Abstract
The vif gene of human immunodeficiency virus type 1 (HIV-1) is essential for viral replication, although the functional target of Vif remains elusive. HIV-1 vif mutant virions derived from nonpermissive H9 cells displayed no significant differences in the amount, ratio, or integrity of their protein composition relative to an isogenic wild-type virion. The amounts of the virion-associated viral genomic RNA and tRNA3Lys were additionally present at normal levels in vif mutant virions. We demonstrate that Vif associates with RNA in vitro as well as with viral genomic RNA in virus-infected cells. A functionally conserved lentivirus Vif motif was found in the double-stranded RNA binding domain of Xenopus laevis, Xlrbpa. The natural intravirion reverse transcriptase products were markedly reduced in vif mutant virions. Moreover, purified vif mutant genomic RNA-primer tRNA complexes displayed severe defects in the initiation of reverse transcription with recombinant reverse transcriptase. These data point to a novel role for Vif in the regulation of efficient reverse transcription through modulation of the virion nucleic acid components.
Lentiviruses are distinct from simpler retroviruses in encoding numerous regulatory proteins that modulate virus infectivity. Vif is a lentiviral protein which modulates viral replication (22, 23, 39) and, consequently, pathogenicity (19). It is believed to act during the late stages of virus assembly by enabling the establishment of integrated provirus in new target cells. Since vif mutant virions show severely impaired infectivity, Vif must regulate one or more of the molecules found in virions. Immunofluorescence analysis of infected cells has demonstrated that Vif and the major structural protein, Gag, colocalize in the cytoplasm (36). Furthermore, Vif cosediments with some of the intracellular preassembly complexes of Gag, but not with the more mature forms of these assembly complexes (38). These data suggest that Vif and Gag may transiently be found together in a complex during virus assembly. However, the failure of Gag and Vif to coimmunoprecipitate tends to argue against a direct interaction of Vif with assembling Gag particles (38), although the involvement of additional bridging molecules cannot be ruled out. Vif is also virtually absent from highly purified human immunodeficiency virus type 1 (HIV-1) virions, further supporting the hypothesis that the role of Vif in infectivity occurs during the preassembly stages of particle formation within the cytoplasm (7).
The requirement for Vif for viral replication exhibits a striking cell-type dependence. For example, Jurkat and SupT1 cells do not require Vif for HIV-1 replication; for H9 cells and primary blood-derived monocytes (PBMCs), however, Vif is essential. In the case of nonhuman lentiviruses, PBMCs derived from the appropriate animals fail to support the replication of vif mutant viruses (14, 33, 42). This finding has led to the concept that specific cellular factors exist which act as inhibitors of lentiviral replication and which Vif must overcome (26, 37). Vif may therefore exist in an intimate molecular relationship with components of the infected cells, where it modulates the biology of the virus.
Analysis of vif mutant virions during entry into target cells has suggested that either the stabilization of the viral nucleoprotein complex is compromised (35), or the processivity of reverse transcription is impaired (30, 40, 44). This type of analysis follows the steps in reverse transcription that coincide with several early-stage events in general, making it difficult to identify the key regulatory event. Studies examining biochemical differences between wild-type and vif mutant virions have provided little compelling evidence for Vif-mediated modification or altered incorporation of the virion-associated proteins Gag, Pol, and Env (1, 11, 32, 44). However, intriguing evidence demonstrated that detergent-treated vif mutant virions are defective in de novo reverse transcription (8, 13), a finding which suggests that Vif may regulate molecules involved in the reverse transcriptase (RT) process itself.
RT is synthesized as part of the Gag-Pol polyprotein, which includes the major Gag structural proteins, the virus-encoded protease, and integrase (IN). Regions of the Gag-Pol polyprotein, particularly RT, are believed to direct the selective incorporation of cellular tRNA3Lys into virions, where it functions as the primer in reverse transcription (27, 28). Placement of the tRNA3Lys molecule on the primer binding site (PBS), which is located near the 5′ end of the viral genome, may be guided by p7NC (3, 18). tRNA itself may also contribute to the processivity of RT. HIV-1 infection has been shown to influence the posttranscriptional modification of tRNAs (15, 24). These modifications of tRNA3Lys may contribute to the synthesis of strong stop DNA, presumably through added contacts between the tRNA and the viral genome in regions outside the PBS (16, 20, 21). Experiments using avian leukosis virus have indicated that the 5′ end of the viral genome and the primer tRNATrp form a complex secondary structure that influences the initiation of reverse transcription (5, 29).
RNA-binding proteins regulate many cellular events, including splicing, translation, and RNA transport (2, 9). Virally encoded proteins, especially those of HIV-1, including Tat, Rev, and p7NC, have contributed substantially to our understanding of RNA-binding protein function. We now report that another HIV-1-encoded protein, Vif, associates with its RNA. Our data indicate that Vif interacts with the viral genomic RNA within the cytoplasm, probably acting in concert with other as yet undetermined cellular factors. Vif appears to direct efficient reverse transcription by affecting the nucleic acid components of the RT complex.
MATERIALS AND METHODS
DNA constructs.
The parental wild-type (WT) virus used for this study was derived from HXB2NEO (7). A deletion mutant resulting in a premature stop codon was generated in the vif open reading frame between nucleotide positions 4702 and 4772 by digestion with NdeI and XmnI, creating blunt ends by using Klenow fragment, and religation with T4 DNA ligase to generate HXB2VifΔNEO (vifΔ). The env deletion vector and the vif mutant vector with an env deletion were created by removing nucleotides 6620 to 7200 of HXB2 by digestion with BglII and religation with T4 ligase.
Cells, DNA transfection, and infection.
COS-7 cells were transfected by the DEAE-dextran method (7). COS-7 cells transfected with HXB2-derived env mutants were co-transfected with a murine leukemia virus Env expression vector, SV-A-MLV-env, in order to pseudotype HIV-1 virions.
Cell culture supernatants containing viral particles were harvested 3 days after transfection, precleared by centrifugation in a Sorvall RT 6000B centrifuge at 3,000 rpm for 30 min, filtered through a 0.2-μm-pore-size membrane, and used to infect Jurkat and H9 cell lines. At 24 h postinfection, cells were grown in the presence of 1.2 mg of G418 per ml for at least 2 weeks before analysis of cellular and viral protein profiles to ensure that all surviving cells contained viral genomes.
Exogenous and endogenous RT assays.
