Abstract
Kaposi’s sarcoma–associated herpesvirus (KSHV) has been implicated in the pathogenesis of Kaposi’s sarcoma (KS) and other malignancies. The cellular origin of KS has been suggested to be either mesenchymal stem cells (MSCs) or endothelial cells. However, receptor(s) for KSHV to infect MSCs remains unknown. By combining bioinformatics analysis and shRNA screening, we identify neuropilin 1 (NRP1) as an entry receptor for KSHV infection of MSCs. Functionally, NRP1 knockout and overexpression in MSCs significantly reduce and promote, respectively, KSHV infection. Mechanistically, NRP1 facilitated the binding and internalization of KSHV by interacting with KSHV glycoprotein B (gB), which was blocked by soluble NRP1 protein. Furthermore, NRP1 interacts with TGF-β receptor type 2 (TGFBR2) through their respective cytoplasmic domains and thus activates the TGFBR1/2 complex, which facilitates the macropinocytosis-mediated KSHV internalization via the small GTPases Cdc42 and Rac1. Together, these findings implicate that KSHV has evolved a strategy to invade MSCs by harnessing NRP1 and TGF-beta receptors to stimulate macropinocytosis.
A systematic screening revealed that NRP1 and TGFBR1/2 mediated KSHV infection through an ingenious mechanism.
INTRODUCTION
Kaposi’s sarcoma–associated herpesvirus (KSHV) is an enveloped double-stranded DNA virus associated with a variety of human malignancies, including Kaposi’s sarcoma (KS) (1), primary effusion lymphoma (2), multicentric Castleman’s disease (3), and recently reported childhood osteosarcoma (4). KS is a multifocal and oligoclonal malignancy with complicated pathogenesis. The origin of spindle-shaped KS cells was originally considered to be endothelial cell lineage (5). However, the heterogeneity of spindle-shaped cells has led to the hypothesis that KS may originate from mesenchymal stem cells (MSCs) or precursors of vascular cells (6). As more and more evidence (7–10) was unearthed, this hypothesis has been gaining ground and forming relatively well-developed perspectives that KS may originate from KSHV-infected pluripotent MSCs and KSHV infection transforms MSCs to KS cells through an mesenchymal-to-endothelial transition process.
MSCs are a group of pluripotent stem cells with multiple differentiation abilities. The initial evidence supporting KSHV infection of MSCs was that primary MSCs were susceptible to infection by both cell-free and cell-associated KSHV in culture (11). The vulnerability of MSCs to KSHV infection was further demonstrated by the report that human MSCs of diverse origins permit persistent KSHV infection (12). Moreover, primary rat embryonic metanephric mesenchymal precursor cells were effectively infected by KSHV and cellular transformation occurred as early as 4 days after the infection (13). Considering that the oral cavity is the major route of KSHV infection and oral MSCs have access to saliva, we investigated the susceptibility of oral MSCs to KSHV infection. We showed that different types of oral MSCs, including periodontal ligament stem cells (PDLSC), dental pulp stem cells (DPSC), and gingiva/mucosa-derived mesenchymal stem cells (GMSC), were highly susceptible to KSHV infection and the infection promotes MSC differentiation (7). These studies suggest that MSCs might be a primary target of KSHV infection in the oral cavity, and KSHV-infected MSCs could be precursors to KS tumor cells. However, how KSHV infects MSCs remains unclear.
KSHV infection is a complicated and multistep process involving the interaction of multiple viral glycoproteins with host receptors. KSHV infection in different target cells except for MSCs have been intensively investigated (14, 15), and the receptors mediating KSHV entry in these cells, including integrins (16), Ephrin receptor family (17–19), Cystine/glutamate transporter (xCT) (20), and Dendritic cell-specific ICAM-3-grabbing non-integrin 1 (DC-SIGN) (21), have been identified. Although MSCs are highly susceptible to KSHV infection, how KSHV enters MSCs has not been explored yet. In particular, the host factors mediating KSHV infection and the mechanism governing KSHV entry into MSCs remain to be unveiled. In this study, we identify neuropilin 1 (NRP1) as a crucial entry receptor for KSHV infection and show that NRP1 takes advantage of transforming growth factor–β receptor type 1/2 (TGFBR1/2)–transduced signaling to promote macropinocytosis to support KSHV entry into MSCs.
RESULTS
Candidate receptors for KSHV were screened by bioinformatics analysis and shRNA screening experiments
To identify potential receptors for KSHV infection of MSCs, we adopted a previously reported screening strategy (22) (Fig. 1A). First, we compared RNA sequencing (RNA-seq) data of primary MSCs and the BJAB cell line and screened 2453 differentially expressed genes (DEGs) that were highly expressed in MSCs but low expressed in BJAB. Similarly, by comparing the RNA-seq data between KS1 (two cases of KS tissue samples that were sequenced in this study) and BJAB and between KS2 {four cases of KS tissue samples from Gene Expression Omnibus [GEO] database [GSE100684; (23)]} and BJAB, we screened 1831 and 2674 DEGs, respectively, that were highly expressed in KS1 and KS2 but lowly expressed in BJAB. These cell lines and tumor samples were chosen because MSCs were susceptible to KSHV, and KS was supposed to derive from MSCs whereas BJAB cells were refractory to cell-free KSHV infection in culture condition (24). We then identified membrane protein genes from these three groups of DEGs and obtained 70 genes shared among them (Fig. 1, B and C). To further narrow down the receptor candidates, these 70 genes were input into KOBAS 3.0 website (25) for an enrichment analysis for items associated with “virus,” which yielded a total of 21 receptor candidates (Fig. 1D, the list of these 21 receptor candidates is shown in fig. S2A).
Fig. 1. Bioinformatics analysis combined with shRNA screening identifies candidate receptors for KSHV infection of MSCs.
(A) Flow charts of bioinformatics analysis for identifying candidate receptors. (B) Venn diagram showing the intersection of three groups of differentially expressed membrane protein genes. (C) Heatmaps showing the expression of the 70 membrane protein genes. (D) KOBAS3.0 enrichment analysis of 70 membrane proteins. Virus-related items are shown here, and the list of specific genes enriched to these items is shown in fig. S2A. KEGG, Kyoto Encyclopedia of Genes and Genomes. (E) shRNA screening for candidate receptors that contribute to KSHV infection. shRNA targeting 21 candidate receptors were transduced to PDLSC by lentivirus. These cells were then infected with KSHV, and the efficiency of infection was determined by flow cytometry. Arrows indicate genes whose knockdown reduces KSHV infection efficiency more than 50% compared to the shRNA control (shCTR). The red line indicated 50% of relative KSHV infection efficiency in shCTR. Data are means ± SEM (n = 3 biological replicates).
The importance of these 21 candidates for KSHV infection was then examined through a loss-of-function strategy. We constructed a short hairpin RNA (shRNA) library containing two or three independent shRNAs targeting each of these candidates to knock down their expression. To eliminate the subjective influence in the experiment, we randomly numbered shRNA and carried out the analysis in a blinded fashion. The knockdown efficiency of each shRNA was verified by quantitative reverse transcription–polymerase chain reaction (RT-qPCR) (fig. S2B). Then, the effect of each shRNA on KSHV infection efficiency was examined in PDLSC transduced with the shRNA. We found that all the shRNAs targeting ephrin receptor A2 (EPHA2), NRP1, and TGFBR2 genes reduced the infection of PDLSC by KSHV to 50% or less (Fig. 1E). We verified the results of shRNA screening with a single guide RNA (sgRNA)–mediated knockout approach and found that KSHV infection was indeed significantly reduced after the knockout of the NRP1, EPHA2, or TGFBR2 gene (Fig. 2, A to C) and that re-expression of these genes could rescue reduced KSHV infection (fig. S3A).
Fig. 2. Loss-of-function and gain-of-function analysis show that EphA2 and NRP1 promote KSHV infection.
(A to C) Knockout of NRP1, EPHA2, and TGFBR2 individually reduces KSHV infection. PDLSC, DPSC, and GMSC transduced with sgRNA targeting NRP1 (A), EPHA2 (B), or TGFBR2 (C) by lentivirus were analyzed for the protein expression by Western blotting (top) or for the KSHV infection efficiency by flow cytometry (bottom). (D) Effect of overexpression of NRP1, EPHA2, and TGFBR1/2 on KSHV infection. 293T cells transfected with expression plasmids for NRP1, EphA2, or TGFBR1 together with TGFBR2 (TGFBR1/2) were analyzed for the protein expression by Western blotting (top) or for the KSHV infection efficiency by flow cytometry (bottom). (E) Endogenous expression levels of NRP1, EPHA2, and TGFBR2 in PDLSC and human umbilical cord endothelial cells (HUVECs). Results are representative of two or three independent experiments. Data are means ± SEM. One-way analysis of variance (ANOVA) was carried out with Tukey’s correction for multiple comparisons (A to C). Comparison between two groups was performed by two-tailed unpaired Student’s t test (D). **P < 0.01; ****P < 0.0001; ns, no significance.
The contribution of EPHA2, NRP1, and TGFBR2 to KSHV infection was further studied using a gain-of-function assay. Overexpression of EPHA2 or NRP1 in both PDLSC and 293T cells increased the susceptibilities to KSHV infection, whereas overexpression of TGFBR2 and TGFBR1 had no effect on KSHV infection (Fig. 2D and fig. S3B). These results suggested that both EPHA2 and NRP1 might serve as receptors for KSHV infection of MSCs. EPHA2 had been reported as a receptor for KSHV to infect endothelial and epithelial cells, which supports the reliability of our screening. EPHA2 was expressed at a very low level in MSCs compared to in endothelial cells, whereas the expression of NRP1 was high in MSCs but was almost undetectable in endothelial cells (Fig. 2E). Compared to NRP1, other reported KSHV receptors including EPHA2, EPHA4, EPHA7, xCT, DC-SIGN, and integrin α3 (ITGA3) also expressed in very low levels in MSCs (fig. S3C). Therefore, we speculated that NRP1 was the primary receptor for KSHV infection of MSCs. To determine whether NRP1 is required for all types of oral MSCs, we performed knockout of NRP1 in DPSC and GMSC in addition to PDLSC and found that NRP1 was important for the KSHV infection of all three types of MSCs (Fig. 2A). Together, our results suggest that NRP1 is an important candidate receptor for KSHV infection of oral MSCs.
