Abstract
Sendai virus (SeV) infection of interferon (IFN)-competent cells is one of the most efficient ways of inducing IFN production. Virus replication is nevertheless largely unaffected, since SeV infection also interfers with IFN action, a prerequisite for the establishment of an antiviral state. This property has been mapped by reverse genetics to the viral C gene, which is also known to act as a promoter-specific inhibitor of viral RNA synthesis. Using luciferase reporter plasmids containing IFN-responsive promoters, we have found that all four C proteins effectively interdict IFN signaling when expressed independently of SeV infection. The C proteins must therefore interact directly with cellular components to carry this out. The C gene in the context of an SeV infection was also found to induce STAT1 instability in some cells, whereas in other cells it apparently acts to prevent the synthesis of STAT1 in response to the virus infection or IFN treatment. The SeV C proteins appear to act in at least two ways to counteract the IFN induced by SeV infection.
Paramyxovirus P genes are remarkable for the complexity of their genetic organization and expression (Fig. 1A). This gene, named for the phosphoprotein that is an essential component of the P/L viral RNA polymerase (vRNAP), contains additional open reading frames (ORFs) that overlap the beginning and the middle of the P protein ORF (the C and V ORFs, respectively). The overlapping ORFs are accessed by a variety of unusual mechanisms of ribosomal choice (reviewed in references 8 and 28) and mRNA editing (41, 42), respectively. The overlapping ORFs are also referred to as “accessory” genes (4) because in each case there is at least one virus that does not contain (or express) the V or C genes.
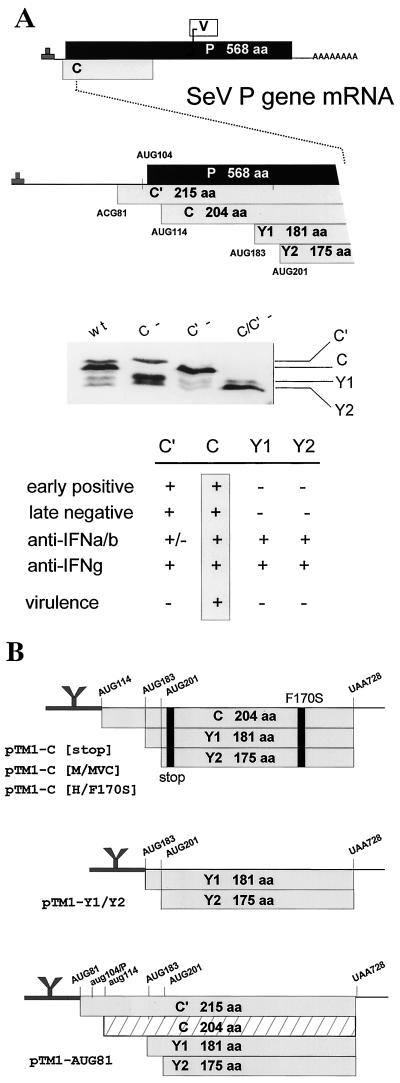
FIG. 1.
(A) The SeV P gene expresses four C proteins. The unedited P gene mRNA is shown at the top and its three ORFs (P, C, and V) are shown as horizontal boxes drawn roughly to scale (above). A blow-up of the 5′ end of the mRNA is shown in the middle, and the five ribosomal start sites in this region are indicated. The numbers refer to positions from the 5′ end of the mRNA and to the first base of the start codon. An immunoblot of the C proteins expressed in BHK cells infected with SeVwt and SeV unable to express C′ or C due to mutation of ACG81 and AUG114 (27) is shown in the middle. The properties of the individual C proteins are summarized at the bottom. (B) Various pTM1-C protein expression plasmids. pTM1 contains a T7 RNAP promoter and generates mRNAs beginning with the EMCV IRES up to the CCAUGG start codon and NcoI restriction site (thick line at left with Y-shaped structure), which permits cap-independent translation. The various C genes cloned into pTM1, as described in the text, are shown schematically. The start codons in lowercase letters are silent (6).
The Sendai virus (SeV) P gene has been extensively studied, in part because SeV RNAP is active in vitro, and it was here that the first property of the C gene was noted. When viral RNA synthesis is reconstituted from purified N-RNA nucleocapsids and P/L-vRNAP (contained in transfected cell extracts), specific ablation of overlapping C gene expression (or its replacement with mutant C genes) increases vRNAP specific activity ca. eightfold (7). C′ and C are equally active in this respect, but Y1 and Y2, the smallest of the four “C” proteins (Fig. 1A), are inactive. How the C proteins reduce vRNAP activity appears to be complex. The inhibitory effect of C can be countered by overexpressing P (and not L) (7, 40), but in glutathione S-transferase pulldown experiments C interacts with L (and not P) (20). C must also be present during P/L expression for inhibition to occur since C added to premade P/L-vRNAP is inactive (7, 19). The inhibitory effects of C are promoter specific in that genomic promoters are considerably more sensitive than antigenomic promoters (4, 40). Remarkably, the negative effects of C on vRNAP activity are accompanied by an equivalent increase in promoter fidelity in that minigenomes with promoter mutations (including those which destroy hexamer genome length) that are lethal in the presence of C gene expression replicate well in its absence (33, 40). C is present in SeV particles at relatively low levels (ca. 40 molecules/genome [42]), and its relative abundance increases greatly (∼50-fold) during intracellular replication. Since the inhibitory effects of C are concentration dependent, C would be expected to act as a brake on RNA synthesis (and as a fidelity factor) late in infection, possibly in preparation for assembling genome nucleocapsids into virions (40).
There is evidence that the C proteins also play a role early in infection. Consistent with their negative effects on vRNA synthesis, infections with recombinant SeV that cannot express C′ or C individually accumulate more viral RNAs than rSeVwt infection, and cytopathic effects are seen earlier (27). However, the simultaneous absence of both C′ and C paradoxically delays viral RNA accumulation (and cytopathic effects), and these viruses form “pinhole” plaques. Thus, in addition to their negative effects on viral RNA synthesis that apparently operate later in infection, C′ and C apparently also provide a positive function without which the onset of viral RNA accumulation is delayed. This positive function of C is not seen when viral RNA synthesis is reconstituted in transfected cells. Here, the early events in the infectious process have been bypassed (by jump-starting viral replication from DNA), and only the later negative effects are detected. Presumably, either C′ or C (but not Y1 or Y2) can supply this early required function in cell culture infections. How C might act at low concentrations early in infection to shorten the latent phase is unclear. The effect may in fact be indirect, e.g., the incoming genomes being initially inactive due to defects stemming from virion assembly in the absence of both C′ and C.
