Abstract
Red mold rice (RMR) is a traditional Chinese medicine prepared using Monascus fermentation. Monascus ruber ( pilosus) and Monascus purpureus have a long history of use as food and medicine. As an economically important starter culture, the relationship between the taxonomy of Monascus and production capabilities of secondary metabolites is crucial for the Monascus food industry. In this study, monacolin K, monascin, ankaflavin, and citrinin production by M. purpureus and M. ruber were genomically and chemically investigated. Our findings suggest that M. purpureus can produce monascin and ankaflavin in a correlated manner, whereas M. ruber produces monascin with minimum ankaflavin. M. purpureus is capable of producing citrinin; however, it is unlikely able to produce monacolin K. In contrast, M. ruber produces monacolin K, but not citrinin. We suggest that the current monacolin K content-related regulation of Monascus food should be revised, and labeling of Monascus species should be considered.
Keywords: Ankaflavin, Citrinin, Monacolin K, Monascin, Monascus purpureus Monascus ruber
1. Introduction
Red mold rice (RMR) is a traditional Chinese medicine prepared using Monascus fermentation. Bioactive compounds, including monacolin K (lovastatin) and pigments, are important functional components of RMR. The Monascus yellow pigments, in particular, monascin and ankaflavin, are the focus of current Monascus functional food research owing to their multiple health benefits [1]. Besides monacolin K and its pigments, the mycotoxin citrinin is also a critical secondary metabolite of Monascus.
Among the Monascus species, only Monascus ruber, Monascus pilosus, and Monascus purpureus have a long history of food andmedicine usage. M. purpureus [2] is distinguished from M. pilosus and M. ruber by colony growth characteristics, pigment production, and molecular genetics [3,4]. M. pilosus was first proposed as a novel species by Hawksworth and Pitt in 1983 and has been proven to be synonymous to M. ruber [5].
Monacolin K is the best-known bioactive ingredient in RMR. Many current Monascus health food regulations are based on the concentrations of monacolin K and citrinin in food [6–8]. Thus, the relationship between the taxonomy of Monascus and the monacolin K and citrinin production capabilities of Monascus is crucial for the regulation of Monascus. However, existing findings remain inconclusive. In this study, monacolin K, azaphilone pigments, and citrinin production by M. purpureus and M. ruber (M. pilosus) was genomically and chemically investigated. Synteny analysis of monacolin K, azaphilone pigment, and citrinin biosynthesis gene clusters was conducted to investigate the biosynthetic capabilities of M. purpureus and M. ruber ( pilosus). Forty-eight Monascus strains were isolated from 23 traditional Chinese medicine RMR samples, and these isolates along with other 41 Monascus strains, were analyzed using high-performance liquid chromatography (HPLC) to explore the relationship between secondary metabolite signatures and Monascus species.
1.1. Hypothesis
The secondary metabolite signatures (including monacolin K, yellow pigment, and citrinin production capability) of M. purpureus and M. ruber (M. pilosus) are species-specific.
2. Materials and methods
2.1. Microorganisms and cultivation
Ninety-five Monascus strains, including 26 reference strains [purchased from the Bioresource Collection and Research Center (BCRC, Taiwan) (Table S1 (https://www.jfda-online.com/cgi/viewcontent.cgi?filename=6=article=3438=context=journal=type=additional)), six strains of M. pilosus, six strains of M. purpureus, and 14 strains of M. ruber], and 69 isolated strains (mostly isolated from the Chinese medicine redmold rice, including eight strains of M. pilosus, 60 strains of M. purpureus, and one strain of M. ruber) were used in this study (Table S1 (https://www.jfda-online.com/cgi/viewcontent.cgi?filename=6=article=3438=context=journal=type=additional)). Monascus str-ains were routinely maintained on potato dextrose agar (PDA; HiMedia Laboratories, India).
2.2. DNA extraction, quantification, polymerase chain reaction (PCR), and sequencing
Eight-day-old Monascus mycelia cultivated in potato dextrose broth (PDB) medium were used for DNA extraction. Mycelia were ground to a powder in liquid nitrogen and vacuum-dried. Monascus mycelium powder (50 mg) was used for DNA extraction using a QIAamp DNA mini kit (QIAGEN GmbH, Germany). DNA concentration was measured using a Qubit™ dsDNA BR assay kit (Life Technologies Corporation, USA) with a Qubit® 2.0 fluorometer (Life Technologies). Partial internal transcribed spacer (ITS)-and β-tubulin-coding genes were amplified and sequenced for phylogenetic analysis [9]. The primer sets used in this study are listed in Table S2 (https://www.jfda-online.com/cgi/viewcontent.cgi?filename=6=article=3438=context=journal=type=additional), and PCR amplifications were performed using EXSEL high-fidelity DNA polymerase (Bersing Ltd., Taiwan).
