Abstract
We characterized a novel 28S rRNA cleavage in cells infected with the murine coronavirus mouse hepatitis virus (MHV). The 28S rRNA cleavage occurred as early as 4 h postinfection (p.i.) in MHV-infected DBT cells, with the appearance of subsequent cleavage products and a decrease in the amount of intact 28S rRNA with increasing times of infection; almost all of the intact 28S rRNA disappeared by 24 h p.i. In contrast, no specific 18S rRNA cleavage was detected in infected cells. MHV-induced 28S rRNA cleavage was detected in all MHV-susceptible cell lines and all MHV strains tested. MHV replication was required for the 28S rRNA cleavage, and mature cytoplasmic 28S rRNA underwent cleavage. In certain combination of cells and viruses, pretreatment of virus-infected cells with interferon activates a cellular endoribonuclease, RNase L, that causes rRNA degradation. No interferon was detected in the inoculum used for MHV infection. Addition of anti-interferon antibody to MHV-infected cells did not inhibit 28S rRNA cleavage. Furthermore, 28S rRNA cleavage occurred in an MHV-infected mouse embryonic fibroblast cell line derived from RNase L knockout mice. Thus, MHV-induced 28S rRNA cleavage was independent of the activation of RNase L. MHV-induced 28S rRNA cleavage was also different from apoptosis-related rRNA degradation, which usually occurs concomitantly with DNA fragmentation. In MHV-infected 17Cl-1 cells, 28S rRNA cleavage preceded DNA fragmentation by at least 18 h. Blockage of apoptosis in MHV-infected 17Cl-1 cells by treatment with a caspase inhibitor did not block 28S rRNA cleavage. Furthermore, MHV-induced 28S rRNA cleavage occurred in MHV-infected DBT cells that do not show apoptotic signs, including activation of caspase-3 and DNA fragmentation. Thus, MHV-induced 28S rRNA cleavage appeared to differ from any rRNA degradation mechanism described previously.
Coronaviruses are enveloped RNA viruses that cause gastrointestinal and upper respiratory tract illnesses in animals and humans. These range, in severity, from very serious neonatal enteritis in domestic animals to the common cold in humans. Although coronavirus infections are usually acute, some coronaviruses cause persistent neurotropic infections in animals (2, 38, 53). Among the coronaviruses, mouse hepatitis virus (MHV) is one of the best characterized in terms of pathogenesis and molecular biology. MHV causes various diseases, including hepatitis, enteritis, and encephalitis in rodents (6, 53). MHV contains a 32-kb-long, positive-sense, single-stranded RNA genome (27, 29, 36) that encodes 11 open reading frames, which are expressed through the production of a genomic-size mRNA and six to eight species of subgenomic mRNAs (26, 30). The identical leader sequence, about 70 nucleotides long, present at the 5′ ends of all MHV mRNAs and each MHV-specific protein, is translated from each subgenomic mRNA. Genomic-size mRNA encodes the most 5′ gene, the 22-kb-long gene 1, which encodes the RNA polymerase function (29). Expression of gene 1 and N protein, which is encoded by the smallest mRNA, mRNA 7, is sufficient for MHV RNA synthesis (24). MHV contains three envelope proteins, S, M, and E. S protein binds to the coronavirus receptor (7) and forms the characteristic coronavirus peplomer. M protein and E protein play an important role in the formation of MHV envelope (4, 23, 52). MHV genomic RNA is associated with N protein, forming a helical nucleocapsid (47).
Extensive morphological, physiological, and biochemical changes occur in coronavirus-infected cells. Some of these changes contribute to the damage of cells and tissues. Progress in molecular biological and biochemical techniques has advanced our knowledge of the intracellular biochemical events of coronavirus replication, whereas the specific basis for the deleterious effects on host cells is not as well understood. Some progress has been made regarding the mechanism of cell death in coronavirus-infected cells; infection of coronavirus transmissible gastroenteritis virus and MHV induces apoptosis in certain cells (1, 3, 8). As found for some lytic viruses (9, 11, 20, 21), host protein translation is inhibited (12, 42, 49, 50) but not completely shut off in MHV-infected cells. Inhibition of host protein synthesis is accompanied by an increase in MHV protein synthesis (42, 43, 44). Specific host mRNAs are degraded in MHV-infected cells, while transcriptional upregulation of some other host mRNAs occurs in MHV-infected cells (25). The mechanism of selective MHV-specific protein synthesis, which occurs concomitantly with host protein inhibition, in infected cells is also poorly characterized, although it has been suggested that MHV mRNAs containing 5′-end leader sequences bind to N protein, forming a complex that may act as a strong translation initiation signal (50, 51).
In this study, we described the specific cleavage of 28S rRNA in MHV-infected cells; cleavage of 28S rRNA in coronavirus-infected cells has not been described previously. There are a few examples of specific 28S rRNA cleavage: interferon (IFN) secretion activates the cytoplasmic endoribonuclease, RNase L, via activation of the 2′,5′-oligoadenylate (2-5A) system; activated RNase L causes 28S and 18S rRNA cleavage in murine cells and human cells (45, 46); in some human and rat tumor cells, apoptosis triggers 28S rRNA cleavage when induced by chemicals like actinomycin D or cyclic AMP (16, 17); porcine reproductive and respiratory syndrome virus (PRRSV) infection results in rRNA degradation that is related to nucleosomal DNA cleavage and apoptosis (48); and rabbit reticulocytes contain a membrane-associated endonuclease activity that cleaves 28S rRNA (54). The present study demonstrates that MHV-induced 28S rRNA cleavage is different from other known rRNA cleavage events. The possible biological significance of 28S rRNA cleavage in MHV infection is discussed.
MATERIALS AND METHODS
Viruses and cells.
The plaque-cloned A59 strain of MHV (26), MHV-JHM (33), and MHV-2 (22) were used. Mouse DBT cells (14) were used for the propagation of virus stocks. DBT cells were maintained in minimal essential medium supplemented with 8% heat-inactivated newborn calf serum. Mouse L929 cells were used for IFN assay. 17Cl-1 cells (1) and mouse embryonic fibroblasts (MEF cells) derived from wild-type (RNase L+/+) and RNase L knockout (RNase L−/−) mice (55) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum.
Northern (RNA) blotting.
Northern blot analysis was performed using [γ-32P]ATP-labeled oligonucleotide probes as previously described (32). Oligonucleotide probe 1 (5′ CTAATCATTCGCTTTACCGG 3′), which specifically binds to nucleotides 1532 to 1551 from the 5′ end of mouse 28S rRNA, was used for the detection of 28S rRNA and its cleavage products. Oligonucleotide probes 2 (5′ ATGCCCCCGGCCGTCCCTCT 3′) and 3 (5′ TAATGATCCTTCCGCAGGTTCACC 3′), which bind to nucleotides 921 to 940 and 1846 to 1870, respectively, from the 5′ end of mouse 18S rRNA, were used to detect 18S rRNA. All hybridizations were performed at 60°C. To detect MHV-specific RNAs, Northern blot analysis was performed using a digoxigenin (DIG)-labeled random-primed probe (Boehringer), corresponding to the 3′ end of MHV genomic RNA, and visualized by using a DIG luminescence detection kit (Boehringer) according to the manufacturer's protocol.
Host protein synthesis analysis.
Intracellular proteins were labeled as described previously (23). Briefly, mock-infected and MHV-infected cells were incubated in methionine-cysteine-free medium for 0.5 h before labeling. Cells were labeled with Tran35S-label (75 μCi/ml; ICN) for 30 min at various times postinfection (p.i.). Labeled cells were lysed in buffer (1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS] in phosphate-buffered saline), and the postnuclear supernatant was separated by SDS-polyacrylamide gel electrophoresis (PAGE).
IFN assay.
