Abstract
Objective
X-linked Dystonia Parkinsonism (XDP) is associated with a SINE-VNTR- Alu (SVA) retrotransposon insertion in an intron of the TAF1 gene that alters gene transcription and splicing. In this study, we determined if the SVA insertion introduces glucocorticoid (GC)-responsive cis-regulatory elements that may contribute to dysregulated TAF1 transcription and XDP disease progression.
Methodology
We performed in silico analysis to identify potential GC receptor (GR) binding sites within the XDP-SVA. We also conducted promoter-reporter assays on HeLa and HEK293T cells to assess the intrinsic promoter activity of three XDP-SVA variants representing different hexameric repeat lengths associated with differences in disease onset. We treated XDP fibroblast cell models with GR agonist (CORT) or antagonist (RU486), then subjected TAF1 and the XDP-associated aberrant transcript, TAF1-32i to gene expression analysis.
Results
A transcription factor binding site search revealed three binding sites for GR within the XDP-SVA—two within the SINE region and one in the Alu region. Promoter-reporter assays showed induction of XDP-SVA promoter activity upon CORT treatment that was dependent on the cell line and XDP-SVA hexamer repeat length. Gene expression analysis showed that baseline TAF1 levels differed between control and patient fibroblast cell lines, and treatment with CORT led to an increasing trend in the expression of the aberrant TAF1-32i transcript but did not reach statistical significance. Treatment with RU486 increased TAF1 mRNA expression only in the control cell lines.
Conclusion
Using reporter assays, the XDP-SVA was shown to exhibit CORT-dependent transcriptional activation. Gene expression analysis also showed that GC signaling may influence TAF1 and TAF1-32i expression, possibly through interaction with the XDP-SVA. Our data provide a potential link between stress and XDP progression.
Keywords: XDP, glucocorticoids, stress, neurodegenerative disease, TAF1
INTRODUCTION
X-linked Dystonia Parkinsonism (XDP) is a neurodegenerative disease that primarily affects males native to the island of Panay, Philippines. It is characterized by the initial appearance of focal dystonia, followed by traits commonly associated with Parkinson’s Disease, including tremors, bradykinesia, masked facies, hypomimia, and drooling.1 At present, the prevalence rate of XDP is unknown2 with the most recent estimate from 2010 placing it at 5.74 in 100,000 on Panay Island, and 0.31 in 100,000 in the Philippines.3 Symptoms manifest late into adulthood, with an average age of onset at 39-40 years,1 making early diagnosis and treatment difficult. Currently, available treatment options include oral medication, chemodenervation, neuroablative surgery, and deep brain stimulation.1 However, these only serve to alleviate symptoms as currently there is no cure. Patient demise usually occurs due to complications in the aerodigestive tract caused by extreme motor dysfunction.1
As an X-linked recessive disorder, XDP patients harbor disease-specific nucleotide changes (DSCs) within the DYT3 gene locus4 and an insertion of a SINE-VNTR-Alu (SVA)-type retrotransposon in the TAF1 locus5 located on the X-chromosome. The SVA insertion contains a hexameric repeat domain whose length appears to inversely correlate with the age of disease onset.6 Furthermore, an aberrant TAF1 transcript, known as TAF1-32i, was found to be exclusively transcribed in XDP patient cells.5 The TAF1-32i transcript is a truncated version of the canonical TAF1 mRNA that terminates ~700 bp before the SVA insertion point and incorporates an intronic segment from the same region. The expression level of the aberrant TAF1-32i transcript is inversely proportional to TAF1 mRNA expression levels.5 Removal of the SVA insertion via CRISPR/Cas9-mediated gene editing in patient-derived neural stem cells abolished TAF1-32i transcript synthesis and rescued the attenuated expression of TAF1 mRNA,5 suggesting that the insertion of the SVA retrotransposon may lead to the synthesis of aberrant transcripts and a decrease in expression of canonical TAF1 that may consequently translate to a reduction of TAF1 protein synthesis. In promoter-luciferase assays, the XDP-SVA has been shown to exhibit intrinsic promoter activity6 as well as cis-regulatory activity via TAF1 promoter repression.7 It has been proposed that the insertion of the SVA retrotransposon provides new binding sites within the locus, which may lead to the transcriptional interference of TAF1 or the binding of trans-acting elements that contribute to disease progression.6
