Abstract
Background & Aims
Non-alcoholic fatty liver disease (NAFLD) contributes to the global epidemic of metabolic syndrome and is considered a prelude to end-stage liver diseases such as cirrhosis and hepatocellular carcinoma. During NAFLD pathogenesis, hepatic parenchymal cells (hepatocytes) undergo both morphological and functional changes owing to a rewired transcriptome. The underlying mechanism is not entirely clear. In the present study, we investigated the involvement of early growth response 1 (Egr1) in NAFLD.
Methods
Quantitative PCR, Western blotting, and histochemical staining were used to assess gene expression levels. Chromatin immunoprecipitation was used to evaluate protein binding to DNA. NAFLD was evaluated in leptin receptor-deficient (db/db) mice.
Results
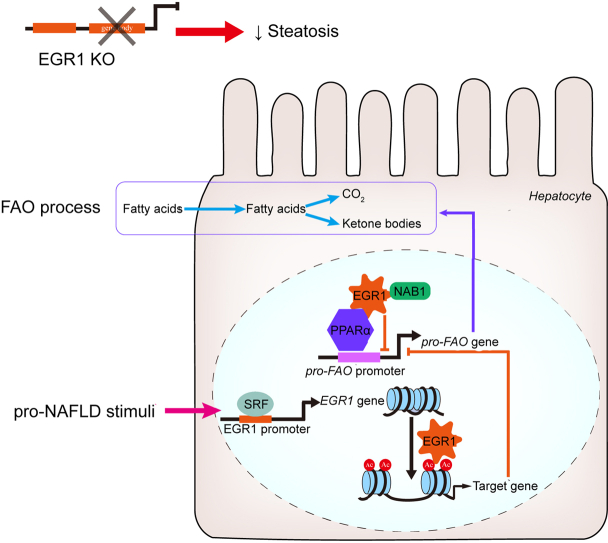
We report here that Egr1 was upregulated by pro-NAFLD stimuli in vitro and in vivo. Further analysis revealed that serum response factor (SRF) was recruited to the Egr1 promoter and mediated Egr1 transactivation. Importantly, Egr1 depletion markedly mitigated NAFLD in db/db mice. RNA sequencing revealed that Egr1 knockdown in hepatocytes, on the one hand, boosted fatty acid oxidation (FAO) and, on the other hand, suppressed the synthesis of chemoattractants. Mechanistically, Egr1 interacted with peroxisome proliferator-activated receptor α (PPARα) to repress PPARα-dependent transcription of FAO genes by recruiting its co-repressor NGFI-A binding protein 1 (Nab1), which potentially led to promoter deacetylation of FAO genes.
Conclusions
Our data identify Egr1 as a novel modulator of NAFLD and a potential target for NAFLD intervention.
Impact and Implications
Non-alcoholic fatty liver disease (NAFLD) precedes cirrhosis and hepatocellular carcinoma. In this paper, we describe a novel mechanism whereby early growth response 1 (Egr1), a transcription factor, contributes to NAFLD pathogenesis by regulating fatty acid oxidation. Our data provide novel insights and translational potential for NAFLD intervention.
Keywords: Non-alcoholic fatty liver disease, Transcriptional regulation, Transcription factor, Hepatocyte
Graphical abstract
Highlights
-
•
SRF mediates Egr1 transactivation by pro-NAFLD stimuli in vitro and in vivo.
-
•
Egr1 levels correlate with steatotic injuries in patients with NAFLD.
-
•
Egr1 depletion in leptin receptor-deficient mice mitigates NAFLD pathologies.
-
•
Egr1 contributes to NAFLD likely by promoting inflammation and suppressing FAO.
-
•
Egr1 and its co-repressor recruit HDAC activity to repress FAO gene transcription.
Introduction
Non-alcoholic fatty liver disease (NAFLD) is a prototypic form of metabolic syndrome that affects 25% of the global population.1 Without effective intervention, some patients with NAFLD will develop cirrhosis, hepatocellular carcinoma, and eventually liver failure. NAFLD shares many common pathological features with alcoholic liver disease (ALD), which include accumulation of lipid droplets, presence of extensive immune infiltrates, and hepatocyte ballooning.2 Indeed, diagnostic criteria for NAFLD are similar to those for ALD except for long-term excessive alcohol consumption. In the past decade, NAFLD has become the primary contributor not only to end-stage liver diseases (e.g. cirrhosis) but also to cardiovascular diseases and diabetes.3 Despite decades of rigorous research, therapeutic strategies for NAFLD remain elusive.4
Transcriptional reprogramming is considered key to the pathologies associated with NAFLD. For instance, increased expression levels of fatty acid transporters, which include members of the fatty acid transport proteins (FATP) family and CD36, are observed in the livers of patients with NAFLD compared with the healthy individuals.5 Rate-limiting enzymes involved in de novo lipogenesis, including fatty acid synthase and acetyl-CoA carboxylase, are found to be upregulated during NAFLD pathogenesis in humans and in experimental animals,6,7 likely owing to elevated activity of sterol response element binding protein (SREBP). On the contrary, genes that encode enzymes participating in fatty acid oxidation (FAO) appear to be lower in patients with NAFLD than in healthy individuals, which can be attributed to deficiencies of the transcription factor peroxisome proliferator-activated receptor α (PPARα).8,9 Thus, enhanced lipid uptake and synthesis in combination with suppressed FAO may help explain hepatic lipid accumulation associated with NAFLD. Upregulation of TRAF3, a key adaptor molecule in the tumour necrosis factor signalling pathway, in hepatocytes by free fatty acids (FAA) as a result of excessive energy influx has been reported.10 Consequently, NF-κB, the master regulator of pro-inflammatory transcription, is activated to program the synthesis of pro-inflammatory mediators.11 Of note, many of the transcription factors are considered ‘versatile’ in coordinating pro-NAFLD transcriptional events. For instance, SREBP1c has been documented to modulate the transcription of pro-inflammatory mediators.[12], [13], [14] On the contrary, there is also evidence to implicate NF-κB in lipid metabolism: NF-κB unorthodoxly represses the transcription of Sorcin, which in turn promotes carbohydrate response element binding protein (ChREBP) nuclear accumulation to influence de novo lipogenesis.15 Recently, it has been demonstrated that multiple networks of transcription factors contribute to the overhaul of hepatic transcriptome during NAFLD pathogenesis.16
Early growth response 1 (Egr1) was independently isolated and characterised by the Siebenlist laboratory17 and the Forsdyke laboratory18 as an early inducible gene in the process of T lymphocyte expansion. Egr1 is a zinc finger transcription factor that plays roles in a wide range of pathophysiological processes including host defence response, carcinogenesis, and organ fibrosis.[19], [20], [21] Previously, Nagi and colleagues have shown that Egr1 expression was upregulated in the livers of rodents fed an ethanol diet to induce ALD.22 Moreover, Egr1 deletion completely abrogated alcohol-induced liver injury in mice likely owing to dampened hepatic inflammation.22 Because of the overall similarity between NAFLD and ALD pathogenesis, we hypothesised that Egr1 might contribute to NAFLD development by regulating transcription in hepatocytes exposed to FAA. We report here that Egr1 expression is regulated by pro-NAFLD stimuli, likely mediated by serum response factor (SRF), in hepatocytes in vivo and in vitro. Importantly, Egr1 regulates transcriptional events relevant to the pathogenesis of NAFLD. Therefore, our data identify Egr1 as a novel modulator of NAFLD and a potential target for NAFLD intervention.
Materials and methods
Animals
All the animal experiments were reviewed and approved by the Liaocheng University Ethics Committee on Humane Treatment of Experimental Animals. The mice were maintained in an SPF environment with 12-h light/dark cycles and libitum access to food and water. NAFLD was induced by four different protocols: (1) db/db mice on a regular chow diet for 12 wk; (2) C57/B6 mice on a high-fat diet (HFD; D09100310, Research Diets) for 12 wk; (3) Apoe-/- mice on a Western diet (D12079B, Research Diets, New Brunswick, NJ, USA) for 7 wk; and (4) C57/B6 mice on choline-deficient, l-amino acid-defined HFD (CDA-HFD; A06071309, Research Diets) for 8 wk as previously described.23 In certain experiments, Egr1-targeting small-hairpin RNA (shRNA) was placed downstream of the human thyroxin-binding globulin (TBG) promoter and packed into AAV8 for tail vein injection into db/db mice.
Cell culture, plasmids, and transient transfection
Human hepatoma cells (HepaRG, Thermo Fisher, San Jose, CA, USA) were maintained in DMEM supplemented with 10% FBS (Hyclone, Logan, UT, USA). Primary murine hepatocytes were isolated as previously described24,25 and seeded at 2 × 105 cells/well for 12-well culture dishes, or 4 × 105 cells/well for 6-well culture dishes, or 4 × 106 cells/well for p100 culture dishes. Cell viability was examined at the time of seeding by trypan blue staining; typical isolation yielded >95% viability. EGR1 promoter-luciferase construct was generated by amplifying genomic DNA spanning the proximal promoter and the first exon of EGR1 gene (-900/+50) and ligating into a pGL3-basic vector (Promega, Madison, WI, USA). Truncation mutants were made using a QuikChange kit (Thermo Fisher Scientific, Waltham, MA, USA) and verified by direct sequencing. Small interfering RNAs (siRNAs) were purchased from Dharmacon (Lafayette, CO, USA). Transient transfections were performed with Lipofectamine 2000 (Thermo Fisher Scientific). Luciferase activities were assayed 24–48 h after transfection using a luciferase reporter assay system (Promega) as previously described.26
RNA isolation and real-time PCR
RNA was extracted with the RNeasy RNA isolation kit (Qiagen, Germantown, MD, USA) as previously described.27,28 Reverse transcriptase reactions were performed using a SuperScript First-strand Synthesis System (Invitrogen, Waltham, MA, USA). Real-time PCRs were performed on an ABI Prism 7500 system using the following primers: mouse Egr1, TCGGCTCCTTTCCTCACTCA and CTCATAGGGTTGTTCGCTCGG; human EGR1, GGTCAGTGGCCTAGTGAGC and GTGCCGCTGAGTAAATGGGA; mouse Il1b, GAAATGCCACCTTTTGACAGTG and TGGATGCTCTCATCAGGACAG; mouse Il6, TGGGGCTCTTCAAAAGCTCC and AGGAACTATCACCGGATCTTCAA; mouse Mcp1, AAAACACGGGACGAGAAACCC and ACGGGAACCTTTATTAACCCCT; and mouse Tnfa, CTGGATGTCAATCAACAATGGGA and ACTAGGGTGTGAGTGTTTTCTGT. Ct values of target genes were normalised to the Ct values of the housekeeping control gene (18s, 5′-CGCGGTTCTATTTTGTTGGT-3′ and 5′-TCGTCTTCGAAACTCCGACT-3′ for both human and mouse genes) using the ΔΔCt method and expressed as relative mRNA expression levels compared with the control group, which is arbitrarily set as 1.
