Summary
Mitochondria produce reactive oxygen species (ROS), which function in signal transduction. Mitochondrial dynamics, encompassing morphological shifts between fission and fusion, can directly impact ROS levels in cancer cells. In this study, we identified an ROS-dependent mechanism for how enhanced mitochondrial fission inhibits triple negative breast cancer (TNBC) cell migration. We found that enforcing mitochondrial fission in TNBC resulted in an increase in intracellular ROS levels and reduced cell migration and the formation of actin-rich migratory structures. Consistent with mitochondrial fission, increasing ROS levels in cells inhibited cell migration. Conversely, reducing ROS levels with either a global or mitochondrially targeted scavenger overcame the inhibitory effects of mitochondrial fission. Mechanistically, we found that the ROS sensitive SHP-1/2 phosphatases partially regulate inhibitory effects of mitochondrial fission on TNBC migration. Overall, our work reveals the inhibitory effects of ROS in TNBC and supports mitochondrial dynamics as a potential therapeutic target for cancer.
Subject areas: Cancer, Cell biology
Graphical abstract

Highlights
-
•
Mitochondrial fission inhibits triple-negative breast cancer cell migration
-
•
Fission drives actin reorganization to inhibit actin-based migratory structures
-
•
Enforcing fission increases the formation of reactive oxygen species (ROS)
-
•
Increased ROS activates SHP1/2 to inhibit TNBC cell migration
Cancer; Cell biology
Introduction
Abnormal mitochondrial morphology, functions, and dynamics underlie malignant transformation and tumor progression. More specifically, cancer cells reprogram mitochondrial energetic pathways to meet the high energy demands of processes that facilitate metastasis, such as growth and migration.1,2 By transitioning along a continuum of fissioned and fused states, mitochondrial morphology can regulate the metabolic state of a cell. Dynamin related protein 1 (Drp1) and mitofusins (Mfn1/2) drive mitochondrial fission and fusion, respectively, with fusion commonly leading to enhanced oxidative phosphorylation (OXPHOS).3,4 In addition to directly impacting metabolism, mitochondrial morphology also directly influences levels of metabolic signaling molecules, such as reactive oxygen species (ROS) and ATP.5,6 However, effects of these metabolic signaling molecules on cell migration remain incompletely defined.
Although originally considered only as a byproduct of OXPHOS, ROS can act as secondary signaling messengers.7,8 Excessive ROS production by mitochondria can lead to oxidative damage or modification of proteins, nucleic acids, lipids, membranes, and organelles, which drives activation of intracellular signaling cascades. Initial work demonstrated increased ROS levels in cancer cells and that elevated ROS levels promote tumor formation and progression.9,10,11 Recent work contradicts these data and demonstrates tumor-suppressive functions for ROS.12,13 This discordance not only suggests context-dependent effects of ROS in cancer but also opens new avenues for further studying effects of ROS and their regulators in tumor formation and metastasis.
Src homology region 2 domain-containing phosphatase 1 (SHP-1) and 2 (SHP-2) are non-transmembrane, cytoplasmic members of the protein tyrosine phosphatase (PTP) superfamily of enzymes that control signaling pathways underlying diverse physiological processes. Initial reports revealed that SHP-1/2 require ROS-dependent oxidation to activate and allow these PTPs to function.14,15 Subsequent studies in solid cancers found that SHP-1/2 act as tumor suppressors, where expression inhibits growth, invasion, and migration.16,17,18,19,20 Moreover, recent studies also showed that SHP-1/2 can localize to the actin cytoskeleton.21,22 However, little is known about effects of SHP-1/2 on migration and the actin cytoskeleton in breast cancer.
In our previous work we found that enhanced mitochondrial fission reduced migration and metastasis of triple-negative breast cancer cells.23,24 Here, we build on this finding and investigate how mitochondrial fission inhibits migration, a key process in metastasis. We discovered that cells with more mitochondrial fission have enhanced levels of ROS compared to cells that display more mitochondrial fusion. Exogenous addition of a mitochondrial-based ROS, H2O2, promoted mitochondrial fission and reduced cell migration. Enhanced mitochondrial fission also drove actin reorganization leading to enhanced stress fiber density and inhibition of the formation of actin-rich migratory structures, the actin-based protrusions at the leading edge of the cell, in both cell culture and mouse animal models. Treatment with the global ROS scavenger, N-acetyl cysteine (NAC), or a mitochondrially targeted ROS scavenger, MitoTEMPO, reversed these phenotypes on migration and actin reorganization. Mechanistically, we found that SHP-1/2 was activated in cells that display enhanced mitochondrial fission, and inhibition of SHP-1/2 overcame inhibitory effects of mitochondrial fission on migration. Together, these data demonstrate that enhanced mitochondrial fission inhibits migration and metastasis at least partially by increasing ROS and activating SHP-1/2.
Results
Enhanced mitochondrial fission inhibits cell migration
We previously showed that enforcing mitochondrial fission, through genetic (stable expression of phosphatidylserine decarboxylase (PISD) or dynamin related protein 1 (Drp1) (Figure S1) or chemical compounds (treatment with L-α-lysophosphatidylethanolamine (LPE)), reduced single-cell migration in a microfluidics device.23,24 As a complementary approach to drive mitochondrial fusion, we first stably expressed MFN2 in both MDA-MB-231 and SUM159 triple-negative breast cancer (TNBC) cells (Figures S1, S2A, and S2B). Stable expression of MFN2 had minimal effects on mitochondrial fission and modestly increased mitochondrial fusion relative to wild-type (WT) cells in MDA-MB-231 cells (Figures S2C and S2D). Consistent with MDA-MB-231 cells, stable expression of MFN2 had only modest effects on mitochondrial fission compared to WT in SUM159 cells. However, stable expression of MFN2 significantly increased mitochondrial fusion compared to WT in SUM159 cells (Figures S2C and S2D).
Next, to investigate effects of enhanced mitochondrial fission on collective cell migration, we performed a wound healing assay. Consistent with our recent findings on single-cell migration, we found that cells with more mitochondrial fission closed the wound significantly less than cells that primarily displayed mitochondrial fusion in both MDA-MB-231 and SUM159 triple-negative breast cancer (TNBC) cells (Figure S3).
Enhanced mitochondrial fission drives actin cytoskeleton reorganization
Cells rely on the actin cytoskeleton for migration, and lamellipodia/membrane ruffles, filopodia, and stress fibers are the most well-known and studied actin-based migratory structures. Lamellipodia/membrane ruffles are sheet-like, actin-rich migratory structures found at the leading edge of the cell; filopodia are the finger-like projections that commonly extend out of the lamellipodia; and stress fibers are contractile, actomyosin bundles. Previous studies have demonstrated that the actin cytoskeleton can regulate mitochondrial morphology and function.25 However, only limited information exists on mitochondrial morphology-based regulation of the actin cytoskeleton. Phalloidin staining revealed dramatic reorganization of the actin cytoskeleton in cells that display mitochondrial fission, where enhanced fission decreased the formation of actin-rich migratory structures (lamellipodia/membrane ruffles) in both MDA-MB-231 (Figure 1A) and SUM159 (Figure S4) cells. Furthermore, cells that display enhanced mitochondrial fission also had increased stress fiber density (stress fiber area divided by total cell area) in both MDA-MB-231 (Figure 1A) and SUM159 (Figure S4) cells. Together, these data suggest that mitochondrial fission inhibits cell migration through actin cytoskeleton reorganization. As cellular adhesion to the ECM and other cells is critical to initiate signaling cascades and forces necessary for migration, we next tested to what extent different mitochondrial morphologies regulate adhesion to human mammary fibroblasts (HMFs). We found no differences in the ability of cells to adhere to HMFs (Figure S5).
Figure 1.
Mitochondrial fission promotes actin cytoskeletal reorganization and reduces actin-based migratory structures
(A) Representative overlaid and zoomed images of immunofluorescence staining of phalloidin (red) and nuclear DAPI (blue) of MDA-MB-231 WT cells, as well as cells stably expressing PISD, Drp1, or MFN2. Arrows point to actin-rich migratory structures. Dashed box represents the area of the zoomed image. Scale bar is 20 μm. Right. Graphs show average number (mean +/- SD) of actin-rich migratory structures per cell in an image (top, n = 6 images) and stress fiber density in an image (bottom, n = 6 images) for each group. ∗ = p<0.05. ∗∗ = p < 0.01. ∗∗∗ = p<0.001.
(B) Representative images of tumors (green) and collagen (gray) formed from MDA-MB-231 WT, PISD, Drp1, or MFN2 cells that express LifeAct-GFP. Scale bar is 50 μm. The box in GFP panel denotes the origin of the inset. Arrows in the inset point to actin-rich migratory structures.
(C) Boxplot and whiskers for the aspect ratio (length/width) of cells in each group demonstrates more elongated morphology of WT and MFN2 cells (n ≥ 103 cells per group). For boxplots and whiskers, the bottom and top of a box define the first and third quartiles, and the band inside the box marks the second quartile (median). The ends of the whiskers represent the 10th and 90th percentiles, respectively. For boxplots and whiskers, the “+” within the box refers to the mean. ∗∗∗ = p< 0.001.
