Abstract
In epithelial cells, planar polarization of subapical microtubule networks is thought to be important for both breaking cellular symmetry and maintaining the resulting cellular polarity. Studies in the Drosophila pupal wing and other tissues have suggested two alternative mechanisms for specifying network polarity. On one hand, mechanical strain and/or cell shape have been implicated as key determinants; on the other hand, the Fat-Dachsous planar polarity pathway has been suggested to be the primary polarizing cue. Using quantitative image analysis in the pupal wing, we reassess these models. We found that cell shape was a strong predictor of microtubule organization in the developing wing epithelium. Conversely, Fat-Dachsous polarity cues do not play any direct role in the organization of the subapical microtubule network, despite being able to weakly recruit the microtubule minus-end capping protein Patronin to cell boundaries. We conclude that any effect of Fat-Dachsous on microtubule polarity is likely to be indirect, via their known ability to regulate cell shape.
INTRODUCTION
The polarity of epithelial cells is essential for their correct function within tissues. There are two principal types of cell polarity—apicobasal and planar, the latter describing the coordinated polarization of cells within the plane of cell sheets (Goodrich and Strutt, 2011; Butler and Wallingford, 2017). One of the manifestations of such polarities is the asymmetry of cytoskeletons such as the microtubule cytoskeleton. Microtubules are polar filaments with highly dynamic plus ends and more stable minus ends. Their preferential directionality within cells leads to biased trafficking of molecules to specific locations by motor proteins moving along individual microtubule filaments (Hirokawa and Takemura, 2005; van Haren and Wittmann, 2019; Masucci et al., 2022). As a result, such directionality of the microtubule cytoskeleton can play roles in both the initial breaking of cellular symmetry as well as the maintenance of polarity once it is established (Shimada et al., 2006; Siegrist and Doe, 2007; Bulgakova et al., 2013; Matis et al., 2014). Many epithelial cells have two microtubule systems with distinct organizations—an apicobasal array with microtubule plus ends pointing basally and a dense network beneath (and parallel to) the apical surface known as the “subapical network” (Figure 1A) (Toya and Takeichi, 2016; Akhmanova and Kapitein, 2022).
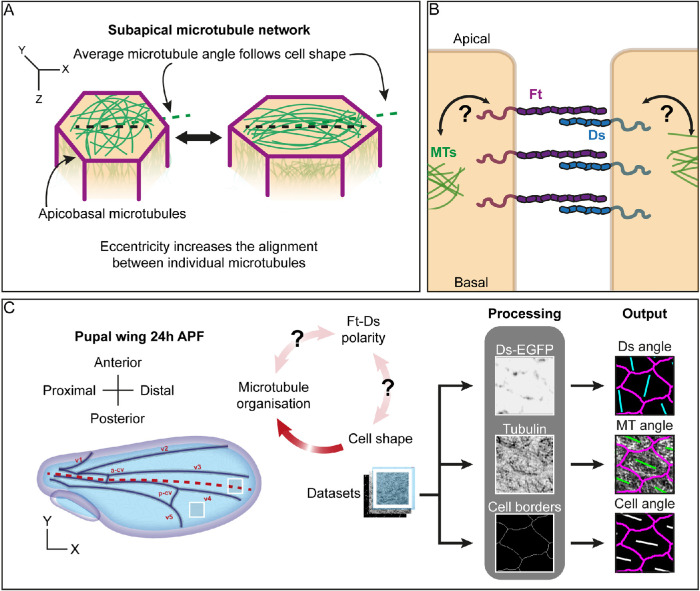
FIGURE 1:
Development of a toolkit to investigate the effects of cell shape and Ft-Ds polarity on the organization of the subapical microtubule network. (A) Cartoon depicting the subapical microtubule network in cells with different elongations. (B) Cartoon of the planar polarized localization of the Fat-Dachsous (Ft-Ds) atypical cadherins at the subapical cell–cell boundary and their possible interaction with the subapical microtubule network (MTs). (C) Diagram of a pupal wing at 24 h APF with the two regions of interest (distal and posterior) highlighted by white squares and the image analysis pipeline. The main longitudinal veins of the wing (v1–5) and the anterior (a-cv) and posterior (p-cv) cross-veins are depicted in black. The combination of software and automated analysis scripts allowed us to extract cell-by-cell data about cell shape (using cell borders) and correlate them with the distribution of proteins at the junctions (e.g., detected with the knock-in Ds-EGFP) and the organization of the subapical microtubule network (detected by immunolabeling against tubulin).
The subapical microtubule network contributes to both morphogenesis and function of epithelia, including such diverse roles as the regulation of cell morphology and adhesion, planar polarity, and positioning of cilia and their coordinated beating (Vladar et al., 2012; Gomez et al., 2016; Herawati et al., 2016; Tateishi et al., 2017; Kimura et al., 2021; Akhmanova and Kapitein, 2022). However, it is unclear what determines the correct organization of subapical microtubule networks, and two competing models have been proposed.
The first model suggests that the microtubules follow the physical constraints set up by mechanical strain (Chien et al., 2015) or by cell shape (Gomez et al., 2016). As a result, the more cells are elongated in the tissue plane, the greater the alignment of microtubules with each other within the subapical network—the overall direction being aligned with the cell’s long axis (Figure 1A) (Gomez et al., 2016). This cell shape–driven microtubule alignment has been shown to be robust in several epithelia, including the Drosophila pupal wing epithelium (Płochocka et al., 2021). It was further proposed that the degree of microtubule alignment but not the overall direction might be affected by the positioning of microtubule minus ends (Płochocka et al., 2021).
The second model suggests that the organization of the subapical network is actively controlled by planar polarity machinery. Planar polarity (also known as planar cell polarity, PCP) is established and maintained by two widely studied systems in animal cells: the “core” Frizzled-dependent pathway and the Fat-Dachsous (Ft-Ds) pathway (Devenport, 2014; Hale et al., 2015). Both pathways rely on the heterophilic binding of membrane proteins that are distributed asymmetrically across the tissue plane in response to upstream patterning signals such as morphogen gradients (Figure 1B) (Lawrence and Casal, 2018). In both invertebrates and vertebrates, there is evidence that these pathways can specify planar polarity on either the same or different axes within a single tissue, depending on the context (Merkel et al., 2014; Zakaria et al., 2014; Vu et al., 2019) (reviewed in Strutt and Strutt, 2021).
Microtubule polarity has been proposed to be regulated by both the core planar polarity pathway in vertebrates (Sepich et al., 2011; Vladar et al., 2012; Chien et al., 2015) and the Ft-Ds pathway in both vertebrates and flies (Harumoto et al., 2010; Li-Villarreal et al., 2015). Specifically, in the Drosophila pupal wing, it has been proposed that the Ft-Ds pathway controls the orientation of microtubules within the subapical network along the proximodistal axis and this provides a polarity cue for the core pathway through the polarized transport of core proteins toward microtubule plus ends at distal cell edges (Shimada et al., 2006; Harumoto et al., 2010; Matis et al., 2014; Olofsson et al., 2014) (reviewed in Strutt and Strutt, 2021). Finally, both cell elongation due to mechanical strain and the core planar polarity pathway have been shown to cooperate in apical microtubule alignment during Xenopus gastrulation (Chien et al., 2015).
Currently, it is unclear whether the Ft-Ds pathway affects the organization of the subapical microtubule network directly or indirectly. In the pupal wing, minus ends of the subapical microtubules are localized at apical junctions in a polarized manner thought to align with Ft-Ds planar polarity (Matis et al., 2014), and Ft-Ds are suggested to be required for the association between the subapical microtubules and adherens junctions (Singh et al., 2018). From this, it has been inferred that Ft-Ds may play a direct role in capturing or nucleating microtubules, although the molecular link between Ft-Ds and microtubules is unknown. At the same time, Ft-Ds might act indirectly on the subapical microtubule network, as there is evidence that they alter cell shape, and in particular cell elongation, by polarizing the distribution of the atypical myosin Dachs (Aigouy et al., 2010; Mao et al., 2011; Bosveld et al., 2012).
Here, we reassess the evidence for these models of how the subapical microtubule network is organized, looking at the Drosophila pupal wing epithelium due to the wealth of acquired knowledge about this system. We have developed a robust pipeline of work where automated image analysis has allowed us to determine characteristics on a cell-by-cell basis (Figure 1C). Similar approaches have been used previously for linking planar polarity and cell orientation (Hirano et al., 2022), but our pipeline introduces microtubule analysis. We identified two regions within pupal wings with distinct relationships between the orientation of cell shape and Ft-Ds polarity, allowing us to uncouple their effects on the organization of subapical microtubule networks. Studying these two regions in wings with normal and altered Ft-Ds function revealed that the Ft-Ds complexes weakly promote localization of the microtubule minus-end capping protein Patronin. Despite this, we found that Ft-Ds polarity cues do not play any substantial role in the subapical microtubule network organization in the developing wing epithelium. Conversely, cell shape was a strong predictor of microtubule organization. Altogether, our findings reconcile the two models of the subapical microtubule network organization by placing cell shape as the primary cue, which might be regulated upstream by Ft-Ds planar polarity in a context-dependent manner.
RESULTS
Divergent axes of Ft-Ds polarity and cell elongation in the pupal wing
To start disentangling the relationship between the Ft-Ds pathway, cell shape, and subapical microtubule organization using automated image analysis, we first quantified the polarity of Ft-Ds in different regions of the pupal wing using our established tool based on principal component analysis (PCA) (Tan et al., 2021). Previous reports have come to conflicting conclusions regarding Ft-Ds polarity in the developing wing; Matis et al. (2014) have reported that polarity is proximodistally oriented at 24 h after puparium formation (APF) when microtubules are also aligned proximodistally following the long axis of the cells (Shimada et al., 2006; Harumoto et al., 2010; Matis et al., 2014; Płochocka et al., 2021). Conversely, Merkel et al. (2014) observed a primarily radial/anteroposterior orientation of Ft-Ds polarity throughout most of the wing at this stage (Merkel et al., 2014).