Functional IFN was detected by the vesicular stomatitis virus (VSV) plaque reduction method as previously described (28). Briefly, confluent DBT cells in 60-mm-diameter dishes were mock infected or infected with MHV-A59 at a multiplicity of infection (MOI) of 10. At 12 h and 24 h p.i., supernatants were harvested and irradiated with UV light (wavelength, 253 nm) for 12 min to inactivate MHV. To 96-well clusters, seeded 24 h earlier with 4 × 104 mouse L929 cells per well and containing 100 μl of complete medium (DMEM supplemented with 10% fetal bovine serum), 50-μl duplicate aliquots of mock-infected or MHV-infected supernatant were added. Each sample was then serially threefold diluted 11 times. After 24 h of incubation at 37°C, 6 × 105 PFU of VSV, in 25 μl of complete medium, was added to each well. After another 24 h incubation at 37°C, L929 cell viability was determined by counting the number of plaques in each dilution. The dilution which yielded a 50% reduction in plaque number was used to determine the IFN concentration in the original supernatant. Calculations were according to IFN concentrations based on National Institutes of Health (NIH) standards for murine IFN-α and IFN-β.
Caspase-3 activity assay.
Presence of activated caspase-3 was detected using a caspase-3/CPP32 calorimetric protease assay kit (BioSource International Inc., Camarillo, Calif.); this assay system measures cleavage of the synthetic tetrapeptide Asp-Glu-Val-Asp (DEVD), linked to the chromophore p-nitroanilide, by activated caspase-3. Cell extracts were prepared by solubilizing mock-infected or MHV-A59-infected DBT cells at 4.5 and 8.5 h p.i. in a buffer consisting of 10 mM Tris-HCl (pH 7.5), 10 mM NaH2PO4, 10 mM Na2HPO4, 150 mM NaCl, and 1% Triton X-100. Cell extracts from DBT cells that were treated continuously in the presence of 50 μM etoposide for 35.5 h were used as a positive control. As a negative control, the 8.5-h-p.i. DBT lysate was incubated with 250 μM Z-DEVD-fmk (Enzyme Systems, Livermore, Calif.), a caspase-3 inhibitor, for 30 min at 37°C. Aliquots of each sample, corresponding to 200 μg of protein, were mixed with the reaction buffer {40 mM HEPES, 200 mM NaCl, 2 mM EDTA, 0.2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 20% glycerol, 10 mM dithiothreitol, 200 μM DEVD–p nitroanilide substrate} and incubated in 96-well clusters. The amount of substrate hydrolyzed was measured at an optical density of 405 nm. Background readings from cell lysates and buffers were subtracted from both mock-infected and MHV-infected samples before determining the fold increase in caspase-3 activation, calculated as (infected lysate + substrate) − (infected lysate − substrate)/(mock lysate + substrate) − (mock lysate − substrate).
RESULTS
Cleavage of 28S rRNA in MHV-infected cells.
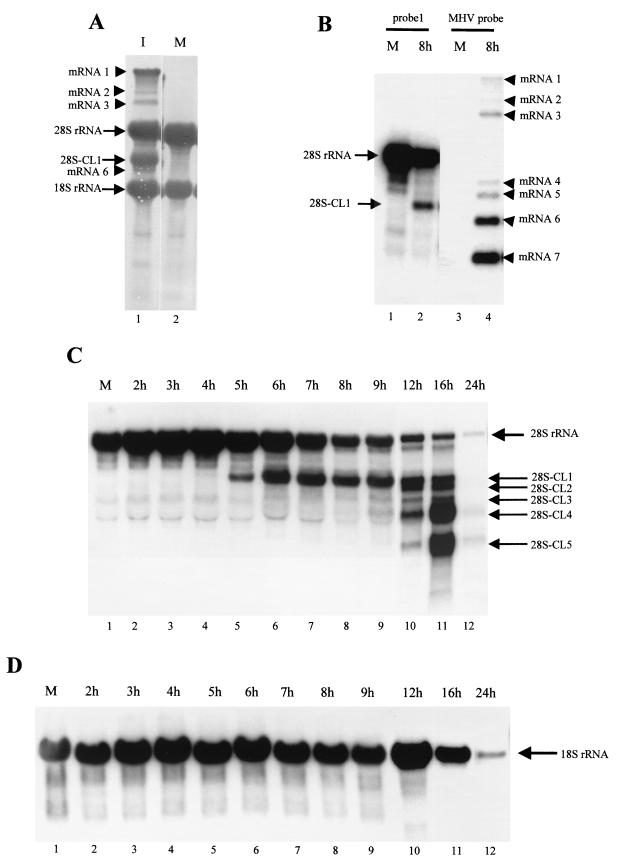
We noticed the appearance of a major band that migrated between the 28S and 18S rRNAs after agarose gel electrophoresis of intracellular RNA from MHV-A59-infected DBT cells. This band was easily detected after ethidium bromide staining of the gels (data not shown) or by methylene blue staining of intracellular RNAs transferred to a nylon membrane (Fig. 1A). MHV subgenomic mRNA 6 migrated slightly faster than this major band, and mRNA 7 comigrated with 18S rRNA. This band appeared to be an RNA of non-MHV origin, as DNase treatment did not affect it and the size differed from that of any of the MHV subgenomic mRNAs. The size and abundance of the band suggested that this RNA may be a cleavage product of 28S rRNA. We tested this possibility by Northern blot analysis of intracellular RNAs from MHV-A59-infected DBT cells, using an oligonucleotide (probe 1) that specifically hybridizes with the mature mouse 28S rRNA at 1.5 kb from the 5′ end (Fig. 1B). This probe specifically hybridized with an intact 28S rRNA and four additional minor bands from uninfected cells; these minor bands most probably represented degraded RNAs that were generated by the normal turnover of 28S rRNAs. The same probe hybridized with intact 28S rRNA and a 3.2-kb-long RNA (28S-CL1) from intracellular RNAs extracted from MHV-A59-infected cells at 8 h p.i. Using two other oligonucleotide probes, each of which hybridized with 28S rRNA at 1.0 and 2.0 kb from the 5′ end, we observed the same 28S rRNA cleavage (data not shown). Judging from the size and amount of this 28S rRNA-related RNA, 28S-CL1 was the cleavage product that we initially noticed (Fig. 1A). Parallel Northern blots of the same RNA samples were probed with the 28S rRNA-specific oligonucleotide probe 1 and an MHV-specific probe that hybridizes with all of the MHV RNAs (Fig. 1B). The 28S-CL1 product differed in size from any MHV mRNA, eliminating the possibility that it was the result of nonspecific hybridization of probe 1 to MHV mRNAs.
FIG. 1.
Characterization of 28S and 18S rRNAs in MHV-A59-infected cells. (A) DBT cells were mock infected (M) or infected with MHV-A59 (I) at an MOI of 10. At 8 h p.i., cytoplasmic RNA was extracted and a portion of the RNA was electrophoresed on a denaturing 1% agarose-formaldehyde gel. The RNA was blotted onto a nylon membrane, which was stained with methylene blue. MHV-A59-specific mRNAs 1, 2, 3, and 6 are indicated by arrowheads. Intact 28S rRNA, 18S rRNA, and the cleaved 28S rRNA product are indicated by arrows. (B) Cytoplasmic RNA was extracted from mock-infected DBT cells (M) or MHV-A59-infected DBT cells at 8 h p.i. (8h). RNAs were electrophoresed on a denaturing 1% agarose-formaldehyde gel. The RNA was blotted onto a nylon membrane, which was cut into two identical halves. One-half was probed with the 5′-end-labeled oligonucleotide probe 1, which specifically binds to mouse 28S rRNA (lanes 1 and 2), and the other half was probed with a random-primed DIG-labeled probe specific for MHV mRNAs (lanes 3 and 4). (C and D) Cytoplasmic RNA was extracted from MHV-A59-infected DBT cells at various times p.i., as shown above the lanes. RNAs were electrophoresed on a denaturing gel and examined by Northern blot analysis, using the 28S rRNA-specific probe 1 to detect 28S rRNA and its cleavage products (C) and a mixture of oligonucleotide probes 2 and 3 to detect 18S rRNA (D). The mock-infected RNA sample (lanes 1 in panels C and D) was extracted at 24 h p.i.
