Abstract
Human hepatitis delta virus (HDV) is a natural subviral agent that uses hepatitis B virus as a helper. Experimentally, HDV can be made to replicate in woodchucks, using woodchuck hepatitis B virus as a helper virus. Also, independent of such helper activity, replication of the HDV RNA genome can be achieved in many mammalian cells. In this study we examined whether such replication could also be achieved in avian cells. We used cotransfection strategies and initially found no detectable genome replication in chicken LMH cells relative to the mammalian cell line Huh7, used as a positive control. We also found that, in contrast to transfected Huh7 cells, the avian cell line was readily and efficiently killed by expression of the delta protein. Three strategies were used to reduce such killing: (i) the delta protein was expressed from a separate expression vector, the amount of which was then reduced as much as 33-fold; (ii) the protein was expressed transiently, using a promoter under tetracycline control; and (iii) the transfected cells were treated with Z-VAD-fmk, a broad-spectrum caspase inhibitor, which reduced cell killing. This last result indicated that cell killing occurred via an apoptotic pathway. After application of these three strategies to reduce cell killing, together with a novel procedure to improve the signal-to-noise ratio in Northern analyses, replication of the HDV genome was then detected in LMH cells. However, even after removal of obvious signs of toxicity, the amount was still >50 times lower than in the Huh7 cells. Our findings explain previous unsuccessful attempts to demonstrate replication of the HDV genome in avian cells and establish the precedent that in certain situations HDV replication can be cytotoxic.
Hepatitis delta virus (HDV) is only found as a natural infection of the liver in humans already infected by hepatitis B virus (HBV). HBV acts as a helper virus in that it provides the envelope proteins needed for the assembly of replicating HDV RNA genomes into new virus particles (16). It was found by Ponzetto et al. that HDV could be experimentally transmitted to woodchucks and could make use of the envelope proteins of woodchuck HBV for its assembly and transmission (23). Following this initial success, there were several reported attempts to achieve a comparable switch to ducklings infected with the duck HBV as a helper (8, 9, 24). The latter studies were not definitive, and a positive outcome could not be confirmed (J. Taylor, unpublished observations).
Independent of the question of which hepadnavirus can act as a helper, it has been believed that replication of just the HDV genome can be achieved in cells from a variety of animal species (e.g., human, chimpanzee, monkey, and mouse) and from tissues other than liver (29). For example, injection of HDV into mice resulted in a low level of HDV genome replication in the absence of any helper virus (21). Some efforts to achieve HDV genome replication in transfected avian cells gave positive results (2, 28), but in retrospect such studies have to be considered controversial in that no steps were taken to distinguish whether the accumulated HDV RNAs were transcribed from RNA or DNA templates (17). Other studies gave negative results (D. Ganem and T.-T. Wu, personal communications). The following studies were therefore undertaken to clarify the limitations to HDV genome replication in avian cells.
As reported here, in LMH cells, an avian cell line, the major restriction was the induction of cell death simply by expression of delta protein. This observation may have additional relevance to the problem of HDV-associated pathogenesis. It has been a controversial question whether or not HDV replication is cytopathic. Some studies have detected little or no cytopathic effect (1), while others have shown that expression of even the small delta protein can inhibit cell growth (6). In contrast, expression of delta protein in insect cells produced both cell cycle arrest (11) and even antiapoptotic effects (12). Our studies show not only that the delta protein can induce cell killing in the avian cells, but also that this killing can be reduced by treatment of cells with a known antiapoptotic agent. This was one of three strategies that we used to reduce the killing induced in avian cells by expression of the delta protein; after such reductions, we were able to see real but minimal amounts of HDV RNA-directed RNA transcription and accumulation in LMH cells.
MATERIALS AND METHODS
Plasmids.
pCMV(D3) is a trimer of HDV cDNA inserted into pcDNA3 (Invitrogen). pJC110 is pcDNA3 with an insert of 1.2 copies of an HDV genome that has a 2-nucleotide (nt) deletion (nt 1434 and 1435) in the open reading frame for the small delta protein. HDV RNA transcribed from this construct will only replicate when small delta protein is provided in trans (15), such as from pTW198, which is a pcDNA3 construct. Green fluorescent protein (GFP) was expressed from a cytomegalovirus (CMV) construct, pGG119, kindly provided by Ketaki Datta (Fox Chase Cancer Center). For expression of small delta protein under tetracycline control, we used pPB106, which is based on vector pBPSTR1 (22).
Cell culture and transfections.
Two cell lines were used, LMH chicken liver cells (14) and Huh7 human hepatoblastoma cells (20). Exponentially growing cells were trypsinized and seeded (105 per well of a 24-well culture dish) at 1 day prior to transfection. For all cotransfections we used a total of 1 μg of plasmid per well in a protocol involving FuGENE 6 (Roche); as needed, we added empty vector to achieve this. Each cotransfection included 0.1 μg of pGG119, a plasmid expressing GFP. For expression of small delta protein in cells transfected with pPB106, we used 1 μg of tetracycline per ml in the growth medium to suppress expression both during and immediately after transfection. Subsequently, to release the transcriptional block, the cultures were washed five times with tetracycline-free medium and then incubated further, as indicated in Fig. 4.
