Abstract
During cell division, the microtubule nucleating and organizing organelle, known as the centrosome, is a critical component of the mitotic spindle. In cells with two centrosomes, each centrosome functions as an anchor point for microtubules, leading to the formation of a bipolar spindle and progression through a bipolar cell division. When extra centrosomes are present, multipolar spindles form and the parent cell may divide into more than two daughter cells. Cells that are born from multipolar divisions are not viable, and hence clustering of extra centrosomes and progression to a bipolar division are critical determinants of viability in cells with extra centrosomes. We combine experimental approaches with computational modeling to define a role for cortical dynein in centrosome clustering. We show that centrosome clustering fails and multipolar spindles dominate when cortical dynein distribution or activity is experimentally perturbed. Our simulations further reveal that centrosome clustering is sensitive to the distribution of dynein on the cortex. Together, these results indicate that dynein’s cortical localization alone is insufficient for effective centrosome clustering and, instead, dynamic relocalization of dynein from one side of the cell to the other throughout mitosis promotes timely clustering and bipolar cell division in cells with extra centrosomes.
INTRODUCTION
Proper formation and maintenance of the dynamic mitotic spindle is necessary to ensure accurate segregation of chromosomes and cell division resulting in two viable daughter cells. Assembly of the spindle is initiated by the nucleation of microtubules at each of two centrosomes, the organelles that function as the dominant microtubule organization center of the cell (Hinchliffe, 2011; Petry, 2016). Microtubule plus ends radiate outward from the centrosome and interact with the cell boundary, other microtubules, and chromosomes (Forth and Kapoor, 2017). In response to mechanical forces on and by the microtubules, the centrosomes are positioned and ultimately become the two poles of a bipolar spindle (Busson et al., 1998; Merdes et al., 2000; Goshima et al., 2005; Kiyomitsu and Cheeseman, 2012; di Pietro et al., 2016; Okumura et al., 2018).
Dynein is the major minus end–directed motor protein in the cell and has many functions. Throughout mitosis, dynein is localized at the kinetochore on chromosomes, where it contributes to chromosome movement and alignment (Bader and Vaughan, 2010), at the spindle poles, where it actively sustains focusing of microtubule minus ends at centrosomes (Merdes et al., 2000), and at the cell cortex, where it contributes to centrosome movement and thus impacts bipolar spindle length, orientation, and positioning (Busson et al., 1998; O’Connell and Wang, 2000; Kiyomitsu and Cheeseman, 2012; di Pietro et al., 2016; Okumura et al., 2018; Mercadante et al., 2021). Dynein and its binding partner dynactin are anchored at the cell cortex by a complex including the proteins LGN, Afadin, and NuMA (Du and Macara, 2004; Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012, 2013; di Pietro et al., 2016; Okumura et al., 2018). Cortical localization of dynein is regulated through phosphorylation of NuMA by the centrosome-localized kinases PLK1 and CDK1 (Kiyomitsu and Cheeseman, 2012; Kotak et al., 2013; Seldin et al., 2013; Sana et al., 2018). Defects in bipolar spindle orientation have been observed when cortical dynein localization is perturbed via enhanced phosphorylation of NuMA or experimental depletion of LGN or Afadin (Kiyomitsu and Cheeseman, 2012; Carminati et al., 2016).
Cells with more than two centrosomes, termed centrosome amplification, are common in cancer. Extra centrosomes induce the formation of multipolar spindles, and cell division can lead to the formation of multiple daughter cells that are nonviable (Kwon et al., 2008). However, cells with centrosome amplification often cluster their extra centrosomes into a functional bipolar spindle (Figure 1A), enabling a bipolar division and giving rise to two viable daughter cells (Kwon et al., 2008). This clustering activity is dependent on both passive and active mechanisms. Passive mechanisms, like contractility of the cortex-associated actin cytoskeleton, contribute broadly to centrosome positioning within the cell (Rhys et al., 2018). This helps to bring centrosomes within proximity of each other, where active mechanisms—those that allow centrosomes to engage and move with respect to each other—become relevant. Both motor-derived forces and cross-linking activity at the centrosomes have been shown to impact active clustering (Quintyne et al., 2005; Barr and Gergely, 2008; Kwon et al., 2008; Fielding et al., 2010; Leber et al., 2010; Ding et al., 2017). While cortical dynein–driven centrosome movement has been demonstrated to impact centrosome and spindle positioning in cells with two centrosomes, whether this activity may be relevant for either passive or active centrosome clustering in cells with centrosome amplification has not been investigated.
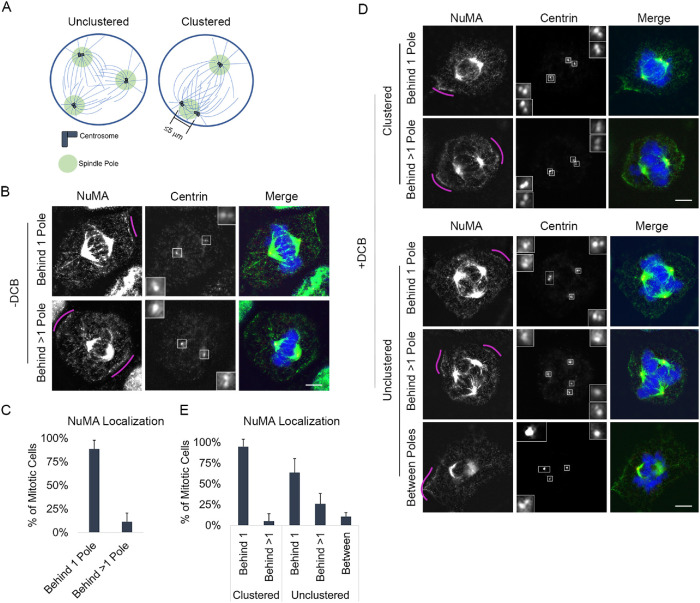
FIGURE 1:
NuMA is primarily enriched in a region behind a single spindle pole, regardless of centrosome number. (A) Schematic of cells with extra centrosomes that are unclustered and form a multipolar spindle (left) or are clustered and form a bipolar spindle (right). Centrosome clustering is defined as a spindle pole with two or more centrosomes that are within 5 µm of each other. (B, C) Representative fixed-cell images and quantification of cortical NuMA enrichment in mitotic RPE cells with two centrosomes. NuMA was characterized as being behind 1 spindle pole or >1 spindle pole. (D, E) Representative fixed-cell images and quantification of cortical NuMA enrichment in mitotic RPE cells following DCB treatment, which induces cytokinesis failure and results in cells with >2 centrosomes. NuMA was characterized as being behind 1 spindle pole or >1 spindle pole or between poles (behind 0 poles). See also Supplemental Figure S1. All RPE cells are stained with antibodies specific for NuMA (green) and centrin-2 (red); DNA is detected with DAPI (blue). In B and D, pink arcs highlight regions of increased cortical NuMA, insets represent 4× enlargements of centrosomes, and scale bars are 5 µm. In C and E, error bars are SD. In E, cells were grouped based on spindle polarity and analysis performed on 50 cells per group for each of three biological replicates.
We exploit a combination of molecular manipulation and computational cell modeling approaches to quantify the impact of cortical dynein–derived forces on centrosome clustering. We utilize image-based approaches to assess how disruption of cortical dynein impacts centrosome clustering and mitotic progression. Informed by previously published work and our own experimental results, we define a computational model of mitotic spindle formation and function to explore the relationship between cortical dynein–derived forces and centrosome movement in cells with centrosome amplification. Our findings implicate cortical dynein in the directed centrosome movement necessary to efficiently cluster centrosomes in cells with centrosome amplification.
RESULTS
Cortical dynein promotes centrosome clustering
In cells with two centrosomes, dynein and its cofactor NuMA are localized to the cell cortex in an asymmetric manner such that enrichment is primarily observed behind one spindle pole at a time (Kiyomitsu and Cheeseman, 2012). To assess whether similar localization is seen in cells with more than two centrosomes, we first induce centrosome amplification in retinal pigment epithelial (RPE) cells through induction of cytokinesis failure by treating cells with dihydrocytochalasin B (DCB). This approach generates tetraploid cells with four centrosomes (Andreassen et al., 1996). We next performed immunofluorescence imaging to assess NuMA localization. NuMA functions to link dynein to the actin cytoskeleton, and its localization mirrors that of dynein at the cell cortex (Kiyomitsu and Cheeseman, 2012; Seldin et al., 2013). Consistent with published work (Kiyomitsu and Cheeseman, 2012; Seldin et al., 2013), we find that cells with two centrosomes primarily exhibit NuMA enrichment behind a single pole (Figure 1, A–C, and Supplemental Figure S1, A and B). Similarly, we find that NuMA is also primarily enriched behind a single spindle pole in cells with centrosome amplification (Figure 1, D and E, and Supplemental Figure S1, A and B). This is true regardless of whether centrosomes are clustered together to form a bipolar spindle (where one or more spindle poles have two or more clustered centrosomes located within 5 µm of each other as in Figure 1A) or remain unclustered to form a multipolar spindle.
Work from our group and others has implicated cortical dynein as a dominant driver of centrosome movement during mitosis in cells with two centrosomes (di Pietro et al., 2016; Mercadante et al., 2021). To test whether cortical dynein–driven centrosome movement is involved in centrosome clustering in cells with centrosome amplification, we perturbed dynein localization or function in cells with experimentally induced centrosome amplification. Two complementary approaches (Supplemental Figure S1, C–E) were used to induce centrosome amplification: overexpression of PLK4 (indPLK4) to induce the biogenesis of extra centrosomes (Godinho et al., 2009) and induction of cytokinesis failure, as described above. Next, cortical dynein motor activity was disrupted using the small molecule inhibitor dynarrestin (Hoing et al., 2018; Mercadante et al., 2021), or dynein localization was perturbed via small interfering RNA (siRNA)-mediated depletion of Afadin or LGN (Supplemental Figure S1, C–E). Individual cells were analyzed for centrosome number, centrosome location, and spindle structure, where clustering was assessed preanaphase and bipolar spindles were considered as those in which one or both spindle poles contained two or more centrosomes located within 5 µm of each other. Cells with centrosome amplification and functional cortical dynein are efficient at clustering centrosomes, such that ∼50% of cells with >2 centrosomes are able to form clustered bipolar spindles (Figure 2, A–D). In contrast, inhibition of dynein (dynarrestin) or dynein delocalization from the cortex (Afadin or LGN depletion) decreased centrosome clustering such that only ∼20% of cells with centrosome amplification exhibit clustered bipolar spindles (Figure 2, A–D).