Protein extraction and Western blot
Whole-cell lysates were obtained by resuspending cell pellets in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris pH7.4, 150 mM NaCl, 1% Triton X-100) with freshly added protease inhibitor (Roche, Basel, Switzerland) as previously described.29 Typically, 100 μl RIPA buffer was used for 1 × 106 cells. Moreover, 30 μg of protein was loaded in each lane and separated by 8% SDS-PAGE gel with all-blue protein markers (Bio-Rad, Hercules, CA, USA). Proteins were transferred to nitrocellulose membranes (Bio-Rad) in a Mini-Trans-Blot Cell (Bio-Rad). The membranes were blocked with 5% fat-free milk powder in TBS at room temperature for half an hour and then incubated with the following primary antibodies at 4 °C overnight: anti-Egr1 (Proteintech, Wuhan, China, 55117-1, 1:500) and anti-β-actin (Sigma, Burlington, MA, USA, A1978, 1:5,000). The next day, the membranes were washed with TBS and incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody (Thermo Fisher, 61-6520, 1:5,000) or anti-mouse secondary antibody (Thermo Fisher, 31464, 1:5,000) for 1 h at room temperature. For densitometrical quantification, densities of target proteins were normalised to those of β-actin. Data are expressed as relative protein levels compared with the control group, which is arbitrarily set as 1.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described before.30,31 Chromatin was cross-linked with 1% formaldehyde for 8 min at room temperature and then sequentially washed with ice-cold PBS, solution I (10 mM HEPES, pH 7.5, 10 mM EDTA, 0.5 mM EGTA, 0.75% Triton X-100), and solution II (10 mM HEPES, pH 7.5, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA). Cells were incubated in lysis buffer (150 mM NaCl, 25 mM Tris pH 7.5, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholate) supplemented with protease inhibitor tablet. DNA was fragmented into 500-bp pieces using a Branson 250 sonicator (Brookfield, CT, USA). Aliquots of lysates containing 100 μg of protein were used for each immunoprecipitation reaction with anti-SRF (Cell Signaling Tech, Danvers, MA, USA, 5147), anti-Egr1 (Cell Signaling Tech, 4154), anti-NGFI-A binding protein 1 (Nab1) (Novus Biologicals, Minneapolis, MN, USA, NBP1-71838), anti-acetyl H3K9 (Millipore, Burlington, MA, USA, 07-352), anti-acetyl H3K27 (Millipore, Burlington, MA, USA 07-360), anti-acetyl H3K14 (Millipore, Burlington, MA, USA, 07-353), anti-acetyl H3K18 (Millipore, 07-354), or IgG followed by adsorption to protein A/G PLUS-agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Precipitated DNA–protein complexes were washed sequentially with RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, 1 mM EDTA), high salt buffer (50 mM Tris, pH 8.0, 500 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, 1 mM EDTA), LiCl buffer (50 mM Tris, pH 8.0, 250 mM LiCl, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, 1 mM EDTA), and TE buffer (10 mM Tris,1 mM EDTA, pH 8.0). DNA–protein cross-link was reversed by heating the samples to 65 °C overnight. Proteins were digested with proteinase K (Sigma), and DNA was phenol/chloroform-extracted and precipitated by 100% ethanol. Precipitated genomic DNA was amplified by real-time PCR.
Human NASH biopsy specimens
Liver biopsies were collected from patients with non-alcoholic steatohepatitis (NASH) referring to the First People's Hospital of Changzhou. Control liver samples were collected from donors without NASH but deemed unsuitable for transplantation. Written informed consent was obtained from participants or families of liver donors. All procedures that involved human samples were approved by the Ethics Committee of the First People's Hospital of Changzhou and adhered to the principles outlined in the Declaration of Helsinki.
Glucose tolerance assays
For the glucose tolerance test (GTT), mice fasted overnight were injected i.p. with 2 g/kg glucose, and blood samples were taken at the indicated intervals. For the insulin tolerance test (ITT), mice were fasted for 6 h and injected i.p. with 0.75 IU/kg soluble insulin, and blood samples were taken at the indicated intervals. For the pyruvate tolerance test, mice were fasted for 6 h and injected i.p. with 2.5 g/kg pyruvate dissolved in PBS, and blood samples were taken at the indicated intervals. Blood glucose was measured using an Accu-Chek compact glucometer (Roche).
Histology
Histological analyses were performed essentially as described before.32 Images were taken using an Olympus IX-70 microscope (Shinjuku, Tokyo, Japan). Quantifications were performed with ImageJ. For each mouse, at least three slides were stained, and at least five different fields were analysed for each slide.
RNA sequencing and data analysis
RNA sequencing (RNA-seq) was performed as previously described.33 Total RNA was extracted using the TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. RNA purity and quantification were evaluated using the NanoDrop 2000 spectrophotometer (Thermo Scientific). RNA integrity was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Then, the libraries were constructed using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer's instructions and sequenced on an Illumina HiSeq X Ten platform, and 150-bp paired-end reads were generated. Raw data (raw reads) of fastq format were firstly processed using Trimmomatic, and the low-quality reads were removed to obtain the clean reads. The clean reads were mapped to the mouse genome (Mus_musculus.GRCm38.99) using HISAT2. FPKM (fragments per kilobase of exon model per million reads mapped) of each gene was calculated using Cufflinks, and the read counts of each gene were obtained using HTSeqcount. Differential expression analysis was performed using the DESeq (2012) R package. A p value <0.05 and fold change >2 or fold change <0.5 were set as the thresholds for significantly differential expression. Hierarchical cluster analysis of differentially expressed genes (DEGs) was performed to demonstrate the expression pattern of genes in different groups and samples. Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEGs were performed respectively using R based on the hypergeometric distribution.
Statistical analysis
For comparison between two groups, a two-tailed t test was performed. For comparison among three or more groups, one-way ANOVA or two-way ANOVA with post hoc Turkey analyses were performed using an SPSS package (IBM Corp., Armonk, NY, USA). The assumptions of normality were checked using the Shapiro–Wilk test, and equal variance was checked using Levene's test; both were satisfied. Values of p less than 0.05 were considered statistically significant (∗). All in vitro experiments were repeated at least three times, and three replicates were estimated to provide 80% power.
Results
Egr1 expression is upregulated by pro-NAFLD stimuli in vitro and in vivo
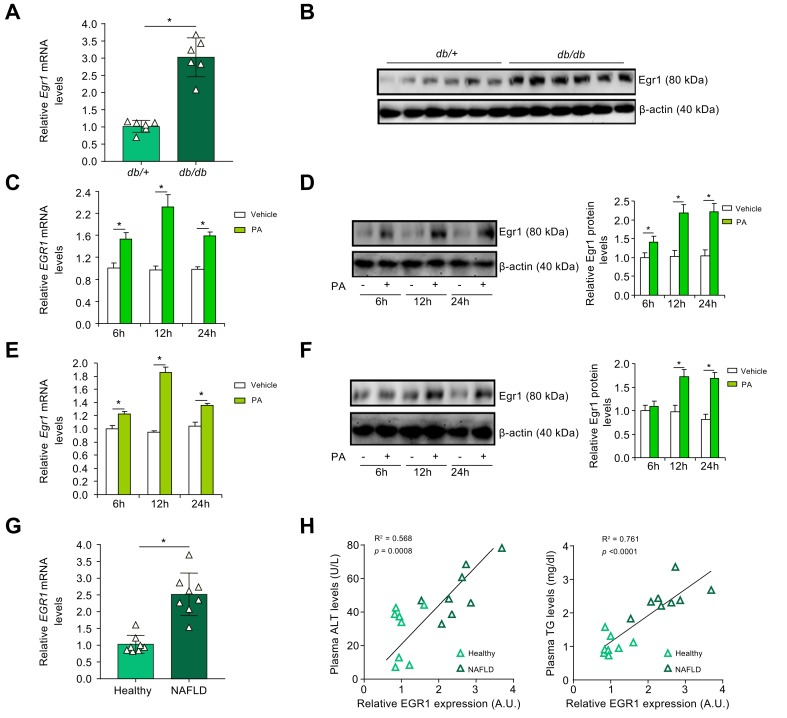
The following experiments were performed to determine whether Egr1 expression was altered during NAFLD pathogenesis in four different animal models. In the livers of 12-wk db/db mice, Egr1 expression at both mRNA levels (Fig. 1A) and protein levels (Fig. 1B) was appreciably higher than that in the livers of db/+ mice. In the second model, male C57/B6 mice were fed an HFD for 12 wk. Compared with the mice fed with the control diet (CD), Egr1 expression was significantly upregulated in the livers of the mice fed the HFD (Fig. S1). In the third model, male Apoe-/- mice were fed a Western diet for 7 wk. Higher expression levels of Egr1 were detected in the livers of mice fed the Western diet than in those fed the CD (Fig. S2). In the fourth model, male C57/B6 mice were fed a CDA-HFD for 8 wk. Again, more Egr1 molecules were detected in the CDA-HFD-fed livers than in the CD-fed livers (Fig. S3).