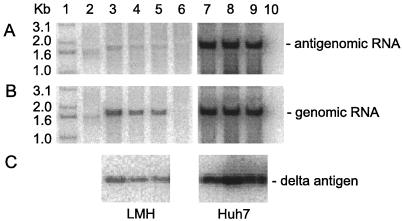
FIG. 4.
Assays of HDV replication in avian and mammalian cells in the presence of tetracycline-controlled expression of small delta protein. Cultures of LMH (lanes 3 to 6) or Huh7 cells (lanes 7 to 10) were cotransfected in the presence of tetracycline with fixed amounts of pGG119, pJC110, and pPB106, expressing the small form of the delta protein under control of tetracycline. Tetracycline was removed after day 1 for 0.5 days (lanes 3 and 7), 3 days (lanes 4 and 8), and 5 days (lanes 5 and 9). Lanes 6 and 10 are untransfected controls. Lane 1 contains 5′-labeled DNA size markers. Lane 2 is a standard of unit-length HDV cDNA (200 pg). At 6 days after transfection, RNA and protein were extracted. RNA was analyzed by Northern blot for antigenomic RNA (A) or genomic RNA (B). Protein was assayed by immunoblot for delta protein (C).
Apoptosis inhibitor.
In some studies we made use of the general apoptosis inhibitor Z-Val-Ala-Asp(OMe)-fluoromethylketone (Z-VAD-fmk) (Enzyme Systems). It was added to the cell growth medium to a final concentration of 40 μM beginning 1 h prior to transfection.
RNA and protein isolation.
At the indicated times after transfection, the medium was removed and the RNA and protein were isolated using a protocol for the Tri reagent (Molecular Research Center).
Northern analysis.
Isolated RNA was glyoxalated prior to electrophoresis into a gel of either 3% (Fig. 1) or 1.5% (Fig. 3 to 5) agarose following electrophoretic transfer and hybridization with a labeled RNA probe. Typically, we first probed to detect antigenomic RNA, and then after quantitation, the filter was stripped and rehybridized to detect genomic RNA. Radioactivity was quantitated using a Fuji imager.
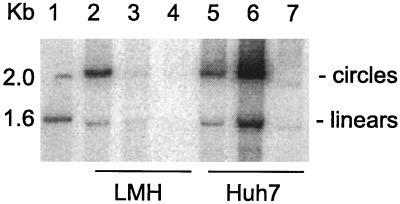
FIG. 1.
Assays of HDV RNA processing in avian cells. LMH (lanes 2 to 4) or Huh7 cells (lanes 5 to 7) were transfected with pCMV(D3) (lanes 2 and 5) or with both pCMV(D3) and pTW198 (lanes 3 and 6) or left untransfected (lanes 4 and 7). After 4 days the RNA was extracted and assayed by Northern analysis to detect genomic RNA. The gel electrophoretic conditions were such as to resolve linear and circular forms of unit-length HDV RNA, as indicated at the right side (4). Lane 1 contains 5′-labeled DNA size markers.
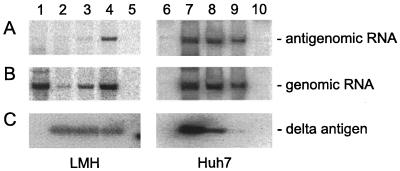
FIG. 3.
Assays of HDV replication in avian and mammalian cells during reduced expression of small delta protein. Cultures of LMH (lanes 1 to 5) or Huh7 cells (lanes 6 to 10) were cotransfected with fixed amounts of pGG119 and pJC110 along with various amounts of pTW198. The ratios of pTW198 to pJC110 are as follows: lanes 1 and 6, 0:1; lanes 2 and 7, 1:1; lanes 3 and 8, 0.3:1; lanes 4 and 9, 0.03:1; lanes 5 and 10, untransfected. At 4 days after transfection, RNA and protein were extracted. RNA was analyzed by Northern blot for antigenomic RNA (A) or genomic RNA (B). Protein was assayed by immunoblot for delta protein (C).
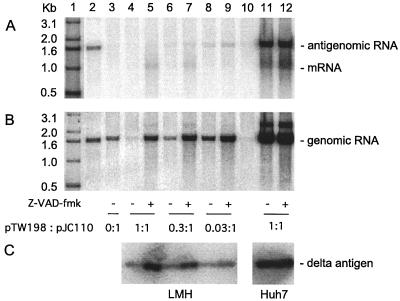
FIG. 5.
