Abstract
Background
Parkinson’s disease (PD) is the second most common neurodegenerative disease with a significant public health burden. It is characterized by the gradual degeneration of dopamine neurons in the central nervous system. Although symptomatic pharmacological management remains the primary therapeutic method for PD, clinical experience reveals significant inter-individual heterogeneity in treatment effectiveness and adverse medication responses. The mechanisms behind the observed interindividual variability may be elucidated by investigating the role of genetic variation in human-to-human variances in medication responses and adverse effects.
Objective
This review aims to explore the impact of gene polymorphism on the efficacy of anti-parkinsonian drugs. The identification of factors associated with treatment effectiveness variability might assist the creation of a more tailored pharmacological therapy with higher efficacy, fewer side outcomes, and cheaper costs.
Methods
In this review, we conducted a thorough search in databases such as PubMed, Web of Science, and Google Scholar, and critically examined current discoveries on Parkinson's disease pharmacogenetics. The ethnicity of the individuals, research methodologies, and potential bias of these studies were thoroughly compared, with the primary focus on consistent conclusions.
Results
This review provides a summary of the existing data on PD pharmacogenetics, identifies its limitations, and offers insights that may be beneficial for future research. Previous studies have investigated the impact of gene polymorphism on the effectiveness and adverse effects of levodopa. The trendiest genes are the COMT gene, DAT gene, and DRD2 gene. However, limited study on other anti-Parkinson's drugs has been conducted.
Conclusion
Therefore, In order to develop an individualized precision treatment for PD, it is an inevitable trend to carry out multi-center, prospective, randomized controlled clinical trials of PD pharmacogenomics covering common clinical anti-PD drugs in large, homogeneous cohorts.
Keywords: Parkinson’s disease, pharmacogenomics, genetic polymorphism, genetic phenotypes, drug response, individualized medicine
1. INTRODUCTION
PD is the second most frequent neurodegenerative disease, affecting approximately 1% of individuals >60 years of age globally [1]. In China, its prevalence rate among people aged over 65 years is about 1.7% [2, 3]. Therefore, as PD is more prevalent in older people, it creates the highest public health burden in the elderly population. It is characterized by a progressive loss of dopamine neurons in the central nervous system. PD manifests clinically as motor symptoms such as bradykinesia, resting tremor, and muscular rigidity, as well as various non-motor symptoms such as autonomic dysfunction, olfactory disturbances, sleep disorders, and cognitive deficits [4].
In recent years, advances in scientific research technologies have provided a new understanding of the occurrence and etiological mechanism of PD. The major pathological manifestations of PD include intracellular aggregation of α-synuclein, which forms Lewy bodies, and loss of dopaminergic neurons, which first occur in the substantia nigra before spreading to other brain parts as the disease progresses [5]. The main mechanism underlying the development of motor symptoms in PD is the loss of dopamine neurons in the substantia nigra. Therefore, exogenous dopamine supplementation is a standard treatment strategy for PD [6].
PD treatment includes both pharmacological therapy and non-pharmacological treatment. Pharmacological therapy remains a crucial treatment method for the current management of PD. Compared with other neurodegenerative diseases, PD symptoms are effectively controlled by drug treatments, thereby improving the quality of life. Based on the mechanism of action, anti-Parkinson's drugs are divided into six categories: dopaminergic drugs, dopamine receptor (DR) agonists, anticholinergic drugs, amantadine, monoamine oxidase (MAO) inhibitors, and catechol-O-methyltransferase (COMT) inhibitors. Unfortunately, these drugs can only alleviate PD symptoms, but cannot delay the progression of the disease, cure it, or reverse its neurodegenerative effects [7].
In clinical settings, patients respond differently to anti-Parkinson's drugs. The following three elements demonstrate the personalized variances in anti-PD medicines [8]. 1) Differences in initial effective dosage: the treatment of anti-PD drugs emphasizes the principle of dose titration. The minimal effective dosage required to induce adequate relief in clinical symptoms varies substantially across people. Some patients achieve considerable relief in symptoms after obtaining a lower dose of medication, whilst others must titrate to a higher dose in order to fulfill their fundamental needs in life. 2) Side effect differences: There is greater individual diversity in the side effects of anti-PD medications. Some people experience no or just slight adverse effects, whilst others experience major side effects such as palpitation, gastrointestinal issues, and impulse control disorder. 3) The disparities in motor problems: most patients who have been on anti-PD medicines for a long time will experience motor complications such as dyskinesia and wearing out, which will have a negative impact on their quality of life. However, the onset of motor problems varies widely between patients, and a small percentage of people who take anti-PD medications for many years still achieve good effectiveness. The mechanism behind this phenomenon remains unclear but many experts suggest a tic basis. In recent decades, mainly due to the development of sequencing technology and increased availability of affordable genetic testing methods, substantial progress has been made in the identification of genetic biomarkers of drug response. Particular gene variants have been associated with drug inefficacy, hypersensitivity, or increased toxicity risk. In addition, there have been efforts to define the role of genetic polymorphisms in optimizing the pharmacotherapy of PD. Studies of the role of genetic variation in human-to-human differences associated with drug responses and adverse reactions may shed light on the mechanisms underlying interindividual variability observed in response to antiparkinsonian agents [9].
Current evidence-based medicine emphasizes drug efficacy in the entire population but ignores individual differences, which makes it less effective in specific subpopulations. Therefore, considering differences in genetic susceptibility, the individualized precision therapy strategy is desirable [9, 10]. The two main research areas in drug response variability are pharmacokinetics and pharmacodynamics. The accumulated to date mainly encompasses three effects of genetic variation on drug response characteristics: affecting serum drug concentration, changing the ability of a drug to cross the blood-brain barrier, and modifying pharmacodynamic characteristics [11]. Pharmacogenomics refers to the study of drug response in various diseases due to genetic variations and the development of new drugs or new drug use methods on this basis. Pharmacogenomic studies have proved that some drugs perform better in certain populations with specific genes, or that the selection of therapeutic drugs based on genes improves the effectiveness of drugs and avoids adverse reactions [12]. Therefore, the ultimate purpose of pharmacogenomics is to identify genetic factors for the different drug responses among individuals, providing a precise personalized medical treatment model. Anti-Parkinson's drugs show 60-90% variability in pharmacokinetics and pharmacodynamics [13]. Transport, metabolic, mechanistic, pathogenic, and polyclonal genes are involved in pharmacogenomics. Recently, increasing evidence suggested that pharmacogenomics affects the efficacy and safety of antiparkinsonian drugs. Therefore, combining personalized treatment of PD with pharmacogenomics could improve the efficacy and safety of drugs.
This is a narrative review comprising a comprehensive summary and novel data compiled from all pharmacogenomic studies published in various databases up to May of this year. To get a better understanding of anti-pharmacogenomics Parkinson's research, we analyzed existing pharmacogenomics research based on the categories of anti-drugs Parkinson's and characterized the study hotspot-polymorphisms in levodopa-related genes in terms of medication efficacy and drug adverse effects. This will serve as a significant reference point for future studies and clinical applications of anti-Parkinson pharmacogenomic research.
2. METHODS
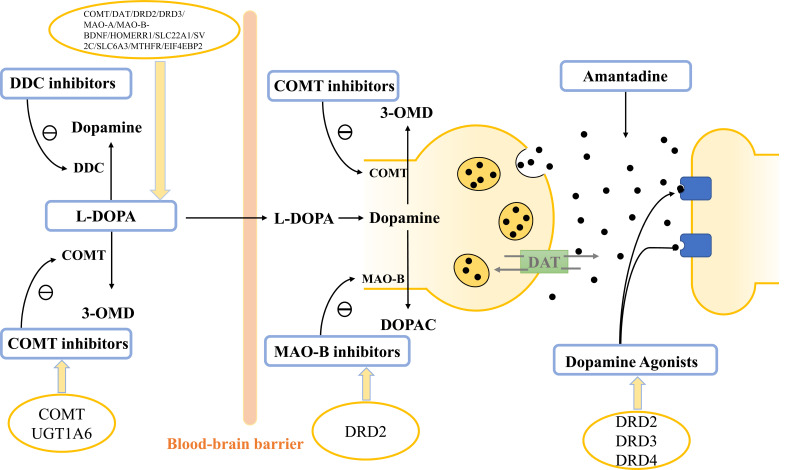
We collected the studies of PD pharmacogenomics published in multiple databases, critically examined current discoveries, enumerated the recent progress of major genes (Table 1), then elaborated on the genetic factors with strong reliability and great clinical value. For a more intuitive understanding of the current research status, we have listed the genetic factors associated with drug response according to the classification of anti-parkinsonian drugs, as shown in Fig. (1).
