Abstract
Cancer treatment is gradually entering an era of precision, with multitude studies in gene testing and immunotherapy. Tumor cells can be recognized and eliminated by the immune system through the expression of tumor-associated antigens, but when the cancer escapes or otherwise suppresses immunity, the balance between cancer cell proliferation and immune-induced cancer cell killing may be interrupted, resulting in tumor proliferation and progression. There has been significant attention to combining conventional cancer therapies (i.e., radiotherapy) with immunotherapy as opposed to treatment alone. The combination of radio-immunotherapy has been demonstrated in both basic research and clinical trials to provide more effective anti-tumor responses. However, the absolute benefits of radio-immunotherapy are dependent on individual characteristics and not all patients can benefit from radio-immunotherapy. At present, there are numerous articles about exploring the optimal models for combination radio-immunotherapy, but the factors affecting the efficacy of the combination, especially with regard to radiosensitivity remain inconclusive. Radiosensitivity is a measure of the response of cells, tissues, or individuals to ionizing radiation, and various studies have shown that the radiosensitivity index (RSI) will be a potential biomarker for predicting the efficacy of combination radio-immunotherapy. The purpose of this review is to focus on the factors that influence and predict the radiosensitivity of tumor cells, and to evaluate the impact and predictive significance of radiosensitivity on the efficacy of radio-immunotherapy combination.
Keywords: Radiotherapy, Immunotherapy, Radiosensitivity, Immune checkpoint blockade, Radiosensitivity index
Introduction
Radiotherapy can provide excellent local control of tumor growth by directly inducing single strand breaks (SSBs) and double strand breaks (DSBs) in DNA, as well as apoptosis and necrosis of tumor cells through the formation of reactive oxygen species (ROS) and free radicals, and is an irreplaceable therapeutic tool in cancer treatment [1]. Radiotherapy also has potent immunomodulatory potential by promoting tumor-specific antigen production and enhancing the initiation and activation of cytotoxic T cells, thereby allowing tumor clearance in immune surveillance. In addition, radiotherapy may induce immunogenic cell death through the release of cytokines, inflammatory mediators, and other immune-related molecules. Despite the activated CD8+ T cells and other immunostimulatory cells can migrate and infiltrate to metastatic sites to act as anti-tumor, but the upregulation of immune-suppressed cells by inflammatory factors may inhibit the anti-tumor effects and lead to tumor progression. This suggests that radiotherapy alone is not sufficient to completely eliminate primary and metastatic tumor lesions [2].
Based on the understanding of cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) and the programmed cell death protein 1/ programmed death-ligand 1 (PD-1/PD-L1) and other pathways in tumor immune microenvironment, immune checkpoint inhibitors (ICIs) can enhance the intrinsic immune response against tumor antigens by promoting T cell activation and function, and have been approved for the treatment of a variety of tumors [3]. However, not all patients derive benefit from this treatment and the effective rate of ICIs alone is only 20-30%, with a majority of patients initially developing primary drug resistance or acquiring secondary drug resistance soon after treatment [4]. Augmented immunotherapy involves increased release of tumor antigens, T cell infiltration, and enhanced antigen presentation. Several mechanisms of immune escape have been postulated to explain the failure of tumor immune attacks. A better understanding of these mechanisms will help us to seek therapeutic strategies to overcome immunotherapy resistance [5].
In recent years, the combination of radiotherapy and immunotherapy (CRI) can increase mutual sensitization and enhance antitumor effects, and their synergistic effects have shown survival benefits in multiple studies [6]. To begin with, radiotherapy triggers the release and presentation of tumor-associated antigens (TAAs), which enhance systemic responses by triggering the recruitment of antigen-presenting cells (APCs), such as macrophages, dendritic cells (DCs), and B cells that enhance T-cell infiltration and promote anti-tumor immune responses in the host [7]. Activation of tumor cells by radiotherapy can reshape the tumor microenvironment to reduce immunotherapy resistance, induce antigen release and cross-presentation of DCs, and trigger the recruitment and activation of APCs, which play a key role in the antitumor immune response [8, 9]. Moreover, radiation promotes the release of cytokines and chemokines, which leads to increased production and recruitment of fibroblast growth factor (FGF), transforming growth factor-β (TGF-β), interleukin 1β (IL-1β) and tumor necrosis factor (TNF), which activate Treg cells, bone marrow-derived suppressor cells and cancer-associated fibroblasts [10, 11]. A recent study indicated that radiation-induced DNA DSBs upregulate PD-L1 expression in tumor cells via ATM/ATR/Chk1 kinase, but immunotherapy can prevent the immunosuppressive effects caused by radiotherapy [12]. In addition, dysfunction of the tumor vascular system can lead to an immunosuppressive microenvironment and induce radioresistance, and immunotherapy creates a potential opportunity to reduce tumor hypoxia and improve radiosensitivity. When tumor cells are activated by immunotherapy, activation of CD8+ T cells and production of interferon (IFN)-γ to normalize tumor vasculature can sensitize tumors to radiation therapy through mechanisms that include normalization of the tumor vascular system and tissue hypoxia [13–15] (Fig. 1). Therefore, the CRI to synergistically counteract the innate and adaptive immunity of cancer cells, as well as to bypass immune tolerance and exhaustion is highly prospective clinically.
Fig. 1.
Mechanism of interaction between radiotherapy and immunotherapy
The identification of biomarker-based approaches is central to the development of clinical strategies for CRI, but most of the previous studies on the efficacy of CRI have focused on the dose, timing, efficacy, and sequence of the combination of the two treatments [13, 16]. There is no standardized choice for the sequence of radiotherapy combined with immunology, so the timing used in different studies currently varies. In multiple preclinical and clinical trials, immunology prior to or concurrent with radiotherapy is the superior choice [17, 18]. There is additional evidence to suggest that sequential therapy and the early use of immunotherapy after radiotherapy can increase the clinical benefits, which is beneficial for newly recruited T cells to destroy tumors [19]. Although preclinical works have shown that immunotherapy has a radiosensitizing effect, the window of opportunity for optimizing this synergy is limited as it includes many confounding factors [20]. Therefore, the optimal sequence of radiotherapy and immunotherapy still needs to be explored through large randomized clinical trials. There is now increasing evidence that the intrinsic radiosensitivity in tumor cells also influences the release of cancer cell antigens and affects antigen-specific T cell activation during the radiation-induced cancer immune cycle [21]. As we all know, the most significant radiobiological factors affecting tumor response to radiotherapy are summarized as the “5 Rs”: DNA damage repair, redistribution in cell cycle, repopulation, reoxygenation, and intrinsic radiosensitivity of cancer cells [22]. Among them, the radiosensitivity of tumor cells is the main determinant of tumor response to radiation [23]. Recently, reactivation of the antitumor immune response has been recognized as the “6th R”, which extends the concept of radiosensitivity beyond the tumor cells themselves and supports improved outcomes when radiotherapy is combined with immunotherapy [24]. In this review, we focus on the radiosensitivity of tumor cells to explore its influencing factors, prediction methods and interactions on the immune system. Furthermore, we explore the predictive value of radiosensitivity to CRI efficacy, which is expected to provide new directions for improving the efficacy of CRI.
The influence factors of tumor radiosensitivity
The intrinsic radiosensitivity of tumor cells is the main determinant of tumor response to radiation, which involves multiple tumor signaling pathways and molecular biological information [23] (Fig. 2). The cellular origin and differentiation of tumor tissues are the main factors affecting the radiosensitivity of tumor cells. Tumors originating from radiosensitive tissues are more sensitive to radiation, while poorly differentiated tumors are less sensitive to radiation [25]. The radiosensitivity of an individual depends to a large extent on biological factors related to epigenetic factors, and the epigenetic mechanisms that determine the selection of metabolic patterns also contribute to the individual radiosensitivity and adaptability of an organism. On the one hand, DNA methylation affects the initial damage process, and on the other hand, methylation shift to ab initio type is associated with further development of protective and repair processes [26]. However, the exact underlying genetic factors that contribute to the inter-individual differences in cellular radiosensitivity are unknown. Understanding the cellular and genetic basis of radiosensitivity and identifying individuals with higher or lower radiosensitivity will facilitate population risk assessment, disease prediction, individualized radiotherapy, and the development of radiation protection standards [27]. Moreover, observations of human tumors have revealed a clear relationship between cell proliferation and cell renewal rates and radiosensitivity. Any tumor with rapidly average growth rate and elevated cell renewal rate is also more sensitive to radiation, and the cellular radiosensitivity differs in different periods, so the redistribution of cell cycle phases within the cell population after irradiation can alter the radiosensitivity.