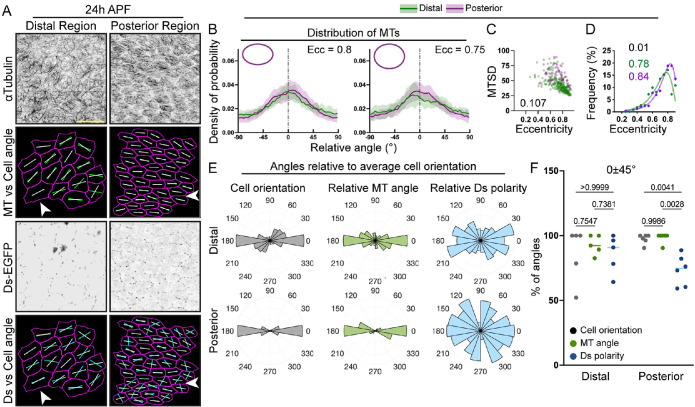
We focused our analysis on two regions that show large discrepancies in the published reports, the distal region of the wing between veins 3 and 4 and the posterior region below vein 4 (v4) at 24 h APF (Figures 1C and 2A). Coarse grain analysis of Ds-EGFP polarity showed a radial/anteroposterior pattern across these regions (Figure 2A), consistent with the previous observations of Merkel et al. (2014). We used our analysis pipeline to map both cell orientation and Ds polarity at a single-cell resolution (Figure 2B, see Materials and Methods). To assess the cell shape polarity—the direction in which cells are elongated within the tissue—we measured cell angles (directions of long axes) of individual cells in both regions (Figure 1C; see Materials and Methods). Cells from the two regions exhibited strong similar magnitudes of Ds polarity as assessed by our PCA method (Figure 2C). At the same time, the cells below v4 (posterior) were more elongated than those above v4 (distal) based on the distributions of their eccentricities (Figure 2D).
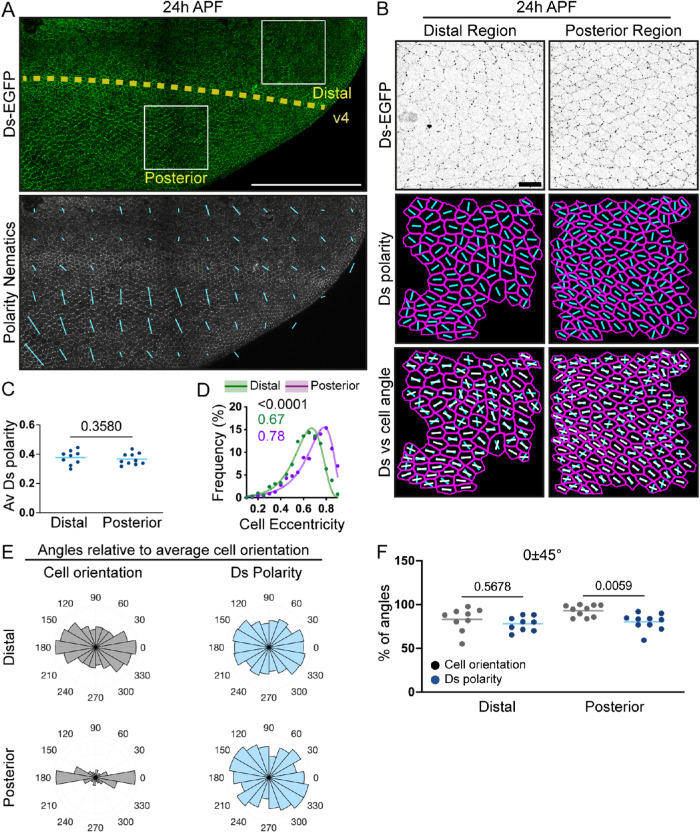
FIGURE 2:
Ds exhibits radial/anteroposterior polarity in the wing independently of cell shape at 24 h APF. (A) Apical view of the distoposterior region of a pupal wing at 24 h APF expressing Ds-EGFP (green with enhanced brightness, top; grayscale, bottom) and showing the coarse grain polarity nematics (6 × 6 cells) of Ds-EGFP (blue lines, bottom). The white dotted line indicates the position of longitudinal vein 4 (v4). Scale bar: 100 µm. Distal is to the right and anterior is up in this and all subsequent images (see Figure 1C). (B) Apical view of cells expressing Ds-EGFP at 24 h APF, located at the distal (left) and posterior (right) regions of the pupal wing. Ds-EGFP distribution (inverted grayscale, top), Ds-EGFP polarity nematics alone (blue lines, middle, cell outlines in magenta) or with cell angle (white lines, bottom) are shown. Scale bar: 10 µm. (C) Average Ds-EGFP polarity (PCA method) in the two analyzed regions. Two-tailed paired t test (for paired regions in nine wings). Total data set N = 9 (distal) and 10 (posterior) wings. (D) Distributions of cell eccentricities in the two regions of interest with the binned data points (dots), the best-fit lognormal distributions (lines), the p value for the probability of the distributions being the same (extra-sum-of-squares F test, black text), and modes of the best-fit distributions (green and magenta text). N = 9 distal and 10 posterior regions. (E) Polar histograms depicting binned cell orientations (gray) and Ds polarity (blue) relative to the average cell orientation in each region in distal (top) and posterior (bottom) regions of pupal wings at 24 h APF. N = cells from 9 distal and 10 posterior regions. (F) Percentage of individual cell angles per wing region in the 0 ± 45° range relative to the average cell orientation for cell orientation and Ds polarity. Lines represent the mean values. Brown-Forsythe and Welch ANOVA. N = 9 distal and 10 posterior regions.
To explore the relationship between cell shape and Ds polarities and directly compare the two regions of the pupal wing, we plotted both cell and Ds polarity angles relative to the average cell angle in each region/wing (Figure 2E). The normalization of cell orientation to the average orientation angle demonstrated high cellular coordination with many cells elongating in the same direction, which is more apparent in the posterior region, potentially due to the higher eccentricities of individual cells (Figure 2, D and E). Next, we calculated the percentages of cell and Ds polarity angles that fell into the quadrant with the most cell angles (i.e., 0 ± 45°) (Harumoto et al., 2010) (Figure 2, E and F). This revealed that cell orientations and Ds polarities were similar in the distal compartment but significantly differed in the posterior (Figure 2F). Therefore, these data demonstrate an uncoupling between cell orientation and Ds polarity in the posterior region below v4 but not in the distal region of the wing. Thus, we found distinct relationships between cell shape and Ft-Ds polarities, depending on the region of the pupal wing. Such an uncoupling in the posterior region indicates that the mechanisms that determine Ft-Ds polarity and cellular elongation within a tissue are, at least partially, independent. Importantly, the differences between cell shapes and Ds polarities in these regions allow us to investigate the effects of both factors on the organization of the subapical microtubule network.
Microtubule organization correlates with cell shape orientation but not Ft-Ds polarity
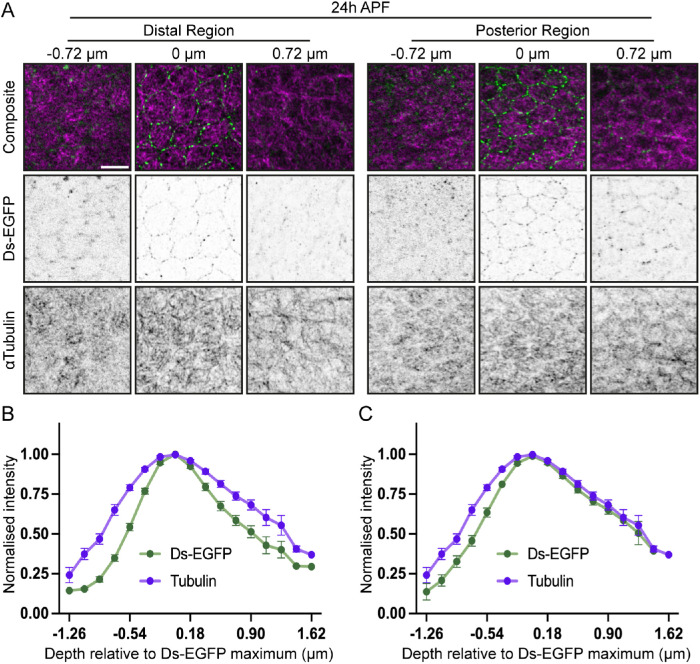
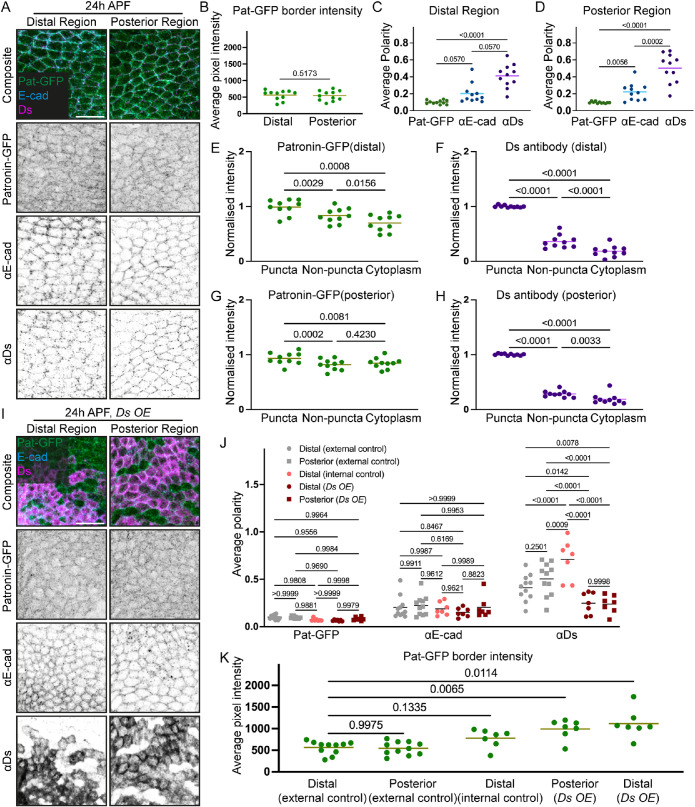
Both Ft-Ds and subapical microtubules are positioned within subapical domains of pupal wing cells (Ma et al., 2003; Shimada et al., 2006; Silva et al., 2006; Harumoto et al., 2010; Matis et al., 2014; Gomez et al., 2016; Płochocka et al., 2021). Therefore, we next employed high-resolution imaging (∼120 nm in XY and ∼350 nm in Z direction) (Wu and Hammer, 2021) to ask whether the subcellular localizations of Ft-Ds and microtubules were consistent with the model whereby Ft-Ds capture microtubules. In the wing, the Ft-Ds complexes localize in “puncta”—discrete accumulations of stable clustered Ft-Ds complexes (Hale et al., 2015). The fixation conditions required to image microtubules (see Materials and Methods) were compatible with the preservation of Ds-EGFP puncta (Figure 3A). Intensity profiles along the apicobasal cell axis showed that Ds-EGFP was indeed found in the same apicobasal plane as the subapical microtubules in both distal and posterior regions (Figure 3, B and C), consistent with the possibility that the Ft-Ds complexes directly affect microtubules.