NRP1 facilitates KSHV binding and internalization
Viral entry is a multistep process, including binding (attachment), internalization, and membrane fusion. To determine at which steps EPHA2, NRP1, and TGFBR2 were involved in the process, the binding, internalization, and fusion assays were performed. Results showed that the knockout of EPHA2, NRP1, or TGFBR2 in PDLSC significantly decreased KSHV internalization and that NRP1 knockout also significantly affected the viral binding, whereas TGFBR2 and EPHA2 knockout has no effect on viral binding (Fig. 3A). Re-expression of these genes could rescue reduced KSHV binding or internalization caused by sgRNAs (fig. S4, A and B). In addition, in 293T cells, overexpression of NRP1 markedly enhanced KSHV binding and internalization, whereas overexpression of EPHA2 only enhanced KSHV internalization (Fig. 3B). The expression of NRP1 only marginally increased the fusion activity, whereas overexpression of EPHA2 markedly enhanced fusion using a previously described cell-based fusion assay (Fig. 3C) (26, 27). This result of EPHA2 is consistent with the finding that EPHA2 interacts with gH/gL of EBV and KSHV to trigger fusion (28). Together, both EPHA2 and NRP1 apparently serve as receptors but contribute to KSHV entry into MSCs through different mechanisms. Since the mechanism of EPHA2 in promoting KSHV infection through interacting with viral gH/gL had been well characterized (17, 28), we decided to focus our investigation on NRP1.
Fig. 3. NRP1 facilitates KSHV binding and internalization.
(A) Binding and internalization assay of KSHV in PDLSC. PDLSC with EPHA2, NRP1, or TGFBR2 knockout were infected with KSHV at 4°C (for binding) for 3 hours or at 37°C (for internalization) for 4 hours followed by relative quantitative real-time PCR analysis for KSHV copy numbers. (B) Binding and internalization assay of KSHV in 293T cells. 293T cells transfected with expression plasmids for EPHA2, NRP1, or TGFBR1/2 were infected with KSHV at 4°C (for binding) for 3 hours or at 37°C (for internalization) for 4 hours followed by relative quantitative real-time PCR analysis for KSHV copy numbers. (C) Cell-based KSHV fusion assay. 293T cells transfected with expression plasmids for pT7-EMCLuc, pRL-SV40, and KSHV gB/gH/gL were cocultured with 293T cells cotransfected with expression plasmid for T7 polymerase and EPHA2, NRP1, or TGFBR1/2. The relative fusion activity was calculated as the ratio of firefly to Renilla luciferase activity 24 hours after coculture, with the empty vector (EV) controls set to 100%. (D) NRP1 colocalizes with KSHV. 293T cells transfected with PCR3.1-NRP1-myc plasmid and PDLSC were infected with biotin-labeled KSHV followed by immunofluorescence analysis. The NRP1 protein was stained using an anti–c-myc antibody (for 293T cells) or anti-NRP1 antibody (for PDLSC). KSHV were stained by streptavidin-Cy3. The colocalization profile on the right was analyzed by ImageJ software with the plot profile analysis function, which describe the intensity change of the fluorescence signal from left to right in the “Enlarge” image. Results are representative of two or three independent experiments. Data are means ± SEM. One-way ANOVA was carried out with Tukey’s correction for multiple comparisons. *P < 0.05; ***P < 0.001; ****P < 0.0001.
To ascertain NRP1-mediated KSHV infection of MSCs, we examined the localization of KSHV and NRP1 in PDLSC and 293T cells during infection. To visualize KSHV virions, we first labeled them with biotin. Labeled virions were then incubated with cells at 4°C for 2 hours plus 37°C for 15 min, and lastly, the location of the biotinylated KSHV virions was indicated with a Streptavidin-Cy3 dye. Confocal microscopy imaging revealed that KSHV colocalized with NRP1 in the cytoplasm (Fig. 3D), which further consolidated the function of NRP1 as a receptor mediating KSHV infection.
NRP1 interacts with KSHV envelope glycoprotein B.
As NRP1 has been reported to play a role in EBV entry into epithelial cells through binding to EBV envelope glycoprotein B(gB) (27), which shares structural and functional similarities with KSHV gB. We therefore investigated whether NRP1 promoted KSHV entry by interacting with KSHV gB. Coimmunoprecipitation assay using the cell lysate from 293T cells cotransfected with the expression vectors for Glutathione S-transferase (GST)-NRP1, and HA-gB showed that the anti–hemagglutinin (HA) agarose beads could precipitated not only HA-gB but also GST-NRP1 from the cell lysate (Fig. 4A). Similarly, the glutathione sepharose beads brought down both gB and NRP1 (Fig. 4B). Moreover, KSHV gB could also interact with endogenous NRP1 proteins in MSCs (fig. S5A). These results indicated that there was a physical interaction between NRP1 and KSHV gB.
Fig. 4. NRP1 interacts with KSHV gB.
(A and B) Coimmunoprecipitation of NRP1 and KSHV gB. Cell lysates from 293T cells cotransfected with the indicated plasmids for 24 hours were immunoprecipitated (IP) with the anti-HA agarose beads (A) or glutathione sepharose beads (B), followed by immunoblotting (IB) analysis for the complex with the indicated antibody. (C) NRP1 pulls down KSHV gB. Cell lysates from 293T cells transfected with pCDNA3.1-HA-gB were incubated with the NRP1 sepharose beads or SA beads at 4°C overnight. After washed five times with wash buffer, the complex remaining on beads was analyzed by IB with the indicated antibody. The NRP1 protein was stained by coomassie brilliant blue. (D) BLI assay. Biotinylated NRP1 proteins were captured onto streptavidin (SA) biosensors and assayed for binding to KSHV gB at the indicated concentrations. The data collected were processed on the Octet Analysis Studio software. The 1:1 fitting model was used to generate the fitting curves and calculate the kinetic parameters. (E) Coimmunoprecipitation of NRP1 and KSHV gB mutants. Cell lysates prepared from 293T cells cotransfected with PEBG-GST-NRP1 and a series of KSHV gB mutants for 24 hours were IP with the anti-HA nanobody agarose beads followed by IB analysis for the complex with the indicated antibodies. (F) Schematic summary of NRP1 and KSHV gB interactions. (G) Soluble NRP1 protein blocks KSHV infection. PDLSC were infected with KSHV that had been preincubated with purified soluble NRP1 protein for 1 hour at 37°C followed by flow cytometry analysis for the KSHV infection efficiency. Bovine serum albumin (BSA; 250 μg/ml) and phosphate-buffered saline (PBS) (CTR) were used as a negative control. Results are representative of two or three independent experiments. Data are means ± SEM. One-way ANOVA was carried out with Tukey’s correction for multiple comparisons. *P < 0.05; **P < 0.01.
To further clarify the gB-NRP1 interaction, we performed an in vitro pull-down assay. We purified the extracellular domain of NRP1 protein and coupled it to streptavidin agarose beads (SA beads) after biotin labeling. Then, the NRP1 beads were incubated with lysates from 293T cells transfected with KSHV gB expression plasmid. Western blotting results showed that NRP1 beads successfully captured gB protein (Fig. 4C). In addition, the kinetics of gB-NRP1 interaction was assayed using biolayer interferometry (BLI), and the results showed that NRP1 bound to the full-length KSHV gB at an affinity of 2.65 × 10−7 nM (Fig. 4D). Therefore, NRP1 can directly bind to KSHV gB.
EBV gB interacts with NRP1 via its N terminus and the CendR motif (27). KSHV gB and EBV gB have high homology in their amino acid sequence, and there are also potential CendR motifs in the KSHV counterpart. Some sites (such as aa116, aa439, and aa440) within the KSHV gB protein sequence conform to R/KXXR/K sequence, where R stands for arginine, K stands for lysine, and X stands for any amino acid (fig. S5B). In vivo, these sites have the potential to be cleaved by furin protease so that R/KXXR/K is exposed at the end of the C terminus, forming the CendR motif (29). To investigate whether these motifs contribute to the interaction between KSHV gB and NRP1 in vitro as it does in EBV, we constructed KSHV gB truncations to expose these CendR motifs. We found that the C terminus of KSHV gB did not participate in the interaction, as gB440-732 failed to interact with NRP1, whereas the truncations exposed to the potential CendR motif, namely, gB27-440, gB27-439, and gB27-116, can interact with NRP1, indicating that the motif exposure was important for gB to bind to NRP1. However, the truncations gB27-435 without exposed CendR motif still retains some ability to bind NRP1 (Fig. 4, E and F), suggesting that, like EBV gB, other site besides the CendR motif was also involved in the binding of KSHV gB to NRP1.
To determine whether the interaction of gB with NRP1 mediated viral entry into MSCs, we infected PDLSC using KSHV preincubated with soluble NRP1 protein for 1 hour and found that soluble NRP1 protein blocked the infection of KSHV in a concentration-dependent manner (Fig. 4G). These results indicate that the interaction between NRP1 and gB is an important step for KSHV infection and suggest that NRP1 is a functional receptor of KSHV entry into MSCs.
KSHV activates TGFBR1/2 complex through interacting between the cytoplasmic domains of NRP1 and TGFBR2
TGFBR2 is a transmembrane receptor that forms a heteromeric protein kinase receptor complex with TGFBR1 to mediate TGF-β signaling (30). Knockout of TGFBR2 or TGFBR1 reduced KSHV infection (Fig. 2C and fig. S6, A and B), especially internalization (Fig. 3A). We therefore hypothesized that TGFBR1/2 complex played a role in KSHV internalization and examined whether TGFBR1/2 interacted with KSHV glycoproteins or NRP1. Results of coimmunoprecipitation assays showed that TGFBR1/2 did not bind KSHV gB, gH/L, or K8.1 (Fig. 5, A to C). However, TGFBR2 interacted with NRP1 through their respective cytoplasmic domains (Fig. 5D), which is consistent with previous reports that NRP1 can bind to TGFBR2 and activates TGF-β signal transduction (31, 32).
Fig. 5. KSHV activates TGFBR1/2 through NRP1 cytoplasmic domain.
