Abstract
The prevalence of seasonal human coronavirus (HCoV) infections in early childhood and adults has not been well analyzed in longitudinal serological studies. Here we analyzed the changes in HCoV (229E, HKU1, NL63, OC43, MERS, and SARS-CoV-2) spike-specific antibody levels in follow-up serum specimens of 140 children at the age of 1, 2, and 3 years, and of 113 healthcare workers vaccinated for Covid-19 with BNT162b2-vaccine. IgG antibody levels against six recombinant HCoV spike subunit 1 (S1) proteins were measured by enzyme immunoassay. We show that by the age of three years the cumulative seropositivity for seasonal HCoVs increased to 38–81% depending on virus type. BNT162b2 vaccinations increased anti-SARS-CoV-2 S1 antibodies, but no increase in seasonal coronavirus antibodies associated with vaccinations. In healthcare workers (HCWs), during a 1-year follow-up, diagnostic antibody rises were seen in 5, 4 and 14% of the cases against 229E, NL63 and OC43 viruses, respectively, correlating well with the circulating HCoVs. In 6% of the HCWs, a diagnostic antibody rise was seen against S1 of HKU1, however, these rises coincided with anti-OC43 S1 antibody rises. Rabbit and guinea pig immune sera against HCoV S1 proteins indicated immunological cross-reactivity within alpha-CoV (229E and NL63) and beta-CoV (HKU1 and OC43) genera.
Subject terms: SARS-CoV-2, RNA vaccines, Antibodies
Introduction
Of the seven human coronaviruses (HCoVs) identified to date, SARS-CoV-2 and the four seasonal coronaviruses, 229E, HKU1, NL63, and OC43, are endemic worldwide causing variable morbidity in different populations. Middle East Respiratory syndrome virus (MERS) is endemic in limited geographic areas such as the Arabic peninsula and SARS after causing a relatively worrisome epidemic in many countries has been extinct since 2003. Seasonal coronaviruses have been estimated to cause 15–30% of the upper respiratory tract infections1, with mild symptoms in the majority of cases, although severe pediatric respiratory infections can occur2–4.
Maternal antibodies may provide immune protection against viral infections during the first six months of life5,6. After the disappearance of maternal antibodies, at the time of increasing human contacts e.g. in the daycare environment, the infants are more susceptible to respiratory virus infections such as those caused by seasonal HCoVs. The prevalence of HCoV infections in early childhood has not been very well characterized. Later in life, during adolescence and adulthood, most individuals are likely (re)exposed to seasonal HCoVs, which leads to most adults having persistent antibody levels against the different HCoVs7,8. SARS-CoV-2 infections induce the production of antibodies especially against SARS-CoV-2 spike protein (S), nucleoprotein (N), or both9,10. The majority of serological studies on seasonal HCoVs have focused on the presence of anti-N antibodies8,11–14. However, a more comprehensive and specific picture on the rate of HCoV infections and reinfections is obtained by analyzing the presence of anti-S antibodies. Cross-protection of pre-existing anti-seasonal HCoV antibodies against SARS-CoV-2 has been under an intensive discussion15–17. Also, the potential role of COVID-19 vaccination in the protection against seasonal HCoV infections has remained uncharacterized.
Here we describe the seropositivity rates and IgG antibody levels against HCoV spike subunit 1 (S1) proteins in sequentially collected serum samples of Finnish children, and in BNT162b2-vaccinated healthcare workers (HCWs) coupled with data on RT-PCR-confirmed circulation of HCoVs in the community. Our data demonstrate increasing seropositivity for HCoV S1-binding IgG antibodies in early childhood, and a good correlation of these antibodies with the corresponding nucleoprotein (N) binding IgG antibodies. Changes in HCoV antibody levels in the follow-up of HCWs match well with the data on circulating HCoVs. BNT162b2-vaccination has no effect on anti-S1 antibody levels against seasonal HCoVs or MERS.
Results
Based on the overall lower amino acid sequence identity (Supplementary Table 1) and on our previous work with SARS-CoV-210, spike protein subunit 1 (S1) was selected as the antigen to set up an enzyme immunoassay (EIA) for the detection of HCoV S binding IgG antibodies. S1 subunits were produced as mFc-fusion (S1-mFc) proteins in HEK-293F cells, purified (Supplementary Fig. 1) and used in EIA.
Seropositivity and HCoV spike-specific IgG antibody levels in children of 1 to 3 years of age
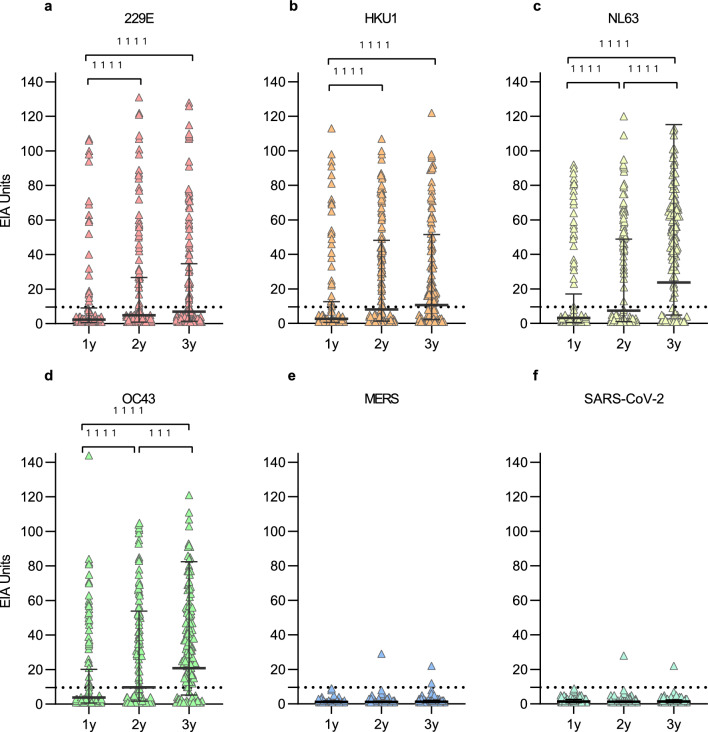
Serum specimens collected in 2009–2013 from 140 healthy children (74 male and 66 female, Table 1) at 1, 2, and 3 years of age were analyzed by EIA for antibodies against S1 proteins of four seasonal HCoVs (229E, HKU1, NL63, OC43), MERS and SARS-CoV-2. The absorbance values were converted to EIA units enabling reliable identification of seropositive samples and to compare antibody levels against different coronavirus species (Fig. 1). An increase in the geometric mean antibody levels between 1- and 2-year samples was significant for all four seasonal HCoVs (p < 0.0001) while the change between 2 and 3 years was significant only for NL63 and OC43 (p < 0.0001 and p = 0.0004, respectively). One participant had low levels of MERS and SARS-CoV-2 S1-binding IgG antibodies at 2 and 3 years of age.
Table 1.