FIGURE 3:
Ft-Ds polarity complexes and the subapical microtubule networks are localized in the same apicobasal position in cells of 24 h APF pupal wings. (A) Subapical localization of Ds-EGFP (green, composite top; inverted grayscale, middle panels) and microtubules stained with antibody against tubulin (magenta, top panels; inverted grayscale, bottom panels) at both the distal (left) and posterior (right) regions of the pupal wing 24 h APF. Images depict the distribution of both markers at different positions across the apicobasal axis relative to the maximum mean intensity of Ds-EGFP (0 µm). Scale bar: 5 µm. (B, C) Normalized intensity of Ds-EGFP and tubulin (mean ± SEM) at the distal (B) and posterior (C) regions along the subapical axis, centered around the maximum mean intensity of Ds-EGFP (0 µm). N = 11 wings.
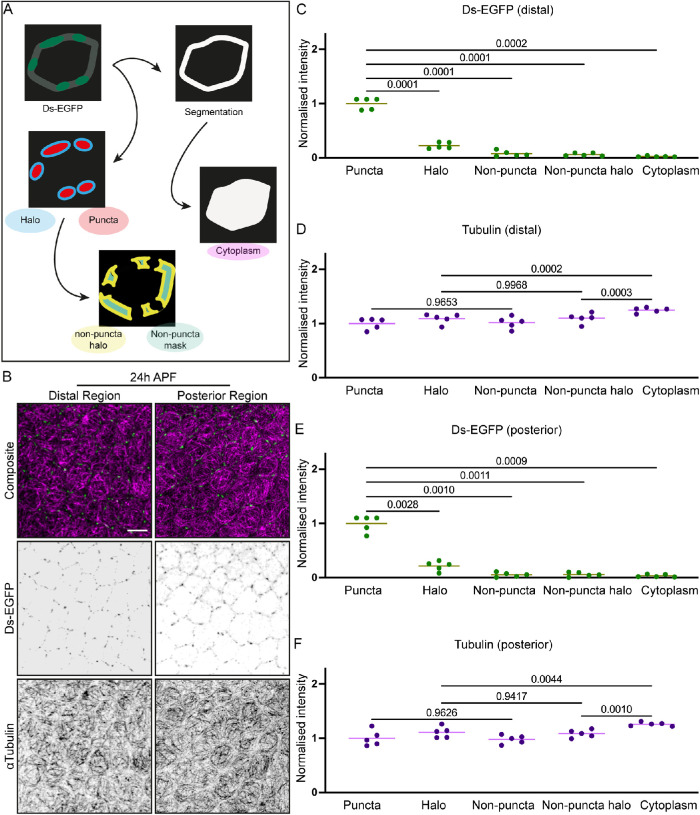
Therefore, we next explored the model that the Ft-Ds complexes capture microtubules. To this end, we used automated image analysis to determine whether there was a correlation between the planar polarized localization of Ds-EGFP puncta and local increases in the density of microtubules in the two regions of interest of the pupal wing (Figure 4, A and B; see Materials and Methods). We compared levels of the tubulin signal within the Ds-EGFP puncta and in the direct vicinity of these puncta (“puncta halo”) to those within nonpuncta regions of the cell boundaries—regions that are largely depleted for Ds-EGFP—and in their vicinity (“non-puncta halo”) and in the cytoplasm (Figure 4A). As expected, Ds-EGFP was predominantly found in puncta and depleted from other cellular areas in both pupal wing regions (Figure 4, B, C, and E). We did not observe an enriched accumulation of tubulin signal in the vicinity of the Ds-EGFP puncta in either pupal wing region (Figure 4, B, D, and F). Thus, there are no detectable local changes in microtubule density depending on Ft-Ds concentrations, making it unlikely that Ft-Ds complexes directly capture or cluster microtubules.
FIGURE 4:
Ft-Ds localization does not correlate with local changes in the density of apical microtubules at 24 h APF. (A) Cartoon illustrating the analysis pipeline for determining the relative signal levels in different subcellular localizations. The automated script thresholds the images to detect Ds puncta and, thus, classify cell boundary pixels from the dilated segmentation masks into puncta and nonpuncta regions. The pixels around the puncta and nonpuncta regions (haloes) are used to examine local tubulin density in the proximity of the Ds puncta. The average signal inside the cell is used as the cytoplasm intensity. (B) Apical view of pupal wings at 24 h APF expressing Ds-EGFP (green, top; inverted grayscale, middle) and stained against tubulin (magenta, top; inverted grayscale, bottom) corresponding to the distal (left) and posterior (right) regions. Scale bar: 5 µm. (C–F) Normalized intensity levels of Ds-EGFP (C, E) and tubulin (D, F) at the distal (C, D) and posterior (E, F) regions of the pupal wing, within specific areas as detected by the analysis. Lines represent the mean values. One-way ANOVA with Tukey’s multiple comparisons test. N = 5 wings.
Next, we asked whether Ft-Ds affect the overall organization of subapical microtubules by testing whether it is the Ft-Ds distribution or cell shape that best correlates with the following two aspects of microtubule organization: the relative alignment of microtubules with each other and the overall direction of the whole subapical network (Figure 1A). We examined these two aspects in individual cells in both distal and posterior regions of pupal wings (Figure 5A) in relation to Ft-Ds polarity and cell elongation.
FIGURE 5:
The organization of subapical microtubule networks in cells with normal expression of Ft-Ds in the pupal wing at 24 h APF. (A) Apical view of cells stained against tubulin (inverted grayscale, top) and expressing Ds-EGFP (inverted grayscale, third row) in the distal (left) and posterior (right) regions of the pupal wing at 24 h APF. Microtubule angles (green lines, second row), cell angles (white lines, second and bottom rows), and Ds polarities (blue lines, bottom row) in individual cells are shown. White arrowheads indicate examples of cells where Ds polarity is parallel to average microtubule and cell angles (left) or perpendicular (right). Scale bar: 5 µm. (B) Microtubule angle distributions (line indicates mean; shading indicates SD) relative to the maximum of the tubulin signal in each cell (0°) binned by cell eccentricity (0.8 ± 0.025, left; 0.75 ± 0.025, right) in the distal (green) and posterior (magenta) regions. N = 14 (0.8, distal), 12 (0.75, distal), 44 (0.8, posterior), and 33 (0.75, posterior) cells (five distal and six posterior wings). (C) Correlation between cell eccentricities and MTSD in the distal (green) and posterior (magenta) regions of pupal wings 24 h APF. Each dot represents an individual cell and the number indicates the p value for the Pearson’s correlations between the two regions tested being different using the bootstrap method. N = 5 distal and 6 posterior regions. (D) Distributions of cell eccentricities in the distal (green) and posterior (magenta) regions of pupal wings 24 h APF with the binned data points (dots), the best-fit lognormal distributions (lines), the p value for the probability of the distributions being the same (extra-sum-of-squares F test, black text), and modes of the best-fit distributions (green and magenta text). N = 5 distal and 6 posterior regions. (E) Polar histograms depicting binned cell orientations (gray), microtubule angles (green), and Ds polarity (blue) relative to the average cell orientation in each region in distal (top) and posterior (bottom) regions of pupal wings at 24 h APF. N = 5 distal and 6 posterior regions. (F) Percentage of individual cell angles in the 0 ± 45° range relative to the average cell orientation for cell orientation, microtubule angle, and Ds polarity. Lines represent the mean values. Kruskal–Wallis with Dunn’s multiple comparisons test. N = 5 distal and 6 posterior regions.
The microtubule angle distribution represents how well the microtubules are aligned with each other within individual cells (Figure 5B) and can be expressed as the microtubule standard deviation (MTSD)—the SD of the best-fit von Mises distribution reflecting the width of the peak (Gomez et al., 2016). MTSD correlated with cell elongation (Figure 5C)—more elongated cells with higher eccentricities had a steeper distribution (smaller MTSD) that reflected more aligned microtubules, with no significant difference between the two regions of the pupal wing examined. Such correlation of the microtubule angle distribution with the cell eccentricity agrees with the reported robustness of microtubule alignment with each other in subapical microtubule networks of epithelial cells (Płochocka et al., 2021).
At the same time, we found that the overall microtubule network direction aligned with the axis of cell orientation (Figure 5, E and F), with an uncoupling from Ds polarity in the posterior region of the developing wing. Additionally, we observed a better alignment of microtubule networks with the orientation of the cells in the posterior region (Figure 5, E and F), which are more elongated than those in the distal region (Figure 5D). Therefore, we conclude that the direction of the microtubule network inside cells depends on the angle of the cell elongation axis, an effect that is particularly strong in the posterior region, rather than Ft-Ds polarity.
Ft-Ds junctional complexes promote localization of the microtubule minus-end binding protein Patronin
Our findings support the overall organization of the subapical microtubule network being robust and depending on the cell shape rather than the distribution of the Ft-Ds complexes. However, they do not exclude changes to the detail of this organization dependent on Ft-Ds activity. The localization of the minus ends in noncentrosomal networks is of particular interest, as it ensures the correct polarized transport by motor proteins (Steinhauer and Kalderon, 2006; Zhelezov et al., 2019).To examine the possibility that Ft-Ds complexes alter the distribution of the minus ends without affecting the overall organization of subapical microtubules, we turned to the minus-end capping protein Patronin, which localizes at apical junctions and exhibits an asymmetrical distribution in the Drosophila embryonic epidermis that depends on the cell elongation (Goodwin and Vale, 2010; Płochocka et al., 2021).