A precise time course analysis of 28S rRNA cleavage during MHV-A59 infection showed that a reduction in the amount of 28S rRNA was detectable at 7 h p.i. (Fig. 1C). Densitometric scanning analysis showed a 60% decrease in the amount of intact 28S rRNA from infected cells by 8 to 9 h p.i. compared to uninfected cells. The amount of 28S rRNA decreased continuously, and 28S rRNA was hardly detectable by 24 h p.i. The first cleavage product, 28S-CL1, appeared as a faint band as early as 4 h p.i., and it increased substantially between 5 and 6 h p.i. 28S-CL1 remained the major rRNA species from 9 to 12 h p.i. Probe 1 also hybridized with four additional RNA bands, 2.8-kb-long 28S-CL2, 2.1-kb-long 28S-CL3, 2.0-kb-long 28S-CL4, and 1.0-kb-long 28S-CL5, from MHV-A59-infected cells; none of these smaller 28S rRNA cleavage products comigrated with MHV mRNAs, and all were clearly visible by 12 h p.i. 28S-CL3 and 28S-CL4 were visible at 9 h p.i., and they accumulated transiently until 16 h p.i. 28S-CL2 and 28S-CL5 appeared around 12 h p.i. Late in infection, 28S-CL4 and 28S-CL5 were the major cleavage products. By 24 h p.i., the cleavage products had virtually disappeared and very little intact 28S rRNA was present.
In contrast to the extensive cleavage of 28S rRNA in MHV-A59-infected cells, Northern blot analysis of 18S rRNA, using two 18S rRNA-specific oligonucleotide probes, showed that 18S rRNA did not undergo cleavage in MHV-infected DBT cells even late in infection (Fig. 1D). The amount of 18S rRNA in MHV-A59-infected cells was similar to that in uninfected cells until 12 h p.i., while the amount of intact 18S rRNA in MHV-A59-infected cells decreased slightly at 16 h p.i. By 24 h p.i. there was a decrease in the total amount of intact 18S rRNA; the mechanism of reduction in the amount of 18S rRNA late in infection is not known. The amount of intact 18S rRNA remained unchanged until 12 h p.i., and Northern blot analysis detected no specific cleavage products; hence, we concluded that 28S rRNA, but not 18S rRNA, underwent specific cleavage in MHV-A59-infected cells.
MHV replication was required for 28S rRNA cleavage.
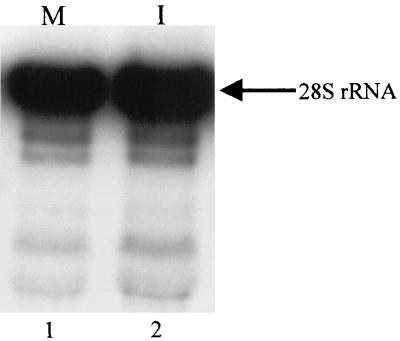
To test whether binding of MHV to MHV receptors, or some unidentified substances other than MHV present in the inoculum, induced 28S rRNA cleavage, the inoculum used in the above experiments was exposed to UV light (wavelength, 253 nm) for 12 min prior to addition to DBT cells. MHV infectivity of UV-irradiated samples was less than 1 PFU/0.2 ml. After incubation for 1 h at 37°C, the inoculum was removed and the cells were incubated for up to 8 h. No 28S rRNA cleavage was detected in cells that underwent this treatment (Fig. 2), demonstrating that binding of MHV to MHV receptors alone or to unidentified substances which may have been present in the inoculum did not induce 28S rRNA cleavage. Induction of 28S rRNA cleavage required MHV replication.
FIG. 2.
Northern blot analysis of 28S rRNA after inoculation of UV-irradiated MHV-A59. DBT cells were mock infected (M) or inoculated (I) with the UV-irradiated MHV-A59 sample. Cytoplasmic RNA was extracted at 8 h p.i., and Northern blot analysis was performed with 5′-end-labeled probe 1. The arrow indicates intact 28S rRNA.
Relationship between 28S rRNA cleavage and other MHV-induced changes in infected cells.
We compared the kinetics of 28S rRNA cleavage with other cellular changes that occurred in MHV-infected cells. MHV-A59 infection in DBT cells induces cell fusion mediated by S protein (7). MHV-A59-induced cell fusion appeared 5 h p.i. Fused cells became 60% and nearly 100% of the total cell population by 6 and 8 h p.i., respectively, but they did not start floating until 18 to 20 h p.i. (data not shown). As shown in Fig. 1C, 28S rRNA cleavage started earlier than the onset of MHV-A59-induced cell fusion.
MHV RNA synthesis peaks at 6 to 7 h p.i. (39). Amounts of MHV RNAs are roughly constant from 8 to 10 h p.i. and then decline at 11 h p.i. (39). A substantial increase in 28S-CL1 in MHV-infected cells preceded the peak of MHV RNA synthesis, and 28S rRNA cleavage continued beyond 12 h p.i. (Fig. 1C).
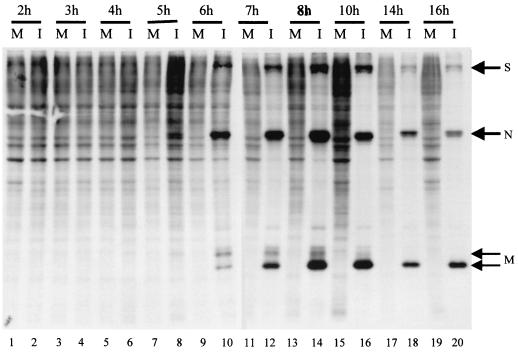
28S rRNA is an integral component of the large subunit of the ribosome; cleavage of 28S rRNA may affect ribosome structure or function and subsequently protein synthesis. Hence we examined the relationship between 28S rRNA cleavage and protein synthesis in MHV-infected cells. MHV-A59-infected DBT cells were labeled with Tran35S-label for 30 min at different times p.i., and cell extracts were analyzed by SDS-PAGE (Fig. 3). Synthesis of S, N, and M proteins was detectable at 5 h p.i.; these MHV structural proteins became major proteins by 6 h p.i. Consistent with previous studies (12, 42, 49, 50), host protein synthesis was inhibited in MHV-infected cells (Fig. 3); inhibition of host protein synthesis was seen around 7 h p.i. and proceeded further. As shown in Fig. 1C, 28S-CL1 appeared as a major cleavage product starting 5 h p.i. These data showed that there was a slight lag period between host protein synthesis inhibition and 28S-CL1 production. Thereafter, both 28S rRNA cleavage and host protein synthesis inhibition continued as infection proceeded.
FIG. 3.
Host protein synthesis inhibition in MHV-A59-infected DBT cells. DBT cells were mock infected (M) or infected with MHV-A59 (I) at an MOI of 10. Culture medium was replaced with methionine-cysteine-free medium 30 min prior to each indicated time point. After 30 min of incubation, Tran35S-label was added to the culture medium at a final concentration of 75 μCi/ml. Intracellular proteins were extracted after 30 min of incubation and analyzed by SDS-PAGE (12% gel). Positions of MHV-A59 S, N, and M structural proteins are indicated by arrows.
Cleavage of 28S rRNA after infection with different MHV strains in different cell lines.
We examined whether 28S rRNA cleavage was confined to a particular cell type or MHV strain. MHV-A59 infection of 17Cl-1 cells also produced all 28S rRNA cleavage products; the kinetics of appearance of cleavage products and reduction in the amount of mature 28S rRNA in MHV-A59-infected 17CL-1 cells were similar to results for MHV-infected DBT cells (data not shown). MHV-JHM-infected DBT cells also induced the same 28S rRNA cleavage products as found in MHV-A59-infected DBT cells, but 28S rRNA cleavage was less prominent in MHV-JHM-infected cells; the amount of intact 28S rRNA remaining in MHV-A59-infected cells was much less than that in MHV-JHM-infected cells at 24 h p.i. (data not shown). MHV-JHM-induced 28S rRNA cleavage was slower than MHV-A59-induced 28S rRNA cleavage, yet 28S-CL1 was clearly detectable at 9 h p.i. in MHV-JHM-infected cells. Cleaved 28S rRNA products were also less abundant than in MHV-A59-infected cells. We detected no major differences in the patterns of 28S rRNA cleavage in MHV-JHM-infected 17CL-1 cells and MHV-JHM-infected DBT cells. Production of infectious MHV from MHV-JHM-infected cells is about 10 times lower than that from MHV-A59-infected cells, and the amount of MHV-specific RNAs in MHV-JHM-infected cells is also lower than that in MHV-A59-infected cells (data not shown). A lower level of MHV-JHM replication efficiency in infected cells may be related to less efficient 28S rRNA cleavage.
