Abstract
Considerable evidence indicates that the semiautonomous organelles mitochondria play key roles in the progression of many neurodegenerative disorders. Mitochondrial DNA (mtDNA) encodes components of the OXPHOS complex but mutated mtDNA accumulates in cells with aging, which mirrors the increased prevalence of neurodegenerative diseases. This accumulation stems not only from the misreplication of mtDNA and the highly oxidative environment but also from defective mitophagy after fission. In this review, we focus on several pivotal mitochondrial proteins related to mtDNA maintenance (such as ATAD3A and TFAM), mtDNA alterations including mtDNA mutations, mtDNA elimination, and mtDNA release-activated inflammation to understand the crucial role played by mtDNA in the pathogenesis of neurodegenerative diseases such as Alzheimer's disease, Parkinson's disease, amyotrophic lateral sclerosis, and Huntington's disease. Our work outlines novel therapeutic strategies for targeting mtDNA.
Keywords: Mitochondrial DNA (mtDNA), mitophagy, reactive oxygen species (ROS), Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, amyotrophic lateral sclerosis
1. INTRODUCTION
Mitochondria are the main powerhouses of cells, providing energy in the form of adenosine triphosphate (ATP). In addition, mitochondria are also involved in the regulation of intracellular calcium homeostasis, synaptic plasticity, apoptosis, and other processes [1-4]. As semiautonomous organelles, mitochondria possess their own DNA called mitochondrial DNA (mtDNA), which is located in the mitochondrial matrix and associated with the inner mitochondrial membrane (IMM). The human mitochondrial genome comprises cyclic DNA containing 16,569 base pairs with two strands of nonhomogeneous mtDNA. mtDNA encodes 13 polypeptides, 2 rRNAs, and 22 tRNAs; the 13 encoded polypeptides are components of the oxidative phosphorylation (OXPHOS) complex, that is, 7 from complex I, 1 from complex III, 3 from complex IV, and 2 from complex V [5].
Distinct from the binding of nuclear DNA and histone proteins, mtDNA and a number of proteins from nucleoids are localized to the IMM. These proteins mainly include mitochondrial transcription factor A (TFAM), mitochondrial single-stranded DNA-binding protein (mtSSB), and the mtDNA helicase Twinkle [6]. As early as 25 years ago, the first pathological mutations were found in mtDNA [7]. In addition to mtDNA mutations, changes in nuclear genes affect DNA maintenance, which in turn harm mtDNA replication, transcription, and translation, resulting in mtDNA deletion and eventually a variety of diseases [8]. Although mtDNA is exclusively inherited in a maternal fashion, most pathological mtDNA mutations are heterogeneous, which means that different mitochondria or even a single mitochondrion in the same cell may contain diverse mtDNA [9]. Mitochondrial mutations have a threshold effect, and clinical symptoms do not emerge until the mutational load exceeds the threshold [10]. Nevertheless, the threshold is low in neurons because their high energy metabolism is highly dependent on mitochondria and because they cannot proliferate.
Neurodegenerative diseases (NDDs) are sporadic or inherited disorders of the central nervous system that lead to a slowly progressive loss of specific neurons and their functions. Based on clinical features, they can be classified as dementia, parkinsonism, and motor neuron disorder. The prevalence of NDDs is increasing as human life expectancy increases [11]. Due to the high energy demand of neurons and the increasing accumulation of oxidatively damaged mtDNA found in neuronal cells in studies related to aging, attention has been focused on the relationship between mitochondria and NDDs [12]. As a result of the extensive use of oxygen and the lack of protection afforded by histones, mtDNA is vulnerable to oxidative damage by reactive oxygen species (ROS), which causes mtDNA mutations that further disrupt mitochondrial function, leading to a vicious cycle [13]. Mutations in mtDNA might accumulate to higher levels within individual cells, resulting in biochemical defects. A higher frequency of biochemically defective cells has been observed in diseases such as Alzheimer’s disease (AD) and Parkinson’s disease (PD) [12, 14], further suggesting the involvement of somatic mutations in the pathogenesis of neurodegeneration. Some of this defective mtDNA can be eliminated by mitophagy after mitochondrial fission, whereas other such DNA will be displaced to the intra- or extracellular compartments, which will trigger inflammation [15, 16].
In this review, we summarize the proteins associated with mtDNA maintenance and mtDNA alterations, including mtDNA mutations caused by replication errors or oxidative damage, mitochondrial fission, and clearance of defective mtDNA by mitophagy, as well as mtDNA release-activated inflammation. At the same time, we illustrate the mtDNA alterations in diseases such as AD, PD, amyotrophic lateral sclerosis (ALS), and Huntington’s disease (HD) and their role in pathogenesis and discuss potential therapeutic strategies according to the nature of these changes.
2. MTDNA MAINTENANCE AND ALTERATION
2.1. mtDNA Maintenance-Related Proteins
Of the approximately 1500 different proteins contained in mitochondria, several hundred are required for mtDNA expression and many are related to mtDNA maintenance [17]. mtDNA maintenance requires the involvement of nuclear DNA (nDNA)-encoded proteins, including those involved in mtDNA synthesis, maintenance of mitochondrial nucleotide pool homeostasis, and mitochondrial dynamics. Defective mtDNA maintenance-related proteins would result in mtDNA depletion or even the deletion of multiple mtDNA sequences [18]. Nucleoids are complexes of mtDNA and proteins. They usually comprise a single copy of mtDNA and the various proteins that play a role in mtDNA maintenance and intermediary metabolism [18].
Of all proteins, TFAM is the most important structural protein in mammalian nucleoids [19, 20], being involved in the packaging of mtDNA into nucleoids and initiation of mtDNA transcription [17]. At the initiation of transcription, TFAM binds to mtDNA promoter regions in a sequence-specific manner and acts as an RNA primer at the beginning of replication [21]. Transcriptional upregulation of TFAM is generally accompanied by an increase in mtDNA, indicating that TFAM mediates mitochondrial biogenesis by linking nuclear transcription to mtDNA [22]. mtDNA can be compacted and packaged into nucleoids due to the binding of mtDNA and TFAM, which force U-turns in the circular molecules [23]. On the one hand, TFAM protects mtDNA from oxidative stress and, on the other, it somewhat reduces mtDNA-mediated inflammation. In cells of TFAM heterozygous (TFAM+/−) mice, reduced TFAM protein levels lead to altered mtDNA packaging and extraction of mtDNA fragments, which culminate in mtDNA stress signaling [24]. The level of TFAM protein is a central regulator of mtDNA copy number, and a meta-analysis showed an average decrease in TFAM protein of about 43% in AD, supporting the belief that a decrease in mtDNA copy number is associated with the TFAM protein level [25].
Another protein engaged in mitochondrial maintenance is the mtDNA-specific helicase Twinkle, which participates in mtDNA replication by unwinding mtDNA prior to polymerase gamma (Pol-γ)-mediated synthesis. In transgenic mice, the mtDNA copy number increases as a result of the overexpression of Twinkle [26]. Moreover, Twinkle mutation will trigger mitochondrial myopathy and NDD due to its contribution to the total mutational load of mtDNA in cells. Pol-γ comprises a p140 subunit encoded by POLG and a p55 subunit encoded by POLG2. Mutations in POLG or POLG2 adversely impact the maintenance of mtDNA, leading to mtDNA depletion, deletion, and mutation. For example, the Chr17: 62492543G>A mutation in POLG2 compromises the stability of the p55 subunit, causing mtDNA depletion and liver failure [27].
Another protein involved in mtDNA maintenance is OPA1, a GTPase with eight isoforms, and its more familiar functions are related to cristae integrity and mitochondrial dynamics [28]. mtDNA maintenance is mainly achieved in two aspects, firstly TM1 and TM2a in OPA1, a short 10-kDa peptide that anchors the nucleoid to the IMM [29], and secondly with other related proteins such as via a TFAM–POL-γ interaction [28, 30]. Previous studies have shown that exon 4b mutations in the OPA1 gene lead to mtDNA deletion and even depletion [31]; its genetic ablation also has similar results [31].
In addition to components of nucleoids such as TFAM, Twinkle, and Pol-γ, other proteins are also involved in mtDNA replication, transcription, and translation, and these are recognized as nucleoid-associated proteins [32]. For example, ATAD3A is a nuclear-encoded mitochondrial protein that spans the inner and outer mitochondrial membranes. As a component of the mitochondrial nucleoid, it plays a role in mitochondrial maintenance [33, 34]. In addition, it exerts functions in maintaining mitochondrial morphology and controlling cholesterol transport. Mutant ATAD3A Drosophila develops severe mitochondrial fission and abnormal cristae, which compromise the integrity of mtDNA [35]. Other nucleoid proteins such as NEK protein kinase also participate in the maintenance of mtDNA by altering the ratio of mtDNA amplification through interactions with SIRT3, ATAD3A, and other proteins [36].
2.2. Causes and Manifestations of mtDNA Mutations
In somatic cells, the levels of mutated mtDNA increase with age [37], and the most common causes of mtDNA mutations are, an error in mtDNA replication [38, 39] and an accumulation of oxidative damage [40, 41]. Most ROS are generated during mitochondrial OXPHOS and ATP production [42], which involve subunit complexes I and III. Under conventional conditions, these ROS can be effectively scavenged by the mitochondrial thioredoxin/peroxiredoxin-3,5 system, glutathione antioxidant system, cytochrome c, and the opening of the mitochondrial permeability transition pore (mPTP). Even though ROS are an important cause of mtDNA mutations, their production is an intrinsic part of OXPHOS and plays a role in several signaling processes. Under physiological conditions, ROS levels are controlled by antioxidant enzyme systems but, when the level of ROS produced by the compromised OXPHOS exceeds the defense capacity, the nearby mtDNA is damaged. In general, it is believed that mtDNA is more susceptible to mutation than nDNA, both on account of the closer proximity of mtDNA to ROS and because mtDNA lacks histone protection. However, it has also been observed that DNA-binding proteins in nucleoids can also protect mtDNA when it is exposed to H2O2 [43]. At the same time, mutations and deletions of mtDNA in turn stimulate the production of new ROS, thereby creating a vicious cycle [12].
ROS cause mtDNA to accumulate oxidative damage, resulting in base modifications including hydroxylation, ring opening and base loss, as well as sugar modifications. Free radical attack also leads to ring opening and single-strand breaks and, most severely, double-strand breaks [44]. Identified using next-generation sequencing, the dominant mutation among the mutation patterns generated by oxidative lesions in mtDNA is the G:C→T:A translocation [45], which mainly arises from oxidative damage-mediated modification of 8-hydroxypurine and formamidopyrimidine. In addition to causing mtDNA mutations, higher levels of ROS are associated with dysregulation of the epigenetic control of mtDNA, which can lead to mtDNA demethylation [46]. This is of interest because mtDNA methylation and hydroxymethylation may play a role in replication and transcription. Furthermore, correlations between mtDNA methylation and gene expression have been reported in mesenchymal stem cells and in peripheral blood samples [46, 47]. For most alkylated and oxidative DNA damage, base excision repair (BER) is the main DNA repair mechanism in mitochondria. In mitochondria, some DNA glycosylases associated with BER, such as uracil-DNA glycosylase and Nth-like DNA glycosylase 1, may help to prevent mtDNA mutations [48].
In recent years, high-throughput DNA sequencing methods have shown that, in addition to oxidative damage, a defective DNA replication process may be one of the sources of mtDNA point mutations, which are closely related to mtDNA Pol-γ [49, 50]. DNA polymerase is a key enzyme involved in DNA synthesis, with the proofreading activity of Pol allowing replication with sufficiently high fidelity to be one of the principal determinants of reliable DNA replication. Studies in mtDNA mutant mice highlight its significance in vivo, with a single point mutation (D257A) introduced into the mouse mtDNA genome eliminating the 3’→5’ nucleic acid exonuclease activity of Pol, and somatic mtDNA mutations accumulate progressively during mitochondrial biogenesis [51]. Recently, it has been reported that mtDNA replication errors can be attributed to the proofreading ability of Pol-γ being diminished by ROS [52]. Thus, oxidation exacerbates mtDNA mutations by affecting Pol-γ, leading to replication errors, which in turn culminate in mtDNA damage.
2.3. mtDNA in Mitochondrial Dynamics
Mitochondrial fission and fusion are essential for the preservation of mitochondrial homeostasis, which involves the maintenance of mtDNA stability and the regulation of mitophagy and apoptosis [53]. Defective copies of mtDNA can be processed by a fusion/fission process. Mitochondria with high mutational load are eliminated by mitophagy after fission, whereas salvageable mitochondria maintain function through fusion to make complementary mutant mtDNA [54, 55]. When the fission/fusion balance is disrupted, excessive mitochondrial division results in reduced mitochondrial content exchange, exacerbating the mtDNA defect [16].
As a major participant in mitochondrial dynamics, OPA1 interacts with other dynamin-like GTPases, such as MFN1, MFN2, and Drp1, and primarily mediates the fusion of the IMM [56, 57]. Dominant optic atrophy can be caused by mutations in OPA1 affecting its GTPase activity with multiple mtDNA deletions accumulating in the postmitotic tissue of these patients [31]. These mitogenomes lacking part of the mtDNA arise due to replication defects and accumulate as a result of OPA1 mutations and ultimately lead to respiratory chain disorders. This demonstrates that mitochondrial fusion is also a key mechanism in mitochondrial maintenance, helping to maintain the integrity of mtDNA. Similar defective mtDNA maintenance is associated with MFN2 mutation [58], further illustrating the important role of mitochondrial fusion in preserving the integrity of the mitochondrial genome.
Mitophagy is also related to mitochondrial dynamics. In general, increased mitochondrial fusion can inhibit mitophagy, either through OPA1 overexpression or DRP1 inhibition [59, 60]. Inhibition of mitochondrial fission in in vitro experiments improves tolerance to a higher mtDNA mutation load but induces overexpression of Parkin to combat mtDNA mutations, which selectively removes mtDNA-mutated mitochondria through fission and mitophagy [61, 62]. Dysfunctional mitochondria are eliminated by Drp1-mediated mitochondrial fission and PTEN-induced kinase-1 (PINK)-dependent mitophagy when mtDNA is altered due to mutations in tyrosyl-DNA phosphodiesterase 1, which is associated with DNA repair, further demonstrating the important role of mitophagy in mtDNA alteration [63].
2.4. mtDNA Elimination via Mitophagy
Mitophagy is a selective form of autophagy that targets damaged mitochondria. Its typical mechanism involves the PINK1/Parkin pathway, which comprises the mitochondrial membrane kinase PINK1 and the cytoplasmic E3 ubiquitin ligase Parkin [4]. Upon sensing a decrease in the mitochondrial membrane potential (ΔΨm), PINK1 autophosphorylates, which allows Parkin activation and the subsequent stimulation of LC3 via the ubiquitinated autophagy adaptor protein SQSTM1 to induce autophagosome formation and digestion by lysosomal enzymes [4]. Besides Parkin/PINK1-dependent mitophagy, hypoxia-triggered mitophagy is characterized by upregulation of BNIP3/Nix levels due to hypoxia-inducible factor 1a transcripts [64] and mitophagy induced by nutrient deprivation involving the phosphoinositide 3-kinase (PI3K) complex [65].
In addition to protection by nucleoid proteins, mitophagy removes mitochondria with damaged mtDNA. Depolarized mitochondria due to mtDNA mutations are unable to integrate with the mitochondrial network after fission [66]. Moreover, when mtDNA mutations are not eliminated by fission and mitophagy, mitochondria will develop a burden in which they switch from OXPHOS to glycolytic metabolism [67]. Mitophagy is repressed in many mtDNA-mutated mitochondria. This results in the accumulation of dysfunctional mitochondria in neurons, which is an important mechanism for the development of NDDs [68]. In contrast, when damaged mitochondria are not promptly removed by mitophagy, mtDNA is released and triggers an inflammatory response, as demonstrated in a model of mice lacking the pro-mitophagy protein Parkin [69].
Accumulation of mutant mtDNA occurs in both Parkin−/− mice and Pink1−/− mice and eventually leads to the loss of dopaminergic neurons [69, 70]. Likewise, the fact that the levels of deleterious mtDNA variants in human cell lines are reduced by Parkin overexpression suggests that selective mitochondrial degradation mechanisms can eliminate mtDNA with deleterious mutations [61]. Many experiments have proven this point. For example, an increase in the mtDNA mutation rate has been found in Parkin knockout mouse somatic cells [61]. A similar phenomenon is observed in rhabdomyosarcoma cells, which accumulate higher levels of mutant m3243G mtDNA compared with adenocarcinoma cells with high Parkin expression [71]. Similarly, overexpression of the granulosa autophagy genes PINK and Parkin significantly increases the heterogeneity of mtDNA compared with the wild-type [72]. However, it remains uncertain whether there is a mechanism for targeting the clearance of mitochondria with mtDNA mutations through mitophagy. For example, the use of Parkin knockout or Parkin mutants has no effect on the purification selection of Drosophila for mtDNA mutations leading to COX1 protein expression [73]. Similarly, when Parkin knockout mice are crossed with mtDNA mutant mice, the total mtDNA mutational load of the mice is also unaltered [74]. However, compared with the elimination of mtDNA mutation, a recent study notably found that mitophagy is not essential for the elimination of mtDNA following the production of double-strand breaks by mitochondria-targeted restriction enzymes [75].
In addition, in a mouse model with deficient mtDNA Pol-γ, mtDNA mutations lead to increased levels of hepatic mitophagy in vivo [76]. Loss of ΔΨm and mTOR inhibition which simulate BCL2 and PINK1/Parkin-mediated pathways are essential for the induction of mtDNA mutation-derived mitophagy [77, 78]. Due to the dependence of mitophagy on a low mitochondrial ΔΨm, certain mtDNA mitochondrial types, such as ATP6, whose mutations can induce mitochondrial hyperpolarization, can evade mitophagy mechanism [79]. Dysfunctional mitochondria caused by mtDNA mutations can evade mitophagy through intercellular mitochondrial transfer, which accounts for the increase in pathological mtDNA copies in NDDs [80].