(A and B) Interaction analysis of TGFBR1/2 complex with KSHV gH/gL or gB. The cell lysates from 293T cells cotransfected with the indicated plasmids were IP with the anti-HA agarose beads followed by IB analysis with the indicated antibodies. (C) Interaction analysis of TGFBR1/2 complex with KSHV glycoprotein K8.1. Cell lysates from 293T cells cotransfected with expression plasmids for TGFBR1/2 and K8.1 were IP with the anti-flag M2 affinity agarose gel followed by IB analysis with the indicated antibodies. (D) Interaction analysis of NRP1 with TGFBR1 or TGFBR2. Cell lysates from 293T cells cotransfected with expression plasmids for cytoplasmic domains of NRP1 and TGFBR1 or TGFBR2 were IP with the glutathione sepharose beads followed by IB analysis with the indicated antibodies. (E) KSHV infection activates SMAD2 which was blocked by inhibitors. PDLSC prestarved for 24 hours were infected with KSHV for different lengths of time followed by IB analysis with the indicated antibodies (left). PDLSC prestarved and pretreated with SB431542 (20 μM) or LY2109761 (10 μM) for 24 hours were mock or infected with KSHV for 30 min followed by IB analysis with the indicated antibodies (right). (F) Knockout of NRP1 impedes KSHV from activating SMAD2. PDLSC with NRP1 knockout or PDLSC with NRP1 re-expression were mock infected or infected with KSHV for 30 min followed by IB analysis with the indicated antibodies. (G) Overexpression of NRP1 activates SMAD2. 293T cells transfected with an expression plasmid for NRP1 or empty vector were treated with dimethyl sulfoxide (DMSO) or 20 μM SB431542 for 24 hours followed by IB analysis with the indicated antibodies. (H) Overexpression of NRP1ΔCTD fails to activate SMAD2. 293T cells were transfected with indicated expression plasmids for 24 hours followed by IB analysis with the indicated antibodies. Results are representative of two independent experiments.
We next investigated whether the signal transduction function of TGFBR1/2 complex was involved in NRP1-mediated KSHV infection. We found that KSHV infection of PDLSC activated TGFBR1/2, revealed by the phosphorylation of Mothers against decapentaplegic homolog (SMAD2) (33), in the early stage of infection (Fig. 5E). The KSHV-mediated TGFBR1/2 activation and SMAD2 phosphorylation were completely blocked by specific inhibitors of TGFBR1 (SB431542) and TGFBR2 (LY2109761) (Fig. 5E). Furthermore, KSHV-mediated TGFBR1/2 activation was NRP1 dependent as the SMAD2 phosphorylation was completely blocked in PDLSC where NRP1 was knocked out and that SMAD2 phosphorylation was rescued with NRP1 re-expression (Fig. 5F). Consistently, overexpression of the full-length NRP1 substantially up-regulated the phosphorylation of SMAD2, which was blocked by SB431542 (Fig. 5G). However, overexpression of the NRP1 mutant lacking the cytoplasmic domain (namely, NRP1 ΔCTD) completely abolished the function of NRP1 in inducing the phosphorylation of SMAD2 (Fig. 5H). Together, these data indicate that KSHV can activate TGFBR1/2 during infection, which relies on the interaction between the cytoplasmic domain of NRP1 and TGFBR2.
Activation of TGFBR1/2 promotes KSHV entry by enhancing macropinocytosis-mediated viral internalization
We then investigated the importance of NRP1-mediated TGFBR1/2 activation in KSHV infection. To address this question, we first examined the effect of deletion of the cytoplasmic domain from NRP1 on KSHV entry into MSCs. Results showed that the full-length NRP1 activated TGFBR1/2 and promoted viral infection, whereas NRP1ΔCTD neither activated TGFBR1/2 nor promoted KSHV infection (Figs. 5H and 6A). Further analysis showed that NRP1ΔCTD still promoted KSHV binding in the same way as the full-length NRP1 but could not promote KSHV internalization (Fig. 6B). These results suggest that the extracellular domain of NRP1 is responsible for recognizing KSHV to facilitate viral adsorption, while the intracellular domain promotes viral entry by activating TGFBR1/2.
Fig. 6. Activation of TGFBR1/2 promotes KSHV entry by enhancing macropinocytosis.
(A and B) Overexpression of NRP1ΔCTD promotes KSHV binding but not KSHV internalization and infection. 293T cells transfected with PCR3.1-NRP1, PCR3.1-NRP1ΔCTD, or empty vector were used for KSHV binding and internalization assay (B) or infected with KSHV for 24 hours followed by flow cytometry analysis (A). (C) TGF-β1 promotes KSHV infection. PDLSC prestarved for 24 hours were infected with KSHV and immediately treated with TGF-β1. The infection efficiency was examined 24 hours after infection by flow cytometry. (D and E) Inhibitors of TGFBR1/2 reduce KSHV infection. PDLSC pretreated with LY2109761 (D) or SB431542 (E) for 24 hours were infected with KSHV for 24 hours, followed by flow cytometry analysis. (F) MβCD and EIPA inhibit KSHV entry. PDLSC pretreated with indicated inhibitors were examined for KSHV internalization. (G) KSHV colocalizes with dextran. PDLSC infected with biotin-labeled KSHV at 4°C for 1 hour were treated with FITC-dextran (1 mg/ml) and transferred to 37°C for another 30 min, followed by immunofluorescence analysis. The colocalization profile on the right was analyzed by ImageJ software with the plot profile analysis function, which describes the intensity change of the fluorescence signal from left to right in the “Enlarge” image. (H) TGF-β enhances macropinocytosis. The procedure of this experiment was described in Results. (I) EIPA blocks the effect of TGF-β1 in promoting KSHV infection. PDLSC prestarved for 16 horus were treated with 20 μM EIPA or DMSO for 45 min, and then the cells were infected with KSHV and treated simultaneously with TGF-β1(100 ng/ml). The KSHV infection efficiency was examined 24 hours after infection by flow cytometry. Results are representative of two or three independent experiments. Data are means ± SEM. One-way ANOVA was carried out with Tukey’s correction for multiple comparisons. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To directly evaluate the effect of TGFBR1/2 activation on KSHV infection, we treated PDLSC with TGF-β1 and found that the treatment significantly stimulated KSHV infection in a dose-dependent manner (Fig. 6C). Consistently, pretreatment of PDLSC with LY2109761 (Fig. 6D) or SB431542 (Fig. 6E) significantly inhibited KSHV infection. Since activation of TGFBR1/2 complex triggers intricate downstream signal transduction, including PI3K-AKT pathway, Rho-(like) guanosine triphosphatase (GTPase) pathway, and ERK/MAP kinase pathway, which may affect KSHV infection at different stages (34–36). We then examined which stage TGFBR1/2 activation is involved in. The internalization assay results showed that TGF-β1 treatment increased the number of KSHV internalization, whereas pretreatment of a TGFBR1 or TGFBR2 inhibitors reduced KSHV internalization (fig. S6C). However, neither TGF-β1 (fig. S6D) nor inhibitor (fig. S6E) treatment after KSHV that had been internalized to cells could affect KSHV infection. Therefore, we conclude that the activation of TGFBR1/2 is critical for KSHV entry rather than for viral gene expression at the postentry stage.
KSHV entry can be mediated by endocytosis or macropinocytosis, depending on the cell type (37–40). Given the paucity of studies on KSHV infection of MSCs, we decided to explore how KSHV enters MSCs. PDLSC were treated with the inhibitor of macropinocytosis (Ethylisopropylamiloride, EIPA), lipid raft [methyl-β-cyclodextrin (MβCD)], or clathrin-mediated endocytosis [chlorpromazine hydrochloride (CPZ)] at the indicated concentration for 45 min and then infected with KSHV for 4 hours followed by relative qRT-PCR analysis for KSHV entry. The results showed that MβCD and EIPA could significantly reduce the KSHV copy number, whereas CPZ had no obvious effect (Fig. 6F), indicating that KSHV entry into MSCs was mediated by lipid raft–dependent macropinocytosis. This conclusion was further supported by the fact that KSHV colocalized with the macropinocytosis marker (dextran) during infection (Fig. 6G). TGF-β was reported to promote macropinocytosis in tumor cells to nonselectively ingest and digest macromolecules and thereby facilitates tumor cells survive and grow in a nutrient-poor tumor microenvironment (41). Therefore, we hypothesized that TGFBR1/2-transduced TGF-β signaling may promote KSHV infection of MSCs by enhancing macropinocytosis-mediated KSHV entry. To test this hypothesis, we conducted two experiments. First, PDLSC pretreated with 20 μM EIPA for 45 min or 20 μM SB431542 for 12 hours were treated with fluorescein isothiocyanate (FITC)–dextran (0.1 mg/ml) for 30 min followed by immunofluorescence analysis. Next, PDLSC prestarved for 16 hours were treated with 20 μM EIPA or phosphate-buffered saline (PBS) for 45 min and then treated simultaneously with FITC-dextran (0.1 mg/ml) and TGF-β1(100 ng/ml) for another 30 min followed by immunofluorescence analysis. The results showed that TGF-β1 could distinctly promote whereas EIPA and SB431542 could distinctly inhibit dextran uptake by MSCs and that TGF-β1 lost its function to promote dextran uptake in the presence of EIPA (Fig. 6H and fig. S6F). Consistently, EIPA was also able to block the function of TGF-β1 in promoting KSHV infection (Fig. 6I). Therefore, these results suggest that TGFBR1/2-transduced TGF-β signaling promotes KSHV entry into cells by enhancing the macropinocytosis level of MSCs.
