Abstract
Biomedical imaging, especially molecular imaging, has been a driving force in scientific discovery, technological innovation, and precision medicine in the past two decades. While substantial advances and discoveries in chemical biology have been made to develop molecular imaging probes and tracers, translating these exogenous agents to clinical application in precision medicine is a major challenge. Among the clinically accepted imaging modalities, magnetic resonance imaging (MRI) and magnetic resonance spectroscopy (MRS) exemplify the most effective and robust biomedical imaging tools. Both MRI and MRS enable a broad range of chemical, biological and clinical applications from determining molecular structures in biochemical analysis to imaging diagnosis and characterization of many diseases and image-guided interventions. Using chemical, biological, and nuclear magnetic resonance properties of specific endogenous metabolites and native MRI contrast-enhancing biomolecules, label-free molecular and cellular imaging with MRI can be achieved in biomedical research and clinical management of patients with various diseases. This review article outlines the chemical and biological bases of several label-free chemically and molecularly selective MRI and MRS methods that have been applied in imaging biomarker discovery, preclinical investigation, and image-guided clinical management. Examples are provided to demonstrate strategies for using endogenous probes to report the molecular, metabolic, physiological, and functional events and processes in living systems, including patients. Future perspectives on label-free molecular MRI and its challenges as well as potential solutions, including the use of rational design and engineered approaches to develop chemical and biological imaging probes to facilitate or combine with label-free molecular MRI, are discussed.
Keywords: Magnetic resonance imaging, Magnetic resonance spectroscopy, Molecular imaging, Label-free, Metabolic imaging, Chemical exchange, Diffusion, Perfusion, Imaging probe
Introduction
The power of noninvasive and anatomically defined diagnosis and assessment of diseases has made medical imaging a highly desirable specialty in modern medicine. While its early development and growth were driven mainly by technological and engineering advances, the explosion of molecular biology and life science research in the 1980s to 2000s, coupled with the need for new real-time visualization tools for scientific discoveries, stimulated cutting-edge innovations in imaging technology and expanded imaging applications beyond clinical care. Different imaging modalities, including optical, ultrasound, X-ray, radio-nuclear, and magnetic resonance imaging (MRI) or even their combinations, have been developed and deployed for in vitro, ex vivo, and in vivo cellular imaging, preclinical live animal imaging, and more recently molecular and functional imaging in patients. These technological advancements have led to the emergence of a new and highly multidisciplinary field of molecular imaging1−3 that specifically explores and visualize biological processes at the cellular and molecular levels using imaging technology.
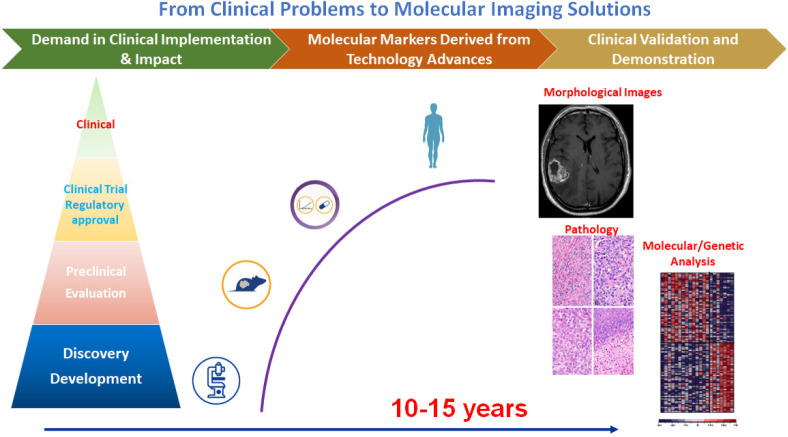
Molecular imaging, in large part, is still centered on the development of novel imaging probes or tracers via chemical biology approaches to target and report specific biomarkers and biological events in vivo.4,5 Typically, these biologically and molecularly targeted probes, whether they are used for optical imaging, MRI, or nuclear imaging as readout methods, are chemically modified and “labeled” with signal emitting or enhancing molecules or radioisotopes to achieve the high sensitivity necessary for molecular imaging of living systems.6,7 Since the inception of molecular imaging two decades ago, tremendous efforts and advances have been made in developing exogenous imaging probes. New knowledge and understandings have been obtained in the areas of rational design of imaging probes, delivery of the probes, and applications toward intended biomedical problems.8,9 On the other hand, translating these innovations and new probes to clinical applications has been challenging with limited success, despite the increasing emphasis on precision medicine demands for molecular imaging approaches to be implemented in critical areas of clinical management of patient care, such as molecular or genetical characterization of diseases, stratification of patients who are suitable for targeted treatment, molecular image-guided interventions and theranostics, and noninvasive and timely monitoring treatment responses or the disease progression.10,11 The gap and major hurdles to the translation of molecular imaging into clinical practices are well recognized. As summarized in Figure 1, while there are rich resources of biological discoveries and development of advanced chemical methods for specific probes in laboratories,12,13 they need to be matched with imageable biomarkers that are clinically relevant for the intended imaging modality. Imaging probes developed and tested in vitro must be stable and deliverable in vivo while not losing or altering their biological and chemical properties and imaging capability. The validation and evaluation of the safety and performance of the developed probes should be done in proper preclinical animal models, which often do not recapitulate human diseases sufficiently. Even if a probe meets the criteria to advance to further investigations in humans, the development of large-scale production of the clinical grade probes, lengthy clinical trials for safety and efficacy, and stringent regulatory processes precede and often limit availability to patients.
Figure 1.
Schematic illustration of the disproportion of scientific discoveries, imaging probe development, and clinical molecular imaging applications highlights the challenges in translating research to clinical applications.
Importantly, in order to move forward with translating and implementing molecular imaging approaches in clinical applications, there are opportunities to take advantage of endogenous biochemicals as imaging reporters and to use existing imaging technologies, particularly those already available in standard clinical care to study and characterize diseases at the molecular and cellular level.14 These endogenous “probes” are present and accumulated in the residing organs or tissue of interest, including specific metabolites, proteins, and cells. Not only do they exhibit unique chemical and biological properties, such as optical, acoustical, or nuclear magnetic properties for imaging, but levels of these endogenous probes change (directly or indirectly) in response to specific biological or disease processes. Therefore, no chemical or biological “labeling” is required to enable these endogenous image-capable probes, while concurrently avoiding toxicity concerns and difficulties involved in site-specific delivery of exogenous agents. Such a “label-free” strategy15 for molecular imaging will be discussed in this review using MRI representative image modality.
MRI, along with its spectroscopic version, MRS, is widely available and arguably the most applied noninvasive radiological imaging modality,16,17 given its substantial advantages in whole-body three-dimensional tomographic capability, superb image resolution, exquisite soft tissue contrast, and radiation-free. MRI-based diagnosis and treatment monitoring are currently applicable in almost all diseases and organ systems, particularly for managing oncological, neurological, and cardiovascular diseases. It is also a widely used robust basic science and preclinical investigative tool in biomedical research and engineering fields, including neurosciences and drug development.18−20 It is worth noting that MRI is one of the few imaging methods fundamentally and technically built on the principles and methods of nuclear magnetic resonance (NMR), which is an essential analytical tool in the chemical and biochemical sciences and discovery. In principle, MRI utilizes a magnetic nucleus (i.e., one with a nonzero nuclear spin), most commonly the hydrogen nucleus (1H) containing a single proton, as the signal source. Since water (H2O) is the most abundant molecule in living systems, most MRI applications depend on the nuclear magnetic and physiological properties of water molecules. Throughout this review, references are made to hydrogen nuclei or protons and their resonant frequencies; however, the same principles apply to nonproton NMR and MRI methods. The exquisite image contrast from MRI manifested by a series of cleverly orchestrated radiofrequency (RF) pulse sequences, can be generated based on the tissue-dependent longitudinal and transverse relaxation times of protons, i.e., T1 and T2, differences, respectively, in spatial distributions of water or other sources of 1H or protons in different organs and tissue types, as well as differences in movement or flow. Because these nuclear magnetic properties of water protons are sensitive to the chemical, biological, and physiological environments of tissues, water molecules can be considered as endogenous probe molecules for in vivo “label-free” MRI to report the normal and abnormal tissues based on disease-induced changes in chemical, biological, and physiological environments. Figure 2 highlights several label-free MRI contrast mechanisms that use intrinsic properties of water molecules to report and even quantitatively measure the physiological and functional status of living systems without using exogenous contrast agents.21−23 In MRI, these often include paramagnetic or superparamagnetic molecules that can alter the nuclear magnetic properties of the surrounding water molecules. Furthermore, intrinsic magnetic properties of some essential biological molecules produced in the body, such as iron-containing hemoglobin and ferritin,24 are sufficient to allow these molecules to serve as endogenous MRI contrast agents, and importantly involve the biological processes in their hosting organs and tissue.
Figure 2.
Labeled vs label-free strategies for generating MRI contrast, namely the change in water MRI signal. The label-free cellular and molecular MRI employs endogenous chemical and biological probes without using exogenous contrast agents and magnetic material labeled probes.
Another well-regarded label-free molecular MRI approach is spatially localized MRS, which directly detects and measures endogenous metabolites in a “virtually” sampled tissue volume selected from anatomic images obtained in the same session as the MRI scans.25−27 Effectively, MRS employs laboratory-based analytic NMR methods are applied to study metabolic diseases and disorders associated with various diseases in both in vivo and ex vivo.
In this article, we will review several label-free molecular MRI methods which have been accepted in biomedical research and clinical practices, focusing on their chemical and biological bases. Examples of these methods, such as MRS, diffusion-weighted imaging (DWI), perfusion-weighted imaging (PWI), and chemical exchange saturation transfer (CEST) imaging, as well as uses of endogenous hemoglobin and ferritin as contrast agents, will be presented to demonstrate the strategies of using label-free endogenous probes to report the molecular, metabolic, physiological and functional events and processes in living systems, including patients. We will also discuss the future perspectives of label-free molecular MRI and the challenges and potential solutions for imaging molecular targets and biomarkers at very low concentrations using in vivo MRI, including rationally designed and engineered chemical and biological imaging probes.
MRS-Based Molecular and Metabolic Imaging
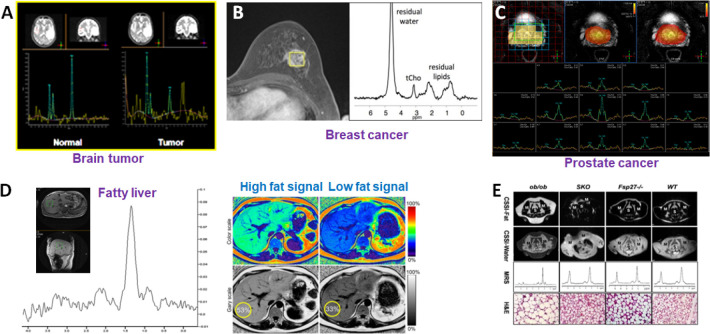
Spatially localized MRS essentially employs the same physics principles and similar hardware as analytical NMR for in vivo detecting and quantifying biomolecules, mostly low molecular weight metabolites.28 However, a gradient system is implemented on MRI scanners for spatially encoding NMR signals. Coupled with MRI to identify the anatomic structures or abnormal tissues, spectra of the NMR signals rising from different molecules appearing at their respective resonant frequencies can be recorded from a volume of interest in the size as small as 0.125 mL using a typical 3 T (T) MRI scanner.29 NMR and MR spectra are typically displayed with x-axis units of chemical shift (in parts per million) rather than frequency units in Hertz, to ensure spectral peak positions are independent of the applied or “carrier” frequency. Both safety concerns and engineering challenges limit the field strength used for preclinical and clinical MRI and MRS to 3T (i.e., 1.5–3T, 64 to 127 MHz) as the current clinical standard in contrast to conventional high-resolution analytical NMR (i.e., 14 to 23 T or 600 MHz to 1 GHz). Notably, 7T whole-body systems have gained recent approval from the US Food and Drug Administration (FDA) for clinical uses, while field strengths of preclinical animal MRI scanners typically fall in the range of 7T (300 MHz) to 11.7T (500 MHz). Granting data acquisition is not disrupted or distorted by the physical and physiological motions and other inevitable in animal and human studies, MRS methods can be applied in almost all organs, but mostly in the brain,30,31 breast,32,33 prostate,34,35 and liver36,37 as examples shown in Figure 3. Importantly, different organs and tissue often show specific or distinctive spectral patterns or metabolite profiles due to different metabolic functions. Changes in metabolite profiles can be observed when the disease alters metabolic functions or produces additional metabolites or compounds.
Figure 3.
Examples of in vivo single-voxel MRS applications in different organs and diseases with corresponding metabolite profiles. (A) The spectra collected from the brain tumor (right) and the contralateral normal region (left) show different spectral patterns. Prepared from unpublished data. (B) Spectrum from a lesion from a breast cancer patient gives choline and lipid signals that can be used to assess the malignancy of the tumor.38 Adapted with permission from ref (38). Copyright 2012 Springer Nature. (C) Multivoxel MRS can also be used for assessing prostate cancer in which spatially distinctive spectra from individual voxels can be extracted and compared. Prepared from unpublished data. (D) MRS signals rising from fat and lipids can be used to map the fat levels in the liver for diagnosis of fatty liver. Prepared from unpublished data. (E) Chemical shift-selective imaging and single-voxel MRS are used to examine the lipid accumulation in the diabetic kidney in a mouse model. Reproduced with permission from ref (61). Copyright 2013 the American Physiological Society.
Running In Vivo NMR Experiments in a 3D Space
Unlike solution NMR which is used in most chemical and biochemical experiments, not all biomolecules in organs, intact tissue, and cells can be detected by MRS in vivo. Usually, only those with millimolar concentrations and above, and proper T1 or T2 relaxation times39 within the limits of the MR hardware can be detected due to the sensitivity and dynamic range of MRS, for example, extracellular low molecular weight metabolites. Many macromolecules that are compartmentalized and restricted in the tissue with very short T2 or long T1 may not be detectable by MRS.40,41 While these “tissue-specific” properties can be a disadvantage in terms of sensitivity under certain data acquisition conditions, resulting in spectra that represent an incomplete picture of molecules present, these properties are also used by MRI physicists and engineers. Data acquisition parameters can be designed and optimized for improving the specificity and resonance assignment to specific molecules, or selectively detecting molecules of interest, particularly when NMR properties such as tissue-specific relaxation times in distinct physiological environments can be determined.
