Abstract
Understanding how viral components collaborate to convert the human immunodeficiency virus type 1 genome from single-stranded RNA into double-stranded DNA is critical to the understanding of viral replication. Not only must the correct reactions be carried out, but unwanted side reactions must be avoided. After minus-strand strong stop DNA (−sssDNA) synthesis, degradation of the RNA template by the RNase H domain of reverse transcriptase (RT) produces single-stranded DNA that has the potential to self-prime at the imperfectly base-paired TAR hairpin, making continued DNA synthesis impossible. Although nucleocapsid protein (NC) interferes with −sssDNA self-priming in reverse transcription reactions in vitro, NC alone did not prevent self-priming of a synthetic −sssDNA oligomer. NC did not influence DNA bending and therefore cannot inhibit self-priming at hairpins by directly blocking hairpin formation. Using DNA oligomers as a model for genomic RNA fragments, we found that a 17-base DNA fragment annealed to the 3′ end of the −sssDNA prevented self-priming in the presence of NC. This implies that to avoid self-priming, an RNA-DNA hybrid that is more thermodynamically stable than the hairpin must remain within the hairpin region. This suggests that NC prevents self-priming by generating or stabilizing a thermodynamically favored RNA-DNA heteroduplex instead of the kinetically favored TAR hairpin. In support of this idea, sequence changes that increased base pairing in the DNA TAR hairpin resulted in an increase in self-priming in vitro. We present a model describing the role of NC-dependent inhibition of self-priming in first-strand transfer.
Human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) synthesizes minus-strand strong stop DNA (−sssDNA) by extending tRNA3Lys bound at the primer binding site (PBS) in the RNA genome (8, 15, 37). To synthesize −sssDNA, RT copies the U5 and R regions; the R region contains two large hairpins known as the poly(A) hairpin and the TAR hairpin (9). When this RNA is copied into DNA, hairpins that correspond to the TAR and poly(A) hairpins of the RNA can form in the nascent −sssDNA; the formation of these hairpins depends on the digestion of the template RNA (17). The RNase H activity of RT cleaves the RNA portion of the RNA-DNA heteroduplex during polymerization, and there is additional RNase H cleavage after −sssDNA synthesis is complete (8, 12, 18, 27, 33). Although it is possible for either the nascent TAR and poly(A) DNA to form hairpins that could self-prime, such self-priming events are not detected when the HIV-1 genome is copied into DNA in infected cells. Because the TAR DNA hairpin forms at the end of R, the likelihood of self-priming is higher for TAR than for the poly(A) hairpin. Instead of self-priming, the 3′ end of the nascent DNA is efficiently transferred to the R sequence on the 3′ end of the template RNA, where synthesis continues. This event is known as the first-strand transfer. Nucleocapsid protein (NC) has been shown to prevent synthesis of self-primed products and promote strand transfer in vitro (17, 23, 26, 30). However, NC does not require a strand transfer acceptor to prevent self-priming. NC can inhibit self-priming of −sssDNA in reverse transcription reactions in vitro in the absence of an acceptor (17; this report). Therefore, the annealing of a strand transfer acceptor cannot be the only mechanism that prevents self-priming. One possibility is that NC interferes with the formation of hairpins by preventing bending of the hairpin DNA (24, 40). However, we present evidence that this is not the mechanism that prevents self-priming. Instead, using DNA oligomers as a model for genomic RNA fragments, we show that NC requires complementary oligonucleotides to prevent self-priming. In in vitro transcription reactions, NC must therefore inhibit self-priming by maintaining RNA-DNA duplexes within TAR that are sufficient to prevent hairpin formation. In experiments performed in vitro in the absence of NC, digestion of the RNA in the RNA-DNA duplex results in loss of the RNA from the heteroduplex and leads to self-priming at the TAR hairpin. However, NC promotes the retention of an RNA-DNA hybrid that inhibits self-priming. For NC to successfully prevent self-priming, the stability of the residual RNA-DNA hybrid must be greater than the stability of the TAR hairpin. Therefore, the template must be subjected to only a limited amount of RNase H digestion. This implies that the extent of RNase H digestion of the RNA genome during reverse transcription directly influences whether or not self-priming occurs at the TAR hairpin. If the RNA genome was extensively digested by RNase H, the stability of the remaining RNA-DNA hybrids would be less than that of the TAR hairpin, and NC would promote hairpin formation instead of RNA-DNA annealing. In addition to affecting the annealing of nucleic acids, NC may protect the RNA-DNA hybrid from RNase H digestion (10, 22, 23, 34; this report), which could affect the digestion of genomic RNA after −sssDNA synthesis.
MATERIALS AND METHODS
Wild-type HIV-1 RT (p66/51) was expressed in Escherichia coli and purified as described previously (5). HIV-1 NC (p7 Zn2+ holoenzyme) was generously provided by Robert Gorelick, Louis Henderson, and Larry Arthur (SAIC Frederick, Frederick, Md.). NC was reconstituted from lyophilized powder in 1× RT binding buffer (50 mM Tris-Cl, [pH 8.0], 80 mM KCl, 1 mM dithiothreitol, 100 μM bovine serum albumin) at a concentration of 30 μM and stored in 4-μl aliquots in 150-μl tubes at −80°C. Fresh aliquots of NC were thawed immediately prior to use. Oligonucleotides were purchased from Life Technologies (Rockville, Md.). HIV-1 sequences were subcloned from the pNL4-3 clone (3; GenBank accession no. AF033819) into the LITMUS 28 plasmid (New England Biolabs, Beverly, Mass.) and sequenced. The R-PBS template RNA was synthesized according to the instructions contained in the Ambion Megashortscript kit (Ambion, Austin, Tex.). In brief, an oligomer containing a T7 promoter modified so that it contained the correct sequence for the 5′ end of the R region (5′-TTACGCCAAGCTACG TAATACGAC TCAC TATAGG TC TC TC TGG T TAGACCAGATCTGAGCCTGGGA-3′) and a second oligomer containing the PBS sequence (5′-AGTCCCTGTTCGGGCGCCA-3′) were used to generate a PCR fragment from the pNL4-3 sequence cloned into LITMUS. The PCR fragment was used as the template for RNA synthesis. RNA was purified by electrophoresis on a 5% denaturing gel, visualized using SYBR Green (Molecular Probes, Eugene, Oreg.), excised, and eluted using an RNaid kit from Bio 101 (Vista, Calif.). RNA was quantitated by both UV spectrophotometry and Ribogreen fluorescence as measured by a Storm 860 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). Oligomers were 32P labeled using [γ-32P]ATP (Amersham Pharmacia, Piscataway, N.J.) and T4 polynucleotide kinase (New England Biolabs).