Mutation of Phe170 of the SeV C gene to Ser (by classical or reverse genetics) strongly reduces virulence in mice. The CF170S mutation was uncovered as one of two mutations (the other in the L gene) in a highly virulent virus strain (SeVM; 50% lethal dose [LD50] of 40) that became avirulent (SeVMVC; LD50 of >800,000) on adaptation for growth in LLC-MK2 cells (21, 22). SeV carrying the CMVC gene in an otherwise wild-type strain Z background (rSeVZ-CMVC) is also avirulent in mice, whereas those carrying the CM gene have not lost virulence (15). This was the first indication that the SeV C gene was essential for virulence in an animal host, and importation of this mutation into human parainfluenza virus type 3 (hPIV3) (CF164S) similarly attenuates virulence in vivo (11). Although rSeVZ-CMVC was avirulent, it grew normally in the mouse respiratory tract at first, but virus was then quickly cleared. The rapidity of the antiviral response in immunologically naive mice suggested that some aspect of innate immunity was involved (15). One such function has recently been identified. Infection of interferon (IFN)-competent cells with SeV, or simian virus 5 (SV5), does not induce an antiviral state. These infections do not affect the secretion of IFN-α/β, but somehow interfere with its receptor-mediated signaling, since they also prevent added IFN from inducing an antiviral state (10, 17). For SeV, the C gene is clearly involved, since viruses which carry the CF170S mutation, or those which cannot specifically express the AUG114-initiated C protein, do not interfere with IFN action (17). Consistent with this, SeVZ–C-minus is avirulent in mice, even though this virus normally expresses C′ and Y1 and overexpresses Y2. The AUG114-initiated C protein is thus specifically required for virulence in mice (16, 26). For SV5, which does not express a C protein, it is the V protein that apparently functions to interdict IFN signaling (9).
The role of SeV gene in countering IFN action has been investigated so far within the context of a natural virus infection, during which C also modulates vRNA synthesis in a promoter-specific fashion (4). Linear paramyxovirus genomes and antigenomes contain promoters for RNA synthesis at their 3′ ends, and C predominantly inhibits antigenome, leader, and mRNA synthesis from the genomic promoter, as opposed to genome and trailer RNAs from the antigenomic promoter. Leader and trailer RNAs have been implicated in the apoptotic response of the host cell to SeV infection (16), and these short RNAs may do this by affecting IFN signaling. The effect of C on IFN signaling may, at least in part, be indirect, acting via its ability to affect the viral RNAs produced (including cytoplasmic double-stranded RNA [23]). However, many viruses are known to directly subvert the IFN system by a wide variety of mechanisms (reviewed in reference 39), and it is equally possible that the C proteins also interact with (and suppress) some component of the signaling pathway leading to an antiviral state. If so, the C gene products should also function when expressed independently of virus infection. We investigate here the ability of various wild-type and mutant C genes expressed by transfection to inhibit IFN signaling to reporter plasmids.
MATERIALS AND METHODS
Cells and viruses.
Murine BF cells (cloned from a primary cell culture of a BALB/c mouse embryo) and mouse embryo fibroblasts (MEFs) (45) were grown as monolayers in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. The generation of rSeV expressing alternate C (and P) proteins has been described elsewhere (14, 15, 16, 17, 27). All SeV stocks were grown in the allantoic cavity of 10-day-old embryonated chicken eggs. Virus present in the allantoic fluids was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining after virus pelleting. Virus titers were determined by plaquing on LLC-MK2 cells. The vesicular stomatitis virus (VSV) stock (Mudd-Summers) was grown in BHK cells. Virus released into the culture medium was clarified by centrifugation to remove cell debris, and the titer was determined by plaquing them in LLC-MK2 cells.
Plasmid DNAs.
The various pTM1 plasmids (13) expressing the various SeV C proteins (pTM1-C) were generated by PCR amplification of the corresponding SeV C gene ORFs, and their insertion between the NcoI (CCATGG) and BamHI sites of pTM1. pTM1 was designed to express T7-RNAP-derived (and thus uncapped) transcripts since its 5′ untranslated region contains the encephalomyocarditis virus (EMCV) internal ribosome entry sequence (IRES) (see Fig. 1B). The primers used included the following: 5′ SeV C′, 5′-CCA TGC CAT GGC TTC GGC TAC ACT-3′ (AUG 81); 5′ SeV C, 5′-GAA GAT CTG CCA TGG CCT CAT TTC-3′ (AUG 114); 5′ SeV Y1, 5′-CCA TGC CAT GGT ATC GGA TTC CTC-3′ (AUG 183); and 3′ SeV C, 5′-CGG GAT CCT CAC TCT TGC ACT ATG-3′ (UGA 726). T7 RNA polymerase was provided by a plasmid expressing a cytoplasmic T7 RNA polymerase under the control of a cytomegalovirus (CMV) promoter (pT7) (36) kindly provided by M. Billeter, Zurich, Switzerland.
The IFN-α/β-responsive reporter plasmid, p(9-27)4tkΔ(−39)lucter (10, 24), referred to here as pISRE-f.luc, contains four tandem repeats of the IFN-inducible gene 9-27 ISRE fused to the firefly luciferase gene. The IFN-γ-responsive reporter plasmid, p(GAS)2tkΔ(−39)lucter (24) (referred to here as pGAS-f.luc) contains a minimal thymidine kinase (TK) promoter and two tandem repeats of the IRF-1 GAS sequences, fused to the firefly luciferase gene. pTK-r.luc., used as a transfection standard, contains the herpes simplex virus TK promoter region upstream of the renilla luciferase gene (Promega).
Transient transfections.