Patronin-GFP was also enriched at the cell junctions in the pupal wing (Figure 6A). There were no differences in the average levels of Patronin-GFP at the cell boundaries between distal and posterior regions (Figure 6B). When its distribution was assessed using PCA, Patronin-GFP appeared to be uniformly distributed along cell boundaries, with the average polarity being lower than that of E-cadherin (Figure 6, C and D). However, we found that there were slightly higher levels of Patronin-GFP within Ds puncta along the cell boundaries in both distal and posterior regions of pupal wings (Figure 6, E–H). This suggests that the Ft-Ds complexes might capture microtubule minus ends associated with Patronin. To strengthen this conclusion, we correlated the intensity of Ds and Patronin-GFP on a puncta-by-puncta basis. We found a mild but significant correlation in both distal and posterior regions—0.06 (p = 0.0002) and 0.09 (p < 0.0001), respectively. This correlation supports the possibility that Ft-Ds complexes could affect subapical microtubule networks via regulation of Patronin localization. Additionally, this highlights the limitations of using the PCA analysis alone—the moderate increase of Patronin-GFP levels within polarized Ds puncta appears to be below its sensitivity to detect polarization at the cell level, probably due to the high levels of unpolarized Patronin-GFP outside of Ds puncta.
FIGURE 6:
Patronin is weakly recruited to cell boundaries by Ft-Ds complexes in the pupal wing at 24 h APF. (A) Apical view of pupal wings at 24 h APF expressing Patronin-GFP (Pat-GFP, green, top; inverted grayscale, second row) and stained against E-cadherin (E-cad, cyan, top; inverted grayscale, third row) and Ds (magenta, top; inverted grayscale, bottom) in the distal (left) and posterior (right) regions. Scale bar: 10 µm. (B) Mean intensity of Patronin-GFP at cell boundaries in cells from distal and posterior regions of pupal wings with normal levels of Ds at 24 h APF. Lines indicate the mean values. Two-tailed paired t test. N = 11. (C, D) Average polarity of Patronin-GFP (Pat-GFP), E-cadherin (E-cad), and Ds in the distal (C) and posterior (D) regions. Friedman test (C) and one-way ANOVA (D) with Dunn’s (C) and Tukey’s (D) tests for multiple comparisons. N = 11 wings. (E–H) Mean intensity levels of Patronin-GFP (E, G), and Ds (F, H) at the distal (E, F) and posterior (G, H) regions of the pupal wing, within specific subcellular areas. One-way ANOVA (E, G, H) and Friedman test (F) and with Tukey’s (E, G, H) and Dunn’s (F) tests for multiple comparisons. N = 11 wings. (I) Apical view of pupal wings at 24 h APF overexpressing Ds from 18 h APF, expressing Patronin-GFP (Pat-GFP, green, top; inverted grayscale, second row) and stained against E-cadherin (E-cad, cyan, top; inverted grayscale, third row) and Ds (magenta, top; inverted grayscale, bottom) in the distal (left) and posterior (right) regions. Scale bar: 10 µm. (J) The average polarities of Patronin-GFP (Pat-GFP), E-cadherin (E-cad), and Ds in the distal and posterior regions with normal Ds levels (external controls), cells without Ds overexpression—adjacent to region with Ds overexpression—in distal regions (internal control) and cells overexpressing Ds (Ds OE) in distal and posterior regions are shown. Two-way ANOVA with Tukey’s tests for multiple comparisons. N = 11 (external control) and 7 (Ds OE) wings. (K) Mean intensities of Patronin-GFP at cell boundaries in cells in the distal and posterior regions with normal Ds levels (external controls), cells without Ds overexpression in distal regions (internal control), and cells overexpressing Ds in distal and posterior regions are depicted. Brown-Forsythe and Welch ANOVA test with Dunnett’s T3 test for multiple comparisons. N = 11 (control) and 7 (Ds OE) wings. Lines represent the mean values in all graphs.
To further investigate the relationship between Ft-Ds and Patronin, we sought to genetically target the Ft-Ds pathway and determine the resulting effects on Patronin localization. Therefore, we disrupted Ft-Ds polarity by temporally inducing Ds overexpression in the posterior compartment of the wing using a hedgehog-Gal4 driver. In this condition, cells in posterior regions overexpress Ds, whereas only cells in the posterior half of the distal regions do, providing an internal control in nearby anterior tissue. The induction from 18 h APF led to an increase in Ds levels at 24 h APF accompanied by reduced Ds polarity in overexpressing posterior and distal regions (Figure 6, I and J). Concomitantly, the polarity of Ds increased in the adjacent distal internal control cells with unmodified Ds levels (Figure 6J), likely due to the nonautonomous propagation of polarity from the overexpression region (Brittle et al., 2012).
We found that neither reduced Ds polarity in the cells that overexpress Ds nor increased Ds polarity in their neighbors (distal internal control region) was accompanied by changes in Patronin-GFP polarities (Figure 6J). However, as shown above, the PCA method is not powerful for detecting the correlation between Ds and Patronin localization. To further examine the interaction between Ds and Patronin-GFP and the consequences of Ds overexpression on its distribution, we next measured the average levels of Patronin-GFP localized at cell boundaries (as there were no apparent Ds puncta when Ds was overexpressed; Figure 6I). Ds overexpression increased the levels of Patronin-GFP at the cell boundaries in both regions, whereas they were not significantly affected in the internal control with normal Ds levels (Figure 6K). This finding supports the recruitment of Patronin to cell boundaries and the potential capture of microtubule minus ends by the Ft-Ds complexes.
Experimental manipulation of Ft-Ds planar polarity does not reorganize microtubules
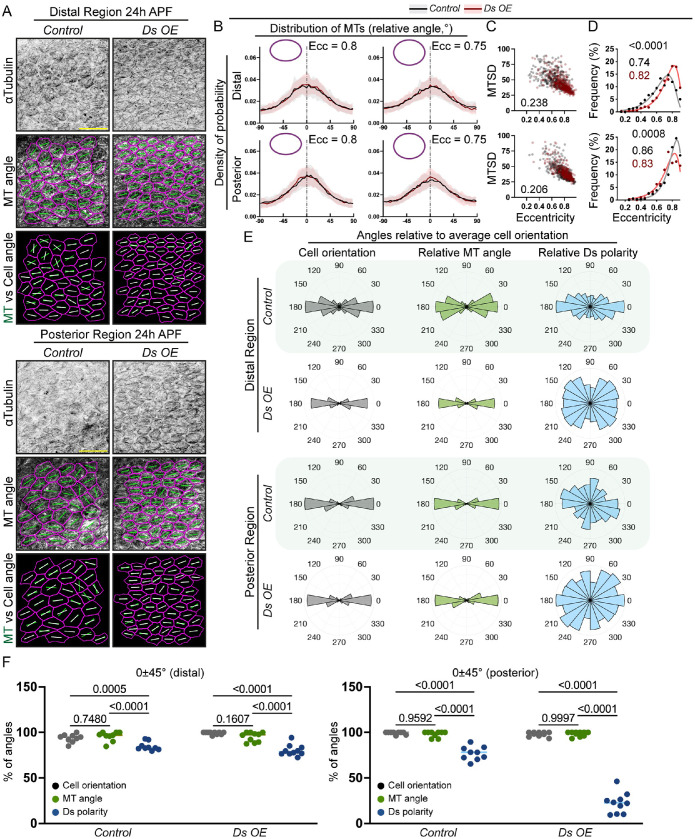
The observed uncoupling of Ft-Ds polarity and microtubule organization in the posterior region suggested that during normal development Ft-Ds are unlikely to override the effects of cell shape on microtubule network organization despite potential effects on the distribution of microtubule minus ends via Patronin. Consistent with this, we found that Ds overexpression did not change the alignment of microtubules with each other in the two wing regions (Figure 7, A and B). The correlation between MTSD and cell eccentricity remained unchanged in both regions when comparing normal and elevated levels of Ds (Figure 7C). Note that Ds overexpression had a mild effect on cell shape, as in the distal region cells with excess Ds were more elongated (Figure 7D), demonstrating that the Ft-Ds pathway can contribute to shaping cells in the pupal wing. Overall microtubule network directions remained aligned with cell angles, regardless of the disrupted Ds polarity produced by the protein overexpression (Figure 7, E and F). Notably, the increase in cell eccentricity distally appeared to increase both the coordination of cell alignment with each other and the alignment of microtubule angles with cell angles (Figure 7E), in agreement with what we observed in pupal wings with an unaltered Ft-Ds pathway (Figure 5, E and F), supporting the primary role of cell shape in organizing the subapical microtubule network.
FIGURE 7:
Ds overexpression does not change microtubule organization at 24 h APF. (A) Apical view of cells in the distal (top group) and posterior (bottom group) regions at 24 h APF stained against tubulin (inverted grayscale, top) without (left) or with Ds overexpression (right). Alongside stainings, microtubule angles (green lines, second and bottom rows) and cell angles (white lines, bottom row) are shown. Scale bar: 10 µm. (B) Microtubule angle distributions (line indicates mean, shading indicates SD) relative to the maximum of the tubulin signal in each cell (0°) binned by cell eccentricity (0.8 ± 0.025, top; 0.75 ± 0.025, bottom) in control cells (black) and cells overexpressing Ds (red) in distal (left) and posterior (right) regions. N = 37 (0.8, distal, control), 42 (0.75, distal, control), 72 (0.8, distal, Ds OE), 52 (0.75, distal, Ds OE), 53 (0.8, posterior, control), 22 (0.75, posterior, control), 46 (0.8, posterior, Ds OE), and 65 (0.75, posterior, Ds OE). (C) Correlations between MTSD and cell eccentricity at the distal (top) and posterior (bottom) regions, comparing the distributions between control (black) and Ds-overexpressing (red) cells. Each dot represents an individual cell, the numbers indicate p values for the Pearson’s correlations between the two genotypes tested being different using the bootstrap method. N = 9 (both regions, control) and 10 (both regions, Ds OE). (D) Distributions of cell eccentricities in control cells (black) or cells overexpressing Ds (red) in the distal (top) and posterior (bottom) regions of pupal wings 24 h APF with the binned data points (dots), the best-fit lognormal distributions (lines), the p values for the probability of the distributions being the same (extra-sum-of-squares F test, black text at top), and modes of the best-fit distributions (black text, middle, and red text). N = 9 (both regions, control) and 10 (both regions, Ds OE). (E) Polar histograms depicting binned cell orientations (gray), microtubule angles (green), and Ds polarity (blue) relative to the average cell orientation in each region in control cells (first and third rows) and cells overexpressing Ds (second and bottom rows) in distal (top two rows) and posterior (bottom two rows) regions of pupal wings at 24 h APF. N = 9 (both regions, control) and 10 (both regions, Ds OE). (F) Percentage of angles in the 0 ± 45° range relative to the average cell orientation for cell orientation, microtubule angle, and Ds polarity. Lines represent the mean values. One-way ANOVA with Dunn’s multiple comparisons test. N = 9 (both regions, control) and 10 (both regions, Ds OE).