Characteristics of the study cohorts and serum sampling intervals.
| Children | COVID-19 vaccinated HCWs | |
|---|---|---|
| N | 140 | 113 |
| Female (%) | 66 (47%) | 104 (92%) |
| Male (%) | 74 (53%) | 9 (8%) |
| Months | Years | |
|---|---|---|
| Age 1st sampling (mean and range) | 13.7 (11.6–16.6) | 43 (25–65) |
| Age 2nd sampling (mean and range) | 25.3 (23.8–27.4) | |
| Age 3rd sampling (mean and range) | 37.5 (36.1–39.6) |
| Sampling interval | Months | Months |
|---|---|---|
| Time between 1st and 2nd sample | 11.6 (7.8–14.7) | 2.3 (1.3–6.2) |
| Time between 2nd and 3rd sample | 12.1 (10.3–14.6) | 8.5 (7.5–10.5) |
Figure 1.
HCoV S1-specific IgG antibody responses in children at 1, 2 and 3 years of age. HCoV S1-specific IgG antibody levels were measured with EIA from sera collected from 140 children at 1, 2, and 3 years of age, during the years 2009 to 2013. Geometric means and geometric standard deviations of antibody levels are shown. Differences between the groups were analyzed using Wilcoxon matched pairs test. Two-tailed P-values < 0.05 were considered statistically significant. P-values are < 0.0001 (1111) and 0.0004 (111). Dashed line indicates the cutoff value for seropositivity.
The rate of antibody positive children increased by age for the four seasonal HCoVs and the antibody levels remained elevated in most of the seropositive children (Table 2). In all age groups the seropositivity was the highest for OC43 with a seropositivity of 31% in 1-year-olds and 81% in 3-year-olds. At 3 years of age the seropositivity rate for 229E (37%) was lower than for the other seasonal HCoVs (59% for HKU1, 76% for NL63, and 81% for OC43). It is noteworthy that the cumulative seropositivity for HKU1 declined between 2 and 3 years of age, while in 229E, NL63 and OC43 the annual and cumulative seropositivity rates increased almost at the same rate (Table 2). A decline in anti-HKU1 S1 antibodies was observed in previously seropositive children whose sequential samples showed a decrease in geometric mean IgG antibody levels (GMALs) from 2 to 3 years (44 to 33 EIA units). For 229E, NL63, and OC43 the mean IgG antibody levels in seropositive children remained relatively stable during the 3-year follow-up. Gender had no significant effect on the HCoV S1 antibodies in the studied children at any time point (Supplementary Fig. 2).
Table 2.
Seroprevalence of IgG antibodies against HCoV S1 in 140 children at ages of 1, 2, and 3 years.
| Age | Seropositivity | Cumulative seropositivity | Antibody level in seropositive (EIA units) | Reinfection rate | |||||
|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | GMAL ± SD | CI 95% | n | % | ||
| 229E | 1y | 20 | 14 | 20 | 14 | 43 ± 2.1 | 30–61 | – | – |
| 2y | 44 | 31 | 45 | 32 | 46 ± 2.0 | 37–57 | 5 | 25 | |
| 3y | 52 | 37 | 53 | 38 | 45 ± 2.0 | 37–55 | 11 | 24 | |
| HKU1 | 1y | 29 | 21 | 29 | 21 | 41 ± 2.1 | 31–55 | – | – |
| 2y | 69 | 49 | 73 | 52 | 44 ± 1.9 | 37–51 | 4 | 14 | |
| 3y | 83 | 59 | 93 | 66 | 33 ± 2.0 | 29–39 | 6 | 8 | |
| NL63 | 1y | 34 | 24 | 34 | 24 | 54 ± 1.6 | 45–64 | – | – |
| 2y | 63 | 45 | 64 | 46 | 55 ± 1.5 | 49–61 | 7 | 21 | |
| 3y | 107 | 76 | 108 | 77 | 54 ± 1.5 | 50–59 | 11 | 17 | |
| OC43 | 1y | 43 | 31 | 43 | 31 | 39 ± 1.8 | 32–47 | – | – |
| 2y | 76 | 54 | 79 | 56 | 42 ± 1.8 | 37–48 | 14 | 33 | |
| 3y | 114 | 81 | 114 | 81 | 38 ± 1.8 | 34–42 | 15 | 19 | |
| Any HCoV | 1y | 88 | 63 | 88 | 63 | 44 ± 1.9 | 39–49 | – | – |
| 2y | 119 | 85 | 120 | 86 | 46 ± 1.8 | 43–49 | 27 | 31 | |
| 3y | 137 | 98 | 137 | 98 | 42 ± 1.9 | 39–45 | 40 | 34 | |
The table presents the seroprevalence and cumulative seroprevalence in numbers (n) and percentages (%) of positive individuals in each age group. The geometric mean titers (GMT) and standard deviations (SD) of positive samples, and the rate of reinfection in previously seropositive children (defined as an increase of > 20 EIA units in sequential samples) are shown.
y, year of age; Seropositive defined as antibody level of > 9.5 EIA units in the sample; n, number of positive samples; GMAL, geometric mean antibody level; SD, geometric standard deviation; CI 95%, 95% confidence interval.
A subset of seropositive 1-year-old or 2-years-old children showed a diagnostic increase in HCoV S1-specific antibody levels in subsequent samples at 2 or 3 years of age, respectively, indicating a likely reinfection or re-exposure (below referred to as reinfection). Altogether 31% (27/88) of the children seropositive for any seasonal HCoV at 1 year showed a diagnostic/significant increase (more than 20 EIA units between sequential samples) in anti-S1 HCoV antibodies by the age of 2 years. A similar rate of likely reinfection for any HCoV (34%) was observed between 2 and 3 years for children who were seropositive at the age of 2 years.