RT/NC assays.
PBS-R template RNA, either wild type or mutant, was mixed with fivefold molar excess [32P]PBS (5′-AGTCCCTGTTCGGGCGCCA-3′), incubated at 65°C, and allowed to cool to 23°C over 30 min. A 20-fold molar excess of RT was added to the annealed template-primer and allowed to bind for 5 min at 37°C. Increasing amounts of NC were added to aliquots of the template-primer-RT reaction and incubated at 37°C for 7 min. Synthesis was initiated with the addition of RT start solution containing deoxynucleoside triphosphates (80 μM, final concentration) and MgCl2 (6 mM, final concentration). Reactions were stopped at the indicated times by the addition of an equal volume of a 90% formamide stop solution containing 1% sodium dodecyl sulfate (SDS), 4 μg of plasmid DNA/ml, bromophenol blue, and xylene cyanol. Reactions were heated to 95°C for 4 min, fractionated by electrophoresis on a denaturing acrylamide gel containing 0.05% SDS, dried under vacuum, and exposed to a PhosphorImager screen (Molecular Dynamics).
Circularization assays.
32P-labeled circularization substrate (5′-GCGAATTCTTTTTTTTTTTTTTTTTTTTGAAGACATAGTCCCTGTTCGGGCGCCAC-3′) was mixed in 1× ligase buffer (New England Biolabs) with increasing concentrations of NC. The bridge primer (5′-AAGAATTCGCGTGGCGCCCG-3′) was incubated with NC in a separate reaction. After 20 min at 37°C, the NC-oligomer reactions were mixed and incubated at 37°C for 8 min. T4 DNA ligase (New England Biolabs) was added in 50-fold molar excess, and the reactions were incubated at 22°C for 30 min. Ligase was inactivated by heating to 65°C, and the reactions were placed on ice. Then 0.3 U of exonuclease VII (Exo VII; Gibco/BRL, Rockville, Md.) was added, and the mixture was incubated for 2 h at 37°C. Reactions were stopped by the addition of an equal volume of formamide loading dye containing 0.1% SDS and fractionated by electrophoresis on a 12% denaturing acrylamide gel. The gels were dried under vacuum and exposed to a PhosphorImager screen.
In the circularization control assays, 15 fmol of an oligomer representing the upstream sequence of the circularization substrate (5′-CGGGCGCCAC-3′) and 1 fmol of the 32P-labeled oligomer representing the downstream sequence of the circularization substrate (5′-[32P]GCGAATTCTT-3′) were mixed in 1× ligase buffer and incubated with increasing concentrations of NC. In a separate reaction, 7.5 fmol of the bridge primer (5′-AAGAATTCGCGTGGCGCCCG-3′), to which the upstream and downstream oligomers could anneal to form a ligatable nick, was coated with NC. After a 20-min incubation at 37°C, the oligomer-NC mixtures were combined and incubation was continued at 37°C for 8 min. T4 DNA ligase (New England Biolabs) was added in 50-fold molar excess and incubated at 22°C for 30 min. The reactions were stopped by the addition of an equal volume of formamide loading dye containing 0.1% SDS and fractionated by electrophoresis on a 20% denaturing acrylamide gel. The gels were dried under vacuum and exposed to a PhosphorImager screen.
Self-priming assays.
Fifteen femtomoles of 32P-labeled 100-nucleotide (nt) DNA complementary to bases 1 to 100 of pNL4-3 (GenBank accession no. AF033819), containing either wild-type or mutant TAR sequences, was mixed with 70-fold excess of the indicated blocking primers in 1× RT binding buffer. The mixtures were heated to 65°C for 10 min and allowed to cool slowly to 22°C. A 20-fold molar excess of RT was added and incubated at 37°C for 5 min. NC was added to the template-primer-RT mix, at a one- to fourfold coating level (assuming 7 nt/NC as onefold) as indicated, and allowed to incubate for 10 min before the addition of RT start solution containing MgCl2 and deoxynucleoside triphosphates. The reactions were incubated at 37°C for 40 min and stopped by the addition of an equal volume of formamide loading dye containing 0.1% SDS. Electrophoresis was performed on a 6% denaturing acrylamide gel. The gels were dried under vacuum and exposed to a PhosphorImager screen.
RESULTS
NC inhibits the synthesis of self-primed products.
