Abstract
Purified retrovirus Gag proteins can assemble in vitro into virus-like particles (VLPs) in the presence of RNA. It was shown previously that a Rous sarcoma virus Gag protein missing only the protease domain forms spherical particles resembling immature virions lacking a membrane but that a similar protein missing the p10 domain forms tubular particles. Thus, p10 plays a role in spherical particle formation. To further study this shape-determining function, we dissected the p10 domain by mutagenesis and examined VLPs assembled within Escherichia coli or assembled in vitro from purified proteins. The results identified a minimal contiguous segment of 25 amino acid residues at the C terminus of p10 that is sufficient to restore efficient spherical assembly to a p10 deletion mutant. Random and site-directed mutations were introduced into this segment of polypeptide, and the shapes of particles formed in E. coli were examined in crude extracts by electron microscopy. Three phenotypes were observed: tubular morphology, spherical morphology, or no regular structure. While the particle morphology visualized in crude extracts generally was the same as that visualized for purified proteins, some tubular mutants scored as spherical when tested as purified proteins, suggesting that a cellular factor may also play a role in shape determination. We also examined the assembly properties of smaller Gag proteins consisting of the capsid protein-nucleocapsid protein (CA-NC) domains with short N-terminal extensions or deletions. Addition of one or three residues allowed CA-NC to form spheres instead of tubes in vitro, but the efficiency of assembly was extremely low. Deletion of the N-terminal residue(s) abrogated assembly. Taken together, these results imply that the N terminus of CA and the adjacent upstream 25 residues play an important role in the polymerization of the Gag protein.
The internal structure of retroviruses is determined by Gag, a polyprotein that is cleaved by the viral protease (PR) late in assembly, leading to virus maturation. While the viral envelope proteins and the viral enzymes are essential for infectivity, Gag alone can form virus-like particles (VLPs) closely resembling real virions in morphology and other physical properties. Immature virions are composed of about 1,500 Gag molecules (34) that form a hollow sphere enclosed by the viral membrane. The molecules are arranged radially in a parallel array, such that the N-terminal end of Gag is outside, in contact with the inner surface of the membrane, while the C-terminal end is inside, in contact with RNA. This arrangement is maintained after maturation. Thus, the N-terminal domain of Gag gives rise to the membrane-associated protein (MA), a middle domain gives rise to the capsid protein (CA) that forms the shell of the mature core, and a domain near the C terminus gives rise to the nucleocapsid protein (NC) in the center of the core. In addition to these three canonical proteins found in all retroviruses, proteolytic processing of Gag also leads to other proteins and short peptides that are specific to the viral genus or species.
The morphology of immature virions is very similar for all retrovirus genera. The protein shell shows characteristic radial density variations that can be seen by cryoelectron microscopy (cryo-EM) (9, 37). The radial density pattern is likely to correspond to positions of globular versus extended polypeptide domains of Gag from the C to the N terminus. In contrast, the morphology of the cores of mature virions is distinctive for different viruses. For example, the cores of lentiviruses, such as human immunodeficiency virus type 1 (HIV-1), have a cone-shaped appearance, while those of C-type viruses, such as Rous sarcoma virus (RSV) and Moloney murine leukemia virus (M-MuLV), have a polyhedral appearance. Neither immature nor mature retroviruses are icosahedral, nor are they strictly uniform in size (9, 34, 37). In some cases mutations in Gag have been reported to lead to the budding of VLPs with tubular morphology (14, 20, 28, 38), but the mechanism for and significance of tube formation are not understood.
Retroviral VLPs can be formed spontaneously in vitro from Gag proteins purified from Escherichia coli (4, 5, 6, 8, 12, 16, 17, 18, 19, 23, 35) or translated in vitro (26, 31, 33). In vitro assembly with purified proteins has been studied most extensively for HIV-1 and RSV. For HIV-1, CA, when alone, assembles into tubes (8, 17) at a high concentration, high ionic strength, and appropriate pH. CA-NC also assembles into tubes but at low ionic strength this process requires the presence of RNA (5, 17). Under some conditions, this protein forms a mixture of tubular and cone-shaped particles resembling authentic mature cores (12). In contrast, longer Gag proteins containing long upstream amino acid sequences in addition to the CA and NC domains generally assemble into spherical particles, although in some cases these particles are much smaller than bona fide immature cores (4, 18, 35). For HIV-1, CA-NC proteins carrying even very short N-terminal extensions form spherical particles instead of tubes, but the assembly process in this case is quite inefficient and the particles are not uniform in size (18, 35). It has been reported recently that the morphology of HIV-1 VLPs is determined not only by the linear structure of the Gag protein but also by the pH of the assembly reaction (19). A protein with the structure ΔMA-CA-NC, where a part of the MA domain is deleted, forms spherical particles at pH 8 but tubular particles at pH 6. This observation, together with a similar pH dependence for monoclonal antibody reactivity to CA, suggests that the underlying mechanism distinguishing between tubular and spherical polymerization may be a conformational change in Gag (19).
Like the equivalent HIV-1 protein, purified RSV CA-NC assembles into tubes in the presence of RNA (5). A longer RSV Gag protein missing only the PR domain, i.e., with the structure MA-p10-CA-NC, gives rise in the presence of RNA to spherical particles of a size and morphology that are indistinguishable from those of immature, detergent-treated RSV particles collected from cells expressing a PR-defective mutant (6). A similar protein with an internal deletion excising the p10 domain of Gag forms tubes instead of spheres in vitro (6). This result suggests that the p10 domain plays a critical role in directing Gag into the spherical mode of polymerization. Using both assembly in vitro and assembly in E. coli cells as assays, we have dissected the p10 domain to locate the minimal sequence necessary for spherical shape determination. In addition, we have analyzed a collection of random and targeted mutations in this minimal segment to try to gain an understanding of the sequence requirements for its function as a shape determinant.