Exogenous RT assays using cell culture supernatants were performed as previously described (7). Endogenous RT assays used virions which were first normalized for viral protein content by immunoblotting and assays of exogenous RT activity. Pelleted virions were resuspended in 50 μl of endogenous RT buffer (40 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 6 mM KCl, 2 mM dithiothreitol [DTT], 0.5 mM dCTP, 0.5 mM dGTP, 0.5 mM dTTP, 20 μCi of [35S]dATP, 0.02% Triton X-100) and incubated at 37°C for 16 h. The reaction products were precipitated with 3 ml of chilled 10% (wt/vol) trichloroacetic acid using tRNA as a carrier. [3H]dTTP bound to GF/C glass microfiber filters after five washes with chilled 5% trichloroacetic acid was quantified with a Beckman LS 6500 scintillation counter.
Immunoblotting.
Virion-associated viral proteins were prepared from cell culture supernatants and analyzed by immunoblotting (7). Membranes were probed with either HIV-1-positive patient serum (diluted 1:200), rabbit polyclonal antiserum against Vif (1:1,000), sheep polyclonal anti-p17MA (1:400), mouse monoclonal anti-RT (1:400; BTI), rabbit polyclonal anti-IN (1:400), goat polyclonal anti-p7NC (1:400), goat polyclonal anti-p6gag (1:1,000), or rabbit polyclonal anti-Vif (1:1,000).
Northern and slot blots for tRNA and genomic RNA.
Virions were pelleted through 30% sucrose and resuspended in TNE (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1 mM EDTA), treated with 1% sodium dodecyl sulfate (SDS), and extracted with phenol-chloroform. Virion-associated nucleic acids were then ethanol precipitated and resuspended in diethyl pyrocarbonate-treated water. Viral genomic RNA was detected from virions by slot blot or Northern blot analysis by using an in vitro-transcribed probe spanning the 5′ long terminal repeat (LTR) and gag regions (BglII-BglII; nucleotide positions 52 to 1675) in the antisense orientation. Hybridization was performed under standard conditions, and blots were subjected to autoradiography.
Viral genomic RNA dimers were analyzed by running viral RNA directly on nondenaturing 1.0% agarose gels in Tris-borate-EDTA (TBE) running buffer. Gels were treated with 10% formaldehyde for 30 min at 70°C, washed twice for 20 min in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and transferred overnight to nylon membranes which were hybridized with the 5′ LTR-gag probe described above.
tRNA3Lyss were obtained from virion-associated nucleic acid preparations similar to those for genomic RNA. RNAs were run on 1% agarose gels containing formaldehyde, washed twice for 20 min in 20× SSC, and transferred overnight to nylon membranes. Membranes were hybridized with a tRNA3Lys-specific primer (5′-TGGCGCCCGAACAGGGAC-3′), which was end labeled with [γ-32P]dATP by using polynucleotide kinase.
RNase protection assay.
Total RNA was isolated from infected H9 cells by the RNA Stat-60 RNA isolation method (Tel-Test, Inc., Friendswood, Tex.). After ethanol precipitation, samples were resuspended in water and stored at −70°C. Typically, 5 μg of total RNA from H9 cells was precipitated in ethanol in the presence of 10 μg of carrier tRNA and resuspended in 30 μl of hybridization buffer [40 mM piperazine-N-N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.4), 1 mM EDTA, and 400 mM NaCl in 80% formamide]. Viral genomic RNA was detected with an in vitro-transcribed probe containing a region of gag (HindIII-PstI; nucleotide positions 1291 to 998) in the antisense orientation. Samples were heated to 85°C for 10 min and hybridized to 2 × 105 cpm of 32P-labelled antisense RNA at 50°C overnight. Samples were then incubated at 25°C for 2 h, followed by a 1-h incubation at 37°C in the presence of 300 μl of RNase digestion buffer (10 mM Tris-HCl [pH 7.5], 300 mM NaCl, 5 mM EDTA, 2-μg/ml RNase T1, 40 μg of RNase A). The RNase digestion was terminated by the addition of SDS (1% final concentration) and proteinase K (300-μg/ml final concentration) and incubated at 37°C for 30 min. Samples were phenol-chloroform extracted and ethanol precipitated in the presence of 10 μg of carrier tRNA, and the dried pellets were resuspended in loading buffer (80% formamide, 10 mM EDTA [pH 8.0]). Samples were analyzed by 6% polyacrylamide–7 M urea–1× Tris-borate-EDTA gels and visualized by autoradiography.
PCR of intravirion nucleic acids.
PCR was performed according to the standard manufacturer's conditions (Perkin-Elmer). PCRs were run at 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s for 30 cycles, followed by a 5-min extension at 72°C. Early RT products or strong stop DNA was detected with primer pairs U5a and U5 (5′-CGTCTGTTGTGTGACTCTGGTAAC-3′ and 5′-CTGCTAGAGATTTTCCACACTGAC-3′, respectively), R (5′-GGCTAACTAGGGAACCCACTGCTT-3′) and U5, and TAR (5′-GGTCTCTCTGGTTAGACCAGATCT-3′) and U5. The first jump was detected with primer pair U3 (5′CACACACAAGGCTACTTCCCT-3′) and U5. Extended minus-strand synthesis into the gag region was detected with primer pair H68 and H71 (5′-GCGAGAGCGTCAGTATTAAGCG-3′) and 5′-TCTGATAATGCTGAAAACATGG-3′, respectively). PCR products were run on 1.6% agarose gels, incubated twice for 20 min in 1.5 M NaCl with 0.5 N NaOH and twice for 20 min in Southern neutralization buffer (1 M Tris [pH 7.4], with 1.5 M NaCl), and transferred overnight to a nylon membrane. Southern blot hybridization was performed with an in vitro-transcribed riboprobe recognizing gag and LTR, as described above. Filters were washed and visualized by autoradiography.
In vitro translation.
In vitro translation of Vif was performed as previously described (12). Vif was in vitro transcribed by using the T7 promoter and translated by using the TNT rabbit reticulocyte lysate system according to the manufacturer's instructions (Promega), in the presence of [35S]cysteine for 90 min at 30°C. Following the TNT reaction, either canine pancreatic microsomal membranes (2 U) (Promega), RNase A (10 μg/ml), or both in combination were added directly to the reaction mixture and incubated for 30 min at 30°C. The sample was diluted with 500 μl of physiological salt buffer (50 mM Tris-acetate [pH 7.5], with 150 mM potassium acetate, 2.5 mM MgOAc, 1 mM DTT), adjusted to 2 mM tetracaine hydrochloride, and incubated at room temperature for 5 min. Samples were layered onto a 1-ml cushion of 500 mM sucrose in physiological salt buffer and sedimented in a TLA 100.4 rotor at 160,000 × g for 15 min at 4°C. Pellets were resuspended in 500 μl of immunoprecipitation buffer (0.15 M NaCl, 0.01 M Tris-HCl [pH 7.4], 1% Triton X-100), while a final concentration of 0.5% Triton X-100 was added to the upper 500 μl of sample supernatants. All samples contained phenylmethylsulfonyl fluoride (PMSF), leupeptin, aprotinin, and antipain. Samples were then immunoprecipitated for 3 h at 4°C with anti-Vif antiserum (12) that had been preabsorbed with protein A-Sepharose. Proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), followed by autoradiography.