Several studies have also looked into the psychosocial aspect of XDP. Measurements of Quality of Life (QoL) through in-depth interviews indicate a pattern of QoL impairment which manifests not only physically but also socially and emotionally.8 Furthermore, case studies have found that depressive symptoms are prevalent among XDP patients, the severity of which are comparable to those seen in patients with Parkinson’s disease (PD), Alzheimer’s disease (AD), and spinocerebellar degeneration.9 As of 2015, the suicide rate among the population of XDP patients is 10.8% which is higher than the 2015 Philippine estimate of 3.59 in 100,000 cases.10 As such, it is also of interest to understand how and why psychosocial factors may influence XDP progression and pathophysiology. One way in which these may be integrated is through the Hypothalamic-Pituitary-Adrenal (HPA) axis which mediates the stress response. The HPA integrates environmental and social stimuli and the neuroendocrine system. The presence of stressors activates the HPA axis that leads to the secretion of glucocorticoids (GC) which then binds to the GC receptor (GR), a member of the steroid receptor superfamily.11 In the presence of a bound ligand, GR forms dimers that translocate to the nucleus and bind to GC response elements (GREs) of target genes to regulate GC-dependent transcription.12 Activation of these target genes leads to the reorganization and modulation of neural circuits, which translate to the phenotypes displayed by stressed individuals.13 One such target is Krüppel-like Factor 9 (KLF9), a transcription factor that is highly expressed in the brain and is involved in various neuronal development processes.14
Stress and dysregulation of the HPA axis have been shown to contribute to the physiology of neurodegenerative diseases. Patients afflicted with PD exhibit increased GC levels indicative of dysregulated GR function, leading to chronic inflammation characteristic of the disease.15 In AD, stress has been shown to contribute to disease progression through the induction of Tau-hyperphosphorylation. Stress-associated GC secretion leads to aggregation of misfolded and hyperphosphorylated variants of the Tau protein which then causes cytoskeletal instability and neuronal damage, ultimately resulting in neurodegeneration.16 However, less is known about the role of stress in XDP.
In this study, we determined whether a relationship between the stress response and XDP-associated dysregulated TAF1 gene transcription exists. We hypothesized that the SVA insertion provides a binding site for GR and that GR binding to the disease-associated XDP-SVA contributes to the dysregulation of TAF1 transcription through a decrease in expression of the canonical transcript and an increase in expression of aberrant transcripts such as TAF1-32i. Towards this end, we tested XDP-SVA promoter activity upon acute hydrocortisone (CORT) treatment via a luciferase reporter assay. We also employed cellular models of XDP for use in gene expression analysis upon GR agonist and antagonist treatment to identify the XDP-specific role of GR on TAF1 and TAF1-32i transcription.
METHODOLOGY
In Silico Analysis
We subjected the full SVA sequence to a transcription factor binding site (TFBS) search using LASAGNA-Search tool 2.0.17 Search parameters were adjusted to identify binding sites for NR3C1 or GR, by selecting the following TFs: vertebrates, JASPAR ID MA0113.1; Homo sapiens, TRANSFAC Accession Number T00337 and T01920; vertebrates, TRANSFAC Accession Number M00205. For the TF Model Input, the option “Use TRANSFAC TFBSs” under “LASAGNA-Aligned Models” was selected. We filtered results to include only those with associated p-values <0.001. We subsequently mapped putative binding sites onto the SVA sequence to identify the location of putative GREs.
Cell Culture
HeLa cells were cultured in Modified Eagle Medium (MEM) with 10% fetal bovine serum (FBS), 1X Penicillin-Streptomycin-Glutamine, and 2.5% Amphotericin B. HEK293T cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 10% FBS, 1X Penicillin-Streptomycin-Glutamine, and 2.5% Amphotericin B. For reporter assays, HeLa and HEK293T cells were seeded in a 96-well plate at a density of 1.0 x 104 cells/well. Upon cell attachment, media was replaced with MEM (HeLa cells) or DMEM (HEK293T) supplemented with 10% charcoalstripped serum (CSS) to remove possible effects of steroid hormones from the media.
Immortalized fibroblast cell lines from XDP patients and ethnically matched controls with Filipino ancestry were obtained from the laboratory of Dr. Christopher Bragg of the Massachusetts General Hospital (USA). The cell lines were generated in a previous study with participants that were evaluated at Massachusetts General Hospital, or in regional clinics in Panay Island affiliated with Jose Reyes Memorial Medical Center (Manila, Philippines). Sample tissue collection and cell line derivation were done with the approval of the institutional review board at both institutions and with the informed consent of all study participants.5 Cell lines were validated through clinical evaluation of the source patients, as well as genetic testing for the seven known haplotype markers of XDP, including the SVA insertion.5 The patient fibroblasts used in this study, with the identifier 35833, were sourced from an XDP patient who had an age of onset of 36 years. The XDP-SVA hexameric domain for these cells was found to have a repeat length of 50. The ethnically matched control fibroblasts had the identifier 36176.5 Primary fibroblasts were cultured in DMEM supplemented with 20% FBS, 1X Penicillin-Streptomycin-Glutamine, and 2.5% Amphotericin B. For gene expression analysis, control and XDP fibroblasts were seeded in 6-well plates at a density of 2.0 x 106 cells/ well three days before hormone treatment. Upon cell attachment, the media was replaced with DMEM with 20% CSS for treatment with hydrocortisone (CORT; Sigma H0888) or 20% FBS for treatment with the GR antagonist RU486 (Sigma M8046). All cell lines were incubated in 5% CO2 at 37°C.