Fig. 2.
The influence factors of tumor intrinsic radiosensitivity
In spite of the many factors (i.e., dose, exposure volume, gender, age, underlying disease, and lifestyle) that may influence individual radiosensitivity and radiosensitivity to cancer, the inherent cellular radiosensitivity is genetically determined and supported by genetic alterations involving DNA damage repair [28, 29]. Genetic alterations in proteins involved in DNA damage repair are responsible for individual differences in radiation response. Genetic mutations in DNA repair response-related genes (i.e., p53, ATM, BRCA1, BRCA2, ERCC1, XRCC3 and Rad51) have also been found to be associated with radiosensitivity in lung cancer correlation [30, 31]. For instance, individuals with pure mutations in ATM have an approximately three-fold increase in radiosensitivity at the cellular, tissue and biological levels compared to average [32]. The development of DNA-based markers is currently underway, and areas for additional research include the role of somatic mutations in DNA damage response genes that affect radiosensitivity. Exposure of cells to extracellular matrix proteins can increase radioresistance by promoting DNA damage repair and activation of the Akt/MAPK signaling pathway [33]. It has been demonstrated that the anti-apoptotic protein nucleolin (C23) can enhance radiosensitivity in non-small cell lung cancer (NSCLC) by affecting the activity of DNA-dependent protein kinase (DNA-PK) [34].
There is growing evidence that viral pathogenic factors are associated with the regulation of cellular radiation response, treatment outcome, and clinical prognosis in patients following radiotherapy, with the regulation of DNA damage repair mechanisms being the most common point of attack [35]. Malignancies with a viral etiology are more immunogenic, such as human papillomavirus (HPV), Epstein-Barr virus (EBV) and other virus types that are more sensitive to anticancer therapy. One work identified a group of Head and neck squamous carcinoma (HNSCC) that may benefit from CRI and showed a significantly improved prognosis in patients with HPV-positive tumors, attributed to increased intrinsic radiosensitivity and possibly to the modulation of cytotoxic T-cell responses in the tumor microenvironment [35]. Recent study indicated that for HPV-positive HNSCC, the virus hijacked cellular mechanisms of DNA repair, altered cell cycle distribution, induced cell proliferation and displayed peculiar hypoxic kinetics during radiation treatment [36]. The mechanism described involves a reduced ability to repair DNA double-strand breaks, accompanied by enhanced radiation-induced G2/M cell cycle arrest [37, 38]. Additionally, excessive expression of immune checkpoints was also strongly associated with radiosensitivity. This finding suggested that high PD-1 expression was significantly associated with the clinical prognosis of HPV/p16-positive HNSCC. Patients in the radioresistant group and HPV/p16-negative group with radioresistant genetic markers could benefit from combination CRI [39]. The central research on EBV-regulated radiation response has focused on LMP-1, which is expressed in most EBV-associated malignancies.LMP-1 inhibits DNA double-strand break repair by inhibiting the phosphorylation and activity of DNA-PKcs, a key enzyme of the NHEJ pathway in nasopharyngeal carcinoma (NPC), and by inhibiting ATM repair of DNA double-strand breaks [40].
In addition to the tumor cells themselves, environmental factors such as oxygenation status may also affect radiosensitivity by further modulating damage induction and cellular responses [41]. Therefore, as a classical regulator of tumor radiation resistance, the elimination of hypoxia may be a potential solution to address radioresistance [42]. Hypoxia inducible factor-1 (HIF-1) remains active in cells that survive radiation therapy and is associated with tumor cell resistance to radiotherapy. It has been suggested that it may modulate tumor radioresistance through reprogramming of glucose metabolism and cell cycle regulation [43]. Tumors contain different proportions of intrinsically radioresistant cancer stem cell (CSC), which are closely associated with tumor hypoxia, and HIF-1α contributes to the development and maintenance of the CSC phenotype [44]. The radioresistance of CSC is characterized by a reduced accumulation of radiation-induced DNA damage and increased activation of anti-apoptotic signaling pathways compared to differentiated tumor cells [45]. Current strategies for predicting normal tissue radiosensitivity are genomics and large-scale prospective studies, and further research is still needed to explore the best predictive methods for radiosensitivity [46].
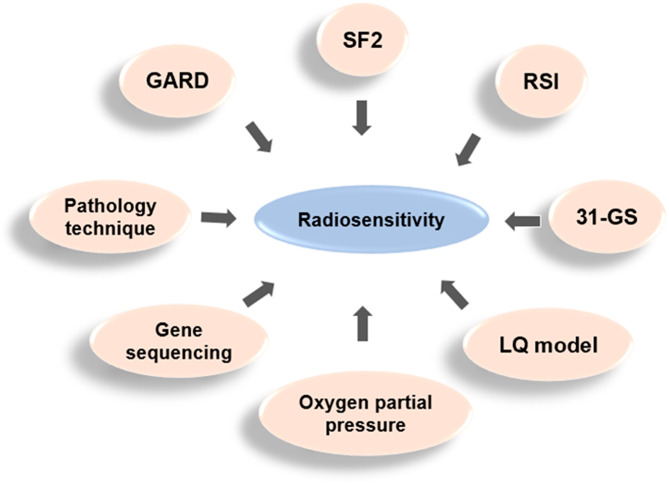
The prediction methods of tumor radiosensitivity
The radiosensitivity of tumor cells is strongly influenced by molecular variation at the genomic, transcriptional and translational levels. Radiosensitivity is a measure of the response of cells, tissues or individuals to ionizing radiation and can be used to predict which individuals will benefit from radiotherapy. Recent advances in gene sequencing technology and microarray technology for high-throughput RNA analysis have driven interest in identifying features that measure the intrinsic radiosensitivity of tumor cells. The development of a successful predictive analysis of radiosensitivity has been a major goal of research, and many genetic markers have been developed to predict the radiosensitivity of tumors [47]. These methods can be broadly divided into two categories: one is the characterization of the surviving fraction of cancer cell lines formed after radiation, which reflects the intrinsic radiosensitivity of cancer cells, but fails to consider the influence of non-malignant cells in the tumor microenvironment, particularly the role of anti-tumor immunity [48]. The second is the prediction of patient progression after radiotherapy. This is dedicated to predicting the clinical outcome of radiotherapy, but cannot be used for cellular level studies and it is difficult to reveal radiobiologically based mechanisms [49]. Nevertheless, how to build a radiosensitivity prediction model has not been discussed systematically in these recent years.
The traditional experimental approach to determine intrinsic radiosensitivity is the survival of tumor cell lines at a single dose of 2 Gy (SF2), but is not applicable for routine use and alternative strategies must be sought. The radiosensitivity index (RSI) is a 10-gene model based on the survival of 48 human cancer cell lines at SF2 radiation and is a measure of clonogenic survival after a given radiation dose [50]. The 10-gene model (AR, cJun, STAT1, PKC, RelA, cABL, SUMO1, CDK1, HDAC1, and IRF1) that hold a crucial role in DNA damage response, histone deacetylation, cell cycle regulation, apoptosis and proliferation [50, 51]. The RSI prediction model is a linear regression algorithm and is independent of the cancer type. RSI is designed to detect intrinsic tumor radiosensitivity independently of cancer type and has been independently validated as a pan-tissue biomarker of radiosensitivity at multiple disease sites [52–54]. The 31-genes were developed by analyzing a panel of NCI-60 cancer cells that were associated with SF2 expression, and its correlation with radiosensitivity has been validated in various malignancies [48]. Similarly, measuring the oxygen partial pressure of a tumor can indicate its level of hypoxia, which can help predict its radiosensitivity [23]. Unfortunately, these parameters, even when used in combination, are insufficient to predict tumor radioresistance for clinical use.
