Abstract
Baculovirus late RNAs are transcribed by a four-subunit RNA polymerase that is virus encoded. The late viral mRNAs are capped and polyadenylated, and we have previously shown that capping is mediated by the LEF-4 subunit of baculovirus RNA polymerase. Here we report studies undertaken to determine the mechanism of 3′-end formation. A globin cleavage/polyadenylation signal, which was previously shown to direct 3′-end formation of viral RNAs in vivo, was cloned into a baculovirus transcription template. In vitro assays with purified baculovirus RNA polymerase revealed that 3′ ends were formed not by a cleavage mechanism but rather by termination after transcription of a T-rich region of the globin sequence. Terminated RNAs were released from ternary complexes and were subsequently polyadenylated. Mutational analyses indicated that the T-rich sequence was essential for termination and polyadenylation, but the poly(A) signal and the GT-rich region of the globin polyadenylation/cleavage signal were not required. Termination was not dependent on ATP hydrolysis, indicating a slippage mechanism.
mRNA 3′-end formation is a complicated process that requires protein-nucleic acid and protein-protein interactions. The 3′ ends of most eukaryotic RNA polymerase II transcripts are produced by a coupled cleavage/polyadenylation reaction that requires multiple factors (reviewed in references 8, 22, and 43). The processing machinery is recruited to transcription complexes by TFIID (9) and is targeted to the phosphorylated carboxy-terminal domain (CTD) of RNA polymerase II during transcription elongation (27). Recent studies have shown that RNA polymerase II itself is an essential factor in the polyadenylation step (20).
Transcription termination by RNA polymerase II occurs downstream of the cleavage site and is less well understood. Termination is defined as the process of disrupting the transcription ternary complex, in which the RNA polymerases cease their elongation function, the nascent transcripts are released, and the RNA polymerases step off from the DNA templates. Disassembly of an RNA polymerase ternary complex is mediated by termination sequences that are recognized by the RNA polymerase or by termination factors (reviewed in references 37 and 39). Failure of the elongating polymerase to stop transcription before reaching an adjacent gene can lead to a reduction in the expression of the downstream gene by destabilizing the assembly of transcription initiation factors on the adjacent promoter (3, 15, 16, 19, 31). This is especially important to the gene expression programs of compact genomes, such as yeast and viruses, which have short intergenic sequences.
Baculoviruses are large double-stranded DNA viruses that have been extensively used as protein expression vectors. The genomes of several baculovirus species have been sequenced and shown to be highly compressed (1, 2, 14). Viral gene expression is regulated in a temporal pattern (4, 11, 30) in which immediate- and delayed-early genes are transcribed by host cell RNA polymerase II, while late and very genes are transcribed by a virus-encoded RNA polymerase (18). The promoters used for overexpression in baculovirus vectors belong to these late-expression categories; therefore, regulation of viral late transcription has received considerable attention in the last decade (reviewed in references 23 and 28).
Baculovirus late and very late mRNAs have typical methyl-7-guanosine caps at their 5′ ends and poly(A) tails at the 3′ ends (32, 41). Baculovirologists have long assumed that the 3′ ends of baculovirus late and very late transcripts were formed by host cleavage and polyadenylation enzymes. This assumption initially arose from transcript mapping analyses showing the presence of AAUAAA motifs near the 3′ end of viral transcripts and was further supported by studies showing that a globin cleavage/polyadenylation signal could be effectively used as a 3′-end processing signal for baculovirus very late genes (41). This conclusion was so well accepted by the baculovirus community that most baculovirus expression vectors were developed with the simian virus 40 cleavage/polyadenylation signal sequences downstream of the polyhedrin promoter (26).
We recently purified the baculovirus RNA polymerase that transcribes late genes and showed that it was a complex of four viral proteins (18). None of these viral proteins has a CTD-like domain, which raises the question of how the cellular machinery could recognize the viral RNA polymerase and its associated transcripts. In addition, we noticed that the AATAAA and AATTAA sequences are not consistently located 10 to 30 bp upstream, as expected for a cleavage/polyadenylation signal. Furthermore, the 3′ noncoding regions of many baculovirus late and very late genes tend to be very AT rich, so even the occurrence of polyadenylation signals in the expected position relative to the 3′ ends could just be coincidental. Together, these observations suggested to us that 3′-end formation of baculovirus late RNA was probably not mediated by the host cleavage/polyadenylation machinery. Rather, we hypothesized that 3′ ends were formed by termination at U-rich sequences, which are present both in the globin signal and in most baculovirus 3′ noncoding regions. This model is also consistent with our previous studies showing that the LEF-4 subunit of viral RNA polymerase is an mRNA capping enzyme (17, 21). Capping enzymes, like the polyadenylation machinery, are targeted to substrates through interactions with the CTD of RNA polymerase II. Thus baculoviruses, which lack this motif, have evolved a unique mechanism for coordinating transcription and 5′-end processing and incorporated the capping function into the RNA polymerase complex.
To test whether the globin cleavage/polyadenylation signal is recognized by baculovirus RNA polymerase as a termination signal, we cloned the sequence into our standard transcription template. In vitro transcription assays revealed that the 3′ ends of late viral mRNAs are formed by termination, which is apparently an intrinsic property of the four-subunit baculovirus RNA polymerase complex. Mutational analyses indicate that the cellular poly(A) signal is not essential for termination or polyadenylation by viral RNA polymerase.
MATERIALS AND METHODS
Construction of DNA template for transcription termination assay.
A SwaI recognition site was inserted in the C-free cassette of Polh/CFS plasmid (42), using a QuikChange site-directed mutagenesis kit (Stratagene) as recommended by the manufacturer. Potential mutant clones were identified by restriction digestion and then verified by DNA sequence analysis. The mutant plasmid was linearized with SwaI and then ligated to a 47-bp synthetic globin sequence. A plasmid containing an insert in the correct orientation was identified by DNA sequence analysis and named Polh/CFS-T. The other mutants in the synthetic globin sequence were constructed using same procedure with the appropriate mutant oligonucleotides.
Baculovirus RNA polymerase purification.