Gradient sedimentation of Vif.
H9 cells (5 × 106) infected with HXB2NEO were washed in phosphate-buffered saline and then lysed with 0.5% Triton X-100 in phosphate-buffered saline containing PMSF, leupeptin, aprotinin, and antipain. Cell lysates were precleared of nuclei by centrifugation at 1,500 × g for 5 min at 4°C and incubated in the presence or absence of RNase A (10 μg/ml) for 30 min at 30°C. Postnuclear supernatants were loaded onto a 15 to 50% (wt/vol) sucrose gradient and centrifuged at 40,000 rpm for 90 min at 4°C in a Beckman SW41Ti rotor. Fractions were collected, precipitated with trichloroacetic acid, resuspended in loading dye, separated by SDS-PAGE (12% polyacrylamide), and analyzed by Western blotting with antiserum against Vif.
In vivo Vif-RNA association.
H9 cells either mock infected or chronically infected with HXB2NEO or HXB2VifΔNEO were washed in phosphate-buffered saline, pelleted, and resuspended in hypotonic lysis buffer (10 mM Tris-HCl [pH 8.0], with 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT) in the presence of protease inhibitors. Cells were incubated on ice for 10 min, homogenized with 20 strokes of a tight-fitting glass Dounce homogenizer, and centrifuged at 1,500 × g for 5 min at 4°C to remove nuclei. Postnuclear supernatants were immunoprecipitated in the presence of 0.5% NP-40 and 10 μg of yeast tRNA per ml with antiserum against Vif (preabsorbed on protein A-Sepharose) for 3 h at 4°C. Precipitated samples were subjected to either cDNA synthesis with avian myeloblastosis virus RT (Invitrogen), followed by PCR, or were submitted directly to PCR. PCRs were run at 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s for 30 cycles, followed by a 5-min extension at 72°C to amplify the env-coding region with primers J10 and J12 (5′-CGCGGGATCCTGTTAAATGGCAGTCTAGCAGAAG-3′ and 5′-CGCGCGCGGAATTCTTAAACAGTAGAAAAATTCCCCTC-3′, respectively). PCR amplification was also performed with the HXB2 construct as a positive control. PCR products were separated on 1.2% agarose gels and visualized by ethidium bromide staining.
In vitro RT reactions.
Supernatants were harvested from H9 cells infected with HXB2NEO(env−) and HXB2VifΔNEO(env−), and virions were pelleted as described above. Total viral RNA was extracted from viral pellets by the guanidinium isothiocyanate method (18) and dissolved in 5 mM Tris buffer (pH 7.5). Total viral RNA was incubated for 15 min at 37°C in 20 μl of RT buffer (50 mM Tris-HCl [pH 7.5], with 60 mM KCl, 3 mM MgCl2, 10 mM DTT) containing 40 ng of purified HIV RT, 10 U of RNasin, and various radioactive α-32P-labeled deoxynucleoside triphosphates (dNTPs). The reaction mixture for measuring total tRNA placement contained 0.2 mM dCTP, 0.2 mM dTTP, 5 μCi of [α-32P]dGTP (Dupont; 3,000 Ci/mmol, 10 mCi/ml), and 0.5 mM ddATP. For experiments assessing the initiation of reverse transcription, the reaction mixture contained only 5 μCi each of [α-32P]dGTP and [α-32P]dCTP (Dupont; 3,000 Ci/mmol, 10 mCi/ml). The reaction products were ethanol precipitated, resuspended, and analyzed on 6% polyacrylamide–7 M urea–1× TBE gels.
RESULTS
Viral growth kinetics and exogenous and endogenous RT.
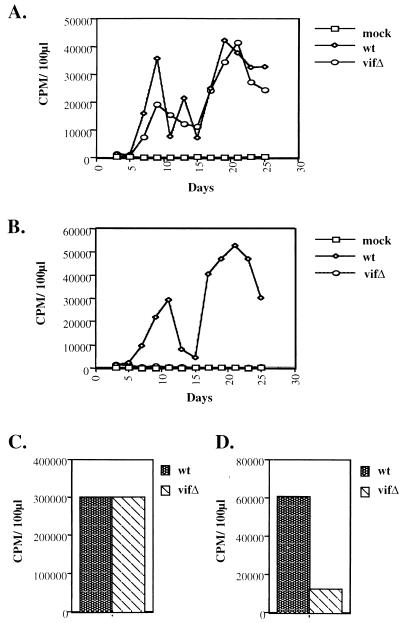
To identify the functional target of Vif, wild-type and vif mutant virions derived from permissive COS-7 cells were used to infect Jurkat and H9 cells. Virion production was monitored by assaying RT activity in the extracellular supernatant every other day for 25 days. The infected Jurkat cells produced comparable levels of wild-type and vif mutant virus (Fig. 1A). In H9 cells, however, the wild-type virus was readily competent for productive infection, but the vif mutant failed to replicate (Fig. 1B).
FIG. 1.
Viral growth curves and exogenous and endogenous RT reactions. Cell culture supernatants from COS-7 cells transfected with wild-type (wt) or vif mutant (vifΔ) viral constructs were used to initiate infections. After infection of Jurkat (A) and H9 T-cell (B) lines, virion production was monitored by RT activity in cell culture supernatants. Exogenous (C) and endogenous (D) RT reactions utilized H9-derived virions (wild type and vif mutant), which were normalized by viral protein content as displayed in Fig. 2. CPM, counts per minute.
Wild-type and vif-mutant virions produced from H9 cells were the focus of further analysis. The activity of virion-associated RT was first examined by measuring exogenous RT activity: virions were lysed with 1% Triton X-100 and added to an exogenous template and primer, and the incorporation of radiolabeled dTTP was determined. When wild-type and vif mutant virions were normalized by virion-associated protein profiles, including RT, they gave indistinguishable levels of exogenous RT activity (Fig. 1C). The same preparation of virions was then monitored for endogenous RT activity. In this case, 0.02% Triton X-100 was used for lysis, and radiolabeled dATP and nonradioactive dGTP, dCTP, and dTTP were added to the reaction. Under these conditions, the vif mutant showed a markedly decreased ability to incorporate radiolabeled nucleotides (Fig. 1D). The endogenous RT assay utilizes viral genomic RNA and tRNA3Lys as template and primer, respectively, a situation distinct from the exogenous RT reaction. Taken together, the data from these two assays suggest a defect in the reverse transcription process in the vif mutant that cannot be attributed to the RT molecules.