Assays of HDV replication in avian and mammalian cells during expression of small delta protein in the presence of the antiapoptotic compound Z-VAD-fmk. Cultures of LMH (lanes 3 to 10) or Huh7 cells (lanes 11 and 12) were cotransfected with fixed amounts of pGG119 and pJC110 along with various amounts of pTW198. The ratios of pTW198 to pJC110 are as follows: lane 3, 0:1; lanes 4, 5, 11, and 12, 1:1; lanes 6 and 7, 0.3:1; lanes 8 and 9, 0.03:1. Lane 1 contains 5′-labeled DNA size markers. Lane 2 is a standard of unit-length HDV cDNA (200 pg). Lane 10 is a sample from untransfected cells. Lanes 5, 7, 9, and 12 correspond to cultures treated, beginning 1 h prior to cotransfection, with 40 μM Z-VAD-fmk. At 4 days after transfection, RNA and protein were extracted. RNA was analyzed by Northern blot for antigenomic RNA (A) or genomic RNA (B). Protein was assayed by immunoblot for delta protein (C).
We found that in transfected avian cells, the low levels of unit-length antigenomic RNA were sometimes very difficult to detect with a standard hybridization protocol. A further complication was cross-hybridization to the abundant 1.8-kb small rRNA, which produced a band with almost the same migration as unit-length HDV RNA. A partial solution to these problems was to prehybridize the probe for 16 h at 65°C in 1/20 the final hybridization volume with 50 μg of RNA from uninfected LMH cells. We used this strategy for the detection of antigenomic RNA (Fig. 3 and 5).
Immunoblot analysis.
Isolated protein was subjected to a standard gel electrophoresis and immunoblot, with the delta protein being detected by a combination of specific rabbit polyclonal antibody, 125I-labeled staphylococcal A protein (DuPont), and quantitation using a Fuji imager.
RESULTS
Transfection of avian cells with wild-type HDV cDNA construct.
We initially set about to determine whether HDV genome replication could be initiated in avian cells transfected with expression vectors containing wild-type HDV sequences. For constructs derived from pSVL, which uses the simian virus 40 late promoter, we did not detect even expression of the delta antigen (data not shown). We therefore switched promoters and made constructs based on vector pcDNA3, which uses a CMV immediate-early promoter. In this way we obtained expression of delta protein with construct pTW198, as judged by immunoblot. Also, we obtained DNA-directed transcription of trimers of wild-type genome RNA with construct pCMV(D3), as judged by Northern analysis; in fact, 4 days after transfection, we detected processed unit-length genomic RNA in LMH cells. Furthermore, with appropriate electrophoretic conditions, we were able to detect both linear and circular RNA conformations (Fig. 1, lane 2) just as in transfected Huh7 cells (lane 5). When we cotransfected cells with both pCMV(D3) and pTW198, which expresses the small delta protein, we detected much more HDV genomic RNAs in the Huh7 cells (lane 6), as expected for enhanced genome replication. In contrast, for the LMH cells, we actually detected a major decrease (lane 3), although the residual amount was still more than the background signal detected for untransfected cells (lane 4). Another concern was that when we hybridized the Northern blot to detect antigenomic RNA, we were unable to detect any indication of unit-length HDV RNAs in the LMH cells, in contrast to the Huh7 cells, which gave a significant signal (data not shown).
Our initial interpretation of these data was that the avian cells could support (i) DNA-directed RNA transcription, (ii) processing from multimers to unit-length species, and (iii) the formation of RNA circles, but they could not achieve RNA-directed transcription to make antigenomic RNA. Even when the cells were cotransfected with a construct, pTW198, to express additional amounts of the small form of the delta protein, we were still unable to detect evidence of RNA-directed transcription in avian cells. Importantly, we noted that expression of the small delta protein from pTW198 produced a decrease in the accumulation of unit-length genomic RNA in transfected avian cells (Fig. 1, lane 3 versus 2). To test the hypothesis that this decrease was somehow caused by toxicity, we carried out the following experiment.
Cytotoxic effect of delta protein expression in avian cells.
Cultures of LMH and Huh7 cells were cotransfected with (i) fixed amounts of pGG119, a plasmid expressing GFP, and (ii) pJC110, a construct which contains 1.2 copies of the HDV genome under control of a CMV promoter, along with (iii) different amounts of pTW198, which expresses the small form of the delta protein. The HDV genome of pJC110 has a 2-nt deletion within the delta protein open reading frame; this mutant HDV does not replicate in mammalian cells unless supported in trans by a construct expressing the wild-type form of the small delta protein (15). The ratios of pTW198 to pJC110 were 0:1, 1:1, 0.3:1, and 0.03:1.
For the LMH cells at 1 day after transfection, the expression of GFP, as monitored by fluorescence microscopy, was detectable in about 25 to 40% of cells, independent of the presence or absence of the small delta protein (data not shown). At days 2, 3, and 4, due to both enhanced expression and cell division, this signal was much stronger in cells cotransfected in the absence of the plasmid expressing small delta protein (data not shown). However, for three parallel cotransfections that included different amounts of pTW198, we saw significant decreases in the number of such GFP-positive cells and also corresponding increases in the release into the medium of GFP-positive cells (data not shown).