(D) Frequency distribution of collagen alignment angle reveals greater alignment of collagen fibers perpendicular to margins of tumors from MFN2 cells. Graphs show the orientation of the collagen fibers relative to the tumor (n ≥ 2 tumors per group, n ≥ 4 images per tumor).
To extend our studies to live cells in an in vivo setting, we stably expressed LifeAct-GFP, a 17-amino acid peptide that binds to filamentous actin (F-actin, the form that underlies actin-based migratory structures) and does not interfere with actin dynamics,26 in wild type (WT), PISD, Drp1, and MFN2 stably expressing cells. We orthotopically implanted MDA-MB-231 WT, PISD, Drp1, or MFN2 cells stably expressing LifeAct-GFP into the fourth mammary fat pad of NSG mice. Although mice injected with PISD and Drp1 TNBC cells had on average smaller tumors at experimental endpoint, we did not find any significant differences in growth kinetics (Figures S6A and S6B) or final tumor weight (Figure S6C). Consistent with phalloidin staining, we found that cells with greater mitochondrial fission had reduced actin-rich migratory structures than TNBC cells that displayed mitochondrial fusion (Figure 1B). In addition, we analyzed the aspect ratio of cells within the tumor, where previous work has shown that a larger aspect ratio is indicative of an invasive, motile phenotype.27 We found that enhanced mitochondrial fission reduced the aspect ratio (length/width) of cells within the orthotopic tumor (Figure 1C). In addition to imaging LifeAct-GFP in cells of the primary tumor, we utilized second harmonic two-photon microscopy to also image the peri-tumoral collagen architecture (Figure 1B). Previous work has demonstrated that collagen fiber alignment perpendicular to the tumor margin defines a more invasive phenotype and a worse prognosis in breast cancer.28 Tumors formed from WT, PISD, or Drp1 cells showed similar collagen alignment (Figure 1D). However, tumors formed from MFN2 stably expressing cells showed greater collagen alignment perpendicular to the tumor margin (Figure 1D), which correlates with the pro-invasive phenotypes, such as enhanced migration, aspect ratio, increase in actin-rich migratory structures and reduction in stress fiber density, that MFN2 cells display. We note that we found no differences in lung metastases (Figure S6A), actin-rich migratory structures or collagen alignment between groups in lung metastases (Figure S7).
Cells that display enhanced mitochondrial fission have greater ROS, and increased ROS inhibits cell migration
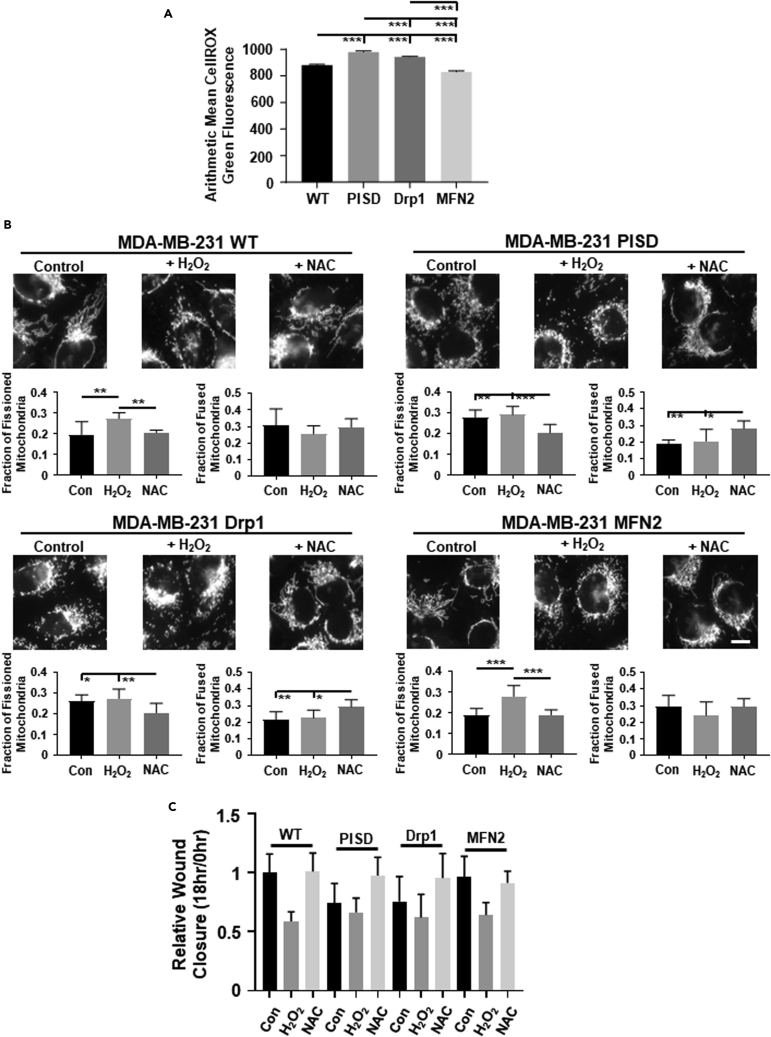
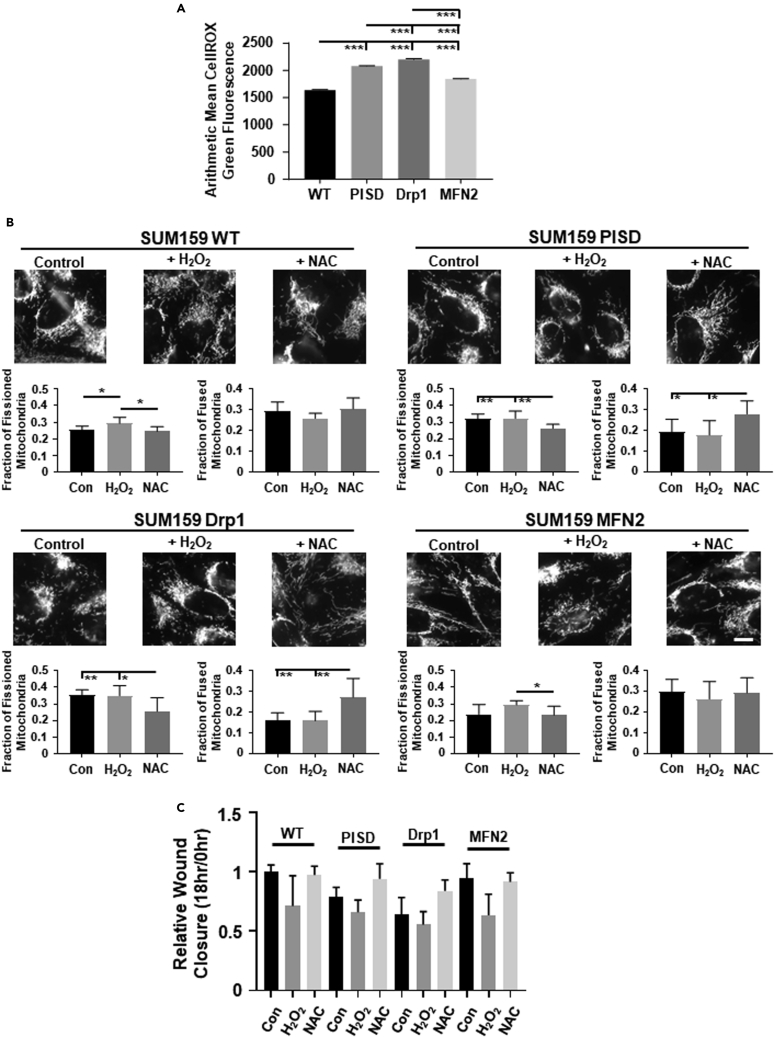
ROS are byproducts of mitochondrial metabolism and can act as secondary signaling messengers,7,8 driving activation of intracellular signaling cascades that can control cellular migration. To investigate to what extent cells with fused versus fissioned mitochondrial morphologies contain different levels of ROS, we stained cells with CellROX green dye. Cells with enhanced mitochondrial fission contained higher levels of ROS than cells with mitochondrial fusion in both MDA-MB-231 (Figures 2A and S8) and SUM159 (Figures 3A and S8) cells. To investigate effects of altering ROS levels in cells, we treated cells with either hydrogen peroxide (H2O2, a key ROS produced by mitochondria) or the global ROS scavenger N-acetyl cysteine (NAC) (Figure S9). We found no significant difference in cell growth by bioluminescence in cells treated with H2O2 or NAC compared to control cells (Figure S10). Treatment of cells with H2O2 significantly enforced mitochondrial fission in MDA-MB-231 (Figures 2B and S11A) and SUM159 (Figures 3B and S11B) cells, resembling the morphology seen in cells that stably express PISD or Drp1. Conversely, treatment of cells with NAC drove a more fused mitochondrial morphology in both MDA-MB-231 (Figure 2B) and SUM159 (Figure 3B) cells, resembling the morphology seen in WT cells and cells that stably express MFN2. Consistent with effects on mitochondrial morphology, we found that treating cells with H2O2 reduced, whereas treatment with NAC increased, the ability of cells to close a wound in MDA-MB-231 (Figures 2C and S12A, and Table S1) and SUM159 (Figures 3C and S12B, and Table S2) cells. Overall, these data suggest that enhanced ROS reduces cell migration.