2.5. mtDNA Release-Activated Inflammation
Dache et al. detected free mitochondria in the blood of healthy people, as well as other mitochondrial components represented by mtDNA (including circular double-stranded or linear single-stranded mtDNA or nucleoids) [81]. Multiple lines of evidence have demonstrated that mtDNA can be displaced to the intra- or extracellular compartments through the release of extracellular vesicles when confronted with stressors [82, 83]. In addition, mtDNA can be recognized by pattern recognition receptors because it has a hypomethylated CpG motif similar to that of bacterial DNA, which will trigger inflammation.
There are several potential routes for mtDNA displacement [84]. The first is associated with mPTP opening, either partial opening of the mPTP due to oxidative stress or permanent opening due to irreversible damage, both of which lead to mtDNA extrusion and the release of other small mitochondrial molecules [85, 86]. This is because, when mitochondrial damage overwhelms mitophagy, apoptosis occurs with activation of the Bax/Bak pathway. Under prolonged stress, the mitochondrial outer membrane forms a pore that reaches the inner membrane, and mtDNA nucleoids are transferred into the cytosol through such structures [87]. Another pathway for mtDNA unloading involves extracellular vesicles of mitochondria. In cases of mild mitochondrial damage, damaged mitochondrial components accumulate adjacent to the mitochondrial membrane and, with the involvement of PINK1 and Parkin, form extracellular vesicles. These extracellular vesicles will later fuse with multivesicular bodies and transport mtDNA and other cellular components to the extracellular space for signaling [88]. FUNDC1 is an outer mitochondrial membrane protein containing an LIR structural domain that interacts with LC3, a key regulator of mitophagy. In mouse models, specific knockdown of the FUNDC1 gene promotes the release of mtDNA into the cytoplasm, enabling the release of the pro-inflammatory cytokine IL-1B, with mitophagy repressed and dysfunctional mitochondria unable to be cleared and accumulated [89]. Therefore, it has been suggested to be a complement to mitophagy.
The inflammatory process, a hallmark of aging, leads to the progression of NDDs. Free mtDNA or TFAM-bound nucleoids will act as damage-associated molecular patterns (DAMPs) to activate inflammation by interacting with one of the following pathways: Toll-like receptors (TLRs), the NLRP3 inflammasome system, or cGAS-STING1-IRF3–mediated signaling [90-92]. mtDNA release can be sensed by the subsequent activation of caspase-1 by TLR9 via NF-κB signaling, which promotes the secretion of IL-1β and IL-18 by macrophages [93]. NLRP3 can sense the hypomethylated CpG motifs in mtDNA. Once oxidized mtDNA is released, it will be preferentially bound as NLRP3 ligand to participate in the activation of caspase-1 and promote the activation of IL-1 and IL-18 [94]. Interestingly, NLRP3 also promotes mPTP opening and thus contributes to the release of mtDNA [95, 96]. Another mtDNA-driven inflammation pathway, cGAS-STING, proceeds as follows. When cGAS binds to mtDNA, its conformation changes, causing the recruitment of STING protein. The subsequent phosphorylation of IRF-3 by TRAF family member-associated NF-κB activator (TANK)-binding kinase 1 triggers the expression of IFN-I and IFN-III and causes inflammation [97, 98]. In addition, ROS directly activates inflammation via NF-κB activation, which regulates the transcription of various pro-inflammatory factors [99].
Notably, in the OPA1 and Drp1 overexpression models, giant mtDNA nucleoids and fragmented mitochondria are found outside the mitochondria, which can trigger an inflammatory response, suggesting that mitochondrial dynamics are also associated with mtDNA-derived inflammation [100]. Naturally, in addition to mtDNA, other substances released from mitochondria upon neuronal death, such as cardiolipin, cytochrome c, and TFAM, may also stimulate inflammation by activating microglia and astrocytes [101].
3. mtDNA ALTERATIONS IN NEURODEGENERATIVE DISEASES
3.1. Alzheimer’s Disease
AD, the most common NDD, is characterized by increasing amounts of neuritic plaques with extracellular amyloid-β (Aβ) accumulation and neurofibrillary tangles with tau hyperphosphorylation in neurons that result in neuron death and a reduced volume of specific regions of the brain, including the hippocampus and frontal cortex [102, 103]. In Table 1 we summarize the mtDNA alterations observed in AD patients reported in the literature. In the hippocampus of AD patients’ brains, TFAM protein levels are reduced by 50%, which is accompanied by lower levels of NRF1, NRF2, and PGC-1α, suggesting impaired mitochondrial biosynthesis [104]. Similarly, TFAM protein levels in the hippocampus of APPswe/PS1dE9 mice are reduced by 20% to 87.5% compared with controls [25]. All of these observations demonstrate that aberrant mtDNA maintenance and biogenesis occur in the pathogenic process of AD.
Table 1.
mtDNA alterations in AD.
| Alteration Type | Observed Change | Analysis Method | Patient Sample | References |
|---|---|---|---|---|
| mtDNA deletion | mtDNA Δ4977 | dilution-PCR | cortex, putamen, and cerebellum | [107] |
| mtDNA deletion | - | real-time PCR and long-extension PCR | putamen, frontal cortex, and substantia nigra | [222] |
| mtDNA deletion | ↑8-OHdG, ↑mtDNA fragmentation, ↓mtDNA | semiquantitative PCR analysis | temporal lobe tissue | [223] |
| mtDNA deletion | ↑8-OHdG and ↓mtDNAΔ4977 deletion | Kinetics PCR | frontal cortex | [110] |
| mtDNA deletion | ↓ mtDNA/ nDNA ratio | real-time PCR | hippocampus and cerebellum | [104] |
| mtDNA deletion | ↓ mtDNA copy number | Exome sequencing | brain tissue | [105] |
| mtDNA deletion | ↓ mtDNA copy number | real-time PCR | blood leukocytes | [224] |
| mtDNA mutation | Frequencies are not increased | RMC assay | brain tissue | [118] |
| mtDNA mutation | ↓mtDNA, ↑mutation frequencies (T→C/A→G and G→A/C→T transitions) |
ultra-sensitive next-generation sequencing |
hippocampus, middle temporal gyrus, and the visual and cerebellar cortex | [108] |
| mtDNA point mutation | T→C conversion | ultra-sensitive next-generation sequencing | hippocampus | [108] |
| mtDNA point mutation | m.7476C→T and m.15812 G→A | PCR sequencing | frontal and parietal associative cortex | [106] |
| mtDNA point mutation | tRNA (Asn) gene variant at nucleotide pair 5075 | PCR sequencing | brain tissue | [225] |
| mtDNA point mutation | tRNA (Asn) gene variant at nucleotide pair 3646 | D-loop sequencing | brain tissue | [226] |
| mtDNA methylation | D-loop↑ and MT-ND1 ↓ | PCR sequencing | entorhinal cortex | [227] |
| mtDNA methylation | D-loop methylation levels: MCI > control group > AD early stages > AD advanced stages | MS-HRM | peripheral blood | [228] |
Abbreviations: AD, Alzheimer’s disease; PCR, polymerase chain reaction; 8-OHdG, 8-oxo-7,8-dihydro-29-deoxyguanosine; RMC assay, a highly sensitive method that discriminates wild-type from mutated DNA based on the cleavage at a specific restriction site (TaqI) composed of all four canonical nucleotide (50 TCGA30); D-loop, displacement-loop; MT-ND1, a gene encoding for NADH-ubiquinone oxidoreductase chain 1, which is a core subunit of complex I; MCI, mild cognitive impairment; MS-HRM, methylation-sensitive high-resolution melting.
The main alterations in mtDNA in AD patients are a decreased mtDNA copy number and increased mtDNA heterogeneity in neurons, as observed in the postmortem brains of AD patients and further confirmed by high-depth whole-exome mtDNA sequencing [105, 106]. Earlier quantitative PCR studies found that a common deletion of mtDNA Δ4977 occurs in AD and accumulates up to 15-fold with age in the frontal cortex [107]. Using the ultra-sensitive next-generation sequencing technique, the frequency of mtDNA point mutations is significantly elevated in the hippocampus of early AD patients compared with controls. mtDNA mutations also lead to a reduction in mitochondrial protein levels and inhibit mitochondrial respiration. For example, an association has been confirmed between the T→C conversion of the gene encoding cytochrome c oxidase subunit III and reduced citrate synthase activity [108]. This mutation is present in up to 15% of AD patients and this process leads to decreased histone acetylation, which supports its importance in the pathogenesis of AD. The clinical study by Chagnon et al. found that mtDNA modifications, such as m.7476C>T and m.15812 G>A, could be significant in the development of early-onset AD [106].
Another critical factor that can moderate mitochondrial pathology in AD is mtDNA methylation. For example, in APP/PS1 mice, an animal model of familial AD, the D-loop region is demethylated and the mtDNA copy number is reduced [108]. The same phenomenon has been observed in patients with late-onset AD. However, increased methylation of the mtDNA D-loop region was observed in brain samples from Braak stage I/II AD patients [109]. It is worth noting that, while mtDNA mutations are important in the development of AD, mtDNA mutations are not specific to AD pathogenesis, because they are also found with aging. Moreover, the incidence of mtDNA mutations is sometimes even lower in AD patients compared with healthy age-matched controls [110]. This may indicate that mtDNA mutations play an early role in the course of AD but that, as the disease progresses, mutated mtDNA is gradually lost with neuronal death.
Oxidative stress occurs in the early stages of AD. As a sensor for oxidative stress caused by elevated ROS production, mtDNA alteration contributes to the positive feedback loop of free radical-induced oxidative damage [111]. This is mainly associated with the pathological process of AD, where the interaction of Aβ peptide aggregation and proteins related to mitochondrial import mechanisms is detrimental to electron transport chain activity, resulting in increased ROS production [112, 113]. Firstly, degraded mtDNA-related proteins such as TFAM and a higher degree of mtDNA oxidative damage have been found in the postmortem AD brain and brain samples of AD transgenic mice [114]. Oxidized bases in nuclear and mtDNA were quantified in AD brains and age-matched controls, and oxidative damage to mtDNA was three times greater than in controls and 10 times greater than that to nDNA [115]. Besides, a higher percentage of cytochrome oxidase-deficient neurons was revealed in postmortem AD brains compared with controls, indicating higher levels of mtDNA mutations. Yan et al. [116] reported that, in AD cell lines, there was reduced cytochrome oxidase activity, increased ROS production, reduced mitochondrial membrane potential, elevated apoptotic pathways, and increased Aβ42 production. However, it is interesting to note that alterations in mtDNA mainly involve transitions, which primarily result from Pol-γ rather than an elevation of G→T/C→A mutations caused by oxidative DNA damage [117]. The impaired BER activity observed in AD patients suggests that mtDNA replication errors are an important cause of mtDNA mutations [117, 118]. Furthermore, mtDNA from AD cytoplasmic hybrid lines display a number of typical AD features such as a calcium homeostasis change, accumulation of Aβ, aggregation of hyperphosphorylated tau, and misfolding of α-synuclein and TDP-43 (transactive response DNA-binding protein of 43 kDa).
The development of AD is also associated with defective mitophagy, with reduced serum levels of Parkin and autophagy-related 5 protein in samples from AD patients compared with controls [113]. In addition, the mRNA and protein levels of the mitophagy proteins PINK1, TERT, and LC3 are reduced in mouse hippocampal cells expressing mAPP and Aβ [113]. Impaired mitophagy is also reflected in altered expression levels of the mitophagy receptor disrupted-in-schizophrenia 1 (DISC1), a protein that protects neurons from Aβ accumulation-induced toxicity by binding to microtubule-associated proteins. Variations in the expression of this receptor have been found in AD patients, AD transgenic mouse models, and Aβ-treated cultured cells [119]. Increased cytoplasmic accumulation of Aβ contributes to reduced levels of Parkin and PINK1, thereby decreasing the numbers of available autophagosomes for efficiently clearing dysfunctional mitochondria [120, 121]. The production of Aβ plaques and phosphorylated tau protein may in fact establish a vicious circle between defective mitophagy and mitochondrial dysfunction, ultimately contributing to neuronal damage. Additionally, Evandro et al. restored neuronal mitophagy by using urea A and actinomycin in a C. elegans model, as well as in the APP/PS1 AD mouse model, which ameliorated cognitive decline, in conjunction with reduced levels of tau phosphorylation and alleviated inflammation induced by microglia [122].
3.2. Parkinson’s Disease
PD is a prevalent age-related NDD with a pathology characterized by the loss of nigrostriatal dopaminergic neurons and the presence of Lewy bodies in neurons [109]. Higher ROS production in PD is associated with suppression of mitochondrial complex I activity due to α-synuclein, a major component of Lewy bodies, and the overexpression of and mutations in Parkin, PINK1, and DJ-1 [123, 124]. mtDNA alterations play a key role in the progression of PD [125, 126] (Table 2), and studies have used rotenone to create experimental animal models that mimic the features of PD through extensive damage to mtDNA [127].
Table 2.
mtDNA alterations in PD.
| Alteration Type | Observed Change | Analysis Method | Patient Sample | References |
|---|---|---|---|---|
| mtDNA deletion | ↓transcription/replication-associated mtDNA molecules and ↓mtDNA copy number | real-time PCR | dopaminergic neurons in the substantia nigra | [229] |
| mtDNA deletion | ↑mtDNA deletion and ↑mtDNA copy number (MT-ND1 and MT-ND4) | quantitative PCR | cholinergic neurons from the substantia nigra | [230] |
| mtDNA deletion | ↓mtDNA copy number and ↑deletion (MT-ND1 and MT-ND4) | long-range PCR and real-time PCR | neurons from the substantia nigra | [135] |
| mtDNA deletion | ↓mtDNA, ↑MT-ND1, and ↑MT-ND4 single and multiple deletions | triplex real-time PCR | single neurons from the substantia nigra | [139] |
| mtDNA deletion | ↓mtDNA copy number | real-time PCR | Venous blood | [133] |
| mtDNA deletion | ↓mtDNA (breakpoints consistently occur in regions of sequence homology) | ultradeep sequencing | dopaminergic neurons in the substantia nigra | [231] |
| mtDNA deletion | ↑mtDNAΔ4977 deletion | PCR sequencing | Striatum | [128] |
| mtDNA deletion | ↑mtDNAΔ4977 deletion | quantitative PCR | brain tissue | [130] |
| mtDNA deletion | ↑8-OHdG and ↑oxidized coenzymeQ-10 | - | CSF | [140] |
| mtDNA deletion | defective mtDNA replication/repair | - | dopaminergic neurons in the substantia nigra | [125] |
| mtDNA point mutation |
4216T>C in MT-ND1 | PCR sequencing | substantia nigra | [232] |
| mtDNA point mutation |
5460G>A in MT-ND2 | PCR sequencing | brain tissue | [226] |
| mtDNA point mutation |
4336T>C in SNP | PCR sequencing | blood | [233] |
| mtDNA point mutation |
4336A>G in MT-TQ | PCR sequencing | brain tissue | [234] |
| mtDNA point mutation |
heteroplasmic variants in genes of complex III (CYTB) and complex IV (COXI and COXII) | PCR sequencing | substantia nigra and frontal cortex | [235] |
| mtDNA point mutation |
↑mtDNA mutation level (G→T or C→A transversions) | high-fidelity PCR protocol followed by TA cloning of the PCR | neurons in the substantia nigra (early-stage PD) | [141] |
| mtDNA methylation | ↓5-methylcytosine in the D-loop region | 454 GS FLX Titanium pyrosequencer | substantia nigra | [227] |
| mtDNA methylation | methylation levels of D-Loop Region and mtDNA copy number show no significantly difference compared with control group | MS-HRM and pyrosequencing techniques; quantitative PCR | Peripheral Blood | [236] |
Abbreviations: PD, Parkinson’s disease; PCR, polymerase chain reaction; MT-ND1, a gene encoding for NADH-ubiquinone oxidoreductase chain 1, which is a core subunit of complex I; CSF, cerebrospinal fluid; EOPD, early-onset PD; LOPD, late onset PD; SNP, a polymorphism in the tRNA gln gene; D-loop, displacement-loop;8-OHdG, 8-oxo-7,8-dihydro-29-deoxyguanosine; MS-HRM, methylation-sensitive high-resolution melting.
PD progression is also associated with the maintenance of mtDNA, with deletions of mtDNA observed in both PD patients and age-matched controls, although higher amounts were identified in PD patients [128]. Among PD patients, the most common mtDNA deletion is the mtDNA 4977-bp deletion, involving the gene encoding the complex I subunit [129]. Furthermore, mtDNA deletion molecules are more severe in the caudate nucleus, putamen, and substantia nigra, which are three regions with high dopamine metabolism, than in the cerebral cortex [130, 131]. Similarly, other studies have found that this accumulation of multiple mtDNA deletions in the substantia nigra is more pronounced with age in PD patients [132, 133]. This is related to nuclear genes that encode proteins associated with mitochondrial maintenance, such as POLG, the gene encoding Pol-γ. Some studies have found that patients with parkinsonism and showing Lewy body pathology have recessive mutations in this gene [134]. Similarly, DAT (dopamine transporter) imaging of patients with mitochondrial disease confirmed that nigrostriatal degeneration occurs only in patients with defective mtDNA maintenance and Pol mutations [135]. Collectively, these studies indicate that PD may be a common manifestation of mtDNA maintenance disorders. Other genes related to mtDNA maintenance, such as c10orf2 and MPV171, have also been examined in other studies of parkinsonism [136, 137]. Another mtDNA alteration associated with PD is related to epigenetics. mtDNA methylation deletion at both CpG and non-CpG sites has been detected in the nigrostriatal D-loop region of PD patients [138]. Considering that methylation may regulate mtDNA replication, this might also be correlated with the deletion of mtDNA. Chen et al. showed that the expression of TFAM could be significantly reduced in PD patients, which leads to D-loop destabilization and impaired mtDNA replication [139]. Cytosine at site 498 and thymine at site 497, which are located between two adjacent TFAM-binding sites, protect against PD. Pyrimidine loss due to TFAM deficiency increases the mtDNA haplogroup K1c in the risk of PD [109].