Role of p10 in spherical conformation.
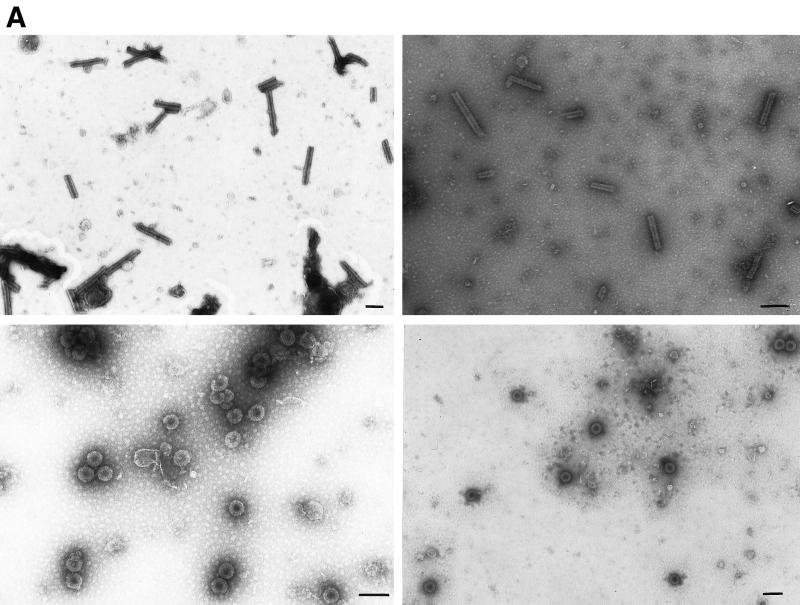
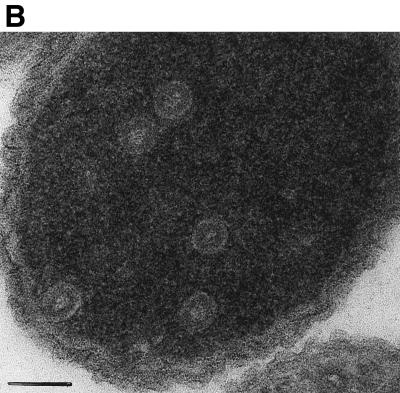
To obtain VLPs for analysis by transmission electron microscopy (TEM), we employed two methods. In the first method, Gag proteins were purified as described previously (5, 6) after expression in E. coli, using ammonium sulfate precipitation and phosphocellulose chromatography as the major steps. To initiate assembly in vitro, purified protein in 0.5 M NaCl, pH 7.5, was mixed with 12% (wt/wt) total E. coli RNA which had been prepared by standard methods (1), and then the mixture was either dialyzed (5, 6) or diluted to 0.1 M NaCl, pH 6.0, and examined by TEM after 30 min. In the second method, crude lysates from cells induced to express viral protein were centrifuged for 5 min at 10,000 × g to remove debris, and then VLPs formed in E. coli and liberated in soluble form were viewed directly by negative staining of the extract with uranyl acetate (5, 6). Previous in vitro assembly studies had shown that VLPs formed by the proteins CA-NC, ΔMBDΔPR, and dp10 (Fig. 1, lines 1, 2, and 5) were either spherical, with a diameter of 60 nm, or tubular, with a diameter of 30 nm and a variable length, which was confirmed in the present study. We constructed a new, shorter protein, p10-CA-NC (Fig. 1, line 3), which also forms spherical VLPs. For these and all other proteins described here, with the exceptions noted below, the morphologies of the VLPs were indistinguishable by the two methods of analysis. This result is shown for a representative pair of proteins, the tube-forming CA-NC and the sphere-forming p10-CA-NC (Fig. 2A). We further confirmed spherical particle assembly for p10-CA-NC in E. coli by thin-section EM (Fig. 2B). Thin-section results showing tubes in E. coli expressing CA-NC were reported previously (5). For all cases examined, the relative efficiencies of particle formation scored by the two methods were qualitatively similar (Table 1).
FIG. 1.
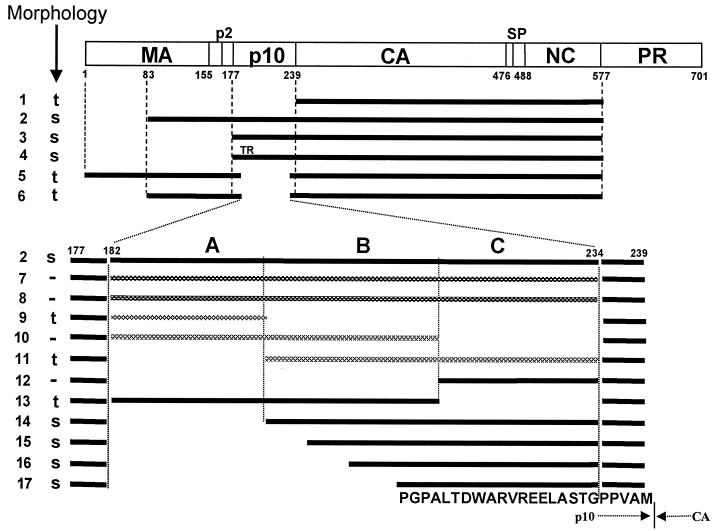
Diagrammatic representation of assembly results. The rectangle shows the structure of the Gag protein, with vertical lines representing cleavage sites and numbers representing the number of amino acid residues from the N terminus, using the standard numbering for the Prague C strain of RSV (32). Horizontal bars indicate the structures of the proteins studied for their assembly properties from both in vitro and particle extraction experiments. Black bars represent viral sense sequences. Cross-hatched bars indicate either antisense RSV sequences or sequences from other retroviruses. Regions A, B, and C in p10 are segments of approximately 20 aa residues each. All constructs were placed into the pET3XC vector by common cloning techniques, propagated in E. coli DH5α cells, confirmed by sequencing, and transformed into BL21(DE3)/pLysS cells for protein expression and purification. ΔMBDΔPR, dp10 (called Δp10.52 in reference 25), and CA-NC have been described previously (5, 6, 25). ΔMBDdp10 combines the deletions of ΔMBDΔPR and dp10. DNA segments encoding the C-terminal 62 aa residues of M-MuLV p12 or HIV-1 MA or various segments of p10 (AB, BC, and C) were amplified by PCR from the appropriate viral clones, using primers encoding a SpeI site, and inserted into the unique SpeI site in ΔMBDdp10. Lines: 1, CA-NC; 2, ΔMBDΔPR; 3, p10-CA-NC; 4, p10TR-CA-NC; 5, dp10; 6, ΔMBDdp10; 7, C-terminal 62 aa of MMLV p12; 8, C-terminal 62 aa of HIV-1 MA (BH10 strain); 9 to 11, insertions of antisense sequences derived from DNA coding for p10; 12, segment C; 13, segment AB; 14, segment BC; 15, segment C plus the last 15 aa of segment B; 16, segment C plus the last 10 aa of segment B; 17, segment C plus the last 5 aa of segment B. The proteins shown in lines 7 to 14 contain the inserted dipeptide Thr-Ser between the wild-type Gly and Pro residues five residues from the C terminus of p10. The proteins shown in lines 15 to 17 contain the wild-type sequence at this location. The 25-aa sequence corresponding to the minimal insertion sufficient for spherical particle formation is shown at the bottom, with the boundary between p10 and CA marked. s, formation of spheres; t, formation of tubes; −, no regular assembly; TR, Thr-Arg insertion. Assembly for all of the proteins shown was tested both in vitro with purified protein and in E. coli lysates, with the same results, shown in the column marked “Morphology.”
