Abstract
Background Patient cohorts generated by machine learning can be enhanced with clinical knowledge to increase translational value and provide a practical approach to patient segmentation based on a mix of medical, behavioral, and social factors.
Objectives This study aimed to generate a pragmatic example of how machine learning could be used to quickly and meaningfully cohort patients using unsupervised classification methods. Additionally, to demonstrate increased translational value of machine learning models through the integration of nursing knowledge.
Methods A primary care practice dataset ( N = 3,438) of high-need patients defined by practice criteria was parsed to a subset population of patients with diabetes ( n = 1233). Three expert nurses selected variables for k-means cluster analysis using knowledge of critical factors for care coordination. Nursing knowledge was again applied to describe the psychosocial phenotypes in four prominent clusters, aligned with social and medical care plans.
Results Four distinct clusters interpreted and mapped to psychosocial need profiles, allowing for immediate translation to clinical practice through the creation of actionable social and medical care plans. (1) A large cluster of racially diverse female, non-English speakers with low medical complexity, and history of childhood illness; (2) a large cluster of English speakers with significant comorbidities (obesity and respiratory disease); (3) a small cluster of males with substance use disorder and significant comorbidities (mental health, liver and cardiovascular disease) who frequently visit the hospital; and (4) a moderate cluster of older, racially diverse patients with renal failure.
Conclusion This manuscript provides a practical method for analysis of primary care practice data using machine learning in tandem with expert clinical knowledge.
Keywords: social determinants of health, phenotypes, primary care, nursing, machine learning, care coordination
Background and Significance
Coordinating transitional care for those with multiple chronic or complex chronic conditions, functional disabilities, and/or social needs 1 requires collaboration with service partners outside the health care sector. For example, persons with housing insecurity may require care from the health, behavioral health, and social service sectors at the time of hospital discharge, yet the fragmented continuum of care adds to treatment burden and jeopardizes the safety of people who already have compromised health. To improve care for people with high medical, behavioral, and/or social needs, such as people experiencing housing insecurity and homelessness in the setting of complex medical needs, it is necessary to prioritize the population receiving care transitions follow-up to those with the greatest need, partner with social sector providers to improve transitional care continuity, and optimize health information technology, such as health information exchange (HIE), to include cross-sector comprehensive shared care plans. 2
Data-driven personalized clinical classification models of high-need persons aid in the development of comprehensive shared care plans and care coordination across care sectors for persons with social needs who are at risk for experiencing poor health outcomes. 3 4 Precision health, defined by the Council for the Advancement of Nursing Science, is “an emerging approach to individualizing health care and includes genomics and other physiological, psychological, environmental, and ethical factors that are central to the development and testing of individualized treatments and prevention strategies for persons, families, and communities.” 5 Highly accurate personalized algorithms, an example of precision health, were employed during the early days of the COVID-19 pandemic to identify patients with the highest mortality risk, which helped to direct limited health system resources to those most in need. 6
Despite the promise of using personalized approaches to improve patient outcomes, relying on algorithms to classify people into clinically actionable cohorts may unintentionally perpetuate health disparities. 7 8 Identifying and stratifying populations for the purpose of enrollment into care coordination programs has historically been achieved through the use of risk score algorithms that are heavily influenced by cost and medical diagnoses. Research suggests that this methodology may be flawed in that it is biased toward patients who have the ability and willingness to incur costs in the medical system and it does not incorporate important patient-centric considerations such as social needs and access to care. 7 As a result, cohorts generated from traditional algorithms (typically based on health utilization data) may be more reflective of high-cost patients, for example, those with medical insurance, than those with significant needs who could benefit from coordinated care such as those algorithms considering the broader constellation of social needs that exert influence on health outcomes. Health systems are particularly susceptible to this offense when predicting important clinical risk factors such as disease onset, the likelihood of hospitalization, and medication adherence 8 9 as the traditional approach to building predictive models utilizes data that capture historical patterns of care that are more likely to be representative of social privilege (e.g., access to childcare and transportation) and structural inequities in health systems (e.g., access to health care providers), rather than revealing the unmet needs of populations who are most vulnerable to harm.
The thoughtful design of precision health algorithms, therefore, requires inclusion of representative voices throughout creation who can bring attention to the contextualized needs of patients, with particular attention being paid to the clinical context to which the algorithms will be applied. 7 9 10 Nurses, who make up the largest number of health care professionals nationwide, are increasingly participating in the design and development of precision health algorithms using machine learning (ML) approaches. 11 12 Nursing has increasingly used ML learning methods to develop a variety of algorithms that address important clinical issues ranging from standardizing nomenclature for machine-identified topic models, through the development of complex care management systems, identifying mortality risk using nurse-generated data, exploring predictors of hospice use, and for addressing important nursing workforce issues such as burnout and staffing. 12 13 14 15 16 However, there remains a need for nursing to lead the expansion of social determinants of health (SDOH) and biopsychosocial features into precision health algorithms to advance health equity and access to care. 12 17
To address these concerns, we have embarked on the preliminary phase of our personalized cross-sector transitional care management project (PC-TCM) and commenced design of an actionable clinical classification model of high-need persons that incorporates relevant medical and behavioral health factors as well as psychosocial phenotypes. 18 The original conceptualization of psychosocial phenotyping is based on the work of Kim and colleagues who sought to more effectively personalize care by identifying key psychological and social features of biological and environmental conditions that influence health outcomes. Our long-term goal is to develop a precision health approach that comprehensively and effectively identifies people with high needs while deemphasizing the costs of their care, an approach that has been exempt from prior studies. Through the development of this model and resulting dissemination of results to cross-sector collaborators, our study makes an important contribution to the literature given its pragmatic approach, consideration of the overlapping influences of SDOH and chronic conditions that can be easily replicated in community-based primary care settings, and reduction of bias through the intentional deemphasis of costs of care as predictors of risk.
