SUMMARY
Distinct locations of different white adipose depots suggest anatomy-specific developmental regulation, a relatively understudied concept. Here, we report a population of Tcf21 lineage cells (Tcf21 LCs) present exclusively in visceral adipose tissue (VAT) that dynamically contributes to VAT development and expansion. During development, the Tcf21 lineage gives rise to adipocytes. In adult mice, Tcf21 LCs transform into a fibrotic or quiescent state. Multiomics analyses show consistent gene expression and chromatin accessibility changes in Tcf21 LC, based on which we constructed a gene-regulatory network governing Tcf21 LC activities. Furthermore, single-cell RNA sequencing (scRNA-seq) identifies the heterogeneity of Tcf21 LCs. Loss of Tcf21 promotes the adipogenesis and developmental progress of Tcf21 LCs, leading to improved metabolic health in the context of diet-induced obesity. Mechanistic studies show that the inhibitory effect of Tcf21 on adipogenesis is at least partially mediated via Dlk1 expression accentuation.
Graphical abstract
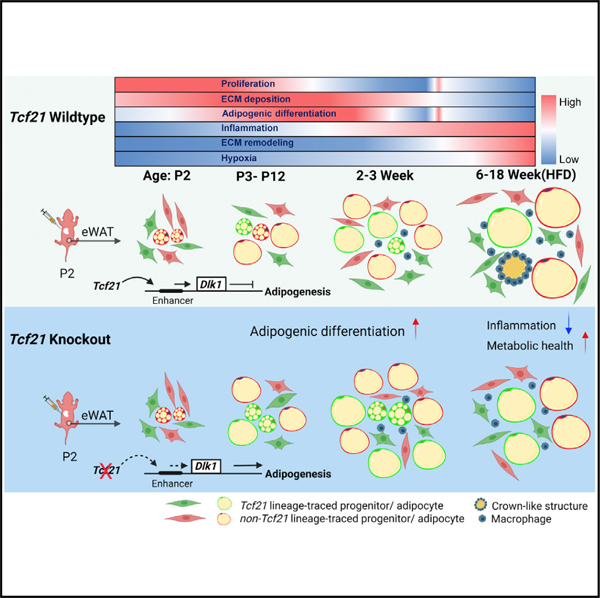
In brief
Liu et al. report that Tcf21, a VAT-specific gene, is exclusive to MPCs. Tcf21 LCs directly contribute to VAT development but have limited adipogenesis in adults because of transcriptional and chromatin accessibility changes. Loss of Tcf21 promotes visceral adipogenesis by reducing Dlk1 expression, improving the metabolic health of obese mice.
INTRODUCTION
White adipose tissue (WAT) is the primary energy reserve and an essential endocrine organ. However, excessive WAT accumulation is a leading cause of obesity and type 2 diabetes. During development and growth, mesenchymal progenitor cells (MPCs) differentiate into white adipocytes before undergoing hypertrophy.1,2 Compared with adipocyte hypertrophy, which can lead to adipocyte hypoxia and death, inflammation, and insulin resistance,3–5 adipocyte hyperplasia is typically associated with improved AT function.6,7
Visceral WAT (VAT) is generally considered more strongly correlated with insulin resistance than subcutaneous WAT (SAT),8,9 rendering VAT a promising therapeutic target. Although SAT and VAT adipogenesis share a core regulatory network,10,11 depot-specific development and expansion have been reported.2,12–15 Moreover, one comparative study identified depot-specific genes, including the VAT-specific Tcf21.13 Tcf21 encodes the basic-helix-loop-helix (bHLH) transcription factor 21 (Tcf21), which is a maker of fibroblasts and similar cells in select visceral organs.16–19
We recently characterized the differentiation of Tcf21 lineage cardiac fibroblasts after myocardial infarction.20,21 However, very little is known about Tcf21 lineage cells (Tcf21 LCs) in VAT. Tcf21 expression manipulation in VAT-derived stromal vascular cells (SVCs) in vitro alters the expression of some extracellular matrix (ECM) remodeling-related genes.22 Nevertheless, the contribution of Tcf21 lineage to VAT development and expansion, and the functional role of Tcf21 in these processes, have not been studied to our knowledge.
Because global Tcf21 knockout mice die perinatally,16–18 studying the function of Tcf21 in VAT, which primarily develops after birth, is extremely challenging.1,15 Here, using lineage tracing (LT) and an inducible cell-type-specific Tcf21 knockout mouse line, coupled with gain-of-function and mechanistic analyses, we show the depot-specific contribution of Tcf21 LCs to VAT development and expansion and report an inhibitory role of Tcf21 in the adipogenesis and developmental progress of Tcf21 LCs.
RESULTS
Tcf21 LCs give rise to visceral adipocytes
To explore the identity of Tcf21 LCs, we generated Tcf21MCM/+; R26tdTomato;PdgfraeGFP mice in which Tcf21 LCs express cytoplasmic tdTomato upon tamoxifen (TM) treatment, and Pdgfra+ cells express nuclear eGFP (Figure 1A). These mice were exposed to TM at 2 months of age (Figure 1B). Fibroblast-like tdTomato+ cells were readily identifiable in VAT (epidydimal VAT [eVAT] shown as a representative) 1 week after TM injection (Figures 1B–1D). Very few tdTomato+ cells were identified in inguinal SAT (iSAT) (Figure 1C). Mesothelial and interstitial Tcf21 LCs were identified in VAT, most of which expressed Pdgfra-eGFP, suggesting MPC identity (Figures 1C–1E). tdTomato was also observed in a few newly differentiated Pdgfra-eGFP+ adipocytes but not in mature adipocytes (Figure 1D), indicating that Tcf21 expression is lost during adipogenesis. Interestingly, no difference in Tcf21 expression, proliferation, or adipogenic efficiency was identified between tdTomato+;Pdgfra-eGFP+ and tdTomato−;Pdgfra-eGFP+ cells (Figures S1A and S1B). This suggests that Tcf21 is expressed in all VAT but not SAT Pdgfrα+ MPCs, which was further supported by re-analysis of recently published single-cell RNA sequencing (scRNA-seq) datasets (Figures S1C and S1D).23,24 Moreover, Tcf21 was specific to Pdgfrα+ MPCs versus other reported VAT progenitor markers, such as Ly6a (Sca1) and WT1, which were also expressed in Pdgfrα− cells (Figure S1C).12
Figure 1. Tcf21 LCs give rise to visceral adipocytes.
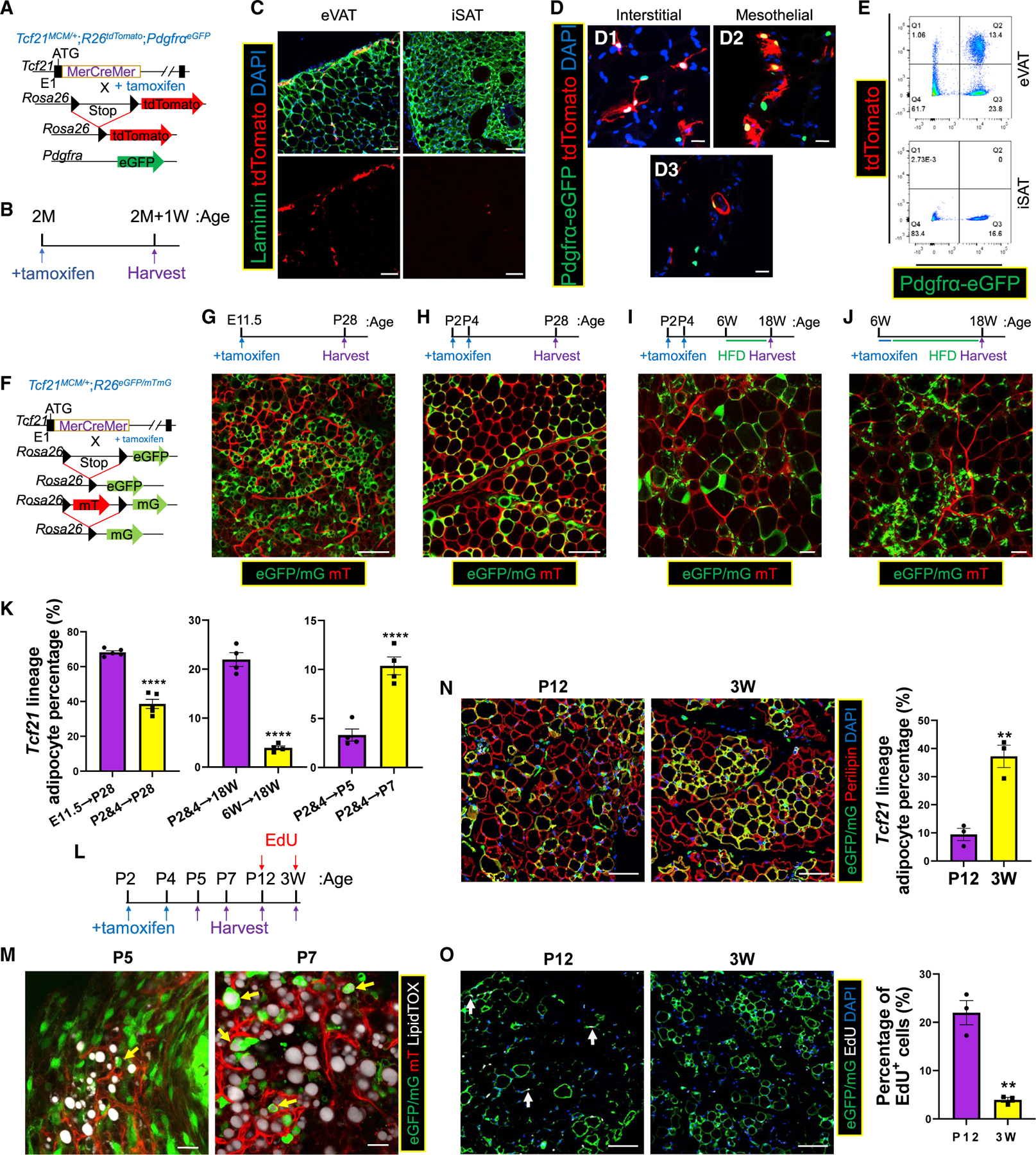
(A and B) Schematics of the generation of Tcf21MCM/+;R26tdTomato;PdgfraeGFP mice (A) and TM treatment and sample collection strategy (B).
(C) eVAT and iSAT were subjected to immunohistochemical staining (IHC), identifying Tcf21 LCs. Scale bar, 20 μm.
(D) IHC images showing Pdgfra-eGFP+ Tcf21 lineage mesothelial (D1) and interstitial (D2) non-adipocytes and differentiating adipocytes (D3). Scale bar, 20 μm.
(E) FC analysis of eVAT and iSAT.
(F) Schematic of the generation of Tcf21 LT mice. (G–J) Tcf21 LT mice were treated with TM at the indicated ages and sampled on the indicated days, followed by whole-mount imaging (G–J). Scale bar, 100 μm.
(K) The fractions of Tcf21 lineage-traced adipocytes in (G)–(J) and (M) were quantified.
n = 5 (G and H); n = 4 (I, J, and M); unpaired t test.
(L) Schematic of TM and EdU treatments in Tcf21 LT mice and sample collection at the indicated time points.
(M) eVAT collected at P5 and P7 was stained with LipidTOX and subjected to whole-mount imaging to identify Tcf21 lineage adipocytes (yellow arrows). Scale bar, 20 μm.
(N and O) Sections of eVAT collected on P12 and P21 were subjected to IHC to quantify the percentage of Tcf21 lineage adipocytes (N) and the percentage of EdU+ Tcf21 LCs (white arrows) (O). Scale bar, 100 μm. n = 3; unpaired t test.
**p < 0.01, ****p < 0.0001. Data are represented as mean ± SEM with individual values plotted. See also Figure S1.
To study the contribution of Tcf21 LCs to VAT development, we generated Tcf21MCM/+;R26eGFP/mTmG LT (Tcf21 LT) mice (Figure 1F). Upon TM treatment, Tcf21 LCs constitutively express cytoplasmic and membrane GFP in these mice. Tcf21 LT mice were treated with TM on embryonic day 11.5 (E11.5) or post-natal day 2 (P2) and P4. P28 eVAT displayed many GFP+ adipocytes (Figures 1G, 1H, and 1K). GFP+ adipocytes were also observed in mesenteric VAT (mVAT) and perirenal VAT (prVAT) but not in iSAT, retroperitoneal AT (rtAT), or pericardial AT (pcAT), indicating that Tcf21 LCs include adipogenic progenitors and exclusively contribute to VAT development (Figures 1H and S1E). TM induction of Tcf21MCM/+; R26tdTomato;PdgfraeGFP mice at E11.5 or P2 and P4 showed the same results (Figure S1F). To study the adipogenic activity of Tcf21 LCs in adult mice, Tcf21 LT mice were treated with TM at P2 and P4 or 6 weeks and subjected to 12 weeks of high-fat diet (HFD) feeding. Compared with mice treated with TM at P2 and P4, far fewer GFP+ adipocytes were observed in eVAT of mice treated with TM at 6 weeks despite similar labeling efficiencies, suggesting less robust adipogenesis of Tcf21 LCs in adult mice (Figures 1I–1K, S1G, and S1H).
To better understand Tcf21 LC activity during eVAT development, Tcf21 LT mice treated with TM at P2 and P4 were sampled at P5, P7, P12, and 3 weeks (Figures 1L). At P5, GFP was expressed in many MPCs and some differentiating multilocular adipocytes (Figures 1M and S1I). At P7 and P12, the proportion of GFP+ adipocytes was elevated; however, the overall percentage of GFP+ adipocytes remained relatively low (Figures 1M, 1N, S1J, and S1K). By 3 weeks, a much larger fraction of adipocytes was of the Tcf21 lineage, suggesting that Tcf21 LC adipogenesis peaks between P12 and 3 weeks (Figures 1N and S1L). To study Tcf21 LC proliferation, Tcf21 LT mice were treated with 5-ethynyl-2’-deoxyuridine (EdU) at P12 or P21, followed by sample collection after 4 h. More EdU+ Tcf21 LCs were present on P12, indicating robust Tcf21 LC proliferation before adipogenesis peaks (Figure 1O).
Multiomics analyses identify the mechanisms governing Tcf21 LC activities
The dynamic activities of Tcf21 LCs during VAT growth and expansion prompted us to study the associated gene expression changes. Wild-type (WT) Tcf21 LT mice were treated with TM on P2 and P4. eVAT Tcf21 LCs isolated at P12, 3 weeks, and 6 months without HFD and after 1 week or 12 weeks with HFD from 6 weeks were subjected to bulk RNA-seq. Principal-component analysis (PCA) and t-distributed stochastic neighbor embedding (t-SNE) analyses show treatment-dependent clustering (Figures S2A and S2B). A total of 5,006 differentially expressed genes (DEGs) were identified (Figure 2A; Table S1).
Figure 2. Multiomics analyses identify the GRN regulating Tcf21 LC activities.