Fig. 1.
Egr1 is upregulated by pro-NAFLD stimuli in vivo and in vitro.
(A and B) Egr1 expression in the liver tissues of 12-wk male db/db mice and db/+ mice was examined by qPCR and Western blotting. N = 6 mice for each group. Data are expressed as mean ± SD. ∗p <0.05, two-tailed Student’s t test. (C and D) HepaRG cells were treated with or without PA (0.2 mM). EGR1 expression was examined by qPCR and Western blotting. N = 3 biological replicates. Data are expressed as mean ± SD. ∗p <0.05, two-tailed Student’s t test. (E and F) Primary hepatocytes were treated with or without PA (0.2 mM). Egr1 expression was examined by qPCR and Western blotting. N = 3 biological replicates. Data are expressed as mean ± SD. ∗p <0.05, two-tailed Student’s t test. (G and H) Egr1 expression levels in the livers of patients with NASH and healthy individuals were examined by qPCR. Linear regression was performed by GraphPad Prism (Dotmatics, Boston, MA, USA). N = 8 cases for each group. Data are expressed as mean ± SD. ∗p <0.05. ALT, alanine transaminase; Egr1, early growth response 1; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; PA, palmitate; qPCR, quantitative PCR; TG, triglyceride.
FFA, including palmitate (PA), are among the best characterised risk factors for NAFLD.34 When primary murine hepatocytes were exposed to PA, a small but significant upregulation of Egr1 expression was detected as early as 6 h after the treatment (Fig. 1C and D). Egr1 expression peaked at 12 h and declined at 24 h. Similar observations were made in the human hepatoma cells (HepaRG): the cells responded to PA treatment by upregulating Egr1 expression fast and transiently (Fig. 1E and F). However, Egr1 expression was unaltered in cells exposed to oleate (Fig. S4), pointing to certain level of selectivity in terms of the responsiveness of Egr1 to nutrients/metabolites. In addition, Egr1 expression could be stimulated in hepatocytes exposed to fructose (Fig. S5), another major risk factor for NAFLD.35
Liver specimens collected from patients with NAFLD and healthy donors were examined for Egr1 expression. As shown in Fig. 1G, Egr1 levels were markedly elevated in individuals diagnosed with NAFLD compared with those in healthy individuals. More importantly, a positive correlation was identified between Egr1 expression and steatotic injuries (as measured by plasma alanine transaminase [ALT] and triglyceride levels) in humans (Fig. 1H). Together, these data support the conclusion that Egr1 expression may be associated with NAFLD pathogenesis.
SRF mediates Egr1 transactivation in hepatocytes
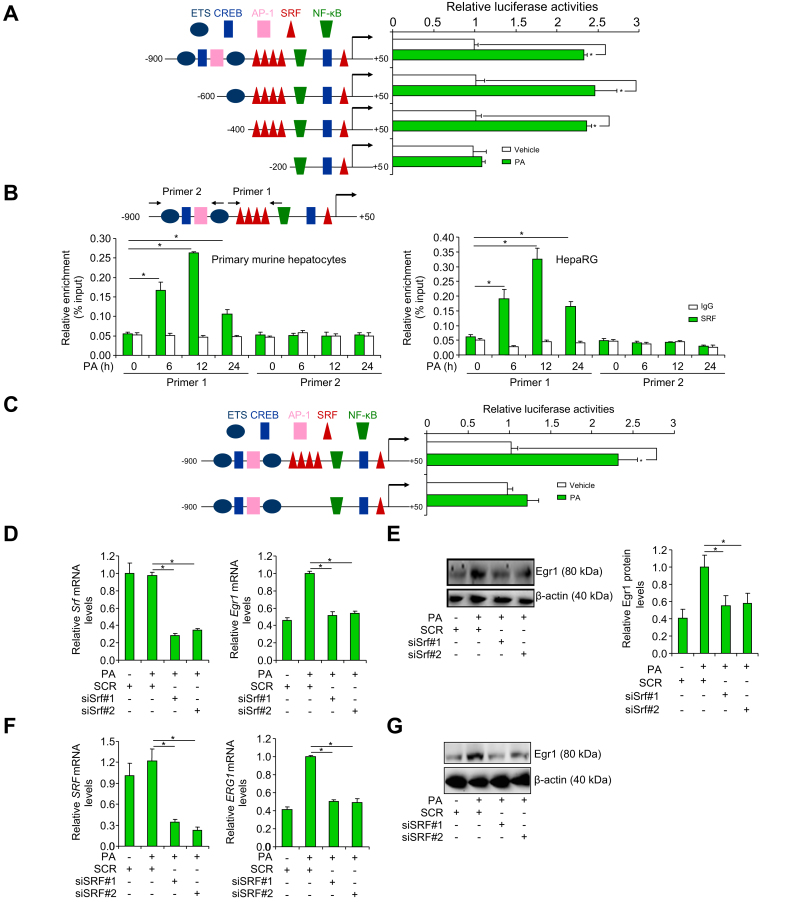
To determine whether the observed upregulation of Egr1 expression occurred at the transcriptional level, an Egr1 promoter-luciferase construct (-900/+50) was transfected into HepaRG cells; PA treatment led to a robust augmentation of the Egr1 promoter activity (Fig. 2A). A host of transcription factors have been identified to bind to the Egr1 promoter and regulate Egr1 gene transcription, which include E26 transformation-specific (ETS), cAMP response element binding protein (CREB), activator protein 1 (AP-1), SRF, and NF-κB (Fig. 2A). Progressive deletions introduced to the Egr1 promoter did not alter its responsiveness to PA treatment unless a string of SRF binding sites located between -400 and -200 relative to the transcription start site (TSS) was removed (Fig. 2A). Of note, PA treatment did not significantly alter SRF expression at either mRNA levels (Fig. S6A) or protein levels (Fig. S6B). Immunofluorescence staining showed that PA treatment altered SRF subcellular localisation in hepatocytes: in the absence of PA, SRF was detected in both the cytoplasm and the nucleus; in the presence of PA, SRF was almost exclusively detected in the nucleus (Fig. S6C). ChIP assay confirmed that PA treatment resulted in enhanced binding of SRF to the Egr1 promoter consistent with its nuclear accumulation (Fig. 2B). Enhanced binding of SRF to the Egr1 promoter was also detected in the livers of the db/db mice compared with the db/+ mice (Fig. S7). Compared with the wild-type Egr1 promoter-luciferase construct, a mutant construct without the SRF motif completely lost the response to PA treatment (Fig. 2C). Finally, knockdown of SRF expression by two different pairs of siRNAs attenuated Egr1 induction by PA treatment in hepatocytes (Fig. 2D–G). Of note, SRF knockdown appeared to significantly dampen Egr1 induction by fructose in both HepaRG cells and murine primary hepatocytes (Fig. S8). Collectively, these data support the conclusion that SRF mediates Egr1 transactivation in hepatocytes.
Fig. 2.
SRF mediates Egr1 transactivation in hepatocytes.
(A) Egr1 promoter-luciferase constructs were transfected into HepaRG cells followed by treatment with PA for 12 h. Luciferase activities were normalised by protein concentration and GFP fluorescence. N = 3 biological replicates. Data are expressed as mean ± SD. ∗p <0.05, two-tailed Student’s t test. (B) Primary hepatocytes and HepaRG cells were treated with or without PA (0.2 mM). ChIP assay was performed with anti-SRF or IgG. N = 3 biological replicates. Data are expressed as mean ± SD. ∗p <0.05, one-way ANOVA with the post hoc Scheffé test. (C) Wild-type and mutant Egr1 promoter-luciferase constructs were transfected into HepaRG cells followed by treatment with PA for 12 h. Luciferase activities were normalised by protein concentration and GFP fluorescence. N = 3 biological replicates. Data are expressed as mean ± SD. ∗p <0.05, two-tailed Student’s t test. (D and E) Primary hepatocytes were transfected with siRNA targeting SRF or SCR followed by treatment with PA (0.2 mM). Egr1 expression was examined by qPCR and Western blotting. N = 3 biological replicates. Data are expressed as mean ± SD. ∗p <0.05, one-way ANOVA with the post hoc Scheffé test. (F and G) HepaRG cells were transfected with siRNA targeting SRF or SCR followed by treatment with PA (0.2 mM). Egr1 expression was examined by qPCR and Western blotting. N = 3 biological replicates. Data are expressed as mean ± SD. ∗p <0.05, one-way ANOVA with the post hoc Scheffé test. AP-1, activator protein 1; ChIP, chromatin immunoprecipitation; CREB, cAMP response element binding protein; Egr1, early growth response 1; ETS, E26 transformation-specific; GFP, green fluorescent protein; PA, palmitate; qPCR, quantitative PCR; SCR, scrambled siRNAs; siRNA, small interfering RNA; SRF, serum response factor.