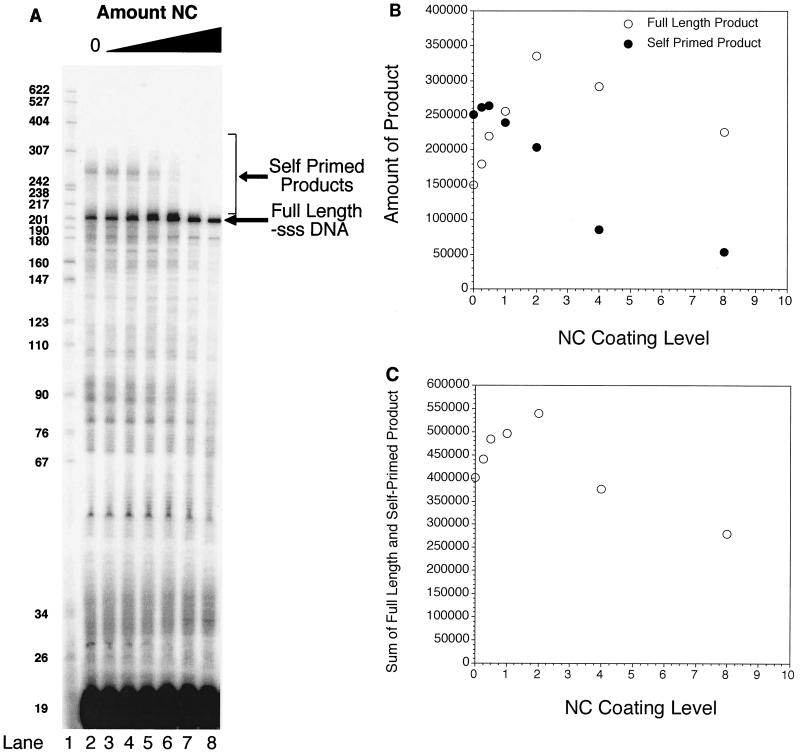
A synthetic RNA encompassing the U5-R region of the HIV-1 genome from the PBS through TAR was synthesized and designated PBS-R RNA. Complete reverse transcription of PBS-R RNA starting from a 19-base DNA primer annealed at the PBS produced a 200-base-long DNA product. A representative reverse transcription experiment is shown in Fig. 1A. Products larger than 200 bases were the result of self-primed synthesis (lanes 2 to 8). NC was added in increasing concentrations in lanes 3 to 8. In calculating the relative amounts of NC and nucleic acid, we have assumed that each NC covers 7 nt (39). Actual NC coating levels of DNA and RNA varied during the course of the reactions because the amounts and sizes of DNA and RNA varied as RT synthesized DNA and degraded RNA.
FIG. 1.
NC inhibits formation of self-primed products during reverse transcription. (A) A 5′ 32P-labeled 19-base DNA containing the PBS sequence was annealed to RNA containing the genomic HIV-1 R-PBS sequence. RT and either no NC (lane 2) or increasing amounts of NC (lanes 3 to 8) were subsequently allowed to bind. Theoretical coating levels of NC/primer-template were as follows: lane 2, none; lane 3, 0.25-fold; lane 4, 0.50-fold; lane 5, 1.0-fold; lane 6, 2.0-fold; lane 7, 4-fold; lane 8, 8-fold. Reverse transcription was initiated, allowed to proceed for 60 min at 37°C, quenched, and visualized on a 6% denaturing polyacrylamide gel. The positions of the 200-base full-length −sssDNA and self-primed products are indicated at the right. Positions of DNA size markers (in bases) are shown in lane 1. (B) Quantitation of full-length and self-primed product. The amount of full-length 200-base product and self-primed product in panel A was quantitated using a PhosphorImager and plotted as a function of NC coating level, where 1 NC/7 bases is a onefold coating level. (C) Sum of full-length and self-primed products in panel A, quantitated using a PhosphorImager and graphed as a function of initial NC coating level.
The addition of NC had significant effects on the synthesis of DNA by HIV-1 RT. Increasing amounts of NC resulted in a corresponding decrease in the amount of the self-primed products (Fig. 1A, lanes 3 to 8). A fourfold excess of NC (lane 7) virtually eliminated self-priming in these assays. The amount of self-primed product was quantitated using a PhosphorImager (Fig. 1B). As NC levels increased from zero (lane 2) to a twofold excess (lane 6), the amount of full-length product increased to more than twice its initial level (Fig. 1B). In reactions containing less than twice the amount of NC required to coat the RNA (lanes 2 to 5), two separate mechanisms increased the amount of 200-base product produced. First, the 200-base DNA, once made, was less likely to be extended because self-priming was inhibited by NC. Second, there was a slight (less than 1.4-fold) increase in the total synthesis of products 200 bases and longer (Fig. 1C). The increase in the amount of large products could have been caused by NC assisting RT through secondary structure, or it could have been the result of increased levels of primer-template annealing facilitated by NC, or both.
As NC levels were increased from two- to eightfold above the amount required to coat the RNA and primer, however, there was a decline in the amount of the full-length (200-base DNA) product produced (Fig. 1A, lanes 6 to 8; quantitated in Fig. 1B and C). High NC coating levels resulted in lower levels of polymerization. Very high levels of NC resulted in a marked decrease in extension by RT (data not shown). This is in agreement with published reports that NC can protect the substrate from RT binding, as well as from enzymatic digestion or modification (22, 33, 34) (see below).
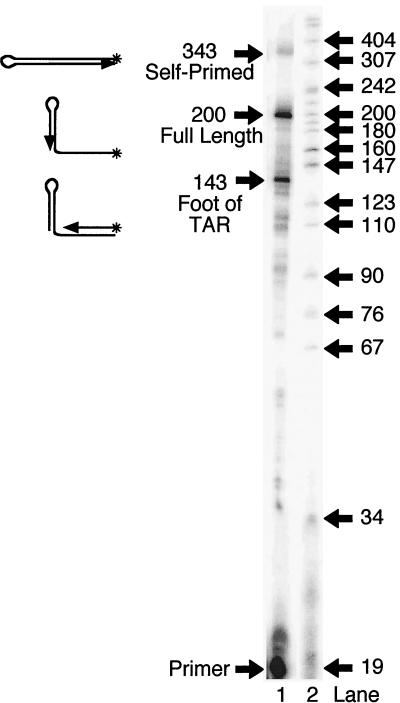
The major self-primed product did not migrate on a denaturing gel to the position corresponding to the expected size of 343 nt. Instead, the major product migrated as though it had an apparent size of 275 nt. We believe that the high level of secondary structure within the newly synthesized DNA prevented the completion of the self-primed product. To test this possibility, the sequence of the 200-base RNA used in Fig. 1A was changed so that base pairing within the poly(A) hairpin was disrupted. The TAR hairpin was still formed, as evidenced by the strong pause seen at 143 bases at the foot of TAR. These sequence changes that disrupted the poly(A) hairpin permitted the synthesis of the full-length 343-base self-primed product (Fig. 2). These data imply that −sssDNA folds into a structure similar to that proposed for viral RNA.
FIG. 2.