Correlation of anti-HCoV S1 and anti-HCoV N antibody levels
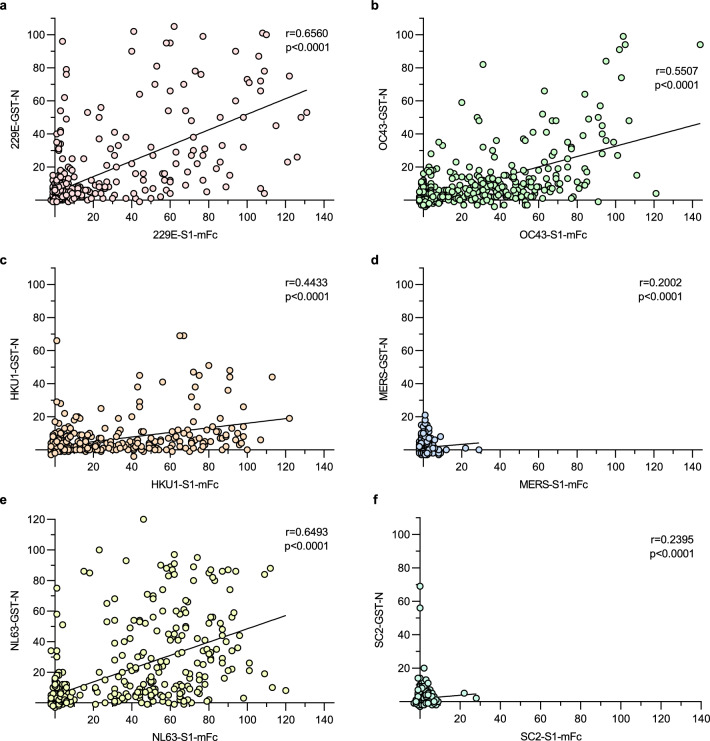
HCoV infections have been shown to induce antibodies for both HCoV S and N proteins9,10. We have previously analyzed anti-HCoV N IgG antibody levels for the same set of serum samples from children (n = 420)18 allowing us to use the existing data to analyze the correlation of N and S1 protein-specific antibody responses. We calculated the correlation coefficients for S1 and N EIA data of each HCoV (Fig. 2). The correlation coefficients of S1 and N IgG antibody levels for 229E and NL63 were relatively high (r = 0.66 and r = 0.65, respectively, p < 0.0001). The rates of correlation for anti-HKU1 S1 and N, and anti-OC43 S1 and N IgG antibodies were somewhat lower, yet significant (r = 0.44 and r = 0.55, respectively, p < 0.0001). However, the negative samples may contribute considerably to the higher rates of correlation since the removal of samples that were negative in both assays weakened the correlation of all assay pairs (Supplementary Fig. 3).
Figure 2.
Correlation of HCoV N and S1 antibody responses. IgG antibody levels for 229E, HKU1, NL63, OC43, MERS, and SARS-CoV-2 S1 and N proteins were measured for 420 serum samples from 1 to 3-year-old children. The correlation of anti-S1 and anti-N antibody levels for each HCoV was analyzed using Spearman’s matched pairs test. The correlation coefficient (r) and two-tailed p-value for each pair is shown.
COVID-19 vaccination has no effect on HCoV S1 IgG antibody levels in adults
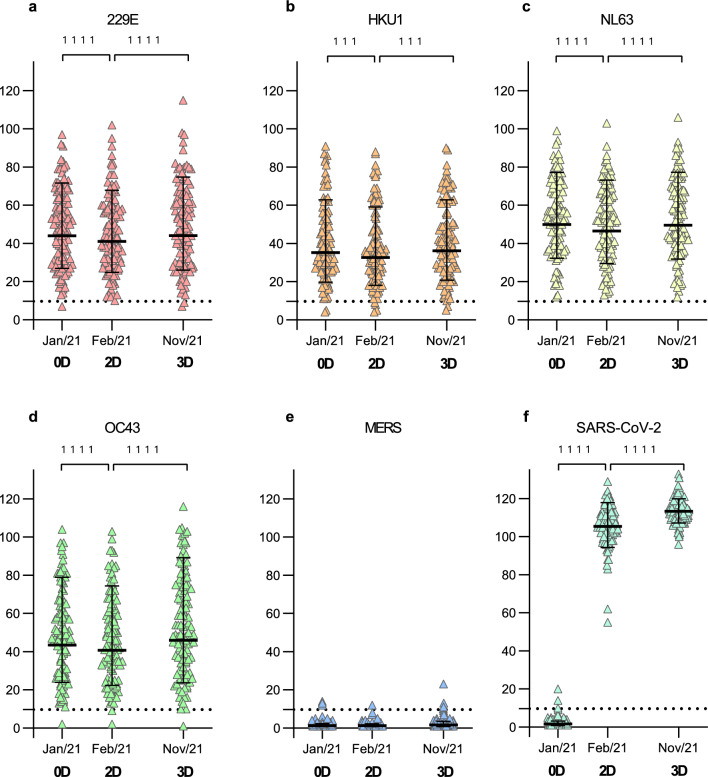
To investigate whether COVID-19 vaccination induces the production of HCoV S1 cross-reactive antibodies, we analyzed sera of COVID-19 vaccinees with HCoV S1 protein-specific EIAs. Sera were collected from 113 HCWs (25–65 years old, Table 1) before vaccination with BNT162b2 (0D; August 2020 to January 2021), three weeks after the second BNT162b2 dose (2D; January to March 2021), and three weeks after the third vaccination dose with BNT162b2 or mRNA-1273 (3D; October 2021 to January 2022). Geometric mean IgG antibody levels (GMALs) for the seasonal HCoV S1 proteins decreased from 0 to 2D but increased from 2 to 3D (44 EIA units at 0D vs. 41 EIA units at 2D vs. 44 EIA units at 3D for 229E, both p < 0.0001; 35 vs. 33 vs. 36 for HKU1, p = 0.0007 and p = 0.0008; 50 vs. 46 vs. 50 for NL63, p < 0.0001; 43 vs. 41 vs. 46, for OC43 p < 0.0001; Fig. 3). GMALs for MERS S1 remained practically negative while two doses of BNT162b2 increased the SARS-CoV-2 S1 GMALs from negative to high levels (GMAL of 1.6 EIA units at 0D vs. 105 at 2D, p < 0.0001). The third mRNA vaccine dose (BNT162b2 or mRNA-1273) further boosted the SARS-CoV-2 antibody levels (GMAL of 105 EIA units at 2D vs. 113 at 3D, p < 0.0001).
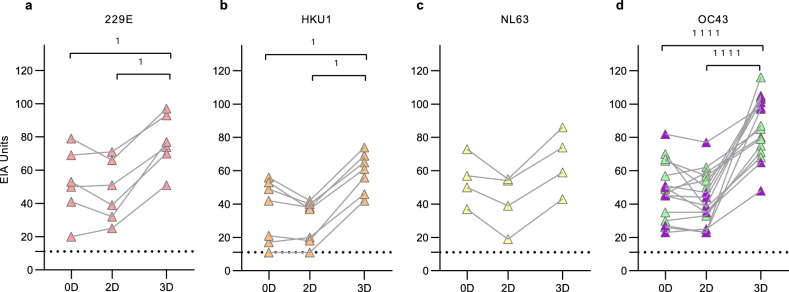
Figure 3.
HCoV S1-specific antibody responses of COVID19 mRNA vaccinated health care workers (HCWs). (a–f) HCoV S1-specific IgG antibody levels were measured with EIA from 339 serum specimens collected before vaccination (0D, Jan/21), and three weeks after second (2D, Feb/21) and third vaccination (3D, Nov/21) from 113 HCWs who received two doses of BNT162b2 with a three-week interval in December 2020 to February 2021 and a third dose of BNT162b2 or mRNA-1273 in September to December 2021. Geometric means and geometric standard deviations of antibody levels are shown. Differences between the groups were analyzed with Wilcoxon matched pairs test. Dashed line indicates the cutoff value for seropositivity. Two-tailed p-values < 0.05 were considered statistically significant. P-values 1111 < 0.0001 and 111 (b) from left to right are 0.0007 and 0.0008.