Since the relationship between radiation dose and survival is nonlinear, various mathematical formulas have been proposed to fit the radiation survival curve. The linear quadratic (LQ) model has become the most popular calculator for analyzing and predicting ionizing radiation response in the laboratory and in the clinic, where the α/β ratio is used to characterize the sensitivity of specific tissue types to segmentation [55]. The LQ model provides a simple equation between cell survival and delivered dose: S = exp (-αD-βD2) [56]. The radiosensitivity of cells is influenced by complex interactions between intrinsic polygenic traits. As the mechanisms and biomarkers of radiosensitivity have become better understood, gene expression classifiers containing few key genes have been used to predict radiosensitivity in specific tumor types or various human cancers [57, 58]. Based on RSI, LQ model, and the time and dose of radiotherapy received by each patient, a team derived a genome-based model for adjusting radiotherapy dose (GARD) on more than 8,000 tumor samples from more than 20 tumor types [59]. The GARD predicts the efficacy of radiotherapy and guides the radiation dose to match the individual tumor radiosensitivity, with higher GARD values associated with better efficacy of radiotherapy. Given that the range of GARD values varies among different types of cancer, the use of RSI alone cannot be a complete representation of the treatment effect, and we need to combine the means of tumor type and genetic testing to determine the appropriate radiotherapy dose for individual patients.
Besides the classical biological mechanisms mentioned above, gene sequencing has further revealed the regulatory role of non-coding RNAs on radiosensitivity, and their high-throughput properties contribute to the study of radiosensitivity mechanisms. A previous study used a gene expression classifier to predict radiosensitivity, which regarded radiosensitivity as a continuous variable, used microarray significance analysis for gene selection, and multiple linear regression model for radiosensitivity prediction [57]. Three new genes (RbAp48, RGS19 and R5PIA) were identified in the gene selection step, and their expression values were correlated with radiosensitivity and were transfected with cancer cell lines. The results established that the RbAp48 gene could induce radiosensitivity 1.5-2 times, and increased the proportion of cells in G2-M phase of cell cycle. In addition, the study also showed that the overexpression of RbAp48 was related to the dephosphorylation of Akt, which suggested that RbAp48 may exert its effects by antagonizing the Ras pathway. This study established that radiosensitivity can be predicted based on gene expression profiles and introduced a genomic approach to identify novel molecular markers of radiosensitivity [60]. Moreover, some traditional pathology techniques remain valid for measuring tumor radiosensitivity. For instance, hematoxylin and eosin staining can be used to identify radiosensitive (i.e., seminoma) or radioresistant (i.e., glioma) tumors [61] (Fig. 3). More advanced pathologic techniques such as DNA methylationome analysis are now used to classify tumors, but have not yet guided the clinical prescription of radiotherapy doses. The current strategy for predicting normal tissue radiosensitivity is genomics and large-scale prospective studies, and further studies are still needed to explore the best predictive methods for radiosensitivity.
Fig. 3.
Common prediction methods for radiosensitivity
The biomarkers of tumor radiosensitivity
Unsatisfactory radiosensitivity has been plagued, and finding biomarkers that predict radiosensitivity could help improve the efficacy of radiotherapy. Chromosomal aberrations and DNA damage, in particular DSB, are among the few cellular markers that have some correlation with cellular radiosensitivity. Signaling pathway molecules involved in the DNA damage response are excellent candidates for the evaluation of radiosensitivity biomarkers, and relevant biomarkers include MRE11, AIMP3, NBN, and BRE, with MRE11 potentially a predictive biomarker for radiotherapy benefit [62]. The current research suggests that γ-H2AX assay as a rapid and sensitive biomarker can be used in epidemiological studies to measure changes in radiosensitivity. The use of γ-H2AX lesion analysis as well as DSB repair gene polymorphisms can be used to assess cellular radiosensitivity, which will assist in population risk assessment, disease prediction, individualized radiotherapy, and the development of radiation protection standards [63]. Additionally, evaluation of the predictive significance of the systemic immune-inflammatory index (SII) on overall survival and radiosensitivity in advanced NSCLC showed favorable radiosensitivity in the low SII group, and higher SII levels were associated with poorer overall survival and radiosensitivity [64]. Cellular radiosensitivity can be assessed by quantifying DSB damage and repair [65]. It has been observed that among the different types of DNA damage, DSBs have the slower and most lethal repair dynamics. Therefore, they are more helpful in explaining clinical radiosensitivity than other types of damage with rapid repair dynamics [66]. The development of DNA-based markers is currently underway and areas for further research include the role of somatic mutations in DNA damage response genes that affect radiosensitivity [67].
The molecular mechanisms involved in the radiation-induced response are complex and the expression levels of genes do not consistently represent the properties of all proteins in the tumor cells. The proteomic approach allows the identification of various proteins involved in the cellular response to ionizing radiation, which may be useful in identifying potential candidates for use as predictive biomarkers. The expression levels of genes do inconsistently represent the nature of all proteins in normal or tumor cells, and therefore direct detection of protein expression may be more effective in determining the complexity of the mechanisms and the large number of molecular signatures involved in the cellular radiation-induced responses. The radiosensitivity of tumors is related to the basal expression levels of intracellular or cell membrane proteins, and the direct detection of protein expression using proteomics studies allows the detection of protein sequences and post-translational modifications stored in genes that can be used for early diagnosis, prognosis and treatment of cancer [68]. Current proteomics technologies can be used to detect and analyze proteomic information using cells, tissues or body fluids, providing a better platform for biomarker research and development [69, 70]. High throughput radio proteomics is the latest tool, where mass spectrometry (MS) is used to analyze and identify unknown proteins by converting protein molecules into gas phase ions through an ionisation source and applying the electromagnetic field of the instrument to separate proteins with a specific mass-to-charge ratio. MS has the advantage of quickly analysis, high sensitivity and resolution. The advantages of MS are its speed, sensitivity and resolution. Proteomics research based on liquid chromatography-mass spectrometry (LC-MS) is now widely used [71]. The intrinsic radiosensitivity of NSCLC is mainly regulated by the signal pathways in the proteoglycans, focal adhesion and the actin cytoskeleton in cancer. Radiosensitivity-specific proteins can guide clinical individualized radiotherapy by predicting radiation response in NSCLC patients [72].
The effect of radiosensitivity on the efficacy of CRI
In the era of immunotherapy, reliable genomic predictors to identify optimal patient populations in CRI are lacking. A comprehensive analysis of radiosensitivity-associated genes and proteins in lung cancer and other solid tumors has been used to identify potential biological predictors of radiosensitivity [73]. There are some evidences that radiosensitivity could predict the effect of radiotherapy and immunotherapy (Table 1). To determine first whether tumor radiosensitivity correlates with immune system activation in all tumor types, Tobin et al. identified 10,240 genotypically distinct solid primary tumors using 12 chemokine genes to define intratumor immune activation and determined that low RSI was significantly associated with elevated immune activation, supporting the association of RSI with immune-related signaling networks in patients’ tumors (using an RSI threshold of 0.3745) [74]. In another study, a total of 12,832 primary tumors from 11 major cancer types were analyzed in relation to DNA repair and immune subtypes in order to determine whether genomic scores of radiosensitivity were associated with immune responses. The results found that RSI was related with various immune-related signatures and predicted responses to PD-1 blockade, emphasizing the promising potential of RSI as a candidate biomarker for CRI [75]. In addition, a study also identified enhanced immune checkpoint interactions in radioresistant tumors, providing a new theoretical basis for radiotherapy and ICIs for the treatment of HNSCC [76]. The RSI-low may be characterized by higher genomic instability and subsequently higher mutational burden, which associated with predicted efficacy of dominant IFN-γ signaling responses and PD-1 blockade. Taken together, RSI-Low tumors may represent a special subgroup and therapeutic target for immunotherapy [75].
Table 1.