Table 1.
Polymorphisms and genomic variants underlying responses to anti-PD drugs.
| Drugs | Gene | SNP | Effect | References |
|---|---|---|---|---|
| L-DOPA | COMT gene | rs4680 | Higher frequency of COMT LL genotype in patients requiring lower L-dopa dosage | [21, 22, 78] |
| No association between COMT polymorphism and the occurrence of LID | [22, 33] | |||
| Higher frequency of COMT LL genotype in patients with dyskinesia | [23, 32] | |||
| Higher frequency of COMT LL/LH genotypes in patients with higher Epworth sleepiness scale scores | [79] | |||
| rs165815 | Higher frequency of COMT C allele in patients with visual hallucinations | [54] | ||
| DAT gene | rs28363170 & rs393795 | Lower frequency of the combination of the two genotypes of the DAT gene in patients with LID | [80] | |
| rs28363170 | Higher frequency of DAT gene VATR sequence in patients with LID | [27] | ||
| Higher frequency of DAT gene VATR sequence in patients with Levodopa-induced hallucinations and psychosis | [27] | |||
| Lower frequency of DAT1 9/9 genotype in patients with LID | [28] | |||
| rs393795 | Higher frequency of DAT C genotype in patients with later occurrence time of LID | [27] | ||
| DRD2 gene | rs1800497 | DRD2/ANKK1 is significantly associated with increased doses of L-dopa | [28] | |
| No association between TaqIA polymorphism and the motor fluctuation | [28] | |||
| Lower frequency of DRD2 13 or 14 alleles in patients with peak-dose dyskinesias | [34, 35] | |||
| Higher frequency of DRD2 14/15 genotype in patients with LID | [36] | |||
| Higher frequency of DRD2 C allele in patients with delayed visual hallucinations | [56] | |||
| DRD3 gene | rs6280 | Higher frequency of DRD3 C genotype in patients with visual hallucinations | [55] | |
| MAO-A gene | rs6323 | No association between MAO-A and the occurrence of LID | [22] | |
| rs1799836 | Higher frequency of A allele and AA genotype in patients LID | [23] | ||
| BDNF gene | rs6265 | No association between BDNF and the occurrence of LID | [22] | |
| Higher frequency of BDNF met allele in patients LID | [81] | |||
| HOMER1 gene | rs4704560 | Lower frequency of HOMER1 G allele in patients with LID | [46] | |
| ACE gene | rs4646994 | Higher frequency of ins/ins genotype in patients with L-DOPA-induced psychosis | [82] | |
| SLC22A1 gene | rs622342 | Higher frequency of the minor C allele in patients requiring higher anti-Parkinsonian drugs dosage | [29] | |
| SV2C gene | rs30196 | Higher frequency of the C allele in patients requiring lower L-dopa dosage | [30] | |
| SLC6A3 gene | rs2836371 | Higher frequency of SLC6A3 9R allele in patients requiring lower L-dopa dosage | [30] | |
| MTHFR gene | rs1801133 | Higher frequency of MTHFR TT677 mutants in patients with lower daily levodopa dose | [83] | |
| Higher frequency of C677T in patients with L-dopa-induced hyperhomocysteinemi | [53] | |||
| Lower frequency of C677T TT homozygosity in patients with L-DOPA resistance | [53] | |||
| EIF4EBP2 gene | rs1043098 | Higher frequency of CC homozygous in patients with the later occurrence of LID and LID peak | [84] | |
| Dopamine Receptor Agonists | DRD2 gene | rs1800497 | No association between Taq1 genotypes and the reaction of pramipexole | [63] |
| Higher frequency of Allele A2 in patients with the sudden onset of sleep in patients taking bromocriptine, pergolide, or cabergoline | [85] | |||
| DRD3 gene | rs6280 | Higher frequency of Ser/ser genotype in patients with a higher response rate of pramipexole | [60] | |
| DRD4 gene | DRD4 48-bp VNTR | No association between the Taq1 genotype and the reaction of pramipexole | [63] | |
| COMT inhibitors | COMT gene | rs4680 | COMT HH genotype prolongs the ON time of patients treated with entacapone | [71] |
| COMT gene rs4608 polymorphism modifies the motor response to COMT inhibitors entacapone | [86] | |||
| UGT1A6 gene | A528G SNP | Higher frequency of A528G SNP with a higher risk of hepatotoxicity induced by entacapone and tolcapone | [72] | |
| MAO Inhibitors | DRD2 gene | rs2283265 & rs1076560 | The two genotypes can significantly improve the motor function of patients treated with rasagiline | [68] |
Abbreviations: L-dopa: levodopa; COMT: catechol-O-methyltransferase; LID: levodopa-induced dyskinesia; DR: dopamine receptor; MAO: monoamine oxidase; DAT: dopamine transporter.
Fig. (1).
Parkinson’s disease drugs and their associated gene polymorphisms.
3. RESULTS-CURRENT ACHIEVEMENTS IN PD PHARMACOGENOMICS
3.1. Pharmacogenetic Studies of Levodopa
Levodopa is one of the most commonly used and effective drugs for PD treatment. Most pharmacogenomic studies have explored polymorphisms associated with its response and adverse effects. The investigated genes were mainly those involved in levodopa metabolism, transport, and excretion, including the catecholamine-o-methyltransferase (COMT) gene, the dopamine receptors (DRD1-5) gene, the dopamine transporter (DAT) gene, the dopa decarboxylase enzyme (DDC) related genes and others [14-16]. Among them, the COMT gene has been confirmed to play a role in levodopa response.
3.1.1. COMT Genes
The COMT gene is localized on chromosome 22q11.1-q11.2. It encodes catecholamine-o-methyltransferase (COMT), which converts levodopa into 3-O-methyldopa in the body [17]. Elevated levels of COMT increase the degradation of L-levodopa, thereby reducing its effectiveness. Conversely, combining levodopa with COMT inhibitors enhances its efficacy. G1947A polymorphism (also called RS4680) in exon 4 of the COMT gene changes valine (Val) to methionine (Met) at amino acid 158 of the COMT enzyme, affecting the thermal instability of the enzyme and decreasing its activity [18]. Studies have found that COMT enzyme activity varies greatly among individuals. Three phenotypes have been characterized: high activity, moderate activity, and low activity in the population, which are determined by co-dominantly inherited COMT-H (high) and COMT-L (low) alleles. Their genotypes include COMT-HH (Val/Val), COMT-HL (Val/Met), and COMT-LL (Met/Met) [19, 20]. Several experiments have confirmed that the COMT genotype affects the efficacy of levodopa. Generally, other conditions held constant, patients with a highly active COMT (COMT-HH) phenotype tend to require a larger dose of levodopa than those with a lowly active COMT (COMT-LL) phenotype. Bialecka and his colleagues divided 95 patients with sporadic PD into two groups based on daily levodopa dose: group 1 received levodopa at dose > 500 mg per day and group 2 received levodopa at dose < 500 mg per day. They found that the COMT-LL genotype was more common in group 2 than in group 1 [21]. A study by Cheshire et al. examined the effect of COMT, MAO-A, and BDNF polymorphisms on levodopa response in 285 patients with PD, establishing that homozygous COMT activity was associated with a higher maximum daily dose of levodopa [22]. These findings were recently corroborated by Sampaio et al. research [23]. Better clinical response to lower L-dopa doses in patients with low COMT activity may be explained by slower catabolism of the drug, more stable serum and CNS (central nervous system) drug concentrations, and lower levels of 3-O-MD. However, some studies showed no significant correlation between COMT gene polymorphism and levodopa use [24, 25].
3.1.2. DR Genes
Dopamine receptor genes DRD1, DRD2, DRD3, DRD4, and DRD5, encode dopamine receptors D1, D2, D3, D4, and D5, respectively. Current studies about DR gene polymorphism and drug response mainly focus on the effects of DRD2 and DRD3 gene polymorphisms on the severity of the side effects of dopamine preparation. Few studies have found that the DRD2 and DRD3 gene polymorphisms are associated with the maximum daily dose limit of levodopa tolerated by patients. DRD2 is located at 11q22-23, with its main genetic variant, TaqIA (RS1800497) [26] associated with movement effects, including movement fluctuations and disorders induced by L-dopa. TaqIA originally belonged to the DRD2 gene family but was later classified as the ANKK1 gene, which lies downstream of DRD2 and overlaps with DRD2. Kaiser et al. [27] found no correlation between DRD2/ANKK1 and the clinical characteristics of patients treated with L-dopa. In contrast, Dos Santos et al. found that DRD2/ANKK1 was significantly associated with increased doses of L-dopa in a 2019 study involving 195 patients with sporadic PD in Brazil [28].