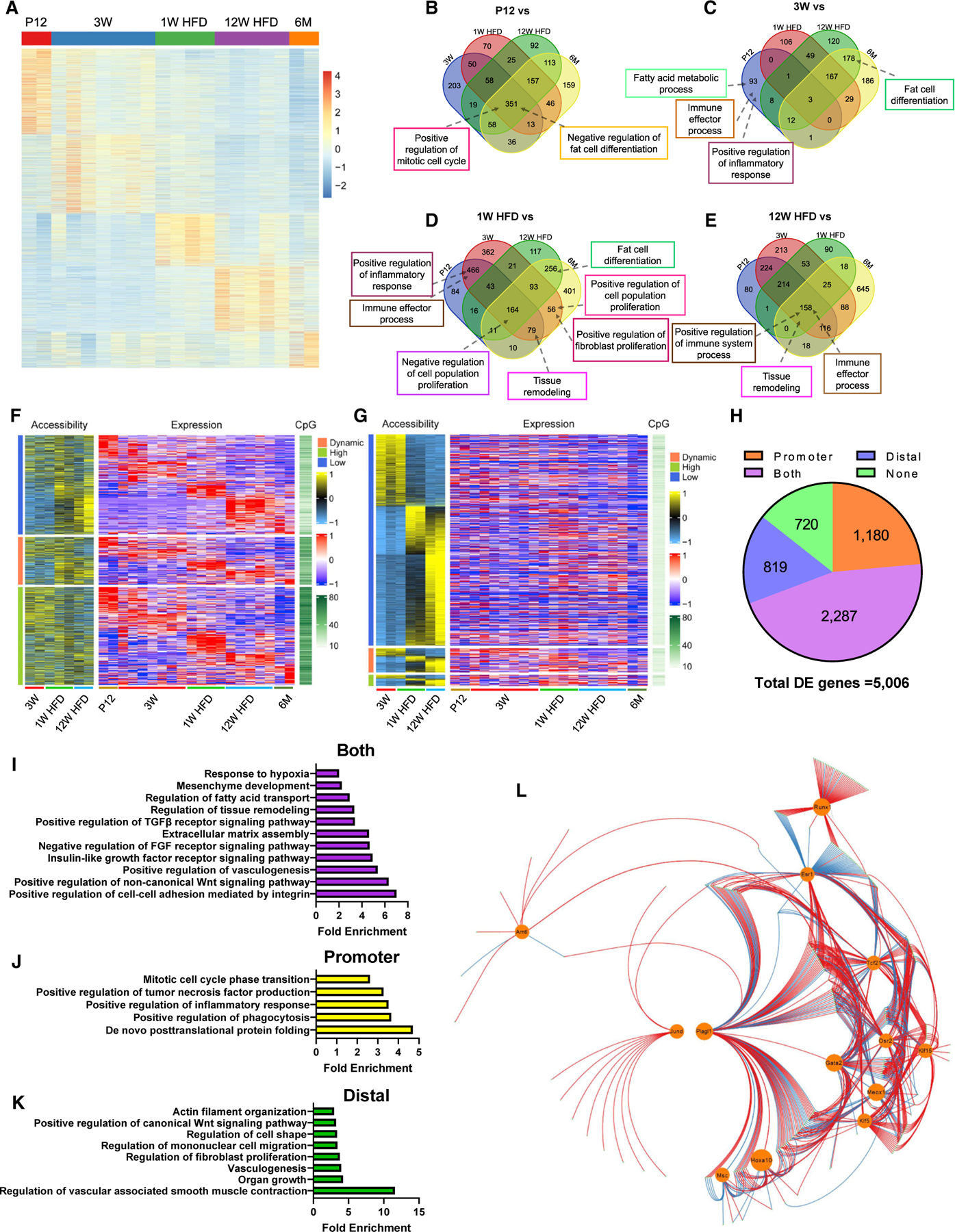
(A–E) WT Tcf21 LCs isolated at the indicated time points were subjected to bulk RNA-seq. (A) A heatmap shows the expression of DEGs. See also Table S1. (B–E) Venn diagrams show the overlap of BPs enriched at P12 (B), 3 weeks (C), 1 week HFD (D), or 12 weeks HFD (E) versus other groups.
(F–H) Tcf21 LCs isolated at the indicated time points were subjected to ATAC-seq. (F) A heatmap shows the PA, CpG density, and expression level of DEGs. Genes were grouped based on PA. See also Table S2. (G) A heatmap shows the accessibility and CpG density of DADs and the expression level of proximal genes. Distal peaks were grouped based on the expression of proximal genes. See also Tables S3 and S4. (H) A pie graph shows the numbers of DEGs with expression strongly correlated with PA, distal region accessibility, both of them, or none of them.
(I–K) Representative BPs enriched in DEGs with expression strongly correlated with PA (I), distal region accessibility (J), or both of them (K), revealed by GO analysis. (L) A GRN was constructed using the top 500 DEGs, including 13 TFs (circled). Red and blue lines indicate positive and negative regulation, respectively. See also Table S5.
n = 2, P12 and 6 months (RNA-seq); n = 7, 3 weeks (RNA-seq); n = 4, 1 week HFD (RNA seq); n = 5, 12 weeks HFD (RNA-seq); n = 3, 3 weeks (ATAC-seq); n = 2, 1 week and 12 weeks HFD (ATAC-seq). See also Figures S2 and S3 and Tables S1, S2, S3, S4, and S5.
Gene set enrichment analysis (GSEA) revealed biological processes (BPs) enriched in individual treatment groups (Figures 2B–2E and S2C–S2H). BPs related to “proliferation” and “negative regulation of fat cell differentiation” were enriched at P12 versus all other groups, together with higher expression of pro-mitotic genes such as Chp2 and Mki67 and anti-adipogenic genes such as Dlk1 and Sirt1 (Figures 2B and S2I). “Fat cell differentiation” and pro-adipogenic genes such as Adipoq and Rxra were enriched at 3 weeks versus 12 weeks HFD and 6 months (Figures 2C and S2I). This supports the observed activities of Tcf21 LCs during eVAT development. Furthermore, “fat cell differentiation” and related genes were enriched at 1 week HFD versus 12 weeks HFD and 6 months (Figures 2D and S2I), and proliferation-related terms and genes were enriched at 1 week HFD versus 3 weeks and 6 months (Figures 2D and S2I). This suggests activation of proliferation and adipogenesis in Tcf21 LCs of adult mice shortly after HFD. However, “negative regulation of cell population proliferation” and anti-mitotic genes such as Bmp7 and Fbxo4 were enriched at 1 week HFD versus all other groups, suggesting co-activation of mechanisms preventing exponential proliferation of Tcf21 LCs following HFD challenge (Figures 2D and S2I).
Interestingly, an increase in enrichment of select inflammation-related terms and genes, such as Tlr4 and Ccr7, was observed as early as 3 weeks, peaking at 12 weeks HFD, suggesting that increased adiposity, including normal development, is associated with elevated inflammation programming in Tcf21 LCs (Figures 2C–2E and S2I). “Negative regulation of cell population proliferation” and related genes were enriched at 6 months versus P12 and 3 weeks (Figures S2G and S2I). Moreover, “fat cell differentiation” and related genes were enriched in all other treatment groups versus 6 months (Figures S2H and S2I). This suggests a relatively quiescent state of Tcf21 LCs in non-obese adult mice. AT fibrosis in obese individuals has been reported.25 Fibroblasts and MPCs are essential players in fibrosis.26 Indeed, “tissue remodeling” was enriched at 1 week HFD and 12 weeks HFD versus other groups (Figures 2D and 2E). Surprisingly, the highest expression of major fibrillar collagen genes (e.g., Col1a1 and Col3a1) and basement collagen genes (e.g., Col4a3 and Col4a4) was observed at P12 and 3 weeks (Figure S2I). In addition, the expression of these genes was not significantly higher at 12 weeks HFD versus 6 months (Figure S2I). This suggests that active ECM protein deposition by Tcf21 LCs occurs during AT development but not with HFD-induced tissue remodeling. Instead, increased ECM remodeling enzyme gene expression (e.g., Mmp12 and Timp1) was identified at 12 weeks HFD (Figure S2I).
To study chromatin remodeling involvement in dynamic gene expression of Tcf21 LCs, Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) was performed using Tcf21 LCs isolated from 3 weeks, 1 week HFD, and 12 weeks HFD mice. Similar to our RNA-seq results, ATAC-seq PCA data showed tight treatment-dependent clustering (Figure S3A). Specifically, 56.87% of 5,006 DEGs exhibited a strong correlation between promoter accessibility (PA) and expression (Figures 2F and S3C; Table S2). Similarly, 63.29% of 1,174 differentially accessible promoters (DAPs) displayed a strong correlation between accessibility and corresponding gene expression (Figures S3B and S3E; Tables S2 and S3). This suggests an important role of PA in Tcf21 LC gene expression regulation. Interestingly, the strongest correlations were observed in genes with generally low PA (Figures 2F and S3C). In addition, we found that DEGs with generally lower PA had lower promoter CpG densities, and DEGs with lower promoter CpG densities displayed stronger correlations between PA and expression (Figures 2F and S3D). It is possible that genes with high promoter CpG density are more heavily regulated by DNA methylation than chromatin accessibility. Accessible regions outside of promoters are enriched with distal regulatory elements such as enhancers. Among the 17,284 DA distal regions (DADs) identified, 38.54% showed strong correlations between accessibility and expression of proximal genes, suggestive of their important function in gene expression regulation (Figures 2G and S3F; Tables S3 and S4).
Interestingly, 4,286 of 5,006 DEGs had a strong correlation between expression and PA and/or between expression and the accessibility of at least one accessible distal region (ADR) (Figure 2H). Subsequent Gene Ontology (GO) enrichment analysis of these genes suggests that genes related to many BPs, such as cell-cell adhesion, insulin growth factor (IGF) signaling, and ECM assembly, are coregulated by promoter and enhancer activities (Figure 2I). However, genes related to BPs such as protein folding, inflammatory response, and proliferation are possibly more heavily regulated by promoter activities, and genes related to BPs such as vascular smooth muscle cell (VSMC) contraction, fibroblast proliferation, and cell migration may be primarily regulated by enhancer activities (Figures 2J and 2K), cumulatively hinting at divergent mechanisms of promoter and enhancer regulation of Tcf21 LCs.
Transcription factors (TFs) bind to target motifs to regulate gene expression. To identify TFs regulating Tcf21 LC gene expression, a gene-regulatory network (GRN) was constructed using the top 500 DEGs, including 13 TF genes, using a newly developed approach (STAR Methods; Figure 2L; Table S5). In total, 331 of 500 DEGs were predicted to be regulated by at least 1 of the 13 TFs. Genes regulated by Tcf21 were subjected to GO enrichment analysis. The results indicate that Tcf21 may be an adipogenesis inhibitor (Figures S3G and S3H). Genes upregulated by 5 other highly and differentially expressed TFs (Gata2, Runx1, Osr2, Klf5, and Plagl1) were also subjected to GO enrichment analysis (Figures S3I–S3M). The analysis suggests that Gata2 promotes organ growth by regulating organ morphogenesis and cell proliferation, which is consistent with its reported pro-proliferative functions.27–29 The results also suggest that Osr2, a known chondrogenesis regulator,30 has an anti-adipogenic role. Our analysis indicates that Klf5 may regulate cell-substrate interactions and glucose import, possibly involving non-canonical Wnt signaling, a pathway with which Klf5 interacts.31 The data also indicate that Runx1, a TF upregulated during inflammation,32 may stimulate an immune response and negatively regulate triglyceride metabolism. Moreover, Plagl1, known to interact with transforming growth factor β (TGF-β) signaling during retina development,33 possibly promotes the response of Tcf21 LCs to TGF-β.
scRNA-seq analysis shows Tcf21 LC heterogeneity
To explore the cell type composition of Tcf21 LCs, we performed scRNA-seq using Tcf21 LCs isolated at P18, 5 weeks, 1 week HFD, and 12 weeks HFD. Pooled clustering analysis (Figures 3A, 3B, S4A, and S4B) and clustering analyses of individual age groups (Figures 3C–3F and S4C–S4F) indicate the heterogeneity of Tcf21 LCs. In general, Tcf21 LCs can be divided into a population expressing genes including Dpp4, Cd55, Ly6c1, Sema3c, and Ccn3 (cluster 1 [C1] in each age group) and a larger population expressing genes including Icam1, Cygb, and Apoe, which can be further sub-divided depending on the age group (C2–C6 for P18, 5 weeks, and 1 week HFD; C2–C8 for 12 weeks HFD) (Figures 3B–3F and S4G–S4J).
Figure 3. scRNA-seq analysis identifies the cell type composition of Tcf21 LCs.
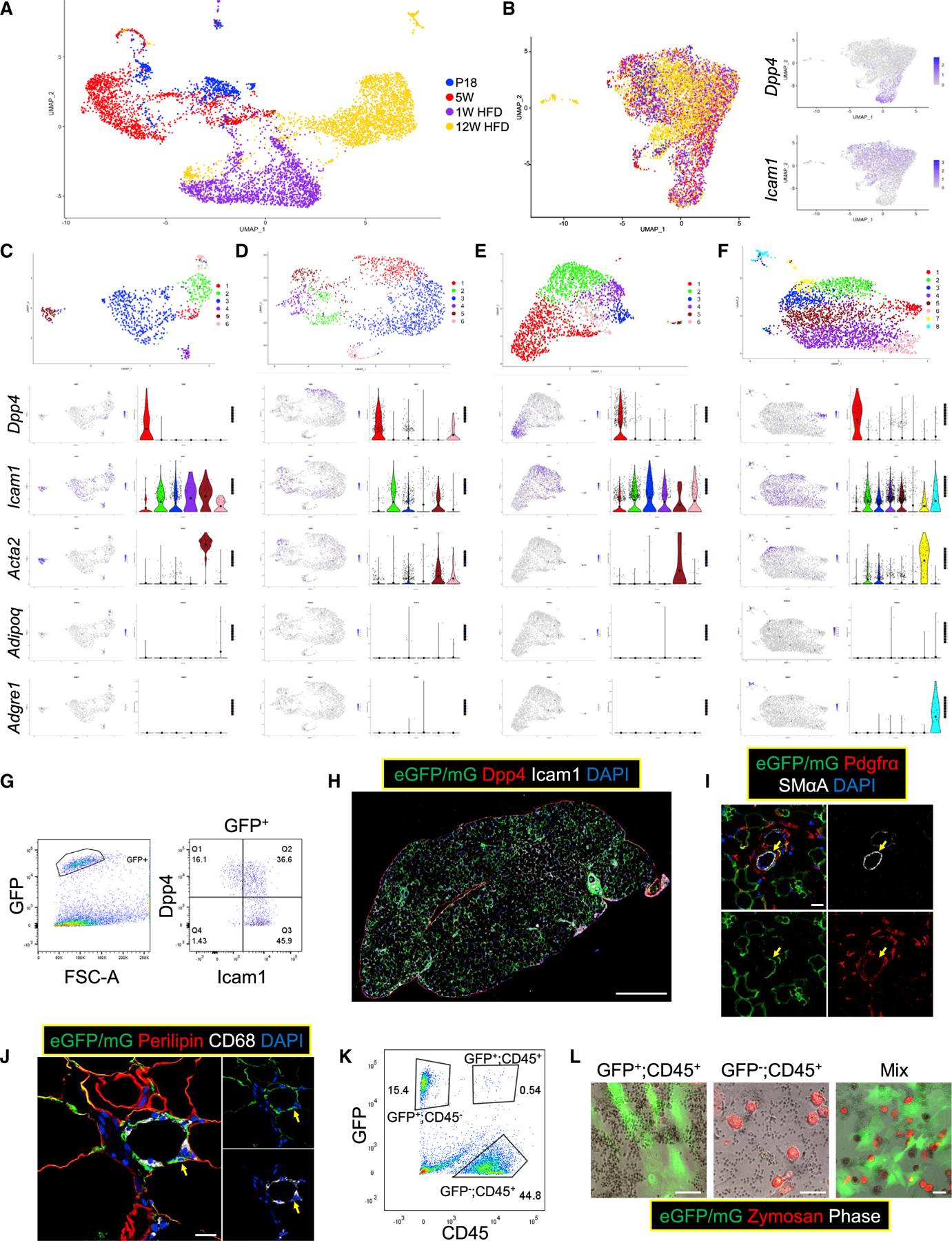
(A and B) scRNAseq was performed using WT Tcf21 LCs isolated at P18 (739 cells), 5 weeks (1,678 cells), 1 week HFD (2,008 cells), and 12 weeks HFD (2,835 cells). Uniform Manifold Approximation and Projection (UMAP) graphs show the clustering of all 7,260 Tcf21 LCs without (A) and with (B) batch correction. The expression of Dpp4 and Icam1 is also shown in (B).