FIG. 2.
Morphology of CA-NC and p10-CA-NC particles assembled in vitro and within E. coli. (A) Electron micrographs of negatively stained particles. Samples were adsorbed onto Formvar- and carbon-coated grids for 2 min and stained with 2% uranyl acetate (pH 5.2) for 20 s. Upper left, CA-NC protein assembled into tubes in vitro; upper right, CA-NC tubes from crude extracts; lower left, p10-CA-NC protein assembled into spheres in vitro; lower right, p10-CA-NC spheres from crude extracts. Bars = 100 nm. (B) Thin-section electron micrograph of VLPs formed within E. coli cells expressing p10-CA-NC protein. Cells were pelleted and fixed for 2 h in 0.1 M sodium maleate (pH 5.2)–3% glutaraldehyde and then washed in 0.1 M sodium cacodylate, pH 7.4. The samples were postfixed for 2 h in 1% OsO4–0.1 M sodium cacodylate (pH 7.4), quickly rinsed in 0.1 M sodium maleate (pH 5.2), and then washed extensively in the same buffer. The samples were then stained with 1% uranyl acetate–0.1 M sodium maleate (pH 6.0), washed in 0.1 M sodium maleate (pH 5.2), and serially dehydrated with 50, 70, 95, and 100% ethanol and 100% propylene oxide. Pellets were embedded in 50% propylene oxide–standard Spurr, and thin sections were stained with 2% uranyl acetate and then Reynold's lead citrate.
TABLE 1.
Characteristics of Gag protein mutants
| Protein | Figure (line) | Shapea
|
Assembly efficiencyb | |
|---|---|---|---|---|
| In vitro | In E. coli | |||
| CA-NC | 1 (1) | t | t | +++ |
| ΔMBDΔPR | 1 (2) | s | s | +++ |
| ΔMBDdp10 | 1 (6) | t | t | ++ |
| dp10 | 1 (5) | t | t | ++ |
| Min0 | 1 (17) | s | s | ++ |
| Min23 (L52F) | 3B | s | t | ++ |
| Min21 (P58T) | 3B | s | t | ++ |
| Min24 (M62V) | 3B | s | t | ++ |
| m-CA-NC | 4 | s | s,t | ± |
| vam-CA-NC | 4 | s | t | ± |
s, spheres; t, tubes.
Relative assembly efficiency based on the average number of VLPs seen with negative-stain TEM. +++, more than 100 particles per grid; ++, 30 to 75 particles per grid; ±, fewer than 10 particles per grid.
The p10 protein is a small proline- and glycine-rich polypeptide with unusual biochemical properties (30) comprising the 62 amino acids (aa) N terminal to CA (Fig. 1, top). In the previously described p10 deletion mutant called dp10 (Fig. 1, line 5), which has also been referred to as Δp10.52 (25), 52 of the 62 residues in p10 are replaced with the dipeptide sequence Thr-Ser, corresponding to a SpeI site, leaving 5 wild-type residues at each end. The dp10 protein includes the 84-aa membrane binding domain at the N terminus of Gag, which represents about one-half of MA. While the membrane binding domain, as well as MA itself and MA-p2-p10, is soluble in E. coli extracts (27; A. Barbera and V. M. Vogt, unpublished observations), previous in vitro assembly experiments indicated that in the context of larger Gag proteins extending through CA, this domain leads to poor solubility of Gag proteins in E. coli, thus complicating studies on in vitro assembly (6). Hence, in order to establish a more tractable system for studying the effects of p10 and to investigate the generality of the effect of p10 deletion on VLP morphology, we combined the deletions of dp10 and ΔMBDΔPR into a single construct, ΔMBDdp10 (Fig. 1, line 6). The resulting purified protein was found to assemble into tubes with an efficiency similar to that of dp10 (Table 1), suggesting that the N-terminal membrane binding domain does not contribute to morphology.