The predictive role of radiosensitivity in radiotherapy and immunotherapy
| Study | Cancer type | Sample Size(n) | Outcome |
|---|---|---|---|
| Tobin et al. (2017) [74] | Breast cancer | 282 | RSI-low status that refer to more radiosensitive tumor (HR 0.58, 95% CI 0.34-1.00; p = 0.05) and 12-CK-high status that refer to more immune-active tumor (HR 0.61,95% CI 0.39–0.96; p = 0.03) were independently related with improved distant metastasis free survival. |
| Dai et al. (2021) [75] | 11 major cancer types | 12,832 primary tumors and 585 metastatic tissues | RSI was significantly associated with immune-related molecular features(p < 0,05). RSI-Low tumors carried more higher portion of follicular T helper cells, T cell gamma delta cells, activated NK cells and M1 macrophages than RSI-High tumors. |
| Grass et al. (2022) [87] | 31 primary tumors types | 10,469 | A weak negative relevance between the RSI and immune score (Pearson’s r = − 0.28; Spearman’s r = − 0.27, P < 0.001). Tumors with high radiosensitivity showed significant enrichment of IFN-related signaling pathways and immune cell infiltration (i.e., CD8+ T cells, activated NK cells, M1-macrophages, q < 0.05). |
RSI, radiosensitivity index; NK, natural killer; IFN, interferon
The molecular mechanisms underlying the biological effects of radiotherapy can affect the response and repair of cells to DSBs, but there is currently limited research on the mechanisms of RSI and immune response [77].Research has found that RSI is associated with various immune-related genomic and molecular characteristics, and low RSI is correlated with dominant response to IFN-γ signaling and predicted efficacy of PD-1 blocking agents [75]. Lower RSI is linked to higher HRD scores and higher TMB, indicating the presence of defective DNA repair mechanisms and potential for response to immune based therapies [78]. Besides, lower RSI is also correlated with higher RNA stemness score, indicating higher degrees of stemness and tumor de-differentiation, which is also related to increased PD-L1 protein expression [79].To further explore the relationship between RSI and immune response, a team used whole transcriptomic and matched proteomic data from 12,832 primary and 585 metastatic tumors and found that RSI was associated with a variety of immune-related genomic and molecular features. Lower RSI was associated with higher homologous recombination deficiency (HRD) scores and higher tumor mutational burden (TMB), suggesting the presence of defective DNA repair mechanisms and response potential to immune-based therapies [78]. HRD scores were correlated with genes involved in homologous repair, including BRCA1, BRCA2, RAD51B, and RAD51C, and alterations in these genes were related to radiosensitivity [80, 81]. Intriguingly, the RSI-Low tumors exhibit both higher microsatellite instability (MSI) and TMB molecular profiles in gastric cancer, which were shown to be subgroups with favorable prognosis after immunotherapy [82]. Furthermore, since RSI genes (STAT1 and IRF1) are downstream of IFN-γ-mediated signaling, RSI correlates better with various immune-related molecular features and phenotypes than other genes and genetic features associated with radiation response [83].
At the same time, the role of the immune system is crucial for tumor radiosensitivity. To explore the relationship between intrinsic tumor radiosensitivity and the immune system, a study has investigated radiation-induced tumor equilibrium and dormancy in animal models, and whether host immune responses contribute to radiation-induced tumor equilibrium [84, 85]. The study developed two mouse models—TUBO (HER2-positive breast cancer) and B16 (melanoma), and has observed four possible tumor responses to radiotherapy. These were non-responsive tumors (non-responsive to radiotherapy); responsive tumors (tumor regression observed within 10 days after radiation); stable tumors (tumors that regress and remain stable and palpable during any 34-60-day observation period); late recurrent tumors (tumor recurrence after 60 days). The inherent cellular radiosensitivity of tumors is frequently hypothesized to explain the observed differences in tumor regeneration rates observed after radiotherapy, and this study determined the radiosensitivity of tumor cells taken from mice that responded variably to radiotherapy. These tumors were surgically removed and digested into single cell suspensions and subjected to 2, 5, or 10 Gy of in vitro irradiation and assessed with clonogenic assays. The results demonstrated that tumor cells with different responses to radiation in vivo exhibited indistinguishable radiosensitivity in vitro. This finding revealed that the degree of tumor cells radiosensitivity was unable to explain the different tumor responses to local radiotherapy, in contrast to immune cells and their cytokines, which have been shown to exhibit a pivotal role in inhibiting tumor cell regeneration in two experimental animal model systems.
Traditional radiosensitivity studies have focused on tumor cells, neglecting the effects of the tumor microenvironment, which consists of stromal and immune cells [86]. To explore the relationship between RSI and its associated unique tumor immune microenvironment, a study used RSI to assess the radiosensitivity of 10,469 primary tumor samples and to assess the immune environmental components of each tumor. The results showed that tumors with high immune cell content were more sensitive to radiation because they were enriched with leukocytes, which are highly sensitive to radiation. Furthermore, tumors estimated to be highly sensitive to radiotherapy exhibited significant enrichment of interferon-related signaling pathways and immune cell infiltration (i.e., CD8+ T cells, activated natural killer cells, M1 macrophages) [87]. In the radiation-induced cancer immune cycle, intrinsic radiosensitivity affects cancer cell antigen release and immune status affects antigen-specific T cell activation [88]. To elucidate the effect of tumor microenvironment on the efficacy of radiotherapy in glioma patients, a study analyzed the differences in the infiltration levels of immune cells. Patients were classified into a radiosensitive (RS) group and a radioresistant (RR) group. The results showed that the level of activated NK cell infiltration was significantly higher in the RS group, whereas the level of macrophage, Treg cell, and resting NK cell infiltration was significantly higher in the RR group, and the immune score and PD-L1 expression levels were significantly higher in the RR group than in the RS group. These results indicated that patients in the RR group had higher immunogenicity, higher TMB and mutational characteristics, which requires more clinical trials to demonstrate [89, 90].
Integrating tumor radiosensitivity and immune status to predict clinical outcomes
In addition to focusing only on intrinsic tumor radiosensitivity, the integration of radiosensitivity features and immune features could predict the clinical outcomes of patients (Table 2). One study has developed independent predictors of radiosensitivity signature (RSS) and an immune signature (IMS) in breast cancer patients treated with radiotherapy. When integrating both signatures, patients with radiosensitive or immune effective tumors gained better disease-specific survival (DSS) from radiotherapy. On the contrary, patients in the other group, defined as radiotherapy resistance and immunodeficient, had significantly lower DSS when they received radiotherapy. Individuals in the radiosensitive and immunodeficient or radiotherapy resistant and immune effective, there was no significant difference in DSS between treatment groups [91]. Another study in the Cancer Genome Atlas (TCGA) dataset showed significantly higher PD-L1 expression in the RR group than in the RS group, and the PD-L1-high-RR group had the worst survival, so the analysis focused on this group of patients. These studies demonstrated that 31 genetic features and PD-L1 expression status as potential predictive markers for radiotherapy. Moreover, patients classified as PD-L1-high-RR exhibit radiotherapy resistance and immunosuppressive TME through multiple mechanisms and may benefit from radiotherapy combined with PD-1/PD-L1 blockers. Therefore, the integration of 31 genetic characteristics and PD-L1 expression status may help to classify the patient population that may benefit most from the combination of radiotherapy and PD-1/PD-L1 blockade in clinical practice [92]. In addition, it has been shown that RSI and PD-L1 status predict clinical outcome in patients with glioblastoma multiforme. The 399 patients were divided into RS and RR groups based on radiosensitivity genetic markers and into PD-L1 high and PD-L1 low groups based on CD274 mRNA expression. Differential and comprehensive analyzes of expression and methylation data were performed. The results demonstrate the potential efficacy of radiotherapy in combination with PD-1/PD-L1 blockade and angiogenesis inhibition in the PD-L1-high-RR group [93].
Table 2.