2.3. Phylogenetic analysis
Geneious R 8.1.6 software (Biomatters Ltd., Auckland, New Zealand) was used for routine sequence management and de novo sequence assemble. MAFFT (ver. 7.017, [10]) plugin of the Geneious software was used for multiple alignments. MEGA X [11] was used for the maximum-likelihood phylogenetic tree construction (1000 bootstrap replicates). The general time-reversible (GTR; [12]) model was used for all phylogenetic analyses.
2.4. Functional genomic analysis
M. ruber NRRL 1597 (accession No. PRJNA196033) and M. ruberFWB13 (accession no.PRJNA433431) and M. ruber CBS 127566 (accession no. PRJEB20852), and M. pilosus MS-1, YDJ-2, YDJ-1, and K104061 (accession no. PRJNA718072; [13]), M. purpureus YY-1 (accession no. PRJNA453427) and M. purpureus NRRL 1596 (BCRC 31542T; accession no. PRJNA204098) whole-genome shotgun assemblies were used for genomic investigations. The M. pilosus pigment (strain BCRC 38072; accession no. KC148521.1), and monacolin K (BCRC 38072; accession no. DQ176595.1), and M. purpureus citrinin biosynthesis gene clusters (strain BCRC 31542) were used as queries for a BLAST search of the corresponding biosynthesis gene cluster sequences (Table S3 (https://www.jfda-online.com/cgi/viewcontent.cgi?filename=6=article=3438=context=journal=type=additional)). Custom discontinuous BLAST searches were performed using the Geneious R 8.1.6. Mauve alignment [14] and LASTZ 1.02 [15] were used for the synteny analysis of citrinin biosynthesis gene cluster sequences. InterProScan 5.20 was used for protein functional analysis and conserved domain prediction [16].
2.5. HPLC analysis of secondary metabolites
The Monascus strains were cultured in PDB medium for metabolite analysis. After eight days of cultivation, mycelia were collected by centrifugation (15 min at 6000×g). HPLC samples were prepared by 95% ethanol extraction of Monascus dry mycelium/pellet at 50 °C for one hour and subsequently filtrated through 0.45 μm PVDF filter. The HPLC analysis method was adopted from a previous report [17]. Isocratic elution (water: acetonitrile: trifluoroacetate = 38:62:0.05 at 1 mL/min) was carried out on a Hitachi D-2000 Elite HPLC system (Hitachi, Japan) with a LUNA C18 column (250 × 4.6 mm, 5 μm particle size, Phenomenex, USA), fluorescence detector (L-2485 FL, Hitachi), and a photodiode array detector (L2455 DAD detector, Hitachi). The 385 nm absorbance chromatogram (yellow pigment) was used as the metabolite signature. Pure monascin and ankaflavin were provided by SunWay Biotech Co. Ltd. (Taipei, Taiwan).
2.6. Statistical analysis
R 4.1.3, using the vegan package, was used for principal component analysis (PCA) and analysis of similarities (ANOSIM) of secondary metabolite signatures. Other data were visualized and analyzed the Prism 6.01 software (GraphPad Software, USA). p < 0.05 was considered as significantly correlated (Pearson’s correlation analysis).
3. Results
3.1. M. purpureus is predominant Monascus species used for traditional Chinese medicine RMR fermentation
The internal transcribed spacer (ITS) region sequence is currently the major molecular marker used for the identification and classification of eukaryotes. β-tubulin gene sequences are also known to be effective molecular taxonomic markers for the classification of fungi [18]. In this study, the taxonomic position of Monascus isolates was investigated using a phylogenetic approach based on both ITS and β-tubulin gene sequences. Penicillium eremophilus (Monascus eremophilus, [5]) was added as an outgroup. Phylogenetic trees based on ITS and βtubulin sequences showed that most of the Monascus strains isolated from RMR belonged to M. purpureus, whereas only one isolate (strain DYH) belonged to M. ruber (M. pilosus). These results indicate that the bioactivity of traditional RMR may largely originate from M. purpureus (Figure S1 (https://www.jfdaonline.com/cgi/viewcontent.cgi?filename=6=article=3438=context=journal=type=additional)).