As explained previously, oxidative stress can cause oxidation of hydroxyl radicals and guanine residues in DNA to produce 8-oxo-2′-deoxyguanosine, which generates mutations by pairing with adenosine. The levels of 8-oxo-2′-deoxyguanosine are higher in the cerebrospinal fluid, serum, and urine of PD patients than in those of controls [140]. In addition, 8-oxo-2′-deoxyguanosine-induced mtDNA mutations (G→T or C→A translocations) are detected at higher levels in the nigrostriatal neurons of patients in the early stages of PD than in those of controls [141]. However, it is interesting to note that mtDNA mutation levels were significantly lower in patients with advanced PD. In addition to the effects of mtDNA mutations caused by ROS, the occurrence of PD is also associated with errors arising from the mtDNA replication and repair machinery [38], which are mainly attributed to mutations in Pol-γ, the enzyme with the nucleotide selectivity and proofreading ability to replicate mtDNA with high fidelity. Several studies have conducted mtDNA sequence analysis of DNA extracted from the striatum or peripheral blood of PD patients, but no specific PD-linked mutations have been identified [142, 143]. It has also not been confirmed that the m.4336A>G/MT-tRNAGln mutation, despite being thought to be associated with PD [142]. Mutations in the leucine-rich repeat kinase 2 (LRRK2) gene are relevant to PD induce degenerative mutations in SH-SY5Y cells and are often accompanied by mtDNA damage [144]. The degeneration of dopaminergic neurons is associated with mtDNA mutations and dysfunctional mitochondrial dynamics [145]. The substantia nigra of PD patients contains very high levels of mtDNA deletion [146]. In addition, Alicia et al. [147] have created a mouse model of PD that reproduces most of the key features of PD by using the mitochondria-targeted restriction enzyme mito-PstI to disrupt mtDNA in dopaminergic neurons.
Given that ROS help to trigger neuroinflammation through dysregulation of cytokine expression [148], we hypothesized that some mtDNA disruptions could lead to increased production of ROS, thereby stimulating the further development of PD. In particular, the production of defective mitophagy and DAMPs is considered to be a major factor in the development of inflammation. Mutations in FBXW7, which participates in mitochondrial autophagy through an interaction with Parkin, are associated with early-onset PD, further confirming the important role of mitochondrial autophagy in the pathogenesis of PD [149]. In aged PD patients, mitochondrial DAMPs have been detected at higher levels in circulating extracellular vesicles, which are characterized by a specific inflammatory feature. CD9, NDUFS3, CRP, and FGF21 are identified as integrating mitochondrial and inflammatory markers in PD, while the clearance of mitochondrial DAMPs (including mtDNA) may provide benefits for PD treatment [150]. In addition, the study by Matheoud et al. revealed that decreased PINK1/Parkin expression in PD is associated with disrupted mitophagy and neuroinflammation via mitochondrial-derived vesicle-mediated antigen delivery [151]. In Parkin−/− mutant mice, mtDNA mutations accumulate with age. Pink and Parkin knockout mice have higher circulating levels of the pro-inflammatory factors IL-6 and INF-β compared with wild-type, which could be attenuated by deletion of STING or administration of INF-α/β receptor-blocking antibodies. This suggests that the prerequisite for the inflammation caused by mtDNA release may be mitochondrial dysfunction in PD [74], which causes inflammation that can be completely rescued by simultaneous loss of STING, along with less nigrostriatal dopaminergic neuronal loss and ameliorated motor deficits [69].
3.3. Huntington’s Disease
HD is an autosomal dominant neurodegenerative disorder that develops due to amplification of the CAG trinucleotide repeat in the Huntingtin gene, which results in the aggregation of mutated Huntington's protein (polyglutamine stretch) in neurons and ultimately leads to chorea (involuntary movements) and cognitive symptoms [152]. SIRT1 and PCG-1, which support mitochondrial biogenesis, are expressed at lower levels in HD patients than in controls, suggesting reduced mitochondrial activity [153]. Alterations in mtDNA in HD are also associated with mtDNA maintenance. For example, the disruption of the homeostasis of the upstream molecule ATAD3A in HD impairs mtDNA maintenance by interfering with TFAM–mtDNA binding. Meanwhile, dimerization of K135 residues in ATAD3A is required for Drp1-mediated mitochondrial fission, which manifests as mitochondrial fragmentation and mtDNA lesions in the mouse striatal cell lines HdhQ7 and Q111 and in HD patient-derived cells, ultimately resulting in bioenergetic defects and cell death [154]. In the striatum of the R6/2 HD fragment mouse model, selective mtDNA depletion is observed with aberrant oligomerization of OPA1 while the fission/fusion balance is not distorted [155].
The proportion of mtDNA heterogeneity in lymphocytes is significantly increased in HD patients compared with healthy individuals. Notably, increased heterogeneity is also associated with progression through the stages of HD and disease severity, including decreases in motor function, cognitive function, and functional capacity [156]. Specifically, with sequencing by targeted amplification of multiplex probes, the location of heterogeneity in HD samples spanned all 13 protein-coding genes in mtDNA. The most common one was identified at m.3244 (n = 21), which is located in the tRNALeu gene, whereas m.3243A>G is the most prevalent pathogenic mtDNA mutation that is a risk factor for aging [156, 157]. Signs of mitochondrial dysfunction are evident in the early stages of HD, accompanied by deleterious effects such as oxidative stress and DNA damage. As a marker of endogenous oxidative stress, 8-oxoguanine has been associated with disease progression in the R6/2 HD model [158]. Cells in an in vitro striatal model of HD exhibit higher levels of the mitochondrial production of ROS and mtDNA damage involving increased lesion frequency and mtDNA deletion, which is also observed in HD skin fibroblasts [159].
One study found that the relative mtDNA/nDNA copy number of leukocytes was significantly higher in patients with symptomatic HD than in controls. Interestingly, mtDNA levels in leukocytes were higher in women with HD than in men with HD, which probably at least somewhat explains the faster rate of HD progression in women [160]. However, the relative mtDNA/nDNA copy number was lower in fibroblasts from HD patients, whereas even the contradictory result of a lower mtDNA/nDNA copy number was observed in leukocytes in another study [161]. Therefore, further investigation is required to determine whether the change in mtDNA is a compensatory mechanism in response to the inefficient mitochondrial respiratory function or whether it is due to lower metabolic activity of mitochondria.
Mitophagy is also impaired in HD, and the mHtt mutant leads to the formation of spherical mitochondria, which also inhibit mitophagy and contribute to impaired mitochondrial clearance [162]. In HD mouse models, both R6/2 mice [163] and STHdhQ111 mice [164] show alterations in mtDNA, with the former exhibiting reduced mtDNA levels in the striatum and the latter showing more mtDNA lesions. These damages can be attributed to the interference of elongated polyQ mutant HTT with mitophagy, such as with the p62-induced binding process of LC3 [76], which ultimately leads to compromised mtDNA quality, manifested by increased heteroplasmy rates. Meanwhile, increased mitochondrial fission is facilitated through mHTT binding to Drp1, thereby promoting the expansion of pathogenic mtDNA heterogeneity in cells [165]. Although the impact of mitochondrial disorders, represented by mtDNA alterations, on the development of the disease is now unclear, because the accumulation of mtDNA defects occurs in the early stages of HD, it is considered a potential biomarker of the disease, while significant neuroinflammation has been observed in the brains of HD patients by PET imaging, implying that microglial activation is associated with the pathological progression [166]. Further examples of mtDNA alteration in HD are summarized in Table 3.
Table 3.
mtDNA alterations in HD.
| Alteration Type | Observed Change | Analysis Method | Patient Sample | References |
|---|---|---|---|---|
| mtDNA deletion | ↓mtDNA/ nDNA relative copy number | real-time quantitative PCR | peripheral leukocytes | [161] |
| mtDNA deletion | mtDNA/ nDNA relative copy number (↑ in leukocytes and ↓ in dermal fibroblasts) | real-time quantitative PCR | leukocytes and dermal fibroblasts | [160] |
| mtDNA deletion | ↓mtDNA (MT-ND2) | real-time PCR | brain and plasma | [158] |
| mtDNA deletion | ↑mtDNA lesion frequency and ↓mtDNA abundance | quantitative PCR | brain tissue | [159] |
| mtDNA mutation | ↑mtDNA heteroplasmies | sensitive mtDNA-targeted sequencing | lymphoblasts and longitudinal blood samples | [156] |
Abbreviations: C9ALS/FTD, amyotrophic lateral sclerosis and frontotemporal dementia with C9ORF72 mutation; PCR, polymerase chain reaction; nDNA, nuclear DNA; MT-ND2, a gene encoding for NADH-ubiquinone oxidoreductase chain 2, which is a core subunit of complex I; mtDNA heteroplasmies (coexistence of mutated and wild-type mtDNA)
3.4. Amyotrophic Lateral Sclerosis
ALS is a progressive NDD with continuous loss of motor neurons in the central nervous system that leads to muscle atrophy, respiratory failure, and, ultimately, death. ALS is characterized by the accumulation of mutant proteins resulting from specific gene mutations (e.g., SOD1, FUS, TDP43, and Sqstm1/p62), with loss of mitochondrial function preceding the onset of the disease [167]. Numerous works of literature have shown that mtDNA alteration is significant in ALS patients (Table 4).
Table 4.
mtDNA alterations in ALS.
| Alteration Type | Observed Change | Analysis Method | Patient Sample | References |
|---|---|---|---|---|
| mtDNA deletion | ↓mtDNA copy number | digital droplet PCR | Prefrontal Cortex in C9ALS/FTD | [237] |
| mtDNA deletion | ↑mtDNA copy number but not statistically significant. | quantitative PCR | peripheral white blood cells |
[172] |
| mtDNA methylation | ↓D-loop region | Bisulfite pyrosequencing | ||
| mtDNA deletion | multiple mitochondrial DNA deletions | long range PCR and Southern blot analysis | muscle | [180] |
| relative mtDNA copy number without finding any depletion | real-time quantitative PCR | |||
| mtDNA methylation | ↓D-loop region (↑mtDNA copy number) |
MS-HRM and quantitative PCR | blood samples | [173] |
Abbreviations: C9ALS/FTD, amyotrophic lateral sclerosis and frontotemporal dementia with C9ORF72 mutation; PCR, polymerase chain reaction; D-loop, displacement-loop; MS-HRM, methylation-sensitive high-resolution melting.
Ladd et al. found reduced levels of mtDNA in postmortem human ALS cervical spinal cord and demonstrated that recombinant human TFAM improved mtDNA gene expression in human neural stem cells [168]. The GGGGCC repeat expansion in C9ORF72 is a common genetic factor contributing to ALS, and DNA damage is greater in motor neurons differentiated from induced pluripotent stem cells of C9ORF72 patients compared with controls in an age-dependent manner, with pharmacological or genetic reduction of oxidative stress ameliorating the DNA damage [169]. Mutations in mtDNA in the mitochondrial genome are responsible for the pathology of ALS. Mutations in superoxide dismutase 1 (SOD1) lead to overproduction of ROS in microglia and abnormal microglial proliferation [170]. An elevation of 8-hydroxy-2-deoxygua- nosine in the spinal motor neurons of transgenic ALS mice results in oxidative damage to mtDNA. In addition, the cortical levels of mtDNA are 30-fold higher in the brains of ALS patients than in control brains [171].
In addition to mtDNA oxidation, mtDNA copy number is increased in ALS patients, particularly in those with SOD1 or C9orf72 mutations. Interestingly, methylation levels in the D-loop region of mtDNA are significantly lower in blood DNA samples from carriers of the SOD1 mutation compared with those from patients with C9orf72 amplification and are negatively correlated with mtDNA copy number [172, 173]. Given the antioxidant role of the SOD1 enzyme, this reduction in methylation, which increases mtDNA replication, probably serves to counteract the burden of oxidative damage.
In patients with sporadic or familial ALS, mislocalization and aggregation of TDP-43 and accumulation of p62 are critical markers of defective mitophagy [174, 175]. The former interacts with the mitophagy receptor prohibitin 2 and voltage-gated anion channel 1 [176], whereas the accumulation of p62 indicates that it is not efficiently autophagically degraded after its binding to misfolded protein aggregates [174, 177]. Patients with late-onset ALS show mutations in the CHCHD10 gene, a nuclear-encoded mitochondrial small protein gene [178]. In the fibroblasts of these patients, in addition to morphological changes in the mitochondria such as the presence of an abnormal cristae structure [179], mtDNA deletions are also observed, which could be secondary to impaired mtDNA repair or restricted clearance of damaged mtDNA [180].
Although the entry of TDP-43 into the mitochondria of ALS patients is evolutionarily conserved [181], once it invades the mitochondria, it releases mtDNA through the mPTP opening and VDAC1 oligomerization independent of the Bax/Bak-dependent process [182]. mtDNA accumulation in the cytoplasm of induced pluripotent stem cell-derived motor neurons and in TDP-43 mutant mice leads to cGAS/STING activation and the subsequent upregulation of NF-κB and IFN. The fact that inflammation can be prevented by pharmacological inhibition or gene deletion of cGAS and its downstream signaling molecule STING further demonstrates the relationship between ALS and the cGAS/STING pathway [182].
4. POTENTIAL THERAPEUTICS RELEVANT TO MTDNA
Because mtDNA alterations occur in the early stages and pathological progresses of NDDs, the targeting of mtDNA has been considered a potential therapeutic strategy for NDDs.
The most compelling approach is to directly target the mutated mtDNA itself for targeted genome editing. Mitochondrially targeted TALENs (transcription activator-like effector nucleases) and mitochondrially targeted zinc-finger nucleases, which have a relatively low risk of interacting with nDNA, are two potential vehicles. The former can not only correct induced mtDNA mutations in mouse models, but also eliminate the m.3243A>G mutation in induced pluripotent stem cells of mitochondrial disease patients [183, 184]; the latter corrected the pathological mtDNA mutation in the m.5024C>T tRNAAla mouse model [185]. Another gene editing tool that is CRISPR-free and can precisely edit mtDNA in vitro is also of interest. This tool is based on a bacterial cytidine deaminase toxin A that catalyzes the C→G to T→A conversion in human mtDNA with high target specificity [186]. However, most mtDNA mutations in NDDs are not specific [184, 187]. Thus, we should carefully consider the exploitation of the balance between healthy and mutant mtDNA to avoid a reduction in normal gene function with mtDNA genome editing.
As described above, oxidative stress can trigger mitochondrial dysfunction, such as mitochondrial gene mutations, and mtDNA alterations contribute to the positive feedback loop of free radical-induced oxidative damage. Therefore, antioxidants are one of the therapeutic approaches for NDDs. Compared with non-targeted cellular antioxidants, antioxidants targeting mitochondria provide better protection against oxidative damage by effectively isolating reactive oxygen intermediates. These antioxidants, including 10-decyltriphenyl-phosphonium, MitoQ, and latrepirdine, have been extensively evaluated in multiple studies in AD and PD models [188, 189]. Recently, MitoQ, a mitochondrially targeted antioxidant, has been tested in a small clinical trial for AD, albeit for its role in endothelial NO production and whether it improves cerebrovascular blood flow in AD patients [188-190]. On the other hand, recent clinical trials for creatine and coenzyme Q10 have not demonstrated any disease-modifying benefit in patients with PD [191, 192], indicating that more targeted antioxidant approaches may be required for PD-related neurodegeneration. Other strategies have been devised to improve mitochondrial function in PD. Enhancing mitophagy presents an effective approach due to growing evidence for its general impairment in PD. In particular, Nip3-like protein X-mediated mitophagy was recently found to restore mitochondrial function and prevent neurodegeneration in the setting of Parkin or PINK1 deficiency, highlighting this pathway as a potential target for therapeutic intervention [193]. Boosting mitochondrial biogenesis is another strategy to replenish neurons with healthy mitochondria. A recent study showed that BG-12 exerts beneficial effects by increasing mitochondrial biogenesis in humans via the transcription factor nuclear factor (erythroid-derived 2)-like 2 (NRF2) [194].
The induction of mitophagy is also a common therapeutic strategy. Several agents targeting mitophagy are in development, such as clomifene, which suppresses mtDNA defects, and hexestrol, which reduces excessive mitochondrial fragmentation while rescuing mtDNA depletion [195]. Furthermore, metformin and resveratrol have been extensively demonstrated to increase mitophagy as modulators of PINK1/ Parkin [196]. There are other PINK1/parkin-independent mitophagy inducers, such as PMI, which is associated with p62 and enhances mitophagy without causing loss of mitochondrial membrane potential [197]. In addition, 3-phos- phate dehydrogenase is a molecular sensor that detects and labels damaged mitochondria because GAPDH is inactivated by mitochondrial ROS and protects cells from damage.