Egr1 depletion attenuates NAFLD in mice
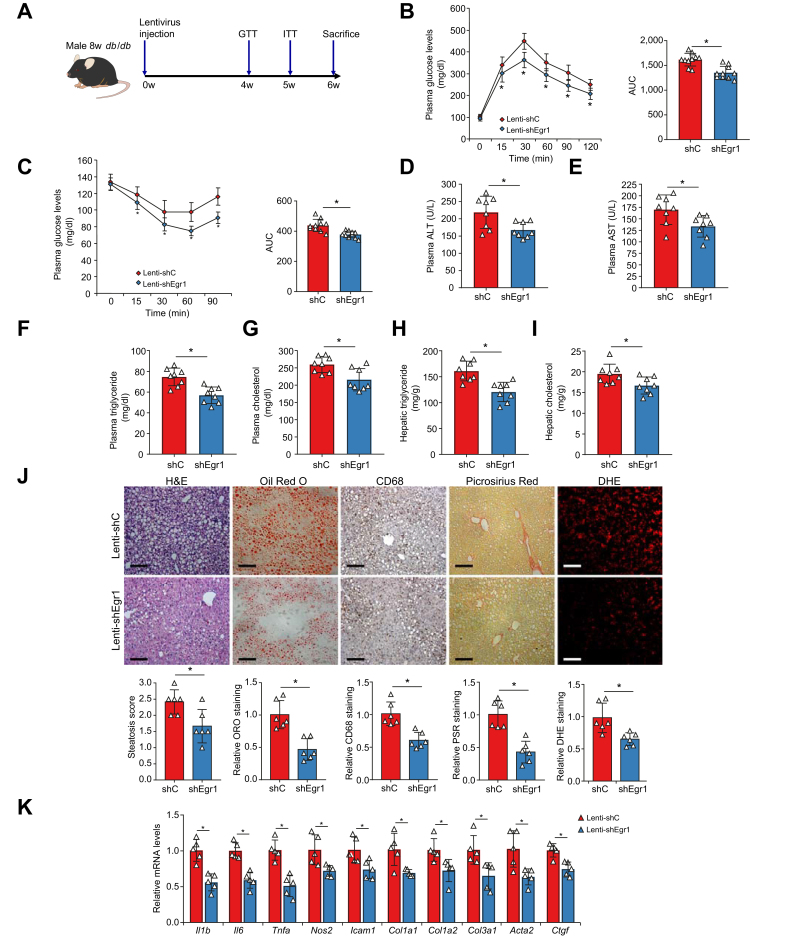
Next, lentivirus carrying shRNA targeting Egr1 (shEgr1) (Lenti-shEgr1) or a control shRNA (shC) (Lenti-shC) was injected into db/db mice to determine whether Egr1 interference would alter NAFLD in vivo (Fig. 3A); quantitative PCR (qPCR) and Western blotting confirmed that Egr1 levels were downregulated by Lenti-shEgr1 injection compared with Lenti-shC injection (Fig. S9). Egr1 silencing did not impact the body weight of the mice (Fig. S10). Nor did Egr1 silencing influence JNK signalling in the liver (Fig. S11). However, insulin sensitivity, as measured by the glucose tolerance test (Fig. 3B) and the insulin tolerance test (Fig. 3C), was greatly improved by Egr1 knockdown. In addition, biochemical analysis of plasma ALT/aspartate aminotransferase (AST) levels (Fig. 3D and E), plasma triglyceride/cholesterol levels (Fig. 3F and G), and hepatic triglyceride/cholesterol levels (Fig. 3H and I) pointed to an amelioration of steatotic injury in the shEgr1-injected mice compared with the shC-injected mice. Histological staining showed that Egr1 knockdown reduced lipid accumulation, immune cell infiltration, deposition of extracellular matrix proteins, and reactive oxygen species (ROS) in the liver (Fig. 3J). Finally, expression levels of pro-inflammatory/fibrogenic mediators were collectively downregulated following Egr1 depletion in the db/db mice (Fig. 3K).
Fig. 3.
Egr1 depletion attenuates NAFLD in mice.
Lenti-shEgr1 or Lenti-shC was injected into db/db mice as described in the Materials and methods section. (A) Scheme of protocol. (B) GTT. (C) ITT. (D) Plasma ALT levels. (E) Plasma AST levels. (F) Plasma triglyceride levels. (G) Plasma total cholesterol levels. (H) Hepatic triglyceride levels. (I) Hepatic total cholesterol levels. (J) Paraffin sections were stained with H&E, ORO, PSR, and anti-CD68. (K) Pro-inflammatory and pro-fibrogenic genes were examined by qPCR. N = 5–10 mice for each group. Data are expressed as mean ± SD. ∗p <0.05, two-tailed Student’s t test. ALT, alanine transaminase; AST, aspartate aminotransferase; DHE, dihydroethidium; Egr1, early growth response 1; GTT, glucose tolerance test; ITT, insulin tolerance test; Lenti-shC, lentivirus carrying a control shRNA; Lenti-shEgr1, lentivirus carrying shRNA targeting Egr1; ORO, Oil Red O; PSR, Picrosirius Red; qPCR, quantitative PCR; shC, control shRNA; shEgr1, shRNA targeting Egr1; shRNA, small-hairpin RNA.
Egr1 knockdown alters transcriptome in hepatocytes
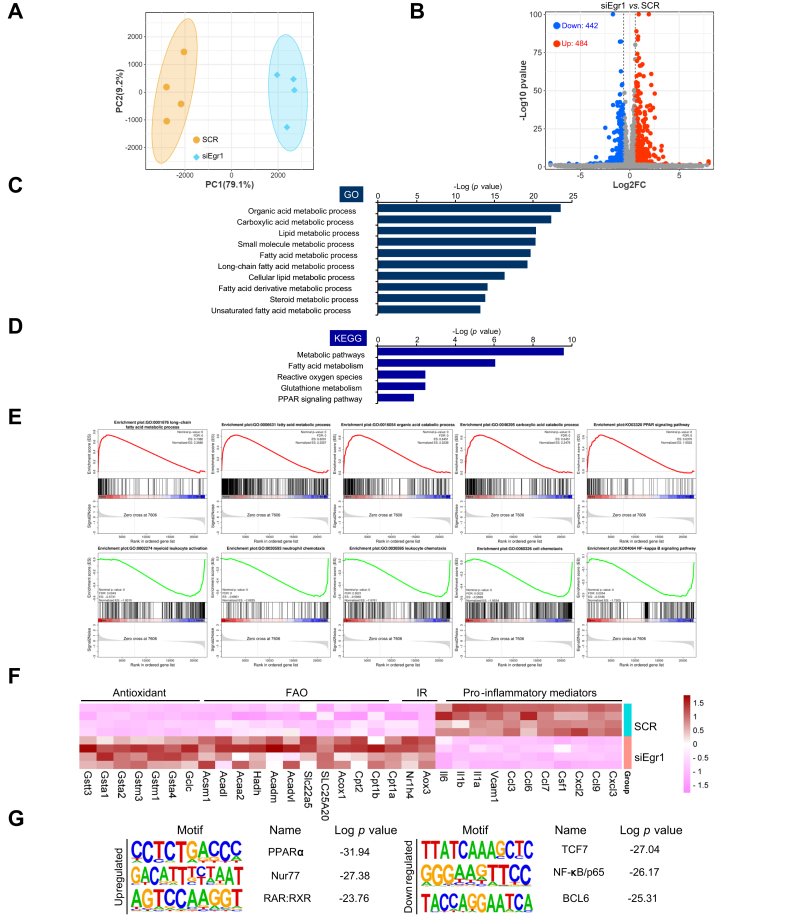
To gain genome-wide perspective on the role of Egr1 in NAFLD pathogenesis, RNA-seq analysis was performed to compare the transcriptome of hepatocytes before and after Egr1 was depleted with siRNAs. As shown in Fig. 4A, Egr1 knockdown significantly altered cellular transcriptome. Using 1.5 × fold change and p <0.05 as thresholds, approximately 1,000 genes were identified to be altered by Egr1 knockdown (Fig. 4B). GO analysis (Fig. 4C) and KEGG analysis (Fig. 4D) indicated that Egr1 knockdown primarily influenced genes involved in cellular metabolism. Further, gene set enrichment analysis (GSEA) illustrated that Egr1 depletion was positively correlated with pathways involved in fatty acid metabolism but inversely correlated with pathways that promote hepatocyte-derived chemoattractive cues (Fig. 4E). Among the top DEGs were pro-inflammatory chemokines/cytokines (downregulated), antioxidants (upregulated), and fatty acid β-oxidation enzymes (upregulated) (Fig. 4F). In congruence, hypergeometric optimisation of motif enrichment (HOMER) analysis revealed that Egr1 deficiency attenuated the activities of pro-inflammatory transcription factors including NF-κB but liberated the activities of metabolic nuclear receptors including PPARα (Fig. 4G). Thus, it is possible that Egr1 might contribute to NAFLD by modulating lipid metabolism and inflammation in hepatocytes.
Fig. 4.
Egr1 knockdown alters transcriptome in hepatocytes.
(A–G) Primary murine hepatocytes were transfected with indicated siRNAs followed by treatment with PA (0.2 mM). RNA-seq was performed and analysed as described in the Materials and methods section. (A) PCA plot. (B) Volcano plot. (C) GO analysis. (D) KEGG analysis. (E) GSEA. (F) Heatmap of differentially expressed genes. (G) HOMER analysis. BCL6, B-cell lymphoma 6; Egr1, early growth response 1; ES, enrichment score; FAO, fatty acid oxidation; FDR, false discovery rate; GO, gene ontology; GSEA, gene set enrichment analysis; HOMER, hypergeometric optimisation of motif enrichment; IR, insulin resistance; KEGG, Kyoto Encyclopedia of Genes and Genomes; PA, palmitate; PC1, principal component 1; PC2, principal component 2; PCA, principal component analysis; PPARα, peroxisome proliferator-activated receptor α; RAR: retinoic acid receptor; RNA-seq, RNA sequencing; RXR, retinoic X receptor; SCR, scrambled siRNAs; siEgr1, Egr1 siRNA; siRNA, small interfering RNA; TCF7, transcription factor 7.