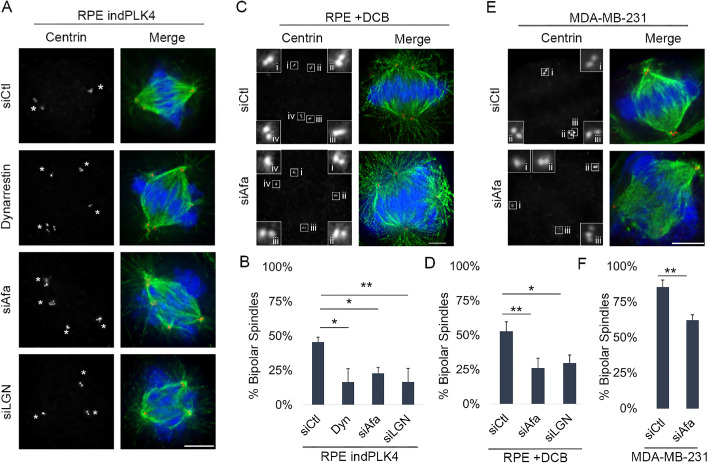
FIGURE 2:
Centrosome clustering is compromised in the absence of cortical dynein activity. Representative fixed-cell images of cell lines with extra centrosomes, through overexpression of PLK4 (A: RPE indPLK4), DCB to induce cytokinesis failure (C: RPE DCB), or preexisting centrosome amplification (E: MDA-MB-231). Cells were treated with the dynein inhibitor dynarrestin, a nontargeting siRNA (siCtl), or a LGN or Afadin-targeting siRNA (siLGN or siAfadin) to disrupt dynein localization at the cortex (see Supplemental Figure S1 for experimental setup and confirmation of Afadin and LGN depletion). Antibodies specific for α-tubulin (green), centrin-2 (red), and DAPI (blue) to detect DNA were utilized to assess mitotic stage and spindle structure. The white asterisks denote spindle poles, insets represent 4× enlargements of individual centrosomes, and scale bars are 5 µm. Quantification of the percentage of mitotic cells (preanaphase) exhibiting clustered bipolar spindles is given: (B: RPE indPLK4), (D: RPE DCB), and (F: MDA-MB-231). The analysis was performed on at least 50 cells per condition from three biological replicates, and error bars are SD. Significance was determined via a Student’s t test when comparing two conditions and a one-way ANOVA with Dunnett’s test for multiple comparisons when comparing multiple conditions to one control; *p < 0.05, **p < 0.01.
To explore whether dynein activity similarly impacts centrosome clustering in cells with preexisting centrosome amplification, we perturbed cortical dynein localization in MDA-MB-231 cells (Supplemental Figure S1F), a well-characterized breast cancer cell line with centrosome amplification (Mittal et al., 2017). Centrosome positioning and spindle structure were assessed as described above. Nearly 45% of mitotic MDA-MB-231 cells have centrosome amplification and/or fragmentation (Supplemental Figure S1F). Of the mitotic cells with more than two centrosomes, ∼80% exhibit centrosome clustering and bipolar spindle formation (Figure 2, E and F). Consistent with our results in cells with induced centrosome amplification, MDA-MB-231 cells exhibit a reduction in centrosome clustering when cortical dynein localization is perturbed, such that only ∼60% of preanaphase cells form a clustered bipolar spindle following Afadin depletion (Figure 2, E and F).
Cells complete multipolar divisions in the absence of cortical dynein
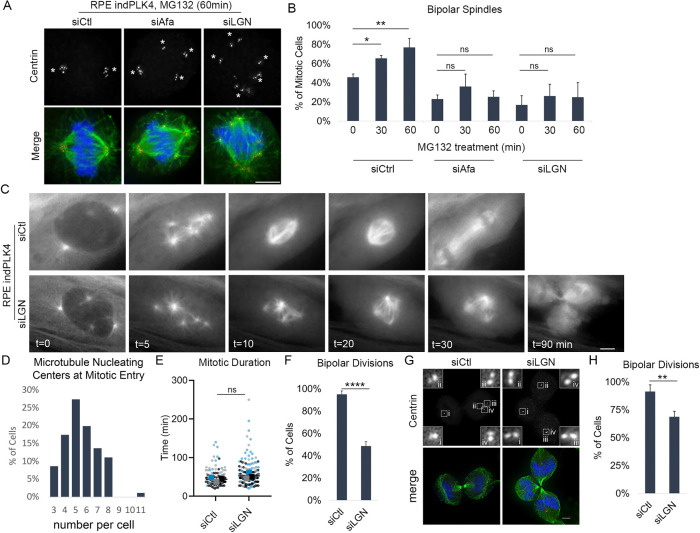
Centrosome clustering is a dynamic process that can take 1 h or longer to achieve (Kwon et al., 2008; Navarro-Serer et al., 2019), and analysis of a single time point in mitosis cannot distinguish between a delay and a deficit in centrosome clustering. To further assess the observed increase in multipolar spindles seen following cortical dynein disruption, cells were additionally treated with the protease inhibitor MG132 before assessment of centrosome positioning and spindle structure. MG132 prevents anaphase onset, providing cells additional time in which to cluster centrosomes. Consistent with previous findings (Rhys et al., 2018), cells with centrosome amplification progress from ∼50 to 60% and then 80% bipolar spindles following 30 or 60 min of MG132-induced mitotic arrest, respectively (siCtl in Figure 3, A and B). In contrast, the percentage of mitotic cells with extra centrosomes and perturbed dynein activity that are able to form a bipolar spindle does not increase with prolonged mitotic duration, remaining primarily multipolar, even after 60 min of MG132 treatment (siAfa and siLGN in Figure 3, A and B).
FIGURE 3:
Cortical dynein activity promotes centrosome clustering and bipolar cell division. (A, B) Representative fixed-cell images and quantification of the frequency of spindle bipolarity in cells induced to have extra centrosomes through overexpression of PLK4 (RPE indPLK4). Cells were treated with nontargeting (siCtl) or Afadin- or LGN-targeting (siAfa or siLGN) siRNA to disrupt cortical dynein localization, subsequently treated as indicated with MG132 to prevent anaphase onset. Antibodies specific for centrin-2 (red), tubulin (green), and DAPI (blue) to detect DNA were utilized to assess spindle structure. (C) Still frames from live cell imaging of RPE indPLK4 cells expressing EGFP-tubulin are shown for both control (siCtl) and LGN (siLGN) depleted cells at the indicated time points. (D) Microtubule-organizing centers in RPE indPLK4 cells were quantified at nuclear envelope breakdown (characterized by loss of EGFP exclusion from the nucleus). (E) Mitotic progression of RPE indPLK4 cells was timed from nuclear envelope breakdown until anaphase B (indicated by rapid elongation of the spindle), and (F) the frequency of mitotic cells progressing through a bipolar division was quantified. (G, H) Representative images and quantification of cells progressing through a bipolar division (determined at anaphase/telophase) in cells with extra centrosomes acquired through DCB-induced cytokinesis failure. The control and LGN depleted RPE DCB cells were stained with antibodies specific for centrin-2 (red) and tubulin (green) and assessed for evidence of bipolar vs. multipolar divisions. Scale bars in A, C, and G are 5 µm, and insets in G represent 4× enlargements of individual centrosomes. All quantifications were performed on at least 50 cells per condition from each of three biological replicates, and error bars are SD. Significance was determined by Student’s t test when comparing two conditions and a one-way ANOVA with Dunnett’s test for multiple comparisons when comparing multiple conditions to one control; *p < 0.05, **p < 0.01, ****p < 0.0001, ns indicates not significant.
Results of these fixed-cell analyses can support two distinct modes in the absence of Afadin/LGN. Either the majority of cells never achieve clustering, or alternatively, dynamic/unstable clustering is achieved while the steady state of clustering remains low. To distinguish between these modes and assess the temporal relationship between spindle dynamics and the mitotic fate of cells following loss of cortical dynein activity, we performed time-lapse imaging of cells stably expressing fluorescently tagged tubulin (EGFP-tubulin; Figure 3C) (Mercadante et al., 2019) and induced cells to have extra centrosomes through induction of PLK4 expression (Figure 3D), as described above. Phase contrast and fluorescence images were captured every 2.5 min throughout mitosis and were used to assess spindle structure, mitotic timing, and the number of progeny resulting from each division. We find that individual mitotic cells with centrosome amplification cluster extra centrosomes and progress through mitosis in ∼45 min. The vast majority (95%) of these cells complete a bipolar division (Figure 3, E and F). Disruption of cortical dynein activity does not alter mitotic timing (Figure 3E). However, consistent with the centrosome clustering defects described above, 50% of cells with centrosome amplification that have disrupted cortical dynein activity exit mitosis with a multipolar division (Figure 3, C and F). To confirm that spindle pole clustering (or lack thereof) in the live cell imaging approaches reflects the behavior of centrosomes, we next performed fixed-cell analysis to quantify the frequency of centrosome clustering and multipolar divisions in anaphase and telophase cells stained for both microtubules and centrosomes. These analyses similarly reveal that clustering is reduced in LGN-depleted anaphase cells in which centrosome amplification has been induced (Figure 3, G and H). Together, these results implicate cortical dynein as necessary for centrosome clustering and bipolar division in cells with centrosome amplification.