3.2. Pigment, monacolin K, and citrinin biosynthesis gene clusters are highly conserved between M. pilosus and M. ruber
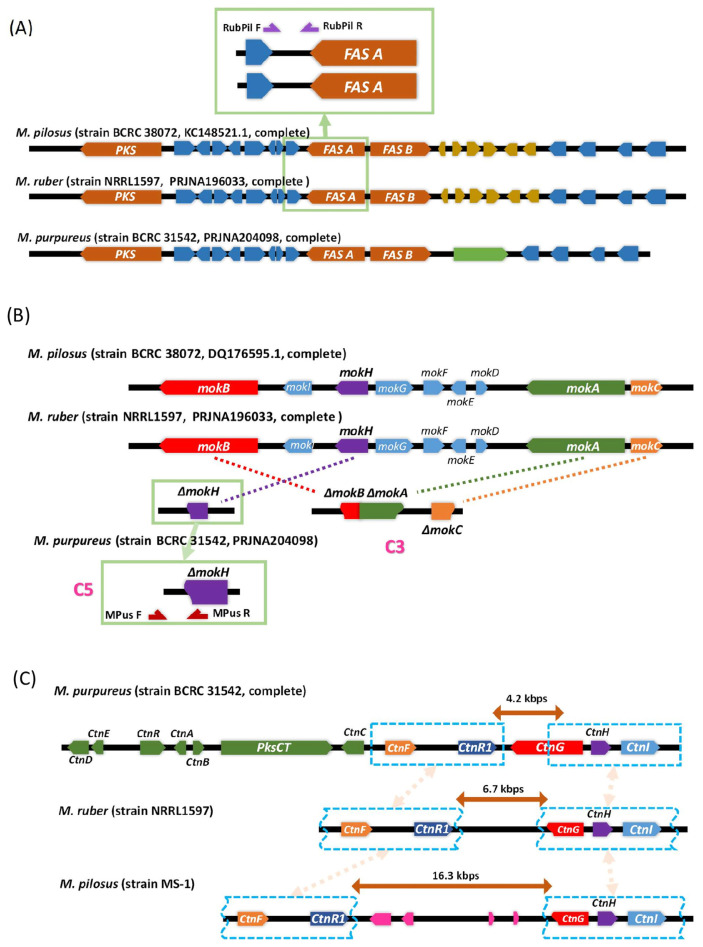
Monascus produces bioactive secondary metabolites including monacolin K, monascin/ankaflavin, and citrinin. However, the ability of the major edible Monascus species, M. purpureus, M. pilosus, and M. ruber to produce these specific secondary metabolites is inconclusive. In this study, synteny analysis of the biosynthesis gene cluster of monacolin K, monascin/ankaflavin, and citrinin was performed to investigate the production capabilities of major secondary metabolites and taxonomic relationships among M. purpureus, M. pilosus, and M. ruber. The sequences of pigment biosynthetic gene clusters were highly conserved (>99.9%) between M. ruber and M. pilosus genomes. In contrast, the pigment biosynthesis gene cluster in the M. purpureus genome showed less homology to its counterparts in M. ruber and M. pilosus (Fig. 1(A)). A conserved region of the pigment biosynthesis gene cluster in M. ruber and M. pilosus strains was confirmed using PCR with the primer set RubPil F/R (Table S2 (https://www.jfda-online.com/cgi/viewcontent.cgi?filename=6=article=3438=context=journal=type=additional)) in all Monascus strains. Despite minor differences in gene cluster structure, all M. purpureus, M. pilosus, and M. ruber species are capable of producing yellow, orange, and red pigments.
Fig. 1.
The pigment, monacolin K, and citrinin biosynthesis gene clusters in M. pilosus, M. ruber NRRL 1597 and M. purpureus BCRC 31542 genomes. These gene clusters were nearly identical between M. pilosus and M. ruber. (A) Pigment biosynthesis gene clusters. Functional pigment biosynthesis gene clusters were present in all three species. M. pilosus (M. ruber)-specific primer set RubPil is targeted to the upstream sequences of FASA gene. (B) Monacolin K biosynthesis gene clusters. Besides lost most part of monacolin K biosynthesis gene cluster sequences, the residual two sequence fragments located at two different chromosomes in M. purpureus (C3 and C5 of M. purpureus YY-1 genome). (C) Citrinin biosynthesis gene clusters. Two types of residual citrinin biosynthesis gene cluster present in the M. pilosus and M. ruber genomes.
The monacolin K (MK) biosynthetic gene cluster was also highly conserved (95.0–99.7%) between M. ruber and M. pilosus. However, all the M. purpureus genomes contained only two fragments of the MK gene cluster. One of the residual MK biosynthesis gene clusters contains only a truncated mokH gene (ΔmokH ), and another residual fragment of the MK gene cluster contains recombined and truncated mokA, mokB, and mokC genes. PCR analysis showed that these two residual gene cluster fragments were highly conserved among M. purpureus strains. Sequence analysis of ΔmokH showed that the Zn (2)-Cys (6) fungal-type transcription factor motif of the mokH protein, which is a key activator of MK biosynthesis, was lost. The presence of ΔmokH in all M. purpureus strains was confirmed using PCR with the event-specific primer set MPus F/MPus R (Fig. 1(B)). The residual MK gene cluster fragments located on different chromosomes also indicated that a large-scale recombination event occurred in the last common ancestor (LCA) of all M. purpureus strains (Fig. 1(B)) and thus, M. purpureus was unable to produce monacolin K.