Reporter Gene Assays
Three XDP-SVA construct variants, bearing 25 (Hex25), 41 (Hex41), or 52 (Hex52) repeats of the hexamer were obtained from the laboratory of Dr. Christopher Bragg. The inserts were subcloned from pGL3-B vectors6 into the pGL4.23 luciferase-reporter vector (Promega), with the XDP-SVA upstream of the luc2 reporter in the anti-sense direction. Briefly, the pGL3-B constructs were digested with SacI and HindIII enzymes and were then subjected to agarose gel electrophoresis. Bands corresponding to the XDP-SVA (~3kb) were extracted using the Wizard SV Gel and PCR Clean-up System (Promega) and ligated into linearized pGL4.23 vectors.
HeLa and HEK293T cells were co-transfected with the pGL4.23 vector constructs and the pRL-TK vector (Promega) which codes for Renilla luciferase under a constitutive promoter. Transfection was performed using X-tremeGENE HP DNA Transfection Reagent (Roche). Transfection with an empty vector and a vector containing a CORT-responsive enhancer sequence for Cyb561 (upstream Cyb561 enhancer, uCE) served as negative and positive controls, respectively. At least 16 hours post-transfection, cells were treated with either 100nM CORT or an equivalent volume of ethanol (vehicle control) for 4 hours, after which the cells were harvested. Luminescence measurements were performed using the Dual Luciferase Assay System (Promega), through the GloMax®-Multi Jr detection system (Promega) for HeLa cells, and Fluoroskan FL microplate luminometer (Thermo Fisher) for HEK293T cells. Relative light unit (RLU) measurements were obtained via normalization of luc2 luminescence against that of Renilla luciferase. Assays were done with four replicates per treatment per construct.
TAF1 and TAF1-32i Gene Expression analysis
Control and XDP fibroblasts (at least 3 replicates per treatment) were treated with 100nM CORT, 50nM RU486, or an equivalent volume of vehicle control for 24 hours before harvesting for RNA extraction using TRIzol Reagent (Invitrogen) following the manufacturer’s protocol.
The RNA extracts were treated with DNAse I (Sigma, AMPD1) and RNAse inhibitor (Applied Biosystems), then converted to cDNA using the ABI High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, 4374967). Quantitative PCR master mixes were prepared using PowerUP SYBR Green (Applied Biosystems, A25742) for GAPDH, KLF9, NR3C1 (GR), and TAF1 or a custom Taqman primer-probe (Thermo Fisher, ID AJWR28J) for TAF1-32i (Table 1). Quantitation of KLF9 transcript levels was used as a positive control as its transcription has been previously shown to be induced by CORT.14 Quantitative PCR was performed using the ABI 7500 Fast sequence detector (Applied Biosystems). Relative gene expression levels across treatments were obtained using the ΔΔCt method, using GAPDH Ct values for normalization.
Table 1.
Primer sequences used for RT-qPCR experiments
| Target | Sequence | |
|---|---|---|
| GAPDH | Forward | 5’-GGCATGGACTGTGGTCATGAG-3 |
| Reverse | 5’-TGCACCACCAACTGCTTAGC-3’ | |
| NR3C1 | Forward | 5’-TATCCTCTGCCTCCCATTCT-3’ |
| Reverse | 5’-CACCTTCCTGTCTCCTGTTTAC-3’ | |
| KLF9 | Forward | 5’-TGGCTGTGGGAAAGTCTATG-3’ |
| Reverse | 5’-GTCTGAGCGGGAGAACTTTT-3’ | |
| TAF1 | Forward | 5’-GGGAGAGCTTTCTGGATGATGTAAA-3’ |
| Reverse | 5’-ACAATCTCCTGGGCAGTCTTAGTAT-3’ | |
| TAF1-32i | Custom Taqman ID AJWR28J (Thermo Fisher) | |
Statistical Analysis
For luciferase assays, data (in relative light units) were log-transformed and analyzed by unpaired sample t-test and Tukey’s post-hoc test (p<0.05; GraphPad Prism 6). For gene expression analysis, data were analyzed using Student’s t-test and Tukey’s post-hoc test (p < 0.05; GraphPad Prism 6).
RESULTS
Identification of GREs within the SVA
LASAGNA-Search 2.0 was used to identify GREs within the XDP-SVA. The tool uses an alignment-based approach to match query sequences to the consensus sequence of a given TFBS. From the search, ten hits were obtained (Table 2). Removing redundant results, three unique hits were obtained. Two GREs mapped to the SINE, 271 and 400bp from the XDP-SVA insertion point, and one GRE mapped to the Alu element, 2,199 base pairs from the insertion point (Figure 1).