Due to the relatively lower external magnetic field strength used in human imaging and inherent heterogeneity of tissue and compartmentalized biomaterials in the sampling volume results in the partial volume effects and line-broadening, the spectral resolution of in vivo MRS is not sufficient to resolve individual groups of protons from each molecule at different chemical shifts as is possible high-resolution analytical NMR. Instead, in vivo MRS often reports detectable metabolites or molecules based on one of their proton resonances resolvable in the spectra.42,43 For example, a typical MR spectrum acquired in the mammalian brain consists of peaks attributed to the resonance of methyl (−CH3) groups of lactate at 1.33 ppm; N-acetyl aspartate (NAA) at 2.01 ppm, which is considered as a marker for neurons; a combined peak attributed to the neurotransmitter glutamate and glutamine (Glx) at 2.1–2.4 ppm creatine and phosphocreatine (Cr) at 3.00–3.05 ppm, a marker of cellular energy metabolism marker; glycerophosphocholine, phosphocholine and free choline (tCho), a marker for cell turnover rate, at 3.19–3.24 ppm, and myo-inositol (mI) at 3.56 and 4.06 ppm, which may be elevated in the presence of neuroinflammation. The signal levels are closely correlated to the concentrations of each metabolite, allowing for quantification of their concentrations using an internal reference, such as H2O. While the number of metabolites and chemicals detectable by in vivo MRS is much smaller than those detected and resolved in high-resolution analytical NMR, simultaneous detection of different metabolites or molecules in vivo in the same scan session is an invaluable and unmatched multiplexing capability. Most clinically available metabolic imaging modalities, such as positron emission tomography (PET), relies on a specific tracer or probe that can only target a single biomarker in each scan, assuming the tracer is available.
In vivo MRS can be performed in a size-selected single voxel, in which a localized sample volume (often ∼1–8 mL) is selected in a well-defined anatomical region to obtain a spectrum of metabolites. In contrast, a multivoxel approach is used when the entire or a significant portion of an anatomical section or slice (thickness of 5 to 20 mm) consisting of multiple voxels is selected for data acquisition. Based on the spectra from each individual voxel, as shown in the application in evaluating prostate diseases (Figure 3C), the signal level or intensity corresponding to different metabolites at specific chemical shifts measured from each voxel can be combined to generate a spatial distribution or image-like maps for each metabolite. Thus, multivoxel MRS or magnetic resonance spectroscopic imaging (MRSI), also known as chemical shift-selective imaging (CSI), yields metabolite maps readily overlaid with corresponding anatomical images,44 similar to the presentation of PET/CT or PET/MRI. As physics and chemistry of in vivo MR methods are originated from NMR, the interest in developing MRS for biomedical applications was one of the primary motivations for the invention and development of clinical MRI scanners in the 1980s. Currently, MRS has many applications in biomedical research and clinical care in the areas highlighted in the following sections.
With direct detection and measurement of metabolites in the diseases affected tissue, MRS provides important biochemical information about the disease process that can be better correlated to the pathology than morphological image features found in structural MRI. To date, clinical applications of noninvasive MRS are mostly for diagnosing central nervous system (CNS) diseases and disorders, particularly neoplasm45 and epilepsy,46 which often require surgical interventions. Since it is a noninvasive and radiation-free method, MRS can be used repeatedly to monitor disease progression and treatment response, circumventing the need for more invasive procedures such as repeated brain biopsy. For example, malignant brain tumors typically exhibit spectral patterns consistent with the loss of neurons as indicated by reduced NAA and Cre levels, high cell turnover rate marked by a sharp increase of tCho, and the presence of necrosis that leads to elevated lactate and lipid levels (Figure 3A).
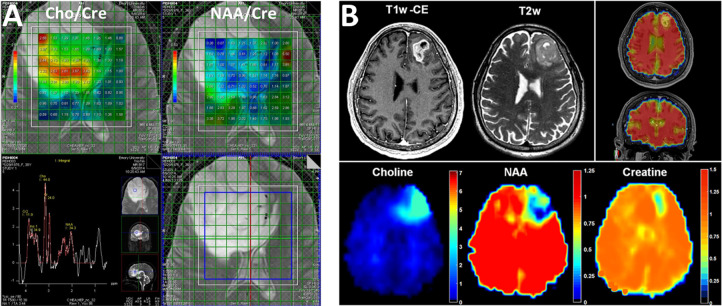
The need for a combination of molecular, functional, and metabolic imaging for better assessment and diagnosis of a disease can be demonstrated in Figure 4, where it is apparent that metabolic alterations in a high-grade brain tumor, highlighted with elevated Cho and a reduction of NAA, are strikingly heterogeneous and cannot be delineated by morphological presentations in the high-resolution anatomic MRI (Figure 4A). For highly infiltrated glioblastoma (GBM), conventional MRI, even with enhancement from injected gadolinium (Gd) contrast agent, can only indicate the tumor boundary while the area with infiltrating cancer cells is only observed by tumor-induced changes in metabolite levels47 (Figure 4B). Since changes in molecular processes and biomarkers always occur prior to structural and morphological changes, molecular and metabolic imaging with MRS provides crucial biochemical information for improving the specificity of diagnosis and characterization of diseases, and early detection. In addition to CNS diseases, MRS has been used in assisting the diagnosis of many other diseases that involve metabolic or biochemical changes, including liver diseases,48 renal dysfunction,49 and muscle degeneration.50,51 With the increasing need for quantitative and multiparametric imaging approaches in precision medicine, integration of MRS in clinical protocols is not only beneficial but also logical and highly feasible, given most current clinical MRI systems are equipped with MRS capabilities.
Figure 4.
Maps of the rations of NAA vs Cre and Cho vs Cre derived from multivoxel MRSI or CSI of a brain tumor (A) show the intratumoral heterogeneity of metabolic changes with highly elevated Cho and reduced NAA in relation to Cre in the specific regions that cannot be localized well in T2-weighted MRI. Composed from unpublished data. (B) Whole-brain CSI performed on a recurring GBM identified areas with tumor cells infiltrating beyond the boundary delineated in T2-weighted (T2w) and T1-weighted contrast-enhanced (T1w-CE) MRI. Reprinted in part with permission from ref (47). Copyright 2016 Society for Neuro-Oncology and Oxford University Press.
Clinically Ready MRS and Its Added Values
The spatially resolved MRS and its combination with high-resolution MRI provides an excellent tool for image-guided intervention and site-specific delivery of treatment. In the oncology clinic, MRS has been used to guide tissue biopsy and tumor resection in diagnosing and treating brain tumors, breast cancer, and prostate cancer. Metabolite maps from MRSI can be used to direct the radiotherapy for infiltrating GBM, as shown in Figure 4B, enabling aggressive and targeted treatment for such lethal tumors by maximizing therapeutic responses while sparing the unaffected brain region. In this case, treatment-targeted regions with cancer cell infiltration are beyond the regions identified by the conventional structural MRI. A recent clinical trial used MRSI to guide the radiotherapy of GBMs with dose escalation to 75 Gy, a unit of ionizing radiation dose, instead of 60 Gy for treating the part of the tumors with elevated Cho/NAA ratio but nonenhancing fluid-attenuated inversion recovery (FLAIR) imaging and contrast-enhanced MRI. The results indicated that this approach is safe and feasible for guiding treatment. More importantly, GBM patients treated with dose-escalated MRSI-guided radiotherapy had improved median overall survival from 16 months with the current standard care to 23 months.52
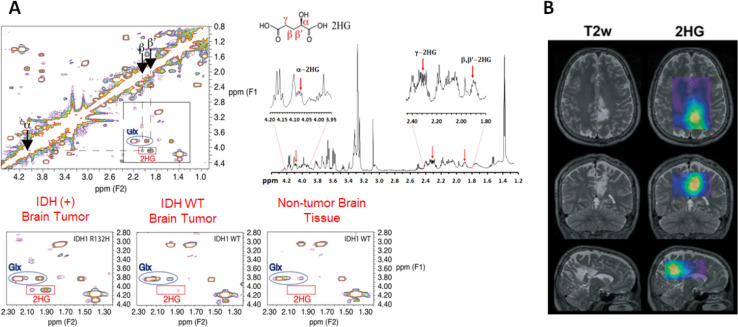
A significant portion of MRS applications and technology development is devoted to providing tools for investigating disease mechanisms and discovering new image-able biomarkers for biomedical research. As MRS is the in vivo analog of laboratory-based analytical NMR, NMR experiments provide excellent approaches and data for bridging in vitro and ex vivo cell or tissue-based mechanistic investigations with in vivo metabolic analysis through visualization-based preclinical and clinical imaging. For example, the original discovery of the MRS biomarker for the isocitrate dehydrogenase 1 (IDH1) mutation in glioma was carried out using high-resolution solid-state NMR analysis of a large number of brain tumor tissue samples.53,54 IDH mutation is a critical genetic alteration in gliomas and is now used as a central component in the criteria for the genetic and molecular classification of gliomas, which has strong implications on outcomes and treatment options. Because of the mutation at the amino acid 132, the mutant IDH then produces a new oncometabolite, D-2-hydroxylglutarate or 2HG, instead of the regular product, α-keto glutarate. 2HG has a concentration level in the MRS detectable range (mM), making 2HG a potential MRS-detectable metabolite biomarker for the IDH mutation. As shown in Figure 5, high-resolution solid-state NMR at 14T (600 MHz) with the assistance of the 2D total correlation spectroscopy (TCOCY) method was able to determine the spectroscopy pattern or “fingerprint” of 2HG in tumor tissue samples collected from glioma patients with pathological and genetic analysis confirmed IDH mutations. Thus, this NMR spectroscopic “fingerprint” of 2HG provided the basis for further developing advanced MRS methods to detect 2HG and characterize IDH1-mutated brain tumors in patients.55−57 In addition, it demonstrates that MRS can directly detect a specific and potent genetic mutation through its corresponding molecular or metabolite marker. Furthermore, MRS has the advantage of enabling the study of metabolic networks and pathways as MRS can detect multiple metabolites simultaneously. This enables the assessment of metabolic changes not only in a single domain but also the interactions between metabolic pathways, a powerful capability for studying metabolic reprogramming in tumors caused by 2HG production that may drive tumor progression, or to report the response to novel drugs developed for targeting IDH mutation and tumor metabolism as shown in recent studies.58,59
Figure 5.
(A) High-resolution ex vivo NMR analysis of intact brain tumor tissue samples revealed resonance signals rising from different protons of the 2HG molecule and their specific chemical shifts. Reproduced with permission from ref (53). Copyright 2012 Springer-Verlag. (B) Spectroscopic editing methods, such as double quantum coherent acquisition, can detect 2HG in patients carrying tumors with IDH mutation. Reproduced with permission from ref (57). Copyright 2015 American Association for Cancer Research.
Readers can find extensive literature reporting the studies on many other diseases using NMR and MRS. For instance, Li et al. applied 1H MRS directly after ketamine administration to measure the glutamine/glutamate ratio.60 The significant increase in this ratio thus reveals a possible mechanism for the antidepressant effect of ketamine. In a preclinical investigation of functions brown and white adipose tissue by Peng et al.,61 fat fractions of these two different types of adipose tissues in ob, seipin, and Fsp27 gene knockout mice were quantified using noninvasive CSI of lipid accumulation combined with MRI, as brown adipose tissue and its content contribute the control of thermogenesis that protects the body from cold and obesity. The tissue-type specific quantitative measurements by CSI revealed that the differences in fat fractions in brown and white adipose tissue in different mouse models were related to the different regulation effects of ob, seipin, or Fsp27 gene.
In addition to metabolic analysis and imaging applications, MRS can be used for noninvasive molecular characterization and measurement of other physiological conditions in deep tissue and organs, such as temperature.62 It is worth noting that Robert Gillies, a brilliant cancer biologist and biophysicist who passed away recently, pioneered the use of phosphorus (31P) MRS and pH dependence of chemical shifts of adenosine triphosphate (ATP) to measure the tissue pH and tumor acidity, a hallmark of tumor biology and the tumor microenvironment.27,63 Gillies’ work not only established an innovative early molecular imaging tool but enabled intense and systematic studies that advanced our understanding of the pathophysiological microenvironment of solid tumors and its roles in initiating and promoting malignant clones, genomic plasticity, and heterogeneity in the early development of cancers.
Molecular and Functional Imaging Using Water Molecules as Probes
Unlike water molecules in the homogeneous solution state, which cannot be differentiated from each other by MRI, those compartmentalized in the different tissue and cellular structures, such as gray and white matters of the brain, collagen of cartilage, and blood vessels, exhibit nuclear magnetic properties that are dependent on biological and physiological conditions in addition to their spatial distribution. Thus, localized changes in those properties as a result of disease-related pathological or physiological disruptions can be revealed by specific MRI pulse sequences or data acquisition schemes. Clinically, diffusion-weighted imaging (DWI)64 and atrial spin-label (ASL) perfusion MRI65 of endogenous water molecules are often used to visualize and quantify tissue-specific pathological and physiological changes that can be correlated to molecular and cellular processes. Since the iron in different biological forms is the most abundant and essential biological metal in living systems,66 its magnetic properties make iron-containing biomolecules endogenous MRI contrast agents, allowing for label-free molecular and functional imaging with MRI. The most notable applications of this approach are on blood oxygenation level-dependent (BOLD)67 imaging that detects tissue oxygenation based on the change of ionic iron (Fe2+ or Fe3+) in oxy- to deoxy-hemoglobin flowing in the capillary and susceptibility weighted imaging (SWI)68 that measures the tissue iron content based on strong susceptibility from deposited and accumulated in the tissue.
