Abstract
Retroviruses are believed to induce tumors by acting as insertional mutagens that activate expression of cellular protooncogenes. Indeed, almost 90% of mouse mammary tumor virus (MMTV)-induced mammary tumors in C3H/He mice show upregulation of Int protooncogenes. We have analyzed three different MMTV variants [MMTV(C3H), MMTV(HeJ), and a genetically engineered MMTV hybrid provirus (HP)] for tumorigenicity in mice from two distinct genetic backgrounds. All three viruses were tumor causing in BALB/cJ mice. However, only MMTV(C3H), but not MMTV(HeJ) or HP, induced mammary tumors in C3H/He mice. All of the viruses were infectious on either background and up-regulated expression of Int genes in tumors they induced. Like HP, MMTV(HeJ) was found to be a genetic recombinant between endogenous Mtv1 provirus and exogenous MMTV(C3H). Sequence comparison of MMTV variants linked the tumorigenicity of MMTV(C3H) to the gag region of the retrovirus.
Many retroviruses carry oncogenes (v-onc) and induce tumors after a short latency period (33). For viruses lacking v-onc genes, tumors arise after an extended latency period and provirus integration near a cellular protooncogene (33). Most tumors induced by retroviruses that lack oncogenes cause hematopoietic malignancies, although a few of these viruses induce carcinomas (33). Mouse mammary tumor virus (MMTV) is a B-type retrovirus that does not have an oncogene but induces mammary carcinomas and, more rarely, T-cell lymphomas (for a review, see reference 35).
Exogenous MMTV is spread via the milk of infected females and is acquired by suckling pups (27). On rare occasions, an exogenous MMTV provirus is inserted into germ or early embryonic cells, thereby becoming a stably inherited endogenous provirus (2, 9). The primary targets for exogenous MMTV are T and B cells located in Peyer's patches of the gastrointestinal tracts of neonatally infected pups (3, 19). MMTV gains access to these cells by traveling through M cells located in the follicle-associated epithelium of the Peyer's patches (18). Both endogenous and exogenous MMTVs encode a superantigen (Sag) in their 3′ long terminal repeat (LTR) (7). In contrast to conventional antigens, Sags stimulate profound T-cell responses, because they are recognized by all T cells that express a particular T-cell receptor Vβ chain (22, 25). Since proliferation increases the number of T cells and because dividing cells are susceptible to retroviral infection, the rate of infection is increased (41). Infection rates of the T and B cells remain high sufficiently long to infect the mammary gland cells when they begin to divide at about 3 to 4 weeks after birth. Overall, the LTR sequences of different MMTVs are highly conserved (4). However, the region encoding the C-terminal segment of Sag is more diverse and is known as the hypervariable region. The amino acid sequence of this region contacts the Vβ chain of the T-cell receptor and thus determines which T cells are affected (45). When recognized as foreign, Sags stimulate specific Vβ+ T-cell proliferation (22, 25) whereas Sags present in the germ line stimulate deletion of the Vβ+ T-cell subset during formation of the immune repertoire (1, 10, 11, 43). The Sag function is indispensable to the MMTV life cycle, because mice that lack Sag-cognate T cells, due to the expression of transgenes (15) or endogenous proviruses (20), cannot be infected with exogenous viruses bearing Sags of the same Vβ specificity. In addition, viruses without functional Sags cannot propagate in vivo (16).
Once proviral DNA is integrated into a chromosome, its expression is regulated by specific sequences within the LTR that cause increased viral transcription in response to glucocorticoid receptor-steroid hormone complexes (44). The increased virion production that occurs during lactation results in a greater number of infected mammary gland cells and more proviral integrations into the genome. MMTV does not encode an oncogene, so mammary tumorigenesis takes place after proviral insertion near specific cellular proto-oncogenes, thereby up-regulating their transcription. Because retroviral integration into the host chromosome occurs at random locations, the more viruses that are produced, the more likely it is that integration near cellular proto-oncogenes will occur. The large majority of MMTV integrations in mammary tumors result in activation of proto-oncogenes that are not normally expressed in the mammary gland (28). Thus, it has been postulated that MMTV-induced mammary tumors result from the expression of proto-oncogenes controlling cellular growth.
It has been shown that exogenous MMTV carried by C3H/HeJ mice [MMTV(HeJ)] cannot efficiently induce mammary tumors in C3H/HeJ mice (30). We have extended these findings and shown that besides MMTV(HeJ), another highly infectious genetically engineered MMTV hybrid provirus (HP) was incapable of efficiently inducing tumors in C3H/He mice. In contrast, both of these viruses were tumorigenic in BALB/cJ mice and integrated into the Wnt-1 and Int-2/Fgf3 loci with high frequency. By comparing the sequences of these viruses to MMTV(C3H), which caused tumors in C3H/He as well as in BALB/cJ mice, we found that a determinant of oncogenicity mapped to the gag gene.
MATERIALS AND METHODS
Mice.
All of the mice used in this study were bred and maintained at the animal facility of The Jackson Laboratory, Bar Harbor, Maine. C3H/HeJ MMTV+ and BALB/cJ mice were obtained from The Jackson Laboratory. Please note that in 1999, C3H/HeJ MMTV+ mice were rederived to improve the overall health status of the distribution colonies, resulting in elimination of exogenous virus (JAX Notes 480:8, 2000). Thus, MMTV-infected C3H/HeJ mice are no longer available from The Jackson Laboratory. MMTV-negative C3H/HeN and MMTV-infected C3H/HeN mice were originally obtained from the National Cancer Institute, Frederick Cancer Research Facility, Frederick, Md. MMTV HP transgenic mice were made on a C3H/HeN genetic background (14).
Cloning and sequencing.