The small GTPases Cdc42/Rac1 of TGF-β signaling downstream effectors contributes to KSHV entry
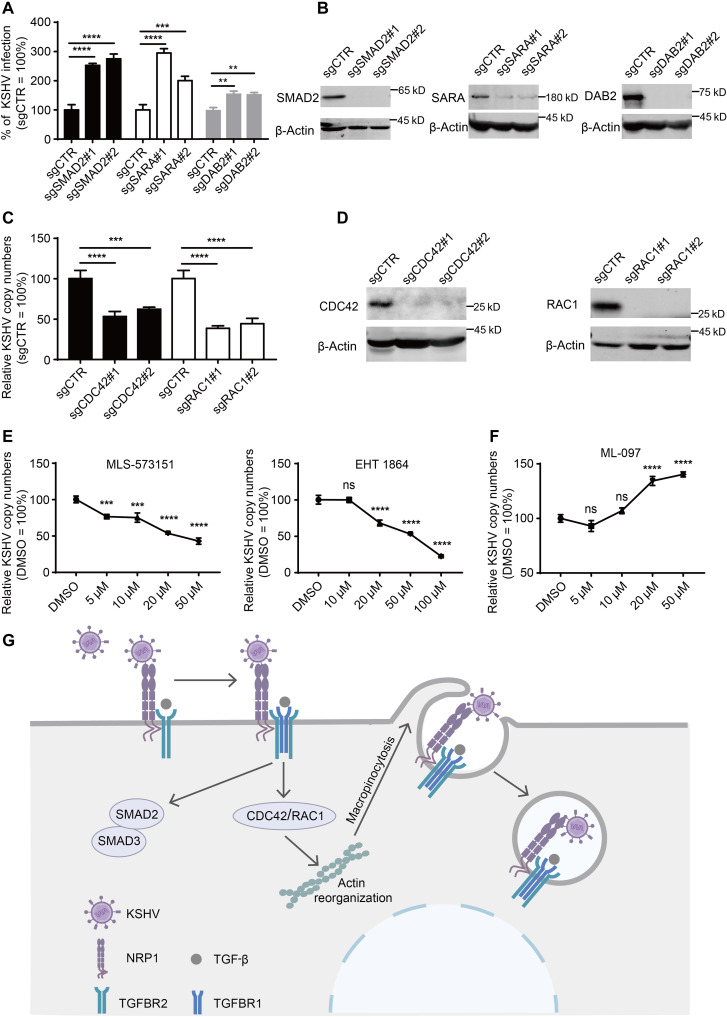
To further explore how TGF-β signaling promotes macropinocytosis and KSHV internalization, we decided to analyze the effect of TGF-β downstream signalings on KSHV infection. Upon activation, the TGFBR1/2 complex can transmit signals to intracellular SMAD proteins to activate the SMAD signaling pathway, and this process requires the cooperation of a collection of adaptor proteins such as Smad anchor for receptor activation (SARA) and Disabled homolog 2 (DAB2) (42). Unexpectedly, knockout of SMAD2, SARA, and DAB2 expression in PDLSC showed that KSHV infection efficiency was increased rather than decreased (Fig. 7, A and B), and re-expression of SMAD2, SARA, and DAB2 could rescue increased KSHV infection (fig. S7A), indicating that the SMAD signaling pathway does not contribute to KSHV infection. In addition to the canonical SMAD signaling, TGFBR1/2 complex can also activate the non-SMAD signaling pathways via other signal transducers. In fibroblasts, TGF-β can activate PAK2 via Rac1 and Cdc42 GTPases, to promote fibroblast morphological transformation. In epithelial cells, TGF-β rapidly induces activation of RhoA and Cdc42 GTPases, resulting in rapid actin reorganization and membrane ruffling (42), which are essential for macropinocytosis. Furthermore, the GTPases Cdc42 and Rac1 are thought to be involved in the initiation of macropinocytosis (43). We therefore examined the effect of Cdc42 and Rac1 on KSHV infection. The results showed that knockout of CDC42 and RAC1 expression in PDLSC result in a significant decrease in the internalization of KSHV (Fig. 7, C and D), which was rescued by re-expression of CDC42 and RAC1 (fig. S7B). Moreover, both the selective GTPase Cdc42 inhibitor (MLS-573151) and the inhibitor of Rac family small GTPases (EHT 1864) significantly inhibited KSHV internalization in a dose-dependent manner (Fig. 7E). Consistently, the pan Ras-related GTPase activator that can activate Rac1, Cdc42, Ras, and Rab7 can also promote KSHV internalization (Fig. 7F). Together, these results suggest that TGF-β signaling promotes macropinocytosis-mediated KSHV internalization through the downstream GTPases Cdc42 and Rac1.
Fig. 7. The small GTPases CDC42/RAC1 of TGF-β signaling downstream contributes to KSHV entry.
(A and B) Knockout of SMAD2, SARA, or DAB2 does not reduce KSHV infection. PDLSC transduced with sgRNA targeting SMAD2, SARA, or DAB2 by lentivirus were analyzed for the protein expression by Western blotting (B) or for the KSHV infection efficiency by flow cytometry (A). (C and D) Knockout of CDC42 or RAC1 can reduce KSHV internalization. PDLSC transduced with sgRNA targeting CDC42 or RAC1 were analyzed for the protein expression by western blotting (D) or infected with KSHV at 37°C for 4 hours followed by relative quantitative real-time PCR analysis for KSHV copy numbers (C). (E) Inhibitor of Cdc42 or Rac1 inhibits KSHV internalization. PDLSC pretreated with MLS-573151 or EHT1864 for 15 min were infected with KSHV at 37°C for 4 hours followed by relative quantitative real-time PCR analysis for KSHV copy numbers. (F) Activator of Cdc42/Rac1 promotes KSHV internalization. PDLSC prestarved for 12 hours were treated with ML-097 for 15 min, and then the cells were infected with KSHV at 37°C for 4 hours followed by relative quantitative real-time PCR analysis for KSHV copy numbers. (G) Model of KSHV entry into MSCs mediated by NRP1 and TGFBR1/2. Results are representative of three independent experiments. Data are means ± SEM. One-way ANOVA was carried out with Tukey’s correction for multiple comparisons. **P < 0.01; ***P < 0.001; ****P < 0.0001.
DISCUSSION
KSHV is a human herpesvirus that infects a variety of target cells including endothelial cells, epithelial cells, fibroblasts, monocytes, macrophages, and MSCs (24, 44, 45). The receptors for KSHV entry into endothelial, epithelial, and mononuclear cells and macrophages, such as EPHA2, integrins, DC-SIGN, and xCT, as well as related entry mechanisms, have been intensively investigated [reviewed in (14, 15)]. In contrast, the receptors for KSHV infection of MSCs had been largely ignored because its pathogenetic significance was not clear until recently when increasing evidence emerged showing that KSHV-infected MSCs may be the origin or progenitor of KS spindle cells (7, 8, 10, 46) and that pediatric osteosarcoma, a MSC-originated malignancy, is reported to be associated with KSHV infection (4). Although numerous KSHV receptors have been identified, the expression of these receptors in MSCs is very low (fig. S3C). Therefore, elucidation of the mechanism governing KSHV entry into MSCs has become a very important task for understanding the pathogenesis of KSHV-associated malignancies. Here, we identify NRP1 as a primary receptor for KSHV to infect MSCs and show that NRP1 takes advantage of TGFBR1/2-transduced signaling to promote macropinocytosis to support KSHV entry into cells. Although NRP1 mediated EBV infection of nasopharyngeal epithelial cells by activating EGFR signaling (27), we found that NRP1 promoted KSHV entry into MSCs by activating the TGFBR1/2 signaling pathway, which reflects the signaling adaptation of NRP1 as a viral receptor according to different viruses and cell types.
NRP1 is a transmembrane receptor involved in angiogenesis, regulation of vascular permeability, and the development of the nervous system. NRP1 recognizes the CendR motif R/KXXR/K on its ligands to cause cellular internalization and vascular leakage (29). Several viruses, such as human T cell lymphotropic virus type 1 (HTLV-1) (47), Epstein-Barr virus (EBV) (27), and severe acute respiratory syndrome coronavirus 2 (48, 49) bind to NRP1 through their CendR motif to enhance their infection. KSHV gB has potential furin cleavage sites, and cleavage of these sites would result in CendR motif exposure. Although our results suggest that the CendR motif exposure enhances the interaction of KSHV gB with NRP1, it is not clear how critical the CendR motif is for KSHV infection under physiological conditions. Both full-length and cleaved forms of gB can be detected on purified virions obtained from induced BCBL-1 cells (50), and full-length KSHV gB can also bind to NRP1 (Fig. 4D), suggesting that gB cleavage may not be always necessary for it to function (51). In addition, we found that NRP1 promotes KSHV infection by enhancing TGFBR1/2 signaling–mediated macropinocytosis. However, studies have shown that NRP1-mediated CendR endocytosis is mechanically different from macropinocytosis (52), again suggesting that the potential CendR motif in KSHV gB may not be involved in KSHV infection. This is likely to be why NRP1 acts as a shared receptor for KSHV and EBV, but the specific mechanism is different.
In this study, we find that NRP1 plays roles in KSHV infection by enhancing viral adherence and promoting internalization but contributes little to membrane fusion (Fig. 3, A to C). Although reports indicate that KSHV binding involves the interaction between KSHV gB and cell surface heparan sulfate moieties (53), it is possible that the interaction of gB with NRP1 adds an additional adhesion of viruses to target cells. This notion is supported by previous observations that NRP1 contributes to the binding of HTLV-1 surface (SU) subunit and virions to target cells (47, 54) and that soluble NRP1 and anti-NRP1–neutralizing antibody were reported to diminish murine cytomegalovirus attachment to the cell membrane surface (55). EPHA2 is recognized as a key receptor for KSHV infection of endothelial cells. EPHA2 is expressed highly in endothelial cells but only at a low level in MSCs, whereas, in contrast, NRP1 is expressed highly in MSCs but not in endothelial cells (Fig. 2E). The expression of NRP1 is also higher than those of other reported KSHV receptors in MSCs (fig. S3C). Therefore, NRP1 is likely the primary functional receptor for KSHV infection of MSCs.
NRP1 also promotes KSHV internalization. During KSHV infection, the virus binds to the extracellular domain of NRP1 via gB, amplifying the TGFBR1/2-transduced TGF-β signaling through the interaction between cytoplasmic domains of NRP1 and TGFBR2. The signal is transmitted down to intracellular small GTPases Cdc42 and Rac1, resulting in rapid actin reorganization and plasma membrane ruffling which wrap around virus particles and form macropinosomes for intracellular release (Fig. 7G). Macropinocytosis is a transient, growth factor–induced, actin-dependent endocytic process that leads to internalization of fluid and membrane into large vacuoles (56). Macropinosomes are also an entry site for intracellular pathogens (e.g., viruses, bacteria, and prions) and a source of metabolites for proliferating cancer cells (43). A collection of viruses has acquired the ability to stimulate macropinocytosis indiscriminately by hijacking a battery of receptors (57). Our findings suggest that KSHV has also evolved strategies to hijacks NRP1 and TGFBRs to enhance macropinocytosis, thereby facilitating viral entry into cells.