Comparison of HCoV S1-specific antibody responses between genders showed similar antibody levels at each time point with the exception of anti-NL63 S1 antibodies, which were slightly, although not significantly higher in female than in male HCWs (GMAL of 37 EIA units for male vs 51 for female at 0D, 33 vs 48 at 2D, and 43 vs. 50 at 3D; Supplementary Fig. 4). Unequal number of the HCWs in the genders (9 males and 104 females) limited the strength of the comparisons between the groups.
Prevalence of HCoV infections in 2020–2021
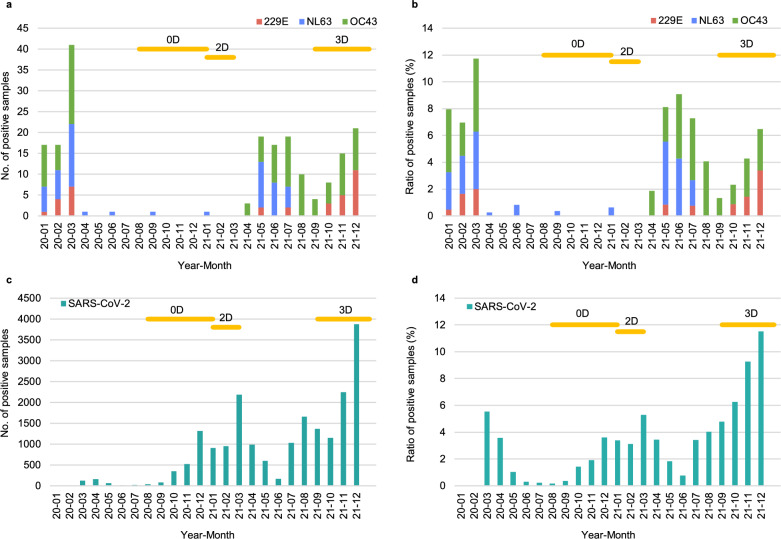
To investigate how the circulation of different HCoVs associated with the changes in HCoV antibody levels in the study population, we analyzed the monthly number and prevalence of HCoV PCR-positive samples in hospitalized patients in Southwest Finland health district in 2020–2021 (Fig. 4). The multiplex RT-qPCR with Allplex Respiratory Panel 3 (Seegene Inc.), which detected 229E, NL63, and OC43 (but not HKU1) RNA, showed the circulation of these viruses in early 2020 (January–March) with a peak of 12% of positive HCoV samples in March 2020 (7 229E-positive, 15 NL63-positive, and 19 OC43-positive out of 349 tested samples). This was followed by 12 months (April 2020 to March 2021) of very low number of positive samples (only 4 NL63-positive samples out of 2356 tested samples). Starting from April 2021 the circulation of OC43 was observed monthly until the end of the study period. NL63 was detected from May to July 2021 but no circulation was seen later in the year. 229E was detected sporadically after May 2021 and occasionally between October and December 2021. Detection of SARS-CoV-2 with laboratory developed RT-qPCR showed higher number of positive cases starting from autumn 2020 with peaks in March and December 2021.
Figure 4.
Monthly prevalence of HCoV RT-qPCR-positive samples in Southwest Finland hospital district in 2020–2021. Multiplex RT-qPCR test (with Allplex Respiratory panel 3, Seegene) including the detection of 229E, NL63, and OC43, or an in-house RT-qPCR for SARS-CoV-2 was used to analyze respiratory samples at the Clinical microbiology unit of Turku University Hospital in 2020 to 2021. Monthly numbers of positive samples for 229E, NL63, OC43 (a), SARS-CoV-2 (c) as well as the ratio of positive samples of tested samples (b and d) are presented. Horizontal orange lines indicate the timelines for serum specimens collected before (0D), three weeks after two doses (2D) and three weeks after the third (3D) mRNA vaccine dose. The number of tested samples for multiplex RT-qPCR varied from 104 to 377 (228 on average) per month and for SARS-CoV-2 RT-qPCR 2280 to 41,300 (22,983 on average) per month.
Congruence of seasonal HCoV occurrence and S1 IgG antibody levels
The absence of serologically observable seasonal HCoV reinfections (defined as > 20 EIA unit increase in antibody levels of sequential samples) between 0 and 2D (Fig. 3a–d) correlated with the low number of seasonal HCoV-positive samples in Autumn 2020 to March 2021 in Southwest Finland health district. The increases in seasonal HCoV S1 IgG antibody levels between the second and third vaccine doses (8 months apart) on the other hand suggested potential circulation of HCoVs and reinfection amongst HCWs (Fig. 5). The PCR-data on the rates of 229E, NL63, and OC43 detections from May 2021 onwards matched well with the serological changes among some of the HCWs on the same period. Diagnostic rises in antibody levels from 2 to 3D indicated a reinfection rate of 5.3% (6/113) for 229E, 6.2% (7/113) for HKU1, 3.5% (4/113) for NL63, and 14.2% (16/113) for OC43. Interestingly, each participant (n = 7) showing an increase in HKU1 S1 antibodies had also an increase in OC43 S1 antibodies (Fig. 5).
Figure 5.
Increases in HCoV S1 binding IgG antibody levels indicate reinfection. HCWs received two doses of BNT162b2 and a third dose of BNT162b2 or mRNA-1273 in 2020–2021. Serum samples were collected before vaccination (0D), 3 weeks after the second (2D), and 3 weeks after the third dose (3D) and analyzed for 229E (a), HKU1 (b), NL63 (c), and OC43 (d) S1 binding IgG antibodies with EIA. An increase of > 20 EIA units between sequential samples was considered as an indication for reinfection or re-exposure. Sequential serum samples of different individuals are connected with lines. In figure (d) purple-marking indicates the antibody levels of individuals who have an increase in antibodies for both OC43 S1 and HKU1 S1 and green-marking those with an increase only in OC43 S1 binding antibodies. Statistical differences between the time points were analyzed using Wilcoxon matched pairs test. Two-tailed p-values < 0.05 were considered statistically significant. P-values from left to right marked with 1 (panels a and b) are 0.031, 0.031, 0.016, and 0.016. 1111 < 0.0001.
