Abstract
Background:
Tumor-only genomic profiling is an important tool in therapeutic management of men with prostate cancer. Since clinically actionable germline variants may be reflected in tumor profiling, it is critical to identify which variants have a higher risk of being germline in origin to better counsel patients and prioritize genetic testing.
Objective:
To determine when variants found on tumor-only sequencing of prostate cancers should prompt confirmatory germline testing.
Design, setting, and participants:
Men with prostate cancer who underwent both tumor and germline sequencing at Memorial Sloan Kettering Cancer Center from January 1, 2015 to January 31, 2020 were evaluated.
Outcome measurements and statistical analysis:
Tumor and germline profiles were analyzed for pathogenic and likely pathogenic (“pathogenic”) variants in 60 moderate- or high-penetrance genes associated with cancer predisposition. The germline probability (germline/germline + somatic) of a variant was calculated for each gene. Clinical and pathologic factors were analyzed as potential modifiers of germline probability.
Results and limitations:
Of the 1883 patients identified, 1084 (58%) had a somatic or germline pathogenic variant in one of 60 cancer susceptibility genes, and of them, 240 (22%) had at least one germline variant. Overall, the most frequent variants were in TP53, PTEN, APC, BRCA2, RB1, ATM, and CHEK2. Variants in TP53, PTEN, or RB1 were identified in 746 (40%) patients and were exclusively somatic. Variants with the highest germline probabilities were in PALB2 (69%), MITF (62%), HOXB13 (60%), CHEK2 (55%), BRCA1 (55%), and BRCA2 (47%), and the overall germline probability of a variant in any DNA damage repair gene was 40%. Limitations were that most of the men included in the cohort had metastatic disease, and different thresholds for pathogenicity exist for somatic and germline variants.
Conclusions:
Of patients with pathogenic variants found on prostate tumor sequencing, 22% had clinically actionable germline variants, for which the germline probabilities varied widely by gene. Our results provide an evidenced-based clinical framework to prioritize referral to genetic counseling following tumor-only sequencing.
Patient summary:
Patients with advanced prostate cancer are recommended to have germline genetic testing. Genetic sequencing of a patient’s prostate tumor may also identify certain gene variants that are inherited. We found that patients who had variants in certain genes, such as ones that function in DNA damage repair, identified in their prostate tumor sequencing, had a high risk for having an inherited cancer syndrome.
Keywords: Advanced prostate cancer, DNA damage repair genes, Germline testing, Tumor-only sequencing
1. Introduction
Tumor sequencing is often performed for men with prostate cancer, particularly to guide therapeutic decisions in advanced disease [1]. Genetic variants in certain DNA damage repair (DDR) genes predict response to poly (ADP-ribose) polymerase inhibitors, and variants can serve as eligibility criteria for clinical trials [2–4]. However, variants identified on tumor-only sequencing without a matched germline subtracted may be acquired (somatic) or inherited (germline) in origin. Most commercial tests are tumor-only sequencing and do not detect whether a variant’s origin is germline or somatic. Identification of the origin of a pathogenic variant found in tumor-only sequencing has important clinical implications for patients and their at-risk family members that go beyond therapeutic actionability. For example, germline BRCA2 or PALB2 pathogenic variants are associated with a high risk of prostate, pancreatic, and male breast cancers, as well as a high risk of breast and ovarian cancers in women [5,6]. For these reasons, the National Comprehensive Cancer Network (NCCN) guidelines recommend referral for germline testing for any variant in the germline upon tumor-only testing with clinical implications, such as pathogenic variants in BRCA1/2, PALB2, PTEN, RB1, and TP53 [1,7]. This approach ensures that most patients are referred for germline genetic testing. When applied in practice, however, this leads to over-referral, since variants in genes such as PTEN and TP53 are seen in close to 50% of prostate tumors [8].
Referring these patients for confirmatory germline testing not only places a large burden to an already stressed system, but could also unnecessarily worry many patients [9]. Universal germline testing in men with high-risk localized or metastatic prostate cancer is also recommended, but in practice remains a challenge, resulting in many men not undergoing the recommended testing [10,11].
Data on specific germline risks after tumor-only sequencing not only are important to prioritize patients for genetic referral, but also provide information for counseling patients based on their pretest germline genetic risk. Most studies have focused on germline risk after tumor profiling across all tumors [12–14], but the risk of a variant being germline may vary widely not just by gene, but also by tumor type [15]. Yet, no study has evaluated the germline risks of variants found on prostate tumor sequencing. To determine when variants found on tumor-only sequencing should prompt germline testing for men with prostate cancer, we leveraged a cohort of 1883 patients with prostate cancer who underwent both tumor and germline sequencing at our institution. Our objective was to define the probability of identifying a germline variant (germline probability) in clinically significant cancer susceptibility genes via prostate cancer tumor-only sequencing, and to identify clinical or pathologic factors that modify the germline probability.
2. Patients and methods
2.1. Study population
Men with prostate cancer who underwent tumor and germline next-generation sequencing with the United States Drug and Food Administration–approved and New York State–certified Memorial Sloan Kettering-Integrated Variant Profiling of Actionable Cancer Targets (MSK-IMPACT) assay under an institutional review board–approved protocol [16] from January 1, 2015, to January 31, 2020 were identified. Patients were offered MSK-IMPACT as per the judgment of their treating physician. Only data from patients consenting to disclosure of germline results were analyzed. Clinical data were collected from the electronic medical records and genetics visit notes, when available. Race and ancestry were self-reported at the time of patient registration or genetic counseling.
2.2. Somatic and germline sequencing and variant interpretation
Tumor and germline (blood) DNA were sequenced using MSK-IMPACT as previously described [17]. All patients underwent a somatic panel analysis of ≥341 genes and a germline panel of ≥76 genes associated with increased cancer predisposition. Separately, we analyzed 60 genes associated with an increased cancer risk and with established clinical guidelines from the American College of Medical Genetics and Genomics (ACMG) and the NCCN for enhanced cancer screening, presuming that any variant in these genes seen on tumor-only sequencing would be of potential clinical significance and warrants confirmatory germline testing as per current guidelines (Supplementary Table 1) [18]. Somatic variants were curated using the OncoKB platform, and only oncogenic or likely oncogenic variants were included in this analysis [19]. If multiple tumor samples were sequenced from the same patient (primary and metastasis, or two or more metastases), only the first tumor sequenced was analyzed. Germline variants were interpreted according to the ACMG-Association for Molecular Pathology (AMP) criteria by a clinical molecular geneticist or pathologist; only pathogenic or likely pathogenic variants were included in this analysis [20]. Genes were classified as of high (relative risk >4) or moderate (relative risk 2–4) penetrance [16]. For MITF and HOXB13, only the hotspot variants MITF E318K and HOXB13 G84E were analyzed in the germline. Variant alleles of low, uncertain, and recessive penetrance identified in APC c.3920T > A (p. I1307K), CHEK2 c.470T > C (p.I157T), and FH c.1431_1433dupAAA (p.K477dup) were similarly not counted in the germline probability of the respective genes. Biallelic inactivation was defined as either a second somatic variant at the locus of a germline variant or a loss of the wild-type allele (loss of heterozygosity [LOH]) in the tumor using the FACETS algorithm, as described previously [21].