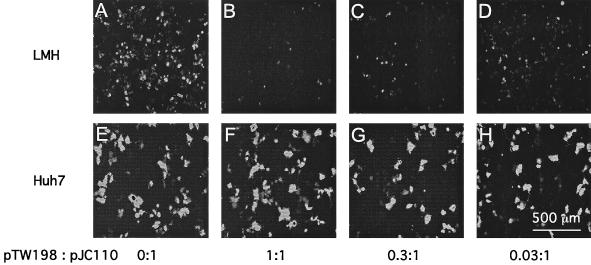
Fluorescence microscopy data for LMH cells at day 4 are shown in Fig. 2A to D. Note that in the absence of delta protein (0:1), a larger number of GFP-positive cells appeared than with the highest amount of delta protein (1:1) (Fig. 2A and B). The number of GFP-positive cells was reduced by about 90%. In Fig. 2C and D we show fluorescence for cells transfected with lower amounts, 0.3:1 and 0.03:1, respectively, of the plasmid expressing small delta protein. We observed less reduction in GFP positivity as the amount of delta antigen expression was reduced. However, even at 0.03:1 (Fig. 2D) there was still some reduction in GFP-positive cells relative to 0:1 (panel A). In contrast to these results with avian cells, no such effects were seen in parallel assays of transfected human liver cell line Huh7 (Fig. 2E to H).
FIG. 2.
Variation in the expression of GFP reveals the toxic effect of delta protein in transfected avian cells. Cultures of LMH cells (A to D) and Huh7 cells (E to H) were cotransfected with fixed amounts of plasmids pGG119 and pJC110 along with variable amounts of pTW198. The ratios of pTW198 to pJC110 are indicated. At 4 days after transfection, fluorescence microscopy, coupled with a charge-coupled device camera, was used to record the levels of GFP fluorescence.
In additional studies with LMH cells, we observed reductions in GFP-positive cells when we expressed the small form of the delta protein but in the absence of pJC110, which expresses the HDV genome. Even the large form of the delta protein (3) and a form with a deletion that inactivated the dimerization domain (18) were able to produce such reductions (data not shown).
These data, along with other observations presented subsequently, support the interpretation that expression of delta protein in avian cells has a toxic effect, leading to death of the cells and their release from the monolayer culture into the medium.
Detection of HDV genome replication in avian cells in the presence of reduced amounts of small delta protein expression.
As part of the experiment described above, we harvested the cells at 4 days after transfection and isolated the RNA and protein. Northern analyses were used to assay for the accumulation of HDV antigenomic RNA (Fig. 3A) and genomic RNA (Fig. 3B). Immunoblots were used to assay for delta protein (Fig. 3C). As previously reported, the nature of the construct pJC110 is such that DNA-directed RNA transcription can make genomic RNA transcripts that may be processed by their two copies of the genomic ribozyme to make unit-length genomic RNAs, which may in turn be further processed to produce circular RNAs (17). We expect that when the small delta protein is provided in trans, there can be RNA-directed RNA transcription leading to increased accumulation of unit-length genomic RNAs together with the appearance of unit-length antigenomic RNAs. This accumulation of processed antigenomic RNAs is thus diagnostic of RNA-directed RNA synthesis.
Consistent with the above explanation, antigenomic RNA accumulation was detected in Huh7 cells in the presence of small delta protein (Fig. 3A, lanes 7 to 9) but not in its absence (lane 6). A similar transfection of LMH cells is shown in lanes 1 to 5. Figure 3A, lane 1, shows that there was no antigenomic RNA accumulation in the absence of small delta protein. The signal was indistinguishable from that of untransfected cells (lane 5). However, we also examined cells transfected with decreasing amounts of pTW198 to express the delta protein (lanes 2 to 4). At 0.3:1 (lane 3) and 0.03:1 (lane 4), the lower amount corresponding to a less toxic effect, we detected signals of antigenomic RNA. Our interpretation is that the signals in lanes 3 and 4 (which correspond to <2% of the signal seen in transfected Huh7 cells) represent RNA-directed replication. Relative to untransfected cells (lane 5), no HDV-specific signal was detected in lane 2, and we consider this to be largely the consequence of the cytotoxicity of the larger amount of small delta protein provided in trans.
In the above Northern analyses to detect antigenomic HDV RNA in transfected avian cells, we initially had problems in that the low signals were relatively close to the background levels of hybridization. In addition, even for untransfected cells, there was also a discrete band at about the same location as HDV RNA, which we consider a cross-reaction with abundant 1.8-kb small rRNA. As described in Materials and Methods, a modification to the Northern analysis procedure was developed that significantly reduced these problems. (This new approach is also used in Fig. 5A.)