Figure 2.
Mitochondrial fission increases reactive oxygen species (ROS) in MDA-MB-231 TNBC cells
(A) Graphs show average amount of ROS (mean +/- SEM) using CellROX Green in MDA-MB-231 WT and PISD, Drp1, or MFN2 stably expressing cells (n ≥ 726 cells per group) from a representative biological replicate. ∗∗∗ = p< 0.001.
(B) Representative zoomed images of WT, PISD, Drp1, and MFN2 cells stably expressing a mitochondrial targeted BFP treated with either vehicle control, hydrogen peroxide (H2O2, 100 μM), or N-acetyl cysteine (NAC, 1 mM). Scale bar is 10 μm. Graphs show the mean fraction +SD of fissioned or fused mitochondria after treatment with vehicle control, H2O2, or NAC (n = 8 images for each treatment group) in MDA-MB-231 WT, PISD, Drp1, and MFN2 cells. ∗ = p<0.05. ∗∗ = p<0.01. ∗∗∗ = p<0.001.
(C) Summary of wound closure of cells after treatment with vehicle control, H2O2, or NAC presented as mean +SD (n = 9) relative to the initial wound size (0 h). Statistical analysis for this graph is in Table S1.
Figure 3.
Mitochondrial fission increases reactive oxygen species (ROS) in SUM159 TNBC cells
(A) Graph show average amount of ROS (mean +/- SEM) using CellROX Green in SUM159 WT and PISD, Drp1, or MFN2 stably expressing cells (n ≥ 4507 cells per group) from a representative biological replicate. ∗∗∗ = p< 0.001.
(B) Representative zoomed images of WT, PISD, Drp1, and MFN2 cells stably expressing a mitochondrial targeted BFP treated with either vehicle control, hydrogen peroxide (H2O2, 100 μM), or N-acetyl cysteine (NAC, 1 mM). Scale bar is 10 μm. Graphs show the mean fraction +SD of fissioned or fused mitochondria after treatment with vehicle control, H2O2, or NAC (n = 8 images for each treatment group) in SUM159 WT, PISD, Drp1, and MFN2 cells. ∗ = p<0.05. ∗∗ = p<0.01.
(C) Summary of wound closure of cells after treatment with vehicle control, H2O2, or NAC presented as mean +SD (n = 9) relative to the initial wound size (0 h). Statistical analysis for this graph is in Table S2.
Increased ROS drive actin cytoskeleton reorganization
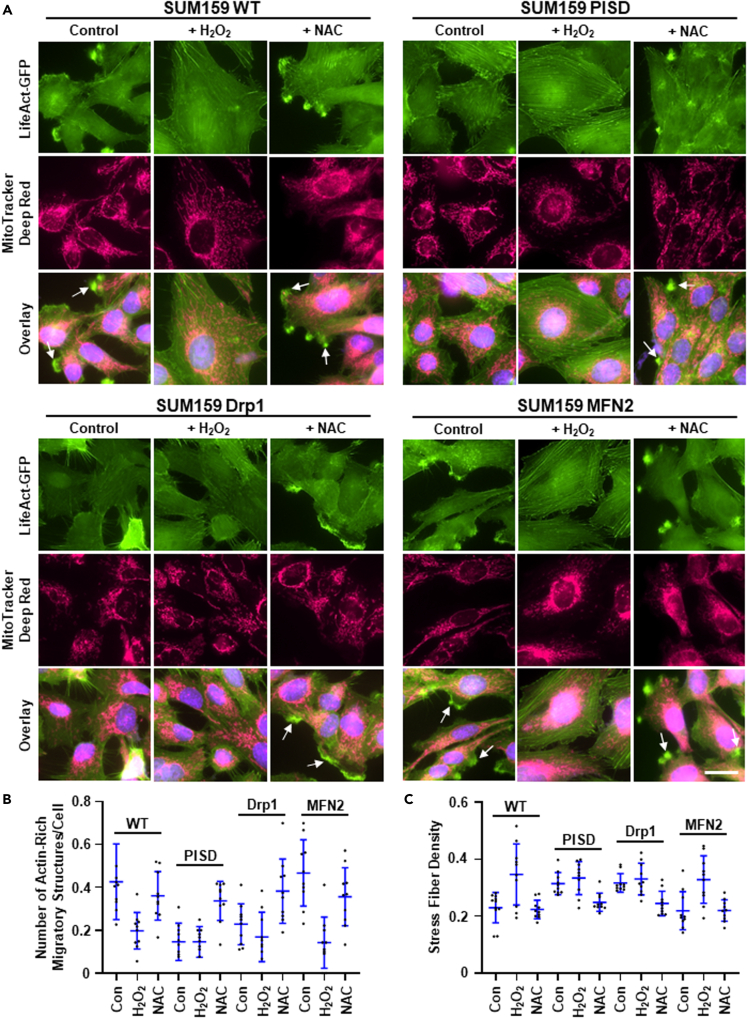
Having established that increased ROS inhibits cell migration, we next investigated effects on the actin cytoskeleton. To directly link mitochondrial morphology, ROS, and the actin cytoskeleton, we stained live TNBC cells that stably expressed LifeAct-GFP with MitoTracker Deep Red. Consistent with our phalloidin data, we found that cells with enhanced mitochondrial fission (PISD and Drp1) had reduced actin-rich migratory structures compared to cells that display increased mitochondrial fusion (WT and MFN2) in MDA-MB-231 (Figures 4A, 4B, and S13, and Table S3) and SUM159 (Figures 5A, 5B, and S14, and Table S4) cells. Treatment of cells showing greater mitochondrial fusion with H2O2 both drove mitochondrial fission and reduced formation of actin-rich migratory structures. However, treatment of cells that already displayed enhanced mitochondrial fission with H2O2 had little effect on the number of actin-rich migratory structures in MDA-MB-231 (Figures 4A, 4B, and S13, and Table S3) and SUM159 (Figures 5A, 5B, and S14, and Table S4) cells. Furthermore, treatment of cells that display enhanced mitochondrial fusion with H2O2 increased stress fiber density in both MDA-MB-231 (Figures 4C and S13, and Table S5) and SUM159 (Figures 5C and S14, and Table S6) cells. Consistent with these data, treatment of cells that already displayed enhanced mitochondrial fusion with NAC had little effect on actin-rich migratory structures. However, NAC treatment of cells with enhanced mitochondrial fission increased formation of actin-rich migratory structures in MDA-MB-231 (Figures 4A, 4B, and S13, and Table S3) and SUM159 (Figures 5A, 5B, and S14, and Table S4) cells. Moreover, treatment of cells that display enhanced mitochondrial fission with NAC reduced stress fiber density in both MDA-MB-231 (Figures 4C and S13, and Table S5) and SUM159 (Figures 5C and S14, and Table S6) cells.
Figure 4.
Enhanced ROS promotes actin reorganization and a less migratory phenotype in MDA-MB-231 TNBC cells
(A) Representative overlaid, zoomed images of immunofluorescence staining of mitochondria (pink) and the nucleus (blue) in MDA-MB-231 WT, PISD, Drp1, and MFN2 cells stably expressing LifeAct-GFP (green) treated with vehicle control, hydrogen peroxide (H2O2, 100 μM), or N-acetyl cysteine (NAC, 1 mM). Arrows point to actin-rich migratory structures. Scale bar is 20 μm. Dot plots show number of actin-rich migratory structures per cell (B) or stress fiber density in each image (C) for each group (n = 10 images per group). Lines depict mean ± SD. Statistical analysis for this graph is in Tables S3 and S4.
Figure 5.
Enhanced ROS promotes actin reorganization and a less migratory phenotype in SUM159 TNBC cells
(A) Representative overlaid, zoomed images of immunofluorescence staining of mitochondria (pink) and the nucleus (blue) in MDA-MB-231 WT, PISD, Drp1, and MFN2 cells stably expressing LifeAct-GFP (green) treated with vehicle control, hydrogen peroxide (H2O2, 100 μM), or N-acetyl cysteine (NAC, 1 mM). Arrows point to actin-rich migratory structures. Scale bar is 20 μm. Dot plots show number of actin-rich migratory structures per cell (B) or stress fiber density in each image (C) for each group (n = 10 images per group). Lines depict mean ± SD. Statistical analysis for this graph is in Tables S5 and S6.