Monolayers of BF cells in 5.5-cm-diameter plates (at 50% confluence) were transfected with a total of 4 μg of DNA and 12 μl of Effectene (Qiagen) according to the manufacturer's instructions. At 24 h posttransfection, the cells were (or were not) infected with 20 PFU of SeV per cell and treated with 1,000 IU of recombinant IFN-α2α1 (43) per ml at 48 h posttransfection. At 4 to 12 h after IFN treatment, cells were harvested and assayed for firefly and renilla luciferase activity (dual-luciferase reporter assay system; Promega). Relative expression levels were calculated by dividing the firefly luciferase values by those of the renilla luciferase. The experiments were repeated several times with reproducible results, and their range is indicated in the figures.
Immunoblotting and immunoprecipitation.
Proteins were separated by SDS-PAGE and transferred to Immobilon-P membranes by semidry transfer. The primary antibody antibodies used included the following: a rabbit polyclonal antiserum to VSV P protein (provided by J. Perrault and D. Summers); a rabbit polyclonal antiserum to SeV P protein isolated from an SDS gel (anti-PSDS; L. Roux, Geneva, Switzerland); a mouse monoclonal antibody to SeV N (N877 [31]); a mouse monoclonal antibody to Stat1 C terminus (Transduction Laboratories, S21120); and a rabbit polyclonal antiserum to actin (provided by G. Gabbiani, Geneva, Switzerland).
The secondary antibodies used were alkaline phosphatase-conjugated goat antibodies specific for either rabbit or mouse immunoglobulin G (Bio-Rad). The immobilized proteins were detected by light-enhanced chemiluminescence (Bio-Rad) and quantified using the Bio-Rad Fluor-S multimager.
To estimate SeV C protein levels, cells were radiolabeled for 2 h with l-[35S]methionine and l-[35S]cysteine (Hartmann Analytic) at 48 h posttransfection (32) and the extracts were precipitated with a rabbit polyclonal antiserum to SeV C protein (provided by Y. Nagai, Tokyo, Japan).
RESULTS
To specifically monitor IFN-α signaling, we used a reporter plasmid with a firefly luciferase gene under the control of a basal promoter containing a tandemized ISRE (pISRE-luc [10, 24]). IFN-competent murine BF cells were transfected with pISRE-luc, along with pT7-RNAP and one of the various pTM1-C (see Materials and Methods). At 48 h posttransfection, the cultures were treated (or not) with IFN-α and harvested 4 h later, and the relative levels of luciferase activity in the cell extracts were determined. To monitor transfection and expression efficiencies, the same cultures were metabolically labeled with [35S]Met (for 2 h at 50 h posttransfection), and the relative levels of the various C proteins made during the pulse were determined by immunoprecipitation.
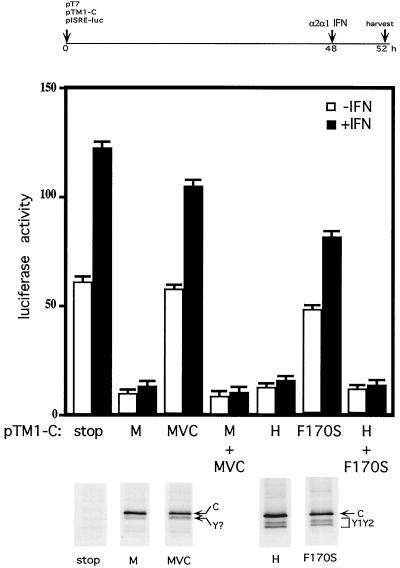
The 5′ end of the SeV P gene contains five ribosomal initiation codons, ACG81 (C′) (5, 18), AUG104 (P), AUG114 (C), AUG185 (Y1), and AUG201 (Y2) (Fig. 1). To limit P gene expression to the C proteins, nucleotides 114 to 730, containing only the C, Y1, and Y2 start codons, were cloned into the expression plasmid pTM1, which contains a T7 RNAP promoter and the EMCV IRES (see Materials and Methods). Five different C genes (all beginning at AUG114/C) were used: Cstop (with a stop codon just downstream of AUG201/Y2) as a negative control; CM and CMVC, which differ only at position 170; and CH and CH/F170S, which again differ only at position 170 (M and H are different strains whose C genes are 85% identical [22]). Cultures transfected with the CM and CMVC genes or with the CH and CH/F170S genes each expressed comparable levels of the AUG114-initiated C protein, the CH proteins being slightly more abundant. No C proteins could be found in the cultures that received Cstop (Fig. 2). Cultures transfected with CH and CH/F170S genes also expressed detectable amounts of the Y1 and Y2 proteins.
FIG. 2.
Effect of wild-type and mutant C gene expression on IFN-α signaling to pISRE-luc. Parallel cultures of BF cells were transfected with 1.5 μg of pT7 (which expresses cytoplasmic T7 RNAP from a CMV promoter), 1 μg of pISRE-luc, and 1.5 μg of one of the various pTM1-C as indicated. When transfections included two pTM1-C plasmids, all other reagents were doubled. At 48 h posttransfection, the cultures were treated or not treated with 1,000 U of IFN-α. The cultures were then metabolically labeled with Tran35S-label at 50 h posttransfection and harvested 2 h later, and the level of luciferase activity in the cell extracts was determined. The averages and ranges of activities from duplicate transfections of each pTM1 constructs and their mixtures are shown. Equal amounts of the various transfected cell extracts were also immunoprecipitated with an excess of anti-C antisera and then separated by SDS-PAGE. An autoradiogram of the gel is shown below. A time line of the experiment is shown above.
As shown in Fig. 2, transfected cultures unable to express the C proteins (Cstop) expressed relatively high levels of luciferase even when not IFN treated, and IFN treatment increased these levels twofold. Expression of the wild-type CM or CH genes strongly reduced both the high endogenous levels and those due to IFN treatment, whereas expression of the mutant CMVC or CH/F170S genes had little or no effect. Moreover, the simultaneous expression of the CM and CMVC proteins or the CH and CH/F170S proteins showed that the wild-type phenotype was dominant, a finding consistent with the F170S substitution being a loss-of-function mutation. Similar experiments in which the reporter gene was under the control of the herpesvirus TK promoter showed no effect of the C proteins on luciferase activity (data not shown). The C proteins therefore do not appear to affect the cellular basal transcription machinery. As C gene expression effectively interdicts this IFN signaling independent of virus infection (i.e., independent of its ability to modulate viral RNA synthesis), the C gene products presumably interact directly with cellular components to carry this out.