(C–F) Cells of each sample were subjected to separate clustering analyses (C, P18; D, 5 weeks; E, 1 week HFD; F, 12 weeks HFD). UMAP graphs and violin plots show the expression of select genes. Large solid circles indicate expression means.
(G and H) FC (G) and IHC (H) identify Dpp4+ and Icam1+ Tcf21 LCs in eVAT at 3 weeks. Scale bar, 100 μm.
(I) IHC shows the presence of smooth muscle alpha actin (SMαA)+;Pdgfrα− Tcf21 lineage VSMCs (yellow arrows) in eVAT at 3 weeks. Scale bar, 20 μm.
(J) IHC images show the presence of Tcf21 lineage CD68+ cells (yellow arrows) in eVAT at 12 weeks HFD. Scale bar, 20 μm.
(K and L) Tcf21 lineage CD45+ cells and non-Tcf21 lineage CD45+ cells were sorted from Tcf21 LT mice at 12 weeks HFD (K) and subjected to a phagocytosis test separately or in a co-culture system (L). Scale bar, 50 μm. See also Figure S4.
GSEA showed that P18 C1 was enriched for “actin filament bundle assembly” and “cell-matrix adhesion” and expressed more Postn, a myofibroblast ECM gene,34 suggesting stronger attachment to the surrounding matrix (Figures 3C and S4K). P18 C2 expressed high Col4a1, a major component of the adipocyte basement membrane, and was enriched for “negative regulation of cell-matrix adhesion,” “regulation of angiogenesis,” and “fatty acid transport” (Figures 3C, S4G, and S4K). This cluster was also enriched for “negative regulation of fat cell differentiation” and expressed anti-adipogenic genes such as Dlk1 (Figures 3C, S4G, and S4K). These cells may represent a mobilized pro-angiogenic population that is halted in the “preadipocyte” state. P18 C3 was enriched for “collagen fibril organization” and expressed high levels of Col1a1, Lox, and Timp1, likely representing a matrix-depositing “fibroblast” population (Figures 3C, S4G, and S4K). P18 C5 was enriched for “smooth muscle contraction” and expressed VSMC markers such as Acta2, Actg2, and Myh11 but was negative for Pdgfra, suggestive of a “VSMC” population (Figures 3C, S4G, and S4K). P18 C6 was enriched for “fatty acid transport” and “fat cell differentiation” and expressed adipogenesis markers, including Pparg, Cebpa, and Adipoq, which may be a differentiating “adipocyte” population absent in other age groups (Figures 3C, S4G, and S4K). P18 C4 expressed elevated levels of the apolipoprotein genes Apod and ApoE and likely consists of cells regulating lipid metabolism (Figure S4G).
Interestingly, 12 weeks HFD Tcf21 LC subpopulations were enriched for many distinct BPs relative to P18 Tcf21 LC counterparts despite some commonly expressed genes. 12 week HFD C1 shared select marker expression with P18 C1 (Figures S4G and S4J) and was enriched for “negative regulation of growth” and “cellular response to decreased oxygen levels,” suggesting hypoxia-induced activity stunting (Figure S4L). “Wound healing” was enriched in 3 related clusters at 12 weeks HFD (C2, C3, and C7), which expressed high levels of Timp1, Lox, and Acta2, respectively, and were likely “myofibroblast”-like populations (Figures S4J and S4L). 12 weeks HFD C8 was enriched for “inflammatory response” and “immune response,” expressed inflammatory markers (e.g., Cd68, Adgre1, and Ptprc), and lacked Pdgfra expression; it resembles macrophages and likely represents an “inflammatory” population (Figures 3F, S4J, and S4L). Moreover, a less significant enrichment of inflammation-related BPs was observed in 12 weeks HFD C2, expressing Ccl2 and Cxcl14 (Figures 3F, S4J, and S4L). Besides, many cells at 12 weeks HFD seemed to be in a “quiescent” state, including C4 (enriched for “negative regulation of response to stimulus”) and C5 (enriched for “cell-matrix adhesion”) (Figure S4L). These data not only show the heterogeneity of Tcf21 LCs but also suggest the dynamic contributions of different Tcf21 LC subpopulations to VAT development and pathological expansion.
Subsequent flow cytometry (FC) of Tcf21 LCs identified Dpp4+ (C1) and Icam+ (non-C1) Tcf21 LCs (Figure 3G). IHC showed Dpp4+ cells primarily located in the mesothelium and Icam+ cells more prevalent interstitially (Figure 3H). Tcf21 lineage VSMC presence was also confirmed by IHC (Figure 3J). In addition, IHC and FC confirmed the presence of Tcf21 LCs expressing CD68 and CD45 (Figures 3K and 3L). However, CD45+ Tcf21 LCs maintained a fibroblast-like morphology and were not phagocytic, suggesting their non-macrophage identity (Figure 3L). The common niche of these Tcf21 LCs with inflammatory cells in crown-like structures (CLSs) might cause their expression of select inflammation marker genes (Figure 3K).
Tcf21 inhibits Tcf21 LC adipogenesis
To test the anti-adipogenic role of Tcf21 suggested by the GRN, we used Tcf21fl/+ mice. Tcf21MCM/+;R26mTmG/mTmG and Tcf21fl/+; R26eGFP/eGFP mice were crossed to generate Tcf21MCM/+; R26eGFP/mTmG (formerly abbreviated as Tcf21 LT; for convenience abbreviated as “WT” hereafter) and Tcf21MCM/fl;R26eGFP/mTmG (knockout [KO]) littermates (Figure 4A). WT and KO littermates were treated with TM on P2 and P4 before sampling at 3 weeks (Figure 4B). The eVAT weight of KO mice was greater than that of the WT despite similar body weights (BWs) and iSAT weights (Figures 4C–4F). Whole-mount imaging of eVAT and enzymatically released adipocytes revealed a higher percentage of Tcf21 lineage adipocytes in KO (>50%) versus WT (~40%) mice (Figures 4G–4I). The average diameter of Tcf21 lineage adipocytes was larger in KO versus WT littermates (Figure 4J), while no difference was observed between non-Tcf21 lineage adipocytes (Figure 4K). Size distribution analysis showed more abundant larger Tcf21 lineage adipocytes in KO versus WT littermates (Figure 4L). These data suggest that loss of Tcf21 promotes Tcf21 LC adipogenesis. Moreover, Tcf21 LC density was lower in KO versus WT eVAT (Figure 4M), likely because of stronger MPC pool depletion. To verify these results in vitro, Tcf21 LCs and non-Tcf21 LCs sorted from the eVAT of 3-week WT and KO littermates were induced for adipogenesis (Figure 4N). More robust adipogenesis was observed in Tcf21 LCs but not non-Tcf21 LCs from KO versus WT mice (Figure 4O). Furthermore, KO Tcf21 LCs showed greater adipogenic efficiency than WT Tcf21 LCs transplanted into the same recipient mice (Figure 4P). Moreover, a retrovirus-mediated gain-of-function study showed that Tcf21 overexpression (OE) inhibited SVC adipogenesis (Figures 4Q–4R).
Figure 4. Tcf21 inhibits Tcf21 LC adipogenesis.
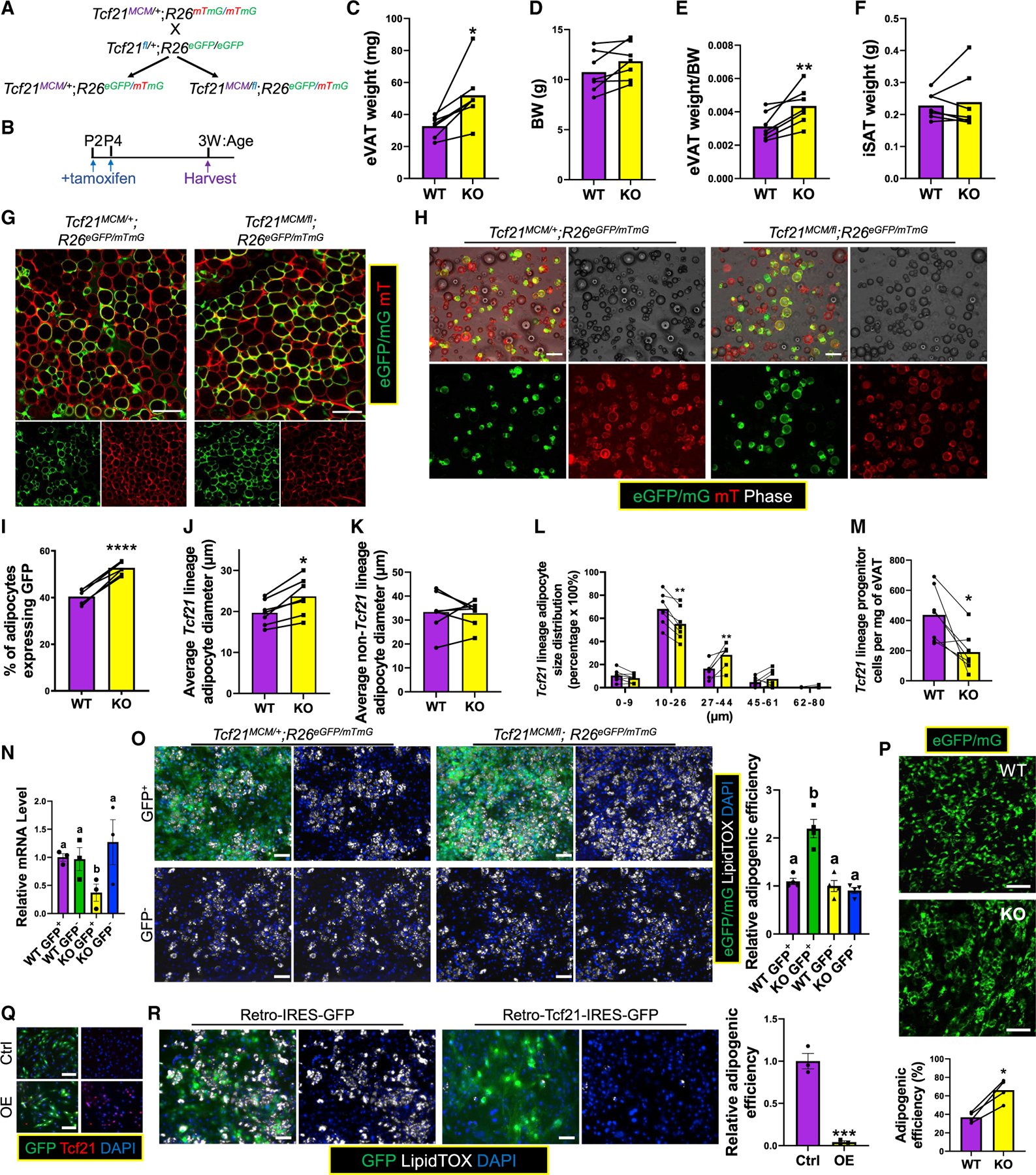
(A and B) Schematics of the generation of WT and KO littermate mice (A) and TM treatments and sample collection strategy (B).
(C–F) eVAT weight (C), BW (D), eVAT weight/BW ratio (E), and iSAT weight (F) were obtained. n = 7; paired t test.
(G) eVAT was subjected to whole-mount imaging. Scale bar, 100 μm. (H–L) Adipocytes released from eVAT (H) were analyzed to calculate the fraction of Tcf21 lineage adipocytes (I), their average size (J) and size distribution (L), and the average size of non-Tcf21 lineage adipocytes (K). Scale bar, 100 μm. n = 7; paired t test.
(M) FC measures the density of Tcf21 LCs in eVAT. n = 7; paired t test.
(N and O) Tcf21 LCs and non-Tcf21 LCs sorted from WT and KO littermates at 3 weeks were measured for Tcf21 expression by real-time PCR (N) and induced for adipogenesis, followed by LipidTOX staining (O). Scale bar, 50 μm. n = 4 (N), n = 3 (O); one-way ANOVA. Different letters indicate significant differences (p < 0.05).
(P) The adipogenic efficiency of transplanted WT and KO Tcf21 LCs was quantified. Scale bar, 100 μm. n = 4; unpaired t test.
(Q and R) SVCs isolated from 3-week C57BL/6 mice were transduced with IRES (internal ribosome entry site) -GFP (control, abbreviated as Ctrl) or Tcf21-IRES-GFP (OE) retroviruses, verified for Tcf21 OE by immunocytochemical staining (ICC) (Q), and induced for adipogenesis followed by LipidTOX staining (R). Scale bar, 50 μm.
*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Data are represented as mean with values of littermates connected (C–M and P) or mean ± SEM with individual values plotted (N and R).
Neonatal Tcf21 deletion improves obese mouse metabolic health
To study the impact of Tcf21 deletion on obese mouse metabolic health, 6-week WT and KO male mice treated with TM at P2 and P4 were subjected to 12 weeks HFD. Higher glucose tolerance and a trend toward higher insulin tolerance were observed in KO mice (Figures 5A and 5B). No differences between WT and KO in BW, eVAT weight, or eVAT weight/BW ratio were observed (Figures 5C–5E). The Tcf21 lineage adipocyte fraction remained higher in KO versus WT mice after HFD treatment (Figures 5F–5H). Interestingly, unlike young mice, Tcf21 lineage and non-Tcf21 lineage eVAT adipocytes were smaller in KO versus WT mice (Figures 5I and 5J). Enhanced Tcf21 lineage adipocyte hyperplasia during development likely underpins the smaller VAT adipocyte size in KO versus WT mice after HFD challenge because no differences in lipid metabolism and thermogenesis gene expression in eVAT adipocytes were identified between from WT and KO mice (Figure 5K). Excess adipocyte hypertrophy can be associated with adipocyte apoptosis, which promotes CLS formation. As expected, more abundant CLSs were observed in WT versus KO eVAT (Figure 5L).
Figure 5. Neonatal deletion of Tcf21 improves obese mouse metabolic health.
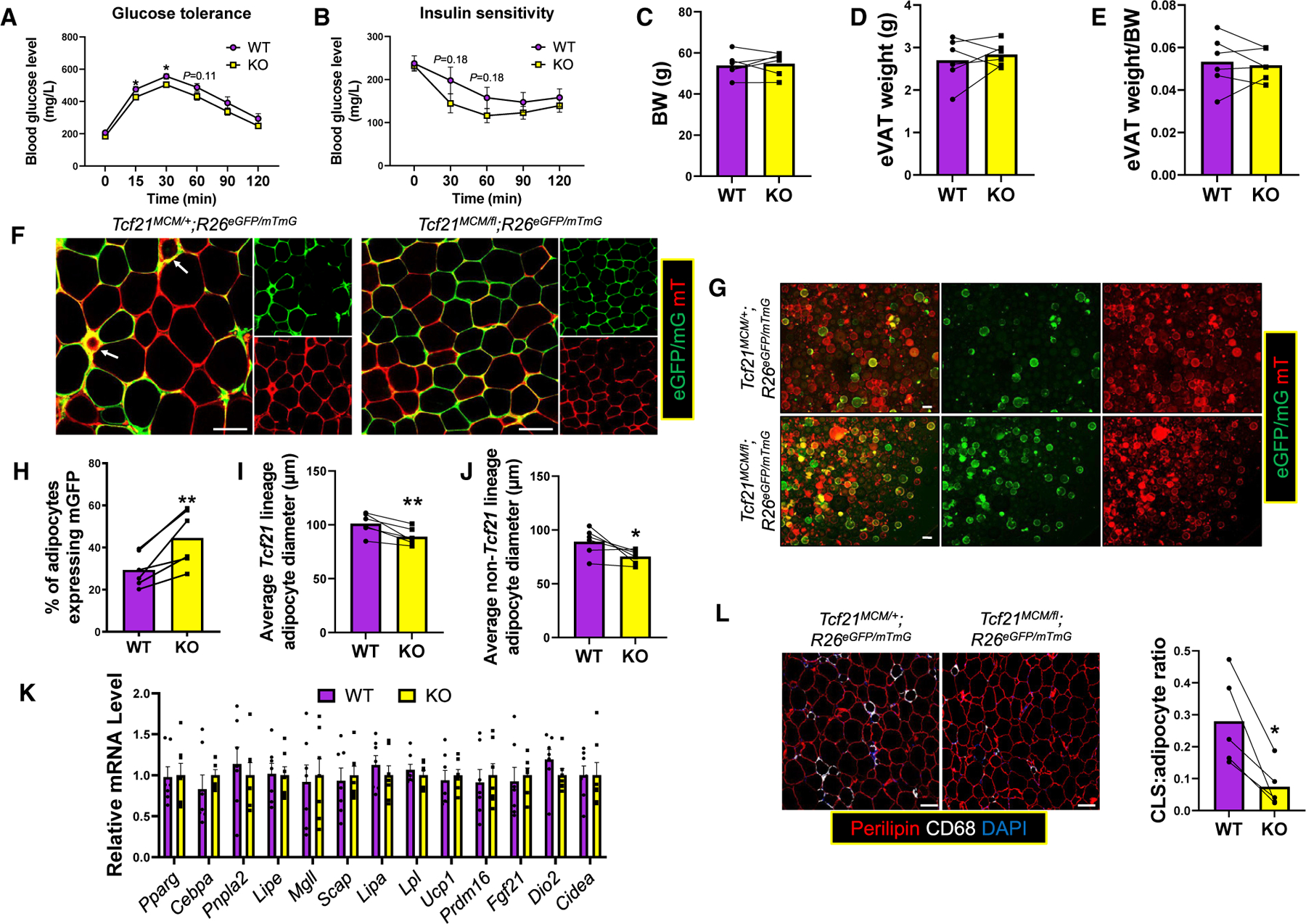
(A and B) WT and KO male littermates treated with TM on P2 and P4 were subjected to 12 weeks HFD starting at 6 weeks of age. Glucose tolerance tests (GTTs; A) and insulin tolerance tests (ITTs; B) were performed. n = 8; unpaired t test.