Objectives
An initial PC-TCM project step builds on our previous work using HIE to alert nurse care coordinators about social and medical complexity for the purpose of prioritization of postacute discharge outreach calls. 19 The current paucity of discrete data on social need available through the HIE led us to explore data available at the participating primary care practice site. Our desire to generate a flexible, translatable, “real world” example of how ML could be used to quickly and meaningfully cohort patients resulted in development of an unsupervised classification model to better understand our data. The purpose of this article is to describe how nursing knowledge informed our methodology, preliminary results, and to discuss how the output could be translated into clinical practice.
Methods
Utilizing primary care practice data, this exploratory study employs data clustering approaches to identify clinically relevant subgroups of high-need individuals that can be addressed as part of a cross-sector care plan. The study is guided by the World Health Organization (WHO) definition of SDOH as “nonmedical factors that influence health outcomes.” 20 According to the WHO, SDOH are influenced by social norms, economic and political policies, and systems that shape the conditions of daily life. 20
Nursing Knowledge Application to the Conceptual Framework
A key aim of our PC-TCM project is to increase the capability to segment the “High-Cost High-Need” population into subsets with specific cross-sector needs. Understanding the limitations of traditional cost-based segmentation algorithms, the team referenced the National Academy of Medicine's (NAM) conceptual model of a starter taxonomy for high-need patients to shape our working definition of “High Need” and to influence the development of our clinical segmentation rules. 1 The NAM's model builds upon the groupings proposed by Joynt and colleagues 21 by layering in behavioral health comorbidities and social risk factors ( Table 1 ). Joynt et al's work provides specific diagnosis-based criteria for identifying clinical and functional groups (i.e., multiple chronic conditions, advancing illness). However, NAM's publication does not share logic for identifying and parameterizing behavioral health and social needs.
Table 1. Adaptation of starter taxonomy for high need patients with features and data sources for variables included in analysis a .
| Multiple chronic b | Major complex chronic b |
|---|---|
| Hypertension | Diabetes |
| Obesity | Cardiovascular |
| Chronic pulmonary disease | Liver disease |
| Hypothyroid | Renal failure |
| Behavioral health factors b | |
| Substance use | |
| Mental health | |
| Social risk factors—extracted from practice electronic health record | |
| Age, sex, childhood illness | |
| Ethnicity, language, race, new refugee | |
| Utilization features—from ADT notification from the health information exchange | |
| Count of hospital visits, flag indicating 2+ hospital visits | |
| Outpatient visit in the past year | |
Abbreviation: ADT, admission, discharge, and transfer.
The original taxonomy 1 segments the high-need population into children with complex needs, disabled, frail elderly, and advancing Illness, but only multiple chronic and major complex chronic are included in this analysis. Behavioral health factors and social risk factors cut across all the segments.
Chronic conditions are extracted from International Classification of Disease (ICD-10 codes).
Given a lack of explicit guidance on a segmentation approach that incorporates a mix of clinical and social indicators, the PC-TCM team initiated a sub-project focused on developing a novel stratification model that integrates these features. Table 1 demonstrates how features available in the practice's table were aligned with the NAM's model for two clinical groups, multiple chronic and major complex chronic. This nurse-led initiative was inspired by the clinical groupings of the NAM starter taxonomy and domains of demographic and social factors proposed by Kim et al. 18 Reflecting on nursing experiences caring for high-need patients, the sub-team considered that a highly parameterized model might not produce meaningful cohorts due to the complex intersection of medical and social needs, implicit bias, and potential membership of single patients in multiple clinical groups (i.e., multiple chronic conditions and frailty). Having collaborated on past data science projects, the sub-team became interested in developing a computer-assisted classification model to facilitate rapid identification of meaningful patient clusters that were reflective of medical and social needs as well as key patient characteristics that could impact care coordination (i.e., language and cultural affinity). The team hypothesized that these clusters could be interpreted and enhanced using nursing knowledge and evidence-based guidelines to produce actionable cohorts and associated care plans for PC-TCM and therefore meet the aim associated with the identification of cross-sector needs.
Setting
The project setting is a Federally Qualified Health Center that is recognized by the National Committee for Quality Assurance, with clinics located in five resource-poor urban neighborhoods in Buffalo, New York, United States. Twenty-eight percent (28%) of the population lives in poverty and 55% report belonging to underrepresented racial and/or ethnic backgrounds. 22 Although there are four large medical centers, two of which are safety net hospitals, access to health care is limited by health-related social needs such as inadequate food, transportation, and housing. The practice services a population of high-need individuals, the majority Medicaid eligible, and a large proportion with immigrant or refugee status. The practice is a partner in the PC-TCM project and requested guidance on how to use a previously existing summary database for clinical quality improvement purposes.