3.1.3. SLC22A1, SLC6A3, and SV2C Genes
The solute carrier (SLC) transporter family is the second-largest membrane protein family in human cells, which is distributed on various biofilm structures in cells, including nuclear and cytoplasmic membranes. SLC susceptibility sites are potential therapeutic targets for various diseases, with SLC22A1 and SLC6A3 genes being clinically significant to PD pharmacogenomics. Becker et al. collected levodopa dosage for 7,983 PD patients aged over 55 years and found a higher dosage of levodopa in SLC22A1 gene carriers than that in the control group [29]. The SLC6A3 gene which encodes the dopamine transporter is highly expressed in dopaminergic neurons in the presynaptic midbrain. Previous studies have found that double-allelic mutations in the SLC6A3 gene cause dopamine transporter deficiency syndrome (DTDS), manifested as PD in infants. In a 2018 study, Altmann et al. conducted multiple regression analysis of levodopa dose and SLC6A3 gene in 224 patients demonstrating that SLC6A3 was associated with low doses of levodopa [30]. Similar results were found in the SV2C genome [30]. In contrast, MTHFR TT677 mutants had a lower daily levodopa dose [31].
It is evident that the studies reviewed on the pharmacogenomics of levodopa differ in the ethnicity of patients, choice of study sites, study endpoints, and methods of analysis. In particular, ethnic differences of study participants may explain the contradictory results of different studies.
3.2. Pharmacogenomics of Levodopa Side Effects
PD patients taking levodopa for a long period may experience various side effects. The most common side effects are dyskinesia, end-use phenomena, visual hallucinations, daytime lethargy, and impulse control disorders.
3.2.1. COMT Gene
Previous studies have found that L-dopa dose is influenced by the COMT genotype. Recent studies have found that COMT polymorphisms are associated with side effects of long-term use of L-dopa. A paper published by Sampaio and his team in 2018 suggested that patients with the COMT-LL phenotype are more prone to levodopa-induced dyskinesia (LID) [23], which was corroborated by de Lau et al.’s research [32]. In other words, patients with low COMT enzyme activity are more likely to develop levodopa-induced dyskinesia than those with high COMT enzyme activity. The low-activity COMT enzyme reduces levodopa degradation, increasing dopamine accumulation in the synaptic cleft, which causes dyskinesia. Cheshire et al. and Watanabe et al. also explored this question [22, 33]. However, both studies found no significant correlation between low-activity COMT enzyme and LID, which may be attributed to individual differences between patients (that is, some patients may have multiple genotypes affecting LID at the same time).
3.2.2. DR Gene
Currently, the DRD2 gene is the main dopamine receptor gene associated with levodopa side effects. It is highly expressed in the basal ganglia, which is a motor-regulating region of the central nervous system and is crucial to PD. Oliveri is the first researcher to study the role of DRD2 polymorphism in LID [34]. In his study, the frequency of both alleles 13 and 14 of DRD2 gene polymorphism was higher in non-dyskinetic than in dyskinetic PD patients. In addition, carrying at least 1 of the 13 or 14 alleles reduced the risk of developing peak-dose dyskinesias by 72% in PD patients compared to PD patients not carrying these alleles. Further, DRD1 polymorphisms were not associated with the risk of developing PD or peak-dose dyskinesias.
Rieck et al. [35] verified this relationship with 199 Brazilian PD patients. Strong and his colleagues found that the DRD2 gene was involved in an early-onset of LID in 92 PD patients and further determined that the early onset PD was due to 14 and 15 DRD2 alleles [36]. Some researchers have also found that DRD3 gene polymorphism is associated with levodopa-induced side effects, but evidence supporting this conclusion is still insufficient.
3.2.3. DAT Genes
The dopamine transporter (DAT, SLC6A3) plays an important role in controlling the intensity and duration of dopaminergic neurotransmission by rapid reuptake DA into presynaptic terminals [37]. The single nucleotide polymorphism (RS393795) in the DAT gene is significantly correlated with the onset time of levodopa-induced motor dysfunction, whereas the C allele of DAT is associated with late-onset LID, which may be regulated by a change in dopamine reuptake rate in synaptic cleft [38]. Sossi et al. have shown that greater DAT expression levels are directly associated with lower dopamine turnover and lower changes in synaptic dopamine concentration in PD patients [39]. Troiano et al. subsequently confirmed decreased DAT in the presynaptic membrane using a DAT-PET (Dopamine transporter-Positron Emission Tomography) [40].
In addition to these three genes implicated in dopamine metabolism, the following genes are also potentially involved in LID. LID is a type of motor complication common in patients with PD during chronic levodopa therapy. It is reported that 40% of patients develop LID four years after receiving levodopa therapy. This risk is even higher among younger patients receiving high doses of levodopa therapy. In addition to the early onset age of PD and higher L-dopa total exposure, other studies have found that LID is more common in patients with a longer course of PD, lower body mass index, and female gender [41-45]. Several researchers have also investigated the potential involvement of genetic factors or individual genetic variations in the development of LID. Analysis of the chi-square correlation between genotype and the presence of multiple levodopa-induced side effects in 205 PD patients by Schumacher-schuh found that rs4704559 G (HOMER1) allele was associated with a lower prevalence of dyskinesia [46]. Contrastingly, a higher prevalence of LID was found in patients with MAO-B [23] (rs1799836, A644G) A allele and AA genotype and in patients with mTOR [23] gene polymorphism after multivariate analysis. A better understanding of the role played by genetic factors in the development of LID may be important to identify patients more likely to develop LID and elucidate the molecular mechanisms underlying this causal link.
With the increasing understanding of the adverse side effects of levodopa, more and more studies have focused on the influence of genotypes on side effects other than LID. Some studies have found an association between COMT gene polymorphism and the Epworth Sleepiness Scale (ESS, a scale designed by Johns MW to assess excessive daytime sleepiness state, facilitating quantitative semi-objective evaluation of the drowsiness state). Sleep disturbances are a well-known disabling nonmotor manifestation of PD, affecting almost 80% of patients [47]. Patients with COMT-LH and COMT-LL genotypes had higher ESS scores than patients with COMT-HH, suggesting that these patients were more likely to experience daytime sleepiness [47]. However, Rissling’s study yielded contradictory results [48]. An SNP C667T (rs1801133) in the MTHFR gene was consistently linked to hyperhomocysteinemia (HHcy), which is defined as elevated serum concentrations of homocysteine (Hcy) exceeding 15 µmol/L [49]. Elevated Hcy levels have been associated with increased cardiovascular, cerebrovascular, and thromboembolic diseases [50-52] due to the L-dopa treatment in several studies. This mutation generates a temperature-labile MTHFR enzyme, which ultimately leads to hyperhomocysteinemia [53]. Regarding visual hallucinations as side effects, current research suggests that the COMT RS165815 C allele is a protective factor against visual hallucinations [54], as the incidence of visual hallucinations in patients with this allele is relatively low. In contrast, the DRD3 RS6280 C genotype is a risk factor for visual hallucinations [55], whereas DRD2 TaqIA C has been associated with delayed visual hallucinations [56]. Other studies have confirmed that OPRK1, HTR2a, and DDC genotypes are associated with the incidence of ICD(impulse control disorder) [57-59].
However, these results have only been confirmed by a few studies focusing on occidental patient samples. It is expected that more studies will be conducted in this area, especially focusing on the Asian population, to support these conclusions.
3.3. Pharmacogenetic Studies on Dopamine Receptor Agonists
Dopamine agonists are one of the most commonly used drugs in PD treatment but have lower efficacy than levodopa. Many studies have shown that dopamine agonists could overcome the shortcomings of levodopa, strengthen levodopa’s curative effect and delay complications. The combination of low-dose levodopa and dopamine receptor agonists is as effective as high-dose levodopa alone, but with a significantly lower incidence of side effects. In the later stages of the disease, due to gradual degeneration and loss of dopaminergic neurons in the nigrostriatal system, exogenous levodopa decarboxylation is no longer converted to dopamine. At this point, levodopa becomes ineffective, while dopamine receptor agonists remain effective. Examples of commonly used dopamine receptor agonists include pramipexole, bromocriptine, and rotigotine. Presently, studies examining the relationship between dopamine receptor agonists and genomics mainly focus on the effect of gene polymorphism on the efficacy of dopamine receptor agonists.