Virion-associated protein and RNA profiles.
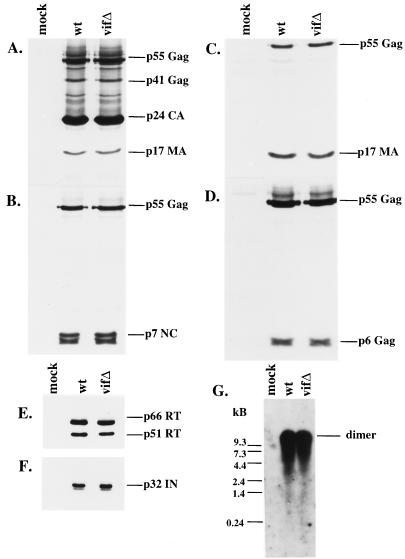
In order to examine the virion-associated protein profiles, we established stable H9 cell lines using wild-type and vif mutant constructs. env was deleted in these constructs in order to prevent the spreading of the wild-type virus in culture; previous studies have established that Env is not the target of Vif (11, 32, 44). These H9 cell lines expressed comparable levels of intracellular viral proteins (data not shown). Pelleted virions were examined by Western blotting, which demonstrated that virion production and release were unaltered in the vif mutant (Fig. 2A to F). Immunoblots were probed with HIV-positive patient serum and with antibodies recognizing p7NC, p17MA, p6gag, RT, and IN. No significant differences were seen in the composition, ratio, or integrity of any of the Gag or Pol proteins examined (Fig. 2A to F).
FIG. 2.
Protein and genomic RNA composition of wild-type (wt) and vif mutant (vifΔ) virions from H9 cells. Protein profiles of virions derived from chronically infected H9 cells expressing wild-type and mutant constructs were evaluated by SDS-PAGE, and immunoblots were stained with HIV-1-positive patient serum (A), antiserum against p7NC (B), antiserum against p17MA (C), antiserum against p6gag (D), a monoclonal antibody against RT (E), or antiserum against IN (F). (G) Virion-associated genomic RNA was hybridized with a probe directed against the 5′ LTR and gag coding regions and visualized by autoradiography.
We then analyzed viral genomic RNA from virions derived from stable H9 cell lines containing wild-type or vif mutant constructs. When the viral RNA was extracted, run on a nondenaturing gel, and then probed for genomic RNA by Northern blotting, the vif mutant virions were found to possess levels of viral genomic RNA in its dimeric form which were indistinguishable from those of the wild type (Fig. 2G).
Natural intravirion RT products.
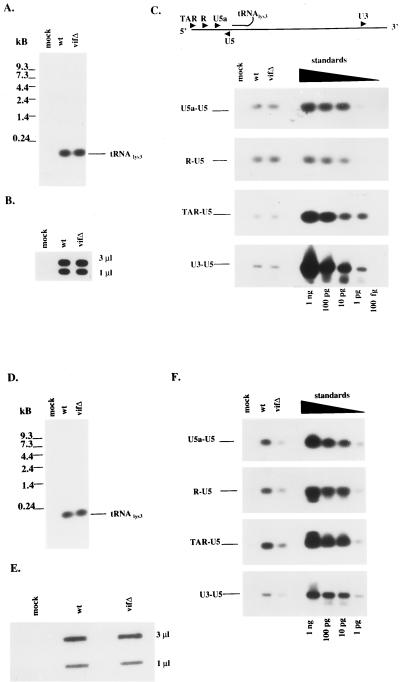
The natural intravirion RT products were then monitored by direct extraction of nucleic acids from virions, followed by a combination of PCR for specific sites in the viral genome and Southern blotting. Our intent was to determine whether vif mutant virions have a defect in reverse transcription that precedes virus entry into target cells and occurs in the absence of any in vitro manipulation. The wild type and vif mutant produced in Jurkat cells were first normalized for protein composition and exogenous RT activity (data not shown) and then examined for virion incorporation of tRNA3Lys and viral genomic RNA (Fig. 3A and B, respectively); in both cases, no distinguishable differences were seen. The existence of RT-directed DNA extensions from the tRNA3Lys primer was then assessed by using PCR primers within the 5′ LTR: U5 and U5a, R and U5, and TAR and U5 for minus-strand strong stop DNA. Additional PCR primers covered the first strand transfer (U3 and U5). PCR products from all four sets of primers were indistinguishable when wild-type and vif mutant virions from Jurkat cells were compared (Fig. 3C). These data suggest that Vif plays no detectable role in the extent of reverse transcription within Jurkat-derived virions, as had been predicted.
FIG. 3.
Intravirion nucleic acid profiles from Jurkat and H9 cells expressing wild-type (wt) or vif mutant (vifΔ) constructs. Nucleic acids were extracted from Jurkat cell-derived virions. (A and B) tRNA3Lys was analyzed by Northern blot hybridization (A), and (B) viral genomic RNA content was analyzed by slot blot analysis (B) with two separate dilutions (3 and 1 μl) of each sample. (C) Natural virion-associated RT products were detected by PCR with specific primers, followed by hybridization with a probe directed against the 5′LTR and gag coding region. The primers are described in Materials and Methods. Control standards were amplified with the indicated primers and the HXB2 construct as the template in serial 10-fold dilutions. Similar analysis was performed for (D) tRNA3Lys (D), virion-associated genomic RNA (E), and natural intravirion RT products (F) from virions derived from H9 cells expressing wild-type or vif mutant constructs.
A similar analysis of intravirion RT products was performed on wild-type and vif mutant virions from H9 cells. Virions were normalized for virion-associated protein content and exogenous RT activity before nucleic acid extraction. Comparable levels of virion incorporation of tRNA3Lys and viral genomic RNA were noted (Fig. 3D and E, respectively). PCR products using the four sets of primers described above were then evaluated by using H9-derived virions. For all DNA-extended products examined, the vif mutant displayed an approximately eightfold reduction in reverse transcription (Fig. 3F). These data argue in favor of an RT-mediated defect in the vif mutant that preceeds the entry of virions into new target cells.
RNA association of Vif.