2.3. Data analysis
The primary aim was to identify variants on tumor-only sequencing that should prompt confirmatory germline testing due to a high probability that the variant was germline in origin. A high germline probability was defined as 10% based on historically suggested thresholds [22]. For each gene, germline probability was defined as the number of germline variants divided by the total number of germline and somatic variants.
For the genes in which more than one patient had a germline variant, we assessed whether the likelihood of a germline variant was modified by tumor location (primary tumor vs metastatic site). For mismatch repair (MMR) genes (MSH2, MSH6, PMS2, and MLH1) associated with Lynch syndrome, we assessed whether germline probability was modified by tumor mutation burden (TMB) ≥10, and microsatellite instability (MSI) status (MSI-high vs MSI-indeterminate/stable) using Fisher’s exact test. Tumor MSI profile was determined by MSIsensor scores [23,24]. To explore whether variant allele frequency (VAF), a measure of the proportion of DNA in the tumor specimen that carries the gene variant, could be used to distinguish pathogenic germline variants and somatic variants, we analyzed the distribution of VAF values for germline and somatic variants. Differences in VAF values between germline and somatic variants were calculated using Welch’s two-sample t test.
All p values were corrected for multiple comparisons using the false discovery rate method [25]. All analyses were conducted using R (version 4.1.0).
3. Results
3.1. Cohort characteristics
Of 2370 men with prostate cancer enrolled in a matched tumor-normal sequencing protocol during the study period, 1883 (79.5%) consented to the disclosure of germline results and were included in this study. Patient demographics are listed in Table 1. Tumor sequencing was performed on 1272 (68%) primary prostate tumors and 611 (32%) metastatic lesions. MSI status was evaluable in 1603 tumors, of which 35 (2.2%) were MSI high and 34 (1.8%) were MSI indeterminate.
Table 1 –
Cohort characteristics
| Characteristic | N | N = 1883a |
|---|---|---|
| Age at diagnosis | 1883 | 62 (56, 68) |
| Tumor location | 1883 | |
| Metastasis | 611 (32%) | |
| Primary | 1272 (68%) | |
| Diagnosis Gleason grade | 1578 | |
| ≤6 | 134 (8.5%) | |
| 7 | 537 (34%) | |
| ≥8 | 907 (57%) | |
| Disease stage at time of tumor sequencing | 1858 | |
| Localized | 502 (27%) | |
| Regional nodal involvement | 264 (14%) | |
| Metastatic | 1092 (59%) | |
| Tumor mutation burden | 1741 | 2.6 (1.0, 3.5) |
| ≥10 | 1741 | 64 (3.7%) |
| MSI | 1603 | |
| High | 35 (2.2%) | |
| Indeterminate/stable | 1568 (98%) | |
| Ashkenazi Jewish Heritage | 1521 | 326 (21%) |
| Family history of cancer | 1883 | 1584 (84%) |
| Family history of prostate cancer | 1883 | 711 (38%) |
| Other cancer Dx | 1883 | 347 (18%) |
| Race | 1760 | |
| White | 1516 (86%) | |
| Black | 144 (8.2%) | |
| Asian | 62 (3.5%) | |
| Other | 38 (2.2%) |
Dx = diagnosis; IQR = interquartile range; MSI = microsatellite instability.
Median (IQR); n (%).
3.2. Germline-focused analysis
Of 1883 patients identified, 1084 men (58%) had at least one germline or somatic variant in one of the 60 genes of potential germline significance (Fig. 1). In 240 (13%) men, 255 germline variants were identified; 15 men had germline variants in more than one gene (Supplementary Table 2). The most frequent variants found in the germline were in DDR genes (191/255; 75%), including BRCA2 (n = 74; 3.9% of total study cohort), CHEK2 (n = 48; 2.5%), ATM (n = 24; 1.3%), BRCA1 (n = 11; 0.6%), and PALB2 (n = 11; 0.6%). Fourteen men (0.7%) had germline variants in genes associated with hereditary renal cell carcinoma (RCC) syndromes (eight in MITF, and one each in VHL, FH, FLCN, and SDHA/B/C).