TGF-β is versatile and its function is highly location- and context-dependent. TGF-β signaling usually plays an immunomodulatory role in immune cells, whereas in the field of tumor research, it is generally accepted that TGF-β signaling has a tumor-suppressive role in normal cells but tumor-promoting and tumor metastasis role in malignant cells (58–60). It is worth noting that both TGF-β ligand and TGF-β receptors are widely expressed in KS tissues (61) and were found to be up-regulated in MSC-originated osteosarcomas (4), suggesting that TGF-β signaling in these tumors may be active. Moreover, in osteosarcoma, TGF-β have the potential to influence the sarcoma’s aggressive clinical behavior (62) and is associated with disease progression (63). Therefore, the TGF-β signaling activated during KSHV infection can not only facilitate viral infection but may also be involved in the pathogenesis of KSHV-related malignancies. Intervention targeting TGF-β signals may be of great significance for the prevention and treatment of these malignancies. TGF-β has also been reported to promote viral replication (64, 65) or increase HSV-1 latency and reactivation (66) but has not been implicated in the early stages of viral entry. Therefore, in this study we report that TGF-β signaling can promote virus entry into cells at the early stage of infection and to elucidate the mechanism.
In conclusion, this study identifies NRP1 as the primary receptor for KSHV infection of MSCs and explores the mechanism that NRP1 mediated KSHV entry by taking advantage of TGFBR1/2-transduced signaling to promote macropinocytosis. This study identifies the receptor that mediates KSHV infection of MSCs and extends the current understanding of the mechanisms of KSHV infection. How NRP1 activates TGFBR1/2 kinase complex and whether EphA2 is involved in NRP1-TGFBR1/2–mediated KSHV infection warrant further investigation and will facilitate a better understanding of the process of KSHV infection, paving the way for the design of efficient therapeutic strategies against KSHV infection and related diseases.
MATERIALS AND METHODS
Ethical guidelines
The human sample collection and the use of MSCs in our research were approved by the Medical Ethics Review Board of Sun Yat-sen University (approval no. 2015-028) and Shihezi University (IIT-2017-004-01 to Y.Z.).
Cell culture
PDLSC, GMSC, and DPSC were isolated as described previously (7) and cultured in complete α-minimal essential medium (Gibco Life Technologies) containing 10% fetal bovine serum (FBS; Gibco) and penicillin-streptomycin solution (C3420-0100, VivaCell, Shanghai, China). The colony-forming unit assessment assay, multiple differentiation ability assays, tissue origin authentication assay, and surface marker identification assay were performed to characterize PDLSC, DPSC, and GMSC, and the results were shown in the Supplementary Materials (fig. S1). The iSLK.219 cells (provided by K. Lan of Wuhan University) and 293T cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% FBS. Human umbilical cord endothelial cells were purchased from the China Center for Type Culture Collection (China) and cultured in endothelial cell growth medium 2 BulletKit (ScienCell). Cells were grown in a humidified 5% CO2 incubator at 37°C.
Plasmids
To construct stable expression vectors, the full-length complementary DNA (cDNA) sequences of NRP1, EPHA2, TGFBR1, and TGFBR2 were PCR-amplified and then integrated into the pMSCV-puro vector. To construct transient expression vectors, the full-length cDNA sequences of NRP1, EPHA2, and TGFBR2 were PCR-amplified and then integrated into the PCR3.1 vector with a myc tag in C terminus. The full-length cDNA of KSHV K8.1 and the truncated form of NRP1 lacking an intracellular segment were integrated into the PCR3.1 vector with a 3× flag tag in C terminus. The full-length cDNA of KSHV gH and gB were PCR-amplified and then integrated into the pCDNA3.1(+) vector with a 3× HA tag in C terminus. For coimmunoprecipitation, those truncated forms of gB were PCR-amplified and then integrated into the pCDNA3.1(+) vector with a 3× HA tag in N terminus. The full-length cDNA of gL was integrated into the pEGFP-C2 vector with a green fluorescent protein (GFP) tag in N terminus. The truncated form of NRP1 was integrated into the pEBG-GST vector a GST tag in N terminus. The cytoplasm domain of TGFBR1 or TGFBR2 was integrated into the PCR3.1 vector with a 3× flag tag in C terminus. As for the rescue plasmids, the full-length cDNAs of NRP1, EPHA2, TGFBR1, TGFBR2, SMAD2, SARA, DAB2, CDC42, and RAC1 were first PCR-amplified and integrated into the pMSCV-puro vector. These plasmids are then mutated at multiple points, synonymously mutating the sequences targeted by the respective sgRNA. The sequences with synonymous mutations have been listed in table. S1.
Reagents and antibodies
The reagents and antibodies used were as follows: recombinant human TGF-β1 protein (240-B, R&D Systems), SB431542 (HY-10431, MedChemExpress), LY2109761(T2123, TargetMol), MβCD (C4555-1G, Sigma-Aldrich, EIPA (E884358-5mg, Macklin), CPZ (C834105-5g, Macklin), FITC-dextran,MW:70K (R-FD-005, Ruixibio), ML-097(HY-118208, MedChemExpress), MLS-573151(HY-113849, MedChemExpress), EHT 1864 (HY-16659, MedChemExpress), anti-NRP1 antibody (ab81321, Abcam), anti-EphA2 antibody (6997, Cell Signaling Technology), anti-GST antibody (ET1611-47, HuaBio), anti-HA nanobody agarose beads (KTSM1305, Alpalife), glutathione sepharose beads (17-0756-01, Cytiva), anti-flag M2 affinity agarose gel (A2220, Sigma-Aldrich), anti–HA-tag mouse monoclonal antibody (M180-3, MBL Beijing Biotech Co. Ltd.), Coomassie Brilliant blue R-250 (821616, MP Biomedicals), anti–flag-tag mouse monoclonal antibody (M185-3L, MBL Beijing Biotech Co. Ltd.), anti-Smad2 antibody (5339T, Cell Signaling Technology), anti–phospho-Smad2 antibody (3108T, Cell Signaling Technology), anti–Myc-tag rabbit monoclonal antibody for Western blot (2278, Cell Signaling Technology), anti–c-myc mouse monoclonal antibody for immunofluorescence (sc-40, Santa Cruz Biotechnology Inc.), anti-TGFBR1 rabbit polyclonal antibody (A16983, ABclonal), anti-TGFBR2 rabbit polyclonal antibody (A1415, ABclonal), anti-CDC42 antibody (10155-1-AP, Proteintech), anti-RAC1 antibody (2465s, Cell Signaling Technology), anti-SARA antibody (22033-1-AP, Proteintech), and anti-DAB2 antibody (10109-2-AP, Proteintech).
Bioinformatics
RNA-seq data of MSCs were obtained in our previous publication (7). RNA-seq data of BJAB cells were from GEO database [GSE82184; (67)]. RNA-seq data of KS2 were from GEO database [GSE100684; (23)]. RNA-seq data of KS1 were obtained by RNA-seq in this study, and the sequencing method was consistent with that described previously (7). Briefly, two KS tissue samples were cut and lysed with TRIzol reagent, and total RNA was extracted following the manufacturer’s manual. RNA concentration was checked using Qubit 3.0 Fluorometer (Life Technologies). For each sample, 2 μg of RNA was used to generate sequencing libraries with the NEBNext Ultra RNA Library Prep Kit for Illumina (#E7530L, NEW ENGLAND BioLabs), referring to the manufacturer’s recommendations. Index codes were added to attribute sequences to each sample. The libraries were sequenced on an Illumina Hiseq 2500 platform, and 150-bp paired-end reads were generated.
The raw data of KS and MSCs were first quality checked and filtered by fastp to obtain the clean reads data with “--cut_front” and “--cut_tail” parameters. The clean reads data were aligned to human genome GRCh38 with -hisat2 and annotated with GENCODE gene annotation (Release 33). Gene expression profiling was exported in transcripts per million (TPM) and read counts by StringTie. The DEGs were screened by setting screening parameters (i.e., TPM of MSCs was over 10, TPM of KS was over 10, TPM of BJAB was less than 10, TPM ratio of MSCs to BJAB was greater than 10, and TPM ratio of KS to BJAB was greater than 10).
shRNA- and sgRNA-mediated gene knockdown/knockout
Target sequences of shRNA and sgRNA are listed in table. S1. For shRNA, oligonucleotides corresponding to the target sequences were synthesized and cloned into the Eco RI/Age I sites in PLKO.1 vector. For sgRNA, oligonucleotides corresponding to the target sequences were synthesized and cloned into Bsm BI site in Cas9-expressing plasmid lentiCRISPR-V2. The constructed shRNA or sgRNA expression vector was packaged into lentivirus and used to infect MSCs. After 48 hours of infection, cells were treated with puromycin (2 μg/ml) for 4 days to remove uninfected cells, and the obtained cells could be used for subsequent experiments. For rescue assay, the rescue plasmids (plasmid construction details have been described above) were transduced to the knockout PDLSC by lentivirus, and expression of individual proteins or KSHV infection was detected after 3 days.
KSHV preparation and infection
KSHV preparation was carried out as previously described (7). Briefly, iSLK.219 cells harboring rKSHV.219 were used for virus preparation. The cells were treated with doxycycline (1 mg/ml) and sodium butyrate (1 mM) and harvested 5 days after induction. The culture supernatants were first centrifuged at 10,000g for 15 min to remove cell debris and then centrifuged at 134,000g for 70 min. Pellet was resuspended in 1/100 volume of PBS and stored at −80°C until use. Virus infection was performed according to an established procedure. Briefly, MSCs (PDLSC, GMSC, and DPSC) were incubated with KSHV in the absence of polybrene. After centrifugation at 2500 rpm for 45 min in room temperature, the cells were incubated at 37°C with 5% CO2 for 4 hours. Then, the inoculum was removed by changing culture medium. For infection of 293T cells, cells were incubated with KSHV at 37°C with 5% CO2 for 4 hours. The inoculum was removed by changing culture medium. Twenty-four hours after infection, the KSHV infection efficiency was analyzed by flow cytometry by measuring the percentage of GFP-positive cells.