Four C proteins.
The various SeV C proteins are not functionally equivalent in all respects. C′ and C (but not Y1 or Y2) enhance RNA synthesis early in infection and inhibit it later on, and only CAUG114 is essential for virulence in mice (Fig. 1A). This latter conclusion is based on rSeV in which expression of individual C proteins was specifically ablated by mutating their start codons (27). Mutation of ACG81 to UCU or of AUG114 to GCG effectively ablates synthesis of C′ or C, respectively. However, we have been unable to completely eliminate Y1 and Y2 synthesis by mutating either AUG183 or AUG201 to seven different non-AUG codons, including ACG (reference 28 and data not shown). This unusual situation appears to be due to the operation of a ribosomal shunt, where codon-anticodon interactions are not as critical for start-site selection, as reported for the hepatitis C virus IRES (37). We have thus prepared rSeVs which do not selectively express C′ (C′-minus), C (C-minus), or C′ and C (C′/C-minus), but not rSeVs specifically unable to express Y1 or Y2. Of these rSeVs, only C′/C-minus is highly debilitated for growth in cell culture. The double mutant also displays a small plaque phenotype and is avirulent for mice. The single mutants C′-minus and C-minus, in contrast, grow more rapidly in cell culture than SeVwt and show a normal plaque phenotype. Nevertheless, despite this similarity of the single mutants in cell culture, C′-minus is as virulent as the SeVwt in mice, whereas C-minus is as avirulent as SeVMVC (27). C and/or Y1/Y2 can thus compensate for the loss of C′ in maintaining virulence, i.e., in sustaining virus multiplication in the mouse respiratory tract. However, C′ plus Y1/Y2 cannot compensate for loss of C, even though C′/C-minus overexpresses Y2. The AUG114-initiated C protein is thus the only one of the four that is essential for virulence in the animal host.
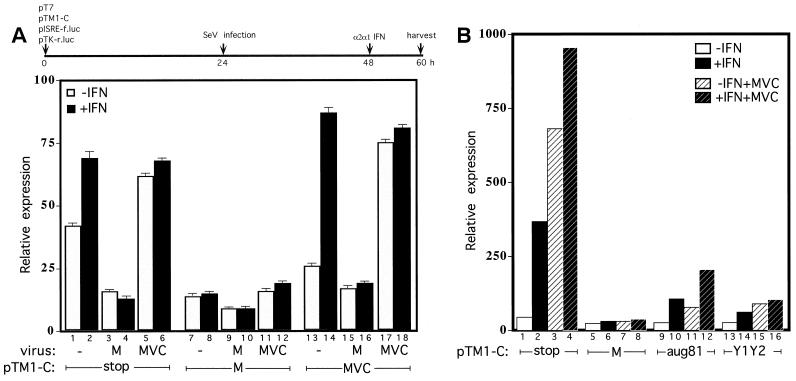
To investigate which of the C proteins could prevent IFN signaling, two additional pTM1-C gene constructs were prepared (Fig. 1B); pTM1-C′ (AUG81), which contains the entire C ORF beginning with AUG81 and which is expected to predominantly expresses C′ and some Y1/Y2, (but not the P fragment or C [6]), and pTM1-Y1/Y2, in which the C gene begins with AUG183/Y1 and which is now in an optimal context for ribosomal initiation. These constructs were tested for their ability to inhibit IFN-induced pISRE-luc activity, alongside the previously used pTM1-CAUG114. Figure 3 shows the various C proteins expressed by transfection and their relative levels, determined as described above. The various C gene constructs again expressed roughly equivalent levels of their respective C proteins (Fig. 3C).
FIG. 3.
Effects of the various C proteins on IFN-α signaling to pISRE-luc and IFN-γ signaling to pGAS-luc. Parallel cultures of BF cells were transfected with 1 μg of pT7 and 1 μg of pISRE-f.luc (A) or 1 μg of pGAS-f.luc (B) and 0.2 μg of pTK-r.luc to control for transfection efficiencies and 1 μg of one of the various pTM1-C as indicated (see also Fig. 1B). At 48 h posttransfection, the cultures were treated (or not) with 1,000 U of IFN-α. The cultures were then metabolically labeled with Tran35S-label at 58 h posttransfection and harvested 2 h later, and the levels of the firefly (f.luc) and renilla (r.luc) luciferase activities in the cell extracts were determined. The averages and ranges of the ratios of firefly to renila activities from duplicate transfections of each pTM1 construct are shown. Equal amounts of the various transfected cell extracts were also immunoprecipitated with an excess of anti-C antisera and separated by SDS-PAGE. (C) An autoradiogram of the gel is also shown. Note that the strain M and H AUG114-initiated C proteins migrate at slightly different rates, as highlighted by a short line which connects the MVC and H lanes. A time line of the experiment is shown above panel A.
The ability of these C gene constructs to inhibit IFN-α signaling to pISRE-luc in BF cells, as well as IFN-γ signaling to an analogous reporter construct containing a tandemized gamma-activated sequence (GAS) element (pGAS-luc), was determined as described in Materials and Methods. IFN-γ is released by activated immune cells and acts through a parallel and partially overlapping Jak-Stat pathway as IFN-α/β to activate IFN-stimulated genes (ISGs) with GAS element's in their promoters (39). Remarkably, the expression of the Y1 and Y2 proteins alone were almost as effective as CAUG114 in interdicting either type of IFN signaling (Fig. 3A and B). The expression of C′ (plus a small amount of Y1; Fig. 3C, lane AUG81) was only partially effective in inhibiting IFN-α signaling (Fig. 3A); however, it was as effective as CAUG114 in inhibiting IFN-γ signaling (Fig. 3B). Thus, with the partial exception of C′, all four C proteins appear to inhibit IFN signaling from both IFN-α/β and IFN-γ receptors. In contrast, similar experiments in which the reporter gene was controlled by the herpesvirus TK promoter showed no effect of the C proteins on luciferase activity (data not shown).
Complementation of the anti-IFN defect of SeVMVC.