In retrovirus Gag proteins other than those of lentiviruses, one or more short proteins or peptides lie between MA and CA. In general, these amino acid sequences are rich in glycine and proline residues. For example, both RSV p10 and M-MuLV p12 are about 30% Gly and Pro. Thus, it seemed possible that the amino acid composition of p10 might be the characteristic responsible for spherical particle formation. To test this hypothesis, we replaced the amino acids deleted in dp10 with a similar-size segment from the C terminus of M-MuLV p12 or, as a control, from the C terminus of HIV-1 MA, which is not rich in Gly or Pro. In each case the foreign sequence was inserted into the SpeI site of ΔMBDdp10. Neither of these chimeric proteins assembled into recognizable structures in vitro (Fig. 1, lines 7 and 8). We conclude that the effect of the p10 domain on particle shape is specific.
In order to delineate the sequences within p10 responsible for the shape-determining function, we originally deleted one-third or two-thirds of p10 in the context of p10-CA-NC. However, protein expression from these two constructs was poor, and the assembly phenotype could not be determined (data not shown). As an alternative approach, and to buffer the Gag protein against possible N-terminus effects, we inserted portions of the p10 coding sequence back into ΔMBDdp10. The experimental logic was to determine the minimal segment of polypeptide sufficient to restore spherical morphology. The central 52-aa core of the p10 domain was divided approximately into thirds, called A, B, and C. Insertion of segment C into ΔMBDdp10 abolished the assembly of regular structures altogether (Fig. 1, line 12). Insertion of segment AB still gave a tubular assembly phenotype (Fig. 1, line 13). Insertion of segment BC restored efficient assembly of spherical particles (Fig. 1, line 14). Since a single restriction site was used for cloning, some of the inserted DNA fragments were inserted in the antisense orientation. One of these abrogated assembly (Fig. 1, line 10), while two others gave rise to tubes (Fig. 1, lines 9 and 11). It should be noted that in all of the insertion mutants shown in Fig. 1, lines 7 to 14, the final protein sequence included an extra dipeptide sequence, Thr-Ser, positioned 6 aa upstream of the start of CA, corresponding to the SpeI cloning site. That this dipeptide insertion itself does not perturb spherical assembly is shown by the phenotype of the BC insertion mutant (Fig. 1, line 14).
To locate the shape-determining function more precisely, a set of serial deletions was constructed. Stretches of 5, 10, and 15 residues were deleted from the N terminus of the BC segment. These constructs were prepared in such a way that the 25 to 35 aa of p10 upstream of CA were entirely wild type, without the added dipeptide sequence from the artificial SpeI site. Each new protein was assayed for particle shape during assembly. In this way, the minimal region sufficient to restore efficient spherical assembly was determined to be the C-terminal 25 residues of p10, which encompass 5 residues from segment B and all of segment C (Fig. 1, line 17). The sequence of this minimal region, including the five residual residues still present in the original dp10 deletion, is PGPALTDWARVREELASTGPPVVAM.
RSV Gag proteins with mutations in p10 were reported to be quantitatively defective in budding in chicken cells (7). In that study, each of several 15- to 20-aa deletions throughout p10 reduced budding about 20-fold. In addition, two more localized mutations, an insertion of Thr-Arg at p10 residue 8 and a Gly-to-Arg substitution at residue 56, resulted in a temperature-sensitive budding phenotype in chicken cells. We built the Thr-Arg insertion mutation into the context of the p10-CA-NC expression plasmid. The resulting protein, called p10TR-CA-NC (Fig. 1, line 4), assembled into spherical particles in E. coli with the same efficiency as the wild-type p10-CA-NC (Table 1).
Random mutagenesis of the minimal region.
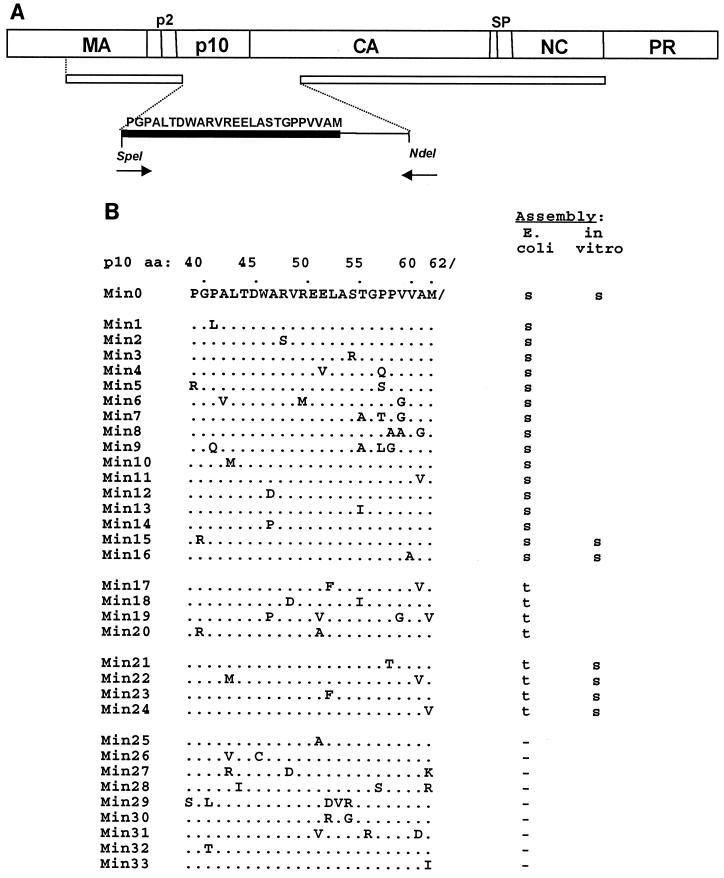
In the absence of structural information to help guide site-directed mutagenesis, we generated a collection of random mutations in the p10 minimal segment, using a partially degenerate primer and amplification by PCR. In the forward primer, a stretch of nucleotides corresponding to the 25 aa of interest was doped such that at each position the nucleotide was correct in 88% of the molecules and mutant in the remainder. The backwards primer was wild type in sequence and started at the natural NdeI site in the CA gene (Fig. 3A). The PCR product was inserted between the unique SpeI and NdeI sites of the plasmid encoding ΔMBDdp10. After transformation into BL21(DE3)/pLysS cells, 125 colonies were picked for analysis. This level of doping resulted in an average of two or three amino acid substitutions per expressed clone, as predicted theoretically.