Table 2.
LASAGNA-search 2.0 results show potential GREs within XDP-SVA sequence
| Name | Sequence | Pos | Strand | Score | p-value | E-value |
|---|---|---|---|---|---|---|
| GR-alpha (T00337) | cagaacaaaatgaaa | 400 | + | 138.5 | 0.000375 | 0.97 |
| GR-alpha (T00337) | cggtgcaagatgtgct | 273 | - | 136.68 | 0.000425 | 1.10 |
| GR-alpha (T00337) | agaacaaaatgaaaa | 401 | + | 127.1 | 0.0009 | 2.34 |
| GR-beta (T01920) | cagaacaaaatgaaa | 400 | + | 138.5 | 0.0008 | 2.08 |
| GR-beta (T01920) | cggtgcaagatgtgct | 273 | - | 136.68 | 0.000825 | 2.15 |
| GR (M00205) | ggtgcaagatgtgct | 273 | - | 16.38 | 5.0E-5 | 0.130 |
| GR (M00205) | ggtgcaagatgtgctt | 272 | - | 12.97 | 0.0004 | 1.04 |
| GR (M00205) | cgttcactcagtgct | 2199 | - | 12.84 | 0.000425 | 1.10 |
| GR (M00205) | ggtgcaagatgtgc | 274 | - | 12.15 | 0.0005 | 1.30 |
| GR (M00192) | gcggtgcaagatgtgcttt | 271 | - | 8.66 | 0.000725 | 1.88 |
Each entry corresponds to a putative GRE. Information on the binding site sequence, position (in bp) within the SVA, and strand are given for each entry, as well as its corresponding score, p-value, and e-value as determined by the alignment algorithm.
Figure 1.
Identification of putative GREs within the XDP-SVA. The map shows the location of the SVA insertion within the TAF1 locus (Xq13.1). LASAGNA-Search 2.0 identified three GREs: two within the SINE, and one within the Alu region. GRE1 is found in the negative strand and is located 271 bp from the XDP-SVA insertion point. GRE2 is found in the positive strand and is located 401 bp from the insertion point. GRE3 is found in the negative strand and is located 2199 bp from the insertion point.
CORT-dependent activity of XDP-SVA luciferase reporter constructs
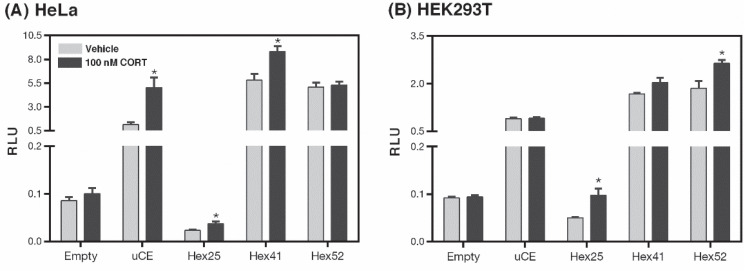
We performed dual luciferase assays on HeLa cells and HEK293T cells following transfection with reporter constructs of varying hexameric repeat lengths within the XDP-SVA-sequence. The XDP-SVA is inserted upstream and anti-sense to the reporter gene in the same way the SVA is inserted into the TAF1 gene in XDP patients. In HeLa cells, CORT treatment increased luciferase activity in the positive control (t(6) = 3.531, p = 0.012) and in XDP-SVA constructs Hex25 (t(6) = 2.601, p = 0.040) and Hex41 (t(6) = 3.295, p = 0.016) (Figure 2A). In HEK293T cells, CORT hormone treatment increased the normalized luciferase activity of cells transfected with Hex25 (t(5) = 3.919, p = 0.011) and Hex52 (t(6) = 3.079, p = 0.022) reporter constructs while cells transfected with the Hex41 construct only showed an increasing pattern of CORT-induced reporter activity that did not reach statistical significance (t(6) = 2.241, p = 0.066) (Figure 2B).
Figure 2.
Luciferase reporter assay to assess CORT-dependent transactivation of XDP-SVA promoter-luciferase constructs. Luciferase reporter constructs containing three variants of the XDP-SVA representing different hexameric domain repeat lengths were transfected into (A) HeLa and (B) HEK293T cells. Induction of luciferase activity (measured in Relative Luminescence Units, RLU) following CORT treatment was observed in HeLa cells transfected with the uCE (positive control), Hex25, and Hex41 constructs, while only HEK293T cells transfected with the Hex25 and Hex52 constructs showed significant CORT-dependent induction of promoter activity. Bars represent the mean of each data set ± standard error of the mean with statistical significance determined through Student’s t-test (*p < 0.05). Each treatment was done with four replicates each.
