Abstract
Adeno-associated virus type 2 (AAV2) has proven to be a valuable vector for gene therapy. Characterization of the functional domains of the AAV capsid proteins can facilitate our understanding of viral tissue tropism, immunoreactivity, viral entry, and DNA packaging, all of which are important issues for generating improved vectors. To obtain a comprehensive genetic map of the AAV capsid gene, we have constructed 93 mutants at 59 different positions in the AAV capsid gene by site-directed mutagenesis. Several types of mutants were studied, including epitope tag or ligand insertion mutants, alanine scanning mutants, and epitope substitution mutants. Analysis of these mutants revealed eight separate phenotypes. Infectious titers of the mutants revealed four classes. Class 1 mutants were viable, class 2 mutants were partially defective, class 3 mutants were temperature sensitive, and class 4 mutants were noninfectious. Further analysis revealed some of the defects in the class 2, 3, and 4 mutants. Among the class 4 mutants, a subset completely abolished capsid formation. These mutants were located predominantly, but not exclusively, in what are likely to be β-barrel structures in the capsid protein VP3. Two of these mutants were insertions at the N and C termini of VP3, suggesting that both ends of VP3 play a role that is important for capsid assembly or stability. Several class 2 and 3 mutants produced capsids that were unstable during purification of viral particles. One mutant, R432A, made only empty capsids, presumably due to a defect in packaging viral DNA. Additionally, five mutants were defective in heparan binding, a step that is believed to be essential for viral entry. These were distributed into two amino acid clusters in what is likely to be a cell surface loop in the capsid protein VP3. The first cluster spanned amino acids 509 to 522; the second was between amino acids 561 and 591. In addition to the heparan binding clusters, hemagglutinin epitope tag insertions identified several other regions that were on the surface of the capsid. These included insertions at amino acids 1, 34, 138, 266, 447, 591, and 664. Positions 1 and 138 were the N termini of VP1 and VP2, respectively; position 34 was exclusively in VP1; the remaining surface positions were located in putative loop regions of VP3. The remaining mutants, most of them partially defective, were presumably defective in steps of viral entry that were not tested in the preliminary screening, including intracellular trafficking, viral uncoating, or coreceptor binding. Finally, in vitro experiments showed that insertion of the serpin receptor ligand in the N-terminal regions of VP1 or VP2 can change the tropism of AAV. Our results provide information on AAV capsid functional domains and are useful for future design of AAV vectors for targeting of specific tissues.
Adeno-associated virus type 2 (AAV2) belongs to the human parvovirus family, which requires a helper virus for productive replication (5, 7, 8). The nonenveloped capsid adopts an icosahedral structure with a diameter of approximately 20 nm. Packaged within the capsid is a single-stranded DNA genome of 4.7 kb that contains two large open reading frames (ORFs), rep and cap (35). Three structural proteins, designated VP1, VP2, and VP3, are encoded in the cap ORF and made from the p40 promoter by use of alternative splicing and alternative start codons. The three proteins share the same ORF and end at the same stop codon. The C-terminal regions common to all three capsid proteins fold into a β-barrel structure that is present in several viruses (31). Their molecular masses are 87, 73, and 62 kDa, and their relative abundances within the capsid are approximately 5, 5, and 90%, respectively (26). Recently, AAV has attracted a significant amount of interest as a vector for gene therapy (6, 26). It has a number of unique advantages that are potentially useful for gene therapy applications, including the ability to infect nondividing cells, a lack of pathogenicity, and the ability to establish long-term gene expression.
Early genetic studies on deletion mutants of AAV revealed that capsid proteins were required for accumulation of single-stranded DNA and production of infectious particles (19, 38). Mutations in the C-terminal region common to all three proteins also abolished virion formation and failed to accumulate single-stranded DNA (32). VP1 was thought to be important for virus infectivity or stability because mutations in the N-terminal region unique to VP1 produced DNA-containing particles with significantly reduced infectivity (19, 38). In vitro assembly studies (33) and capsid initiation codon mutagenesis studies (25) suggested that both VP2 and VP3 were required for capsid formation and production of infectious particles, and either VP1 or VP2 was required for nuclear localization of VP3. Recently, Hoque et al. (19b) have shown that the VP2 N-terminal residues 29 to 34 are sufficient for nuclear translocation and suggested that the major function of VP2 is to translocate VP3 into the nucleus. A recent insertional mutation study on AAV capsid protein revealed that mutations in the capsid gene could affect AAV capsid assembly and infection (30). Since the crystal structure of AAV was still unavailable, the functional domains of the AAV capsid proteins were mostly predicted based on information derived from other related autonomous parvoviruses, canine parvovirus (CPV), feline panleukopenia virus, and B19, whose crystal structures were available (1, 2, 40, 41). Sequence comparison of AAV to these viruses revealed a few conserved functional domains (9, 10), but the exact functions of these domains were not clear.
While certain groups of cells cannot be transduced by AAV (22, 27), AAV can transduce a wide variety of tissues, including brain, muscle, liver, lung, vascular endothelial, and hematopoietic cells (12–14, 16, 21, 45, 48). Recently, Summerford and Samulski (37) reported that heparan sulfate proteoglycan is the primary cellular receptor for AAV, and their group further revealed that the binding site lies within VP3 (30). In addition, human fibroblast growth factor receptor 1 and αvβ5 integrin were identified as coreceptors for AAV (28, 36). Attempts to alter the AAV capsid also have been made in order to expand the tropism of AAV. Yang et al. (47) showed improved infectivity of hematopoietic progenitor cells by generating a chimeric recombinant AAV (rAAV) having the single-chain antibody against human CD34 protein. Girod et al. (15) showed that insertion of the L14 epitope into the capsid coding region can expand the tropism of this virus to cells nonpermissive for AAV infection that bear the L14 receptor. However, in both cases the normal AAV tropism was not disrupted. Ideally, for the purpose of retargeting, the normal AAV receptor binding would need to be modified so that rAAV infects only targets bearing the receptors for the engineered epitope.
In this study, we used site-directed mutagenesis to mutate the capsid ORF. Initially, 48 alanine scanning mutations were made in which two to five charged amino acids in the AAV capsid ORF were mutated to alanine residues by site-directed mutagenesis. We reasoned that since the mutations were an average of 15 to 20 amino acids (aa) apart and spanned the whole capsid gene, some of them would inevitably fall in or near the functional domains of AAV capsid. In addition, over 40 substitution and insertion mutations were made in a search for regions that could tolerate insertions for the purpose of retargeting AAV vectors. By analyzing these mutants, we obtained a preliminary functional map of the AAV capsid protein. Our results identified critical regions within the capsid that were potentially responsible for receptor binding, DNA packaging, capsid formation, and infectivity. In addition, we identified sites that were suitable for epitope insertions that might be useful for targeted gene delivery.
MATERIALS AND METHODS
Cell culture.
Low-passage-number (passages 27 to 38) HFK 293 cells (17) and HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, penicillin (100 U/ml), and streptomycin (100 U/ml) at 37°C and 5% CO2. IB3 cells were propagated as described elsewhere (34).
Construction of AAV capsid mutant plasmids.
Plasmid pIM45 (previously called pIM29-45 [23]) was used as the template for all mutant constructions. Mutagenesis was achieved by using the Stratagene site-directed mutagenesis kit according to the supplier's manual. For each mutant, we designed two PCR primers which contained the sequence of alanine substitution or insertion plus a unique endonuclease restriction site flanked by 15 to 20 homologous bp on each side of the substitution or insertion. The restriction site was designed to facilitate subsequent DNA sequencing of the mutants and for potential insertion of tags or foreign epitopes. The PCR products were digested with endonuclease DpnI to eliminate the parental plasmid template and were propagated in Escherichia coli XL-Blue (Stratagene). Miniprep DNAs were extracted from ampicillin-resistant colonies and were screened by restriction endonuclease digestion. Positive clones were sequenced in the capsid ORF region. The capsid ORF was then subcloned back into the pIM45 backbone with SmaI and SphI to eliminate background mutations. The same mutagenesis strategy was used for peptide substitution and insertion mutant constructions.
Production of rAAV particles.
To produce rAAV with mutant capsid proteins, we transfected 293 cells with three plasmids: (i) pIM45, which supplied either wild-type (wt) or mutant capsid proteins (23); (ii) pXX6, which contained the adenovirus (Ad) helper genes (46); and (iii) pTRUF5, which contains the green fluorescent protein (gfp) gene driven by the cytomegalovirus (CMV) promoter and flanked by the AAV terminal repeats (22). In some experiments, pTRUF5 was substituted with CBA-AT, a recombinant AAV plasmid that contains the human α1-antitrypsin (hAAT) gene under the control of the CMV–β-actin promoter. The plasmids were mixed at a 1:1:1 molar ratio. Plasmid DNAs used for transfection were purified by the QIAGEN Maxi-prep kit according to the supplier's manual.
The transfections were carried out as follows. 293 cells were split 1:2 the day before the transfection so that they could reach 75% confluency the next day. Ten 15-cm-diameter plates were transfected at 37°C, using calcium phosphate as described elsewhere (51), and incubated at 37°C. Forty-eight hours after transfection, cells were harvested by centrifugation at 1,140 × g for 10 min, the pellets were resuspended in 10 ml of lysis buffer (0.15 M NaCl, 50 mM Tris-HCl [pH 8.5]), and viruses were released by freezing and thawing three times. The crude rAAV lysates were treated with Benzonase (pure grade; Nycomed Pharma A/S) at a final concentration of 50 U/ml at 37°C for 30 min. The crude lysates were clarified by centrifugation at 3,700 × g for 20 min, and the supernatant was subjected to further purification by iodixanol step gradient and heparan sulfate affinity purification as previously described (51).
To determine whether any of the mutants were temperature sensitive, the transfections were done in six-well dishes as duplicates at 39.5 and 32°C. Viruses were resuspended in 250 μl of lysis buffer. All crude rAAV preparations were stored at −80°C until their titers were determined.
Gel electrophoresis, immunoblotting, and immunoprecipitation.