The predictive role of radiosensitivity and immune gene signatures for clinical outcome of patients
| Study | Cancer type | Sample Size(n) | Outcome |
|---|---|---|---|
| Cui et al. (2018) [91] | Breast cancer | 1439 |
Patients treated with radiotherapy had significantly better DSS in the immune-effective group (HR 0.46; P = 0.0076). Both radiosensitivity and immune signatures could predict the benefit from radiotherapy (Pinteraction=0.007 and 0.005). |
| Jang et al. (2018) [92] | Lower grade glioma | 511 | Patients classified as the PD-L1-high-radioresistant group showed a detrimental effect on OS rate and may benefit most from radiotherapy combined with immunotherapy (HR: 1.96; CI: 1.01–3.80; p = 0.047). |
| Jang et al. (2020) [93] | Glioblastoma | 399 | PD-L1-high-radioresistant group could potentially benefit from radiotherapy combined with immunotherapy and angiogenesis inhibition (HR, 1.70, 95%CI, 1.03–2.81; p = 0.037). |
| Dai et al. (2021) [94] | Head and neck squamous cell carcinoma | 288 | The survival rate and B cell count of the radioresistant and PD-L1-high group were lower than those of the other groups (p < 0.05). |
| Sun et al. (2021) [97] | Head and neck squamous cell carcinoma | 392 | Only patients in the radiosensitive-immune group had better OS after receiving radiotherapy (HR 0.194, 95%CI 0.788–0.480; p < 0.001). |
DSS, disease specific survival; OS, overall survival
Tumor radiosensitivity is also governed by other features of cancer, including tumor microenvironment dynamics, nutrient utilization, and multiple cellular complexes. A study showed that the RR-PD-L1-high group had depleted B cells and had a significantly lower survival rate than the other groups, which predicted the prognosis of patients with locally advanced HNSCC [94]. Some evidence points to the possibility that the pathways associated with radiosensitivity may also modulate the immunogenicity of tumors and predict their response to immunotherapy. For example, inactivation of the DNA repair mechanism may trigger an immune response and impair tumor growth by triggering the release of neoantigens, and the therapeutic efficacy of immunotherapy can be predicted by the presence of these DNA repair defects [95]. Several common regulators of DNA repair and immune checkpoints have been identified, such as PARP inhibitors capable of DNA repair proficiency and radiosensitization of tumor cells [96]. Several studies have demonstrated that the combined stratification of intrinsic radiosensitivity and immune status is superior to considering intrinsic radiosensitivity or immune status separately, and can therefore be used in preclinical evaluations to select patients or to determine whether radiation sensitizers and immunotherapy should be used together [97]. With respect to whether immunotherapy modulates tumor intrinsic radiosensitivity, increasing evidence supports the idea that DNA repair defects modulate tumor immune checkpoints, but whether the immune checkpoints in turn modulate DNA repair pathways remains unclear, and this potential new mechanism by which immunotherapy modulates tumor intrinsic radiosensitivity still deserves further exploration in the future.
Future Prospect
Since intrinsic radiosensitivity and immune status affect the initial and effective phases of the radiation-induced cancer immune cycle, respectively, it is necessary to consider radiation in combination with immunity when selecting patients who may benefit from radiotherapy. Moreover, the prognostic value of RSI has been validated using multiple independent datasets, such as those used to predict the prognosis of patients treated with radiation for breast, pancreatic, glioblastoma, esophageal, and metastatic colorectal cancers [51, 98–100]. Despite the recognized differences in tumor radiosensitivity in preclinical and clinical settings, radiation dose prescriptions are not currently individualized in the field of radiation oncology based on the biology of the patient’s tumor. However, individualized adjustment of radiation dose based on patient tumor radiosensitivity is a promising strategy for effective radiotherapy, and radiosensitivity indices are expected to be potential biomarkers for combination radiotherapy and immunotherapy.
Conclusion
In this review, we first present the mechanisms underlying the interaction between radiotherapy and immunotherapy, where radiotherapy serves as an essential adjunct to immunotherapy by providing a source of danger signals, antigens and activation of innate immunity. Similarly, immunotherapy can sensitize tumors to subsequent radiotherapy, reducing the radiation dose required to eradicate them. We next describe the effect of tumor cell radiosensitivity and the method to predict it. The biological effects of radiation are mediated by a complex network of signaling pathways, and advances in genomics can be used to guide radiotherapy alone or in combination, and the commercialization of genomic-based tools will be important to facilitate its implementation. Furthermore, radiosensitivity holds favorable promise for the predictive role and clinical application of radiation-free combination, and future clinical investigations will need to emphasize the implementation of preclinical and translational discovery data in the development of new clinical trials to demonstrate reproducibility in the patient setting and to help optimize the efficacy of their combination therapy. In summary, the radiosensitivity of tumor cells can help predict the efficacy of CRI and the integration of immune status with radiosensitivity can also help better predict clinical outcome. In the future, the treatment of CRI should rely on the mining and detection of multiple biomarkers to achieve precision oncology.
Acknowledgements
None.
Abbreviations
- APCs
Antigen-presenting cells
- CRI
Combination of radiotherapy and immunotherapy
- CSC
Cancer stem cell
- CTLA-4
Cytotoxic T lymphocyte-associated antigen 4
- DCs
Dendritic cells
- DNA-PK
DNA-dependent protein kinase
- DSBs
Double strand breaks
- DSS
Disease-specific survival
- FGF
Fibroblast growth factor
- GARD
Adjusting radiotherapy dose
- HIF-1
Hypoxia inducible factor-1
- HNSCC
Head and neck squamous carcinoma
- HPV
Human papillomavirus
- HRD
Homologous recombination deficiency
- ICIs
Immune checkpoint inhibitors
- IFN
Interferon
- IL-1β
Interleukin 1β
- IMS
Immune signature
- LC-MS
Liquid chromatography-mass spectrometry
- LQ
Linear quadratic
- MS
Mass spectrometry
- MSI
Microsatellite instability
- NK
Natural killer
- NPC
Nasopharyngeal carcinoma
- NSCLC
Non-small cell lung cancer
- PD-1
Programmed death protein-1
- PD-L1
Programmed death-ligand 1
- ROS
Reactive oxygen species
- RR
Radioresistant
- RS
Radiosensitive
- RSI
Radiosensitivity index
- RSS
Radiosensitivity signature
- SII
Systemic immune-inflammatory index
- SSBs
Single strand breaks
- TAAs
Tumor-associated antigens
- TCGA
The Cancer Genome Atlas
- TGF-β
Transforming growth factor-β
- TMB
Tumor mutational burden
- TNF
Tumor necrosis factor
Author contributions
ZG performed data analysis and manuscript preparation. QZ and YX re-verified the data and polished the language. LW designed the study and revised the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by National Natural Science Foundation of China (Grant number 82172865), Start-up fund of Shandong Cancer Hospital (Grant number 2020-B14), Clinical Research Special Fund of Wu Jieping Medical Foundation (Grant number 320.6750.2021-02-51 and 320.6750.2021-17-13).
Data Availability
Not applicable.
Declarations
Conflict of interests
The authors declare that they have no competing interests.
Ethics approval
Not applicable.