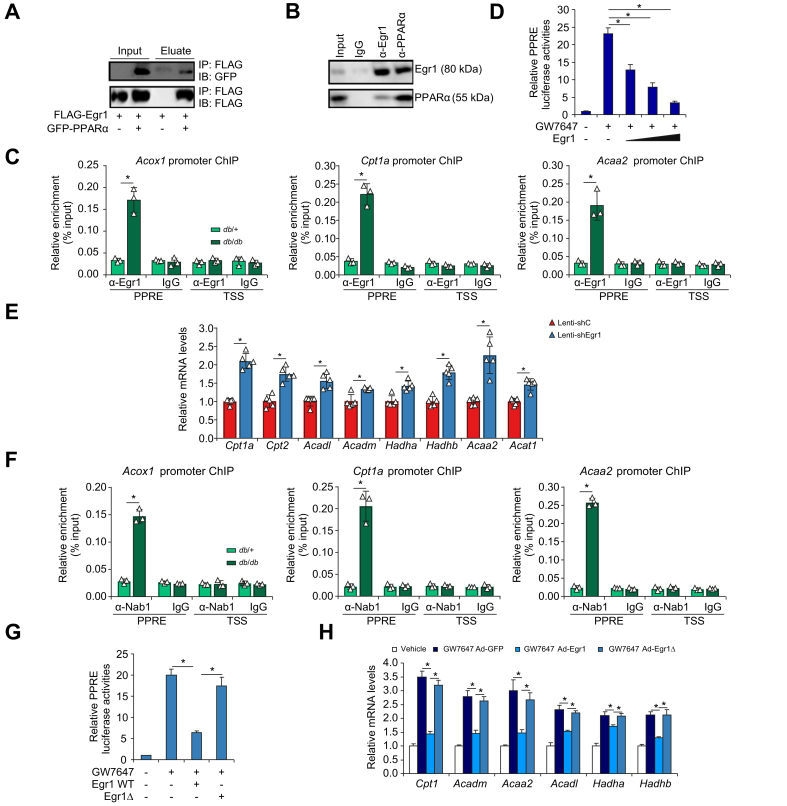
Erg1 interacts with PPARα to repress FAO transcription
Bioinformatic analysis offered compelling evidence for a potential interplay between Egr1 and PPARα. Co-immunoprecipitation assays showed that (1) ectopically expressed Egr1 and PPARα formed a complex in HEK293 cells (Fig. 5A) and (2) endogenous Egr1 and PPARα interacted with each other in the murine livers (Fig. 5B). Importantly, ChIP assay detected strong association of Egr1 with FAO gene promoters, all known PPARα targets, surrounding the PPAR response element (PPRE) in the db/db mice compared with the db/+ mice (Fig. 5C). Overexpression of Egr1 dose-dependently repressed PPARα activity measured by a reporter fused to six tandem repeats of PPRE (Fig. 5D). On the contrary, Egr1 knockdown significantly upregulated FAO genes in the db/db mice as measured by qPCR (Fig. 5E). Consistently, plasma ketone body (β-hydroxybutyrate) levels were higher in the shEgr1 mice than in the shC mice (Fig. S12). The hepatokine fibroblast growth factor 21 (FGF21) is a well-established PPARα target implicated in FAO.36,37 However, FGF21 levels were not significantly altered by Egr1 knockdown in db/db mice or in primary hepatocytes (Fig. S13). Because FAO is a key physiological process during fasting, Egr1 expression was examined in the livers isolated from mice subjected to 12 h of fasting followed by 12 h of refeeding. Egr1 expression was mildly but significantly downregulated in the liver following fasting but returned to basal levels following refeeding (Fig. S14).
Fig. 5.
Erg1 interacts with PPARα to repress FAO transcription.
(A) FLAG-tagged Egr1 and GFP-tagged PPARα were transfected into HEK293 cells. Immunoprecipitation was performed with indicated antibodies. (B) Liver lysates from db/db mice were immunoprecipitated with indicated antibodies. (C) ChIP assay was performed with anti-Egr1 or IgG using liver lysates from db/db mice and db/+ mice. N = 3 mice for each group. Data are expressed as mean ± SD. ∗p <0.05, one-way ANOVA with the post hoc Scheffé test. (D) A PPRE reporter was transfected into HepaRG cells with increasing doses of Egr1 followed by treatment with GW7647 (0.1 μM). Luciferase activities were normalised by protein concentration and GFP fluorescence. N = 3 biological replicates. Data are expressed as mean ± SD. ∗p <0.05, one-way ANOVA with the post hoc Scheffé test. (E) Lenti-shEgr1 or Lenti-shC was injected into db/db mice. Gene expression levels were examined by qPCR. N = 5 mice for each group. Data are expressed as mean ± SD. ∗p <0.05, two-tailed Student’s t test. (F) ChIP assay was performed with anti-Nab1 or IgG using liver lysates from db/db mice and db/+ mice. N = 3 mice for each group. Data are expressed as mean ± SD. ∗p <0.05, one-way ANOVA with the post hoc Scheffé test. (G) A PPRE reporter was transfected into HepaRG cells with indicated Egr1 vectors followed by treatment with GW7647 (0.1 μM). Luciferase activities were normalised by protein concentration and GFP fluorescence. N = 3 biological replicates. Data are expressed as mean ± SD. ∗p <0.05, one-way ANOVA with the post hoc Scheffé test. (H) HepaRG cells were transduced with Ad-Egr1, Ad-Egr1Δ, or Ad-GFP followed by treatment with GW7647 (0.1 μM). FAO genes were examined by qPCR. N = 3 biological replicates. Data are expressed as mean ± SD. ∗p <0.05, one-way ANOVA with the post hoc Scheffé test. Ad-Egr1, adenovirus carrying Egr1; Ad-Egr1Δ, adenovirus carrying Egr1 mutant; Ad-GFP, adenovirus carrying a control vector; ChIP, chromatin immunoprecipitation; Erg1, early growth response 1; FAO, fatty acid oxidation; GFP, green fluorescent protein; IB, immunoblotting; IP, immunoprecipitation; Lenti-shC, lentivirus carrying a control shRNA; Lenti-shEgr1, lentivirus carrying shRNA targeting Egr1; Nab1, NGFI-A binding protein 1; PPARα, peroxisome proliferator-activated receptor α; PPRE, PPAR response element; qPCR, quantitative PCR; shRNA, small-hairpin RNA; TSS, transcription start site; WT, wild-type.
Previous studies indicate that Egr1 relies on co-repressor Nab1 to repress transcription.38 Indeed, Nab1 was detected on the FAO gene promoters, in a similar fashion as Egr1, in the db/db mice compared with the db/+ mice (Fig. 5F). Consistently, the Egr1 mutant that lacks the Nab1 interaction domain (Egr1Δ)39 lost the ability to repress PPARα activity (Fig. 5G) and PPARα-dependent transactivation of FAO genes (Fig. 5H). In addition, Nab1 levels were found to be higher in the patients with NAFLD than in healthy individuals and correlated with steatotic injuries (Fig. S15).
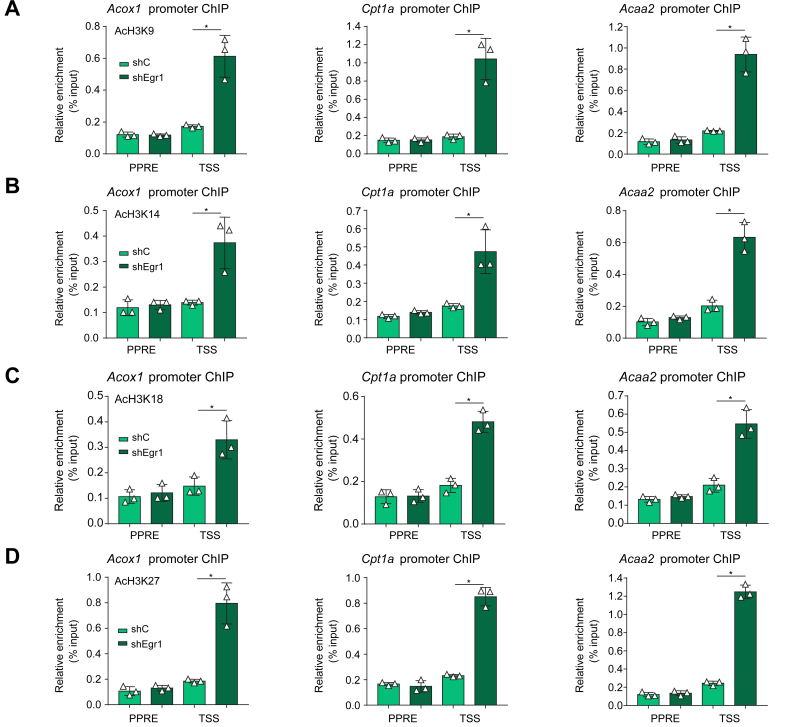
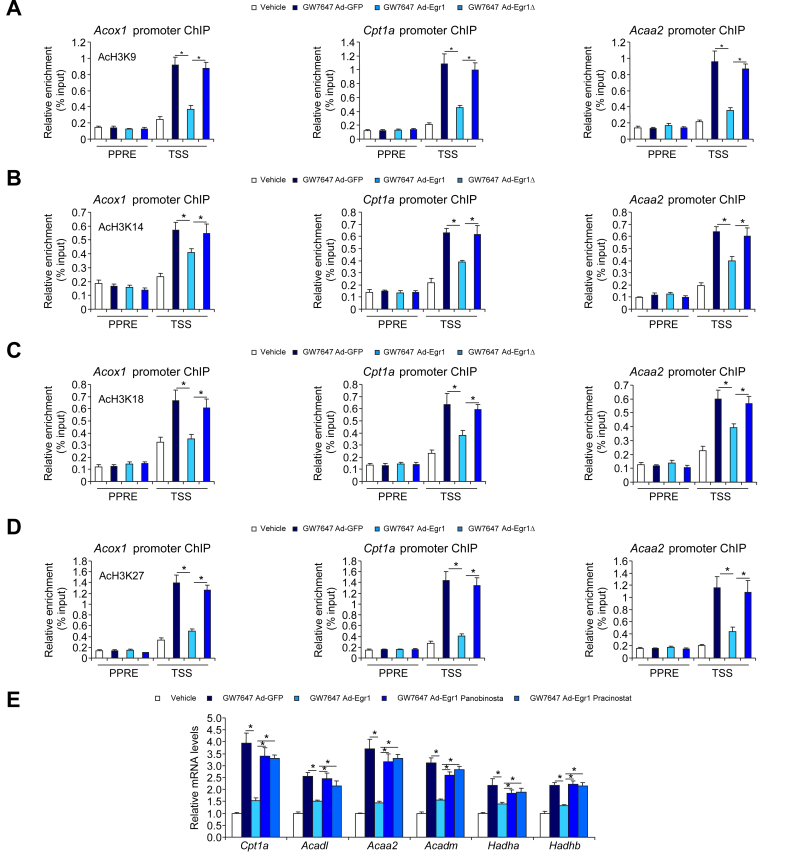
Erg1 regulates FAO transcription by promoting histone deacetylation
Histone acetylation is considered a prototypical marker for actively transcribed chromatin. ChIP assay demonstrated that Egr1 deficiency significantly augmented acetylation of histone H3K9 (Fig. 6A), H3K14 (Fig. 6B), H3K18 (Fig. 6C), and H3K27 (Fig. 6D) on FAO gene promoters consistent with the upregulation of these genes. In HepaRG cells, exposure to the PPARα agonist evoked accumulation of acetylated histones on the FAO gene promoters, which was suppressed by wild-type Egr1 but not by Egr1Δ (Fig. 7A–D). These observations suggest that Erg1 might recruit histone deacetylases (HDACs), likely via Nab1, to the FAO promoters to repress transcription. To authenticate this hypothesis, two pan-HDAC inhibitors (HDACis), were exploited. As shown in Fig. 7E, repression of PPARα-induced FAO gene expression by Egr1 was partially relieved by co-treatment of either HDACi.
