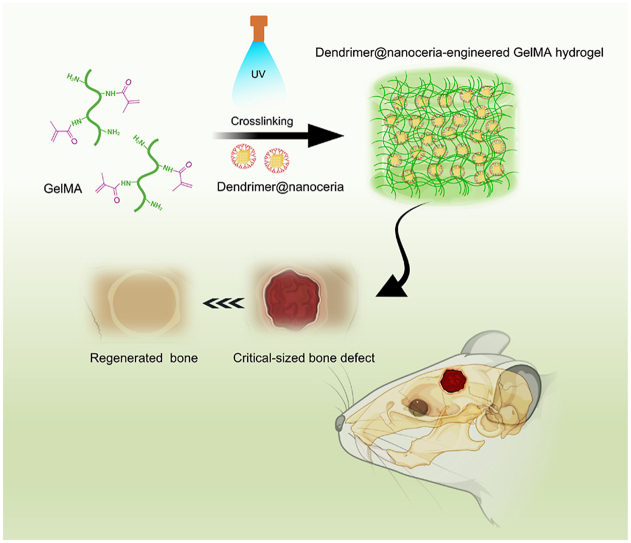
Abstract
Bone defects in patients entail the microenvironment that needs to boost the functions of stem cells (e.g., proliferation, migration, and differentiation) while alleviating severe inflammation induced by high oxidative stress. Biomaterials can help to shift the microenvironment by regulating these multiple events. Here we report multifunctional composite hydrogels composed of photo-responsive Gelatin Methacryloyl (GelMA) and dendrimer (G3)-functionalized nanoceria (G3@nCe). Incorporation of G3@nCe into GelMA could enhance the mechanical properties of hydrogels and their enzymatic ability to clear reactive oxygen species (ROS). The G3@nCe/GelMA hydrogels supported the focal adhesion of mesenchymal stem cells (MSCs) and further increased their proliferation and migration ability (vs. pristine GelMA and nCe/GelMA). Moreover, the osteogenic differentiation of MSCs was significantly stimulated upon the G3@nCe/GelMA hydrogels. Importantly, the capacity of G3@nCe/GelMA hydrogels to scavenge extracellular ROS enabled MSCs to survive against H2O2-induced high oxidative stress. Transcriptome analysis by RNA sequencing identified the genes upregulated and the signalling pathways activated by G3@nCe/GelMA that are associated with cell growth, migration, osteogenesis, and ROS-metabolic process. When implanted subcutaneously, the hydrogels exhibited excellent tissue integration with a sign of material degradation while the inflammatory response was minimal. Furthermore, G3@nCe/GelMA hydrogels demonstrated effective bone regeneration capacity in a rat critical-sized bone defect model, possibly due to an orchestrated capacity of enhancing cell proliferation, motility and osteogenesis while alleviating oxidative stress.
Keywords: Bone regeneration, Dendrimer, Nanoceria, Hydrogel, Stem cell activation, ROS scavenging
Graphical abstract
1. Introduction
Bone defects occasioned by an infection, trauma or enduring inflammation can result in a serious burden on a patient's physical and mental health. Repair and regeneration of critical bone defects is a challenging process that remains a serious clinical task [1,2]. Bone biomaterials are important in bone defects because they function as a link between native bone tissues, and cells and also steer the functional regeneration of bone tissues [3]. To facilitate the new bone formation, tissue engineering based on scaffolds and stem cells has also been derived as an alternative approach [4,5]. Being the most favourable biomatrices, hydrogels possess immense scientific interest. Apart from acting as a cell-friendly microenvironment, the inner porous morphology of hydrogels maintains and facilitates nutrient and gas exchange which supports endogenous cell growth and could act as a delivery platform for the controlled release of several bioactive molecules [[6], [7], [8]]. Naturally derived gelatin-based Gelatin Methacryloyl (GelMA) hydrogel is one such candidate widely used in various tissue regeneration applications due to its versatile cell supportiveness, controllable properties, and enzymatic disintegration [[9], [10], [11]]. Although GelMA hydrogels have likenesses with native bone extracellular matrix (ECM), they lack osteogenic factors or other inorganic components that could endorse bone mineralization, hence GelMA hydrogels in their native form are not suitable for in situ bone regeneration [12]. Moreover, the native bone comprises of multifaceted structure (micro/nanoscale) which guides cell growth and differentiation.
Several exertions have been done to modulate the degradation tendency, and mechanical performance as well as advance biological functionalities of microscale GelMA, especially by engineering using bioactive nanoscale materials [12,13]. For example, Z. Wu et al. engineered GelMA using lithium-modified bioglass for functional bone regeneration under diabetic conditions [14]. Here GelMA regulated cellular activities such as cell adhesion and proliferation while the nanomaterial integrated supported osteogenesis, angiogenesis, and immunomodulation. Halloysite (HNT)-incorporated GelMA hydrogels were recently reported by Huang et al. to promote bone regeneration [15]. The study showed that the HNT-based composite improved bone cell proliferation and differentiation in vitro and enhanced bone regeneration in a rat calvarial defect model in vivo. The researchers attributed the improved bone regeneration to the unique structure and properties of HNTs, which can serve as bioactive and biocompatible platforms for engineering bones.
Similarly, Q. Ou and colleagues used nanosilver-incorporated HNT to engineer GelMA hydrogels to form a hydrogel with superior bioactivity [16]. Moreover, the incorporation of nanomaterials rendered GelMA hydrogel with addition properties such as antibacterial activity, and immunomodulatory property which supported regeneration of infected bone tissues. In another study, ultrathin nano silicates were incorporated into GelMA to design a highly stiff GelMA hydrogel which could support osteogenesis even in the absence of osteoinductive factors due to the supreme bioactivity of nanomaterial incorporated [17]. In some cases, GelMA hydrogel has been directly functionalized with drugs such as alendronate, without the use of nanomaterials, for supporting osteogenesis. Liu et al. conducted a study using this approach [18]. Although such hydrogels have bioactive properties due to the modulation of the physiochemical properties of GelMA, their applications in bone tissue engineering are limited by factors such as the need for multiple functionalization steps, concerns over biodegradability, and high cost. All these studies point out that bone tissue engineering (BTE) using GelMA hydrogels requires engineered nanomaterials as functional units, which should greatly improve the physicochemical properties, osteogenic capacity, biocompatibility etc., and renders other superior properties to the hydrogel [12].