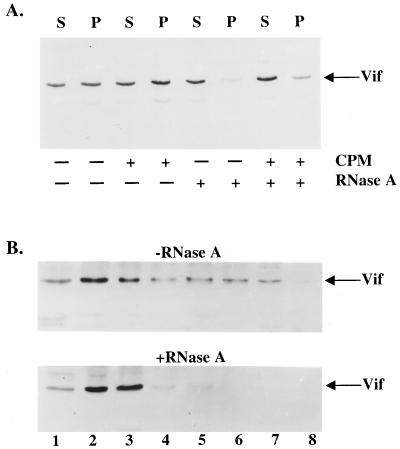
We next examined the association of Vif with RNA as a means of evaluating the possibility that Vif could regulate the RT complex through an association with viral genomic RNA or tRNA3Lys. We therefore translated Vif in vitro by using a rabbit reticulocyte lysate and assessed its ability to be pelleted by a 160,000 × g centrifugation. We also posttranslationally treated Vif-containing lysates with RNase A, supplemented the lysates with canine pancreatic microsomes, or exposed them to a combination of both treatments. This assay served as a preliminary means of assessing the ability of Vif to associate with a complex containing membranes, as previously suggested (12), or RNA. In the absence of RNase A or microsome treatment, approximately 50 to 60% of Vif was pelletable, as previously shown (12) (Fig. 4A). When lysates were treated with RNase A, irrespective of whether canine pancreatic microsomes were present, the ability of Vif to pellet was severely impaired. The presence of canine pancreatic microsomes did not appear to significantly influence Vif sedimentation.
FIG. 4.
RNA association with HIV-1 Vif in vitro and in vivo. (A) HIV-1 Vif was in vitro translated with rabbit reticulocyte lysates, followed by incubation with canine pancreatic microsomes (CPM), RNase A, or both in combination. Vif-containing lysates were subjected to centrifugation at 160,000 × g through a sucrose cushion in order to separate samples as supernatants (S) and pellets (P). (B) H9 cells infected with HXB2NEO (wild type) were lysed in the presence of 0.5% Triton X-100, and postnuclear supernatants were incubated in the presence (bottom panel) or absence (top panel) of RNase A. Lysates were layered on top of a 15 to 50% sucrose gradient and subjected to centrifugation, and fractions were collected. Fraction numbers are labeled, with fraction 1 being the top of the gradient.
Bearing in mind that Vif participates in the coordination of a cell-type-specific event, we were interested to assess its ability to associate with RNA in virus-infected cells. Infected H9 cells were lysed with 0.5% Triton X-100, and postnuclear supernatants containing Vif were then treated with RNase A or directly centrifuged without RNase A pretreatment through a sucrose velocity gradient. In the untreated sample, Vif was present in fractions 1 to 7 with slightly higher protein levels in fractions 2 and 3 than in other fractions (Fig. 4B). After RNase A treatment, a much lower level of Vif was seen in the fractions closer to the bottom of the gradient (fractions 4 to 7), with a corresponding increase of Vif in fractions 2 and 3. These data suggest that Vif associates with RNA either directly or through interaction with other molecules.
Association of Vif with viral genomic RNA.
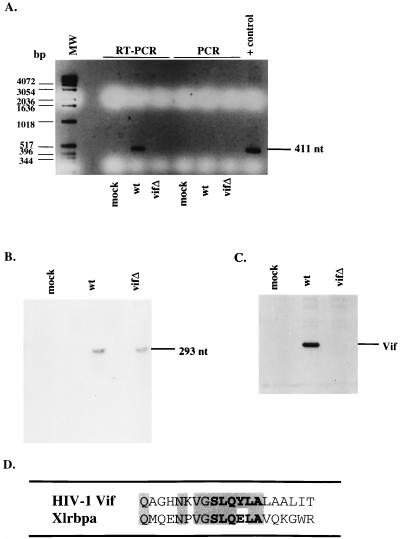
To determine whether Vif is associated with viral genomic RNA, we attempted to immunoprecipitate RNA from virus-infected H9 cells by using an anti-Vif antibody. Postnuclear supernatants from mock-, wild-type-, or vif mutant-infected cells were immunoprecipitated with antiserum against Vif, and the resulting precipitates were subjected to either RT-PCR or PCR using primers to the viral genome. Whereas viral genomic RNA was successfully amplified from the cell lysate containing Vif, the vif mutant lysate failed to give a detectable product (Fig. 5A). In addition, when lysates were used in PCR, no products were obtained, confirming that the amplification in the wild type was RNA. RNase protection was performed with cell lysates expressing the wild-type or vif mutant viral genome demonstrating the comparable abundance of intracellular viral genomic RNA (Fig. 5B). Additionally, cell lysates were probed in an immunoblot with antiserum to Vif demonstrating the expression of Vif protein in the wild type, but not mock or vif mutant-infected cells (Fig. 5C).
FIG. 5.
Association of Vif with viral genomic RNA. (A) H9 cell lysates from mock-infected cells or cells expressing the wild type (wt) or vif mutant (vifΔ) were subjected to immunoprecipitation with antiserum against Vif. Immunoprecipitated pellets were either directly PCR amplified with primers directed against the HIV-1 genome, or cDNA synthesis was performed first with avian myeloblastosis virus RT, followed by PCR. Direct PCR amplification were performed on the HXB2 construct as a positive control. MW, molecular weight markers; nt, nucleotides. (B) RNA of H9 cell lysates from mock-infected cells or cells expressing the wild type or vif mutant were subjected to RNase protection with a probe to the HIV-1 genome. (C) Western blot analysis of cell lysates was performed with antiserum against Vif. (D) Amino acid sequence alignment of HIV-1 Vif and Xlrbpa. Residues in boldface display the highly conserved Vif sequence among lentiviruses. Shaded residues display sequence identity between HIV-1 Vif and Xlrbpa.
Although the vif open reading frame is conserved within the lentivirus family, the overall amino acid sequence identity among various Vif proteins is low. Lentiviral Vifs, which are highly basic proteins of 192 to 251 amino acids, do share one common feature: a highly conserved sequence motif, SLQXLA. Since Vif appears to associate with viral genomic RNA, it is interesting to note that the same amino acid motif appears in the Xenopus laevis RNA-binding protein Xlrbpa (10). In fact, this motif is present in the double-stranded RNA binding region of the molecule (34). HIV-1 Vif and Xlrbpa show amino acid sequence identity in 7 of 8 residues or, when extended, 9 of 14 residues encompassing this motif (Fig. 5D).