Summary Table
The summary table provided by the project primary care practice for their roster of current patients (i.e., those with an outpatient visit in the past year) included demographic information, annual utilization counts (inpatient or emergency visits based on admission, discharge, and transfer information from the HIE) and selected medical and social conditions of importance to the practice. The practice extracted a de-identified dataset of unique individuals containing 25 risk factor indicators for all adult patients with at least two conditions ( n = 3,438). The contents of the summary table were at the discretion of the practice and included features that were important to practice, and this may introduce selection bias. The extracted data were determined to not qualify as human subjects research by the Institutional Review Board.
Data Acquisition, Cleaning, and Exploration
The summary table was provided by the practice, downloaded from a secure folder, and imported into R version 4.1.3 using R Studio version 2022.02.1 Build 461. The dataframe underwent extensive exploratory analysis and cleaning to prepare for analysis. First steps included removal of the total line and normalizing variable names. Because missing values were rare (less than 1% of language, race, ethnicity, and 3% of secondary insurance), we chose to recode them as “Unknown” as opposed to censoring the affected records or imputing the values. All condition variables were recoded to binary (1/0) and dummy variables were created to collapse some data points. For example, the lang_diverse variable was created such that English language was coded as 0 and all others coded as 1. Similarly, the race_diverse variable was created such that White was coded as 0 and all others coded as 1. English language and White race were selected as the standard (0) as they are the most prevalent in the greater Buffalo area according to Census data, 22 therefore, the presence of positive indicators for nondominant language and race were felt to be potential key differentiators. Frequencies were visualized in the form of bar plots for age, gender, race, ethnicity, language, practice location, primary insurance, secondary insurance, count of inpatient visits, and medical conditions to help determine which to select for input into the model. Finally, a correlation matrix was generated to help detect strong relationships between variables.
Nursing Knowledge Application to Variable and Sample Selection
Our initial aim was to develop clusters that centered around intrapersonal and social features. As such, the following risk factor indicators were initially considered: age, sex (female = 1), language spoken (lang_diverse, 1 = non-English), race (race_diverse, 1 = not White), number of hospitalizations (hosp_visits), number of conditions (number_of_yes), and specific conditions of substance use disorder (SUD) and mental health diagnoses. However, further consideration of the importance of preserving a holistic patient view led to expanding the list of conditions to include any condition flags found in at least 3% of the primary care practice population to include more features examining medical complexity in addition to social and behavioral risks.
The analysis excluded specific variables for a variety of reasons. The variable “appt_in_12mos” indicates whether a patient on the roster had an appointment within the 12 months prior to its generation; we omitted it as the value was always set to “Yes” and therefore provided no classification value. We chose to exclude the location number (loc_number) from the model as we wanted to permit patients in clusters to span across physical practice locations. Conditions affecting fewer than 3% of the adult population were excluded to prevent the generation of clusters with very small volumes of patients with rare disorders that were not appropriate for care coordination programs. We excluded primary and secondary insurance given that we intentionally set out to design a model that was not focused on cost or primary payment source. The roster contained two variables pertaining to hospital-based utilization; we chose to keep the discrete count of visits and omit the flag for 2 or more visits (2 + _hosp_visits) so that we could compute the mean number of visits for patients in each cluster. Finally, we observed that the ethnicity variable stratified patients on the basis of Hispanic/Latino heritage and its values were different from those in the race variable. As explained above, we chose to collapse race into a binary variable using White as the comparison group and all other races as the reference group. As such, we omitted the ethnicity variable. The final list of excluded variables includes: appt_in_12_months (always “Yes”), loc_number (classifier), ethnicity (given that we opted for a binary race variable), prim_ins and sec_ins (cost- and payer-centric classifiers), 2 + _hosp_visits (duplicative to hospital encounter counts), and conditions where <3% of the population was flagged.
Consistent with our desire to produce a machine-generated model whose outputs could be rapidly translated into clinical practice, we decided to limit the input data to patients with a diagnosis of diabetes. The top three most prevalent chronic conditions in the population included hypertension, diabetes, and obesity; of these, diabetes involves the widest range of clinical complexities and array of interventions based on disease subtypes. Additionally, there is a strong positive correlation between diabetes, hypertension, and obesity.