We used a nonfusogenic MHV strain, MHV-2, to test whether MHV-induced cell fusion was required for 28S rRNA cleavage. The same 28S rRNA cleavage products accumulated in MHV-2-infected DBT cells (data not shown). Kinetics of 28S rRNA cleavage in MHV-2-infected DBT cells was similar to that of MHV-JHM-infected DBT cells. These data demonstrated that MHV-induced 28S rRNA cleavage was not restricted to any particular MHV strain or MHV-susceptible cell line.
Evidence for cleavage of mature cytoplasmic 28S rRNA in MHV-infected cells.
Mature cytoplasmic 28S rRNA is generated from a precursor 45S rRNA; 45S rRNA undergoes specific cleavages to produce 28S rRNA, 18S rRNA, and other small rRNAs in the nucleolus (37). After processing, 28S rRNA associates with ribosomal proteins to form the large subunit, which is then transported to the cytoplasm (37). Accumulation of 28S rRNA cleavage products in MHV-infected cells may be the result of cleavage of mature cytoplasmic 28S rRNA or of aberrant rRNA processing, which takes place in the nucleolus. To examine the possibility that mature cytoplasmic 28S rRNA is cleaved in MHV-infected cells, we first determined the time taken by the precursor rRNA to be transported from the nucleolus to the cytoplasm. DBT cells at 50% confluency were incubated in the presence of [3H]uridine (60 μCi/ml). After 16 h of incubation, the culture medium was replaced with growth medium lacking [3H]uridine. The labeled rRNAs were chased in medium without [3H]uridine. At various times during this chase period, intracellular RNA was extracted and analyzed on a 1% agarose-formaldehyde gel. Labeled mature 28S and 18S rRNAs were detected in the cytoplasm after a 5.5- to 12-h chase (data not shown). To test whether mature cytoplasmic 28S rRNA was cleaved in MHV-infected cells, 50% confluent DBT cells were incubated in the presence of [3H]uridine (100 μCi/ml). After 16 h of incubation, the culture medium was replaced with growth medium lacking [3H]uridine and chased for another 13 h; processing and transportation of 28S rRNA were completed during this chase period. Cells were then mock infected or infected with MHV-A59. After 1 h of virus adsorption, the inoculum was removed and cells were incubated in a medium containing actinomycin D (5 μg/ml) to prevent transcription of host RNAs, including rRNAs. Cytoplasmic RNA was extracted at 8 h p.i. and analyzed by electrophoresis on a 1% denaturing agarose-formaldehyde gel (Fig. 4). Both 28S and 18S rRNAs were seen in mock-infected cells, while 28S-CL1 and a reduced level of 28S rRNA were detected in MHV-infected cells. Since the majority of intact rRNAs were radiolabeled and transported to the cytoplasm prior to MHV infection, and only cytoplasmic RNAs were extracted in this experiment, rRNAs detected in this study should represent cytoplasmic rRNAs that were made prior to MHV infection. Reduction of intact 28S rRNA and the presence of 28S-CL1 in MHV-infected cells demonstrated that MHV infection induced cleavage of mature cytoplasmic 28S rRNA that was made prior to MHV infection. Thus, cytoplasmic 28S rRNA, which is part of the large subunit, was cleaved during MHV infection.
FIG. 4.
Evidence for the cleavage of mature cytoplasmic 28S rRNA in MHV-infected DBT cells. DBT cells at a low confluency were labeled with [3H]uridine for 16 h and then chased, in a medium lacking isotope, for 13 h. Cells were then mock infected (M) or infected with MHV-A59 (I) at an MOI of 10. After adsorption for 1 h, cells were incubated in the presence of actinomycin D (5 μg/ml), and cytoplasmic RNA was extracted at 8 h p.i. Cytoplasmic RNAs were electrophoresed on a 1% denaturing gel. The gel was washed and enhanced prior to autoradiography. The gel was exposed at −80°C for 60 days. The arrows indicate intact 28S rRNA, 28S-CL1, and 18S rRNA.
Independence of MHV-induced 28S rRNA cleavage from 2-5A system-mediated rRNA degradation.
In certain combination of viruses and cells, pretreatment of cells with IFN and subsequent viral infection result in rRNA degradation (45). This rRNA degradation is mediated by the 2-5A system (45, 46), in which IFN upregulates the enzyme 2-5A synthetase, which synthesizes 2-5A molecules. These 2-5A moieties are highly unstable and are rapidly degraded by phosphatases. In the presence of viral double-stranded RNAs, 2-5A binds and activates the endoribonuclease, RNase L. RNase L activation is localized and serves to cleave viral mRNAs, thus inhibiting viral replication and limiting viral spread. At high 2-5A concentrations, RNase L activation leads to extensive degradation of rRNAs (31).
We examined whether the 28S rRNA cleavage in MHV-infected cells was due to activation of the 2-5A system. RNase L-mediated rRNA degradation usually requires treatment of cells with IFN prior to virus infection (45). MHV-induced 28S rRNA cleavage occurred in the absence of IFN pretreatment; therefore, it was less likely that the conventional 2-5A system was mediating MHV-induced 28S rRNA cleavage. However, the MHV sample used for virus inoculation could contain IFN secreted from infected cells. Subsequent infection using that inoculum would then expose the cells to IFN during virus adsorption. Such a brief exposure of cells to IFN and the subsequent replication of MHV may be sufficient to activate the 2-5A system in the infected cell. We tested this possibility by examining the production of IFN from MHV-infected DBT cells, as all MHV stocks used for virus inoculation were grown in DBT cells. A classical biological assay for IFN, which uses the susceptibility of VSV replication in IFN-pretreated L929 cells (28), was used to detect biologically active IFN in MHV-infected supernatants. VSV replication in L929 cells was very sensitive to pretreatment with a mixture of IFN-α and IFN-β NIH standards, as no VSV plaques formed at moderate dilutions of these NIH standards. In contrast, supernatant from MHV-infected and mock-infected cells failed to protect L929 cells from VSV replication (data not shown), suggesting that MHV-infected culture fluid contained no or an undetectable level of biologically active IFN.
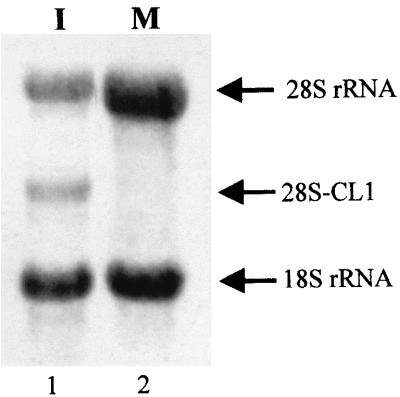
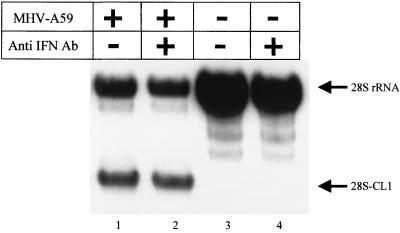
Next we examined the possibility that the autocrine pathway of IFN action (40) could mediate MHV-induced 28S rRNA cleavage. If very low levels of IFN mediate MHV-induced 28S rRNA cleavage through the autocrine pathway, then neutralizing the putative IFN which is secreted into MHV-infected culture fluid should block this pathway. After adsorption of MHV-A59, DBT cells were incubated in the presence of an anti-mouse IFN-α and -β rabbit antibody mixture (20% IFN-α–80% IFN-β; 30 U/ml; NIAID catalog no. G024-501-568). This amount of anti-IFN antibodies can neutralize 3,000 U of IFN. This concentration of antibodies was more than enough to neutralize any IFN that might be produced from MHV-infected cells, because the IFN bioassay showed that less than 3 U of IFN per ml may be secreted by MHV-infected cells. Northern blot analysis of intracellular RNAs that were extracted at 8 h p.i. showed that incubation of MHV-infected cells with anti-IFN antibodies did not block 28S rRNA cleavage (Fig. 5). Taken together, these data demonstrated that IFN was not involved in 28S rRNA cleavage in MHV-infected cells.
FIG. 5.