All procedures were carried out at 4°C. Nuclear extracts and RNA polymerases were prepared according to the protocol of Guarino et al. (18). Ten-liter aliquots of infected SF-9 cells were collected to prepare nuclear extracts. Pooled nuclear extracts were treated with 0.1% polymin P to remove nucleic acid. The soluble fraction was twice precipitated with 50% ammonium sulfate. Precipitated proteins were then resuspended in column buffer (50 mM Tris [pH 7.9], 100 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol [DTT]), loaded onto a 5-ml heparin (Pharmacia) column, and eluted with a step gradients of 5 ml each at 10, 20, 30, 40, 50, and 100% 1 M KCl buffer. Fractions with transcription activity were pooled and precipitated with an equal volume of saturated ammonium sulfate at 4°C overnight. The ammonium sulfate precipitate was collected by centrifugation, resuspended in column buffer, and applied to a Mono Q HR 5/5 column (Pharmacia). The bound proteins were eluted with a 20-ml linear gradient from 100 to 550 mM KCl. Fractions with transcription activity were pooled and diluted to 80 mM KCl in the buffer. RNA polymerase aggregates at low salt; precipitated material was collected by ultracentrifugation, resuspended into 200 μl of 2 M KCl-Tris buffer, and filtered through a Superose 6 column in 2 M KCl-Tris buffer. Fractions were individually dialyzed against RNA polymerase storage buffer (50 mM Tris [pH 7.9], 400 mM KCl, 0.1 mM EDTA, 1 mM DTT, 50% glycerol), assayed for transcription activity, and stored at −20°C.
In vitro transcription assays.
In vitro transcription assays for transcription termination reactions were carried out using conditions previously described (42), with some modifications. Briefly, 50 μl of transcription reaction mixture contains 0.2 pmol of purified RNA polymerase, 50 mM Tris (pH 7.9), 100 mM KCl, 2 mM MgCl2, 5 mM DTT, 0.1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 1 mM each ATP and UTP, 20 μM GTP, 5 μCi of [α-32P]GTP, 8 U of RNasin, 0.2 U of inorganic pyrophosphatase, 0.2 pmol of Polh/CFS or Polh/CFS-T. Reaction mixtures were incubated at 30°C for 15 min. The reactions were stopped by adding 150 μl of stop buffer (20 mM Tris [pH 7.5], 0.1% sodium dodecyl sulfate [SDS], 1 mM EDTA, 400 mM NaCl, 50 μg of tRNA/ml). RNA transcripts were extracted once with an equal volume of phenol-chloroform and precipitated by adding a 2.5× volume of 100% ethanol at −80°C for 30 min. After centrifugation at 4°C, the RNA was resuspended into sequence stop buffer (36) and resolved on a 6% polyacrylamide–8 M urea gel. The gel was dried and exposed on Kodak film or PhosphorImager plates.
Northern blot analysis.
After in vitro transcription, RNA transcripts were resolved on a 6% polyacrylamide–8 M urea gel and transferred to nylon membrane with 0.6× Tris-borate-EDTA (TBE) buffer at 16 V at 4°C for 16 h. The RNA transcripts were then cross-linked to the nylon membrane using a Stratagene UV cross-linker. The membrane was prehybridized in hybridization buffer (6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 10 mM EDTA, 5× Denhardt's solution, 0.5% SDS, 100 μg of sheared calf thymus DNA/ml, 25% formamide) at room temperature for 2 h, and then a biotin-labeled oligonucleotide probe (10 ng/ml) was added to hybridize for 16 h. After hybridization, the membrane was rinsed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS, washed in 0.2× SSC–0.1% SDS for 15 min twice, and then washed in 2× SSC for 5 min once at room temperature. The hybridization signals were detected by alkaline phosphatase-conjugated streptavidin (Gibco) and developed with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate as substrates.
Synthesis of RNA templates for in vitro cleavage assays.
PCR was performed using Pfu DNA polymerase with Polh/CFS or Polh/CFS-T plasmid as DNA template and two oligonucleotides (GCGGTACCATTTAGTGACACTATAGAAGTAT TT TAGTGT TT TTGTAAT TTGTAATAAAAAAAT TATAAATGGG and GGCCTCGAGCTCCATACCCTTCCTCCATCTATACCACCC) as primers. The underlined sequence corresponds to the SP6 promoter; the remaining sequences hybridize to the C-free cassette. PCR products were inserted into a pUC18 plasmid previously linearized with SmaI. The resulting plasmids were called SP6/CFS and SP6/CFS-T. The RNA templates were synthesized using SP6 RNA polymerase and purified on an agarose-TBE gel.
RNA 3′ RACE.
Transcription reactions were performed as described above but incubated at 30°C for 60 min. Then the RNA transcripts were precipitated with 5 μg of glycogen and 2 volumes of ethanol at −80°C for 30 min. RNA pellets were resuspended into diethylpyrocarbonate-treated sterile water. The RNA samples were then used to make the first-strand cDNA using Superscript II reverse transcriptase (Gibco) with the oligo(dT) adapter primer (AP primer; GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT) as recommended by the manufacturer. Reverse transcriptase reactions were performed in the absence of dGTP. Then the first-strand cDNA was amplified by PCR with Taq DNA polymerase (Promega), an abridged universal amplification primer (AUAP primer; GGCCACGCGTCGACTAGTAC) corresponding to sequence beyond the oligo(dT) of the AP primer, and a 3′ RACE (rapid amplification of 3′ cDNA ends) primer (TGGAGGGGATATGGAAAGGGAAAGGAG) corresponding to the upstream region of the C-free cassette. The PCR products were ligated with a linear T vector (Promega). The 3′-end sequences of RNA transcripts were determined by sequencing using the primers that hybridized to sequences on the T vector flanking the insert.
RNA 5′ RACE.
After in vitro transcription, RNA transcripts were resolved on a denaturing 6% acrylamide–8 M urea gel. The 130-nucleotide (nt) transcripts were excised, crushed in buffer, and purified by centrifugation through a spin column. RNA transcripts were then precipitated by the addition of 5 μg of glycogen and 2 volumes of ethanol at −80°C for 30 min. The RNA pellets were resuspended in diethylpyrocarbonate-treated sterile water. The RNA samples were then used to make the first-strand cDNA using Superscript II reverse transcriptase (Gibco) in the absence of dGTP, using a 3′ primer (CTCCATACCCTTCCTCCAT) that hybridized to the C-free cassette. The cDNA samples were purified on a Sephadex G-50 spin column to remove the free nucleotides. The first-strand cDNAs were tailed using terminal deoxynucleotidyltransferase (Gibco) in the presence of 1 mM dGTP. The dG-tailed cDNAs were amplified with Taq DNA polymerase (Promega) with 3′ long primer (GGCCTCGAGCTCCATACCCTTCCTCCATCTATACCACCC) and 5′ RACE primer (GGCCACGCGTCGACTAGTACCCCCCCCCCCCCCCCC). The PCR products were ligated with linearized T vector (Promega). The 5′ ends of the RNA transcripts were determined by sequencing using the primers that annealed to sequences on the T vector flanking the insert.