Consent for publication
All authors give their consent to publish this manuscript.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Park SS, Dong H, Liu X, et al. PD-1 restrains Radiotherapy-Induced Abscopal Effect. Cancer Immunol Res. 2015;3(6):610–9. doi: 10.1158/2326-6066.CIR-14-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chicas-Sett R, Zafra-Martin J, Morales-Orue I et al. Immunoradiotherapy as an effective therapeutic strategy in Lung Cancer: from Palliative Care to curative intent. Cancers (Basel). 2020; 12(8). [DOI] [PMC free article] [PubMed]
- 3.Das S, Johnson DB. Immune-related adverse events and anti-tumor efficacy of immune checkpoint inhibitors. J Immunother Cancer. 2019;7(1):306. doi: 10.1186/s40425-019-0805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schoenfeld AJ, Hellmann MD. Acquired Resistance to Immune Checkpoint inhibitors. Cancer Cell. 2020;37(4):443–55. doi: 10.1016/j.ccell.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kon E, Benhar I. Immune checkpoint inhibitor combinations: current efforts and important aspects for success. Drug Resist Updat. 2019;45:13–29. doi: 10.1016/j.drup.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Kunos CA, Coleman CN. Current and future initiatives for Radiation Oncology at the National Cancer Institute in the era of Precision Medicine. Int J Radiat Oncol Biol Phys. 2018;102(1):18–25. doi: 10.1016/j.ijrobp.2017.02.225. [DOI] [PubMed] [Google Scholar]
- 7.Pointer KB, Pitroda SP, Weichselbaum RR. Radiotherapy and immunotherapy: open questions and future strategies. Trends Cancer. 2022;8(1):9–20. doi: 10.1016/j.trecan.2021.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Taieb J, Chaput N, Ménard C, et al. A novel dendritic cell subset involved in tumor immunosurveillance. Nat Med. 2006;12(2):214–9. doi: 10.1038/nm1356. [DOI] [PubMed] [Google Scholar]
- 9.Sharabi AB, Nirschl CJ, Kochel CM, et al. Stereotactic Radiation Therapy augments Antigen-Specific PD-1-Mediated Antitumor Immune responses via Cross-Presentation of Tumor Antigen. Cancer Immunol Res. 2015;3(4):345–55. doi: 10.1158/2326-6066.CIR-14-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanpouille-Box C, Diamond JM, Pilones KA, et al. TGFβ is a Master Regulator of Radiation Therapy-Induced Antitumor Immunity. Cancer Res. 2015;75(11):2232–42. doi: 10.1158/0008-5472.CAN-14-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasi P, Paolini A, Nano R, Di Liberto R, Capelli E. Effects of single or combined treatments with radiation and chemotherapy on survival and danger signals expression in glioblastoma cell lines. Biomed Res Int. 2014;2014:453497. doi: 10.1155/2014/453497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato H, Niimi A, Yasuhara T, et al. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat Commun. 2017;8(1):1751. doi: 10.1038/s41467-017-01883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharabi AB, Lim M, DeWeese TL, Drake CG. Radiation and checkpoint blockade immunotherapy: radiosensitisation and potential mechanisms of synergy. Lancet Oncol. 2015;16(13):e498–509. doi: 10.1016/S1470-2045(15)00007-8. [DOI] [PubMed] [Google Scholar]
- 14.Ma Y, Adjemian S, Mattarollo SR, et al. Anticancer chemotherapy-induced intratumoral recruitment and differentiation of antigen-presenting cells. Immunity. 2013;38(4):729–41. doi: 10.1016/j.immuni.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Zheng X, Fang Z, Liu X, et al. Increased vessel perfusion predicts the efficacy of immune checkpoint blockade. J Clin Invest. 2018;128(5):2104–15. doi: 10.1172/JCI96582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williamson CW, Sherer MV, Zamarin D, et al. Immunotherapy and radiation therapy sequencing: state of the data on timing, efficacy, and safety. Cancer. 2021;127(10):1553–67. doi: 10.1002/cncr.33424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dovedi SJ, Adlard AL, Lipowska-Bhalla G, et al. Acquired resistance to fractionated radiotherapy can be overcome by concurrent PD-L1 blockade. Cancer Res. 2014;74(19):5458–68. doi: 10.1158/0008-5472.CAN-14-1258. [DOI] [PubMed] [Google Scholar]
- 18.Golden EB, Demaria S, Schiff PB, Chachoua A, Formenti SC. An abscopal response to radiation and ipilimumab in a patient with metastatic non-small cell lung cancer. Cancer Immunol Res. 2013;1(6):365–72. doi: 10.1158/2326-6066.CIR-13-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antonia SJ, Villegas A, Daniel D, et al. Overall survival with Durvalumab after Chemoradiotherapy in Stage III NSCLC. N Engl J Med. 2018;379(24):2342–50. doi: 10.1056/NEJMoa1809697. [DOI] [PubMed] [Google Scholar]
- 20.Azad A, Yin Lim S, D’Costa Z, et al. PD-L1 blockade enhances response of pancreatic ductal adenocarcinoma to radiotherapy. EMBO Mol Med. 2017;9(2):167–80. doi: 10.15252/emmm.201606674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39(1):1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 22.Steel GG, McMillan TJ, Peacock JH. The 5Rs of radiobiology. Int J Radiat Biol. 1989;56(6):1045–8. doi: 10.1080/09553008914552491. [DOI] [PubMed] [Google Scholar]
- 23.Gerweck LE, Vijayappa S, Kurimasa A, Ogawa K, Chen DJ. Tumor cell radiosensitivity is a major determinant of tumor response to radiation. Cancer Res. 2006;66(17):8352–5. doi: 10.1158/0008-5472.CAN-06-0533. [DOI] [PubMed] [Google Scholar]
- 24.Boustani J, Grapin M, Laurent PA, Apetoh L, Mirjolet C. The 6th R of Radiobiology: reactivation of Anti-Tumor Immune Response. Cancers (Basel). 2019; 11(6). [DOI] [PMC free article] [PubMed]
- 25.Szumiel I. Intrinsic radiation sensitivity: cellular signaling is the key. Radiat Res. 2008;169(3):249–58. doi: 10.1667/RR1239.1. [DOI] [PubMed] [Google Scholar]
- 26.Kravets AP, Sokolova DA. Epigenetic factors of individual radiosensitivity and adaptive capacity. Int J Radiat Biol. 2020;96(8):999–1007. doi: 10.1080/09553002.2020.1767819. [DOI] [PubMed] [Google Scholar]
- 27.Jeggo P, Lavin MF. Cellular radiosensitivity: how much better do we understand it? Int J Radiat Biol. 2009;85(12):1061–81. doi: 10.3109/09553000903261263. [DOI] [PubMed] [Google Scholar]
- 28.Schuster B, Hecht M, Schmidt M et al. Influence of gender on Radiosensitivity during Radiochemotherapy of Advanced rectal Cancer. Cancers (Basel). 2021; 14(1). [DOI] [PMC free article] [PubMed]
- 29.Klement RJ. The influence of ketogenic therapy on the 5 R’s of radiobiology. Int J Radiat Biol. 2019;95(4):394–407. doi: 10.1080/09553002.2017.1380330. [DOI] [PubMed] [Google Scholar]
- 30.Lee YS, Oh JH, Yoon S, et al. Differential gene expression profiles of radioresistant non-small-cell lung cancer cell lines established by fractionated irradiation: tumor protein p53-inducible protein 3 confers sensitivity to ionizing radiation. Int J Radiat Oncol Biol Phys. 2010;77(3):858–66. doi: 10.1016/j.ijrobp.2009.12.076. [DOI] [PubMed] [Google Scholar]
- 31.Jin JY, Wang W, Ten Haken RK, et al. Use a survival model to correlate single-nucleotide polymorphisms of DNA repair genes with radiation dose-response in patients with non-small cell lung cancer. Radiother Oncol. 2015;117(1):77–82. doi: 10.1016/j.radonc.2015.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thariat J, Chevalier F, Orbach D, et al. Avoidance or adaptation of radiotherapy in patients with cancer with Li-Fraumeni and heritable TP53-related cancer syndromes. Lancet Oncol. 2021;22(12):e562–e74. doi: 10.1016/S1470-2045(21)00425-3. [DOI] [PubMed] [Google Scholar]
- 33.Sandfort V, Koch U, Cordes N. Cell adhesion-mediated radioresistance revisited. Int J Radiat Biol. 2007; 83(11–12): 727 – 32. [DOI] [PubMed]
- 34.Xu JY, Lu S, Xu XY, et al. Knocking Down Nucleolin expression enhances the radiosensitivity of Non-Small Cell Lung Cancer by influencing DNA-PKcs activity. Asian Pac J Cancer Prev. 2015;16(8):3301–6. doi: 10.7314/APJCP.2015.16.8.3301. [DOI] [PubMed] [Google Scholar]