KSHV binding and internalization
The protocols for binding and internalization assay refer to previous publications (26, 27). Briefly, cells were seeded at cell culture plates for 24 hours. KSHV for binding and internalization assay was pretreated with deoxyribonuclease I (2270A, Takara) to remove the exposed KSHV genome. To measure virus binding, cells were washed with ice-cold PBS twice and then were incubated with KSHV for 3 hours at 4°C to allow cell surface binding. The cells were then washed three times with ice-cold PBS to remove unbound virus, followed by collection to extract genome DNA. To measure virus internalization, cells were washed with PBS twice and then were incubated with KSHV for 4 hours at 37°C to allow internalization and then washed three times with PBS to remove unbound virus. To remove the surface-bound virus, the cell was resuspended in 200 μl of trypsin-EDTA and proteinase K for 5 min, and the cell pellets were then washed three times with PBS. KSHV genome DNA was extracted from collected cells using a Hipure tissue DNA Mini Kit (Magen, D2121-03) as recommended by the manufacturer’s instructions. The copy number of KSHV bound to the cell surface or internalized into cells was measured using qPCR for detection of the ORF73 gene of the KSHV genome. qPCR for the glyceraldehyde phosphate dehydrogenase (GAPDH) DNA was used for cell counting estimation. The relative KSHV copy number was expressed as a ratio of the copy number of the KSHV genome to the GAPDH DNA.
Cell fusion assay
The virus-free, cell-based fusion assay was carried out as previously described (26, 27). Effector human embryonic kidney (HEK) 293T cells were transiently transfected with expression plasmid (pT7EMCLuc, pRL-SV40, KSHV gB, gH, and gL). Target cells (HEK-293T) were transfected with expression plasmid for pCAGT7 (expressing T7 RNA polymerase) together with EPHA2, NRP1, TGFBR1/2, or empty vector. Twenty-four hours later, effector cells (2.5 × 105 per 24-well plate) were cocultured with target cells (2.5 × 105 per 24-well plate) for 24 hours. To quantify fusion, the relative fusion activity, calculated as the ratio of firefly luciferase activity to Renilla luciferase activity, was measured by a GloMax 96 Microplate Luminometer with a dual-luciferase reporter assay system (E1960, Promega).
Protein expression and purification
For NRP1 construction, NRP1 (22-644) with N-terminal CD5 signal peptide and C-terminal 8*His Tag was cloned into mammalian expression vector pcDNA3.1(+). The plasmids were transfected to Expi293F cell lines with polyethylenimine (Polysciences, catalog no. 24765). The supernatant of medium was harvested after 6-day culture by centrifugation, filtered with 0.22-μm filter membrane, and applied to gravity column loaded with Ni Sepharose excel resin (Cytiva, catalog no. 17371201). The resin was washed with PBS with 30 mM imidazole and eluted with PBS with 300 mM imidazole. Then, concentrated elution was applied to a Superdex 200 Increase 10/300 GL column (Cytiva, catalog no. 28990944) for size exclusion chromatography (SEC). The peak fraction was pooled and stored at −80°C for further use. To purify the full-length KSHV gB, KSHV gB coding sequence (CDS) fused with 6*His Tag was cloned into mammalian expression vector PCR3.1-3*flag. As for purification, the full-length KSHV gB could be purified as membrane protein as previously reported (68). Briefly, the plasmid was transfected to Expi293F, and cell was harvested after 3-day culture by centrifugation and resuspended in PBS with 10% glycerol plus 1× protease inhibitor cocktail (Yaesen, catalog no. 20123ES10). Then, the suspension was lysed by pressure cracking at 850 MPa. The lysed suspension was further centrifuged for clarity at 2000g for 30 min. The membrane extraction was collected from the clarified supernant through ultracentrifuge at 120,000g for 2 hours and resuspended in PBS with 10% glycerol plus 1.5% n-dodecyl-β-d-maltopyranoside (DDM; Macklin, catalog no. D806359). The resuspended membrane extraction was filtered with 0.22-μm filter membrane and applied to gravity column loaded with Ni Sepharose excel resin as previously described. The resin was washed with PBS with 30 mM imidazole plus 5% glycerol and 0.1% DDM and eluted with PBS with 300 mM imidazole plus 5% glycerol and 0.1% DDM. The full-length KSHV gB was further purified from the elution through SEC.
Pull-down assay
For pulldown assay, NRP1 agarose beads were first prepared. Briefly, biotin (A39257, Thermo Fisher Scientific) was labeled to soluble NRP1 protein according to the instructions followed by enrichment of labeled protein with SA beads (16-126, Millipore). For the mechanism of biotin labeling, please refer to the reagent specification, which states that the biotin reagent is a biotin derivative with N-hydroxysuccinimide (NHS) esters. NHS-activated biotin reacts with primary amine group (−NH2) efficiently in alkaline buffer to form a stable amide bond. Proteins generally have several primary amines in the side chain of lysine (K) residues and the N terminus of each polypeptide that are available as targets for labeling with NHS-activated biotin reagents. The beads were washed three times with lysis buffer to remove unbound protein. After preparation of NRP1 beads, 293T cells transfected with KSHV gB were lysed in immunoprecipitation (IP) lysis buffer, and the lysate was collected and incubated with NRP1 beads for 6 hours at 4°C. After washing five times with lysis buffer to remove unbound proteins, the sample was suspended in 2× SDS sample buffer and boiled for 10 min at 95°C. The complex was then analyzed by Western blotting with the indicated antibodies.
Blocking assay
For the soluble NRP1 protein blocking assays, KSHV was preincubated with soluble NRP1 protein at indicated concentration or bovine serum albumin (BSA; as a negative control) at 37°C for 1 hour, and then the KSHV was used to infect PDLSC. The percentage of KSHV infected cells were determined by flow cytometry 24 hours after infection.
BLI assay
To determine the affinity between NRP1 and KSHV gB ex vivo, we performed BLI assay to determine the binding between these two proteins, as previously described (69). Briefly, the soluble NRP1 protein was biotinylated using the Sulfo-NHS-LC-LC-biotin kit (Thermo Fisher Scientific, catalog no. A35358), and kinetic buffer (KB) was prepared as PBS with 0.1% Tween 20. Then, the SA biosensors were incubated in the KB for 15 min, after which, biotinylated NRP1 at 5 μg/ml was captured on the sensors for 120 s, and full-length KSHV gB protein diluted from 4 μM to 500 nM was applied to the biosensors for 300-s association, followed by 600-s dissociation in KB. The data collected were processed on the Octet Analysis Studio software. The 1:1 fitting model was used to generate the fitting curves and calculate the kinetic parameters.
KSHV labeling with biotin
For fluorescent staining of KSHV, the virus particles were first labeled with 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethyleneglycol)-2000] [DSPE-PEG (2000)] biotin (Sigma-Aldrich, 880129P-10MG) to facilitate subsequent reactions with streptavidin-Cy3. Briefly, the DSPE-PEG (2000) biotin powder was dissolved in anhydrous ethanol to form a biotin storage solution (5 mg/ml). The biotin storage solution was then added to the concentrated KSHV virus to achieve a final concentration of biotin (0.1 mg/ml). After incubation at room temperature for 2 hours, the labeled KSHV and excess biotin were separated by illustra NAP Columns (Cytiva, 17085301) according to the manufacturer’s instructions. The labeled KSHV was stored at 4°C and used as soon as possible.
Immunofluorescence confocal microscopy
293T cells were seeded on coverslips in 48-well plates and then transfected with the PCR3.1-NRP1-myc plasmids for 24 hours. For PDLSC, cells were seeded on coverslips in 48-well plates without transfection. Both 293T and PDLSC cells were then infected with the labeled KSHV at 4°C for 2 hours and then transferred to 37°C for 15 min, followed by briefly washing with PBS twice, fixed with 4% paraformaldehyde in PBS for 15 min, and permeabilized with 0.1% Triton X-100 in PBS for 10 min. After blocking with 1% BSA in PBS, 293T cells were stained with an anti–c-myc mouse monoclonal antibody (Santa Cruz Biotechnology, sc-40), and PDLSC were stained with an anti-NRP1 rabbit monoclonal antibody (Abcam, ab81321) for 4°C overnight and washed with PBS three times, followed by incubation with Alexa Fluor 488–labeled goat antibody to mouse or rabbit IgG. After wash with PBS three times, cells were stained with streptavidin-Cy3 in PBS (1:1000 dilution; Invitrogen, 434315) at 4°C for 15 min, followed by nuclei staining with Hoechst 33342. After wash with PBS three times, cells were mounted with ProLong Diamond (Invitrogen, P36961). The confocal images were acquired using a Nikon Eclipse Ni-E confocal laser scanning microscope.
Acknowledgments
Funding: This work was supported by National Key R&D Program of China (2022YFC3400900), National Natural Science Foundation of China (82030046, 81830090, 82202510 and 81621004), Guangdong Science and Technology Department (2020B1212030004), and The Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2019BT02Y198).
Author contributions: Experimental design: Z.-Z.L. Experimental perform: Z.-Z.L., C.S., X.Z., Y.P., and N.Z. Provision of materials (MSCs and KS tissue samples): Y.W. and Y.Z. Project guidance: Y.Y. and M.-S.Z. Manuscript writing and revision: Z.-Z.L., Y.Y., and M.-S.Z.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data of KS1 have been deposited in the NCBI (National Center for Biotechnology Information) GEO database (GSE201321).