RESULTS
Baculovirus RNA polymerase terminates within the synthetic globin sequence.
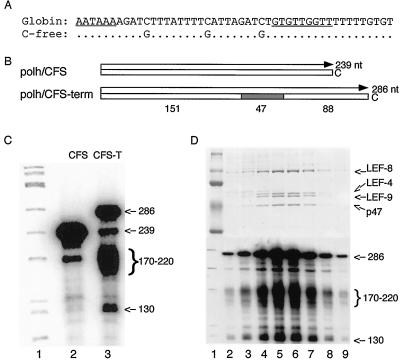
Previous studies have shown that a synthetic globin cleavage/polyadenylation signal directs 3′-end formation in vivo as well as the native polyhedrin signal (41). To determine whether purified baculovirus RNA polymerase recognizes this globin signal in vitro, a 47-nt C-free version (Fig. 1A) was inserted into the template region of Polh/CFS, the transcription template used for purification of viral RNA polymerase (18, 42). The resulting construct, Polh/CFS-T, should direct the synthesis of a 286-nt transcript if the signal is not recognized by the baculovirus RNA polymerase (Fig. 1B). Alternatively, if RNA polymerase terminates transcription or cleaves the RNA transcripts within the globin signal, then the RNA products should be approximately 88 nt shorter than those transcribed from the standard Polh/CFS template.
FIG. 1.
The globin cleavage/polyadenylation signal is recognized by purified baculovirus RNA polymerase. (A) Sequence of the globin cleavage/polyadenylation signal. To allow transcription in the absence of CTP, the three C's on the nontemplate strand of the globin sequence were changed to G. (B) Schematic diagram of transcription templates Polh/CFS and Polh/CFS-T. (C) RNA transcription pattern. Purified RNA polymerase (0.2 pmol) was incubated with 0.2 pmol of the indicated DNA template using standard transcription reaction conditions at 30°C for 15 min. Lane 1, φX174/HinfI marker; lane 2, Polh/CFS as template; lane 3, Polh/CFS-T as template. Sizes of the transcripts produced from Polh/CFS (left) and Polh/CFS-T (right) are indicated. (D) Transcription termination activity copurified with baculovirus RNA polymerase. RNA polymerase was filtered through a Superose 6 size exclusion column. Fractions were collected and dialyzed individually against polymerase storage buffer (50 mM Tris [pH 7.9], 400 mM KCl, 0.1 mM EDTA, 1 mM DTT, 50% glycerol). Fractions across the peak of RNA polymerase were analyzed by SDS-PAGE followed by staining with Coomassie brilliant blue (top). The same fractions were assayed for transcription termination using 2 μl from each fraction and 1.0 μg of Polh/CFS-T as template. The RNA transcripts were resolved on a 6% polyacrylamide–8 M urea gel and exposed to X-ray film. Lane 1 contains protein (top) or DNA (bottom) molecular markers.
In vitro transcription reactions were performed in parallel with purified RNA polymerase and either the standard Polh/CFS template or Polh/CFS-T. The transcription products were resolved on a denaturing polyacrylamide gel to compare the sizes of the transcription products. As usual, in vitro transcription with Polh/CFS template produced two transcripts (Fig. 1C, lane 2). The major band was 239 nt, which corresponds to the full-length transcript that initiates at TAAG and stalls at the first C in the nontemplate strand. The minor one was approximately 50 nt shorter than the full-length RNA transcripts. The origin of this shorter transcript is unknown, although we have speculated that it is a pause product (42). In vitro transcription reactions with Polh/CFS-T generated four different RNA products (Fig. 1C, lane 3). The size of the largest band, 286 nt, corresponds to the size predicted for initiation at the polyhedrin promoter and stalling at the first C on the nontemplate strand. The minor band at 239 nt is probably equivalent to the pause product seen with Polh/CFS. The shorter two bands were unique to the transcription reaction with Polh/CFS-T. The major product was a heterogeneously sized band of approximately 170 to 220 nt. There was also a less abundant transcript of 130 nt.
To confirm that this transcription pattern was due to intrinsic properties of baculovirus RNA polymerase and not to contaminants in the enzyme preparation, we assayed for copurification of this transcription pattern with RNA polymerase (Fig. 1D). The last step in the purification protocol is a Superose 6 gel exclusion column in the presence of 2 M KCl to disrupt nonspecific interactions with other proteins (18). Aliquots of the individual fractions from the Superose column were analyzed by polyacrylamide gel electrophoresis (PAGE) to confirm the purity of the enzyme preparation (Fig. 1D, top). The corresponding fractions were also assayed for transcription activity on Polh/CFS-T. As expected, the total transcription activity increased and decreased concomitantly with the peak of RNA polymerase. The pattern of transcripts produced was identical in every fraction, indicating that minor contaminants probably do not contribute to the pattern observed (Fig. 1D, bottom). The peaks of protein and transcription activity also exactly copurified with the guanylyltransferase activity of the RNA polymerase (data not shown).
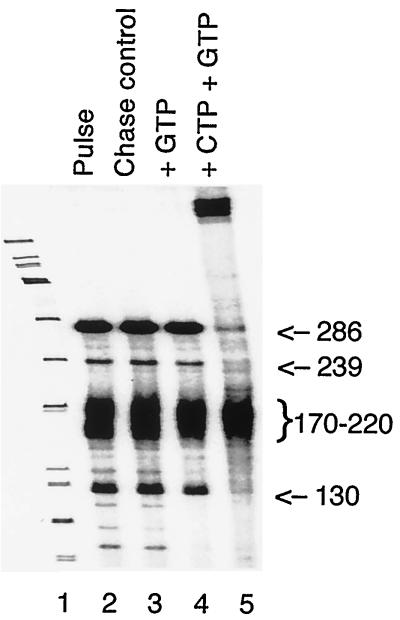
We considered the possibility that the two smaller RNAs were transcriptional pause products resulting from the low concentration of GTP used in the in vitro transcription reactions; [α-32P]GTP is used as the radiolabel, and so the GTP concentration is 50 times lower than the concentration of other nucleotides. To test this, reaction products from a 15-min reaction (Fig. 2, lane 2) were chased with 1 mM GTP (lane 4). All four of the major bands, including the 130- and 170- to 220-nt transcripts, were resistant to the GTP chase (lane 4). This indicates they are not transcriptional pause products due to limiting GTP concentration. Other minor products, however, were chased by the addition of GTP, confirming that polymerase was still active and able to extend paused products. Addition of 1 mM CTP and 1 mM GTP chased the 130-nt product as well as the 239- and 286-nt transcripts (lane 5), indicating that these transcripts were in stable ternary complexes that stalled at the first C on the nontemplate strand. The 170- to 220-nt fragments, however, were not extended, indicating that these transcripts were no longer associated with RNA polymerase ternary complexes.