FIG. 3.
Results of random and site-directed mutagenesis. (A) Mutagenesis strategy. A schematic diagram shows the PCR strategy for the partial-randomization experiment. The top bar represents the Gag polyprotein. The second, narrower bar denotes the ΔMBDdp10 protein; the gap is at the SpeI site. The forward primer starts at the corresponding SpeI site; the reverse primer starts at the natural NdeI site for the CA gene. The sequence that is shown (thick black bar) was partially randomized. The methionine at the end of this sequence is the C-terminal residue of the wild-type p10 protein. (B) Mutant sequences and assembly phenotypes. Dots denote the wild-type sequence, with changes from the wild-type sequence shown by letters. The results are for particles observed in crude lysates and for some particles assembled in vitro from purified proteins. Blanks indicate that the protein was not tested. s, spheres; t, tubes; −, no assembly.
A crude extract from each of the transformants was tested for expression of Gag protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The results showed that 60 clones did not express sufficient protein to be identifiable as an obvious band on stained gels, or they expressed a protein that was smaller than the expected size (∼47 kDa), due to premature termination. For the 65 clones that did express the correct product, particles in the crude lysates were visualized by TEM. Since the wild-type sequence restores spherical particle formation to ΔMBDdp10, we were most interested in mutations that altered the assembly phenotype back to tubes, indicating that the function of the 25-aa segment was perturbed. In total, 18 of the mutant proteins still had efficient spherical assembly, 15 had tubular assembly, and 32 lacked assembly altogether. DNA sequences were determined for all clones giving rise to tubes or spheres and for a subset of the clones that did not lead to assembly. The mutant clones were assigned numbers with the prefix Min, to show that they had been created in the minimal p10 segment required to promote efficient spherical assembly, with the wild-type segment being called Min0 (Fig. 3B).
Inspection of the collection of unique amino acid sequences and their phenotypes (Fig. 3B) does not show an obvious pattern. For instance, each of the prolines in the minimal region was mutated in at least one clone, without changing spherical assembly. In fact, three of the four prolines were changed in one mutant, Min9, without changing spherical particle formation. Nevertheless, the prolines could not be changed arbitrarily, since mutant Min21 (P58T) had a tubular assembly phenotype and mutant Min32 (P40T) did not assemble at all. Similarly, the percentages of all the nonconservative changes were similar for the collections of mutants that formed spheres or tubes did not assemble. We conclude from these results that since the minimal segment of p10 is sensitive to mutation, the amino acid sequence must fold into an ordered structure, either by itself or in conjunction with other sequences in CA.
Since most of the mutants had more than one amino acid substitution, we next used site-directed mutagenesis to isolate individual mutations and thus try to determine which were responsible for the phenotype. Twelve single amino acid mutants were constructed in this way. In the collection of all single amino acid mutants, including those obtained in the original screen, 10 assembled into spheres in E. coli, 3 assembled into tubes, and 3 failed to assemble into regular structures. The three tube-forming mutants, Min21 (P58T), Min23 (M62V), and Min24 (L52F), have conservative substitutions, consistent with the notion that subtle changes of a structural element are enough to perturb the Gag polymerization pathway.
The three single mutants and one double mutant, Min22 (L42M and A61V), that gave rise to tubular particles in crude extracts were also tested for assembly phenotype in vitro, after purification of each protein. Surprisingly, each of these mutant proteins assembled in vitro into spherical rather than into tubular particles (Fig. 3B). This finding of altered assembly phenotypes was unexpected, since all other Gag proteins examined previously (all the proteins shown in Fig. 1) had the same phenotype in vitro and in E. coli. As a control we also tested two single mutants (Min15 and Min16) that formed spheres in E. coli; both of them showed the same spherical phenotype as purified proteins. Several further experiments were carried out to try to gain evidence for the underlying difference between assembly in E. coli and in vitro (data not shown). First, a concentrated E. coli lysate without Gag protein was added to the in vitro assembly reaction with several purified mutant proteins. No change from spheres to tubes was observed. Second, for the same mutants, particles in crude lysates were diluted in high-salt (0.5 M NaCl) lysis buffer in order to break apart any preformed VLPs and then dialyzed against in vitro assembly buffer (0.1 M NaCl, pH 6.0). The VLPs observed in this experiment had the same tubular shape as those in the original crude extracts. Third, purified Gag proteins were assembled in vitro at pH 7.0, which is similar to the intracellular pH. Although the assembly was less efficient at this pH, the VLPs observed still had the same spherical shape as seen at pH 6.0.
We interpret these results to mean that some factor or condition in E. coli favors tubular assembly for mutant proteins that are intrinsically able to polymerize in either a spherical or a tubular mode. The propensity to form one shape or another may be keyed by alternative conformations of Gag, as suggested for HIV in vitro assembly of Gag proteins (19). The more subtle mutations in the 25-aa stretch of p10 may, then, be viewed as balancing the protein between two alternative conformations, such that small changes in the environment lead to different polymerization modes. We speculate that once assembly has been nucleated in either mode, proteins will adapt their conformation to add to the polymerized lattice in the same fashion.
Importance of residues at the immediate N terminus of CA.