The citrinin biosynthesis gene cluster was firstly cloned from M. purpureus [19] (accession no. EU309474.1). M. purpureus strains shared highly conserved citrinin biosynthesis gene clusters (>99.9% among strains NRRL1596, YY-1, and Gen-Bank EU309474.1). However, only two parts of the citrinin biosynthesis gene cluster were present in both M. pilosus MS-1 and M. ruber NRRL 1597 genomes (Fig. 1(C)). One residual citrinin biosynthesis gene sequence contained ctnF and ctnR1, and the other contained ctnH, ctnI, and a truncated ctnG gene. The loss of the major citrinin synthesis gene, pksCT, indicates that M. pilosus and M. ruber are not capable of producing citrinin. Remarkably, the residual citrinin gene sequences were nearly identical between M. pilosus MS-1 and M. ruber NRRL 1597. These two regions of citrinin gene sequence also highly conserved (92.69–96.30% nucleotide similarity) among M. purpureus, M. pilosus, and M. ruber. However, the insert sequences between the two residual citrinin gene cluster sequences are different in M. pilosus MS-1 (approximately 16.3 kbps, designated as insert ρ) and M. ruber NRRL 1597 (approximately 6.7 kbps, designated as insert π). The insert sequence π contained no ORF, and insert ρ contained four predicted ORFs with unknown functions. The sequence of insert π shares no homology to insert ρ, and both insert π and ρ have no homology to any other region of the M. purpureus, M. pilosus, and M. ruber genomes (Fig. 2).
Fig. 2.
Residual citrinin biosynthesis gene clusters in M. pilosus (strain MS-1) and M. ruber (strain NRRL 1597) genomes. (A) Mauve alignment analysis revealed two types of insert sequences flanked by highly conserved residual citrinin biosynthesis gene cluster sequences. π: The insert sequence region of citrinin biosynthesis gene cluster in M. ruber NRRL 1597. ρ: The insert sequence region of citrinin biosynthesis gene cluster in M. pilosus MS-1. Shaded region in red: sequence similarity of conserved region. Blank: regions with no homology. (B) Schematic diagram of residual citrinin biosynthesis gene clusters. Core citrinin biosynthesis gene pksCT was not found in both genomes.
3.3. M. pilosus and M. ruber shared similar signature of yellow pigments production, differing from M. purpureus
Ninety-fiveMonascus strains, including 14 strains of M. pilosus, 66 strains of M. purpureus, 15 strains of M. ruber, and 27 RMR samples (Chinese herbal medicine fermented by M. purpureus) were used for secondary metabolite analysis. Peak areas of the absorbance spectrum at 385 nm (major absorbance wavelengths of monascin and ankaflavin) were selected for metabolite signature comparison (Fig. 3(A)). PCA analysis of seven peak areas of the absorbance spectrum (385 nm) indicated that the production of citrinin and ankaflavin was closely related to M. purpureus, whereas M. pilosus was characterized by low-level pigmentation (Fig. 3(B)). Among M. purpureus, M. pilosus, and M. ruber, only M. purpureus produced higher amounts of ankaflavin, which was significantly correlated with monascin in both the mycelium (Pearson’s r = 0.9017, p < 0.0001; Fig. 4(A)) and RMR (Pearson’s r = 0.9393, p < 0.0001; Fig. 4(B)). In contrast, M. ruber ( pilosus) had a higher p10.9 peak area which also significantly correlated with the peak area of monascin (Pearson’s r = 0.9363, p < 0.0001; Fig. 5). ANOSIM analysis also suggested that the metabolite signatures of M. purpureus and M. ruber ( pilosus) were significantly different (R = 0.8517; p = 0.001).
Fig. 3.
M. pilosus and M. ruber shared common HPLC absorbance chromatogram signature at 385 nm that was different from that of M. purpureus. (A) Comparison of absorbance chromatograms (385 nm) and selected peaks. p10.9: Unidentified peak with 10.9 min retention time. pMS: peak of monascin. pAK: peak of ankaflavin. RMR-AB: Red mold rice AB which purchased from a Chinese herbal medicine pharmacy (prepared with M. purpureus). (B) PCA analysis of 385 nm absorbance chromatogram (peak area of seven peaks including citrinin, monascin, ankaflavin, p10.9, p12.5, p25, and p30). Citrinin and ankaflavin production were correlated to M. purpureus and differentiate it from M. ruber ( pilosus).