Analysis of aberrant and canonical TAF1 transcript expression in XDP fibroblasts
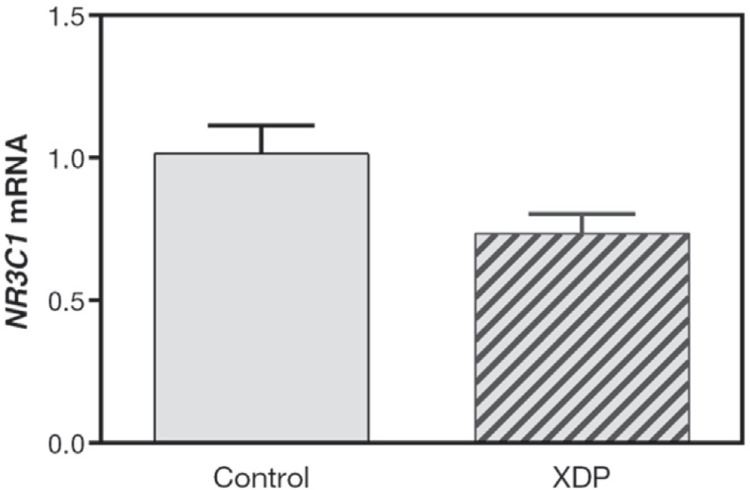
XDP fibroblasts and ethnically matched controls were used to determine the effect of hormone treatment on the expression of TAF1 and TAF1-32i transcripts. Before hormone treatment, we quantified NR3C1 (GR) mRNA expression in these cell lines and found that NR3C1 mRNA levels did not differ between control and XDP fibroblasts (Figure 3). Gene expression fold change values for three target genes, KLF9 (positive control), TAF1, and TAF1-32i were measured in CORT-treated control and XDP fibroblasts (Figure 4). The established CORT-responsive gene KLF9 exhibited CORT-dependent induction in XDP fibroblasts but not in control-derived cells (t(5) = 8.315, p = 0.0004) (Figure 4A). The expression level of TAF1 mRNA was not significantly affected by CORT treatment in both control and XDP fibroblasts. However, baseline TAF1 mRNA level is significantly reduced in XDP relative to control fibroblasts (t(5) = 4.409, p = 0.007) (Figure 4B). As expected, we did not detect TAF1-32i transcript expression in both vehicle- and CORT-treated control fibroblasts (data not shown). CORT treatment of XDP fibroblasts led to an increasing trend in the expression of the aberrant TAF1-32i transcript but did not reach statistical significance (t(5) = 2.545, p = 0.0516) (Figure 4C).
Figure 3.
Baseline expression of NR3C1 in XDP fibroblasts. Untreated control and XDP fibroblasts were harvested for analysis of NR3C1 (GR) mRNA. The expression level of NR3C1, normalized to GAPDH mRNA, did not significantly differ across control and XDP fibroblasts (p = 0.0718, Student’s t-test). Bars represent the fold induction ± standard error of the mean relative to control fibroblasts. Three to four replicates were used for each setup.
Figure 4.
Analysis of TAF1 and TAF1-32i expression in CORT-treated control and XDP fibroblasts. Control and XDP fibroblasts were treated with 100nM CORT or an equivalent volume of 100% ethanol (vehicle control) for 24 hours before harvesting for analysis of mRNA. Fold induction values were obtained using the ΔΔCt method with GAPDH as a housekeeping control. (A) KLF9 mRNA levels, used as a positive control for CORT treatment, increased significantly in XDP fibroblasts (p = 0.0004, Student’s t-test), but not in control fibroblasts (p = 0.1224). (B) TAF1 transcript levels differ across vehicle-treated control and XDP fibroblasts (p = 0.007, Student’s t-test). CORT treatment did not significantly change TAF1 expression levels in either control or XDP fibroblasts (Control, p = 0.0833; XDP, p = 0.7448, Student’s t-test). (C) The apparent increase in TAF1-32i expression levels upon CORT treatment of XDP fibroblasts approaches significance (p = 0.0516, Student’s t-test). Bars represent the fold induction ± standard error of the mean relative to vehicle control with statistical significance determined through Student’s t-test (*p < 0.001 for statistically significant effects of CORT within a cell line, and #p < 0.01 for statistically significant effects between the same hormone treatment). All treatments were done with three to four replicates.
Expression of TAF1 and TAF1-32i transcripts in control and XDP fibroblasts was also observed after treatment with the GR-specific antagonist RU486. Treatment with RU486 did not affect KLF9 mRNA expression in the control and XDP fibroblasts (Figure 5A). We observed an increase in TAF1 mRNA expression with RU486 treatment of control cells (t(5) = 3.597, p = 0.007) (Figure 5B) while TAF1 and aberrant TAF1-32i transcript expression were not affected by RU486 treatment in XDP fibroblasts (Figure 5B and 5C).
Figure 5.