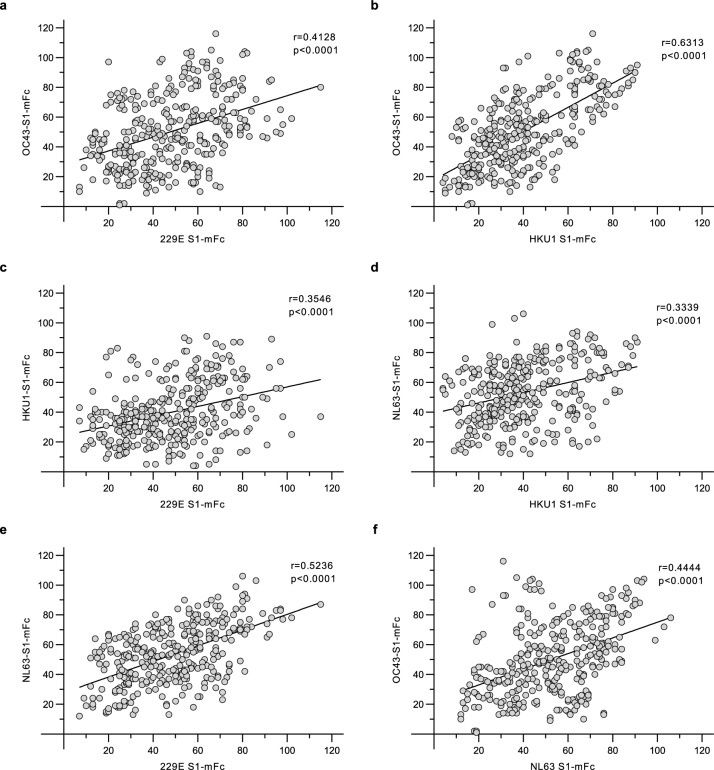
Correlation of HCoV S1-binding antibody levels
To evaluate the presence of homologous and potentially cross-reactive anti-S1 antibodies, we used the data from 339 serum samples of COVID-19 vaccinated HCWs to analyze the correlation of seasonal HCoV S1-binding IgG antibodies. The highest rates of correlation were observed for anti-229E and anti-NL63 S1 antibodies (r = 0.524, p < 0.0001), and anti-HKU1 and anti-OC43 S1 antibodies (r = 0.631, p < 0.0001) but also other assay pairs showed moderate correlation coefficients (r > 0.334, p < 0.0001 for other anti-HCoV S1 antibody level pairs) (Fig. 6).
Figure 6.
Correlation of seasonal HCoV S1 binding antibody responses. Pairwise correlation of 229E, HKU1, NL63, and OC43 S1 binding IgG antibody levels were analyzed from the data of 113 HCWs (n = 339 serum samples) with Spearman’s matched pairs test. The correlation coefficient (r) and two-tailed p-value for each pair is shown.
In children (420 serum samples, Supplementary Fig. 5), a moderate correlation was observed for anti-HKU1 S1 and anti-OC43 S1 binding antibodies (r = 0.589, p < 0.0001) and for anti-229E S1 and anti-NL63 S1 binding antibodies (r = 0.512, p < 0.0001) while the correlation coefficient values of other anti-HCoV S1 pairs was lower (r < 0.26). Pairwise comparison of S1 amino acid sequences (Supplementary Table 1) showed that the pairs of 229E and NL63, as well as HKU1 and OC43 shared more identical amino acids (50% and 59%, respectively) in comparison to other sequence pairs (10–22%), which may contribute to sequence identity-related immunological correlation.
The correlation coefficient values for anti-SARS-CoV-2 S1 antibodies and other anti-HCoV S1 antibodies after COVID-19-vaccination were low at 2D and 3D time points (Supplementary Fig. 6). Despite the low correlation coefficient values, likely due to mainly seronegative specimens, the correlation of anti-SARS-CoV-2 S1 antibodies was statistically significant (p < 0.05) with anti-OC43 antibodies at 2D (r = 0.3033, p = 0.0011), and with anti-MERS antibodies both at 2D and 3D (r = 0.2462, p = 0.0086 and r = 0.2098, p = 0.0257, respectively).
Homologous and cross-reactive S1 antibodies in immunized animals
The data from HCoV reinfections in HCWs as well as the correlations between anti-S1 antibody levels suggested potential cross-reactivity between HCoV S1-specific antibodies. To validate further the antigens chosen for EIA, we immunized rabbits and guinea pigs with the HCoV S1 proteins without mFc-fusion (except for OC43 for which S1 with mFc was used due to unsuccessful production of OC43 S1 without mFc, Supplementary Fig. 1). Each animal was immunized with one antigen. Antigens without mFc-fusion were used in immunization to avoid formation of antibodies against the mFc domain. The specificity of immune sera was examined by immunofluorescence (IF) assay for homologous and cross-reactive antibodies using 229E, OC43, and NL63 virus-infected cells (Huh7 or LLC-MK2). Virus-infected cells were detected with high IF titers (1:1000 to 1:5000) by the corresponding anti-S1 sera of immunized rabbits and guinea pigs (Table 3, Supplementary Fig. 7). In addition to homologous reactivity of sera, cross-reactivity was also observed. Cells infected with 229E were recognized by anti-HKU1 S1 rabbit serum (titer 50) and by both rabbit and guinea pig anti-NL63 S1 sera (titer 200; Table 3, Supplementary Fig. 8). OC43-infected cells were recognized by rabbit and guinea pig anti-HKU1 S1 sera (titer 200, Supplementary Fig. 9) and NL63-infected cells were recognized by rabbit anti-OC43 S1-mFc sera (titer 200, Supplementary Fig. 10).
Table 3.
Cross-reactive and homologous titers of serum from HCoV S1 immunized rabbit and guinea pig.
| Serum | 229E IF titer | NL63 IF titer | OC43 IF titer |
|---|---|---|---|
| Rb anti-229E S1 | 5000 | < 50 | < 50 |
| Gp anti-229E S1 | 1000 | < 50 | < 50 |
| Rb anti-HKU1 S1 | < 50 | < 50 | 200 |
| Gp anti-HKU1 S1 | 50 | < 50 | 200 |
| Rb anti-NL63 S1 | 200 | 5000 | < 50 |
| Rb anti-NL63 S1 | 200 | 5000 | < 50 |
| Rb anti-OC43 S1 | < 50 | 200 | 5000 |
| Gp anti-OC43 S1 | < 50 | < 50 | 5000 |
| Rb anti-SARS-CoV-2 S1 | < 50 | < 50 | < 50 |
| Gp anti-SARS-CoV-2 S1 | < 50 | < 50 | < 50 |
Titers for sera of one rabbit and one guinea pig immunized with three doses of each of the indicated HCoV S1 antigens were determined using immunofluorescence assay. Huh7 cells were infected with 229E or OC43 virus and LLC-MK2 cells with NL63 virus and labelled with a dilution series (1:50 to 1:25,000) of sera against each HCoV S1. Serum titer > 1:50 was considered positive. Of note, infectious HKU1 is globally not available.
Rb, rabbit; Gp, guinea pig; S1, spike subunit 1; IF titer, immunofluorescence titer; < 50, IF titer lower than 1:50.
Significant values are in bold.
Discussion
In this study we investigated the seroprevalence of HCoV spike-specific antibodies by analyzing anti-HCoV IgG antibodies in sequential serum samples collected from 140 children between 1 and 3 years of age and from BNT162b2-vaccinated HCWs in three time points: before the vaccination and after two and three vaccine doses. Spike subunits 1 (S1) were chosen as antigens for the EIA since they are genetically more distant from each other compared to the whole spike proteins or the S2 subunits. The spike protein is the main structural protein on the coronavirus surface, and it is responsible for the binding and fusion of the virus with the target cell, and due to its immunogenicity, it is a good antigen for serological studies. Dimeric S1-mFc fusion proteins were chosen as antigens for serological assays due to their enhanced stability and reliable functionality in EIA10.