Figure 3B shows the corresponding analysis to detect genomic RNA. Again this was detected for LMH in lanes 3 and 4, and now a signal was also detected in lane 2. The unit-length genomic RNA was more abundant than the corresponding antigenomic RNA, but since such RNAs could arise by the processing of both RNA- and DNA-directed transcripts, the assay was not diagnostic for genome replication. In fact, in Fig. 3B, lane 1, we detected relatively large amounts of unit-length genomic RNA accumulated in LMH cells in the total absence of delta protein; thus, we infer that in this case, all HDV RNA transcription was DNA directed. (Somehow, this level of processing and accumulation was not achieved in the Huh7 cells under the same conditions [Fig. 3B, lane 6]. Apparently LMH cells differ from mammalian cells in that the requirement for delta protein in either HDV RNA processing [13] or stabilization of processed transcripts [17] was less stringent.) Note in Fig. 3B, lanes 2 to 4, that the accumulation of unit-length genomic RNA in LMH cells increased as the amount of cotransfected plasmid expressing small delta protein decreased. Again, this observation supports the interpretation of significant toxicity associated with expression of the delta protein.
The immunoblot analyses to detect delta protein provided independent evidence of the induced death of transfected LMH cells. As we decreased the amount of pTW198 used in the cotransfection by 3- and 33-fold, we did not detect a corresponding decrease in the amount of expressed protein per culture. In Fig. 3C, consider lanes 2 to 4, which correspond to 1:1, 0.3:1, and 0.03:1 ratios of pTW198, respectively. Since lanes 2 to 4 indicate roughly similar amounts of delta protein per sample, we can deduce that after transfection with 33 times more plasmid per culture (lane 2 versus lane 4), there is no increase in the amount of delta protein detected; we interpret this as a consequence of loss of transfected cells due to toxicity. In contrast to this, for the cotransfected Huh7 cells, the amount of expressed protein increased as the amount of transfected pTW198 increased (Fig. 3C, lanes 7 to 9).
We deduce that relative to an amount of small delta protein that causes what we interpret as toxicity in LMH cells, a 33-times-greater amount had no detectable effect in Huh7 cells. Thus, the above studies support the preliminary interpretation that toxicity is the major block for HDV genome replication in LMH cells.
Detection of HDV genome replication in avian cells in the presence of tetracycline-controlled expression of small delta protein.
As an additional strategy for reducing cytotoxic effects in avian cells while encouraging HDV genome replication, we made use of a more controlled expression of the small delta protein. To do this, the coding region of the small delta protein was first inserted into a TET-off vector to produce pPB106. For this vector, DNA-directed RNA transcription is suppressed in the presence of tetracycline. Cotransfections of LMH and Huh7 cells were carried out using this construct along with pJC110, to express a mutant genomic RNA, and pGG119, to express GFP. Following cotransfection, expression of small delta protein was suppressed by the presence of tetracycline (1 μg/ml) in the growth medium. Then, at day 1, the tetracycline was removed for 0.5, 3, or 5 days. Finally, at day 6, cells were harvested and analyses were made, as before, for antigenomic RNA, genomic RNA, and delta protein, with the results as shown in Fig. 4. Note that antigenomic RNA (Fig. 4A), which is diagnostic of genome replication, was detected in all the transfected LMH cells (lanes 3 to 5), although the amount obtained was still at least 50 times less than achieved for comparable cotransfections of Huh7 cells (lanes 7 to 9).
Even under these conditions of controlled expression of delta protein, we detected what is interpreted as toxicity in LMH but not in Huh7 cells. Specifically, in Huh7 cells, as the time period in the absence of tetracycline was increased, we detected more delta protein and more genomic and antigenomic RNAs (lanes 7 to 9). This was all as expected. In contrast, in LMH cells, the increased expression time in the absence of tetracycline led to some decreases in the amounts of delta protein and of the HDV RNAs. Our interpretation is that some toxicity was associated with even transient expression of the delta protein in LMH cells. The amount of toxicity was nevertheless much lower than detected in Fig. 2, in that we were unable to detect any major difference in the expression of GFP by fluorescence microscopy at day 6 (data not shown).
Suppression of cell killing using an antiapoptotic agent.
The above studies suggest that expression of delta protein in the avian cell line was associated with cell death. We reasoned that if this death occurred via apoptosis, then it might be feasible to suppress it with a known antiapoptotic agent (25–27). Furthermore, if such suppression were achieved, then the extent of HDV genome replication might be enhanced.
The experimental design was largely as for Fig. 3. Cotransfections were done with different amounts of plasmid pTW198, which expresses the delta protein. In addition, we also tested the effect of adding Z-VAD-fmk (40 μM), beginning 1 h prior to transfection and extending to the end of the experiment. At various times after cotransfection, we monitored the cultures by fluorescence microscopy for the expression of GFP. It was readily apparent that Z-VAD-fmk greatly suppressed the cytotoxic effects associated with delta protein expression, especially for the 1:1 ratio, in LMH cells. As judged by the GFP signal, cell death was substantially reduced, although it was not eliminated (data not shown).
As before, at 4 days after cotransfection, we analyzed HDV antigenomic RNA, genomic RNA, and delta protein, with the results as shown in Fig. 5A to C, respectively.