(C–E) The BW (C), eVAT weight (D), and eVAT weight/BW ratio (E) were obtained. n = 8; paired t test.
(F) eVAT was subjected to whole-mount imaging. Scale bar, 100 μm.
(G–J) Adipocytes released from eVAT (G) were analyzed to calculate the percentage of Tcf21 lineage adipocytes (H) and the average sizes of Tcf21 lineage adipocytes (I) and non-Tcf21 lineage adipocytes (J). n = 6; paired t test. Scale bar, 100 μm.
(K) The expression of select genes in adipocytes isolated from HFD-treated WT and KO mice was measured. n = 7; unpaired t test.
(L) eVAT sections were analyzed by IHC to identify CLSs (CD68+;Perilipin−) and calculate the CLS/adipocyte ratio. n = 5; paired t test. Scale bar, 100 μm.
*p < 0.05, **p < 0.01. Data are represented as mean ± SEM (A, B, and K) or mean with values of littermates connected (C–J and L).
Loss of Tcf21 alters Tcf21 LC gene expression and chromatin accessibility
To explore the role of Tcf21 in gene expression regulation in Tcf21 LCs, we performed bulk RNA-seq on Tcf21 LCs from eVAT of KO mice that were littermates of some WT mice sequenced in earlier experiments (3 weeks, 1 week HFD, and 12 weeks HFD). Interestingly, compared with WT littermate counterparts, cells of KO mice clustered closer to cells of the next age group in PCA, suggesting advanced development (Figure 6A). Because of the more robust adipogenic activity of Tcf21 LCs and higher Tcf21 expression at 3 weeks (Figure 6B), Tcf21 LCs of 3-week WT and KO mice were compared, identifying 651 DEGs (376 upregulated in WT and 275 upregulated in KO mice) (Figure 6C). Consistent with this, KO 3-week Tcf21 LC gene expression clustered closer to that of WT 1 week HFD relative to WT 3-week Tcf21 LCs, further suggesting that Tcf21 deletion promotes Tcf21 LC developmental progress. Moreover, DEGs upregulated in WT Tcf21 LCs included those regulating cell adhesion, proliferation, and transcription and, more importantly, genes inhibiting adipogenesis (Figure 6D). DEGs downregulated in WT Tcf21 LCs included those regulating ECM remodeling and promoting responses to stimuli (Figure 6E).
Figure 6. Tcf21 inhibits Tcf21 LC adipogenic efficiency through promoting Dlk1 expression.
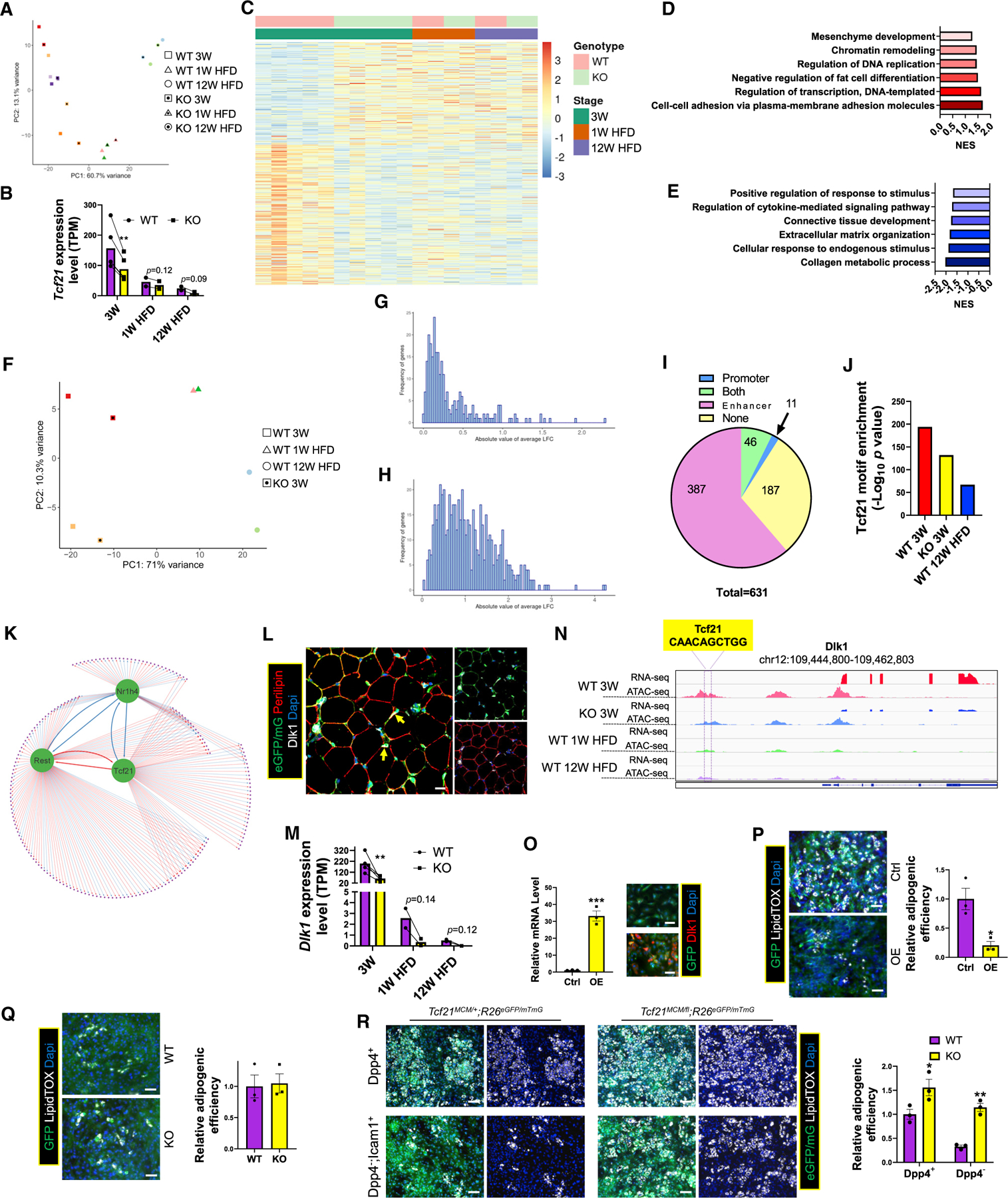
(A) Tcf21 LCs from eVAT of KO mice of the indicated groups were subjected to bulk RNA-seq and PCA. Samples of the same color indicate littermates.
(B) A bar graph shows Tcf21 expression in the indicated groups. n = 5 for 3 weeks; n = 2 for 1 week HFD and 12 weeks HFD; paired t test. Data are represented as mean with values of littermates connected.
(C) A heatmap shows the expression of DEGs between Tcf21 LCs of WT and KO 3-week mice in cells of the indicated groups.
(D and E) Graphs show the NES (normalized enrichment score) of representative BPs enriched in DEGs that were upregulated (D) or downregulated (E) in 3-week WT versus KO mice.
(F) Tcf21 LCs isolated from eVAT of the indicated groups were subjected to ATAC-seq and PCA. Samples of the same color indicate littermates.
(G–I) The absolute LFC in promoter (G) and distal region (H) accessibility of DEGs between Tcf21 LCs of 3-week WT and KO mice are plotted. A pie graph (I) shows the numbers of DEGs with corresponding DAPs, DADs, DAPs and DADs, or none. See also Table S6.
(J) The enrichment of the Tcf21 motif in ADRs in Tcf21 LCs of the indicated sample groups.
(K) A GRN was constructed using direct target genes of Tcf21 and 2 Tcf21-targeted TFs. See also Table S7.
(L) eVAT of P12 Tcf21 LT mice was subjected to IHC to identify Dlk1+ Tcf21 LCs (yellow arrows). Scale bar, 20 μm.
(M) The expression of Dlk1 in eVAT Tcf21 LCs of the indicated groups was measured by RNA-seq. n = 5 for 3 weeks; n = 2 for 1 week HFD and 12 weeks HFD; paired t test.
(N) The Integrative Genomics Viewer view shows the expression and chromatin accessibility of Dlk1 in Tcf21 LCs of the indicated sample groups.
(O and P) SVCs isolated from 3-week C57BL/6 mice were transduced with IRES-GFP (Ctrl) or Dlk1-IRES-GFP (OE) retroviruses, verified for Dlk1 OE by real-time PCR and ICC (O), and induced for adipogenesis followed by LipidTOX staining (P). n = 3; unpaired t test. Scale bar, 20 μm.
(Q) Tcf21 LCs sorted from 3-week WT and KO mice were transduced with Dlk1-IRES-GFP retroviruses and induced for adipogenesis, followed by LipidTOX staining. n = 3; unpaired t test. Scale bar, 20 µm.
(R) Dpp4+ and Dpp4−;Icam1+ Tcf21 LCs isolated from 3-week WT and KO mice were induced for adipogenesis and stained with LipidTOX. n = 3; unpaired t test. Scale bar, 50 μm.
*p < 0.05, **p < 0.01, ***p < 0.001. Data are represented as mean with values of littermates connected (B and M) or mean ± SEM with individual values plotted (O–R). See also Figure S5 and Tables S6 and S7.
KO 3-week Tcf21 LCs were also subjected to ATAC-seq. Similar to the RNA-seq data, KO 3-week Tcf21 LCs clustered closer to WT 1 week HFD Tcf21 LCs in PC1 of PCA, which accounted for 71% of differences (Figure 6F). The log2 fold change (LFC) of PA and ADR for the 631 DEGs (20 mitochondrial genes were removed) were calculated (Figures 6G and 6H). In total, 433 DEGs had corresponding differences in at least 1 ADR, while only 57 DEGs had corresponding PA differences (Figure 6I; Table S6). In addition, significant Tcf21 motif enrichment in ADR (Figure 6J), but not in promoters, was observed. Moreover, Tcf21 motif enrichment in ADR was less pronounced in KO versus WT 3-week Tcf21 LCs. However, both were more pronounced than that in WT 12 weeks HFD Tcf21 LCs, showing a positive correlation between Tcf21 motif enrichment and Tcf21 expression (Figure 6J). This suggests that Tcf21 mainly targets enhancers, and differential enhancer activity likely mediates differential gene expression between WT and KO Tcf21 LCs. To identify direct Tcf21 target genes from the 631 DEGs, a Tcf21-regulated GRN was constructed (Figure 6K; Table S7). Forty positively and 25 negatively regulated genes by Tcf21 were identified, including 2 TF genes, Nr1h4 and Rest.
Tcf21 inhibits VAT MPC adipogenesis at least partially by promoting Dlk1 expression
To identify genes through which Tcf21 inhibits adipogenesis, 65 direct Tcf21 target genes were screened, identifying Dlk1, a negative regulator of adipogenesis.35,36 Dlk1 was also a core enrichment gene for the BP “negative regulation of fat cell differentiation,” which was enriched in WT versus KO 3-week Tcf21 LCs (Figure 6D) and in Tcf21-targeted genes, as predicted by a GRN constructed using WT Tcf21 LC data (Figure S3G). Dlk1 was also expressed in a halted Tcf21 lineage preadipocyte population (Figures 6D, S4G, and S4K). In eVAT, Dlk1 was expressed in interstitial Tcf21 LCs but not adipocytes (Figure 6L). Dlk1 expression in Tcf21 LCs was negatively correlated with mouse adiposity and consistently lower in KO versus WT littermates (Figure 6M). In the WT 3-week Tcf21 LC ATAC-seq data, a typical “peak-dip-peak” TF motif in a putative enhancer upstream of Dlk1 was identified, where a Tcf21 motif was located at the “dip.” (Figure 6N). However, in KO 3-week, WT 1-week, and 12 weeks HFD mice, such a “peak-dip-peak” pattern was less pronounced, and the flanking areas were less accessible, suggesting that Tcf21 promotes Dlk1 expression by targeting this putative enhancer (Figure 6N).
To verify the effect of Dlk1 on adipogenesis, eVAT SVCs of C57BL/6 mice were transduced with Dlk1 OE or control retroviruses, showing that Dlk1 OE inhibited adipogenesis (Figures 6O and 6P). Moreover, Dlk1 OE and knockdown abolished the difference in adipogenesis between WT and KO Tcf21 LCs (Figures 6Q, S5A, and S5B). Interestingly, Dlk1 knockdown failed to abolish the difference in adipogenesis between Tcf21-overexpressing and control cells, suggesting the presence of other Tcf21-targeted anti-adipogenic effector gene(s), which possibly only occurs when Tcf21 is supraphysiologically expressed (Figure S5C). Because of the higher Dlk1 expression in some interstitial Tcf21 LCs (Figures S4G–S4J), Tcf21 LCs were separated by expression of Dpp4, a mesothelial Tcf21 LC marker, and Icam1, an interstitial Tcf21 LC marker. While Dpp4+ and Dpp4−Icam1+ cells of KO mice had stronger adipogenesis than WT controls, a more significant difference was identified in the Dpp4−Icam1+ population (Figure 6R). These data strongly suggest that Tcf21 inhibits adipogenesis, at least partially, by promoting Dlk1 expression.
Loss of Tcf21 accelerates Tcf21 LC differentiation progress
These data combined suggest accelerated development of KO Tcf21 LCs. To further test this, we performed scRNA-seq on Tcf21 LCs from KO mice that were littermates of tested WT mice (P18 and 5 weeks) (Figures 7A and S6A). Consistent with bulk RNA-seq data, Tcf21 and Dlk1 expression in Tcf21 LCs was negatively correlated with adiposity and was lower in KO versus WT littermates (Figures 7B and 7C). Because of the lower population of Tcf21 LCs in eVAT of KO mice, fewer KO cells were obtained for scRNA-seq. Nevertheless, Tcf21 LCs of KO mice were also mainly composed of mesothelial and interstitial populations (Figures S6B–S6G). Pseudotime analysis using Monocle 2 showed that, at P18, KO cells were developmentally more advanced compared with the WT (Figures 7D and 7E). RNA velocity generated a similar result (Figure 7F). Pseudotime analysis using Monocle 3 also showed accelerated KO versus WT development in a branch of cells enriched for Dlk1 expression (Figures S7A–S7G). In addition, decreased Dlk1 expression was observed along the pseudotime trajectory (Figures S7E–S7G).