FIG. 2.
Terminated transcripts are released from ternary complexes. After a 15-min transcription reaction, parallel transcription reactions were stopped immediately (lane 2) or chased for 5 min at 30°C with 1 mM GTP (lane 4), 1 mM GTP plus 1 mM CTP (lane 5), or an equivalent volume of transcription buffer (lane 3). Sizes are indicated in nucleotides.
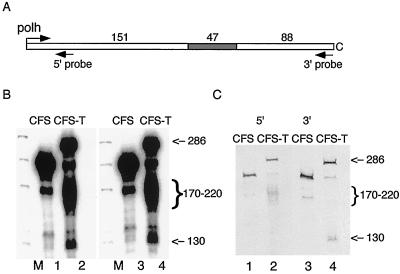
Northern blot analyses were performed to confirm this result. Parallel in vitro transcription reactions with both templates were performed and separated by acrylamide gel electrophoresis. The reaction products were transferred to nylon membrane and exposed to film to locate the positions of the reaction products (Fig. 3B). Blots were then separately probed with biotin-labeled oligonucleotides that corresponded to unique sequences that mapped to either the 5′ or 3′ region of the C-free cassette. Both probes hybridized to the full-length transcripts, as expected. The 170- to 220-nt transcripts hybridized with only the 5′ probe, while the 130-nt band hybridized with the 3′ probe but not the 5′ probe (Fig. 3C).
FIG. 3.
Mapping of terminated transcripts. (A) Schematic diagram of Polh/CFS-T with the relative positions of the 5′ and 3′ probes indicated. (B) Transcription reactions. RNA transcripts were resolved on a 6% polyacrylamide–8 M urea gel, transferred to a nylon membrane, and detected by exposure to X-ray film. Lanes: M, φX174/HinfI marker; 1 and 3, Polh/CFS as template; 2 and 4, Polh/CFS-T as template. (C) Northern blot analysis. The nylon membrane was hybridized with biotin-labeled probes and detected using alkaline phosphatase-conjugated streptavidin and a chromogenic substrate. Sizes are indicated in nucleotides.
Purified RNA polymerase has polyadenylation, but not cleavage, activity.
The results of the Northern blot analysis combined with the CTP chase data indicate that the 5′ ends of the 170- to 220-nt transcripts mapped to the polyhedrin promoter, and so the 3′ ends must map to positions within, or downstream of, the globin sequence. The 3′ ends of the 130-nt transcripts, on the other hand, must correspond to the last nucleotide of the C-free cassette. This positions the 5′ ends of 130-nt transcripts at the extreme left end of the globin cassette, apparently upstream of the 3′ ends of the 170- to 220-nt transcripts. This would suggest that these two bands were not produced by cleavage of the full-length transcript. Also arguing against a cleavage hypothesis is the fact that the relative molar amounts of the two products are not equivalent. The 170- to 220-nt fragments were present at an approximately fivefold molar excess compared to the 130-nt band.
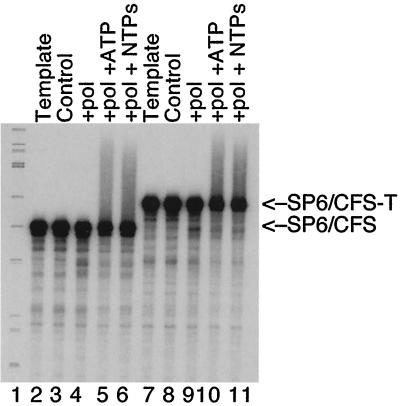
To validate this conclusion, we assayed directly for posttranscriptional RNA cleavage activity associated with RNA polymerase. The Polh/CFS and Polh/CFS-T cassettes were cloned under the control of the SP6 promoter, and RNA transcripts were synthesized using SP6 RNA polymerase. Transcripts made by SP6 RNA polymerase are identical to the baculovirus RNA polymerase-derived RNAs except for an additional G at the 5′ end. SP6 RNA polymerase-derived RNA transcripts were incubated with purified baculovirus RNA polymerase for 15 min at 30°C in the standard in vitro transcription buffer (Fig. 4). RNAs were then analyzed for cleavage activity on a 6% polyacrylamide—8 M urea denaturing gel. RNAs were stable in both the presence and absence of RNA polymerase, indicating that they were not cleaved by the enzyme and also did not self-cleave. Addition of ATP alone or ATP plus GTP and CTP resulted in the formation of longer, heterogeneously sized products, suggesting the RNA substrates were polyadenylated (Fig. 4, lanes 5, 6, 10, and 11). The polyadenylation reaction was apparently not sequence dependent, since the two transcripts were elongated to similar extents.
FIG. 4.
Cleavage and polyadenylation activities of purified baculovirus RNA polymerase. The two C-free cassettes were cloned under the control of the SP6 promoter, and RNA was produced in vitro using SP6 RNA polymerase. Ten femtomoles of each RNA was analyzed directly or incubated at 30°C for 15 min in transcription buffer with RNA polymerase and nucleotides as indicated. The samples were then were extracted with phenol and chloroform, precipitated with ethanol, and resolved on 6% polyacrylamide–8 M urea gel. Lanes: 1, φX174/HinfI marker; 2 and 7, SP6 RNA transcripts; 3 and 8, buffer only; 4 and 9, RNA polymerase; 5 and 10, RNA polymerase plus 1 mM ATP; 6 and 11, RNA polymerase plus 1 mM ATP and UTP plus 20 μM GTP.
This polyadenylation activity of the purified RNA polymerase suggests that heterogeneity of the 170- to 220-nt transcripts was due to polyadenylation of the terminated transcripts. To test this, we performed a time course experiment with Polh/CFS-T plasmid (Fig. 5). If the terminated transcripts were polyadenylated, they should be elongated with the extended reaction time. Alternatively, if heterogeneity was due to random termination or cleavage, the sizes should not change with time of incubation.