Fig. 1 –
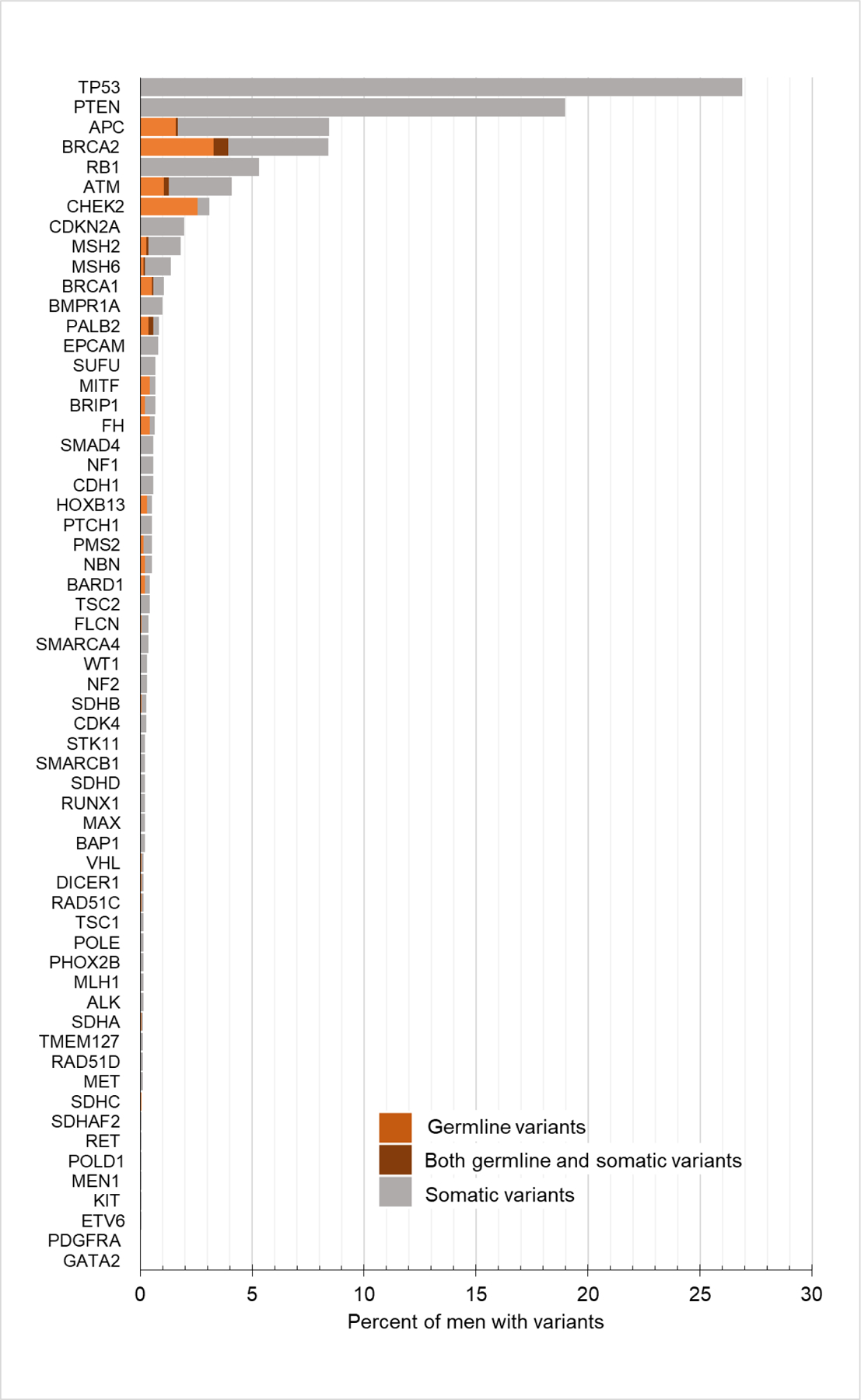
Distribution of germline and somatic variants in men undergoing prostate tumor sequencing.
The overall probability of a variant in a DDR gene being germline was 44% (191 germline variants of 433 total variants); those with the highest germline probability were PALB2 (number of germline variant/number of total variants, 11/16, 69%), CHEK2 (32/58, 55%), BRCA1 (11/20, 55%), BRCA2 (74/158, 47%), NBN (4/10, 40%), RAD51C (1/3, 33%), ATM (24/77, 31%), and BRIP1 (4/13, 33%; Fig. 2). The germline probabilities of DNA MMR genes were 30% (3/10) for PMS2, 21% (7/34) for MSH2, 15% (4/26) for MSH6, and 0% (0/3) for MLH1. The germline probability of a variant in HOXB13, a gene associated with hereditary prostate cancer [26], was 60% overall, but all were in the common hotspot variant HOXB13 G84E. The overall germline probability of a variant in a gene associated with hereditary RCC syndromes was 33% (14/43). The common founder variants BRCA1 c.68_69delAG (p.E23*fs), BRCA1 c.5266dupC (p.Q1756*fs), BRCA2 c.5946delT (p.S1982*fs), CHEK2 c.1283C > T (p.S428F), and APC c.3920T > A (p. I1307K) were identified in 77 (4.1%) patients, and all were germline.
Fig. 2 –
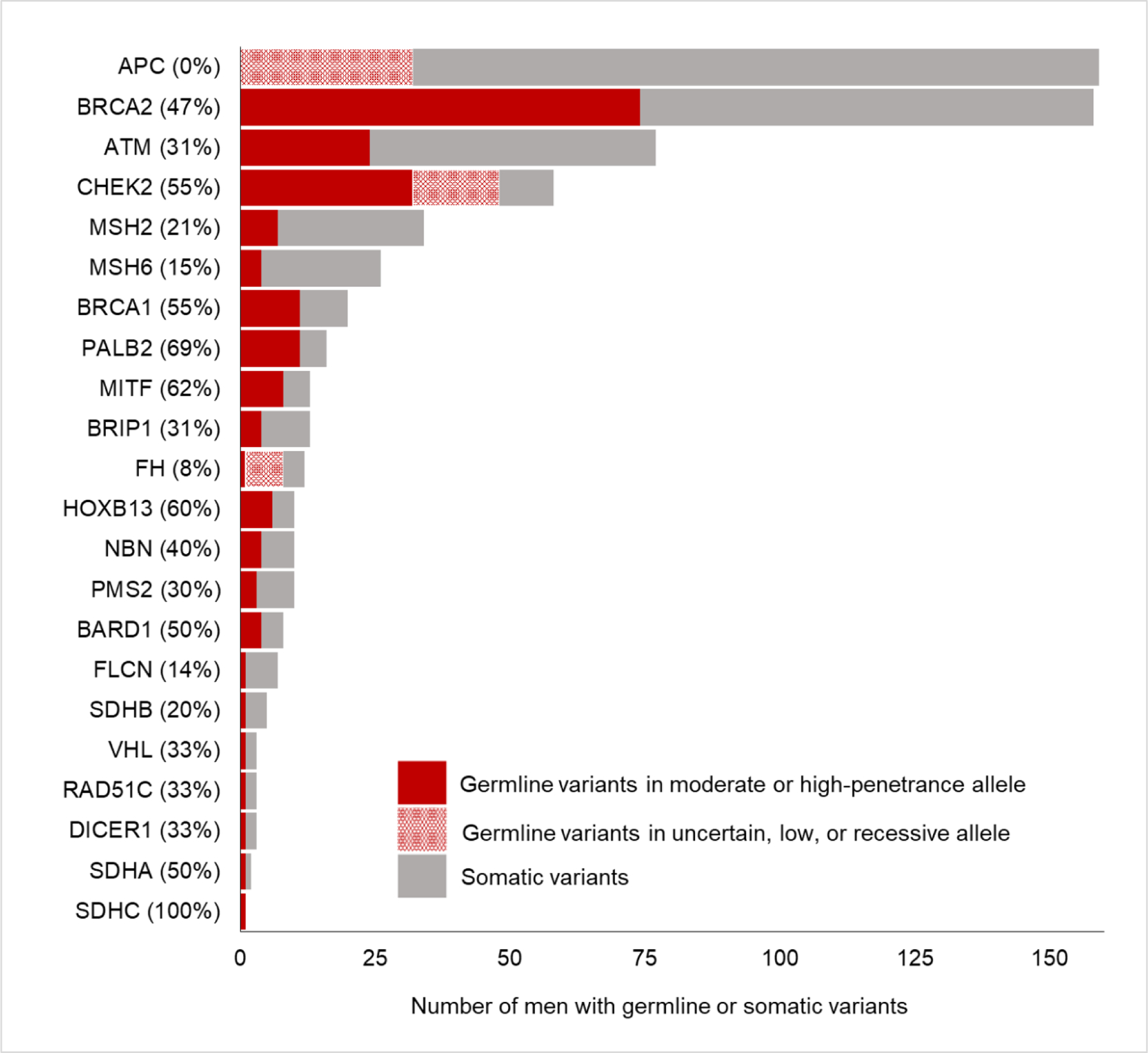
Germline probability in 22 genes in which pathogenic germline variants were identified. Germline variants in uncertain, low, or recessive alleles in APC, CHEK2, and FH were not included in the numerator of the germline probability calculation: APC low-penetrance allele variant c.3920T > A (p.I1307K). CHEK2 variant allele of uncertain penetrance c.470T > C (p.I157T), and FH recessive allele variant c.1431_1433dupAAA (p.K477dup).