- 35.Rödel F, Martin D, Balermpas P, et al. Modulation of radiation sensitivity and antitumor immunity by viral pathogenic factors: implications for radio-immunotherapy. Biochim Biophys Acta Rev Cancer. 2019;1871(1):126–37. doi: 10.1016/j.bbcan.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 36.Swick AD, Chatterjee A, De Costa AM, Kimple RJ. Modulation of therapeutic sensitivity by human papillomavirus. Radiother Oncol. 2015;116(3):342–5. doi: 10.1016/j.radonc.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rieckmann T, Tribius S, Grob TJ, et al. HNSCC cell lines positive for HPV and p16 possess higher cellular radiosensitivity due to an impaired DSB repair capacity. Radiother Oncol. 2013;107(2):242–6. doi: 10.1016/j.radonc.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 38.Kimple RJ, Smith MA, Blitzer GC, et al. Enhanced radiation sensitivity in HPV-positive head and neck cancer. Cancer Res. 2013;73(15):4791–800. doi: 10.1158/0008-5472.CAN-13-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyu X, Zhang M, Li G, Jiang Y, Qiao Q. PD-1 and PD-L1 expression predicts Radiosensitivity and Clinical Outcomes in Head and Neck Cancer and is Associated with HPV infection. J Cancer. 2019;10(4):937–48. doi: 10.7150/jca.27199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu J, Tang M, Li H, et al. EBV-LMP1 suppresses the DNA damage response through DNA-PK/AMPK signaling to promote radioresistance in nasopharyngeal carcinoma. Cancer Lett. 2016;380(1):191–200. doi: 10.1016/j.canlet.2016.05.032. [DOI] [PubMed] [Google Scholar]
- 41.Martinive P, Defresne F, Bouzin C, et al. Preconditioning of the tumor vasculature and tumor cells by intermittent hypoxia: implications for anticancer therapies. Cancer Res. 2006;66(24):11736–44. doi: 10.1158/0008-5472.CAN-06-2056. [DOI] [PubMed] [Google Scholar]
- 42.Dewhirst MW. A potential solution for eliminating hypoxia as a cause for radioresistance. Proc Natl Acad Sci U S A. 2018;115(42):10548–50. doi: 10.1073/pnas.1814212115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harada H, Inoue M, Itasaka S, et al. Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate towards tumour blood vessels. Nat Commun. 2012;3:783. doi: 10.1038/ncomms1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linge A, Lohaus F, Löck S, et al. HPV status, cancer stem cell marker expression, hypoxia gene signatures and tumour volume identify good prognosis subgroups in patients with HNSCC after primary radiochemotherapy: a multicentre retrospective study of the German Cancer Consortium Radiation Oncology Group (DKTK-ROG) Radiother Oncol. 2016;121(3):364–73. doi: 10.1016/j.radonc.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 45.Price JM, Prabhakaran A, West CML. Predicting tumour radiosensitivity to deliver precision radiotherapy. Nat Rev Clin Oncol. 2023;20(2):83–98. doi: 10.1038/s41571-022-00709-y. [DOI] [PubMed] [Google Scholar]
- 46.Chua ML, Rothkamm K. Biomarkers of radiation exposure: can they predict normal tissue radiosensitivity? Clin Oncol (R Coll Radiol) 2013;25(10):610–6. doi: 10.1016/j.clon.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 47.Du Z, Zhang X, Tang Z. More evidence for prediction model of radiosensitivity. Biosci Rep. 2021; 41(4). [DOI] [PMC free article] [PubMed]
- 48.Kim HS, Kim SC, Kim SJ, et al. Identification of a radiosensitivity signature using integrative metaanalysis of published microarray data for NCI-60 cancer cells. BMC Genomics. 2012;13:348. doi: 10.1186/1471-2164-13-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li G, Jiang Y, Lyu X, et al. Gene signatures based on therapy responsiveness provide guidance for combined radiotherapy and chemotherapy for lower grade glioma. J Cell Mol Med. 2020;24(8):4726–35. doi: 10.1111/jcmm.15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eschrich SA, Pramana J, Zhang H, et al. A gene expression model of intrinsic tumor radiosensitivity: prediction of response and prognosis after chemoradiation. Int J Radiat Oncol Biol Phys. 2009;75(2):489–96. doi: 10.1016/j.ijrobp.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eschrich SA, Fulp WJ, Pawitan Y, et al. Validation of a radiosensitivity molecular signature in breast cancer. Clin Cancer Res. 2012;18(18):5134–43. doi: 10.1158/1078-0432.CCR-12-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rink L, Skorobogatko Y, Kossenkov AV, et al. Gene expression signatures and response to imatinib mesylate in gastrointestinal stromal tumor. Mol Cancer Ther. 2009;8(8):2172–82. doi: 10.1158/1535-7163.MCT-09-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eschrich S, Zhang H, Zhao H, et al. Systems biology modeling of the radiation sensitivity network: a biomarker discovery platform. Int J Radiat Oncol Biol Phys. 2009;75(2):497–505. doi: 10.1016/j.ijrobp.2009.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khan MT, Yang L, More E, et al. Developing Tumor Radiosensitivity Signatures using LncRNAs. Radiat Res. 2021;195(4):324–33. doi: 10.1667/RADE-20-00157.1. [DOI] [PubMed] [Google Scholar]
- 55.Daguenet E, Khalifa J, Tolédano A, et al. To exploit the 5 ‘R’ of radiobiology and unleash the 3 ‘E’ of immunoediting: ‘RE’-inventing the radiotherapy-immunotherapy combination. Ther Adv Med Oncol. 2020;12:1758835920913445. doi: 10.1177/1758835920913445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pérez-Romasanta LA, González-Del Portillo E, Rodríguez-Gutiérrez A, Matías-Pérez Á. Stereotactic radiotherapy for Hepatocellular Carcinoma, Radiosensitization Strategies and Radiation-Immunotherapy Combination. Cancers (Basel). 2021; 13(2). [DOI] [PMC free article] [PubMed]
- 57.Torres-Roca JF, Eschrich S, Zhao H, et al. Prediction of radiation sensitivity using a gene expression classifier. Cancer Res. 2005;65(16):7169–76. doi: 10.1158/0008-5472.CAN-05-0656. [DOI] [PubMed] [Google Scholar]
- 58.Amundson SA, Do KT, Vinikoor LC, et al. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer drug screen. Cancer Res. 2008;68(2):415–24. doi: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
- 59.Scott JG, Berglund A, Schell MJ, et al. A genome-based model for adjusting radiotherapy dose (GARD): a retrospective, cohort-based study. Lancet Oncol. 2017;18(2):202–11. doi: 10.1016/S1470-2045(16)30648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meng L, Xu J, Ye Y, et al. The combination of Radiotherapy with Immunotherapy and potential predictive biomarkers for treatment of Non-Small Cell Lung Cancer Patients. Front Immunol. 2021;12:723609. doi: 10.3389/fimmu.2021.723609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Avril D, Foy JP, Bouaoud J, Grégoire V, Saintigny P. Biomarkers of radioresistance in head and neck squamous cell carcinomas. Int J Radiat Biol. 2022: 1–11. [DOI] [PubMed]
- 62.Forker LJ, Choudhury A, Kiltie AE. Biomarkers of Tumour Radiosensitivity and Predicting Benefit from Radiotherapy. Clin Oncol (R Coll Radiol) 2015;27(10):561–9. doi: 10.1016/j.clon.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 63.Mumbrekar KD, Goutham HV, Vadhiraja BM, Bola Sadashiva SR. Polymorphisms in double strand break repair related genes influence radiosensitivity phenotype in lymphocytes from healthy individuals. DNA Repair (Amst) 2016;40:27–34. doi: 10.1016/j.dnarep.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 64.Li X, Hu P, Liu J, Zhang J, Liu Q. Systemic immune-inflammation index predicted overall survival and radiosensitivity in advanced non-small-cell lung cancer. Future Oncol. 2020;16(5):103–15. doi: 10.2217/fon-2019-0761. [DOI] [PubMed] [Google Scholar]
- 65.Taneja N, Davis M, Choy JS, et al. Histone H2AX phosphorylation as a predictor of radiosensitivity and target for radiotherapy. J Biol Chem. 2004;279(3):2273–80. doi: 10.1074/jbc.M310030200. [DOI] [PubMed] [Google Scholar]
- 66.Popanda O, Ebbeler R, Twardella D, et al. Radiation-induced DNA damage and repair in lymphocytes from breast cancer patients and their correlation with acute skin reactions to radiotherapy. Int J Radiat Oncol Biol Phys. 2003;55(5):1216–25. doi: 10.1016/S0360-3016(02)04415-2. [DOI] [PubMed] [Google Scholar]
- 67.Price JM, Prabhakaran A, West CML. Predicting tumour radiosensitivity to deliver precision radiotherapy. Nat Rev Clin Oncol. 2022. [DOI] [PubMed]