While numerous GelMA nanocomposites have shown promise in BTE , many of them lack ROS scavenging properties, which can restrict their effectiveness to only osteogenic properties. As a result, it is crucial to integrate ROS scavenging properties into pristine GelMA, while maintaining osteogenic potential to enhance their efficiency in regenerating bone tissues. ROS-responsive GelMA hydrogels can reduce oxidative stress, providing a better microenvironment for cell proliferation and differentiation [19]. Over the years, nanoceria (nCe) incorporated GelMA hydrogels have been extensively studied for their potential applications in regenerative medicine. The nCe has emerged as a strong candidate in biomedical research because of its exceptional multi-enzymatic properties owing to the ability to shift between oxidation states (Ce3+ & Ce4+) by an auto-sequential redox cycle [20,21]. Since bioactive and biocompatible, it is one of the potent candidates for handling health conditions related to oxidative stress and is also known to be efficacious in a wide range of inflammatory diseases as well as in cancer therapy [[21], [22], [23]]. Moreover, numerous studies have already demonstrated the therapeutic potential of nCe in BTE [[24], [25], [26]]. Previous studies using nCe and GelMA have primarily relied on the intrinsic antioxidant property of nCe to achieve targeted regenerative potential, especially for soft tissues like skin [27,28]. However, nCe alone may not provide the necessary microenvironmental cues when it comes to hard tissues like bones. In recent years,bioinspired materials based on dendrimers also have lately attracted extensive scientific interest because of their unique biocompatibility, chelation with proteins, and provision of a cell microenvironment that benefits tissue regeneration [[29], [30], [31]]. Dendrimer-engineered nanomaterials are very desirable to be deployed in the development of functional biomaterials for bone regeneration because of their highly defined chemical structure in nanometer dimension and globular form [32,33].
To address the existing limitations, we propose an innovative approach to develop a hybrid nanomaterial based on nCe, which is surface functionalized with macromolecular structures called dendrimers (G3). For this, first, we synthesized nCe and functionalized it with 3rd generation polyamidoamine (PAMAM) dendrimer to generate G3@nCe with nCe as the core and amine groups (-NH2) from G3 as the periphery [34,35]. The resulting G3@nCe retains the intrinsic properties of nCe, including ROS responsiveness, crystallinity, and stability, while also providing additional features to enhance bioactivity. The combination of superior bioactivity and antioxidant property of G3@nCe can lead to synergistic effects, where the properties of each component enhance the properties of the other. When incorporated into the GelMA matrix by photogelation, G3@nCe led to enhanced interfacial interactions and stability of resultant hydrogels characterized by improved mechanical properties and swelling kinetics, because of their cationic nature and symmetrical geometry. The immobilization capacity of dendrimers enables G3@nCe to function as a “nano-reservoir” that can immobilize and release a range of bioactive molecules (including growth factors and ECM proteins) in a controlled and sustained manner, providing superior bioactivity compared to nCe. Moreover, their unique architecture allows for high surface area which can enhance the stability and activity of immobilized proteins. Also due to the cationic nature, dendrimers interact with negatively charged cell membranes, enhancing their cellular uptake. The increased cellular uptake of G3@nCe leads to its better distribution and localization within the targeted cells or tissues.
For this reason, G3@nCe/GelMA served as a bioactive platform which offers superior microenvironmental cues for modulating cellular responses (proliferation, migration, and osteogenic differentiation) compared to pristine GelMA and nCe/GelMA. G3@nCe/GelMA also has antioxidant properties which benefit cells under oxidative stress. The transcriptome analysis by RNA sequencing revealed that G3@nCe/GelMA upregulated various genes related to cell growth, migration, osteogenesis, and ROS metabolic process and revealed the involvement of the canonical Wnt signalling pathway in stimulating osteogenesis. We believe that the design of G3@nCe/GelMA hydrogel that can successfully orchestrate cellular responses will meet multiple requirements to ensure the successful restoration of bone defects and could provide insightful ideas for developing future bone biomaterials. In addition, our findings suggest that this approach to engineer GelMA hydrogels using dendrimer-functionalized nanomaterials is not only universal but also can be extended to other tissue platforms, providing a unique opportunity to impart new biological properties and functions to regenerate diseased and injured tissues.
2. Experimental section
2.1. Synthesis of GelMA
GelMA was made according to previous reports with slight modifications [36]. Briefly, a 10% (w/v) uniform gelatin solution was made by dissolving gelatin (Bovine skin type B, Bloom 300, Sigma Aldrich) in distilled water (DW) at 50°C. Then gradually add 0.6 ml of methacrylic anhydride (MA, 94%, Sigma Aldrich) per gram of gelatin and continue stirring for 1 h. This mixture was then centrifugated at 3500 rpm for 2 min and purified by dialyzing against warm DW for 7 days using 12–14 kDa tubular dialysis membrane (CelluSep, Regenerated Cellulose Tubular Membrane (T4) MFPI). The pH of the solution was then adjusted to 7.4 using 1 molar (1 M) sodium bicarbonate (NaHCO3, Sigma Aldrich) before being snap-frozen in liquid nitrogen and lyophilized for 1 week (ilshin Lab Co. Ltd, Korea). The freeze-dried GelMA prepolymer was stored at −20°C before use. Further characterization of GelMA was done using Fourier transform infrared spectroscopy (FTIR, Varian 640-IR, Australia). The degree of methacrylation (DOM) was found by comparing the free –NH2 groups in GelMA before and after functionalization using ninhydrin assay following a slightly modified protocol [36]. For this gelatin and GelMA solutions were made in DW at 50°C until the solutions become clear. Ninhydrin reagent (Sigma Aldrich) was made in sodium citrate monobasic (Sigma Aldrich) or glycerol (Sigma Aldrich) mixture and made into a final concentration of 2.5 mg/ml. Then 50 μl of gelatin or GelMA was mixed with 950 μl of ninhydrin in an Eppendorf tube and warmed in a water bath for 15 min. DW is used as the blank solution. After 45 min of cooling, the absorbance was measured at 570 nm with a plate reader (Molecular Devices, USA). A linear regression line was plotted from the gelatin dilution series using the average absorbance value. The absorbance value at 570 nm corresponds to the concentration of amine groups in free form. The decrease in free amine concentration is due to the successful methacryloyl substitution. The average absorbance value of GelMA corresponds to A% gelatin concentration on the standard curve. Then the DOM was calculated as (100-A) %.