SeVMVC infection of BF cells is particularly adept at inducing an antiviral state for subsequent VSV growth and at elevating STAT1 levels. This is not because SeVMVC infection produces more IFN than SeVM but presumably because SeVMVC has lost the ability to interfere with IFN action (17). We would therefore expect SeVMVC infection itself to stimulate transfected pISRE-luc, whereas SeVM infection should have the opposite effect. BF cells were therefore transfected as described above [plus pTK-luc(ren) to control for transfection efficiency] and infected with either SeVM or SeVMVC or mock infected at 24 h posttransfection. The cultures were then IFN treated (or not) at 48 h posttransfection, and the luciferase levels were determined at 60 h posttransfection. As shown in Fig. 4A, transfected cultures without C protein expression (Cstop) again expressed relatively high levels of firefly luciferase without IFN treatment. However, this expression was in fact further stimulated by SeVMVC infection and inhibited by SeVM infection. SeVM infection also inhibited the additional signaling due to IFN-α treatment, whereas SeVMVC infection again had no effect. In contrast, SeVM and SeVMVC infection equally stimulated luciferase activity from constructs that contained the IFN-β promoter (consistent with the ability of SeVM and SeVMVC to equally induce IFN production) and had no effect on constructs that contained the herpesvirus TK promoter (data not shown).
FIG. 4.
Effect of SeV infection and expression of the various C proteins on pISRE-luc activity. Parallel cultures of BF cells were transfected as described in Fig. 3. At 24 h posttransfection, the cultures were either not infected or infected with SeVM or SeVMVC, and treated (or not treated) with 1,000 U of IFN-α at 48 h posttransfection as indicated. The cultures were harvested at 60 h posttransfection, and the levels of the firefly (f.luc) and renilla (r.luc) luciferase activities in the cell extracts were determined. The averages and ranges of the ratios of activities from duplicate transfections of each pTM1 construct are shown in panel A. The experiment in panel B was carried out similarly to that in panel A, except that only SeVMVC infection was used, as indicated. A time line of both experiments is shown above.
Since SeV infection affected pISRE-luc expression as expected, we next examined whether the C proteins expressed by transfection could act in trans to prevent the SeVMVC infection from stimulating pISRE-luc, thus complementing this defect in SeVMVC. Natural SeV infections should activate pISRE-luc via an endogenous mix of many IFN-α/β types (many of which are more powerful than the single recombinant IFN-α that we added), and they may also induce alternative pathways activated by double-stranded RNA (23). Furthermore, the various C proteins expressed by transfection are now operating in the context of a natural virus infection, in which the resident C gene is inactive due to the F170S mutation. As shown in Fig. 4B, pTM1-CM expression effectively prevents SeVMVC infection (as well as does IFN treatment) from stimulating pISRE-luc, whereas pTM1-CMVC expression was without effect. Moreover, Y1/Y2 expression alone or expression of C′ (AUG81) were almost as effective as CAUG114 in preventing SeVMVC infection from activating pISRE-luc. Thus, all of the four C proteins can apparently complement this defect. Since SeVMVC contains two mutations relative to SeVM (LE2050A and CF170S), the results of Fig. 4 suggest that the LE2050A mutation of SeVMVC does not contribute greatly to the inability of this virus to interfere with IFN action.
rSeV infections of murine NIH 3T3 cells.
Murine BF cells have relatively low endogenous levels of STAT1 (as determined by immunoblotting). Their infection with SeVwt does not alter these STAT1 levels, and subsequent VSV replication occurs normally. In contrast, infection of BF cells with rSeV which carry the CF170S mutation (and are avirulent in mice) leads to an antiviral state in which VSV replication is restricted and STAT1 levels increase dramatically (17). This increase in STAT1 presumably results from the activation of ISGs by the virus infection and the failure of the mutant C genes of these avirulent viruses to interdict IFN signaling (as STAT1 is also an ISG). Infection of cells with the paramyxovirus SV5 similarly leads to the interdiction of IFN signaling to pISRE-luc (in human diploid fibroblasts but not in murine BF cells), and STAT1 levels in this case are strongly decreased (10). Rubulaviruses such as SV5, however, do not contain C genes, and Didcock et al. (9) have provided evidence that these effects are due to the V protein (another product of the P gene), which somehow targets STAT1 for proteosome-mediated degradation.
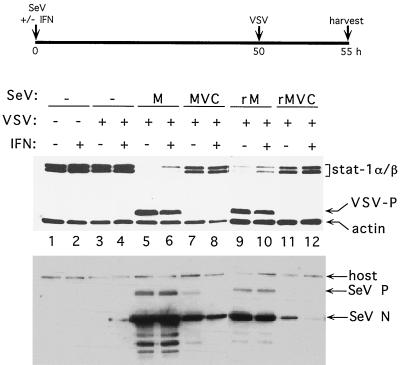
We have examined the effects of SeV infection on STAT1 levels in NIH 3T3 MEFs, which contain elevated endogenous STAT1 levels relative to BF cells (ca. 10-fold). This disparity does not affect the relative abilities of the various SeVs to replicate in the MEFs, which occur in the following order: M > rM > MVC > rMVC (Fig. 5), a result identical to that in BF cells (17). The elevated STAT1 levels, however, may have restricted VSV replication, since VSV clearly does not replicate in naive MEFs (Fig. 5, lanes 3 and 4), in contrast to BF cells. However, when MEFs are infected with SeVM (lanes 5 and 6) or rSeVZ-CM (lanes 9 and 10), STAT1 levels decreased dramatically, and the block on VSV replication was reversed. In fact, VSV replication here was almost as robust as in naive BF cells (data not shown), even when these cultures were also IFN-α treated (lanes 6 and 10). In contrast, infection with SeVMVC (lanes 7 and 8) or rSeVZ-CMVC (lanes 11 and 12) had little or no effect on these parameters. Two conclusions can be drawn. (i) The permissiveness of murine cells to VSV replication is strongly determined by the endogenous levels of STAT1. This was not unexpected, since STAT1 is known to be highly important in combating VSV infections of mice. Targeted disruption of this gene lowers the LD50 of VSV by 3 logs (12, 30). (ii) Similar to SV5 infection of human diploid fibroblasts (10), SeV infection of MEFs strongly reduces STAT1 levels, presumably by inducing STAT1 instability. This reduction, and the accompanying permissiveness of these cells to VSV replication, requires an active C gene as defined by the F170S mutation. We note, however, that these effects are being measured 50 h post-SeV infection and thus that many upstream events may contribute to STAT1 instability.