In the presence of RNA, HIV-1 CA-NC assembles into tubes in vitro (5, 17). However, CA-NC carrying a few extra N-terminal residues forms spheres instead of tubes, although the assembly products in this case are heterogenous in size and the assembly is not efficient (18, 35). A model explaining this result (35) invokes the hypothetical malformation of the β-hairpin at the N terminus of CA, a feature seen in the three-dimensional structure of the N-terminal domain of CA (10, 15). The hairpin is stabilized by a salt bridge between the amino-terminal residue Pro1 and Asp51, both of which are highly conserved residues in CA proteins of diverse retroviruses. It was thus hypothesized that an intact hairpin and salt bridge are involved in the CA-CA interactions leading to tube formation (35). In contrast, extensions of the CA N terminus, which would prevent formation of the salt bridge and thus presumably abrogate the formation of the hairpin or cause its malformation or displacement, would promote a structural change that allows CA to polymerize into spheres. A β-hairpin almost identical to that in HIV CA is found in the N-terminal CA domain of RSV (22).
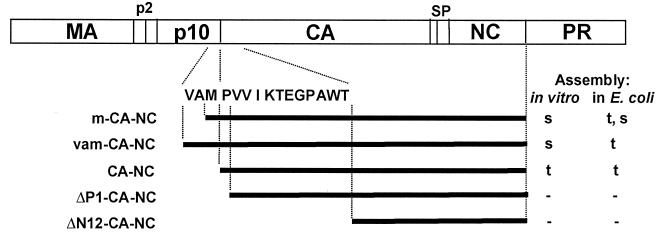
To examine the importance of the amino terminus of RSV CA in tube formation, we constructed two CA-NC-related proteins with very short extensions directly upstream of CA. These new proteins were called m-CA-NC and vam-CA-NC; “m” and “vam” indicate the addition of Met and Val-Ala-Met, respectively. These residues occur naturally at this position in Gag, comprising the C terminus of p10. In addition to codons for Met or Val-Ala-Met, the constructs encoding these proteins also contained an initiating AUG coding for an additional Met residue. The m-CA-NC and vam-CA-NC proteins were purified and tested for the ability to assemble in vitro. Both assembled into spheres (Fig. 4). However, the efficiency of assembly was extremely low (Table 1), with only occasional particles visible on an EM grid. In contrast, the same proteins assembled into tubes within E. coli. This difference in assembly phenotype in the two environments is reminiscent of the differences seen for several of the single mutants described above. For the shorter protein, a caveat in the interpretation of the assembly phenotype comes from N-terminal sequence analysis that we carried out, which showed a mixture of two ends (data not shown). The predominant species of m-CA-NC started with the sequence PVV, i.e., corresponding to CA-NC itself, implying that two Met residues were removed. The minor species, representing about 10% of the ends, started with the expected sequence MPVV, corresponding to a protein with just the initiating Met removed. Since CA-NC by itself forms tubes, the tubes visualized in E. coli are most easily explained as the product of polymerization of CA-NC. The absence of any tubes visualized in vitro might reflect the poisoning of tubular assembly in vitro by the minor species with one Met residue. Although we did not determine the N-terminal sequence of vam-CA-NC, presumably in this case three or four amino acid residues preceded CA-NC, and thus the observation of tubular VLPs in crude extracts and rare spherical VLPs in vitro is most likely explained by whatever factor also causes some of the point mutants to form tubes in the environment of the E. coli cell.
FIG. 4.
Effects of small additions and deletions at the N terminus of CA-NC. Assembly phenotypes of proteins with minimal additions or deletions at the N terminus of CA-NC are shown. ΔP1, deletion of Pro1 residue; ΔN12, deletion of the N-terminal 12 residues of CA. All proteins had an additional initiating Met aside from the sequence shown. VAM/PVVIKTEGPAWT is the sequence of the C-terminal 3 residues of p10 and the N-terminal 12 residues of CA. Mutants with changes in this region are not drawn to scale relative to the rest of the CA-NC protein. s, spheres; t, tubes; −, no assembly.
In a different set of experiments, we created constructs designed to delete the first residue (Pro1) or the first 12 residues of the CA domain of CA-NC. In the former construct, an initiating Met residue was used to replace Pro1. Neither of these mutant proteins assembled into regular structures in vitro or in E. coli (Fig. 4). Since an initiating Met residue is poorly removed in overexpressed proteins if followed by a residue with a large side chain, the mutant CA-NC protein possibly had a mixture of the N termini Met-Val-Val and Val-Val.
In summary, the results for some three dozen Gag proteins assembled in E. coli or in vitro lead to the conclusion that the N-terminal sequence of CA and the immediately upstream 25-aa residues in p10 are important elements in the polymerization of RSV Gag proteins. While CA-NC proteins extended N terminally by a only few amino acid residues formed rare spherical particles, for efficient assembly of spherical particles the stretch of the C-terminal 25 residues of p10 was crucial. In a collection of 33 proteins with mutations in this segment of polypeptide, 9 mutants did not assemble into regular structures in E. coli, 8 formed tubular particles, and 16 formed spherical particles like those of the wild-type sequence. The lack of any obvious pattern in this collection of mutants suggests that it is conformation, and not sequence per se, that is important for promoting spherical assembly. The simplest hypothesis to account for these observations is that the 25-aa segment folds into a structure that interacts specifically with CA. Given that mutation of Pro1 in RSV CA-NC abrogates assembly and that short N-terminal extensions of CA-NC lead to at least some spherical particles, it seems likely that this hypothetical interaction is with the N-terminal domain of CA and, in particular, perhaps with the β-hairpin found there (22).