Viral RNA components of the vif mutant virions display a defect in initiation of reverse transcription.
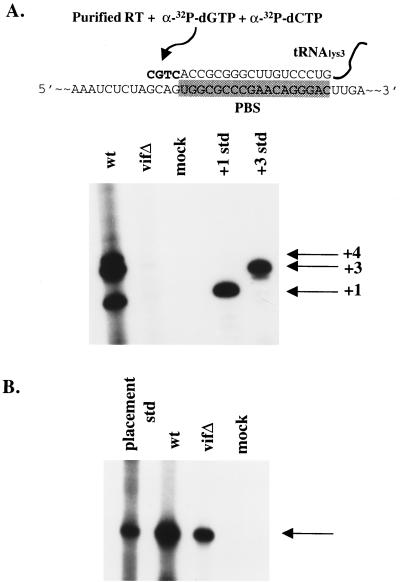
It is possible that Vif influences reverse transcription by regulating the viral RNA, but not viral proteins. To test this idea, total viral RNAs (tRNA3Lys and genomic RNA) were extracted from virions devoid of proteins. Initiation of reverse transcription with purified viral RNAs, recombinant HIV-1 RT, and limited concentrations of dCTP and dGTP was then performed. Previous studies have shown that primer tRNA3Lys exists in unextended and 2-base extended forms within HIV-1 virions (17). The wild type and vif mutant produced in H9 cell lines were first normalized for viral genomic RNA content (data not shown). When extended in vitro, RNAs extracted from wild-type virions showed 1-base (+1), 3-base (+3), and 4-base (+4) extended tRNA3Lys forms (Fig. 6A, lane 1), consistent with previous reports (17). The vif mutant RNAs were markedly impaired in their ability to produce the extended products (Fig. 6A, lane 2). This defect in the initiation of reverse transcription could not be attributed to a defect in the placement of tRNA3Lys onto the viral genomic RNA, since a placement assay revealed only a modest reduction in primer placement (Fig. 6B). This type of analysis utilizes only the RNA components derived from vif mutant virions, which supports the notion that Vif regulates the viral RNA.
FIG. 6.
Primer placement and initiation of reverse transcription. (A) In vitro initiation of reverse transcription was assessed with [32P]dCTP and [32P]dGTP from H9-derived virion-associated nucleic acids. Positive controls for +1- and +3-base extensions are indicated. The diagram depicts viral genomic RNA with bound tRNA3Lys on the PBS (shaded box). Addition of recombinant RT and dCTP and dGTP results in +1-, +3-, and +4-base extensions. Std, standard. (B) In vivo tRNA3Lys primer placement for H9-derived wild-type (wt) and vif mutant (vifΔ) virions. An arrow indicates the primer placement product for a 6-base extension.
DISCUSSION
Previous efforts to determine the target for Vif activity have not yielded definitive results. It was relatively certain that Vif must affect components found within the virion, although analysis of virion protein content from wild-type and vif mutant producer cells revealed indistinguishable levels of Gag, Pol, and Env proteins (1, 11, 32, 44). In addition, studies using vif mutant virions from semipermissive cell lines (CEM) showed no differences in viral RNA incorporation (13, 44) or dimerization of RNA when compared to wild-type virions (13). In the present study, we also observed no differences in the virion protein content for the vif mutant (Fig. 2). Viral genomic RNA and primer tRNA3Lys were packaged normally in the vif mutant virions.
Several investigators have shown that Vif plays a regulatory role in reverse transcription during the entry phase of the virus's life cycle (6, 8, 30, 35, 40, 44). However, an alteration in the appearance of RT products after viral entry into target cells does not clearly indicate whether penetration, uncoating, virus core stability, or reverse transcription itself is being affected. Previous reports have demonstrated defects in the ability of vif mutant virions to undergo endogenous reverse transcription (8, 13). These studies suggested the existence of alterations in virion components involved in the regulation of reverse transcription; however, the target molecule(s) of this regulation remained uncertain. Since partial RT products have been demonstrated within virions (25, 43), we decided to examine the contribution of Vif to the presence of natural intravirion cDNAs. This analysis revealed that Vif contributed to the enhancement of natural intravirion RT products in a cell type-dependent manner (Fig. 3). A reduction in RT products was evident in vif mutant virions derived from H9 cells, but not from Jurkat cells, a finding which correlates with their replication growth kinetics (Fig. 1). These data were consistent with the defect observed in the endogenous RT reaction (Fig. 1D), underscoring the notion that virion-associated components must regulate reverse transcription. Since RT itself failed to display an enzymatic defect when a heterologous template primer was used (Fig. 1C) and no difference was observed in the virion-associated proteins (Fig. 2), it remained possible that either the primer tRNA3Lys or the viral genomic RNA would be the likely target.
When we examined the contribution of Vif to virion-associated RNAs, we saw no differences in the levels of virion-incorporated viral genomic RNA or tRNA3Lys. However, use of purified virion-associated RNAs as a template and primer for the initiation of reverse transcription resulted in a drastic reduction in the +1-, +3-, and +4-base-extended forms of tRNA3Lys in the vif mutant, as compared to the wild type (Fig. 6A). Since the vif mutant showed a less dramatic, but still approximately fivefold reduction in the assay of placement of tRNA3Lys on the viral genome, this finding imply an altered binding affinity of the primer to its template. It remains possible that the structure of the RNA complex at the 5′ end of the viral genome may be altered by Vif in such a way that it could indirectly influence the accessibility of the primer tRNA3Lys in annealing to the primer binding site. Whether the placement of tRNA3Lys on to the viral genome is truly altered remains uncertain, since the tRNA placement assay used in these studies relies not only on tRNA placement, but also on the ability of RT to perform a 6-nucleotide extension from the primer. From the data provided, it's not clear whether the RT-mediated extension defect in the vif mutant could be completely independent of a defect in tRNA3Lys placement. Although the details of Vif's role in this reaction remain to be determined, these findings provided clear evidence that the vif mutant RNAs differed from wild-type RNAs in some manner that influenced the initiation of reverse transcription.
In eukaryotic cells, several types of posttranscriptional regulation involve the recognition of RNA by proteins. RNA-binding proteins can often be divided into distinct classes on the basis of their amino acid sequence motifs (2, 41). We have already noted that the highly conserved and essential Vif motif SLQYLA shows a high degree of homology (8 out of 10 residues identical) to the double-stranded RNA-binding protein from X. laevis, XlrbpA (Fig. 5D). This motif in XlrbA has been shown to participate directly in binding of RNA (34). In addition, XlrbpA associates with ribosomes and hnRNPs (10). The double-stranded RNA-binding motif (DSRM) of XlrbpA extends over a 65- to 70-amino-acid sequence and includes many basic and hydrophobic residues. Although Vif lacks additional sequence homology with the DSRM outside the SLQYLA motif, it contains a high percentage of basic residues, with a predicted pI of 10.7 (31).