Analysis of TAF1 and TAF1-32i expression in control and XDP fibroblast cells treated with GR-specific antagonist RU486. Control and XDP fibroblasts were treated with 50nM RU486 or an equivalent volume of vehicle control for 24 hours before harvesting for analysis of mRNA. Fold induction values were obtained using the ΔΔCt method with GAPDH as a housekeeping control. (A) No significant change in KLF9 mRNA levels is observed upon RU486 treatment. (B) TAF1 expression levels in control fibroblasts increase with RU486 treatment (p = 0.007). No effect of RU486 treatment is seen in XDP fibroblasts. (C) RU486 treatment did not alter TAF1-32i expression levels in XDP fibroblasts. Bars represent the mean fold induction ± standard error of the mean relative to vehicle control with statistical significance determined through Student’s t-test (*p < 0.01). All treatments were done with four to five replicates.
DISCUSSION
In this study, we aimed to establish the possible effects that the stress hormone CORT, as mediated by GR, may have on XDP. We hypothesized that this could occur via the interaction of GR with the XDP-SVA, a retrotransposon insertion previously identified to be a causal mutation of the disease. Through in silico analysis, three candidate GREs were identified. Indirect evidence for GR association with the SVA sequence was provided by reporter assays, which revealed CORT-responsiveness of the XDP-SVA in human cell lines. Gene expression analysis showed that NR3C1 (GR) mRNA levels in XDP and control fibroblasts were not significantly different. Control fibroblasts treated with CORT showed a decreasing but non-significant trend in TAF1 mRNA expression. Conversely, treatment of the control fibroblasts with the GR antagonist RU486 increased TAF1 expression. In XDP fibroblasts, CORT treatment did not alter TAF1 mRNA expression and led to an increasing but non-significant trend of TAF1-32i expression. However, treatment of the XDP fibroblasts with RU486 did not affect TAF1-32i transcript levels. Overall, the results suggest that GR signaling, through association with the XDP-SVA, could enhance the dysregulation of TAF1 leading to the production of TAF1-32i. This in turn contributes to the disease phenotype.
GR is predicted to bind to the XDP-SVA
A representative XDP-SVA sequence was used as an input for LASAGNA-Search 2.0 to identify possible GR binding sites within the SVA sequence. For the search, all factors named 'GR' were selected. Results returned ten possible binding sites of which only three sites were non-redundant. Mapping the putative GREs to the XDP-SVA sequence revealed two unique sites in the SINE region and one in the Alu region. Sequences similar to the SINE and Alu have been found to bind other nuclear hormone receptors such as estrogen and progesterone receptors. Of these, only the Alu sequences have been tested in vitro for nuclear receptor binding.18 While this type of analysis provided a starting point in establishing the association of GRs to the SVA, the results are not definitive in terms of direct GR binding to the SVA. This must be tested experimentally through electrophoretic mobility shift assays, mutational analysis of GR binding sequence, or GR chromatin immunoprecipitation assays. Nevertheless, this in silico analysis provides a possible association between the stress-dependent gene regulatory consequences of the SVA insertion and the production of aberrant transcription products from the TAF1 locus.
The XDP-SVA confers GC responsiveness in human cell lines
We performed luciferase reporter assays in vitro to determine the GC response in the context of the XDP-SVA. The XDP-SVA reporter constructs have proven intrinsic promoter activity.6 We wanted to test if the XDP-SVA would be responsive to CORT treatment. We made use of different variants of the XDP-SVA to test for possible effects of hexameric repeat length on reporter activity induction. In HeLa and HEK293T cell lines used in the promoterreporter assays, baseline promoter activity represented by the vehicle treatment showed that only constructs Hex41 and Hex52 have increased reporter activity relative to the empty vector control, consistent with previously published studies that used the same reporter constructs in human neuroblastoma (SH-SY5Y) and human osteosarcoma epithelial (U2OS) cells.6 Similar to the results we obtained for Hex25, an XDP-SVA promoter-luciferase construct with 35 hexameric repeat length (Hex35) did not exhibit promoter activity in SH-SY5Y cells.6 However, Hex35 exhibited promoter activity in U2OS cells, higher than what was observed for Hex41 and Hex52. They also tested promoter activity of the XDP-SVA with no hexameric repeats (ΔHex) and observed luciferase activity equal to the empty vector in SH-SY5Y cells, but higher than the empty vector in U2OS cells.6 These results suggest that SVA insertion confers intrinsic promoters outside of the hexameric repeat domain. Moreover, the previously demonstrated lack of a consistent trend between hexameric repeat lengths and promoter activity across cell lines was also reflected in our findings, indicating that the XDP-SVA confers promoter activity in a cell-specific manner.
Comparing reporter assay results across treatments, it appears that CORT treatment enhances the intrinsic promoter activity of XDP-SVA. This suggests that GR may associate with the SVA to mediate CORT-dependent transcriptional regulation. However, the CORT-dependent induction was only observed in two of three XDP-SVA promoter-reporter constructs used. Whether GR directly binds the identified GREs within the SVA or indirectly associates through the recruitment of TFs remains to be seen.