The data presented here are based exclusively on EM, a technique that is necessarily qualitative or, at best, semiquantitative. While we have noted gross quantitative differences in assembly efficiency, for example, for comparisons of assembly of m-CA-NC and p10-CA-NC, we have not reported lesser quantitative differences because of uncertainty about their significance. However, the major conclusions from this study do not rest on quantitative differences but rather on the obvious distinctions in the shapes of tubes and spheres. All visual assays by EM were carried out at least twice with independent protein preparations or with cell extracts from independently induced E. coli cultures. With the single exception of the protein m-CA-NC, there was no protein for which particles of both shapes were observed together or particles of one shape were observed once and particles of another shape were observed in a different experiment. To verify that both tubes and spheres would be identifiable in the same extract, we prepared mixtures of two crude extracts, one containing tubes and the other containing spheres, at ratios of 1:1, 10:1, and 1:10. Both types of particles were readily visible in all of these mixtures (data not shown). The observation of two shapes for the m-CA-NC protein is a special case, given that the protein preparation was found to be composed of molecules with two different N-terminal amino acid sequences. Thus, overall, the scoring of particle shapes of the various mutants should be reliable. The scoring of lack of assembly, which by nature is a negative result, arguably is less convincing, since assembly might take place with some alteration in conditions. However, for these mutants, at least two independent experiments were also carried out, with the same phenotype being recorded for each.
While the roles of several RSV Gag domains during assembly are understood at least in part, the role of the p10 domain has remained largely unknown. The Gag proteins of most retroviruses include one or two small proteins derived from amino acid sequences between MA and CA, and at least one of these proteins is typically very rich in Pro and Gly, suggesting the lack of an overall globular fold. This prediction is consistent with radial density measurements of immature HIV and M-MuLV particles, which imply that part of the Gag sequence between the membrane binding domain of MA and the N-terminal domain of CA is in an extended conformation (9, 37). RSV p10 is just C terminal to the sequence PPPY, comprising the core of the late domain, which plays a key role in a late step in budding (29, 36), probably by interacting with one of the members of the cellular family of WW proteins (13). Three previous studies have investigated the role of p10 in RSV assembly in vivo in transfected cells. Krishna et al. (25) reported that deletion of part or most of p10 modestly reduced assembly in COS cells and gave rise to particles that appeared somewhat smaller by rate zonal sedimentation. Dupraz and Spahr (7) showed that several partial deletions of p10 reduced the budding of virus particles as much as 20-fold in chicken cells. In addition, they found that two of five more subtle mutations in p10 gave a temperature-sensitive budding phenotype. On the other hand, T. Cairns and R. Craven (personal communication) found that budding from transfected COS cells was not significantly impaired either in the dp10 mutant or in another mutant deleting 31 aa residues of p10, in the context of an otherwise infectious viral clone expressed in quail cells. In addition, they found that a mutant with a deletion of 26 residues from the N-terminal portion of p10 retains infectivity. The reason for the apparent discrepancies among these results remains unclear, but they might be due to differences in virus strain or cell type. In none of these cases was the morphology of the budding particles determined.
What is the significance of the two shapes of VLPs? The spherical particles are closely similar to immature virions without a membrane, as shown for HIV-1 (19) and RSV (R. Kingston, personal communication) by cryo-EM. That is, the in vitro-assembled particles show the characteristic radial density profile first noted for HIV-1 (9) and M-MuLV (37), in which the most prominent features are the densities of the two structurally distinct domains of CA. We hypothesize that the tubular particles are analogues of mature cores, but the logic for this hypothesis is indirect, relying on comparisons with HIV-1 assembly in vitro. In HIV-1, under some conditions CA-NC forms a mixture of tubes and cones (12), and thus the protein-protein contacts in these two structures are probably similar. Mature lentivirus cores are conical. To date, no in vitro assembly system has produced mature cores of C-type retroviruses, like RSV or M-MuLV, which appear to have an irregular polyhedral shape (37; R. Kingston, personal communication). An alternative hypothesis is that tubular particles represent the polymerization of Gag in a manner like that observed for some mutants expressed in vivo. For example, baculovirus expression of an HIV-1 Gag protein with the structure MA-CA leads to the massive appearance of membrane-enclosed tubular structures on the surfaces of insect cells (14, 28). Similarly, deletion of the M-MuLV p12 domain leads to budding of both tubular and spherical particles (38). Since p12 carries the M-MuLV late domain sequence, PPPY, believed to interact with a cellular protein, the alternative morphologies recorded in that study may be related to interaction or lack of interaction with a cellular factor. Conceivably, the discordance we have observed for some RSV p10 mutants which showed spherical assembly in vitro but tubular assembly in E. coli is also accounted for by a factor present in E. coli cells.
What mechanisms underlie the difference in the shapes of tubes and spheres? It has been known for several decades that capsid proteins of plants and bacteriophages can polymerize into diverse regular shapes. For example, cowpea chlorotic mosaic virus capsid protein forms either spheres, sheets, or tubes, depending on pH and ionic strength (2). The different polymerization modes observed in retroviral Gag proteins are likely to reflect subtle differences in the conformation of the protein, in particular the CA domain, since this part of Gag is generally considered to be the major locus of interactions between Gag molecules in assembly. This notion is based in part on the observation that HIV CA forms dimers in vitro via interactions between C-terminal domains (11). However, the nature of CA-CA interaction in RSV is less clear, since RSV CA remains monomeric even at concentrations as high as 20 mg/ml (22). The first detailed model proposed to explain the tubular and spherical polymerization modes in vitro (35) invoked the presence or absence of the β-hairpin at the N terminus of mature CA (10, 15). However, both reduction of pH and deletion of the spacer peptide between HIV CA and NC can change the in vitro assembly product from spheres to tubes (19). This recent result suggests that the alternative assembly modes reflect more global conformational changes or, alternatively, that there is some communication between the C-terminal and the N-terminal domains of CA. The published solution structures of retroviral CA proteins (3, 21) show little evidence for interaction between the two CA domains. However, structures of larger Gag proteins, encompassing CA with both upstream and downstream sequences, have yet to be determined.
Acknowledgments
We thank Stephen Campbell and Yu Ma for helpful advice, Mark Berryman for help with thin sectioning, Wes Sundquist for communication of unpublished results, and Marc Johnson and Deborah Lynn for critical reading of the manuscript.
This work was supported by USPHS grant CA-20081.