K-Means Clustering
After parsing out a subset of persons with diabetes ( n = 1,233) from the dataset, we used the selected variables to develop a classification model that incorporated medical and social features. Moreover, we considered the characteristics of our data when determining the best type of ML for our project. Given the manageable size of the sample, the limited number of input features, and the lack of a predefined grouping variable, we chose to employ the K-means clustering technique. This algorithm is a popular unsupervised ML approach that is highly effective at identifying patterns not necessarily discernable through manual analytic work. 23
K-means clustering works by sorting cases into groups that most closely match their combination of characteristics. An optimized model will minimize differences between members in the same cluster while maximizing differences between clusters. Each cluster is defined by a centroid (central features) and all cases whose features most closely match its parameters. Therefore, it is important to set the number of clusters appropriately to achieve optimization. Given the pilot nature of our work, we favored speed to conceptual model over statistical precision when setting cluster count. As such, we used the “elbow method” to set cluster N . The elbow method involves creating a visualization to illustrate the cut point where increasing the number of clusters generated yields no additional value. It can be used to determine the amount of variation in the data explained by the model and compute the marginal gain or loss resulting from increasing or decreasing the number of clusters. 24 Based on visualizing an elbow diagram from our data, we chose four centroids to define four distinct patient cohorts (clusters) representing the natural groupings of data features. 23
The output of a K-means model is simply a number indicating the cluster (centroid) to which each case most closely aligns. We applied the cluster numbers back to our table containing diabetic patient data and then profiled each cluster. This involved counting the number of patients in each cluster, determining the average age, number of hospital-based encounters, and risk factor count. We also computed prevalence rates (percentages) for the medical and social features.
Nursing Knowledge Application to Clinical Relevance
Initial cluster profiles helped begin conceptualization of the ways in which each cluster could be described in clinical terms as distinct cohorts. However, we recognized the importance of contextualizing the results to promote better translation and adoption into practice. As such, we enhanced the machine-generated clusters using nursing knowledge within a psychosocial phenotyping framework 18 to further describe cohorts in a clinically meaningful way. Registered Nurses with PhD degrees in medical anthropology and nursing; MS degrees and content expertise in gerontology, business administration and health informatics; and certification in hospice and palliative care and health data analysis determined the clinical relevance of the data features. 4 5 18 As opposed to traditional, diagnosis-based segmentation models, our nurse-enhanced model resulted in cluster profiles that differentiated patients by a mix of medical, behavioral, and social needs as well as key priorities for cross-sector care coordination.
Results
Descriptive Statistics
The total adult population meeting the practice's criteria for high need (those with two or more risk factors) was 3,438 with a minimum age of 20 and the maximum age truncated at 90, with an average age of 50. Fifty-six percent (56%) of the adult population were female, and 49% spoke English, with other common languages being Burmese, Bangla/Bengali, Nepali, and Karen. A third of the adult population were described as Asian (34%), 27% were Black/African American, and 19% were White. Eleven conditions were present in at least 3% of the adult population ( Fig. 1 ) with hypertension (50%), childhood illness (40%), obesity (39%), and diabetes (36%) topping the list. The subset of cases with diabetes ( n = 1233) was entered into the model.
Fig. 1.
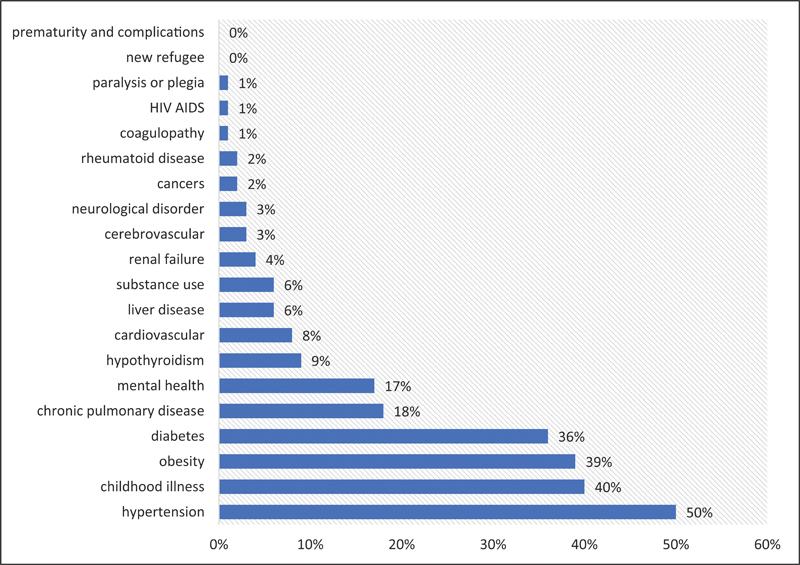
Prevalence of selected conditions in the adult population ( N = 3,438) a . a Conditions with greater than 3% prevalence were entered into analysis.
Data Clusters and Phenotype Description
The enhanced model ( N = 1,233 unique individuals) yielded four distinct patient cohorts that differed by factors such as comorbidities, primary language, gender, and hospital utilization patterns ( Table 2 ). Together, these characteristics help create profiles of the underlying patients that can facilitate a deeper understanding of their clinical conditions, cultural preferences, social roles, and patterns of interaction with health care systems. Each variable used in the model was summarized to allow for comparison. Mean_age, hosp_visits, and number_of_yes are summarized as the mean for the cluster, and the remaining variables are summarized as percentages (sum of flags/total patients). After application of nursing knowledge, clusters were grouped into descriptive psychosocial phenotypes.