Effect of anti-mouse IFN antibodies on MHV-induced 28S rRNA cleavage. DBT cells were mock infected (lanes 3 and 4) or infected with MHV-A59 (lanes 1 and 2) at an MOI of 10. After virus adsorption for 1 h, cells were incubated in the absence (lanes 1 and 3) or presence of 30 U of rabbit anti-mouse IFN antibodies (Ab) per ml (lanes 2 and 4). Cytoplasmic RNA was extracted at 8 h p.i., and 28S rRNA cleavage was examined by Northern blot analysis using 5′-end-labeled probe 1.
We tested yet another possibility, that MHV infection directly activates RNase L in the absence of IFN. We used RNase L+/+ and RNase L−/− MEF cells to examine this possibility. If RNase L plays a vital role in 28S rRNA cleavage in MHV-infected cells, 28S rRNA cleavage should not occur in MHV-infected RNase L−/− cells. We conducted a [32P]2-5A cross-linking assay (35) to confirm the absence of RNase L in RNase L−/− cells; our data unambiguously showed the lack of RNase L in RNase L−/− cells and the presence of RNase L in RNase L+/+ cells (data not shown).
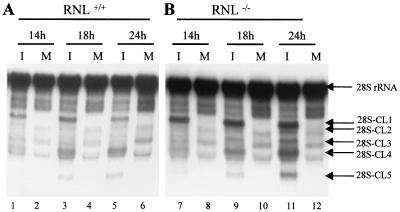
One-step MHV-A59 growth curve analysis showed that MHV replicated with similar kinetics, with maximum virus titer at about 24 h p.i. in both RNase L+/+ and RNase L−/− cell lines. The maximum MHV titer in both cell lines was about 5 to 10 times lower than that in DBT cells. Immunofluorescence studies using an anti-N protein monoclonal antibody showed that in both cell lines approximately 15% cells supported MHV replication after MHV inoculation at an MOI of 10 (data not shown); we do not know why only 15% of cells supported MHV replication. Both cell lines showed no apparent cytopathic effects, including cell fusion, after MHV-A59 infection. To determine whether MHV-induced 28S rRNA cleavage occurred in the absence of RNase L expression, RNase L+/+ and RNase L−/− cells were infected with MHV-A59 at an MOI of 10. Intracellular RNA was extracted at various times p.i., and 28S rRNA cleavage was examined by Northern blot analysis. Cleavage of 28S rRNA occurred in both cell lines (Fig. 6). The sizes of the 28S rRNA cleavage products in both cell lines were the same as those found in MHV-infected DBT cells. The amount of intact 28S rRNA did not decrease drastically in either type of MEF cells after MHV infection. This was not surprising, as only 15% of each cell type was infected with MHV; 28S rRNA cleavage did not occur in uninfected cells.
FIG. 6.
MHV-induced 28S rRNA cleavage in RNase L+/+ (A) and RNase L−/− (B) MEF cells. RNase L+/+ (RNL+/+) and RNase L−/− (RNL−/−) MEF cells were mock infected (M) or infected with MHV-A59 (I) at an MOI of 10. Cytoplasmic RNA was extracted at the indicated times and electrophoresed on a 1% denaturing gel. Cleavage of 28S rRNA was examined by Northern blot analysis using 5′-end-labeled probe 1.
Blocking apoptosis did not affect MHV-induced 28S rRNA cleavage.
There have been some reports of rRNA degradation occurring in apoptotic cells. Treatment of certain tumor cells with chemical inducers of apoptosis, e.g., actinomycin D and cyclic AMP, causes 28S rRNA degradation coincident with DNA fragmentation, which is a characteristic change found in apoptotic cells (16, 17). PRRSV infection of susceptible cells causes apoptosis, characterized by DNA fragmentation, concomitant with cleavage of both 18S and 28S rRNAs (48).
Apoptosis is induced in MHV-infected 17Cl-1 cells but not in MHV-infected DBT cells (1). Nevertheless 28S rRNA cleavage occurred in both DBT cells (Fig. 1) and 17Cl-1 cells (data not shown). Furthermore, a DNA ladder, representing apoptotic DNA fragmentation, is detected at about 24 h p.i. in MHV-infected 17Cl-1 cells (1), while significant 28S rRNA cleavage occurred much earlier, around 5 h p.i. (data not shown). These differences signify that 28S rRNA cleavage detected in MHV-infected cells and rRNA degradation associated with apoptosis are not identical, yet MHV-induced 28S rRNA cleavage and apoptosis may be related. A possible relationship between MHV-induced 28S rRNA cleavage and apoptosis was examined.
First we examined whether induction of apoptosis in DBT cells resulted in 28S rRNA cleavage, i.e., whether 28S rRNA cleavage is a typical change that occurs in apoptotic DBT cells. DBT cells at 50% confluency were incubated in medium containing 2% serum for 18 h. After addition of 50 μM etoposide, cells were incubated for 33 h, and then internucleosomal DNA (13) and intracellular RNA were extracted. Etoposide treatment of DBT cells resulted in DNA ladder formation, demonstrating that DBT cells can die by apoptosis. However, no specific cleavage of either 18S or 28S rRNA occurred (data not shown), indicating that 28S rRNA cleavage was not a common apoptotic change in DBT cells that underwent apoptosis.
Although MHV-infected DBT cells showed no apoptotic signs, certain steps of the apoptotic process may occur in MHV-infected DBT cells but apoptosis may be blocked. Caspase-3 plays a central role in caspase-dependent apoptosis (5, 34). We wondered whether caspase-3 was activated in MHV-infected DBT cells; caspase-3 may be activated early in MHV-infected DBT cells, and activated caspase-3 may trigger 28S rRNA cleavage. To examine this possibility, DBT cells were mock infected or infected with MHV-A59 at an MOI of 10, and cell lysates were prepared at 4.5 and 8.5 h p.i. Etoposide-treated DBT cells were used as a positive control. Caspase-3 activity assay demonstrated a fourfold increase in caspase-3 activation in etoposide-treated DBT cells, with no caspase-3 activation in MHV-infected DBT cells at 4.5 and 8.5 h p.i. These data suggested that 28S rRNA cleavage was upstream of the activation of caspase-3 or not regulated by caspase-3.
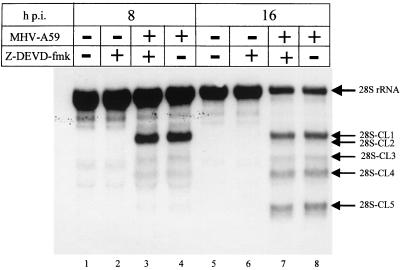
MHV-infected 17Cl-1 cells die by apoptosis (1). Treatment of MHV-infected 17Cl-1 cells with the irreversible, cell-permeable caspase-3 inhibitor Z-DEVD-fmk inhibits MHV-induced apoptosis without suppressing MHV growth (1). If MHV-induced 28S rRNA cleavage is an event downstream of caspase-3 activation, then Z-DEVD-fmk treatment of MHV-infected 17Cl-1 cells would inhibit MHV-induced 28S rRNA cleavage. For 2 h prior to MHV-A59 infection and after MHV-A59 infection, 17Cl-1 cells were incubated with 80 μM Z-DEVD-fmk, because apoptosis is blocked in MHV-infected 17CL-1 cells under this experimental condition (1). Control samples were incubated with dimethyl sulfoxide, which was used to dissolve Z-DEVD-fmk. As expected, a DNA ladder assay at 24 h p.i. showed that Z-DEVD-fmk treatment blocked apoptosis, while MHV-infected cells that were incubated with dimethyl sulfoxide showed clear DNA fragmentation (data not shown). Northern blot analysis of intracellular RNAs that were extracted at 8 and 16 h p.i. showed that Z-DEVD-fmk treatment had no effect on MHV-induced 28S rRNA cleavage (Fig. 7). Inhibition of caspase-3 activity in MHV-infected 17Cl-1 cells did not block or delay 28S rRNA cleavage, indicating that MHV-induced 28S rRNA cleavage was not an event downstream of caspase-3 activation.
FIG. 7.