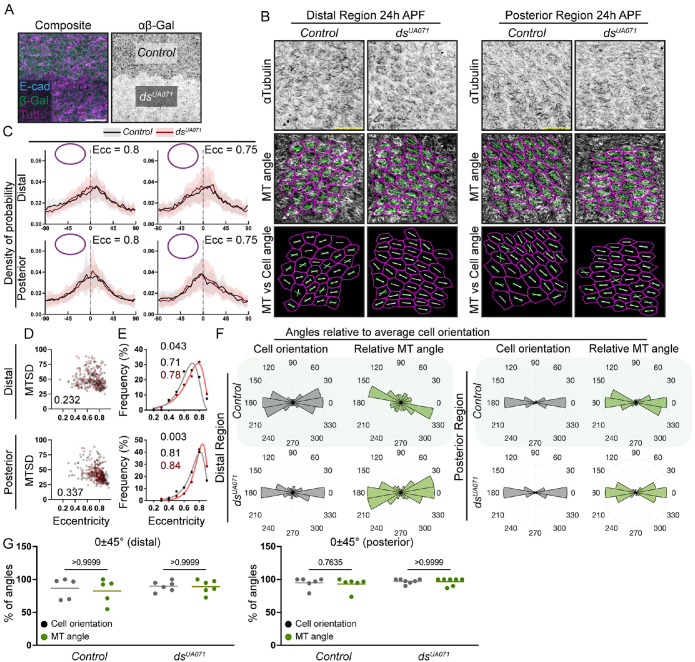
Finally, we disrupted Ft-Ds polarity by removing Ds using the strong loss-of-function dsUA071 allele and flippase recognition target (FRT)-mediated mitotic recombination with an arm-LacZ transgene on the homologous chromosome to generate homozygous clonal tissue (Adler et al., 1998; Strutt and Strutt, 2002; Germani et al., 2018). We compared the microtubule organization in the control tissue with wild-type ds against the mutant clonal tissue, as detected by the presence or absence of β-galactosidase, respectively (Figure 8A). In ds clones in both distal and posterior regions, microtubules remained aligned with each other according to the cell eccentricity (Figure 8, B and C), and in both regions the correlations between MTSD and cell eccentricity were unaffected (Figure 8D). Concurrently, clone cells increased their eccentricities in both regions (Figure 8E). Thus, both loss and overexpression of Ds can increase cellular elongation, suggesting that it is loss of the polarity rather than the levels of Ft-Ds complexes that contributes to the cell shape phenotype. The microtubule angle in the mutant cells remained coupled with the cell angle, regardless of the region, with the same percentages of angles in 0 ± 45° quadrants (Figure 8, F and G).
FIGURE 8:
Microtubule organization does not change in ds mutant tissue at 24 h APF. (A) Representative apical views of a clonal border in the posterior region of a pupal wing at 24 h APF stained against β-galactosidase (green, left; inverted grayscale, right) whose absence indicates the strong mutant dsUA071/dsUA071 clone tissue. Stainings against E-cadherin (E-cad, blue) and tubulin (Tub, magenta) are shown on the left. Scale bar: 10 µm. (B) Apical view of cells in the distal (top group) and posterior (bottom group) regions at 24 h APF stained against tubulin (inverted grayscale, top) in control (left) or dsUA071 mutant (right) cells. Microtubule angles (green lines, second and bottom rows) and cell angles (white lines, bottom row) are shown. Scale bar: 10 µm. (C) Microtubule angle distributions (line indicates mean, shading indicates SD) relative to the maximum of the tubulin signal in each cell (0°) binned by cell eccentricity (0.8 ± 0.025, left; 0.75 ± 0.025, right) in control cells (green) and dsUA071 mutant cells (red) in distal (top) and posterior (bottom) regions. N = 16 (0.8, distal, control), 18 (0.75, distal, control), 24 (0.8, distal, dsUA071), 16 (0.75, distal, dsUA071), 27 (0.8, posterior, control), 19 (0.75, posterior, control), 21 (0.8, posterior, dsUA071), and 26 (0.75, posterior, dsUA071). (D) Correlation between MTSD and cell eccentricity at the distal (top) and posterior (bottom) regions, comparing the distributions between control (black) and dsUA071 (red) mutant cells. Each dot represents an individual cell, and the numbers indicate p values for the Pearson’s correlations between the two genotypes tested being different using the bootstrap method. N = 5 (distal, control), 6 (distal, dsUA071), 6 (posterior, control), and 7 (distal, dsUA071). (E) Distributions of cell eccentricities in control (black) or dsUA071 mutant (red) cells in the distal (top) and posterior (bottom) regions of pupal wings 24 h APF with the binned data points (dots), the best-fit lognormal distributions (lines), the p values for the probability of the distributions being the same (extra-sum-of-squares F test, black text top), and modes of the best-fit distributions (black text, middle, and red text). N = 5 (distal, control), 6 (distal, dsUA071), 6 (posterior, control), and 7 (distal, dsUA071). (F) Polar histograms depicting binned and cell orientations (gray) and microtubule angles (green) relative to the average cell orientation in each region in control (top rows) and dsUA071 mutant (bottom rows) cells in distal (left two columns) and posterior (right two columns) regions of pupal wings at 24 h APF. N = 5 (distal, control), 6 (distal, dsUA071), 6 (posterior, control), and 7 (distal, dsUA071). (G) Percentage of angles in the 0 ± 45° range relative to the average cell orientation for cell orientation, microtubule angle, and Ds polarity. One-way ANOVA with Dunn’s multiple comparisons test. Lines represent the mean values. N = 5 (distal, control), 6 (distal, dsUA071), 6 (posterior, control), and 7 (distal, dsUA071).
To summarize, while we found evidence for the Ft-Ds pathway affecting both cell shape and the localization of the microtubule minus-end capping protein Patronin, these effects do not produce detectable changes in the organization of the subapical microtubule network. Specifically, they do not influence how well the microtubules align with each other inside cells and the direction in which the overall network orients. On the basis of our experimental data, we concluded that cell shape is the primary cue for the direction and alignment of the subapical microtubule network in Drosophila pupal wings.
DISCUSSION
Our work has revealed three key findings. First, we found that the cell shape polarity (the direction in which cells elongate) and Ft-Ds planar polarity are uncoupled in Drosophila pupal wings. Second, assisted by this uncoupling of two polarities, we demonstrated that subapical microtubule networks orient themselves in agreement with the cell shape rather than planar polarity established by the Ft-Ds complexes. Finally, we found that the Ft-Ds complexes promote the recruitment of the microtubule minus-end capping protein Patronin to cell boundaries, although any contribution of this recruitment to the organization of subapical microtubule networks in the pupal wing was not detectable.
Cell shape is ultimately determined by a tug-of-war between tissue-scale forces such as shear stress and the forces generated internally within the cell by the cytoskeleton (Mao and Baum, 2015). Ft-Ds planar polarity is established by the developmental program, namely, by opposing expression gradients of Ds and the Golgi-localized kinase Four-jointed (reviewed in Strutt and Strutt, 2021). In turn, Ft-Ds complexes are reported to actively control cell shape by asymmetrically localizing the atypical myosin Dachs, which creates anisotropic junctional tension promoting cellular elongation (Mao et al., 2011; Bosveld et al., 2012). As a result, the Ft-Ds pathway promotes the alignment of cell angles in such epithelia as the Drosophila abdomen (Mangione and Martín-Blanco, 2018). At the same time, cell elongation is also influenced by the axis of mechanical forces in an epithelium—indeed, tissue-scale anisotropic stress applied to epithelial monolayers leads to cellular elongation in the direction of the applied force in both Drosophila and vertebrate cells (Aigouy et al., 2010; Wyatt et al., 2015; Duda et al., 2019; Nestor-Bergmann et al., 2019).
Interestingly, we find that depending on the region of the pupal wing, the cell shape and Ft-Ds polarity vectors either align (in the distal region) or not (in the posterior region below vein 4) (Figure 2, E and F). We also observed that reduced Ds polarity due to either its loss or overexpression led to an increase in cell elongation (Figures 7C and 8D). Such an uncoupling between cell shape and Ds polarity could be caused by several factors. First, although we did not observe gross differences in the strength of Ds polarity between the two studied regions (Figure 2C), it is possible that due to the presence of additional yet unidentified factors, the degrees to which the Ft-Ds complexes activate Dachs differ between the two regions. Note that as in other tissues, Ft-Ds promote polarized recruitment of Dachs to cell junctions in the pupal wing (Merkel et al., 2014). Lowered activation of orthogonally oriented Dachs in the posterior region would allow the proximodistal global tension to override the Dachs-dependent internal forces and align the cell elongation pattern along the vector of tissue-scale stress. Second, dissipation of the external stress that originates from contraction of the wing hinge due to the viscoelastic properties of epithelial cells may lead to a nonuniform distribution of stress in the pupal wing. Indeed, such a nonuniform stress distribution is suggested by the differences in the anisotropy of the response to laser ablation experiments (Etournay et al., 2015). However, the reported anisotropy of the recoil in response to laser cuts was smaller in the posterior region than distal (Etournay et al., 2015), which makes it unlikely that the lack of alignment of cell elongation with the Ft-Ds polarity can be solely explained by greater tissue-scale stress in this region. It is also possible that other factors such as the core planar polarity pathway or adherens junctions contribute to determining the cell shape and elongation direction. Such combinatorial regulation of cell shape agrees with the observation that the clones of cells that lack ft have either normal or irregular geometries depending on the clone position in pupal wings (Ma et al., 2008). Dissecting the contributions of individual factors and interactions between them would be essential for understanding how cells are shaped in epithelia and how the tissue’s ultimate form is established.