Centrosome clustering is sensitive to dynein activity at the cortex
To further manipulate cortical dynein and explore the extent to which it impacts centrosome clustering in cells with centrosome amplification, we developed a force balance model that tracks centrosome movement and spindle formation, taking into account major motor-derived forces during mitosis. This model was informed by our previous work (Mercadante et al., 2021) and modified to account for the presence of >2 centrosomes. To simulate a cell that has rounded in mitosis, the cell boundary is defined as a rigid circle with a diameter of 30 µm. Dynamic microtubules are short at mitotic entry and elongate through mitotic progression (Figure 4A). Motor proteins Eg5, HSET, and dynein push or pull on the microtubule (and hence exert force on the centrosome the microtubule is attached to). These motor-dependent forces, along with forces associated with microtubules pushing on the cell cortex, are evaluated at 0.5 s intervals to determine centrosome movement in the mitotic cell (Supplemental Figure S2A). Motor-dependent force generation is dependent on distances between model entities and modulated by altering the stochastic binding probability of a motor; increased binding probability equates to increased motor activity (Supplemental Figure S2B). Additional model details are given in Materials and Methods.
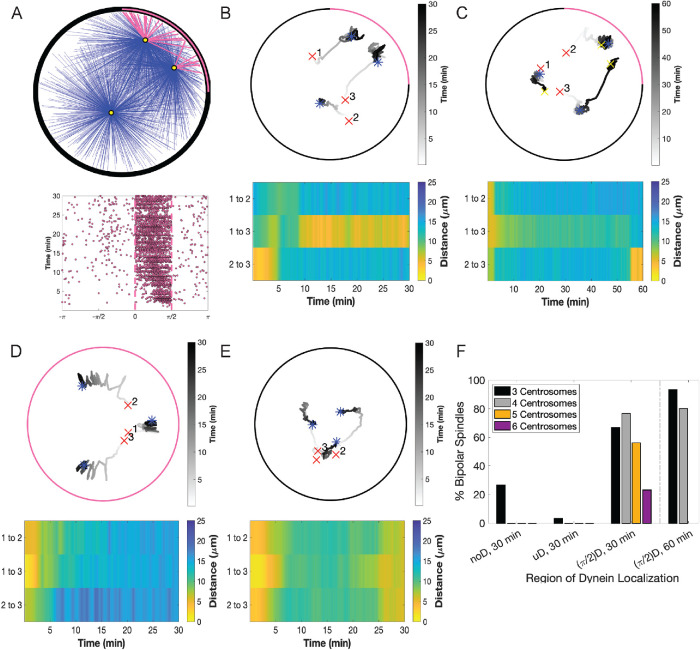
FIGURE 4:
Computational modeling suggests that restricted cortical dynein localization drives cen- trosome clustering. (A) Simulation results where cortical dynein is enriched in the region from 0 to π/2 (region shown with a pink arc in A, top, and dashed lines in A, bottom). Top, Simulation at t = 30 min showing centrosomes in yellow, microtubules bound to cortical dynein in pink, and all other microtubules in blue. Bottom, Plot of microtubules binding to cortical dynein on the boundary of the cell (−π to π) from the simulation shown above. Each dot indicates an individual microtubule binding to dynein. (B–E) Top, Trace of centrosome movement over time through the duration of a simulation, where a red “x” indicates initial centrosome position, a blue “*” indicates centrosome position at 30 min, and a yellow “x” (C only) indicates centrosome position at 60 min. Numbers mark individual centrosomes, and grayscale indicates time. The pink on the cell boundary indicates the region of high dynein activity (where 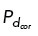 = 0.5; elsewhere
= 0.5; elsewhere 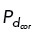 = 0.01). Here, B and C have dynein enrichment in the region 0 to π/2, D has uniform dynein on the entire cell boundary, and E has no dynein. (B–E) Bottom, Heat map representing the pairwise distances between all centrosome pairs indicated in the corresponding traces in the top panel. (F) The percentage of simulations that achieve bipolar spindles (each of the two poles having one or more clustered centrosomes) when cortical dynein is absent (noD), distributed uniformly across the cell boundary (uD), or enriched in the region from 0 to π/2 ((π/2)D); duration of simulations is t = 30 or 60 min as indicated. Data are an average over 30 simulations, and data for five and six centrosomes are shown for only t = 30 min.
= 0.01). Here, B and C have dynein enrichment in the region 0 to π/2, D has uniform dynein on the entire cell boundary, and E has no dynein. (B–E) Bottom, Heat map representing the pairwise distances between all centrosome pairs indicated in the corresponding traces in the top panel. (F) The percentage of simulations that achieve bipolar spindles (each of the two poles having one or more clustered centrosomes) when cortical dynein is absent (noD), distributed uniformly across the cell boundary (uD), or enriched in the region from 0 to π/2 ((π/2)D); duration of simulations is t = 30 or 60 min as indicated. Data are an average over 30 simulations, and data for five and six centrosomes are shown for only t = 30 min.
Our immunofluorescence analysis showed that regardless of centrosome number, dynein primarily localized in a region behind a single spindle pole (Figure 1E and Supplemental Figure S1B). To assess the functional relevance of the NuMA/dynein distribution on centrosome clustering, we manipulated the distribution of cortical dynein activity in our model. The NuMA/dynein distribution was mimicked by permitting microtubules on centrosomes to bind to cortical dynein with probability 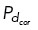 = 0.5 on one fixed quadrant (angular region of π/2), while having a much smaller binding probability,
= 0.5 on one fixed quadrant (angular region of π/2), while having a much smaller binding probability, 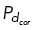 = 0.01, everywhere else (Figure 4, A and B, and Supplemental Figure S3, A, D, and G). We then assessed the movement of three, four, five, and six centrosomes to reflect the three to six centrosomes seen, on average, in our experimentally induced systems (Supplemental Figure 1, D and E; Figure 3D). Figure 4B is a representative simulation with three centrosomes. Initially, centrosomes numbered 2 and 3 start close together but move apart within a few minutes, whereas centrosomes 1 and 3 start further apart and cluster together around t = 10 min and remain clustered up until t = 30 min. We define this as sustained clustering: centrosomes that have a pairwise distance of ≤5 µm that is maintained for the duration of the simulation after initial centrosome separation (after t = 10 min).
= 0.01, everywhere else (Figure 4, A and B, and Supplemental Figure S3, A, D, and G). We then assessed the movement of three, four, five, and six centrosomes to reflect the three to six centrosomes seen, on average, in our experimentally induced systems (Supplemental Figure 1, D and E; Figure 3D). Figure 4B is a representative simulation with three centrosomes. Initially, centrosomes numbered 2 and 3 start close together but move apart within a few minutes, whereas centrosomes 1 and 3 start further apart and cluster together around t = 10 min and remain clustered up until t = 30 min. We define this as sustained clustering: centrosomes that have a pairwise distance of ≤5 µm that is maintained for the duration of the simulation after initial centrosome separation (after t = 10 min).
This simulation is also classified as having formed a bipolar spindle where the sustained clustering of centrosomes 1 and 3 is one pole and the other pole is centrosome 2. Similarly, Supplemental Figure S3A shows the case where four centrosomes move and after t = 10 min, there is sustained clustering of three centrosomes that form a spindle pole close to the dynein-enriched region with the second pole as a single centrosome 15 µm away. Simulations with five and six centrosomes also form a bipolar spindle, with one pole having a single centrosome and the other pole consisting of the rest of the centrosomes clustered together (Supplemental Figure S3, D and G). In contrast, cases where the three (or four) centrosomes each have pairwise distances apart in the range of 7–20 µm from each other after t = 10 min and do not cluster are shown in Figure 4, D and E (and Supplemental Figure S3, B and C, for four centrosomes), corresponding to the case of uniform dynein (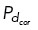 = 0.5 everywhere on the cortex) and no dynein (
= 0.5 everywhere on the cortex) and no dynein (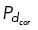 = 0.01 everywhere on the cortex), respectively. Similarly, in the cases of uniform dynein or no dynein, a bipolar spindle is not formed for both five and six centrosomes (Supplemental Figure S3, E, F, H, and I).
= 0.01 everywhere on the cortex), respectively. Similarly, in the cases of uniform dynein or no dynein, a bipolar spindle is not formed for both five and six centrosomes (Supplemental Figure S3, E, F, H, and I).
Owing to the inherent stochasticity of motor and microtubule dynamics, we ran 30 simulations (unless otherwise stated) to quantify the frequency of sustained clustering and bipolar spindle formation. We find that in simulations with cortical dynein primarily in a single quadrant or region of π/2, ∼65% of cells with three centrosomes and ∼75% of cells with four centrosomes formed bipolar spindles within 30 min (Figure 4F). In simulations with three centrosomes where dynein is uniformly distributed along the cortex or removed entirely, bipolarity is greatly reduced to ∼5 and ∼25%, respectively. We observe an even larger reduction in bipolarity in simulations with four, five, and six centrosomes in the case of uniform dynein and removal of dynein (Figure 4F). In this case, the simulations are not able to form a bipolar spindle and always have multiple distinct poles (Supplemental Figure S3, B, C, E, F, H, and I). The reduction in bipolar spindle formation in the case of no dynein is consistent with our biological results in RPE cells with centrosome amplification and perturbed dynein localization (Figure 2, B, D, and F). Given that our biological experiments indicate that the frequency of centrosome clustering is sensitive to mitotic duration (when cortical dynein is not perturbed), we next sought to determine whether centrosome clustering in our model is similarly sensitive to simulation duration. Consistent with our biological results, extension of the simulations from 30 to 60 min increases the frequency of centrosome clustering from ∼65 to ∼90% with three centrosomes (Figures 3, A and B, and 4, C and F) and from ∼75 to ∼80% with four centrosomes (Figure 4F).
To further investigate the functional relationship between dynein and centrosome clustering, we quantified cortical dynein–derived forces and centrosome position over time in simulations where dynein was enriched in an angular region of π/2, or alternatively, distributed uniformly along the cortex. These data reveal that as cortical dynein engages and pulls on centrosome-anchored microtubules (as indicated by peaks in dynein-derived force felt by each centrosome; red bars, Supplemental Figure S5, A and B), a corresponding decrease in the distance between the centrosomes and the region of dynein enrichment was observed (blue trace, Supplemental Figure S5, A and B). In simulations with three centrosomes, where dynein is restricted to a cortical region equal to π/2, two centrosomes generally exhibit sustained clustering and form a pole closer to the region of dynein enrichment. The remaining centrosome does not interact with cortical dynein and remains ∼17 µm from the dynein-enriched region, forming a bipolar spindle (Supplemental Figures S5, C and D, and S6A).