Crude or purified rAAV samples were analyzed on sodium dodecyl sulfate (SDS)–10% polyacrylamide gels. The samples were mixed with sample buffer and boiled at 100°C for 5 min before loading. For immunoblotting, the proteins were transferred to a Nitro-bond membrane at 4°C, and the membrane was probed with monoclonal antibody (MAb) B1, directed against the capsid proteins (43). The capsid bands were visualized by peroxidase-coupled secondary antibodies using ECL (enhanced chemiluminescence detection) (Amersham) as suggested by the supplier.
For immunoprecipitation, heparan column-purified rAAV samples were diluted in 10 volumes of NETN buffer (0.1 M NaCl, 1 mM EDTA, 20 mM Tris-HCl [pH 7.5], 0.5% Nonidet P-40) and incubated overnight at 4°C in the presence of a MAb to the hemagglutinin (HA) epitope conjugated to Sepharose beads (BAbCo). For a negative control, MAb AU1-conjugated beads (BAbCo) were used. AU1 is a commonly used epitope, DTYRYI. After incubation, the samples were centrifuged for 5 min at 17,600 × g at 4°C. The beads were washed three times with 1 ml of NETN for 10 min at room temperature and resuspended in protein loading buffer. After centrifugation, the supernatant was precipitated with 15% trichloroacetic acid on ice for 1 h and centrifuged for 45 min at 4°C, and the pellet was resuspended in loading buffer. The samples then were boiled in sample buffer and analyzed by Western blotting with MAb B1 as described above.
Virus titers.
The infectious titers of rAAV-containing wt and mutant capsids were measured at two temperatures, 39.5 and 32°C, for the alanine scanning mutants and at 37°C for all other mutants by using the fluorescent cell assay, which detects expression of the gfp gene. This was done essentially as described previously by Zolotukhin et al. (51). Briefly, 293 cells were seeded in a 96-well dish the day before infection so that they would reach about 75% confluence the next day. Serial dilutions of wt and mutant rAAV-GFP crude preparations were added to the cells in the presence of Ad5 at a multiplicity of infection (MOI) of 10. The cells and viruses were incubated at 37°C (or 32° and 39.5°C) for 48 h, and the titers were determined by counting the number of green cells with the fluorescence microscope. For each mutant, the infections were done twice and the average was taken. For mutants that contained a packaged CBA-AT gene, infectivity was measured by the infectious center assay on 293 cells as previously described (51) and by enzyme-linked immunosorbent assay (ELISA) measurement of hAAT secreted into culture media from infected cells as described elsewhere (34).
To determine the rAAV physical particle titer, we used the A20 ELISA kit (American Research Bioproducts). The crude rAAV stocks were serially diluted and incubated with the A20 kit plate. The readings that fell into the linear range were taken, and the titers were read off the standard according to the manufacturer's instructions. The A20 antibody detects both full and empty particles (44).
To determine the titer of rAAV physical particles that were full (i.e., contained DNA), we used the quantitative competitive PCR (QC-PCR) assay as described previously (51). The crude rAAV stocks (100 μl) were digested first with DNase I to eliminate contaminating unpackaged DNA in 50 mM Tris-HCl (pH 7.5)–10 mM MgCl2 for 1 h at 37°C and then incubated with proteinase K (Boehringer) in 10 mM Tris HCl (pH 8.0)–10 mM EDTA–1% SDS for 1 h at 37°C. Viral DNA was extracted twice in phenol-chloroform and once with chloroform and then precipitated by ethanol in the presence of glycogen (10%). The DNA was washed with ethanol, dried, and dissolved in 100 μl of H2O, and 1 μl of the viral DNA was used for QC-PCR. Serial dilutions of the internal standard plasmid DNA with a deletion of GFP were included in the reaction, and the PCR products were separated by 2% agarose gel electrophoresis. The densities of the target and competitor bands in each lane were measured using ZERO-Dscan image analysis system software (version 1.0; Scanalytics) to determine the DNA concentration of the virus stock.
Heparan column binding assay.
The ability of mutants to bind to heparan sulfate was tested essentially as previously described (51). Crude rAAV preparations containing wt or mutant capsids were first subjected to iodixanol gradient purification. The 40% layer was then collected and loaded onto a 1-ml preequilibrated heparan column at room temperature (immobilized on cross-linked 4% beaded agarose; Sigma H-6508). The flowthrough fraction, wash (3 column volumes), and 1 M NaCl eluate were collected, and equivalent amounts of each sample were mixed with SDS sample buffer and electrophoresed on SDS-polyacrylamide gels. The yield of capsid proteins in each fraction was monitored with MAb B1 by Western blotting and ECL detection.
EM.
Electron microscopy (EM) was done in the ICBR EM lab of the University of Florida. Iodixanol gradient and heparan column-purified wt or mutant GFP-rAAVs were desalted and concentrated by using a Centricon 10 filter (Amicon). About a 5-μl drop of the virus sample was spotted onto carbon-coated grids and left for 1 min at room temperature. Excess fluid was drawn off, and the sample was washed three times with phosphate-buffered saline; 5 μl of 1% uranyl acetate was added for 10 s, and the grid was dried at room temperature for 10 min before viewing under EM.
RESULTS
Generation of AAV capsid mutations.
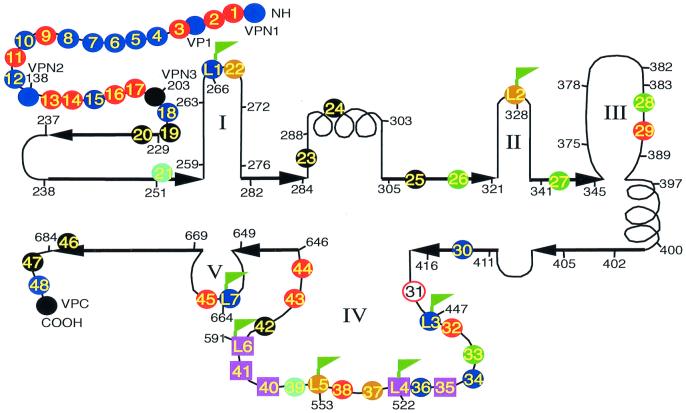
We began our studies by using alanine scanning site-directed mutagenesis in the hope that some of the mutants would be temperature sensitive (11). The mutants were constructed in the noninfectious AAV plasmid, pIM45, which contains all of the AAV DNA sequence except the AAV terminal repeats. There are approximately 60 charged clusters in the AAV capsid gene. Some of the clusters are overlapping; in those cases, only one cluster was chosen. For the initial round of mutagenesis, 48 sites, named mut1 to mut48, were targeted. These were spaced approximately equally over the capsid gene, with 12 mutants exclusively in VP1, 5 in VP2, and the rest in VP3 (Fig. 1). With the exceptions noted below, in each cluster, all charged amino acids were converted to alanine. The mutations were created so that they also contained a restriction site at the site of mutation to facilitate confirmation of the mutant sequence and subsequent insertion of foreign epitopes (Table 1). In addition, after sequence comparison of AAV serotypes 1 to 6, several other positions were targeted. mut28 and mut35 were made at positions where extra amino acids were found in AAV4 by sequence comparison with AAV2. mut32 was made by replacing TTT with AAA since TTT was not conserved among other AAV serotypes at aa 454. Finally, in mut29 and mut31, only one Arg residue was changed to Ala, and in mut45 and mut48, only one Lys was changed to Ala. The positions of the alanine scanning mutants and the specific amino acid substitutions are summarized in Table 1 and Fig. 1.
FIG. 1.
Distribution of alanine scanning and HA epitope insertion mutants. Positions of the alanine scanning mutants (colored circles or squares) and the HA insertion mutants (flagged circles or squares) are shown on a diagram of the putative secondary structure of the AAV capsid protein adapted from a comparison of parvovirus capsid sequences by Chapman and Rossmann (9). Some important amino acid positions and mutant positions are illustrated by numbers with short lines. Heavy arrows represent putative β sheets, and helices represent putative α helices. The five putative loop regions are numbered I to V. The colors of the circles indicate the phenotypes of the mutants as shown below: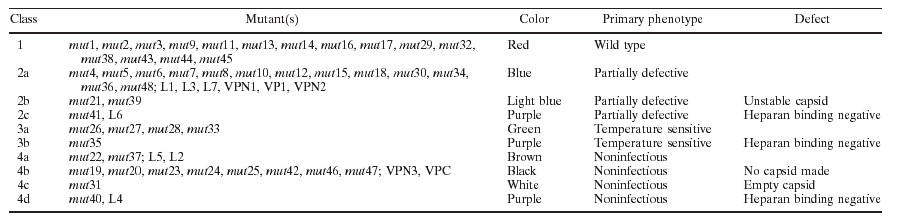
TABLE 1.