Variants in tumor suppressor genes TP53, PTEN, and RB1 were detected in 506 (26.9%), 357 (19.0%), and 100 (5.3%) tumors, respectively, and all were exclusively somatic variants (Fig. 1). APC somatic and germline variants were also common in prostate cancers (159, 8.4%); however, all identified germline variants were low-penetrance I1307K. No high-risk germline APC variants diagnostic of familial adenomatous polyposis were identified.
Consistent with the increasing complexity of metastatic lesions relative to primary prostate cancers was the higher frequency of identifying somatic alterations (69% vs 42%, p < 0.001) [8]. A subanalysis was performed to determine whether germline probability changed with the site of tumor used for sequencing. To do so, we evaluated the germline probabilities stratified by site (primary vs metastatic) sequenced for 13 genes in which more than one patient had a germline variant (Fig. 3). In seven genes (BRCA2, CHEK2, BRCA1, PALB2, MITF, NBN, and PMS2), the germline probabilities of a variant found in a primary tumor were numerically higher than in a metastatic tumor; however, this difference was not statistically significant (Supplementary Table 3).
Fig. 3 –
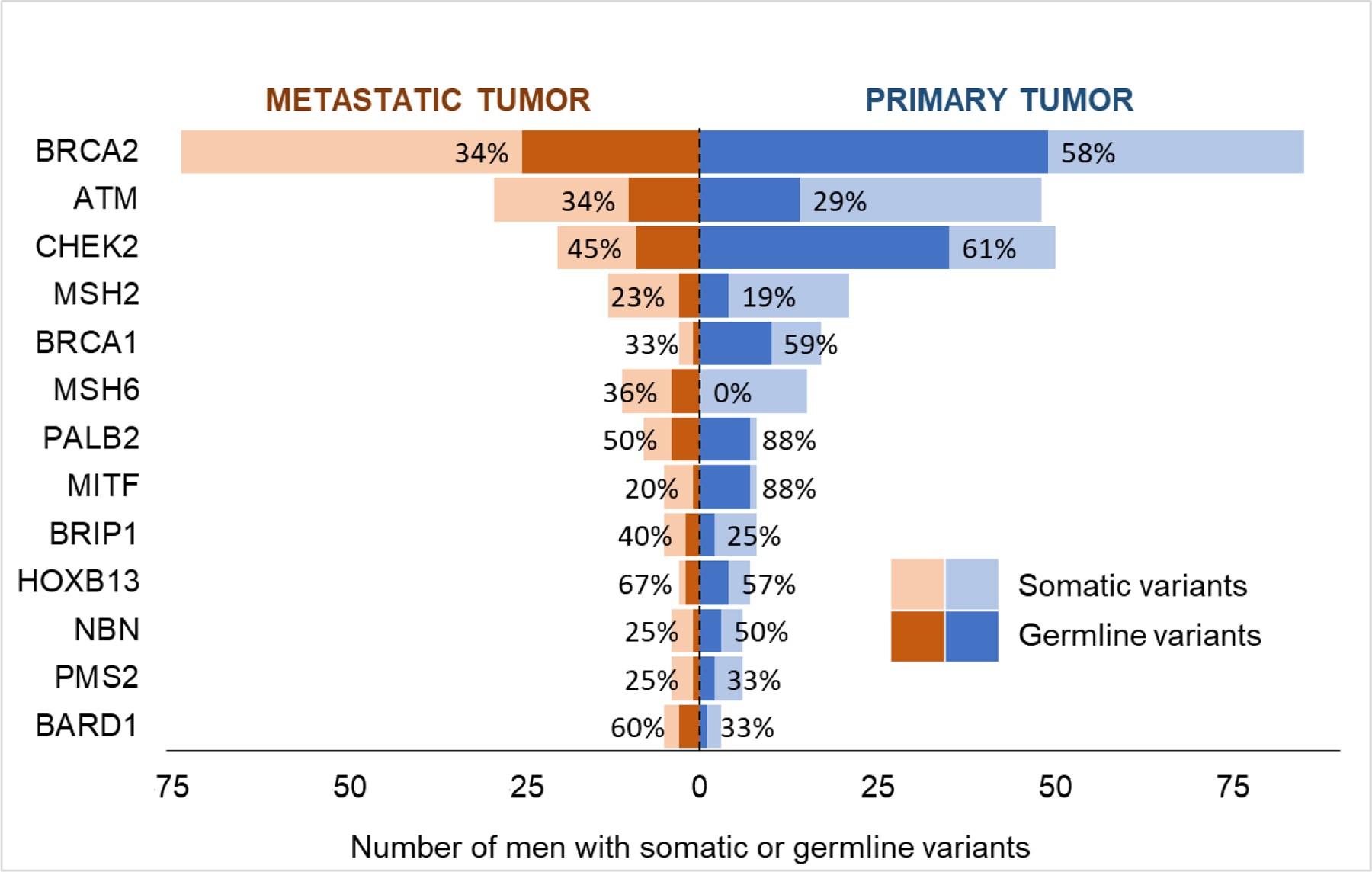
Comparison of germline probability in primary versus metastatic tumor sequencing. Only 13 genes, in which at least two germline variants in moderate- or high-penetrance alleles were identified, were included. Patients who had both germline and somatic variants in a specific gene were classified as having a germline variant only for the purpose of the analysis presented in this figure. Percentages represent the germline probability.
3.3. VAF analysis
To determine whether VAF could be used to distinguish germline from somatic variants, we evaluated the distribution of VAF in tumors for somatic and germline variants (Fig. 4). As expected, the median VAF was 0.50 (interquartile range [IQR] 0.47–0.59) for germline variants and 0.24 (IQR 0.13–0.36) for somatic variants—significantly higher in germline variants than in somatic variants (difference = 0.27, 95% confidence interval 0.23–0.30, p < 0.001). Setting a cutoff of tumor-detected VAF of ≥0.3 would have detected all germline variants and filtered out 63% of somatic variants.