Probing Cellularity and Tissue Integrity with DWI
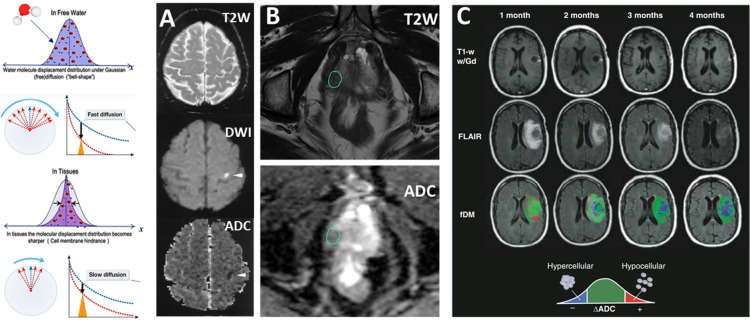
DWI is established and well-integrated into the clinical protocols in brain, abdominal, and pelvis imaging. This is the method for detecting and quantifying the movement of the water molecules inside tissues and cells, typically by a pair of magnetic field gradients (often described by the b value, which depends on the strength, duration, and spacing of these pulsed gradients) together with RF pulses. The measured DWI signal in tissues to obtain the apparent diffusion constant (ADC, mm2/s). ADC is lower than that of the solution because the random walk-type of Brownian dynamics movement of water molecules is restricted and hindered by cell membranes and intercellular boundaries. When cells within a tissue become deformed, swollen, necrotic, or apoptotic, or experience membrane disruption, the movement of water molecules is less restricted, resulting in an increase in the ADC values. DWI can be used to quantitatively characterize tissue properties such as cellularity. The sensitivity and accuracy of DWI are dependent on the maximum gradient strength delivered by an MRI scanner, with high b-values up to 4000 being possible in preclinical animal and research-only human scanners. When the directionally applied gradient scheme is used in DWI, directions of the water diffusion in the given tissue compartment can also be calculated from the images obtained from diffusion tensor imaging (DTI),69 allowing for quantitative assessment of the orientation-specific microscopic structures of the fibrous tissue, such as axons in brain white matter70 and cartilage of the tendons.71
Clinically, DWI is widely used for imaging and quantitative assessment of tissue degeneration and damage, especially in detecting mild traumatic brain injury (mTBI) and early ischemic lesions in stroke patients, as shown in Figure 6A. In this example, areas undergoing ischemia-induced tissue damage were visualized by increased diffusivity in DWI but reduced ADC as the result of cytotoxicity from the edema. Generally, ADC decreases even without a noticeable change in T2-weighted images at the earliest times from stroke onset. A substantial decrease in ADC is believed to represent the most severe ischemic tissue damage with the highest risk of cell death, a factor critical to the clinical decision on treatment options.72 In oncological applications, DWI is readily used for characterizing the cellularity of the tumors based on the measurement of ADC. In the case of patients with suspected prostate cancer, the lesion not only showed typical T2 signal decreases but has low ADC, indicating a high cellularity solid tumor73 (Figure 6B). When used to assess tumor responses to the treatment, ADC can be considered as a surrogate marker for cancer cell apoptosis. In the GBM patient shown in Figure 6C, who received bevacizumab antiangiogenesis treatment over a period of four months, the regional spatial distribution of ADC change, termed functional diffusion map (fDM),74 showed initial response to the treatment but recurred and progressed. Even though both contrast-enhanced T1 weighted MRI and FLAIR imaging looked improved, time-dependent fDM maps showed a rapid increase in the hypercellularity volume, indicating the tumor’s growth and treatment failure75 (Figure 6C).
Figure 6.
Diffusion properties of water are used to quantitatively assess the integrity of the cell and tissue in (A) ischemic brain. Adapted with permission from ref (72). Copyright 1999 American Academy of Neurology. (B) Change of ADC in a prostate lesion (circled), and (C) quantitative measurement of diffusion properties in a brain tumor that responded to the treatment before relapsing. Adapted with permission from ref (75). Copyright 2010 Wiley-Liss, Inc.
Tracking Magnetically Labeled Water Molecules to Measure Blood Perfusion
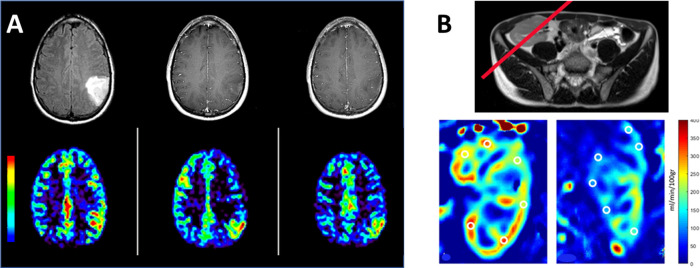
Different from DWI, perfusion-weighted imaging looks for the movement of water molecules in the microvascular space, particularly capillary. Both blood flow and volume perfused into the tissue are important physiological and functional parameters that are important to the health and functions of organs and tissues. Absolute quantification of these two hemodynamics parameters noninvasively with imaging is not trivial. PET imaging using the H2O labeled with the short half-time radioisotope O15 as a tracer is the “gold standard” and the most accurate deep tissue imaging method. In the case of ASL perfusion-weighted imaging, water molecules in the flowing arterial blood are “magnetically labeled” by one or a series of RF pulses to invert out of phase from their original alignment or orientation in the external magnetic field.76 As these RF-labeled water molecules flow into the region of imaging, they are distinguishable from those residing in the region by readout RF pulses at particular times. The volume and rate of blood perfusing into the targeted organ or tissue can be calculated from the amount of these RF-labeled water molecules based on time-dependent or time series data collected from ASL perfusion imaging.77 While bolus tracking of the intravenously injected Gd-contrast agent with dynamic susceptibility contrast-enhanced (DSC) MRI can be easily implemented in clinical routes for measuring blood perfusion, modeling the kinetics of the injected contrast agent that can vary its contrast effect from changes of T1, T2, and T2* relaxation times depending on the tissue compartmentalization is difficult to derive accurate and absolute measurements of hemodynamic parameters. In comparison, ASL perfusion MRI with endogenous water molecules as a tracer alleviates such complicated and uncertain agent-tissue interaction problems. In the situation in which the concern over the clearance and safety of the Gd-based contrast agents,78 such as patients with abnormal renal functions, ASL perfusion MRI provides an excellent alternative if motion artifacts and acquisition time with the spin-label scheme designed for the flow and magnetic properties of water in the blood are manageable by the performance of the gradient and RF systems of the MRI scanners and blood flow kinetics in the vasculature of the organs, such as brain79 and kidney,80 where ASL perfusion MRI has been used with examples shown in Figure 7. More importantly, because the measurement of the amount of endogenous water flowing into the tissue is affected by the concentration-dependent change of contrast or signal intensity in the DSC MRI, the regional cerebral blood flow (rCBF) rate and regional cerebral blood volume (rCBV), when used in a brain, can be calculated accurately. In addition, ASL perfusion MRI can be repeated easily, which is a significant advantage as patient motion or other clinical situations can cause the scan to fail. In this case, repeated injection of contrast agent and the interference from the first bolus injection limit the effectiveness of DSC perfusion MRI.
Figure 7.
ASL perfusion MRI from a patient (A) with a brain tumor that is contrast-enhanced in T1 weighted imaging (top row) and increased rCBF (bottom row). Composed from unpublished data. (B) In patients receiving renal transplants, ASL perfusion MRI provides functional and quantitative assessments of the renal filtration, showing a normal blood perfusion map (left) and impaired blood perfusion of the kidney (right). T2 weighted image was used to place the slice (red line) where the labeling RF was applied. Reprinted with permission from ref (80). Copyright 2022 Springer Nature.
Iron Containing Endogenous MRI Contrast Agents
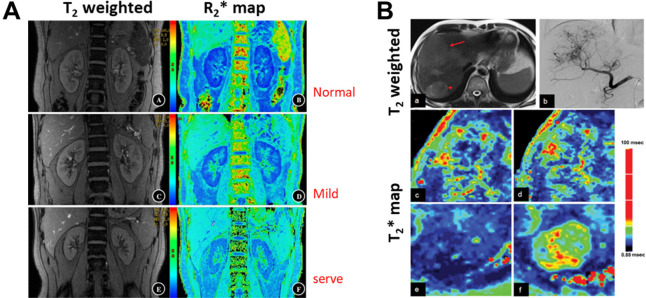
Using biological iron to gain biochemical and molecular information can be traced back to the early development and applications of NMR and MRI. Depending on its biochemical forms and the tissue environment, the presence of iron or iron-containing molecules can cause surrounding protons (or waters) changes in chemical shift, T1, T2, and T2 relaxation times as well as the phase of signals. The change in the ratio of blood paramagnetic Fe3+ heme-containing deoxyhemoglobin to diamagnetic oxyhemoglobin with Fe2+ heme and the difference in T2* shortening effect from to leads to the BOLD contrast is pronounced enough to be captured by the susceptibility weighted imaging. Advances in MRI technology and engineering, such as high field scanners, powerful gradient systems and rapid acquisition sequences, coupled with the BOLD contrast, led to the development of functional MRI (fMRI) that revolutionized the field of neuroscience.81 fMRI is now an essential tool in studying the fundamental brain functions and assisting diagnosis of brain diseases and disorders and image-guided neurosurgery. However, oxygen-sensitive BOLD contrast can be readily applied to investigate other diseases and disorders based on the change in the tissue oxygenation modulated by the amount of oxyhemoglobin delivered by the circulating blood. A decrease in the T2* (or increase in R2*) values, which can be spatially mapped in the entire imaging volume or organ by BOLD MRI, indicates an increase in deoxyhemoglobin and a decrease in tissue oxygen partial pressure. Such a noninvasive ability to assess and quantify tissue oxygenation level is essential in understanding and treating many diseases, given the central role of tissue oxygenation in physiology, including cell signaling, metabolism, and energy and functional outputs. In addition to applications in CNS diseases, BOLD MRI has been extensively investigated in kidney functions in renal diseases that are associated with hypoxia,82 such as ischemic kidney injury, chronic kidney disease (CKD), kidney transplantation, ureteral obstruction, diabetic nephropathy, kidney tumors. Current clinical practice in diagnosing renal hypoxia uses a microelectrode inserted into the kidneys to measure the partial pressure of oxygen. This method not only has the risk of the side effect commonly related to invasive procedures but also limits the measurement of the tissue oxygenation level in a small region and cannot be repeated routinely for patient follow-up. Using BOLD MRI, the kidney functions of patients with CKD or renal transplant can be examined in the cortex and medulla, respectively. For example, Li et al. showed that early renal impairment in patients with CKD causes hypoxia, as observed by reductions in the R2* value of the medulla and cortex83 (Figure 8A). They also compared with the current clinical standard of renal function test, which is the blood test of the creatinine level to estimate glomerular filtration rate (eGFR) that is not sensitive to the renal functional change in the early stage of the disease. They reported a significant positive correlation between the reduction of the R2* value of the medulla and the lower, although the correlation is weak in the cortex. Using medullary R2* as an imaging biomarker, they demonstrated a sensitivity of 92.31% and a specificity of 85.19% in differentiating the severity of renal function impairment, respectively.
Figure 8.
Multiecho T2 weighted GRE MRI of kidneys were used to derive BOLD R2*, i.e., 1/T2*, maps (A) of a normal individual (top row), a patient with IgA glomerulonephritis and mild renal impairment (middle row) and a patient with IgA glomerulonephritis and moderate to severe renal impairment (bottom row). The gradual increase of R2* (blue to green) indicates lower oxygenation correlated with the aggravation of renal impairment. Reprinted with permission from ref (83). Copyright 2018 Springer Nature. (B) BOLD MRI with carbogen challenge in a patient with diffuse liver masses in the right posterior lobe shown in T2 weighted MRI and angiography (arrows in the top row). Compared with T2* mapping (left panel) prior to carbogen breathing, the T2* value of the larger lesion is mildly increased (right panel), suggesting that the tumor has heterogeneous hypoxic regions. Reprinted with permission from ref (84). Copyright 2015 Spandidos Publications.
The sensitivity of BOLD-based MRI measurement of tissue oxygenation can be further enhanced by inhaling carbogen gas that can promote the increase of the level of deoxyhemoglobin in the blood. In the study reported by Zhang et al., carbogen gas-challenged BOLD MRI is used to assess the oxygenation changes of liver tumors as the result of the tumor vascular embolic treatment with transarterial chemoembolization (TACE) in patients with hepatocellular carcinoma (HCC).84 Hypoxia is a key characteristic of the tumor microenvironment and is strongly associated with poor responses to treatment. A higher level of oxygen in the tumor tissue may improve the treatment response of these tumors. Carbogen (95% O2 and 5% CO2) has been clinically applied during radiation therapy to raise oxygen tension and sensitize the radiation-induced reactive oxygen species in the anoxic region. Inhaling 5% CO2 mixed with oxygen reduces oxygen-induced vasoconstriction. With 15 min of carbogen-challenge monitored by BOLD MRI, their results, highlighted in Figure 8B, revealed that TACE treatment led to a decreased BOLD response of HCC compared with untreated HCC as the treatment can block the tumor vessels, enabling imaging assessment of the treatment-related tumor tissue oxygenation changes.
The magnetic susceptibility contrast used for BOLD imaging is further amplified when biological iron becomes accumulated in the forms of clusters or packed in the iron storage proteins, i.e., ferritin, allowing for the detection of abnormal presence or accumulation of iron in the tissue. Susceptibility weighted imaging (SWI), along with quantitative susceptibility mapping (QSM) based on the large phase change induced by iron that also can be derived from MRI, are normally used in clinical MRI protocols to examine the vasculature of the venous blood vessels (Figure 9A), detect the microbleeding in the brains (Figure 9B) with microvascular diseases that are not detectable in conventional MRI, and abnormal iron-deposition in specific brain regions.85 In particular, SWI provides the imaging biomarker for the early diagnosis of Parkinson’s disease (PD) based86 on the signal intensity changes in deep brain nuclei, such as the SN, the putamen (PUT), the globus pallidus (GP), the thalamus (TH), the red nucleus (RN) and the caudate nucleus (CN) as shown in Figure 9C. Iron overloading in those regions may lead to reactive iron species that can damage surrounding neurons. In the case of multiple sclerosis (MS), which is an autoimmune disease affecting the CNS, MS lesions cause neuroinflammation and activation of microglia which preferentially take up iron. Thus, hyperintense signal in QSM in MS as seen in Figure 9D is considered as a biomarker for microglial activation and subsequent demylination.87
Figure 9.