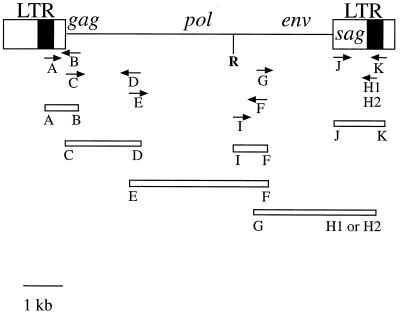
Sequences of exogenous MMTV(C3H) and MMTV(HeJ) and endogenous Mtv1 proviruses were obtained using a panel of overlapping plasmids (Fig. 1). Except for pAB, all plasmids were cloned from viral RNA templates. The viral RNA templates were isolated from the milk-filled stomachs of MMTV-infected C3H/HeJ, C3H/HeN, or MMTV-negative C3H/HeN pups as previously described (17). Viral cDNA was prepared by using SuperScript II reverse transcriptase in the buffer supplied by the manufacturer (GIBCO/BRL, Gaithersburg, Md.) and a (dT)15 primer. The amplified DNA was cloned into a vector using the PCRScript cloning kit (Stratagene, La Jolla, Calif.) and subsequently sequenced. A single-copy pACYC177 vector (New England Biolabs, Beverly, Mass.) was used to clone MMTV(C3H) gag (pCD plasmid) to avoid problems with the poison sequences (5). Since primer A was specific for the Sag hypervariable region located in the U3 LTR (not present in RNA), high-molecular-weight DNA was isolated from spleens of 2- to 3-month-old MMTV-infected C3H/HeJ and C3H/HeN mice and used as a template for the inserts of plasmids pAB. MMTV(C3H) and MMTV(HeJ) were sequenced using plasmids pAB, pCD, pEF, pGH1, and pJK. Mtv1 was sequenced using pHP, which consists of Mtv1 from the beginning of the 5′ LTR to the EcoRI site in the pol gene (37) and the pIF, pJK, and pGH2 plasmids. The three latter plasmids were cloned using reverse transcription-PCR products obtained from RNA isolated from the milk of MMTV-negative C3H/HeN females (a small amount of Mtv1 is produced in the milk of MMTV-negative C3H/He mice [data not shown]). The following primers were used: forward MMTV(C3H) Sag-specific primer A, 5′GACAGTGGCTGGACTAATAGAACATT3′ (nucleotides [nt] 897 to 922, according to the numbering system of Brandt-Carlson et al. [4]); reverse gag-specific primer B, 5′CTCCTTCTTCGGGAAACCAAG3′ (nt 151 to 131 from the start codon in gag); forward gag-specific primer C, 5′ATGGGGGTCTCGGGCTCAAAAGGG3′ (nt 1 to 24 from the start codon in gag); reverse gag-specific primer D, 5′GGGACTGCCCCTTTACAAGTTTTTTGA3′ (nt 1888 to 1762 from the start codon in gag); forward gag-specific primer E, 5′GATGGGAATCCACTTCCTCCC3′ (nt 1720 to 1740 from the start codon in gag); reverse env-specific primer F, 5′GGACCCAGATTGGTGTTTCGGCAT3′ (nt 24 to 1 from to the start codon in env); forward env-specific primer G, 5′ATGCCGAAACACCAATCTGGG3′ (nt 1 to 21 from the start codon in env); reverse MMTV(C3H) Sag-specific primer H1, 5′AATGTTCTATTAGTCCAGCCACTGTC3′ (reverse of primer A); reverse Mtv1 Sag-specific primer H2, 5′GAAAGCTAAGGGCAAAGCCTT3′ (nt 945 to 925 according to the numbering system of Brandt-Carlson et al. [4]); forward pol-specific primer I, 5′GGGAAATGCCTATGCCTATGCAGATTC3′ (nt 5999 to 6019 according to BR6 provirus); forward LTR-specific primer J, 5′GAACTCCCGAGAGTGTCCTACAC3′ (nt 27 to 49 according to the numbering system of Brandt-Carlson et al. [4]); reverse LTR-specific primer K, 5′GGACTGTTGCAAGTTTACTC3′ (nt 1204 to 1185 according to the numbering system of Brandt-Carlson et al. [4]). Primers A and H1 are specific for the hypervariable region of MMTV(C3H) sag, whereas primer H2 is specific for the hypervariable region of Mtv1 sag.
FIG. 1.
Diagram of the MMTV provirus with the primers used (see Materials and Methods) to PCR amplify different regions of MMTV(HeJ), HP, and MMTV(C3H). Solid box, Sag hypervariable region. R, EcoRI.
Two or three independently isolated clones of each plasmid were sequenced.
Southern blot analyses.
Mammary gland tumors were excised from the surrounding normal tissue, and DNA was isolated as previously described (17). Twenty micrograms of each DNA sample was digested with indicated restriction enzymes and electrophoresed on 0.8% agarose gels. After transfer to nylon, the blots were hybridized with 32P-labeled probe (see Fig. 3A), washed, and exposed to Kodak XAR-5 film using Cronex Lightning-Plus intensifying screens.
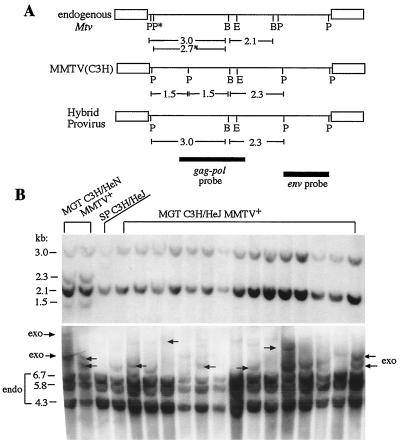
FIG. 3.
Mammary tumors of MMTV(HeJ)-infected C3H/HeJ mice contain a recombinant MMTV. (A) Map of endogenous (endo) proviruses present in C3H/He and BALB/c mice. Also shown are the maps of exogenous (exo) MMTV(C3H) and HP. The presence of the 2.3- or 2.3- and 1.5-kb fragments is characteristic of newly integrated copies of HP or MMTV(C3H), respectively. Abbreviations: E, EcoRI; P, PstI; B, BglII; P∗, PstI site present in Mtv9 provirus inherited by BALB/c but not C3H/He mice. Filled bars, two probes used for hybridization. (B) Mammary tumors of C3H/HeJ mice are not induced by wild-type exogenous MMTV(C3H). Top panel, high-molecular-weight DNAs from mammary gland tumors of MMTV(HeJ)-infected C3H/HeJ mice and from the spleen of a C3H/HeJ mouse were digested with PstI and BglII and subjected to Southern blot analysis with the hybridization gag-pol probe depicted in panel A. DNAs isolated from mammary tumors induced by MMTV(C3H) in C3H/HeN MMTV+ mice are shown for comparison (MGT C3H/HeN MMTV+). Bottom panel, the same DNA samples were digested with EcoRI endonuclease and hybridized with the env probe depicted in panel A. The 6.7-, 5.8-, and 4.3-kb fragments correspond to endogenous Mtv proviruses present in C3H/He mice. Arrows show newly integrated exogenous MMTVs. SP, splenic DNA of a C3H/HeJ mouse.
RNA isolation and Northern blot analysis.
RNA was isolated from mammary gland tumors or normal mammary glands in accordance with a protocol published elsewhere (6). Twenty micrograms of total RNA was subjected to electrophoresis on a 1% formaldehyde gel, transferred to a nylon membrane, and hybridized with a Wnt-1 (29) or Int-2/Fgf-3 (32) probe.
RNase T1 protection analysis.
RNA was isolated from lactating mammary glands and milk of the indicated mice as previously described (14, 17); 40- and 5-μg samples were used for RNase T1 protection analysis, respectively, with the MMTV(C3H) Sag-specific probe (14). In order to compare virus load in MMTV-infected C3H/HeJ and C3H/HeN mice, the ratios of exogenous to endogenous viral RNA expression levels were quantified using a phosphorimager (Bio-Imaging analyzer BAS 1000 MacBas; Fuji Photo Film Co., Ltd.).
Mammary gland tumorigenesis.
Mammary gland tumor incidence was monitored by weekly palpation of the animals. Tumor-bearing mice were sacrificed, and DNA isolated from a portion of each tumor was subjected to Southern blot analysis to confirm the viral etiology (17, 23).
Nucleotide sequence accession numbers.