Fig. 4.
Monascin and ankaflavin production are significantly correlated in M. purpureus. (A) Monascin and ankaflavin production are significantly correlated in M. purpureus (20 strains, triplicated test) cultivated in PDB medium. (B) Monascin and ankaflavin production were also significantly correlated in RMRs (23 samples).
Fig. 5.
Besides monascin and ankaflavin production, absorbance chromatogram (385 nm) also revealed specific metabolite signature of M. ruber ( pilosus). (A) M. pilosus (ruber) and M. purpureus could be differentiated with the ratio of peak area of monascin (pMS) to p10.9. Purple circle: M. purpureus strains. Pink triangle: M. pilosus/ruber strains. Pearson’s r for the correlation analysis between peak area of pMS and p10.9 in M. pilosus/ruber strains. (B) Peak area ratios of p10.9 to pMS were significantly different between M. purpureus and M. ruber ( pilosus).
3.4. Average Nucleotide Identity (ANI) and the distribution of insert sequences among M. pilosus and M. ruber strains suggest that the strains are synonymous
ANI analysis is an alternative method for DNA–DNA hybridization based on genome sequences, and is generally recognized as a standard for taxonomic investigation of microorganisms [20,21]. ANI analysis among M. purpureus, M. pilosus, and M. ruber genomes showed that ANI between M. purpureus and M. ruber are 94.22–94.30%, while ANI between M. purpureus and M. pilosus are 94.20–94.29%. These interspecies ANI were significantly lower than intraspecies ANI (M. purpureus to M. purpureus, 99.89%; M. pilosus to M. pilosus 99.92–99.98%; M. ruber to M. ruber 99.86–99.89%; Figure S2 (https://www.jfda-online.com/cgi/viewcontent.cgi?filename=6=article=3438=context=journal=type=additional)). However, the interspecies ANI between M. pilosus and M. ruber are 99.87–99.98%, which is not significantly lower than intraspecies ANI.
The insert sequences π (in the M. ruber NRRL 1596 genome) and ρ (in the M. pilosus MS-1 genome) of M. ruber and M. pilosus strains were further investigated by partial PCR amplification and sequencing (Fig. 6 (A)). Although the DNA sequences of π and ρ inserts are highly conserved (89.9–99.99% mean pairwise identity) among Monascus strains, only one type (π or ρ) of insert sequence exists in Monascus genome and not correlate with specific Monascus species (Fig. 6 (B)). M. pilosus K104061, MS-1, YDJ-1, and M. ruber FWB-13 genomes had ρ type insert sequences (99.7%, 99.7%, 99.7%, and 99.9% identical sites, respectively). M. pilosus YDJ-2 and M. ruber CBS 127566 genomes have a π-type insert sequence (with 99.8% and 99.4% identical sites, respectively; Fig. 6 (C) and (D)).
Fig. 6.
The distribution of π and ρ insert sequences in M. pilosus and M. ruber strains. (A) Primer sets designed for π and ρ insert sequences PCR and sequencing. Yellow arrow: predicted genes in insert sequences. White box: regions for PCR amplification. Green box: PCR and sequenced region. (B) The distribution of π and ρ insert sequences in M. pilosus and M. ruber strains irrelevant to species.
4. Discussion
M. ruber, M. pilosus, and M. purpureus have a long history of food and medicinal usage. As an economically important starter culture, the relationship between the taxonomy of Monascus and the production capabilities of secondary metabolites is crucial for food and regulation in Monascus.
M. pilosus and M. ruber have been proposed to be synonymous in several previous studies [5,9]. Barbosa et al. [5] also reported that pigmentation is not a robust taxonomic key for distinguishing M. pilosus from M. ruber. In our study, synteny analysis showed that M. pilosus and M. ruber share highly conserved and complete monacolin K and azaphilone pigment biosynthesis gene clusters, which were significantly different from those of M. purpureus (Fig. 1 and Fig. 6). Although the pigment synthesis gene cluster in the M. purpureus genome is complete, all M. purpureus strains have only two highly conserved residual monacolin K biosynthesis gene cluster sequences in which all residual genes appeared to be nonfunctional (Fig. 1(B)). Kwon et al. [22] reported that most part of monacolin K biosynthesis gene clusters were lost in M. purpureus NRRL 1596 and M. purpureus YY-1. Our findings suggest that all M. purpureus strains shared a highly conserved residual monacolin K biosynthesis gene cluster with the disrupted mokH gene (confirmed using PCR and sequencing analysis), which indicates that the disruption of the monacolin K biosynthesis gene cluster is species-specific. Chen et al. [23] showed that mokH is essential for monacolin K biosynthesis. Higa et al. [24] reported that M. purpureus NBRC 4478 lacks a complete monacolin K gene sequence, consistent with our findings. The strain-specific disruption of the monacolin K biosynthesis gene cluster suggests that M. purpureus lost its monacolin K-producing capability.