(A) Cerebral venous blood vessels are highlighted in the QSM of the brain. (B) SWI revealed the microbleeds that cannot be clearly delineated in T2 and T2* weighted MRI. A, B: Adapted with permission from ref (85). Copyright 2021 Radiological Society of North America. (C) Iron deposition in the thalamus of a PD patient led to hyperintense signals in QSM and R2* map. Reprinted with permission from ref (86). Copyright 2016 PLosOne. (D) Enhanced MS lesions in QSM due to iron uptake by microglia are correlated to the inflammation-related hyperintensity shown in the T2 weighted image. Reprinted with permission from ref (87). Copyright 2018 American Society of Neuroradiology.
CEST Based Molecular Imaging
Although chemical exchange saturation transfer (CEST) is an established NMR method to study slow exchange or reaction typically involved in biological molecules, such as confirmational changes in proteins, the feasibility of using the CEST contrast for MRI was only demonstrated in the 2000s in pursuing a new molecular imaging approach.88 The principle of CEST NMR is simple: a detectable NMR signal change in one type of proton is generated indirectly via the magnetization transfer between it and another type of proton, when the exchange rate of these protons is within the dynamic range of NMR frequency used in the measurement.89 Because conventional MRI typically uses water as the signal source, the diamagnetic effect of biomolecules with functional groups having strong electron shielding, such as amide (−NH2), amine (−NH), or even hydroxyl (−OH) in proteins or peptides, can affect the surrounding water molecules through their hydrogen bonds. These water molecules are “magnetically” different from bulky water, but can undergo slow exchange with bulky water. In CEST MRI, water protons are protons showing detectable signal change, and water-exchanging protons, referred to as exchangeable protons, are the protons on other molecules serving as a “recipient” of magnetization modulation, often saturation achieved by applying frequency selective RF irradiation pulses. The advantage of CEST MRI lies in the ability to amplify the signal of exchangeable protons (∼mM) for more than 1,000 times i.e., a few percent of signal change of water ([H+] = 110 M), so that the signal changes can be readily detected by MRI. Detailed descriptions of CEST MRI can be found in several excellent review papers.90−92 As NMR-rooted techniques, CEST MRI and MRS share many common physics and chemistry principles and practices. They are also complementary and can be used in combination, as it was demonstrated by Liu et al. in their work on using CEST MRI and MRS to investigate mannitol accumulation following the osmotic blood–brain barrier (BBB), in which mannitol in the brain can be directly measured by both CEST and MRS signals.93 Their results suggested that MRS had low spatial resolution but high specificity, whereas those by CEST MRI had high spatial resolution and high sensitivity. Quantifying CEST images is much more complicated than MRS as signals are from exchangeable protons with an exchange rate that is dependent on various chemical and biochemical conditions. One common approach is to plot the water signal intensity as a function of saturation frequency to obtain a Z-spectrum, which can be used to directly visualize the position and relative intensity of the CEST effect from a CEST agent. For in vivo applications, using a Z-spectrum to quantify the CEST effect from the exchangeable protons is affected by the T1 and T2 relaxation properties of water in specific tissue environments and the presence of macromolecules. Therefore, a CEST effect is generally measured by the magnetization transfer ratio asymmetry (MTRasym), which is defined by MTRasym = (S–Δω – S+Δω)/S0, where S+Δω and S–Δω are the MRI signals at offsets +Δω and −Δω when an RF saturation pulse (B1) is applied with a period of time (Tsat). S0 is the signal acquired without the RF saturation. Since this approach is based on the assumption that the water saturation profile is perfectly asymmetric without CEST agents, there are increasing effects to improve the methods of quantifying CEST images. The Web site (https://www.cest-sources.org) and review articles by Zaiss et al.94,95 provided in-depth descriptions of CEST theory, mathematical modeling using Bloch-McConnell equations, and quantifying Z-spectral data, along with several examples.
CEST Imaging of Changes in Tissue Properties
CEST MRI provides an effective way to detect endogenous biomolecules with weak diamagnetic properties indirectly through the water. CEST MRI is sensitive enough to detect these molecules through the amplification strategy mentioned above. Therefore, exchangeable protons in the mM concentration range would result in a few percent change of water signal that makes it readily detectable. As such, a wide variety of biomolecules with such or similar diamagnetic properties have been explored and discussed in our recent review papers.96−98 Given its capability of chemically selective imaging, CEST MRI has become an attractive molecular imaging method increasingly explored for investigating the disease-associated changes in the tissue that are not informed through structural MRI and MRS. While DWI provides structural and mechanical properties of the cells and tissue, tissue properties and their changes probed by CEST imaging are more relevant to tissue biological and physiological conditions.
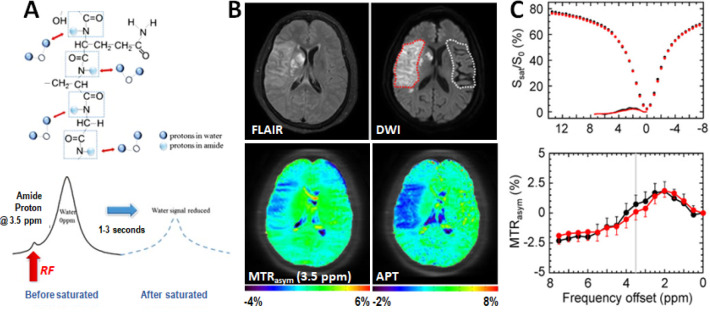
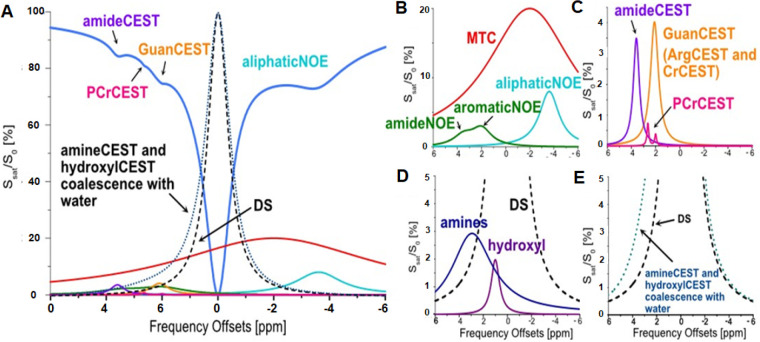
Amide proton (−NH) is the most widely used CEST proton investigated in clinical applications. Not only it is abundant in proteins and peptides and well separated from the bulk water signals in the CEST spectrum, but the exchange of amide proton hydrogen-bound water with the bulk water is also very sensitive to the physiological environment and conditions of the tissue, such as pH and temperature. Amide proton transfer weighted (APTw) CEST imaging was first demonstrated by Zhou et al. in detecting pH changes in ischemic brain tissue based on amide proton signals from intracellular proteins using the rat models of stroke.99 Subsequently, it has been advanced in the clinical studies of stroke patients (Figure 10). Since then, APTw CEST imaging and its strategy have been applied to assess abnormal tissue’s molecular and physiological conditions in various diseases, including tumors and Alzheimer’s disease. Because of the high concentration of mobile proteins in tumors due to the rapid turnover of cancer cells, the higher APTw signals at 3.5 ppm are commonly observed in various types of tumors, including glioma,100,101 breast cancer,102 and head and neck cancer.103 APTw imaging is an effective way to examine the intratumoral or even intracellular pH.104 A recent consensus paper on the technical aspects of APTw MRI summarized the previously published 3T clinical APTw studies,105 providing specific recommendations for pulse sequences, acquisition protocols, and data processing methods. APT imaging takes advantage of the slow magnetic transfer effect between the amide protons of mobile proteins on the hydrogen-bound water and bulk water to capture this process to report the tissue damage.106−108
Figure 10.
(A) Schematic illustration of the CEST imaging of slow exchange of hydrogen-bound water on amide protons and bulk water. An example of APT imaging of an acute stroke patient with right MCA occlusion106 (B) with conventional FLAIR and DWI images (top) showing the ischemic region. MTR asymmetry analysis and APT (bottom) showed much clearer ischemic contrasts than MTRasym (3.5 ppm). (C) Z-spectra and MTRasym spectra were obtained from the contralateral normal (black) and the ischemic lesion (red). B, C: Adapted with permission from ref (106). Copyright 2017 International Society for Magnetic Resonance in Medicine and Wiley.
CEST Imaging of Metabolites
In addition to the mobile proteins that satisfy the CEST MRI detection scheme or conditions, some endogenous CEST-detectable biomolecules include glucose and amino-containing amino acids, such as l-arginine and cytosine and nucleosides and nucleotides, such as deoxycytodine. Therefore, CEST imaging approaches can be expanded to noninvasive imaging of metabolites directly to probe the disease-altered metabolism.109 For example, glutamate (Glu) is the most abundant excitatory neurotransmitter in the brain, and altered glutamate levels are found to be highly related to many neurodegenerative disorders, such as AD, Parkinson’s disease (PD), as well as epilepsy, in which the glutamate level decline. Based on amino protons resonant at 3 ppm downfield from the resonance of water, glutamate can generate a highly pH-sensitive CEST signal.110 It should be noted that the so-called gluCEST signal at 3 ppm is attributed to a blend of endogenous agents, not just glutamate solely, even though the concentration of glutamate is typically much higher than other compounds (cytoplasm concentration is ∼5–10 mM in neuron cells). It is instrumental in identifying and localizing epileptic foci for proper diagnosis and presurgical planning,111 which is difficult with conventional morphological MRI. gluCEST has also been applied to monitor AD, another major neuro-degenerative disease requiring multibiomarkers for diagnosis. Results showed a strong correlation between the progression of AD and the reduction of the Glu level.112 However, the exchange of amino protons from glutamate is too fast at physiological pH, making it unsuitable as a CEST agent at 3T or lower field strengths. Nowadays, most clinical gluCEST MRI studies are conducted using 7T MRI scanners.
Creatine (Cr) and phosphocreatine (PCr) are other types of endogenous CEST-sensitive metabolites that have been exclusively studied in recent years. Coupling together, Cr and PCr are essential for energetic homeostasis in skeletal and cardiac muscles. The concentration of PCr is approximately 30 mM in the skeletal muscle and 5–6 mM in the brain. Thus, their intracellular concentrations are relatively high, making them desirable endogenous CEST contrast agents. Cr has a single CEST signal peak at 2 ppm, whereas PCr has two CEST peaks at 1.8 and 2.5 ppm, respectively, all rising from guanidinium protons. Cr-weighted and PCr-weighted CEST MRI have been demonstrated in the skeletal muscle,113 myocardium,114 and brain.115 In addition to Cr and PCr, other important brain metabolites, including the neurotransmitter γ-aminobutyric acid (GABA)116 and myoinositol117 have been shown detectable by CEST MRI.
Aside from the CEST effect raised from exchangeable protons, saturation transfer with water can also be achieved using nonexchangeable protons if those protons can rapidly exchange their magnetization with surrounding exchangeable protons via the intramolecular nuclear Overhauser effect (NOE), which successively exchanges with water molecules to generate CEST contrast.118 Because of the underlying mechanism of a two-step exchange, this effect is often called the relayed NOE (rNOE)t.90 In order to obtain fast enough exchange between nonexchangeable and exchangeable protons, both must be a part of the same macromolecules, such as mobile protons or glycogen. The rNOE of aliphatic protons has been reported in the frequency range from −1.6 to −3.5 ppm. Several studies have revealed that an altered rNOE effect is associated with cancer,119 stroke,120 kidney diseases,121 and, recently, glycogen disorders,122 suggesting the potential clinical applications of rNOE MRI.
Taken together, many endogenous molecules can be used as CEST contrast agents for label-free molecular imaging when a proper CEST imaging technique is used (Figure 11). Compared to traditional MRS, signals from endogenous CEST molecules often lack enough specificity. Current models for interpreting and quantifying CEST signals are oversimplified, while biological, physiological, and chemical conditions in the tissue are heterogeneous, requiring much more complex and multivariant analysis. Furthermore, changes in the biochemical environment and altered metabolism due to disease-associated anatomical, pathological, and functional changes present even more challenges to CEST MRI. The desirable single metabolite-targeted CEST imaging typically can be contaminated by other biomolecules as current metabolite-specific CEST pulse sequences are not specific enough. In this context, efforts have been made to develop sophisticated data acquisition schemes or methodologies and CEST imaging sequences for measuring exchange rates more accurately and quantitatively123 or filtering out interfering protons in a specific exchange rate range to “purify” the measured CEST signal.124,125 It should be noted that many exogenous agents are inherently CEST contrast agents and are already entering the phases of clinical trials.109
Figure 11.
(A) Schematic representation of brain Z-spectrum with typical contributions including direct water saturation (DS), magnetization transfer contrast (MTC), amideNOE, aromaticNOE, aliphaticNOE, amideCEST, CrCEST, PCrCEST, hydroxylCEST, and amineCEST. The DS component is also included in the amineCEST and hyroxylCEST line shapes for reference only. The CEST contributions are plotted in B–E. (B) The non-CEST saturation transfer processes contribute to the brain Z-spectrum. MTC is a strong and broad signal centering around −3.5 to −3 ppm. Amide and aromatic NOE peaks are distributed from 2 to 5 ppm, while aliphaticNOE centers are at −3.5 to −3 ppm. (C) The CEST signals from the protons with slow to intermediate exchange rates (e.g., ArgCEST and CrCEST at 2 ppm, amideCEST at 3.5 ppm, and PCrCEST at 2 and 2.5–2.6 ppm). (D) The peak locations of the amine protons from glutamate (3 ppm) and protein (2.7 ppm), as well as hydroxyl protons (1 ppm). (E) The amine and hydroxyl CEST signal coalesce with the water peak due to higher exchange rates (>1000 s–1). A–E: Reproduced with permission from ref (115). Copyright 2022 John Wiley and Sons, Ltd.