Figure 7. Loss of Tcf21 accelerates Tcf21 LC developmental progress.
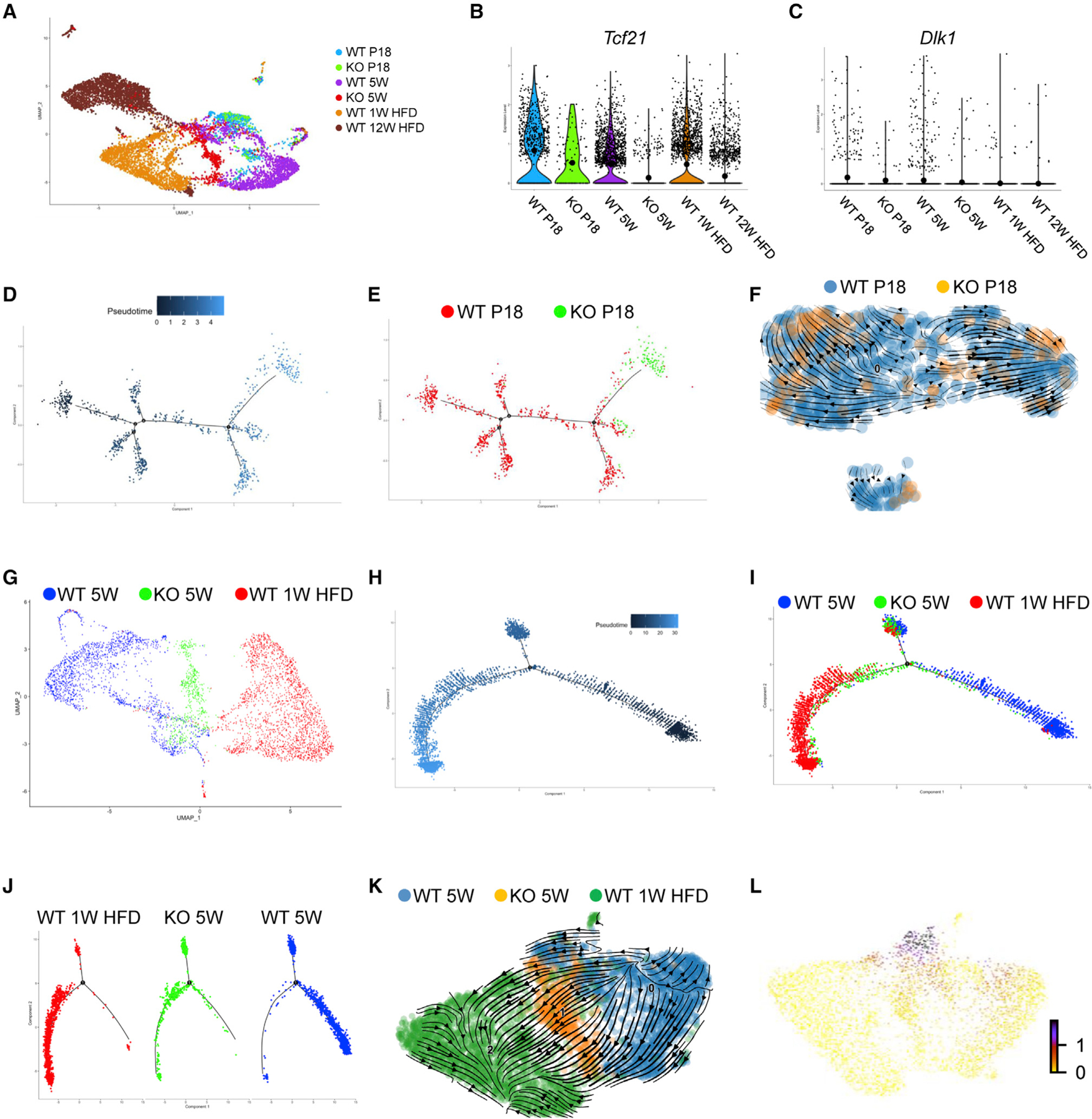
(A) Tcf21 LCs isolated from WT and KO mice at P18, 5 weeks, 1 week HFD (WT only), and 12 weeks HFD (WT only) were subjected to scRNA-seq. The UMAP graph shows the clustering of Tcf21 LCs from different sample groups, which include 7,260 WT cells, 122 KO cells at P18, and 555 KO cells at 5 weeks.
(B and C) Violin plots show the expression of Tcf21 (B) and Dlk1 (C) in Tcf21 LCs of different samples.
(D and E) Monocle 2 pseudotime analysis of WT and KO Tcf21 LCs at P18.
(F) RNA velocity analysis of WT and KO Tcf21 LCs at P18.
(G) A UMAP graph shows the clustering of Tcf21 LCs of WT and KO 5-week mice and WT 1 week HFD mice.
(H and J) Monocle 2 pseudotime analysis of Tcf21 LCs of WT and KO 5-week mice and WT 1 week HFD mice.
(K and L) RNA velocity analysis of Tcf21 LCs of WT and KO 5-week mice and WT 1 week HFD mice (K) and a UMAP showing the expression of Dlk1 on the RNA velocity trajectory (L).
See also Figures S6 and S7.
Tcf21 LCs from WT 5-week, KO 5-week, and WT 1-week HFD mice were also combined for pseudotime analysis. Monocle 2 and RNA velocity showed a “WT 5 weeks to KO 5 weeks to WT 1 week HFD” developmental order (Figures 7G–7K). Moreover, decreased Dlk1 expression along the RNA velocity trajectory was identified (Figure 7L). Monocle 3 pseudotime analysis revealed more progressive development of KO versus WT 5-week cells within a cluster primarily comprising cells of 5-week WT and KO mice, including cells expressing Dlk1 (Figures S7H–S7N). Similar to the branch of Dlk1+ cells identified at P18, a decrease in Dlk1 expression was identified along the pseudotime trajectory (Figures S7L–S7N). Together, these results further suggest that loss of Tcf21 promotes the developmental progress of Tcf21 LCs, which is at least partially attributed to reduced Dlk1 expression.
DISCUSSION
Despite some reports of Tcf21 expression in VAT, the roles of Tcf21 and Tcf21 LCs in VAT development and expansion have not been carefully studied. Here, using multiple mouse lines and various approaches, we report the identity of Tcf21 LCs in VAT, describe the dynamic gene expression and chromatin accessibility of Tcf21 LCs during development, and identify the inhibitory role of Tcf21 in adipogenesis of VAT PCs.
The identified contribution of Tcf21 LCs to VAT development suggests that MPCs in VAT and some other visceral organs18,37,38 share a developmental origin. The absence of Tcf21 LCs in rtAT and pcAT suggests a distinct developmental ancestry of MPCs in these adipose depots despite their proximity to visceral organs where Tcf21 LCs are present. Gene expression profiling of Tcf21 LCs from WT mice of different developmental stages showed precisely regulated gene expression associated with Tcf21 LC dynamic activities. Besides downregulation of adipogenic genes, select fibrosis and inflammation gene expression was concomitantly elevated in Tcf21 LCs of adult mice. This likely contributes to their reduced adipogenic activity because inflammatory and fibrotic signaling often inhibit adipogenesis.39–41 These results are consistent with a recent study showing elevated inflammatory programs in many VAT-resident cell types in obese mice.42 However, unchanged collagen gene expression and elevated ECM remodeling gene expression in Tcf21 LCs of obese mice suggest that Tcf21 LCs are not a major source of collagen accumulating around adipocytes in obese mice25 but do contribute to ECM remodeling.
Interestingly, we found that, while promoters and enhancers may coregulate genes related to many BPs, genes related to some BPs appear to be primarily regulated by one of the mechanisms, which may deserve further investigation. Moreover, the expression of genes with generally accessible promoters appears to be more heavily regulated by DNA methylation rather than PA. This is likely due to the ability of DNA methylation to regulate TF binding and transcription activity without affecting PA.43,44
Another interesting finding is the identification of a relatively complex cell type composition of Tcf21 LCs by scRNA-seq. VAT Tcf21 LCs are mainly composed of mesothelial and interstitial populations, sharing certain gene signatures with some MPC populations identified in mouse AT in recent scRNA-seq studies.24,45 Here we found that cell-matrix adhesion genes are more highly expressed in mesothelial Tcf21 LCs of young mice, likely enabling them to anchor to the mesothelial membrane. Reduced cell-matrix adhesion gene expression in interstitial Tcf21 LCs likely enables their migration to different locations and execute more diverse BPs and gene expression. Interestingly, in obese mice, mesothelial Tcf21 LCs seem to suffer from hypoxia, and many interstitial Tcf21 LCs are in a fibrotic, inflammatory, or quiescent state, which, taken together, likely underpins the reduced adipogenic activity of Tcf21 LCs in these mice.
Using the constructed GRN, we predicted an anti-adipogenic role of Tcf21. We also validated the inhibitory role of Tcf21 in adipogenesis, which is contrary to a recent report of a pro-adipogenic role of Tcf21 in immortalized chicken cells.46 Several etiologies likely underpin this discrepancy, including species (mouse versus chicken) and cell (in vivo and primary in vitro [here] versus Immortalized) differences, among others. Here, Tcf21 deletion resulted in more abundant Tcf21 lineage adipocytes, leading to improved metabolic health after HFD treatment. This is likely attributed to the larger quantity of VAT adipocytes in KO mice, which can store more lipids without excess hypertrophy. A similar phenomenon was observed in a recent study promoting de novo adipogenesis in WAT by enhancing Pparg expression.7 Our results further indicate that increased adipocyte hyperplasia in VAT during development alone can ameliorate obesity-induced metabolic syndrome in adulthood. Another interesting observation is that Tcf21 LCs of KO mice are developmentally more advanced compared with the WT. Our mechanistic study shows that Tcf21 mainly targets enhancers to regulate gene expression, including Dlk1. Dlk1 encodes an inhibitory non-canonical Notch1 ligand and inhibits many differentiation processes, including adipogenesis, through Notch-related and -unrelated mechanisms.36,47–50 Our data indicate that Tcf21 functions as a rate-limiting factor in Tcf21 LC adipogenic regulation through Dlk1. Our conclusion is further supported by the observation that the difference in adipogenesis between WT and KO Tcf21 LCs is mainly due to the interstitial population, which expresses a higher level of Dlk1. Moreover, because of the inhibitory effect of Dlk1 on the differentiation of other mesodermal lineages and the switch from a pro-adipogenic to a pro-fibrotic niche environment in adult VAT, the reduction of Tcf21 and Dlk1 expression in Tcf21 LCs of adult mice may facilitate their differentiation into other non-adipocyte cell types, such as myofibroblast, which requires additional investigation.
In conclusion, Tcf21 LCs actively contribute to VAT development but transition into quiescent and fibrotic states in adult mice, which is epigenetically regulated. Moreover, Tcf21 negatively regulates adipogenesis during VAT development, at least partially, through Dlk1 expression stimulation.
Limitations of the study
Because of the lack of a chromatin immunoprecipitation sequencing (ChIP)-grade antibody recognizing the endogenous level of mouse Tcf21, our study does not include experiments validating predicted Tcf21 binding or the causal link between Tcf21 deficiency and chromatin accessibility. Our stringent approach has enabled us to confidently identify some Tcf21-targeted genes but may also omit other genes regulated by Tcf21. Even though our results show that Dlk1 is a mediator of the inhibitory effect of Tcf21 on adipogenesis, it does not rule out the possible existence of other effector genes. Another limitation was the exclusion of non-Tcf21 LCs in the scRNA-seq experiment, which hampered our ability to understand the indirect impact of Tcf21 deficiency on these cell types.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xing Fu (xfu1@agcenter.lsu.edu).
Materials availability
All new unique reagents generated in this study will be shared in accordance with the relevant material transfer agreements.
Data and code availability
Raw bulk RNAseq, bulk ATACseq, and scRNAseq datasets have been deposited at Gene Expression Omnibus (GEO: GSE196569) and are publicly available as of the date of publication.
All original code for bulk sequencing data analysis has been deposited at GitHub (https://doi.org/10.5281/zenodo.7258615) and is publicly available as of the date of publication.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal models
All animal experimentation was approved by the Louisiana State University’s Institutional Animal Care and Use Committee. Tcf21MCM/+;R26tdTomato;PdgfraeGFP lineage-tracing mice (Figure 1A) in which Tcf21 LCs express cytoplasmic tdTomato upon tamoxifen (TM) treatmen and Pdgfra+ cells express nuclear eGFP were generated through crossing Tcf21MCM,38 R26tdTomato (Jackson Laboratories, stock no. 007914), and PdgfraeGFP (Jackson Laboratories, stock no. 007669) mice. Tcf21MCM/+;R26eGFP/mTmG lineage-tracing mice (Figure 1F) in which Tcf21 LCs express cytoplasmic and membrane GFP were generated through crossing Tcf21MCM,38 R26eGFP,51 and R26mTmG (Jackson Laboratories, stock no. 007676) mice. Tcf21MCM/fl;R26eGFP/mTmG KO lineage-tracing mice (Figure 4A) in which Tcf21 LCs express cytoplasmic and membrane GFP and Tcf21 is deleted in Tcf21 LCs were generated through crossing Tcf21MCM/+;R26mTmG/mTmG and Tcf21fl/+;R26eGFP/eGFP37 mice, which also generated Tcf21MCM/+;R26eGFP/mTmG littermates for WT and KO comparison studies. Breeding and lactating mice were fed with a breeder diet (LabDiet #5015). After weaning, mice were fed with a regular diet containing 13.384% calories from fat (LabDiet #5001). For HFD challenges, mice were fed with a diet containing 60.3% calories from fat (Teklad #TD.06414) starting at 6W of age for 1 week or 12 weeks. To induce the activity of the MerCreMer, mice were treated with TM (MilliporeSigma, T5648) dissolved in corn oil (MP Biomedicals, 901,414) through gavage (adult mice) or intragastric injection (neonatal mice). Experimentalists remained blinded to the genotypes or treatments until the completion of data analysis. Only male mice were used.
Primary cell cultures
Primary cells used in this study include stromal vascular cells and Tcf21 LCs isolated from eVAT of male Tcf21MCM/+;R26eGFP/mTmG, Tcf21MCM/fl;R26eGFP/mTmG, and WT C57BL/6 mice. Cells were maintained in DMEM with 10% bovine growth serum at 37°C and 5% CO2.
METHOD DETAILS
TM treatments
To induce the activity of the MerCreMer in adult male mice, mice were treated with TM (MilliporeSigma, T5648) dissolved in corn oil (MP Biomedicals, 901,414) through gavage at a dosage of 75 mg/kg BW/d starting at 6-week age for 3 days except for Figures 1A–1E, in which only a single dose was given. To induce the activity of the MerCreMer in neonatal male mice, pups were treated with TM through intragastric injection at a dosage of 100 μg/day on P2 and P4. To induce the activity of the MerCreMer in fetuses, on E11.5, pregnant female mice were treated with a single dose of TM at 75 mg/kg BW.
Cesarean section
We noticed that pregnant mice treated with TM often suffer from dystocia. To extract fetal mice, pregnant mice treated with TM were euthanized on gestation day 19.5 by cervical dislocation. The uterus was removed and placed onto a sterile drape or Petri dish. Pups were dissected from the yolk sac and amnion, followed by cutting of the umbilical cord. Pups were then transferred to a clean prewarmed dry paper towel to clean the amniotic fluid and secretions, especially the nasal area and simultaneously stimulated for breathing by gently tapping their chest area. Pups were placed on a heating pad (37°C) with a moist paper towel in between until regular breath was seen. Pups were rubbed with bedding material from the cage housing the foster dam and mixed with pups from the natural litter of the foster dam.