We detected somewhat higher seroprevalence rates for anti-HCoV S1 antibodies as compared to the seroprevalence rates of anti-HCoV N antibodies in the same child population18. This observation is well in line with previous studies showing a high seroprevalence for binding antibodies against the whole HCoV S proteins19 and somewhat more variation in seroprevalence for HCoV N antibodies3,12,13. Also, based on the cumulative seroprevalence, the waning of anti-S1 antibodies as shown by the present study and previous publications was slower compared to the waning of anti-N antibodies8,18. Despite the relatively high correlation coefficient values for anti-N and anti-S1 antibodies for four seasonal HCoVs, our data shows that in some individuals HCoV infections induce the production of IgG antibodies only against one of these antigens, an observation similar to the one seen in SARS-CoV-2 infections9,10.
The PCR data suggested a low rate of circulation of HCoVs from April 2020 to March 2021, which is well in line with the reports on limited circulation of endemic seasonal respiratory viruses20,21 due to “lockdown” of the society and other measures used to prevent the spread of the COVID19 pandemic. The data also showed that two doses of BNT162b2 did not increase antibody levels against other HCoV S1 proteins indicating practically a complete lack of immunological cross-reactivity. The gradual release of COVID-19 restrictions in the spring of 2021 in Finland22 resulted in the circulation of seasonal HCoVs followed by an increase in anti-HCoV S1 antibody levels in the autumn 2021. Furthermore, the reinfection events of OC43, seen as a diagnostic increase in the anti-OC43 S1 antibodies between 2 and 3D serum specimens, matched well with the circulation of OC43 during the year 2021. In epidemiological studies the occurrence of HKU1 has been lower than that of other seasonal HCoVs23,24. Thus, the high seroprevalence for HKU1 in this study may be due to cross-reactive antibodies induced by OC43 infections. In our analysis all individuals showing an increase in anti-HKU1 antibodies also had an increase in anti-OC43 antibodies indicating that immunological cross-reactivity between OC43 and HKU1 may affect the anti-HKU1 antibody positivity rate. Similar immunological cross-reactivity between OC43 and HKU1 spike protein was suggested by our analyses of rabbit and guinea pig immune sera.
Data from immunized animals, antibody level correlation coefficient values and HCW serum analyses with indications of seasonal HCoV reinfections suggest significant cross-reactivity between anti-HKU1 and anti-OC43 S1 antibodies and anti-229E and anti-NL63 S1 antibodies, respectively. Sequence identities between the above HCoV S1 protein pairs is higher than those of other S1 protein pairs and similar cross-reactivity of antibodies exist with the corresponding anti-N antibody pairs18,23,25. Immunizations with OC43 S1 were done with mFc-fusion of the S1 and the effect of mFc-specific antibodies into cross-reactivity cannot be completely ruled out, although our data indicates no such reactivity. Immunological cross-reactivity between HKU1 and OC43 or 229E and NL63 may also provide cross-protection even though we have no experimental evidence for that. The level of cross-reactive antibodies as well as the cross-reactive HCoV pairs would likely be greater if the full-length spike protein or the S2 subunit were used as the antigens due to higher sequence similarities in these protein sequences. For example, OC43 and HKU1 S2-specific antibody responses have been described to correlate with antibody responses induced by SARS-CoV-2 infection26,27. However, since our prevaccination sera were devoid of binding activity to SARS-CoV-2 S1 protein and COVID-19 vaccination did not induce any increase in seasonal HCoV S1-specific antibody responses, we consider it quite unlikely that there is humoral cross-protection induced by seasonal HCoV infections against SARS-CoV-2 and vice versa.
Our data shows that the number of children with HCoV S1-binding IgG antibodies increases by age, reaching 40–80% by the age of three years. In adults these values are nearly 100% against all the seasonal HCoVs indicating a high rate of circulation of 229E, NL63 and OC43 viruses. Twelve-month follow-up of vaccinated adults showed that increased circulation of HCoVs matched relatively well with changes in anti-HCoV antibody levels. These observations provide further information on the characteristics of humoral immune responses of coronavirus infections which appear to be extremely common respiratory pathogens also among the adults.
Materials and methods
Study participants
Infant participants were part of the STEPS study28, a prospective observational birth-cohort study in the Southwest Finland health district. Serum samples were collected at 13.7 (1y), 25.3 (2y) and 37.5 (3y) months of age from 140 children (53% male and 47% female; n = 420 samples) during visits to a study clinic in 2009 – 2013, and the same serum specimens have been used in our previous study concentrating on anti-HCoV nucleoprotein antibodies18. The Ethics Committee of the Southwest Finland health district approved the study (decision no. 16/180/2008). Parents of participating children gave their written, informed consent.
Healthcare workers (HCWs, n = 113, 92% female and 8% male) aged 25–65 years (average 43 years) had obtained two doses of BNT162b2 with a three-week (21 days for each participant) dosing interval between the first two doses in December 2020–February 2021 and a BNT162b2 or an mRNA-1273 as the third vaccine 9 months (7.7–10.3 months post the 2nd dose, 8.5 months as an average) later in September–December 2021. The participants were randomly selected for this study from a larger cohort29. Serum samples from the vaccinees were collected 0–129 days (average 22.7 days) before the vaccination (0D) in August 2020 to January 2021, 3 weeks (17–49 days, average 24.2 days) after the second dose (2D) February to March 2021, and 3 weeks (15–44 days, average 24.4 days) after the third dose (3D) in September to December 2021. Ethical permission for this study was obtained from the Southwest Finland health district (ETMK 19/1801/2020, EudraCT 2021-004419-14). All participants signed a written informed consent at the enrollment for the study.
Recombinant HCoV spike protein S1 antigen production
Codon-optimized genes encoding the spike protein subunit 1 (S1) of 229E (GenBank accession KY621348.1, residues 1–565), HKU1 (KY674943.1, residues 1–750), NL63 (KY554967.1, residues 1–717), OC43 (MN306053.1, 1–760), SARS-CoV-2 (NC_045512.2, residues 16–541), and MERS (JX869059.2, residues 1–747) were obtained from GeneUniversal. The HCoV S1 genes were cloned into a vector with a C-terminal mouse IgG2a Fc part (mFc) tagged with polyhistidine (His) tail, and a vector with only the C-terminal His tail. HCoV S1-mFc-His (referred to as HCoV S1-mFc) proteins, the EIA antigens, and the HCoV S1-His (HCoV S1 without mFc) proteins were produced in HEK-293F cells, and purified as described previously10.