Fig. 6.
Erg1 regulates FAO transcription by promoting histone deacetylation.
(A–D) Lenti-shEgr1 or Lenti-shC was injected into db/db mice. ChIP assays were performed with anti-acetyl H3K9 (A), anti-acetyl H3K14 (B), anti-acetyl H3K18 (C), and anti-acetyl H3K27 (D). N = 3 mice for each group. Data are expressed as mean ± SD. ∗p <0.05, one-way ANOVA with the post hoc Scheffé test. ChIP, chromatin immunoprecipitation; Erg1, early growth response 1; FAO, fatty acid oxidation; Lenti-shC, lentivirus carrying a control shRNA; Lenti-shEgr1, lentivirus carrying shRNA targeting Egr1; PPAR, peroxisome proliferator-activated receptor; PPRE, PPAR response element; shC, control shRNA; shEgr1, shRNA targeting Egr1; shRNA, small-hairpin RNA.
Fig. 7.
Erg1 regulates FAO transcription via Nab1-dependent recruitment of HDACs.
(A–D) HepaRG cell were transduced with Ad-Egr1, Ad-Egr1Δ, or Ad-GFP followed by treatment with GW7647 (0.1 μM). ChIP assays were performed with anti-acetyl H3K9 (A), anti-acetyl H3K14 (B), anti-acetyl H3K18 (C), and anti-acetyl H3K27 (D). N = 3 biological replicates. Data are expressed as mean ± SD. ∗p <0.05, one-way ANOVA with the post hoc Scheffé test. (E) HepaRG cell were transduced with Ad-Egr1 or Ad-GFP followed by treatment with GW7647 (0.1 μM) in the presence or absence of two pan-HDAC inhibitors (panobinostat [0.1 μM] and pracinostat [0.5 μM]). FAO genes were examined by qPCR. N = 3 biological replicates. Data are expressed as mean ± SD. ∗p <0.05, one-way ANOVA with the post hoc Scheffé test. Ad-Egr1, adenovirus carrying Egr1; Ad-Egr1Δ, adenovirus carrying Egr1 mutant; Ad-GFP, adenovirus carrying a control vector; ChIP, chromatin immunoprecipitation; Erg1, early growth response 1; FAO, fatty acid oxidation; GFP, green fluorescent protein; HDAC, histone deacetylase; Nab1, NGFI-A binding protein 1; PPAR, peroxisome proliferator-activated receptor; PPRE, PPAR response element; qPCR, quantitative PCR.
Discussion
Stimuli that promote NAFLD pathogenesis elicit profound changes in hepatocytes.40 In the course of NAFLD development and progression, multiple transcription factors form extensive regulatory networks to influence transcriptome in hepatocytes.41 We describe here a novel finding that Egr1, a transcription factor previously reported to regulate the pathogenesis of ALD, appears to be upregulated during NAFLD development in mice and in hepatocytes exposed to FFA. Moreover, single-gene and genome-wide profiling of expression patterns in hepatocytes indicate that Egr1 knockdown may dampen NAFLD pathogenesis. Thus, it is possible that Egr1 might represent a novel biomarker and a potential therapeutic target for NAFLD.
Our data indicate that SRF mediates PA-induced transactivation of Egr1 in hepatocytes by binding to the Egr1 promoter. Although SRF has been implicated in the regulation of multiple pathophysiological processes related to NAFLD, including inflammation,42 production of ROS,43 and lipid metabolism,44 no direct evidence exists to link SRF to NAFLD pathogenesis, probably owing to a lack of suitable genetic tools. Sun et al.45 have previously made an attempt to specifically delete SRF in hepatocytes by crossing the Srff/f mice to the Alb-Cre mice. The resulting SRF conditional knockout mice were born at sub-Mendelian ratio, and the surviving mice displayed retarded growth compared with the wild-type littermates, which could be attributed to disruption of glucose and fat metabolism. These abnormalities, although preclude the analysis of NAFLD pathogenesis, suggest that SRF may be indispensable for the maintenance of physiological homoeostasis of the liver. Of interest, a study by Jin et al.46 has found that nuclear accumulation of SRF in skeletal muscle cells exposed to PA correlated with a gene signature of insulin resistance. Our observation that PA treatment promoted migration of SRF into the nucleus in hepatocytes echoes that by Jin et al.46 and suggests SRF might be a nutrition sensor that contributes to the regulation of insulin response.
A key finding of the present study is that Egr1 contributes to NAFLD pathogenesis by suppressing fatty acid β-oxidation in hepatocytes. Egr1 and its co-repressor repress FAO gene transcription by recruiting an HDAC activity. Concordantly, treatment with two pan-HDACis relieved the repression of FAO gene transcription. However, the identity of the specific HDAC(s) mediating repression of FAO gene transcription by Egr1 remains obscure. As such, whether HDACi administration could bring about any beneficial effects in established NAFLD models in vivo awaits to be tested. It should be noted that existing literature alludes to distinct roles for different HDACs in NAFLD pathogenesis. There is strong evidence to suggest that HDAC3, a class I HDAC, protects against NAFLD by promoting lipid metabolism, although this effect is likely deacetylase activity independent.47 It has also been reported that HDAC5, a class IIa HDAC, promotes FAO and safeguards against liver steatosis in a PPARα-dependent manner.48 On the contrary, both HDAC11,49 the sole class IV HDAC, and HDAC1,50 the ubiquitously expressed class I HDAC, appear to suppress FAO in the skeletal muscle and liver, respectively. Future studies should aim to unravel the precise epigenetic mechanism whereby Egr1 (re)programs cellular metabolism.
We show here that Egr1 knockdown in hepatocytes results in downregulation of several pro-inflammatory mediators. This is consistent with a previous report by Mackman and colleagues, in which it was found that the same set of pro-inflammatory mediators including IL-6 and monocyte chemoattractant protein 1 (MCP1) was repressed in the lungs and kidneys of Egr1 knockout mice compared with those of the wild-type mice in a model of lipopolysaccharide-induced endotoxaemia.51 In addition, Cho et al.52 have presented evidence to show that Egr1 deletion blunted pulmonary inflammation in IL-13 transgenic mice. Further, it has been documented that Egr1 deletion attenuates atherosclerotic lesions in Ldlr-/- mice fed a Western diet.53 Because atherosclerosis is considered a prototypical pathology of chronic inflammation, it is tempting to speculate that Egr1 might be a master regulator of inflammatory response. However, a recent study by the Gardini laboratory indicated that Egr1 is able to bind to non-classic motifs located on the enhancers and repress the transcription of several pro-inflammatory genes in macrophages.54 More studies are certainly warranted to reconcile this discrepancy.
Our RNA-seq data show that Egr1 knockdown is associated with upregulation of a panel of antioxidant genes, suggesting that Egr1 might contribute to NAFLD pathogenesis by altering redox homoeostasis. There has been abundant evidence that connects Egr1 expression/activity and intracellular ROS levels. Egr1 expression can be upregulated by a long list of oxygen-free radicals in vascular smooth muscle cells,55 osteoblasts,56 myelocytic cells,57 and tubular epithelial cells.58 The precise mechanism whereby Egr1 regulates cell-specific redox status is not entirely clear. Recently, Pang et al.59 have made an interesting observation that Egr1 suppresses macrophage phagocytosis by reducing the expression of P62, thus depriving Nrf2, the master regulator of anti-oxidative transcription, a key co-factor.59 Because increased ROS production and/or impaired ROS clearance is a hallmark event in NAFLD pathogenesis,60 targeting Egr1 could certainly help restore redox homoeostasis in the liver.