Because uncontrolled production of DAMPs leads to neuroinflammation, a viable therapeutic option would be the reduction of neuroinflammation by targeting DAMPs and their downstream signaling molecules. Resveratrol alleviates microglial activation by inhibiting TLR4/NF-κB/STAT, as well as cytokine release upon Aβ stimulation, and is currently being used in clinical trials of AD [198]. In addition, a selective NLRP3 inhibitor has also been used to eliminate IL-1β to attenuate the immune response to Aβ and the propagation of neuroinflammation in an AD model [199]. As for PD, a promising immunotherapeutic approach is the use of anti-inflammatory drugs and immunosuppressants to inhibit the release of pro-inflammatory cytokines and accelerate the clearance of α-synuclein and thereby ameliorate the inflammation triggered by the accumulation of α-synuclein in PD [200]. Considering the presence of DNA released via the mPTP to activate cGAS/STING in AD, PD, and ALS, a series of cGAS and STING inhibitors have recently been developed with the potential to target this pathway and ultimately ameliorate the symptoms of neuronal degeneration in patients [201, 202].
CONCLUSION AND FUTURE PERSPECTIVE
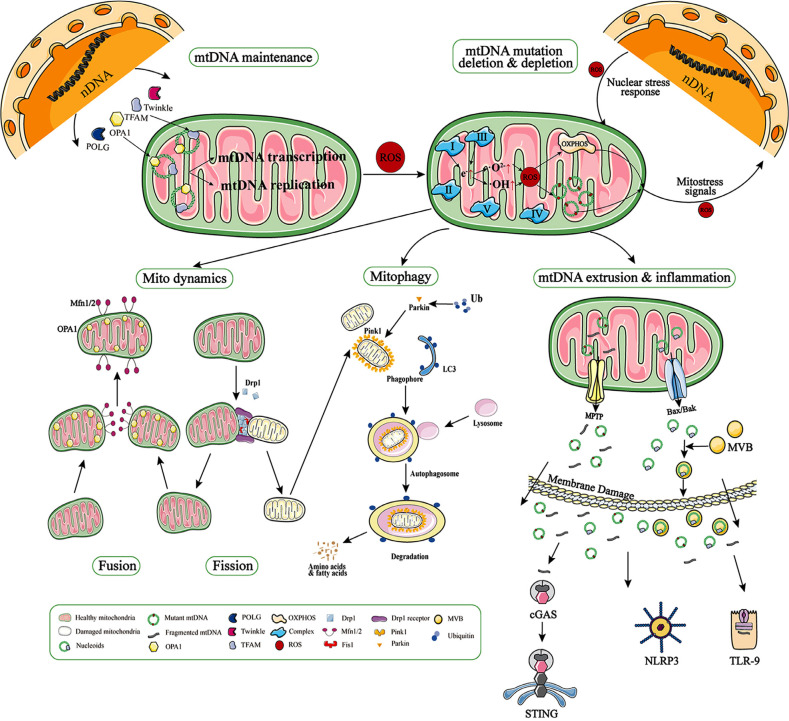
mtDNA is of widespread interest because of its specific structure and important function, and its mutations are associated with a number of diseases [203]. Because the proteins that it encode are important components of oxidative phosphorylation, mtDNA mutation leads to inefficient ATP supply and increased production of ROS in the mitochondria of affected tissues, which in turn results in further effects on mtDNA. Our review shows that, although oxidative stress, represented by ROS, is associated with mtDNA mutation, the mtDNA maintenance-related proteins TFAM and Pol-γ can cause mutations in mtDNA, and even this effect is more prominent than oxidative stress. When mtDNA is damaged, the dysfunctional mitochondria can be eliminated by mitophagy, which is also related to fusion/fission [204]. When mitophagy is impaired or insufficient to clear damaged mitochondria, mtDNA may escape to the cytoplasm and body fluids. In turn, as important DAMPs, mtDNA triggers cGAS/STING-mediated inflammation, and this is now considered to be possibly complementary to impaired mitophagy [15] (Fig. 1).
Fig. (1).
mtDNA maintenance and alteration. The proteins related to mtDNA maintenance (e.g., TFAM, POLG, Twinkle, OPA1) are encoded by nDNA and contribute to mtDNA maintenance by participating in nucleoid formation, mtDNA replication, and mitochondrial dynamics. Impaired mitochondrial maintenance results in point mutations, deletion, or even depletion of the mitochondrial genome. Another cause of mtDNA mutations is oxidative stress triggered by ROS generated by electron leakage from complexes I and III. mtDNA mutations and deletions stimulate the production of new ROS, creating a vicious cycle. Damaged mtDNA accumulates in the mitochondria and causes clinical symptoms when the mutational load exceeds a threshold. In the meantime, damaged mitochondria can be rescued by fusion with other mitochondria or by fission and mitophagy. When the damage exceeds the mitochondrial quality control, the mPTP will remain open and the mtDNA breaks and is extruded into the cytoplasm, while nucleoids are extruded with the activation of Bax/Bak. These nucleoids and mtDNA fragments can further be extruded into the extracellular space via MVBs (multivesicular bodies) recognized by DNA-binding receptors within cGAS and TLR-9, ultimately triggering inflammation.
The mitochondrial cascade hypothesis suggests that the accumulation of mtDNA modifications and mutations in brain somatic cells with age determines the disease phenotype [126, 205]. Although mtDNA haplogroups are usually specific, they may impact the pathogenesis of more than one disease. There is a correlation between the impact of the mtDNA haplogroup B5 on NDD development and mitochondrial defects [206], the severity of which depends on the frequency of mtDNA mutations [207]. As a result of mtDNA haplotypes, certain human haplotypes are more likely than other human populations to develop specific types of NDDs during their lifetime [67].
With increasing human life expectancy, the contribution of mtDNA to the pathogenesis of multifactorial degenerative diseases will further increase. Further understanding of mtDNA alterations is necessary to comprehend the mechanism of NDD progression and to effectively conduct early diagnosis and treatment. In recent years, it has been observed that circulating cell-free mtDNA concentrations can be correlated with NDDs. For example, circulating cell-free mtDNA has been observed at low levels in the cerebrospinal fluid of asymptomatic patients with AD, suggesting that mtDNA alterations may be a precursor to AD injury [208]. In addition, its levels are also a valid physiological indicator for differentiating idiopathic PD and PD related to PRKN/PINK1 mutations [126]. In addition to cell-free mtDNA, the D-loop methylation level of mtDNA in blood samples is of interest as a potential biomarker, with about 25% reduced mtDNA methylation observed in AD patients [209]. Furthermore, Gonzalez-Hunt, C P et al. revealed that the level of mtDNA damage may serve as a powerful and sensitive readout of altered LRRK2 kinase activity in LRRK2 PD [210]. However, these results have not been replicated in some studies, so further research with a large sample is needed to determine whether mtDNA can be a stable biomarker for investigating the progression of NDDs [211].
In addition, because DAMPs and dysfunctional mtDNA exacerbate inflammation by triggering innate immune responses, which contribute to the onset and pathological processing of NDDs, therapeutic strategies have been aimed at reducing cytosolic mtDNA accumulation and inhibiting molecules of the downstream signaling pathway of DAMP [212, 213]. On the other hand, healthy mtDNA is able to activate the mesenchymal stem cell pathway, which transfers healthy extracellular mitochondria via vesicles to damaged cells, thereby revitalizing the metabolic activity of nearby cells [214-217]. In our recent study, transplanted mitochondria led to the reuptake of extracellular mtDNA, resulting in improved mtDNA homeostasis [218]. Therefore, it is necessary to generate effective mtDNA-targeting approaches for NDDs.
Moreover, some computerized methods based on chaos game representation and lacunarity analysis have been proposed for differentiating among AD, PD, and aging by characterizing mutated mtDNA [219, 220]. However, it is important to note that, although next-generation sequencing methods improve accuracy, the data may still be contaminated with nuclear mitochondrial DNA sequences (NUMTs), which can lead to a false estimate of mtDNA heterogeneity [221]. In future studies, it will be highly valuable to further elucidate the relationship between mitosis and mtDNA alterations in the somatic cells of NDD patients and to look for specific mtDNA mutations. It would be very exciting if these prove to be beneficial in treatment. On this basis, it would be exciting to work with tools for mtDNA gene manipulation, such as TALEN and Zinc-finger nucleases, or to develop better tools for targeting mitophagy to achieve the desired therapeutic effect in patients with NDDs.
ACKNOWLEDGEMENTS
Declared none.
LIST OF ABBREVIATIONS
- PD
Parkinson’s Disease
- PCR
Polymerase Chain Reaction
- CSF
Cerebrospinal Fluid
- EOPD
Early-onset PD
- LOPD
Late Onset PD
- SNP
A polymorphism in the tRNA gln gene
- D-loop
Displacement-loop
- 8-OHdG
8-oxo-7,8-dihydro-29-deoxyguanosine
- MS-HRM
Methylation-sensitive high-resolution Melting
AUTHORS’ CONTRIBUTIONS
D.S., M.H. and X.Z. made substantial contributions to the conception and design of the study. D.S., M.H., B.W., X.Y., Z.W. and X.Z. searched the data and participated in drafting the article. D.S., M.H. and B.W. created the figure and tables. All authors revised and approved the final manuscript.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
This work was supported by the National Natural Science Foundation of China (Grant No. 81700977 to X. Y.; Grant No. 81500858 to X. Z.) and the Japanese KAKENHI Grant No. 22K09927 (Grants-in-Aid for Scientific Research to Z.W.).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Modesti L., Danese A., Angela Maria Vitto V., Ramaccini D., Aguiari G., Gafà R., Lanza G., Giorgi C., Pinton P. Mitochondrial Ca2+ signaling in health, disease and therapy. Cells. 2021;10(6):1317. doi: 10.3390/cells10061317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh L.N., Kao S.H., Wallace D.C. Unlocking the complexity of mitochondrial DNA: A key to understanding neurodegenerative disease caused by injury. Cells. 2021;10(12):3460. doi: 10.3390/cells10123460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rone M.B., Fan J., Papadopoulos V. Cholesterol transport in steroid biosynthesis: Role of protein–protein interactions and implications in disease states. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2009;1791(7):646–658. doi: 10.1016/j.bbalip.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang B., Huang M., Shang D., Yan X., Zhao B., Zhang X. Mitochondrial behavior in axon degeneration and regeneration. Front. Aging Neurosci. 2021;13:650038. doi: 10.3389/fnagi.2021.650038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Figueira T.R., Barros M.H., Camargo A.A., Castilho R.F., Ferreira J.C.B., Kowaltowski A.J., Sluse F.E., Souza-Pinto N.C., Vercesi A.E. Mitochondria as a source of reactive oxygen and nitrogen species: From molecular mechanisms to human health. Antioxid. Redox Signal. 2013;18(16):2029–2074. doi: 10.1089/ars.2012.4729. [DOI] [PubMed] [Google Scholar]
- 6.Garrido N., Griparic L., Jokitalo E., Wartiovaara J., van der Bliek A.M., Spelbrink J.N. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell. 2003;14(4):1583–1596. doi: 10.1091/mbc.e02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holt I.J., Harding A.E., Morgan-Hughes J.A. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331(6158):717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 8.Hällberg B.M., Larsson N.G. Making proteins in the powerhouse. Cell Metab. 2014;20(2):226–240. doi: 10.1016/j.cmet.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Alston C.L., Rocha M.C., Lax N.Z., Turnbull D.M., Taylor R.W. The genetics and pathology of mitochondrial disease. J. Pathol. 2017;241(2):236–250. doi: 10.1002/path.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filograna R., Mennuni M., Alsina D., Larsson N.G. Mitochondrial DNA copy number in human disease: The more the better? FEBS Lett. 2021;595(8):976–1002. doi: 10.1002/1873-3468.14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heemels M.T. Neurodegenerative diseases. Nature. 2016;539(7628):179. doi: 10.1038/539179a. [DOI] [PubMed] [Google Scholar]
- 12.Klein H.U., Trumpff C., Yang H.S., Lee A.J., Picard M., Bennett D.A., De Jager P.L. Characterization of mitochondrial DNA quantity and quality in the human aged and Alzheimer’s disease brain. Mol. Neurodegener. 2021;16(1):75. doi: 10.1186/s13024-021-00495-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nissanka N., Moraes C.T. Mitochondrial DNA damage and reactive oxygen species in neurodegenerative disease. FEBS Lett. 2018;592(5):728–742. doi: 10.1002/1873-3468.12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buneeva O., Fedchenko V., Kopylov A., Medvedev A. Mitochondrial dysfunction in Parkinson’s disease: Focus on mitochondrial DNA. Biomedicines. 2020;8(12):591. doi: 10.3390/biomedicines8120591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Gaetano A., Solodka K., Zanini G., Selleri V., Mattioli A.V., Nasi M., Pinti M. Molecular mechanisms of mtDNA-mediated inflammation. Cells. 2021;10(11):2898. doi: 10.3390/cells10112898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishra P., Chan D.C. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 2014;15(10):634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gustafsson C.M., Falkenberg M., Larsson N.G. Maintenance and expression of mammalian mitochondrial DNA. Annu. Rev. Biochem. 2016;85(1):133–160. doi: 10.1146/annurev-biochem-060815-014402. [DOI] [PubMed] [Google Scholar]
- 18.El-Hattab A.W., Craigen W.J., Scaglia F. Mitochondrial DNA maintenance defects. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863(6):1539–1555. doi: 10.1016/j.bbadis.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 19.Zhao M., Wang Y., Li L., Liu S., Wang C., Yuan Y., Yang G., Chen Y., Cheng J., Lu Y., Liu J. Mitochondrial ROS promote mitochondrial dysfunction and inflammation in ischemic acute kidney injury by disrupting TFAM-mediated mtDNA maintenance. Theranostics. 2021;11(4):1845–1863. doi: 10.7150/thno.50905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao M., Liu S., Wang C., Wang Y., Wan M., Liu F., Gong M., Yuan Y., Chen Y., Cheng J., Lu Y., Liu J. Mesenchymal stem cell-derived extracellular vesicles attenuate mitochondrial damage and inflammation by stabilizing mitochondrial DNA. ACS Nano. 2021;15(1):1519–1538. doi: 10.1021/acsnano.0c08947. [DOI] [PubMed] [Google Scholar]
- 21.Campbell C.T., Kolesar J.E., Kaufman B.A. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim. Biophys. Acta. Gene Regul. Mech. 2012;1819(9-10):921–929. doi: 10.1016/j.bbagrm.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Picca A., Lezza A.M.S. Regulation of mitochondrial biogenesis through TFAM–mitochondrial DNA interactions. Mitochondrion. 2015;25:67–75. doi: 10.1016/j.mito.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Rubio-Cosials A., Solà M. U-turn DNA bending by human mitochondrial transcription factor A. Curr. Opin. Struct. Biol. 2013;23(1):116–124. doi: 10.1016/j.sbi.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 24.West A.P., Khoury-Hanold W., Staron M., Tal M.C., Pineda C.M., Lang S.M., Bestwick M., Duguay B.A., Raimundo N., MacDuff D.A., Kaech S.M., Smiley J.R., Means R.E., Iwasaki A., Shadel G.S. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520(7548):553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang I., Chu C.T., Kaufman B.A. The mitochondrial transcription factor TFAM in neurodegeneration: Emerging evidence and mechanisms. FEBS Lett. 2018;592(5):793–811. doi: 10.1002/1873-3468.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song L., Shan Y., Lloyd K.C.K., Cortopassi G.A. Mutant Twinkle increases dopaminergic neurodegeneration, mtDNA deletions and modulates Parkin expression. Hum. Mol. Genet. 2012;21(23):5147–5158. doi: 10.1093/hmg/dds365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoff K.E., DeBalsi K.L., Sanchez-Quintero M.J., Longley M.J., Hirano M., Naini A.B., Copeland W.C. Characterization of the human homozygous R182W POLG2 mutation in mitochondrial DNA depletion syndrome. PLoS One. 2018;13(8):e0203198. doi: 10.1371/journal.pone.0203198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Del Dotto V., Fogazza M., Lenaers G., Rugolo M., Carelli V., Zanna C. OPA1: How much do we know to approach therapy? Pharmacol. Res. 2018;131:199–210. doi: 10.1016/j.phrs.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 29.Kukat C., Davies K.M., Wurm C.A., Spåhr H., Bonekamp N.A., Kühl I., Joos F., Polosa P.L., Park C.B., Posse V., Falkenberg M., Jakobs S., Kühlbrandt W., Larsson N.G. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc. Natl. Acad. Sci. USA. 2015;112(36):11288–11293. doi: 10.1073/pnas.1512131112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elachouri G., Vidoni S., Zanna C., Pattyn A., Boukhaddaoui H., Gaget K., Yu-Wai-Man P., Gasparre G., Sarzi E., Delettre C., Olichon A., Loiseau D., Reynier P., Chinnery P.F., Rotig A., Carelli V., Hamel C.P., Rugolo M., Lenaers G. OPA1 links human mitochondrial genome maintenance to mtDNA replication and distribution. Genome Res. 2011;21(1):12–20. doi: 10.1101/gr.108696.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hudson G., Amati-Bonneau P., Blakely E.L., Stewart J.D., He L., Schaefer A.M., Griffiths P.G., Ahlqvist K., Suomalainen A., Reynier P., McFarland R., Turnbull D.M., Chinnery P.F., Taylor R.W. Mutation of OPA1 causes dominant optic atrophy with external ophthalmoplegia, ataxia, deafness and multiple mitochondrial DNA deletions: A novel disorder of mtDNA maintenance. Brain. 2008;131(2):329–337. doi: 10.1093/brain/awm272. [DOI] [PubMed] [Google Scholar]