3.3.1. DRD2 and DRD3 Gene
The DRD2 and DRD3 gene polymorphisms may influence drug efficacy and tolerance. Together with his team, Liu found that DRD3 Ser/Gly polymorphism influenced the efficacy of dopamine receptor agonists in 30 Chinese PD patients [60], which was corroborated by Xu’s study [61]. To investigate the association between Dopamine receptor D type 2 (DRD2) dinucleotide short tandem repeat (CA(n)-STR) and Dopamine receptor D type 3 (DRD3) Ser9Gly polymorphisms and different doses of Dopamine receptor agonists (DAs) in PD patients, professor Xu recruited 168 idiopathic PD patients and 182 controls. Further exploration showed no association between DRD2 CA(n)-STR polymorphism and DA dosage. Among patients with three different DRD3 Ser9Gly genotypes (Ser/Ser, Ser/Gly, and Gly/Gly), patients carrying Gly/Gly genotype used higher doses of DAs than those with Ser/Gly and Ser/Ser genotypes. DRD3 Ser9Gly Ser/Ser genotype has a higher response rate to dopamine agonists and requires a smaller dose. In a recent study of patients in Brazil, the presence of the TTCTA haplotype, derived from five DRD2 SNPs, was also linked with a high risk of dyskinesia [62].
3.3.2. DRD4 Gene
Sleep attacks in PD were initially reported to occur only with particular dopamine agonists, pramipexole, and ropinirole. The DRD4 48-bp VNTR short/short variant was significantly associated with sleep attacks without warning signs [63].
In general, research in this area is insufficient. Considering the complexity of nervous system diseases, comorbidities in elderly patients, multiple treatment regimens, lack of dose-response relation for many drugs, and other non-genetic factors affecting treatment results, as well as various problems that may be encountered in the design and implementation of PD pharmacogenomics conclusive studies, confirming the clinical utility of pharmacogenomics is challenging. Therefore, the positive results obtained for some candidate genes studied to date indicate that pharmacogenomics of PD warrants more extensive studies involving more uniform, large patient groups to minimize the effect of nongenetic factors.
3.4. Pharmacogenetic Studies on Other Anti-PD Drugs
Until now, levodopa has been the most effective treatment for patients with PD. However, with long-term use, the effect of levodopa gradually decreases and contributes to various motor complications. Therefore, for patients with advanced PD, combination therapy including levodopa and other drugs is routinely indicated. Monoamine oxidase (MAO) inhibitor drugs, such as selegiline and rasagiline, could reduce the metabolic degradation of levodopa, thus they are often concurrently administered with it [64]. In later stages of the disease requiring levodopa, adjunctive monoamine oxidase B inhibitors reduce ‘off’ time and may improve gait and freezing [65]. Monoamine oxidase is an extra-mitochondrial protein found in almost all human tissues [66]. This enzyme regulates neurotransmitter metabolism in the brain and other systems. Therefore, its inhibition can boost neurotransmitter levels in the brain. MAO inhibitors can be divided into two categories according to their pharmacological effects. MAO-A is primarily involved in the oxidative metabolism of tyramine, whereas inhibition of MAO-B primarily reduces the metabolism of dopamine and β-phenylethylamine [67]. Because of their role in levodopa metabolism, selective MAO-B inhibitors are clinically used for PD treatment drugs. Few pharmacogenetic studies have evaluated the effect of DRD2 gene polymorphisms on the clinical response to rasagiline. The results of a large study including 692 PD patients indicated that two SNPs of the DRD2 gene, rs2283265, and rs1076560, were significantly correlated with improved motor function in response to 12-week management with rasagiline after controlling for placebo effects [68].
COMT inhibitors are concomitantly used with levodopa to inhibit levodopa metabolism. PD treatment is based on replacing lost DA with levodopa, which can pass the blood-brain barrier. Administration of COMT inhibitors blocks methylation of levodopa to 3-methyldopa (3-OMD) through the inhibition of the COMT enzyme, thereby preventing levodopa degradation through this major peripheral metabolic pathway and improving its clinical potency efficacy. Presently, COMT inhibitors commonly used in clinical practice include entacapone, tolcapone, and opicapone. Tolcapone and entacapone have mainly been used with patients with more advanced diseases, including chronic motor fluctuations. Whereas tolcapone is associated with hepatotoxicity, entacapone has a short half-life in plasma and requires dosing with every administration of levodopa [69]. However, compared with other COMT inhibitors, opicapone, a third-generation COMT inhibitor, not only reduces the risk of toxicity but also improves COMT inhibitory efficacy and peripheral tissue selectivity [70].
In general, the COMT inhibitors such as entacapone and tolcapone are concomitantly used with levodopa, as they could inhibit the metabolism of levodopa. The effect of genetic variants in the COMT gene on clinical response to entacapone has been evaluated in a clinical trial including 33 PD patients. The results demonstrated that entacapone-treated patients with Val/Val genotype showed a higher levodopa concentration [71]. Besides, the A528G SNP of the UGT1A6 gene is associated with the hepatotoxicity of entacapone and tolcapone in a large European study of 409 PD patients [72].
4. DISSCUSION
The present review gives an overview of the published data on PD pharmacogenetics, shows their limitations and gives insights that may be useful to future studies. Previous studies have explored the effect of gene polymorphism on the efficacy and side effects of levodopa. The efficacy and side effects of levodopa may be connected to the genetic variation of various genes involved in metabolism and transport in vivo, such as the catechol-o-methyltransferase (COMT) gene, the dopamine receptor (DRD 1-5) gene, and the dopamine transporter (SLCA3) gene, among others. Multiple studies have shown that patients with the COMT-LL genotype require less levodopa but are at greater risk for levodopa-induced dyskinesia; Lower frequency of DRD2 13 or 14 alleles in patients with peak-dose dyskinesias; Higher frequency of DAT gene VATR sequence in patients with LID, and so on.
The role of many genetic polymorphisms in drug response has been demonstrated by only one or a few studies and is inconclusive, for instance: MAO-A gene, BDNF gene, HOMER1 gene, MTHFR gene, etc. In addition, compared with dopamine agents, pharmacogenomic investigations of other antiparkinson medicines, such as COMT inhibitors, MAO inhibitors, dopamine receptor agonists, anticholinergic pharmaceuticals, and amantadine, have been restricted, and firm findings have not been achieved. In brief, pharmacogenomic investigations have demonstrated that gene polymorphisms are strongly related to disparities in the efficacy and side effects of several anti-PD medications, including levodopa and rasagiline. However, due to the paucity of prospective large-scale clinical studies in China, it cannot be utilized to guide clinical application.
As we all know, PD is a neurodegenerative disease manifesting as impaired motor function [73]. Pharmacological treatment of symptoms has shown excellent but highly variable efficacy in PD patients. It can be shown that the efficacy, side effects, and problems of anti-PD medications vary greatly depending on the person, and clinical medication must focus on tailored exact therapy [9, 10]. Unfortunately, appropriate assistance is unavailable due to a lack of professional consensus or recommendations.
Thus, studies identifying factors associated with this variability would contribute to more personalized treatment of PD. In the last decade, pharmacogenetic studies in PD have shed light on the role of genetic factors in drug response and the adverse effects of anti-PD drugs [74]. However, results on the link between investigated genes and drug response phenotypes have been inconclusive. The inability to replicate findings among these studies may be due to the small sample size used and the diverse genetic backgrounds of patients. Each genetic variant has a small contribution to the inter-individual variability in drug response [16]. This effect could be missed if a small number of subjects are analyzed. In addition, different ethnicities in the investigated cohort could be another source of variability among different studies [75]. Therefore, genotype groups and genetic backgrounds should be considered when evaluating the effect of genetic factors in pharmacogenetic studies. It is important to note that drug responses or adverse effects are associated with multiple factors, including genetic variants, drug interactions, concomitant therapy, and environmental factors [76]. The effect of a certain genetic variant would be diluted if the other risk factors in the investigated cohort are not adequately considered. Besides, the large heterogeneity of PD in clinical settings is another important issue that should be taken into account. PD manifests as a heterogeneous clinical syndrome, with motor and nonmotor symptoms and variable rates of disease progression [77]. On one hand, although significant strides have been made to improve clinical instruments for accurately assessing the main outcomes of the disease, some characteristics, such as motor fluctuation and some non-motor symptoms, still lack clear definitions and precise scales. On the other hand, identifying PD subtypes may help understand the underlying disease mechanism and design better clinical trials for pharmacogenetic studies. Moreover, most pharmacogenetic studies in PD to date are cross-sectional or retrospective. Given that PD is a progressive disease, longitudinal studies are needed to better identify the effect of genetic factors on drug response and the onset of adverse events [16].