Effects of ROS on actin cytoskeleton and migration are mitochondrial-dependent
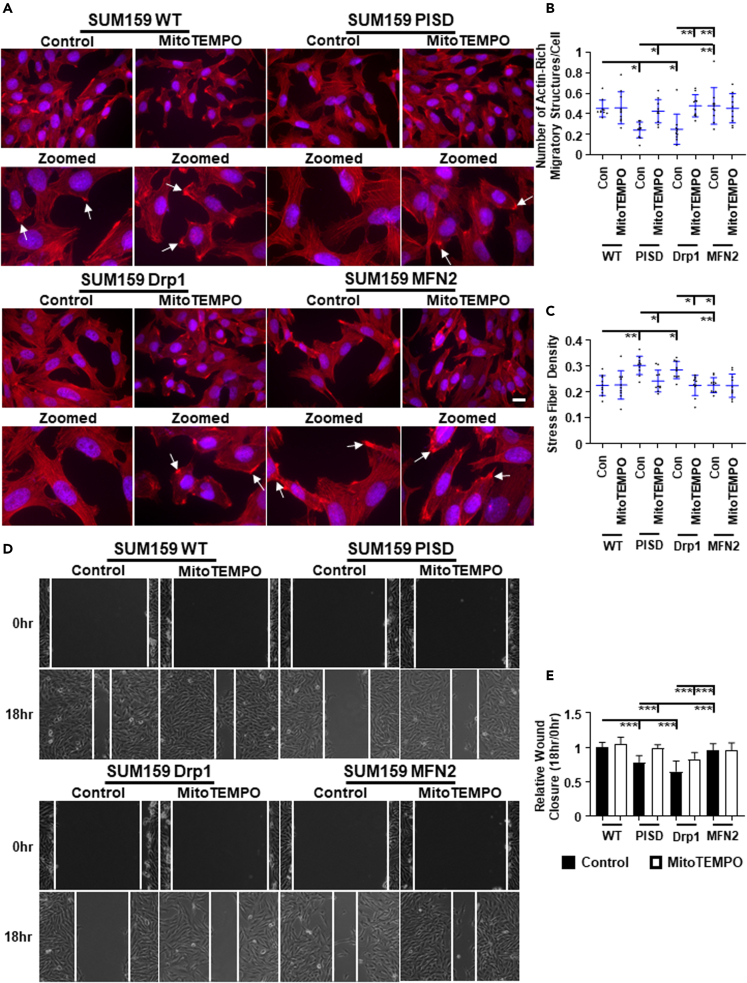
As H2O2 is a key ROS produced by mitochondria and NAC is a global ROS scavenger, we next investigated direct effects of mitochondrial ROS on actin cytoskeleton reorganization. We treated cells with MitoTEMPO, a mitochondrially targeted ROS scavenger,29 or vehicle control and then visualized actin with phalloidin staining. We found no significant difference in cell growth by bioluminescence in cells treated with MitoTEMPO compared to control cells (Figure S15). In cells with increased mitochondrial fusion, MitoTEMPO had little effect on the actin cytoskeleton in both MDA-MB-231 (Figures 6A–6C) and SUM159 (Figures 7A–7C) cells. However, in cells showing enhanced mitochondrial fission, MitoTEMPO drove actin cytoskeleton reorganization, promoted actin-rich migratory structures, and reduced stress fiber density in both MDA-MB-231 (Figures 6A–6C) and SUM159 (Figures 7A–7C) cells.
Figure 6.
Mitochondrial-generated ROS inhibits cell migration and actin-based migratory structures in MDA-MB-231 TNBC cells
(A) Representative overlaid and zoomed images of immunofluorescence staining of phalloidin (red) and nuclear DAPI (blue) of WT, PISD, Drp1, or MFN2 stably expressing cells treated with vehicle control or the mitochondrially targeted ROS scavenger MitoTEMPO (50 μM). Arrows point to actin-rich migratory structures. Dot plots show number of actin-rich migratory structures per cell (B) or stress fiber density in each image (C) for each group (n = 10 images per group). Lines depict mean ± SD. ∗ = p < 0.05. ∗∗ = p < 0.01. ∗∗∗ = p < 0.001.
(D) Representative images of wound healing cell migration assay in MDA-MB-231 TNBC cells displaying different mitochondrial morphologies after treatment with vehicle control or MitoTEMPO (50 μM) at the initial time (0 h) and 18 h after the wound was made. Scale bar is 100 μm.
(E) Summary of wound closure of cells after treatment with vehicle control or MitoTEMPO (50 μM) presented as mean +SD (n = 9) relative to the initial wound size (0 h). ∗∗∗ = p < 0.001.
Figure 7.
Mitochondrial-generated ROS inhibits cell migration and actin-based migratory structures in SUM159 TNBC cells
(A) Representative overlaid and zoomed images of immunofluorescence staining of phalloidin (red) and nuclear DAPI (blue) of WT, PISD, Drp1, or MFN2 stably expressing cells treated with vehicle control or the mitochondrially targeted ROS scavenger MitoTEMPO (50 μM). Arrows point to actin-rich migratory structures. Dot plots show number of actin-rich migratory structures per cell (B) or stress fiber density in each image (C) for each group (n = 10 images per group). Lines depict mean ± SD. ∗ = p < 0.05. ∗∗ = p < 0.01.
(D) Representative images of wound healing cell migration assay in SUM159 TNBC cells displaying different mitochondrial morphologies after treatment with vehicle control or MitoTEMPO (50 μM) at the initial time (0 h) and 18 h after the wound was made. Scale bar is 100 μm.
(E) Summary of wound closure of cells after treatment with vehicle control or MitoTEMPO (50 μM) presented as mean +SD (n = 9) relative to the initial wound size (0 h). ∗∗∗ = p < 0.001. (n = 10 images per group).
As MitoTEMPO caused significant reorganization of the actin cytoskeleton, we next investigated effects of mitochondrial ROS on cell migration. Consistent with phalloidin staining, we found that treatment with MitoTEMPO increased wound closure by cells with greater mitochondrial fission in both MDA-MB-231 (Figures 6D and 6E) and SUM159 (Figures 7D and 7E) cells. Conversely, MitoTEMPO had modest effects on the migratory capability of cells that displayed enhanced mitochondrial fusion.
Mitochondrial ROS inhibits cell migration by activating SHP-1/2
We next investigated a potential mechanism linking mitochondrial fission to suppression of TNBC cell migration. Previous studies have shown that Src homology region 2 domain-containing phosphatases 1 (SHP-1) and 2 (SHP-2) require ROS-dependent oxidation to activate and can interact with the actin cytoskeleton.21,22 Moreover, high mRNA levels of both SHP-1 and -2 correlate with better distant metastasis-free survival in TNBC patients (Figure S16). Therefore, we next identified whether SHP-1/2 are critical for the inhibitory effect of mitochondrial fission on migration in TNBC. To first determine if SHP-1/2 are active in cells with enhanced mitochondrial fission, we performed a Western blot for STAT3, a known downstream target of these phosphatases.30,31 We found decreased STAT3 phosphorylation in cells that display primarily mitochondrial fission and increased STAT3 phosphorylation in cells that display mitochondrial fusion (Figure S17), demonstrating that SHP-1/2 are more active in cells that display enhanced mitochondrial fission. To determine effects of active SHP-1/2 on TNBC migration, we next performed a wound healing assay in the presence of NSC87877, a dual SHP-1/2 inhibitor, or vehicle control. We found no significant difference in cell growth by bioluminescence in cells treated with NSC87877 compared to control cells (Figure S15). We found that in cells that display primarily mitochondrial fusion, inhibition of SHP-1/2 had no significant effect on MDA-MB-231 (Figures 8A and 8B) and SUM159 (Figures 8C and 8D) migration. However, in cells with greater mitochondrial fission, inhibition of SHP-1/2 significantly increased the migratory capability of these cells (Figures 8A–8D). Consistent with effects on migration, phalloidin staining after treatment with NSC87877 revealed increased actin-rich migratory structures in both MDA-MB-231 (Figure S18) and SUM159 (Figure S19) cells. We did not identify any effects of NSC87877 on mitochondrial morphology in either cell line (Figures S20 and S21). Together, these data suggest that enhanced mitochondrial fission reduces TNBC cell migration by activating SHP-1/2.
Figure 8.
Inhibition of SHP-1/2 promotes cell migration
Representative images of wound healing cell migration assay in MDA-MB-231 (A) and SUM159 (C) TNBC cells displaying different mitochondrial morphologies after treatment with vehicle control or the SHP-1/2 inhibitor NSC87877 (25 μM) at the initial time (0 h) and 18 h after the wound was made. Scale bar is 100 μm. Summary of wound closure of MDA-MB-231 (B) and SUM159 (D) cells after treatment with vehicle control or NSC87877 (25 μM) presented as mean +SD (n = 9) relative to the initial wound size (0 h). ∗ = p < 0.05. ∗∗ = p < 0.01. ∗∗∗ = p < 0.001.
Discussion
Here we provide evidence that mitochondrial fission inhibits TNBC cell migration through a ROS-SHP-1/2 mechanism. Using in vitro and in vivo studies, we demonstrate that enforcing mitochondrial fission not only increases endogenous ROS but also drives actin cytoskeleton reorganization to a less migratory state by reducing actin-rich migratory structures. Moreover, scavenging excess ROS in TNBC cells can reverse many inhibitory effects of mitochondrial fission. We also found that inhibitory effects of mitochondrial fission on migration and the actin cytoskeleton result in part from increased activation of the non-receptor tyrosine phosphatases SHP-1/2, based on data showing that inhibition of SHP-1/2 partially recovered effects on migration, formation of actin-rich migratory structures, and stress fiber density. These results highlight the importance of mitochondrial dynamics in TNBC and suggest mitochondria as a central node in regulation of the actin cytoskeleton.