Table 2. Clusters of patients within the diabetes cohort ( N = 1,233 cases) .
| Cluster | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Count (percent of cohort) | 727 (59%) | 412 (33%) | 21 (2%) | 73 (6%) |
| Mean age | 51 | 56 | 53 | 63 |
| Mean hospital visit count | 0.7 | 3.1 | 8 | 3.5 |
| Mean risk factor count | 2.7 | 3.8 | 5.1 | 4.7 |
| Female | 56% | 57% | 29% | 51% |
| Racially diverse | 97% | 66% | 76% | 74% |
| Non-English speakers | 80% | 25% | 29% | 53% |
| Substance use disorder | 0% | 0% | 100% | 0% |
| Mental health disorder | 4% | 24% | 29% | 15% |
| Hypertension | 54% | 76% | 67% | 89% |
| Cardiovascular disease | 1% | 18% | 33% | 18% |
| Renal failure | 0% | 0% | 10% | 100% |
| Obesity disorder | 25% | 44% | 33% | 27% |
| Chronic pulmonary disease | 3% | 28% | 14% | 21% |
| Hypothyroidism | 6% | 9% | 5% | 11% |
| Liver disease | 3% | 2% | 19% | 3% |
| Childhood illness | 55% | 29% | 24% | 29% |
| Phenotype | Large cluster of racially diverse female, non-English speakers with low medical complexity, history of childhood illness | Large cluster of English speakers. Significant comorbidities include obesity and respiratory disease. Least diverse group | Small cluster of male patients with substance use disorder who frequently visit the hospital. Significant comorbidities include mental health issues, liver and cardiovascular disease | Moderate cluster of older, racially diverse patients with renal failure. May not speak English. Significant comorbidities include hypertension, respiratory disease, hypothyroidism |
| Priorities | Attention to potential language barrier to prevent comorbidities | Management of comorbidities and avoidance of hospital use | Substance use intervention and treatment to avoid hospital encounters | Attention to comorbidities and renal failure to avoid hospital encounters |
The first cluster can be described as a phenotype representing a cohort (59%) of racially diverse (97%), non-English speakers (80%) ( n = 727). The mean age is 51, 56% were female, and the majority (55%) have experienced childhood illness. Social priorities for individuals within the first cluster include attention to potential language barriers during health promotion activities.
The second largest cluster includes 412 (33%) persons who are, on average, 56 years old. Fifty-seven percent (57%) of these individuals are female, with an average of four risk factors, and three hospital visits in a year. This cohort has the lowest rate of racial diversity (66%) and the highest percentage of English speakers (75%). The priorities of those individuals in cluster 2 include attention to management of comorbidities of obesity and respiratory disease and avoidance of hospital use.
The third cluster includes 21 individuals (2%) who are 53 years old, on average, primarily male (71%), and 76% racially diverse. Twenty-nine percent (29%) of individuals in cluster 3 are non-English speakers, 100% have SUD, and 29% have at least one mental health diagnosis. Those individuals represented in cluster 3 experienced an average of eight hospital visits and had an average of five risk factors. This cluster represents a small cohort of male patients with SUD who frequently visit the hospital. Although we profiled these individuals in the context of diabetes, higher social priorities for these patients include SUD intervention and treatment to avoid hospital encounters.
The fourth cluster represents 73 (6%) individuals from the population. Half are female and are slightly older, averaging 63 years old. One hundred percent (100%) of these individuals have renal disease, 74% are racially diverse, and 53% are non-English speakers. Those persons in this phenotype have an average of five risk factors and three to four hospital visits. This phenotype represents a moderate cohort of older, racially diverse patients with renal failure who may not speak English. Significant comorbidities include hypertension, respiratory disease, and hypothyroidism. Social priorities include attention to management of comorbidities, avoidance of hospital use, and coordination of services related to kidney disease.
Clinical Application
Clinical application of these results must consider the context of the primary care practice which provided the high needs report and requested guidance in how to use the information more effectively. 10 It is important to remember that the high-need population was defined by having two or more conditions and is only 15% of the practice population. Targeting this group for immediate postdischarge outreach and assessment by nurses can stabilize them in the community by rapid deployment of the right services. The average number of hospital visits range from 0.7 to 8 per year, indicating that there is a need for transitional care coordination. We enhanced the machine-generated clusters using nursing knowledge within a psychosocial phenotyping framework to identify cohorts that were clinically meaningful. Table 2 demonstrates how clinical expertise is used to interpret or curate the statistical findings to write descriptive statements about the cohort characteristics. The phenotype description then allows identification of clinical and cross-sector priorities. The cohort number can be applied back to the dataset to segment the diabetes population into actionable groups and flag clinical and psychosocial needs for the primary care practice, resulting in the ability to quickly create plans of care that consider each patient's clinical and psychosocial phenotype ( Table 3 ). Knowing the differences in social factors between the groups is essential in developing a care team and plan to address their very different needs.