2.2. Synthesis of G3@nCe
For the synthesis of G3@nCe, first, nCe was made using a hydrothermal process with slight modifications from previous reports [37,38]. In brief, 2.6 g of cerium (III) nitrate hexahydrate (Ce(NO3)3·6H2O, 99%, Sigma Aldrich) was dissolved in 60 ml DW and pH was adjusted to basic (8.0) using ammonium hydroxide solution (NH4OH, 28.0–30.0%, Sigma Aldrich). Separately 22 mg of Hexadecyltrimethylammonium bromide (CTAB, 98%, Sigma Aldrich) was dissolved in DW and used as a surfactant. To obtain hydrothermally processed nCe, the Teflon vessel was transferred to an autoclave and thermal-treated at 140°C for 24 h. The unreacted surfactants were removed completely by repeated washing and drying at 450°C for 3 h. Before functionalizing with G3, first, the surface of nCe was modified independently by a carboxylation reaction. For this, a known amount of nCe was treated with citric acid (99.5%, Sigma Aldrich). After that, the pH was adjusted to 5.0 with 1 M sodium hydroxide (NaOH, 97.0%, Sigma Aldrich). The obtained carboxylated nCe were thoroughly washed using DW and dried in a freeze-dryer. For G3 conjugation, a calculated amount of carboxylated nCe was dispersed in DW by ultrasonication and an equimolar mixture of N-(3-dimethylamino-propyl)-N′-ethylcarbodiimide hydrochloride (EDC, 98% Sigma Aldrich), and N-hydroxysuccinimide (NHS, 98% Sigma Aldrich) was added to the solution by continuous stirring and allowed stirring for 30 min for activation. Finally, the PAMAM dendrimer ethylenediamine core, generation 3.0 solution (20 wt% in methanol, Sigma Aldrich) was gradually added to the mixture and allowed to stir for 24 h at room temperature (RT). The obtained G3@nCe was then washed with DW several times to remove impurities, then freeze-dried and stored in a vacuum desiccator.
2.3. Characterization of G3@nCe
The nanosized G3@nCe was first observed using Transmission electron microscopy (TEM, JEOL-7100). The characteristic infrared spectra were then analyzed using FTIR. The X-ray diffraction (XRD) measurements were performed to analyze the changes in the crystalline structure of nCe after functionalization using a Rigaku Ultima IV powder diffractometer with Cu-Kα radiation. The Zeta potential of particles was determined with the Malvern Zetasizer device (ZEN3600; Malvern). The surface chemistry of G3@nCe was investigated using X-ray photoelectron spectroscopy (XPS, ESCA2000, Thermo VG, U.K.). MagicPlot software was used to deconvolve the peaks of Ce (3d) orbital satellites. Further experiments were carried out using bovine serum albumin (BSA) as the model protein to confirm the protein immobilizing capacity of G3@nCe. For the test, different amounts of BSA were well dispersed in 1 ml of PBS along with a calculated amount of G3@nCe by sonication. After that, the mixture was placed in a 37°C water bath for 24 h. The nanoparticle-BSA dispersion was centrifuged at 15,000 rpm for 5 min, and the absorbance values of BSA in the supernatant at an absorbance maximum of 280 nm were measured using a UV–Vis spectrophotometer (Biochrom UV libra S22), and the corresponding loading quantity was determined using the BSA standard curve.
The antioxidant effects of synthesized G3@nCe were then confirmed using various assays. First, the peroxidase-mimic catalytic activity was investigated using the redox chemistry between 3,3′,5,5′-tetramethylbenzidine (TMB, ≥99%, Sigma Aldrich) and hydrogen peroxide (H2O2; 30 wt% in H2O, Sigma Aldrich). The TMB solution was prepared using acetate buffer solution (pH 4.1) and added with 1 mM H2O2. A known volume of G3@nCe at various concentrations was added to the prepared TMB solution and incubated for 30 min at RT. Following that, a broad wavelength scan of UV–vis spectroscopy (Cary Varian UV, USA) was used to record the representative absorbance peak at 652 nm. The superoxide anion (O2.) radical scavenging capacity was determined using the superoxide dismutase (SOD) assay kit (Cell Biolabs, Inc.). Following the manufacturer's protocol, a specific volume of various concentrations of G3@nCe was added to the working solution for 1 h at 37°C. The absorbance of the solution was measured at 490 nm after 1 h using a microplate reader (Varioskan LUX, Thermo Scientific). The SOD inhibition percentage was calculated according to the protocol. The ability of G3@nCe to bleach the stable 1,1-diphenyl-2-picrylhydrazyl (DPPH) radical was used to assess its DPPH radical scavenging capacity (DPPH Assay kit, Dojindo). According to the manufacturer's protocol, a definite volume of various concentrations of G3@nCe was incubated with a working solution for 30 min in the dark at 25°C. After 30 min, the absorbance of the mixture was measured using a microplate reader at 517 nm. The inhibition percentage of DPPH was calculated by following the protocol. The autocatalytic or self-regeneration property of G3@nCe was evaluated based on the shift in the UV spectrum of G3@nCe under continuous exposure to H2O2. An equimolar amount of H2O2 and G3@nCe were used for the tests. The UV absorbance curves were plotted before and after adding H2O2. After adding H2O2, G3@nCe was kept under dark conditions and the UV absorbance curves were observed on the 7th and 14th days. The spectrum shifts from lower to higher value over time confirms the autocatalytic property of G3@nCe.