FIG. 5.
Effect of SeV infection on STAT1 levels and induction of an antiviral state. Parallel cultures of NIH 3T3 mouse embryo fibroblasts were infected with either SeVM, SeVMVC, SeVZ-CM (rM), or SeVZ-CMVC (rMVC) or were not infected with SeV and then concomitantly treated (or not) with 1,000 U of IFN-α as indicated. Most cultures were also superinfected with VSV at 50 h post-SeV infection, as indicated. The cultures were harvested at 55 h postinfection, and the relative levels of the cellular STAT1 and VSV P proteins were determined by immunoblotting. An anti-actin antibody was included to control for the amount of cellular material loaded on each lane. Equal samples of the various cell extracts were used to determine the relative levels of the SeV N and P proteins present by immunoblotting.
DISCUSSION
The SeV C gene in the context of a natural SeV infection functions to prevent the infected cell from establishing an antiviral state in response to the virus infection (or added IFN), at least in part by interdicting IFN action (17). Using reporter genes to specifically assess and IFN-α/β and IFN-γ signaling and plasmids that express C genes independent of virus infection, we found that expression of wild-type CM or CH genes effectively blocked IFN signaling to pISRE-luc induced either by exogenous IFN, double-stranded RNA (data not shown), or SeVMVC infection. C genes carrying the F170S mutation, in contrast, were largely inactive. Thus, in addition to whatever effects the C proteins might have indirectly on IFN action by modulating viral RNA synthesis during intracellular replication (Fig. 1), they presumably also interact directly with cellular component(s) to interdict IFN signaling to pISRE-luc. In contrast to the anti-IFN effects of the SV5 V gene which are specific to cells of human origin, the SeV C genes acts equally well in human and mouse cells (data not shown).
An important question now is whether C gene expression independent of SeV infection is sufficient to prevent IFN from blocking VSV replication in BF cells. Unfortunately, one limitation of these transfection experiments is that we were unable to determine this, since only ca. 15% of the BF cells were productively transfected. The IFN-induced antiviral state is known to be remarkably complex (39), and the interdiction of IFN signaling to transfected pISRE-luc is not the same as preventing IFN from inducing an anti-VSV state. This distinction may be germane, particularly to the action of the Y proteins. In contrast to their ability to effectively prevent IFN signaling to pISRE-luc, even in the context of SeVMVC infection (Fig. 4B), Y protein expression alone in the context of SeVZ–C′/C-minus infection is clearly unable to prevent IFN action from blocking VSV replication (17). The interdiction of IFN signaling to pISRE/luc and pGAS-luc that we measure may not extend to other aspects of IFN action, e.g., Y protein expression alone (in the context of SeVZ–C′/C-minus infection) is also unable to prevent the elevation of STAT1 in BF cells. Elevated STAT1 levels may then be an additional requirement for an effective antiviral state in these murine cells. Many of these questions regarding what the various C proteins can and cannot do should be addressed with uninfected, inducible C-expressing cells. However, although cell lines in which high levels of Y1 and Y2 are expressed in response to tetracycline are easy to establish, we have been unable to prepare cells expressing CAUG114, presumably because even low levels of this protein are toxic (data not shown).
In SV5-infected cells, the V protein of this rubulavirus is thought to interdict IFN signaling to pISRE-luc by targeting STAT1 for proteasome-mediated degradation (9). The action of the SeV C gene on STAT1 levels appears to be more complex. In murine BF cells with low endogenous STAT1 levels, SeV-CF170S infection dramatically increases STAT1, whereas SeV-Cwt infection has no effect. Here, Cwt functions to prevent the increase of this ISG in response to the viral infection. In MEFs with high endogenous STAT1 levels, in contrast, Cwt functions to destabilize STAT1, and CF170S has lost this property. While these two effects appear to be quite different, they are, of course, two sides of the same coin. In both cases the SeV C gene prevents the induction of a cellular antiviral state by removing this key player in IFN signaling, regardless of the STAT1 status of the uninfected host cell.
How might this effect of C on IFN action operate? IFN-γ and IFN-α/β bind to their respective cell surface receptors and activate distinct but related signal transduction pathways, culminating in the activation of an overlapping set of ISGs. Upon binding of IFN-α/β to IFN-α/β receptors, their associated protein tyrosine kinases (Jak1 and tyk2) phosphorylate Y701 of STAT1 and Y690 of STAT2. STAT1 and STAT2 then heterodimerize, translocate to the nucleus, and associate with p48 to form ISG factor 3 (ISGF3). ISGF3 activates the transcription of genes containing ISREs within their promoters. The full activity of STAT1 also depends on phosphorylation of S727 via the mitogen-activated protein kinase pathway. Upon binding of IFN-γ to IFN-γ receptors, their associated protein tyrosine kinases (Jak1 and Jak2) similarly phosphorylate Y701 of STAT1. Phosphorylated STAT1 also homodimerizes to form gamma-activated factor (GAF), which activates the transcription of genes containing GAS elements within their promoters. Phosphorylation of STAT1 on S727 is also required for the full activity of GAF (for a review of IFN signaling, see reference 39).
Our experiments offer little insight into how the primary effect(s) of C might operate on IFN action, since our reporter gene measurements are made at relatively late times (>24 h) posttransfection, and there is considerable cycling and cross-talk in the IFN signaling pathways. However, Komatsu et al. (25) have recently reported that SeV infection of HeLa cells specifically prevented phosphorylation of STAT1 Y701 by 2 h postinfection. Assuming that this effect is due to the C proteins, they must be acting here at relatively low concentrations, as there are few (ca. 40) C proteins per genome in virions (44), and little primary translation has occurred by 2 h postinfection. After genome amplification, however, there are >300 C molecules per genome intracellularly, and the number of genomes/cell has increased by ca. 2 logs. The C proteins are thus considerably more abundant late in infection, and at higher concentrations they may act as well to destabilize STAT1 (when STAT1 levels are elevated). In analogy to the different effects of C on viral RNA synthesis at early and late times of infection, C may be acting differently to counteract the IFN system as its intracellular concentration increases during the infection. C may act first by preventing the formation of activated STAT1 and then by reducing its abundance intracellularly. The SeV C gene may be a useful addition in nonretrovirus vectors that are sensitive to IFN action.