GCs can alter TAF1 and TAF1-32i expression in an XDP cell model
Following confirmation of expression of endogenous GR in XDP and control fibroblasts, we conducted expression analysis to determine the effects of CORT and the GR-specific antagonist RU486 on TAF1 and TAF1-32i transcript levels. We observed lower expression of TAF1 mRNA in vehicle-treated XDP fibroblasts relative to control cells maintained in steroid-stripped media. This is in agreement with previous studies that found reduced expression of the functional TAF1 transcript in XDP cell lines compared to corresponding control cells.5 However, the reduced expression of TAF1 mRNA in XDP versus control fibroblasts was not observed when the cells were maintained in full serum as in the RU486-treated cells, providing further evidence that steroid hormones may be involved in TAF1 transcriptional regulation.
CORT treatment of control fibroblasts led to a decreasing but non-significant trend in TAF1 mRNA level (t(6) = 2.075, p = 0.0833). Conversely, RU486 treatment significantly upregulated TAF1 expression in control fibroblasts (t(5) = 3.597, p = 0.007). In addition, we also observed an increasing trend in the expression of the aberrant TAF1-32i transcript in CORT-treated XDP-fibroblasts; however, this increase did not reach statistical significance (t(5) = 2.545, p = 0.0516). These data suggest that in the context of the wild-type TAF1 gene, GCs can negatively regulate the expression of the canonical TAF1 transcript through transcriptional repressor activity of GR. In the presence of the SVA as in XDP cells, the CORT-dependent negative regulation of TAF1 expression is lost and the effect of CORT shifts towards positive regulation of the aberrant TAF1-32i transcript expression, possibly through GR association with the SVA as established by the CORT-dependent transactivation assay. The SVA sequence may sequester GR from the TAF1 cis-regulatory elements where it would normally bind. Concurrently, GR binding to the SVA could lead to the recruitment of trans-acting elements, resulting in faulty transcription and transcript processing, hence producing aberrant transcripts. Further experiments involving GR chromatin immunoprecipitation assays in control and XDP cells are needed to confirm this mechanism of GR-dependent regulation.
Although these results establish a possible link between stress as mediated by GR and TAF1 dysregulation, we performed expression analysis only on one clonal population of XDP and control fibroblasts. TAF1 and TAF1-32i expression in cell lines from other probands and unaffected controls should be interrogated to definitively ascribe the transcriptional effects to the SVA insertion, and not to other genetic variations. Alternatively, neural stem cells (NSCs) which may more accurately recapitulate XDP neuropathology, may be employed as in vitro models. Prolonged or iterative GC treatment may also be employed to better observe the effect of chronic stress on TAF1 transcriptional regulation. Lastly, further studies should also consider how the decrease in TAF1 expression and the emergence of aberrant transcripts as observed in XDP cell lines lead to disease neuropathology (i.e. progressive loss of neurons in the striatum) and clinical manifestations.
CONCLUSION
Overall, our results suggest that acute stress, through CORT-dependent GR transactivation of the XDP-SVA, could enhance the dysregulation of TAF1 expression in XDP patients and enhance the production of aberrant transcripts such as TAF1-32i, which in turn may contribute to the disease phenotype. Our study provides a new facet for the management of XDP cases, taking into account psychological and social factors in the form of stress. It may also provide a basis for new treatment avenues, wherein GC levels may be manipulated to reduce the production of aberrant transcripts which may consequently delay disease onset.
Acknowledgments
The authors thank Dr. Christopher Bragg (Massachusetts General Hospital) for providing the cell lines and the XDP-SVA constructs, and Weaverly Lee for subcloning the XDP-SVA into the vectors used for the luciferase assays.
Statement of Authorship
Both authors certified fulfillment of ICMJE authorship criteria.
Authors Contribution Statement
SEDC and PB developed the methodology; verified results; synthesized the study data; reviewed and edited the manuscript; and prepared the data presentation. SEDC implemented computer code and supporting algorithms; conducted the research and investigation process; curated data; and prepared the original draft of the manuscript. PB formulated the research goals and aims; provided the study materials; supervised and managed the research activity planning and execution; and acquired financial support for the project.
Author Disclosure
Both authors declared no conflict of interest.
Funding Source
The research received funding support from the University of the Philippines-National Institute of Molecular Biology and Biotechnology.