The regulatory role of the actin cytoskeleton on mitochondrial function is well appreciated.25 However, only limited information exists about the reciprocal regulation of the actin cytoskeleton by mitochondrial dynamics. Activation of signaling cascades leads to actin cytoskeleton reorganization and underlies cell migration. Different levels of oxidative stress confer discordant outcomes in cancer cells; where mild oxidative stress promotes activation of cell signaling cascades and high levels cause cell death.32 Our observations suggest that increased ROS levels lead to actin cytoskeleton reorganization and reduced cell migration in TNBC cells. Here, we found that (1) cells with enhanced mitochondrial fission produced increased levels of ROS; (2) exogenous treatment with ROS or enforcing mitochondrial fission promoted actin cytoskeleton reorganization to a state that reduced formation of actin-rich migratory structures and promoted stress fiber density; and (3) treatment with a mitochondrially targeted ROS scavenger promoted formation of actin-rich migratory structures and cell migration, and reduced stress fiber density. Although we recognize that mitochondria form multiple ROS species and we consider ROS a general output from mitochondria for this manuscript, these observations, combined with others,33 suggest a novel ROS/actin-based mechanism by which mitochondrial dynamics controls TNBC cell migration.
Controversial reports exist on the function of ROS in cancer. Recent work demonstrated that increased levels of ROS produced by targeting critical antioxidant pathways reduced tumor initiation, proliferation, a stem-like phenotype, and metastasis.34,35,36,37 In support of these data, our current study demonstrates that increased levels of ROS limit cell migration and characteristics that define breast tumor progression in mice. Although we found that mitochondrial fission reduces actin-rich migratory structures and collagen alignment in the primary tumor, we found no differences in actin-rich migratory structures, collagen alignment, or metastasis between groups that displayed more mitochondrial fission or fusion. This result may be because we euthanized mice at a relatively early time point (3 weeks) after implantation to facilitate actin and collagen imaging by two photon microscopy. However, we showed that scavenging ROS can overcome the inhibitory effects of ROS on characteristics that define these processes in vitro. These data support an inhibitory function of ROS in TNBC and are consistent with the failure to identify patient benefit after reducing ROS with antioxidants or targeting major contributors to ROS production.38,39 Together, these data not only suggest clinical caution when utilizing antioxidant therapy but also that methods to enforce mitochondrial fission may be useful for TNBC.
We hypothesize that enhanced mitochondrial fission inhibits cell migration partially because of increased ROS levels. Indeed, we found that cells showing enhanced mitochondrial fission contain increased levels of ROS, which is consistent with previous studies.40,41 Mechanistically, we found that the ROS-dependent tyrosine phosphatases SHP-1/2 are important for this mitochondrial-dependent inhibition on migration. Intriguingly, SHP-1 and SHP-2 demonstrate discordant functions in cancer, where SHP-1 generally functions as a tumor suppressor and SHP-2 typically promotes oncogenic phenotypes.42,43 However, outcomes data show that increased expression of SHP-1 and SHP-2 correlate with better distant metastasis-free survival. In support of this data, we found decreased STAT3 phosphorylation in cells that displayed increased mitochondrial fission, suggesting increased SHP-1/2 activation in these cells. Furthermore, pharmacologic inhibition of SHP-1/2 partially reversed mitochondrial fission-dependent inhibition of migration in TNBC. Therefore, we propose that (i) an increase in oxidative stress promotes SHP-1/2 activation to inhibit cell migration; and (ii) elevated levels of SHP-1/2 might benefit TNBC patients. Our results support further mechanistic and prognostic studies of SHP-1/2 in TNBC.
Our data support an inhibitory function for mitochondrial fission in TNBC, consistent with previous work demonstrating that mitochondrial fusion promotes cell migration.44,45,46,47 As mitochondrial fission correlates with increased levels of ROS,40,41 we propose that mitochondrial fission inhibits tumorigenesis and tumor progression by regulating redox signaling. More specifically, the increase in ROS induced by mitochondrial fission activates SHP-1/2 to reduce the polymerization of actin-rich migratory structures. Therefore, data from our study provide strong rational for increasing mitochondrial fission or ROS as a therapeutic approach for TNBC. In summary, this study not only clarifies conflicting data on ROS but also suggest the potential benefit of enforcing mitochondrial fission in TNBC.
Limitations of the study
Our study uncovers an ROS-based mechanism that regulates TNBC cell migration. In this study, we utilized an immune-compromised mouse model to determine effects of ROS on actin-rich migratory structures in primary tumors and metastases. Although this model is important in pre-clinical studies, the immune system is important in cancer initiation and progression. Furthermore, other cell types of the microenvironment, such as fibroblasts, are instrumental in cancer progression. Therefore, more studies focused on these complex interactions are needed in the future to address the pathophysiological significance of ROS pathways in breast cancer.
In addition, here we investigated effects of genes that alter mitochondrial morphology within an overexpression context. To better understand the effects of mitochondria in TNBC cell migration and the actin cytoskeleton, future studies will need to expand this work by investigating effects of these mitochondrial morphology genes after knockdown or knockout and after rescue. These studies will further solidify mitochondrial morphology as a central node in the regulation of TNBC cell migration.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PISD (OTI4G5) Mouse mAb | ThermoFisher | Cat# MA5-26861;RRID:AB_2723486 |
| Drp1 (D6C7) Rabbit mAb | Cell Signaling Technology | Cat# 8570;RRID:AB_10950498 |
| Phospho-Drp1 (Ser616) (D9A1) Rabbit mAb | Cell Signaling Technology | Cat# 4494;RRID:AB_11178659 |
| MFN2-2 (D1E9) Rabbit mAb | Cell Signaling Technology | Cat# 11925;RRID:AB_2750893 |
| Phospho-STAT3 (Tyr705) (D3A7) XP® Rabbit mAb | Cell Signaling Technology | Cat# 9145;RRID:AB_2491009 |
| STAT3 (79D7) Rabbit mAb | Cell Signaling Technology | Cat# 4904;RRID:AB_331269 |
| β-actin (D6A8) Rabbit mAb | Cell Signaling Technology | Cat# 8457;RRID:AB_10950489 |
| Chemicals, peptides, and recombinant proteins | ||
| Hydrogen Peroxide (30% in Water) | Fisher BioReagents | Cat# BP2633500;CAS: 7722-84-1 |
| N-acetyl cysteine | Sigma-Aldrich | Cat# A7250;CAS: 616-91-1 |
| NSC87877 | Cayman Chemical | Cat# 14908;CAS: 56990-57-9 |
| MitoTracker Deep Red FM | ThermoFisher | Cat# M22426 |
| Hoechst 33342 | ThermoFisher | Cat# H1399 |
| Texas Red-X Phalloidin | ThermoFisher | Cat# T7471 |
| ProLong™ Gold with DAPI | ThermoFisher | Cat# P36930 |
| L-α-lysophosphatidylethanolamine | Avanti Polar Lipids | Cat# 860081;CAS: 97281-40-8 |
| Leflunomide | Cayman Chemical | Cat# 14860;CAS: 75706-12-6 |
| MitoTEMPO | Cayman Chemical | Cat# 16621;CAS: 1569257-94-8 |
| CellTracker Green CMFDA | ThermoFisher | Cat# C2925 |
| CellROX Green | ThermoFisher | Cat# C10444 |
| D-luciferin, Potassium Salt | Promega | Cat# E1605 |
| Polybrene | Sigma-Aldrich | Cat# TR-1003-G |
| Critical commercial assays | ||
| RNeasy Mini Kit | QIAGEN | Cat# 217004 |
| QuantiTect Reverse Transcrition Kit | QIAGEN | Cat# 205311 |
| QuantiTect SYBR Green PCR Kit | QIAGEN | Cat# 204143 |
| Halt™ Protease Inhibitor Cocktail | ThermoFisher | Cat# 78429 |
| Halt™ Phosphatase Inhibitor Single-Use | ThermoFisher | Cat# 78428 |
| Immobilon®-P Transfer Membrane | Millipore Sigma | Cat# IPVH00010 |
| NuPAGE MOPS SDS Running Buffer | Life Technologies | Cat# NP0001 |
| Pierce™ BCA Protein Assay Kit | ThermoFisher | Cat# 23225 |
| Experimental models: Cell lines | ||
| MDA-MB-231 | ATCC | Cat# CRM-HTB-26;RRID:CVCL_0062 |
| SUM159 | Gift from Dr. Stephen Ethier | N/A; RRID:CVCL_5423 |
| Experimental models: Organisms/strains | ||
| NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG™) mice | The Jackson Laboratory | Cat# 005557;RRID:IMSR_JAX:005557 |
| Oligonucleotides | ||
| KiCqStart® SYBR® Green Primers - PISD | Millipore Sigma | Cat# KSPQ12012; H_PISD_1 |
| KiCqStart® SYBR® Green Primers – Drp1 | Millipore Sigma | Cat# KSPQ12012; H_DNM1L_1 |
| KiCqStart® SYBR® Green Primers – MFN2 | Millipore Sigma | Cat# KSPQ12012; H_MFN2_1 |
| KiCqStart® SYBR® Green Primers – β-actin | IDT | Forward: 5’-TGT-ACG-TTG-CTA-TCC-AGG-CTG-TGC-3’; Reverse: 5’-CGG-TGA-GGA-TCT-TCA-TGA-GGT-AGT-C-3’ |
| Recombinant DNA | ||
| pLV.PGK.MFN2.P2A.hygromycin | VectorBuilder | NM_014874.4 |
| pLenti.PGK.LifeAct-GFP.W | Addgene | Cat# 51010;RRID:Addgene_51010 |
| Software and algorithms | ||
| cellSens | Olympus | https://www.olympus-lifescience.com/en/software/cellsens/ |
| MATLAB | MathWorks | https://www.mathworks.com/products/matlab.html |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Graphpad Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| Fluorobrite DMEM | ThermoFisher | Cat# A1896701 |
| 35mm glass bottom dish wih 20 mm micro-well #1.5 cover glass | Cellvis | Cat# D35-20-1.5-N |
Resource availability
Lead contact
Information and requests for resources should be directed to and be fulfilled by the Lead Contact, Gary D. Luker (gluker@med.umich.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cell culture
We purchased MDA-MB-231 cells from the ATCC (Manassas, VA) and cultured cells in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin (Pen/Strep) (Thermo Fisher Scientific, Waltham, MA), and 1% GlutaMAX (Thermo Fisher Scientific, Waltham, MA). We obtained SUM159 cells from Dr. Stephen Ethier and cultured cells in F-12 media supplemented with 10% fetal bovine serum, 1% Pen/Strep, 1% glutamine, 5 μg/mL hydrocortisone, and 1 μg/mL insulin. We authenticated all cells by analysis of short tandem repeats and characterized cells as free ofMycoplasma at the initial passage, and cultured cells in an anti-mycoplasma prophylactic (2.5 mg/mL, Plasmocin, InvivoGen, San Diego, CA). We used all cells within 3 months after resuscitation and maintained all cells at 37°C in a humidified incubator with 5% CO2.