2.4. Fabrication of G3@nCe/GelMA hydrogel
The G3@nCe/GelMA hydrogels were fabricated following a UV polymerization. Specifically, 10% GelMA (w/v) was uniformly mixed in DW and added with 0.5% (w/v) of photoinitiator 2-hydroxy-1-[4-(2-hydroxyethoxy)- phenyl]-2-methyl-1-propanone (Irgacure-2959, 98% Sigma Aldrich) which is dissolved in warm DW beforehand. G3@nCe was then mixed with the GelMA prepolymer solution at a final concentration of 500 μg/ml with proper sonication to prevent particle agglomeration. The mixture was then transferred to a rectangular-shaped Teflon mould and crosslinked using a UV light (Omnicure S2000, Lumen Dynamics, Canada) for 5 min. The resultant hydrogels were punched out from the mould using a biopsy punch and kept in PBS for further material characterization. Hydrogels intended for cell culture were directly formed inside cell culture plates under sterile conditions and were additionally incubated in PBS and growth media to remove the unreacted photo-initiator. For in vivo studies the hydrogel samples were formed in sterile Teflon blocks under a clean bench and punched to a suitable size using a biopsy punch. The punched hydrogel discs are then washed gently with PBS and kept in sterile conditions until implantation.
2.5. Characterization of G3@nCe/GelMA hydrogel
Firstly, we examined the microarchitecture and element composition of G3@nCe/GelMA hydrogels using a scanning electron microscope (JEOL-SEM 3000, Hitachi, Japan) at a 10 keV operating voltage and an ultradry EDS detector to observe the porous morphology and chemical composition (Thermo Fisher, USA). The hydrogels were freeze-dried and frozen-fractured in liquid nitrogen for this purpose, and the cross-sections were observed after sputter-coating with Pt (IB-3 Eiko, Japan). Next, the stress versus strain curves and respective compressive modulus values of G3@nCe/GelMA hydrogels were also measured. A single-column testing system was also used to measure the results (3344, Instron, USA). Each hydrogel sample was subjected to a uniform compression load (10 mm diameter x 4 mm height) at a constant strain of 15% and a deformation rate of 0.1 mm/min using a static load of 10 N.. To evaluate the swelling performance of hydrogel constructs, disc-shaped hydrogel constructs (10 mm × 4 mm) were prepared and directly soaked in DW, 1X PBS, and minimum essential medium (αMEM) as growth medium before being incubated at 37°C in an incubator for 24 h. Later constructs were gently blotted with KimWipe to remove residual solutions, and weight values were recorded. The swollen constructs were then lyophilized after being frozen at −80°C. The hydrogel swelling ratio was calculated using the formula:
where Wswollen and Wdry are the mass of swollen and dry hydrogels respectively. Next, the hydroxyapatite-forming ability of G3@nCe/GelMA hydrogels was tested using simulated body fluid (2x SBF) at 37°C. All the required chemicals used for preparing SBF were obtained from Sigma Aldrich and used directly without additional purification. For mimicking acellular mineralization, the fabricated hydrogel constructs (10 mm × 4 mm) were immersed at 37°C for 3 days. The apatite-like granule formation on hydrogel constructs was then observed using SEM (JEOL JSM 6510, Japan).
2.6. Adhesion, proliferation, and migration of rMSCs
For in vitro studies, rat bone marrow mesenchymal stem cells (rMSCs) were harvested and maintained according to the procedures described in a previous study [39]. Cells sustained at 4−5 passages were used for all experiments. The optimal G3@nCe/GelMA hydrogels needed for in vitro studies were first chosen by performing a cytotoxicity check using cell counting kit-8 (CCK-8, Dojindo Molecular Technologies, Inc.) by culturing 2 × 104 rMSCs on top of various hydrogels using α-MEM media (HyClone, USA) supplemented with 10% fetal bovine serum (FBS, Corning, USA) and 1% penicillin/streptomycin antibiotic (PS, Gibco, USA) for 24 h. The early responses of rMSCs with the hydrogels were assessed by analyzing the expression of focal adhesion protein vinculin. For this study, 1 × 104 rMSCs were seeded on hydrogel samples in 24-well plates and cells were fixed with 4% paraformaldehyde solution (PFA, Tech & Innovation, South Korea) after 6 h of culture, followed by permeabilization of cell membrane with 0.05% Triton-X (Sigma Aldrich) for 10 min and blocking with 1% BSA (Solmate) for 30 min at RT. Each step involved twice washing samples using phosphate buffer solution (PBS, Tech and Innovation, Korea). Finally, the cells are marked with anti-vinculin (Abcam, USA) primary antibody and stained using TRITC-conjugated secondary antibody (Santacruz, USA), followed by staining AlexaFluor-488 conjugated Phalloidin for cytoskeleton and 4,6-diamidino-2-phenylindole (DAPI, Thermofisher, USA) for nuclei. Cells were imaged using a confocal laser scanning microscope (ZEISS-LSM 700, Germany).
After confirming the focal adhesion, the proliferation of rMSC on hydrogel samples was evaluated at different time points. For this, hydrogel samples were first made inside 24 well plates and 1 × 104 rMSCs were seeded on each sample and cultured for up to 5 days. At different culturing times, the proliferation of cells was measured using CCK-8 by measuring mean optical density (OD) at 450 nm using a microplate reader (Molecular Devices). The cell morphology was then examined using a fluorescence microscope (Olympus IX71, Japan) by staining the cytoskeleton with Phalloidin and the nuclei with DAPI. To confirm the rMSC proliferation, immunofluorescence of the Ki67 proliferation marker was performed. For this brief, 3 × 104 rMSCs were cultured with hydrogels in 24-well plates for 24 h. Following washing and fixing, cells were stained with rabbit anti-human Ki67 (Abcam, USA) overnight at 4°C and then with goat anti-rabbit Alexa 488 (Abcam, USA) at RT for 2 h. The nuclei were stained with DAPI, the cell nuclei were detected using a fluorescence microscope, and the number of Ki67-positive cells was quantified using ImageJ. For confirming the directional behaviour of rMSCs under the influence of G3@nCe/GelMA, an in vitro wound healing assay was performed. Initially, a 2-well silicone insert (Ibidi) is attached inside a 24-well plate and a 70 μl suspension of 3 × 105 cells was seeded to attain a confluent layer of rMSC after 24 h. Then carefully remove the insert using forceps and washed with PBS to remove the debris. The complete media is then replaced by serum-free media and culture with hydrogels for up to 24 h. After live staining at different time points, the fluorescent images were obtained and quantified the wound closure by using ImageJ. All experiments were performed in triplicate.