Nucleotide sequences have been submitted to the GenBank nucleotide sequence database and have been assigned accession numbers AF228552 for MMTV(C3H) provirus, AF228551 for MMTV(HeJ) provirus, and AF228550 for Mtv1 provirus.
RESULTS
Tumor occurrence in C3H substrains.
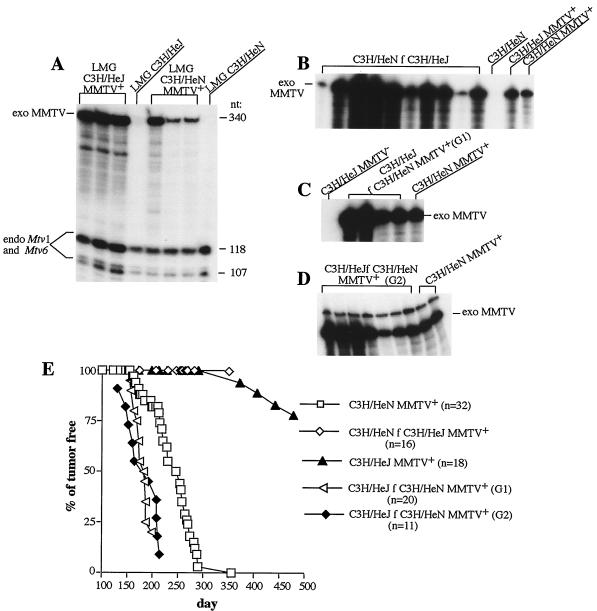
Approximately 50 years ago, the high-tumor-incidence, MMTV-infected C3H mouse strain was divided between the National Institutes of Health (C3H/HeN) and The Jackson Laboratory (C3H/HeJ). In 1973, it was reported that C3H/HeJ mice demonstrated a drastic decrease in mammary tumor incidence compared to the infected C3H/HeN strain (30; D. M. Richardson, JAX Notes 413:1–3, 1973). We have repeated these experiments and confirmed the results. Indeed, 50% of MMTV-infected C3H/HeN mice developed tumors after approximately 250 days whereas only 10% of the C3H/HeJ mice developed tumors after 350 days (Fig. 2E). Exogenous MMTV was present in C3H/HeJ mice, since they showed deletion of Sag-cognate Vβ14+ T cells characteristic of MMTV(C3H) infection. Three- to four-month-old C3H/HeJ mice had only about 3.5% CD4+/Vβ14+ T cells among peripheral T cells, in contrast to 7.5% in virus-free C3H/HeJ mice. Furthermore, RNA isolated from the C3H/HeJ mammary tumors contained large amounts of virus-specific RNA (data not shown). Quantitative analysis of the viral transcripts in the mammary glands of MMTV-infected C3H/HeN and C3H/HeJ mice ruled out differences in the virus load between these two strains; if anything, there was a slight increase in MMTV(HeJ) expression compared to MMTV(C3H) expression (a factor of 1.2) (Fig. 2A; expression of endogenous Mtv proviruses was used as an internal control).
FIG. 2.
Mammary tumor incidence in C3H/HeN and C3H/HeJ mice infected with either MMTV(C3H) or MMTV(HeJ). (A) C3H/HeJ and C3H/HeN mice are equally infected with exogenous MMTVs. RNA was isolated from the lactating mammary glands (LMG) of MMTV-infected C3H/HeJ (C3H/HeJ MMTV+) and C3H/HeN (C3H/HeN MMTV+) females after the first pregnancy and subjected to RNase T1 protection analysis with a probe specific for MMTV(C3H) Sag (14). A 340-nt full-length protection fragment corresponds to expression of integrated exogenous (exo) MMTV [MMTV(C3H) or MMTV(HeJ)] provirus. The smaller fragments of 118 and 107 nt observed with RNA from infected and uninfected LMG of C3H/He mice correspond to RNA produced by Mtv1 and Mtv6 endogenous (endo) proviruses (17). Normalization by expression of endogenous Mtv1 and Mtv6 proviruses showed that, if anything, there was a slight increase in MMTV(HeJ) expression compared to MMTV(C3H). LMG C3H/HeJ and LMG C3H/HeN, RNAs isolated from uninfected C3H/HeJ and C3H/HeN mice, respectively. (B) Tumor-susceptible C3H/HeN mice became infected with exogenous MMTV(HeJ). MMTV-free C3H/HeN mice were foster nursed by MMTV(HeJ)-infected C3H/HeJ females (C3H/HeN f C3H/HeJ). Foster mothers were 6-month-old breeding females. RNA was isolated from their milk after the second pregnancy and subjected to RNase T1 protection analysis with the MMTV(C3H) Sag-specific probe. C3H/HeN, milk RNA of MMTV-negative C3H/HeN mice; C3H/HeJ MMTV+, milk RNA of MMTV(HeJ)-infected C3H/HeJ mice; C3H/HeN MMTV+, milk RNA of MMTV(C3H)-infected C3H/HeN mice. (C) MMTV-negative C3H/HeJ mice became infected with exogenous MMTV(C3H). MMTV-negative C3H/HeJ mice were foster nursed on MMTV(C3H)-infected C3H/HeN milk (C3H/HeJ f C3H/HeN MMTV+, generation 1 [G1]). RNA was isolated from their milk after the first pregnancy and subjected to RNase T1 protection analysis with the MMTV(C3H) sag-specific probe. C3H/HeJ MMTV−, milk RNA of MMTV-negative C3H/HeJ mice; C3H/HeN MMTV+, milk RNA of MMTV(C3H)-infected C3H/HeN mice; exo MMTV, full-length protection. (D) MMTV(C3H)-infected C3H/HeJ mice passed infectious virus to the generation 2 (G2) animals. MMTV(C3H)-infected C3H/HeJ females were mated with C3H/HeJ males to produce the G2 infected females. The females were bred, and RNA isolated from their milk after the first pregnancy was subjected to RNase T1 protection analysis with the MMTV(C3H) sag-specific probe. (E) Mammary tumorigenesis in MMTV-infected C3H/HeJ and C3H/HeN females. MMTV-infected C3H/He mice were bred and monitored for mammary tumors. n represents the number of mice used. All C3H/HeN MMTV+ mice developed tumors by 350 days.
It has been suggested that such a radical decrease in tumor incidence in C3H/HeJ mice is due to mutations in MMTV itself resulting in attenuation of the virus [(MMTV(HeJ) in C3H/HeJ mice versus MMTV(C3H) in C3H/HeN mice] (30). To confirm this, we fostered MMTV-negative C3H/HeN mice on MMTV-infected C3H/HeJ milk and monitored them for mammary gland tumors. C3H/HeN mice became infected with MMTV(HeJ), since they secreted a large amount of this virus into the milk, as was determined by RNase T1 protection analysis (Fig. 2B). These fostered mice were bred and monitored for mammary gland tumors together with MMTV-infected C3H/HeN mice (C3H/HeN MMTV+) as a control. At the time when 100% of the C3H/HeN MMTV+ females developed tumors, no tumors were detected in their littermates nursed on C3H/HeJ MMTV+ milk (Fig. 2E). Therefore, MMTV(HeJ) did not cause tumors in tumor-susceptible C3H/HeN mice, at least within 350 days (Fig. 2E).