- 68.Lacombe J, Azria D, Mange A, Solassol J. Proteomic approaches to identify biomarkers predictive of radiotherapy outcomes. Expert Rev Proteomics. 2013;10(1):33–42. doi: 10.1586/epr.12.68. [DOI] [PubMed] [Google Scholar]
- 69.Zhang H, Liu T, Zhang Z, et al. Integrated Proteogenomic characterization of Human High-Grade Serous Ovarian Cancer. Cell. 2016;166(3):755–65. doi: 10.1016/j.cell.2016.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang B, Wang J, Wang X, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513(7518):382–7. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li X, Wang W, Chen J. Recent progress in mass spectrometry proteomics for biomedical research. Sci China Life Sci. 2017;60(10):1093–113. doi: 10.1007/s11427-017-9175-2. [DOI] [PubMed] [Google Scholar]
- 72.Zhu X, Wang Y, Jiang C, et al. Radiosensitivity-specific Proteomic and Signaling Pathway Network of Non-Small Cell Lung Cancer (NSCLC) Int J Radiat Oncol Biol Phys. 2022;112(2):529–41. doi: 10.1016/j.ijrobp.2021.08.041. [DOI] [PubMed] [Google Scholar]
- 73.Kim EH, Park AK, Dong SM, Ahn JH, Park WY. Global analysis of CpG methylation reveals epigenetic control of the radiosensitivity in lung cancer cell lines. Oncogene. 2010;29(33):4725–31. doi: 10.1038/onc.2010.223. [DOI] [PubMed] [Google Scholar]
- 74.Strom T, Harrison LB, Giuliano AR, et al. Tumour radiosensitivity is associated with immune activation in solid tumours. Eur J Cancer. 2017;84:304–14. doi: 10.1016/j.ejca.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dai YH, Wang YF, Shen PC, et al. Radiosensitivity index emerges as a potential biomarker for combined radiotherapy and immunotherapy. NPJ Genom Med. 2021;6(1):40. doi: 10.1038/s41525-021-00200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li G, Jiang Y, Li G, Qiao Q. Comprehensive analysis of radiosensitivity in head and neck squamous cell carcinoma. Radiother Oncol. 2021;159:126–35. doi: 10.1016/j.radonc.2021.03.017. [DOI] [PubMed] [Google Scholar]
- 77.Masuda Y, Kamiya K. Molecular nature of radiation injury and DNA repair disorders associated with radiosensitivity. Int J Hematol. 2012;95(3):239–45. doi: 10.1007/s12185-012-1008-y. [DOI] [PubMed] [Google Scholar]
- 78.Knijnenburg TA, Wang L, Zimmermann MT et al. Genomic and molecular Landscape of DNA damage Repair Deficiency across the Cancer Genome Atlas. Cell Rep. 2018; 23(1): 239 – 54.e6. [DOI] [PMC free article] [PubMed]
- 79.Malta TM, Sokolov A, Gentles AJ et al. Machine learning identifies stemness features Associated with Oncogenic Dedifferentiation. Cell 2018; 173(2): 338 – 54.e15. [DOI] [PMC free article] [PubMed]
- 80.Baeyens A, Thierens H, Claes K, et al. Chromosomal radiosensitivity in BRCA1 and BRCA2 mutation carriers. Int J Radiat Biol. 2004;80(10):745–56. doi: 10.1080/09553000400017937. [DOI] [PubMed] [Google Scholar]
- 81.Zhong X, Luo G, Zhou X, et al. Rad51 in regulating the radiosensitivity of non-small cell lung cancer with different epidermal growth factor receptor mutation status. Thorac Cancer. 2016;7(1):50–60. doi: 10.1111/1759-7714.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goodman AM, Sokol ES, Frampton GM, Lippman SM, Kurzrock R. Microsatellite-stable tumors with high mutational Burden Benefit from Immunotherapy. Cancer Immunol Res. 2019;7(10):1570–73. doi: 10.1158/2326-6066.CIR-19-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shao L, Hou W, Scharping NE, et al. IRF1 inhibits Antitumor Immunity through the Upregulation of PD-L1 in the Tumor cell. Cancer Immunol Res. 2019;7(8):1258–66. doi: 10.1158/2326-6066.CIR-18-0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liang H, Deng L, Chmura S, et al. Radiation-induced equilibrium is a balance between tumor cell proliferation and T cell-mediated killing. J Immunol. 2013;190(11):5874–81. doi: 10.4049/jimmunol.1202612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weichselbaum RR, Liang H, Deng L, Fu YX. Radiotherapy and immunotherapy: a beneficial liaison? Nat Rev Clin Oncol. 2017;14(6):365–79. doi: 10.1038/nrclinonc.2016.211. [DOI] [PubMed] [Google Scholar]
- 86.Byrne NM, Tambe P, Coulter JA. Radiation Response in the Tumour Microenvironment: predictive biomarkers and future perspectives. J Pers Med. 2021; 11(1). [DOI] [PMC free article] [PubMed]
- 87.Grass GD, Alfonso JCL, Welsh E, et al. The Radiosensitivity Index Gene signature identifies distinct Tumor Immune Microenvironment characteristics Associated with susceptibility to Radiation Therapy. Int J Radiat Oncol Biol Phys. 2022;113(3):635–47. doi: 10.1016/j.ijrobp.2022.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Darragh LB, Oweida AJ, Karam SD. Overcoming resistance to Combination Radiation-Immunotherapy: a focus on contributing pathways within the Tumor Microenvironment. Front Immunol. 2018;9:3154. doi: 10.3389/fimmu.2018.03154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yan D, Zhao Q, Du Z, et al. Development and validation of an immune-related gene signature for predicting the radiosensitivity of lower-grade gliomas. Sci Rep. 2022;12(1):6698. doi: 10.1038/s41598-022-10601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jang BS, Han W, Kim IA. Tumor mutation burden, immune checkpoint crosstalk and radiosensitivity in single-cell RNA sequencing data of breast cancer. Radiother Oncol. 2020;142:202–09. doi: 10.1016/j.radonc.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 91.Cui Y, Li B, Pollom EL, Horst KC, Li R. Integrating radiosensitivity and Immune Gene Signatures for Predicting Benefit of Radiotherapy in breast Cancer. Clin Cancer Res. 2018;24(19):4754–62. doi: 10.1158/1078-0432.CCR-18-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jang BS, Kim IA. A radiosensitivity gene signature and PD-L1 predict the clinical outcomes of patients with lower grade glioma in TCGA. Radiother Oncol. 2018;128(2):245–53. doi: 10.1016/j.radonc.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 93.Jang BS, Kim IA. A radiosensitivity gene signature and PD-L1 Status Predict Clinical Outcome of patients with Glioblastoma Multiforme in the Cancer Genome Atlas dataset. Cancer Res Treat. 2020;52(2):530–42. doi: 10.4143/crt.2019.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dai D, Guo Y, Shui Y, et al. Combination of Radiosensitivity Gene signature and PD-L1 Status predicts clinical outcome of patients with locally Advanced Head and Neck squamous cell carcinoma: a study based on the Cancer Genome Atlas dataset. Front Mol Biosci. 2021;8:775562. doi: 10.3389/fmolb.2021.775562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Germano G, Lamba S, Rospo G, et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature. 2017;552(7683):116–20. doi: 10.1038/nature24673. [DOI] [PubMed] [Google Scholar]
- 96.Alotaibi M, Sharma K, Saleh T, et al. Radiosensitization by PARP inhibition in DNA repair proficient and deficient tumor cells: proliferative recovery in senescent cells. Radiat Res. 2016;185(3):229–45. doi: 10.1667/RR14202.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun C, Zhang M, Qiao Q, Wang Y. Integrating intrinsic radiosensitivity and Immune Status for Predicting benefits of Radiotherapy in Head and Neck squamous cell carcinoma. Med Sci Monit. 2021;27:e932126. doi: 10.12659/MSM.932126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ahmed KA, Chinnaiyan P, Fulp WJ, et al. The radiosensitivity index predicts for overall survival in glioblastoma. Oncotarget. 2015;6(33):34414–22. doi: 10.18632/oncotarget.5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Strom T, Hoffe SE, Fulp W, et al. Radiosensitivity index predicts for survival with adjuvant radiation in resectable pancreatic cancer. Radiother Oncol. 2015;117(1):159–64. doi: 10.1016/j.radonc.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ahmed KA, Fulp WJ, Berglund AE, et al. Differences between Colon Cancer Primaries and Metastases using a molecular assay for Tumor Radiation Sensitivity Suggest Implications for potential oligometastatic SBRT patient selection. Int J Radiat Oncol Biol Phys. 2015;92(4):837–42. doi: 10.1016/j.ijrobp.2015.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.