Table 3. Process for integrating nursing knowledge into translation of phenotype to social and medical care plans.
| Process Step | Description | ||||
|---|---|---|---|---|---|
| 1. Select Population | High-need primary care practice patients ( N = 3,438) | ||||
| 2. Inform selection of variables based on nursing knowledge | Included variables | Age, sex, language spoken, race, number of hospital visits, number of conditions, conditions found in at least 3% of population including psychosocial characteristics available in dataset | |||
| Excluded variables | Always yes (appointment in 12 months), irrelevant classifiers (clinic number; insurance); highly correlated variables (ethnicity; two or more hospital visits), conditions found in <3% of population | ||||
| 3. Parse out subset condition | Diabetes sub-population ( n = 1,233) | ||||
| 4. Perform cluster | Cluster 1: 59% (727 patients) |
Cluster 2: 33% (412 patients) |
Cluster 3: 2% (21 patients) |
Cluster 4: 6% (73 patients) |
|
| 5. Apply nursing knowledge | Review summary statistics of clinical and social variables in each cluster to develop psychosocial phenotype description and recommend care plan elements | ||||
| 6. Describe psychosocial phenotype | Large cluster of racially diverse female, non-English speakers with low medical complexity, history of childhood illness | Large cluster of English speakers. Significant comorbidities include obesity and respiratory disease. Least diverse group. | Small cluster of male patients with substance use disorder who frequently visit the hospital. Significant comorbidities include mental health issues, liver and cardiovascular disease | Moderate cluster of older, racially diverse patients with renal failure. May not speak English. Significant comorbidities include hypertension, respiratory disease, hypothyroidism | |
| 7. Draft social care plan | Culturally sensitive interventions in the patient's preferred language | Weight and comorbidity management to promote hospital avoidance | Referral for SUD treatment, screening, and assistance with SDOH needs including homelessness, community health worker/peer advocate | Complex care coordination for multiple chronic conditions, culturally sensitive interventions aimed at avoidance of hospital encounters | |
| 8. Align medical care plan | Antidiabetic agent, A1c, health promotion, patient teaching, surgical or nonsurgical weight loss interventions for population experiencing obesity (patients experiencing obesity comprise 31% [ n = 381] and occur in each cluster). | ||||
Abbreviations: SDOH, social determinants of health; SUD, substance use disorder.
Segmenting patients into meaningful cohorts for care coordination programs can be a challenging and time-consuming process for health care organizations. This is particularly true among resource-constrained practices that lack time to manually produce cohorts or sophisticated applications to automate their generation. Additionally, existing segmentation models tend to rely heavily on clinical condition diagnoses and omit social factors, resulting in patient groups that lack person-centric dimensionality. Our machine-generated, nurse-enhanced clustering algorithm demonstrates direct and meaningful application to nursing practice by quickly and efficiently producing distinct and specific patient cohorts as well as informing characteristics of the resources required to manage their needs. For example, although our sample population consisted of patients with diabetes, our model identified a niche population of males with SUD whose behavioral health needs are likely more critical than the management of blood sugar and related lifestyle changes. Traditional segmentation models that do not incorporate social factors may have failed to identify this subgroup, or more importantly, may have failed to inform the practice of their most critical needs. Our simple clustering algorithm, created with open-source software and a limited set of features, was able to generate multi-dimensional patient profiles that deliver actionable information to prospective care coordinators.
Discussion
In this study we used data science approaches to develop a precision health approach for integrating medical and SDOH factors into interpretable models for the personalization of care of high-need persons with multiple chronic illness and/or social needs. Using psychosocial phenotyping, 18 nursing knowledge, and through focus on clinical and social needs, in lieu of cost of care need, this preliminary project underscores the possibility of generating a flexible, translatable, “real world” example of how ML could be used to quickly and meaningfully cohort patients into actionable phenotypes. Psychosocial phenotyping is an emerging field with significant promise for addressing an array of social and behavioral needs of vulnerable groups. 25 26 To the best of our knowledge, our study is the first to develop phenotypes of persons with “high needs” combining chronic health conditions with social needs. In addition, our work intentionally avoids using health utilization costs to classify high-need individuals. This is important because there is very little research investigating disease conditions within the social environment without attention to the cost of care despite the fact that it is well known that SDOH and health conditions (and outcomes) are inextricably linked. 25 Moreover, a recent scoping review of predictive models and other data-science oriented research found that merely 20% of included studies were conducted within a social or community context. 25
Most similar to our work, Byrne and colleagues 27 have predicted housing instability and homelessness within the Veterans Health Administration using social history (e.g., branch of service, service use, and diagnosis) for personalized interventions, but their models are fit to a narrow population with a specific need, and do not take into account medical complexity. Burgermaster and Rodriguez 28 used sophisticated analytic approaches to define 20 different phenotypes predicting elevated weight (body mass index [BMI] ≥ 25 kg/m 2 ) and personalizing behavioral interventions based on psychosocial-behavioral characteristics. Their work yielded both positive and negative associations of elevated BMI, which has important clinical implications for personalizing care. However, their models do not take into account the impact of the intersection of SDOH and chronic conditions on health outcomes. 28 Recently SDOH phenotypes predicting maternal health morbidity from income, stress, and immigration status show promise for improving maternal health outcomes, but the models were limited to a population of healthy individuals rather than those with chronic illness. 29 Thus, our study makes an important contribution to the literature given its pragmatic approach, consideration of the overlapping influences of SDOH and chronic conditions that can be easily replicated in community-based primary care settings, and reduction of bias through the intentional de-emphasis of costs of care as a predictor of risk.