2.7. Osteogenic differentiation of rMSCs
To assess the role of G3@nCe/GelMA in promoting osteogenesis, we performed osteogenic differentiation of rMSCs and their relative gene expressions were analyzed using quantitative real-time polymerase chain reaction (qRT-PCR). A suspension of 5 × 104 rMSCs was seeded on each sample and cultured with differentiation media (DM) composed of αMEM, 100 nM dexamethasone, 10 mM glycerophosphate, and 50 g/ml ascorbic acid for 3 and 7 days. After extracting the differentiated cells from hydrogels, Total mRNA was collected with a Ribospin kit (GeneAll, Korea) following the manufacturer's protocol. First, the cDNA was synthesized using AccuPower PCR premix (Bioneer, Korea) and the reverse transcription was executed with the help of a thermal cycler (HID Veriti Thermal Cycler, Applied Biosystems). For qRT-PCR, SensiMi SYBR Hi-ROX kit (Bioline) with added MgCl2 (Bioline) was used and the reaction was executed using StepOne plus software (Applied Biosystems). The change in the fold of the osteogenic genes was determined by the comparative Ct method (2−ΔΔCt) and normalized to GAPDH (housekeeping gene). The expression of bone-associated genes such as collagen type I (Col-I), Runt-related transcription factor-2 (RunX-2), alkaline phosphatase (ALP) and osteopontin (OPN) was confirmed in this way. The primers used are listed in Table S1.
Following qRT-PCR, rMSC differentiation and mineralization were confirmed using alkaline phosphatase (ALP) and alizarin red staining (ARS). For this rMSCs were seeded on hydrogels at a density of 3 × 104 cells per well and cultured using DM and staining was conducted on the 7th and 14th day. The cells were washed with PBS and fixed with 4% PFA for 30 min at RT before using an ALP staining kit (Sigma, FAST BCIP/NBT tablet) according to the manufacturer's instructions. The fixed cells were stained with ALP solution for 1 h at 37°C. The optical images were obtained after removing the excess stain. For ARS, the fixed cells were stained at RT for 2 h using 1% alizarin-red solution (Sigma Aldrich) with a pH of 4.2. The cells were then thoroughly washed with PBS and dried at RT. For quantification, the calcium deposits on hydrogels from the 14th day of osteogenic differentiation were dissolved in 10% cetylpyridinium chloride (CPC, Sigma Aldrich), and then the mean OD value of the solution was measured at 562 nm using an iMark microplate reader (Bio-Rad, USA). Next, the expression of RunX2 was analyzed by immunostaining on the 7th day of osteogenic differentiation. For this, cells were first fixed with 4% PFA and marked with RunX2 antibody (Santacruz, USA) at 4°C overnight and then further stained with TRITC-conjugated secondary antibody (Santacruz, USA) at RT for 2 h. Actin was stained using Phalloidin and nuclei using DAPI and the relative expression of RunX2 was quantified using ImageJ.
2.8. Oxidative stress assay and intracellular ROS production
For confirming the ROS scavenging effects of hydrogels under high ROS conditions, first, we performed an oxidative stress assay using H2O2. For this brief, 3 × 104 rMSCs were seeded and cultured with hydrogels. Then, H2O2 of various concentrations (0.1 mM, 0.5 mM and 1 mM) were introduced and cultured for 24 h to challenge the rMSCs under a pathologic oxidative stress microenvironment. Then CCK-8 assay was performed to quantify the survival of rMSCs. Following this, the live and dead cells were marked using a calcein AM and ethidium homodimer-1-based kit (Thermo Fisher Scientific, USA). Further, the intracellular ROS levels in rMSC under oxidative stress conditions were then analyzed using the Image-iT live green ROS detection kit (Invitrogen, USA). For this, briefly, 3 × 104 cells per well were first seeded and cultured with hydrogels for 24 h. After aspirating media and hydrogels, high oxidative stress conditions were enabled by treating 0.1 mM H2O2 for 3 h. After carefully removing H2O2, cells were gently washed with warm Hanks Balanced Salt Solution (HBSS with Ca/Mg, Welgene) and labelled with enough 25 μM carboxy-2′,7′-dichlorodihydrofluorescein diacetate (Carboxy-H2DCFDA) working solution at 37°C in dark for 30 min. The mean fluorescence values were then calculated using a plate reader (Ex. 495 nm; Em. 529 nm). For imaging, the fluorescently stained samples were mounted in warm HBSS and imaged immediately. For performing and confirming osteogenic differentiation of rMSC on G3@nCe/GelMA hydrogels under ROS conditions, the same steps were followed as mentioned in the previous section with an additional induction of oxidative stress conditions using 0.05 mM H2O2 during differentiation.
2.9. QuantSeq 3′ mRNA-sequencing and data analysis
For QuantSeq 3′ mRNA sequencing the 5 × 104 rMSCs were cultured on hydrogels for 24 h afterwards the total RNA was collected using Ribospin-II (Geneall, Korea) based on manufacturers' instructions. The Agilent TapeStation 4000 system (Agilent Technologies) was used to evaluate the quality of isolated RNA, and the ND-2000 Spectrophotometer (Thermo Inc., USA) was used to quantify the RNA amount. The QuantSeq 3′mRNA-Seq Library Prep Kit (Lexogen, Inc.) was used to build libraries for samples. After total RNA preparation, reverse transcription was carried out using an oligo-dT primer that had an Illumina-compatible sequence at its 5′ end. A random primer with an Illumina-compatible linker sequence at its 5′end launched second strand synthesis after the RNA template had degraded. By using magnetic beads, the double-stranded library was made completely free of reaction by-products. The entire adaptor sequences necessary for cluster creation were added to the library through amplification. Using NextSeq 550 (Illumina, Inc., USA), single-end 75 sequencing for high-throughput sequencing was carried out and QuantSeq 3′mRNA-Seq reads were aligned using Bowtie2 analyzing tool [40]. For aligning to the genome and transcriptome, Bowtie2 indices were either produced from the representative transcript sequences or the genome assembly sequence. The transcripts were collated, abundances calculated, and differential gene expression was discovered using the alignment file. Using coverage in Bedtools counts from single and multiple alignments were used to discover which genes were differentially expressed [41]. The TMM + CPM normalization method was used with EdgeR within R (R Development Core Team, 2020) using Bioconductor to process the Read Count data [42]. Gene classification was based on searches in the Medline and Database for Annotation, Visualization, and Integrated Discovery (DAVID) databases respectively [43]. The Excel-based differentially expressed gene analysis tool (ExDEGA, ExDEGA Graphic Plus, Ebiogen Inc., Korea) was used for data analysis and visualization. The transcription factor enrichment analysis (TFEA) was done by orthogonal omics integration using ChIP-X Enrichment Analysis Version 3 (ChEA3) [44] based on the submitted gene sets and the top 15 enriched transcription factors (TF) were considered.