In reciprocal experiments, MMTV-free C3H/HeJ mice were foster nursed by C3H/HeN MMTV+ females. These MMTV(C3H)-infected C3H/HeJ mice (generation G1) then were mated with C3H/HeJ males to produced infected offspring (generation G2). All of the animals from both the G1 and G2 generations became MMTV infected and produced MMTV in their milk (Fig. 2C and D). The generation G1 and G2 females were bred and observed for mammary tumors. MMTV(C3H)-infected C3H/HeJ mice in both generations were even more susceptible to MMTV(C3H)-induced tumors than were C3H/HeN mice. Fifty percent of MMTV(C3H)-infected C3H/HeJ mice developed mammary tumors by 183 days, whereas 50% of C3H/HeN females infected with the same virus developed mammary tumors by 250 day (Fig. 2E). Therefore, C3H/HeJ mice are genetically susceptible to MMTV-induced mammary tumors and the change in tumor incidence and latency in this substrain must be due to the occurrence of a new MMTV.
MMTV-induced mammary tumors in C3H/HeJ mice contain recombinant virus.
In addition to endogenous loci, MMTV-induced mammary tumors always demonstrate newly acquired exogenous proviruses (8). We sought to determine whether the newly integrated exogenous MMTV proviruses present in MMTV-infected C3H/HeJ mammary tumors differ from those found in C3H/HeN MMTV+ tumors. Tumor DNA was isolated and subjected to Southern blot restriction fragment length polymorphism analysis and compared with splenic DNA (Fig. 3A) (17, 37). Splenic DNA digested with BglII and PstI endonucleases and hybridized with the gag-pol probe yielded two bands of 3.0 and 2.1 kb corresponding to the germ line-encoded Mtv proviruses present in C3H/He mice (Fig. 3) (23). Two additional bands of 1.5- and 2.3-kb corresponding to exogenous MMTV(C3H) proviruses were obvious in the tumor DNA of C3H/HeN MMTV+ mice but not in tumor DNA of C3H/HeJ mice (Fig. 3B, top). To confirm that mammary tumors in C3H/HeJ mice were induced by exogenous MMTV, we digested the same DNA samples with EcoRI endonuclease, which cleaves MMTV proviral DNA at a single internal site (Fig. 3A). Since the tumors are clonal, each provirus yields two virus-host junction fragments, the lengths of which depend upon the site of integration within the host DNA. The new fragments generally differ in size from the EcoRI fragments derived from the few endogenous Mtv proviruses present in C3H/He mice (8, 9). As a hybridization probe, we used a 1.8-kb PstI fragment of cloned MMTV DNA that contains the viral env gene and detects only EcoRI fragments derived from the 3′ portion of MMTV proviruses (Fig. 3A). This probe also detects three EcoRI fragments (6.7, 5.8, and 4.5 kb) derived from endogenous Mtv proviruses present in C3H/He mice (23). Almost all of the MMTV-infected C3H/HeJ mammary tumors exhibited multiple additional proviruses located at different sites in the host genome (Fig. 3B, bottom). Based on these results, we concluded that mammary tumors in C3H/HeJ mice were induced by exogenous MMTV; however, this virus [MMTV(HeJ)] was different from MMTV(C3H).
Genetically engineered MMTV HP is not tumorigenic in tumor-susceptible C3H/HeN mice.
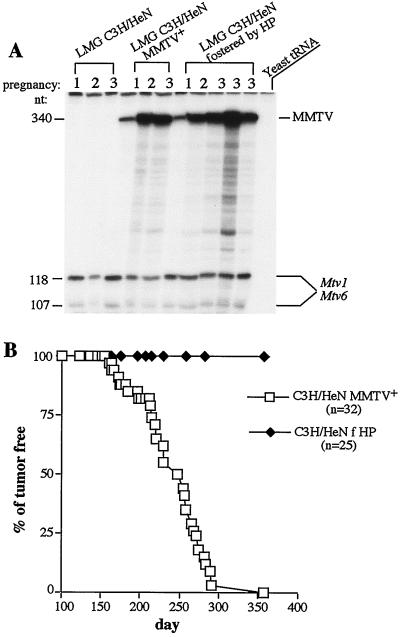
Previously, we produced transgenic mice on a C3H/HeN genetic background with a genetically engineered MMTV HP (37) of which the 5′ half was derived from the endogenous Mtv1 provirus and the 3′ half was derived from exogenous MMTV(C3H) (Fig. 3A) (14). Transgenic females shed virus into the milk, and nontransgenic mice foster nursed by them became infected with the virus, suggesting that HP was infectious (14). We have also tested whether HP amplification within the mammary gland was similar to that of exogenous wild-type MMTV(C3H) by analyzing viral RNA production in the mammary gland after each subsequent pregnancy. HP reached the same level of amplification as wild-type MMTV(C3H) in the mammary glands of infected females (Fig. 4A; all RNA samples were normalized by expression of endogenous Mtv1/Mtv6 proviruses). However, when we monitored mice infected with HP for mammary gland tumors, we discovered that this virus did not cause tumors on a C3H/HeN background. Whereas 100% of C3H/HeN females infected with MMTV(C3H) developed tumors after approximately 350 days, C3H/HeN mice infected with HP developed no tumors during this period (Fig. 4B).
FIG. 4.
Hybrid MMTV produced by HP transgenic mice does not cause tumors in C3H/HeN mice. (A) C3H/HeN mice were equally infected with HP or MMTV(C3H). C3H/HeN mice were foster nursed on HP transgenic milk together with their littermates fostered by C3H/HeN MMTV+ mothers. RNA was isolated from their lactating mammary glands (LMG) after the first (lanes 1), second (lanes 2), and third (lanes 3) pregnancies and subjected to RNase T1 protection analysis with the MMTV(C3H) Sag-specific probe. MMTV, full-length protection. LMG C3H/HeN, RNA isolated from the lactating mammary glands of MMTV-negative C3H/HeN mice. Quantitation analysis was performed as described in the legend to Fig. 2, and no difference in viral RNA expression was observed between MMTV(C3H)- and HP-infected C3H/HeN females. (B) Mammary gland tumor incidence in C3H/HeN mice infected with HP. C3H/HeN mice fostered by HP transgenic (C3H/HeN f HP) or MMTV(C3H)-infected C3H/HeN females (C3H/HeN MMTV+) were bred and monitored for mammary gland tumors. n, number of animals used.