Summary of all mutants
| Mutanta | Typeb | Amino acid positionsb | Class | Phenotypec |
|---|---|---|---|---|
| mut11 | Ala sub | 9-13 DWLED-AWLAA | 1 | wt |
| mut21 | Ala sub | 24-28 KLKPG-ALAPG | 1 | wt |
| mut32 | Ala sub | 33-37 KPKER-APAAA | 1 | wt, surface |
| mut42 | Ala sub | 39-43 KDDSR-AAASA | 2a | pd, hep+ |
| mut53 | Ala sub | 63-67 EPVNE-APVNA | 2a | pd, hep+ |
| mut62 | Ala sub | 67-71 EADAA-AAAAA | 2a | pd, hep+ |
| mut72 | Ala sub | 74-78 EHDKA-AHAAA | 2a | pd, hep+ |
| mut82 | Ala sub | 76-80 DKAYD-AAAYA | 2a | pd, hep+ |
| mut91 | Ala sub | 84-88 DSGDN-ASGAN | 1 | wt |
| mut102 | Ala sub | 95-99 HADAE-AAAAA | 2a | pd, hep+ |
| mut112 | Ala sub | 102-107 ERLKED-AALAAA | 1 | wt |
| mut122 | Ala sub | 122-126 KKRVL-AAAVL | 2a | pd, hep+ |
| mut132 | Ala sub | 142-146 KKRPV-AAAPV | 1 | wt |
| mut141 | Ala sub | 152-156 EPDSS-APASS | 1 | wt |
| mut152 | Ala sub | 168-172 RKRLN-AAALN | 2a | pd, hep+ |
| mut162 | Ala sub | 178-182 GDADS-GAAAS | 1 | wt |
| mut171 | Ala sub | 180-184 DSVPD-ASVPA | 1 | wt |
| mut182 | Ala sub | 216-220 EGADG-AGAAG | 2a | pd, hep+ |
| mut191 | Ala sub | 228-232 WHCDS-WACAS | 4b | ni, no capsid |
| mut202 | Ala sub | 235-239 MGDRV-MGAAV | 4b | ni, no capsid |
| mut214 | Ala sub | 254-258 NHLYK-NALYA | 2b | pd, unstable capsid |
| mut224 | Ala sub | 268-272 NDNHY-NANAY | 4a | ni, full particle |
| mut234 | Ala sub | 285-289 NRFHC-NAFAC | 4b | ni, no capsid |
| mut242 | Ala sub | 291-295 FSPRD-FSPAA | 4b | ni, no capsid |
| mut252 | Ala sub | 307-311 RPKRL-APAAL | 4b | ni, no capsid |
| mut262 | Ala sub | 320-324 VKEVT-VAAVT | 3a | hs |
| mut271 | Ala sub | 344-348 TDSEY-TASAY | 3a | hs |
| mut282 | Ala ins | 384-385 AAA | 3a | cs |
| mut291 | Ala sub | 389 R-A | 1 | wt |
| mut302 | Ala sub | 415-419 FEDVP-FAAVP | 2a | pd, hep+ |
| mut314 | Ala sub | 432 R-A | 4c | ni, empty particle |
| mut322 | Ala sub | 454-456 TTT-AAA | 1 | wt |
| mut332 | Ala sub | 469-472 DIRD-AIAA | 3a | hs |
| mut342 | Ala sub | 490-494 KTSAD-ATSAA | 2a | pd, hep+ |
| mut352 | Ala ins | 509 AAAA | 3b | cs, hep−, surface |
| mut361 | Ala sub | 513-517 RDSLV-AASLV | 2a | pd, hep+ |
| mut372 | Ala sub | 527-532 KDDEEK-AAAAA | 4a | ni, full particle |
| mut382 | Ala sub | 547-551 SEKTN-SAATN | 1 | wt |
| mut392 | Ala sub | 553-557 DIEKV-AIAAV | 2b | pd, unstable capsid |
| mut402 | Ala sub | 561-565 DEEEI-AAAAI | 4d | ni, hep−, full particle, surface |
| mut412 | Ala sub | 585-588 RGNR-AGAA | 2c | pd, hep−, surface |
| mut422 | Ala sub | 607-611 QDRDV-QAAAV | 4b | ni, no capsid |
| mut432 | Ala sub | 624-628 TDGHF-TAGAF | 1 | wt |
| mut441 | Ala sub | 637-641 FGLKH-FGLAA | 1 | wt |
| mut452 | Ala sub | 665 K-A | 1 | wt |
| mut462 | Ala sub | 681-683 EIE-AAA | 4b | ni, no capsid |
| mut472 | Ala sub | 689-693 ENSKR-ASSAA | 4b | ni, no capsid |
| mut481 | Ala sub | 706 K-A | 2a | pd, hep+ |
| L1 | HA ins | 266 | 2a | pd, A20−, A20 epitope−, surface |
| L2 | HA ins | 328 | 4a | ni, A20+, surface |
| L3 | HA ins | 447 | 2a | pd, hep+, surface |
| L4 | HA ins | 522 | 4d | ni, hep−, surface |
| L5 | HA ins | 553 | 4a | ni, A20+, surface |
| L6 | HA ins | 591 | 2c | pd, hep−, surface |
| L7 | HA ins | 664 | 2a | pd, hep+, surface |
| VPN1 | HA, AU ins | 1 | 2a | pd, hep+, surface |
| VP1 | HA ins, Ser sub | 34 | 2a | pd, hep+, surface |
| VPN2d | HA, Ser ins | 138 | 2a | pd, hep+, surface |
| VPN3 | HA, Ser ins | 203 | 4b | ni, no capsid |
| VPC | HA, Ser, AU, His ins | 735 | 4b | ni, no capsid |
| mut1subser1 | Ser sub | 10 | 4a | ni, A20+ |
| mut2subser2 | Ser sub | 24 | 4a | ni, A20+ |
| mut3subser3 | Ser sub | 34 | 2a | pd, hep+ |
| mut9subser4 | Ser sub | 84 | 4a | ni, A20+ |
| mut14subser5 | Ser sub | 150 | 4a | ni, A20+ |
| mut16subser6 | Ser sub | 178 | 4b | ni, no capsid |
| mut19subser7 | Ser sub | 224 | 4b | ni, no capsid |
| mut32subser8 | Ser sub | 454 | 4b | ni, no capsid |
| mut37subser9 | Ser sub | 526 | 4b | ni, no capsid |
| mut39subser10 | Ser sub | 553 | 4b | ni, no capsid |
| mut40subser11 | Ser sub | 562 | 4b | ni, no capsid |
| mut41subser12 | Ser sub | 590 | 4b | ni, no capsid |
| mut44subser13 | Ser sub | 638 | 4b | ni, no capsid |
| mut45subser14 | Ser sub | 664 | 4b | ni, no capsid |
| mut46subser15 | Ser sub | 682 | 4b | ni, no capsid |
| mut4subflg2 | FLAG sub | 39 | 4a | ni, A20+ |
| mut8subflg3 | FLAG sub | 76 | 4a | ni, A20+ |
| mut16subflg4 | FLAG sub | 178 | 4a | ni, A20+ |
| mut32subflg5 | FLAG sub | 454 | 4a | ni, A20+ |
| mut37subflg6 | FLAG sub | 526 | 4a | ni, A20+ |
| mut38subflg7 | FLAG sub | 547 | 4a | ni, A20+ |
| mut40subflg8 | FLAG sub | 562 | 4b | ni, no capsid |
| mut44subflg9 | FLAG sub | 638 | 4b | ni, no capsid |
| mut45subflg10 | FLAG sub | 664 | 4b | ni, no capsid |
| mut46subflg11 | FLAG sub | 682 | 4b | ni, no capsid |
Superscripts 1 to 4 indicate that a restriction site was introduced as a result of the alanine substitution mutation: 1, NheI; 2, EagI; 3, HpaI; 4, MluI.
Ala sub, alanine substitution mutant; Ala ins, string of alanine residues inserted after the indicated amino acid; HA, AU, His, or Ser ins, insertion of the HA, AU, His, or Ser epitope immediately after the indicated amino acid of wt cap; Ser or FLAG sub, substitution of the Ser or FLAG epitope for the wt AAV capsid sequence beginning immediately after the indicated AAV amino acid residue. Amino acid tags: HA, YPYDVPDYA; AU, DTYRYI; HIS, HHHHHH; Ser, FVFLI; FLAG, DYKDDDDK.
pd, partially defective for infectivity, between 1 to 3 logs lower than wt; cs and hs, cold sensitive and heat sensitive, respectively; ni, noninfectious, 5 logs lower than wt; hep+ mutant bound to a heparan column; hep−, mutant did not bind to heparan sulfate; no capsid, mutant was A20 ELISA negative and MAb B1 negative; A20+, mutant could be detected with A20 antibody; surface, position was present on the surface of the capsid.
The serpin insertion in VPN2 was KFNKPFVFLI.
Infectious titer assays reveal four general classes of mutants.
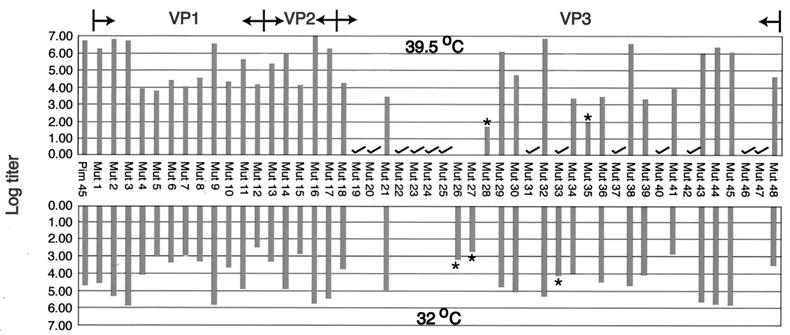
To determine the effect of each mutation on viral infectivity, we used either wt pIM45 or a mutant pIM45 plasmid to complement the growth of pTRUF5. pTRUF5 is a recombinant AAV plasmid that contains the gfp gene under the control of a CMV enhancer-promoter (22). The resulting recombinant TRUF5 virus contained either wt or mutant capsid proteins and could be titered for infectivity by counting green fluorescent cells in the presence of an Ad5 coinfection. We had shown previously that the fluorescent cell assay produced titers within two- to threefold of those obtained with a conventional infectious center assay (51). Initially, each mutant was grown and titered at either 39.5 or 32°C to determine if any of the mutants were temperature sensitive. The experiments were done twice, and there was no significant variation in titer. On the basis of these titers, the mutants could be grouped into four classes (Fig. 2; Table 1). Class 1 contained mutants that have an infectious titer similar to the wt titer (less than 1 log difference; for example, mut1 and mut2). Class 2 contained partially defective mutants with infectious titers 2 to 3 logs lower than the wt titer (for example, mut4 and mut5). Class 3 contained temperature-sensitive mutants; three of these (mut26, mut27, and mut33) were heat sensitive, and two (mut28 and mut35) were cold sensitive. Class 4 consisted of 12 noninfectious mutants, whose titers were more than 5 logs lower than the wt titer.
FIG. 2.
Infectious titers of virus stocks containing wt and mutant capsid proteins. The GFP fluorescent cell assay was used to titer virus stocks of wt and mutant virus stocks containing the pTRUF5 genome. 293 cells were transfected with wt or mutant pIM45 complementing plasmid in the presence of pTRUF5 and pXX6 at 39.5 and 32°C. Cells were collected 48 h posttransfection and then frozen and thawed three times. The crude lysate was used to infect 293 cells at 39.5 and 32°C with Ad5 (MOI = 10). The log value of the average infectious titer (infectious particles/milliliter) that was obtained from two independent experiments is shown. There was no significant difference between experiments. The distribution of mutants unique to VP1, VP2, or VP3 is shown at the top. Asterisks indicate temperature-sensitive mutants; noninfectious mutants are indicated by check marks.