Fig. 4 –
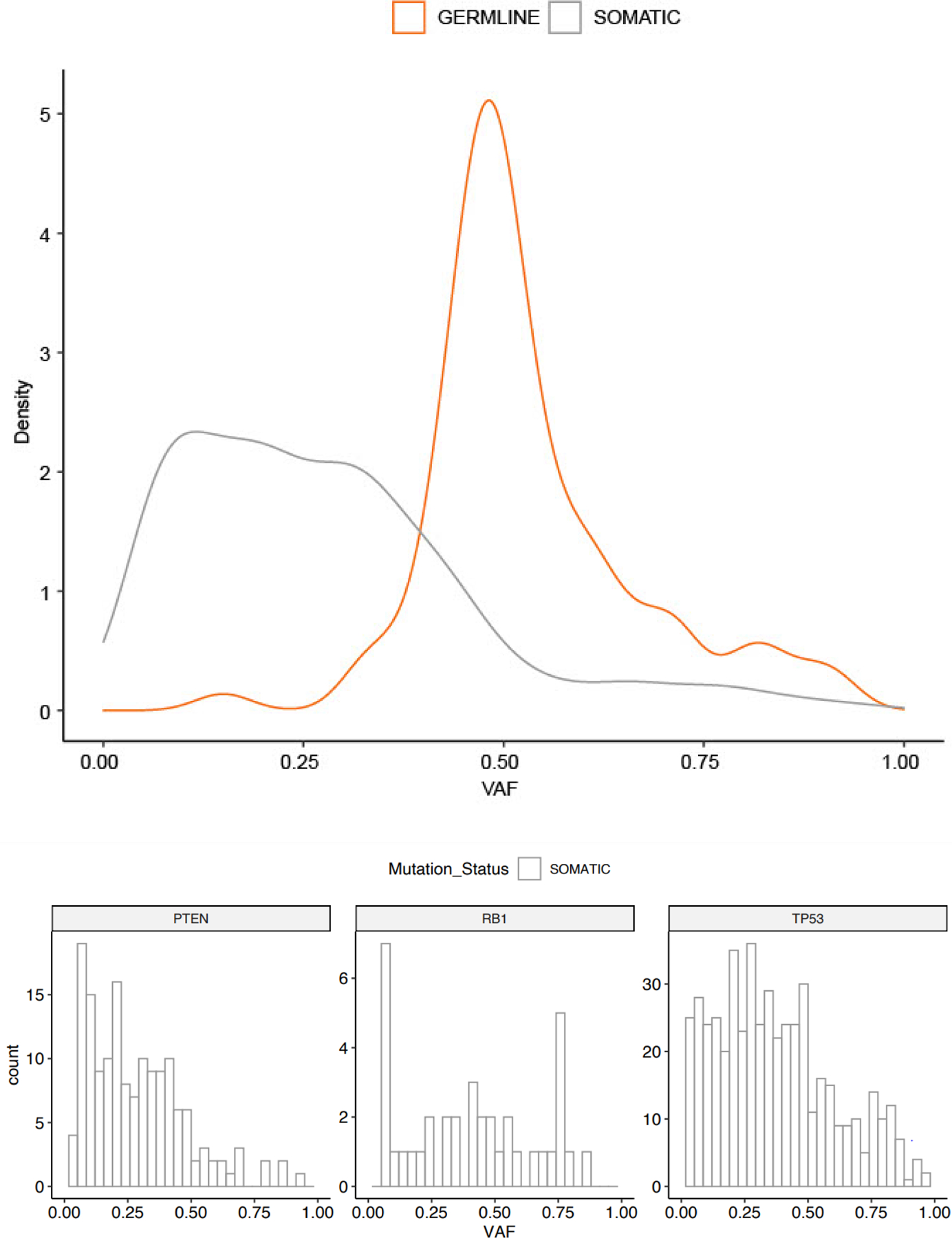
Distribution of variant allele frequency of germline and somatic variants: (A) variants in 22 genes in which both germline and somatic variants were identified and (B) variants in TP53, PTEN, and RB1 in which only somatic variants were identified. VAF = variant allele frequency.
The mean tumor-detected VAF values of variants in TP53, PTEN, and RB1 (all common, somatic-only variants) were 0.37 (standard deviation, 0.23), 0.29 (0.19), and 0.41 (0.26), respectively (Fig. 4). VAF ≥0.5 was detected in 25% of TP53 variants, 12% of PTEN variants, and 39% of RB1 variants.
3.4. TMB and MSI analysis
We then evaluated the TMB and MSI phenotype of tumors of men with germline variants in MMR genes (MSH2, MSH6, and PMS2). Six of 14 (43%) men with MMR germline variants had MSI-high tumors and eight of 12 (67%) men with available TMB score had TMB ≥10, with prevalence varying significantly by gene. Whereas five of the seven (71%) men with germline variants in MSH6 had an MSI-high tumor, only one of four (25%) men with MSH2 germline variants and zero of three men with PMS2 germline variants had MSI-high tumors. Conversely, among men with MSI-high tumors or TMB ≥10, only 6/35 (17%) and 8/64 (13%) had an MMR germline variant. There was no statistically significant difference in the germline probabilities of MMR genes in men with and without TMB ≥10 or MSI-high tumors (Supplementary Table 4). These data suggest that the MSI and TMB phenotypes of prostate tumors in men with tumor-detected MMR variants do not predict germline probabilities.
3.5. Biallelic inactivation
To infer the potential contribution of germline variants in prostate cancer pathogenesis, we evaluated biallelic inactivation through second somatic alteration or LOH in the tumor. Of men with germline variants, 108 had tumors with available LOH status of whom 61 (57%) had biallelic inactivation. Biallelic inactivation in DDR genes was found in 59 (60%) tumors (Fig. 5), with BRCA2 having the highest rate of inactivation at 83% followed by PALB2 at 78%. There was no statistical difference in biallelic loss of DDR genes in primary versus metastatic tumors (25/37 [68%] primary vs 36/64 [56%] metastatic tumors, p = 0.3). No biallelic inactivation was seen in tumors of patients with germline variants in inherited RCC syndrome genes.