Effect of the caspase inhibitor Z-DEVD-fmk on MHV-induced 28S rRNA cleavage. Mouse 17Cl-1 cells were incubated in serum-free DMEM (lanes 1, 4, 5, and 8) or 80 μM Z-DEVD-fmk (lanes 2, 3, 6 and 7) for 2 h prior to MHV infection. Cells were then mock infected (lanes 1, 2, 5, and 6) or infected with MHV-A59 (lanes 3, 4, 7, and 8) at an MOI of 10. After adsorption, cells were incubated in the presence (lanes 2, 3, 6, and 7) or absence (lanes 1, 4, 5, and 8) of 80 μM Z-DEVD-fmk. Cytoplasmic RNA was extracted at the indicated times. Cleavage of 28S rRNA was examined by Northern blot analysis using 5′-end-labeled probe 1.
In summary, triggering apoptosis by etoposide treatment in DBT cells did not cause any 28S rRNA cleavage. Conversely, blocking apoptosis by Z-DEVD-fmk treatment in MHV-infected 17Cl-1 cells did not block MHV-induced 28S rRNA cleavage. Also, no activation of caspase-3 was detected in MHV-infected DBT cells. These data strongly indicated either that 28S rRNA degradation was occurring upstream of caspase-3 activation or that apoptosis induction and 28S rRNA cleavage occurred via two independent pathways.
DISCUSSION
Comparison of MHV-induced 28S rRNA cleavage and other 28S rRNA cleavage mechanisms.
In this study, we report a novel MHV-induced 28S rRNA cleavage. The MHV-induced rRNA cleavage occurred only in 28S rRNA, not in 18S rRNA; generation of 28S rRNA cleavage products of specific sizes argues against a random RNase activation, which would result in smeared bands of degraded rRNAs, with no preference for 18S rRNA or 28S rRNA. The cleavage products appeared around 4 h p.i., with further cleavage products appearing with increasing times of infection. Mature cytoplasmic 28S rRNA, a part of the 60S large ribosomal subunit, underwent cleavage. UV-inactivated MHV failed to induce 28S rRNA cleavage, demonstrating that the binding of virus to the cell surface receptor or unidentified substances which may be present in the inoculum did not induce 28S rRNA cleavage. Specific cleavage of 28S rRNA required ongoing MHV replication. Currently, however, we do not know which step of the viral life cycle or specific viral factor(s) causes this 28S rRNA cleavage. MHV infection to all susceptible cell lines, using several different MHV strains, induced 28S rRNA cleavage; MHV-induced 28S rRNA cleavage was independent of virus-induced cytopathic effect. Kyuwa et al. reported a 50% decrease in intact 28S rRNA at 12 h p.i. in MHV-JHM-infected J774.1 BALB/c monocytic cells (25). Their data are consistent with our observation of reduced amount of intact 28S rRNA with increasing times of MHV infection.
One of the known mechanisms of rRNA degradation is through the activation of RNase L, a cellular endoribonuclease, activated by the 2-5A system. We showed that MHV-induced 28S rRNA cleavage was independent of the 2-5A system and RNase L activation. Namely, IFN was undetectable in the inoculum used for MHV infection. Also, neutralization of putative IFN in the culture fluid from MHV-infected cells by anti-IFN antibodies did not affect MHV-induced 28S rRNA cleavage. Furthermore, MHV infection of RNase L−/− MEF cells induced a pattern of 28S rRNA cleavage similar to that induced in RNase L+/+ MEF cells.
Apoptosis-associated rRNA cleavage is another known mechanism of rRNA degradation; DNA fragmentation and RNA fragmentation are usually temporally linked in this type of rRNA cleavage (16, 17). The only report of rRNA degradation during viral infection is during PRRSV infection or expression of the PRRSV p25 gene product (48). In both cases, rRNA degradation is coincident with DNA fragmentation and other morphological features of apoptosis. MHV-induced 28S rRNA cleavage differed from apoptosis-related rRNA degradation and PRRSV-induced rRNA degradation. MHV-induced 28S rRNA cleavage occurred in DBT cells that do not undergo apoptosis (1), and no activated caspase-3 was detected in MHV-infected DBT cells. MHV-infected 17Cl-1 cells undergo caspase-dependent apoptosis, where DNA fragmentation is detected at about 24 h p.i. (1), whereas MHV-induced 28S rRNA cleavage was detected much earlier. Treatment of MHV-infected 17Cl-1 cells with the caspase-3 inhibitor Z-DEVD-fmk did not affect 28S rRNA cleavage, whereas this treatment did inhibit DNA fragmentation; hence, inhibition of apoptosis did not block 28S rRNA cleavage. These data argue against the involvement of an activated caspase-3 and the triggering of apoptosis in inducing 28S rRNA cleavage. MHV-induced 28S rRNA cleavage was probably an event independent of apoptosis or, if related, was upstream of the activation of caspase-3 and a very early event in MHV infection.
Wreschner et al. (54) have reported the presence in rabbit reticulocyte lysates of a membrane-bound RNase M which is inactivated during maturation of reticulocytes to erythrocytes. RNase M cleaves 28S rRNA. No such RNase has been found in DBT or 17Cl-1 cells, and since these are immortalized, transformed cell lines, the presence of a developmentally regulated RNase is questionable. Specific 28S rRNA cleavage and DNA fragmentation are seen in rat brains during traumatic brain injury (10), and the authors concluded that the rRNA fragmentation in their system may be related to either necrosis or apoptosis. Cleavage of 28S rRNA by RNase M or during rat brain injury generated 28S rRNA cleavage products that differed in size from the 28S rRNA cleavage products found in MHV-infected cells. Thus, MHV-induced 28S rRNA specific cleavage appeared to be different from other reported rRNA degradations.
The mechanism of MHV-induced 28S rRNA cleavage.
Although RNase L was not responsible for MHV-induced 28S rRNA cleavage, it is possible that such a cleavage was the result of activation of another RNase of cellular or viral origin. Others have reported the degradation of few cellular mRNAs during MHV infection (12, 25, 49). Therefore, it is conceivable that the RNase responsible for cleaving 28S rRNA may also degrade these host mRNAs, as the reduction of cellular mRNAs appears to be, at least in part, responsible for the host protein synthesis inhibition in MHV-infected cells (12). MHV-encoded RNase activity has not been demonstrated. Another possibility is that the structure of 60S ribosome may be altered by binding of unidentified MHV factors or host factors which are induced by infection in MHV-infected cells. This putative structural alteration may allow a cellular or viral RNase to access specific regions of 28S rRNA, resulting in specific cleavage of 28S rRNA.
MHV-induced 28S rRNA cleavage and protein translation.
28S rRNA is an integral component of 60S ribosome, whose major function is protein translation. Hence, MHV-induced 28S rRNA cleavage may affect protein synthesis. Indeed, consistent with previous studies (12, 42, 49, 50), host protein translation was severely inhibited but not completely shut off in MHV-infected cells (Fig. 3). It is tempting to speculate that ribosomes containing the cleaved 28S rRNA may not be able to form polysomes or may be functionally inactive in protein synthesis. In that case, MHV-specific protein synthesis may take place on polysomes containing intact 28S rRNAs. This possibility is consistent with the finding that the amount of 80S monosome which is not involved in protein translation increased in MHV-infected cells (12), and both MHV and host proteins were poorly synthesized late in MHV infection, when only a minute amount of intact 28S rRNA was detected (Fig. 1C). Alternatively, most host protein synthesis does not occur on ribosomes containing cleaved 28S rRNA, while MHV-specific proteins are preferentially synthesized on ribosomes containing the cleaved 28S rRNA species. 28S-CL1, the first cleavage product, appeared early in infection and remained a major stable rRNA species until 12 h p.i. (Fig. 1C). It is conceivable that ribosomes containing 28S-CL1 are functional but structurally altered to better translate the increasing amounts of MHV-specific mRNAs, which begin to accumulate from 5 h p.i. Tahara et al. reported that chimeric mRNAs containing the MHV leader sequence upstream of the human α-globin region translate more efficiently than the authentic human α-globin mRNA in extracts from MHV-infected cells, whereas this translational enhancement is not seen in extracts from uninfected cells (50). Recent studies on translation analysis of bovine coronavirus (BCV) mRNAs also showed that the presence of BCV leader sequence in chimeric mRNAs increases their translation activity in BCV-infected cells (41). Since MHV N protein binds to the leader sequence (51), Tahara et al. speculated that binding of N protein to the 5′ end of the MHV leader sequence may augment translation efficiency in MHV-infected cells (51). Presence of MHV leader sequence at the 5′ ends of MHV mRNAs and N protein binding may allow MHV protein synthesis in ribosomes containing cleaved 28S rRNAs. There is yet another possibility, that 28S rRNA cleavage does not affect the translational activity of ribosomes; ribosomes containing cleaved 28S rRNAs may be biologically active for translation of both host and MHV proteins. In that case, the inhibition of host protein synthesis in MHV-infected cells is mediated by some other, unidentified mechanism(s).