HCoV anti-S1 and anti-N enzyme immunoassays
The presence of HCoV S1 binding IgG antibodies (anti-HCoV-S1) in the serum samples was analyzed with an enzyme immunoassay (EIA) using HCoV-S1-mFc proteins. EIA was conducted with 1:300 dilution of serum samples and the quantification of antigen-binding IgG was done with measurement of absorbance at wavelength of 450 nm as described previously10,18. The protocol was adjusted slightly to optimize the assay for the detection of lower IgG concentrations without saturation in the higher IgG concentrations: coating concentration for S1-mFc proteins was 2.0 µg/ml and the dilution of horseradish peroxidase (HRP) conjugated anti-human antibodies 1:6000 (Dako A/S).
The optical density (OD) value of each sample was converted into EIA units by comparing it to the OD values of positive (marked as 100 EIA units) and negative (marked as 0) control sample pools measured by SARS-CoV-2 S1 EIA. To allow the calculation of geometric means the samples with an EIA unit value lower than 1 were marked as 1. The cutoff value was defined as the mean of anti-SARS-CoV-2 S1 EIA results from the negative child serum samples (n = 417, one child showed detectable antibodies and therefore it was excluded in the calculations) plus five standard deviations (SD). The inter-assay variation was controlled with an additional control sample pool and observed to remain < 10% except for anti-MERS S1 for which no positive control was available. Based on the limited inter-assay variation we considered a > 20 EIA unit increase in antibody levels to indicate a reinfection.
Immunization of rabbits and guinea pigs
A total of five rabbits (Hsdlf:NZW, female, 13–14 weeks of age) and five guinea pigs (HsdDhl:DH, female, 12–14 weeks of age) obtained from Envigo (The Netherlands), were acclimatized for 2 weeks before being immunized. A rabbit and a guinea pig were immunized with three 50 µg-doses of either 229E S1-His, HKU1 S1-His, NL63 S1-His, or OC43 S1-mFc-His protein or four 50 µg-doses of SARS-CoV-2 S1-His protein. Each animal was immunized with one antigen. Immunizations were carried out with a 2-week dosing interval and the proteins were administered with adjuvant system 03 (AS03, GlaxoSmithKline) intramuscularly (rabbits) or subcutaneously (guinea pigs).
Both rabbits and guinea pigs were sampled for serum before the first immunization (0-sample) and at the end of the immunization 7–10 days after the last booster dose. The rabbits were additionally sampled at the time of each immunization. The animals were anesthetized with isoflurane (guinea pigs) or with subcutaneous injection of medetomidine and ketamine injection (rabbits) and euthanized by cardiac puncture. Project Authorization Board of Finland approved the procedures and protocols (license numbers: ESAVI/16751/2020 and ESAVI/20869/2020) in accordance with the 2010/EU/63 EU Directive and Finnish national legislation (497/2013 and 564/2013) on the protection of animals used for scientific purposes.
Indirect immunofluorescent (IF) assay with cells infected with 229E, NL63, and OC43
LLC-MK2 (rhesus monkey kidney) cells and Huh7 (human hepatoma) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Lonza BioWhittaker) supplemented with 10% FBS (Biowest Nuaille), 1% l-glutamine (ThermoFischer Scientific), and 1% penicillin/streptomycin (Lonza). For the infections the concentration of FBS in the medium was reduced to 2%. Infections of Huh7 cells with 229E (GenBank Accession OK662398) and OC43 (OK662397) and the following fixing and permeabilization for IF were done as described previously18. LLC-MK2 cells were infected with 0.1 FFU/ml of NL63 (AY567487.2, a kind gift from Prof. van der Hoek) for 48 h followed by fixing the cells with 4% paraformaldehyde and permeabilization with 0.1% Triton-X-100 in PBS. An infectious virus for HKU1 is globally not available and thus HKU1 virus infected cells could not be used in IF.
Rabbit and guinea pig sera were diluted 1:50, 1:200, 1:1000, 1:5000, and 1:25,000 into PBS supplemented with 3% bovine serum albumin (BSA). Dilution series was added onto virus- or mock-infected cells in duplicates and incubated for 1 h. For double-staining of 229E-infected, OC43-infected, and NL63-infected cells the guinea pig serum dilutions were supplemented with a 1:1000 dilution of polyclonal rabbit anti-229E N-GST, rabbit anti-OC43 N-GST, or rabbit anti-NL63 sera and the rabbit serum dilutions were supplemented with corresponding guinea pig anti-HCoV N-GST sera (Huttunen et al., unpublished data), which had been prepared in serial immunizations as described above. The antibodies were detected with 1:1000 dilution of secondary antibodies goat anti-rabbit IgG 488/564 and/or goat anti-guinea pig IgG 488 (Thermo Scientific) and 1:2500 dilution of DAPI for 1 h. Antibody-binding to infected cells was imaged on EVOS FL Auto Fluorescence Inverted Microscope (Life Technologies).
Statistical and sequence analysis
Statistical analysis of EIA results was conducted using GraphPad Prism 8 software as the platform. Wilcoxon matched pair signed-rank test corrected with Pratt’s method was used for the analysis of pairwise differences between the groups. Spearman’s correlation test was used to analyze the correlation of results between two different HCoV S1 type specific assays. Two-tailed p-values < 0.05 were considered significant. Exact p-values are shown in the figures and marked according to their significance with stars: p-value of < 0.05 *, < 0.002 **, < 0.0002 ***, and < 0.00001 ****.
Amino acid sequences of the spike, spike subunits 1 and 2, and nucleoprotein for the HCoV isolates used in this study were aligned using ClustalW30 in MEGA 1131 platform and compared pairwise for sequence identity.
Ethics statement
All experiments were performed in accordance with relevant guidelines and regulations. The Ethics Committee of the Southwest Finland health district approved the study (decision no. 16/180/2008). Parents of participating children gave their written, informed consent. All methods are reported in accordance with ARRIVE guidelines (https://arriveguidelines.org) for the reporting of animal experiments. Procedures and protocols concerning animal experiments were approved by the Project Authorization Board of Finland (license numbers: ESAVI/16751/2020 and ESAVI/20869/2020) in accordance with the 2010/EU/63 EU Directive and Finnish national legislation (497/2013 and 564/2013) on the protection of animals used for scientific purposes.
Supplementary Information
Acknowledgements
Anne-Mari Pieniniemi and Anne Suominen are gratefully acknowledged for their excellent technical help. We thank Joonas Khabbal, Varpu Laine and personnel of Central Animal Laboratory for their expert animal care. We thank all the children and their parents for participation in the STEPS study as well as the participants of the COVID-19 vaccination study. We thank professors Lia van der Hoek for the kind gift of NL63 (Amsterdam) and Varpu Marjomäki for the kind gift of OC43 virus isolate. This work was supported by the Jane and Aatos Erkko Foundation (grant numbers 3067-84b53 and 5360-cc2fc to IJ), the Sigrid Juselius Foundation (to IJ and LK), the Medical Research Council of the Academy of Finland (grant numbers 297329 and 336410 to IJ) and the Foundation for Pediatric Research, Finland (to VP).