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S7
Legend for table S1
Other Supplementary Material for this manuscript includes the following:
Table S1
REFERENCES AND NOTES
- 1.P. S. Moore, Y. Chang, Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and those without HIV infection. N. Engl. J. Med. 332, 1181–1185 (1995). [DOI] [PubMed] [Google Scholar]
- 2.E. Cesarman, Y. Chang, P. S. Moore, J. W. Said, D. M. Knowles, Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332, 1186–1191 (1995). [DOI] [PubMed] [Google Scholar]
- 3.J. Soulier, L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, F. Sigaux, Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86, 1276–1280 (1995). [PubMed] [Google Scholar]
- 4.Q. Chen, J. Chen, Y. Li, D. Liu, Y. Zeng, Z. Tian, A. Yunus, Y. Yang, J. Lu, X. Song, Y. Yuan, Kaposi's sarcoma herpesvirus is associated with osteosarcoma in Xinjiang populations. Proc. Natl. Acad. Sci. U.S.A. 118, e2016653118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.L. Cancian, A. Hansen, C. Boshoff, Cellular origin of Kaposi's sarcoma and Kaposi's sarcoma-associated herpesvirus-induced cell reprogramming. Trends Cell Biol. 23, 421–432 (2013). [DOI] [PubMed] [Google Scholar]
- 6.S. Gurzu, D. Ciortea, T. Munteanu, I. Kezdi-Zaharia, I. Jung, Mesenchymal-to-endothelial transition in Kaposi sarcoma: A histogenetic hypothesis based on a case series and literature review. PLOS ONE 8, e71530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Y. Li, C. Zhong, D. Liu, W. Yu, W. Chen, Y. Wang, S. Shi, Y. Yuan, Evidence for Kaposi Sarcoma originating from mesenchymal stem cell through KSHV-induced mesenchymal-to-endothelial transition. Cancer Res. 78, 230–245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.W. Chen, Y. Ding, D. Liu, Z. Lu, Y. Wang, Y. Yuan, Kaposi's sarcoma-associated herpesvirus vFLIP promotes MEndT to generate hybrid M/E state for tumorigenesis. PLOS Pathog. 17, e1009600 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Y. Ding, W. Chen, Z. Lu, Y. Wang, Y. Yuan, Kaposi's sarcoma-associated herpesvirus promotes mesenchymal-to-endothelial transition by resolving the bivalent chromatin of PROX1 gene. PLOS Pathog. 17, e1009847 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.J. Naipauer, S. Rosario, S. Gupta, C. Premer, O. Méndez-Solís, M. Schlesinger, V. Ponzinibbio, V. Jain, L. Gay, R. Renne, H. L. Chan, L. Morey, D. Salyakina, M. Abba, S. Williams, J. M. Hare, P. J. Goldschmidt-Clermont, E. A. Mesri, PDGFRA defines the mesenchymal stem cell Kaposi's sarcoma progenitors by enabling KSHV oncogenesis in an angiogenic environment. PLOS Pathog. 15, e1008221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.C. H. Parsons, B. Szomju, D. H. Kedes, Susceptibility of human fetal mesenchymal stem cells to Kaposi sarcoma-associated herpesvirus. Blood 104, 2736–2738 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.M. S. Lee, H. Yuan, H. Jeon, Y. Zhu, S. Yoo, S. Shi, B. Krueger, R. Renne, C. Lu, J. U. Jung, S. J. Gao, Human mesenchymal stem cells of diverse origins support persistent infection with Kaposi's sarcoma-associated herpesvirus and manifest distinct angiogenic, invasive, and transforming phenotypes. MBio 7, e02109–e02115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.T. Jones, F. Ye, R. Bedolla, Y. Huang, J. Meng, L. Qian, H. Pan, F. Zhou, R. Moody, B. Wagner, M. Arar, S. J. Gao, Direct and efficient cellular transformation of primary rat mesenchymal precursor cells by KSHV. J. Clin. Investig. 122, 1076–1081 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.E. van der Meulen, M. Anderton, M. J. Blumenthal, G. Schäfer, Cellular receptors involved in KSHV infection. Viruses 13, 118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.S. J. Dollery, Towards understanding KSHV fusion and entry. Viruses 11, 1073 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.S. M. Akula, N. P. Pramod, F. Z. Wang, B. Chandran, Integrin α3β1 (CD 49c/29) is a cellular receptor for Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 108, 407–419 (2002). [DOI] [PubMed] [Google Scholar]
- 17.A. S. Hahn, J. K. Kaufmann, E. Wies, E. Naschberger, J. Panteleev-Ivlev, K. Schmidt, A. Holzer, M. Schmidt, J. Chen, S. König, A. Ensser, J. Myoung, N. H. Brockmeyer, M. Stürzl, B. Fleckenstein, F. Neipel, The ephrin receptor tyrosine kinase A2 is a cellular receptor for Kaposi's sarcoma-associated herpesvirus. Nat. Med. 18, 961–966 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.J. Chen, X. Zhang, S. Schaller, T. S. Jardetzky, R. Longnecker, Ephrin receptor A4 is a new Kaposi's sarcoma-associated herpesvirus virus entry receptor. MBio 10, e02892-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A. K. Großkopf, S. Schlagowski, B. F. Hörnich, T. Fricke, R. C. Desrosiers, A. S. Hahn, EphA7 functions as receptor on BJAB cells for cell-to-cell transmission of the Kaposi's sarcoma-associated herpesvirus and for cell-free infection by the related rhesus monkey rhadinovirus. J. Virol. 93, e00064-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.J. A. Kaleeba, E. A. Berger, Kaposi's sarcoma-associated herpesvirus fusion-entry receptor: Cystine transporter xCT. Science 311, 1921–1924 (2006). [DOI] [PubMed] [Google Scholar]
- 21.G. Rappocciolo, F. J. Jenkins, H. R. Hensler, P. Piazza, M. Jais, L. Borowski, S. C. Watkins, C. R. Rinaldo Jr., DC-SIGN is a receptor for human herpesvirus 8 on dendritic cells and macrophages. J. Immunol. 176, 1741–1749 (2006). [DOI] [PubMed] [Google Scholar]
- 22.J. Chen, K. Sathiyamoorthy, X. Zhang, S. Schaller, B. E. Perez White, T. S. Jardetzky, R. Longnecker, Ephrin receptor A2 is a functional entry receptor for Epstein-Barr virus. Nat. Microbiol. 3, 172–180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.F. Y. Tso, A. V. Kossenkov, S. J. Lidenge, O. Ngalamika, J. R. Ngowi, J. Mwaiselage, J. Wickramasinghe, E. H. Kwon, J. T. West, P. M. Lieberman, C. Wood, RNA-Seq of Kaposi's sarcoma reveals alterations in glucose and lipid metabolism. PLOS Pathog. 14, e1006844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R. Renne, D. Blackbourn, D. Whitby, J. Levy, D. Ganem, Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 72, 5182–5188 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D. Bu, H. Luo, P. Huo, Z. Wang, S. Zhang, Z. He, Y. Wu, L. Zhao, J. Liu, J. Guo, S. Fang, W. Cao, L. Yi, Y. Zhao, L. Kong, KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 49, W317–W325 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.H. Zhang, Y. Li, H. B. Wang, A. Zhang, M. L. Chen, Z. X. Fang, X. D. Dong, S. B. Li, Y. du, D. Xiong, J. Y. He, M. Z. Li, Y. M. Liu, A. J. Zhou, Q. Zhong, Y. X. Zeng, E. Kieff, Z. Zhang, B. E. Gewurz, B. Zhao, M. S. Zeng, Ephrin receptor A2 is an epithelial cell receptor for Epstein-Barr virus entry. Nat. Microbiol. 3, 1–8 (2018). [DOI] [PubMed] [Google Scholar]
- 27.H. B. Wang, H. Zhang, J. P. Zhang, Y. Li, B. Zhao, G. K. Feng, Y. du, D. Xiong, Q. Zhong, W. L. Liu, H. du, M. Z. Li, W. L. Huang, S. W. Tsao, L. Hutt-Fletcher, Y. X. Zeng, E. Kieff, M. S. Zeng, Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat. Commun. 6, 6240 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.J. Chen, S. Schaller, T. S. Jardetzky, R. Longnecker, Epstein-Barr Virus gH/gL and Kaposi's sarcoma-associated herpesvirus gH/gL bind to different sites on EphA2 to trigger fusion. J. Virol. 94, e01454-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.T. Teesalu, K. N. Sugahara, V. R. Kotamraju, E. Ruoslahti, C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. U.S.A. 106, 16157–16162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.J. L. Wrana, L. Attisano, J. Cárcamo, A. Zentella, J. Doody, M. Laiho, X. F. Wang, J. Massague, TGFβ signals through a heteromeric protein kinase receptor complex. Cell 71, 1003–1014 (1992). [DOI] [PubMed] [Google Scholar]
- 31.Y. Glinka, S. Stoilova, N. Mohammed, G. J. Prud'homme, Neuropilin-1 exerts co-receptor function for TGF-beta-1 on the membrane of cancer cells and enhances responses to both latent and active TGF-β. Carcinogenesis 32, 613–621 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Y. Cao, A. Szabolcs, S. K. Dutta, U. Yaqoob, K. Jagavelu, L. Wang, E. B. Leof, R. A. Urrutia, V. H. Shah, D. Mukhopadhyay, Neuropilin-1 mediates divergent R-Smad signaling and the myofibroblast phenotype. J. Biol. Chem. 285, 31840–31848 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.M. Macías-Silva, S. Abdollah, P. A. Hoodless, R. Pirone, L. Attisano, J. L. Wrana, MADR2 is a substrate of the TGFbeta receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell 87, 1215–1224 (1996). [DOI] [PubMed] [Google Scholar]