Noninfectious (class 4) mutants and temperature-sensitive (class 3) mutants were defective in packaging DNA or in forming stable virus particles.
To determine the probable causes for the different defective mutants, we focused first on class 3 and 4 mutants. For convenience, we ignored the fact that the temperature-sensitive mutants had low infectivity when grown at the partially restrictive temperature of 37°C (data not shown), and viral preparations for all class 3 and 4 mutants were made at 37°C. To determine if these mutants were able to make capsids, we used the A20 ELISA. The A20 antibody recognizes only intact AAV particles (43) and is useful for determining the physical particle titer irrespective of whether the capsids contain DNA (18). Eight of sixteen mutants that were tested were negative by ELISA reading (Table 2), indicating that they were unable to make capsids or that the capsids were unstable even in crude lysate preparations. All of these were class 4 (noninfectious) mutants and were classified as class 4b (Table 1; Fig. 1).
TABLE 2.
Determination of physical particle titer and DNA-containing particle titer of class 2 and 3 mutants
| Constructa | A20 ELISAb | QC-PCRc |
|---|---|---|
| pIM45 (wt) | +++ | +++ |
| mut19 | − | − |
| mut20 | − | − |
| mut22 | ++ | ++ |
| mut23 | − | − |
| mut24 | − | − |
| mut25 | − | − |
| mut26 (hs) | NDd | ND |
| mut27 (hs) | + | ND |
| mut28 (cs) | + | ND |
| mut31 | ++ | − |
| mut33 (hs) | ++ | + |
| mut35 (cs) | ++ | ++ |
| mut37 | ++ | + |
| mut40 | ++ | ++ |
| mut42 | − | − |
| mut46 | − | ND |
| mut47 | − | ND |
hs, heat sensitive; cs, cold sensitive.
+++, >1012 particles/ml; ++, >1011 particles/ml; +, >1010 particles/ml; −, <108 particles/ml, which was the limit of detection by A20 ELISA.
+++, >1011 full particles/ml; ++, >1010 full particles/ml; +, >109 full particles/ml; −, <108 full particles/ml.
ND, not done.
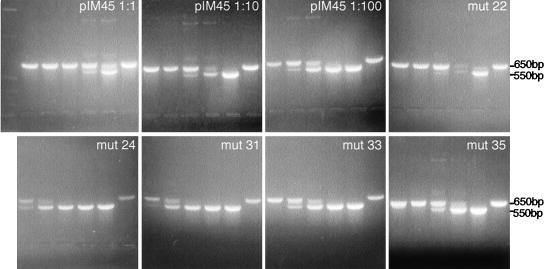
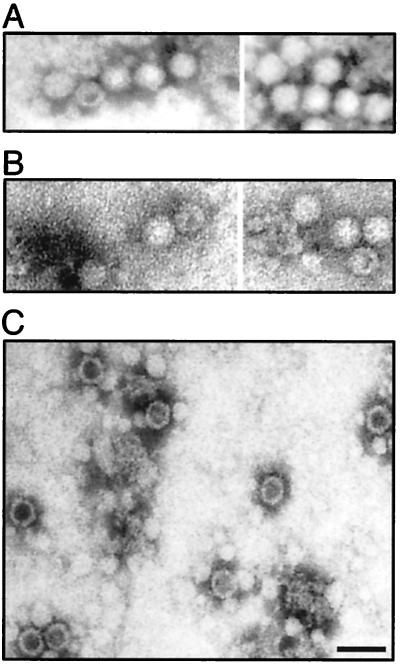
QC-PCR assays also were performed on most of the class 3 and 4 mutants. The QC-PCR assay measures the titer of AAV particles that contain DNase-resistant rAAV genomes (Fig. 3). We have shown previously that it provides physical particle titers that are equivalent to those obtained by dot blot assay but has better sensitivity at low particle titers (51). As expected, mutants that were negative for the synthesis of AAV particles by A20 ELISA were also negative by QC-PCR assay (Table 2; Fig. 3). Most of the remaining mutants, which were positive for A20 particles, were also positive for packaged viral DNA in the QC-PCR assay (Fig. 3; Table 2). This group of noninfectious mutants (mut22 and mut37) were called class 4a (Table 1; Fig. 1). Their defect was not in packaging but rather in the binding, internalization, or uncoating steps of the viral entry process. One A20-positive mutant (mut31) was an exception in that it was A20 positive but DNA negative by QC-PCR assay. This meant that mut31 formed intact virus particles that were empty. To confirm this, mut31 was examined by EM (Fig. 4), and it did indeed make empty particles. In contrast, the partially defective class 2 mutant, mut4, produced particles similar to wt particles. mut31 was assigned to class 4c (Fig. 1; Table 1).
FIG. 3.
QC-PCR assay of wt and mutant virus stocks to determine the DNA-containing particle titers. Crude viruses were treated with DNase to digest unpackaged DNA and then treated with proteinase K to release the packaged DNA. The viral DNA was extracted with phenol-chloroform, precipitated with ethanol, and dissolved in water. Equal amounts of viral DNA were incubated with (from left to right in each panel) 100 fg, 1 pg, 10 pg, 100 pg, 1 ng, or none of the pTRUF5 plasmid DNA containing a deletion in the gfp gene and amplified by PCR. The PCR products were separated on 2% agarose gels and viewed by ethidium bromide staining. The arrangement of lanes in each panel is the same. Results for wt pIM45 viral DNA at three dilutions (1:1, 1:10, and 1:100) are also shown (top left three panels). Molecular markers were included in the left lane of the top left panel.
FIG. 4.
EM analysis of wt (A) and mutant (mut4 [B] and mut31 [C]) rAAVs. The viruses were purified by iodixanol step gradient centrifugation and heparan column chromatography as described elsewhere (51), concentrated in a Centricon 10, and negatively stained with 1% uranyl acetate. Bar = 40 nm. Although the iodixanol step gradient might be expected to remove empty particles, these particles apparently accumulate at the 25 to 40% interface, and a significant fraction are recovered during this purification step.
Some mutants are defective for binding the viral receptor.
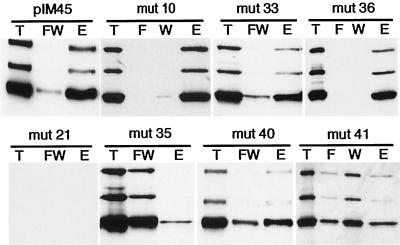
One potential cause for the reduced infectivity of class 2, 3, or 4 mutants might be that they were unable to bind the viral cell surface receptor, the first step of the infectious cycle. Heparan sulfate proteoglycan has been identified as the primary cell surface receptor for AAV (37). To test whether these mutants could bind heparan, we developed a heparan column binding assay (Materials and Methods). Iodixanol-purified wt or mutant rAAVs were passed through a heparan agarose column, and the AAV capsid proteins in the starting material and the bound (eluate) and unbound (flowthrough and wash) fractions were monitored by Western blotting using MAb B1, which recognizes all three capsid proteins (Fig. 5; Table 3). As expected, wt AAV had a high affinity for the heparan column, since little capsid protein was detected in the flowthrough and wash fractions, and most of the capsid protein was detected in the eluate. The same was true of most of the mutants tested (Fig. 5; Table 3). Two mutants, however, mut35 and mut41, bound poorly to heparan (Fig. 5). A third mutant, mut40, which is located about 20 aa away from mut41, also bound with reduced affinity (Fig. 5). This suggested that the primary defect in these mutants was their inability to bind to heparan sulfate proteoglycan. We classified mut35 as class 3b (temperature sensitive and heparan binding negative), mut41 as class 2c (partially defective and heparan binding negative), and mut40 as class 4d (noninfectious and heparan binding negative) (Fig. 1; and Table 1).
FIG. 5.
Heparan binding properties of mutant viruses. Iodixanol gradient-purified virus stocks were loaded onto a heparan column. Equivalent volumes of the starting, 40% iodixanol material (T), flowthrough (F), wash (W), and eluted (E) fractions were separated on SDS–10% acrylamide gels and Western blotted with MAb B1. In some cases, the flowthrough and wash fractions were pooled (FW) and loaded together.
TABLE 3.
Heparan column binding properties of class 2, 3, and 4 mutantsa
| Construct | Heparan binding | Construct | Heparan binding | |
|---|---|---|---|---|
| pIM45 | + | mut27 | 0 | |
| mut4 | + | mut28 | + | |
| mut5 | + | mut30 | + | |
| mut6 | + | mut31 | + | |
| mut7 | + | mut32 | + | |
| mut8 | + | mut33 | + | |
| mut10 | + | mut34 | + | |
| mut11 | + | mut35 | − | |
| mut12 | + | mut36 | + | |
| mut13 | + | mut37 | + | |
| mut14 | + | mut39 | 0 | |
| mut15 | + | mut40 | − | |
| mut18 | + | mut41 | − | |
| mut20 | 0 | mut43 | + | |
| mut21 | 0 | mut46 | 0 | |
| mut22 | + | mut48 | + | |
| mut25 | 0 |
+, mutant virus bound to a heparan column with the same affinity as wt pIM45 virus; −, virus bound with at least a threefold-lower affinity; 0, no protein signal detected by Western blotting.
Three class 4b mutants, mut20, mut25, and mut46, could not be detected by Western analysis (Table 3). This was consistent with the fact that they made no capsid that was detectable with the A20 antibody (Table 2). Additionally, mut27, a temperature-sensitive mutant, and two class 2 mutants, mut21 and mut39, did not give any Western signal with MAb B1 (Fig. 5; Table 3). The heat-sensitive mutant, mut27, was presumably unstable at the nonpermissive temperature used for growing this virus. mut21 and mut39 were partially defective when assayed in crude extracts (Fig. 2). The fact that they could not be detected by capsid antibody after iodixanol centrifugation suggests that these capsids were also unstable during purification. These mutants were assigned to class 2b on the basis of their capsid instability (Table 1; Fig. 1). The rest of the mutants in class 2 that bind to heparan were classified as class 2a, partially defective, and heparan binding positive (Tables 1 and 3; Fig. 1). The nature of their defect was not clear but presumably was due to some step in the infectious process that occurs after viral attachment to the cell surface.
Regions tolerating alanine substitutions do not tolerate other kinds of substitutions.