Taken as a whole, PD pharmacogenomics research has become a local and worldwide hotspot, but there are still obstacles in clinical application and directing the customized precision treatment of PD, which may be attributable to the following causes. 1) Most pharmacogenomic studies of Parkinson's disease concentrate on levodopa rather than other regularly prescribed anti-PD medications. 2) The function of the majority of gene polymorphisms is not entirely understood. 3) Existing pharmacogenomics researches on Parkinson's disease concentrate mostly on PD patients in western nations. It must be determined if the research results have reference value for PD patients in China.
CONCLUSION AND FUTURE PERSPECTIVE
In order to create personalised precision treatments for PD, it is important to conduct large, homogenous, multi-center, prospective, randomized controlled clinical studies of PD pharmacogenomics encompassing common therapeutic anti-PD medicines. Additionally, to better apply the research results of pharmacogenomics to guide the clinical diagnosis and treatment of PD, we need to incorporate the probable genetic variations of all patients with suspected PD into the diagnostic chain (clinical examination, MRI, LP, DatScan, FDG-PET) for better drug selection. The identification of a genetic variant associated with drug response in PD is a significant step preceding its use in clinical practice. Therefore, the biological functional effect of identified genetic variants and their interactions with other genetic or environmental factors should be explored further. Finally, cost-effectiveness analyses should be conducted to assess the translation of research evidence into clinical practice.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the BRZ and JLP contributed to the design of the manuscript. YC and DDS contributed to the polishing of the manuscript. This work was financial support by Zhejiang Provincial Program for Medical and Health Science Co-sponsored by Province and Ministry.
LIST OF ABBREVIATIONS
- COMT
Catechol-O-methyltransferase
- DAs
Dopamine Receptor Agonists
- DAT
Dopamine Transporter
- DR
Dopamine Receptor
- DRD2
Dopamine Receptor D Type 2
- MAO
Monoamine Oxidase
- PD
Parkinson’s Disease
- SLC
Solute Carrier
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
This work was financially supported by Zhejiang Provincial (Grant no. LQ18E020002) Program for Medical and Health Science Co-sponsored by Province and Ministry (Application number 2023569421).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.deRijk M.C., Tzourio C., Breteler M.M.B., Dartigues J.F., Amaducci L., Lopez P.S., Manubens B.J.M., Alperovitch A., Rocca W.A. Prevalence of parkinsonism and Parkinson’s disease in Europe: The EUROPARKINSON collaborative study. J. Neurol. Neurosur. PS. 1997;62(1):10–15. doi: 10.1136/jnnp.62.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Z., Roman G., Hong Z., Wu C., Qu Q., Huang J., Zhou B., Geng Z., Wu J., Wen H., Zhao H., Zahner G.E.P. Parkinson’s disease in China: Prevalence in beijing, xian, and shanghai. Lancet. 2005;365(9459):595–597. doi: 10.1016/S0140-6736(05)70801-1. [DOI] [PubMed] [Google Scholar]
- 3.Qi S., Yin P., Wang L., Qu M., Kan G.L., Zhang H., Zhang Q., Xiao Y., Deng Y., Dong Z., Shi Y., Meng J., Chan P., Wang Z. Prevalence of Parkinson’s Disease: A Community‐Based Study in China. Mov. Disord. 2021;36(12):2940–2944. doi: 10.1002/mds.28762. [DOI] [PubMed] [Google Scholar]
- 4.Wolters E.Ch., Braak H. Parkinson’s disease: Premotor clinico-pathological correlations. J. Neural Transm. Suppl. 2006;(70):309–319. doi: 10.1007/978-3-211-45295-0_47. [DOI] [PubMed] [Google Scholar]
- 5.Rocha E.M., De Miranda B., Sanders L.H. Alpha-synuclein: Pathology, mitochondrial dysfunction and neuroinflammation in Parkinson's disease. Neurobiol. Dis. 2018;109(Pt B):249. doi: 10.1016/j.nbd.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Fahn S. The history of dopamine and levodopa in the treatment of Parkinson’s disease. Mov. Disord. 2008;23(S3) Suppl. 3:S497–S508. doi: 10.1002/mds.22028. [DOI] [PubMed] [Google Scholar]
- 7.Armstrong M.J., Okun M.S. Diagnosis and treatment of Parkinson disease. JAMA. 2020;323(6):548–560. doi: 10.1001/jama.2019.22360. [DOI] [PubMed] [Google Scholar]
- 8.Corvol J.C., Poewe W. Pharmacogenetics of Parkinson’s disease in clinical practice. Mov. Disord. Clin. Pract. (Hoboken) 2017;4(2):173–180. doi: 10.1002/mdc3.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cacabelos R. Parkinson’s disease: From pathogenesis to pharmacogenomics. Int. J. Mol. Sci. 2017;18(3):551. doi: 10.3390/ijms18030551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payami H. The emerging science of precision medicine and pharmacogenomics for Parkinson’s disease. Mov. Disord. 2017;32(8):1139–1146. doi: 10.1002/mds.27099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalliokoski A., Niemi M. Impact of OATP transporters on pharmacokinetics. Br. J. Pharmacol. 2009;158(3):693–705. doi: 10.1111/j.1476-5381.2009.00430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roden D.M., McLeod H.L., Relling M.V., Williams M.S., Mensah G.A., Peterson J.F., Van Driest S.L. Pharmacogenomics. Lancet. 2019;394(10197):521–532. doi: 10.1016/S0140-6736(19)31276-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cacabelos R., Cacabelos P., Aliev G. Genomics of schizophrenia and pharmacogenomics of antipsychotic drugs. Open J. Psychiatr. 2013;3(1):46–139. doi: 10.4236/ojpsych.2013.31008. [DOI] [Google Scholar]
- 14.Schumacher-Schuh A.F., Rieder C.R.M., Hutz M.H. Parkinson’s disease pharmacogenomics: New findings and perspectives. Pharmacogenomics. 2014;15(9):1253–1271. doi: 10.2217/pgs.14.93. [DOI] [PubMed] [Google Scholar]
- 15.Redenšek S., Dolžan V. The role of pharmacogenomics in the personalization of Parkinson’s disease treatment. Pharmacogenomics. 2020;21(14):1033–1043. doi: 10.2217/pgs-2020-0031. [DOI] [PubMed] [Google Scholar]
- 16.Politi C., Ciccacci C., Novelli G., Borgiani P. Genetics and treatment response in Parkinson’s disease: An update on pharmacogenetic studies. Neuromol. Med. 2018;20(1):1–17. doi: 10.1007/s12017-017-8473-7. [DOI] [PubMed] [Google Scholar]
- 17.Scanlon P.D., Raymond F.A., Weinshilboum R.M. Catechol-O-methyltransferase: Thermolabile enzyme in erythrocytes of subjects homozygous for allele for low activity. Science. 1979;203(4375):63–65. doi: 10.1126/science.758679. [DOI] [PubMed] [Google Scholar]
- 18.McLeod H.L., Fang L., Luo X., Scott E.P., Evans W.E. Ethnic differences in erythrocyte catechol-O-methyltransferase activity in black and white Americans. J. Pharmacol. Exp. Ther. 1994;270(1):26–29. [PubMed] [Google Scholar]
- 19.Emin Erdal M., Herken H., Yilmaz M., Bayazit Y.A. Significance of the catechol-O-methyltransferase gene polymorphism in migraine. Brain Res. Mol. Brain Res. 2001;94(1-2):193–196. doi: 10.1016/S0169-328X(01)00219-4. [DOI] [PubMed] [Google Scholar]