2.10. Biocompatibility of G3@nCe/GelMA hydrogel
The in vivo biocompatibility of G3@nCe/GelMA hydrogel was evaluated by performing blood and tissue compatibility studies using SD rats. The Animal Care and Use Committee at Dankook University, Republic of Korea approved all animal experiments (Approval no: DKU-18-032). First, the blood compatibility of hydrogels was analyzed using an in vitro haemolysis assay. For this, blood was collected from male Sprague Dawley (SD) rats (5 weeks old) in a sampling tube without coagulation. Then red blood cells (RBC) were obtained by centrifuging whole blood at 3000 rpm for 5 min and repeatedly washing with PBS 3 times. The pure RBCs obtained were diluted to 5% (v/v). Then 500 μl of hydrogel and 500 μl of 5% RBCs were added into a 2 ml mini centrifuge tube and mixed uniformly and kept at 37°C for 1 h. Then the samples are centrifuged at 3000 rpm for 10 min. A microplate reader was used to measure the absorbance of the clear supernatant at 540 nm. The negative control is PBS, and the positive control is 0.1% Triton-X. All tests were performed three times, and the percentage of hemolysis was calculated using the formula:
where AS, AP, AT are the absorbance values of the sample, PBS, and Triton-X respectively.
In vivo, tissue compatibility and immune responses were then assessed using subcutaneous implantation of hydrogels in male SD rats (5 weeks old). All rats were first sedated by an intramuscular injection of Xylazine (10 mg/kg body weight) and Ketamine HCl (80 mg/kg body weight). Hydrogel constructs of a specific size (10 mm × 3 mm) were made and sterilized using UV irradiation before use. The dorsal skin was clean-shaven and sterilized with ethanol/povidone-iodine rub and an incision (2 cm) was made to form subcutaneous pouches. The experimental groups were randomly allocated, and the hydrogel constructs were implanted in each rat (n = 5 per group). Following implantation, the incision was sutured using a polypropylene suture (Prolene, B. Braun, Germany). For examining the impact of implantation in host tissues, rats were sacrificed following 2 and 4 weeks of implantation and the hydrogel-integrated tissue samples were retrieved. This time point was chosen for the study because shorter periods of implantation might not be sufficient to observe the complete range of tissue reactions and will not provide information about the long-term biocompatibility of the material. Tissue samples were fixed in neutral buffered formalin (10% NBF) before being dehydrated in a series of ethanol solutions. After embedding the samples in paraffin, thin slices (5 μm) were cut with a microtome (Leica, USA) and stained with hematoxylin and eosin (H&E) to confirm tissue responses. The involvement of monocytes/macrophages and host response to implanted hydrogel constructs at implantation sites was then assessed through immunohistochemistry (IHC) of CD68 and CD3 (Santacruz, USA). All samples were imaged with a confocal microscope (CLSM; Zeiss LSM 700, Germany) and were quantified using ImageJ.
2.11. Implantation of G3@nCe/GelMA hydrogels in critical-sized bone defects
12 weeks aged healthy male SD rats (SJ Bio, Korea) were used for inducing critical-sized calvaria bone defects in vivo. The Animal Care and Use Committee at Dankook University, Republic of Korea approved all animal experiments (Approval no: DKU-18-032). Rats were sustained in an optimum environment and maintained under sedation throughout surgery by an intramuscular administration of ketamine and xylazine. The hydrogel constructs that exactly fit the defects was prepared and sterilized by UV before use. The dorsal skin above the cranium was clean-shaven and sterilized with ethanol/povidone-iodine rub. Then, using a surgical blade, a linear sagittal midline skin cut was made over the skull, and the incised skin flap was opened to properly locate the defect site. Then, under sterile saline solution flow, 5 mm diameter critically sized circular full thickness calvarial bone defects were created at the centre of each parietal bone by drilling. The experimental groups were randomly allocated, and 2 hydrogel constructs were implanted per animal (n = 5 per group). Pristine GelMA hydrogel was used as the control group. Following implantation, the defects were first sewed with an absorbable suture (4-0 Vicryl, Germany) inside and then with a non-absorbable suture (Dafilon, B. Braun, Germany) outside. The animals were then sustained in separate cages in an optimum environment and observed to see any signs of inflammation or infection. The animals were sacrificed 12 weeks after the operation to collect bone samples from the defect site and surrounding bone.
After being fixed in 10% NBF for 24 h at RT, the specimens were prepared for micro-computed tomography (μCT), histology, and immunohistochemistry. The first μCT scan was done to check for neo-bone formation. A μCT scanning machine was used to scan all the specimens (Skyscan, Belgium). The reconstructed images were formed and were used to examine the neo-bone formation over the region of interest such as volume over total volume (BV/TV) and bone surface density (BSD) using CTAn Skyscan software. 3D images were produced and visualized using software (CTvol Skyscan software). The harvested samples were prepared for histology after μCT analysis. NBF-fixed bone samples were decalcified (RapidCal, BBC Chemical Co, USA), dehydrated with a series of ethanol solutions, and then embedded in paraffin for slicing. For histology, 5 mm thick tissue sections were prepared at the central region of the circular defects using a microtome (Leica, Germany), and the tissue slices were stained with H&E staining for new bone formation and then Masson-Trichome staining (MT) to check collagen deposition. For IHC, tissue samples were stained with primary antibodies related to bone formation in vivo (Col-1, OCN, OPN, and CD31) overnight at 4°C, followed by 2 h of treatment with Alexa Flour 594-conjugated secondary antibodies at RT. The nuclei were then counterstained with DAPI (Invitrogen) and images were captured using a confocal laser scanning microscope. ImageJ was used to quantify relative expressions to compare groups.