In contrast, when dynein is uniformly distributed on the cortex, we observe cortical dynein acting on microtubules associated with all centrosomes; all centrosomes end up at a distance of ∼5 µm from the cortex but are not able to cluster (Supplemental Figures S5, B and C, and S6B). In the case of no dynein, centrosomes do not cluster and pairwise distances between centrosomes stabilizes at ∼8 µm apart (Supplemental Figures S5, D and E, and S6C). These data indicate that centrosome movement is responsive to cortical dynein–derived forces but that restricted localization of dynein along the cortex is necessary to promote centrosome clustering and spindle bipolarity.
Oscillations in cortical dynein enrichment impact centrosome movement
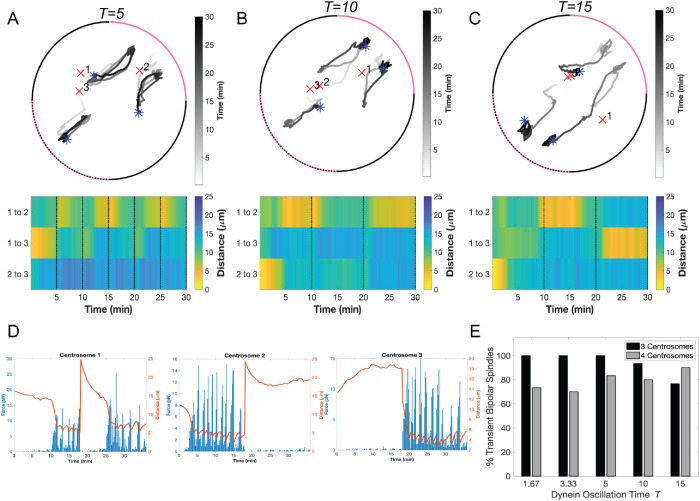
In cells with two centrosomes, cortical dynein localization is primarily behind one spindle pole at a time and oscillates from behind one spindle pole to the other every few minutes (Kiyomitsu and Cheeseman, 2012). To determine whether dynamic cortical dynein localization influences centrosome clustering in cells with centrosome amplification, we mimicked the observed biological oscillations by moving the π/2 angular region of enrichment to the opposite side of the cortex every t minutes (t = 5, 10, and 15 in Figure 5, A–C, and t = 1.67 and 2.33 in Supplemental Figure S7, A and B). In contrast to static dynein enrichment in an angular region of π/2, we do not observe sustained clustering for periods of 20 or 25 min (comparing Figure 4B with Figure 5, A–C). Instead, centrosomes with microtubules bound to cortical dynein moved toward the region of dynein activity, effectively dragging similarly engaged centrosomes toward each other, and when dynein activity oscillates to the opposing quadrant, previously engaged centrosomes do not sustain their close proximity to each other (Figure 5, A–C; Supplemental Figure S7, A and B).
FIGURE 5:
Oscillatory redistribution of cortical dynein activity in computational modeling enhances centrosome clustering. (A–C) Top, Traces of centrosome movement over time from a simulation with dynein-enriched regions (with 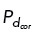 = 0.5) oscillating between the upper-right (0 to π/2, solid pink arc) and the lower-left (−π to −π/2, dashed pink arc) quadrants with periods of t = 5 min (A), 10 min (B), or 15 min (C). Initial centrosome position indicated by a red “x,” and final centrosome position indicated by a blue asterisk. Grayscale indicates time. (A–C) Bottom, Heat map representing the pairwise distances between all centrosomes from the corresponding simulation above. Black dotted lines indicate a time point when the quadrant of cortical dynein enrichment is updated. (D) Plots depicting the magnitude of cortical dynein–derived forces over time (blue bars, scale on left axis) and the distance between each centrosome and the midpoint of dynein localization (red trace, scale on right axis) from the simulation shown in C, where cortical dynein is redistributed at t = 15 min. (E) Quantification of transient bipolar spindle formation (each of the two poles having one or more clustered centrosomes) for simulations with either three or four centrosomes. Data are for 30 simulations.
= 0.5) oscillating between the upper-right (0 to π/2, solid pink arc) and the lower-left (−π to −π/2, dashed pink arc) quadrants with periods of t = 5 min (A), 10 min (B), or 15 min (C). Initial centrosome position indicated by a red “x,” and final centrosome position indicated by a blue asterisk. Grayscale indicates time. (A–C) Bottom, Heat map representing the pairwise distances between all centrosomes from the corresponding simulation above. Black dotted lines indicate a time point when the quadrant of cortical dynein enrichment is updated. (D) Plots depicting the magnitude of cortical dynein–derived forces over time (blue bars, scale on left axis) and the distance between each centrosome and the midpoint of dynein localization (red trace, scale on right axis) from the simulation shown in C, where cortical dynein is redistributed at t = 15 min. (E) Quantification of transient bipolar spindle formation (each of the two poles having one or more clustered centrosomes) for simulations with either three or four centrosomes. Data are for 30 simulations.
We do observe transient clustering where centrosomes are ≤5 µm apart for several minutes and move apart only after the region of dynein localization oscillates to the other side of the cortex. For example, in Figure 5C, in the first 15 min period where dynein is enriched in the upper right quadrant of the cortex, centrosomes 1 and 2 exhibit clustering, and cortical dynein–induced forces on these centrosomes is largest for t = 10–15 min (Figure 5D). At t = 15 min, dynein is then enriched in the lower left quadrant and we observe the pairwise distances between centrosomes 1 and 2 increasing whereas the pairwise distances of centrosomes 1 and 3 decrease and they cluster (with increased cortical dynein–induced forces acting on these centrosomes after t = 20 min; Figure 5D). To determine whether changes in pairwise centrosome distance is a consequence of loss of cortical dynein behind the clustered pair or instead results from dynein-dependent pulling from the opposing side, we performed simulations in which microtubules bind to asymmetric cortical dynein until t = 15 min and then dynein is removed for the duration of the simulation (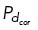 = 0.01 everywhere for t ≥ 15 min). Assessment of dynein-derived forces and corresponding centrosome movement reveals that centrosomes cluster toward the region of enriched dynein but begin separating after dynein is removed at t = 15 min (Supplemental Figure S7, C and D), indicating that loss of clustering is a passive event.
= 0.01 everywhere for t ≥ 15 min). Assessment of dynein-derived forces and corresponding centrosome movement reveals that centrosomes cluster toward the region of enriched dynein but begin separating after dynein is removed at t = 15 min (Supplemental Figure S7, C and D), indicating that loss of clustering is a passive event.
To quantify the propensity for a bipolar spindle to form, we recorded the number of simulations where we observe transient centrosome clustering at least one time, sustained for at least 1.5 min. The frequency of transient centrosome clustering in simulations with cortical dynein oscillations is >75% for simulations with three or four centrosomes, regardless of the period of dynein oscillations (Figure 5E). In the case of three centrosomes, this propensity to form a bipolar spindle is much higher than the ∼60% of simulations achieving centrosome clustering when cortical dynein activity was stably enriched in one quadrant of the cortex (comparing to Figure 4F). Together, these results suggest that centrosome movement and clustering are driven by cortical dynein activity.
DISCUSSION
In this work, we combine biological experimentation with computational modeling to inform novel dynein-dependent mechanisms driving centrosome clustering in cells with extra centrosomes. Our simulations indicate that cortical dynein–dependent forces are responsible for actively directing movement of centrosomes toward the cell cortex (Figure 4; Supplemental Figure S5). When dynein localization along the cell cortex is primarily in a region behind one of the poles of the mitotic spindle (Figure 1), dynein-dependent forces serve to bring individual centrosomes within close proximity of each other, near the region of cortical dynein activity (Figures 4 and 5; Supplemental Figures S3 and S7).
We propose that the directed movement of centrosomes toward a common region on the cell cortex is indicative of passive centrosome clustering. The centrosome(s) not near the region of cortical dynein activity do not cluster and instead are held at a stable distance from clustered centrosomes by other microtubule-derived forces (i.e., binding to dynein at spindle poles, Eg5, and/or HSET and pushing on the cell cortex; Supplemental Figure S6), resulting in a clustered bipolar spindle structure (Figures 4 and 5; Supplemental Figures S3 and S7). Centrosome clustering in this model is dependent on cortical dynein activity and is sensitive to the distribution of dynein on the cell cortex, as either removing cortical dynein activity from simulations or defining dynein to be uniformly distributed on the cortex precludes centrosome clustering (Figure 4; Supplemental Figure S4). Our biological data support this model and demonstrate that cortical dynein contributes to centrosome clustering in cells with centrosome amplification, such that loss of cortical dynein results in primarily unclustered spindles and multipolar divisions (Figures 2 and 3). This model is further supported by recent work from the Kyomitsu group that describes loss of spindle pole clustering in HCT116 cells with extra centrosomes when NuMA is acutely depleted during mitosis (van Toorn et al., 2023).
Our modeling framework provides an opportunity to exploit temporal and spatial regulation of cortical dynein activity in ways not easily achieved in biological systems. Through assessing progressively smaller angular regions of high cortical dynein activity from π to π/8, we find that the frequency of clustering is maximized at π/2 and decreases as the region of dynein activity is increased above or decreased below π/2 (Supplemental Figure S4E; results for three and four centrosomes). Importantly, our calculated range of dynein cortical distribution that is effective for centrosome clustering is consistent with the range of NuMA cortical distribution that has been described in different experimental systems: from ∼1/4 of the cortex in human epithelial cells (Kiyomitsu and Cheeseman, 2012; Figure 1) to ∼1/3 of the cortex in mouse keratinocytes (Seldin et al., 2013) and in human cells with exogenously expressed tagged NuMa (Kiyomitsu and Cheeseman, 2012). While our experimental approach to reduce NuMA/dynein localization to the cortex disrupts centrosome clustering in a manner consistent with that seen in our computational model, experimental tethering of NuMA at discrete regions of the cortex, such as that done by Kiyomitsu and colleagues (Okumura et al., 2018), would be necessary to further validate the causal relationship between NuMA/dynein cortical positioning and centrosome clustering.