Thus far, it is unclear whether Vif has specificity for the HIV-1 genomic RNA. However, we have provided evidence that Vif affects viral nucleic acid components to influence the initiation of reverse transcription. Vif of FIV has recently been shown to localize to the nucleus, which raises the interesting possibility that Vif may traffic with the viral RNA out of the nucleus to the cytoplasm (4). Although it appears unlikely that Vif regulates the movement or targeting of the viral genomic RNA itself, since the vif mutant fails to influence viral protein translation and RNA packaging, its position within the nucleus may provide the initial contact point with viral genomic RNA. The association of Vif with RNA from postnuclear supernatants (Fig. 4B) and its colocalization with Gag proteins in infected cells (35) are consistent with the possibility that Vif is recruited to the same cytoplasmic location as the Gag assembling complex, where it may aid in an RNA regulatory step in viral assembly. The virtual absence of Vif from virions suggests that its association with viral RNA, however, may be transient. Vif may function by associating with the viral genomic RNA prior to or during the binding of the RNA with the cytoplasmic viral assembly complex. From this position, we speculate that Vif regulates the viral genomic RNA-tRNA3Lys complex in a yet-to-be-determined manner. Once this alteration is achieved, Vif may be removed from this complex, leaving the RNA in a state that would promote efficient initiation of reverse transcription. Since Vif functions in a strict cell-type-dependent fashion, it may be working in concert with other cellular molecules. It has been argued that Vif is necessary to overcome an inhibitory factor present in H9 and Hut-78 cell lines (26, 37). Although this factor has yet to be identified, it would be intriguing to see whether it functions with Vif in connection with the virion-associated nucleic acids. Future studies of Vif are likely to provide insight into the regulatory steps of reverse transcription.
ACKNOWLEDGMENTS
We thank Dolph Hatfield, Richard Markham, and Jef Boeke for useful discussions of the manuscript and Larry Arthur for providing antiserum to p7NC.
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: antiserum to HIV-1 Vif (catalog no. 2221), antiserum to HIV-1 IN (catalog no. 756), and antiserum to p17MA (catalog no. 286).
REFERENCES
- 1.Bouyac M, Rey F, Nascimbeni M, Courcoul M, Sire J, Blanc D, Clavel F, Vigne R, Spire B. Phenotypically Vif− human immunodeficiency virus type 1 is produced by chronically infected restrictive cells. J Virol. 1997;71:2473–2477. doi: 10.1128/jvi.71.3.2473-2477.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 3.Cen S, Huang Y, Khorchid A, Darlix J-L, Wainberg M A, Kleiman L. The role of Pr55gag in annealing of tRNA3Lys to human immunodeficiency virus type 1 genomic RNA. J Virol. 1999;73:4485–4488. doi: 10.1128/jvi.73.5.4485-4488.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterji U, Grant C K, Elder J H. Feline immunodeficiency virus Vif localizes to the nucleus. J Virol. 2000;74:2533–2540. doi: 10.1128/jvi.74.6.2533-2540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cobrinik D, Soskey L, Leis J. A retroviral RNA secondary structure required for efficient initiation of reverse transcription. J Virol. 1988;62:3622–3630. doi: 10.1128/jvi.62.10.3622-3630.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courcoul M, Patience C, Rey F, Blanc D, Harmache A, Sire J, Vigne R, Spire B. Peripheral blood mononuclear cells produce normal amounts of defective Vif− human immunodeficiency virus type 1 particles which are restricted for the preretrotranscription steps. J Virol. 1995;69:2068–2074. doi: 10.1128/jvi.69.4.2068-2074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dettenhofer M, Yu X F. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J Virol. 1999;73:1460–1467. doi: 10.1128/jvi.73.2.1460-1467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dornadula G, Yang S, Pomerantz R J, Zhang H. Partial rescue of the Vif-negative phenotype of mutant human immunodeficiency virus type 1 strains from nonpermissive cells by intravirion reverse transcription. J Virol. 2000;74:2594–2602. doi: 10.1128/jvi.74.6.2594-2602.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Draper D E. Protein-RNA recognition. Annu Rev Biochem. 1995;64:593–620. doi: 10.1146/annurev.bi.64.070195.003113. [DOI] [PubMed] [Google Scholar]
- 10.Eckmann C R, Jantsch M F. Xlrbpa, a double-stranded RNA-binding protein associated with ribosomes and heterogeneous nuclear RNPs. J Cell Biol. 1997;138:239–253. doi: 10.1083/jcb.138.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouchier R A M, Simon J H M, Jaffe A B, Malim M H. Human immunodeficiency virus type 1 Vif does not influence expression or virion incorporation of gag-, pol-, and env-encoded proteins. J Virol. 1996;70:8263–8269. doi: 10.1128/jvi.70.12.8263-8269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goncalves J, Jallepalli P, Gabuzda D H. Subcellular localization of the Vif protein of human immunodeficiency virus type 1. J Virol. 1994;68:704–712. doi: 10.1128/jvi.68.2.704-712.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goncalves J, Korin Y, Zack J, Gabuzda D. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J Virol. 1996;70:8701–8709. doi: 10.1128/jvi.70.12.8701-8709.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harmache A, Russo P, Guiguen F, Vitu C, Vignoni M, Bouyac M, Hieblot C, Pepin M, Vigne R, Suzan M. Requirement of caprine arthritis encephalitis virus vif gene for in vivo replication. Virology. 1996;224:246–255. doi: 10.1006/viro.1996.0526. [DOI] [PubMed] [Google Scholar]