Fig. 5 –
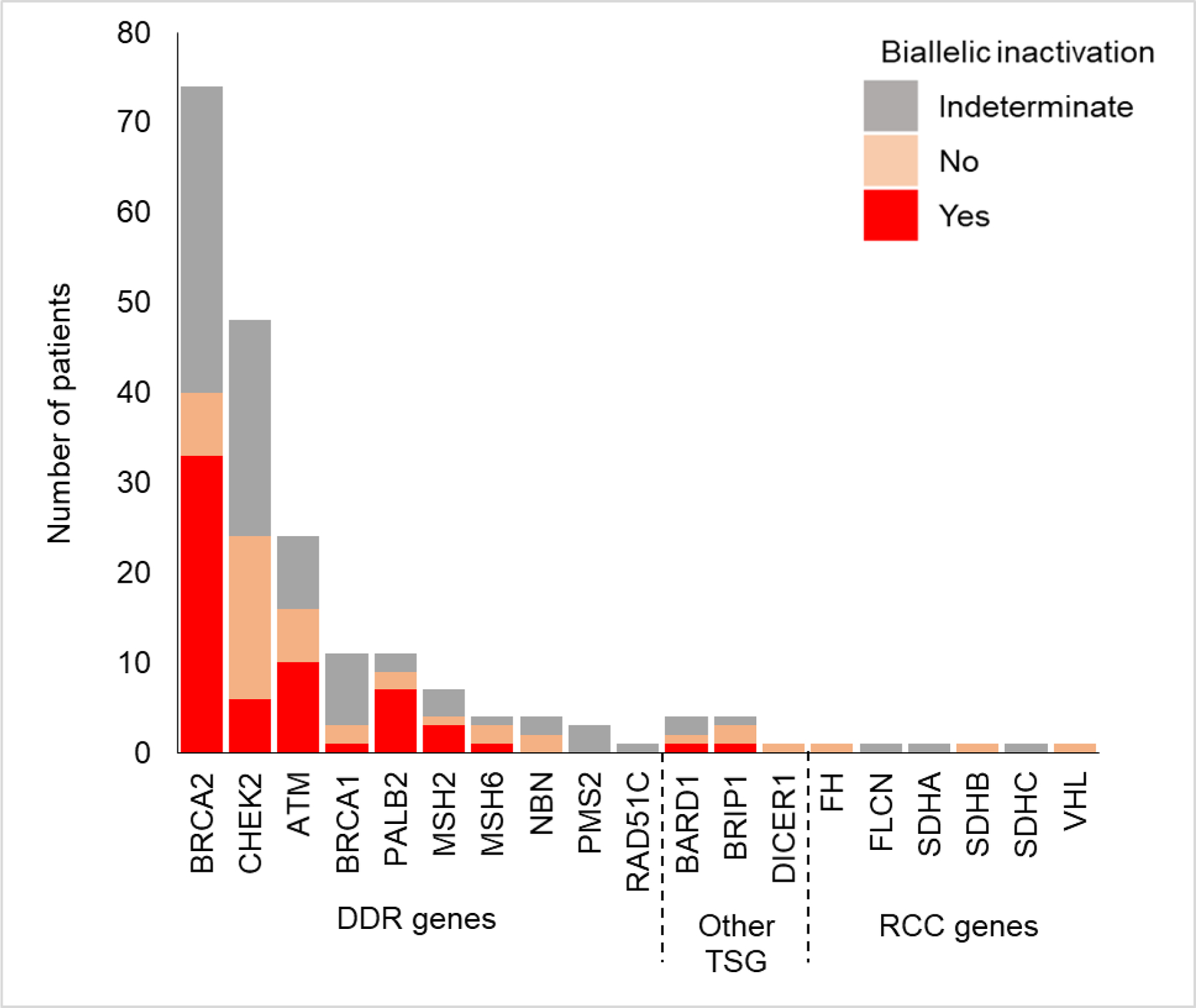
Biallelic inactivation in tumors of patients with germline variants. Biallelic inactivation was evaluated by inferring loss of heterozygosity or the presence of a somatic variant in the same gene in the tumor. DDR = DNA damage repair; RCC = renal cell carcinoma; TSG = tumor suppressor gene.
3.6. Role of personal and family history in germline testing
We examined whether confirmatory germline testing should be restricted to men who met the current NCCN guidelines for germline genetic testing based on a personal history of non–prostate cancer(s) or a family history of cancer. Of 212 men with moderate- or high-penetrance germline variants, 107 (51%) met germline testing criteria based on a family or personal history of an additional primary cancer associated with the respective gene (Supplementary Table 5). Notable, however, is that 14 men with germline variants in genes associated with hereditary RCC syndromes would not have met the criteria for testing.
4. Discussion
In this study of 1883 men who underwent both prostate tumor and germline sequencing, we found that the probabilities of a variant seen in tumor-only testing being germline varied widely by gene, and that clinical or pathologic factors alone did not reliably predict gene-specific germline probabilities. Using this evidence, we provide a framework to identify those at the highest risk of a hereditary syndrome after tumor-only testing (Fig. 6). Overall, 40% of tumor-detected variants in DDR genes were found to be germline in origin, ranging from 15% in MSH6 to 69% in PALB2, surpassing the predetermined 10% probability used historically to indicate clinically meaningful pretest probabilities [22]. The results support confirmatory germline testing for a tumor-detected variant in any DDR gene, not just BRCA1 and BRCA2, and provide evidence for physicians and genetic counselors to educate patients. Conversely, while 40% of tumors had TP53, PTEN, or RB1 variants, all were somatic, representing the potential for a significant number of referrals of men who lack any clinically significant germline variants. Accordingly, we recommend limiting confirmatory testing for these genes to men who meet the personal or family history criteria suggestive of associated cancer syndromes. Finally, we identified variants in genes associated with nonprostate hereditary cancer syndromes, such as VHL, which, although rare, had a high germline probability. These genes are typically not tested in prostate cancer germline panels, making it important for ordering providers to specifically refer these patients for confirmatory germline testing.