Cells with induced or naturally occurring centrosome amplification exhibit a range of centrosome numbers and may have different regions of cortical dynein enrichment. In our model, we fix dynein enrichment to an angular region of π/2 and explore dynein-induced centrosome movement in simulated cells with two to six centrosomes. Cortex-directed centrosome movement corresponds with peaks in dynein activity (Supplemental Figure S5) for simulations with two and more than two centrosomes (Supplemental Figures S5 and S8; Mercadante et al., 2021). Our base model of mitotic spindle formation with supernumerary centrosomes reproduces critical features of passive centrosome clustering and is robust to centrosome number—revealing similar frequency and dynamics of spindle pole clustering with three to six centrosomes with fixed dynein enrichment as well as perturbations to dynein localization (Figure 4 and Supplemental Figure S3). We do note a few interesting features. Simulations with four centrosomes created a bipolar spindle that is predominantly asymmetric (one pole having three centrosomes and the other having one centrosome; this is consistent with previous experimental studies utilizing DLD-1 and RPE-1 p53−/− cells (Baudoin et al., 2020). In the case of five and six centrosomes, the fraction of simulated cells that form a bipolar spindle at t = 30 min is less than that observed in our experiments, as well as in simulations with three and four centrosomes. However, many of the multipolar spindles observed were close to being bipolar, with one of the centrosomes being within 1 µm of our criteria to change classification from three to two poles. Future work can consider additional forces or modify criteria for bipolarity classifications in simulated cells with larger centrosome numbers. Overall, these results show that the propensity to form a bipolar spindle is sensitive to the region of dynein localization, whereas formation of a bipolar spindle with a given region of dynein enrichment is not sensitive to centrosome numbers between two and four. Our model considers centrosomes as a microtubule-nucleating and -organizing center and does not distinguish between a centriole pair/centrosome and individual centrioles. As such, the ability of our model to reflect clustering activity is dependent on the number of microtubule-organizing centers and irrespective of centriole number. In this way, our simulations are relevant to understanding dynein’s role in clustering extra spindle poles that arise either from centrosome overduplication or from centrosome fragmentation.
Previous studies of mitotic cell division utilizing different modeling approaches have been valuable in understanding and informing force-derived centrosome clustering mechanisms in cells with centrosome amplification (Chatterjee et al., 2020; Goupil et al., 2020; Miles et al., 2022). In particular, these models have provided insight into chromosome-dependent centrosome clustering mechanisms that implicated kinetochore microtubule–derived torque (Miles et al., 2022) and have highlighted that there must exist a delicate balance between attraction forces for efficient centrosome clustering to occur, including centrosome-cortex forces (Chatterjee et al., 2020). Our results further expand on the latter observation by identifying that the centrosome-cortex force must correspond to a region on the cell cortex, either fixed or dynamically changing, for efficient clustering to occur via dynein motor activation (Figure 4).
Chromosomes and chromosome-derived forces have been implicated in centrosome clustering in two distinct ways. First, chromosomes form a physical barrier that segments the cell, thereby restricting centrosome movement (Goupil et al., 2020). Second, chromosomes form stable interactions, via kinetochores, with bundles of microtubules that are in turn anchored at the centrosomes (DeLuca et al., 2006). The bioriented configuration and associated forces of paired kinetochores enforce a bipolar geometry where centrosomes are positioned along the spindle axis (Leber et al., 2010; Tanaka, 2010; Chatterjee et al., 2020; Miles et al., 2022). Similarly, cell shape and actin-dependent cortical contractility impact centrosome clustering by restricting the space within which centrosomes can move (Kwon et al., 2008; Rhys et al., 2018). Dynein is functionally linked to the actin cell cortex through the proteins NuMA/LGN/Afadin (di Pietro et al., 2016), suggesting that cortical dynein and actin-dependent contractility act in concert to drive passive clustering by bringing centrosomes into proximity with each other. Once centrosomes are within ∼8 µm of each other, activities by static cross-linkers and motor proteins (i.e., HSET) at spindle poles engage centrosome-associated microtubules and drive robust and sustained centrosome clustering (Quintyne et al., 2005; Kwon et al., 2008; Rhys et al., 2018). Our simulations lack both chromosome-derived forces and cross-linking activity, explaining why when cortical dynein activity is turned off or redistributed on the cell cortex, previously clustered centrosomes begin to move apart (Figure 5).
Our results indicate that cortical localization of dynein alone is not sufficient for centrosome clustering and instead specific enrichment of dynein behind a single spindle pole is critical to drive clustering. Because cortical dynein localization is negatively regulated by centrosome-localized kinases (Kiyomitsu and Cheeseman, 2012), this suggests an iterative process to achieve and sustain centrosome clustering where dynein pulls centrosomes toward a common point on the cell cortex and then is itself inhibited by the centrosomes clustered nearby. In this way, we propose that regional enrichment of cortical dynein can both drive and be driven by centrosome clustering. Additional mechanisms such as kinase activity, nonmotor activity, and tension from kinetochore–microtubule interactions would then allow for sustained clustering when regions of dynein localization are dynamic. While existing in vivo data have not described dynein oscillations between more than two positions, it is reasonable to assume that, at least early in mitosis before a spindle structure has been established, dynein may localize to and even oscillate between >2 cortex locations. Our model would predict that, due to a lack of sustained and repetitive dynein activity pulling toward a consistent cortex region, oscillations between >2 cortex locations may disrupt centrosome clustering in a manner consistent with that seen with very quick oscillations between two locations. If true, this would further implicate the critical nature of the iterative process by which centrosomes both drive and respond to cortical dynein distribution to establish and maintain a bipolar spindle structure.
The presence of centrosome amplification is a hallmark of cancer and is associated with drug resistance, tumor progression, and poor patient prognosis (D’Assoro et al., 2002; Fukasawa, 2005; Mittal et al., 2020). Owing to the requirement of cancer cells with centrosome amplification to cluster their centrosomes to remain proliferative, prevention of centrosome clustering in cancer cells is believed to be a promising therapeutic approach (Kwon et al., 2008; Leber et al., 2010; Godinho and Pellman, 2014; Sabat-Pospiech et al., 2019). The motor protein HSET and proteins involved in the formation and maintenance of cell–cell junctions, cortical contractility, and kinetochore–microtubule interactions have been implicated as potential targets to limit centrosome clustering in cancer cells (Kwon et al., 2008, 2015; Hebert et al., 2012; Rhys et al., 2018). Dynein had previously been implicated in centrosome clustering, although this role was believed to be associated with its function at spindle poles (Quintyne et al., 2005). Our data now indicate that both cortical dynein activity and its dynamic and asymmetric localization behind spindle poles are critical for efficient centrosome clustering. Through simulations and molecular manipulations in cells with experimentally induced centrosome amplification or cancer cells with preexisting centrosome amplification, we show that perturbation of dynein’s ATP-driven motor activity or kinase-sensitive cortical localization impacts centrosome clustering. These results suggest that inhibition of either feature of dynein (activity or dynamic localization) may be of therapeutic interest in cancers with a high frequency of centrosome amplification.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Modeling
In this work, we optimize and expand upon our previously described two-dimensional model with dynamic microtubules and stochastic motor-dependent force generation to capture the centrosome movement in cells with centrosome amplification (Mercadante et al., 2021). We allow microtubule–motor protein interactions with Eg5 and HSET on antiparallel microtubules, capturing the dominant roles of these proteins in mitosis. Dynein is localized at the cell cortex and spindle poles to account for its functions in pole focusing and spindle dynamics, respectively (Supplemental Figure S2A) (Vaisberg et al., 1993; Sharp et al., 2000; Ferenz et al., 2009, 2010; van Heesbeen et al., 2014; Mann and Wadsworth, 2019; Loncar et al., 2020). Kinesin-5, Eg5 in mammalian cells, is a homotetrameric motor protein with two motor domains on either side of an elongated stalk (Kashina et al., 2009). Each of the motor domains binds to a microtubule and walks toward the plus end (Bodrug et al., 2020). When the microtubules are antiparallel, as they are in the interpolar region of the spindle, this movement causes microtubule sliding in opposite directions and drives centrosome separation and early spindle formation (Mayer et al., 1999; Kapoor et al., 2000). Kinesin-14, HSET in mammalian cells, is a minus end–directed dimeric motor protein with two motor heads on one end of the molecule and nonmotor microtubule-binding domains on the other (Braun et al., 2009; Fink et al., 2009). At the interpolar region of the spindle, HSET facilitates antiparallel microtubule–microtubule sliding to help maintain mitotic spindle length (Fink et al., 2009). HSET movement opposes that of Eg5, resulting in an inward force between spindle poles (Mountain et al., 1999; Sharp et al., 2000). Dynein is the major minus end–directed motor protein in mammalian cells. While dynein at the cell cortex is essential for spindle positioning and orientation, it has additional essential roles within the spindle that are required for the maintenance of spindle structure. Dynein activity in the interpolar region of the spindle, where interpolar microtubules overlap, counteracts that of Eg5, with one of dynein’s two motor heads walking along each microtubule. This movement pulls spindle poles together, antagonizing centrosome separation and bipolar spindle formation (Ferenz et al., 2009; Raaijmakers et al., 2013; Raaijmakers and Medema, 2014). Additionally, dynein motor activity on parallel microtubules is critical for maintaining microtubule minus end focusing at spindle poles, where loss of dynein results in splayed poles and barrel-like spindles (Echeverri et al., 1996; Goshima et al., 2005).
Dynamic microtubules.