Regardless of the mechanism that establishes the cell shape polarity, we found that it is this cell shape that the subapical microtubule network follows. Namely, we demonstrated that the overall direction of microtubules follows the cell’s long axis, whereas the alignment of microtubules with each other is consistent with the cell eccentricity in agreement with the robustness of this alignment previously reported (Płochocka et al., 2021). A similar role for cell shape in organizing microtubule polarity independently of Ft-Ds polarity in the Drosophila larval epidermis has also been suggested (Pietra et al., 2020).
The instructive role of the cell shape in the organization of the subapical microtubule network is consistent with the model whereby microtubules “passively” sense the cell shape constraints due to their dynamic instability (Gomez et al., 2016). Specifically, as the frequency of microtubule catastrophe in the cytoplasm is low in vivo (Rogers et al., 2002; Komarova et al., 2009), microtubules grow until they reach the cell boundary, where they either undergo a catastrophe or “zip” with the boundary to continue growing along it depending on the angle of approach (Gomez et al., 2016). In this scenario, any role of the Ft-Ds complexes in the organization of the subapical microtubule network is indirect through their effects on the cell shape. If by recruiting Dachs and thus increasing the anisotropy of junctional tension, the Ft-Ds pathway is able to override other cues such as the global tissue stress, the microtubules will follow this new shape, aligning according to Ft-Ds localization—we suggest that such a scenario may be observed in Drosophila wing discs (Mao et al., 2011; Matis et al., 2014).
Finally, we found that the Ft-Ds complexes promote the localization of Patronin as it is enriched within Ds puncta in wings with normal Ft-Ds function and its levels are elevated at cell boundaries following Ds overexpression. This recruitment did not appear to have a detectable effect on the organization of the subapical microtubule network, which correlated with the cell shape alone in the pupal wing. Previously, it was suggested that the localization of microtubule minus ends influences how well the microtubules align with each other within a cell (Płochocka et al., 2021). While it does not appear to be the case in pupal wings studied here, it might occur in other tissue contexts where the Ft-Ds complexes exert stronger effects, for example, in the Drosophila abdomen, where both the Ft-Ds pathway and Patronin have major contributions to shaping the tissue during morphogenesis (Mangione and Martín-Blanco, 2018; Panzade and Matis, 2021).
Altogether, our findings highlight the central role of cell shape/elongation in the organization of the subapical microtubules. We conclude that while the Ft-Ds pathway is likely to participate in regulating cell shape, it is not the only contributing factor and can be overridden by other cues. These findings were enabled by our simultaneous analysis on a cell-by-cell basis of three parameters—cell shape, planar cell polarity, and microtubule organization—from microscopy images. The example of planar polarity–microtubule–cell shape relationships studied here shows that such multifactorial image analyses and integration of relevant parameters aids the dissection of relationships between protein distribution, subcellular organization, and tissue morphogenesis.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Fly genetics and husbandry
Drosophila melanogaster flies were raised on standard cornmeal/agar/molasses media at 18°C or 25°C unless otherwise specified. Constructs are detailed in Table 1. To express constructs of interest, the GAL4/UAS system was used (Brand and Perrimon, 1993), with the hedgehog-GAL4 (hh-Gal4) driver. Prepupa individuals were selected and scored against selection markers and aged for 24 h at 25°C before dissecting.
TABLE 1:
Key reagents and tools.
| Reagent/resource | Source | ID/catalogue number |
|---|---|---|
| Drosophila strains | ||
| ds-EGFP | Brittle et al., 2012 | FLYB: FBti0202074 |
| ft-EGFP | Hale et al., 2015 | |
| Ubi-p63E-Patronin.A.GFP[3M] | Wang et al., 2013 | BDSC:55129; FLYB:FBst0055129; RRID:BDSC_55129 |
| P[w+, UAS-FRT-stop-FRT-ds] | This study | |
| y w hsFLP | Bloomington Drosophila Stock Center (Golic and Lindquist, 1989) | BDSC:7; FLYB:FBti0002044; RRID:BDSC_7 |
| hh-GAL4 | Tanimoto et al., 2000 | FLYB: FBal0121962 |
| dsUA071 | Adler et al., 1998 | BDSC:41784; FLYB:FBal0089339; RRID:BDSC_41784 |
| FRT40 | Bloomington Drosophila Stock Center (Xu and Rubin, 1993) | FLYB: FBti0002071 |
| y w P[w+, UbxFLP]1 | Bloomington Drosophila Stock Center (Emery et al., 2005) | BDSC:42718; FLYB:FBti0150334; RRID:BDSC_42718 |
| P[w+, arm-lacZ] FRT40 | Bloomington Drosophila Stock Center | BDSC:7371; RRID:BDSC_7371 |
| Antibodies | ||
| Mouse monoclonal anti–α-tubulin, 1:1000 | Sigma-Aldrich | Cat#SC-32293 |
| Mouse monoclonal anti-armadillo, 1:100 | Developmental Studies Hybridoma Bank | Cat#N2 7A1 |
| Rat monoclonal anti–E-cad, 1:200 | Developmental Studies Hybridoma Bank | Cat#DCAD2 |
| Rabbit anti-Ds affinity purified, 1:50 | Strutt and Strutt, 2002 | N/A |
| Software | ||
| FV10-ASW | Olympus | N/A |
| Zen | Zeiss | N/A |
| Fiji (ImageJ) | https://fiji.sc | N/A |
| Tissue Analyzer | Benoit Aigouy (Aigouy et al., 2010). | N/A |
| QuantifyPolarity | Sara Tan (Tan et al., 2021). | N/A |
| MATLAB R2019b | Mathworks | N/A |
| GraphPad Prism Version 7 | GraphPad Software | N/A |
| Office Excel 16 | Microsoft | N/A |
| Illustrator 20 | Adobe | N/A |
| Custom scripts for MATLAB | N. Bulgakova (https://github.com/nbul) | N/A |
| Other | ||
| FV1000 confocal microscope | Olympus | N/A |
| LSM 880 Airyscan Microsope | Zeiss | N/A |
Induction and recombination experiments.
hsFLP was used to excise an FRT-Stop-FRT cassette to allow expression of Ds in the posterior compartment under control of hh-GAL4 (hh-GAL4/UAS-FRT-Stop-FRT-ds). Heat shocks were performed for 2 h at 37°C at 18 h APF, and pupa were left to age for 6 h at 25°C before dissection. P[w+, UAS-FRT-Stop-FRT-ds] was generated by cloning the ds coding sequence in the vector pUASt with insertion of a removable stop cassette between the promoter and the coding sequence and random integration into the genome by P-element transgenesis.
dsUA071 clones were generated using UbxFLP in the X chromosome and FRT40 with P[w+, arm-lacZ] FRT40 to mark cells where recombination did not occur.
Dissection and immunostaining
Pupal wings were dissected as described previously (Strutt, 2001). Pupae without cuticles were fixed with paraformaldehyde in phosphate-buffered saline [PBS] (PP) and wings dissected away, with 10% PP for a total of 60 min in assays involving microtubule imaging and 4% PP for 40 min for all other labeling. Pupal wings were blocked for 60 min in PBS containing 0.3% Triton X-100 (PBST) and 10% normal goat serum (NGS). Wings were incubated with primary antibodies and 10% NGS PBST overnight at 4°C, washed 10 times in PBST, incubated overnight with secondary antibodies and 10% NGS PBST overnight at 4°C, and washed in PBST again. Antibodies are listed in Table 1. Finally, wings were equilibrated in Vectashield (Vector Labs; H-1000) and mounted in the same media.
Microscopy
Images were acquired at room temperature (20–22°C). The subapical domain of the cell was identified using the localization of both tubulin and E-cadherin (see Figure 3). Pupal wings were oriented along the proximodistal axis using v4 as a reference. For analysis of Ds-EGFP and Patronin-GFP distribution and polarity, an upright confocal microscope (FV1000; Olympus) using a 60× 1.42 NA oil PlanApoN objective lens was used. Sixteen-bit depth images were taken at a magnification of 12.8 pixels/μm with 0.15 (Ds-EGFP) or 0.38 (Pat-GFP) µm between z-sections. Images of the subapical microtubule network were acquired using a Zeiss Airyscan microscope and the 63× objective lens. Z-stacks consisted of seven sections with 23.5 pixels/μm in XY resolution and 0.185 µm distance between sequential z-sections. All processing was done at 6.5 power in ZEN software. Representative images used for figure preparation are the average projections of the region of interest/analysis. Figures were assembled using Adobe CS3 Photoshop and Illustrator (http://www.adobe.com). The processing of images shown in the figures involved adjusting gamma settings.
Image processing
The QuantifyPolarity graphic user interface was previously described (Tan et al., 2021), and our custom automated image analysis scripts were written for MATLAB and can be found at www.github.com/nbul.
Planar polarity (QuantifyPolarity).
To measure polarity, wing images were aligned along the proximodistal wing axis based on wing vein orientation, and border masks were generated using Tissue Analyzer (Aigouy et al., 2010). On a cell-by-cell basis, the polarity magnitude of membrane proteins was computed using PCA in QuantifyPolarity (Tan et al., 2021). In this algorithm, a cell is first transformed onto a regular shape depending on the number of cell vertices. Then, intensities of individual pixels are normalized, and the polarity angle is calculated as the angle that produces the largest variance of normalized intensities. Finally, the normalized intensities are converted into pseudo–XY-coordinates and the eigenvalues of the covariation matrix of these transformed coordinates are used to compute the polarity magnitude. Importantly, this approach produces cell shape–independent measures of polarity angle and magnitude. The full description of the algorithm and the instructions for downloading are available in Tan et al. (2021). The same methodology was applied to measure the PCA component of Patronin-GFP and the distributions of E-cadherin and Ds as revealed by immunolabeling. Polarity angles were extracted and drawn over the segmentation masks using our custom automated script in MATLAB (https://github.com/nbul/Cytoskeleton/tree/master/PCP-MT).