2.12. Statistical analysis
GraphPad Prism software was used to generate all statistical analyses (version 8.4.3). All data are presented as mean standard deviation (SD). ANOVA or the Student's t-test were used to assess significant differences between groups, and any ∗p value less than 0.05 was considered statistically significant.
3. Results
3.1. Physicochemical characterization of G3@nCe
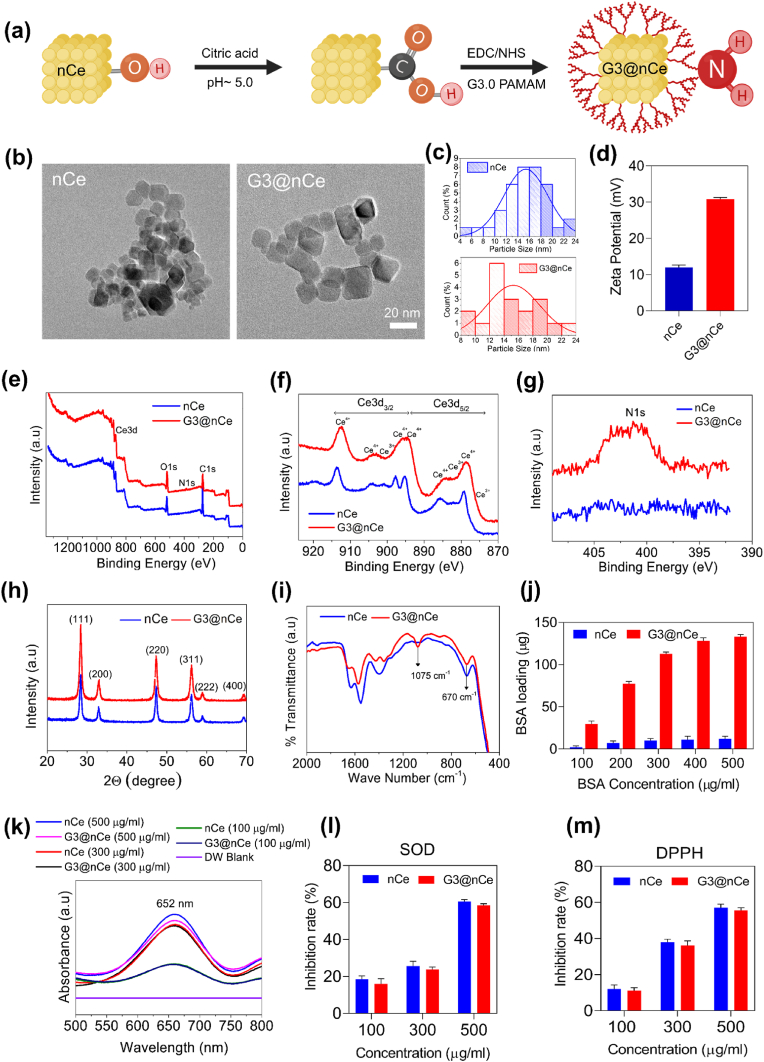
G3@nCe was synthesized using nCe and PAMAM dendrimer following a reaction involving carboxylation and EDC-NHS activation as indicated in (Fig. 1a). For this nCe was first synthesized by a hydrothermal reaction following the previous studies. The TEM images of nCe before and after functionalization displayed a cube-like morphology (Fig. 1b). The particle size distribution was also calculated based on the TEM images (Fig. 1c). This variation in particle size distribution is owing to the surface functionalization of nCe with G3. It was observed that after the surface functionalization of nCe, its zeta potential increased significantly from +12.01 mV to +30.8 mV (Fig. 1d). Next, we performed the XPS analysis of G3@nCe to validate the elemental composition and chemical bonding. Fig. 1e shows the full-width XPS curves which provide detailed information on the chemical composition of G3@nCe. Out of the major peaks we focused on the Ce3d and N1s peaks as it is more relevant to the proposed nanomaterial. Fig. 1f verified the distribution of Ce3+ and Ce4+ valence states in G3@nCe and the percentage of Ce3+ and Ce4+ in G3@nCe was found to be 16.42% and 83.57% respectively (Fig. S1). Fig. 1g shows the characteristic N1s peak in G3@nCe owing to the presence of nitrogen-containing NH2 groups from G3, which is absent in nCe. The XRD peaks of both nCe and G3@nCe showed similar characteristic peaks indicating a cubic fluorite phase (JCPDS card no: 81–0792) (Fig. 1h). XRD results also confirm that the surface functionalization doesn't affect the crystal structure of nCe. The FTIR spectra showed a characteristic C–N stretching peak in G3@nCe at 1075 cm−1. Similar peaks were absent in nCe, indicating the existence of plentiful amine groups in G3@nCe. The characteristic peak corresponding to Ce–O was observed at 670 cm−1 (Fig. 1i).
Fig. 1.
Synthesis and characterization of G3@nCe (a) Schematic illustration of the steps involved in the synthesis of G3@nCe. (b) TEM images showing the cubical particle morphology. Scale bar: 20 nm. (c) Size distribution curves obtained from representative TEM images (d) Zeta potential measurement. (e) Full-width XPS analysis showing characteristic peaks in G3@nCe. (f) XPS analysis at Ce3d region. (g) XPS analysis at N1s region. (h) Characterization by XRD. (i) FTIR spectra of G3@nCe showing characteristic peaks. (j) The protein loading capacity of G3@nCe evaluated using BSA as a model protein. (k) Oxidase-like activity of G3@nCe evaluated using TMB assay at 652 nm. (l) Superoxide radical scavenging assay. (m) DPPH radical scavenging assay. Data reported as mean ± SD (n = 3).
The delocalization or loading of biomolecules on implantable biomaterials is crucial for improving their bioactivity [45,46]. Being abundant in plasma, we used albumin as the model protein for our protein loading studies. Fig. 1j shows the amount of BSA loaded by G3@nCe and nCe. The G3@nCe showed a significantly higher amount of protein loading at all concentrations compared to nCe. Being highly cationic G3@nCe is expected to retain the attached proteins while the non-covalently attached proteins may readily desorb from the surface in the case of nCe [34]. The presence of detrimental ROS in the tissue microenvironment leads to the impairment of biological functions and holds a negative role in bone tissue remodelling. Designing biomatrices with superior ROS scavenging properties is hence considered to be a good approach to solving such problems [47]. So next we evaluated the multi-enzymatic properties of nanoparticles using the oxidase-like activity assay, SOD radical scavenging assay, DPPH radical scavenging assay (Fig. 1k-m), and self-regeneration assay (Fig. S2). In all assays, G3@nCe also exhibited radical scavenging properties like nCe confirming that surface functionalization does not interfere with the innate ROS scavenging properties of nCe.