Biological significance of MHV-induced 28S rRNA cleavage.
What is the biological significance of a specific 28S rRNA cleavage occurring early in MHV infection? It has been proposed that 28S rRNAs may serve as cytoplasmic “biosensors” regulating cellular processes (18, 19). In apoptosis-related 28S rRNA cleavage, the cleavages occur at specific sites within rRNA D domains (17). Although the biological function of D domains of 28S rRNA has not been established, Houge and Doskeland speculated that various proapoptotic signal transduction pathways, e.g., those involving phosphorylation and/or proteolysis, can convert the D domain from a passive to an active state (15). If a sufficient number of ribosomes are in the active state, the threshold for apoptosis is exceeded. Secondary to the postulated D-domain modulations, apoptotic rRNA cleavage may inactivate the D domains or liberate the cleaved 28S rRNA fragments, which may have additional biological effects (15). Iordanov et al. described other cellular responses to changes in the status of 28S rRNA, reporting that damage at a specific loop of the 28S rRNA, or binding of peptidyltransferase inhibitors to the adjacent peptidyltransferase center of the 28S rRNA, induced a ribotoxic stress response (19). This response involves the activation of the stress-activated protein kinase/c-Jun NH2-terminal kinase, the p38 mitogen-activated protein kinase, and the transcriptional induction of immediate-early genes such as c-fos and c-jun (18, 19). The signal transduction cascade promotes either cell recovery and survival after cellular damage or apoptotic cell death (reference 19 and references therein). MHV-induced 28S rRNA cleavage may have a similar biological consequence as ribotoxic stress response; MHV-induced 28S rRNA cleavage may activate a signal transduction pathway(s), which may result in the alteration of the cellular environment. The altered environment may suppress MHV replication as a cellular countermeasure against MHV infection. Alternatively, MHV may have evolved to trigger a signal transduction pathway to enhance its replication; such an altered cellular environment, triggered by the MHV-induced 28S rRNA cleavage-mediated transduction pathway, may offer a better environment for efficient MHV replication.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI29984 (to S.M.) and CA44059 (to R.H.S.) from the National Institutes of Health.
We thank Chun-Jen Chen and Gunnar Houge for valuable information and suggestions for etoposide-induced apoptosis in DBT cells and for the Northern blot analyses of 28S rRNA, respectively. We also thank Samuel Baron and Joyce Poast for the anti-IFN antibodies and invaluable help with the IFN assays.
REFERENCES
- 1.An S, Chen C-J, Yu X, Leibowitz J L, Makino S. Induction of apoptosis in murine coronavirus-infected cultured cells and demonstration of E protein as an apoptosis inducer. J Virol. 1999;73:7853–7859. doi: 10.1128/jvi.73.9.7853-7859.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey O, Pappenheimer A M, Cheever F S, Daniels J B. A murine virus (JHM) causing disseminated encephalomyelitis with extensive destruction of myelin. II. Pathology. J Exp Med. 1949;90:195–205. doi: 10.1084/jem.90.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belyavskyi M, Belyavskaya E, Levy G A, Leibowitz J L. Coronavirus MHV-3-induced apoptosis in macrophages. Virology. 1998;250:41–49. doi: 10.1006/viro.1998.9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos E C W, Luytjes W, van der Meulen H, Koerten H K, Spaan W J M. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen G M. Caspases: the executioners of apoptosis. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Compton S R, Barthold S W, Smith A L. The cellular and molecular pathogenesis of coronaviruses. Lab Anim Sci. 1993;43:15–28. . (Erratum, 43:203.) [PubMed] [Google Scholar]
- 7.Dveksler G S, Pensiero M N, Cardellichio C B, Williams R K, Jiang G-S, Holmes K V, Dieffenbach C W. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J Virol. 1991;65:6881–6891. doi: 10.1128/jvi.65.12.6881-6891.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eleouet J-F, Chilmonczyk S, Besnardeau L, Laude H. Transmissible gastroenteritis coronavirus induces programmed cell death in infected cells through a caspase-dependent pathway. J Virol. 1998;72:4918–4924. doi: 10.1128/jvi.72.6.4918-4924.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Etchison D, Milburn S C, Edery I, Sonenberg N, Hershey J W B. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eukaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- 10.Fan L, Yakovlev A G, Faden A L. Site-specific cleavage of 28S rRNA as a marker of traumatic brain injury. J Neurotrauma. 1999;16:357–364. doi: 10.1089/neu.1999.16.357. [DOI] [PubMed] [Google Scholar]
- 11.Her L S, Lund E, Dahlberg J E. Inhibition of Ran guanosine triphosphate-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science. 1997;276:1845–1848. doi: 10.1126/science.276.5320.1845. [DOI] [PubMed] [Google Scholar]
- 12.Hilton A, Mizzen L, Macintyre G, Cheley S, Anderson R. Translational control in murine hepatitis virus infection. J Gen Virol. 1986;67:923–932. doi: 10.1099/0022-1317-67-5-923. [DOI] [PubMed] [Google Scholar]
- 13.Hinshaw V S, Olsen C W, Dybdahl-Sissoko N, Evans D. Apoptosis: a mechanism of cell killing by influenza A and B viruses. J Virol. 1994;68:3667–3673. doi: 10.1128/jvi.68.6.3667-3673.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirano N, Fujiwara K, Hino S, Matsumoto M. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch Gesamte Virusforsch. 1974;44:298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- 15.Houge G, Doskeland S O. Divergence towards a dead end? Cleavage of the divergent domains of ribosomal RNA in apoptosis. Experientia. 1996;52:963–967. doi: 10.1007/BF01920105. [DOI] [PubMed] [Google Scholar]
- 16.Houge G, Doskeland S O, Boe R, Lanotte M. Selective cleavage of 28S rRNA variable regions V3 and V13 in myeloid leukemia cell apoptosis. FEBS Lett. 1993;315:16–20. doi: 10.1016/0014-5793(93)81123-h. [DOI] [PubMed] [Google Scholar]
- 17.Houge G, Robaye B, Eikhom T S, Golstein J, Mellgren G, Gjertsen B T, Lanotte M, Doskeland S O. Fine mapping of 28S rRNA sites specifically cleaved in cells undergoing apoptosis. Mol Cell Biol. 1995;15:2051–2062. doi: 10.1128/mcb.15.4.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iordanov M S, Paranjape J M, Zhou A, Wong J, Williams B R, Meurs E F, Silverman R H, Magun B E. Activation of p38 mitogen-activated protein kinase and c-Jun NH2-terminal kinase by double-stranded RNA and encephalomyocarditis virus: involvement of RNase L, protein kinase R, and alternative pathways. Mol Cell Biol. 2000;20:617–627. doi: 10.1128/mcb.20.2.617-627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iordanov M S, Pribnow D, Magun J L, Dinh T-H, Pearson J A, Chen S L-Y, Magun B E. Ribotoxic stress response: activation of the stress-activated protein kinase JNK1 by inhibitors of the peptidyl transferase reaction and by the sequence-specific RNA damage to the α-sarcin/ricin loop in the 28S rRNA. Mol Cell Biol. 1997;17:3373–3381. doi: 10.1128/mcb.17.6.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katze M G, DeCorato D, Krug R. Cellular mRNA translation is blocked at both initiation and elongation after infection by influenza virus and adenovirus. J Virol. 1986;60:1027–1039. doi: 10.1128/jvi.60.3.1027-1039.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katze M G, Krug R. Metabolism and expression of RNA polymerase II transcripts in influenza virus-infected cells. Mol Cell Biol. 1984;4:2198–2206. doi: 10.1128/mcb.4.10.