- 32.Peter B., Falkenberg M. TWINKLE and other human mitochondrial DNA helicases: Structure, function and disease. Genes (Basel) 2020;11(4):408. doi: 10.3390/genes11040408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He J., Cooper H.M., Reyes A., Di Re M., Sembongi H., Litwin T.R., Gao J., Neuman K.C., Fearnley I.M., Spinazzola A., Walker J.E., Holt I.J. Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic Acids Res. 2012;40(13):6109–6121. doi: 10.1093/nar/gks266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baudier J. ATAD3 proteins: brokers of a mitochondria-endoplasmic reticulum connection in mammalian cells. Biol. Rev. Camb. Philos. Soc. 2018;93(2):827–844. doi: 10.1111/brv.12373. [DOI] [PubMed] [Google Scholar]
- 35.Harel T., Yoon W.H., Garone C., Gu S., Coban-Akdemir Z., Eldomery M.K., Posey J.E., Jhangiani S.N., Rosenfeld J.A., Cho M.T., Fox S., Withers M., Brooks S.M., Chiang T., Duraine L., Erdin S., Yuan B., Shao Y., Moussallem E., Lamperti C., Donati M.A., Smith J.D., McLaughlin H.M., Eng C.M., Walkiewicz M., Xia F., Pippucci T., Magini P., Seri M., Zeviani M., Hirano M., Hunter J.V., Srour M., Zanigni S., Lewis R.A., Muzny D.M., Lotze T.E., Boerwinkle E., Gibbs R.A., Hickey S.E., Graham B.H., Yang Y., Buhas D., Martin D.M., Potocki L., Graziano C., Bellen H.J., Lupski J.R. Recurrent de novo and biallelic variation of ATAD3A, encoding a mitochondrial membrane protein, results in distinct neurological syndromes. Am. J. Hum. Genet. 2016;99(4):831–845. doi: 10.1016/j.ajhg.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peres de Oliveira A., Basei F.L., Slepicka P.F., de Castro Ferezin C., Melo-Hanchuk T.D., de Souza E.E., Lima T.I., dos Santos V.T., Mendes D., Silveira L.R., Menck C.F.M., Kobarg J. NEK10 interactome and depletion reveal new roles in mitochondria. Proteome Sci. 2020;18(1):4. doi: 10.1186/s12953-020-00160-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reynolds J.C., Bwiza C.P., Lee C. Mitonuclear genomics and aging. Hum. Genet. 2020;139(3):381–399. doi: 10.1007/s00439-020-02119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeBalsi K.L., Hoff K.E., Copeland W.C. Role of the mitochondrial DNA replication machinery in mitochondrial DNA mutagenesis, aging and age-related diseases. Ageing Res. Rev. 2017;33:89–104. doi: 10.1016/j.arr.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zekonyte U., Bacman S.R., Moraes C.T. DNA‐editing enzymes as potential treatments for heteroplasmic mtDNA diseases. J. Intern. Med. 2020;287(6):685–697. doi: 10.1111/joim.13055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson L.V. Oxidative stress, mitochondria and mtDNA-mutator mice. Exp. Gerontol. 2006;41(12):1220–1222. doi: 10.1016/j.exger.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su B.C., Pan C.Y., Chen J.Y. Antimicrobial peptide TP4 induces ROS-mediated necrosis by triggering mitochondrial dysfunction in wild-type and mutant p53 glioblastoma cells. Cancers (Basel) 2019;11(2):171. doi: 10.3390/cancers11020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu M., Bogoyevitch M.A., Jans D.A. Subversion of host cell mitochondria by RSV to favor virus production is dependent on inhibition of mitochondrial complex I and ROS generation. Cells. 2019;8(11):1417. doi: 10.3390/cells8111417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guliaeva N.A., Kuznetsova E.A., Gaziev A.I. Proteins associated with mitochondrial DNA protect it against the action of X-rays and hydrogen peroxide. Biofizika. 2006;51(4):692–697. [PubMed] [Google Scholar]
- 44.Napolitano G., Fasciolo G., Venditti P. Mitochondrial management of reactive oxygen species. Antioxidants (Basel) 2021;10(11):1824. doi: 10.3390/antiox10111824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yasui M., Kanemaru Y., Kamoshita N., Suzuki T., Arakawa T., Honma M. Tracing the fates of site-specifically introduced DNA adducts in the human genome. DNA Repair (Amst.) 2014;15:11–20. doi: 10.1016/j.dnarep.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 46.Sanyal T., Bhattacharjee P., Bhattacharjee S., Bhattacharjee P. Hypomethylation of mitochondrial D-loop and ND6 with increased mitochondrial DNA copy number in the arsenic-exposed population. Toxicology. 2018;408:54–61. doi: 10.1016/j.tox.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 47.Yu D., Du Z., Pian L., Li T., Wen X., Li W., Kim S.J., Xiao J., Cohen P., Cui J., Hoffman A.R., Hu J.F. Mitochondrial DNA hypomethylation is a biomarker associated with induced senescence in human fetal heart mesenchymal stem cells. Stem Cells Int. 2017;2017:1–12. doi: 10.1155/2017/1764549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szczepanowska K., Trifunovic A. Origins of mtDNA mutations in ageing. Essays Biochem. 2017;61(3):325–337. doi: 10.1042/EBC20160090. [DOI] [PubMed] [Google Scholar]
- 49.Qian Y., Kachroo A.H., Yellman C.M., Marcotte E.M., Johnson K.A. Yeast cells expressing the human mitochondrial DNA polymerase reveal correlations between polymerase fidelity and human disease progression. J. Biol. Chem. 2014;289(9):5970–5985. doi: 10.1074/jbc.M113.526418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kauppila J.H.K., Stewart J.B. Mitochondrial DNA: Radically free of free-radical driven mutations. Biochim. Biophys. Acta Bioenerg. 2015;1847(11):1354–1361. doi: 10.1016/j.bbabio.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., Bohlooly-Y M., Gidlöf S., Oldfors A., Wibom R., Törnell J., Jacobs H.T., Larsson N.G. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 52.Anderson A.P., Luo X., Russell W., Yin Y.W. Oxidative damage diminishes mitochondrial DNA polymerase replication fidelity. Nucleic Acids Res. 2020;48(2):817–829. doi: 10.1093/nar/gkz1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan D.C. Mitochondrial Dynamics and Its Involvement in Disease. Annu. Rev. Pathol. 2020;15(1):235–259. doi: 10.1146/annurev-pathmechdis-012419-032711. [DOI] [PubMed] [Google Scholar]
- 54.Carelli V., Maresca A., Caporali L., Trifunov S., Zanna C., Rugolo M. Mitochondria: Biogenesis and mitophagy balance in segregation and clonal expansion of mitochondrial DNA mutations. Int. J. Biochem. Cell Biol. 2015;63:21–24. doi: 10.1016/j.biocel.2015.01.023. [DOI] [PubMed] [Google Scholar]
- 55.Aryaman J., Bowles C., Jones N.S., Johnston I.G. Mitochondrial network state scales mtDNA genetic dynamics. Genetics. 2019;212(4):1429–1443. doi: 10.1534/genetics.119.302423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee J.E., Westrate L.M., Wu H., Page C., Voeltz G.K. Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 2016;540(7631):139–143. doi: 10.1038/nature20555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 2003;278(10):7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- 58.Pellino G., Faggioli R., Galuppi A., Leon A., Fusco C., Tugnoli V., Suppiej A. Mitofusin 2: The missing link between mtDNA maintenance defects and neurotransmitter disorders. Mitochondrion. 2021;61:159–164. doi: 10.1016/j.mito.2021.09.011. [DOI] [PubMed] [Google Scholar]
- 59.Twig G., Elorza A., Molina A.J.A., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., Alroy J., Wu M., Py B.F., Yuan J., Deeney J.T., Corkey B.E., Shirihai O.S. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27(2):433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gomes L.C., Benedetto G.D., Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011;13(5):589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suen D.F., Narendra D.P., Tanaka A., Manfredi G., Youle R.J. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc. Natl. Acad. Sci. USA. 2010;107(26):11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hashimoto M., Bacman S.R., Peralta S., Falk M.J., Chomyn A., Chan D.C., Williams S.L., Moraes C.T. MitoTALEN: A general approach to reduce mutant mtDNA loads and restore oxidative phosphorylation function in mitochondrial diseases. Mol. Ther. 2015;23(10):1592–1599. doi: 10.1038/mt.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghosh A., Bhattacharjee S., Chowdhuri S.P., Mallick A., Rehman I., Basu S., Das B.B. SCAN1-TDP1 trapping on mitochondrial DNA promotes mitochondrial dysfunction and mitophagy. Sci. Adv. 2019;5(11):eaax9778. doi: 10.1126/sciadv.aax9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao A., Jiang J., Xie F., Chen L. Bnip3 in mitophagy: Novel insights and potential therapeutic target for diseases of secondary mitochondrial dysfunction. Clin. Chim. Acta. 2020;506:72–83. doi: 10.1016/j.cca.2020.02.024. [DOI] [PubMed] [Google Scholar]
- 65.Dombi E., Mortiboys H., Poulton J. Modulating mitophagy in mitochondrial disease. Curr. Med. Chem. 2019;25(40):5597–5612. doi: 10.2174/0929867324666170616101741. [DOI] [PubMed] [Google Scholar]
- 66.Twig G., Shirihai O.S. The interplay between mitochondrial dynamics and mitophagy. Antioxid. Redox Signal. 2011;14(10):1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herst P.M., Rowe M.R., Carson G.M., Berridge M.V. Functional mitochondria in health and disease. Front. Endocrinol. (Lausanne) 2017;8:296. doi: 10.3389/fendo.2017.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Corti O., Blomgren K., Poletti A., Beart P.M. Autophagy in neurodegeneration: New insights underpinning therapy for neurological diseases. J. Neurochem. 2020;154(4):354–371. doi: 10.1111/jnc.15002. [DOI] [PubMed] [Google Scholar]
- 69.Sliter D.A., Martinez J., Hao L., Chen X., Sun N., Fischer T.D., Burman J.L., Li Y., Zhang Z., Narendra D.P., Cai H., Borsche M., Klein C., Youle R.J. Parkin and PINK1 mitigate STING-induced inflammation. Nature. 2018;561(7722):258–262. doi: 10.1038/s41586-018-0448-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Matheoud D., Cannon T., Voisin A., Penttinen A.M., Ramet L., Fahmy A.M., Ducrot C., Laplante A., Bourque M.J., Zhu L., Cayrol R., Le Campion A., McBride H.M., Gruenheid S., Trudeau L.E., Desjardins M. Intestinal infection triggers Parkinson’s disease-like symptoms in Pink1 −/− mice. Nature. 2019;571(7766):565–569. doi: 10.1038/s41586-019-1405-y. [DOI] [PubMed] [Google Scholar]
- 71.Malena A., Pantic B., Borgia D., Sgarbi G., Solaini G., Holt I.J., Spinazzola A., Perissinotto E., Sandri M., Baracca A., Vergani L. Mitochondrial quality control: Cell-type-dependent responses to pathological mutant mitochondrial DNA. Autophagy. 2016;12(11):2098–2112. doi: 10.1080/15548627.2016.1226734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kandul N.P., Zhang T., Hay B.A., Guo M. Selective removal of deletion-bearing mitochondrial DNA in heteroplasmic Drosophila. Nat. Commun. 2016;7(1):13100. doi: 10.1038/ncomms13100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma H., Xu H., O’Farrell P.H. Transmission of mitochondrial mutations and action of purifying selection in Drosophila melanogaster. Nat. Genet. 2014;46(4):393–397. doi: 10.1038/ng.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pickrell A.M., Huang C.H., Kennedy S.R., Ordureau A., Sideris D.P., Hoekstra J.G., Harper J.W., Youle R.J. Endogenous parkin preserves dopaminergic substantia nigral neurons following mitochondrial DNA mutagenic stress. Neuron. 2015;87(2):371–381. doi: 10.1016/j.neuron.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moretton A., Morel F., Macao B., Lachaume P., Ishak L., Lefebvre M., Garreau-Balandier I., Vernet P., Falkenberg M., Farge G. Selective mitochondrial DNA degradation following double-strand breaks. PLoS One. 2017;12(4):e0176795. doi: 10.1371/journal.pone.0176795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun N., Yun J., Liu J., Malide D., Liu C., Rovira I.I., Holmström K.M., Fergusson M.M., Yoo Y.H., Combs C.A., Finkel T. Measuring in vivo mitophagy. Mol. Cell. 2015;60(4):685–696. doi: 10.1016/j.molcel.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marin J.J.G., Hernandez A., Revuelta I.E., Gonzalez-Sanchez E., Gonzalez-Buitrago J.M., Perez M.J. Mitochondrial genome depletion in human liver cells abolishes bile acid-induced apoptosis: Role of the Akt/mTOR survival pathway and Bcl-2 family proteins. Free Radic. Biol. Med. 2013;61:218–228. doi: 10.1016/j.freeradbiomed.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 78.Yang X., Zhang R., Nakahira K., Gu Z. Mitochondrial DNA mutation, diseases, and nutrient-regulated mitophagy. Annu. Rev. Nutr. 2019;39(1):201–226. doi: 10.1146/annurev-nutr-082018-124643. [DOI] [PubMed] [Google Scholar]
- 79.Knorre D.A. Intracellular quality control of mitochondrial DNA: evidence and limitations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020;375(1790):20190176. doi: 10.1098/rstb.2019.0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dickson-Murray E., Nedara K., Modjtahedi N., Tokatlidis K. The Mia40/CHCHD4 oxidative folding system: Redox regulation and signaling in the mitochondrial intermembrane space. Antioxidants (Basel) 2021;10(4):592. doi: 10.3390/antiox10040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Al Amir Dache Z., Otandault A., Tanos R., Pastor B., Meddeb R., Sanchez C., Arena G., Lasorsa L., Bennett A., Grange T., El Messaoudi S., Mazard T., Prevostel C., Thierry A.R. Blood contains circulating cell‐free respiratory competent mitochondria. FASEB J. 2020;34(3):3616–3630. doi: 10.1096/fj.201901917RR. [DOI] [PubMed] [Google Scholar]
- 82.Pérez-Treviño P., Velásquez M., García N. Mechanisms of mitochondrial DNA escape and its relationship with different metabolic diseases. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866(6):165761. doi: 10.1016/j.bbadis.2020.165761. [DOI] [PubMed] [Google Scholar]
- 83.Picca A., Calvani R., Coelho-Junior H.J., Marzetti E. Cell death and inflammation: the role of mitochondria in health and disease. Cells. 2021;10(3):537. doi: 10.3390/cells10030537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao Y., Liu B., Xu L., Yu S., Fu J., Wang J., Yan X., Su J. ROS-induced mtDNA release: The emerging messenger for communication between neurons and innate immune cells during neurodegenerative disorder progression. Antioxidants. 2021;10(12):1917. doi: 10.3390/antiox10121917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bernardi P. Why F-ATP Synthase remains a strong candidate as the mitochondrial permeability transition pore. Front. Physiol. 2018;9:1543. doi: 10.3389/fphys.2018.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Urbani A., Giorgio V., Carrer A., Franchin C., Arrigoni G., Jiko C., Abe K., Maeda S., Shinzawa-Itoh K., Bogers J.F.M., McMillan D.G.G., Gerle C., Szabò I., Bernardi P. Purified F-ATP synthase forms a Ca2+-dependent high-conductance channel matching the mitochondrial permeability transition pore. Nat. Commun. 2019;10(1):4341. doi: 10.1038/s41467-019-12331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Riley J.S., Quarato G., Cloix C., Lopez J., O’Prey J., Pearson M., Chapman J., Sesaki H., Carlin L.M., Passos J.F., Wheeler A.P., Oberst A., Ryan K.M., Tait S.W.G. Mitochondrial inner membrane permeabilisation enables mt DNA release during apoptosis. EMBO J. 2018;37(17):e99238. doi: 10.15252/embj.201899238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sugiura A., McLelland G.L., Fon E.A., McBride H.M. A new pathway for mitochondrial quality control: mitochondrial‐derived vesicles. EMBO J. 2014;33(19):2142–2156. doi: 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li W., Li Y., Siraj S., Jin H., Fan Y., Yang X., Huang X., Wang X., Wang J., Liu L., Du L., Chen Q. FUN14 Domain‐containing 1–mediated mitophagy suppresses hepatocarcinogenesis by inhibition of inflammasome activation in mice. Hepatology. 2019;69(2):604–621. doi: 10.1002/hep.30191. [DOI] [PubMed] [Google Scholar]