Destabilizing the poly(A) hairpin sequence allows completion of self-primed products. The sequence of the poly(A) RNA hairpin was changed so that base pairing between the arms of the hairpin was disrupted. 5′ 32P-labeled PBS DNA was annealed to the RNA, start solution and RT were added, and the mixture was incubated for 60 min at 37°C, as for Fig. 1A. The reaction was loaded in lane 1, and DNA markers were loaded in lane 2. Sizes of the markers are indicated in bases.
Does NC binding influence DNA bending?
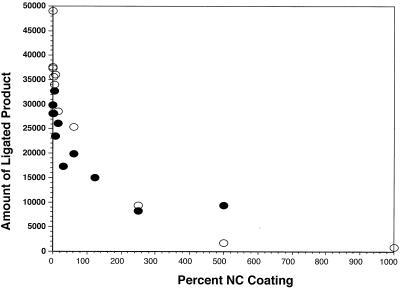
Clearly, NC is able to prevent self-priming from the TAR hairpin. How is this accomplished? NC can promote the annealing of thermodynamically stable structures (36). This would suggest that the addition of NC to an oligomer capable of forming a stable hairpin should actually promote hairpin formation. One possibility is that NC could unfold single-stranded DNA and hold it in a linear form, in a manner similar to the adenovirus DNA binding protein (40). This would also be consistent with reports that NC can unfold secondary structures in RNA and DNA, leading to increased RT polymerization rates through regions containing secondary structure (19, 38). To test this possibility, two substrates were created. The first was a linear substrate constructed of three synthetic DNA oligonucleotides, two of which were 10 bases long and were designed to anneal to a 20-base oligonucleotide, creating a double-stranded DNA with a nick that could be sealed by T4 DNA ligase. The second substrate contained a 56-base oligonucleotide designed such that its ends annealed to the same 20-base DNA used to make the first substrate. The 56-base oligonucleotide and the 20-base oligonucleotide were annealed to form a circular substrate with the same ligatable nick present in the linear substrate. In this assay, the ability of ligase to form the circular product depended on the ability of both ends of the linear 56-base DNA to anneal to the same 20-base DNA, forming a circle. If NC binding produced a rigid linear DNA structure, formation of a ligatable circle would be inhibited relative to the ligation of the linear substrate with an identical nick. DNA components of both linear and circular substrates were first coated with NC and then mixed together and allowed to anneal. Increasing amounts of NC were added to the substrates, and the amount of circularized substrate was quantitated and compared with the amount of ligation obtained using linear substrate. To ensure that the ligation product was a circle, the completed ligation reactions were digested with Exo VII, which can digest single-stranded DNA from both 5′ and 3′ exposed ends. The only ligated product resistant to Exo VII was the circular 56-base DNA, which migrated on a sequencing gel at an apparent size of 70 nt (data not shown). As can be seen in Fig. 3, both the linear and circular substrates were ligated with the same efficiency by T4 DNA ligase in the presence of NC. Although increasing levels of NC were able to interfere with T4 DNA ligase, presumably by blocking access to the nick, the degree of inhibition was the same for both substrates. This shows that NC does not affect the ability of the 56-base DNA to form a circle and implies that NC does not prevent self-priming by preventing TAR DNA from forming a hairpin. These results also show that NC can protect DNA from modification by ligase. Substantial (i.e., 60 to 70%) protection of the ends of the DNA was obtained at only a one- to twofold NC coating level (in these experiments, twofold is 1 NC/3.5 bp). These results are similar to those reported by Lapadat-Tapolsky et al. (22), who showed that interior of the DNA is completely protected from restriction enzyme digestion by twofold excess NC (1 NC/ 4 bp).
FIG. 3.
NC does not inhibit circularization of DNA. The effects of NC on the formation of a linear and circular ligation product were compared. The 5′ end of the circularization substrate (●) and the 5′ end of the downstream 10-base DNA of the linear substrate (○) were 32P labeled as described in Materials and Methods. Both DNA substrates, which contained the same ligatable nick, were incubated with increasing concentrations of NC. The reactions were incubated with ligase and fractionated by electrophoresis, and the ligated products were quantitated using a PhosphorImager as described in Materials and Methods.
NC requires an annealed oligomer to block TAR self-priming.
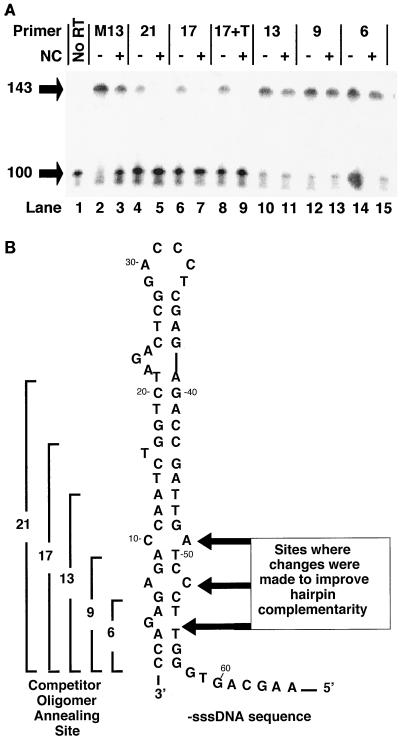
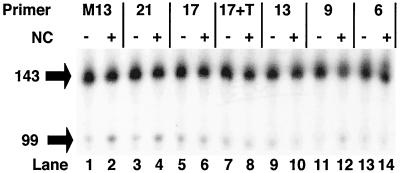
Since NC does not directly inhibit the formation of DNA hairpins, it must block the self-priming of the TAR hairpin by another mechanism. We noticed that in reactions in vitro where −sssDNA was synthesized by copying an RNA template, NC blocked self-priming. However, for NC to inhibit self-priming in reactions containing synthetic −sssDNA in the absence of RNA, it was necessary to include an oligonucleotide complementary to the sequence that forms the hairpin. This indicated that both NC and an oligomer (either DNA or RNA) were required to block self-priming. Either NC alone or an oligomer alone was not capable of blocking self-priming of DNA hairpin. To demonstrate this, a 32P-labeled synthetic 100-nt −sssDNA oligomer, consisting of the entire TAR sequence plus part of the poly(A) hairpin, was synthesized. This oligomer could self-prime by forming a hairpin involving the TAR sequence and be extended by RT to produce a product of 143 nt (Fig. 4A). A 70-fold excess of unlabeled DNA oligomers of increasing length was included in the experiments shown in Fig. 4A, lanes 2 to 15. The unlabeled DNA oligomers were intended to mimic the RNA fragments remaining after RNase H digestion