FIG. 5.
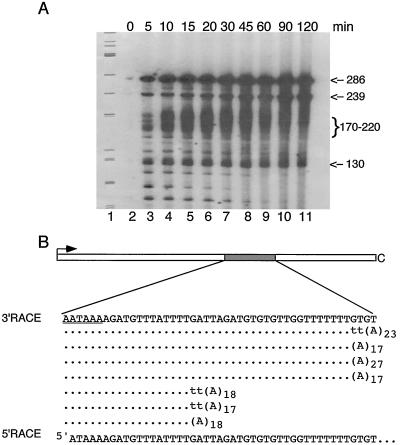
Terminated transcripts are polyadenylated. (A) Time course of transcription and polyadenylation. In vitro transcription reactions with Polh/CFS-T and purified RNA polymerase were stopped at the times indicated above the lanes. RNA transcripts were resolved on a 6% polyacrylamide–8 M urea gel. Lane 1, φX174/HinfI marker. Positions of the relevant transcripts are indicated in nucleotides on the right. (B) Sequences of cDNA clones. RNA 3′ RACE and cDNA sequencing were used to determine the sites of transcription termination. The sequence corresponds to the nontemplate strand of the globin cleavage/polyadenylation signal in Polh/CFS-T; the poly(A) signal is underlined. The viral RNA polymerase terminates at the end of two T-rich sequences in the globin sequence. Three clones contained two nontemplated T residues before the poly(A) tails.
Analysis of the time course experiment revealed that all four reaction products were synthesized by 5 min of incubation. The amount of product increased somewhat between 5 and 10 min of incubation, but the four bands were relatively constant in abundance at the later time points. The reaction products were constant with respect to size, with the exception of the 170- to 220-nt transcripts, which were elongated with increasing time. At later times the heterogeneous transcripts ranged in size from 200 to >400 nt, with an average length of approximately 250 nt.
Significantly, the amount of full-length product did not decrease with time, and there was no evidence of cleavage of this product. This supports our conclusion that the shorter RNAs are not produced by cleavage of the full-length transcript. Thus, these data suggest a model in which the 170- to 220-nt transcripts arise by termination of transcription, followed by posttranscriptional polyadenylation.
Viral RNA polymerase terminates at a T-rich region in the globin cleavage/polyadenylation sequence.
To determine the site of transcription termination, the 3′ ends of the terminated transcripts were determined by 3′ RACE using oligo(dT) for the initial reverse transcription reaction. Sequencing of cDNA clones revealed the presence of a poly(A) tail, confirming that the transcripts were indeed polyadenylated. The 3′ ends of these transcripts were mapped to the two T-rich regions of the synthetic globin sequence (Fig. 5B). Several clones had two additional, nontemplated T residues before the poly(A) tail. This suggests that baculovirus RNA polymerase may terminate by a slippage mechanism.
We also performed RNA 5′ RACE on the transcription products to determine the origin of the 130-nt product. Sequence analysis revealed that the 5′ end of this transcript mapped to the poly(A) signal AATAAA (Fig. 4B). Therefore, the 130- and 170- to 220-nt transcripts could not possibly arise by cleavage of the full-length product, since their 5′ and 3′ ends overlap. Presumably, the 130-nt product is produced by internal initiation of transcription, although there is not a consensus baculovirus late promoter motif (A/GTAAG) near the putative transcriptional start site. The poly(A) signal is followed by an AG dinucleotide, and it is possible that the resulting sequence ATAAAAG is read as a transcription start site in this in vitro system.
To confirm that the 130-nt transcripts were due to internal initiation and not to cleavage of the full-length transcript, we used the construct SP6/Polh-T as a transcription template with purified RNA polymerase (Fig. 6A). This plasmid has the same C-free termination cassette as Polh/CFS-T but has a bacteriophage SP6 promoter instead of the baculovirus polyhedrin promoter. We found that 130-nt transcripts were transcribed by purified viral RNA polymerase from this template (Fig. 6A, lane 3). Transcripts were not synthesized from a similar plasmid in which the AATAAA motif was changed to AGGAAA, confirming that this sequence directed internal initiation of transcription.
FIG. 6.
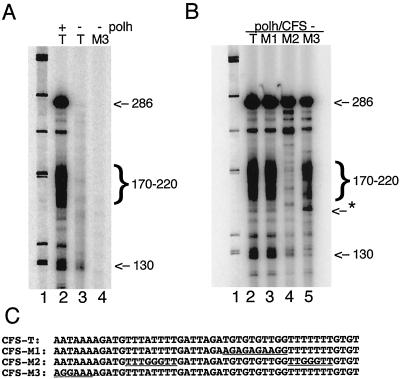
Mutational analysis of globin cleavage/polyadenylation cassette. (A) Nonspecific transcription initiation at the globin poly(A) signal. Standard in vitro transcription reactions were carried out with 0.2 pmol of purified RNA polymerase and 0.2 pmol of the indicated templates at 30°C for 15 min. Lanes: 1, φX174/HinfI marker; 2, Polh/CFS-T; 3, SP6/CFS-T; 4, SP6/Polh-M3 as template in which AATAAA was changed to AGGAAA in the C-free cassette. Sizes are indicated in nucleotides. (B) Sequence requirements for termination. Lanes: 1, φX174/HinfI marker; 2, RNA transcripts synthesized from Polh/CFS-T plasmid; 3, RNA transcripts from Polh/CFS-M1 plasmid; 4, RNA transcripts from Polh/CFS-M2 plasmid; 5, RNA transcripts from Polh/CFS-T3 plasmid. (C) Sequences of the globin cleavage/polyadenylation cassette and three mutant versions. The relevant substituted regions are underlined.
Oligo(T) is the major determinant for transcription termination.
The synthetic globin sequence has two major sequence features of RNA polymerase II cleavage/polyadenylation signals: an AAUAAA motif located 10 to 30 bases upstream of the cleavage site and a GU-rich sequence 20 to 40 bases downstream. To determine whether these sequence features are essential for transcription termination by baculovirus RNA polymerase, the hexanucleotide AATAAA in the globin C-free cassette was changed to AGGAAA (Polh/CFS-M3) and the T residues in the GT-rich sequence were changed to A's (Polh/CFS-M1). The ability of these mutant versions to direct transcription termination was tested by in vitro transcription. The termination activity of viral RNA polymerase was not affected either by substitutions in the GT-rich sequence (Figure 6B, lane 3) or by the poly(A) signal mutation (Fig. 6B, lane 5). This indicates that neither of these sequence features is a major determinant for termination in vitro.