Despite the advances proffered by our study, major limitations exist that dampen enthusiasm. First, lentiviral delivery of shRNA, although driven by a hepatocyte-specific (TBG) promoter, may non-specifically target cell lineages other than hepatocytes. Thus, the observed phenotype could be accounted for by Egr1 in macrophages or sinusoidal endothelial cells. New mouse strains with lineage-specific manipulation of Egr1 expression would help clarify this issue. Second, the regulatory role of Egr1 in NAFLD pathogenesis was investigated in a single mouse model, the genetically predisposed db/db model, considered not ideal in terms of recapitulating the human NAFLD-NASH pathology owing to low level of liver fibrosis and low incidence of hepatocellular carcinoma.61 Future studies should exploit multiple and preferably humanised NAFLD-NASH models to validate the regulatory role of Egr1.
In summary, our data provide novel mechanistic insight on Egr1 upregulation during NAFLD pathogenesis. More importantly, our data suggest that Egr1 regulates a transcriptional programme in hepatocytes that might contribute to NAFLD. Further studies should be conducted to provide genetic evidence that links Egr1 to NAFLD in experimental animals and in humans to justify targeting Egr1 for NAFLD intervention.
Financial support
This work was supported by grants from the National Natural Science Foundation of China (82170592, 82200684, and 81900513), the Natural Science Foundation of Jiangsu Province (BK20221032), and Liaocheng University (318012118).
Authors’ contributions
Conceived the project: ZWF, ZLL, QHW. Designed experiments: all authors. Performed experiments, collected data, and analysed data: YG, XLM, XYS, LYL, AQZ, XZ, ZLL, ZWF. Secured funding: ZLL, ZWF. Drafted the manuscript: YX. Edited and finalised the manuscript: all authors.
Data availability statement
The data that support the findings of this study are available upon reasonable request.
Conflicts of interest
The authors delare no conflict of interest.
Please refer to the accompanying ICMJE disclosure forms for further details.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2023.100724.
Contributor Information
Qinghua Wang, Email: 20214132092@stu.suda.edu.cn.
Zilong Li, Email: lzl1114@cpu.edu.cn.
Zhiwen Fan, Email: fanzhiwenfff@126.com.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Cotter T.G., Rinella M. Nonalcoholic fatty liver disease 2020: the state of the disease. Gastroenterology. 2020;158:1851–1864. doi: 10.1053/j.gastro.2020.01.052. [DOI] [PubMed] [Google Scholar]
- 2.Anstee Q.M., Reeves H.L., Kotsiliti E., Govaere O., Heikenwalder M. From NASH to HCC: current concepts and future challenges. Nat Rev Gastroenterol Hepatol. 2019;16:411–428. doi: 10.1038/s41575-019-0145-7. [DOI] [PubMed] [Google Scholar]
- 3.Godoy-Matos A.F., Silva Júnior W.S., Valerio C.M. NAFLD as a continuum: from obesity to metabolic syndrome and diabetes. Diabetol Metab Syndr. 2020;12:60. doi: 10.1186/s13098-020-00570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdelmalek M.F. Nonalcoholic fatty liver disease: another leap forward. Nat Rev Gastroenterol Hepatol. 2021;18:85–86. doi: 10.1038/s41575-020-00406-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu L., Baker S.S., Liu W., Tao M.H., Patel R., Nowak N.J., et al. Lipid in the livers of adolescents with nonalcoholic steatohepatitis: combined effects of pathways on steatosis. Metabolism. 2011;60:1001–1011. doi: 10.1016/j.metabol.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Dorn C., Riener M.O., Kirovski G., Saugspier M., Steib K., Weiss T.S., et al. Expression of fatty acid synthase in nonalcoholic fatty liver disease. Int J Clin Exp Pathol. 2010;3:505–514. [PMC free article] [PubMed] [Google Scholar]
- 7.Xie Z., Li H., Wang K., Lin J., Wang Q., Zhao G., et al. Analysis of transcriptome and metabolome profiles alterations in fatty liver induced by high-fat diet in rat. Metabolism. 2010;59:554–560. doi: 10.1016/j.metabol.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 8.Croci I., Byrne N.M., Choquette S., Hills A.P., Chachay V.S., Clouston A.D., et al. Whole-body substrate metabolism is associated with disease severity in patients with non-alcoholic fatty liver disease. Gut. 2013;62:1625–1633. doi: 10.1136/gutjnl-2012-302789. [DOI] [PubMed] [Google Scholar]
- 9.Fujita K., Nozaki Y., Wada K., Yoneda M., Fujimoto Y., Fujitake M., et al. Dysfunctional very-low-density lipoprotein synthesis and release is a key factor in nonalcoholic steatohepatitis pathogenesis. Hepatology. 2009;50:772–780. doi: 10.1002/hep.23094. [DOI] [PubMed] [Google Scholar]
- 10.Wang P.X., Zhang X.J., Luo P., Jiang X., Zhang P., Guo J., et al. Hepatocyte TRAF3 promotes liver steatosis and systemic insulin resistance through targeting TAK1-dependent signalling. Nat Commun. 2016;7 doi: 10.1038/ncomms10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Beek M., Oravecz-Wilson K.I., Delekta P.C., Gu S., Li X., Jin X., et al. Bcl10 links saturated fat overnutrition with hepatocellular NF-κB activation and insulin resistance. Cell Rep. 2012;1:444–452. doi: 10.1016/j.celrep.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reboldi A., Dang E.V., McDonald J.G., Liang G., Russell D.W., Cyster J.G. Inflammation. 25-Hydroxycholesterol suppresses interleukin-1-driven inflammation downstream of type I interferon. Science. 2014;345:679–684. doi: 10.1126/science.1254790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Im S.S., Yousef L., Blaschitz C., Liu J.Z., Edwards R.A., Young S.G., et al. Linking lipid metabolism to the innate immune response in macrophages through sterol regulatory element binding protein-1a. Cell Metab. 2011;13:540–549. doi: 10.1016/j.cmet.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oishi Y., Spann N.J., Link V.M., Muse E.D., Strid T., Edillor C., et al. SREBP1 contributes to resolution of pro-inflammatory TLR4 signaling by reprogramming fatty acid metabolism. Cel Metab. 2017;25:412–427. doi: 10.1016/j.cmet.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniel P.V., Dogra S., Rawat P., Choubey A., Khan A.S., Rajak S., et al. NF-κB p65 regulates hepatic lipogenesis by promoting nuclear entry of ChREBP in response to a high carbohydrate diet. J Biol Chem. 2021;296 doi: 10.1016/j.jbc.2021.100714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez-Schindler J., Vargas-Fernández E., Karrer-Cardel B., Ritz D., Schmidt A., Handschin C. Characterization of regulatory transcriptional mechanisms in hepatocyte lipotoxicity. Sci Rep. 2022;12 doi: 10.1038/s41598-022-15731-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright J.J., Gunter K.C., Mitsuya H., Irving S.G., Kelly K., Siebenlist U. Expression of a zinc finger gene in HTLV-I- and HTLV-II-transformed cells. Science. 1990;248:588–591. doi: 10.1126/science.2110381. [DOI] [PubMed] [Google Scholar]
- 18.Siderovski D.P., Blum S., Forsdyke R.E., Forsdyke D.R. A set of human putative lymphocyte G0/G1 switch genes includes genes homologous to rodent cytokine and zinc finger protein-encoding genes. DNA Cel Biol. 1990;9:579–587. doi: 10.1089/dna.1990.9.579. [DOI] [PubMed] [Google Scholar]
- 19.Wang B., Guo H., Yu H., Chen Y., Xu H., Zhao G. The role of the transcription factor EGR1 in cancer. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.642547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerji R., Saroj S.D. Early growth response 1 (EGR1) activation in initial stages of host–pathogen interactions. Mol Biol Rep. 2021;48:2935–2943. doi: 10.1007/s11033-021-06305-0. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharyya S., Fang F., Tourtellotte W., Varga J. Egr-1: new conductor for the tissue repair orchestra directs harmony (regeneration) or cacophony (fibrosis) J Pathol. 2013;229:286–297. doi: 10.1002/path.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMullen M.R., Pritchard M.T., Wang Q., Millward C.A., Croniger C.M., Nagy L.E. Early growth response-1 transcription factor is essential for ethanol-induced fatty liver injury in mice. Gastroenterology. 2005;128:2066–2076. doi: 10.1053/j.gastro.2005.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumoto M., Hada N., Sakamaki Y., Uno A., Shiga T., Tanaka C., et al. An improved mouse model that rapidly develops fibrosis in non-alcoholic steatohepatitis. Int J Exp Pathol. 2013;94:93–103. doi: 10.1111/iep.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong W., Zhu Y., Zhang Y., Fan Z., Zhang Z., Fan X., et al. BRG1 links TLR4 trans-activation to LPS-induced SREBP1a expression and liver injury. Front Cel Dev Biol. 2021;9 doi: 10.3389/fcell.2021.617073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan Z., Kong M., Miao X., Guo Y., Ren H., Wang J., et al. An E2F5-TFDP1-BRG1 complex mediates transcriptional activation of MYCN in hepatocytes. Front Cel Dev Biol. 2021;9 doi: 10.3389/fcell.2021.742319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lv F., Shao T., Xue Y., Miao X., Guo Y., Wang Y., et al. Dual regulation of tank binding kinase 1 (TBK1) by BRG1 in hepatocytes contributes to ROS production. Front Cel Dev Biol. 2021;9 doi: 10.3389/fcell.2021.745985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong M., Dong W., Zhu Y., Fan Z., Miao X., Guo Y., et al. Redox-sensitive activation of CCL7 by BRG1 in hepatocytes during liver injury. Redox Biol. 2021;46 doi: 10.1016/j.redox.2021.