- 20.Lachman H.M., Papolos D.F., Saito T., Yu Y.M., Szumlanski C.L., Weinshilboum R.M. Human catechol-O-methyltransferase pharmacogenetics: Description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics. 1996;6(3):243–250. doi: 10.1097/00008571-199606000-00007. [DOI] [PubMed] [Google Scholar]
- 21.Białecka M., Droździk M., Kłodowska-Duda G., Honczarenko K., Gawrońska-Szklarz B., Opala G., Stankiewicz J. The effect of monoamine oxidase B (MAOB) and catechol-O-methyltransferase (COMT) polymorphisms on levodopa therapy in patients with sporadic Parkinson’s disease. Acta Neurol. Scand. 2004;110(4):260–266. doi: 10.1111/j.1600-0404.2004.00315.x. [DOI] [PubMed] [Google Scholar]
- 22.Cheshire P., Bertram K., Ling H., O’Sullivan S.S., Halliday G., McLean C., Bras J., Foltynie T., Storey E., Williams D.R. Influence of single nucleotide polymorphisms in COMT, MAO-A and BDNF genes on dyskinesias and levodopa use in Parkinson’s disease. Neurodegener. Dis. 2014;13(1):24–28. doi: 10.1159/000351097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sampaio T.F., dos Santos E.U.D., de Lima G.D.C., dos Anjos R.S.G., da Silva R.C., Asano A.G.C., Asano N.M.J., Crovella S., de Souza P.R.E. MAO-B and COMT genetic variations associated with levodopa treatment response in patients with Parkinson’s disease. J. Clin. Pharmacol. 2018;58(7):920–926. doi: 10.1002/jcph.1096. [DOI] [PubMed] [Google Scholar]
- 24.Contin M., Martinelli P., Mochi M., Riva R., Albani F., Baruzzi A. Genetic polymorphism of catechol-O-methyltransferase and levodopa pharmacokinetic-pharmacodynamic pattern in patients with Parkinson’s disease. Mov. Disord. 2005;20(6):734–739. doi: 10.1002/mds.20410. [DOI] [PubMed] [Google Scholar]
- 25.Lee M.S., Lyoo C.H., Ulmanen I., Syvänen A.C., O. Rinne. J. Genotypes of catechol-O-methyltransferase and response to levodopa treatment in patients with Parkinson’s disease. Neurosci. Lett. 2001;298(2):131–134. doi: 10.1016/S0304-3940(00)01749-3. [DOI] [PubMed] [Google Scholar]
- 26.Grandy D.K., Litt M., Allen L., Bunzow J.R., Marchionni M., Makam H., Reed L., Magenis R.E., Civelli O. The human dopamine D2 receptor gene is located on chromosome 11 at q22-q23 and identifies a TaqI RFLP. Am. J. Hum. Genet. 1989;45(5):778–785. [PMC free article] [PubMed] [Google Scholar]
- 27.Kaiser R., Hofer A., Grapengiesser A., Gasser T., Kupsch A., Roots I., Brockmöller J. L -Dopa-induced adverse effects in PD and dopamine transporter gene polymorphism. Neurology. 2003;60(11):1750–1755. doi: 10.1212/01.WNL.0000068009.32067.A1. [DOI] [PubMed] [Google Scholar]
- 28.dos Santos E.U.D., Sampaio T.F., Tenório dos Santos A.D., Bezerra Leite F.C., da Silva R.C., Crovella S., Asano A.G.C., Asano N.M.J., de Souza P.R.E. The influence of SLC6A3 and DRD2 polymorphisms on levodopa-therapy in patients with sporadic Parkinson’s disease. J. Pharm. Pharmacol. 2019;71(2):206–212. doi: 10.1111/jphp.13031. [DOI] [PubMed] [Google Scholar]
- 29.Becker M.L., Visser L.E., van Schaik R.H.N., Hofman A., Uitterlinden A.G., Stricker B.H.C. OCT1 polymorphism is associated with response and survival time in anti-Parkinsonian drug users. Neurogenetics. 2011;12(1):79–82. doi: 10.1007/s10048-010-0254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altmann V., Schumacher-Schuh A.F., Rieck M., Callegari-Jacques S.M., Rieder C.R.M., Hutz M.H. Influence of genetic, biological and pharmacological factors on levodopa dose in Parkinson’s disease. Pharmacogenomics. 2016;17(5):481–488. doi: 10.2217/pgs.15.183. [DOI] [PubMed] [Google Scholar]
- 31.Vuletić V., Rački V., Papić E., Peterlin B. A systematic review of Parkinson’s disease pharmacogenomics: Is there time for translation into the clinics? Int. J. Mol. Sci. 2021;22(13):7213. doi: 10.3390/ijms22137213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Lau L.M.L., Verbaan D., Marinus J., Heutink P., van Hilten J.J. Catechol-O-methyltransferase Val158Met and the risk of dyskinesias in Parkinson’s disease. Mov. Disord. 2012;27(1):132–135. doi: 10.1002/mds.23805. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe M., Harada S., Nakamura T., Ohkoshi N., Yoshizawa K., Hayashi A., Shoji S. Association between catechol-O-methyltransferase gene polymorphisms and wearing-off and dyskinesia in Parkinson’s disease. Neuropsychobiology. 2003;48(4):190–193. doi: 10.1159/000074637. [DOI] [PubMed] [Google Scholar]
- 34.Oliveri R.L., Annesi G., Zappia M., Civitelli D., Montesanti R., Branca D., Nicoletti G., Spadafora P., Pasqua A.A., Cittadella R., Andreoli V., Gambardella A., Aguglia U., Quattrone A. Dopamine D2 receptor gene polymorphism and the risk of levodopa-induced dyskinesias in PD. Neurology. 1999;53(7):1425–1430. doi: 10.1212/WNL.53.7.1425. [DOI] [PubMed] [Google Scholar]
- 35.Rieck M., Schumacher-Schuh A.F., Altmann V., Francisconi C.L.M., Fagundes P.T.B., Monte T.L., Callegari-Jacques S.M., Rieder C.R.M., Hutz M.H. DRD2 haplotype is associated with dyskinesia induced by levodopa therapy in Parkinson’s disease patients. Pharmacogenomics. 2012;13(15):1701–1710. doi: 10.2217/pgs.12.149. [DOI] [PubMed] [Google Scholar]
- 36.Strong J.A., Dalvi A., Revilla F.J., Sahay A., Samaha F.J., Welge J.A., Gong J., Gartner M., Yue X., Yu L. Genotype and smoking history affect risk of levodopa-induced dyskinesias in Parkinson’s disease. Mov. Disord. 2006;21(5):654–659. doi: 10.1002/mds.20785. [DOI] [PubMed] [Google Scholar]
- 37.Kurzawski M., Białecka M., Droździk M. Pharmacogenetic considerations in the treatment of Parkinson’s disease. Neurodegener. Dis. Manag. 2015;5(1):27–35. doi: 10.2217/nmt.14.38. [DOI] [PubMed] [Google Scholar]
- 38.Kaplan N., Vituri A., Korczyn A.D., Cohen O.S., Inzelberg R., Yahalom G., Kozlova E., Milgrom R., Laitman Y., Friedman E., Rosset S., Hassin-Baer S. Sequence variants in SLC6A3, DRD2, and BDNF genes and time to levodopa-induced dyskinesias in Parkinson’s disease. J. Mol. Neurosci. 2014;53(2):183–188. doi: 10.1007/s12031-014-0276-9. [DOI] [PubMed] [Google Scholar]
- 39.Sossi V., de la Fuente-Fernández R., Schulzer M., Troiano A.R., Ruth T.J., Stoessl A.J. Dopamine transporter relation to dopamine turnover in Parkinson’s disease: A positron emission tomography study. Ann. Neurol. 2007;62(5):468–474. doi: 10.1002/ana.21204. [DOI] [PubMed] [Google Scholar]
- 40.Troiano A.R., de la Fuente-Fernandez R., Sossi V., Schulzer M., Mak E., Ruth T.J., Stoessl A.J. PET demonstrates reduced dopamine transporter expression in PD with dyskinesias. Neurology. 2009;72(14):1211–1216. doi: 10.1212/01.wnl.0000338631.73211.56. [DOI] [PubMed] [Google Scholar]
- 41.Bjornestad A., Forsaa E.B., Pedersen K.F., Tysnes O.B., Larsen J.P., Alves G. Risk and course of motor complications in a population-based incident Parkinson’s disease cohort. Parkinsonism Relat. Disord. 2016;22:48–53. doi: 10.1016/j.parkreldis.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Stocchi F., Olanow C.W. Continuous dopaminergic stimulation in early and advanced Parkinson’s disease. Neurology. 2004;62(1, Supplement 1 Suppl. 1):S56-S63. doi: 10.1212/WNL.62.1_suppl_1.S56. [DOI] [PubMed] [Google Scholar]