However, transcription of the hexanucleotide mutant produced two alterations in the pattern of transcription products (Fig. 6B, lane 5). The 130-nt product was not produced. This was expected from the results presented in Fig. 6A, showing that the sequence surrounding the poly(A) signal could function as a weak initiator. Also a transcript of approximately 160 nt was produced; this is apparently a transcription pause product, because it could be chased by the addition of 1 mM GTP (data not shown).
Sequence analysis of 3′ RACE products indicated that termination occurred at the two T-rich sequences of the synthetic globin C-free cassette (Fig. 5B). Therefore, we also constructed mutant versions of Polh/CFS-T to test whether T-rich sequences are required for transcription termination. Both of the T-rich sequences were changed to TTGGGTT to make the double mutant Polh/CFS-M2, in which both of the T-rich sequences were disrupted by G residues. In vitro transcription reactions were then performed to compare the transcription patterns between Polh/CFS-T and the double mutant Polh/CFS-M2. Termination was completely abolished during transcription of this mutant template (Fig. 6B, lane 4). This result confirms our hypothesis that the globin termination cassette was able to function in 3′-end formation because it contained T-rich sequences, not because it was recognized by cellular cleavage/polyadenylation enzymes.
The mutations in the T-rich regions also resulted in a dramatic decrease in the production of the nonspecific initiation product at 130 nt. This suggests that the ability of the ATAAAAG sequence to serve as an initiator is strongly influenced by, and probably dependent on, the presence of a T-rich sequence immediately downstream.
Transcription termination by baculovirus RNA polymerase is an energy-uncoupled process.
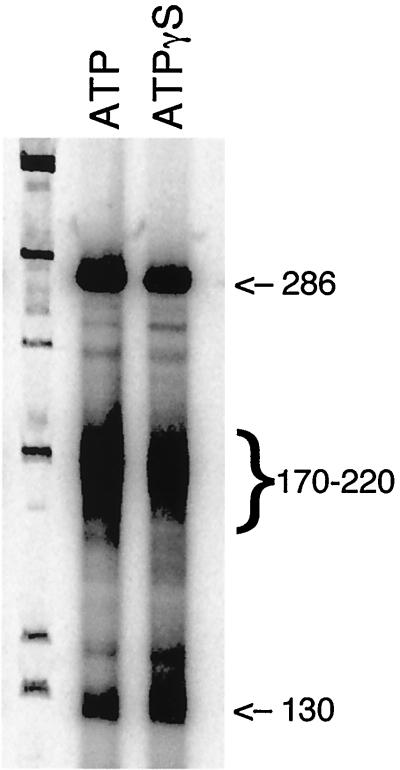
In some systems, termination of transcription requires ATP to provide the energy to break the hydrogen bonds between the template and the transcript. Previous data from our lab have shown that transcription initiation and elongation are independent of ATP hydrolysis and are able to proceed in the presence of 1 mM ATPγS (B. Xu and L. A. Guarino, unpublished data). To test whether transcription termination is coupled to ATP hydrolysis, 1 mM ATPγS was substituted for ATP in a transcription assay with Polh/CFS-T plasmid. There were no differences in either the transcription or termination patterns in parallel reactions containing ATP or ATPγS (Fig. 7). These data indicate that transcription termination by baculovirus RNA polymerase does not require ATP hydrolysis.
FIG. 7.
ATP hydrolysis is not required for transcription termination by baculovirus RNA polymerase. Purified RNA polymerase (0.2 pmol) was incubated with 0.2 pmol of Polh/CFS-T plasmid at 30°C for 60 min in standard transcription buffer containing 1 mM ATP (lane 2) or 1 mM ATPγs to replace ATP. Lane 1, φX174/HinfI marker. Sizes are indicated in nucleotides.
DISCUSSION
The data presented here indicate that purified viral RNA polymerase recognizes a T-rich region within a globin cleavage/polyadenylation sequence as a termination signal. This contradicts the prevailing view of baculovirus 3′-end formation, which holds that cellular enzymes process viral transcripts by a cleavage and polyadenylation mechanism. We questioned this assumption based on recent reports showing that transcription and 3′-end processing were coordinated through interactions between the polyadenylation machinery and the CTD of RNA polymerase II (27). This emerging model, coupled with our finding that the viral RNA polymerase lacked a CTD-like domain, suggested to us that 3′-end formation was an intrinsic property of the RNA polymerase and occurred independently of host machinery.
The model invoking cellular machinery in the formation of viral 3′ ends was based on a study showing that a synthetic globin cleavage/polyadenylation cassette could substitute for the native 3′ noncoding region of the polyhedrin gene (41). This globin sequence contains the two major determinants of a cleavage/polyadenylation signal: a hexanucleotide AAUAAA and a downstream GU-rich sequence. In addition, the signal contains a U-rich sequence, which we predicted would serve as a termination signal. To test this hypothesis, we cloned the globin sequence into a transcription template and found that this sequence induced termination of transcription at the predicted site. The terminated transcripts were released from ternary complexes and were polyadenylated.
We found that the peak of termination activity exactly coincided with the peak RNA polymerase protein as well as the peaks of transcription and guanylyltransferase activities. This suggests that these activities are intrinsic to the baculovirus RNA polymerase, although we cannot exclude the possibility that minor contaminants in our enzyme preparation influence transcription termination. If there are contaminating activities, they are more likely to be viral proteins than host proteins. Cellular cleavage enzymes are unlikely to contribute significantly to the activities detected here since our data indicate that the mechanism of 3′ end formation is termination and not cleavage. Host polyadenylation enzymes are also unlikely to contribute to polyadenylation because they preferentially add adenylates to CA residues, which were not present in the C-free sequence. In addition, most eukaryotic poly(A) polymerases recognize the AAUAAA sequence, which was not essential for the poly(A) polymerase activity of the viral RNA polymerase. Adenylates were added to exogenously transcribed RNAs that lacked the globin termination cassette as efficiently as an RNA containing this sequence.
Only 50% of the transcribing RNA polymerases terminated in the T-rich region on the globin sequence (Fig. 1). This is similar to the in vivo result in which the synthetic globin sequence was inserted upstream of the native polyhedrin termination sequence (41). Northern blotting analyses indicated that about half of the RNA polymerases read through the synthetic globin sequence in vivo and terminated downstream at the authentic polyhedrin signal. This suggests that efficient termination may require additional sequence features that are not present in the globin cassette. Additional in vivo and in vitro analyses will be necessary to fully delineate the sequence requirements for transcription and polyadenylation.