Synteny analysis also revealed that only M. purpureus had a complete citrinin biosynthesis gene cluster. M. pilosus and M. ruber shared highly conserved residual gene cluster sequences in which major citrinin biosynthesis genes, such as pksCT, were lost (Fig. 6 (A), confirmed using PCR and sequencing analysis). Chen et al. [25] showed that, among Monascus species, pksCT was only present in M. purpureus and Monascus sanguineus, consistent with our findings. Shimizu et al. [26] reported that disruption of the pksCT gene abolished citrinin production. Higa et al. [24] also reported that M. pilosus NBRC 4520 and M. ruber NBRC 4483 lacked the complete citrinin gene sequence, consistent with our findings. Since all strains of M. ruber ( pilosus) share two types of residual citrinin biosynthesis gene clusters and all pksCT genes were missing, we suggest that M. ruber ( pilosus) is unable to produce citrinin.
Monacolin K, citrinin, and red pigments have been reported to possess antimicrobial activities [27]. These antimicrobial secondary metabolites may provide Monascus ecological competitiveness at the cost of a metabolic burden. Pigments are also a common UV protection strategy for terrestrial fungi, and may thus be conserved among M. purpureus, M. pilosus, and M. ruber. Loss of monacolin K or citrinin biosynthesis in M. purpureus and M. ruber/M. pilosus may represent a trade-off between resource competitiveness and metabolic burden during the evolution of these species.
The highly conserved pigment, citrinin, and residual monacolin K biosynthesis gene cluster in M. purpureus genomes suggests that major recombination events of secondary metabolite biosynthesis gene clusters occurred during the speciation of M. purpureus, indicating that the evolution of these major secondary metabolite gene cluster sequences may correlate with Monascus taxonomy. The residual monacolin K and citrinin gene clusters in M. purpureus and M. ruber ( pilosus) also suggest that these three Monascus species share a common ancestor capable of producing both monacolin K and citrinin. The production of secondary metabolites is an important adaptation strategy that is usually species-specific fashion [28]. In this study, metabolite analysis revealed that the signature of M. purpureus was capable of producing monascin and ankaflavin in a correlated manner (median MS to AK ratio of all M. purpureus was 2.016), whereas M. ruber ( pilosus) produced monascin with minimum ankaflavin. M. purpureus is also capable of producing citrinin, but not monacolin K. In contrast, M. ruber ( pilosus) produces monacolin K, but not citrinin.
M. purpureus, M. pilosus, and M. ruber were phylogenetically closely related (Figure S1 (https://www.jfda-online.com/cgi/viewcontent.cgi?filename=6=article=3438=context=journal=type=additional)). Our findings indicate that monacolin K, azaphilone pigment, and citrinin biosynthesis gene sequences, as well as production capabilities, are conserved within species of M. purpureus and M. ruber ( pilosus). However, the monacolin K and citrinin production capabilities of M. purpureus and M. ruber ( pilosus) have been inconclusive in previous studies [29–32]. These controversies may arise from incorrect species identification caused by unreliable morphological taxonomic keys and internal transcribed spacer (ITS) sequence-based phylogenetic analysis. For example, M. ruber M7 is a well-known strain with a fully sequenced genome. Our current knowledge of azaphilone pigment biosynthesis and regulation in Monascus has been elucidated through comprehensive studies of M. ruber M7 [31,33]. Synteny analysis revealed that M. ruber M7 has an M. purpureus-type pigment [31] and complete citrinin (accession KT781075.1, approximately 42.8 kbps, >99.9% identity to M. purpureus YY-1 with 99.55% coverage) biosynthesis gene clusters, which indicate that M. ruber M7 may belong to M. purpureus.
Monacolin K and citrinin contents in Monascus products are legally regulated in many countries. The U.S. Food and Drug Administration (FDA) has determined that red yeast rice products containing more than trace amounts of monacolin K are unapproved new drugs and cannot be sold legally as dietary supplements [34]. Citrinin is particularly crucial for Monascus products because it is a major safety concern for Monascus. Our investigation helps to clarify the production capabilities of important secondary metabolites and their relation to Monascus species, which will augment Monascus product regulation, such as labeling or approval of Monascus species used for production.