of the template. They either had no complementarity to the 100-base DNA (lanes 2 and 3, M13 primer) or could base pair with sequences at the 3′ end of TAR (Fig. 4B). The length of the complementary segment was either 21, 17, 13, 9, or 6 nt, as indicated in Fig. 4B. In the absence of a complementary oligonucleotide (Fig. 4A, lanes 2 and 3), the addition of NC caused a slight reduction in the formation of 143-base self-primed product, which is most likely due to inhibition of polymerization, similar to what was seen in Fig. 1A. Likewise, in lanes 10 to 15, the inclusion of short complementary oligonucleotides also had little or no effect on the ability to self-prime, either in the presence or in the absence of NC. In lanes 4 to 7, however, the self-priming of the 100-base DNA was significantly reduced by the addition of oligomer and NC. In the absence of NC, self-priming at the TAR hairpin resulted in the formation of significant amounts of self-primed product (lanes 4 and 6), even in the presence of a 70-fold excess of competing unlabeled oligomer. In the presence of NC, however (lanes 5 and 7), the annealing of either the 17- or the 21-base DNA blocked self-priming. This implies that annealing of the 17- or 21-base DNA produced structures that were thermodynamically more stable than the hairpin, whereas the oligomers 13 bases and smaller did not. In this case, NC was acting to promote annealing, not simply protecting the annealed structure from RNase H, because the DNA oligomers are not susceptible to RNase H.
FIG. 4.
NC blocks self-priming by promoting the annealing of complementary oligomers that are more stable than the hairpin. (A) A synthetic oligonucleotide comprising the 3′ 100 bases of the −sssDNA was end labeled, heated, and slowly cooled in the presence of either a 21-nt M13 DNA primer (lanes 2 and 3) or DNA competitor of decreasing length, complementary to the 3′ end of the −sssDNA (lanes 4 to 15; see panel B). The length of each oligonucleotide is indicated at the top, and the site of annealing to the hairpin is shown in panel B. 17+T has the same sequence as 17, with the addition of a 7-nt unannealed tail at the 3′ end (see text). Each reaction was performed both in the presence and in the absence of enough NC to coat the primer-template at 7 bases/NC, as indicated at the top. Positions of migration of the 100-nt −sssDNA and the 143-nt self-primed product are shown at left. Lane 1 contains unmodified, labeled 100-base DNA. (B) The −sssDNA sequence which folds into a structure resembling the TAR hairpin. The lengths and sites of annealing of the competitor DNA oligomers discussed above are shown at the left. The sequence of the competitor DNA oligomer used to block self-priming is identical to the genomic RNA sequence. The sites of modifications of the hairpin discussed in the text are indicated by arrows at the right.
The oligomer 17+Tail (17+T) was included in the experiments in Fig. 4A as a control. This oligomer is identical to the 17-base DNA except for the addition of seven T residues at the 3′ end. The 3′ tail will not anneal to the template. This oligonucleotide was used to test the possibility that the decrease in production of self-primed products observed for the reactions containing NC was due, at least in part, to an increase in extendable 3′ ends created by annealing perfectly complementary competitor oligomers to the TAR hairpin. In addition to interfering with hairpin formation, the short, fully annealed oligomers created an additional site for RT to bind. The presence of the unannealed 3′ tail in 17+T should have interfered with the ability of RT to bind and extend the 3′ end of this oligomer. However, the results obtained with the 17+T oligonucleotide were indistinguishable from those seen with the 17-base DNA. This demonstrated that increasing in the amount of extendable substrate did not influence the ability of NC to inhibit self-priming at the TAR hairpin.
Improvements in base pairing within the TAR hairpin prevent NC from blocking TAR self-priming.
If there is a competition between the formation of the TAR hairpin and oligomers annealing to TAR that is affected by NC, then changes to either the length of the oligomers or the extent of base pairing in TAR should change whether NC can block self-priming. It is clear from Fig. 4A that changing the length of the oligomer annealed to TAR determines whether or not NC can block self-priming. To test the possibility that stabilizing the TAR hairpin could promote self-priming, we constructed a second hairpin based on the TAR sequence but with an increase in base pairing between the arms of the hairpin. Sequence changes (Fig. 4B) were introduced on the 5′ arm of the TAR hairpin so that the competitor oligomers used in the experiments shown in Fig. 4A could be used in the self-priming assays with increased TAR base pairing. The mutant TAR sequence contained three changes, eliminating a single-nucleotide bubble at base 4 by the addition of a complementary base at position 55, and eliminating the mismatches between bases 7 to 52 and 10 to 49 by making changes at positions 49 and 52 (Fig. 4B). These three changes abolished the ability of NC to block the formation of the hairpin, even in the presence of the complementary 21-base DNA (Fig. 5). This shows that the ability of NC to block self-priming at the TAR hairpin depends on the fact that the hairpin is imperfect.
FIG. 5.
Oligomers cannot block self-priming of more stable mutant TAR hairpin. A 99-base DNA containing the changes in the TAR hairpin sequence indicated in Fig. 4B was subjected to the same conditions as for Fig. 4A. Sizes of the 99-base starting material and the 143-base self-primed product are indicated at the left.