Mouse studies
The University of Michigan IACUC approved all animal procedures (protocol 00008822). The animals used in this study received humane care in compliance with the principles of laboratory animal care formulated by the National Society for Medical Research and Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences and published by the National Institute of Health (Publication no NIH 85-23, revised 1996).
We established orthotopic tumor xenografts in the fourth inguinal mammary fat pads of 8-28-week-old female NSG mice (n = 20 mice total) as described previously,48 implanting 2 × 105 MDA-MB-231 wild type (WT), PISD, Drp1, or MFN2 cells stably expressing LifeAct-GFP cells. All injected cells stably expressed click beetle green (CBG) luciferase for bioluminescence imaging. We quantified tumor burden the day after (Day 1), 10 days (Day 11), and 20 days (Day 21) after implantation by bioluminescence imaging as previously described.49 When tumors reached 5-6 mm in diameter (Day 24), we resected orthotopic tumors to allow mice additional time to develop metastases. We weighed resected tumors and then immediately visualized collagen and actin in orthotopic tumors ex vivo by two-photon microscopy. We used 880 nm excitation and collected second harmonic signal from fibrillary collagen and emission from LifeAct-GFP with 410–460 nm and 495–540 nm filters, respectively.50 We continued to monitor metastatic burden by bioluminescence imaging one and 10 days after resection. Eleven days after resection, we euthanized mice and then immediately visualized collagen and actin in lung metastases ex vivo by two-photon microscopy as described above. We analyzed collagen alignment using custom MATLAB code as described previously.50 To calculate aspect ratios of cells in the primary tumor, we used Fluoview (Olympus) to analyze length and width of individual cells within each plane of a Z-stack. Briefly, we drew perpendicular lines across each cell and, based upon the line intensity profile, measured the long and short axis of the cells. For consistency, we used the longer value as the length and the smaller as the width for each cell.
Method details
Vectors and cell lines
We used MDA-MB-231 and SUM159 cells stably expressing phosphatidylserine decarboxylase (PISD) or dynamin related protein 1 (Drp1) as well as mitochondrially-targeted BFP (Mito-BFP) to visualize mitochondrial morphology as used in our recent study.24 For stable expression of mitofusin 2 (MFN2), we designed and purchased a MFN2 vector from VectorBuilder (Chicago, IL, USA). This pLV-based vector contains a human PGK promoter, and we added a hygromycin selection marker after the MFN2 open reading frame (NM_014874.4) separated by a P2A sequence (MFN2-hygromycin). For direct visualization of the actin cytoskeleton, we used LifeAct-GFP as used in our previous study.50 We produced recombinant lentiviral vectors for MFN2-hygromycin and LifeAct-GFP as previously described51 and generated cells as we have done previously.24 Briefly, we produced stocks of lentivirus by transient transfection of HEK293T cells. MDA-MB-231 and SUM159 cells were transduced at a multiplicity of infection (MOI) of 2 in the presence of 8 μg/mL polybrene.
Fluorescence microscopy
We visualized effects of ROS, ROS scavenging, SHP-1/2 inhibition on mitochondrial morphology, and mitochondrial morphology on actin cytoskeleton in live cells as described previously.24 Briefly, we seeded 2 × 104 cells onto 35-mm dishes with a 20-mm glass bottom insert (Cellvis, Mountain View, CA) in FluoroBrite DMEM media (ThermoFisher Scientific), containing 10% FBS, 1% Pen/Strep, 1% GlutaMAX, and 1% Sodium Pyruvate (ThermoFisher Scientific, Waltham, MA). We also added 5 μg/mL hydrocortisone and 1 μg/mL insulin for the SUM159 cells. Two and three days after seeding, we changed media to FluoroBrite DMEM as described above containing either hydrogen peroxide (H2O2; 100 μM) as a source of ROS, N-acetyl cysteine (NAC; 1 mM) to globally scavenge ROS, or vehicle control. For SHP-1/SHP-2 inhibition, we added NSC87877 (Cayman Chemical; 25 μM) or vehicle control 24 hours before visualization. We acquired all images four days after seeding on an Olympus IX73 microscope with a DP80 CCD camera using cellSens software (Olympus). For mitochondria and actin co-staining, on day four we co-stained dishes with MitoTracker Deep Red FM (ThermoFisher Scientific) and Hoechst 33342 (ThermoFisher Scientific) for 30 mins in the incubator and acquired images as described above. We analyzed mitochondrial morphology, actin-rich migratory structures, and stress fiber density as described in our previous publications.24,50 For mitochondrial morphological analysis we used MATLAB to skeletonize mitochondria and measure total pixel area and major axis length for all mitochondria within an image. We separated mitochondria into fused or fissioned groups by area and major axis length using a user-defined cutoff that we applied to every cell group analyzed in each experiment. We categorized an identified mitochondrion that had total area/majoraxislength >2 as networked and did not include these mitochondria in the analysis.24 We counted actin-rich migratory structures by hand for each image using accepted visual presentations of these structures, and analyzed stress fiber density (stress fiber area divided by total cell size) with our custom MATLAB code as described in our previous publication.50 Briefly, we identified stress fibers through measuring connected ridges in the horizontal and vertical directions using a user-defined peak height and size cutoff that we applied to every cell group analyzed in each experiment. We also identified individual cells and, using the regionprops command, calculated stress fiber and total cell pixel area.
To visualize the actin cytoskeleton only, we seeded 2.0 × 104 cells on glass cover slips in the appropriate FluoroBrite media within a 6-well plate. We changed media two and three days after seeding. Four days after seeding we fixed cells with 4% formaldehyde for 15 minutes at room temperature (RT) and then washed three times with PBS for 5 minutes each. We permeabilized cells with 0.1% Triton X-100 in PBS for 3 minutes at RT, washed with PBS for 5 minutes, and blocked with 10% goat serum in 0.05% Triton X-100 in PBS for 1 hour at RT. Next, we incubated cells with a Texas Red-X phalloidin (ThermoFisher Scientific) for actin for 1 hour at RT in the dark. We washed the slides and mounted with medium containing an anti-fade reagent and DAPI (ProLong™ Gold; ThermoFisher Scientific). We counted actin-rich migratory structures by hand for each image using accepted visual presentations of these structures, and analyzed stress fiber density (stress fiber area divided by total cell size) with our custom MATLAB code as described in our previous publication.50
Wound healing assays
We performed wound healing assays as previously described.50 Briefly, we seeded 2 x 104 cells into 35 mm plastic dishes in the appropriate medium described above. For initial effects of morphology on wound healing, two and three days after seeding, we changed dishes to media containing either L-α-lysophosphatidylethanolamine (LPE; Avanti Polar Lipids Inc., Alabaster, AL, USA; 50 μM) to drive fragmentation, leflunomide (Lef; Cayman Chemical Company, Ann Arbor, MI, USA; 50 μM) to drive fusion, or vehicle control as described previously.24 For ROS-mediated effects on wound healing, both two and three days after seeding, we changed dishes to media containing either hydrogen peroxide (H2O2, 100 μM), N-acetyl cysteine (NAC, 1 mM), or vehicle control as described above. Once cells formed confluent monolayers four days after seeding, we created a linear scratch with a 200 μL pipette tip. After the scratch, we washed dishes once with phosphate-buffered saline (PBS), added fresh medium, and placed the plate into the incubator. For MitoTEMPO (Cayman Chemical; 50 μM) and NSC87877 (Cayman Chemical; 25 μM) assays, we added these compounds or vehicle controls 24 hours before and after we made the wound. We acquired images on an Olympus IX73 microscope with a DP80 CCD camera using cellSens software (Olympus) at the time points indicated in the figure. We quantified wound healing results as:[1 − (wound width at 18 hours/wound width at 0 hour)] × 100.