Fig. 6 –
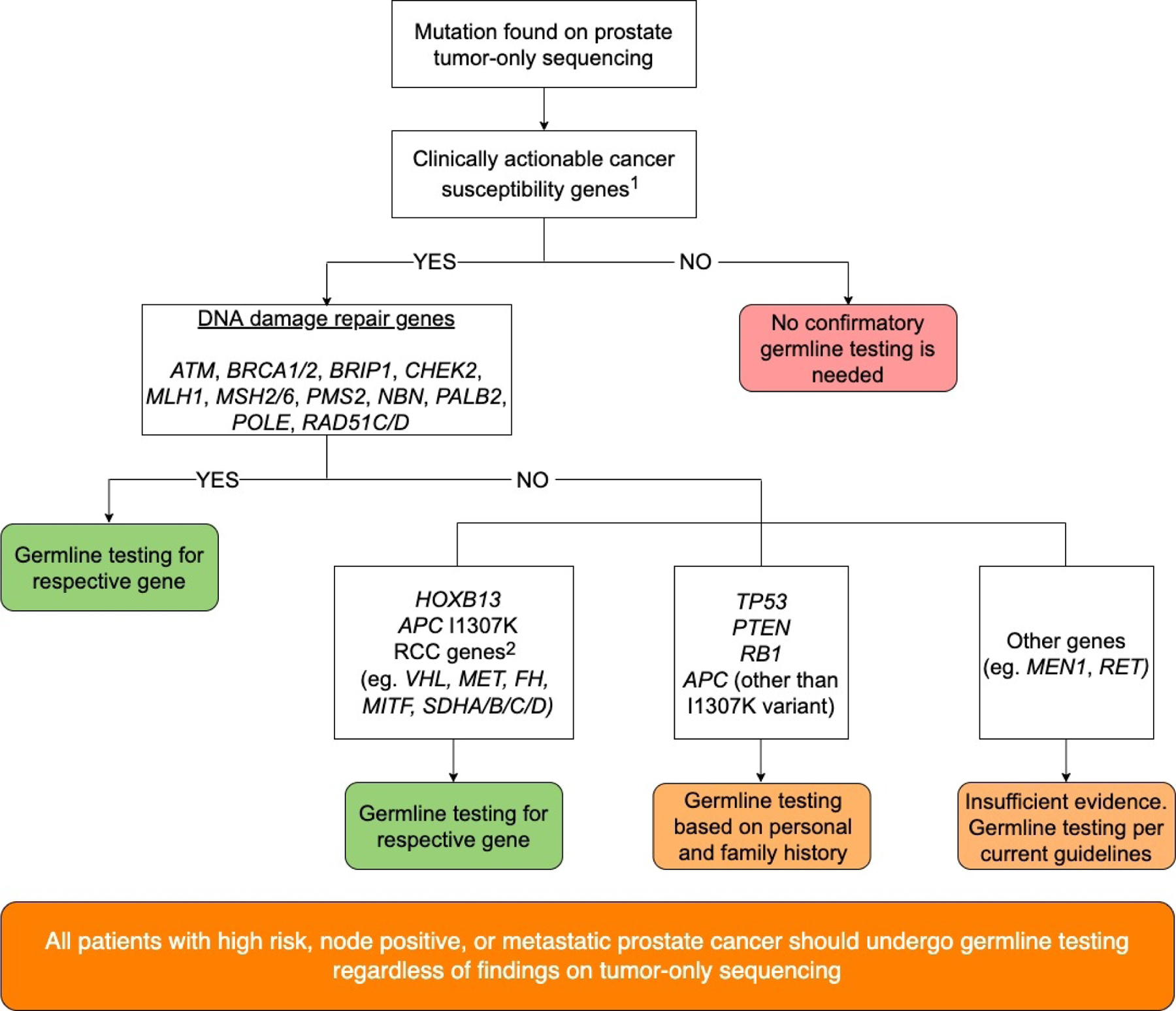
Risk stratification for confirmatory germline genetic testing following prostate tumor-only sequencing. a Clinically actionable genes were defined as cancer susceptibility genes with moderate or high penetrance or those with published medical management guidelines (see Supplementary Table 1). b RCC genes are genes associated with hereditary renal cell carcinoma (RCC) syndromes.
In addition to gene-based probabilities, we investigated clinical and pathologic factors that could further refine germline probabilities. We found that VAF can be informative in screening men for germline variants. In DDR genes and HOXB13, the higher the VAF in the tumor, the more likely the variant to be of germline origin. Nevertheless, there are limitations of establishing a VAF threshold for clinical significance, including the potential for undetected germline mosaicism [27], reliance on the purity of the tumor analyzed [28], and the effect of tumor heterogeneity [29]. Furthermore, even at a VAF of >0.5, which raises concerns for germline origin, variants in TP53, PTEN, and RB1 are still more likely to be somatic. Therefore, we do not recommend routine use of VAF as a screening test for germline testing. Similarly, the site of sequencing (primary or metastatic) had comparable germline probabilities and was not useful as a predictive factor. Interestingly, the presence of an MSI-high tumor phenotype did not affect the germline probabilities of variants in MMR genes. This is an important distinction for therapeutic decision-making since it is the MSI-high phenotype and not the presence of a somatic or germline variant in an MMR-associated gene that determines the likelihood of a response to PD-1 inhibitors. We recommend that if a variant in an MMR gene is found on prostate tumor-only sequencing, patients should undergo germline testing regardless of tumor phenotype.
To explore the contribution of germline variants in prostate tumor pathogenesis, we analyzed tumors for LOH or second somatic variants. We found that the genes most associated with prostate cancer risk (BRCA2, ATM, and PALB2) had a high rate of biallelic loss, whereas other genes such as CHEK2 and BRCA1 had biallelic loss in <50% of tumors. This should be explored in larger studies—for example, the role of BRCA1 or CHEK2 in pathogenesis may be less than previously thought and could have important implications for treatment selection [30,31]. Not surprisingly, in RCC risk genes, we found no LOH, indicating that these are likely unrelated to prostate cancer risk.
Many guidelines recommend universal germline testing for men with high-risk or metastatic prostate cancer, and this study highlights the importance of such testing given the high rate of variants identified, especially in DDR genes. Tumor-only testing remains widespread; however, most eligible men do not undergo germline testing and increasing the awareness of genetic testing remains important [10,11]. Our study offers a risk stratification framework for germline referral specifically for men with prostate cancer. Evidence from this large cohort study can help clinicians best counsel patients who have tumor-only results. In particular, we strongly encourage confirmatory germline testing if a variant in DDR genes such as BRCA2 or PALB2 is present, while reassuring patients with TP53, PTEN, or RB1 alterations in the tumor that the likelihood of an underlying heritable syndrome is low. Clinicians and genetic counselors should also be aware of non–prostate cancer hereditary cancer syndrome variants, such as VHL or FLCN, which can be revealed in tumor-only testing and warrant confirmatory testing.
Other approaches, such as automated germline referral process for patients undergoing tumor-only testing, have been tested in pan-cancer populations and appear feasible [12]. Guidelines also recommend routine confirmatory testing of any variant in tumor-only testing that could be clinically significant if found in the germline [1,7]. Our data suggest that these recommendations, at least for prostate cancer, could be refined to exclude commonly mutated somatic genes and to emphasize the high probability in DDR genes.