- 15.Hatfield D, Feng Y X, Lee B J, Rein A, Levin J G, Oroszlan S. Chromatographic analysis of the aminoacyl-tRNAs which are required for translation of codons at and around the ribosomal frameshift sites of HIV, HTLV-1, and BLV. Virology. 1989;173:736–742. doi: 10.1016/0042-6822(89)90589-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Shalom A, Li Z, Wang J, Mak J, Wainberg M A, Kleiman L. Effects of modifying the tRNA3Lys anticodon on the initiation of human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1996;70:4700–4706. doi: 10.1128/jvi.70.7.4700-4706.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Wang J, Shalom A, Li Z, Khorchid A, Wainberg M A, Kleiman L. Primer tRNA3Lys on the viral genome exists in unextended and two-base extended forms within mature human immunodeficiency virus type 1. J Virol. 1997;71:726–728. doi: 10.1128/jvi.71.1.726-728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Khorchid A, Gabor J, Wang J, Li X, Darlix J-L, Wainberg M A, Kleiman L. The role of nucleocapsid and U5 stem/A-rich loop sequences in tRNA3Lys genomic placement and initiation of reverse transcription in human immunodeficiency virus type 1. J Virol. 1998;72:3907–3915. doi: 10.1128/jvi.72.5.3907-3915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoshima Y, Kohmoto M, Ikeda Y, Yamada H, Kawaguchi Y, Tomonaga K, Miyazawa Y, Kai C, Umemura T, Mikami T. Roles of the auxiliary genes and AP-1 binding site in the long terminal repeat of feline immunodeficiency virus in the early stage of infection in cats. J Virol. 1996;70:8518–8526. doi: 10.1128/jvi.70.12.8518-8526.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isel C, Marquet R, Keith G, Ehresmann C, Ehresmann B. Modified nucleotides of tRNAlys3 modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J Biol Chem. 1993;268:25269–25272. [PubMed] [Google Scholar]
- 21.Isel C, Lanchy J M, Le Grice S F, Ehresmann C, Ehresmann B, Marquet R. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNAlys3. EMBO J. 1996;15:917–924. [PMC free article] [PubMed] [Google Scholar]
- 22.Kan N C, Franchini G, Wong-Staal F, DuBois G C, Robey W G, Lautenberger J A, Papas T S. Identification of HTLV-III/LAV sor gene product and detection of antibodies in human sera. Science. 1986;231:1553–1555. doi: 10.1126/science.3006245. [DOI] [PubMed] [Google Scholar]
- 23.Lee T H, Coligan J E, Allan J S, McLane M F, Groopman J E, Essex M. A new HTLV-III/LAV protein encoded by a gene found in cytopathic retroviruses. Science. 1986;231:1546–1549. doi: 10.1126/science.3006243. [DOI] [PubMed] [Google Scholar]
- 24.Li Z, Shalom A, Huang Y, Mak J, Arts E, Wainberg M A, Kleiman L. Multiple forms of tRNA(Lys3) in HIV-1. Biochem Biophys Res Commun. 1996;227:530–540. doi: 10.1006/bbrc.1996.1541. [DOI] [PubMed] [Google Scholar]
- 25.Lori F, di Marzo Veronese F, De Vico A L, Lusso P, Reitz M S, Jr, Gallo R C. Viral DNA carried by human immunodeficiency virus type 1 virions. J Virol. 1992;66:5067–5074. doi: 10.1128/jvi.66.8.5067-5074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madani N, Kabat D. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J Virol. 1998;72:10251–10255. doi: 10.1128/jvi.72.12.10251-10255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mak J, Kleiman L. Primer tRNAs for reverse transcription. J Virol. 1997;71:8087–8095. doi: 10.1128/jvi.71.11.8087-8095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marquet R, Isel C, Ehresmann C, Ehresman B. tRNA as primer of reverse transcriptases. Biochimie. 1995;77:113–124. doi: 10.1016/0300-9084(96)88114-4. [DOI] [PubMed] [Google Scholar]
- 29.Miller J T, Ge Z, Morris S, Das K, Leis J. Multiple biological roles associated with the Rous sarcoma virus 5′ untranslated RNA U5-IR stem and loop. J Virol. 1997;71:7648–7656. doi: 10.1128/jvi.71.10.7648-7656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nascimbeni M, Bouyac M, Rey F, Spire B, Clavel F. The replicative impairment of Vif− mutants of human immunodeficiency virus type 1 correlates with an overall defect in viral DNA synthesis. J Gen Virol. 1998;79:1945–1950. doi: 10.1099/0022-1317-79-8-1945. [DOI] [PubMed] [Google Scholar]
- 31.Oberste M S, Gonda M A. Conservation of amino-acid sequence motifs in lentivirus Vif proteins. Virus Genes. 1992;6:95–102. doi: 10.1007/BF01703760. [DOI] [PubMed] [Google Scholar]
- 32.Ochsenbauer C, Wilk T, Bosch V. Analysis of vif-defective human immunodeficiency virus type 1 (HIV-1) virions synthesized in ‘non-permissive’ T lymphoid cells stably infected with selectable HIV-1. J Gen Virol. 1997;78:627–635. doi: 10.1099/0022-1317-78-3-627. [DOI] [PubMed] [Google Scholar]
- 33.Park I W, Myrick K, Sodroski J. Effects of vif mutants on cell-free infectivity and replication of simian immunodeficiency virus. J Acquir Immune Defic Syndr. 1994;7:1228–1236. [PubMed] [Google Scholar]
- 34.Ryter J M, Schultz S C. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon J H M, Malim M H. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J Virol. 1996;70:5297–5305. doi: 10.1128/jvi.70.8.5297-5305.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon J H M, Fouchier R A M, Southerling T E, Guerra C B, Grant C K, Malim M H. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J Virol. 1997;71:5259–5267. doi: 10.1128/jvi.71.7.5259-5267.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon J H M, Gaddis N C, Fouchier R A M, Malim M H. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat Med. 1998;4:1397–1400. doi: 10.1038/3987. [DOI] [PubMed] [Google Scholar]
- 38.Simon J H M, Carpenter E A, Fouchier R A M, Malim M H. Vif and the p55Gag polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J Virol. 1999;73:2667–2674. doi: 10.1128/jvi.73.4.2667-2674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sodroski J, Goh W C, Rosen C, Tartar A, Portetelle D, Burny A, Haseltine W. Replicative and cytopathic potential of HTLV-III/LAV with sor gene deletions. Science. 1986;231:1549–1553. doi: 10.1126/science.3006244. [DOI] [PubMed] [Google Scholar]
- 40.Sova P, Volsky D J. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. J Virol. 1993;67:6322–6326. doi: 10.1128/jvi.67.10.6322-6326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.St Johnston D, Brown N H, Gall J G, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci USA. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomonaga K, Norimine J, Shin Y-S, Fukasawa M, Miyazawa T, Adachi A, Toyosaki T, Kawaguchi Y, Kai C, Mikami T. Identification of a feline immunodeficiency virus gene which is essential for cell-free virus infectivity. J Virol. 1992;66:6181–6185. doi: 10.1128/jvi.66.10.6181-6185.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trono D. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses. J Virol. 1992;66:4893–4900. doi: 10.1128/jvi.66.8.4893-4900.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Schwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]