As mentioned above, hPIV1 and SeV are very closely related; they are virtually indistinguishable by serology (38). Nevertheless, even within this virus pair, P gene expression varies. hPIV1 is the only paramyxovirus without a V ORF (or an mRNA editing site), and although hPIV1 also expresses four C proteins, here the GUG67-initiated C′ protein is clearly the most abundantly expressed of the four (29, 34). This non-AUG start site lies in a particularly favorable context for ribosomal initiation (including positions +5 and +6), and GUG67, in contrast to SeV ACG81, is almost as efficient a start site as AUG at this position, but paradoxically this does not compromise leaky scanning to AUG104/P and AUG114/C of hPIV1 (2, 3). This conservation of the four C protein start sites (and the unusual mechanisms involved in their initiation by ribosomes) suggests that all four proteins play a role in virus replication, presumably because the various C proteins are not functionally equivalent in all respects. Other respiroviruses (h/bPIV3) and the morbilliviruses express but a single C protein (1, 35), and it is satisfying that this protein appears to be the homologue of SeV CAUG114, the only one of the four that retains all of this C gene's known activities (Fig. 1A).
Although highly speculative, the C′ and the Y1 and Y2 start sites may have been added to h/mPIV1 during evolution to express proteins which have retained only a subset of the activities of CAUG114. The C′, C, and Y1 and Y2 proteins are initiated by different mechanisms (non-AUG initiation, leaky scanning, and ribosomal shunting, respectively), and their relative efficiencies of expression may be differentially affected by cellular stress (46). The variety of ribosomal initiation possibilities may ensure that one or more aspects of their ability to interfere with the IFN action is particularly resistant to cellular responses that normally limit translational initiation, a hallmark of the antiviral state (23, 26, 39). Cellular responses that favor the ribosomal shunt would relieve the negative effects of the C gene on viral RNA synthesis, without affecting their ability to interfere with some aspects of IFN action.
ACKNOWLEDGMENTS
We thank Rick Randall, Aberdeen, Scotland, for the pISRE-luc and pGAS-luc reporter plasmids; Martin Billeter, Zurich, Switzerland, for the pT7 RNAP; and Jean-Baptiste Marq for excellent technical assistance.
This work was supported by a grant from the Swiss National Science Fund.
REFERENCES
- 1.Bellini W J, Englund G, Rozenblatt S, Arnheiter H, Richardson C D. Measles virus P gene codes for two proteins. J Virol. 1985;53:908–919. doi: 10.1128/jvi.53.3.908-919.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeck R, Kolakofsky D. Positions +5 and +6 can be major determinants of the efficiency of non-AUG initiation codons for protein synthesis. EMBO J. 1994;13:3608–3617. doi: 10.1002/j.1460-2075.1994.tb06668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeck R, Curran J, Matsuoka Y, Compans R, Kolakofsky D. The parainfluenza virus type 1 P/C gene uses a very efficient GUG codon to start its C′ protein. J Virol. 1992;66:1765–1768. doi: 10.1128/jvi.66.3.1765-1768.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cadd T, Garcin D, Tapparel C, Itoh M, Homma M, Roux L, Curran J, Kolakofsky D. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J Virol. 1996;70:5067–5074. doi: 10.1128/jvi.70.8.5067-5074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curran D, Kolakofsky Ribosomal initiation from an ACG codon in the Sendai virus P/C mRNA. EMBO J. 1988;7:245–251. doi: 10.1002/j.1460-2075.1988.tb02806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curran J, Kolakofsky D. Scanning independent ribosomal initiation of the Sendai virus Y proteins in vitro and in vivo. EMBO J. 1989;8:521–526. doi: 10.1002/j.1460-2075.1989.tb03406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curran J, Marq J-B, Kolakofsky D. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology. 1992;189:647–656. doi: 10.1016/0042-6822(92)90588-g. [DOI] [PubMed] [Google Scholar]
- 8.Curran J, Latorre P, Kolakofsky D. Translational gymnastics on the Sendai virus P/C mRNA. Semin Virol. 1998;8:351–357. [Google Scholar]
- 9.Didcock L, Young D F, Goodbourn S, Randall R E. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J Virol. 1999;73:9928–9933. doi: 10.1128/jvi.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Didcock D F, Young L, Goodbourn S, Randall R E. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J Virol. 1999;73:3125–3133. doi: 10.1128/jvi.73.4.3125-3133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durbin A P, McAuliffe J M, Collins P L, Murphy B R. Mutations in the C, D, and V open reading frames of human parainfluenza virus type 3 attenuate replication in rodents and primates. Virology. 1999;261:319–330. doi: 10.1006/viro.1999.9878. [DOI] [PubMed] [Google Scholar]
- 12.Durbin J E, Hackenmiller R, Simon M C, Levy D E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 13.Elroy-Stein O, Fuerst T R, Moss B. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc Natl Acad Sci USA. 1989;86:6126–6130. doi: 10.1073/pnas.86.16.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcin D, Pelet T, Calain P, Roux L, Curran J, Kolakofsky D. A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: generation of a novel copy-back nondefective interfering virus. EMBO J. 1995;14:6087–6094. doi: 10.1002/j.1460-2075.1995.tb00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcin D, Itoh M, Kolakofsky D. A point mutation in the Sendai virus accessory C proteins attenuates virulence for mice, but not virus growth in cell culture. Virology. 1997;238:424–431. doi: 10.1006/viro.1997.8836. [DOI] [PubMed] [Google Scholar]
- 16.Garcin D, Taylor G, Tanebayashi K, Compans R, Kolakofsky D. The short sendai virus leader region controls induction of programmed cell death. Virology. 1998;243:340–353. doi: 10.1006/viro.1998.9063. [DOI] [PubMed] [Google Scholar]
- 17.Garcin D, Latorre P, Kolakofsky D. Sendai virus C proteins counteract the interferon mediated induction of an antiviral state. J Virol. 1999;73:6559–6565. doi: 10.1128/jvi.73.8.6559-6565.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta K C, Patwardhan S. ACG, the initiator codon for Sendai virus C protein. J Biol Chem. 1988;263:8553–8556. [PubMed] [Google Scholar]