- 90.Contis A., Mitrovic S., Lavie J., Douchet I., Lazaro E., Truchetet M.E., Goizet C., Contin-Bordes C., Schaeverbeke T., Blanco P., Rossignol R., Faustin B., Richez C., Duffau P. Neutrophil-derived mitochondrial DNA promotes receptor activator of nuclear factor κB and its ligand signalling in rheumatoid arthritis. Rheumatology (Oxford) 2017;56(7):1200–1205. doi: 10.1093/rheumatology/kex041. [DOI] [PubMed] [Google Scholar]
- 91.Yoo S.M., Park J., Kim S.H., Jung Y.K. Emerging perspectives on mitochondrial dysfunction and inflammation in Alzheimer’s disease. BMB Rep. 2020;53(1):35–46. doi: 10.5483/BMBRep.2020.53.1.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang X., Wang R., Hu D., Sun X., Fujioka H., Lundberg K., Chan E.R., Wang Q., Xu R., Flanagan M.E., Pieper A.A., Qi X. Oligodendroglial glycolytic stress triggers inflammasome activation and neuropathology in Alzheimer’s disease. Sci. Adv. 2020;6(49):eabb8680. doi: 10.1126/sciadv.abb8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mottis A., Herzig S., Auwerx J. Mitocellular communication: Shaping health and disease. Science. 2019;366(6467):827–832. doi: 10.1126/science.aax3768. [DOI] [PubMed] [Google Scholar]
- 94.Zhong Z., Liang S., Sanchez-Lopez E., He F., Shalapour S., Lin X., Wong J., Ding S., Seki E., Schnabl B., Hevener A.L., Greenberg H.B., Kisseleva T., Karin M. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature. 2018;560(7717):198–203. doi: 10.1038/s41586-018-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nakahira K., Haspel J.A., Rathinam V.A.K., Lee S.J., Dolinay T., Lam H.C., Englert J.A., Rabinovitch M., Cernadas M., Kim H.P., Fitzgerald K.A., Ryter S.W., Choi A.M.K. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011;12(3):222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vince J.E., De Nardo D., Gao W., Vince A.J., Hall C., McArthur K., Simpson D., Vijayaraj S., Lindqvist L.M., Bouillet P., Rizzacasa M.A., Man S.M., Silke J., Masters S.L., Lessene G., Huang D.C.S., Gray D.H.D., Kile B.T., Shao F., Lawlor K.E. The mitochondrial apoptotic effectors BAX/BAK activate caspase-3 and -7 to trigger NLRP3 inflammasome and caspase-8 driven IL-1β activation. Cell Rep. 2018;25(9):2339–2353.e4. doi: 10.1016/j.celrep.2018.10.103. [DOI] [PubMed] [Google Scholar]
- 97.White M.J., McArthur K., Metcalf D., Lane R.M., Cambier J.C., Herold M.J., van Delft M.F., Bedoui S., Lessene G., Ritchie M.E., Huang D.C.S., Kile B.T. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159(7):1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sun J., Zhou Y., Xu B., Li J., Zhang L., Li D., Zhang S., Wu J., Gao S., Ye D., Mei W. STING/NF-κB/IL-6-mediated inflammation in microglia contributes to spared nerve injury (SNI)-induced pain initiation. J. Neuroimmune Pharmacol. 2022;3-4:453–469. doi: 10.1007/s11481-021-10031-6. [DOI] [PubMed] [Google Scholar]
- 99.Li S., Li H., Zhang Y.L., Xin Q.L., Guan Z.Q., Chen X., Zhang X.A., Li X.K., Xiao G.F., Lozach P.Y., Cui J., Liu W., Zhang L.K., Peng K. SFTSV infection induces BAK/BAX-dependent mitochondrial dna release to trigger NLRP3 inflammasome activation. Cell Rep. 2020;30(13):4370–4385.e7. doi: 10.1016/j.celrep.2020.02.105. [DOI] [PubMed] [Google Scholar]
- 100.Rodríguez-Nuevo A., Díaz-Ramos A., Noguera E., Díaz-Sáez F., Duran X., Muñoz J.P., Romero M., Plana N., Sebastián D., Tezze C., Romanello V., Ribas F., Seco J., Planet E., Doctrow S.R., González J., Borràs M., Liesa M., Palacín M., Vendrell J., Villarroya F., Sandri M., Shirihai O., Zorzano A. Mitochondrial DNA and TLR9 drive muscle inflammation upon Opa1 deficiency. EMBO J. 2018;37(10):e96553. doi: 10.15252/embj.201796553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bajwa E., Pointer C.B., Klegeris A. The role of mitochondrial damage-associated molecular patterns in chronic neuroinflammation. Mediators Inflamm. 2019;2019:1–11. doi: 10.1155/2019/4050796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mangialasche F., Solomon A., Winblad B., Mecocci P., Kivipelto M. Alzheimer’s disease: clinical trials and drug development. Lancet Neurol. 2010;9(7):702–716. doi: 10.1016/S1474-4422(10)70119-8. [DOI] [PubMed] [Google Scholar]
- 103.Lashley T., Schott J.M., Weston P., Murray C.E., Wellington H., Keshavan A., Foti S.C., Foiani M., Toombs J., Rohrer J.D., Heslegrave A., Zetterberg H. Molecular biomarkers of Alzheimer’s disease: progress and prospects. Dis. Model. Mech. 2018;11(5):dmm031781. doi: 10.1242/dmm.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sheng B., Wang X., Su B., Lee H., Casadesus G., Perry G., Zhu X. Impaired mitochondrial biogenesis contributes to mitochondrial dysfunction in Alzheimer’s disease. J. Neurochem. 2012;120(3):419–429. doi: 10.1111/j.1471-4159.2011.07581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wei W., Keogh M.J., Wilson I., Coxhead J., Ryan S., Rollinson S., Griffin H., Kurzawa-Akanbi M., Santibanez-Koref M., Talbot K., Turner M.R., McKenzie C.A., Troakes C., Attems J., Smith C., Al Sarraj S., Morris C.M., Ansorge O., Pickering-Brown S., Ironside J.W., Chinnery P.F. Mitochondrial DNA point mutations and relative copy number in 1363 disease and control human brains. Acta Neuropathol. Commun. 2017;5(1):13. doi: 10.1186/s40478-016-0404-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chagnon P., Gee M., Filion M., Robitaille Y., Belouchi M., Gauvreau D. Phylogenetic analysis of the mitochondrial genome indicates significant differences between patients with Alzheimer disease and controls in a French-Canadian founder population. Am. J. Med. Genet. 1999;85(1):20–30. doi: 10.1002/(SICI)1096-8628(19990702)85:1<20:AID-AJMG6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 107.Corral-Debrinski M., Horton T., Lott M.T., Shoffner J.M., McKee A.C., Beal M.F., Graham B.H., Wallace D.C. Marked changes in mitochondrial DNA deletion levels in Alzheimer brains. Genomics. 1994;23(2):471–476. doi: 10.1006/geno.1994.1525. [DOI] [PubMed] [Google Scholar]
- 108.Xu Y., Xu L., Han M., Liu X., Li F., Zhou X., Wang Y., Bi J. Altered mitochondrial DNA methylation and mitochondrial DNA copy number in an APP/PS1 transgenic mouse model of Alzheimer disease. Biochem. Biophys. Res. Commun. 2019;520(1):41–46. doi: 10.1016/j.bbrc.2019.09.094. [DOI] [PubMed] [Google Scholar]
- 109.Antonyová V., Kejík Z., Brogyányi T., Kaplánek R., Pajková M., Talianová V., Hromádka R., Masařík M., Sýkora D., Mikšátková L., Martásek P., Jakubek M. Role of mtDNA disturbances in the pathogenesis of Alzheimer’s and Parkinson’s disease. DNA Repair (Amst.) 2020;91-92:102871. doi: 10.1016/j.dnarep.2020.102871. [DOI] [PubMed] [Google Scholar]
- 110.Lezza A.M.S., Mecocci P., Cormio A., Real M.F., Cherubini A., Cantatore P., Senin U., Gadaleta M.N. Mitochondrial DNA 4977 bp deletion and OH 8 dG levels correlate in the brain of aged subjects but not Alzheimer’s disease patients. FASEB J. 1999;13(9):1083–1088. doi: 10.1096/fasebj.13.9.1083. [DOI] [PubMed] [Google Scholar]
- 111.Nunomura A., Perry G., Aliev G., Hirai K., Takeda A., Balraj E.K., Jones P.K., Ghanbari H., Wataya T., Shimohama S., Chiba S., Atwood C.S., Petersen R.B., Smith M.A. Oxidative damage is the earliest event in Alzheimer disease. J. Neuropathol. Exp. Neurol. 2001;60(8):759–767. doi: 10.1093/jnen/60.8.759. [DOI] [PubMed] [Google Scholar]
- 112.Devi L., Prabhu B.M., Galati D.F., Avadhani N.G., Anandatheerthavarada H.K. Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer’s disease brain is associated with mitochondrial dysfunction. J. Neurosci. 2006;26(35):9057–9068. doi: 10.1523/JNEUROSCI.1469-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Reddy P.H., Yin X., Manczak M., Kumar S., Pradeepkiran J.A., Vijayan M., Reddy A.P. Mutant APP and amyloid beta-induced defective autophagy, mitophagy, mitochondrial structural and functional changes and synaptic damage in hippocampal neurons from Alzheimer’s disease. Hum. Mol. Genet. 2018;27(14):2502–2516. doi: 10.1093/hmg/ddy154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Reddy P.H., McWeeney S., Park B.S., Manczak M., Gutala R.V., Partovi D., Jung Y., Yau V., Searles R., Mori M., Quinn J. Gene expression profiles of transcripts in amyloid precursor protein transgenic mice: up-regulation of mitochondrial metabolism and apoptotic genes is an early cellular change in Alzheimer’s disease. Hum. Mol. Genet. 2004;13(12):1225–1240. doi: 10.1093/hmg/ddh140. [DOI] [PubMed] [Google Scholar]
- 115.Wang J., Xiong S., Xie C., Markesbery W.R., Lovell M.A. Increased oxidative damage in nuclear and mitochondrial DNA in Alzheimer’s disease. J. Neurochem. 2005;93(4):953–962. doi: 10.1111/j.1471-4159.2005.03053.x. [DOI] [PubMed] [Google Scholar]
- 116.Yan M.H., Wang X., Zhu X. Mitochondrial defects and oxidative stress in Alzheimer disease and Parkinson disease. Free Radic. Biol. Med. 2013;62:90–101. doi: 10.1016/j.freeradbiomed.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hoekstra J.G., Hipp M.J., Montine T.J., Kennedy S.R. Mitochondrial DNA mutations increase in early stage Alzheimer disease and are inconsistent with oxidative damage. Ann. Neurol. 2016;80(2):301–306. doi: 10.1002/ana.24709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Soltys D.T., Pereira C.P.M., Rowies F.T., Farfel J.M., Grinberg L.T., Suemoto C.K., Leite R.E.P., Rodriguez R.D., Ericson N.G., Bielas J.H., Souza-Pinto N.C. Lower mitochondrial DNA content but not increased mutagenesis associates with decreased base excision repair activity in brains of AD subjects. Neurobiol. Aging. 2019;73:161–170. doi: 10.1016/j.neurobiolaging.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 119.Wang Z.T., Lu M.H., Zhang Y., Ji W.L., Lei L., Wang W., Fang L.P., Wang L.W., Yu F., Wang J., Li Z.Y., Wang J.R., Wang T.H., Dou F., Wang Q.W., Wang X.L., Li S., Ma Q.H., Xu R.X. Disrupted-in-schizophrenia-1 protects synaptic plasticity in a transgenic mouse model of Alzheimer’s disease as a mitophagy receptor. Aging Cell. 2019;18(1):e12860. doi: 10.1111/acel.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Reddy P.H., Oliver D.M.A. Amyloid beta and phosphorylated tau-induced defective autophagy and mitophagy in Alzheimer’s disease. Cells. 2019;8(5):488. doi: 10.3390/cells8050488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Morton H., Kshirsagar S., Orlov E., Bunquin L.E., Sawant N., Boleng L., George M., Basu T., Ramasubramanian B., Pradeepkiran J.A., Kumar S., Vijayan M., Reddy A.P., Reddy P.H. Defective mitophagy and synaptic degeneration in Alzheimer’s disease: Focus on aging, mitochondria and synapse. Free Radic. Biol. Med. 2021;172:652–667. doi: 10.1016/j.freeradbiomed.2021.07.013. [DOI] [PubMed] [Google Scholar]
- 122.Fang E.F., Hou Y., Palikaras K., Adriaanse B.A., Kerr J.S., Yang B., Lautrup S., Hasan-Olive M.M., Caponio D., Dan X., Rocktäschel P., Croteau D.L., Akbari M., Greig N.H., Fladby T., Nilsen H., Cader M.Z., Mattson M.P., Tavernarakis N., Bohr V.A. Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat. Neurosci. 2019;22(3):401–412. doi: 10.1038/s41593-018-0332-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Reeve A.K., Ludtmann M.H., Angelova P.R., Simcox E.M., Horrocks M.H., Klenerman D., Gandhi S., Turnbull D.M., Abramov A.Y. Aggregated α-synuclein and complex I deficiency: exploration of their relationship in differentiated neurons. Cell Death Dis. 2015;6(7):e1820. doi: 10.1038/cddis.2015.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dodson M.W., Guo M. Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson’s disease. Curr. Opin. Neurobiol. 2007;17(3):331–337. doi: 10.1016/j.conb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 125.Tzoulis C., Schwarzlmüller T., Biermann M., Haugarvoll K., Bindoff L.A. Mitochondrial DNA homeostasis is essential for nigrostriatal integrity. Mitochondrion. 2016;28:33–37. doi: 10.1016/j.mito.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 126.Park J.S., Davis R.L., Sue C.M. Mitochondrial dysfunction in Parkinson’s disease: new mechanistic insights and therapeutic perspectives. Curr. Neurol. Neurosci. Rep. 2018;18(5):21. doi: 10.1007/s11910-018-0829-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li N., Ragheb K., Lawler G., Sturgis J., Rajwa B., Melendez J.A., Robinson J.P. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J. Biol. Chem. 2003;278(10):8516–8525. doi: 10.1074/jbc.M210432200. [DOI] [PubMed] [Google Scholar]
- 128.Ikebe S., Tanaka M., Ohno K., Sato W., Hattori K., Kondo T., Mizuno Y., Ozawa T. Increase of deleted mitochondrial DNA in the striatum in Parkinson’s disease and senescence. Biochem. Biophys. Res. Commun. 1990;170(3):1044–1048. doi: 10.1016/0006-291X(90)90497-B. [DOI] [PubMed] [Google Scholar]
- 129.Gezen-Ak D., Alaylıoğlu M., Genç G., Şengül B., Keskin E., Sordu P., Güleç Z.E.K., Apaydın H., Bayram-Gürel Ç., Ulutin T., Yılmazer S., Ertan S., Dursun E. Altered transcriptional profile of mitochondrial DNA-encoded OXPHOS subunits, mitochondria quality control genes, and intracellular ATP levels in blood samples of patients with Parkinson’s disease. J. Alzheimers Dis. 2020;74(1):287–307. doi: 10.3233/JAD-191164. [DOI] [PubMed] [Google Scholar]
- 130.Wei Soong N., Hinton D.R., Cortopassi G., Arnheim N. Mosaicism for a specific somatic mitochondrial DNA mutation in adult human brain. Nat. Genet. 1992;2(4):318–323. doi: 10.1038/ng1292-318. [DOI] [PubMed] [Google Scholar]
- 131.Kraytsberg Y., Kudryavtseva E., McKee A.C., Geula C., Kowall N.W., Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat. Genet. 2006;38(5):518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 132.Basu S., Xie X., Uhler J.P., Hedberg-Oldfors C., Milenkovic D., Baris O.R., Kimoloi S., Matic S., Stewart J.B., Larsson N.G., Wiesner R.J., Oldfors A., Gustafsson C.M., Falkenberg M., Larsson E. Accurate mapping of mitochondrial DNA deletions and duplications using deep sequencing. PLoS Genet. 2020;16(12):e1009242. doi: 10.1371/journal.pgen.1009242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen S-H., Kuo C-W., Lin T-K., Tsai M-H., Liou C-W. Dopamine therapy and the regulation of oxidative stress and mitochondrial DNA copy number in patients with Parkinson’s disease. Antioxidants (Basel) 2020;9(11):1159. doi: 10.3390/antiox9111159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hsieh P.C., Wang C.C., Tsai C.L., Yeh Y.M., Lee Y.S., Wu Y.R. POLG R964C and GBA L444P mutations in familial Parkinson’s disease: Case report and literature review. Brain Behav. 2019;9(5):e01281. doi: 10.1002/brb3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tzoulis C., Tran G.T., Schwarzlmüller T., Specht K., Haugarvoll K., Balafkan N., Lilleng P.K., Miletic H., Biermann M., Bindoff L.A. Severe nigrostriatal degeneration without clinical parkinsonism in patients with polymerase gamma mutations. Brain. 2013;136(8):2393–2404. doi: 10.1093/brain/awt103. [DOI] [PubMed] [Google Scholar]
- 136.Kiferle L., Orsucci D., Mancuso M., Lo Gerfo A., Petrozzi L., Siciliano G., Ceravolo R., Bonuccelli U. Twinkle mutation in an Italian family with external progressive ophthalmoplegia and parkinsonism: A case report and an update on the state of art. Neurosci. Lett. 2013;556:1–4. doi: 10.1016/j.neulet.2013.09.034. [DOI] [PubMed] [Google Scholar]
- 137.Garone C., Rubio J.C., Calvo S.E., Naini A., Tanji K., DiMauro S., Mootha V.K., Hirano M. MPV17 Mutations causing adult-onset multisystemic disorder with multiple mitochondrial DNA deletions. Arch. Neurol. 2012;69(12):1648–1651. doi: 10.1001/archneurol.2012.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fonseca Cabral G., Schaan A.P., Cavalcante G.C., Sena-dos-Santos C., de Souza T.P. Souza Port’s, N.M.; dos Santos Pinheiro, J.A.; Ribeiro-dos-Santos, Â.; Vidal, A.F. Nuclear and mitochondrial genome, epigenome and gut microbiome: Emerging molecular biomarkers for Parkinson’s disease. Int. J. Mol. Sci. 2021;22(18):9839. doi: 10.3390/ijms22189839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen C., Vincent A.E., Blain A.P., Smith A.L., Turnbull D.M., Reeve A.K. Investigation of mitochondrial biogenesis defects in single substantia nigra neurons using post-mortem human tissues. Neurobiol. Dis. 2020;134:104631. doi: 10.1016/j.nbd.2019.104631. [DOI] [PubMed] [Google Scholar]