We wanted to determine whether the class 1 mutants defined positions in the capsid genes that were truly nonessential for capsid function. To test this, we constructed a series of mutants in which either the serpin receptor ligand, FVFLI (50), or the FLAG antibody epitope, DYKDDDDKYK, was substituted for capsid sequences at many of the class 1 mutant positions (Table 4). A number of class 2 and class 4 mutants were tried as well. The serpin substitution (5 aa) was the same size as the largest alanine substitutions. The FLAG epitope is highly charged, as were many of the substituted wt sequences. As expected, substitutions at class 2 (partially defective) or class 4 (nonviable) positions did not produce infectious virus (Table 4). Surprisingly, although many of the class 1 serpin or FLAG substitutions produced some physical particles detectable with the A20 antibody, only one of the substitutions, serpin at aa 34 (the mut3 position), produced infectious virus particles in substantial yield (Table 4). Most infectious titers were reduced by 5 logs or more, and particle titers (as judged by A20 ELISA) were reduced or undetectable as well. Thus, although modification of charged residues in class 1 mutants to alanine was permissible, these regions of the capsid were nevertheless essential for capsid formation and were sensitive to other kinds of substitutions.
TABLE 4.
Substitution of serpin or FLAG epitopes at capsid positions that tolerated alanine substitutions
| Mutant | Titera
|
|
|---|---|---|
| Infectious | Physical particle | |
| mut1subser1 | − | + |
| mut2subser2 | − | + |
| mut3subser3 | 1 log lower | + |
| mut9subser4 | − | + |
| mut14subser5 | − | + |
| mut16subser6 | − | − |
| mut19subser7 | − | − |
| mut32subser8 | − | − |
| mut37subser9 | − | − |
| mut39subser10 | − | − |
| mut40subser11 | − | − |
| mut41subser12 | − | − |
| mut44subser13 | − | − |
| mut45subser14 | − | − |
| mut46subser15 | − | − |
| mut4subflg2 | − | + |
| mut8subflg3 | − | + |
| mut16subflg4 | − | + |
| mut32subflg5 | − | + |
| mut37subflg6 | − | + |
| mut38subflg7 | − | + |
| mut40subflg8 | − | − |
| mut44subflg9 | − | − |
| mut45subflg10 | − | − |
| mut46subflg11 | − | − |
Either a serpin peptide sequence or the FLAG sequence was substituted for the AAV capsid sequence at the positions used previously for alanine scanning mutagenesis (Fig. 2). Infectious titers were determined by GFP fluorescent cell assay. −, infectious virus could not be detected. Physical particle titers were judged by A20 ELISA. +, particles were detectable; −, particles were not detectable.
Putative loop regions and the N-terminal regions of VP1 and VP2 are able to accept insertions of foreign epitopes.
We also chose several other sites for insertion of foreign sequences. For these mutants, we chose to insert the less charged HA epitope, YPVDVPDYA. The target positions for insertion were the N-terminal regions of the three capsid proteins, VP1, VP2, and VP3, the C terminus of the cap ORF and seven positions (mutants L1 to L7) that were believed to be in loop regions of the capsid protein based on an alignment of the AAV capsid sequence to that of CPV (9). Since these sites were suspected to be on the surface of the capsid, insertions at these sites might not affect capsid assembly or stability (Fig. 1). Mutations in the loop regions had been targeted successfully before by Girod et al. (15), who were able to insert the L14 ligand at aa 587 without significant loss in infectivity.
Insertions at the N termini of VP1 (VPN1) and VP3 (VPN3) and the C terminus of the cap ORF (VPC) were not well tolerated (Table 5). To eliminate the possibility that the defect in these mutants was due to the HA tag, other tags such as AU, His, and Myc were also inserted at the N termini of VP1 and VP3 and the C terminus of cap, and they also were not tolerated at those positions (Table 1 and data not shown). Insertions at three of the putative loop regions were also not viable (Table 5, mutants L2, L4, and L5). Mutants L4 (aa 522) and L5 (aa 553) were interesting in that they produced a significant yield of physical particles that were not infectious.
TABLE 5.
HA insertion mutants
| Mutant | Position | Titer
|
|
|---|---|---|---|
| Infectiousa | Physical particleb | ||
| L1 | aa 266 | ++ | + |
| L2 | aa 328 | − | + |
| L3 | aa 447 | ++ | ++ |
| L4 | aa 522 | − | ++ |
| L5 | aa 553 | − | ++ |
| L6 | aa 591 | ++ | ++ |
| L7 | aa 664 | ++ | ++ |
| VPN1 | aa 1 | + | ++ |
| VP1 | aa 34 | +++ | ++ |
| VPN2 | aa 138 | +++ | +++ |
| VPN3 | aa 203 | − | − |
| VPC | C terminus | − | − |
Determined by GFP fluorescence cell assay. +++, 1 log lower than wt; ++, 2 logs lower; +, 3 logs lower; −, at least 5 logs lower.
Determined by A20 ELISA. +, 4 logs lower than wt pIM45; ++, 2 to 3 logs lower; +++, 1 log lower; −, undetectable.
However, HA insertions were well tolerated at aa 34 within the N-terminal region of VP1, at the N terminus of VP2, and within three of the putative loop regions, loop I (mutant L1), loop IV (mutants L3 and L6), and loop V (mutant L7) (Table 5; Fig. 1).
Some HA insertion positions are on the capsid surface.
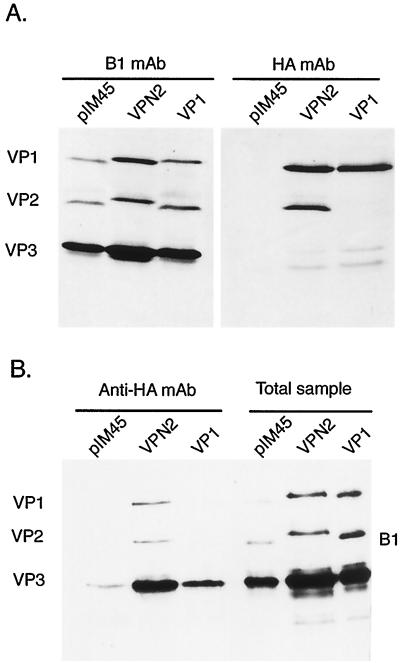
To determine whether the HA insertion mutants contained the HA sequence exposed on the surface of the capsid, we used batch immunoprecipitation with HA MAb-conjugated beads. In each case virus was purified by iodixanol density centrifugation and heparan column chromatography to remove any soluble capsid protein that might be present in crude viral preparations. As expected, insertion of the HA tag at the N terminus of VP2 (mutant VPN2) produced a slight increase in the molecular weight of VP2 and VP1 compared to wt protein, pIM45 (Fig. 6A, 1 mAb). Western blotting with the HA MAb confirmed that the HA tag was present in both VP1 and VP2 (Fig. 6A, HA mAb). In the case of the VP1 mutant (HA insertion at aa 34 in VP1), only VP1 had a higher molecular weight and only VP1 contained the HA tag (Fig. 6A), as expected. When the viable insertions, VPN2 (HA insertion at the N terminus of VP2) and VP1 (insertion at aa 34), were treated with HA MAb-conjugated beads, substantial amounts of both viruses were precipitated (Fig. 6B, HA mAb). This demonstrated that in both cases the HA epitope was on the surface of the virus particle and accessible to the antibody. Control wt virus particles (Fig. 6B, pIM45), were not precipitated with HA MAb to any significant extent. The amount of virus in the starting material was monitored by Western blotting with B1 or HA MAb.
FIG. 6.
Immunoprecipitation analysis of VP1 and VPN2 HA insertion mutants to determine the accessibility of the HA epitope. (A) Western blot analysis of iodixanol gradient-purified viruses with either B1 (left) or HA (right) MAb. (B) Iodixanol gradient and heparan column-purified viruses were precipitated with HA antibody coupled to agarose beads. The bound virus (Anti-HA mAb lanes) was eluted with SDS sample buffer and detected by Western blotting using MAb B1. For comparison, virus that had not been treated with HA MAb (Total sample) was also Western blotted with the B1 antibody.
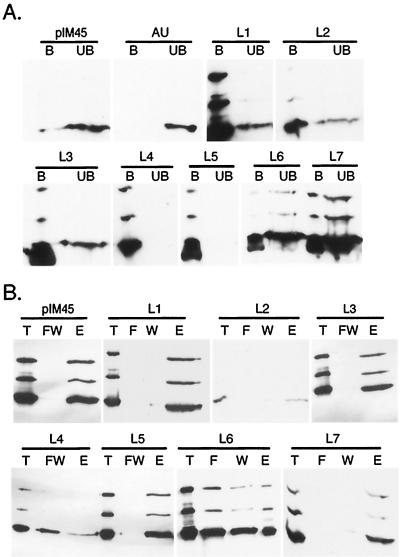
The putative loop HA insertion mutants, L1 to L7, were also incubated with HA MAb-conjugated beads. Although the insertions in some of these mutants produced noninfectious virus, they all produced sufficient A20 antibody-positive virus particles to test for the presence of the HA tag on the surface of the capsid. When this was done, all of the L-series insertions were shown to be in the immunoprecipitate (bound fraction) compared to the wt (pIM45) control (Fig. 7A). This demonstrated that each of these insertions at putative loop sites resulted in the HA epitope being on the surface of the capsid.
FIG. 7.
Properties of HA insertion mutants. (A) Immunoprecipitation of HA loop insertion mutants to determine whether HA is exposed on the capsid surface. Iodixanol gradient and heparan column-purified viruses were incubated with HA MAb beads as described for Fig. 6. The antibody bound (B) and unbound (UB) fractions were separated on SDS–10% gels and detected by Western blotting with MAb B1. As a negative control, AU MAb was used in the panel marked AU. The pIM45 panel contained recombinant virus made with the wt helper plasmid. (B) Heparan binding properties of wt and HA loop insertion mutants. The virus samples were treated as described for Fig. 5. Virus in the starting material (T), flowthrough (F), wash (W), combined flowthrough and wash (FW), or eluate (E) was detected by Western blotting with MAb B1. pIM45 is virus with wt capsid.