In conclusion, our findings suggest that M. purpureus can produce monascin and ankaflavin in a correlated fashion, whereas M. ruber ( pilosus) produces monascin with minimum ankaflavin. M. purpureus is also capable of producing citrinin, but not monacolin K. In contrast, M. ruber ( pilosus) produces monacolin K, but not citrinin. These results confirm our hypothesis that the secondary metabolite signatures of M. purpureus and M. ruber (M. pilosus) are species-specific. Since M. purpureus is the predominant species used for traditional RMR preparation, the current monacolin K (lovastatin) content-related regulation of Monascus food should be revised. The common bioactive Monascus metabolite monascin is a good alternative functional component for Monascus product regulation, and labeling of Monascus species should be considered.
Supplementary Information
Acknowledgements
Part of this study is kindly funded by Taiwan Ministry of Science and Technology (project 104-2311-B-143-001, 105-2320-B-143-001 and 106-2314-B-143-001).
Funding Statement
Part of this study is kindly funded by Taiwan Ministry of Science and Technology (project 104-2311-B-143-001, 105-2320-B-143-001 and 106-2314-B-143-001).
References
- 1. Lin CH, Lin TH, Pan TM. Alleviation of metabolic syndrome by monascin and ankaflavin: the perspective of Monascus functional foods. Food Funct. 2017;8:2102. doi: 10.1039/c7fo00406k. [DOI] [PubMed] [Google Scholar]
- 2. Went FAFC. Le champignon de l’ang-quac: une nouvelle thelebolee. Ann Sci Nat. 1895;81:1–18. [Google Scholar]
- 3. Hawksworth DL, Pitt JI. A new taxonomy for Monascus species based on cultural and microscopical characters. Aust J Bot. 1983;31:51–61. [Google Scholar]
- 4.Pitt JI, Hocking AD. Fungi and food spoilage. 2nd ed. USA: Springer Nature; 2009. [Google Scholar]
- 5. Barbosa RN, Leong SL, Vinnere-Pettersson O, Chen AJ, Souza-Motta CM, Frisvad JC, et al. Phylogenetic analysis of Monascus and new species from honey, pollen and nests of stingless bees. Stud Mycol. 2017;86:29–51. doi: 10.1016/j.simyco.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Commossion Europrean. Commission Regulation (EU) No 432/2012 establishing a list of permitted health claims made on foods, other than those referring to the reduction of disease risk and to children’s development and health Text with EEA relevance. OJEU. 2012;L136:1–40. [Google Scholar]
- 7.China State Food and Drug Administration. Kuo shih yao jian shu No. [2010]2. State food and drug administration, people; Republic of China: 2010. [accessed July, 13, 2022]. Available at, http://www.gov.cn/gzdt/2010-01/11/content_1507641.htm. [Google Scholar]
- 8.Taiwan Food and Drug Administration. Specifications requirement for Monascus health food. Taiwan food and drug administration, Taiwan. 2007. [accessed July, 13, 2022]. Available at, https://law.moj.gov.tw/LawClass/LawAll.aspx?pcode=L0040077.
- 9. Park HG, Elena KS, Jong SC. Phylogenetic relationships of Monascus species inferred from the ITS and the partial β-tubulin gene. Bot Bull Acad Sinica. 2004;45:325–30. [Google Scholar]
- 10. Katoh K, Misawas K, Kuma KI, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar S, Stecher G, Li M, Knyaz C, Tamura K, Mega X. Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tavaré S, Miura RM. Lectures on mathematics in the life sciences. USA: American Mathematical Society; 1986. Some probabilistic and statistical problems in the analysis of DNA sequences; pp. 57–86. [Google Scholar]
- 13. Dai WH, Shao YC, Chen FS. Production of Monacolin K in Monascus pilosus: comparison between industrial strains and analysis of its gene clusters. Microorganisms. 2021;9:747. doi: 10.3390/microorganisms9040747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, et al. Human–mouse alignments with BLASTZ. Genome Res. 2003;13:103–7. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mitchell AL, Teresa KA, Babbitt PC, Blum M, Bork P, Bridge A, et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019;47:D351–60. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee CL, Chen WP, Wang JJ, Pan TM. A simple and rapid approach for removing citrinin while retaining monacolin K in red mold rice. J Agric Food Chem. 2007;55:11101–8. doi: 10.1021/jf071640p. [DOI] [PubMed] [Google Scholar]