Author contributions
P.K., and I.J. designed the study. P.K, M.H., and S.M. performed the experiments. M.H., P.K., and M.W. collected and analyzed the data. L.T., L.I., P.A.T, J.L., and V.P. performed recruitment and sample and participant data collection. E.Y., and S.T. contributed to the methodology and A.I., A.P., R.N., and O.R. provided key reagents. P.K., L.K., and I.J. supervised the study. P.K., L.K., and I.J. wrote the manuscript. All authors participated in the editing of the text and approved the final version of the manuscript.
Data availability
The datasets generated and/or analysed during the current study are available in the supplementary info file or in the NCBI GenBank repository (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers KY621348.1, KY674943.1, KY554967.1, MN306053.1, NC_045512.2, JX869059.2, OK662398, OK662397, and AY567487.2.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Laura Kakkola, Olli Ritvos and Ilkka Julkunen.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-35471-3.
References
- 1.Desforges M, et al. Human coronaviruses and other respiratory viruses: Underestimated opportunistic pathogens of the central nervous system? Viruses. 2019;12:14. doi: 10.3390/v12010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talbot HKB, et al. Coronavirus infection and hospitalizations for acute respiratory illness in young children. J. Med. Virol. 2009;81:853–856. doi: 10.1002/jmv.21443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang AT, et al. A systematic review of antibody mediated immunity to coronaviruses: Kinetics, correlates of protection, and association with severity. Nat. Commun. 2020;11:4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Self WH, et al. Respiratory viral detection in children and adults: Comparing asymptomatic controls and patients with community-acquired pneumonia. J. Infect. Dis. 2016;213:584–591. doi: 10.1093/infdis/jiv323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albrecht M, Arck PC. Vertically transferred immunity in neonates: Mothers, mechanisms and mediators. Front. Immunol. 2020;11:555. doi: 10.3389/fimmu.2020.00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shook LL, et al. Durability of anti-spike antibodies in infants after maternal COVID-19 vaccination or natural infection. JAMA. 2022;327:1087–1089. doi: 10.1001/jama.2022.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorse GJ, Patel GB, Vitale JN, O’Connor TZ. Prevalence of antibodies to four human coronaviruses is lower in nasal secretions than in serum. Clin. Vaccine Immunol. 2010;17:1875–1880. doi: 10.1128/CVI.00278-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edridge AWD, et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- 9.Seow J, et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jalkanen P, et al. A combination of N and S antigens with IgA and IgG measurement strengthens the accuracy of SARS-CoV-2 serodiagnostics. J. Infect. Dis. 2021 doi: 10.1093/infdis/jiab222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dijkman R, et al. The dominance of human coronavirus OC43 and NL63 infections in infants. J. Clin. Virol. 2012;53:135–139. doi: 10.1016/j.jcv.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dijkman R, et al. Human coronavirus NL63 and 229E seroconversion in children. J. Clin. Microbiol. 2008;46:2368–2373. doi: 10.1128/JCM.00533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao X, Guo X, Esper F, Weibel C, Kahn JS. Seroepidemiology of group I human coronaviruses in children. J. Clin. Virol. 2007;40:207–213. doi: 10.1016/j.jcv.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanchard EG, Miao C, Haupt TE, Anderson LJ, Haynes LM. Development of a recombinant truncated nucleocapsid protein based immunoassay for detection of antibodies against human coronavirus OC43. J. Virol. Methods. 2011;177:100–106. doi: 10.1016/j.jviromet.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sagar M, et al. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Investig. 2021;131:e143380. doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waterlow NR, et al. How immunity from and interaction with seasonal coronaviruses can shape SARS-CoV-2 epidemiology. Proc. Natl. Acad. Sci. 2021;118:e2108395118. doi: 10.1073/pnas.2108395118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nasrallah GK. Do preexisting antibodies against seasonal coronaviruses have a protective role against SARS-CoV-2 infections and impact on COVID-19 severity? EBioMedicine. 2022;76:103831. doi: 10.1016/j.ebiom.2022.103831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolehmainen P, et al. Serological follow-up study indicates high seasonal coronavirus infection and reinfection rates in early childhood. Microbiol. Spectr. 2022;10:e01967–e2021. doi: 10.1128/spectrum.01967-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou W, Wang W, Wang H, Lu R, Tan W. First infection by all four non-severe acute respiratory syndrome human coronaviruses takes place during childhood. BMC Infect. Dis. 2013;13:433. doi: 10.1186/1471-2334-13-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chow EJ, Uyeki TM, Chu HY. The effects of the COVID-19 pandemic on community respiratory virus activity. Nat. Rev. Microbiol. 2022 doi: 10.1038/s41579-022-00807-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groves HE, et al. The impact of the COVID-19 pandemic on influenza, respiratory syncytial virus, and other seasonal respiratory virus circulation in Canada: A population-based study. Lancet Reg. Heal. Am. 2021;1:100015. doi: 10.1016/j.lana.2021.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Government adopts plan to lift COVID-19 restrictions. https://valtioneuvosto.fi/en/-//10616/government-adopts-plan-to-lift-covid-19-restrictions (Accessed 28 Oct 2022).
- 23.Gao X, et al. Antibody against nucleocapsid protein predicts susceptibility to human coronavirus infection. J. Infect. 2015;71:599–602. doi: 10.1016/j.jinf.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan CM, et al. Examination of seroprevalence of coronavirus HKU1 infection with S protein-based ELISA and neutralization assay against viral spike pseudotyped virus. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2009;45:54–60. doi: 10.1016/j.jcv.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trivedi SU, et al. Development and evaluation of a multiplexed immunoassay for simultaneous detection of serum IgG antibodies to six human coronaviruses. Sci. Rep. 2019;9:1390. doi: 10.1038/s41598-018-37747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dispinseri S, et al. Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat. Commun. 2021;12:2670. doi: 10.1038/s41467-021-22958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen-Contant P, et al. S Protein-reactive IgG and memory B cell production after human SARS-CoV-2 infection includes broad reactivity to the S2 subunit. MBio. 2020;11:e01991–e2020. doi: 10.1128/mBio.01991-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagström H, et al. Cohort profile: Steps to the healthy development and well-being of children (the STEPS Study) Int. J. Epidemiol. 2013;42:1273–1284. doi: 10.1093/ije/dys150. [DOI] [PubMed] [Google Scholar]
- 29.Jalkanen P, et al. COVID-19 mRNA vaccine induced antibody responses against three SARS-CoV-2 variants. Nat. Commun. 2021;12:3991. doi: 10.1038/s41467-021-24285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li K-B. ClustalW-MPI: ClustalW analysis using distributed and parallel computing. Bioinformatics. 2003;19:1585–1586. doi: 10.1093/bioinformatics/btg192. [DOI] [PubMed] [Google Scholar]
- 31.Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. (2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the supplementary info file or in the NCBI GenBank repository (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers KY621348.1, KY674943.1, KY554967.1, MN306053.1, NC_045512.2, JX869059.2, OK662398, OK662397, and AY567487.2.