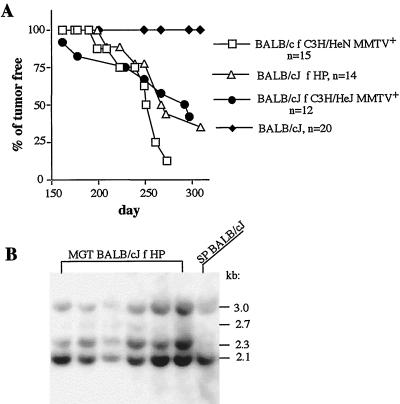
To ensure that the HP can cause tumors, a tumor-susceptible strain of mice, BALB/cJ, was foster nursed on HP-containing milk, bred, and monitored for mammary tumors. Fifty percent of BALB/cJ mice infected with HP developed mammary tumors by 267 days (Fig. 5A and reference 37). To confirm that these tumors were induced by HP, we isolated their DNA and subjected it to Southern blot analysis, which allows distinction between endogenous and exogenous MMTVs as described above. The endogenous BALB/cJ loci (except for Mtv6) yielded fragments of 3.0, 2.7, and 2.1 kb, while digestion and hybridization of integrated HP yielded fragment of 2.3 kb (Fig. 3A). BALB/cJ mammary tumors were induced by HP, since all of the tumors contained the 2.3-kb fragment characteristic of integrated HP (Fig. 5B). We also fostered BALB/cJ mice on C3H/HeJ milk and monitored them for mammary tumors. Fifty percent of BALB/cJ females infected with MMTV(HeJ) developed mammary tumors by 292 days (Fig. 5A), and all of the tumors were induced by MMTV(HeJ) (data not shown). Thus, both MMTV(HeJ) and HP were capable of causing mammary tumors in BALB/cJ mice but not in tumor-susceptible C3H/HeN mice.
FIG. 5.
BALB/cJ mice are susceptible to mammary tumors induced by both MMTV(HeJ) and HP. (A) BALB/cJ mice were fostered by MMTV-infected C3H/HeJ females (BALB/c f C3H/HeJ MMTV+), HP transgenic females (BALB/c f HP), or MMTV(C3H)-infected C3H/HeN females (BALB/cJ f C3H/HeN MMTV+); bred; and monitored for mammary gland tumors. n, number of animals used. BALB/cJ, uninfected mice. (B) BALB/cJ mammary tumors were induced by HP. DNA isolated from mammary tumors developed in BALB/cJ mice fostered on HP milk was subjected to Southern blot analysis as described in the legend to Fig. 3. SP BALB/cJ, splenic DNA of a BALB/cJ mouse.
Both MMTV(HeJ) and HP are capable of up-regulating expression of Int genes.
In contrast to MMTV(C3H), the two other viruses, MMTV(HeJ) and HP, did not cause tumors on the C3H/HeN background even though both were infectious. In over 80% of C3H/He MMTV-induced mammary tumors, at least one of the additional MMTV genomes is integrated near the cellular Wnt-1 gene (29) and in approximately 10% of the tumors integration occurs near the Int-2/Fgf-3 locus (12, 31). Proviral integrations are found at several locations near these genes, but insertions always leave the protein-coding domain intact. In most tumors, the transcriptional orientation of the proviruses is directed away from the Int genes, an indication that the proviral DNA enhancers up-regulate expression of the oncogenes. The induction of Int transcription by MMTV provirus insertion is believed to be an early step in the transformation process.
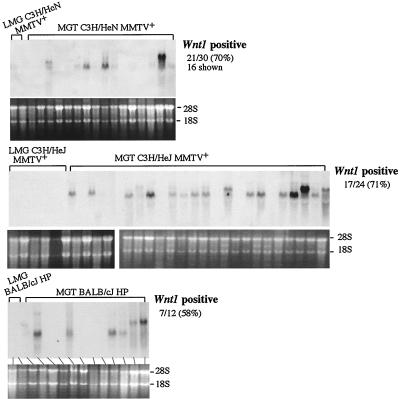
It had been shown previously that HP-induced mammary tumors in BALB/c mice exhibit induced or altered expression of different int genes (24, 36), suggesting that HP is capable of up-regulating int gene expression. We also analyzed RNA isolated from tumors induced by HP or MMTV(HeJ) for the expression of Wnt-1 and Int-2/Fgf-3 transcripts. Expression of Wnt-1 was detected in 71% of MMTV(HeJ)-infected C3H/HeJ mammary tumors and in 58% of HP-infected BALB/cJ mouse mammary tumors (Fig. 6). Expression of Int-2/Fgf-3 was detected in 25% of C3H/HeJ tumors and in 16% of BALB/cJ mammary tumors induced by HP (data not shown). Thus, both HP and MMTV(HeJ) are capable of integrating next to and up-regulating the expression of Int genes.
FIG. 6.
Both HP and MMTV(HeJ) are capable of upregulating expression of the Wnt-1 gene. RNA was isolated from mammary tumors induced by MMTV(C3H) (top panel), MMTV(HeJ) (middle panel), and HP (bottom panel) in MMTV(C3H)-infected C3H/HeN (C3H/HeN MMTV+), MMTV(HeJ)-infected C3H/HeJ (C3H/HeJ MMTV+), and HP-infected BALB/cJ (BALB/cJ HP) mice, respectively, and used for Northern blot analysis with a Wnt-1-specific probe (29). Fragments with molecular weights that were higher than predicted reflect initiation of the transcripts in the LTR. The RNA samples run on a 1% formaldehyde gel were stained with ethidium bromide before blotting to verify their integrity and equal loading (bottom of each blot). 28S and 18S indicate rRNA bands. LMG, RNA isolated from lactating mammary glands of MMTV-infected mice; MGT, mammary gland tumors.
MMTV(HeJ) is a genetic recombinant between endogenous Mtv1 and exogenous MMTV(C3H).
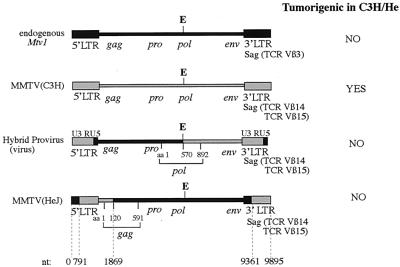
Based on our Southern blot data, we hypothesized that MMTV(HeJ) is a recombinant virus. Of five different endogenous MMTVs present in the C3H/HeN genome, Mtv1 is the only one that can be copackaged with exogenous MMTV in the mammary gland to give rise to a new recombinant (17). Thus, we have cloned and sequenced Mtv1, MMTV(C3H), and MMTV(HeJ). MMTV(HeJ) was found to be a recombinant between Mtv1 and MMTV(C3H) (Fig. 7). Interestingly, almost the entire genome of the virus was derived from Mtv1, except for the Sag hypervariable region and the first 360 bp of the gag region, which were from MMTV(C3H). Knowing the sequences of different viruses, it was possible to compare the phenotypes they produced and to deduce which gene might be responsible for tumor resistance in C3H/He mice. The presence of the MMTV(C3H) gag gene in the context of the provirus was found to correlate with tumorigenicity in the mammary glands of C3H/He mice. Although the entire pro and pol genes in MMTV(HeJ) and pro and part of the pol gene (until the EcoRI site) in HP were of Mtv1 origin, we think that it is unlikely that these genes can contribute to tumorigenesis. First, the majority of amino acid changes in the pro and pol genes of Mtv1 were conservative relative to those of pro and pol of MMTV(C3H). Second, the nonconservative differences in the pol gene of Mtv1 relative to the pol gene of MMTV(C3H) were found downstream of the EcoRI site (after amino acid [aa] 571), where the pol gene in nontumorigenic HP is of MMTV(C3H) origin.