Termination by the viral RNA polymerase was independent of ATP hydrolysis. This suggests that the energy necessary for transcription termination of baculovirus RNA polymerase comes from an intrinsic feature of this viral RNA polymerase, possibly involving a conformational change of the RNA polymerase and an interaction between the components of the elongation complex. Transcription termination can be divided into two categories: intrinsic termination such as the rho-independent transcription termination by bacterial RNA polymerase (reviewed in reference 34), and protein factor-dependent termination, such as the rho-dependent transcription of Escherichia coli RNA polymerase (reviewed in reference 34) and the termination of vaccinia virus RNA polymerase (7) and RNA polymerase II (reviewed in reference 43). Our data suggest that transcription termination by baculovirus RNA polymerase is similar to rho-independent transcription termination of E. coli RNA polymerase.
RNA 3′ RACE and cDNA sequencing data revealed that baculovirus RNA polymerase terminated after transcription of two T-rich regions on the nontemplate strand downstream of the poly(A) signal AAUAAA. Mutational analyses suggest that the T-rich sequence is the major element specifying transcription termination in the globin cassette. Similar T-rich sequences are present downstream of most, or possibly all, baculovirus late genes (41), and we suggest that this element, and not the AAUAAA sequence, specifies 3′-end formation. However, oligo(T) is unlikely to be the only determinant because T-rich sequences can be found within the open reading frames of several baculovirus late and very late genes. If oligo(T) were sufficient to specify termination of transcription, then expression of these late and very late genes would be decreased by untimely termination of RNA polymerase during elongation. It is possible that an antitermination factor is required to prevent premature termination at T-rich sequences within open reading frames. If true, we hypothesize that this termination factor is not part of the core RNA polymerase and was lost during purification.
Mutational analyses suggested that the poly(A) AAUAAA signal is not essential for transcription termination. These poly(A) signals have been noted in the 3′ untranslated regions of most, if not all, mRNAs of baculovirus late and very late genes (41), suggesting that they have been conserved for some purpose. Although our data do not support an essential role for this signal, they also do not exclude an accessory role for the poly(A) signal in transcription termination. For example, the poly(A) signal could be a positioning element for termination. Once RNA polymerase passes this signal, the antitermination factor could be released or inactivated, causing RNA polymerase to terminate at the nearest downstream T-rich sequence. This type of role would not be detected in our experimental system containing RNA polymerase alone but would be essential for transcription in vivo. The role of the poly(A) signal in transcription termination is so far inconclusive based on our data and others (38, 41).
Purified RNA polymerase was able to polyadenylate nascent transcripts as well as exogenously added templates, although the processivity of this reaction was fairly low (Fig. 4). Vaccinia virus also uses a mechanism of transcription termination at oligo(T), followed by polyadenylation for the formation of early viral 3′ ends (12, 35). In this case, polyadenylation is mediated by a poly(A) polymerase that is distinct from the RNA polymerase. The enzyme is a heterodimer, consisting of VP55 and VP39 proteins (12). VP55, the catalytic subunit, can add 30 to 35 adenylates to RNA 3′-end-related primers in a rapid reaction. After this burst of polyadenylation, the processivity suddenly turns into a slow, nonprocessive mode. VP39 can convert this slow reaction back to a rapid semiprocessive addition of adenylates (13). Thus, it is possible that polyadenylation by the baculovirus polymerase is stimulated by a dissociable factor that was lost during purification.
Poly(A) polymerases have been isolated from a number of sources, and conserved motifs are beginning to emerge (5, 6, 10, 12, 24, 25, 29, 33, 40). Comparisons of the amino acid sequences of poly(A) polymerases from the poxviruses, yeast, plants, and mammals have not provided a strong match to any of the baculovirus RNA polymerase subunits. In addition, we have been unable to demonstrate that poly(A) polymerase activity is associated with any of the individual subunits. Previously we were able to map the guanylyltransferase and RNA triphosphatase activities of RNA polymerase to the LEF-4 subunit by expressing this enzyme as a single subunit (17, 21). Unfortunately, LEF-4 is the only one of the RNA polymerase subunits that is soluble when expressed as a single subunit in bacteria. Thus, we have been unable to identify the protein responsible for the poly(A) polymerase activity.
ACKNOWLEDGMENTS
We thank Wen Dong for technical assistance and help with the purification of baculovirus RNA polymerase.
This research was supported by grant MCB-9874532 from the National Science Foundation.