DISCUSSION
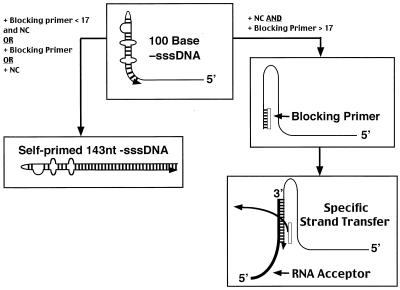
Self-priming of the DNA copied from the 5′ end of the HIV-1 RNA genome requires the formation of a TAR hairpin in the newly synthesized −sssDNA. Because NC impedes self-priming of −sssDNA in vitro, we considered the possibility that NC directly interferes with hairpin formation by forcing the DNA to adopt a rigid, linear structure, in a manner similar to that of the adenovirus DNA binding protein (40), but we found that this was not the case. There was other evidence to support the idea that NC did not constrain the RNA in a rigid linear structure. Although NC could block self-priming from −sssDNA generated by RT copying an RNA template in vitro, NC alone could not prevent the self-priming of synthetic −sssDNA. NC by itself was not sufficient to block self-priming of −sssDNA; there was an additional component in the reactions containing an RNA template that was necessary for NC to inhibit self-priming. The additional component is a complementary oligonucleotide. Substituting DNA oligomers for the RNase H-susceptible RNA in our model reactions, we found that a 17-base DNA primer annealed to the 3′ end of the TAR hairpin was sufficient to block self-priming of −sssDNA in the presence of NC (Fig. 6). NC is a nucleic acid chaperone which facilitates the formation of the most stable nucleic acid structures; under these circumstances, NC promotes the annealing of a thermodynamically more stable −sssDNA-blocking primer duplex even in the presence of the kinetically favored hairpin competitor (36). Formation of the hairpin in the −sssDNA is a first-order, unimolecular reaction, whereas the equilibrium for the RNA-DNA duplex is dependent on the concentrations of both the RNA and DNA, a second-order reaction. As a result, in the absence of NC, the equilibrium of the RNA-DNA exchange favors hairpin formation, in spite of the fact that the hairpin is less thermodynamically stable. Essentially, displacement of the more stably annealed RNA fragment in favor of the less stable DNA hairpin formation is driven by the high local concentration of complementary strands in the hairpin DNA. NC may facilitate the aggregation of nucleic acids, which could affect the local concentrations of the oligonucleotides relative to the hairpin itself; this could reduce the kinetic advantage of hairpin formation (25, 32). It has been proposed that NC destabilizes secondary structures in RNA and DNA, which could facilitate the progression of RT through the genome (20, 36). However, more recent reports have demonstrated that NC does not unwind secondary structures in RNA, suggesting that NC binding has little or no effect on RNA folding (7, 16). After RT extends the DNA hairpin, the hairpin becomes increasingly stable due to the additional base pairing; thus this process, once begun, is not readily reversible.
FIG. 6.
Blocking primer prevents self-priming of −sssDNA and promotes specific strand transfer. The 100-base −sssDNA forms a hairpin similar to the TAR RNA hairpin, which can self-prime and be extended by RT. NC promotes the annealing of a blocking primer 17 bases or longer to the 3′ end of the −sssDNA, preventing self-priming. The blocking primer is released in the presence of NC only if it is displaced by an acceptor which has greater complementarity to the −sssDNA. During reverse transcription, the blocking primer(s) would be a fragment or fragments of the RNA genome that remained annealed to the −sssDNA after synthesis was complete.
Combining our data with data from the literature, we can propose a model describing how the polymerase and RNase H activities of RT collaborate with NC and the TAR hairpin to produce efficient, selective strand transfer (Fig. 6). During DNA synthesis, RNase H cuts relatively infrequently, leaving relatively large RNA oligomers (11, 33), one or more of which may serve as blocking primers in the presence of NC. Fu and Taylor showed that HIV-1 RT leaves a 14- to 18-base RNA oligomer annealed to the end of a newly synthesized DNA strand in the absence of NC (14). NC could protect the RNA oligomers from being displaced by the formation of the hairpin by the same mechanism we observed with the DNA blocking primers (Fig. 6). NC may also block hairpin formation by limiting the extent of RNase H digestion; it has been shown by other investigators that NC can protect RNA from RNase A (10, 28, 33). However, because RT generates genomic RNA fragments of the correct size (14 to 18 nt) at the 3′ end of the −sssDNA even in the absence of NC (14), NC-mediated protection of the RNA may not be necessary.
Transfer of the completed −sssDNA to the 3′ R region RNA is required for the continuation of reverse transcription. This step must be performed without releasing the TAR hairpin sequence in a fashion that would allow self-priming. In this context, it is important to remember that hairpin formation is kinetically favored. The ability of NC to promote the thermodynamically favored event means that as long as the complementarity between the −sssDNA and the RNA acceptor is greater than that of the −sssDNA and the digested RNA, transfer will occur (as illustrated in Fig. 6). Long regions of complementarity are known to promote more efficient strand transfer (4, 21). Since NC promotes the formation of the thermodynamically favored duplex, the extensive base pairing between the nascent DNA and the strand transfer acceptor is favored over either the annealing of the digested RNA or hairpin formation. Thus, targeted and efficient strand transfer is accomplished.
Although in some cases NC altered the degree of RT pausing at secondary structures, NC had either a neutral or inhibitory effect on polymerization in our assays. Other investigators have also reported that NC either had no effect on or inhibited polymerization by RT (29, 31, 33). High concentrations of NC reduced the numbers of initiation events, and the amount of full-length product formed after a given initiation event, in a dose-dependent manner. However, lower levels of NC can promote the annealing of the primer-template, increasing the availability of the nucleic acid substrate and offsetting the inhibition of polymerization, as has been reported for primer tRNA (2, 6, 13, 23). NC not only promotes annealing of primer-template but also inhibits the formation of self-primed products that would prevent completion of the full-length HIV genome (17). An appropriate amount of NC can block self-priming while only slightly inhibiting reverse transcription. As shown in Fig. 1B, almost 90% of the self-primed products were eliminated at a fourfold NC coating level, while the amount of full-length product (200-base DNA) was reduced by only 15%. An eightfold excess of NC over the amount needed to coat the RNA virtually eliminated self-priming but allowed the synthesis of 65% of the maximal amount of full-length DNA product. In our assays we selected conditions where NC displayed the ability to inhibit self-priming while still allowing high levels of polymerization to occur.