FIG. 7.
Structure and tumorigenic features of different MMTVs. Exogenous MMTV(C3H), MMTV(HeJ), and HP and endogenous Mtv1 proviruses were cloned and sequenced as described in Materials and Methods. The HP pol gene is chimeric; its first 570 aa were derived from Mtv1, and the rest are from exogenous MMTV(C3H). The recombination break point in MMTV(HeJ) occurred in the gag gene between nt 1845 and 1869 [nt 1845 is specific for MMTV(C3H), whereas nt 1869 is specific for Mtv1] and in LTR between nt 9333 and 9361 [nt 9333 is specific for Mtv1, whereas nt 9361 is specific for MMTV(C3H)]. The hypervariable region of sag is nt 9430 to 9530. As a result, the first 120 aa of the gag gene and the hypervariable region of the sag gene in MMTV(HeJ) are derived from MMTV(C3H). Although the original DNA construct used to make HP transgenic mice has a 5′ LTR derived from Mtv1 and a 3′ LTR derived from MMTV(C3H), upon infection of cells with the resulting virus, MMTV(C3H)-specific information in the U3 region of the 3′ LTR is present in the newly synthesized 5′ LTR. TCR, T-cell receptor.
The Gag protein is the precursor to the internal structural proteins of all retroviruses. All Gags are organized in the same order, from the amino terminus to the carboxyl terminus, with domains that are cleaved inside the viral particle to yield the matrix (MA), capsid (CA), and nucleocapsid (NC) proteins. The MA protein is located beneath the viral membrane and initiates virus assembly, the CA protein forms the mature virion core, and the NC protein (small basic protein) coats the viral RNA in a sequence-independent manner. We found three regions of nonconservative amino acid changes; two are located within putative bipartite nuclear localization domains of the MA protein (aa 174 to 191 and 231 to 248), and one lies inside the Zn2+ binding finger motif CX2CX4HX4C of the NC protein (aa 527 to 540) (Fig. 8).
FIG. 8.
Amino acid sequence comparison of Mtv1 and MMTV(C3H) gag genes. Proteins: aa 2 to 100, MA protein; aa 271 to 497, CA protein; aa 498 to 591, NC protein. In MMTV(HeJ), the first 120 aa are derived from MMTV(C3H) and the rest are from Mtv1. The first two boxes (aa 174 to 192 and 231 to 148) show predicted nuclear localization motifs; the third box (aa 527 to 540) shows the Zn2+ motif.
DISCUSSION
Our studies provide an explanation for the major change in mammary tumor incidence and latency occurring in the MMTV-infected C3H/HeJ mouse strain maintained at The Jackson Laboratory (30; D. M. Richardson, JAX Notes 413:1–3, 1973) (Fig. 2E). Only 30% of breeding MMTV-infected C3H/HeJ females developed tumors by 500 days, in contrast to the 97% of MMTV+ C3H/HeN mice that developing tumors by 290 days (Fig. 2E). However, C3H/HeJ mice are genetically susceptible to MMTV-induced tumors because they developed high-incidence mammary tumors when fostered by C3H/HeN MMTV+ females (Fig. 2E), as has been previously reported (30). Furthermore, tumor-susceptible C3H/HeN mice do not develop mammary tumors when foster nursed on MMTV-infected C3H/HeJ milk (Fig. 2E). Therefore, our data concur with those of Outzen et al. (30) and indicate that the attenuated tumor incidence in the C3H/HeJ substrain is due to the occurrence of a new exogenous MMTV [MMTV(HeJ)].
We have demonstrated that MMTV(HeJ) is a genetic recombinant between endogenous nonpathogenic Mtv1 and exogenous MMTV(C3H). Almost the entire provirus consists of Mtv1 sequences, except for a small portion of the gag gene and the Sag hypervariable region, which were derived from exogenous MMTV(C3H) (Fig. 7). Retention of the Vβ14+ T-cell-specific Sag in the context of MMTV(HeJ) is not surprising, because exogenous virus with Mtv1-derived Vβ3+ T-cell-specific Sag would not be able to mediate infection in C3H/He mice (they lack Vβ3+ T cells due to expression of endogenous Mtv1/Mtv6 proviruses). Another MMTV variant, genetically engineered HP, that has the 5′ half (including the 5′ LTR, gag, and part of the pol gene) from Mtv1 and the 3′ half (including the rest of the pol gene, env, and the 3′ LTR with Sag) from MMTV(C3H), is also not tumorigenic on the C3H/He background (Fig. 4B). However, both MMTV(HeJ) and HP caused mammary tumors in BALB/cJ mice (Fig. 5A). Nucleotide sequence comparison of tumor-inducing MMTV(C3H) and nontumorigenic MMTV(HeJ) and HP made it possible to map the tumor attenuation region to gag of Mtv1.
The change in tumor incidence and latency in C3H/HeJ MMTV+ mice was investigated by Outzen et al. in 1985 (30). According to them, only 37% of the C3H/HeJ breeding females had detectable MMTV antigens in their milk samples during the first lactation and the percentage of positive milk samples increased with parity to 63% in the second lactation and to 74% in the third lactation (30). In addition, Outzen et al. demonstrated that C3H/HeJ mice fostered by C3H/HeOuJ MMTV+ females (a high tumor incidence substrain of C3H/He mice similar to the C3H/HeN substrain) had lower transmission of exogenous MMTV(OuJ) through milk since the percentage of exogenous MMTV antigen-positive milk samples at the third parity declined from more than 83% in the first generation to less than 67% in the third generation (30). Based on these results, the authors concluded that the C3H/HeJ host inhibited the milk-borne transmission of exogenous MMTV. Although we have not analyzed as many mice for milk production as did Outzen et al., we have consistently seen the same level of virus production by MMTV-infected C3H/HeJ and C3H/HeN mice. The most likely explanation for the discrepancy between their and our results is the sensitivity of the assays used to detect virus production. Outzen et al. tested milk samples by means of an immunodiffusion assay (IDA) for the presence of MMTV antigen (30). Because the IDA is relatively insensitive, it usually detects only large quantities of antigen (>50 μg) (30). Indeed, only 89% of MMTV-infected C3H/OuJ mice had MMTV antigen-positive milk as determined by IDA (30). Nevertheless, all of them developed mammary tumors by 300 days (30 and our own data). We have used the much more sensitive RNase T1 protection assay to detect virus in the milk of infected mice. Using this approach, we have shown that there is always virus produced by C3H/HeN MMTV+ or C3H/HeJ MMTV+ mice and that the level of production does not show dramatic differences between the different age- and pregnancy-matched mice of these two strains (Fig. 2 and 4).