- 34.H. Raghu, N. Sharma-Walia, M. V. Veettil, S. Sadagopan, A. Caballero, R. Sivakumar, L. Varga, V. Bottero, B. Chandran, Lipid rafts of primary endothelial cells are essential for Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8-induced phosphatidylinositol 3-kinase and RhoA-GTPases critical for microtubule dynamics and nuclear delivery of viral DNA but dispensable for binding and entry. J. Virol. 81, 7941–7959 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.N. Sharma-Walia, H. H. Krishnan, P. P. Naranatt, L. Zeng, M. S. Smith, B. Chandran, ERK1/2 and MEK1/2 induced by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) early during infection of target cells are essential for expression of viral genes and for establishment of infection. J. Virol. 79, 10308–10329 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.P. P. Naranatt, S. M. Akula, C. A. Zien, H. H. Krishnan, B. Chandran, Kaposi's sarcoma-associated herpesvirus induces the phosphatidylinositol 3-kinase-PKC-zeta-MEK-ERK signaling pathway in target cells early during infection: Implications for infectivity. J. Virol. 77, 1524–1539 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.H. Raghu, N. Sharma-Walia, M. V. Veettil, S. Sadagopan, B. Chandran, Kaposi's sarcoma-associated herpesvirus utilizes an actin polymerization-dependent macropinocytic pathway to enter human dermal microvascular endothelial and human umbilical vein endothelial cells. J. Virol. 83, 4895–4911 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.N. Kerur, M. V. Veettil, N. Sharma-Walia, S. Sadagopan, V. Bottero, A. G. Paul, B. Chandran, Characterization of entry and infection of monocytic THP-1 cells by Kaposi's sarcoma associated herpesvirus (KSHV): Role of heparan sulfate, DC-SIGN, integrins and signaling. Virology 406, 103–116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.N. Inoue, J. Winter, R. B. Lal, M. K. Offermann, S. Koyano, Characterization of entry mechanisms of human herpesvirus 8 by using an Rta-dependent reporter cell line. J. Virol. 77, 8147–8152 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.S. M. Akula, P. P. Naranatt, N. S. Walia, F. Z. Wang, B. Fegley, B. Chandran, Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) infection of human fibroblast cells occurs through endocytosis. J. Virol. 77, 7978–7990 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Y. F. Zhang, Q. Li, P. Q. Huang, T. Su, S. H. Jiang, L. P. Hu, X. L. Zhang, Y. Sun, H. Pan, X. M. Yang, J. Li, Y. Z. Gai, L. Zhu, L. L. Yao, D. X. Li, Y. W. Sun, Z. G. Zhang, D. J. Liu, Y. L. Zhang, H. Z. Nie, A low amino acid environment promotes cell macropinocytosis through the YY1-FGD6 axis in Ras-mutant pancreatic ductal adenocarcinoma. Oncogene 41, 1203–1215 (2022). [DOI] [PubMed] [Google Scholar]
- 42.K. Tzavlaki, A. Moustakas, TGF-β signaling. Biomolecules 10, 487 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.P. E. Marques, S. Grinstein, S. A. Freeman, SnapShot:Macropinocytosis. Cell 169, 766–766.e1 (2017). [DOI] [PubMed] [Google Scholar]
- 44.J. T. Bechtel, Y. Liang, J. Hvidding, D. Ganem, Host range of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 77, 6474–6481 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D. J. Blackbourn, E. Lennette, B. Klencke, A. Moses, B. Chandran, M. Weinstein, R. G. Glogau, M. H. Witte, D. L. Way, T. Kutzkey, B. Herndier, J. A. Levy, The restricted cellular host range of human herpesvirus 8. AIDS 14, 1123–1133 (2000). [DOI] [PubMed] [Google Scholar]
- 46.J. Naipauer, E. A. Mesri, The Kaposi's sarcoma progenitor enigma: KSHV-induced MEndT-EndMT axis. Trends Mol. Med. 29, 188–200 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.S. Lambert, M. Bouttier, R. Vassy, M. Seigneuret, C. Petrow-Sadowski, S. Janvier, N. Heveker, F. W. Ruscetti, G. Perret, K. S. Jones, C. Pique, HTLV-1 uses HSPG and neuropilin-1 for entry by molecular mimicry of VEGF165. Blood 113, 5176–5185 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.J. L. Daly, B. Simonetti, K. Klein, K. E. Chen, M. K. Williamson, C. Antón-Plágaro, D. K. Shoemark, L. Simón-Gracia, M. Bauer, R. Hollandi, U. F. Greber, P. Horvath, R. B. Sessions, A. Helenius, J. A. Hiscox, T. Teesalu, D. A. Matthews, A. D. Davidson, B. M. Collins, P. J. Cullen, Y. Yamauchi, Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370, 861–865 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.L. Cantuti-Castelvetri, R. Ojha, L. D. Pedro, M. Djannatian, J. Franz, S. Kuivanen, F. van der Meer, K. Kallio, T. Kaya, M. Anastasina, T. Smura, L. Levanov, L. Szirovicza, A. Tobi, H. Kallio-Kokko, P. Österlund, M. Joensuu, F. A. Meunier, S. J. Butcher, M. S. Winkler, B. Mollenhauer, A. Helenius, O. Gokce, T. Teesalu, J. Hepojoki, O. Vapalahti, C. Stadelmann, G. Balistreri, M. Simons, Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370, 856–860 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.A. Baghian, M. Luftig, J. B. Black, Y. X. Meng, C. P. Pau, T. Voss, P. E. Pellett, K. G. Kousoulas, Glycoprotein B of human herpesvirus 8 is a component of the virion in a cleaved form composed of amino- and carboxyl-terminal fragments. Virology 269, 18–25 (2000). [DOI] [PubMed] [Google Scholar]
- 51.A. Kopp, E. Blewett, V. Misra, T. C. Mettenleiter, Proteolytic cleavage of bovine herpesvirus 1 (BHV-1) glycoprotein gB is not necessary for its function in BHV-1 or pseudorabies virus. J. Virol. 68, 1667–1674 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.H. B. Pang, G. B. Braun, T. Friman, P. Aza-Blanc, M. E. Ruidiaz, K. N. Sugahara, T. Teesalu, E. Ruoslahti, An endocytosis pathway initiated through neuropilin-1 and regulated by nutrient availability. Nat. Commun. 5, 4904 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.S. M. Akula, F. Z. Wang, J. Vieira, B. Chandran, Human herpesvirus 8 interaction with target cells involves heparan sulfate. Virology 282, 245–255 (2001). [DOI] [PubMed] [Google Scholar]
- 54.D. Ghez, Y. Lepelletier, S. Lambert, J. M. Fourneau, V. Blot, S. Janvier, B. Arnulf, P. M. van Endert, N. Heveker, C. Pique, O. Hermine, Neuropilin-1 is involved in human T-cell lymphotropic virus type 1 entry. J. Virol. 80, 6844–6854 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.R. K. Lane, H. Guo, A. D. Fisher, J. Diep, Z. Lai, Y. Chen, J. W. Upton, J. Carette, E. S. Mocarski, W. J. Kaiser, Necroptosis-based CRISPR knockout screen reveals Neuropilin-1 as a critical host factor for early stages of murine cytomegalovirus infection. Proc. Natl. Acad. Sci. U.S.A. 117, 20109–20116 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.J. Mercer, A. Helenius, Virus entry by macropinocytosis. Nat. Cell Biol. 11, 510–520 (2009). [DOI] [PubMed] [Google Scholar]
- 57.J. Mercer, A. Helenius, Gulping rather than sipping: Macropinocytosis as a way of virus entry. Curr. Opin. Microbiol. 15, 490–499 (2012). [DOI] [PubMed] [Google Scholar]
- 58.A. V. Ark, J. Cao, X. Li, TGF-β receptors: In and beyond TGF-β signaling. Cell. Signal. 52, 112–120 (2018). [DOI] [PubMed] [Google Scholar]
- 59.S. Li, J. Zhang, S. Qian, X. Wu, L. Sun, T. Ling, Y. Jin, W. Li, L. Sun, M. Lai, F. Xu, S100A8 promotes epithelial-mesenchymal transition and metastasis under TGF-β/USF2 axis in colorectal cancer. Cancer Commun. (London, England) 41, 154–170 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Y. Yuan, H. Li, W. Pu, L. Chen, D. Guo, H. Jiang, B. He, S. Qin, K. Wang, N. Li, J. Feng, J. Wen, S. Cheng, Y. Zhang, W. Yang, D. Ye, Z. Lu, C. Huang, J. Mei, H. F. Zhang, P. Gao, P. Jiang, S. Su, B. Sun, S. M. Zhao, Cancer metabolism and tumor microenvironment: Fostering each other? Sci. China Life Sci. 65, 236–279 (2022). [DOI] [PubMed] [Google Scholar]
- 61.I. F. Ciernik, B. H. Krayenbühl Ciernik, C. J. Cockerell, J. D. Minna, A. F. Gazdar, D. P. Carbone, Expression of transforming growth factor β and transforming growth factor β receptors on AIDS-associated Kaposi's sarcoma. Clin. Cancer Res. 1, 1119–1124 (1995). [PubMed] [Google Scholar]
- 62.A. Franchi, L. Arganini, G. Baroni, A. Calzolari, R. Capanna, D. Campanacci, P. Caldora, L. Masi, M. L. Brandi, G. Zampi, Expression of transforming growth factor β isoforms in osteosarcoma variants: Association of tgfβ1 with high-grade osteosarcomas. J. Pathol. 185, 284–289 (1998). [DOI] [PubMed] [Google Scholar]
- 63.P. Kloen, M. C. Gebhardt, A. Perez-Atayde, A. E. Rosenberg, D. S. Springfield, L. I. Gold, H. J. Mankin; TGF-β3 is related to disease progression , Expression of transforming growth factor-β (TGF-β) isoforms in osteosarcomas. Cancer 80, 2230–2239 (1997). [PubMed] [Google Scholar]
- 64.K. L. McCann, F. Imani, Transforming growth factor β enhances respiratory syncytial virus replication and tumor necrosis factor alpha induction in human epithelial cells. J. Virol. 81, 2880–2886 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.J. D. Gibbs, D. M. Ornoff, H. A. Igo, J. Y. Zeng, F. Imani, Cell cycle arrest by transforming growth factor beta1 enhances replication of respiratory syncytial virus in lung epithelial cells. J. Virol. 83, 12424–12431 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.S. J. Allen, K. R. Mott, S. L. Wechsler, R. A. Flavell, T. Town, H. Ghiasi, Adaptive and innate transforming growth factor beta signaling impact herpes simplex virus 1 latency and reactivation. J. Virol. 85, 11448–11456 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.S. Thakker, R. C. Strahan, A. N. Scurry, T. Uppal, S. C. Verma, KSHV LANA upregulates the expression of epidermal growth factor like domain 7 to promote angiogenesis. Oncotarget 9, 1210–1228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.R. S. Cooper, E. R. Georgieva, P. P. Borbat, J. H. Freed, E. E. Heldwein, Structural basis for membrane anchoring and fusion regulation of the herpes simplex virus fusogen gB. Nat. Struct. Mol. Biol. 25, 416–424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.C. Sun, Y. F. Kang, Y. T. Liu, X. W. Kong, H. Q. Xu, D. Xiong, C. Xie, Y. H. Liu, S. Peng, G. K. Feng, Z. Liu, M. S. Zeng, Parallel profiling of antigenicity alteration and immune escape of SARS-CoV-2 Omicron and other variants. Signal Transduct. Target. Ther. 7, 42 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S7
Legend for table S1
Table S1