GTT and ITT
GTT and ITT were performed using our established protocol52 as follows. Briefly, mice were transferred to a clean cage with new bedding and drinking water and no food or feces in the hopper or bottom of the cage. After 16-h fasting for GTT or 6-h fasting for ITT, the weight of the mice was measured for glucose (1 g/kg BW) or insulin (1 unit/kg BW) dosage calculation. Baseline blood glucose level was then measured with a glucose meter by cutting the tail tip with a sterile scissor. Right after the baseline measuring, the mice were intraperitoneally (i.p.) injected with calculated amount of either D-glucose (VWR, BDH9230) or insulin (Sigma, I6634). Blood glucose levels were measured at 15, 30, 60, 90, and 120 min after glucose or insulin injection.
Viral packaging
Mouse Tcf21 (BC053525) and Dlk1 (BC052159) were cloned from pCMV-SPORT6-Tcf21 (Horizon #MMM1013–202770706) and pIRESh-GFPII-SM-Dlk1 into MIGR1 retroviral plasmid which carries an IRES-GFP cassette (Addgene #27490)53 to make MIGR1-Tcf21 and MIGR1-Dlk1 plasmids, respectively. Platinum-E (PE, Cell Biolabs # RV-101) cells were transfected with empty MIGR1, MIGR1-Tcf21, and MIGR1-Dlk1 plasmids to produce Retro-IRES-GFP, Retro-Tcf21-IRES-GFP, or Retro-Dlk1-IRES-GFP retroviruses, respectively. Lenti-shGFP and Lenti-shDlk1 lentiviruses were produced in 293T cells (ATCC #CRL-3216) using pLKO.1 GFP shRNA (Addgene #30323) and pLKO.1 mouse Dlk1 shRNA (Sigma # TRCN0000095446), respectively.54
Cell isolation
Cell isolation was performed using our established protocol55 as follows. Briefly, AT freshly dissected from mice was finely minced into 1–2 mm3 pieces and digested in DMEM containing 0.75 U/ml collagenase D (Roche, 11,088,866,001), 1.0 U/ml Dispase II (Roche, 10,165,859,001), and 1 mM CaCl2 at 37°C for 10 min with gentle agitation. Released floating adipocytes were transferred to a 1.5 mL centrifuge tube containing room temperature PBS with wide-bore pipette tips for quantification analysis or RNA extraction. The slurry was further digested for another 5–15 min. Digested tissue was centrifuged at 4°C and 500 g for 10 min. The cell pellet was then resuspended in HBSS, filtered with 100 μm and 40 μm cell strainers, followed by centrifugation. Finally, the isolated cells were resuspended in HBSS for FACS or a culture medium for in vitro experiments.
Quantification of the percentage of GFP+ adipocytes and adipocyte diameter
Adipocytes released from tissue samples by enzymatic digestion were placed on glass slides and imaged using an ECHO Revolve fluorescence microscope. The numbers of GFP+ and GFP− adipocytes and their diameters were quantified using ImageJ. The numbers of GFP+ and GFP− adipocytes were used to calculate the percentage of GFP+ adipocytes in total adipocytes.
Cell culture
Isolated cells were resuspended in DMEM with 10% bovine growth serum and 1% antibiotic mixture containing 10,000 U/ml penicillin, 10 mg/mL streptomycin, and 25 μg/mL amphotericin B. Cells were cultured on 6-well plates for 48 h, followed by subculturing on 96-well plates at a density of 1 × 104 cells/well. Twenty-four hours later, adipogenic differentiation was induced using an induction medium containing 1 μg/mL insulin, 0.25 mM dexamethasone, 0.125 mM IBMX, and 10 μM Rosiglitazone for 3 days, followed by a maintaining medium containing 1 μg/mL insulin only for another 3 days. For experiments involving viral transduction, cells were seeded on 96-well plates at a density of 5 × 103 cells/well. Overnight viral transduction was performed 12 h after seeding, followed by adipogenic differentiation induction.
Cell transplantation
Fifty thousand Tcf21 LCs isolated from 3W WT or KO mice were mixed with 30 μL Matrigel (Corning #356255) and transplanted subcutaneously into the chest area of the age-matched recipient mice. To avoid the influence of different recipient mice on the adipogenesis of transplanted cells, Matrigel containing WT and KO Tcf21 LCs were transplanted symmetrically to the left and right chests of the same recipient mice. Transplants were removed for imaging 2 weeks after transplantation.
IHC
AT samples were fixed in 4% paraformaldehyde (PFA) overnight, rinsed in PBS for 30 min, and processed for paraffin embedding using a Leica TP1020 Automatic Benchtop Tissue Processor. Paraffin-embedded tissue samples were cut into 5 μm sections. Deparaffinized and rehydrated tissue sections were subjected to antigen retrieval by heating sections in sodium citrate buffer (pH 6.0) for 20 min before blocking in a buffer containing TBS, 5% goat serum, and 0.2% Triton X-100 for 2 h. Blocked sections were stained with primary antibodies (See key resources table) 1:100 or 1:200 diluted in blocking buffer at 4°C overnight, rinsed 3 × 5 min in TBS containing 0.2% Triton X-100, and then incubated in appropriate fluorophore-conjugated secondary antibodies (See key resources table) 1:500 diluted in blocking buffer for 1 h at room temperature. Sections were then rinsed 3 × 5 min in TBS containing 0.2% Triton X-100 and mounted in a mounting medium containing DAPI. Images were captured using an inverted Leica SP8 confocal microscope.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Rabbit anti Laminin antibody | Abcam | Cat# ab11575, RRID:AB_298179 |
| Rabbit anti RFP antibody | Rockland | Cat# 600–401-379, RRID: AB_2209751 |
| Chicken anti-GFP antibody | Abcam | Cat# ab13970, RRID:AB_300798 |
| Rabbit anti-GFP antibody | Rockland | Cat# 600–401-215L, RRID:AB_2612813 |
| Goat anti-Perilipin-1 antibody | Abcam | Cat# ab61682, RRID:AB_944751 |
| Goat anti-Dpp4 antibody | R & D Systems | Cat# AF954, RRID:AB_355739 |
| Mouse anti-Icam1 antibody | Novus | Cat# NBP2–22541 |
| Goat anti-Pdgfrα antibody | Novus | Cat# AF1062, RRID: AB_2236897 |
| Mouse anti-α-Smooth Muscle Actin antibody | Cell Signaling Technology | Cat# 48938, RRID:AB_2799347 |
| Rabbit anti-CD 68 antibody | Cell Signaling Technology | Cat# 97778, RRID:AB_2928056 |
| Rabbit anti-Tcf21 antibody | Sigma-Aldrich | Cat# HPA013189, RRID:AB_10601215 |
| Rabbit anti-Dlk1 antibody | R & D Systems | Cat# MAB8634 |
| Rat anti-CD16/CD32 antibody | Thermo Fisher Scientific | Cat# 14–0161-82, RRID:AB_467133 |
| Alexa Fluor 488 donkey anti-chicken secondary antibody | Jackson ImmunoResearch Labs | Cat# 703–545-155, RRID:AB_2340375 |
| Alexa Fluor 488 donkey anti-rabbit secondary antibody | Invitrogen | Cat# A-21206, RRID:AB_2535792 |
| Alexa Fluor 555 donkey anti-rabbit secondary antibody | Invitrogen | Cat# A-31572, RRID:AB_162543 |
| Alexa Fluor 555 donkey anti-goat secondary antibody | Invitrogen | Cat# A-21432, RRID:AB_141788 |
| Alexa Fluor 647 donkey anti-mouse secondary antibody | Invitrogen | Cat# A-31571, RRID:AB_162542 |
| Alexa Fluor 647 donkey anti-rabbit secondary antibody | Invitrogen | Cat# A-31573, RRID:AB_2536183 |
| BV421 Rat anti-mouse Icam1 antibody | BioLegend | Cat# 116141, RRID:AB_2876428 |
| PE/Cy7 Rat anti-mouse CD45 antibody | BioLegend | Cat# 103114, RRID:AB_312979 |
| APC Rat anti-mouse Dpp4 antibody | BioLegend | Cat# 137807, RRID:AB_10663403 |
| APC Rabbit anti-mouse Dlk1 antibody | R & D Systems | Cat# FAB8634A, RRID:AB_2890004 |
| PE/Cyanine7 anti-mouse CD140a Antibody | BioLegend | Cat# 135911, RRID: AB_2715973 |
| APC anti-mouse CD140a Antibody | BioLegend | Cat# 135908, RRID:AB_2043970 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Retro-IRES-GFP | This manuscript | N/A |
| Retro-Tcf21-IRES-GFP | This manuscript | N/A |
| Retro-Dlk1-IRES-GFP | This manuscript | N/A |
| Lenti-shGFP | This manuscript | N/A |
| Lenti-shDlk1 | This manuscript | N/A |
|
| ||
| Biological samples | ||
|
| ||
| Mouse adipose tissue | This manuscript | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Tamoxifen | Sigma | Cat# T5648 |
| Coin oil | MP Biomedicals | Cat# 901414 |
| Collagenase D | Roche | Cat# 11088882001 |
| Dispase II | Roche | Cat# 04942078001 |
| DMEM, high glucose | Corning | Cat# 10–013-CV |
| Bovine growth serum | VWR | Cat# 76324–910 |
| HBSS | VWR | Cat# 02–0121-0500 |
| BSA | VWR | Cat# 97061–420 |
| Penicillin-Streptomycin | Thermo Fisher Scientific | Cat# 15140122 |
| Amphotericin B | Thermo Fisher Scientific | Cat# 15290026 |
| Glucose | VWR | Cat# BDH9230 |
| Insulin | Sigma | Cat# I6634 |
| IBMX | Sigma | Cat# I7018 |
| Dexamethasone | Sigma | Cat# D4902 |
| Rosiglitazone | Sigma | Cat# R2408 |
| Polybrene | Sigma | Cat# TR-1003-G |
| TRIzol | Invitrogen | Cat# 15596018 |
| Antifade Mounting Medium with DAPI | Vector Laboratories | Cat# H-1200–10 |
| AMPure XP Reagent beads | Beckman Coulter | Cat# A63881 |
| Sodium Chloride | Sigma | Cat# 59222C |
| Digitonin | Sigma | Cat# 300410 |
| Tween 20 | Promega | Cat# H5152 |
| Tris | VWR | Cat# 97062 |
| Magnesium Chloride | Sigma | Cat# M1028 |
| DMF | Sigma | Cat# D4551 |
| HCS LipidTOX™ Deep Red | Thermo Fisher Scientific | Cat# H34477 |
| SYBR Green | BIO-RAD | Cat# 1725274 |
| PFA | Electron Microscopy Sciences | Cat# 15714 |
| Red Zymosan Bioparticles | Invitrogen | Cat# P35364 |
| Matrigel | Corning | Cat# 356255 |
|
| ||
| Critical commercial assays | ||
|
| ||
| iScript™ cDNA Synthesis Kit | BIO-RAD | Cat# 1708891 |
| miRNeasy Micro Kit | QIAGEN | Cat# 217084 |
| MinElute Reaction Cleanup Kit | QIAGEN | Cat# 28206 |
| Click-iT™ Plus EdU Alexa Fluor™ 647 Flow Cytometry Assay Kit | Invitrogen | Cat# C10643 A |
| Phusion® High-Fidelity PCR Master Mix with HF Buffer | NEB | Cat# M0531L |
| Ultra™ II DNA Library Prep Kit for Illumina | NEB | Cat# E7645S |
| Multiplex Oligos for Illumina® (Dual Index Primers Set 1) | NEB | Cat# E7600S |
| Multiplex Oligos for Illumina® (Dual Index Primers Set 2) | NEB | Cat# E7780S |
| Multiplex Oligos for Illumina® (Index Primers Set 1) | NEB | Cat# E7335S |
| Multiplex Oligos for Illumina®(Index Primers Set 2) | NEB | Cat# E7500S |
| Illumina Tagment DNA Enzyme and Buffer Large Kit | Illumina | Cat# 20034198 |
| Single Cell/Low Input RNA Library Prep Kit for Illumina | NEB | Cat# E6420S |
| Qubit™ dsDNA HS and BR Assay Kits | Invitrogen | Cat# Q32854 |
| High Sensitivity RNA ScreenTape | Agilent | Cat# 5067–5579 |
| D5000 ScreenTape | Agilent | Cat# 5067–5588 |
| Chromium Single Cell 3′ Reagent Kits v3 | 10X Genomics | Cat# 1000157 |
|
| ||
| Deposited data | ||
|
| ||
| Raw sequencing data | GEO | GSE196569 |
| Code for bulk sequencing data analysis | GitHub | https://doi.org/10.5281/zenodo.7258615 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Tcf21MCM/+;R26eGFP/mTmG Tcf21 LCs | Primary isolation | N/A |
| Tcf21MCM/fl;R26eGFP/mTmG Tcf21 LCs | Primary isolation | N/A |
| C57BL/6 SVCs | Primary isolation | N/A |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: Tcf21MCM; B6.129-Tcf21tm3.1(cre/Esr1*)Eno | Acharya et al.38 | N/A |
| Mouse: R26tdTomato; B6.Cg-Gt(ROSA) 26Sortm14(CAG-tdTomato)Hze/J | Jackson Laboratories | Cat# 007914 |
| Mouse: R26eGFP; FVB.Cg-Gt(ROSA) 26Sortm1(CAG-lacZ,-EGFP)Glh/J | Yamamoto et al.51 | N/A |
| Mouse: R26mTmG; B6.129(Cg)-Gt(ROSA) 26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | Jackson Laboratories | Cat# 007676 |
| Mouse: PdgfraeGFP; B6.129S4-Pdgfratm11(EGFP)Sor/J | Jackson Laboratories | Cat# 007669 |
| Mouse: Tcf21fl; B6.129-Tcf21floxed | Maezawa et al.37 | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S8 for Oligonucleotides | N/A | |
|
| ||
| Recombinant DNA | ||
|
| ||
| pCMV-SPORT6-Tcf21 | Horizon | Cat#MMM1013–202770706 |
| pIRESh-GFPII-SM-Dlk1 | Horizon | Cat# 108932 |
| MIGR1 | Addgene | Cat# 27490 RRID: Addgene_27490 |
| MIGR1-Tcf21 | This manuscript | N/A |
| MIGR1-Dlk1 | This manuscript | N/A |
| pLKO.1 GFP shRNA | Addgene | Cat# 30323 RRID: Addgene_30323 |
| pIRESh-GFPII-SM-Dlk1 | Addgene | Cat# 108932 RRID: Addgene_108932 |
| Software and algorithms | ||
| GraphPad Prism | GraphPad Software, Inc | https://www.graphpad.com |
| FlowJo | FlowJo, LLC | https://www.flowjo.com |
| ImageJ | Fiji | https://fiji.sc/ |
| RStudio | R Studio | https://www.rstudio.com/ |
|
| ||
| Other | ||
|
| ||
| Breeder diet | LabDiet | Cat# 5015 |
| Regular diet | LabDiet | Cat# 5001 |
| High-fat diet | Envigo | Cat# TD 06414 |
ICC and LipidTOX staining of cells
For ICC, cells grown in 96-well plates were fixed in ice-old methanol for 10 min, rinse rinsed 3 times in TBS with 0.1% Triton X-100, incubated in blocking buffer (TBS, 0.1% Triton X-100, and 3% BSA), and then incubated with primary antibodies (See key resources table) 1:100 or 1:200 diluted in blocking buffer at 4°C overnight. Cells were then rinsed in TBS with 0.1% Triton X-100 3 × 5 min and stained with corresponding secondary antibodies (See key resources table) 1:500 diluted in blocking buffer for 1 h at room temperature. Stained plates were then rinsed and covered in a mounting medium containing DAPI. For LipidTOX staining of cells, cells were fixed in 4% PFA for 10 min at room temperature, rinsed 3 × 5 min in TBS with 0.1% Triton X-100, and stained with LipidTOX for 30 min at room temperature. Stained cells were rinsed and covered in a mounting medium containing DAPI. Images were captured using an ECHO revolve fluorescence microscope. Adipogenic efficiency was quantified by dividing the LipidTOX+ area by the DAPI+ area.