- 19.Horikami S M, Curran J, Kolakofsky D, Moyer S A. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J Virol. 1992;66:4901–4908. doi: 10.1128/jvi.66.8.4901-4908.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horikami S M, Hector R E, Smallwood S, Moyer S A. The Sendai virus C protein binds the L polymerase protein to inhibit viral RNA synthesis. Virology. 1997;235:261–270. doi: 10.1006/viro.1997.8702. [DOI] [PubMed] [Google Scholar]
- 21.Itoh M, Hotta H, Homma M. Increased induction of apoptosis by a sendai virus mutant is associated with attenuation of mouse pathogenicity. J Virol. 1998;72:2927–2934. doi: 10.1128/jvi.72.4.2927-2934.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itoh M, Isegawa Y, Hotta H, Homma M. Isolation of an avirulent mutant of Sendai virus with two amino acid mutations from a highly virulent field strain through adaptation to LLC-MK2 cells. J Gen Virol. 1997;78:3207–3215. doi: 10.1099/0022-1317-78-12-3207. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs B L, Langland J O. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology. 1996;219:339–349. doi: 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- 24.King P, Goodbourn S. STAT1 is inactivated by a caspase. J Biol Chem. 1998;273:8699–8704. doi: 10.1074/jbc.273.15.8699. [DOI] [PubMed] [Google Scholar]
- 25.Komatsu T, Takeuchi K, Yokoo J, Tanaka Y, Gotoh B. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J Virol. 2000;74:2477–2480. doi: 10.1128/jvi.74.5.2477-2480.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar A, Commane M, Flickinger T W, Horvath C M, Stark G R. Defective TNFα-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science. 1997;278:1630–1632. doi: 10.1126/science.278.5343.1630. [DOI] [PubMed] [Google Scholar]
- 27.Latorre P, Cadd T, Itoh M, Curran J, Kolakofsky D. The various sendai virus C proteins are not functionally equivalent and exert both positive and negative effects on viral RNA accumulation during the course of infection. J Virol. 1998;72:5984–5993. doi: 10.1128/jvi.72.7.5984-5993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Latorre P, Kolakofsky D, Curran J. Sendai virus Y proteins are initiated by a ribosomal shunt. Mol Cell Biol. 1998;18:5021–5031. doi: 10.1128/mcb.18.9.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuoka Y, Curran J, Pelet T, Kolakofsky D, Ray R, Compans R W. The P gene of human parainfluenza virus type 1 encodes P and C proteins but not a cysteine-rich V protein. J Virol. 1991;65:3406–3410. doi: 10.1128/jvi.65.6.3406-3410.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meraz M A, White J M, Sheehan K C, Bach E A, Rodig S J, Dighe A S, Kaplan D H, Riley J K, Greenlund A C, Campbell D, Carver-Moore K, DuBois R N, Clarks R, Aguet M, Schreiber R D. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 31.Orvell C, Grandien M. The effects of monoclonal antibodies on biological activities of structural proteins of Sendai virus. J Immunol. 1982;129:2779–2787. [PubMed] [Google Scholar]
- 32.Pelet T, Curran J, Kolakofsky D. The P gene of bovine parainfluenza virus 3 expresses all three reading frames from a single mRNA editing site. EMBO J. 1991;10:443–448. doi: 10.1002/j.1460-2075.1991.tb07966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelet T, Delenda C, Gubbay O, Garcin D, Kolakofsky D. Partial characterization of a Sendai virus replication promoter and the rule of six. Virology. 1996;224:405–414. doi: 10.1006/viro.1996.0547. [DOI] [PubMed] [Google Scholar]
- 34.Power U F, Ryan K W, Portner A. The P genes of human parainfluenza virus type 1 clinical isolates are polycistronic and microheterogeneous. Virology. 1992;189:340–343. doi: 10.1016/0042-6822(92)90712-x. [DOI] [PubMed] [Google Scholar]
- 35.Radecke F, Billeter M A. The nonstructural C protein is not essential for multiplication of Edmonston B strain measles virus in cultured cells. Virology. 1996;217:418–421. doi: 10.1006/viro.1996.0134. [DOI] [PubMed] [Google Scholar]
- 36.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dotsch C, Christiansen G, Billeter M A. Rescue of measles viruses from cloned DNA. EMBO J. 1995;14:5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reynolds J E, Kaminski A, Kettinen H J, Grace K, Clarke B E, Carroll A R, Rowlands D J, Jackson R J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan K W, Portner A, Murti K G. Antibodies to paramyxovirus nucleoproteins define regions important for immunogenicity and nucleocapsid assembly. Virology. 1993;193:376–384. doi: 10.1006/viro.1993.1134. [DOI] [PubMed] [Google Scholar]
- 39.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 40.Tapparel C, Hausmann S, Pelet T, Curran J, Kolakofsky D, Roux L. Inhibition of Sendai virus genome replication due to promoter-increased selectivity: a possible role for the accessory C proteins. J Virol. 1997;71:9588–9599. doi: 10.1128/jvi.71.12.9588-9599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas S M, Lamb R A, Paterson R G. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell. 1988;54:891–892. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidal S, Curran J, Kolakofsky D. A stuttering model for paramyxovirus P mRNA editing. EMBO J. 1990;9:2017–2022. doi: 10.1002/j.1460-2075.1990.tb08330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weber H, Valenzuela D, Luijber G, Gubler M, Weissmann C. Single amino acid changes that render human IFN-alpha 2 biologically active on mouse cells. EMBO J. 1987;6:591–598. doi: 10.1002/j.1460-2075.1987.tb04795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada H, Hayata T, Omata-Yamada T, Taira H, Mizumoto K, Iwasaki K. Association of the Sendai virus C protein with nucleocapsids. Arch Virol. 1990;113:245–253. doi: 10.1007/BF01316677. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y L, Reis L F, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams B R, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6995–7006. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yueh A, Schneider R J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]