Completion of the full-length DNA hairpin was dependent on the DNA sequence. The wild-type HIV-1 DNA sequence caused RT to pause, producing predominantly a 275-base self-primed product instead of the expected 343-base product. The pause may be caused by secondary structure within the self-primed DNA template. This proposal was supported by the finding that sequence changes within the poly(A) hairpin eliminated the pause, leading to the formation of the 343-base product.
Introducing mutations that stabilized the structure of the TAR hairpin resulted in the efficient production of self-primed products that could not be inhibited by a 21-base blocking primer and NC. The increased stability of the mutated hairpin made hairpin formation the favored reaction both kinetically and thermodynamically. Significantly, this indicates that a more stable hairpin is likely to be detrimental to the virus, presenting the possibility that in mutants in which the TAR hairpin is more stable than the strand transfer intermediate, NC may favor hairpin formation over RNA-DNA heteroduplex formation. Normally, this is not an issue; however, if mutations are introduced into either the upstream or the downstream R region (but not in both), this may diminish the stability of the acceptor RNA-donor DNA duplex required for strand transfer, interfering with the strand transfer reaction. In support of this idea, at least some mutations that both increase TAR stability and block viral replication if they are present only in one TAR element have no discernible effect on HIV-1 replication when introduced into both the upstream and downstream TAR elements (Jared Clever, personal communication).
Although strand transfer is possible in vitro with unstructured complementary regions as short as 2 to 7 bases (1), the ability to form the TAR hairpin would restrict the use of such short regions as strand transfer acceptors at the end of R. HIV-1 has relatively long R regions compared to other retroviruses (8, 35). Longer R regions promote more efficient strand transfer (4, 21). Long R regions may be necessary for efficient strand transfer in the presence of the TAR structure. In retroviral genomes that have highly structured elements in the R region, R is long. This is true for HIV-1 and for two viruses with more complex structures in R, HIV-2 and human T-cell leukemia virus type 1 (HTLV-1). In all three viral genomes, R is longer than the structural element (TAR in HIV-1 and HIV-2; Rex response element for HTLV-1). A long R region would ensure that, in the strand transfer reaction, the strand transfer intermediate is more stable than any structure that would be created from −sssDNA.
ACKNOWLEDGMENTS
We are grateful to Robert Gorelick, Louis Henderson, and Larry Arthur for the gift of the HIV-1 NC used in this study and for helpful discussions. We are grateful to Hilda Marusiodis for preparing the manuscript.
Research in S. Hughes' laboratory was sponsored by the National Cancer Institute, DHHS, under contract with ABL, and by the National Institute of General Medical Sciences.
REFERENCES
- 1.Allain B, Lapadat-Tapolsky M, Berlioz C, Darlix J L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barat C, Schatz O, Le Grice S, Darlix J L. Analysis of the interactions of HIV1 replication primer tRNA(Lys,3) with nucleocapsid protein and reverse transcriptase. J Mol Biol. 1993;231:185–190. doi: 10.1006/jmbi.1993.1273. [DOI] [PubMed] [Google Scholar]
- 3.Benn S, Rutledge R, Folks T, Gold J, Baker L, McCormick J, Feorino P, Piot P, Quinn T, Martin M. Genomic heterogeneity of AIDS retroviral isolates from North America and Zaire. Science. 1985;230:949–951. doi: 10.1126/science.2997922. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout B, van Wamel J, Klaver B. Requirements for DNA strand transfer during reverse transcription in mutant HIV-1 virions. J Mol Biol. 1995;252:59–69. doi: 10.1006/jmbi.1994.0475. [DOI] [PubMed] [Google Scholar]
- 5.Boyer P L, Tantillo C, Jacobo-Molina A, Nanni R G, Ding J, Arnold E, Hughes S H. Sensitivity of wild-type human immunodeficiency virus type 1 reverse transcriptase to dideoxynucleotides depends on template length; the sensitivity of drug-resistant mutants does not. Proc Natl Acad Sci USA. 1994;91:4882–4886. doi: 10.1073/pnas.91.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cen S, Huang Y, Khorchid A, Darlix J L, Wainberg M A, Kleiman L. The role of Pr55gag in the annealing of tRNA3Lys to human immunodeficiency virus type 1 genomic RNA. J Virol. 1999;73:4485–4488. doi: 10.1128/jvi.73.5.4485-4488.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan B, Weidemaier K, Yip W T, Barbara P F, Musier-Forsyth K. Intra-tRNA distance measurements for nucleocapsid protein-dependent tRNA unwinding during priming of HIV reverse transcription. Proc Natl Acad Sci USA. 1999;96:459–464. doi: 10.1073/pnas.96.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 9.Das A T, Klaver B, Klasens B I, van Wamel J L, Berkhout B. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J Virol. 1997;71:2346–2356. doi: 10.1128/jvi.71.3.2346-2356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeStefano J J. Interaction of human immunodeficiency virus nucleocapsid protein with a structure mimicking a replication intermediate. Effects on stability, reverse transcriptase binding, and strand transfer. J Biol Chem. 1996;271:16350–16356. [PubMed] [Google Scholar]
- 11.DeStefano J J, Buiser R G, Mallaber L M, Myers T W, Bambara R A, Fay P J. Polymerization and RNase H activities of the reverse transcriptases from avian myeloblastosis, human immunodeficiency, and Moloney murine leukemia viruses are functionally uncoupled. J Biol Chem. 1991;266:7423–7431. [PubMed] [Google Scholar]