In a majority of mammary tumors, the MMTV proviruses are integrated into the host's genome near one of the Int protooncogenes activating their expression (29, 31). The oncogenic properties of the Wnt-1 gene were proven in transgenic mice, where expression of this gene in the mammary epithelium induced mammary adenocarcinoma development in both males and females (40). However, the median latency of mammary tumor formation in female Wnt-1 transgenic mice was 5 months of age, with >80% of mice developing tumors by 7 months (40). In addition, transgenic females rarely developed more than one tumor per mouse and never developed more than three tumors per mouse (36, 40). This relatively long latency period before tumor development and the stochastic nature of mammary tumors in Wnt-1 transgenic mice argue that Wnt-1 contributes to, but is not sufficient for, tumorigenesis in these mice. Interestingly, Wnt-1 transgenic mice infected with exogenous MMTV demonstrated a dramatic increase in the number of mammary tumors (36). At 4 months of age, infected female breeders showed >5 mammary tumors per mouse, with some animals developing 10 tumors (36). Analysis of provirus-containing tumors for induced or altered expression of known Wnt genes showed activation of Int-2/Fgf-3 in 39% and Hst/Fgf-4 in 3% of the MMTV-infected Wnt-1 transgenic tumors (36). Therefore, it was suggested that cooperation between different oncogenes is an important step in mammary gland tumor induction. Our data indicate that in addition to cooperation between oncogenes, viral genes also contribute to mammary tumorigenesis.
The original virus in the progenitor C3H/He strain was highly tumorigenic. Even if there were two different viruses in the original C3H/He stock, why is it that the attenuated MMTV(HeJ) was selected or became prominent in the C3H/HeJ strain? Because mammary tumors are not required for the virus life cycle, selection for a tumor-attenuated MMTV would be unusual unless a less tumorigenic virus had a competitive advantage over the more tumorigenic ones. Tumor-causing MMTV(C3H) is infectious in C3H/HeJ mice (Fig. 2C and D), suggesting that the selection and retention of a less tumorigenic MMTV(HeJ) required the presence of a heritable, selective pressure operating over many generations. It could be related to some mutation, such as Tlr4Lps-d, which occurred during the same time period when mammary tumor incidence changed in the C3H/HeJ substrain (34, 38, 39, 42). The Tlr4Lps-d mutation is carried homozygously in the C3H/HeJ substrain. This gene was originally named for its ability to increase resistance to lipopolysaccharide toxicity (Lps) and was recently shown to be the Toll-like receptor 4 gene (Tlr4), a member of the neonate Il-1/Toll receptor family (21, 26). Studies are ongoing to determine why and how the new recombinant tumor-attenuated virus was selected in C3H/HeJ mice.
ACKNOWLEDGMENTS
L.M.H. and Y.A. contributed equally to this work.
This work was supported by PHS grants CA65795 to T.V.G. and CA45954 to S.R.R. and by a grant from The Jackson Laboratory to T.V.G. This work was also supported by a grant (CA34196) from the National Cancer Institute to The Jackson Laboratory.
We are thankful to A. Chervonsky and D. Roopenian for helpful discussion.
REFERENCES
- 1.Acha Orbea H, Shakhov A N, Scarpellino L, Kolb E, Muller V, Vessaz Shaw A, Fuchs R, Blochlinger K, Rollini P, Billotte J, Sarafidou M, MacDonald H R, Diggelmann H. Clonal deletion of V beta 14-bearing T cells in mice transgenic for mammary tumour virus. Nature. 1991;350:207–211. doi: 10.1038/350207a0. [DOI] [PubMed] [Google Scholar]
- 2.Benvelzen P, Hilgers J. Murine mammary tumor virus. In: Klein G, editor. Viral oncology. New York, N.Y: Raven Press; 1980. pp. 311–355. [Google Scholar]
- 3.Beutner U, Kraus E, Kitamura D, Rajewsky K, Huber B T. B cells are essential for murine mammary tumor virus transmission, but not for presentation of endogenous superantigens. J Exp Med. 1994;179:1457–1466. doi: 10.1084/jem.179.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandt-Carlson C, Butel J S, Wheeler D. Phylogenetic and structural analysis of MMTV LTR ORF sequences of exogenous and endogenous origins. Virology. 1993;185:171–185. doi: 10.1006/viro.1993.1113. [DOI] [PubMed] [Google Scholar]
- 5.Brookes S, Placzek M, Moore R, Dixon M, Dickson C, Peters G. Insertion elements and transitions in cloned mouse mammary tumour virus DNA: further delineation of the poison sequences. Nucleic Acids Res. 1986;14:8231–8245. doi: 10.1093/nar/14.21.8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chirgwin J M, Prxybyla A E, MacDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 7.Choi Y, Kappler J W, Marrack P. A superantigen encoded in the open reading frame of the 3′ long terminal repeat of the mouse mammary tumor virus. Nature. 1991;350:203–207. doi: 10.1038/350203a0. [DOI] [PubMed] [Google Scholar]
- 8.Cohen J C, Shank P R, Morris V L, Cardiff R, Varmus H E. Integration of the DNA of mouse mammary tumor virus in virus-infected normal and neoplastic tissue of the mouse. Cell. 1979;16:333–345. doi: 10.1016/0092-8674(79)90010-2. [DOI] [PubMed] [Google Scholar]
- 9.Cohen J C, Varmus H E. Endogenous mammary tumour virus DNA varies among wild mice and segregates during inbreeding. Nature. 1979;278:418–423. doi: 10.1038/278418a0. [DOI] [PubMed] [Google Scholar]
- 10.Dyson P J, Knight A M, Fairchild S, Simpson E, Tomonari K. Genes encoding ligands for deletion of V beta 11 T cells cosegregate with mammary tumour virus genomes. Nature. 1991;349:531–532. doi: 10.1038/349531a0. [DOI] [PubMed] [Google Scholar]
- 11.Frankel W N, Rudy C, Coffin J M, Huber B T. Linkage of Mls genes to endogenous mammary tumour viruses of inbred mice. Nature. 1991;349:526–528. doi: 10.1038/349526a0. [DOI] [PubMed] [Google Scholar]
- 12.Gallahan D, Callahan R. Mammary tumorigenesis in feral mice: identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors. J Virol. 1987;61:66–74. doi: 10.1128/jvi.61.1.66-74.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golovkina T V. A novel mechanism of resistance to mouse mammary tumor virus infection. J Virol. 2000;74:2752–2759. doi: 10.1128/jvi.74.6.2752-2759.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golovkina T V, Chervonsky A, Prescott J C, Janeway C A, Ross S R. The mouse mammary tumor virus envelope gene product is required for superantigen presentation to T cells. J Exp Med. 1994;179:439–446. doi: 10.1084/jem.179.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golovkina T V, Chervonsky A V, Dudley J P, Ross S R. Transgenic mouse mammary tumor virus superantigen expression prevents viral infection. Cell. 1992;69:637–645. doi: 10.1016/0092-8674(92)90227-4. [DOI] [PubMed] [Google Scholar]