References
- 1.Rosales RL. X-linked dystonia parkinsonism: Clinical phenotype, genetics and therapeutics. J Mov Disord. 2010;3(2):32-8. PMID: . PMCID: . 10.14802/jmd.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diestro JDB, Pasco PMD, Lee LV. Validation of a screening questionnaire for X-linked dystonia parkinsonism: The first phase of the population-based prevalence study of X-linked dystonia parkinsonism in Panay. Neurol Clin Neurosci. 2017;5(3):79-85. 10.1111/ncn3.12113. [DOI] [Google Scholar]
- 3.Lee LV, Rivera C, Teleg RA, et al. The unique phenomenology of sex-linked dystonia parkinsonism (XDP, DYT3, “Lubag”). Int J Neurosci. 2011;121 Suppl 1:3-11. PMID: . 10.3109/00207454.2010.526728. [DOI] [PubMed] [Google Scholar]
- 4.Evidente VGH, Nolte D, Niemann S, et al. Phenotypic and molecular analyses of X-linked dystonia-parkinsonism (“lubag”) in women. Archives of Neurology. 2004;61(12):1956-9. PMID: . 10.1001/archneur.61.12.1956. [DOI] [PubMed] [Google Scholar]
- 5.Aneichyk T, Hendriks WT, Yadav R, et al. Dissecting the causal mechanism of X-linked dystonia-parkinsonism by integrating genome and transcriptome assembly. Cell. 2018;172(5):897-909.e21. PMID: . PMCID: . 10.1016/j.cell.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bragg DC, Mangkalaphiban K, Vaine CA, et al. Disease onset in X-linked dystonia-parkinsonism correlates with expansion of a hexameric repeat within an SVA retrotransposon in TAF1. Proc Natl Acad Sci U S A. 2017;114(51):E11020-8. PMID: . PMCID: . 10.1073/pnas.1712526114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pozojevic J, Algodon SM, Cruz JN, et al. Transcriptional alterations in X-linked dystonia parkinsonism caused by the SVA retrotransposon. Int J Mol Sci. 2022;23(4):2231. PMID: . . 10.3390/ijms23042231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alcachupas A, Bellosillo K, Catolico WR, et al. Biopsychosocial aspect of patients with X-linked dystonia-parkinsonism: Its implications on quality of life. Cureus. 2022;14(1):e21699. PMID: . PMCID: . 10.7759/cureus.21699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morigaki R, Nakataki M, Kawarai T, et al. Depression in X-linked dystonia-parkinsonism: A case-control study. Parkinsonism Relat Disord. 2013;19(9):844-6. PMID: . 10.1016/j.parkreldis.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 10.Jamora RD, Rae F. Cenina A, A. Teleg R, V. Lee L. Suicidality among patients with sex-linked dystonia-parkinsonism (XDP). Acta Medica Philipp. 2015;49(1):20-3. 10.47895/amp.v49i1.1009. [DOI] [Google Scholar]
- 11.Heitzer MD, Wolf IM, Sanchez ER, Witchel SF, DeFranco DB. Glucocorticoid receptor physiology. Rev Endocr Metab Disord. 2007;8(4):321-30. PMID: . 10.1007/s11154-007-9059-8. [DOI] [PubMed] [Google Scholar]
- 12.Timmermans S, Souffriau J, Libert C. A general introduction to glucocorticoid biology. Front Immunol. 2019;10:1545. PMID: . PMCID: . 10.3389/fimmu.2019.01545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tost H, Champagne FA, Meyer-Lindenberg A. Environmental influence in the brain, human welfare and mental health. Nat Neurosci. 2015;18(10):1421-31. PMID: . 10.1038/nn.4108. [DOI] [PubMed] [Google Scholar]
- 14.Bagamasbad P, Ziera T, Borden SA, et al. Molecular basis for glucocorticoid induction of the Kruppel-like factor 9 gene in hippocampal neurons. Endocrinology. 2012;153(11):5334-45. PMID: . PMCID: . 10.1210/en.2012-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrero M-T, Estrada C, Maatouk L, Vyas S. Inflammation in Parkinson's disease: Role of glucocorticoids. Frontiers in neuroanatomy. 2015;9:32. PMID: . PMCID: . 10.3389/fnana.2015.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okawa Y, Ishiguro K, Fujita SC. Stress-induced hyperphosphorylation of tau in the mouse brain. FEBS letters. 2003;535(1-3):183-9. PMID: . 10.1016/s0014-5793(02)03883-8. [DOI] [PubMed] [Google Scholar]
- 17.Lee C, Huang CH. LASAGNA-Search 2.0: Integrated transcription factor binding site search and visualization in a browser. Bioinformatics. 2014;30(13):1923-5. PMID: . 10.1093/bioinformatics/btu115. [DOI] [PubMed] [Google Scholar]
- 18.Jacobsen BM, Jambal P, Schittone SA, Horwitz KB. ALU repeats in promoters are position-dependent co-response elements (coRE) that enhance or repress transcription by dimeric and monomeric progesterone receptors. Mol Endocrinol. 2009;23(7):989-1000. PMID: . PMCID: . 10.1210/me.2009-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]