Microtubules emanate from a centrosome and are initialized with uniformly distributed random lengths ℓ and angles α (ℓ ∈ U [0, 0.5] µm and α ∈ U [0, 2π], respectively). At each time step, new microtubules are randomly nucleated a rate MTnuc. Microtubules are modeled as semirigid filaments with a constant bending rigidity κ. Microtubule plus ends are dynamic and have constant growth velocity vg and shrinking velocity that is vb for microtubules bound to dynein and vs for all other shrinking microtubules. Microtubule dynamic instability for each microtubule i is defined by a constant rescue frequency (k1) and a length-dependent catastrophe frequency (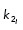 ). We use a stochastic Monte Carlo method to determine microtubule dynamic instability; a random number n ∈ U [0, 1] is generated and if
). We use a stochastic Monte Carlo method to determine microtubule dynamic instability; a random number n ∈ U [0, 1] is generated and if 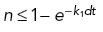 or
or 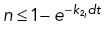 , the microtubule will undergo rescue or catastrophe, respectively (Supplemental Figure S2B). Shrinking microtubules that do not undergo rescue will depolymerize completely and no longer be considered in the system when ℓi ≤ vgdt. Throughout the simulation, the microtubule length ℓ and angle α are updated based on the state of the microtubule (Supplemental Figure S2B).
, the microtubule will undergo rescue or catastrophe, respectively (Supplemental Figure S2B). Shrinking microtubules that do not undergo rescue will depolymerize completely and no longer be considered in the system when ℓi ≤ vgdt. Throughout the simulation, the microtubule length ℓ and angle α are updated based on the state of the microtubule (Supplemental Figure S2B).
Initialization and algorithm.
Centrosomes are initialized in random positions within 7.5 µm from the cell center, to simulate positioning following nuclear envelope breakdown at the start of mitosis. At each time step, new microtubules are nucleated for each centrosome at a rate MTnuc. Microtubule dynamics and microtubule binding to motor proteins are determined and updated by a set of stochastic rules based on distances and probabilities (Supplemental Figure S2B). The force on each microtubule is calculated and summed to determine the total force on centrosome c. The position of centrosome c is updated based on Eq. 4. The length and angle of each microtubule is updated based on its state, and this process is repeated until t = 30 min. Parameters are highlighted in Supplemental Table S1.
Stochastic microtubule–motor interactions.
Microtubule dynamic instability results in changes in microtubule length and positioning at every time step. As such, microtubule interactions with motor proteins are transient, with motor interactions depending on proximity to a microtubule. In addition to proximity, we consider each motor protein population (Eg5, HSET, cortical dynein, and dynein at spindle poles) to have a distinct binding probability (Supplemental Table S1). If a distance argument and probability are satisfied, the microtubule will bind to the motor protein. For example, a microtubule tip within a distance 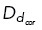 from the cell boundary will then bind to cortical dynein with probability
from the cell boundary will then bind to cortical dynein with probability 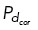 (implemented by choosing
(implemented by choosing 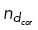 ∈ U [0, 1] and allowing binding to occur if
∈ U [0, 1] and allowing binding to occur if 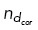 <
< 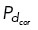 ). Otherwise, the microtubule will continue to grow and push against the cell boundary with a length-dependent force:
). Otherwise, the microtubule will continue to grow and push against the cell boundary with a length-dependent force:
| 1 |
where fstall is the stall force of the microtubule and κ is the bending rigidity. Interpolar microtubules i, j nucleated from centrosomes c, k, that are within a distance DEg5 or DHSET will have a probability of binding to Eg5 (PE) and/or HSET (PH ) and generating force. Using a Monte Carlo method, if a random number nEg5, nHSET is less than PE, PH , binding of Eg5 and/or HSET occurs, respectively. We allow each microtubule from centrosome c to have Eg5 and/or HSET binding on up to two microtubules from centrosome k. Microtubules also interact with spindle pole dynein near opposing centrosomes when a microtubule gets within a distance 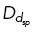 and satisfies the probability
and satisfies the probability 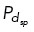 (Supplemental Figure S2B).
(Supplemental Figure S2B).
Motor-dependent force generation and centrosome movement.
We consider motor-dependent forces to be stochastic, where force by motor m is generated if both a distance argument between the two interacting structures (microtubule–microtubule, microtubule–cortex, or microtubule–spindle pole) and a motor-specific binding probability Pm are satisfied (Supplemental Figure S2B). Individual motor forces on the ith microtubule, fm, are calculated using a standard force–velocity relationship (Svoboda and Block, 1994):
| 2 |
where fo,m is the stall force of motor m, vo,m is the walking velocity of motor m, vc is the velocity of centrosome c that microtubule i emanates from, and ui is the unit vector in the direction of microtubule i. The total force by all motors m bound to the Nc,m microtubules nucleated from centrosome c is calculated by
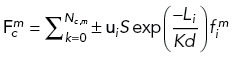 |
3 |
where the sign indicates the direction of the force (cortical dynein, dynein at spindle poles, and Eg5 are [–] while HSET is [+]). The exponential term accounts for increased drag-dependent force as the centrosome approaches a boundary, that is, the cell cortex or opposing centrosome (Aponte-Rivera and Zia, 2016). Therefore, this term is dependent on the distance, Li, between the centrosome and the point of force application on microtubule i and the distance, d, either between centrosomes or between the centrosome and the cell cortex. K is a constant scaling factor, and S is an additional motor-dependent scaling. For dynein-dependent forces, where only one microtubule is bound, S = 1. For HSET and Eg5-derived forces, where two antiparallel microtubules are interacting, S = a(1+Oi,j)C, where a is a constant that depends on the angle between interacting microtubules, Oi,j is the overlap distance between interpolar microtubules, and C is a constant scaling factor to account for both active and passive cross-linking activity at antiparallel microtubule overlap regions (Mollinari et al., 2002; Peterman and Scholey, 2009; Shimamoto et al., 2015; Reinemann et al., 2018; Lamson et al., 2019; Edelmaier et al., 2020).
All motor-dependent and non–motor-dependent forces on all microtubules emanating from centrosome c are summed to determine the total force on the centrosome, and the position of centrosome c is updated as
| 4 |
where 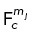 is the total force generated on centrosome c by motor mj (cortical dynein, HSET, Eg5, or spindle pole dynein),
is the total force generated on centrosome c by motor mj (cortical dynein, HSET, Eg5, or spindle pole dynein), 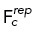 is the repulsive force on centrosome c when within a distance Dr of another centrosome,
is the repulsive force on centrosome c when within a distance Dr of another centrosome, 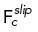 is the total slipping force on centrosome c, vc is the velocity of centrosome c, and ξ is a constant drag coefficient.
is the total slipping force on centrosome c, vc is the velocity of centrosome c, and ξ is a constant drag coefficient.
All parameters were optimized to match experimental results. Additionally, all parameters that were modified from our previous publication (Mercadante et al., 2021) were retested in simulations with two centrosomes and asymmetric dynein localization to confirm that appropriate bipolar spindle length and dynamics were maintained (Supplemental Figure S8). In this work, we set the probability of HSET binding to be higher than that of Eg5 and set cortical dynein binding to be higher than that for spindle pole dynein (see Supplemental Table S1 for exact values). These values were chosen to achieve appropriate clustering frequencies and velocities of centrosome movement that are comparable to that seen in our cell culture model. We note that a higher probability could correspond to a higher local level of expression and/or activity; we do not account for any spatial variation of motor binding other than the geometric constraints highlighted in Supplemental Figure S2B.
Modulating and assessing cortical dynein activity.
To spatially and temporally regulate cortical dynein localization and activity within the model, we specify which microtubules are able to bind to cortical dynein with probability 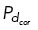 based on the position of the microtubule end. When cortical dynein is uniformly distributed, we allow all microtubules to have an equal probability of binding to dynein and generating force. We remove cortical dynein by setting the probability
based on the position of the microtubule end. When cortical dynein is uniformly distributed, we allow all microtubules to have an equal probability of binding to dynein and generating force. We remove cortical dynein by setting the probability 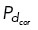 = 0.01, preventing most microtubules from binding and generating force. With dynein localized in a specific region as shown in Figure 4, A and D, we allow only those microtubules whose end falls within the upper-right quadrant of the cell to bind to cortical dynein at a probability
= 0.01, preventing most microtubules from binding and generating force. With dynein localized in a specific region as shown in Figure 4, A and D, we allow only those microtubules whose end falls within the upper-right quadrant of the cell to bind to cortical dynein at a probability 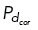 . We allow all other microtubules throughout the cell to have a probability
. We allow all other microtubules throughout the cell to have a probability 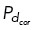 = 0.01 under the assumption that cortical dynein is unlikely to be entirely absent from this region. When regions of dynein localization oscillate in time, we change the position requirement of microtubule ends to bind to cortical dynein, dependent on the period of dynein oscillations, T. Specifically, microtubules in either the upper-right or lower-left quadrant bind to cortical dynein with probability
= 0.01 under the assumption that cortical dynein is unlikely to be entirely absent from this region. When regions of dynein localization oscillate in time, we change the position requirement of microtubule ends to bind to cortical dynein, dependent on the period of dynein oscillations, T. Specifically, microtubules in either the upper-right or lower-left quadrant bind to cortical dynein with probability 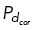 while all other microtubules have a small probability
while all other microtubules have a small probability 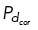 = 0.01 of binding. Peaks in the traces defining the pairwise distances between centrosomes were determined by the MATLAB function “findpeaks” (MathWorks, 2023), with significant peaks defined as those having a prominence greater than 1 SD of the average peak prominence.
= 0.01 of binding. Peaks in the traces defining the pairwise distances between centrosomes were determined by the MATLAB function “findpeaks” (MathWorks, 2023), with significant peaks defined as those having a prominence greater than 1 SD of the average peak prominence.
Computation and code availability.
All computational modeling and model analysis was performed in MATLAB utilizing GPUs on a cluster. Thirty simulations of each set of parameters were run with three centrosomes for 30 min of mitosis unless otherwise specified. Code will be made available upon request.