Growth and adhesion assays
We analyzed effects of H2O2, NAC, MitoTEMPO, and NSC87877 on cell growth for the length of time indicated in the figures using bioluminescence imaging for CBG with medium binning and 30 second exposure as previously described.49 Cells were incubated with 150 μg/mL luciferin for 5 minsat 37°C before bioluminescence imaging.
For adhesion assays, we seeded 1 × 105 HMFs expressing mCherry into a 24-well plate 48 hours before the assay as described previously.50 After 48 hours, we seeded 2.5 × 105MDA-MB-231 and SUM159 wild type (WT), PISD, Drp1, or MFN2 stably expressing cells, stained with CellTracker Green CMFDA (ThermoFisher Scientific) following the manufacturers protocol, onto the HMFs and incubated at 37°C for 15 minutes. We removed non-adherent cells with PBS and visualized adherent cancer cells by fluorescence imaging. We present data as the number of adherent cells to total number of cells seeded relative to matched WT cells.
Flow cytometry
To determine total amount of ROS in cells displaying different mitochondrial morphologies or in cells treated with ROS or ROS scavenging compound, we stained cells with CellROX Green (ThermoFisher Scientific) and analyzed cells following the manufacturers protocol, analyzing at least 10,000 total events for each group.52
Western blotting
We lysed cells using RIPA buffer and detected amounts of target proteins in total cell lysates by western blotting as described previously53 after seeding cells as described in the wound healing assays section. For mitochondrial morphology proteins, we probed total cell lysates for PISD (ThermoFisher Scientific), Drp1 (Cell Signaling Technology), pDrp1S616 (Cell Signaling Technology), MFN2 (Cell Signaling Technology), and β-actin (Cell Signaling Technology). For SHP1/2 activity, we probed total cell lysates for phospho-STAT3 Tyr705 (Cell Signaling Technology) followed by total STAT3 (Cell Signaling Technology) as a loading control. We quantified relative intensities of bands with ImageJ (National Institutes of Health, Bethesda, MD, USA).
Quantification and statistical analysis
Data represent experiments repeated independently three times. For experiments comparing multiple groups, we used one-way ANOVA and Tukey’s multiple comparisons test. We considered p < 0.05 as statistically significant (∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001). We prepared bar graphs (mean values +SD or SEM as denoted in figure legends), dot plots, and boxplots and whiskers using GraphPad Prism 8. For dot plot, bars denote the mean ± SD as identified in the figure legends. For boxplots and whiskers, the bottom and top of a box define the first and third quartiles, and the band inside the box marks the second quartile (median). The ends of the whiskers represent the 10th and 90th percentiles, respectively. For boxplots and whiskers, the “+” within the box refers to the mean.
Acknowledgments
We thank Sean Linkes for assistance with Flow cytometry experiments and interpretation. We acknowledge support from the Flow Cytometry Core and the Center for Molecular Imaging Core of the University of Michigan Medical School’s Biomedical Research Core Facilities supported by US National Institutes of Health grant P30CA046592. We acknowledge funding from US National Institutes of Health grants R01CA238042, U01CA210152, R01CA238023, R33CA225549, U24 CA237683, and R37CA222563. We also acknowledge funding from the W.M. Keck Foundation.
Author contributions
Conceptualization, B.A.H. and G.D.L.; Methodology, B.A.H., J.M.B., S.R., and G.D.L.; Investigation, B.A.H., A.Z., J.M.B., A.B., A.F., S.R., and A.C.C.; Formal analysis, B.A.H., A.Z., J.M.B., A.B., A.F., and S.R.; Visualization, B.A.H., A.Z., and J.M.B.; Writing – Original Draft, B.A.H. and G.D.L.; Writing – Review and Editing, B.A.H., A.Z., J.M.B., A.B., A.F., S.R., A.C.C., and G.D.L.; Funding Acquisition, G.D.L.; Supervision, G.D.L. The authors read and approved the final manuscript.
Declaration of interests
GDL has received research funding from InterAx Biotech AG and Polyphor (now part of Spexis). The remaining authors declare no competing interests.
Published: May 3, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106788.
Supplemental information
Data and code availability
-
•
All data has been included in main figures or supplemental information. All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
References
- 1.Ward P.S., Thompson C.B. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshida G.J. Metabolic reprogramming: the emerging concept and associated therapeutic strategies. J. Exp. Clin. Cancer Res. 2015;34:111. doi: 10.1186/s13046-015-0221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liesa M., Palacín M., Zorzano A. Mitochondrial dynamics in mammalian health and disease. Physiol. Rev. 2009;89:799–845. doi: 10.1152/physrev.00030.2008. [DOI] [PubMed] [Google Scholar]
- 4.Roth K.G., Mambetsariev I., Kulkarni P., Salgia R. The mitochondrion as an emerging therapeutic target in cancer. Trends Mol. Med. 2020;26:119–134. doi: 10.1016/j.molmed.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Picard M., Burelle Y. Mitochondria: starving to reach quorum?: insight into the physiological purpose of mitochondrial fusion. Bioessays. 2012;34:272–274. doi: 10.1002/bies.201100179. [DOI] [PubMed] [Google Scholar]
- 6.Wallace D.C., Fan W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010;10:12–31. doi: 10.1016/j.mito.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan L.B., Chandel N.S. Mitochondrial reactive oxygen species and cancer. Cancer Metab. 2014;2:17. doi: 10.1186/2049-3002-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J., Wang X., Vikash V., Ye Q., Wu D., Liu Y., Dong W. ROS and ROS-mediated cellular signaling. Oxid. Med. Cell. Longev. 2016;2016:4350965. doi: 10.1155/2016/4350965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szatrowski T.P., Nathan C.F. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991;51:794–798. [PubMed] [Google Scholar]
- 10.Gao P., Zhang H., Dinavahi R., Li F., Xiang Y., Raman V., Bhujwalla Z.M., Felsher D.W., Cheng L., Pevsner J., et al. HIG-dependent antitumorigenic effect of antioxidants in vivo. Cancer Cell. 2007;12:230–238. doi: 10.1016/j.ccr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinberg F., Hamanaka R., Wheaton W.W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G.M., Budinger G.R.S., Chandel N.S. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. USA. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris I.S., Treloar A.E., Inoue S., Sasaki M., Gorrini C., Lee K.C., Yung K.Y., Brenner D., Knobbe-Thomsen C.B., Cox M.A., et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell. 2015;27:211–222. doi: 10.1016/j.ccell.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 13.Piskounova E., Agathocleous M., Murphy M.M., Hu Z., Huddlestun S.E., Zhao Z., Leitch A.M., Johnson T.M., DeBerardinis R.J., Morrison S.J. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527:186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michalek R.D., Nelson K.J., Holbrook B.C., Yi J.S., Stridiron D., Daniel L.W., Fetrow J.S., King S.B., Poole L.B., Grayson J.M. The requirement of reversible cysteine sulfenic acid formation for T cell activation and function. J. Immunol. 2007;179:6456–6467. doi: 10.4049/jimmunol.179.10.6456. [DOI] [PubMed] [Google Scholar]
- 15.Crump K.E., Juneau D.G., Poole L.B., Haas K.M., Grayson J.M. The reversible formation of cysteine sulfenic acid promotes B-cell activation and proliferation. Eur. J. Immunol. 2012;42:2152–2164. doi: 10.1002/eji.201142289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yo K., Iwata S., Hashizume Y., Kondo S., Nomura S., Hosono O., Kawasaki H., Tanaka H., Dang N.H., Morimoto C. SHP-2 inhibits tyrosine phosphorylation of Cas-L and regulates cell migration. Biochem. Biophys. Res. Commun. 2009;382:210–214. doi: 10.1016/j.bbrc.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Bard-Chapeau E.A., Li S., Ding J., Zhang S.S., Zhu H.H., Princen F., Fang D.D., Han T., Bailly-Maitre B., Poli V., et al. Ptpn11/Shp2 acts as a tumor suppressor in hepatocellular carcinogenesis. Cancer Cell. 2011;19:629–639. doi: 10.1016/j.ccr.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan L.C., Shiau C.W., Tai W.T., Hung M.H., Chu P.Y., Hsieh F.S., Lin H., Yu H.C., Chen K.F. SHP-1 is a negative regulator of epithelial-mesenchymal transition in hepatocellular carcinoma. Oncogene. 2015;34:5252–5263. doi: 10.1038/onc.2014.445. [DOI] [PubMed] [Google Scholar]
- 19.Joo M.K., Park J.J., Yoo H.S., Lee B.J., Chun H.J., Lee S.W., Bak Y.T. Epigenetic regulation and anti-tumorigenic effects of SH2-containing protein tyrosine phosphatase 1 (SHP1) in human gastric cancer cells. Tumour Biol. 2016;37:4603–4612. doi: 10.1007/s13277-015-4228-y. [DOI] [PubMed] [Google Scholar]