Consider first the accumulation of delta protein in the LMH cells (Fig. 5C, lanes 4 to 9). For each of the three amounts of delta protein construct transfected, the additional treatment with Z-VAD-fmk produced a dramatic increase in the amount of delta protein accumulated per culture. Lanes 4, 6, and 8 contain much less than the corresponding treated samples in lanes 5, 7, and 9, respectively. More specifically, lane 4, which corresponds to cells transfected with the highest amount, 1:1, the treatment with Z-VAD-fmk caused an eightfold increase in the amount of accumulated protein (lane 5). Our interpretation is that Z-VAD-fmk suppressed what would otherwise be a loss of protein per culture due to cell death.
The treatments with Z-VAD-fmk increased the amounts not only of accumulated delta protein but also of genomic RNA (Fig. 5B, lanes 4 to 9).
When we assayed the RNA for antigenomic species, we were now able to detect, in addition to the unit-length RNA, a species of about 1 kb, which is the delta protein mRNA produced by the plasmid pTW198 (Fig. 5A). The amounts of this species were significantly increased by Z-VAD-fmk treatment (as seen in lanes 5 and 7). In contrast, the amounts of unit-length antigenomic RNA, which are diagnostic of RNA-directed RNA synthesis, showed only modest changes due to Z-VAD-fmk treatment. In lanes 8 and 9, at 0.03:1, and lanes 6 and 7, at 0.3:1, we detected about a twofold increase. For lane 5, at 1:1 and with Z-VAD-fmk treatment, we saw, for the first time with this amount of delta protein, a detectable level of genome replication.
Our interpretation of these experiments with LMH cells is that treatment with the antiapoptotic compound Z-VAD-fmk had a major effect on what we have described as delta protein-associated killing. We thus infer that the cell killing probably occurred via apoptosis. Consistent with this interpretation, we found that Z-VAD-fmk treatment of transfected Huh7 cells had no effect on the accumulation of antigenomic RNA, genomic RNA, or delta protein, even at the highest ratio, 1:1, of the plasmid expressing delta protein (Fig. 5A to C, lanes 11 and 12).
In the above studies, Z-VAD-fmk treatment clearly suppressed the obvious symptoms of what we have interpreted as LMH cell killing (as judged by quantitation of GFP-positive cells and accumulation of DNA-directed transcripts and of delta protein). Nevertheless, we were still unable to achieve levels of HDV genome replication comparable to what can be achieved in Huh7 cells. One explanation might be that there was still some less obvious toxic effect(s) of small delta protein expression. A second interpretation might be that these avian cells, for reasons independent of any toxic effects, might be unable to efficiently replicate the HDV genome.
The most important finding from these studies with the inhibitor is that they provide evidence, albeit indirect, that the cytotoxic effect of delta protein in LMH cells is associated with apoptosis.
DISCUSSION
The present studies make clear that independent of genome replication, expression of delta protein produced toxicity in the chicken LMH cell line. Not only the small form of the delta protein did this. Toxicity was also observed with the large form and with a deleted version of the small form that is known to be unable to make dimers (18). Three strategies enabled us to reduce this toxicity and go on and detect, for the LMH cells, low levels of HDV genome replication: (i) the overall expression of delta protein was reduced 33-fold; (ii) the expression was made only transient by use of a tetracycline-controlled promoter; and (iii) we used an anticaspase agent, Z-VAD-fmk, to suppress the toxic effect. Since this anticaspase agent is known to be antiapoptotic (25–27), we infer that the toxic effect of the delta protein involved induction of apoptosis.
In other studies we used the quail tumor cell line QT-6 (19). As with the LMH cells, we observed toxicity induced by expression of the delta protein that could be blocked by the antiapoptoic agent. However, in contrast to the LMH results, we were unable to find any conditions under which we could detect the accumulation of even trace amounts of antigenomic RNAs, indicative of RNA-directed RNA synthesis and processing (data not shown). These negative results are in agreement with unpublished studies by Don Ganem (personal communication).
Unlike the toxicity in avian cells, we detected no such effect in human Huh7 cells. In this respect our results are compatible with those of Guilhot et al., who made mice transgenic for both the small and large delta proteins and saw no toxic effects (10). Nevertheless, our results with the avian cells, in which the toxic effects were so extensive, make us more receptive to the possibility that under certain other conditions of expression in mammalian cells there may be toxic effects. For example, Cole et al. expressed a series of increasing amounts of the small delta protein in human HeLa and HepG2 cells and saw effects on both cell growth and, at the highest amounts, cell toxicity (5). Maybe even in the liver of an HDV-infected human, overexpression of small delta protein could have cytostatic and/or cytotoxic effects. This may be a significant part of the morbidity and mortality associated with HDV infection (7). This possibility leads us to speculate that a patient with a fulminating HDV infection might profit from infusion with an antiapoptotic agent such as Z-VAD-fmk.