Future Development and Perspectives of Label-Free Molecular MRI
As the field of molecular imaging grows further, the concepts and approaches of using endogenous biological and chemical conditions and specific molecules to enable label-free molecular MRI and examples highlighted in the review have pointed to an essential direction for future development that can address the unmet need of filling the gap between the scientific discoveries and the translational and clinical needs. A broad range of opportunities and challenging areas need multidisciplinary efforts, including, but not limited to, these areas. First, using endogenous biomolecules as probes for the chemical and biological events in vivo can be limited by intrinsically low probe concentrations. Furthermore, background interferences from other residing biomolecules with similar biological and chemical properties can dampen the sensitivity and specificity of the detection and quantification of the probe. These two most critical standards in molecular imaging can be improved by continuous advances in the engineering and technology of MRI scanners. For example, increasing the field strength of the magnetic field, or B0, from the current clinical standard of 3T to 7T naturally should more than double the sensitivity and, in some cases, specificity by increasing the spectral resolution in the chemical shift domain in the MRS applications or broadening the dynamic range and magnetic transfer to capture more CEST effects and molecules in CEST MRI. Notably, 7T whole-body MRI scanners have gained FDA approval recently in certain clinical applications. With the increased sensitivity, dynamic range and T1 relaxation time in the high field strength, heteronuclear imaging with low natural abundance stable isotope 31P and 23Na MRI, which has been shown to provide exquisite information to many diseases,126,127 may become clinically feasible in the near future. In addition, parallel data acquisition approaches are well-integrated into the clinical application,128 shortening the otherwise long scan time needed for sufficient signals. Given the rapid advances in data science and artificial intelligence in medical imaging, machine-learning, more recently, deep-learning methods are increasingly used to improve the sensitivity and specificity.129,130 For example, noises and signals in MRS and MRI data can be classified and separated, followed by removing noises to reconstruct the “signal-only” spectra and images.128 Implementation of parallel data acquisition can be coupled with intelligent signal-selective data combination approaches to further improve the signal-to-noise ratio.131 For chemically and molecularly targeted imaging, novel and sophisticatedly designed RF or B1 systems, typically transmission and/or receiving coils, can deliver well-controlled pulses, such as B1, accurately tuned to the frequency of the targeted molecules.132 However, these engineering and customized improvements need to be guided by the understanding of the imaging contrast mechanism and, more importantly, the biological and chemical properties of the probes that lead to the NMR properties and MRI contrast. The other area that warrants specific attentions and in-depth investigations is changes in chemical and magnetic properties of endogenous molecules or probes in the tissue environment or physiological conditions that are different from those documented in the solution or cultural medium conditions in vitro. A simple example is that T1 or T2 relaxation times of metabolites in the brain gray matter are different from the white matter that is composed of dense axon fibers, leading to the tissue compartment-related apparent difference that should be taken into account when quantifying the level of neuro-metabolites, such as Glu and GABA. Indeed, those changes in the probes provide information on the biological environment if they can be detectable and measurable by a specific MRI method. However, there are significant gaps of knowledge on how and what differences in the physiological and biological environment make changes to the MRI properties of the probes, even though they are well characterized in vitro. The data and parameters derived from further investigations in this area are essential to improve the modeling and quantification of endogenous biomarkers and the optimization of imaging methods. More importantly, these pieces of knowledge are necessary for appropriate interpretations of the data and results for accurate diagnosis of clinical conditions that these molecular imaging methods are intended for. In this regard, there are still needs in various levels of preclinical investigations using both in vitro cell or tissue culture-based experiments and proper animal models of diseases. Lastly, as label-free molecular imaging approaches and capabilities can be readily available for clinical applications, there are still steps of clinical validation and evaluation before being implemented and integrated into clinical practices. The paradigm-shifting changes often need to show substantial improvements in clinical benefits. Therefore, the chemically and molecularly targeted imaging approaches, data acquisition technology, and interpretation of the results, which may require in-depth knowledge of chemistry and biology, need to be introduced and accepted by the clinical communities that manage different diseases and healthcare problems. On the other hand, having many label-free molecular imaging capabilities available on clinically approved scanner platforms provide a significant advantage in accelerating clinical validations and trials.
Certainly, MRI-based molecular imaging for precision medicine can be empowered substantially with the development of exogenous probes with chemical biology and materials engineering that can overcome the limitations of label-free molecular imaging. Given the rich chemistry and wealthy molecular biology approaches already available and continuously developed, the development of molecular imaging probes and molecularly targeted contrast agents for MRI should remain an exciting and growing research area. Within the same topic of using endogenous biological and physiological relevant molecules as probes as we discussed in the label-free molecular MRI, using exogenously introduced nonradioactive isotope “labeled” versions of endogenous metabolites, such as 13C-glucose, 13C-pyruvate, and 13C-lactate, enables hyperpolarized (HP) 13C MRS to study fast metabolic conversions in vivo with sensitivity ≤4 orders of magnitude higher than conventional MRS signals. Thus, there are increasing needs and opportunities to rationally design and synthesize novel 13C and 15N “labeled” hyperpolarized molecular probes based on different substrates for detection and quantifying biochemical and metabolic processes and the biomolecules involved using HP-MRS.133 Similarly, without chemical labeling, molecules with the stable isotope deuterium (D, i.e., 2H) instead of the proton, can be directly used for MRI tuned to the frequency of 2H not proton 1H. Indeed, d-glucose and its derivatives, such as 3-oxy-methyl-d-glucose (3OMG) have been applied for MRI of glucose uptake and consumption in animals and humans through its exchangeable hydroxyl proton groups by CEST (glucoCEST)134,135 or glucose-based chemical exchange-sensitive spin-lock (glucoCESL),136 which are being tested clinically now.137,138 Arguably, the most accessible and cost-effective stable-isotope labeled molecule is deuterium oxide (D2O). While D2O has been explored for measuring blood flow and perfusion in normal organs139 and tumors,140 no clinical application of D2O MRI has been reported, likely due to the low sensitivity (i.e., ∼6.5-times lower than 1H) and limited availability of 2H-tuned MRI hardware. However, D2O can be used to generate negative contrast in the 1H MRI due to hypointensity caused by replacing a fraction of H2O with D2O (also known as the proton replace effect). “Dark Water” MRI was demonstrated to measure cerebral perfusion in rat brains and track the spatial distribution of endovascularly infused drug solutions.141 This approach can be directly used on clinical 3T MRI scanners, as demonstrated by our study in a dog model (Figure 12), to accurately visualize the perfusion territory of intra-arterial drug delivery and predict the region with BBB opening.142
Figure 12.
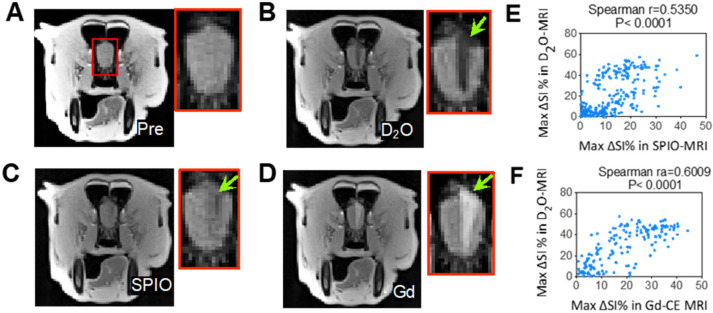
3T D2O MRI can guide the intervention for the hyperosmotic opening of BBB in a dog. (A) T2w image preinjection. (B) D2O MRI contrast-enhancement map. (C) SPIO MRI contrast-enhancement map. (D) Gd-contrast-enhancement map after the administration of mannitol. (E) Correlation between SPIO- and D2O-MRI. The Spearman coefficient (r) = 05350. (F) Correlation between Gd- and D2O- MRI. The Spearman coefficient (r) = 0.6009. Reprinted with permission from ref (142). Copyright 2021 Ivyspring International Publisher.
Using endogenous biomolecules for molecular imaging also inspires many innovations in developing exogenous imaging probes. For example, using iron-storage proteins (e.g., ferritin) for T2* imaging or QSM quantification for iron in vivo led to the development of MRI reporter genes that can be incorporated into the therapeutic cells for MRI tracking of the cell transplants and other cell-based therapies.143,144 On the other hand, many drugs and therapeutic agents are inherently CEST contrast agents, including some already entering the phases of clinical trials.98 Thus, these compounds can be used as theranostic agents that can include image-guided drug delivery in their clinical applications to improve the efficacy and timely assess treatment responses, as demonstrated in a number of previous studies.145−147
Conclusion
Modern MRI technology not only offers a primary clinical imaging modality for disease diagnosis, prognosis, and treatment monitoring but also enables label-free yet chemically and molecularly selective molecular imaging tools for scientific discovery, preclinical investigation, and biomarker-specific noninvasive imaging characterization of the diseases in clinical management of patients. Combining advanced MRI contrast generation and acquisition strategies with endogenous molecules as imaging probes allows timely clinical translation and adoption of molecular imaging approaches to precision medicine-focused patient care. Notably, the approaches developed and demonstrated for label-free molecular imaging with MRI also facilitate innovation and development of chemically and biologically modified or engineered imaging probes that can expand and advance molecular imaging capabilities.
Glossary
Vocabulary
- Label-Free
Molecules, e.g., imaging probes, are used in their original forms without chemical modifications or covalent attachment of other molecules or isotopes.
- Molecular Imaging
A noninvasive imaging approach that enables visualization of chemical, biological and physiological processes at the molecular level.
- Magnetic resonance imaging (MRI)
As a noninvasive medical imaging technology, MRI generates detailed images of almost every internal structure in a living system, such as the human body, including the organs, bones, muscles and blood vessels, and biological properties or even functions of the tissue.
- Proton relaxation time
The relaxation times of protons, typically described as longitudinal (T1) or transverse (T2) relaxation times, are the magnetic properties of the tissue and basic parameters used for MRI to generate the image contrast between different types of tissue.
- Endogenous probes
Small biochemical or molecules presented or found within cells or tissue emit signals or produce contrast detectable by the corresponding imaging modality. These probes can be then used to report the biological or physiological or metabolic processes in vivo by imaging.
- Chemical exchange saturation transfer (CEST)
It is a chemical phenomenon or effect that is used to generate MRI contrast in which a molecule containing either exchangeable protons or exchangeable molecules are selectively saturated by radiofrequency selected irradiation, then indirectly detected through the exchange patterns, e.g., water signals or molecules.
- Theranostics
Approaches and materials that combine diagnostic and therapeutic capabilities or functions are considered as “theranostics”.
This work is supported in parts by the grants from NIH (R01CA203388–04, R01AG067736–02) to HM, (R01CA261974, R33HL161756) to GL. CCF is supported, in part, by NIH grant DP2NS127704–01.
The authors declare no competing financial interest.