- 43.Gilgun-Sherki Y., Djaldetti R., Melamed E., Offen D. Polymorphism in candidate genes: Implications for the risk and treatment of idiopathic Parkinson’s disease. Pharmacogenomics J. 2004;4(5):291–306. doi: 10.1038/sj.tpj.6500260. [DOI] [PubMed] [Google Scholar]
- 44.Zappia M., Annesi G., Nicoletti G., Arabia G., Annesi F., Messina D., Pugliese P., Spadafora P., Tarantino P., Carrideo S., Civitelli D., De Marco E.V., Cirò-Candiano I.C., Gambardella A., Quattrone A. Sex differences in clinical and genetic determinants of levodopa peak-dose dyskinesias in Parkinson disease: An exploratory study. Arch. Neurol. 2005;62(4):601–605. doi: 10.1001/archneur.62.4.601. [DOI] [PubMed] [Google Scholar]
- 45.Fahn S. Parkinson disease, the effect of levodopa, and the ELLDOPA trial. Earlier vs Later L-DOPA. Arch. Neurol. 1999;56(5):529–535. doi: 10.1001/archneur.56.5.529. [DOI] [PubMed] [Google Scholar]
- 46.Schumacher-Schuh A.F., Altmann V., Rieck M., Tovo-Rodrigues L., Monte T.L., Callegari-Jacques S.M., Medeiros M.S., Rieder C.R.M., Hutz M.H. Association of common genetic variants of HOMER1 gene with levodopa adverse effects in Parkinson’s disease patients. Pharmacogenomics J. 2014;14(3):289–294. doi: 10.1038/tpj.2013.37. [DOI] [PubMed] [Google Scholar]
- 47.Shen Y., Huang J.Y., Li J., Liu C.F. Excessive daytime sleepiness in Parkinson’s disease. Chin. Med. J. (Engl.) 2018;131(8):974–981. doi: 10.4103/0366-6999.229889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rissling I., Frauscher B., Kronenberg F., Tafti M., Stiasny-Kolster K., Robyr A.C., Körner Y., Oertel W.H., Poewe W., Högl B., Möller J.C. Daytime sleepiness and the COMT val158met polymorphism in patients with Parkinson disease. Sleep. 2006;29(1):108–111. [PubMed] [Google Scholar]
- 49.Ying Y., Lin S., Kong F., Li Y., Xu S., Liang X., Wang C., Han L. Ideal cardiovascular health metrics and incidence of ischemic stroke among hypertensive patients: A prospective cohort study. Front. Cardiovasc. Med. 2020;7:590809. doi: 10.3389/fcvm.2020.590809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fallest-Strobl P.C., Koch D.D., Stein J.H., McBride P.E. Homocysteine: A new risk factor for atherosclerosis. Am. Fam. Physician. 1997;56(6):1607–1612, 1615-1616. [PubMed] [Google Scholar]
- 51.House D.J., Brosnan E.M., Brosnan T.J. Characterization of homocysteine metabolism in the rat kidney. Biochem. J. 1997;328(1):287–292. doi: 10.1042/bj3280287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anniwaer J., Liu M., Xue K., Maimaiti A., Xiamixiding A. Homocysteine might increase the risk of recurrence in patients presenting with primary cerebral infarction. Int. J. Neurosci. 2019;129(7):654–659. doi: 10.1080/00207454.2018.1517762. [DOI] [PubMed] [Google Scholar]
- 53.De Bonis M.L., Tessitore A., Pellecchia M.T., Longo K., Salvatore A., Russo A., Ingrosso D., Zappia V., Barone P., Galletti P., Tedeschi G. Impaired transmethylation potential in Parkinson’s disease patients treated with l-Dopa. Neurosci. Lett. 2010;468(3):287–291. doi: 10.1016/j.neulet.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 54.Redenšek S., Flisar D., Kojović M., Gregorič Kramberger M., Georgiev D., Pirtošek Z., Trošt M., Dolžan V. Dopaminergic pathway genes influence adverse events related to dopaminergic treatment in Parkinson’s disease. Front. Pharmacol. 2019;10:8. doi: 10.3389/fphar.2019.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goetz C.G., Burke P.F., Leurgans S., Berry-Kravis E., Blasucci L.M., Raman R., Zhou L. Genetic variation analysis in parkinson disease patients with and without hallucinations: Case-control study. Arch. Neurol. 2001;58(2):209–213. doi: 10.1001/archneur.58.2.209. [DOI] [PubMed] [Google Scholar]
- 56.Makoff A., Graham J., Arranz M., Forsyth J., Li T., Aitchison K., Shaikh S., Grünewald R. Association study of dopamine receptor gene polymorphisms with drug-induced hallucinations in patients with idiopathic Parkinson’s disease. Pharmacogenetics. 2000;10(1):43–48. doi: 10.1097/00008571-200002000-00006. [DOI] [PubMed] [Google Scholar]
- 57.Cormier-Dequaire F., Bekadar S., Anheim M., Lebbah S., Pelissolo A., Krack P., Lacomblez L., Lhommée E., Castrioto A., Azulay J.P., Defebvre L., Kreisler A., Durif F., Marques-Raquel A., Brefel-Courbon C., Grabli D., Roze E., Llorca P.M., Ory-Magne F., Benatru I., Ansquer S., Maltête D., Tir M., Krystkowiak P., Tranchant C., Lagha-Boukbiza O., Lebrun-Vignes B., Mangone G., Vidailhet M., Charbonnier-Beaupel F., Rascol O., Lesage S., Brice A., Tezenas du Montcel S., Corvol J.C., Grp B-P.S. Suggestive association between OPRM1 and impulse control disorders in Parkinson’s disease. Mov. Disord. 2018;33(12):1878–1886. doi: 10.1002/mds.27519. [DOI] [PubMed] [Google Scholar]
- 58.Zainal Abidin S., Tan E.L., Chan S.C., Jaafar A., Lee A.X., Abd Hamid M.H.N., Abdul Murad N.A., Pakarul Razy N.F., Azmin S., Ahmad Annuar A., Lim S.Y., Cheah P.S., Ling K.H., Mohamed Ibrahim N. DRD and GRIN2B polymorphisms and their association with the development of impulse control behaviour among Malaysian Parkinson’s disease patients. BMC Neurol. 2015;15(1):59. doi: 10.1186/s12883-015-0316-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kraemmer J., Smith K., Weintraub D., Guillemot V., Nalls M.A., Cormier-Dequaire F., Moszer I., Brice A., Singleton A.B., Corvol J.C. Clinical-genetic model predicts incident impulse control disorders in Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry. 2016;87(10):1106–1111. doi: 10.1136/jnnp-2015-312848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y.Z., Tang B.S., Yan X.X., Liu J., Ouyang D.S., Nie L.N., Fan L., Li Z., Ji W., Hu D.L., Wang D., Zhou H.H. Association of the DRD2 and DRD3 polymorphisms with response to pramipexole in Parkinson’s disease patients. Eur. J. Clin. Pharmacol. 2009;65(7):679–683. doi: 10.1007/s00228-009-0658-z. [DOI] [PubMed] [Google Scholar]
- 61.Xu S., Liu J., Yang X., Qian Y., Xiao Q. Association of the DRD2 CA n -STR and DRD3 Ser9Gly polymorphisms with Parkinson’s disease and response to dopamine agonists. J. Neurol. Sci. 2017;372:433–438. doi: 10.1016/j.jns.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 62.Paus S., Grünewald A., Klein C., Knapp M., Zimprich A., Janetzky B., Möller J.C., Klockgether T., Wüllner U. TheDRD2 TaqIA polymorphism and demand of dopaminergic medication in Parkinson’s disease. Mov. Disord. 2008;23(4):599–602. doi: 10.1002/mds.21901. [DOI] [PubMed] [Google Scholar]
- 63.Paus S., Seeger G., Brecht H.M., Köster J., El-Faddagh M., Nöthen M.M., Klockgether T., Wüllner U. Association study of dopamine D2, D3, D4 receptor and serotonin transporter gene polymorphisms with sleep attacks in Parkinson’s disease. Mov. Disord. 2004;19(6):705–707. doi: 10.1002/mds.20134. [DOI] [PubMed] [Google Scholar]