In our studies we were able to get low levels of HDV genome replication in the LMH cells. There is no question that to get even such low levels of replication in LMH cells (only 2% relative to replication in Huh7 cells) the virus has to achieve a precarious balance; the small protein has many roles which make it essential for the support of genome replication, and yet at the same time, expression of this protein is toxic to the cell. Perhaps this balance can only be achieved in LMH cells when replication is 50 times less than in Huh7 cells. (Maybe it cannot be achieved at all in QT-6.) Alternatively, there may be a factor(s) other than toxicity that limits genome replication. For example, in previous studies with mutagenesis of the HDV genome, we found that in many cases what seemed to be a small change in the HDV genome could reduce the ability of that genome to replicate and accumulate in transfected Huh7 cells by 100-fold (30). Thus, it should not be unreasonable to suggest that during evolution, numerous “small genetic differences” between avian and mammalian cells might also have a major impact on HDV replication. It should be noted that we did find that avian cells were able to process nonreplicating DNA-directed HDV multimeric RNA transcripts, to make unit-length linear and circular species, just as well as mammalian cells. Furthermore, there was no indication that the HDV RNA species produced in the avian cells were subsequently less stable than those produced in mammalian cells (Fig. 1). In fact, it was striking that in the total absence of the delta protein, the nonreplicating processed HDV genomic RNA accumulated to greater amounts in LMH than in Huh7 cells (Fig. 3B, lanes 1 and 6).
Finally, we consider that our results may have implications for both prior (8, 9, 24) and any future attempts to achieve a switch for HDV from its replication in mammalian cells using a mammalian hepadnavirus as helper to replication in ducklings in the presence of duck HBV as helper. If our results with the chicken and quail cell lines are an indicator of what happens in the duck, then initiation of genome replication might be either absent or at too low a level to be of use in the spread of the virus within the liver.
ACKNOWLEDGMENTS
This work was supported by grants AI-26522 and CA-06927 from the NIH and by an appropriation from the Commonwealth of Pennsylvania.
Jon Boyd and the Microscopy Facility assisted with the fluorescence imaging. We thank Werner Paulus for the TET vector and Preetha Biswas for the derived construct, pPB106. Thanks to Don Ganem for his personal communication regarding the QT-6 cells. Thanks to Ting-Ting Wu for preliminary experiments. Special thanks to Ju-Tao Guo and Christoph Seeger for experimental suggestions. Editorial comments were given by Glenn Rall, William Mason, and Severin Gudima.
REFERENCES
- 1.Bichko V V, Taylor J M. Redistribution of the delta antigens in cells replicating the genome of hepatitis delta virus. J Virol. 1996;70:8064–8070. doi: 10.1128/jvi.70.11.8064-8070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chao M. Ph.D. thesis. Philadelphia: University of Pennsylvania; 1991. [Google Scholar]
- 3.Chao M, Hsieh S-Y, Taylor J. Role of two forms of the hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J Virol. 1990;64:5066–5069. doi: 10.1128/jvi.64.10.5066-5069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen P-J, Kalpana G, Goldberg J, Mason W, Werner B, Gerin J, Taylor J. Structure and replication of the genome of hepatitis δ virus. Proc Natl Acad Sci USA. 1986;83:8774–8778. doi: 10.1073/pnas.83.22.8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole S, Gowans E J, Macnaughton T B, Hall P M, Burrell C J. Direct evidence for cytotoxicity associated with expression of hepatitis delta virus antigen. Hepatology. 1991;13:845–851. [PubMed] [Google Scholar]
- 6.Cole S M, Macnaughton T B, Gowans E J. Differential roles for HDAg-p24 and -p27 in HDV pathogenesis. In: Hadziyannis S J, Taylor J M, Bonino F, editors. Hepatitis delta virus: molecular biology, pathogenesis, and clinical issues. New York, N.Y: Wiley-Liss; 1993. pp. 131–138. [PubMed] [Google Scholar]
- 7.Fattovich G, Giustina G, Christensen E, Pantalena M, Zagni I, Realadi G, Schalm S W. Influence of hepatitis delta virus infection on morbidity and mortality in compensated cirrhosis type B. Gut. 2000;46:420–426. doi: 10.1136/gut.46.3.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forzani B, Rapicetta M, Smedile A, Hele C, Morace G, Di Rienzo A M, Buonavoglia C, Avanzini L, Gerin J L, Verme G, Ponzetto A. Delta virus transmission to the pekin duck. J Med Virol. 1987;21:41A. [PubMed] [Google Scholar]
- 9.Frieman J, Williams G, Dimitrakis M, Holmes M, Cossart Y. Transmission of hepatitis delta virus to the pekin duck. J Med Virol. 1987;21:42A. [Google Scholar]