This study has several limitations. Classifications used for somatic variants by OncoKB [32] and germline variants by the ACMG-AMP [20] were not uniform. Criteria for classifying germline variants as pathogenic or likely pathogenic were more stringent than those for classifying somatic variants as oncogenic or likely oncogenic. The result may have led to an underestimation of the fraction of germline variants that were classified as pathogenic or likely pathogenic. We also excluded current variants of unknown significance that may have clinical implications in the future. Only single nucleotide variants or small indels were assessed, limiting the interpretation of findings as large structural variants were not included. The study population was from a single tertiary cancer center with a high proportion of patients with Ashkenazi Jewish ancestry (21%) and metastatic disease. Prevalence of germline variants, especially founder variants, may have been influenced by characteristics of this population and may not be generalizable to the overall population. Finally, tumor-only testing may fail to detect germline variants due to somatic events that may mask germline variants, such as deletions that encompass the pathogenic germline variant [15,33,34]. Therefore, patients with a personal or family history suggestive of an inherited variant should undergo guideline-based genetic testing even if the suspected variant was not found on tumor-only sequencing [15].
5. Conclusions
We report an in-depth, germline-focused analysis of tumor sequencing in a large cohort of men with prostate cancer. The probability of a variant being germline in origin varied widely by gene; while DDR genes had a high overall probability, other commonly altered genes, such as TP53, PTEN, and RB1, had a very low probability. These data provide evidence to refine current guidelines for reflex confirmatory germline testing of variants found on tumor-only testing.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial disclosures: Maria I. Carlo certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
In our analysis of men who underwent prostate cancer tumor sequencing, the possibility of a variant being germline in origin varied by gene. Reflex germline testing of DNA damage repair genes should be emphasized given their very high probability of being germline in origin.
References
- [1].National Comprehensive Cancer Network. Prostate cancer (version 2.2021)
- [2].De Bono J, Mateo J, Fizazi K, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med 2020;382:2091–102. [DOI] [PubMed] [Google Scholar]
- [3].Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015;33:244–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Clarke N, Wiechno P, Alekseev B, et al. Olaparib combined with abiraterone in patients with metastatic castration-resistant prostate cancer: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol 2018;19:975–86. [DOI] [PubMed] [Google Scholar]
- [5].Evans DG, Susnerwala I, Dawson J, Woodward E, Maher ER, Lalloo F. Risk of breast cancer in male BRCA2 carriers. J Med Genet 2010;47:710–1. [DOI] [PubMed] [Google Scholar]
- [6].Van Asperen CJ, Brohet RM, Meijers-Heijboer EJ, et al. Cancer risks in BRCA2 families: estimates for sites other than breast and ovary. J Med Genet 2005;42:711–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].National Comprehensive Cancer Network. Genetic/familial high-risk assessment: breast, ovarian, and pancreatic (version 1.2022) [DOI] [PubMed]
- [8].Abida W, Armenia J, Gopalan A, et al. Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making. JCO Precis Oncol 2017;2017:PO.17.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hoskovec JM, Bennett RL, Carey ME, et al. Projecting the supply and demand for certified genetic counselors: a workforce study. J Genet Couns 2018;27:16–20. [DOI] [PubMed] [Google Scholar]
- [10].Loeb S, Li R, Sanchez Nolasco T, et al. Barriers and facilitators of germline genetic evaluation for prostate cancer. Prostate 2021;81:754–64. [DOI] [PubMed] [Google Scholar]
- [11].Loeb S, Byrne N, Walter D, et al. Knowledge and practice regarding prostate cancer germline testing among urologists: gaps to address for optimal implementation. Cancer Treat Res Commun 2020;25:100212. [DOI] [PubMed] [Google Scholar]
- [12].Clark DF, Maxwell KN, Powers J, et al. Identification and confirmation of potentially actionable germline mutations in tumor-only genomic sequencing. JCO Precis Oncol 2019;3:PO.19.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Meric-Bernstam F, Brusco L, Daniels M, et al. Incidental germline variants in 1000 advanced cancers on a prospective somatic genomic profiling protocol. Ann Oncol 2016;27:795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Schrader KA, Cheng DT, Joseph V, et al. Germline variants in targeted tumor sequencing using matched normal DNA. JAMA Oncol 2016;2:104–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mandelker D, Donoghue M, Talukdar S, et al. Germline-focussed analysis of tumour-only sequencing: recommendations from the ESMO Precision Medicine Working Group. Ann Oncol 2019;30:1221–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mandelker D, Zhang L, Kemel Y, et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA 2017;318:825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hampel H, Bennett RL, Buchanan A, Pearlman R, Wiesner GL. A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: referral indications for cancer predisposition assessment. Genet Med 2015;17:70–87. [DOI] [PubMed] [Google Scholar]
- [19].Chakravarty D, Gao J, Phillips SM, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol 2017;2017:PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].National Institute for Health and Care Excellence. Familial breast cancer: classification, care and managing breast cancer and related risks in people with a family history of breast cancer. Clinical guideline [CG164] [PubMed]
- [23].Niu B, Ye K, Zhang Q, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2014;30:1015–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Middha S, Zhang L, Nafa K, et al. Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data. JCO Precis Oncol 2017;2017:PO.17.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 1995;57:289–300. [Google Scholar]
- [26].Ewing CM, Ray AM, Lange EM, et al. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med 2012;366:141–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Batalini F, Peacock EG, Stobie L, et al. Li-Fraumeni syndrome: not a straightforward diagnosis anymore-the interpretation of pathogenic variants of low allele frequency and the differences between germline PVs, mosaicism, and clonal hematopoiesis. Breast Cancer Res 2019;21:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Shin HT, Choi YL, Yun JW, et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat Commun 2017;8:1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].DeLeonardis K, Hogan L, Cannistra SA, Rangachari D, Tung N. When should tumor genomic profiling prompt consideration of germline testing? J Oncol Pract 2019;15:465–73. [DOI] [PubMed] [Google Scholar]
- [30].Li S, Silvestri V, Leslie G, et al. Cancer risks associated with BRCA1 and BRCA2 pathogenic variants. J Clin Oncol 2022;40:1529–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Abida W, Campbell D, Patnaik A, et al. Non-BRCA DNA damage repair gene alterations and response to the PARP inhibitor rucaparib in metastatic castration-resistant prostate cancer: analysis from the phase II TRITON2 study. Clin Cancer Res 2020;26:2487–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].OncoKB curation standard operating procedure, version 2.1 2021. OncoKB.org
- [33].Jonsson P, Bandlamudi C, Cheng ML, et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature 2019;571:576–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Terraf P, Pareja F, Brown DN, et al. Comprehensive assessment of germline pathogenic variant detection in tumor-only sequencing. Ann Oncol 2022;33:426–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