Apicobasal intensity.
Only the pupal wing regions with cells in the same z-plane across the entire field were used for imaging. Average intensity was measured in each z-section for both Ds-EGFP and tubulin signals. Each z-section measurement was normalized to the maximum average intensity for this protein in every Z-stack, and the different samples were vertically aligned with respect to the maximum tubulin signal distribution. Average intensity profiles across all the samples were then calculated for both Ds-EGFP and tubulin signals.
Proximity analysis.
A MATLAB script (https://github.com/nbul/Proximity) was used to quantify the spatial proximity between tubulin or Patronin-GFP signals and the Ds puncta. The masks generated using Tissue Analyzer (Aigouy et al., 2010) were cleared of any cells contacting image borders and dilated using a disk-shaped structuring element with a radius of 3 pixels. The average signal intensities within cells defined by these dilated masks were used to calculate the cytoplasm intensity from average intensity projections of Z-stacks with tubulin, Patronin-GFP, or Ds signals. These average intensity projections were adjusted so that 0.5% of the pixels with the lowest intensities were set to black and the 0.5% of the pixels with the highest intensities were set to white to normalize the variability in signal between images and data sets but maintain the relative differences between different regions within each image. Then, Ds puncta were defined by thresholding images with the average projection of the Ds signal using a global threshold calculated by Otsu’s method and multiplied by an empirically determined factor (= 2.5). The overlap between the dilated masks and thresholded Ds puncta was used to determine the puncta and nonpuncta boundary regions and calculate their respective average intensities. Images with isolated puncta and nonpuncta regions were then dilated using a disk-shaped structuring element with a radius of 8 pixels. Following subtraction of undilated puncta and nonpuncta regions, these images were used to calculate the average intensities of puncta and nonpuncta haloes (areas directly adjacent to cell boundary regions with Ds puncta or depleted of Ds).
Microtubules.
For the analysis of the subapical microtubule network, we employed a protocol previously developed by us (Gomez et al., 2016; Płochocka et al., 2021). The average projections of images with tubulin signals were adjusted so that 0.5% of the pixels with the lowest intensities were set to black and the 0.5% of the pixels with the highest intensities were set to white to normalize the variability in signal between images and increase the contrast. Masks generated using Tissue Analyzer (Aigouy et al., 2010) were used to isolate tubulin signals in individual cells. Then, the magnitude of the tubulin signal according to its direction (gradient of the signal) in each cell was calculated by convolving the tubulin signal using two 5 × 5 Sobel operators (Gomez et al., 2016). The resulting distributions of tubulin signals were aligned to their maxima at 0 and averaged for cells with specified eccentricities (0.750 ± 0.025 and 0.800 ± 0.025) to produce average profiles of microtubule angle distributions in cells (as in Figure 5) (Płochocka et al., 2021). At the same time, the unaltered distributions were fitted with the Von Mises distribution and the estimated mean and SD of the fitted curve in each cell. The mean was used as the main direction of the subapical microtubule network in this cell and the SD as the measure of the microtubule alignment with each other (MTSD). Finally, the cell directions were calculated by fitting the pixel coordinates of each cell isolated using masks to an ellipse and obtaining the direction of the long axis of the best-fit ellipse. The MTSD was plotted against cell eccentricity for cells with a MTSD <90, excluding cells with unfittable distributions of tubulin signal. The discarded cells (cells/% of total) were as follows: Figure 5C, 8/7% (distal) and 5/2% (posterior); Figure 7C, 16/4% (distal control), 9/3% (distal Ds OE), 11/3% (posterior control), and 9/2% (posterior Ds OE); Figure 8D, 11/8% (distal control), 7/4% (distal Ds), 4/2% (posterior control), and 2/1% (posterior Ds). The MATLAB code used in this study is available at https://github.com/nbul/Cytoskeleton/tree/master/PCP-MT.
Angle visualization and quantification.
Data about directions of cell elongation (the direction of the long axis of the best-fit ellipse), overall directions of microtubule networks, and Ds polarity angle (transferred from QuantifyPolarity) were plotted on image masks produced using Tissue Analyzer (Aigouy et al., 2010). The length of plotted lines does not reflect polarity magnitudes or length of elongation but is fixed at half of the average long cell axis. Average cell orientation within each biological replicate/image was used to normalize the Ds polarity angle, individual cell orientations, and microtubule angles for each cell in polar histograms with nine bins in 0–180° format. Data were mirrored to ease visualization. Normalized angle data were also used to separate angles into quadrants: 0 ± 45° and 90 ± 45°. The plotting and calculations were performed within the script for the microtubule organization analysis (https://github.com/nbul/Cytoskeleton/tree/master/PCP-MT).
Statistical analysis
Data sets were subjected to the D’Agostino and Pearson test to determine normality (Shapiro–Wilk test was used for small data sets). PCA polarity magnitude and average intensity in different regions were tested with one-way analysis of variance (ANOVA) (normal distribution) or Friedman test (nonnormal distribution), with multiple comparisons performed with Tukey’s or Dunn’s tests, respectively. Data sets in different regions were paired when belonging to the same biological replicate (individual). The curve fitting of cell eccentricities was compared with an extra-sum-of-squares F test. Average border intensity of Pat-GFP was compared with Brown-Forsythe and Welch ANOVA test and Dunnett’s T3 test for multiple comparisons (comparisons with Ds OE) or paired t test (regions from the control genotype). The relationships between MTSD and cell eccentricity were tested between regions or genotypes by comparing their respective Pearson’s correlations using the bootstrap method (Wilcox, 2009). Percentages of angles in the proximodistal quadrant (0 ± 45°) for PCP polarity, average microtubule orientation or cell angle, were compared using either one-way ANOVA (normal distribution; Figure 2), Kruskal–Wallis (nonnormal distribution) or two-way ANOVA (polarity data in Figure 6). Multiple comparisons were performed with Brown-Forsythe and Welch (for ANOVAs), Dunn’s (for Kruskal–Wallis), or Šidák tests.
Data availability
The generated Drosophila strains are available upon request. All in-house scripts are available at https://github.com/nbul/.
Acknowledgments
We acknowledge the Bloomington Drosophila Stock Center for fly stocks and the Developmental Studies Hybridoma Bank for antibodies. We thank the technical staff of the Wolfson Light Microscopy Facility and the Drosophila Facility at the University of Sheffield. All imaging work was performed at the Wolfson Light Microscopy Facility, using the FV1000 Olympus and Airyscan Zeiss microscopes. This work was supported by a grant from the Biotechnology and Biological Sciences Research Council, UK Research and Innovation (BB/SOO7342/1) to D.S. and N.A.B. and a Wellcome Trust Senior Fellowship (210630/Z/18/Z) to D.S.
Abbreviations used:
- a-cv
anterior crossvein
- ANOVA
analysis of variance
- APF
after pupa formation
- arm
armadillo
- Ds
dachsous
- EGFP
enhanced green fluorescent protein
- FLP
flippase
- FRT
flippase recognition target
- Ft
fat
- hh
hedgehog
- β-Gal
β-galactosidase
- MTs
microtubules
- MTSD
microtubule angle standard deviation
- NA
numeric aperture
- NGS
normal goat serum
- OE
overexpression
- p-cv
posterior crossvein
- Pat
patronin
- PBS
phosphate-buffered saline
- PBST
PBS with Triton X-100
- PCA
principal component analysis
- PP
paraformaldehyde in PBS
- UAS
upstream activating sequence
- Ubx
ultrabithorax
- v4
wing vein 4
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-09-0442) on February 3, 2023.