We also checked whether these loop insertions affected heparan binding of the mutant capsids. Interestingly, two loop insertion mutants, L4 and L6, were found to bind heparan columns with reduced affinity (Fig. 7B), which probably accounted for the lower infectivity of these mutants in the standard fluorescent cell assay. The L4 and L6 insertions were near the heparan-binding-negative mutants mut35, mut40, and mut41 (Fig. 1). All five of these heparan-binding-negative mutants were located between aa 509 and 591, suggesting that this region within the AAV capsid constitutes the heparan binding domain of the capsid protein.
Changing the tropism of AAV.
To determine whether we could change the tropism of rAAV by inserting a novel receptor ligand into the capsid, we constructed two mutant plasmids that contained a serpin receptor ligand. In one case the serpin ligand FVFLI (50) was substituted for the AAV capsid sequence immediately after aa 34. In the second mutant an expanded serpin receptor ligand, KFNKPFVFLI (50), was inserted at the N terminus of VP2, aa 138 (Table 1). The mutant capsid plasmids were then used to package CBA-AT, an rAAV genome that contained the hAAT gene under the control of a hybrid CMV–β-actin promoter. As seen with the HA insertion mutants described above, the serpin mutants produced rAAV viral titers that were slightly (sixfold) lower in infectivity when titered by the infectious center assay on 293 cells (data not shown). However, when equal amounts of wt or mutant virus (as determined on 293 cells) were infected into IB3 cells, both mutant viruses showed substantially higher infectivity than wt (Fig. 8). The VP2 serpin insertion was 15-fold more infectious, and the VP1 substitution mutant was approximately 62-fold more active. This suggested that IB3 cells, a lung epithelial cell line believed to express the serpin receptor, were a much better target for the serpin-tagged chimeric rAAVs than wt and that the tropism of the mutant rAAVs had been changed. Because both mutants retained the wt heparan binding region, we also infected IB3 cells in the presence of heparan sulfate to see if they continued to use heparan sulfate proteoglycan for viral entry. When this was done, both wt and mutant infectivity dropped to barely detectable levels (Fig. 8). Taken together, these findings suggested that the serpin-tagged viruses continued to use heparan sulfate proteoglycan as the primary receptor and were using an alternative coreceptor, presumably the serpin receptor.
FIG. 8.
Infection of IB3 cells with wt and mutant viruses containing a serpin ligand insertion. IB3 cells (1.5 × 105 per 15-mm well) were infected with Ad5 for 60 min at an MOI of 10 and washed twice with medium. The cells then were infected for 60 min at an MOI of 400 with rAAV containing a genome that expressed the hAAT gene under the control of a CMV–β-actin hybrid promoter. Following infection, the cells were washed with medium and incubated at 37°C. At 72 h postinfection, medium samples were taken to determine the AAT concentration by ELISA. All experiments were done in triplicate, and the average for each experiment is shown. WT indicates that rAAV containing a wt AAV capsid (grown by complementation with pIM45) was used. VP1 virus was grown by complementation with a mutant plasmid containing the serpin ligand sequence (FVFLI) substituted for the AAV capsid sequence after aa 34 of the cap ORF. VP2 virus contained a serpin insertion (KFNKPFVFLI) at the N terminus of VP2, aa 138 of the cap ORF. In the +HS samples, rAAV infection was done in the presence of soluble heparan sulfate at a concentration of 2 mg/ml.
DISCUSSION
In this study we describe the phenotypes of 93 AAV2 capsid mutants at 59 different positions within the capsid ORF. Several classes of mutants were analyzed, including epitope tag or peptide ligand insertion mutants, alanine scanning mutants, and epitope substitution mutants. From this, we could identify some eight separate phenotypes (Fig. 1; Table 1).
Noninfectious mutants.
The bulk of the mutants that were noninfectious either were unable to assemble capsids or the capsids were unstable. These mutants (class 4b) were located predominantly but not exclusively in what are likely to be β-strand structures in the capsid proteins (Fig. 1). Two of these mutants were insertions at the N- and C-terminal residues of VP3, suggesting that both ends of VP3 play a role that is important for capsid assembly or stability. We note that Ruffing et al. (32) have previously characterized deletions of the C terminus of the capsid ORF, and these deletions also were noninfectious.
One noninfectious mutant, mut31, produced viable capsids that were empty. This mutant, which consists of a single amino acid substitution (R432A), was apparently defective in packaging viral DNA and is located in putative loop IV (Fig. 1). It is not clear what the mechanism of viral DNA packaging is. Ruffing et al. (33) demonstrated that empty capsids could assemble in the absence of viral DNA. Some studies have suggested that packaging is an active process that requires interaction of Rep proteins with capsid proteins (42) or possibly is coupled with DNA replication (49). Further studies with mut31 may be helpful in understanding the mechanism of packaging.
Most of the remaining noninfectious mutants (Fig. 1, class 4a) were capable of assembling capsids and packaging DNA. These are likely to be defective in some aspect of viral entry or uncoating and will require further study to uncover the mechanism of the defect.
Receptor binding mutants.
Two of the noninfectious mutants, mut40 and L4, were apparently noninfectious because they were unable to bind to heparan sulfate (Fig. 1, class 4d). Heparan sulfate proteoglycan is believed to be the primary cell surface receptor for AAV (37). Three other mutants also were identified as defective for binding heparan sulfate, two partially defective mutants (Fig. 1, class 2c) and one temperature-sensitive mutant (class 3b). Together, the five mutants were distributed into two clusters in loop IV that were separated by 40 aa. The first cluster spanned aa 509 to 520 (mut35 and L4); the second was between aa 561 and 591 (mut40, mut41, and L6). Mutants L4 and L6 consisted of HA epitope insertions into the two heparan binding clusters. These were found to be capable of being immunoprecipitated by HA MAb, confirming that these positions were on the surface of the capsid. We note also that Girod et al. (15) reported that insertion of the L14 epitope at aa 587, the position of our heparan-negative mut41 mutant, was capable of targeting the virus to the L14 receptor, thus confirming that this region is on the surface of the capsid. A heparan-negative insertion mutant also was reported by Rabinowitz et al. (30) while this report was in preparation; it fell near the first cluster at aa 522. Taken together, analyses of these mutants suggest that the putative loop IV region contains two blocks of residues that are on the surface of the capsid and involved in heparan sulfate binding.
A heparan binding motif which consists of a negatively charged amino acid cluster of the type XBBBXXBX (where B is a basic amino acid and X is any amino acid) has been identified in several receptors and viruses (19a). Regions containing these clusters also appear to be sensitive to spacing changes. Although no heparan binding consensus motif of this kind was found in our heparan binding mutants, there were basic amino acids near these domains. mut35, an insertion at aa 509, was near basic amino acids K507 and H509. Interestingly, K507 is conserved in AAV1, -2, -3, -4, and -6 and in AAV5 is an R. H509 is present only in AAV2 and -3. AAV1, -2, and -3 are known to bind to heparan sulfate, while AAV4 and -5 do not. Additionally, L4, an insertion at aa 520, was near basic amino acids H526 and K527, and L6, an insertion at aa 591, was near R585 and R588. H526 and K527 are conserved except for AAV4 and -5, while R585 and R588 are unique to AAV2. For all of these mutants, the insertions could have disrupted local conformation that hindered normal heparan binding. For mut41, R-to-A substitutions at aa 585 and 588 might contribute directly to reduced heparan binding. Finally, mut40 did not affect either basic amino acids or spacing within the capsid protein.
Capsid regions that are on the surface of the virus particle.
In addition to the heparan binding clusters, several other regions were also present on the capsid surface. These include four of the five putative loop regions (mutants L1 to L7), the N terminus of VP2 (mutant VPN2), and a region within the N terminus of VP1 at amino acid 34 (mutant VP1). HA epitope insertions at these positions were all capable of being immunoprecipitated with anti-HA antibody (Fig. 6 and 7). We note that the L1 insertion mutant at aa 266 had the peculiar phenotype of being partially viable (Table 1) but was not detectable with the A20 MAb, an antibody that recognizes a conformational epitope that is present only in intact viral particles. A nearby capsid-forming mutant made by Girod et al. (15) at aa 261 was also negative for A20 antibody binding. This suggests that at least part of the epitope for the A20 MAb consists of amino acids between 261 and 266 and confirms that this region is on the surface of the intact particle.
Of the positions identified as being on the surface of the capsid, we found six that potentially are capable of accepting foreign epitope or ligand insertions for retargeting the viral capsid to alternative receptors. These are the N-terminal region of VP1 (near aa 34), the N terminus of VP2 (aa 138), the loop I region (aa 266), the loop IV region (near aa 447 and 591), and the loop V region (aa 664). All of these locations were capable of tolerating an HA (or serpin) insertion and produced recombinant virus titers that were within 1 to 2 logs of the wt value. Furthermore, HA epitope insertions at these positions were capable of being immunoprecipitated with anti-HA antibody (Fig. 6 and 7). Two of these positions, when tested with a serpin ligand insertion or substitution, produced virus that was much more infectious on IB3 cells than wt virus. Curiously, both serpin mutants were still inhibited by soluble heparan sulfate, suggesting that heparan sulfate proteoglycan was still the primary receptor for these mutants and that the serpin receptor was being used as an alternative coreceptor. It is conceivable that one or both of these capsid positions is involved in binding to one or both of the proteins that normally act as coreceptors for wt virus, fibroblast growth factor (28), or integrin αvβ5 (36). This would explain their partial defect on 293 cells and the recovery of infectivity on IB3 cells. Further studies will be needed to test this possibility.
Mutants with unstable capsids and temperature-sensitive phenotypes.
Three mutants, mut21, mut27, and mut39, were found to have capsids that were unstable when purified through an iodixanol gradient. Iodixanol is an iso-osmotic gradient purification method that appears to be gentler than CsCl centrifugation (51). Thus, these mutants appear to be particularly sensitive to capsid denaturation. mut21 and mut27 are in putative β sheets, and mut39 is in loop IV. It is worth noting that Rabinowitz et al. (30) also isolated an unstable capsid mutant at aa 247 that is near the mut21 position, aa 254. mut27 is also one of five temperature-sensitive mutants isolated during this study. The temperature-sensitive mutants and the unstable capsid mutants should prove useful in future studies for identifying steps in the capsid assembly or the infection process.
Viable and partially defective mutants.