- 140.Isobe C., Abe T., Terayama Y. Levels of reduced and oxidized coenzymeQ-10 and 8-hydroxy-2′-deoxyguanosine in the cerebrospinal fluid of patients with living Parkinson’s disease demonstrate that mitochondrial oxidative damage and/or oxidative DNA damage contributes to the neurodegenerative process. Neurosci. Lett. 2010;469(1):159–163. doi: 10.1016/j.neulet.2009.11.065. [DOI] [PubMed] [Google Scholar]
- 141.Lin M.T., Cantuti-Castelvetri I., Zheng K., Jackson K.E., Tan Y.B., Arzberger T., Lees A.J., Betensky R.A., Beal M.F., Simon D.K. Somatic mitochondrial DNA mutations in early parkinson and incidental lewy body disease. Ann. Neurol. 2012;71(6):850–854. doi: 10.1002/ana.23568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Simon D.K., Mayeux R., Marder K., Kowall N.W., Beal M.F., Johns D.R. Mitochondrial DNA mutations in complex I and tRNA genes in Parkinson’s disease. Neurology. 2000;54(3):703–709. doi: 10.1212/WNL.54.3.703. [DOI] [PubMed] [Google Scholar]
- 143.Giannoccaro M.P., La Morgia C., Rizzo G., Carelli V. M itochondrial DNA and primary mitochondrial dysfunction in P arkinson’s disease. Mov. Disord. 2017;32(3):346–363. doi: 10.1002/mds.26966. [DOI] [PubMed] [Google Scholar]
- 144.Sanders L.H., Laganière J., Cooper O., Mak S.K., Vu B.J., Huang Y.A., Paschon D.E., Vangipuram M., Sundararajan R., Urnov F.D., Langston J.W., Gregory P.D., Zhang H.S., Greenamyre J.T., Isacson O., Schüle B. LRRK2 mutations cause mitochondrial DNA damage in iPSC-derived neural cells from Parkinson’s disease patients: Reversal by gene correction. Neurobiol. Dis. 2014;62:381–386. doi: 10.1016/j.nbd.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu Z., Ye Q., Wang F., Guo Y., Cui R., Wang J., Wang D. Overexpression of thioredoxin reductase 1 can reduce DNA damage, mitochondrial autophagy and endoplasmic reticulum stress in Parkinson’s disease. Exp. Brain Res. 2021;239(2):475–490. doi: 10.1007/s00221-020-05979-5. [DOI] [PubMed] [Google Scholar]
- 146.Gambardella S., Limanaqi F., Ferese R., Biagioni F., Campopiano R., Centonze D., Fornai F. ccf-mtDNA as a potential link between the brain and immune system in neuro-immunological disorders. Front. Immunol. 2019;10:1064. doi: 10.3389/fimmu.2019.01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pickrell A.M., Pinto M., Hida A., Moraes C.T. Striatal dysfunctions associated with mitochondrial DNA damage in dopaminergic neurons in a mouse model of Parkinson’s disease. J. Neurosci. 2011;31(48):17649–17658. doi: 10.1523/JNEUROSCI.4871-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cheng A.N., Cheng L.C., Kuo C.L., Lo Y.K., Chou H.Y., Chen C.H., Wang Y.H., Chuang T.H., Cheng S.J., Lee A.Y.L. Mitochondrial Lon-induced mtDNA leakage contributes to PD-L1–mediated immunoescape via STING-IFN signaling and extracellular vesicles. J. Immunother. Cancer. 2020;8(2):e001372. doi: 10.1136/jitc-2020-001372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yang Y., Zhou X., Liu X., Song R., Gao Y., Wang S. Implications of FBXW7 in neurodevelopment and neurodegeneration: molecular mechanisms and therapeutic potential. Front. Cell. Neurosci. 2021;15:736008. doi: 10.3389/fncel.2021.736008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Picca A., Guerra F., Calvani R., Marini F., Biancolillo A., Landi G., Beli R., Landi F., Bernabei R., Bentivoglio A., Lo Monaco M., Bucci C., Marzetti E. Mitochondrial signatures in circulating extracellular vesicles of older adults with Parkinson’s disease: results from the exosomes in Parkinson’s disease (EXPAND) study. J. Clin. Med. 2020;9(2):504. doi: 10.3390/jcm9020504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Matheoud D., Sugiura A., Bellemare-Pelletier A., Laplante A., Rondeau C., Chemali M., Fazel A., Bergeron J.J., Trudeau L.E., Burelle Y., Gagnon E., McBride H.M., Desjardins M. Parkinson’s disease-related proteins PINK1 and parkin repress mitochondrial antigen presentation. Cell. 2016;166(2):314–327. doi: 10.1016/j.cell.2016.05.039. [DOI] [PubMed] [Google Scholar]
- 152.Gusella J.F., Wexler N.S., Conneally P.M., Naylor S.L., Anderson M.A., Tanzi R.E., Watkins P.C., Ottina K., Wallace M.R., Sakaguchi A.Y., Young A.B., Shoulson I., Bonilla E., Martin J.B. A polymorphic DNA marker genetically linked to Huntington’s disease. Nature. 1983;306(5940):234–238. doi: 10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- 153.Askeland G., Dosoudilova Z., Rodinova M., Klempir J., Liskova I., Kuśnierczyk A., Bjørås M., Nesse G., Klungland A., Hansikova H., Eide L. Increased nuclear DNA damage precedes mitochondrial dysfunction in peripheral blood mononuclear cells from Huntington’s disease patients. Sci. Rep. 2018;8(1):9817. doi: 10.1038/s41598-018-27985-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhao Y., Sun X., Hu D., Prosdocimo D.A., Hoppel C., Jain M.K., Ramachandran R., Qi X. ATAD3A oligomerization causes neurodegeneration by coupling mitochondrial fragmentation and bioenergetics defects. Nat. Commun. 2019;10(1):1371. doi: 10.1038/s41467-019-09291-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hering T., Kojer K., Birth N., Hallitsch J., Taanman J-W., Orth M. Mitochondrial cristae remodelling is associated with disrupted OPA1 oligomerisation in the Huntington’s disease R6/2 fragment model. Exp. Neurol. 2017;288:167–175. doi: 10.1016/j.expneurol.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 156.Wang Y., Guo X., Ye K., Orth M., Gu Z. Accelerated expansion of pathogenic mitochondrial DNA heteroplasmies in Huntington’s disease. Proc. Natl. Acad. Sci. USA. 2021;118(30):e2014610118. doi: 10.1073/pnas.2014610118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Williams S.L., Mash D.C., Züchner S., Moraes C.T. Somatic mtDNA mutation spectra in the aging human putamen. PLoS Genet. 2013;9(12):e1003990. doi: 10.1371/journal.pgen.1003990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Disatnik M.H., Joshi A.U., Saw N.L., Shamloo M., Leavitt B.R., Qi X., Mochly-Rosen D. Potential biomarkers to follow the progression and treatment response of Huntington’s disease. J. Exp. Med. 2016;213(12):2655–2669. doi: 10.1084/jem.20160776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Siddiqui A., Rivera-Sánchez S., Castro M.R., Acevedo-Torres K., Rane A., Torres-Ramos C.A., Nicholls D.G., Andersen J.K., Ayala-Torres S. Mitochondrial DNA damage Is associated with reduced mitochondrial bioenergetics in Huntington’s disease. Free Radic. Biol. Med. 2012;53(7):1478–1488. doi: 10.1016/j.freeradbiomed.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jędrak P., Krygier M., Tońska K., Drozd M., Kaliszewska M., Bartnik E., Sołtan W., Sitek E.J., Stanisławska-Sachadyn A., Limon J., Sławek J., Węgrzyn G., Barańska S. Mitochondrial DNA levels in Huntington disease leukocytes and dermal fibroblasts. Metab. Brain Dis. 2017;32(4):1237–1247. doi: 10.1007/s11011-017-0026-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Petersen M.H., Budtz-Jørgensen E., Sørensen S.A., Nielsen J.E., Hjermind L.E., Vinther-Jensen T., Nielsen S.M.B., Nørremølle A. Reduction in mitochondrial DNA copy number in peripheral leukocytes after onset of Huntington’s disease. Mitochondrion. 2014;17:14–21. doi: 10.1016/j.mito.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 162.Rasola A., Bernardi P. Mitochondrial permeability transition in Ca2+-dependent apoptosis and necrosis. Cell Calcium. 2011;50(3):222–233. doi: 10.1016/j.ceca.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 163.Hering T., Birth N., Taanman J.W., Orth M. Selective striatal mtDNA depletion in end-stage Huntington’s disease R6/2 mice. Exp. Neurol. 2015;266:22–29. doi: 10.1016/j.expneurol.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 164.Quintanilla R.A., Jin Y.N., von Bernhardi R., Johnson G.V.W. Mitochondrial permeability transition pore induces mitochondria injury in Huntington disease. Mol. Neurodegener. 2013;8(1):45. doi: 10.1186/1750-1326-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shirendeb U.P., Calkins M.J., Manczak M., Anekonda V., Dufour B., McBride J.L., Mao P., Reddy P.H. Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum. Mol. Genet. 2012;21(2):406–420. doi: 10.1093/hmg/ddr475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Crotti A., Glass C.K. The choreography of neuroinflammation in Huntington’s disease. Trends Immunol. 2015;36(6):364–373. doi: 10.1016/j.it.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vicencio E., Beltrán S., Labrador L., Manque P., Nassif M., Woehlbier U. Implications of selective autophagy dysfunction for ALS pathology. Cells. 2020;9(2):381. doi: 10.3390/cells9020381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ladd A.C., Brohawn D.G., Thomas R.R., Keeney P.M., Berr S.S., Khan S.M., Portell F.R., Shakenov M.Z., Antkowiak P.F., Kundu B., Tustison N., Bennett J.P. RNA-seq analyses reveal that cervical spinal cords and anterior motor neurons from amyotrophic lateral sclerosis subjects show reduced expression of mitochondrial DNA-encoded respiratory genes, and rhTFAM may correct this respiratory deficiency. Brain Res. 2017;1667:74–83. doi: 10.1016/j.brainres.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 169.Lopez-Gonzalez R., Lu Y., Gendron T.F., Karydas A., Tran H., Yang D., Petrucelli L., Miller B.L., Almeida S., Gao F.B. Poly(GR) in C9ORF72-related ALS/FTD compromises mitochondrial function and increases oxidative stress and DNA damage in iPSC-derived motor neurons. Neuron. 2016;92(2):383–391. doi: 10.1016/j.neuron.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Barber S.C., Shaw P.J. Oxidative stress in ALS: Key role in motor neuron injury and therapeutic target. Free Radic. Biol. Med. 2010;48(5):629–641. doi: 10.1016/j.freeradbiomed.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 171.Kok J.R., Palminha N.M., Dos Santos Souza C., El-Khamisy S.F., Ferraiuolo L. DNA damage as a mechanism of neurodegeneration in ALS and a contributor to astrocyte toxicity. Cell. Mol. Life Sci. 2021;78(15):5707–5729. doi: 10.1007/s00018-021-03872-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Stoccoro A., Smith A.R., Mosca L., Marocchi A., Gerardi F., Lunetta C., Cereda C., Gagliardi S., Lunnon K., Migliore L., Coppedè F. Reduced mitochondrial D-loop methylation levels in sporadic amyotrophic lateral sclerosis. Clin. Epigenetics. 2020;12(1):137. doi: 10.1186/s13148-020-00933-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Stoccoro A., Mosca L., Carnicelli V., Cavallari U., Lunetta C., Marocchi A., Migliore L., Coppedè F. Mitochondrial DNA copy number and D-loop region methylation in carriers of amyotrophic lateral sclerosis gene mutations. Epigenomics. 2018;10(11):1431–1443. doi: 10.2217/epi-2018-0072. [DOI] [PubMed] [Google Scholar]
- 174.Walker C., El-Khamisy S.F. Perturbed autophagy and DNA repair converge to promote neurodegeneration in amyotrophic lateral sclerosis and dementia. Brain. 2018;141(5):1247–1262. doi: 10.1093/brain/awy076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Huang C., Yan S., Zhang Z. Maintaining the balance of TDP-43, mitochondria, and autophagy: a promising therapeutic strategy for neurodegenerative diseases. Transl. Neurodegener. 2020;9(1):40. doi: 10.1186/s40035-020-00219-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Davis S.A., Itaman S., Khalid-Janney C.M., Sherard J.A., Dowell J.A., Cairns N.J., Gitcho M.A. TDP-43 interacts with mitochondrial proteins critical for mitophagy and mitochondrial dynamics. Neurosci. Lett. 2018;678:8–15. doi: 10.1016/j.neulet.2018.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ichimura Y., Kominami E., Tanaka K., Komatsu M. Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy. 2008;4(8):1063–1066. doi: 10.4161/auto.6826. [DOI] [PubMed] [Google Scholar]
- 178.Johnson J.O., Glynn S.M., Gibbs J.R., Nalls M.A., Sabatelli M., Restagno G., Drory V.E., Chiò A., Rogaeva E., Traynor B.J. Mutations in the CHCHD10 gene are a common cause of familial amyotrophic lateral sclerosis. Brain. 2014;137(12):e311. doi: 10.1093/brain/awu265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhou Z.D., Saw W.T., Tan E.K. Mitochondrial CHCHD-containing proteins: Physiologic functions and link with neurodegenerative diseases. Mol. Neurobiol. 2017;54(7):5534–5546. doi: 10.1007/s12035-016-0099-5. [DOI] [PubMed] [Google Scholar]
- 180.Bannwarth S., Ait-El-Mkadem S., Chaussenot A., Genin E.C., Lacas-Gervais S., Fragaki K., Berg-Alonso L., Kageyama Y., Serre V., Moore D.G., Verschueren A., Rouzier C., Le Ber I., Augé G., Cochaud C., Lespinasse F., N’Guyen K., de Septenville A., Brice A., Yu-Wai-Man P., Sesaki H., Pouget J., Paquis-Flucklinger V. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain. 2014;137(8):2329–2345. doi: 10.1093/brain/awu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wang P., Deng J., Dong J., Liu J., Bigio E.H., Mesulam M., Wang T., Sun L., Wang L., Lee A.Y.L., McGee W.A., Chen X., Fushimi K., Zhu L., Wu J.Y. TDP-43 induces mitochondrial damage and activates the mitochondrial unfolded protein response. PLoS Genet. 2019;15(5):e1007947. doi: 10.1371/journal.pgen.1007947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yu C.H., Davidson S., Harapas C.R., Hilton J.B., Mlodzianoski M.J., Laohamonthonkul P., Louis C., Low R.R.J., Moecking J., De Nardo D., Balka K.R., Calleja D.J., Moghaddas F., Ni E., McLean C.A., Samson A.L., Tyebji S., Tonkin C.J., Bye C.R., Turner B.J., Pepin G., Gantier M.P., Rogers K.L., McArthur K., Crouch P.J., Masters S.L. TDP-43 triggers mitochondrial DNA release via mPTP to activate cGAS/STING in ALS. Cell. 2020;183(3):636–649.e18. doi: 10.1016/j.cell.2020.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yang Y., Wu H., Kang X., Liang Y., Lan T., Li T., Tan T., Peng J., Zhang Q., An G., Liu Y., Yu Q., Ma Z., Lian Y., Soh B.S., Chen Q., Liu P., Chen Y., Sun X., Li R., Zhen X., Liu P., Yu Y., Li X., Fan Y. Targeted elimination of mutant mitochondrial DNA in MELAS-iPSCs by mitoTALENs. Protein Cell. 2018;9(3):283–297. doi: 10.1007/s13238-017-0499-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bacman S.R., Kauppila J.H.K., Pereira C.V., Nissanka N., Miranda M., Pinto M., Williams S.L., Larsson N.G., Stewart J.B., Moraes C.T. MitoTALEN reduces mutant mtDNA load and restores tRNAAla levels in a mouse model of heteroplasmic mtDNA mutation. Nat. Med. 2018;24(11):1696–1700. doi: 10.1038/s41591-018-0166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gammage P.A., Viscomi C., Simard M.L., Costa A.S.H., Gaude E., Powell C.A., Van Haute L., McCann B.J., Rebelo-Guiomar P., Cerutti R., Zhang L., Rebar E.J., Zeviani M., Frezza C., Stewart J.B., Minczuk M. Genome editing in mitochondria corrects a pathogenic mtDNA mutation in vivo. Nat. Med. 2018;24(11):1691–1695. doi: 10.1038/s41591-018-0165-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mok B.Y., de Moraes M.H., Zeng J., Bosch D.E., Kotrys A.V., Raguram A., Hsu F., Radey M.C., Peterson S.B., Mootha V.K., Mougous J.D., Liu D.R. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature. 2020;583(7817):631–637. doi: 10.1038/s41586-020-2477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pereira C.V., Bacman S.R., Arguello T., Zekonyte U., Williams S.L., Edgell D.R., Moraes C.T. mitoTev‐TALE: a monomeric DNA editing enzyme to reduce mutant mitochondrial DNA levels. EMBO Mol. Med. 2018;10(9):e8084. doi: 10.15252/emmm.201708084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zinovkin R.A., Zamyatnin A.A. Mitochondria-targeted drugs. Curr. Mol. Pharmacol. 2019;12(3):202–214. doi: 10.2174/1874467212666181127151059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bharadwaj P.R., Bates K.A., Porter T., Teimouri E., Perry G., Steele J.W., Gandy S., Groth D., Martins R.N., Verdile G. Latrepirdine: molecular mechanisms underlying potential therapeutic roles in Alzheimer’s and other neurodegenerative diseases. Transl. Psychiatry. 2013;3(12):e332. doi: 10.1038/tp.