There are important limitations to our study that warrant mentioning. The dataset used in this example is based on historical data rather than prospective data and comes from a single primary care practice that contained internally defined risk factors. Therefore, the findings of this study cannot be generalized to the broader population and historical data will always contain some bias. However, our approach is scalable and relevant to a variety of clinical settings, whether it is a community-based organization working with limited, local data and resources (i.e., manual analysis), or a large organization with extensive resources for conducting sophisticated statistical analysis.
Our next steps are to explore other, more sophisticated clustering models, including PAM (partitioning around medoids), hierarchical clustering, and fuzzy k-means. Additionally, we would like to explore the use of other data science methods for developing psychosocial phenotypes such as decision-tree, latent class analysis, or random forest. In the future, we intend to further explore how unstructured data may be used in refining our phenotypes, including the use of natural language processing approaches. 25 26 Additionally, we intend to apply the principles of distributive justice 8 when evaluating and deploying our models into clinical practice, recognizing that a multi-pronged approach is necessary to comprehensively identify algorithmic bias. According to Rajkomar and colleagues, 8 principles of distributive justice help ensure fairness in algorithms through the consideration of equal outcomes (i.e., assurance of equal benefit in outcomes), equal performance (i.e., equally accurate models), and equal allocation (i.e., demographic parity). This process requires input from key stakeholders, particularly marginalized voices.
Finally, we plan to collaborate with interdisciplinary team members at the primary care practice to continue translational research in two ways: first, we want to better understand barriers and accelerants to implementing ML-based cohorts into clinical practice. Next, we want to understand the degree to which ML-generated cohorts “fit” the population by direct stakeholder query: were the cohorts truly clinically actionable? Is this technique associated with improved patient and care coordination process outcomes?
Conclusion
The present study provides an example of a pragmatic approach for the rapid development of classification models for segmenting populations into actionable cohorts of high-need individuals using data that are typically available in a primary care setting. This preliminary work makes an important contribution to the literature by demonstrating the application of nursing knowledge into ML approaches for advancing ways of knowing in learning health systems to guide clinical practice.
Clinical Relevance Statement
We enhanced the machine-generated clusters using nursing knowledge within a psychosocial phenotyping framework to identify cohorts that were clinically meaningful, resulting in the ability to quickly create plans of care that consider each patient's clinical and psychosocial phenotype.
Multiple-Choice Questions
-
How do historical data contribute to algorithmic bias?
Historical data only capture encounters and do not reflect barriers to care.
Historical data were collected for purposes other than algorithm development and may contain racial and gender biases introduced during data entry.
Historical data do not necessarily represent the population, which can lead to flawed assumptions when applied to a new population.
All of the above
Correct Answer: The correct answer is option d.
-
Which of the following is TRUE about k-means clustering algorithms?
They minimize differences between data points in each cluster
They maximize differences between data points in each cluster
They minimize differences between clusters
They divide cases equally among clusters
Correct Answer: The correct answer is option a.
-
In this project, when was nursing knowledge employed?
During selection of the features to be included in the analysis.
During interpretation of significant features in the clusters.
During translation of the cohorts into care plans.
All of the above.
Correct Answer: The correct answer is option d.
Funding Statement
Funding Research reported in this publication was supported, in part, by the Agency for Healthcare Research and Quality under Award number: R01 HS028000-01.
Conflict of Interest None declared.
Protection of Human and Animal Subjects
The study was performed in compliance with the World Medical Association Declaration of Helsinki on Ethical Principles for Medical Research Involving Human Subjects and was reviewed by the University at Buffalo Institutional Review Board and the extracted data were determined to not qualify as human subject research.
References
- 1.Long P AM, Milstein A, Anderson G, Apton K L, Dahlberg M.Effective care for high-need patientsNational Academy of Medicine; 2017. Accessed March 20, 2023 at:https://nam.edu/HighNeeds/highNeedPatients.html [PubMed]
- 2.Sullivan S S, Mistretta F, Casucci S, Hewner S. Integrating social context into comprehensive shared care plans: a scoping review. Nurs Outlook. 2017;65(05):597–606. doi: 10.1016/j.outlook.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gambhir S S, Ge T J, Vermesh O, Spitler R. Toward achieving precision health. Sci Transl Med. 2018;10(430):eaao3612. doi: 10.1126/scitranslmed.aao3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corwin E, Redeker N S, Richmond T S, Docherty S L, Pickler R H. Ways of knowing in precision health. Nurs Outlook. 2019;67(04):293–301. doi: 10.1016/j.outlook.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hacker E D, McCarthy A M, DeVon H. Precision health: emerging science for nursing research. Nurs Outlook. 2019;67(04):287–289. doi: 10.1016/j.outlook.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Gao Y, Cai G-Y, Fang W. Machine learning based early warning system enables accurate mortality risk prediction for COVID-19. Nat Commun. 2020;11(01):5033. doi: 10.1038/s41467-020-18684-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Obermeyer Z, Powers B, Vogeli C, Mullainathan S.Dissecting racial bias in an algorithm used to manage the health of populations Science 2019366(6464):447–453. [DOI] [PubMed] [Google Scholar]