The two largest classes of mutants isolated were either wt (class 1) or partially defective (class 2a) with no identifiable defect (Fig. 1). Both class 1 and class 2a mutants were distributed either in the VP1 and VP2 unique regions or in the predicted loop regions of the capsid protein. We naively assumed that class 1 mutant positions, which produced viable capsids after substitution of two to five alanine residues, were regions that were nonessential for capsid assembly or stability and therefore should accommodate other kinds of substitutions. However, when serpin or FLAG epitopes were substituted at many of these sites, most of the mutants were nonviable, with the exception of aa 34 in VP1. Indeed, many of these viruses were negative for capsid assembly and should also be useful for identifying possible intermediates in capsid assembly.
Ruffing et al. (33) showed previously that VP1 and VP2 but not VP3 contained nuclear localization signals (NLS), and three putative NLS are located in the VP1/VP2 region at aa 121 to 125, 141 to 145, and 167 to 171. Hoque et al. (19b) have shown that aa 167 to 172 were sufficient to target VP2 to the nucleus, although their experiments did not rule out possible redundancy with the other two putative NLS sequences. All three of these putative signals were targeted with alanine scanning mutants (mut12, mut13, and mut15) in our study. Two of these mutants, mut12 and mut15, were partially defective, and the inactivation of an NLS may be the reason for their phenotype (19b, 33). We note that mut15 should have eliminated the NLS identified by Hoque and colleagues. The fact that mut15 was only partially defective suggests that there may be an alternative, redundant NLS sequences that are used by the capsid proteins. The third mutant (mut13) was classified as viable, but it also showed a lower than wt titer (Fig. 1).
Molecular computer graphics construction of an AAV model and structural localization of mutant residues.
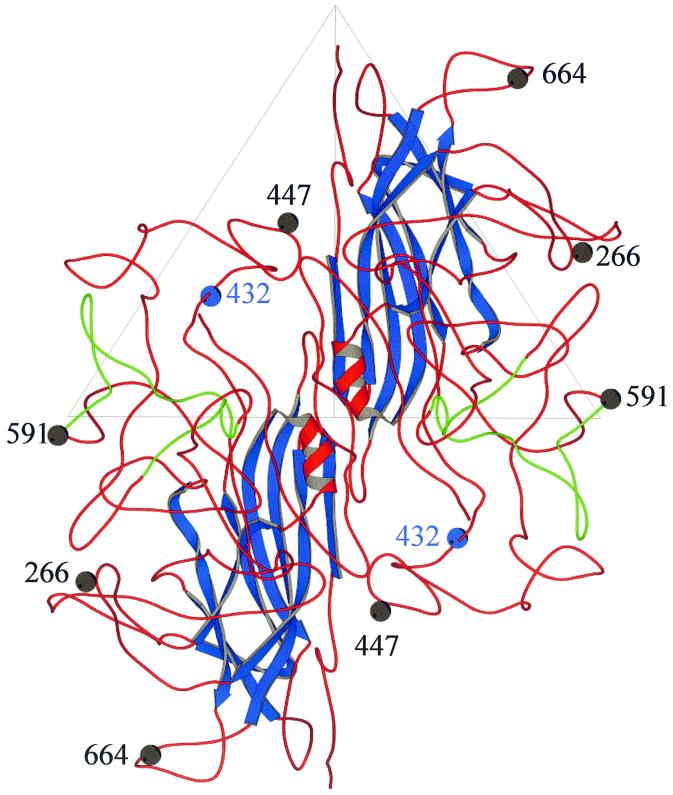
Because the AAV crystal structure is not available, the atomic coordinates of CPV VP2 (PDB accession no. 4DPV) were interactively mutated using the program O (20) to generate a homology-based model of the AAV capsid, using modifications of the alignments of the AAV major capsid protein (VP3) with the VP2 capsid protein of CPV (9, 15). The mutations were followed by refinement constrained with standard geometry in the O database. The model provided a means for preliminary structural identification of the heparan receptor attachment sites in the surface depression (dimple) near the twofold icosahedral axes of the capsid, surface loop regions which can tolerate foreign peptide sequence insertions, and a possible explanation for the phenotype of mut31 (Fig. 9).
FIG. 9.
Ribbon diagrams of a dimer of the AAV VP3 model built based on structural alignments with the VP2 capsid protein of CPV. The view is down an icosahedral twofold axis. The strands of the β-barrel motif are colored blue, and the portion of VP3 in green indicates the heparan binding region. The rest of VP3 is depicted in red. The blue ball identifies the location of residue R432 mutated to an alanine in mut31. The gray balls identify the location of residues 266, 477, 591, and 664 (which had HA insertions in mutants L1, L3, L6, and L7, respectively). The large triangle indicates an icosahedral asymmetric unit.
The topographic location of the putative heparan binding region is consistent with regions that have been suggested as being involved in host cellular factor(s) recognition and implicated in tissue tropism and in vivo pathogenicity for other parvoviruses (3, 4, 24, 39). It is of interest that the putative heparan binding site is adjacent to a region of the AAV capsid that contains a peptide insert when the AAV VP3 sequence is compared to that of CPV VP2 and the VP2 of most of the other autonomous parvovirus sequences (9). Also a similar insertion of peptide sequences compared to CPV (although not in a homologous region of the VP2 to that observed in AAV) is present in the capsid of Aleutian mink disease parvovirus and minute virus of mice, proximal to residues in the dimple depression which are implicated in tissue tropism (24). Thus, these insertions may be capsid surface adaptations that enable the capsids to recognize different receptors during infection. In the case of AAV, its dimple peptide insertion, which is absent in the other parvoviruses, may enable it to recognize heparan sulfate, which has not been implicated in cellular infectivity by any other parvovirus.
The model also clearly shows that regions of the capsid which tolerated the insertions of the HA epitope (i.e., at residues 266, 447, 591, and 664) are on the surface loops present between the β strands of the β-barrel motif (Fig. 9). The β-barrel motif forms the core contiguous shell of parvovirus capsids, while the surface loops make up the surface decorations, dictating the strain-specific biological properties of the members. The observation that these surface regions can tolerate foreign peptide insertion is an indication that they are not involved in the interactions that govern capsid assembly.
Finally, the model provides a possible explanation for the observation that mut31 (R432A) is able to form only empty particles. In the unassembled VP3 monomer, the side chain of R432, points toward the interior of the capsid and would most likely be in contact with DNA. If recognition and encapsidation of AAV DNA precede final capsid assembly and involve oligomeric intermediates, then R432 contacts with DNA may be essential for initiating capsid assembly around a nascent DNA strand.
In summary, we have reported a preliminary analysis of mutants at 59 positions within the AAV2 capsid ORF. We have identified regions in the capsid proteins that affect infectivity, capsid formation, capsid stability, DNA packaging, and receptor binding. These mutants should be valuable for defining the functional domains of AAV capsid proteins and for dissecting the molecular mechanism of viral entry. Additionally, we have defined a number of regions in the capsid gene at which foreign ligands can be inserted and have demonstrated that insertion of a foreign receptor ligand at some of these positions can change the tropism of the virus. This is the first step in the development of the next generation of AAV vectors, which can be targeted to specific cellular receptors or tissues.
ACKNOWLEDGMENTS
We thank J. Kleinschmidt for kindly providing MAbs A20 and B1. We also thank R. J. Samulski for providing plasmid pXX6. We acknowledge the Vector Core Laboratory at the Powell Gene Therapy Center, University of Florida Medical School, for technical assistance on rAAV production. We thank Corrine Abernathy, Daniel Lackner, and Eric Kolbrener for help on this project.
This work was supported by grants PO1 HL59412, PO1 HL51811, and PO1 NS36302 from the National Institutes of Health.