- 20.Wen L.Z., Ding K., Wang Z.R., Ding C.H., Lei S.J., Liu J.P., Yin C., Hu P.F., Ding J., Chen W.S., et al. SHP-1 acts as a tumor suppressor in hepatocarcinogenesis and HCC progression. Cancer Res. 2018;78:4680–4691. doi: 10.1158/0008-5472.CAN-17-3896. [DOI] [PubMed] [Google Scholar]
- 21.Varone A., Amoruso C., Monti M., Patheja M., Greco A., Auletta L., Zannetti A., Corda D. The phosphatase Shp1 interacts with and dephosphorylates cortactin to inhibit invadopodia function. Cell Commun. Signal. 2021;19:64. doi: 10.1186/s12964-021-00747-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacGillivray M., Herrera-Abreu M.T., Chow C.W., Shek C., Wang Q., Vachon E., Feng G.S., Siminovitch K.A., McCulloch C.A.G., Downey G.P. The protein tyrosine phosphatase SHP-2 regulates interleukin-1-induced ERK activation in fibroblasts. J. Biol. Chem. 2003;278:27190–27198. doi: 10.1074/jbc.M213083200. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y.C., Humphries B., Brien R., Gibbons A.E., Chen Y.T., Qyli T., Haley H.R., Pirone M.E., Chiang B., Xiao A., et al. Functional isolation of tumor-initiating cells using microfluidic-based migration identifies phosphatidylserine decarboxylase as a key regulator. Sci. Rep. 2018;8:244. doi: 10.1038/s41598-017-18610-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humphries B.A., Cutter A.C., Buschhaus J.M., Chen Y.C., Qyli T., Palagama D.S.W., Eckley S., Robison T.H., Bevoor A., Chiang B., et al. Enhanced mitochondrial fission suppresses signaling and metastasis in triple-negative breast cancer. Breast Cancer Res. 2020;22:60. doi: 10.1186/s13058-020-01301-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Illescas M., Peñas A., Arenas J., Martín M.A., Ugalde C. Regulation of mitochondrial function by the actin cytoskeleton. Front. Cell Dev. Biol. 2021;9:795838. doi: 10.3389/fcell.2021.795838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keren K., Pincus Z., Allen G.M., Barnhart E.L., Marriott G., Mogilner A., Theriot J.A. Mechanism of shape determination in motile cells. Nature. 2008;453:475–480. doi: 10.1038/nature06952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conklin M.W., Eickhoff J.C., Riching K.M., Pehlke C.A., Eliceiri K.W., Provenzano P.P., Friedl A., Keely P.J. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 2011;178:1221–1232. doi: 10.1016/j.ajpath.2010.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trnka J., Blaikie F.H., Smith R.A.J., Murphy M.P. A mitochondria-targeted nitroxide is reduced to its hydroxylamine by ubiquinol in mitochondria. Free Radic. Biol. Med. 2008;44:1406–1419. doi: 10.1016/j.freeradbiomed.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 30.Huang T.T., Su J.C., Liu C.Y., Shiau C.W., Chen K.F. Alteration of SHP-1/p-STAT3 signaling: a potential target for anticancer therapy. Int. J. Mol. Sci. 2017;18:1234. doi: 10.3390/ijms18061234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tai W.T., Cheng A.L., Shiau C.W., Huang H.P., Huang J.W., Chen P.J., Chen K.F. Signal transducer and activator of transcription 3 is a major kinase-independent target of sorafenib in hepatocellular carcinoma. J. Hepatol. 2011;55:1041–1048. doi: 10.1016/j.jhep.2011.01.047. [DOI] [PubMed] [Google Scholar]
- 32.Reuter S., Gupta S.C., Chaturvedi M.M., Aggarwal B.B. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic. Biol. Med. 2010;49:1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luanpitpong S., Talbott S.J., Rojanasakul Y., Nimmannit U., Pongrakhananon V., Wang L., Chanvorachote P. Regulation of lung cancer cell migration and invasion by reactive oxygen species and caveolin-1. J. Biol. Chem. 2010;285:38832–38840. doi: 10.1074/jbc.M110.124958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S., et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohta T., Iijima K., Miyamoto M., Nakahara I., Tanaka H., Ohtsuji M., Suzuki T., Kobayashi A., Yokota J., Sakiyama T., et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 36.Singh A., Boldin-Adamsky S., Thimmulappa R.K., Rath S.K., Ashush H., Coulter J., Blackford A., Goodman S.N., Bunz F., Watson W.H., et al. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–7984. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diehn M., Cho R.W., Lobo N.A., Kalisky T., Dorie M.J., Kulp A.N., Qian D., Lam J.S., Ailles L.E., Wong M., et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Casas A.I., Nogales C., Mucke H.A.M., Petraina A., Cuadrado A., Rojo A.I., Ghezzi P., Jaquet V., Augsburger F., Dufrasne F., et al. On the clinical pharmacology of reactive oxygen species. Pharmacol. Rev. 2020;72:801–828. doi: 10.1124/pr.120.019422. [DOI] [PubMed] [Google Scholar]
- 39.Forman H.J., Zhang H. Targeting oxidative stress in disease: promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021;20:689–709. doi: 10.1038/s41573-021-00233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu T., Robotham J.L., Yoon Y. Increased production of reactive oxygen species in hyperglycemic conditions requires dynamic change of mitochondrial morphology. Proc. Natl. Acad. Sci. USA. 2006;103:2653–2658. doi: 10.1073/pnas.0511154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu T., Sheu S.S., Robotham J.L., Yoon Y. Mitochondrial fission mediates high glucose-induced cell death through elevated production of reactive oxygen species. Cardiovasc. Res. 2008;79:341–351. doi: 10.1093/cvr/cvn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J., Zhang F., Niu R. Functions of Shp2 in cancer. J. Cell Mol. Med. 2015;19:2075–2083. doi: 10.1111/jcmm.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varone A., Spano D., Corda D. Shp1 in solid cancers and their therapy. Front. Oncol. 2020;10:935. doi: 10.3389/fonc.2020.00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caino M.C., Ghosh J.C., Chae Y.C., Vaira V., Rivadeneira D.B., Faversani A., Rampini P., Kossenkov A.V., Aird K.M., Zhang R., et al. PI3K therapy reprograms mitochondrial trafficking to fuel tumor cell invasion. Proc. Natl. Acad. Sci. USA. 2015;112:8638–8643. doi: 10.1073/pnas.1500722112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rivadeneira D.B., Caino M.C., Seo J.H., Angelin A., Wallace D.C., Languino L.R., Altieri D.C. Survivin promotes oxidative phosphorylation, subcellular mitochondrial repositioning, and tumor cell invasion. Sci. Signal. 2015;8:ra80. doi: 10.1126/scisignal.aab1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caino M.C., Seo J.H., Aguinaldo A., Wait E., Bryant K.G., Kossenkov A.V., Hayden J.E., Vaira V., Morotti A., Ferrero S., et al. A neuronal network of mitochondrial dynamics regulates metastasis. Nat. Commun. 2016;7:13730. doi: 10.1038/ncomms13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu M.J., Chen Y.S., Kim M.R., Chang C.C., Gampala S., Zhang Y., Wang Y., Chang C.Y., Yang J.Y., Chang C.J. Epithelial-Mesenchymal transition directs stem cell polarity via regulation of mitofusin. Cell Metab. 2019;29:993–1002.e6. doi: 10.1016/j.cmet.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 48.Luker K.E., Mihalko L.A., Schmidt B.T., Lewin S.A., Ray P., Shcherbo D., Chudakov D.M., Luker G.D. In vivo imaging of ligand receptor binding with Gaussia luciferase complementation. Nat. Med. 2011;18:172–177. doi: 10.1038/nm.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cavnar S.P., Rickelmann A.D., Meguiar K.F., Xiao A., Dosch J., Leung B.M., Cai Lesher-Perez S., Chitta S., Luker K.E., Takayama S., Luker G.D. Modeling selective elimination of quiescent cancer cells from bone marrow. Neoplasia. 2015;17:625–633. doi: 10.1016/j.neo.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Humphries B.A., Buschhaus J.M., Chen Y.C., Haley H.R., Qyli T., Chiang B., Shen N., Rajendran S., Cutter A., Cheng Y.H., et al. Plasminogen activator inhibitor 1 (PAI1) promotes actin cytoskeleton reorganization and glycolytic metabolism in triple-negative breast cancer. Mol. Cancer Res. 2019;17:1142–1154. doi: 10.1158/1541-7786.MCR-18-0836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith M.C.P., Luker K.E., Garbow J.R., Prior J.L., Jackson E., Piwnica-Worms D., Luker G.D. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 2004;64:8604–8612. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- 52.Buschhaus J.M., Rajendran S., Humphries B.A., Cutter A.C., Muñiz A.J., Ciavattone N.G., Buschhaus A.M., Cañeque T., Nwosu Z.C., Sahoo D., et al. Effects of iron modulation on mesenchymal stem cell-induced drug resistance in estrogen receptor-positive breast cancer. Oncogene. 2022;41:3705–3718. doi: 10.1038/s41388-022-02385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luker K., Gupta M., Luker G. Bioluminescent CXCL12 fusion protein for cellular studies of CXCR4 and CXCR7. Biotechniques. 2009;47:625–632. doi: 10.2144/000113126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data has been included in main figures or supplemental information. All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.