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keck J G, Soe L H, Makino S, Stohlman S, Lai M M C. RNA recombination of murine coronaviruses: recombination between fusion-positive mouse hepatitis virus A59 and fusion-negative mouse hepatitis virus 2. J Virol. 1988;62:1989–1998. doi: 10.1128/jvi.62.6.1989-1998.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K-H, Narayanan K, Makino S. Assembled coronavirus from complementation of two defective interfering RNAs. J Virol. 1997;71:3922–3931. doi: 10.1128/jvi.71.5.3922-3931.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim K H, Makino S. Two murine coronavirus genes suffice for viral RNA synthesis. J Virol. 1995;69:2313–2321. doi: 10.1128/jvi.69.4.2313-2321.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyuwa S, Cohen M, Nelson G W, Tahara S M, Stohlman S A. Modulation of cellular macromolecular synthesis by coronavirus: implications for pathogenesis. J Virol. 1994;68:6815–6819. doi: 10.1128/jvi.68.10.6815-6819.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai M M C, Brayton P R, Armen R C, Patton C D, Pugh C, Stohlman S A. Mouse hepatitis virus A59: mRNA structure and genetic localization of the sequence divergence from hepatotropic strain MHV-3. J Virol. 1981;39:823–834. doi: 10.1128/jvi.39.3.823-834.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai M M C, Stohlman S A. RNA of mouse hepatitis virus. J Virol. 1978;26:236–242. doi: 10.1128/jvi.26.2.236-242.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langford M P, Weigent D A, Stanton G J, Baron S. Virus plaque reduction assay for interferon: microplaque reduction assays. In: Petska S, editor. Methods in enzymology. New York, N.Y: Academic Press; 1981. pp. 330–346. [DOI] [PubMed] [Google Scholar]
- 29.Lee H-J, Shieh C-K, Gorbalenya A E, Koonin E V, La Monica N, Tuler J, Bagdzhadzhyan A, Lai M M C. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leibowitz J L, Wilhemsen K C, Bond C W. The virus-specific intracellular RNA species of two murine coronaviruses: MHV-A59 and MHV-JHM. Virology. 1981;114:39–51. doi: 10.1016/0042-6822(81)90250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X-L, Blackfoot J A, Hassel B A. RNase L mediates the antiviral effect of interferon through a selective reduction in viral RNA during encephalomyocarditis virus infection. J Virol. 1998;72:2752–2759. doi: 10.1128/jvi.72.4.2752-2759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makino S, Joo M, Makino J K. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J Virol. 1991;65:6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makino S, Shieh C-K, Keck J G, Lai M M C. Defective interfering particles of murine coronavirus: mechanism of synthesis of defective viral RNAs. Virology. 1988;163:104–111. doi: 10.1016/0042-6822(88)90237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholson D W, Thornberry N A. Caspases: killer proteases. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 35.Nolan-Sorden N L, Lesiak K, Bayard B, Torrence P F, Silverman R H. Photochemical crosslinking in oligonucleotide-protein complexes between a bromine-substituted 2-5A analog and 2-5A-dependent RNase by ultraviolet lamp or laser. Anal Biochem. 1990;184:298–304. doi: 10.1016/0003-2697(90)90684-2. [DOI] [PubMed] [Google Scholar]
- 36.Pachuk C J, Bredenbeek P J, Zoltick P W, Spaan W J M, Weiss S R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis virus, strain A59. Virology. 1989;171:141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raue H A, Planta R J. Ribosome biogenesis in yeast. Prog Nucleic Acid Res Mol Biol. 1991;41:89–129. doi: 10.1016/s0079-6603(08)60007-0. [DOI] [PubMed] [Google Scholar]
- 38.Rowe C L, Baker S C, Nathan M J, Fleming J O. Evolution of mouse hepatitis virus: detection and characterization of spike deletion variants during persistent infection. J Virol. 1997;71:2959–2969. doi: 10.1128/jvi.71.4.2959-2969.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawicki S G, Sawicki D L. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J Virol. 1986;57:328–334. doi: 10.1128/jvi.57.1.328-334.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sen G C, Lengyel P. The interferon system: a bird's eye view of its biochemistry. J Biol Chem. 1992;267:5017–5020. [PubMed] [Google Scholar]
- 41.Senanayake S D, Brian D A. Translation from the 5′ untranslated region (UTR) of mRNA 1 is repressed but that from the 5′ UTR of mRNA 7 is stimulated in coronavirus-infected cells. J Virol. 1999;73:8003–8009. doi: 10.1128/jvi.73.10.8003-8009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siddell S, Wege H, Barthel A, ter Meulen V. Coronavirus JHM: intracellular protein synthesis. J Gen Virol. 1981;53:145–155. doi: 10.1099/0022-1317-53-1-145. [DOI] [PubMed] [Google Scholar]
- 43.Siddell S, Wege H, Barthel A, ter Meulen V. Intracellular protein synthesis and the in vitro translation of coronavirus JHM mRNA. Adv Exp Med Biol. 1981;142:193–207. doi: 10.1007/978-1-4757-0456-3_17. [DOI] [PubMed] [Google Scholar]
- 44.Siddell S, Wege H, Barthel A, ter Meulen V. Coronavirus JHM: cell-free translation of structural protein p60. J Virol. 1980;33:10–17. doi: 10.1128/jvi.33.1.10-17.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silverman R H. 2-5A-dependent RNase L: a regulated endoribonuclease in the interferon system. In: D'Alessio G, Riordan J F, editors. Ribonucleases: structure and function. New York, N.Y: Academic Press; 1997. pp. 515–551. [Google Scholar]
- 46.Silverman R H, Skehel J J, James T C, Wreschner D H, Kerr I M. rRNA cleavage as an index of ppp(A2′p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J Virol. 1983;46:1051–1055. doi: 10.1128/jvi.46.3.1051-1055.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sturman L S, Holmes K V, Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J Virol. 1980;33:449–462. doi: 10.1128/jvi.33.1.449-462.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suarez P, Diaz-Guerra M, Prieto C, Esteban M, Castro J M, Nieto A, Ortin J. Open reading frame 5 of porcine reproductive and respiratory syndrome virus as a cause of virus-induced apoptosis. J Virol. 1996;70:2876–2882. doi: 10.1128/jvi.70.5.2876-2882.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tahara S M, Bergmann C C, Nelson G W, Anthony R P, Dietlin T A, Kyuwa S, Stohlman S A. Effects of mouse hepatitis virus infection on host cell metabolism. Adv Exp Med Biol. 1994;342:111–116. doi: 10.1007/978-1-4615-2996-5_18. [DOI] [PubMed] [Google Scholar]
- 50.Tahara S M, Dietlin T A, Bergmann C C, Nelson G W, Kyuwa S, Anthony R P, Stohlman S A. Coronavirus translation regulation: leader affects mRNA efficiency. Virology. 1994;202:621–630. doi: 10.1006/viro.1994.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tahara S M, Dietlin T A, Nelson G W, Stohlman S A, Manno D J. Mouse hepatitis virus nucleocapsid protein as a translational effector of viral mRNAs. Adv Exp Med Biol. 1998;440:313–318. doi: 10.1007/978-1-4615-5331-1_41. [DOI] [PubMed] [Google Scholar]
- 52.Vennema H, Godeke G-J, Rossen J W A, Voorhout W F, Horzinek M C, Opstelten D-J E, Rottier P J M. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wege H, Siddell S, ter Meulen V. The biology and pathogenesis of coronaviruses. Curr Top Microbiol Immunol. 1982;99:165–200. doi: 10.1007/978-3-642-68528-6_5. [DOI] [PubMed] [Google Scholar]
- 54.Wreschner D, Melloul D, Herzberg M. Interaction between membrane functions and protein synthesis in reticulocytes: specific cleavage of 28S ribosomal RNA by a membrane constituent. Eur J Biochem. 1978;85:233–240. doi: 10.1111/j.1432-1033.1978.tb12231.x. [DOI] [PubMed] [Google Scholar]
- 55.Zhou A, Paranjape J, Brown T L, Nie H, Naik S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C, Silverman R H. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]