Cell culture
Cell lines were maintained at 37°C with 5% CO2. hTERT-immortalized RPE cells were obtained from and authenticated by the American Type Culture Collection. RPE cells expressing EGFP-tubulin were generated by viral transduction of L304-EGFP-Tubulin, a gift from Weiping Han (Singapore Bioimaging Consortium; Addgene plasmid #64060; http://n2t.net/addgene:64060; RRID:Addgene 64060; Yang et al., 2013). RPE cells expressing the tet-inducible PLK4, a gift from Neil Ganem (Boston University), and MDA-MB-231 cells, a gift from Catherine Whittington (Worcester Polytechnic Institute), were maintained in DMEM. RPE p53 deficient cells, a gift from Meng-Fu Bryan Tsou (Memorial Sloan Kettering Cancer Center), were maintained in DMEM F-12. Cell culture medium was supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. DNA stain (4′,6-diamidino-2-phenylindole [DAPI]) was used to monitor and confirm the absence of mycoplasma contamination.
Induction of centrosome amplification and perturbation of cortical dynein activity
Disruption of cortical localization of dynein was achieved through depletion of Afadin or LGN using lipid-based transfection of 50 nM Horizon ON-TARGET plus pools of siRNA (Afadin target sequences: 5′-ugagaaaccucua guugua-3′, 5′-ccaaaugguuuacaagaau-3′, 5′-guuaagggcccaagacaua-3′, 5′- acuugagcggcaucgaaua-3′, LGN target sequences: 5′-gaacuaacagcacgacuua-3′, 5′-cuucagggaugcaguuaua-3′, 5′-acagugaaauucuugcuaa-3′, 5′-ugaaggguucuuugacuua-3′). Horizon nontargeting siRNA pool (siCtl) was used as a negative control for siRNA experiments (5′-ugguuuacaugucgacuaa-3′, 5′- ugguuuacauguuguguga-3′, 5′-ugguuuacauguuuucuga-3′, 5′-gguuuacauguuuuccua-3′). Knockdown ef- ficiency was confirmed using Afadin (F:5′-gtgggacagcattaccgaca-3′, R:5′tcatcggcttcaccattcc-3′), LGN (F:5′-gtgaccacccgtctgtcg-3′, R:5′-ttcagcaacatttctcccgc-3′), and GAPDH (F:5′-ctagctggcccgatttctcc- 3′, R:5′-cgcccaatacgaccaaatcaga-3′)-specific primers. Inhibition of cortical dynein activity was achieved with exposure to 25 µM dynarrestin for 1 h. Four hours posttransfection with siRNA, RPE indPLK4 cells were induced to express PLK4 and amplify centrosome biogenesis by the addition of 2 µg/ml doxycycline 4 h posttransfection with siRNA. PLK4 induction was sustained for 48 h until cell fixation. Alternatively, 24 h after siRNA transfection, RPE p53-/- cells were treated with 1.5 µg/ml DCB for 24 h to induce cytokinesis failure and generate tetraploid cells with four centrosomes. Cells were washed out of DCB and cultured for an additional 24 h before tetraploid cells were allowed to progress to mitosis. These experimental timelines are summarized in Supplemental Figure S1. Where relevant, 20 µM MG132 was added to the media for the final 30 min or 1 h of culture before fixation.
Fluorescence imaging and analysis
Images were captured with a Zyla sCMOS (Oxford Instruments, Belfast, UK) camera mounted on a Nikon Ti-E microscope (Nikon, Tokyo, Japan). For live cell imaging of RPE cells expressing EGFP-tubulin, a 20× CFI Plan Fluor objective was used to capture images every 2.5 min for the duration of mitosis (Mercadante et al., 2019). Analysis of mitotic timing and cell fate was performed on at least 50 mitotic cells. Mitotic duration was quantified as the time between nuclear envelope breakdown (determined by loss of GFP exclusion from the nucleus) to anaphase B (determined by rapid elongation of the spindle). Phase contrast images were used to assess the number of progeny resulting from each division.
For analysis of cortex-localized NuMA, cells were rinsed briefly in phosphate-buffered saline (PBS) and then incubated in PHEM buffer (60 mM PIPES pH 6.9, 25 mM HEPES pH 6.9, 10 mM EGTA pH 7.0, 2 mM MgCl2) with 0.3% Triton X-100 (TX-100) for 5 min. Cells were fixed in warmed 3.7% paraformaldehyde supplemented with 30 mM sucrose for 15 min at room temperature and then permeabilized in 0.1% TX-100 in PBS for 5 min. Cells were blocked in 3% PBS–bovine serum albumin (BSA) with 0.05% Tween-20 for 15 min and then incubated in primary antibody (NuMa: Abcam ab109262, Cambridge, UK; centrin-2: Sigma 04-1624) diluted in blocking buffer overnight at 4°C. Cells were washed briefly and incubated in secondary antibody diluted in TBS (Tris-buffered Saline)-BSA containing 0.2 µg/ml DAPI. NuMA distribution was determined by measuring the intensity of NuMA staining along a line drawn from the center of a cell, through the spindle pole, and across the cell boundary. The ratio of NuMA staining intensity along the line at the cell boundary to NuMA staining intensity in the cytoplasm was calculated, with a ratio ≥1.2 indicating cortical enrichment. Enrichment scores were calculated for the cortical region behind each spindle within the cell (a cell with three spindle poles would have three enrichment scores) (Supplemental Figure S1A), and the number of cells with cortical enrichment behind one, more than one, or no spindle poles in each condition was was quantified (Supplemental Figure S1B). NuMA cortical enrichment scores were consistent with visual inspection, and qualitative assessment of NuMA distribution with respect to spindle pole position was performed for additional replicates. For analysis of spindle morphology, cells were fixed in ice-cold methanol for 10 min at –20°C. Blocking, primary (α-tubulin: Abcam ab18251, Cambridge, UK; centrin: Millipore 04-1624, Burlington, MA) and secondary antibody dilutions were prepared in TBS-BSA. DNA was detected with the addition of 0.2 µg/ml DAPI to the secondary antibody dilution. Images of individual mitotic cells were captured using a 60× Plan Apo oil immersion objective and 0.3 µm z-stacks through the depth of the cell. NuMA localization was assessed as described previously (Seldin et al., 2013). Clustered bipolar spindles were defined as those in which microtubules were organized into two spindle poles and in which one or both spindle poles contained two or more centrosomes positioned within 5 µm of each other. Statistical analysis between two conditions was determined using a two-tailed Student’s t test. For multiple comparisons, a one-way analysis of variance (ANOVA) was performed and Dunnett’s test post-hoc was used for simultaneous comparison between each test condition and a control.
Supplementary Material
Acknowledgments
Results in this paper were obtained in part using a high-performance computing system acquired through National Science Foundation (NSF) Major Research Instrumentation Program (MRI) Grant DMS-1227943 to Worcester Polytechnic Institute. This work was supported by NSF Graduate Research Fellowship Program (GRFP) to D.L.M. and National Institutes of Health R01 GM140465-01 to A.L.M. and S.D.O.
Abbreviations used:
- BSA
bovine serum albumin
- CDK1
cyclin dependent kinase 1
- DAPI
4′,6-diamidino-2-phenylindole
- DCB
dihydrocytochalasin B
- EGFP
enhanced green fluorescent protein
- GPU
Graphics processing unit
- LGN
Leucine-Glycine-Asparagine repeat protein
- MT
microtubule
- NuMA
nuclear mitotic apparatus protein
- PBS
phosohate buffered saline
- PHEM
pipes hepes EGTA MgCl 2
- PLK1
polo like kinase 1
- PLK4
polo like kinase 4
- RPE
retinal pigment epithelial cell
- SD
standard deviation
- TBS
Tris buffered saline
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-07-0296) on April 5, 2023.
