Dear Editor,
China has represented an unprecedented and unique landscape for COVID-19 propagation, its population being mostly vaccinated with whole virus inactivated vaccines, but infection-naive at the beginning of the 2022-2023 Winter wave. China's vaccination rate has been unfortunately low in people aged 80 and above (42.3%) [1]. Understanding whether China-released data are reliable, and exactly which sublineages circulate in China is of relevance for the evolution of the pandemic.
In order to better assess the consistency of China-released data with ex-China surveillance implemented by several countries at the end of 2022 [2,3], we conducted a search of the GISAID database [4] for all SARS-CoV-2 sequences sampled in China between September 1 2022 and January 31, 2023. We excluded sequences labeled as “imported” in metadata. We also searched all SARS-CoV-2 sequences labelled as deposited from the rest of the world during the same time interval and labelled as incoming from China in metadata: the latter data could not be assigned to individual provincial divisions within China. We monitored Omicron sublineages which are fast-growing in the rest of the World (BQ.1.*, XBB.1.*, BN.1.3 and CH.1.1.*), while leaving all other lineages within the “others” group.
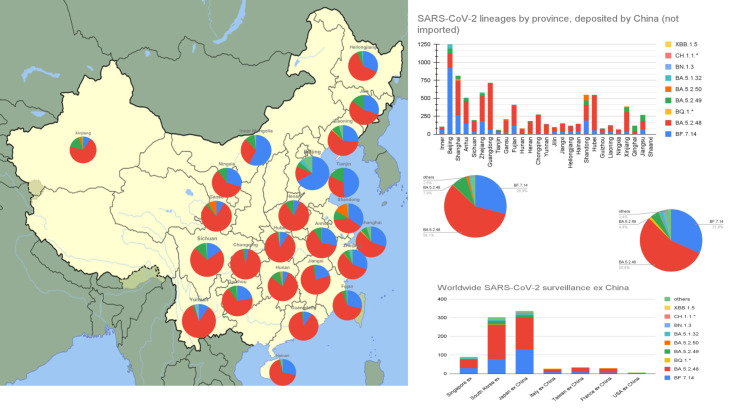
Fig. 1 shows that the 2 cohorts (8380 sequences reported from 23 China provincial divisions, and 827 sequences reported from surveillance of Chinese travelers to the rest of the world) largely overlap, with great dominance of the novel autochthonous BA.5-derived PANGOLIN lineages BA.5.2.48 (58.1% vs. 55.6%) and BF.7.14 (29.0% vs. 31.4%). Three more novel autochthonous SARS-CoV-2 lineages have been also detected in China but in very low prevalence (BA.5.2.49 7.9% vs. 4.4%, BA.5.2.50 1.4% vs 0.2%, BA.5.1.32 0.9% vs. 2.3%). BQ.1.* was detected in only 13 cases (0.2%) and 11 cases (1.3%), while BN.1.3 was detected in just 6 (0.1%) and 5 cases (0.6%) respectively. CH.1.1 was detected only in 3 cases from foreign surveillance, and XBB.1.5 was not detected.
Fig. 1.
China-released sequences of local COVID-19 cases stratified by province compared to data from surveillance of Chinese passengers fled abroad (September 1 2022-January 31 2023).
The concordance between China-released and foreign data is good news for transparency, and provides coverage of almost all of China's provincial divisions, such sharing from China is still negligible in a country peaking at an estimate of 1 million cases and 5,000 deaths per day at the end of 2022. By comparison, the GISAID EpiCoV™ database has collected 14.8 million SARS-CoV-2 genomes from the rest of the world in 3 years.
From pandemic evolution in the rest of the world, we have learnt how the virus mutates to escape Spike-based vaccine-elicited and infection-elicited immunities. This is not the case yet in China's first wave, where, despite unrestricted and massive passenger traffic between China and the rest of the world, autochthonous lineages are outperforming those currently dominant in the western world [5]. Vice versa, the lack of relevant detection of such China-originated lineages outside China is likely attributable to their poor immune escape scores. The cumulative infection attack rate (that is the proportion of population who has been infected) in Beijing has been estimated at 92.3% since November 1 2022 to January 31, 2023 [6]. In such quick burst, China has soon developed its peculiar immune escaping variant soup, with derivatives of BF.7.14 (BF.7.14.1-7), BA.5.2.48 (DY.1-4) and BA.5.2.49 (DZ.1-2). Interestingly, no additional mutations within the Spike receptor binding domain have been reported in Chinese lineages so far. In addition to founder effects, differences in former exposure to variants, type of vaccine, and eventually host genetics are likely contributors to the ongoing geographical restriction.
Experience from a putative former coronavirus pandemic suggests that the most severe wave is not necessarily among the first ones [7], and the impressive decline in genomic sequencing rates across the world (from 5% in November 2021 to 1% in December 2022) represents a serious concern [8].
Funding
None.
Data sharing statement
the original SARS-CoV-2 sequences are available from GISAID.org. The dataset used to generate Fig. 1 is available at https://docs.google.com/spreadsheets/d/e/2PACX-1vRdallgY8 × 5ZAwDRM7o2PvR7iAP120yuSZojcT378oUG1StpM8NLFdzv0A6E37w_ZlNwWuj7mN7H67N/pubhtml
Declaration of Competing Interest
We declare we have no conflict of interest related to this manuscript.
Acknowledgements
We gratefully acknowledge all data contributors, i.e., the Authors and their Originating laboratories responsible for obtaining the specimens, and their Submitting laboratories for generating the genetic sequence and metadata and sharing via the GISAID Initiative, on which this research is based.
References
- 1.The press conference of the Joint Prevention and Control Mechanism of the State Council on December 14, 2022 introduced the relevant situation of medical and health services and drug production and supply. Accessed online at http://www.nhc.gov.cn/xwzb/webcontroller.do?titleSeq=11497&gecstype=1 on January 16, 2023.
- 2.Novazzi F, Giombini E, Rueca M, Baj A, Fabeni L, Genoni A, et al. Genomic surveillance of SARS-CoV-2 positive passengers on flights from China to Italy, December 2022. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2023;28 doi: 10.2807/1560-7917.ES.2023.28.2.2300008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pung R, Kucharski AJ, Marc Ho ZJ, Lee VJ. Patterns of infection among travellers to Singapore arriving from mainland China. 2023:2023.01.16.23284584.
- 4.Khare S, Gurry C, Freitas L, Schultz MB, Bach G, Diallo A, et al. GISAID's Role in Pandemic Response. China CDC Wkly. 2021;3:1049–1051. doi: 10.46234/ccdcw2021.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Jiang S, Jiang S, Li X, Ai J, Lin K, et al. Neutralization of SARS-CoV-2 BQ.1.1 and XBB.1.5 by Breakthrough Infection Sera from Previous and Current Waves in China. 2023:2023.02.07.527406. [DOI] [PMC free article] [PubMed]
- 6.Leung K, Lau EHY, Wong CKH, Leung GM, Wu JT. Estimating the transmission dynamics of SARS-CoV-2 Omicron BF.7 in Beijing after the adjustment of zero-COVID policy in November - December 2022. Nat Med. 2023 doi: 10.1038/s41591-023-02212-y. [DOI] [PubMed] [Google Scholar]
- 7.Vijgen L, Keyaerts E, Moës E, Thoelen I, Wollants E, Lemey P, et al. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. Journal of virology. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.SARS-CoV-2 sequencing intensity. Accessed online at https://cov-spectrum.org/explore/World/AllSamples/Past6M on January 18, 2022.