REFERENCES
- 1.Ausubel F M, et al., editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 2.Bancroft J B. The self-assembly of spherical plant viruses. Adv Virus Res. 1970;16:99–134. doi: 10.1016/s0065-3527(08)60022-6. [DOI] [PubMed] [Google Scholar]
- 3.Berthet-Colominas C, Monaco S, Novelli A, Sibal G, Mallet F, Cusack S. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 1999;18:1124–1136. doi: 10.1093/emboj/18.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell S, Vogt V M. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupraz P, Spahr P-F. Analysis of deletions and thermosensitive mutations in Rous sarcoma virus gag protein p10. J Virol. 1993;67:3826–3834. doi: 10.1128/jvi.67.7.3826-3834.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrlich L S, Agresta B E, Carter C A. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J Virol. 1992;66:4874–4883. doi: 10.1128/jvi.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller S D, Wilk T, Gowen B E, Kräusslich H-G, Vogt V M. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 10.Gamble T L, Vajdos F F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 11.Gamble T L, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 12.Ganser B K, Li S, Klishko V Y, Finch J T, Sundquist W I. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 13.Garnier L, Wills J W, Verderame M F, Sudol M. WW domains and retrovirus budding. Nature (London) 1996;381:744–745. doi: 10.1038/381744a0. [DOI] [PubMed] [Google Scholar]
- 14.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 15.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 16.Grättinger M, Hohenberg H, Thomas D, Wilk T, Müller B, Kräusslich H-G. In vitro assembly properties of wild-type and cyclophilin-binding defective human immunodeficiency virus capsid proteins in the presence and absence of cyclophilin A. Virology. 1999;257:247–260. doi: 10.1006/viro.1999.9668. [DOI] [PubMed] [Google Scholar]
- 17.Gross I, Hohenberg H, Kräusslich H-G. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 18.Gross I, Hohenberg H, Huckhagel C, Kräusslich H-G. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J Virol. 1998;72:4798–4810. doi: 10.1128/jvi.72.6.4798-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross I, Hohenberg H, Wilk T, Wiegers K, Grättinger M, Müller B, Fuller S, Kräusslich H-G. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000;19:103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hockley D J, Nermut M V, Grief C, Jowett J B M, Jones I M. Comparative morphology of Gag protein structures produced by mutants of the gag gene of human immunodeficiency virus type 1. J Gen Virol. 1994;75:2985–2997. doi: 10.1099/0022-1317-75-11-2985. [DOI] [PubMed] [Google Scholar]
- 21.Jin Z, Jin L, Peterson D L, Lawson C L. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J Mol Biol. 1999;286:83–93. doi: 10.1006/jmbi.1998.2443. [DOI] [PubMed] [Google Scholar]
- 22.Kingston R, Eisenmesser E Z, Fitzon-Ostendorp T, Schatz G W, Vogt V M, Post C B, Rossmann M G. Structure and self-association of the Rous sarcoma virus capsid protein. Structure. 2000;8:617–628. doi: 10.1016/s0969-2126(00)00148-9. [DOI] [PubMed] [Google Scholar]
- 23.Klikova M, Rhee S S, Hunter E, Ruml T. Efficient in vivo and in vitro assembly of retroviral capsids from Gag precursor proteins expressed in bacteria. J Virol. 1995;69:1093–1098. doi: 10.1128/jvi.69.2.1093-1098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovari L C, Momany C A, Miyagi F, Lee S, Campbell S, Vuong B, Vogt V M, Rossmann M G. Crystals of Rous sarcoma virus capsid protein show a helical arrangement of protein subunits. Virology. 1997;238:79–84. doi: 10.1006/viro.1997.8807. [DOI] [PubMed] [Google Scholar]
- 25.Krishna N K, Campbell S, Vogt V M, Wills J W. Genetic determinants of Rous sarcoma virus particle size. J Virol. 1998;72:564–577. doi: 10.1128/jvi.72.1.564-577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lingappa J R, Hill R L, Wong M L, Hegde R S. A multistep ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J Cell Biol. 1997;136:567–581. doi: 10.1083/jcb.136.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McConnell J M, Fushman D, Cahill S M, Zhou W, Wolven A, Wilson C B, Nelle T D, Resh M D, Wills J, Cowburn D. Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus. J Mol Biol. 1998;279:921–928. doi: 10.1006/jmbi.1998.1788. [DOI] [PubMed] [Google Scholar]
- 28.Morikawa Y, Hockley D J, Nermut M V, Jones I M. Roles of matrix, p2, and N-terminal myristoylation in human immunodeficiency virus type 1 Gag assembly. J Virol. 2000;74:16–23. doi: 10.1128/jvi.74.1.16-23.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pepinsky R B, Vogt V M. Purification and properties of a fifth major viral gag protein from avian sarcoma and leukemia viruses. J Virol. 1983;45:648–658. doi: 10.1128/jvi.45.2.648-658.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakalian M, Parker S D, Weldon R A, Jr, Hunter E. Synthesis and assembly of retrovirus Gag precursors into immature capsids in vitro. J Virol. 1996;70:3706–3715. doi: 10.1128/jvi.70.6.3706-3715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz D E, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983;32:853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- 33.Spearman P, Ratner L. Human immunodeficiency virus type 1 capsid formation in reticulocyte lysates. J Virol. 1996;70:8187–8194. doi: 10.1128/jvi.70.11.8187-8194.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogt V M, Simon M N. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J Virol. 1999;73:7050–7055. doi: 10.1128/jvi.73.8.7050-7055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Schwedler U K, Stemmler T L, Klishko V Y, Li S, Albertine K H, Davis D R, Sundquist W I. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeager M, Wilson-Kubalek E M, Weiner S G, Brown P O, Rein A. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc Natl Acad Sci USA. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan B, Campbell S, Bacharach E, Rein A, Goff S P. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J Virol. 2000;74:7250–7260. doi: 10.1128/jvi.74.16.7250-7260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]