References
- Weissleder R. Molecular Imaging: Exploring the Next Frontier. Radiology 1999, 212 (3), 609–614. 10.1148/radiology.212.3.r99se18609. [DOI] [PubMed] [Google Scholar]
- Weissleder R.; Mahmood U. Molecular Imaging. Radiology 2001, 219 (2), 316–333. 10.1148/radiology.219.2.r01ma19316. [DOI] [PubMed] [Google Scholar]
- Herschman H. R. Molecular Imaging: Looking at Problems, Seeing Solutions. Science 2003, 302 (5645), 605–608. 10.1126/science.1090585. [DOI] [PubMed] [Google Scholar]
- Danthi S. N.; Pandit S. D.; Li K. C. A Primer on Molecular Biology for Imagers: Vii. Molecular Imaging Probes1. Academic Radiology 2004, 11, 1047–1054. 10.1016/j.acra.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Ottobrini L.; Ciana P.; Biserni A.; Lucignani G.; Maggi A. Molecular Imaging: A New Way to Study Molecular Processes in vivo. Mol. Cell. Endocrinol. 2006, 246 (1–2), 69–75. 10.1016/j.mce.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Boyce M.; Bertozzi C. R. Bringing Chemistry to Life. Nat. Methods 2011, 8 (8), 638–642. 10.1038/nmeth.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks K. M.; Nolan G. P. Chemical Labeling Strategies for Cell Biology. Nat. Methods 2006, 3 (8), 591–596. 10.1038/nmeth906. [DOI] [PubMed] [Google Scholar]
- Bengel F. M.; Gambhir S. S.. Clinical Molecular Imaging and Therapy—Moving Ahead Together; Springer, 2005; Vol. 32, pp S323–S323. [DOI] [PubMed] [Google Scholar]
- Miller J. C.; Thrall J. H. Clinical Molecular Imaging. Journal of the American College of Radiology 2004, 1 (1), 4–23. 10.1016/S1546-1440(03)00025-5. [DOI] [PubMed] [Google Scholar]
- Jaffer F. A.; Weissleder R. Molecular Imaging in the Clinical Arena. Jama 2005, 293 (7), 855–862. 10.1001/jama.293.7.855. [DOI] [PubMed] [Google Scholar]
- Kipper M. S. Steps in Moving Molecular Imaging to the Clinic. J. Nucl. Med. 2008, 49 (6), 58N–60N. [PubMed] [Google Scholar]
- Conti P. S.; McEwan A. J.; Pomper M. G. Molecular Imaging: The Future of Modern Medicine. J. Nucl. Med. 2008, 49 (6), 16N–20N. [PubMed] [Google Scholar]
- Seto B.; McLaughlin A.; Li K.; Menkens A.; Sullivan D.; Pettigrew R. Advancing the Boundaries of Molecular Imaging. Biol. Psychiatry 2006, 59 (10), 881–882. 10.1016/j.biopsych.2005.07.037. [DOI] [PubMed] [Google Scholar]
- Golman K.; Ardenkjaer-Larsen J. H.; Petersson J. S.; Mansson S.; Leunbach I. Molecular Imaging with Endogenous Substances. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (18), 10435–10439. 10.1073/pnas.1733836100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R.; Ma C.; Li Y.; Zhao Y.; Wang T.; Li Y.; El Fakhri G.; Liang Z.-P.. High-Resolution Label-Free Molecular Imaging of Brain Tumor. In 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC); IEEE, 2021; pp 3049–3052. 10.1109/EMBC46164.2021.9630623. [DOI] [PubMed]
- Harisinghani M. G.; O’Shea A.; Weissleder R. Advances in Clinical MRI Technology. Science Translational Medicine 2019, 11 (523), eaba2591 10.1126/scitranslmed.aba2591. [DOI] [PubMed] [Google Scholar]
- Edelman R. R.Clinical Magnetic Resonance Imaging; Saunders Elsevier, 2006. [Google Scholar]
- Hormuth D. A.; Sorace A. G.; Virostko J.; Abramson R. G.; Bhujwalla Z. M.; Enriquez-Navas P.; Gillies R.; Hazle J. D.; Mason R. P.; Quarles C. C.; et al. Translating Preclinical MRI Methods to Clinical Oncology. Journal of Magnetic Resonance Imaging 2019, 50 (5), 1377–1392. 10.1002/jmri.26731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha L.; Horvath I.; Ferreira S.; Lemos J.; Costa P.; Vieira D.; Veres D. S.; Szigeti K.; Summavielle T.; Máthé D.; et al. Preclinical Imaging: An Essential Ally in Modern Biosciences. Molecular diagnosis & therapy 2014, 18 (2), 153–173. 10.1007/s40291-013-0062-3. [DOI] [PubMed] [Google Scholar]
- Koo V.; Hamilton P.; Williamson K. Non-Invasive in vivo Imaging in Small Animal Research. Analytical Cellular Pathology 2006, 28 (4), 127–139. 10.1155/2006/245619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtea C.; Laurent S.; Vander Elst L.; Muller R.. Molecular Imaging I; Springer: Berlin, 2008. [Google Scholar]
- Wahsner J.; Gale E. M.; Rodriguez-Rodriguez A.; Caravan P. Chemistry of MRI Contrast Agents: Current Challenges and New Frontiers. Chem. Rev. 2019, 119 (2), 957–1057. 10.1021/acs.chemrev.8b00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.-D.; Paudel R.; Liu J.; Ma C.; Zhang Z.-S.; Zhou S.-K. MRI Contrast Agents: Classification and Application. International journal of molecular medicine 2016, 38 (5), 1319–1326. 10.3892/ijmm.2016.2744. [DOI] [PubMed] [Google Scholar]
- Hahn P.; Granick S.; Bale W. F.; Michaelis L. Ferritin. 6. Conversion of Inorganic and Hemoglobin Iron into Ferritin Iron in the Animal Body. Storage Function of Ferritin Iron as Shown by Radioactive and Magnetic Measurements. J. Biol. Chem. 1943, 150, 407–4127. 10.1016/S0021-9258(18)72163-7. [DOI] [Google Scholar]
- Faghihi R.; Zeinali-Rafsanjani B.; Mosleh-Shirazi M.-A.; Saeedi-Moghadam M.; Lotfi M.; Jalli R.; Iravani V. Magnetic Resonance Spectroscopy and Its Clinical Applications: A Review. Journal of Medical Imaging and Radiation Sciences 2017, 48 (3), 233–253. 10.1016/j.jmir.2017.06.004. [DOI] [PubMed] [Google Scholar]
- Hwang J.-H.; Choi C. S. Use of in Vivo Magnetic Resonance Spectroscopy for Studying Metabolic Diseases. Experimental Molecular Medicine 2015, 47 (2), e139 10.1038/emm.2014.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies R. J.; Morse D. L. In Vivo Magnetic Resonance Spectroscopy in Cancer. Annu. Rev. Biomed Eng. 2005, 7, 287–326. 10.1146/annurev.bioeng.7.060804.100411. [DOI] [PubMed] [Google Scholar]
- Serkova N. J.; Hasebroock K. M.; Kraft S. L. Magnetic Resonance Spectroscopy of Living Tissues. Tumor Biomarker Discovery 2009, 520, 315–327. 10.1007/978-1-60327-811-9_22. [DOI] [PubMed] [Google Scholar]
- Tognarelli J. M.; Dawood M.; Shariff M. I.; Grover V. P.; Crossey M. M.; Cox I. J.; Taylor-Robinson S. D.; McPhail M. J. Magnetic Resonance Spectroscopy: Principles and Techniques: Lessons for Clinicians. Journal of clinical and experimental hepatology 2015, 5 (4), 320–328. 10.1016/j.jceh.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares D.; Law M. Magnetic Resonance Spectroscopy of the Brain: Review of Metabolites and Clinical Applications. Clinical radiology 2009, 64 (1), 12–21. 10.1016/j.crad.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Aseel A.; McCarthy P.; Mohammed A. Brain Magnetic Resonance Spectroscopy to Differentiate Recurrent Neoplasm from Radiation Necrosis: A Systematic Review and Meta-Analysis. Journal of Neuroimaging 2023, 33, 189. 10.1111/jon.13080. [DOI] [PubMed] [Google Scholar]
- Mountford C.; Ramadan S.; Stanwell P.; Malycha P. Proton MRS of the Breast in the Clinical Setting. NMR in Biomedicine 2009, 22 (1), 54–64. 10.1002/nbm.1301. [DOI] [PubMed] [Google Scholar]
- Sharma U.; Jagannathan N. R. Magnetic Resonance Imaging (MRI) and MR Spectroscopic Methods in Understanding Breast Cancer Biology and Metabolism. Metabolites 2022, 12 (4), 295. 10.3390/metabo12040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Lisse U. G.; Scherr M. K. Proton MR Spectroscopy of the Prostate. European journal of radiology 2007, 63 (3), 351–360. 10.1016/j.ejrad.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Stamatelatou A.; Scheenen T. W.; Heerschap A. Developments in Proton MR Spectroscopic Imaging of Prostate Cancer. Magnetic Resonance Materials in Physics, Biology and Medicine 2022, 35 (4), 1–21. 10.1007/s10334-022-01011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perman W. H.; Balci N. C.; Akduman I. Review of Magnetic Resonance Spectroscopy in the Liver and the Pancreas. Topics in Magnetic Resonance Imaging 2009, 20 (2), 89–97. 10.1097/RMR.0b013e3181c422f1. [DOI] [PubMed] [Google Scholar]
- Gursan A.; Prompers J. J. Magnetic Resonance Imaging and Spectroscopy Methods to Study Hepatic Glucose Metabolism and Their Applications in the Healthy and Diabetic Liver. Metabolites 2022, 12 (12), 1223. 10.3390/metabo12121223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley J. K.; Redpath T. W.; Bolan P. J.; Gilbert F. J. In Vivo Proton Magnetic Resonance Spectroscopy of Breast Cancer: A Review of the Literature. Breast Cancer Res. 2012, 14 (2), 207. 10.1186/bcr3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzola-Aldamizetxebarria S.; Fernández-Méndez L.; Padro D.; Ruíz-Cabello J.; Ramos-Cabrer P. A Comprehensive Introduction to Magnetic Resonance Imaging Relaxometry and Contrast Agents. ACS Omega 2022, 7 (42), 36905–36917. 10.1021/acsomega.2c03549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landheer K.; Gajdošík M.; Treacy M.; Juchem C. Concentration and Effective T2 Relaxation Times of Macromolecules at 3T. Magn. Reson. Med. 2020, 84 (5), 2327–2337. 10.1002/mrm.28282. [DOI] [PubMed] [Google Scholar]
- Murali-Manohar S.; Borbath T.; Wright A. M.; Soher B.; Mekle R.; Henning A. T2 Relaxation Times of Macromolecules and Metabolites in the Human Brain at 9.4 T. Magn. Reson. Med. 2020, 84 (2), 542–558. 10.1002/mrm.28174. [DOI] [PubMed] [Google Scholar]
- Novotny E.; Ashwal S.; Shevell M. Proton Magnetic Resonance Spectroscopy: An Emerging Technology in Pediatric Neurology Research. Pediatric research 1998, 44 (1), 1–10. 10.1203/00006450-199807000-00001. [DOI] [PubMed] [Google Scholar]
- Vigneron D. B. Magnetic Resonance Spectroscopic Imaging of Human Brain Development. Neuroimaging Clinics 2006, 16 (1), 75–85. 10.1016/j.nic.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Brateman L. Chemical Shift Imaging: A Review. American Journal of Roentgenology 1986, 146 (5), 971–980. 10.2214/ajr.146.5.971. [DOI] [PubMed] [Google Scholar]
- Horská A.; Barker P. B. Imaging of Brain Tumors: MR Spectroscopy and Metabolic Imaging. Neuroimaging Clinics 2010, 20 (3), 293–310. 10.1016/j.nic.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J. W.; Kuzniecky R. I. Utility of Magnetic Resonance Spectroscopic Imaging for Human Epilepsy. Quantitative imaging in medicine and surgery 2015, 5 (2), 313. 10.3978/j.issn.2223-4292.2015.01.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordova J. S.; Shu H. K.; Liang Z.; Gurbani S. S.; Cooper L. A.; Holder C. A.; Olson J. J.; Kairdolf B.; Schreibmann E.; Neill S. G.; Hadjipanayis C. G.; Shim H. Whole-brain spectroscopic MRI biomarkers identify infiltrating margins in glioblastoma patients. Neuro-oncology 2016, 18 (8), 1180–1189. 10.1093/neuonc/now036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qayyum A. MR Spectroscopy of the Liver: Principles and Clinical Applications. Radiographics 2009, 29 (6), 1653. 10.1148/rg.296095520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban R. S. MRS of the Kidney. Investigative Radiology 1989, 24 (12), 988–992. 10.1097/00004424-198912000-00014. [DOI] [PubMed] [Google Scholar]
- Krššák M.; Lindeboom L.; Schrauwen-Hinderling V.; Szczepaniak L. S.; Derave W.; Lundbom J.; Befroy D.; Schick F.; Machann J.; Kreis R.; et al. Proton Magnetic Resonance Spectroscopy in Skeletal Muscle: Experts’ Consensus Recommendations. NMR in Biomedicine 2021, 34 (5), e4266 10.1002/nbm.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Gu Y.; Yu X. Assessing Tissue Metabolism by Phosphorous-31 Magnetic Resonance Spectroscopy and Imaging: A Methodology Review. Quantitative imaging in medicine and surgery 2017, 7 (6), 707. 10.21037/qims.2017.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh K.; Mellon E. A.; Gurbani S. S.; Weinberg B. D.; Schreibmann E.; Sheriff S. A.; Goryawala M.; de le Fuente M.; Eaton B. R.; Zhong J.; et al. A Multi-Institutional Pilot Clinical Trial of Spectroscopic MRI-Guided Radiation Dose Escalation for Newly Diagnosed Glioblastoma. Neuro-oncology advances 2022, 4 (1), vdac006. 10.1093/noajnl/vdac006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina J.; Carroll A.; Wang L.; Yu Q.; Mancheno D. E.; Wu S.; Liu F.; Ahn J.; He M.; Mao H.; et al. Detection of “Oncometabolite” 2-Hydroxyglutarate by Magnetic Resonance Analysis as a Biomarker of IDH1/2 Mutations in Glioma. Journal of molecular medicine 2012, 90 (10), 1161–1171. 10.1007/s00109-012-0888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkhaled A.; Jalbert L. E.; Phillips J. J.; Yoshihara H. A.; Parvataneni R.; Srinivasan R.; Bourne G.; Berger M. S.; Chang S. M.; Cha S.; et al. Magnetic Resonance of 2-Hydroxyglutarate in IDH1-Mutated Low-Grade Gliomas. Science translational medicine 2012, 4 (116), 116ra115–116ra115. 10.1126/scitranslmed.3002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C.; Ganji S. K.; DeBerardinis R. J.; Hatanpaa K. J.; Rakheja D.; Kovacs Z.; Yang X.-L.; Mashimo T.; Raisanen J. M.; Marin-Valencia I.; et al. 2-Hydroxyglutarate Detection by Magnetic Resonance Spectroscopy in IDH-Mutated Patients with Gliomas. Nature medicine 2012, 18 (4), 624–629. 