- 16.Golovkina T V, Dudley J P, Ross S R. B and T cells are required for mouse mammary tumor virus spread within the mammary gland. J Immunol. 1998;161:2375–2382. [PubMed] [Google Scholar]
- 17.Golovkina T V, Jaffe A, Ross S R. Coexpression of exogenous and endogenous mouse mammary tumor virus RNA in vivo results in viral recombination and broadens the virus host range. J Virol. 1994;68:5019–5026. doi: 10.1128/jvi.68.8.5019-5026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golovkina T V, Shlomchik M, Hannum L, Chervonsky A. Organogenic role of B lymphocytes in mucosal immunity. Science. 1999;286:1965–1968. doi: 10.1126/science.286.5446.1965. [DOI] [PubMed] [Google Scholar]
- 19.Held H, Shakhov A N, Izui S, Waanders G A, Scarpellino L, MacDonald H R, Acha-Orbea H. Superantigen-reactive CD4+ T cells are required to stimulate B cells after infection with mouse mammary tumor virus. J Exp Med. 1993;177:359–366. doi: 10.1084/jem.177.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Held W, Waanders G, Shakhov A N, Scarpellino L, Acha-Orbea H, MacDonald H R. Superantigen-induced immune stimulation amplifies mouse mammary tumor virus infection and allows virus transmission. Cell. 1993;74:529–540. doi: 10.1016/0092-8674(93)80054-i. [DOI] [PubMed] [Google Scholar]
- 21.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 22.Kappler J W, Staerz U, White J, Marrack P C. Self-tolerance eliminates T cells specific for Mls-modified products of the major histocompatibility complex. Nature. 1988;332:35–40. doi: 10.1038/332035a0. [DOI] [PubMed] [Google Scholar]
- 23.Kozak C, Peters G, Pauley R, Morris V, Michalides R, Dudley J, Green M, Davisson M, Prakash O, Vaidya A, Hilgers J, Verstraeten A, Hynes N, Diggelmann H, Peterson D, Cohen J C, Dickson C, Sarkar N, Nusse R, Varmus H, Callahan R. A standardized nomenclature for endogenous mouse mammary tumor viruses. J Virol. 1987;61:1651–1654. doi: 10.1128/jvi.61.5.1651-1654.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee F S, Lane T F, Kuo A, Shackleford G M, Leder P. Insertional mutagenesis identifies a member of the Wnt gene family as a candidate oncogene in the mammary epithelium of int-2/Fgf-3 transgenic mice. Proc Natl Acad Sci USA. 1995;92:2268–2272. doi: 10.1073/pnas.92.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacDonald H R, Schneider R, Lees R K, Howe R C, Acha Orbea H, Festenstein H, Zinkernagel R M, Hengartner H. T-cell receptor V beta use predicts reactivity and tolerance to Mls1a-encoded antigens. Nature. 1988;332:40–45. doi: 10.1038/332040a0. [DOI] [PubMed] [Google Scholar]
- 26.Medzhitov R, Preston-Hurlburt P, Janeway C A., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 27.Nandi S, McGrath C M. Mammary neoplasia in mice. Adv Cancer Res. 1973;17:353–414. [Google Scholar]
- 28.Nusse R. The int genes in mammary tumorigenesis and in normal development. Trends Genet. 1988;4:291–295. doi: 10.1016/0168-9525(88)90172-2. [DOI] [PubMed] [Google Scholar]
- 29.Nusse R, Varmus H. Mammary tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 30.Outzen H C, Corrow D, Shultz L D. Attenuation of exogenous murine mammary tumor virus virulence in the C3H/HeJ mouse substrain bearing the Lps mutation. JNCI. 1985;75:917–923. doi: 10.1093/jnci/75.5.917. [DOI] [PubMed] [Google Scholar]
- 31.Peters G, Brookes S, Smith R, Dickson C. Tumorigenesis by mouse mammary tumor virus: evidence for a common region for provirus integration in mammary tumors. Cell. 1983;33:369–377. doi: 10.1016/0092-8674(83)90418-x. [DOI] [PubMed] [Google Scholar]
- 32.Peters G, Lee A E, Dickson C. Concerted activation of two potential proto-oncogenes in carcinomas induced by mouse mammary tumour virus. Nature. 1986;320:628–631. doi: 10.1038/320628a0. [DOI] [PubMed] [Google Scholar]
- 33.Rosenberg N, Jolicoeur P. Retroviral pathogenesis. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 475–586. [PubMed] [Google Scholar]
- 34.Rosenstreich D L, Glode L M. Difference in B cell mitogen responsiveness between closely related strains of mice. J Immunol. 1975;115:777–780. [PubMed] [Google Scholar]
- 35.Salmons B, Ginzburg W H. Current perspectives in the biology of mouse mammary tumor virus. Virus Res. 1987;8:81–102. doi: 10.1016/0168-1702(87)90022-0. [DOI] [PubMed] [Google Scholar]
- 36.Shackleford G M, MacArthur C A, Kwan H C, Varmus H E. Mouse mammary tumor virus infection accelerates mammary carcinogenesis in Wnt-1 transgenic mice by insertional activation of int-2/Fgf-3 and hst/Fgf-4. Proc Natl Acad Sci USA. 1993;90:740–744. doi: 10.1073/pnas.90.2.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shackleford G M, Varmus H E. Construction of a clonable, infectious and tumorigenic mouse mammary tumor virus provirus and a derivative genetic vector. Proc Natl Acad Sci USA. 1988;85:9655–9659. doi: 10.1073/pnas.85.24.9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skidmore B J, Chiller J M, Weigle W O, Riblet R, Watson J. Immunologic properties of bacterial lipopolysaccharide (LPS). III. Genetic linkage between the in vitro mitogenic and in vivo adjuvant properties of LPS. J Exp Med. 1976;143:143–150. doi: 10.1084/jem.143.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sultzer B M. Genetic control of host responses to endotoxin. Infect Immun. 1972;5:107–113. doi: 10.1128/iai.5.1.107-113.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsukamoto A S, Grosschedl R, Guzman R C, Parslow T, Varmus H E. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell. 1988;55:619–625. doi: 10.1016/0092-8674(88)90220-6. [DOI] [PubMed] [Google Scholar]
- 41.Varmus H E, Padgett T, Heasley S, Simon G, Bishop J M. Cellular functions are required for the synthesis and integration of avian sarcoma virus-specific DNA. Cell. 1977;11:307–319. doi: 10.1016/0092-8674(77)90047-2. [DOI] [PubMed] [Google Scholar]
- 42.Watson J, Riblet R. Genetic control of responses to bacterial lipopolysaccharides in mice. I. Evidence for a single gene that influences mitogenic and immunogenic responses to lipopolysaccharides. J Exp Med. 1974;140:1147–1161. doi: 10.1084/jem.140.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woodland D L, Happ M P, Gollob K J, Palmer E. An endogenous retrovirus mediating deletion of alpha beta T cells? Nature. 1991;349:529–530. doi: 10.1038/349529a0. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto K. Steroid receptor regulated transcription of specific genes and gene networks. Annu Rev Genet. 1985;19:209–252. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]
- 45.Yazdanbakhsh K, Park C G, Winslow G M, Choi Y. Direct evidence for the role of COOH terminus of mouse mammary tumor virus superantigen in determining T cell receptor Vβ specificity. J Exp Med. 1993;178:737–741. doi: 10.1084/jem.178.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]