REFERENCES
- Andreassen PR, Martineau SN, Margolis RL (1996). Chemical induction of mitotic checkpoint override in mammalian cells results in aneuploidy following a transient tetraploid state. Mutat Res 372, 181–194. [DOI] [PubMed] [Google Scholar]
- Aponte-Rivera C, Zia RN (2016). Simulation of hydrodynamically interacting particles confined by a spherical cavity. Phys Rev Fluids 1, 023301. [Google Scholar]
- Bader JR, Vaughan KT (2010). Dynein at the kinetochore: timing, interactions, and functions. Semin Cell Dev Biol 21, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr AR, Gergely F (2008). MCAK-independent functions of ch-Tog/XMAP15 in microtubule plus-end dynamics. Mol Cell Biol 28, 7199–7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudoin C, Soto K, Martin O, Nicholson JM, Chen J, Cimini D (2020). Asymmetric clustering of centrosomes defines the early evolution of tetraploid cells. eLife 9, e54565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodrug T, Wilson-Kubalek EM, Nithianantham S, Thompson AF, Alfieri A, Gaska I, Major J, Debs G, Inagaki S, Gutierrez P, et al. (2020). The kinesin-5 tail domain directly modulates the machanochemical cycle of the motor domain for anti-parallel microtubule sliding. eLife 9, e51131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M, Drummond DR, Cross RA, McAnish AD (2009). The kinesin-14 Klp2 organizes microtubules into parallel bundles by an ATP-dependent sorting mechanism. Nat Cell Biol 11, 724–730. [DOI] [PubMed] [Google Scholar]
- Busson S, Dujardin D, Moreau A, Dompierrre J, De Mey JR (1998). Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr Biol 8, 541–544. [DOI] [PubMed] [Google Scholar]
- Carminati M, Gallini S, Pirovano L, Alfieri A, Bisi S, Mapelli M (2016). Concomitant binding of Afadin to LGN and F-actin directs planar spindle orientation. Nat Struct Mol Biol 23, 155–163. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Sarker A, Khodjakov A, Mogilner A, Paul R (2020). Mechanics of multi-centrosomal clustering in bipolar mitotic spindles. Biophys J 119, 434–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Assoro AB, Lingle WL, Salisbury JL (2002). Centrosome amplification and the development of cancer. Oncogene 21, 6146–6153. [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED (2006). Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127, 969–982. [DOI] [PubMed] [Google Scholar]
- Ding Z, Huang C, Jiao X, Wu D, Huo L (2017). The role of TACC3 in mitotic spindle organization. Cytoskeleton 74, 369–378. [DOI] [PubMed] [Google Scholar]
- di Pietro F, Echard A, Morin X (2016). Regulation of mitotic spindle orientation: an integrated view. EMBO Rep 17, 1106–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Macara IGM (2004). Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell 119, 503–516. [DOI] [PubMed] [Google Scholar]
- Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB (1996). Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex is chromosome alignment and spindle organization during mitosis. J Cell Biol 132, 617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmaier C, Lamson AR, Gergley ZR, Ansari S, Blackwell R, McIntosh JR, Glaser MA, Betterton MD (2020). Mechanisms of chromosome biorientation and bipolar spindle assembly analyzed by computational modeling. eLife 9, e48787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenz NP, Gable A, Wadsworth P (2010). Mitotic functions of kinesin-5. Semin Cell Dev Biol 21, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenz NP, Paul R, Fagerstrom C, Mogilner A, Wadsworth P (2009). Dynein antagonizes Eg5 by crosslinking and sliding antiparallel microtubules. Curr Biol 19, 1833–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding AB, Lim S, Montgomery K, Dobreva I, Dedhar S (2010). A critical role of integrin-linked kinase, ch-TOG and TACC3 in centrosome clustering in cancer cells. Oncogene 30, 521–534. [DOI] [PubMed] [Google Scholar]
- Fink G, Hajdo L, Skowronek KJ, Reuther C, Kasprzak AA, Diez S (2009). The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat Cell Biol 11, 717–723. [DOI] [PubMed] [Google Scholar]
- Forth S, Kapoor TM (2017). The mechanics of microtubule networks in cell division. J Cell Biol 216, 1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K (2005). Centrosome amplification, chromosome instability, and cancer development. Cancer Lett 8, 6–19. [DOI] [PubMed] [Google Scholar]
- Godinho SA, Kwon M, Pellman D (2009). Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev 28, 85–98. [DOI] [PubMed] [Google Scholar]
- Godinho SA, Pellman D (2014). Causes and consequences of centrosome abnormalities in cancer. Philos Trans R Soc B Biol Sci 369, 20130467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Nedelec F, Vale RD (2005). Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J Cell Biol 171, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goupil A, Nano M, Letort G, Gemble S, Edwards F, Goundiam O, Gogendeau D, Pennetier C, Basto R (2020). Chromsomes function as a barrier to mitotic spindle bipolarity in polyploid cells. J Cell Biol 219, e201908006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AM, DuBoff B, Casaletto JB, Gladden AB, McClatchey AI (2012). Merlin/ERM proteins establish cortical asymmetry and centrosome position. Genes Dev 26, 2709–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchliffe EH. (2011). The centrosome and bipolar spindle assembly. Cell Cycle 10, 3841–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoing S, Yeh TY, Baumann M, Martinex NE, Habenberger P, Kremer L, Drexler HC, Kuchler P, Reinhardt P, Choidas A, et al. (2018). Dynarrestin, a novel inhibitor of cytoplasmic dynein. Cell Chem Biol 25, 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ (2000). Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol 150, 975–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM (2009). A bipolar kinesin. Nature 379, 270–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM (2012). Chromosome- and spindle-pole derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol 14, 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Busso C, Gonczy P (2013). NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function. EMBO J 32, 2517–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Busso C, Gonczy PJ (2012). Cortical dynein is critical for proper spindle positioning in human cells. Cell Biol 199, 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J, Godinho S, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D (2008). Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev 22, 2189–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M, Bagonis M, Danuser G, Pellman D (2015). Direct microtubule-binding by Myosin-10 orients centrosomes toward retraction fibers and subcortical actin clouds. Dev Cell 34, 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamson AR, Edelmaier CJ, Glaser MA, Betterton MD (2019). Theory of cytoskeletal reorganization during cross-linker-mediated mitotic spindle assembly. Biophys J 116, 1719–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber B, Maier B, Fuchs F, Chi J, Riffel P, Anderhub S, Wagner L, Ho AD, Salisbury JL, Boutros M, Kramer A (2010). Proteins required for centrosome clustering in cancer cells. Cancer 2, 33ra38. [DOI] [PubMed] [Google Scholar]
- Loncar A, Rincon SA, Ramirez ML, Paoletti A, Tran PT (2020). Kinesin-14 family proteins and microtubule dynamics define S. pombe mitotic and meiotic spindle assembly, and elongation. J Cell Sci 133, jcs240234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann BJ, Wadsworth P (2019). Kinesin-5 regulation and function in mitosis. Trends Cell Biol 29, 66–79. [DOI] [PubMed] [Google Scholar]
- MathWorks (2023). findpeaks: find local maxima. Available at https://www.mathworks.com/help/signal/ref/findpeaks.html.
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ (1999). Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 286, 971–974. [DOI] [PubMed] [Google Scholar]
- Mercadante DL, Crowley EA, Manning AL (2019). Live cell imaging to assess the dynamics of metaphase timing and cell fate following mitotic spindle perturbations. J Vis Exp 2019, 151. [DOI] [PubMed] [Google Scholar]
- Mercadante DL, Manning AL, Olson SD (2021). Modeling reveals cortical dynein-dependent fluctuations in bipolar spindle length. Biophys J 120, 3192–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW (2000). Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol 149, 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles CE, Zhu J, Mogilner A (2022). Mechanical torque promotes bipolarity of the mitotic spindle through multi-centrosomal clustering. Bull Math Biol 84, 29. [DOI] [PubMed] [Google Scholar]
- Mittal K, Choi DH, Ogden A, Donthamsetty S, Melton BD, Gupta MV, Pannu V, Cantuaria G, Varambally S, Reid MD, et al. (2017). Amplified centrosomes and mitotic index display poor concordance between patient tumors and cultured cancer cells. Sci Rep 7, 43984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal K, Kaur J, Jaczko M, Wei G, Toss MS, Rakha EA, Janssen EAM, Soiland H, Kucuk O, Reid MD, et al. (2020). Centrosome amplification: a quantifiable cancer cell trait with prognostic value in solid malignancies. Cancer Metastasis Rev 40, 319–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinari C, Kleman J, Jiang W, Schoehn G, Hunter T, Margolis RL (2002). PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol 157, 1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain V, Simerly C, Howard L, Ando A, Schatten G, Compton D (1999). The kinesin-related protein, Hset, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol 147, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Serer B, Childers EP, Hermance NM, Mercadante D, Manning AL (2019). Aurora A inhibition limits centrosome clustering and promotes mitotic catastrophe in cells with supernumerary centrosomes. Oncotarget 10, 1649–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell CB, Wang Y (2000). Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol Biol Cell 11, 1765–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura M, Natsume T, Kanemaki MT, Kiyomitsu T (2018). Dynein-dynactin-NuMA clusters generate cortical spindle-pulling forces as a multi-arm ensemble. eLife 7, e36559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman EJG, Scholey JM (2009). Mitotic microtubule crosslinkers: insights from mechanistic studies. Curr Biol 19, R1089–R1094. [DOI] [PubMed] [Google Scholar]
- Petry S (2016). Mechanisms of mitotic spindle assembly. Annu Rev Biochem 85, 659–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS (2005). Spindle multipolarity is prevented by centrosomal clustering. Science 307, 127–129. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JA, Medema RH (2014). Function and regulation of dynein in mitotic chromosome segregation. Chromosoma 123, 407–422. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JA, Tanenbaum ME, Medema RH (2013). Systematic dissection of dynein regulators in mitosis. J Cell Biol 201, 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinemann DN, Norris SR, Ohi R, Lang MJ (2018). Processive kinesin-14 HSET exhibits directional flexibility depending on motor traffic. Curr Biol 28, 2356–2362.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhys AD, Monteiro P, Smith C, Vaghela M, Arnandis T, Kato T, Leitinger B, Sahai E, McAinsh A, Charras G, Godinho SA (2018). Loss of E-cadherin provides tolerance to centrosome amplification in epithelial cancer cells. J Cell Biol 217, 195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabat-Pospiech D, Fabian-Kolpanowicz K, Prior IA, Coulson JM, Fielding AB (2019). Targeting centrsome amplification, and Achilles’ heel of cancer. Biochem Soc Trans 47, 1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana S, Keshri R, Rajeevan A, Kapoor S, Kotak S (2018). Plk1 regulates spindle orientation by phosphorylating NuMA in human cells. Life Sci Alliance 1, e201800223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin L, Poulson ND, Foote HP, Lechler T (2013). NuMA localization, stability, and function in spindle orientation involve 4.1 and Cdk1 interactions. Mol Biol Cell 24, 3651–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Rogers GC, Scholey JM (2000). Microtubule motors in mitosis. Nature 407, 41–47. [DOI] [PubMed] [Google Scholar]
- Shimamoto Y, Forth S, Kapoor TM (2015). Measuring pushing and braking forces generated by ensembles of kinesin-5 crosslinking two microtubules. Dev Cell 34, 669–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Block SM (1994). Force and velocity measured for single kinesin molecules. Cell 77, 773–784. [DOI] [PubMed] [Google Scholar]
- Tanaka TU (2010). Kinetochore-microtubule interactions: steps towards bi-orientation. EMBO J 29, 4070–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisberg EA, Koonce MP, McIntosh RJ (1993). Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol 123, 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heesbeen RGHP, Tanenbaum ME, Medema RH (2014). Balanced activity of three mitotic motors is required for bipolar spindle assembly and chromosome segregation. Cell Rep 8, 948–956. [DOI] [PubMed] [Google Scholar]
- van Toorn M, Gooch A, Boerner S, Kiyomitsu T (2023). NuMA deficiency causes micronuclei via checkpoint-insensitive k-fiber minus-end detachment from mitotic spindle poles. Curr Biol 33, 572–580.e2. [DOI] [PubMed] [Google Scholar]
- Yang W, Guo X, Thein S, Xu F, Sugii S, Baas PW, Radda GK, Han W (2013). Regulation of adipogenesis by cytoskeleton remodelling is facilitated by acetyltransferase MEC-17-dependent acetylation of alpha-tubulin. Biochem J 449, 605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.