Whole-mount staining and imaging
eVAT and associated organs from neonatal mice were fixed in 1% PFA at 4°C overnight, rinsed 3 times in TBS with 0.1% Triton X-100, and stained with LipidTOX for 1 h at room temperature, followed by imaging. eVAT from adult mice were fixed in 1% PFA at 4°C overnight and imaged without staining. Imaging was done using an inverted Leica SP8 confocal microscope.
FACS
Tcf21 LCs were sorted on a FACSaria II system (BD Biosciences). Gates were made based on WT eGFP–control. Sorted eGFP+ Tcf21 LCs were used for in vitro experiments or subjected to RNAseq or ATACseq analyses. For FACS involving staining, cells isolated through tissue digestion were blocked in HBSS containing 1% BSA and an anti-mouse CD16/32 antibody on ice for 30 min, then stained with fluorophore-conjugated primary antibodies diluted in HBSS containing 1% BSA (See key resources table) on ice for 30 min, rinsed in HBSS containing 1% BSA, and sorted on a FACSaria II system. Sorted cells were cultured for in vitro experiments.
In vivo EdU proliferation assay
Mice were treated with EdU at a dosage of 50 mg/kg BW through i.p. injections. Four hours after EdU injections, mice were sacrificed, and samples were collected. EdU detection was carried out after IHC staining using the Click-iT Plus Alexa Fluor 647 Picolyl Azide Toolkit (C10643) from Thermo Fisher Scientific. GFP+ cells with DAPI signal were identified as Tcf21 LCs and were included in the quantification analysis.
Red Zymosan Bioparticle phagocytosis test
Red Zymosan Bioparticle phagocytosis test was performed using pHrodo Red Zymosan Bioparticles Conjugate for Phagocytosis (Invitrogen) following the manufacturer’s manual. Briefly, Tcf21 lineage CD45+ cells and non-Tcf21 lineage CD45+ cells were seeded onto 96 well plates separately or together at a 1:1 ratio and cultured for 24 h. Red Zymosan Bioparticles were then added to the cell culture. Images were captured using an ECHO revolve fluorescence microscope after 3 h of incubation.
Real-time PCR
cDNA was synthesized using an iScript cDNA synthesis kit (Bio-Rad). Real-time PCR (RT-PCR) was carried out using a CFX RT-PCR detection system (Bio-Rad) with SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). After amplification, a melting curve (0.01 °C/s) was used to confirm product purity, and agarose gel electrophoresis was performed to confirm that only a single product of the correct size was amplified. Relative mRNA content was normalized to 18S rRNA content. Primers used can be found in Table S8.
Bulk RNAseq library construction and sequencing
Total RNA was extracted from 2,000–5,000 FACS-sorted Tcf21 LCs using the miRNeasy Micro Kit (cat# 217084) from Qiagen. cDNA libraries were constructed using the NEBNext single cell/low input RNA library prep kit for Illumina (E6420) from New England BioLabs. The cDNA libraries were sequenced on the Illumina Hi-seq platform using 150 bp paired-end sequencing. Around 30 million read pairs were obtained for each sample.
ATACseq library construction and sequencing
The ATACseq was performed following our established protocol.21,56 Briefly, 5,000 FACS-sorted Tcf21 LCs were lysed to isolate the nuclei. Isolated nuclei were then incubated with the Tn5 transposase (TDE1, Illumina) and tagmentation buffer at 37°C for 30 min with shaking on a thermomixer at 500 g. Tagmentated DNA was purified using MinElute Reaction Cleanup Kit (Qiagen). PCR was performed to amplify the ATACseq libraries using Illumina TrueSeq primers and multiplex by indexes primers. After the PCR reaction, libraries were purified with the 1.1X AMPure beads (Beckman). The concentration of the sequencing libraries was determined by using Qubit dsDNA HS Assay Kit (Life Technologies). The size of the sequencing libraries was determined using a High Sensitivity D5000 Assay kit with a Tapestation 4200 system (Agilent). ATACseq libraries were sequenced on the Illumina Hi-seq platform using 150 bp paired-end sequencing. Around 50 million read pairs were obtained for each sample.
Bulk RNAseq data processing
RNAseq reads were filtered to remove low-quality reads and trimmed to remove adaptors by TrimGalore-0.6. The quality of reads after filtering was assessed by fastQC, followed by aligning reads to the mouse genome (MM10) by STAR (2.5.3a) with parameters used in Encode project.57 The expression quantification (transcripts per million, TPM) of individual genes was subsequently obtained by RSEM with gene annotations downloaded from Ensembl. Genes with a TPM >0.1 in at least one sample were deemed as expressed genes, otherwise unexpressed genes. PCA and t-SNE were carried out using R packages, DESeq2 and Rtsne, respectively. All differential gene expression analyses except the comparison between WT and KO samples from 3W were performed using DESeq258 from R directly or OneStopRNAseq.59 The differential expression of a gene was considered to be significant when the absolute value of the estimated shrunken LFC in TPM is over 0.585 and the adjusted p value is less than 0.05. For analysis comparing WT and KO samples, to account for batch effects from factors such as maternal traits and litter size, the LFC was first calculated for each gene between each pair of WT and KO littermates, followed by a one-sample t test to identify differentially expressed genes between WT and KO at 3W. To classify genes by expression level, a Z score for each gene in each sample was first obtained by standardizing the TPM values of all genes in the sample to have a mean of 0 and a variance of 1. Then, genes with a Z score below (above) zero in all sample groups were deemed to have constantly low (high) expression, with remaining genes classified as having dynamic expression. Z score was used to demonstrate gene expression changes for each gene in all relevant heatmaps.
ATACseq data processing
Sequencing reads of all samples underwent adapter removal and low-quality reads filtering using TrimGalore-0.6, followed by quality assessment using FastQC. Reads were then aligned to the mouse reference genome MM10 using Bowtie 2.3 with the following options: – very-sensitive -X 2000 – no-mixed – no-discordant. Only unique alignments within each sample were retained in subsequent analyses. Moreover, alignments resulting from PCR duplicates or located in mitochondria were excluded. The mouse genome was tiled with consecutive non-overlapping 300bp bins. The accessibility of each of the 300bp bins was assessed by the number of fragments per million mapped (FPM) that was aligned to the bin. There was a total of 8,406,114 bins with at least one aligned fragment in at least one sample. Pairwise Poisson distance between samples was calculated using package PoiClaClu in R. Same as in RNAseq data processing, the PCA and t-SNE were also done in R.
Peak calling and enrichment of genomic features in peaks
ATACseq peaks were called separately for each sample by MACS260 with the following options: – keep-dup all – nolambda – nomodel. Peaks in individual samples were subsequently merged using bedtools (https://bedtools.readthedocs.io/en/latest/). The accessibility of each merged peak in individual samples was accessed by the number of ATACseq fragments mapped to the peak region (FPM, fragments per million mapped). The annotations of genomic features, including transcription start sites (TSS), transcription end sites (TES), exons, introns, and CpG islands, were downloaded from Ensembl and UCSC genome browser. Promoters were defined as 500bp up- and downstream of the TSS of each annotated gene (TSS ±500bp). Intergenic regions were defined as genomic regions before the TSS of the first gene and after the TES of the last gene in each chromosome, and in-between the TES and TSS of two consecutive genes. Peaks that did not overlap with annotated promoters were deemed as distal peaks. The enrichment of transcriptional factor motifs in peaks was evaluated using HOMER (http://homer.ucsd.edu/homer/motif/).
Assessment of promoter and distal region accessibility
Promoter and distal region accessibilities in each sample were assessed by the number of ATACseq fragments (FPM) mapped to the defined promoter/distal region. To classify genes by promoter accessibility, a Z score for each promoter in each sample was first obtained by standardizing the FPM values in every sample to have a mean of 0 and a variance of 1 across all promoters. Promoters with Z-scores below zero in all samples were deemed to have constantly low accessibility, while those with z-scores above zero in all samples were deemed to have constantly high accessibility. The remaining promoters were considered to have dynamic accessibility. The differential accessibility analysis for promoters and distal regions was performed by the DESeq2 R package. The differential accessibility of a promoter or distal region was considered to be significant when the absolute value of the estimated shrunken LFC in FPM is over 0.585 and the adjusted p value is less than 0.05.
Assessment of the correlation between promoter/distal region accessibility and gene expression
The gene expression and accessibility of promoters and distal regions (i.e., distal peaks) at a stage were determined by averaging over samples (i.e., replicates) from that stage. Pearson correlation (cor) between promoter(distal) region accessibility and corresponding gene expression across studied stages were calculated. A cor > 0.5 was considered a strong positive correlation. Raw correlation data are included in Tables S2 and S4. To better visualize the expression and accessibility changes in the heatmap of individual genes/promoters/distal regions across samples, the data of accessibility and expression were standardized such that each row (i.e., gene/promoter/distal region) has a mean of 0 and a standard deviation of 1 (Z score). Z score was used to demonstrate accessibility changes for each ATAC peak in all relevant heatmaps.
Assessment of promoter CpG density
CpG density of annotated promoters (TSS ±500bp) was assessed by function CpGDensityByRegion in R package BSgenome. In our analysis, when categorization of the promoter CpG density was needed, it was done as follows: low (<25), medium (≥25 and ≤75), and high (>75).
GSEA and gene ontology enrichment analyses
GSEA of bulk RNAseq data was performed using OneStopRNAseq.59 GSEA of scRNAseq data was performed using clusterProfiler.61 Gene ontology enrichment analysis was performed on http://geneontology.org/.
Prediction of enhancers
Since enhancers can regulate the expression of genes that are far away from them in base-pair distance, it is very challenging to identify enhancers of genes. To predict enhancers for each gene, a set of putative enhancers was first determined, which included all ATAC peaks within the ±100 kb region of the gene except those overlapping with its promoters. Then, the putative enhancers with a high correlation (cor > 0.8) between their accessibility and the expression of the gene across studied samples were predicted as the enhancers of the gene.
Prediction of TF binding
The prediction of TF bindings in promoters and enhancers using ATAC data was performed with TOBIAS, which has shown superior performance over other methods for bias correction and footprinting, according to a recent study.62 Briefly, The TF binding score for individual TFs at each of their motif-appearing locations was first obtained by running TOBIAS for each sample. For each TF, the mean (μ) and standard deviation (σ) among binding scores within the 0.75 quantiles for the TF in a sample were subsequently calculated. A threshold t determined by μ+σ was then used to predict bindings, with a score > t deemed as binding and no binding otherwise.
Gene regulatory network reconstruction
The reconstruction of gene regulatory networks is essential to identify regulating TFs for each (target) gene, for which we developed a two-step approach to leverage information carried by both ATAC and RNA data. In the first step, a candidate set of regulating TFs were composed for each gene by including all TFs with a predicted binding in promoters and predicted enhancers of the gene. The prediction of TF bindings and enhancers of genes was made, as introduced above. In the second step, this candidate set was tailored to retain only TFs whose corresponding gene expression has a high correlation (absolute cor > 0.85) with that of the target gene across the involved samples. The TFs remained in the set were predicted as the regulating TFs of the target gene with positive/negative correlation indicating expression promotion/inhibition. The network topological analysis and visualization were done in Cytoscape63 with additional plugins: NetAnalyzer64 and yFiles.65
ScRNAseq library construction, sequencing, and data analysis
scRNAseq was performed using FACS-sorted Tcf21 LCs. Libraries were constructed using Chromium Single Cell 3′ Reagent Kits v3 (10x Genomics). The cDNA libraries were sequenced on the Illumina Hi-seq platform using 150 bp paired-end sequencing at a depth of more than 20,000 read pairs/cell. Raw sequencing data were processed using Cell Ranger 6.0.0 (10X Genomics) and MM10 mouse genome. Processed data were analyzed using Seurat (version 4.0.4) in R. Only cells with detected expression of the reporter gene were selected. Cells with mitochondrial gene expression greater than 5% of total gene expression were removed from the analysis. The resulting dataset was then normalized with a global-scaling normalization method (“LogNormalize”) with default parameters. Highly variable features were identified by directly calculating the mean-variance relationship using the FindVariableFeatures function (Seurat). Data were subjected to linear transformation to alter the relative expression of each gene with a mean equal to 0 and a standard deviation equal to 1 (Z score). For batch correction, Seurat function FindIntegrationAnchors was used to find pairwise anchors between batches, followed by integrating all datasets by the IntegrateData function. PCA was then performed to linearly reduce the dimensionality, followed by the elbow plot evaluation. The first 50 PCs were selected to cluster cells in a graph structure by drawing potential edges between cells according to the different gene expression patterns using k-nearest neighbor graph. Edges surrounded clusters were further refined with modularity optimization tools using the Findclusters function (Seurat). Then, the UMAP embedding parameters were determined, and cells were embedded in the 2-dimension graphs. The obtained clusters were subsequently compared with each other using FindAllMarkers function (Seurat) to identify differentially expressed genes among clusters, where only genes detected in at least 25% of all cells were analyzed and the Mann-Whitney Wilcoxon test with setting minimal average LFC as 0.25 was used. GSEA was performed using clusterProfiler v4.2.2 in R.61 Pseudotime analysis and trajectory reference were accomplished by using Monocle (versions 2&3) in R. For RNA velocity analysis, spliced and unspliced transcripts were first identified and quantified in each cell using Velocyto.66 The obtained data were then used to run scVelo67 in Python to infer relationships among cells based on RNA velocity.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were analyzed using GraphPad Prism 9 (GraphPad Software, Inc.) unless otherwise stated. Two-tailed unpaired or paired t-tests were used to determine the significance of differences between the means of 2 groups. Significance levels are as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. One-way ANOVA followed by multiple comparisons was used to determine the significance of differences among means of more than 2 groups. Different letters indicate significant differences (p < 0.05). n indicates the number of mice.
Supplementary Material
Highlights.
Tcf21 is specifically expressed in VAT MPCs giving rise to visceral adipocytes
Tcf21 LCs developmentally undergo notable transcriptional and chromatin remodeling
Loss of Tcf21 promotes visceral adipogenesis and improves metabolic health
Tcf21 inhibits adipogenesis of VAT MPCs by activating Dlk1 expression
ACKNOWLEDGMENTS
This work was supported by NIH/NIDDK under grant 1R15DK122383 (to X.F.), NIH/NIGMS P20GM130555 (to X.F.), NIH/NHLBI under 1R01HL157519 (to X.F.), and the Louisiana Board of Regents under grant BOR.Fu.LEQSF(2019–22)-RD-A-01 (to X.F.). We thank Dr. Michelle Tallquist (University of Hawaii) for sharing Tcf21MCM mice and Dr. Susan Quaggin (Northwestern University) for sharing Tcf21 fl/fl mice. MIGR1 was a gift from Warren Pear (Addgene plasmid 27490; http://n2t.net/addgene:27490; RRID: Addgene_27490). pLKO.1 GFP shRNA was a gift from David Sabatini (Addgene plasmid 30323; http://n2t.net/addgene:30323; RRID: Addgene_30323). pIRESh-GFPII-SM-Dlk1 was a gift from Anne Ferguson-Smith (Addgene plasmid 108932; http://n2t.net/addgene:108932; RRID: Addgene_108932).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112166.