- 18. Einaxa E, Voigta K. Oligonucleotide primers for the universal amplification of β-tubulin genes facilitate phylogenetic analyses in the regnum Fungi. Org Divers Evol. 2003;3:185–94. [Google Scholar]
- 19. Li YP, Xu Y, Huang ZB. Isolation and characterization of the citrinin biosynthetic gene cluster from Monascus aurantiacus. J Biotechnol Lett. 2012;34:131–6. doi: 10.1007/s10529-011-0745-y. [DOI] [PubMed] [Google Scholar]
- 20. Yoon SH, Ha SM, Lim JM, Kwon S, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Leeuwenhoek. 2017;110:1281–6. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 21. Zhuang KW, Liu Y, Dai YL, Xu JR, Li WJ, Ming H, et al. Nocardia huaxiensis sp. nov., an actinomycete isolated from human skin. Int J Syst Evol Microbiol. 2021;71(8):2021. doi: 10.1099/ijsem.0.004970. [DOI] [PubMed] [Google Scholar]
- 22. Kwon HJ, Balakrishnan B, Kim YK. Some Monascus purpureus genomes lack the monacolin K biosynthesis locus. J Appl Biol Chem. 2016;59:45–7. [Google Scholar]
- 23. Chen YP, Yuan GF, Hsieh SY, Lin YS, Wang WY, Liaw LL, et al. Identification of the mokH gene encoding transcription factor for the upregulation of monacolin K biosynthesis in Monascus pilosus. J Agric Food Chem. 2010;58:287–93. doi: 10.1021/jf903139x. [DOI] [PubMed] [Google Scholar]
- 24. Higa Y, Kim YS, Amin AU, Huang M, Ono N, Kanaya S. Divergence of metabolites in three phylogenetically close Monascus species (M. pilosus, M. ruber, and M. purpureus) based on secondary metabolite biosynthetic gene clusters. BMC Genom. 2020;21:679. doi: 10.1186/s12864-020-06864-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen YP, Tseng CP, Chien IL, Wang WY, Liaw LL, Yuan GF. Exploring the distribution of citrinin biosynthesis related genes among Monascus species. J Agric Food Chem. 2008;56:11767–72. doi: 10.1021/jf802371b. [DOI] [PubMed] [Google Scholar]
- 26. Shimizu T, Kinoshita H, Ishihara S, Sakai K, Nagai S, Nihira T. Polyketide synthase gene responsible for citrinin biosynthesis in Monascus purpureus. Appl Environ Microbiol. 2005;71:3453–7. doi: 10.1128/AEM.71.7.3453-3457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gyetvai Á, Emri T, Takács K, Dergez T, Fekete A, Pesti M, et al. Lovastatin possesses a fungistatic effect against Candida albicans, but does not trigger apoptosis in this opportunistic human pathogen. FEMS Yeast Res. 2006;6:1140–8. doi: 10.1111/j.1567-1364.2006.00097.x. [DOI] [PubMed] [Google Scholar]
- 28. Thynne E, Mead OL, Chooi YH, McDonald MC, Solomon PS. Acquisition and loss of secondary metabolites shaped the evolutionary path of three emerging phytopathogens of wheat. Genome Biol Evol. 2019;11:890–905. doi: 10.1093/gbe/evz037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tien AJ, Chueh TH, Hsia CP, Chien CT. Monascus adlay and monacolin K attenuates arterial thrombosis in rats through the inhibition of ICAM-1 and oxidative stress. Kidney Blood Press Res. 2016;41:815–27. doi: 10.1159/000452584. [DOI] [PubMed] [Google Scholar]
- 30. Marič A, Skočaj M, Likar M, Sepčić K, Cigić IK, Grundner M, et al. Comparison of lovastatin, citrinin and pigment production of different Monascus purpureus strains grown on rice and millet. J Food Sci Technol. 2019;56:3364–73. doi: 10.1007/s13197-019-03820-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen WP, Chen RF, Liu QP, He Y, He K, Ding XL, et al. Orange, red, yellow: biosynthesis of azaphilone pigments in Monascus fungi. Chem Sci. 2017;8:4917–25. doi: 10.1039/c7sc00475c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang C, Zhu QQ, Zhang H, Zhang N, Xue L, Yang ZY, et al. Effects on the sporulation and secondary metabolism yields of Monascus purpureus with mokH gene deletion and overexpression. Fungal Biol. 2020;124:661–70. doi: 10.1016/j.funbio.2020.03.010. [DOI] [PubMed] [Google Scholar]
- 33. Chen WP, Feng YL, Molnár I, Chen FS. Nature and nurture: confluence of pathway determinism with metabolic and chemical serendipity diversifies Monascus azaphilone pigments. Nat Prod Rep. 2019;36:561–72. doi: 10.1039/c8np00060c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gordon RY, Cooperman T, Obermeyer W, David JB. Marked variability of monacolin levels in commercial red yeast rice products: Buyer beware. Arch Intern Med. 2010;170:1722–7. doi: 10.1001/archinternmed.2010.382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.