- 64.Seppi K., Weintraub D., Coelho M., Perez-Lloret S., Fox S.H., Katzenschlager R., Hametner E.M., Poewe W., Rascol O., Goetz C.G., Sampaio C. The movement disorder society evidence-based medicine review update: Treatments for the non-motor symptoms of Parkinson’s disease. Mov. Disord. 2011;26(Suppl. 3):S42–S80. doi: 10.1002/mds.23884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shoulson I., Oakes D., Fahn S., Lang A., Langston J.W., LeWitt P., Olanow C.W., Penney J.B., Tanner C., Kieburtz K., Rudolph A. Impact of sustained deprenyl (selegiline) in levodopa-treated Parkinson’s disease: A randomized placebo-controlled extension of the deprenyl and tocopherol antioxidative therapy of parkinsonism trial. Ann. Neurol. 2002;51(5):604–612. doi: 10.1002/ana.10191. [DOI] [PubMed] [Google Scholar]
- 66.Mahmood I. Clinical pharmacokinetics and pharmacodynamics of selegiline. An update. Clin. Pharmacokinet. 1997;33(2):91–102. doi: 10.2165/00003088-199733020-00002. [DOI] [PubMed] [Google Scholar]
- 67.Foley P., Gerlach M., Youdim M.B.H., Riederer P. MAO-B inhibitors: Multiple roles in the therapy of neurodegenerative disorders? Parkinsonism Relat. Disord. 2000;6(1):25–47. doi: 10.1016/S1353-8020(99)00043-7. [DOI] [PubMed] [Google Scholar]
- 68.Masellis M., Collinson S., Freeman N., Tampakeras M., Levy J., Tchelet A., Eyal E., Berkovich E., Eliaz R.E., Abler V., Grossman I., Fitzer-Attas C., Tiwari A., Hayden M.R., Kennedy J.L., Lang A.E., Knight J., Investigators A. Dopamine D2 receptor gene variants and response to rasagiline in early Parkinson’s disease: A pharmacogenetic study. Brain. 2016;139(7):2050–2062. doi: 10.1093/brain/aww109. [DOI] [PubMed] [Google Scholar]
- 69.Kaakkola S. In: In: Basic Aspects of Catechol-O-Methyltransterase and the Clinical Applications of Its Inhibitors. Nissinen E., editor. Vol. 95. San Diego: Elsevier Academic Press Inc; 2010. Problems with the present inhibitors and a relevance of new and improved comt inhibitors in parkinson’s disease. pp. 207–225. [DOI] [PubMed] [Google Scholar]
- 70.Kiss L.E., Soares-da-Silva P. Medicinal chemistry of catechol O-methyltransferase (COMT) inhibitors and their therapeutic utility. J. Med. Chem. 2014;57(21):8692–8717. doi: 10.1021/jm500572b. [DOI] [PubMed] [Google Scholar]
- 71.Corvol J.C., Bonnet C., Charbonnier-Beaupel F., Bonnet A.M., Fiévet M.H., Bellanger A., Roze E., Meliksetyan G., Ben Djebara M., Hartmann A., Lacomblez L., Vrignaud C., Zahr N., Agid Y., Costentin J., Hulot J.S., Vidailhet M. The COMT Val158Met polymorphism affects the response to entacapone in Parkinson’s disease: A randomized crossover clinical trial. Ann. Neurol. 2011;69(1):111–118. doi: 10.1002/ana.22155. [DOI] [PubMed] [Google Scholar]
- 72.Acuña G., Foernzler D., Leong D., Rabbia M., Smit R., Dorflinger E., Gasser R., Hoh J., Ott J., Borroni E., To Z., Thompson A., Li J., Hashimoto L., Lindpaintner K. Pharmacogenetic analysis of adverse drug effect reveals genetic variant for susceptibility to liver toxicity. Pharmacogenomics J. 2002;2(5):327–334. doi: 10.1038/sj.tpj.6500123. [DOI] [PubMed] [Google Scholar]
- 73.Bloem B.R., Okun M.S., Klein C. Parkinson’s disease. Lancet. 2021;397(10291):2284–2303. doi: 10.1016/S0140-6736(21)00218-X. [DOI] [PubMed] [Google Scholar]
- 74.Kalinderi K., Papaliagkas V., Fidani L. Pharmacogenetics and levodopa induced motor complications. Int. J. Neurosci. 2019;129(4):384–392. doi: 10.1080/00207454.2018.1538993. [DOI] [PubMed] [Google Scholar]
- 75.Sauerbier A., Aris A., Lim E.W., Bhattacharya K., Ray Chaudhuri K. Impact of ethnicity on the natural history of Parkinson disease. Med. J. Aust. 2018;208(9):410–414. doi: 10.5694/mja17.01074. [DOI] [PubMed] [Google Scholar]
- 76.Ciccacci C., Borgiani P. Pharmacogenomics in Parkinson’s disease: Which perspective for developing a personalized medicine? Neural Regen. Res. 2019;14(1):75–76. doi: 10.4103/1673-5374.243706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Greenland J.C., Williams-Gray C.H., Barker R.A. The clinical heterogeneity of Parkinson’s disease and its therapeutic implications. Eur. J. Neurosci. 2019;49(3):328–338. doi: 10.1111/ejn.14094. [DOI] [PubMed] [Google Scholar]
- 78.Bialecka M., Kurzawski M., Klodowska-Duda G., Opala G., Tan E.K., Drozdzik M. The association of functional catechol-O-methyltransferase haplotypes with risk of Parkinson’s disease, levodopa treatment response, and complications. Pharmacogenet. Genomics. 2008;18(9):815–821. doi: 10.1097/FPC.0b013e328306c2f2. [DOI] [PubMed] [Google Scholar]
- 79.Frauscher B., Högl B., Maret S., Wolf E., Brandauer E., Wenning G.K., Kronenberg M.F., Kronenberg F., Tafti M., Poewe W. Association of daytime sleepiness with COMT polymorphism in patients with Parkinson disease: A pilot study. Sleep. 2004;27(4):733–736. doi: 10.1093/sleep/27.4.733. [DOI] [PubMed] [Google Scholar]
- 80.Purcaro C., Vanacore N., Moret F., Di Battista M.E., Rubino A., Pierandrei S., Lucarelli M., Meco G., Fattapposta F., Pascale E. DAT gene polymorphisms (rs28363170, rs393795) and levodopa-induced dyskinesias in Parkinson’s disease. Neurosci. Lett. 2019;690:83–88. doi: 10.1016/j.neulet.2018.10.021. [DOI] [PubMed] [Google Scholar]
- 81.Foltynie T., Cheeran B., Williams-Gray C.H., Edwards M.J., Schneider S.A., Weinberger D., Rothwell J.C., Barker R.A., Bhatia K.P. BDNF val66met influences time to onset of levodopa induced dyskinesia in Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry. 2009;80(2):141–144. doi: 10.1136/jnnp.2008.154294. [DOI] [PubMed] [Google Scholar]
- 82.Lin J.J., Yueh K.C., Lin S.Z., Harn H.J., Liu J.T. Genetic polymorphism of the angiotensin converting enzyme and l-dopa-induced adverse effects in Parkinson’s disease. J. Neurol. Sci. 2007;252(2):130–134. doi: 10.1016/j.jns.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 83.Gorgone G., Currò M., Ferlazzo N., Parisi G., Parnetti L., Belcastro V., Tambasco N., Rossi A., Pisani F., Calabresi P., Ientile R., Caccamo D. Coenzyme Q10, hyperhomocysteinemia and MTHFR C677T polymorphism in levodopa-treated Parkinson’s disease patients. Neuromol. Med. 2012;14(1):84–90. doi: 10.1007/s12017-012-8174-1. [DOI] [PubMed] [Google Scholar]
- 84.Martín-Flores N., Fernández-Santiago R., Antonelli F., Cerquera C., Moreno V., Martí M.J., Ezquerra M., Malagelada C. MTOR pathway-based discovery of genetic susceptibility to L-DOPA-induced dyskinesia in Parkinson’s disease patients. Mol. Neurobiol. 2019;56(3):2092–2100. doi: 10.1007/s12035-018-1219-1. [DOI] [PubMed] [Google Scholar]
- 85.Rissling I., Körner Y., Geller F., Stiasny-Kolster K., Oertel W.H., Möller J.C. Preprohypocretin polymorphisms in Parkinson disease patients reporting “sleep attacks”. Sleep. 2005;28(7):871–875. doi: 10.1093/sleep/28.7.871. [DOI] [PubMed] [Google Scholar]
- 86.Bonifácio M.J., Palma P.N., Almeida L., Soares-da-Silva P. Catechol-O-methyltransferase and its inhibitors in Parkinson’s disease. CNS Drug Rev. 2007;13(3):352–379. doi: 10.1111/j.1527-3458.2007.00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]