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
REFERENCES
- 1.Wang QA, Tao C, Gupta RK, and Scherer PE (2013). Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med 19, 1338–1344. 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeffery E, Church CD, Holtrup B, Colman L, and Rodeheffer MS (2015). Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nat. Cell Biol 17, 376–385. 10.1038/ncb3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gustafson B, Hammarstedt A, Hedjazifar S, and Smith U (2013). Restricted adipogenesis in hypertrophic obesity: the role of WISP2, WNT, and BMP4. Diabetes 62, 2997–3004. 10.2337/db13-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trayhurn P, and Wood IS (2004). Adipokines: inflammation and the pleiotropic role of white adipose tissue. Br. J. Nutr 92, 347–355. 10.1079/bjn20041213. [DOI] [PubMed] [Google Scholar]
- 5.Veilleux A, Caron-Jobin M, Noël S, Laberge PY, and Tchernof A (2011). Visceral adipocyte hypertrophy is associated with dyslipidemia independent of body composition and fat distribution in women. Diabetes 60, 1504–1511. 10.2337/db10-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim SM, Lun M, Wang M, Senyo SE, Guillermier C, Patwari P, and Steinhauser ML (2014). Loss of white adipose hyperplastic potential is associated with enhanced susceptibility to insulin resistance. Cell Metab 20, 1049–1058. 10.1016/j.cmet.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao M, Vishvanath L, Busbuso NC, Hepler C, Shan B, Sharma AX, Chen S, Yu X, An YA, Zhu Y, et al. (2018). De novo adipocyte differentiation from Pdgfrbeta(+) preadipocytes protects against pathologic visceral adipose expansion in obesity. Nat. Commun 9, 890. 10.1038/s41467-018-03196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Després JP, and Lemieux I (2006). Abdominal obesity and metabolic syndrome. Nature 444, 881–887. [DOI] [PubMed] [Google Scholar]
- 9.Verboven K, Wouters K, Gaens K, Hansen D, Bijnen M, Wetzels S, Stehouwer CD, Goossens GH, Schalkwijk CG, Blaak EE, and Jocken JW (2018). Abdominal subcutaneous and visceral adipocyte size, lipolysis and inflammation relate to insulin resistance in male obese humans. Sci. Rep 8, 4677. 10.1038/s41598-018-22962-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta RK, Mepani RJ, Kleiner S, Lo JC, Khandekar MJ, Cohen P, Frontini A, Bhowmick DC, Ye L, Cinti S, and Spiegelman BM (2012). Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metab 15, 230–239. 10.1016/j.cmet.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosen ED, Walkey CJ, Puigserver P, and Spiegelman BM (2000). Transcriptional regulation of adipogenesis. Genes Dev 14, 1293–1307. [PubMed] [Google Scholar]
- 12.Chau YY, Bandiera R, Serrels A, Martínez-Estrada OM, Qing W, Lee M, Slight J, Thornburn A, Berry R, McHaffie S, et al. (2014). Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat. Cell Biol 16, 367–375. 10.1038/ncb2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen P, Levy JD, Zhang Y, Frontini A, Kolodin DP, Svensson KJ, Lo JC, Zeng X, Ye L, Khandekar MJ, et al. (2014). Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell 156, 304–316. 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joe AWB, Yi L, Even Y, Vogl AW, and Rossi FMV (2009). Depot-specific differences in adipogenic progenitor abundance and proliferative response to high-fat diet. Stem Cell 27, 2563–2570. 10.1002/stem.190. [DOI] [PubMed] [Google Scholar]
- 15.Han J, Lee JE, Jin J, Lim JS, Oh N, Kim K, Chang SI, Shibuya M, Kim H, and Koh GY (2011). The spatiotemporal development of adipose tissue. Development 138, 5027–5037. 10.1242/dev.067686. [DOI] [PubMed] [Google Scholar]
- 16.Acharya A, Baek ST, Huang G, Eskiocak B, Goetsch S, Sung CY, Banfi S, Sauer MF, Olsen GS, Duffield JS, et al. (2012). The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development 139, 2139–2149. 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quaggin SE, Vanden Heuvel GB, and Igarashi P (1998). Pod-1, a mesoderm-specific basic-helix-loop-helix protein expressed in mesenchymal and glomerular epithelial cells in the developing kidney. Mech. Dev 71, 37–48. [DOI] [PubMed] [Google Scholar]
- 18.Quaggin SE, Schwartz L, Cui S, Igarashi P, Deimling J, Post M, and Rossant J (1999). The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development 126, 5771–5783. [DOI] [PubMed] [Google Scholar]
- 19.Shen YC, Shami AN, Moritz L, Larose H, Manske GL, Ma Q, Zheng X, Sukhwani M, Czerwinski M, Sultan C, et al. (2021). TCF21(+) mesenchymal cells contribute to testis somatic cell development, homeostasis, and regeneration in mice. Nat. Commun 12, 3876. 10.1038/s41467-021-24130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu X, Khalil H, Kanisicak O, Boyer JG, Vagnozzi RJ, Maliken BD, Sargent MA, Prasad V, Valiente-Alandi I, Blaxall BC, and Molkentin JD (2018). Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. J. Clin. Invest 128, 2127–2143. 10.1172/JCI98215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Sun J, Liu Q, Dodlapati S, Ming H, Wang L, Li Y, Li R, Jiang Z, Francis J, and Fu X (2022). The landscape of accessible chromatin in quiescent cardiac fibroblasts and cardiac fibroblasts activated after myocardial infarction. Epigenetics 17, 1020–1039. 10.1080/15592294.2021.1982158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akama T, and Chun T-H (2018). Transcription factor 21 (TCF21) promotes proinflammatory interleukin 6 expression and extracellular matrix remodeling in visceral adipose stem cells. J. Biol. Chem 293, 6603–6610. 10.1074/jbc.RA117.000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong H, Sun W, Shen Y, Baláz M, Balázová L, Ding L, Löffler M, Hamilton B, Klöting N, Blüher M, et al. (2022). Identification of a regulatory pathway inhibiting adipogenesis via RSPO2. Nat. Metab 4, 90–105. 10.1038/s42255-021-00509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merrick D, Sakers A, Irgebay Z, Okada C, Calvert C, Morley MP, Percec I, and Seale P (2019). Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science 364, eaav2501. 10.1126/science.aav2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcelin G, Silveira ALM, Martins LB, Ferreira AV, and Clément K (2019). Deciphering the cellular interplays underlying obesity-induced adipose tissue fibrosis. J. Clin. Invest 129, 4032–4040. 10.1172/jci129192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kendall RT, and Feghali-Bostwick CA (2014). Fibroblasts in fibrosis: novel roles and mediators. Front. Pharmacol 5, 123. 10.3389/fphar.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamata M, Okitsu Y, Fujiwara T, Kanehira M, Nakajima S, Takahashi T, Inoue A, Fukuhara N, Onishi Y, Ishizawa K, et al. (2014). GATA2 regulates differentiation of bone marrow-derived mesenchymal stem cells. Haematologica 99, 1686–1696. 10.3324/hae-matol.2014.105692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong Q, Dalgin G, Xu H, Ting CN, Leiden JM, and Hotamisligil GS (2000). Function of GATA transcription factors in preadipocyte-adipocyte transition. Science 290, 134–138. 10.1126/science.290.5489.134. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Huynh H, Zuo H, Salminen M, and Wan Y (2016). Gata2 is a rheostat for mesenchymal stem cell fate in male mice. Endocrinology 157, 1021–1028. 10.1210/en.2015-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y, Lan Y, Liu H, and Jiang R (2011). The zinc finger transcription factors Osr1 and Osr2 control synovial joint formation. Dev. Biol 352, 83–91. 10.1016/j.ydbio.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McConnell BB, Kim SS, Yu K, Ghaleb AM, Takeda N, Manabe I, Nusrat A, Nagai R, and Yang VW (2011). Krüppel-like factor 5 is important for maintenance of crypt architecture and barrier function in mouse intestine. Gastroenterology 141, 1302–1313.e1–6. 10.1053/j.gastro.2011.06.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarroll CS, He W, Foote K, Bradley A, McGlynn K, Vidler F, Nixon C, Nather K, Fattah C, Riddell A, et al. (2018). Runx1 deficiency protects against adverse cardiac remodeling after myocardial infarction. Circulation 137, 57–70. 10.1161/circulationaha.117.028911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma L, Cantrup R, Varrault A, Colak D, Klenin N, Götz M, McFarlane S, Journot L, and Schuurmans C (2007). Zac1 functions through TGFbetaII to negatively regulate cell number in the developing retina. Neural Dev 2, 11. 10.1186/1749-8104-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanisicak O, Khalil H, Ivey MJ, Karch J, Maliken BD, Correll RN, Brody MJ, J Lin SC, Aronow BJ, Tallquist MD, and Molkentin JD (2016). Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat. Commun 7, 12260. 10.1038/ncomms12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smas CM, and Sul HS (1993). Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 73, 725–734. 10.1016/0092-8674(93)90252-l. [DOI] [PubMed] [Google Scholar]
- 36.Mitterberger MC, Lechner S, Mattesich M, Kaiser A, Probst D, Wenger N, Pierer G, and Zwerschke W (2012). DLK1(PREF1) is a negative regulator of adipogenesis in CD105⁺/CD90⁺/CD34⁺/CD31⁻/FABP4⁻ adipose-derived stromal cells from subcutaneous abdominal fat pats of adult women. Stem Cell Res 9, 35–48. 10.1016/j.scr.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Maezawa Y, Onay T, Scott RP, Keir LS, Dimke H, Li C, Eremina V, Maezawa Y, Jeansson M, Shan J, et al. (2014). Loss of the podocyte-expressed transcription factor Tcf21/Pod1 results in podocyte differentiation defects and FSGS. J. Am. Soc. Nephrol 25, 2459–2470. 10.1681/ASN.2013121307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acharya A, Baek ST, Banfi S, Eskiocak B, and Tallquist MD (2011). Efficient inducible Cre-mediated recombination in Tcf21 cell lineages in the heart and kidney. Genesis 49, 870–877. 10.1002/dvg.20750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang N, Li Y, Shu T, and Wang J (2019). Cytokines and inflammation in adipogenesis: an updated review. Front. Med 13, 314–329. 10.1007/s11684-018-0625-0. [DOI] [PubMed] [Google Scholar]
- 40.Ruan H, Hacohen N, Golub TR, Van Parijs L, and Lodish HF (2002). Tumor necrosis factor-alpha suppresses adipocyte-specific genes and activates expression of preadipocyte genes in 3T3-L1 adipocytes: nuclear factor-kappaB activation by TNF-alpha is obligatory. Diabetes 51, 1319–1336. 10.2337/diabetes.51.5.1319. [DOI] [PubMed] [Google Scholar]
- 41.Choy L, and Derynck R (2003). Transforming growth factor-beta inhibits adipocyte differentiation by Smad3 interacting with CCAAT/enhancer-binding protein (C/EBP) and repressing C/EBP transactivation function. J. Biol. Chem 278, 9609–9619. 10.1074/jbc.M212259200. [DOI] [PubMed] [Google Scholar]
- 42.Sárvári AK, Van Hauwaert EL, Markussen LK, Gammelmark E, Marcher AB, Ebbesen MF, Nielsen R, Brewer JR, Madsen JGS, and Mandrup S (2021). Plasticity of epididymal adipose tissue in response to diet-induced obesity at single-nucleus resolution. Cell Metab 33, 437–453.e5. 10.1016/j.cmet.2020.12.004. [DOI] [PubMed] [Google Scholar]
- 43.Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, et al. (2017). Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356, eaaj2239. 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu H, Wang G, and Qian J (2016). Transcription factors as readers and effectors of DNA methylation. Nat. Rev. Genet 17, 551–565. 10.1038/nrg.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hepler C, Shan B, Zhang Q, Henry GH, Shao M, Vishvanath L, Ghaben AL, Mobley AB, Strand D, Hon GC, and Gupta RK (2018). Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. Elife 7, e39636. 10.7554/eLife.39636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X, Cheng B, Liu C, Du Z, Zhang H, Wang N, Wu M, Li Y, Cao Z, and Li H (2019). A novel regulator of preadipocyte differentiation, transcription factor TCF21, functions partially through promoting LPL expression. Front. Physiol 10, 458. 10.3389/fphys.2019.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sánchez-Solana B, Nueda ML, Ruvira MD, Ruiz-Hidalgo MJ, Monsalve EM, Rivero S, García-Ramírez JJ, Díaz-Guerra MJM, Baladrón V, and Laborda J (2011). The EGF-like proteins DLK1 and DLK2 function as inhibitory non-canonical ligands of NOTCH1 receptor that modulate each other’s activities. Biochim. Biophys. Acta 1813, 1153–1164. 10.1016/j.bbamcr.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 48.Lin GL, and Hankenson KD (2011). Integration of BMP, Wnt, and notch signaling pathways in osteoblast differentiation. J. Cell. Biochem 112, 3491–3501. 10.1002/jcb.23287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Traustadóttir GÁ, Lagoni LV, Ankerstjerne LBS, Bisgaard HC, Jensen CH, and Andersen DC (2019). The imprinted gene Delta like non-canonical Notch ligand 1 (Dlk1) is conserved in mammals, and serves a growth modulatory role during tissue development and regeneration through Notch dependent and independent mechanisms. Cytokine Growth Factor Rev 46, 17–27. 10.1016/j.cytogfr.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Zhao L, Smas C, and Sul HS (2010). Pref-1 interacts with fibronectin to inhibit adipocyte differentiation. Mol. Cell Biol 30, 3480–3492. 10.1128/mcb.00057-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto M, Shook NA, Kanisicak O, Yamamoto S, Wosczyna MN, Camp JR, and Goldhamer DJ (2009). A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis 47, 107–114. 10.1002/dvg.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu X, Zhu M, Zhang S, Foretz M, Viollet B, and Du M (2016). Obesity Impairs Skeletal muscle regeneration through inhibition of AMPK. Diabetes 65, 188–200. 10.2337/db15-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, Pendergast AM, Bronson R, Aster JC, Scott ML, and Baltimore D (1998). Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood 92, 3780–3792. [PubMed] [Google Scholar]
- 54.Li Y, Li C, Liu Q, Wang L, Bao AX, Jung JP, Dodlapati S, Sun J, Gao P, Zhang X, et al. (2022). Loss of Acta2 in cardiac fibroblasts does not prevent the myofibroblast differentiation or affect the cardiac repair after myocardial infarction. J. Mol. Cell. Cardiol 171, 117–132. 10.1016/j.yjmcc.2022.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu X, Zhao JX, Zhu MJ, Foretz M, Viollet B, Dodson MV, and Du M (2013). AMP-activated protein kinase α1 but not α2 catalytic subunit potentiates myogenin expression and myogenesis. Mol. Cell Biol 33, 4517–4525. 10.1128/mcb.01078-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ming H, Sun J, Pasquariello R, Gatenby L, Herrick JR, Yuan Y, Pinto CR, Bondioli KR, Krisher RL, and Jiang Z (2021). The landscape of accessible chromatin in bovine oocytes and early embryos. Epigenetics 16, 300–312. 10.1080/15592294.2020.1795602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li R, Hu K, Liu H, Green MR, and Zhu LJ (2020). OneStopRNAseq: a web application for comprehensive and efficient analyses of RNA-seq data. Genes 11, 1165. 10.3390/genes11101165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-seq (MACS). Genome Biol 9, R137. 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bentsen M, Goymann P, Schultheis H, Klee K, Petrova A, Wiegandt R, Fust A, Preussner J, Kuenne C, Braun T, et al. (2020). ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat. Commun 11, 4267. 10.1038/s41467-020-18035-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Assenov Y, Ramírez F, Schelhorn SE, Lengauer T, and Albrecht M (2008). Computing topological parameters of biological networks. Bioinformatics 24, 282–284. 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 65.Wiese R, Eiglsperger M, and Kaufmann M (2004). yFiles — visualization and automatic layout of graphs. In Graph Drawing Software, Jünger M and Mutzel P, eds. (Springer Berlin Heidelberg; ), pp. 173–191. 10.1007/978-3-642-18638-7_8. [DOI] [Google Scholar]
- 66.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bergen V, Lange M, Peidli S, Wolf FA, and Theis FJ (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol 38, 1408–1414. 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw bulk RNAseq, bulk ATACseq, and scRNAseq datasets have been deposited at Gene Expression Omnibus (GEO: GSE196569) and are publicly available as of the date of publication.
All original code for bulk sequencing data analysis has been deposited at GitHub (https://doi.org/10.5281/zenodo.7258615) and is publicly available as of the date of publication.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