REFERENCES
- 1.Agbandje M, Kajigaya S, McKenna R, Young N S, Rossmann M G. The structure of human parvovirus B19 at 8 Å resolution. Virology. 1994;203:106–115. doi: 10.1006/viro.1994.1460. [DOI] [PubMed] [Google Scholar]
- 2.Agbandje M, McKenna R, Rossmann M G, Strassheim M L, Parrish C R. Structure determination of feline panleukopenia virus empty particles. Proteins. 1993;16:155–171. doi: 10.1002/prot.340160204. [DOI] [PubMed] [Google Scholar]
- 3.Agbandje-McKenna M, Llamas-Saiz A L, Wang F, Tattersall P, Rossmann M G. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure. 1998;6:1369–1381. doi: 10.1016/s0969-2126(98)00137-3. [DOI] [PubMed] [Google Scholar]
- 4.Barbis D P, Chang S F, Parrish C R. Mutations adjacent to the dimple of the canine parvovirus capsid structure affect sialic acid binding. Virology. 1992;191:301–308. doi: 10.1016/0042-6822(92)90192-r. [DOI] [PubMed] [Google Scholar]
- 5.Berns K I, Bohenzky R A. Adeno-associated viruses: an update. Adv Virus Res. 1987;32:243–306. doi: 10.1016/s0065-3527(08)60479-0. [DOI] [PubMed] [Google Scholar]
- 6.Berns K I, Giraud C. Adenovirus and adeno-associated virus as vectors for gene therapy. Ann N Y Acad Sci. 1995;772:95–104. doi: 10.1111/j.1749-6632.1995.tb44735.x. [DOI] [PubMed] [Google Scholar]
- 7.Buller R M, Janik J E, Sebring E D, Rose J A. Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. J Virol. 1981;40:241–247. doi: 10.1128/jvi.40.1.241-247.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casto B C, Armstrong J A, Atchison R W, Hammon W M. Studies on the relationship between adeno-associated virus type 1 (AAV-1) and adenoviruses. II. Inhibition of adenovirus plaques by AAV; its nature and specificity. Virology. 1967;33:452–458. doi: 10.1016/0042-6822(67)90120-1. [DOI] [PubMed] [Google Scholar]
- 9.Chapman M S, Rossmann M G. Structure, sequence, and function correlations among parvoviruses. Virology. 1993;194:491–508. doi: 10.1006/viro.1993.1288. [DOI] [PubMed] [Google Scholar]
- 10.Chiorini J A, Yang L, Liu Y, Safer B, Kotin R M. Cloning of adeno-associated virus type 4 (AAV4) and generation of recombinant AAV4 particles. J Virol. 1997;71:6823–6833. doi: 10.1128/jvi.71.9.6823-6833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham B C, Wells J A. High resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 12.Fisher K J, Gao G P, Weitzman M D, DeMatteo R, Burda J F, Wilson J M. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J Virol. 1996;70:520–532. doi: 10.1128/jvi.70.1.520-532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher-Adams G, Wong K K, Jr, Podsakoff G, Forman S J, Chatterjee S. Integration of adeno-associated virus vectors in CD34+ human hematopoietic progenitor cells after transduction. Blood. 1996;88:492–504. [PubMed] [Google Scholar]
- 14.Flotte T R, Afione S A, Conrad C, McGrath S A, Solow R, Oka H, Zeitlin P L, Guggino W B, Carter B J. Stable in vivo expression of the cystic fibrosis transmembrane conductance regulator with an adeno-associated virus vector. Proc Natl Acad Sci USA. 1993;90:10613–10617. doi: 10.1073/pnas.90.22.10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girod A, Ried M, Wobus C, Lahm H, Leike K, Kleinschmidt J, Deleage G, Hallek M. Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2. Nat Med. 1999;5:1438. doi: 10.1038/71021. [DOI] [PubMed] [Google Scholar]
- 16.Gnatenko D, Arnold T E, Zolotukhin S, Nuovo G J, Muzyczka N, Bahou W F. Characterization of recombinant adeno-associated virus-2 as a vehicle for gene delivery and expression into vascular cells. J Investig Med. 1997;45:87–98. [PubMed] [Google Scholar]
- 17.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 18.Grimm D, Kern A, Pawlita M, Ferrari F, Samulski R, Kleinschmidt J. Titration of AAV-2 particles via a novel capsid ELISA: packaging of genomes can limit production of recombinant AAV-2. Gene Ther. 1999;6:1322–1330. doi: 10.1038/sj.gt.3300946. [DOI] [PubMed] [Google Scholar]
- 19.Hermonat P L, Labow M A, Wright R, Berns K I, Muzyczka N. Genetics of adeno-associated virus: isolation and preliminary characterization of adeno-associated virus type 2 mutants. J Virol. 1984;51:329–339. doi: 10.1128/jvi.51.2.329-339.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Hileman R E, Fromm J R, Weiler J M, Linhardt R J. Glycosaminoglycan-protein interactions: definition of consensus sites in glycosaminoglycan binding proteins. Bioessays. 1998;2:156–167. doi: 10.1002/(SICI)1521-1878(199802)20:2<156::AID-BIES8>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 19b.Hoque M, Ishizu K, Matsumoto A, Han S I, Arisaka F, Takayama M, Suzuki K, Kato K, Kanda T, Watanabe H, Handa H. Nuclear transport of the major capsid protein is essential for adeno-associated virus capsid formation. J Virol. 1999;73:7912–7915. doi: 10.1128/jvi.73.9.7912-7915.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones T A, Zou J Y, Cowan S W, Kjeldgaard Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 21.Kaplitt M G, Leone P, Samulski R J, Xiao X, Pfaff D W, O'Malley K L, During M J. Long-term gene expression and phenotypic correction using adeno-associated virus vectors in the mammalian brain. Nat Genet. 1994;8:148–154. doi: 10.1038/ng1094-148. [DOI] [PubMed] [Google Scholar]
- 22.Klein R L, Meyer E M, Peel A L, Zolotukhin S, Meyers C, Muzyczka N, King M A. Neuron-specific transduction in the rat septohippocampal or nigrostriatal pathway by recombinant adeno-associated virus vectors. Exp Neurol. 1998;150(2):183–194. doi: 10.1006/exnr.1997.6736. [DOI] [PubMed] [Google Scholar]
- 23.McCarty D M, Christensen M, Muzyczka N. Sequences required for coordinate induction of adeno-associated virus p19 and p40 promoters by Rep protein. J Virol. 1991;65:2936–2945. doi: 10.1128/jvi.65.6.2936-2945.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna R, Olson N H, Chipman P R, Baker T S, Booth T F, Christensen J, Aasted B, Fox J M, Bloom M E, Wolfinbarger J B, Agbandje-McKenna M. Three-dimensional structure of Aleutian mink disease parvovirus: implications for disease pathogenicity. J Virol. 1999;73:6882–6891. doi: 10.1128/jvi.73.8.6882-6891.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muralidhar S, Becerra S P, Rose J A. Site-directed mutagenesis of adeno-associated virus type 2 structural protein initiation codons: effects on regulation of synthesis and biological activity. J Virol. 1994;68:170–176. doi: 10.1128/jvi.68.1.170-176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 27.Ponnazhagan S, Mukherjee P, Wang X S, Qing K, Kube D M, Mah C, Kurpad C, Yoder M C, Srour E F, Srivastava A. Adeno-associated virus type 2-mediated transduction in primary human bone marrow-derived CD34+ hematopoietic progenitor cells: donor variation and correlation of transgene expression with cellular differentiation. J Virol. 1997;71:8262–8267. doi: 10.1128/jvi.71.11.8262-8267.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat Med. 1999;5:71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- 29.Rabinowitz J E, Samulski J. Adeno-associated virus expression systems for gene transfer. Curr Opin Biotechnol. 1998;9:470–475. doi: 10.1016/s0958-1669(98)80031-1. [DOI] [PubMed] [Google Scholar]
- 30.Rabinowitz J E, Xiao W, Samulski R J. Insertional mutagenesis of AAV2 capsid and the production of recombinant virus. Virology. 1999;265:274–285. doi: 10.1006/viro.1999.0045. [DOI] [PubMed] [Google Scholar]
- 31.Rossmann M G. The canyon hypothesis. Hiding the host cell receptor attachment site on a viral surface from immune surveillance. J Biol Chem. 1989;264:14587–14590. [PubMed] [Google Scholar]
- 32.Ruffing M, Heid H, Kleinschmidt J A. Mutations in the carboxy terminus of adeno-associated virus 2 capsid proteins affect viral infectivity: lack of an RGD integrin-binding motif. J Gen Virol. 1994;75:3385–3392. doi: 10.1099/0022-1317-75-12-3385. [DOI] [PubMed] [Google Scholar]
- 33.Ruffing M, Zentgraf H, Kleinschmidt J A. Assembly of viruslike particles by recombinant structural proteins of adeno-associated virus type 2 in insect cells. J Virol. 1992;66:6922–6930. doi: 10.1128/jvi.66.12.6922-6930.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song S, Morgan M, Ellis T, Poirier A, Chesnut K, Wang J, Brantly M, Muzyczka N, Byrne B J, Atkinson M, Flotte T R. Sustained secretion of human alpha-1-antitrypsin from murine muscle transduced with adeno-associated virus vectors. Proc Natl Acad Sci USA. 1998;95:14384–14388. doi: 10.1073/pnas.95.24.14384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srivastava A, Lusby E W, Berns K I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Summerford C, Bartlett J S, Samulski R J. AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat Med. 1999;5:78–82. doi: 10.1038/4768. [DOI] [PubMed] [Google Scholar]
- 37.Summerford C, Samulski R J. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tratschin J D, Miller I L, Carter B J. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J Virol. 1984;51:611–619. doi: 10.1128/jvi.51.3.611-619.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tresnan D B, Southard L, Weichert W, Sgro J Y, Parrish C R. Analysis of the cell and erythrocyte binding activities of the dimple and canyon regions of the canine parvovirus capsid. Virology. 1995;211:123–132. doi: 10.1006/viro.1995.1385. [DOI] [PubMed] [Google Scholar]
- 40.Tsao J, Chapman M S, Agbandje M, Keller W, Smith K, Wu H, Luo M, Smith T J, Rossmann M G, Compans R W, et al. The three-dimensional structure of canine parvovirus and its functional implications. Science. 1991;251:1456–1464. doi: 10.1126/science.2006420. [DOI] [PubMed] [Google Scholar]
- 41.Tsao J, Chapman M S, Wu H, Agbandje M, Keller W, Rossmann M G. Structure determination of monoclinic canine parvovirus. Acta Crystallogr B. 1992;48:75–88. doi: 10.1107/s0108768191009539. [DOI] [PubMed] [Google Scholar]
- 42.Weger S, Wendland M, Kleinschmidt J A, Heilbronn R. The adeno-associated virus type 2 regulatory proteins Rep78 and Rep68 interact with the transcriptional coactivator PC4. J Virol. 1999;73:260–269. doi: 10.1128/jvi.73.1.260-269.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wistuba A, Kern A, Weger S, Grimm D, Kleinschmidt J A. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J Virol. 1997;71:1341–1352. doi: 10.1128/jvi.71.2.1341-1352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wistuba A, Weger S, Kern A, Kleinschmidt J A. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J Virol. 1995;69:5311–5319. doi: 10.1128/jvi.69.9.5311-5319.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao X, Li J, Samulski R J. Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao X, Li J, Samulski R J. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Q, Mamounas M, Yu G, Kennedy S, Leaker B, Merson J, Wong-Staal F, Yu M, Barber J R. Development of novel cell surface CD34-targeted recombinant adeno-associated virus vectors for gene therapy. Hum Gene Ther. 1998;9:1929–1937. doi: 10.1089/hum.1998.9.13-1929. [DOI] [PubMed] [Google Scholar]
- 48.Zhou S Z, Cooper S, Kang L Y, Ruggieri L, Heimfeld S, Srivastava A, Broxmeyer H E. Adeno-associated virus 2-mediated high efficiency gene transfer into immature and mature subsets of hematopoietic progenitor cells in human umbilical cord blood. J Exp Med. 1994;179:1867–1875. doi: 10.1084/jem.179.6.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou X, Muzyczka N. In vitro packaging of adeno-associated virus DNA. J Virol. 1998;72:3241–3247. doi: 10.1128/jvi.72.4.3241-3247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ziady A G, Perales J C, Ferkol T, Gerken T, Beegen H, Perlmutter D H, Davis P B. Gene transfer into hepatoma cell lines via the serpin enzyme complex receptor. Am J Physiol. 1997;273(2 Pt. 1):G545–G552. doi: 10.1152/ajpgi.1997.273.2.G545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zolotukhin S, Byrne B J, Mason E, Zolotukhin I, Potter M, Chesnut K, Summerford C, Samulski R J, Muzyczka N. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]