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leather T.; Jenkinson M. D.; Das K.; Poptani H. Magnetic Resonance Spectroscopy for Detection of 2-Hydroxyglutarate as a Biomarker for IDH Mutation in Gliomas. Metabolites 2017, 7 (2), 29. 10.3390/metabo7020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andronesi O. C.; Loebel F.; Bogner W.; Marjanska M.; Vander Heiden M. G.; Iafrate A. J.; Dietrich J.; Batchelor T. T.; Gerstner E. R.; Kaelin W. G.; et al. Treatment Response Assessment in IDH-Mutant Glioma Patients by Noninvasive 3D Functional Spectroscopic Mapping of 2-Hydroxyglutarate. Clin. Cancer Res. 2016, 22 (7), 1632–1641. 10.1158/1078-0432.CCR-15-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy A. R.; Najac C.; Viswanath P.; Lakhani A.; Subramani E.; Batsios G.; Radoul M.; Gillespie A. M.; Pieper R. O.; Ronen S. M. MR-Detectable Metabolic Biomarkers of Response to Mutant IDH Inhibition in Low-Grade Glioma. Theranostics 2020, 10 (19), 8757. 10.7150/thno.47317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batsios G.; Viswanath P.; Subramani E.; Najac C.; Gillespie A. M.; Santos R. D.; Molloy A. R.; Pieper R. O.; Ronen S. M. PI3K/mTOR Inhibition of IDH1Mutant Glioma Leads to Reduced 2HG Production That Is Associated with Increased Survival. Sci. Rep. 2019, 9 (1), 1–15. 10.1038/s41598-019-47021-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.; Demenescu L. R.; Colic L.; Metzger C. D.; Heinze H.-J.; Steiner J.; Speck O.; Fejtova A.; Salvadore G.; Walter M. Temporal Dynamics of Antidepressant Ketamine Effects on Glutamine Cycling Follow Regional Fingerprints of AMPA and NMDA Receptor Densities. Neuropsychopharmacology 2017, 42 (6), 1201–1209. 10.1038/npp.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X. G.; Ju S.; Fang F.; Wang Y.; Fang K.; Cui X.; Liu G.; Li P.; Mao H.; Teng G. J. (2013). Comparison of brown and white adipose tissue fat fractions in ob, seipin, and Fsp27 gene knockout mice by chemical shift-selective imaging and 1H-MR spectroscopy. American Journal of Physiology. Endocrinology and Metabolism 2013, 304 (2), E160 10.1152/ajpendo.00401.2012. [DOI] [PubMed] [Google Scholar]
- Lutz N. W.; Bernard M. Contactless Thermometry by MRI and MRS: Advanced Methods for Thermotherapy and Biomaterials. Iscience 2020, 23 (10), 101561. 10.1016/j.isci.2020.101561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies R. J.; Raghunand N.; Karczmar G. S.; Bhujwalla Z. M. MRI of the Tumor Microenvironment. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine 2002, 16 (4), 430–450. 10.1002/jmri.10181. [DOI] [PubMed] [Google Scholar]
- Le Bihan D; Breton E; Lallemand D; Grenier P; Cabanis E; Laval-Jeantet M MR Imaging of Intravoxel Incoherent Motions: Application to Diffusion and Perfusion in Neurologic Disorders. Radiology 1986, 161 (2), 401–407. 10.1148/radiology.161.2.3763909. [DOI] [PubMed] [Google Scholar]
- Detre J. A.; Wang J.; Wang Z.; Rao H. Arterial Spin-Labeled Perfusion MRI in Basic and Clinical Neuroscience. Current opinion in neurology 2009, 22 (4), 348–355. 10.1097/WCO.0b013e32832d9505. [DOI] [PubMed] [Google Scholar]
- Chifman J.; Laubenbacher R.; Torti S. V. A Systems Biology Approach to Iron Metabolism. A Systems Biology Approach to Blood 2014, 844, 201–225. 10.1007/978-1-4939-2095-2_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-G.; Ogawa S. Biophysical and Physiological Origins of Blood Oxygenation Level-Dependent fMRI Signals. Journal of Cerebral Blood Flow & Metabolism 2012, 32 (7), 1188–1206. 10.1038/jcbfm.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. C. Use of Magnetic Resonance Imaging to Monitor Iron Overload. Hematology/Oncology Clinics 2014, 28 (4), 747–764. 10.1016/j.hoc.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser P. J.; Pajevic S.; Pierpaoli C.; Duda J.; Aldroubi A. In Vivo Fiber Tractography Using DR-MRI Data. Magn. Reson. Med. 2000, 44 (4), 625–632. . [DOI] [PubMed] [Google Scholar]
- Lazar M. Mapping Brain Anatomical Connectivity Using White Matter Tractography. NMR in Biomedicine 2010, 23 (7), 821–835. 10.1002/nbm.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya J. G. Techniques and Applications of in Vivo Diffusion Imaging of Articular Cartilage. Journal of magnetic resonance imaging 2015, 41 (6), 1487–1504. 10.1002/jmri.24767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M.; Albers G. W. Applications of diffusion-perfusion magnetic resonance imaging in acute ischemic stroke. Neurology 1999, 52 (9), 1750–1756. 10.1212/WNL.52.9.1750. [DOI] [PubMed] [Google Scholar]
- Tamada T.; Ueda Y.; Ueno Y.; Kojima Y.; Kido A.; Yamamoto A. Diffusion-Weighted Imaging in Prostate Cancer. J. Magn. Magn. Mater. 2022, 35 (4), 533–547. 10.1007/s10334-021-00957-6. [DOI] [PubMed] [Google Scholar]
- Moffat B. A.; Chenevert T. L.; Lawrence T. S.; Meyer C. R.; Johnson T. D.; Dong Q.; Tsien C.; Mukherji S.; Quint D. J.; Gebarski S. S.; et al. Functional Diffusion Map: A Noninvasive MRI Biomarker for Early Stratification of Clinical Brain Tumor Response. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (15), 5524–5529. 10.1073/pnas.0501532102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellingson B. M.; Malkin M. G.; Rand S. D.; Connelly J. M.; Quinsey C.; LaViolette P. S.; Bedekar D. P.; Schmainda K. M. Validation of Functional Diffusion Maps (fDMs) as a Biomarker for Human Glioma Cellularity. Journal of Magnetic Resonance Imaging 2010, 31 (3), 538–548. 10.1002/jmri.22068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. S.; Detre J. A.; Leigh J. S.; Koretsky A. P. Magnetic Resonance Imaging of Perfusion Using Spin Inversion of Arterial Water. Proc. Natl. Acad. Sci. U. S. A. 1992, 89 (1), 212–216. 10.1073/pnas.89.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsop D. C.; Detre J. A.; Golay X.; Günther M.; Hendrikse J.; Hernandez-Garcia L.; Lu H.; MacIntosh B. J.; Parkes L. M.; Smits M.; et al. Recommended Implementation of Arterial Spin-Labeled Perfusion MRI for Clinical Applications: A Consensus of the Ismrm Perfusion Study Group and the European Consortium for ASL in Dementia. Magn. Reson. Med. 2015, 73 (1), 102–116. 10.1002/mrm.25197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhave G.; Lewis J. B.; Chang S. S. Association of Gadolinium Based Magnetic Resonance Imaging Contrast Agents and Nephrogenic Systemic Fibrosis. Journal of urology 2008, 180 (3), 830–835. 10.1016/j.juro.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller S.; Zaharchuk G.; Thomas D. L.; Lovblad K.-O.; Barkhof F.; Golay X. Arterial Spin Labeling Perfusion of the Brain: Emerging Clinical Applications. Radiology 2016, 281 (2), 337–356. 10.1148/radiol.2016150789. [DOI] [PubMed] [Google Scholar]
- Radovic T.; Jankovic M. M.; Stevic R.; Spasojevic B.; Cvetkovic M.; Pavicevic P.; Gojkovic I.; Kostic M. Detection of impaired renal allograft function in paediatric and young adult patients using arterial spin labelling MRI (ASL-MRI). Sci. Rep. 2022, 12 (1), 828. 10.1038/s41598-022-04794-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel S. A.; Song A. W.; McCarthy G. Functional Magnetic Resonance Imaging. Yale J. Biol. Med. 2009, 82 (4), 233. [Google Scholar]
- Hall M. E.; Jordan J. H.; Juncos L. A.; Hundley W. G.; Hall J. E. BOLD Magnetic Resonance Imaging in Nephrology. International journal of nephrology and renovascular disease 2018, 11, 103. 10.2147/IJNRD.S112299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.; Liu H.; Li X.; Zhou L.; Wang R.; Zhang Y. Application of BOLD-MRI in the Classification of Renal Function in Chronic Kidney Disease. Abdominal Radiology 2019, 44 (2), 604–611. 10.1007/s00261-018-1750-6. [DOI] [PubMed] [Google Scholar]
- Zhang L. J.; Zhang Z.; Xu J.; Jin N.; Luo S.; Larson A. C.; Lu G. M. Carbogen Gas-Challenge Blood Oxygen Level-Dependent Magnetic Resonance Imaging in Hepatocellular Carcinoma: Initial Results. Oncology Letters 2015, 10 (4), 2009–2014. 10.3892/ol.2015.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller S.; Haacke E. M.; Thurnher M. M.; Barkhof F. Susceptibility-Weighted Imaging: Technical Essentials and Clinical Neurologic Applications. Radiology 2021, 299 (1), 3–26. 10.1148/radiol.2021203071. [DOI] [PubMed] [Google Scholar]
- Langkammer C.; Pirpamer L.; Seiler S.; Deistung A.; Schweser F.; Franthal S.; Homayoon N.; Katschnig-Winter P.; Koegl-Wallner M.; Pendl T.; Stoegerer E. M.; Wenzel K.; Fazekas F.; Ropele S.; Reichenbach J. R.; Schmidt R.; Schwingenschuh P. Quantitative Susceptibility Mapping in Parkinson’s Disease. PloS One 2016, 11 (9), e0162460 10.1371/journal.pone.0162460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y.; Nguyen T. D.; Pandya S.; Zhang Y.; Rúa S. H.; Kovanlikaya I.; Kuceyeski A.; Liu Z.; Wang Y.; Gauthier S. A. Combining Quantitative Susceptibility Mapping with Automatic Zero Reference (QSM0) and Myelin Water Fraction Imaging to Quantify Iron-Related Myelin Damage in Chronic Active MS Lesions. American Journal of Neuroradiology 2018, 39 (2), 303–310. 10.3174/ajnr.A5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward K. M.; Aletras A. H.; Balaban R. S. A New Class of Contrast Agents for MRI Based on Proton Chemical Exchange Dependent Saturation Transfer (CEST). J. Magn. Reson. 2000, 143 (1), 79–87. 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- Forsén S.; Hoffman R. A. Study of Moderately Rapid Chemical Exchange Reactions by Means of Nuclear Magnetic Double Resonance. J. Chem. Phys. 1963, 39 (11), 2892–2901. 10.1063/1.1734121. [DOI] [Google Scholar]
- van Zijl P. C.; Yadav N. Chemical Exchange Saturation Transfer (CEST): What Is in a Name and What Isn’t?. Magn Reson Med. 2011, 65, 927–948. 10.1002/mrm.22761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. M.; Pollard A. C.; Pagel M. D. Clinical Applications of Chemical Exchange Saturation Transfer (CEST) MRI. Journal of Magnetic Resonance Imaging 2018, 47 (1), 11–27. 10.1002/jmri.25838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.; Song X.; Chan K. W.; McMahon M. T. Nuts and Bolts of Chemical Exchange Saturation Transfer MRI. NMR Biomed 2013, 26 (7), 810–828. 10.1002/nbm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Chu C.; Zhang J.; Bie C.; Chen L.; Aafreen S.; Xu J.; Kamson D. O.; van Zijl P. C.; Walczak P.; et al. Label-Free Assessment of Mannitol Accumulation Following Osmotic Blood–Brain Barrier Opening Using Chemical Exchange Saturation Transfer Magnetic Resonance Imaging. Pharmaceutics 2022, 14 (11), 2529. 10.3390/pharmaceutics14112529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss M.; Bachert P. Chemical Exchange Saturation Transfer (CEST) and MR Z-Spectroscopy in Vivo: A Review of Theoretical Approaches and Methods. Physics in Medicine & Biology 2013, 58 (22), R221. 10.1088/0031-9155/58/22/R221. [DOI] [PubMed] [Google Scholar]
- Zaiss M.; Jin T.; Kim S. G.; Gochberg D. F. Theory of Chemical Exchange Saturation Transfer MRI in the Context of Different Magnetic Fields. NMR in Biomedicine 2022, 35 (11), e4789 10.1002/nbm.4789. [DOI] [PubMed] [Google Scholar]
- Han Z.; Liu G. Sugar-Based Biopolymers as Novel Imaging Agents for Molecular Magnetic Resonance Imaging. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology 2019, 11 (4), e1551 10.1002/wnan.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo D. L.; Carella A.; Corrado A.; Pirotta E.; Mohanta Z.; Singh A.; Stabinska J.; Liu G.; McMahon M. T. A Snapshot of the Vast Array of Diamagnetic CEST MRI Contrast Agents. NMR Biomed 2022, e4715 10.1002/nbm.4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.; Han Z.; Liu G. Repurposing Clinical Agents for Chemical Exchange Saturation Transfer Magnetic Resonance Imaging: Current Status and Future Perspectives. Pharmaceuticals (Basel) 2021, 14 (1), 11. 10.3390/ph14010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.; Payen J.-F.; Wilson D. A.; Traystman R. J.; Van Zijl P. C. Using the Amide Proton Signals of Intracellular Proteins and Peptides to Detect pH Effects in MRI. Nature medicine 2003, 9 (8), 1085–1090. 10.1038/nm907. [DOI] [PubMed] [Google Scholar]