- 8.Rajkomar A, Hardt M, Howell M D, Corrado G, Chin M H. Ensuring fairness in machine learning to advance health equity. Ann Intern Med. 2018;169(12):866–872. doi: 10.7326/M18-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gervasi S S, Chen I Y, Smith-McLallen A. The potential for bias in machine learning and opportunities for health insurers to address it. Health Aff. 2022;41(02):212–218. doi: 10.1377/hlthaff.2021.01287. [DOI] [PubMed] [Google Scholar]
- 10.Romero-Brufau S, Wyatt K D, Boyum P, Mickelson M, Moore M, Cognetta-Rieke C. Implementation of artificial intelligence-based clinical decision support to reduce hospital readmissions at a regional hospital. Appl Clin Inform. 2020;11(04):570–577. doi: 10.1055/s-0040-1715827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.American Association of Colleges of Nursing Nursing fact sheet. Accessed January 8, 2023 at:https://www.aacnnursing.org/news-Information/fact-sheets/nursing-fact-sheet
- 12.Douthit B J, Walden R L, Cato K. Data science trends relevant to nursing practice: a rapid review of the 2020 literature. Appl Clin Inform. 2022;13(01):161–179. doi: 10.1055/s-0041-1742218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korach Z T, Cato K D, Collins S A. Unsupervised machine learning of topics documented by nurses about hospitalized patients prior to a rapid-response event. Appl Clin Inform. 2019;10(05):952–963. doi: 10.1055/s-0039-3401814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sullivan S S, Hewner S, Chandola V, Westra B L. Mortality risk in homebound older adults predicted from routinely collected nursing data. Nurs Res. 2019;68(02):156–166. doi: 10.1097/NNR.0000000000000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sullivan S S, Casucci S, Li C S. Eliminating the surprise question leaves home care providers with few options for identifying mortality risk. Am J Hosp Palliat Care. 2020;37(07):542–548. doi: 10.1177/1049909119892830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan S S, Bo W, Li C S, Xu W, Chang Y P. Predicting hospice transitions in dementia caregiving dyads: an exploratory machine learning approach. Innov Aging. 2022;6(06):igac051. doi: 10.1093/geroni/igac051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobensack M, Song J, Scharp D, Bowles K H, Topaz M. Machine learning applied to electronic health record data in home healthcare: a scoping review. Int J Med Inform. 2023;170:104978. doi: 10.1016/j.ijmedinf.2022.104978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim M T, Radhakrishnan K, Heitkemper E M, Choi E, Burgermaster M. Psychosocial phenotyping as a personalization strategy for chronic disease self-management interventions. Am J Transl Res. 2021;13(03):1617–1635. [PMC free article] [PubMed] [Google Scholar]
- 19.Hewner S, Sullivan S S, Yu G. Reducing emergency room visits and in-hospitalizations by implementing best practice for transitional care using innovative technology and big data. Worldviews Evid Based Nurs. 2018;15(03):170–177. doi: 10.1111/wvn.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization [WHO]. Social determinants of health. Accessed February 24, 2022 at:https://www.who.int/health-topics/social-determinants-of-health#tab=tab_1
- 21.Joynt K E, Figueroa J F, Beaulieu N, Wild R C, Orav E J, Jha A K.Segmenting high-cost Medicare patients into potentially actionable cohorts Healthc (Amst) 20175(1–2):62–67. [DOI] [PubMed] [Google Scholar]
- 22.United States Census Bureau Quick Facts: Buffalo City, New York. Accessed December 14, 2022 at:https://www.census.gov/quickfacts/buffalocitynewyork%20accessed%201/5/2023
- 23.Tan P-N, Steinbach M, Kumar V. Data mining cluster analysis: basic concepts and algorithms. Introduction Data Mining. 2013;487:533. [Google Scholar]
- 24.Bholowalia P, Kumar A. EBK-means: a clustering technique based on elbow method and k-means in WSN. Int J Comput Appl. 2014;105(09):17–24. [Google Scholar]
- 25.Bompelli A, Wang Y, Wan R.Social and behavioral determinants of health in the era of artificial intelligence with electronic health records: a scoping review. Health Data Science 2021;2021; [DOI] [PMC free article] [PubMed]
- 26.Patra B G, Sharma M M, Vekaria V. Extracting social determinants of health from electronic health records using natural language processing: a systematic review. J Am Med Inform Assoc. 2021;28(12):2716–2727. doi: 10.1093/jamia/ocab170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byrne T, Montgomery A E, Fargo J D. Predictive modeling of housing instability and homelessness in the Veterans Health Administration. Health Serv Res. 2019;54(01):75–85. doi: 10.1111/1475-6773.13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgermaster M, Rodriguez V A. Psychosocial-behavioral phenotyping: a novel precision health approach to modeling behavioral, psychological, and social determinants of health using machine learning. Ann Behav Med. 2022;56(12):1258–1271. doi: 10.1093/abm/kaac012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erickson E N, Carlson N S. Maternal morbidity predicted by an intersectional social determinants of health phenotype: a secondary analysis of the NuMoM2b dataset. Reprod Sci. 2022;29(07):2013–2029. doi: 10.1007/s43032-022-00913-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