- 12.DeStefano J J, Mallaber L M, Fay P J, Bambara R A. Quantitative analysis of RNA cleavage during RNA-directed DNA synthesis by human immunodeficiency and avian myeloblastosis virus reverse transcriptases. Nucleic Acids Res. 1994;22:3793–3800. doi: 10.1093/nar/22.18.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Y X, Campbell S, Harvin D, Ehresmann B, Ehresmann C, Rein A. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J Virol. 1999;73:4251–4256. doi: 10.1128/jvi.73.5.4251-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu T B, Taylor J. When retroviral reverse transcriptases reach the end of their RNA templates. J Virol. 1992;66:4271–4278. doi: 10.1128/jvi.66.7.4271-4278.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goff S P. Retroviral reverse transcriptase: synthesis, structure, and function. J Acquir Immune Defic Syndr. 1990;3:817–831. [PubMed] [Google Scholar]
- 16.Gregoire C J, Gautheret D, Loret E P. No tRNA3Lys unwinding in a complex with HIV NCp7. J Biol Chem. 1997;272:25143–25148. doi: 10.1074/jbc.272.40.25143. [DOI] [PubMed] [Google Scholar]
- 17.Guo J, Henderson L E, Bess J, Kane B, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes S H, Hostomsky Z, Le Grice S F, Lentz K, Arnold E. What is the orientation of DNA polymerases on their templates? J Virol. 1996;70:2679–2683. doi: 10.1128/jvi.70.5.2679-2683.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji X, Klarmann G J, Preston B D. Effect of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein on HIV-1 reverse transcriptase activity in vitro. Biochemistry. 1996;35:132–143. doi: 10.1021/bi951707e. [DOI] [PubMed] [Google Scholar]
- 20.Khan R, Giedroc D P. Recombinant human immunodeficiency virus type 1 nucleocapsid (NCp7) protein unwinds tRNA. J Biol Chem. 1992;267:6689–6695. [PubMed] [Google Scholar]
- 21.Klaver B, Berkhout B. Premature strand transfer by the HIV-1 reverse transcriptase during strong-stop DNA synthesis. Nucleic Acids Res. 1994;22:137–144. doi: 10.1093/nar/22.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapadat-Tapolsky M, De Rocquigny H, Van Gent D, Roques B, Plasterk R, Darlix J L. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. . (Erratum, 21:2024.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lapadat-Tapolsky M, Gabus C, Rau M, Darlix J L. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J Mol Biol. 1997;268:250–260. doi: 10.1006/jmbi.1997.0978. [DOI] [PubMed] [Google Scholar]
- 24.Lapadat-Tapolsky M, Pernelle C, Borie C, Darlix J L. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 1995;23:2434–2441. doi: 10.1093/nar/23.13.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Cam E, Coulaud D, Delain E, Petitjean P, Roques B P, Gerard D, Stoylova E, Vuilleumier C, Stoylov S P, Mely Y. Properties and growth mechanism of the ordered aggregation of a model RNA by the HIV-1 nucleocapsid protein: an electron microscopy investigation. Biopolymers. 1998;45:217–229. doi: 10.1002/(SICI)1097-0282(199803)45:3<217::AID-BIP4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Quan Y, Arts E J, Li Z, Preston B D, de Rocquigny H, Roques B P, Darlix J L, Kleiman L, Parniak M A, Wainberg M A. Human immunodeficiency virus type 1 nucleocapsid protein (NCp7) directs specific initiation of minus-strand DNA synthesis primed by human tRNA3Lys in vitro: studies of viral RNA molecules mutated in regions that flank the primer binding site. J Virol. 1996;70:4996–5004. doi: 10.1128/jvi.70.8.4996-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palaniappan C, Fuentes G M, Rodriguez-Rodriguez L, Fay P J, Bambara R A. Helix structure and ends of RNA/DNA hybrids direct the cleavage specificity of HIV-1 reverse transcriptase RNase H. J Biol Chem. 1996;271:2063–2070. [PubMed] [Google Scholar]
- 28.Peliska J A, Balasubramanian S, Giedroc D P, Benkovic S J. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 29.Raja A, DeStefano J J. Kinetic analysis of the effect of HIV nucleocapsid protein (NCp) on internal strand transfer reactions. Biochemistry. 1999;38:5178–5184. doi: 10.1021/bi9828019. [DOI] [PubMed] [Google Scholar]
- 30.Rein A, Henderson L E, Levin J G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez-Rodriguez L, Tsuchihashi Z, Fuentes G M, Bambara R A, Fay P J. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J Biol Chem. 1995;270:15005–15011. doi: 10.1074/jbc.270.25.15005. [DOI] [PubMed] [Google Scholar]
- 32.Stoylov S P, Vuilleumier C, Stoylova E, De Rocquigny H, Roques B P, Gerard D, Mely Y. Ordered aggregation of ribonucleic acids by the human immunodeficiency virus type 1 nucleocapsid protein. Biopolymers. 1997;41:301–312. doi: 10.1002/(SICI)1097-0282(199703)41:3<301::AID-BIP5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 33.Suo Z, Johnson K A. Effect of RNA secondary structure on RNA cleavage catalyzed by HIV-1 reverse transcriptase. Biochemistry. 1997;36:12468–12476. doi: 10.1021/bi971218+. [DOI] [PubMed] [Google Scholar]
- 34.Tanchou V, Gabus C, Rogemond V, Darlix J L. Formation of stable and functional HIV-1 nucleoprotein complexes in vitro. J Mol Biol. 1995;252:563–571. doi: 10.1006/jmbi.1995.0520. [DOI] [PubMed] [Google Scholar]
- 35.Temin H M. Structure, variation and synthesis of retrovirus long terminal repeat. Cell. 1981;27:1–3. doi: 10.1016/0092-8674(81)90353-6. [DOI] [PubMed] [Google Scholar]
- 36.Tsuchihashi Z, Brown P O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitcomb J M, Hughes S H. Retroviral reverse transcription and integration: progress and problems. Annu Rev Cell Biol. 1992;8:275–306. doi: 10.1146/annurev.cb.08.110192.001423. [DOI] [PubMed] [Google Scholar]
- 38.Wu W, Henderson L E, Copeland T D, Gorelick R J, Bosche W J, Rein A, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.You J C, McHenry C S. HIV nucleocapsid protein. Expression in Escherichia coli, purification, and characterization. J Biol Chem. 1993;268:16519–16527. [PubMed] [Google Scholar]
- 40.Zijderveld D C, Stuiver M H, van der Vliet P C. The adenovirus DNA binding protein enhances intermolecular DNA renaturation but inhibits intramolecular DNA renaturation. Nucleic Acids Res. 1993;21:2591–2598. doi: 10.1093/nar/21.11.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]