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong M., Dong W., Xu H., Fan Z., Miao X., Guo Y., et al. Choline kinase alpha is a novel transcriptional target of the Brg1 in hepatocyte: implication in liver regeneration. Front Cel Dev Biol. 2021;9 doi: 10.3389/fcell.2021.705302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen B., Dong W., Shao T., Miao X., Guo Y., Liu X., et al. A KDM4-DBC1-SIRT1 axis contributes to TGF-b induced mesenchymal transition of intestinal epithelial cells. Front Cel Dev Biol. 2021;9 doi: 10.3389/fcell.2021.697614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen B., Zhu Y., Chen J., Feng Y., Xu Y. Activation of TC10-like transcription by lysine demethylase KDM4B in colorectal cancer cells. Front Cel Dev Biol. 2021;9 doi: 10.3389/fcell.2021.617549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan Z., Kong M., Dong W., Dong C., Miao X., Guo Y., et al. Trans-activation of eotaxin-1 by Brg1 contributes to liver regeneration. Cell Death Dis. 2022;13:495. doi: 10.1038/s41419-022-04944-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong W., Kong M., Liu H., Xue Y., Li Z., Wang Y., et al. Myocardin-related transcription factor A drives ROS-fueled expansion of hepatic stellate cells by regulating p38-MAPK signalling. Clin Transl Med. 2022;12:e688. doi: 10.1002/ctm2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X., Dong W., Kong M., Ren H., Wang J., Shang L., et al. Down-regulation of CXXC5 de-represses MYCL1 to promote hepatic stellate cell activation. Front Cel Dev Biol. 2021;9 doi: 10.3389/fcell.2021.680344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feldstein A.E., Werneburg N.W., Canbay A., Guicciardi M.E., Bronk S.F., Rydzewski R., et al. Free fatty acids promote hepatic lipotoxicity by stimulating TNF-α expression via a lysosomal pathway. Hepatology. 2004;40:185–194. doi: 10.1002/hep.20283. [DOI] [PubMed] [Google Scholar]
- 35.Jensen T., Abdelmalek M.F., Sullivan S., Nadeau K.J., Green M., Roncal C., et al. Fructose and sugar: a major mediator of non-alcoholic fatty liver disease. J Hepatol. 2018;68:1063–1075. doi: 10.1016/j.jhep.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inagaki T., Dutchak P., Zhao G., Ding X., Gautron L., Parameswara V., et al. Endocrine regulation of the fasting response by PPARα-mediated induction of fibroblast growth factor 21. Cel Metab. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 37.Badman M.K., Pissios P., Kennedy A.R., Koukos G., Flier J.S., Maratos-Flier E. Hepatic fibroblast growth factor 21 is regulated by PPARα and is a key mediator of hepatic lipid metabolism in ketotic states. Cel Metab. 2007;5:426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 38.Russo M.W., Sevetson B.R., Milbrandt J. Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc Natl Acad Sci USA. 1995;92:6873–6877. doi: 10.1073/pnas.92.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu C., Adamson E., Mercola D. Transcription factor EGR-1 suppresses the growth and transformation of human HT-1080 fibrosarcoma cells by induction of transforming growth factor beta 1. Proc Natl Acad Sci USA. 1996;93:11831–11836. doi: 10.1073/pnas.93.21.11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parthasarathy G., Revelo X., Malhi H. Pathogenesis of nonalcoholic steatohepatitis: an overview. Hepatol Commun. 2020;4:478–492. doi: 10.1002/hep4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang B., Kang B., Roh T.Y., Seong R.H., Kim W. The chromatin accessibility landscape of nonalcoholic fatty liver disease progression. Mol Cell. 2022;45:343–352. doi: 10.14348/molcells.2022.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin C., Hindes A., Burns C.J., Koppel A.C., Kiss A., Yin Y., et al. Serum response factor controls transcriptional network regulating epidermal function and hair follicle morphogenesis. J Invest Dermatol. 2013;133:608–617. doi: 10.1038/jid.2012.378. [DOI] [PubMed] [Google Scholar]
- 43.Kong M., Chen X., Lv F., Ren H., Fan Z., Qin H., et al. Serum response factor (SRF) promotes ROS generation and hepatic stellate cell activation by epigenetically stimulating NCF1/2 transcription. Redox Biol. 2019;26 doi: 10.1016/j.redox.2019.101302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenwald M., Efthymiou V., Opitz L., Wolfrum C. SRF and MKL1 independently inhibit Brown adipogenesis. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun K., Battle M.A., Misra R.P., Duncan S.A. Hepatocyte expression of serum response factor is essential for liver function, hepatocyte proliferation and survival, and postnatal body growth in mice. Hepatology. 2009;49:1645–1654. doi: 10.1002/hep.22834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin W., Goldfine A.B., Boes T., Henry R.R., Ciaraldi T.P., Kim E.Y., et al. Increased SRF transcriptional activity in human and mouse skeletal muscle is a signature of insulin resistance. J Clin Invest. 2011;121:918–929. doi: 10.1172/JCI41940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Z., Feng D., Fang B., Mullican S.E., You S.H., Lim H.W., et al. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol Cel. 2013;52:769–782. doi: 10.1016/j.molcel.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu X., Li J., Lv S., Yu J., Jiang J., Yao J., et al. HDAC5 integrates ER stress and fasting signals to regulate hepatic fatty acid oxidation. J Lipid Res. 2018;59:330–338. doi: 10.1194/jlr.M080382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Donde H., Ghare S., Joshi-Barve S., Zhang J., Vadhanam M.V., Gobejishvili L., et al. Tributyrin inhibits ethanol-induced epigenetic repression of CPT-1A and attenuates hepatic steatosis and injury. Cell Mol Gastroenterol Hepatol. 2020;9:569–585. doi: 10.1016/j.jcmgh.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pawlinski R., Pedersen B., Kehrle B., Aird W.C., Frank R.D., Guha M., et al. Regulation of tissue factor and inflammatory mediators by Egr-1 in a mouse endotoxemia model. Blood. 2003;101:3940–3947. doi: 10.1182/blood-2002-07-2303. [DOI] [PubMed] [Google Scholar]
- 52.Cho S.J., Kang M.J., Homer R.J., Kang H.R., Zhang X., Lee P.J., et al. Role of early growth response-1 (Egr-1) in interleukin-13-induced inflammation and remodeling. J Biol Chem. 2006;281:8161–8168. doi: 10.1074/jbc.M506770200. [DOI] [PubMed] [Google Scholar]
- 53.Albrecht C., Preusch M.R., Hofmann G., Morris-Rosenfeld S., Blessing E., Rosenfeld M.E., et al. Egr-1 deficiency in bone marrow-derived cells reduces atherosclerotic lesion formation in a hyperlipidaemic mouse model. Cardiovasc Res. 2010;86:321–329. doi: 10.1093/cvr/cvq032. [DOI] [PubMed] [Google Scholar]
- 54.Trizzino M., Zucco A., Deliard S., Wang F., Barbieri E., Veglia F., et al. EGR1 is a gatekeeper of inflammatory enhancers in human macrophages. Sci Adv. 2021;7:eaaz8836. doi: 10.1126/sciadv.aaz8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hasan R.N., Schafer A.I. Hemin upregulates Egr-1 expression in vascular smooth muscle cells via reactive oxygen species ERK-1/2-Elk-1 and NF-κB. Circ Res. 2008;102:42–50. doi: 10.1161/CIRCRESAHA.107.155143. [DOI] [PubMed] [Google Scholar]
- 56.Nose K., Ohba M. Functional activation of the egr-1 (early growth response-1) gene by hydrogen peroxide. Biochem J. 1996;316:381κ3. doi: 10.1042/bj3160381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Datta R., Taneja N., Sukhatme V.P., Qureshi S.A., Weichselbaum R., Kufe D.W. Reactive oxygen intermediates target CC(A/T)6GG sequences to mediate activation of the early growth response 1 transcription factor gene by ionizing radiation. Proc Natl Acad Sci USA. 1993;90:2419–2422. doi: 10.1073/pnas.90.6.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bek M.J., Reinhardt H.C., Fischer K.G., Hirsch J.R., Hupfer C., Dayal E., et al. Up-regulation of early growth response gene-1 via the CXCR3 receptor induces reactive oxygen species and inhibits Na+/K+-ATPase activity in an immortalized human proximal tubule cell line. J Immunol. 2003;170:931–940. doi: 10.4049/jimmunol.170.2.931. [DOI] [PubMed] [Google Scholar]
- 59.Pang Z., Xu Y., Zhu Q. Early growth response 1 suppresses macrophage phagocytosis by inhibiting NRF2 activation through upregulation of autophagy during Pseudomonas aeruginosa infection. Front Cel Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.773665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Z., Tian R., She Z., Cai J., Li H. Role of oxidative stress in the pathogenesis of nonalcoholic fatty liver disease. Free Radic Biol Med. 2020;152:116–141. doi: 10.1016/j.freeradbiomed.2020.02.025. [DOI] [PubMed] [Google Scholar]
- 61.Farrell G., Schattenberg J.M., Leclercq I., Yeh M.M., Goldin R., Teoh N., et al. Mouse models of nonalcoholic steatohepatitis: toward optimization of their relevance to human nonalcoholic steatohepatitis. Hepatology. 2019;69:2241–2257. doi: 10.1002/hep.30333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available upon reasonable request.