- Togao O.; Yoshiura T.; Keupp J.; Hiwatashi A.; Yamashita K.; Kikuchi K.; Suzuki Y.; Suzuki S. O.; Iwaki T.; Hata N.; et al. Amide Proton Transfer Imaging of Adult Diffuse Gliomas: Correlation with Histopathological Grades. Neuro-oncology 2014, 16 (3), 441–448. 10.1093/neuonc/not158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z.; Hu S.; Huang F.; Wang X.; Guo L.; Quan X.; Wang S.; Zhou J. MR Imaging of High-Grade Brain Tumors Using Endogenous Protein and Peptide-Based Contrast. Neuroimage 2010, 51 (2), 616–622. 10.1016/j.neuroimage.2010.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dula A. N.; Arlinghaus L. R.; Dortch R. D.; Dewey B. E.; Whisenant J. G.; Ayers G. D.; Yankeelov T. E.; Smith S. A. Amide Proton Transfer Imaging of the Breast at 3T: Establishing Reproducibility and Possible Feasibility Assessing Chemotherapy Response. Magn. Reson. Med. 2013, 70 (1), 216–224. 10.1002/mrm.24450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L.; Li C.; Luo X.; Zhou J.; Zhang C.; Zhang Y.; Chen M. Differentiation of Malignant and Benign Head and Neck Tumors with Amide Proton Transfer-Weighted MR Imaging. Molecular Imaging and Biology 2019, 21 (2), 348–355. 10.1007/s11307-018-1248-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüre J. R.; Shrestha M.; Breuer S.; Deichmann R.; Hattingen E.; Wagner M.; Pilatus U. The Ph Sensitivity of Apt-CEST Using Phosphorus Spectroscopy as a Reference Method. NMR in Biomedicine 2019, 32 (11), e4125 10.1002/nbm.4125. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Zaiss M.; Knutsson L.; Sun P. Z.; Ahn S. S.; Aime S.; Bachert P.; Blakeley J. O.; Cai K.; Chappell M. A.; et al. Review and Consensus Recommendations on Clinical APT-Weighted Imaging Approaches at 3T: Application to Brain Tumors. Magn. Reson. Med. 2022, 88, 546. 10.1002/mrm.29241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo H. Y.; Zhang Y.; Burton T. M.; Jiang S.; Zhao Y.; van Zijl P. C.; Leigh R.; Zhou J. Improving the Detection Sensitivity of P H-Weighted Amide Proton Transfer MRI in Acute Stroke Patients Using Extrapolated Semisolid Magnetization Transfer Reference Signals. Magn. Reson. Med. 2017, 78 (3), 871–880. 10.1002/mrm.26799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo H. Y.; Lee D. H.; Zhang Y.; Zhao X.; Jiang S.; Chen M.; Zhou J. Insight into the Quantitative Metrics of Chemical Exchange Saturation Transfer (CEST) Imaging. Magn. Reson. Med. 2017, 77 (5), 1853–1865. 10.1002/mrm.26264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.; Heo H. Y.; Knutsson L.; van Zijl P. C.; Jiang S. APT-Weighted MRI: Techniques, Current Neuro Applications, and Challenging Issues. Journal of Magnetic Resonance Imaging 2019, 50 (2), 347–364. 10.1002/jmri.26645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg J. M.; Pagel M. D. Assessments of Tumor Metabolism with CEST MRI. NMR in Biomedicine 2019, 32 (10), e3943 10.1002/nbm.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai K.; Haris M.; Singh A.; Kogan F.; Greenberg J. H.; Hariharan H.; Detre J. A.; Reddy R. Magnetic Resonance Imaging of Glutamate. Nature medicine 2012, 18 (2), 302–306. 10.1038/nm.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis K. A.; Nanga R. P. R.; Das S.; Chen S. H.; Hadar P. N.; Pollard J. R.; Lucas T. H.; Shinohara R. T.; Litt B.; Hariharan H.; et al. Glutamate Imaging (GluCEST) Lateralizes Epileptic Foci in Nonlesional Temporal Lobe Epilepsy. Science translational medicine 2015, 7 (309), 309ra161–309ra161. 10.1126/scitranslmed.aaa7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haris M.; Nath K.; Cai K.; Singh A.; Crescenzi R.; Kogan F.; Verma G.; Reddy S.; Hariharan H.; Melhem E. R.; et al. Imaging of Glutamate Neurotransmitter Alterations in Alzheimer’s Disease. NMR in biomedicine 2013, 26 (4), 386–391. 10.1002/nbm.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.; Barker P. B.; Weiss R. G.; van Zijl P. C.; Xu J. Creatine and Phosphocreatine Mapping of Mouse Skeletal Muscle by a Polynomial and Lorentzian Line-Shape Fitting CEST Method. Magn. Reson. Med. 2019, 81 (1), 69–78. 10.1002/mrm.27514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.; Nguyen C.; Chen Y.; Shaw J. L.; Deng Z.; Xie Y.; Dawkins J.; Marbán E.; Li D. Optimized Cest Cardiovascular Magnetic Resonance for Assessment of Metabolic Activity in the Heart. Journal of cardiovascular magnetic resonance 2017, 19 (1), 1–7. 10.1186/s12968-017-0411-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.; Chung J. J.; Jin T. Chemical Exchange Saturation Transfer Imaging of Creatine, Phosphocreatine, and Protein Arginine Residue in Tissues. NMR in Biomedicine 2022, e4671 10.1002/nbm.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G.; Zhang T.; Dai Z.; Yi M.; Jia Y.; Nie T.; Zhang H.; Xiao G.; Wu R. A Potential Magnetic Resonance Imaging Technique Based on Chemical Exchange Saturation Transfer for in vivo γ-Aminobutyric Acid Imaging. PloS one 2016, 11 (10), e0163765 10.1371/journal.pone.0163765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haris M.; Cai K.; Singh A.; Hariharan H.; Reddy R. In Vivo Mapping of Brain Myo-Inositol. Neuroimage 2011, 54 (3), 2079–2085. 10.1016/j.neuroimage.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. K.; Huang A.; Xu J.; Edden R. A.; Schär M.; Hua J.; Oskolkov N.; Zacà D.; Zhou J.; McMahon M. T.; et al. Nuclear Overhauser Enhancement (NOE) Imaging in the Human Brain at 7 T. Neuroimage 2013, 77, 114–124. 10.1016/j.neuroimage.2013.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.; Hong X.; Zhao X.; Gao J. H.; Yuan J. APT-Weighted and NOE-Weighted Image Contrasts in Glioma with Different RF Saturation Powers Based on Magnetization Transfer Ratio Asymmetry Analyses. Magn. Reson. Med. 2013, 70 (2), 320–327. 10.1002/mrm.24784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-Y.; Wang F.; Afzal A.; Xu J.; Gore J. C.; Gochberg D. F.; Zu Z. A New NOE-Mediated MT Signal at around– 1.6 ppm for Detecting Ischemic Stroke in Rat Brain. Magn. Reson. Imaging 2016, 34 (8), 1100–1106. 10.1016/j.mri.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Han Z.; Chen G.; Li Y.; Zhang J.; Xu J.; van Zijl P. C.; Zhang S.; Liu G. CEST MRI of Sepsis-Induced Acute Kidney Injury. NMR in Biomedicine 2018, 31 (8), e3942 10.1002/nbm.3942. [DOI] [PubMed] [Google Scholar]
- Zhou Y.; van Zijl P. C.; Xu X.; Xu J.; Li Y.; Chen L.; Yadav N. N. Magnetic Resonance Imaging of Glycogen Using Its Magnetic Coupling with Water. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (6), 3144–3149. 10.1073/pnas.1909921117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P. Z. Quasi–Steady-State CEST (QUASS CEST) Solution Improves the Accuracy of Cest Quantification: QUASS CEST MRI-Based Omega Plot Analysis. Magn. Reson. Med. 2021, 86 (2), 765–776. 10.1002/mrm.28744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.; Xu X.; Zeng H.; Chan K. W. Y.; Yadav N.; Cai S.; Schunke K. J.; Faraday N.; van Zijl P. C. M.; Xu J. Separating Fast and Slow Exchange Transfer and Magnetization Transfer Using Off-Resonance Variable-Delay Multiple-Pulse (VDMP) MRI. Magn Reson Med. 2018, 80 (4), 1568–1576. 10.1002/mrm.27111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Abecassis B.; Vinogradov E.; Wijnen J. P.; van Harten T.; Wiegers E. C.; Hoogduin H.; van Osch M. J.; Ercan E. The Use of Variable Delay Multipulse Chemical Exchange Saturation Transfer for Separately Assessing Different CEST Pools in the Human Brain at 7T. Magn. Reson. Med. 2022, 87 (2), 872–883. 10.1002/mrm.29005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulborn K. R. Quantitative Sodium MR Imaging: A Review of Its Evolving Role in Medicine. Neuroimage 2018, 168, 250–268. 10.1016/j.neuroimage.2016.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaric O.; Juras V.; Szomolanyi P.; Schreiner M.; Raudner M.; Giraudo C.; Trattnig S. Frontiers of Sodium MRI Revisited: From Cartilage to Brain Imaging. Journal of Magnetic Resonance Imaging 2021, 54 (1), 58–75. 10.1002/jmri.27326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton J.; Franson D.; Seiberlich N. Recent Advances in Parallel Imaging for MRI. Progress in nuclear magnetic resonance spectroscopy 2017, 101, 71–95. 10.1016/j.pnmrs.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji B.; Hosseini Z.; Wang L.; Zhou L.; Tu X.; Mao H. Spectral Wavelet-Feature Analysis and Classification Assisted Denoising for Enhancing Magnetic Resonance Spectroscopy. NMR in Biomedicine 2021, 34 (6), e4497 10.1002/nbm.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal Z.; Nguyen D.; Thomas M. A.; Jiang S. Deep Learning Can Accelerate and Quantify Simulated Localized Correlated Spectroscopy. Sci. Rep 2021, 11 (1), 8727. 10.1038/s41598-021-88158-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung D.; Risk B. B.; Owusu-Ansah M.; Zhong X.; Mao H.; Fleischer C. C. Optimized Truncation to Integrate Multi-Channel MRS Data Using Rank-R Singular Value Decomposition. NMR Biomed 2020, 33 (7), e4297 10.1002/nbm.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennecke A.; Khakzar K. M.; German A.; Herz K.; Fabian M. S.; Liebert A.; Blümcke I.; Kasper B. S.; Nagel A. M.; Laun F. B.; et al. 7 Tricks for 7T CEST: Improving the Reproducibility of Multipool Evaluation Provides Insights into the Effects of Age and the Early Stages of Parkinson’s Disease. NMR in Biomedicine 2022, e4717 10.1002/nbm.4717. [DOI] [PubMed] [Google Scholar]
- Saito Y.; Yatabe H.; Tamura I.; Kondo Y.; Ishida R.; Seki T.; Hiraga K.; Eguchi A.; Takakusagi Y.; Saito K.; et al. Structure-Guided Design Enables Development of a Hyperpolarized Molecular Probe for the Detection of Aminopeptidase N Activity in vivo. Science advances 2022, 8 (13), eabj2667 10.1126/sciadv.abj2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker-Samuel S.; Ramasawmy R.; Torrealdea F.; Rega M.; Rajkumar V.; Johnson S. P.; Richardson S.; Goncalves M.; Parkes H. G.; Arstad E.; et al. In Vivo Imaging of Glucose Uptake and Metabolism in Tumors. Nat. Med. 2013, 19 (8), 1067–1072. 10.1038/nm.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. W.; McMahon M. T.; Kato Y.; Liu G.; Bulte J. W.; Bhujwalla Z. M.; Artemov D.; van Zijl P. C. Natural D-Glucose as a Biodegradable MRI Contrast Agent for Detecting Cancer. Magn Reson Med. 2012, 68 (6), 1764–1773. 10.1002/mrm.24520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T.; Mehrens H.; Hendrich K. S.; Kim S. G. Mapping Brain Glucose Uptake with Chemical Exchange-Sensitive Spin-Lock Magnetic Resonance Imaging. J. Cereb Blood Flow Metab 2014, 34 (8), 1402–1410. 10.1038/jcbfm.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.; Sehgal A. A.; Yadav N. N.; Laterra J.; Blair L.; Blakeley J.; Seidemo A.; Coughlin J. M.; Pomper M. G.; Knutsson L.; et al. d-Glucose Weighted Chemical Exchange Saturation Transfer (glucoCEST)-Based Dynamic Glucose Enhanced (DGE) MRI at 3T: Early Experience in Healthy Volunteers and Brain Tumor Patients. Magn Reson Med. 2020, 84 (1), 247–262. 10.1002/mrm.28124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz K.; Lindig T.; Deshmane A.; Schittenhelm J.; Skardelly M.; Bender B.; Ernemann U.; Scheffler K.; Zaiss M. T1rho-Based Dynamic Glucose-Enhanced (DGErho) MRI at 3T: Method Development and Early Clinical Experience in the Human Brain. Magn Reson Med. 2019, 82 (5), 1832–1847. 10.1002/mrm.27857. [DOI] [PubMed] [Google Scholar]
- Detre J. A.; Subramanian V.; Mitchell M.; Smith D.; Kobayashi A.; Zaman A.; Leigh Jr J. Measurement of Regional Cerebral Blood Flow in Cat Brain Using Intracarotid 2H2O and 2H NMR Imaging. Magn. Reson. Med. 1990, 14 (2), 389–395. 10.1002/mrm.1910140223. [DOI] [PubMed] [Google Scholar]
- Kim S. G.; Ackerman J. J. Quantification of Regional Blood Flow by Monitoring of Exogenous Tracer Via Nuclear Magnetic Resonance Spectroscopy. Magn. Reson. Med. 1990, 14 (2), 266–282. 10.1002/mrm.1910140212. [DOI] [PubMed] [Google Scholar]
- Wang F. N.; Peng S. L.; Lu C. T.; Peng H. H.; Yeh T. C. Water Signal Attenuation by D2O Infusion as a Novel Contrast Mechanism for 1H Perfusion MRI. NMR in Biomedicine 2013, 26 (6), 692–698. 10.1002/nbm.2914. [DOI] [PubMed] [Google Scholar]
- Chen L.; Liu J.; Chu C.; Han Z.; Yadav N.; Xu J.; Bai R.; Staedtke V.; Pearl M.; Walczak P.; et al. Deuterium Oxide as a Contrast Medium for Real-Time MRI-Guided Endovascular Neurointervention. Theranostics 2021, 11 (13), 6240. 10.7150/thno.55953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Cheng E. C.; Long Jr R. C.; Yang S.-H.; Wang L.; Cheng P.-H.; Yang J.; Wu D.; Mao H.; Chan A. W. Noninvasive Monitoring of Embryonic Stem Cells in Vivo with MRI Transgene Reporter. Tissue Engineering Part C: Methods 2009, 15 (4), 739–747. 10.1089/ten.tec.2008.0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulc D. A.; Lee X. A.; Cheng H.-Y. M.; Cheng H.-L. M. Bright Ferritin—a Reporter Gene Platform for on-Demand, Longitudinal Cell Tracking on MRI. iScience 2020, 23 (8), 101350. 10.1016/j.isci.2020.101350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock L. L.; Li Y.; Mao X.; Chen H.; Staedtke V.; Bai R.; Ma W.; Lin R.; Li Y.; Liu G.; et al. One-Component Supramolecular Filament Hydrogels as Theranostic Label-Free Magnetic Resonance Imaging Agents. ACS Nano 2017, 11 (1), 797–805. 10.1021/acsnano.6b07196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Jablonska A.; Li Y.; Cao S.; Liu D.; Chen H.; Van Zijl P. C.; Bulte J. W.; Janowski M.; Walczak P.; et al. Label-Free CEST MRI Detection of Citicoline-Liposome Drug Delivery in Ischemic Stroke. Theranostics 2016, 6 (10), 1588. 10.7150/thno.15492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Chen H.; Xu J.; Yadav N. N.; Chan K. W.; Luo L.; McMahon M. T.; Vogelstein B.; Van Zijl P. C.; Zhou S.; et al. Cest Theranostics: Label-Free MR Imaging of Anticancer Drugs. Oncotarget 2016, 7 (6), 6369. 10.18632/oncotarget.7141. [DOI] [PMC free article] [PubMed] [Google Scholar]