REFERENCES
- Adler PN, Charlton J, Liu J (1998). Mutations in the cadherin superfamily member gene dachsous cause a tissue polarity phenotype by altering frizzled signaling. Development 125, 959–968. [DOI] [PubMed] [Google Scholar]
- Aigouy B, Farhadifar R, Staple DB, Sagner A, Röper JC, Jülicher F, Eaton S (2010). Cell flow reorients the axis of planar polarity in the wing epithelium of Drosophila. Cell 142, 773–786. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Kapitein LC (2022). Mechanisms of microtubule organization in differentiated animal cells. Nat Rev Mol Cell Biol 23, 541–558. [DOI] [PubMed] [Google Scholar]
- Bosveld F, Bonnet I, Guirao B, Tlili S, Wang Z, Petitalot A, Marchand R, Bardet PL, Marcq P, Graner F, Bellaïche Y (2012). Mechanical control of morphogenesis by Fat/Dachsous/Four-jointed planar cell polarity pathway. Science 336, 724–727. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Brittle A, Thomas C, Strutt D (2012). Planar polarity specification through asymmetric subcellular localization of Fat and Dachsous. Curr Biol 22, 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgakova NA, Grigoriev I, Yap AS, Akhmanova A, Brown NH (2013). Dynamic microtubules produce an asymmetric E-cadherin-Bazooka complex to maintain segment boundaries. J Cell Biol 201, 887–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MT, Wallingford JB (2017). Planar cell polarity in development and disease. Nat Rev Mol Cell Biol 18, 375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien YH, Keller R, Kintner C, Shook DR (2015). Mechanical strain determines the axis of planar polarity in ciliated epithelia. Curr Biol 25, 2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devenport D (2014). The cell biology of planar cell polarity. J Cell Biol 207, 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda M, Kirkland NJ, Khalilgharibi N, Tozluoglu M, Yuen AC, Carpi N, Bove A, Piel M, Charras G, Baum B, Mao Y (2019). Polarization of myosin II refines tissue material properties to buffer mechanical stress. Dev Cell 48, 245–260.e247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery G, Hutterer A, Berdnik D, Mayer B, Wirtz-Peitz F, Gaitan MG, Knoblich JA (2005). Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell 122, 763–773. [DOI] [PubMed] [Google Scholar]
- Etournay R, Popović M, Merkel M, Nandi A, Blasse C, Aigouy B, Brandl H, Myers G, Salbreux G, Jülicher F, Eaton S (2015). Interplay of cell dynamics and epithelial tension during morphogenesis of the Drosophila pupal wing. eLife 4, e07090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germani F, Bergantinos C, Johnston LA (2018). Mosaic analysis in Drosophila. Genetics 208, 473–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic KG, Lindquist S (1989). The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59, 499–509. [DOI] [PubMed] [Google Scholar]
- Gomez JM, Chumakova L, Bulgakova NA, Brown NH (2016). Microtubule organization is determined by the shape of epithelial cells. Nat Commun 7, 13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV, Strutt D (2011). Principles of planar polarity in animal development. Development 138, 1877–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin SS, Vale RD (2010). Patronin regulates the microtubule network by protecting microtubule minus ends. Cell 143, 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale R, Brittle AL, Fisher KH, Monk NA, Strutt D (2015). Cellular interpretation of the long-range gradient of Four-jointed activity in the Drosophila wing. eLife 4, e05789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harumoto T, Ito M, Shimada Y, Kobayashi TJ, Ueda HR, Lu B, Uemura T (2010). Atypical cadherins Dachsous and Fat control dynamics of noncentrosomal microtubules in planar cell polarity. Dev Cell 19, 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herawati E, Taniguchi D, Kanoh H, Tateishi K, Ishihara S, Tsukita S (2016). Multiciliated cell basal bodies align in stereotypical patterns coordinated by the apical cytoskeleton. J Cell Biol 214, 571–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S, Mii Y, Charras G, Michiue T (2022). Alignment of cell long axis by unidirectional tension acts cooperatively with Wnt signalling to establish PCP. Development 149, dev200515. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R (2005). Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci 6, 201–214. [DOI] [PubMed] [Google Scholar]
- Kimura T, Saito H, Kawasaki M, Takeichi M (2021). CAMSAP3 is required for mTORC1-dependent ependymal cell growth and lateral ventricle shaping in mouse brains. Development 148, dev195073. [DOI] [PubMed] [Google Scholar]
- Komarova Y, De Groot CO, Grigoriev I, Gouveia SM, Munteanu EL, Schober JM, Honnappa S, Buey RM, Hoogenraad CC, Dogterom M, et al. (2009). Mammalian end binding proteins control persistent microtubule growth. J Cell Biol 184, 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence PA, Casal J (2018). Planar cell polarity: two genetic systems use one mechanism to read gradients. Development 145, dev168229. [DOI] [PubMed] [Google Scholar]
- Li-Villarreal N, Forbes MM, Loza AJ, Chen J, Ma T, Helde K, Moens CB, Shin J, Sawada A, Hindes AE, et al. (2015). Dachsous1b cadherin regulates actin and microtubule cytoskeleton during early zebrafish embryogenesis. Development 142, 2704–2718. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Ma D, Amonlirdviman K, Raffard RL, Abate A, Tomlin CJ, Axelrod JD (2008). Cell packing influences planar cell polarity signaling. Proc Natl Acad Sci USA 105, 18800–18805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Yang CH, McNeill H, Simon MA, Axelrod JD (2003). Fidelity in planar cell polarity signalling. Nature 421, 543–547. [DOI] [PubMed] [Google Scholar]
- Mangione F, Martín-Blanco E (2018). The Dachsous/Fat/Four-Jointed pathway directs the uniform axial orientation of epithelial cells in the Drosophila abdomen. Cell Rep 25, 2836–2850. [DOI] [PubMed] [Google Scholar]
- Mao Y, Baum B (2015). Tug of war—the influence of opposing physical forces on epithelial cell morphology. Dev Biol 401, 92–102. [DOI] [PubMed] [Google Scholar]
- Mao Y, Tournier AL, Bates PA, Gale JE, Tapon N, Thompson BJ (2011). Planar polarization of the atypical myosin Dachs orients cell divisions in Drosophila. Genes Dev 25, 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci EM, Relich PK, Lakadamyali M, Ostap EM, Holzbaur ELF (2022). Microtubule dynamics influence the retrograde biased motility of kinesin-4 motor teams in neuronal dendrites. Mol Biol Cell 33, ar52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matis M, Russler-Germain DA, Hu Q, Tomlin CJ, Axelrod JD (2014). Microtubules provide directional information for core PCP function. eLife 3, e02893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel M, Sagner A, Gruber FS, Etournay R, Blasse C, Myers E, Eaton S, Jülicher F (2014). The balance of prickle/spiny-legs isoforms controls the amount of coupling between core and fat PCP systems. Curr Biol 24, 2111–2123. [DOI] [PubMed] [Google Scholar]
- Nestor-Bergmann A, Stooke-Vaughan GA, Goddard GK, Starborg T, Jensen OE, Woolner S (2019). Decoupling the roles of cell shape and mechanical stress in orienting and cueing epithelial mitosis. Cell Rep 26, 2088–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson J, Sharp KA, Matis M, Cho B, Axelrod JD (2014). Prickle/spiny-legs isoforms control the polarity of the apical microtubule network in planar cell polarity. Development 141, 2866–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzade S, Matis M (2021). The microtubule minus-end binding protein Patronin is required for the epithelial remodeling in the Drosophila abdomen. Front Cell Dev Biol 9, 682083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietra S, Ng K, Lawrence PA, Casal J (2020). Planar cell polarity in the larval epidermis of Drosophila and the role of microtubules. Open Biol 10, 200290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Płochocka AZ, Ramirez Moreno M, Davie AM, Bulgakova NA, Chumakova L (2021). Robustness of the microtubule network self-organization in epithelia. eLife 10, e59529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC, Sharp DJ, Vale RD (2002). Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol 158, 873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepich DS, Usmani M, Pawlicki S, Solnica-Krezel L (2011). Wnt/PCP signaling controls intracellular position of MTOCs during gastrulation convergence and extension movements. Development 138, 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Yonemura S, Ohkura H, Strutt D, Uemura T (2006). Polarized transport of Frizzled along the planar microtubule arrays in Drosophila wing epithelium. Dev Cell 10, 209–222. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ (2007). Microtubule-induced cortical cell polarity. Genes Dev 21, 483–496. [DOI] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H (2006). The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol 16, 2081–2089. [DOI] [PubMed] [Google Scholar]
- Singh A, Saha T, Begemann I, Ricker A, Nüsse H, Thorn-Seshold O, Klingauf J, Galic M, Matis M (2018). Polarized microtubule dynamics directs cell mechanics and coordinates forces during epithelial morphogenesis. Nat Cell Biol 20, 1126–1133. [DOI] [PubMed] [Google Scholar]
- Steinhauer J, Kalderon D (2006). Microtubule polarity and axis formation in the Drosophila oocyte. Dev Dyn 235, 1455–1468. [DOI] [PubMed] [Google Scholar]
- Strutt DI (2001). Asymmetric localization of frizzled and the establishment of cell polarity in the Drosophila wing. Mol Cell 7, 367–375. [DOI] [PubMed] [Google Scholar]
- Strutt H, Strutt D (2002). Nonautonomous planar polarity patterning in Drosophila: dishevelled-independent functions of frizzled. Dev Cell 3, 851–863. [DOI] [PubMed] [Google Scholar]
- Strutt H, Strutt D (2021). How do the Fat-Dachsous and core planar polarity pathways act together and independently to coordinate polarized cell behaviours? Open Biol 11, 200356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SE, Tan W, Fisher KH, Strutt D (2021). QuantifyPolarity, a new tool-kit for measuring planar polarized protein distributions and cell properties in developing tissues. Development 148, dev198952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto H, Itoh S, ten Dijke P, Tabata T (2000). Hedgehog creates a gradient of DPP activity in Drosophila wing imaginal discs. Mol Cell 5, 59–71. [DOI] [PubMed] [Google Scholar]
- Tateishi K, Nishida T, Inoue K, Tsukita S (2017). Three-dimensional organization of layered apical cytoskeletal networks associated with mouse airway tissue development. Sci Rep 7, 43783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toya M, Takeichi M (2016). Organization of non-centrosomal microtubules in epithelial cells. Cell Struct Funct 41, 127–135. [DOI] [PubMed] [Google Scholar]
- van Haren J, Wittmann T (2019). Microtubule plus end dynamics—do we know how microtubules grow? Cells boost microtubule growth by promoting distinct structural transitions at growing microtubule ends. Bioessays 41, e1800194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar EK, Bayly RD, Sangoram AM, Scott MP, Axelrod JD (2012). Microtubules enable the planar cell polarity of airway cilia. Curr Biol 22, 2203–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu HT, Mansour S, Kücken M, Blasse C, Basquin C, Azimzadeh J, Myers EW, Brusch L, Rink JC (2019). Dynamic polarization of the multiciliated planarian epidermis between body plan landmarks. Dev Cell 51, 526–542.e526. [DOI] [PubMed] [Google Scholar]
- Wang H, Brust-Mascher I, Civelekoglu-Scholey G, Scholey JM (2013). Patronin mediates a switch from kinesin-13-dependent poleward flux to anaphase B spindle elongation. J Cell Biol 203, 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox RR (2009). Comparing Pearson correlations: dealing with heteroscedasticity and nonnormality. Commun Stat Simul Comput 38, 2220–2234. [Google Scholar]
- Wu X, Hammer JA (2021). ZEISS Airyscan: optimizing usage for fast, gentle, super-resolution imaging. Methods Mol Biol 2304, 111–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt TP, Harris AR, Lam M, Cheng Q, Bellis J, Dimitracopoulos A, Kabla AJ, Charras GT, Baum B (2015). Emergence of homeostatic epithelial packing and stress dissipation through divisions oriented along the long cell axis. Proc Natl Acad Sci USA 112, 5726–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Rubin GM (1993). Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117, 1223–1237. [DOI] [PubMed] [Google Scholar]
- Zakaria S, Mao Y, Kuta A, de Sousa CF, Gaufo GO, McNeill H, Hindges R, Guthrie S, Irvine KD, Francis-West PH (2014). Regulation of neuronal migration by Dchs1-Fat4 planar cell polarity. Curr Biol 24, 1620–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelezov G, Alfred V, Bulgakova NA, Chumakova L (2019). The walkoff effect: cargo distribution implies motor type in bidirectional microtubule bundles. bioRxiv, 831024. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The generated Drosophila strains are available upon request. All in-house scripts are available at https://github.com/nbul/.