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Perez Ortiz J.M., Swerdlow R.H. Mitochondrial dysfunction in Alzheimer’s disease: Role in pathogenesis and novel therapeutic opportunities. Br. J. Pharmacol. 2019;176(18):3489–3507. doi: 10.1111/bph.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Beal M.F., Oakes D., Shoulson I., Henchcliffe C., Galpern W.R., Haas R., Juncos J.L., Nutt J.G., Voss T.S., Ravina B., Shults C.M., Helles K., Snively V., Lew M.F., Griebner B., Watts A., Gao S., Pourcher E., Bond L., Kompoliti K., Agarwal P., Sia C., Jog M., Cole L., Sultana M., Kurlan R., Richard I., Deeley C., Waters C.H., Figueroa A., Arkun A., Brodsky M., Ondo W.G., Hunter C.B., Jimenez-Shahed J., Palao A., Miyasaki J.M., So J., Tetrud J., Reys L., Smith K., Singer C., Blenke A., Russell D.S., Cotto C., Friedman J.H., Lannon M., Zhang L., Drasby E., Kumar R., Subramanian T., Ford D.S., Grimes D.A., Cote D., Conway J., Siderowf A.D., Evatt M.L., Sommerfeld B., Lieberman A.N., Okun M.S., Rodriguez R.L., Merritt S., Swartz C.L., Martin W.R.W., King P., Stover N., Guthrie S., Watts R.L., Ahmed A., Fernandez H.H., Winters A., Mari Z., Dawson T.M., Dunlop B., Feigin A.S., Shannon B., Nirenberg M.J., Ogg M., Ellias S.A., Thomas C.A., Frei K., Bodis-Wollner I., Glazman S., Mayer T., Hauser R.A., Pahwa R., Langhammer A., Ranawaya R., Derwent L., Sethi K.D., Farrow B., Prakash R., Litvan I., Robinson A., Sahay A., Gartner M., Hinson V.K., Markind S., Pelikan M., Perlmutter J.S., Hartlein J., Molho E., Evans S., Adler C.H., Duffy A., Lind M., Elmer L., Davis K., Spears J., Wilson S., Leehey M.A., Hermanowicz N., Niswonger S., Shill H.A., Obradov S., Rajput A., Cowper M., Lessig S., Song D., Fontaine D., Zadikoff C., Williams K., Blindauer K.A., Bergholte J., Propsom C.S., Stacy M.A., Field J., Mihaila D., Chilton M., Uc E.Y., Sieren J., Simon D.K., Kraics L., Silver A., Boyd J.T., Hamill R.W., Ingvoldstad C., Young J., Thomas K., Kostyk S.K., Wojcieszek J., Pfeiffer R.F., Panisset M., Beland M., Reich S.G., Cines M., Zappala N., Rivest J., Zweig R., Lumina L.P., Hilliard C.L., Grill S., Kellermann M., Tuite P., Rolandelli S., Kang U.J., Young J., Rao J., Cook M.M., Severt L., Boyar K. A randomized clinical trial of high-dosage coenzyme Q10 in early Parkinson disease: no evidence of benefit. JAMA Neurol. 2014;71(5):543–552. doi: 10.1001/jamaneurol.2014.131. [DOI] [PubMed] [Google Scholar]
- 192.Kieburtz K., Tilley B.C., Elm J.J., Babcock D., Hauser R., Ross G.W., Augustine A.H., Augustine E.U., Aminoff M.J., Bodis-Wollner I.G., Boyd J., Cambi F., Chou K., Christine C.W., Cines M., Dahodwala N., Derwent L., Dewey R.B., Jr, Hawthorne K., Houghton D.J., Kamp C., Leehey M., Lew M.F., Liang G.S.L., Luo S.T., Mari Z., Morgan J.C., Parashos S., Pérez A., Petrovitch H., Rajan S., Reichwein S., Roth J.T., Schneider J.S., Shannon K.M., Simon D.K., Simuni T., Singer C., Sudarsky L., Tanner C.M., Umeh C.C., Williams K., Wills A.M. Effect of creatine monohydrate on clinical progression in patients with Parkinson disease: a randomized clinical trial. JAMA. 2015;313(6):584–593. doi: 10.1001/jama.2015.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Koentjoro B., Park J.S., Sue C.M. Nix restores mitophagy and mitochondrial function to protect against PINK1/Parkin-related Parkinson’s disease. Sci. Rep. 2017;7(1):44373. doi: 10.1038/srep44373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hayashi G., Jasoliya M., Sahdeo S., Saccà F., Pane C., Filla A., Marsili A., Puorro G., Lanzillo R., Brescia Morra V., Cortopassi G. Dimethyl fumarate mediates Nrf2-dependent mitochondrial biogenesis in mice and humans. Hum. Mol. Genet. 2017;26(15):2864–2873. doi: 10.1093/hmg/ddx167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Delerue T., Tribouillard-Tanvier D., Daloyau M., Khosrobakhsh F., Emorine L.J., Friocourt G., Belenguer P., Blondel M., Arnauné-Pelloquin L. A yeast-based screening assay identifies repurposed drugs that suppress mitochondrial fusion and mtDNA maintenance defects. Dis. Model. Mech. 2019;12(2):dmm.036558. doi: 10.1242/dmm.036558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wang Y., Liu N., Lu B. Mechanisms and roles of mitophagy in neurodegenerative diseases. CNS Neurosci. Ther. 2019;25(7):cns.13140. doi: 10.1111/cns.13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gureev A.P., Sadovnikova I.S., Starkov N.N., Starkov A.A., Popov V.N. p62-Nrf2-p62 mitophagy regulatory loop as a target for preventive therapy of neurodegenerative diseases. Brain Sci. 2020;10(11):847. doi: 10.3390/brainsci10110847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Rahman M.H., Akter R., Bhattacharya T., Abdel-Daim M.M., Alkahtani S., Arafah M.W., Al-Johani N.S., Alhoshani N.M., Alkeraishan N., Alhenaky A., Abd-Elkader O.H., El-Seedi H.R., Kaushik D., Mittal V. Resveratrol and neuroprotection: impact and its therapeutic potential in Alzheimer’s disease. Front. Pharmacol. 2020;11:619024. doi: 10.3389/fphar.2020.619024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Coll R.C., Robertson A.A.B., Chae J.J., Higgins S.C., Muñoz-Planillo R., Inserra M.C., Vetter I., Dungan L.S., Monks B.G., Stutz A., Croker D.E., Butler M.S., Haneklaus M., Sutton C.E., Núñez G., Latz E., Kastner D.L., Mills K.H.G., Masters S.L., Schroder K., Cooper M.A., O’Neill L.A.J. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 2015;21(3):248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Savitt D., Jankovic J. Targeting α-synuclein in Parkinson’s disease: Progress towards the development of disease-modifying therapeutics. Drugs. 2019;79(8):797–810. doi: 10.1007/s40265-019-01104-1. [DOI] [PubMed] [Google Scholar]
- 201.Haag S.M., Gulen M.F., Reymond L., Gibelin A., Abrami L., Decout A., Heymann M., van der Goot F.G., Turcatti G., Behrendt R., Ablasser A. Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559(7713):269–273. doi: 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
- 202.Vincent J., Adura C., Gao P., Luz A., Lama L., Asano Y., Okamoto R., Imaeda T., Aida J., Rothamel K., Gogakos T., Steinberg J., Reasoner S., Aso K., Tuschl T., Patel D.J., Glickman J.F., Ascano M. Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice. Nat. Commun. 2017;8(1):750. doi: 10.1038/s41467-017-00833-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Russell O.M., Gorman G.S., Lightowlers R.N., Turnbull D.M. Mitochondrial diseases: Hope for the future. Cell. 2020;181(1):168–188. doi: 10.1016/j.cell.2020.02.051. [DOI] [PubMed] [Google Scholar]
- 204.Yoo S.M., Jung Y.K. A Molecular approach to mitophagy and mitochondrial dynamics. Mol. Cells. 2018;41(1):18–26. doi: 10.14348/molcells.2018.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Swerdlow R.H. Mitochondria and mitochondrial cascades in Alzheimer’s disease. J. Alzheimers Dis. 2018;62(3):1403–1416. doi: 10.3233/JAD-170585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hwang I.W., Kwon B.N., Kim H.J., Han S.H., Lee N.R., Lim M.H., Kwon H.J., Jin H.J. Assessment of associations between mitochondrial DNA haplogroups and attention deficit and hyperactivity disorder in Korean children. Mitochondrion. 2019;47:174–178. doi: 10.1016/j.mito.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 207.DeBrosse S., Parikh S. Neurologic disorders due to mitochondrial DNA mutations. Semin. Pediatr. Neurol. 2012;19(4):194–202. doi: 10.1016/j.spen.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 208.Moya G.E., Rivera P.D., Dittenhafer-Reed K.E. Evidence for the Role of Mitochondrial DNA Release in the Inflammatory Response in Neurological Disorders. Int. J. Mol. Sci. 2021;22(13):7030. doi: 10.3390/ijms22137030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Stoccoro A., Siciliano G., Migliore L., Coppedè F. Decreased methylation of the mitochondrial D-Loop region in late-onset Alzheimer’s disease. J. Alzheimers Dis. 2017;59(2):559–564. doi: 10.3233/JAD-170139. [DOI] [PubMed] [Google Scholar]
- 210.Gonzalez-Hunt C.P., Thacker E.A., Toste C.M., Boularand S., Deprets S., Dubois L., Sanders L.H. Mitochondrial DNA damage as a potential biomarker of LRRK2 kinase activity in LRRK2 Parkinson’s disease. Sci. Rep. 2020;10(1):17293. doi: 10.1038/s41598-020-74195-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zambrano K., Barba D., Castillo K., Robayo P., Argueta-Zamora D., Sanon S., Arizaga E., Caicedo A., Gavilanes A.W.D. The war against Alzheimer, the mitochondrion strikes back! Mitochondrion. 2022;64:125–135. doi: 10.1016/j.mito.2022.03.003. [DOI] [PubMed] [Google Scholar]
- 212.Matsui H., Ito J., Matsui N., Uechi T., Onodera O., Kakita A. Cytosolic dsDNA of mitochondrial origin induces cytotoxicity and neurodegeneration in cellular and zebrafish models of Parkinson’s disease. Nat. Commun. 2021;12(1):3101. doi: 10.1038/s41467-021-23452-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Onyango I., Bennett J., Stokin G. Regulation of neuronal bioenergetics as a therapeutic strategy in neurodegenerative diseases. Neural Regen. Res. 2021;16(8):1467–1482. doi: 10.4103/1673-5374.303007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Caicedo A., Zambrano K., Sanon S., Luis Vélez J., Montalvo M., Jara F., Moscoso S.A., Vélez P., Maldonado A., Velarde G. The diversity and coexistence of extracellular mitochondria in circulation: A friend or foe of the immune system. Mitochondrion. 2021;58:270–284. doi: 10.1016/j.mito.2021.02.014. [DOI] [PubMed] [Google Scholar]
- 215.Caicedo A., Zambrano K., Sanon S., Gavilanes A.W.D. Extracellular mitochondria in the cerebrospinal fluid (CSF): Potential types and key roles in central nervous system (CNS) physiology and pathogenesis. Mitochondrion. 2021;58:255–269. doi: 10.1016/j.mito.2021.02.006. [DOI] [PubMed] [Google Scholar]
- 216.Nakamura Y., Park J.H., Hayakawa K. Therapeutic use of extracellular mitochondria in CNS injury and disease. Exp. Neurol. 2020;324:113114. doi: 10.1016/j.expneurol.2019.113114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhang X., Hu D., Shang Y., Qi X. Using induced pluripotent stem cell neuronal models to study neurodegenerative diseases. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866(4):165431. doi: 10.1016/j.bbadis.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Zhang W., Gu G.J., Shen X., Zhang Q., Wang G.M., Wang P.J. Neural stem cell transplantation enhances mitochondrial biogenesis in a transgenic mouse model of Alzheimer’s disease–like pathology. Neurobiol. Aging. 2015;36(3):1282–1292. doi: 10.1016/j.neurobiolaging.2014.10.040. [DOI] [PubMed] [Google Scholar]
- 219.Zaia A., Maponi P., Zannotti M., Casoli T. Biocomplexity and fractality in the search of biomarkers of aging and pathology: Mitochondrial DNA profiling of Parkinson’s disease. Int. J. Mol. Sci. 2020;21(5):1758. doi: 10.3390/ijms21051758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zaia A., Maponi P., Di Stefano G., Casoli T. Biocomplexity and fractality in the search of biomarkers of aging and pathology: Focus on mitochondrial DNA and Alzheimer’s disease. Aging Dis. 2017;8(1):44–56. doi: 10.14336/AD.2016.0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Santibanez-Koref M., Griffin H., Turnbull D.M., Chinnery P.F., Herbert M., Hudson G. Assessing mitochondrial heteroplasmy using next generation sequencing: A note of caution. Mitochondrion. 2019;46:302–306. doi: 10.1016/j.mito.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Krishnan K.J., Ratnaike T.E., De Gruyter H.L.M., Jaros E., Turnbull D.M. Mitochondrial DNA deletions cause the biochemical defect observed in Alzheimer’s disease. Neurobiol. Aging. 2012;33(9):2210–2214. doi: 10.1016/j.neurobiolaging.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 223.de la Monte S.M., Luong T., Neely T.R., Robinson D., Wands J.R. Mitochondrial DNA damage as a mechanism of cell loss in Alzheimer’s disease. Lab. Invest. 2000;80(8):1323–1335. doi: 10.1038/labinvest.3780140. [DOI] [PubMed] [Google Scholar]
- 224.Liou C.W., Chen S.H., Lin T.K., Tsai M.H., Chang C.C. Oxidative stress biomarkers and mitochondrial DNA copy number associated with APOE4 allele and cholinesterase inhibitor therapy in patients with Alzheimer’s disease. Antioxidants (Basel) 2021;10(12):1971. doi: 10.3390/antiox10121971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hutchin T.P., Heath P.R., Pearson R.C.A., Sinclair A.J. Mitochondrial DNA mutations in Alzheimer’s disease. Biochem. Biophys. Res. Commun. 1997;241(2):221–225. doi: 10.1006/bbrc.1997.7793. [DOI] [PubMed] [Google Scholar]
- 226.Shoffner J.M., Brown M.D., Torroni A., Lott M.T., Cabell M.F., Mirra S.S., Beal M.F., Yang C.C., Gearing M., Salvo R., Watts R.L., Juncos J.L., Hansen L.A., Crain B.J., Fayad M., Reckord C.L., Wallace D.C. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993;17(1):171–184. doi: 10.1006/geno.1993.1299. [DOI] [PubMed] [Google Scholar]
- 227.Blanch M., Mosquera J.L., Ansoleaga B., Ferrer I., Barrachina M. Altered mitochondrial DNA methylation pattern in Alzheimer disease–related pathology and in Parkinson disease. Am. J. Pathol. 2016;186(2):385–397. doi: 10.1016/j.ajpath.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 228.Stoccoro A., Baldacci F., Ceravolo R., Giampietri L., Tognoni G., Siciliano G., Migliore L., Coppedè F. Increase in mitochondrial D-loop region methylation levels in mild cognitive impairment individuals. Int. J. Mol. Sci. 2022;23(10):5393. doi: 10.3390/ijms23105393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Grünewald A., Rygiel K.A., Hepplewhite P.D., Morris C.M., Picard M., Turnbull D.M. Mitochondrial DNA depletion in respiratory chain–deficient p arkinson disease neurons. Ann. Neurol. 2016;79(3):366–378. doi: 10.1002/ana.24571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Bury A.G., Pyle A., Elson J.L., Greaves L., Morris C.M., Hudson G., Pienaar I.S. Mitochondrial DNA changes in pedunculopontine cholinergic neurons in Parkinson disease. Ann. Neurol. 2017;82(6):1016–1021. doi: 10.1002/ana.25099. [DOI] [PubMed] [Google Scholar]
- 231.Nido G.S., Dölle C., Flønes I., Tuppen H.A., Alves G., Tysnes O.B., Haugarvoll K., Tzoulis C. Ultradeep mapping of neuronal mitochondrial deletions in Parkinson’s disease. Neurobiol. Aging. 2018;63:120–127. doi: 10.1016/j.neurobiolaging.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 232.Richter G., Sonnenschein A., Grünewald T., Reichmann H., Janetzky B. Novel mitochondrial DNA mutations in Parkinson’s disease. J. Neural Transm. (Vienna) 2002;109(5-6):721–729. doi: 10.1007/s007020200060. [DOI] [PubMed] [Google Scholar]
- 233.Huerta C., Castro M.G., Coto E., Blázquez M., Ribacoba R., Guisasola L.M., Salvador C., Martínez C., Lahoz C.H., Alvarez V. Mitochondrial DNA polymorphisms and risk of Parkinson’s disease in Spanish population. J. Neurol. Sci. 2005;236(1-2):49–54. doi: 10.1016/j.jns.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 234.Egensperger R., Kösel S., Schnopp N.M., Mehraein P., Graeber M.B. Association of the mitochondrial tRNAA4336G mutation with Alzheimer’s and Parkinson’s diseases. Neuropathol. Appl. Neurobiol. 1997;23(4):315–321. doi: 10.1111/j.1365-2990.1997.tb01301.x. [DOI] [PubMed] [Google Scholar]
- 235.Coxhead J., Kurzawa-Akanbi M., Hussain R., Pyle A., Chinnery P., Hudson G. Somatic mtDNA variation is an important component of Parkinson’s disease. Neurobiol. Aging. 2016;38:217.e1–217.e6. doi: 10.1016/j.neurobiolaging.2015.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Stoccoro A., Smith A.R., Baldacci F., Del Gamba C., Lo Gerfo A., Ceravolo R., Lunnon K., Migliore L., Coppedè F. Mitochondrial D-loop region methylation and copy number in peripheral blood DNA of Parkinson’s disease patients. Genes (Basel) 2021;12(5):720. doi: 10.3390/genes12050720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Alvarez-Mora M.I., Podlesniy P., Riazuelo T., Molina-Porcel L., Gelpi E., Rodriguez-Revenga L. Reduced mtDNA copy number in the prefrontal cortex of C9ORF72 patients. Mol. Neurobiol. 2022;59(2):1230–1237. doi: 10.1007/s12035-021-02673-7. [DOI] [PubMed] [Google Scholar]