- 10.Guilhot S, Huang S-N, Xia Y-P, La Monica N, Lai M M C, Chisari F V. Expression of hepatitis delta virus large and small antigens in transgenic mice. J Virol. 1994;68:1052–1058. doi: 10.1128/jvi.68.2.1052-1058.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang S B, Park K J. Cell cycle arrest mediated by hepatitis delta antigen. FEBS Lett. 1999;449:41–44. doi: 10.1016/s0014-5793(99)00394-4. [DOI] [PubMed] [Google Scholar]
- 12.Hwang S B, Park K J, Kim Y S. Overexpression of hepatitis delta antigen protects insect cells from baculovirus-induced cytolysis. Biochem Biophys Res Commun. 1998;244:652–658. doi: 10.1006/bbrc.1998.8317. [DOI] [PubMed] [Google Scholar]
- 13.Jeng K-S, Su P-Y, Lai M M C. Hepatitis delta antigen enhances the ribozyme activities of hepatitis delta virus RNA in vivo. J Virol. 1996;70:4205–4209. doi: 10.1128/jvi.70.7.4205-4209.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawaguchi T, Nomura K, Hirayama Y, Kitagawa T. Establishment and characterization of a chicken hepatocellular carcinoma cell line, LMH. Cancer Res. 1987;47:4460–4464. [PubMed] [Google Scholar]
- 15.Kuo M Y-P, Chao M, Taylor J. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J Virol. 1989;63:1945–1950. doi: 10.1128/jvi.63.5.1945-1950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai M M C. The molecular biology of hepatitis delta virus. Annu Rev Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 17.Lazinski D W, Taylor J M. Expression of hepatitis delta virus RNA deletions: cis and trans requirements for self-cleavage, ligation, and RNA packaging. J Virol. 1994;68:2879–2888. doi: 10.1128/jvi.68.5.2879-2888.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazinski D W, Taylor J M. Relating structure to function in the hepatitis delta virus antigen. J Virol. 1993;67:2672–2680. doi: 10.1128/jvi.67.5.2672-2680.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moscovici C, Moscovici M, Jiminez H, Lai M M C, Hayman M J, Vogt P K. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell. 1977;11:95–103. doi: 10.1016/0092-8674(77)90320-8. [DOI] [PubMed] [Google Scholar]
- 20.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 21.Netter H J, Kajino K, Taylor J. Experimental transmission of human hepatitis delta virus to the laboratory mouse. J Virol. 1993;67:3357–3362. doi: 10.1128/jvi.67.6.3357-3362.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulus W, Baur I, Boyce F M, Breakefield X O, Reeves S A. Self-contained, tetracycline-regulated retroviral vector system for gene delivery to mammalian cells. J Virol. 1996;70:62–67. doi: 10.1128/jvi.70.1.62-67.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponzetto A, Cote P J, Popper H, Hoyer B H, London W T, Ford E C, Bonino F, Purcell R H, Gerin J L. Transmission of the hepatitis B virus-associated δ agent to the eastern woodchuck. Proc Natl Acad Sci USA. 1984;81:2208–2212. doi: 10.1073/pnas.81.7.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponzetto A, Rapicetta M, Forzani B, Smedile A, Hele C, Morace G, di Rienzo A M, Palladina P, Gerin J L, Verme G. Hepatitis delta virus infection in Pekin ducks chronically infected by the duck hepatitis B virus. Prog Clin Biol Res. 1987;234:47–49. [PubMed] [Google Scholar]
- 25.Pronk G J, Ramer K, Amiri P, Williams L T. Requirement of an ICE-like protease for induction of apoptosis and ceramide generation by REAPER. Science. 1996;271:808–810. doi: 10.1126/science.271.5250.808. [DOI] [PubMed] [Google Scholar]
- 26.Slee E A, Zhu H, Chow S C, MacFarlane M, Nicholson D W, Cohen G M. Benzoylcarbonyl-Val-Ala-Asp (OMe) fluoromethylketone (Z-VAD.FMK) inhibits apoptosis by blocking the processing of CPP32. Biochem J. 1996;315:21–24. doi: 10.1042/bj3150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart S A, Poon B, Song J Y, Chen I S Y. Human immunodeficiency virus type I Vpr induces apoptosis through caspase activation. J Virol. 2000;74:3105–3111. doi: 10.1128/jvi.74.7.3105-3111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor J. HDV as a precedent for RNA-directed RNA synthesis by RNA polymerase II. In: Dinter-Gottlieb G, editor. The unique hepatitis delta virus. R. G. Austin, Tex: Landes, Co.; 1995. pp. 47–54. [Google Scholar]
- 29.Taylor J M. The structure and replication of hepatitis delta virus. Annu Rev Microbiol. 1992;46:253–276. doi: 10.1146/annurev.mi.46.100192.001345. [DOI] [PubMed] [Google Scholar]
- 30.Wu T-T, Netter H J, Lazinski D W, Taylor J M. Effects of nucleotide changes on the ability of hepatitis delta virus to transcribe, process, and accumulate unit-length, circular RNA. J Virol. 1997;71:5408–5414. doi: 10.1128/jvi.71.7.5408-5414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]