REFERENCES
- 1.Ahrens C H, Russell R L Q, Funk C J, Evans J T, Harwood S H, Rohrmann G F. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology. 1997;229:381–399. doi: 10.1006/viro.1997.8448. [DOI] [PubMed] [Google Scholar]
- 2.Ayres M D, Howard A C, Kuzio J, Lopéz-Ferber M, Possee R D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 3.Bateman E, Paule M R. Promoter occlusion during ribosomal RNA transcription. Cell. 1988;54:985–993. doi: 10.1016/0092-8674(88)90113-4. [DOI] [PubMed] [Google Scholar]
- 4.Blissard G W, Rohrmann G F. Baculovirus diversity and molecular biology. Annu Rev Entomol. 1990;35:127–155. doi: 10.1146/annurev.en.35.010190.001015. [DOI] [PubMed] [Google Scholar]
- 5.Cao G J, Sarkar N. Identification of the gene for Escherichia coli poly(A) polymerase. Proc Natl Acad Sci USA. 1992;89:10380–10384. doi: 10.1073/pnas.89.21.10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao G J, Pogliano J, Sarkar N. Identification of the coding region for a second poly(A) polymerase in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:11580–11585. doi: 10.1073/pnas.93.21.11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christen L M, Sanders M, Wiler C, Niles E G. Vaccinia virus nucleoside triphosphate phosphohydrolase I is an essential viral early gene transcription termination factor. Virology. 1998;245:360–371. doi: 10.1006/viro.1998.9177. [DOI] [PubMed] [Google Scholar]
- 8.Colgan D F, Manley J L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 9.Dantonel J, Murthy K G K, Manley J L, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 10.Das Gupta J, Li Q, Thomas A B, Hunt A G. Characterization of a cDNA encoding a novel plant poly(A) polymerase. Plant Mol Biol. 1998;37:729–734. doi: 10.1023/a:1006000213403. [DOI] [PubMed] [Google Scholar]
- 11.Friesen P D, Miller L K. Temporal regulation of baculovirus RNA overlapping early and late transcripts. J Virol. 1985;54:392–400. doi: 10.1128/jvi.54.2.392-400.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gershon P D, Ahn B Y, Garfield M, Moss B. Poly(A) polymerase and a dissociable polyadenylation stimulatory factor encoded by vaccinia virus. Cell. 1991;66:1269–1278. doi: 10.1016/0092-8674(91)90048-4. [DOI] [PubMed] [Google Scholar]
- 13.Gershon P D, Moss B. Simulation of poly(A) tail elongation by the VP39 subunit of the vaccina virus-encoded poly(A) polymerase. J Biol Chem. 1993;268:2203–2210. [PubMed] [Google Scholar]
- 14.Gomi S, Majima K, Maeda S. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J Gen Virol. 1999;80:1323–1337. doi: 10.1099/0022-1317-80-5-1323. [DOI] [PubMed] [Google Scholar]
- 15.Greger I H, Proudfoot N J. Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 1998;17:4771–4779. doi: 10.1093/emboj/17.16.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greger I H, Demarchi F, Giacca M, Proudfoot N J. Transcriptional interference perturbs the binding of Sp1 to the Hiv-1 promoter. Nucleic Acids Res. 1998;26:1294–1300. doi: 10.1093/nar/26.5.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guarino L A, Jin J, Dong W. Guanylyltransferase activity of the LEF-4 subunit of baculovirus RNA polymerase. J Virol. 1998;72:10003–10010. doi: 10.1128/jvi.72.12.10003-10010.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guarino L A, Xu B, Jin J, Dong W. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J Virol. 1998;72:7985–7991. doi: 10.1128/jvi.72.10.7985-7991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson S L, Ryan K, Sollner-Webb B. The promoter-proximal rDNA terminator augments initiation by preventing disruption of the stable complex caused by polymerase read-in. Genes Dev. 1989;3:212–223. doi: 10.1101/gad.3.2.212. [DOI] [PubMed] [Google Scholar]
- 20.Hirose Y, Manley J L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 21.Jin J, Dong W, Guarino L A. The LEF-4 subunit of baculovirus RNA polymerase has 5′-RNA triphosphatase and ATPase activities. J Virol. 1998;72:10011–10019. doi: 10.1128/jvi.72.12.10011-10019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keller W. No end yet to messenger RNA 3′ processing. Cell. 1995;81:829–832. doi: 10.1016/0092-8674(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 23.King L A, Possee R D. The baculovirus expression system. A laboratory guide. London, England: Chapman & Hall; 1992. [Google Scholar]
- 24.Li Q, Das Gupta J, Hunt A G. Polynucleotide phosphorylase is a component of a novel plant poly(A) polymerase. J Biol Chem. 1998;273:17539–17543. doi: 10.1074/jbc.273.28.17539. [DOI] [PubMed] [Google Scholar]
- 25.Lingner J, Kellermann J, Keller W. Cloning and expression of the essential gene for poly(A) polymerase from S. cerevisiae. Nature. 1991;354:496–498. doi: 10.1038/354496a0. [DOI] [PubMed] [Google Scholar]
- 26.López-Ferber M, Sisk W P, Possee R D. Baculovirus transfer vectors. Methods Mol Biol. 1995;39:25–63. doi: 10.1385/0-89603-272-8:25. [DOI] [PubMed] [Google Scholar]
- 27.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson D S, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 28.Miller L K. The baculoviruses. New York, N.Y: Plenum Press; 1997. [Google Scholar]
- 29.Ohnacker M, Minvielle-Sebastia L, Keller W. The Schizosaccharomyces pombe pla1 gene encodes a poly(A) polymerase and can functionally replace its Saccharomyces cerevisiae homologue. Nucleic Acids Res. 1996;24:2585–2591. doi: 10.1093/nar/24.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors: a laboratory manual. W. H. New York, N.Y: Freeman & Company; 1992. [Google Scholar]
- 31.Proudfoot N J. Transcriptional interference and termination between duplicated alpha-globin gene constructs suggests a novel mechanism for gene regulation. Nature. 1986;322:562–565. doi: 10.1038/322562a0. [DOI] [PubMed] [Google Scholar]
- 32.Qin J C, Weaver R F. Capping of viral RNA in cultured Spodoptera frugiperda cells infected with Autographa californica nuclear polyhedrosis virus. J Virol. 1982;43:234–240. doi: 10.1128/jvi.43.1.234-240.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raabe T, Bollum F J, Manley J L. Primary structure and expression of bovine poly(A) polymerase. Nature. 1991;353:229–234. doi: 10.1038/353229a0. [DOI] [PubMed] [Google Scholar]
- 34.Richardson J P. Transcription termination. Crit Rev Biochem Mol Biol. 1993;28:1–30. doi: 10.3109/10409239309082571. [DOI] [PubMed] [Google Scholar]
- 35.Rohrmann G, Yuen L, Moss B. Transcription of vaccinia virus early genes by enzymes isolated from vaccinia virions terminates downstream of a regulatory sequence. Cell. 1986;46:1029–1035. doi: 10.1016/0092-8674(86)90702-6. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Uptain S M, Kane C M, Chamberlin M J. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 38.van Oers M M, Vlak J M, Voorma H O, Thomas A A M. Role of the 3′ untranslated region of baculovirus p10 mRNA in high-level expression of foreign genes. J Gen Virol. 1999;80:2253–2262. doi: 10.1099/0022-1317-80-8-2253. [DOI] [PubMed] [Google Scholar]
- 39.von Hippel P H. An integrated model of the transcription complex in elongation, termination, and editing. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 40.Wahle E, Martin G, Schilz E, Keller W. Isolation and expression of cDNA clones encoding mammalian poly(A) polymerase. EMBO J. 1991;10:4251–4257. doi: 10.1002/j.1460-2075.1991.tb05003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westwood J A, Jones I M, Bishop D H. Analyses of alternative poly(A) signals for use in baculovirus expression vectors. Virology. 1993;195:90–99. doi: 10.1006/viro.1993.1349. [DOI] [PubMed] [Google Scholar]
- 42.Xu B, Yoo S, Guarino L A. Differential transcription of baculovirus late and very late promoters: fractionation of nuclear extracts by phosphocellulose chromatography. J Virol. 1995;69:2912–2917. doi: 10.1128/jvi.69.5.2912-2917.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ end in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]