3.2. Physicochemical characterization of G3@nCe/GelMA hydrogel
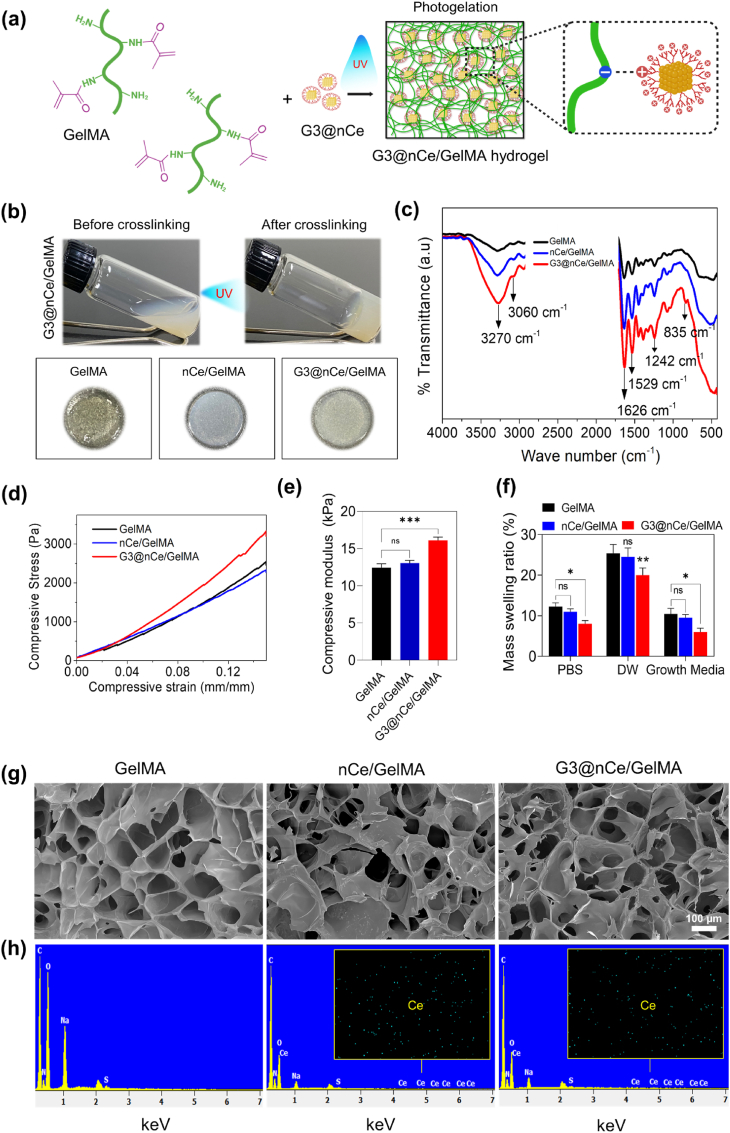
GelMA was prepared by introducing the methacrylate groups on the gelatin chain following a chemical reaction between amine functional groups in gelatin with MA [9,12]. The successful functionalization was further confirmed by performing FTIR (Fig. S3b) and the DOM is calculated to be 84% by ninhydrin assay (Fig. S3c). Next, we prepared G3@nCe/GelMA hydrogels with a combination of 10% (w/v) GelMA polymer and 500 μg/ml of G3@nCe (Fig. 2a). This concentration of nanoparticles was chosen for the study as it is the optimal amount at which cells exhibited no observable toxicity compared to pristine GelMA (Fig. S4). Fig. 2b shows the images of G3@nCe/GelMA before and after UV cross-linking and the images of different hydrogel groups after photogelation. Following fabrication, the hydrogel samples were characterized by FTIR (Fig. 2c). The stretching vibrations of hydroxy groups in GelMA were observed at 3200-3600 cm−1 and the stretching bonds of C O at 1626 cm−1. N–H deformation bonds appeared at 1242 cm−1 and 1529 cm−1. In the case of G3@nCe/GelMA, an additional peak was observed at 835 cm−1. Since most of the characteristic peaks from G3@nCe overlapped with GelMA peaks, it was not easily distinguishable. Next, we experimented with the mechanical properties of the G3@nCe/GelMA hydrogel by evaluating the stress versus strain curves and the compressive modulus.
Fig. 2.
Physicochemical characterization of G3@nCe/GelMA hydrogel. (a) Graphical illustration showing the photo-encapsulation of G3@nCe into GelMA hydrogel and existing supramolecular interactions between GelMA and G3@nCe. (b) Images showing G3@nCe/GelMA before and after cross-linking and images of different hydrogel groups after photogelation. (c) FTIR spectra of hydrogels showing characteristic peaks. (d) Stress versus strain curves of hydrogels under compression. (e) Compressive modulus measured for various hydrogels. (f) Swelling behaviour of hydrogels in PBS, DW and growth media. (g) SEM images showing honeycomb-like morphology by the cross-section of freeze-dried hydrogels. Scale bar: 100 μm. (h) Elemental mapping of freeze-dried hydrogel performed using EDS. The presence of ceria detected in both G3@nCe/GelMA and nCe/GelMA are also shown in the EDS graph. Data reported as mean ± SD (n = 3; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
It is recognized that stem cells tend to differentiate towards osteogenic lineage under the influence of a stiff microenvironment [48,49]. Pristine GelMA is known to have inferior mechanical properties when compared to its composites; therefore improving the mechanical performances of GelMA by incorporating various nanomaterials provides it with remarkable properties for bone tissue regeneration [12]. Fig. 2d displays stress versus strain curves of various hydrogels under compression mode and the compressive modulus of GelMA, nCe/GelMA, and G3@nCe/GelMA was observed to be 12.44 kPa, 13.04 kPa and 16.10 kPa respectively (Fig. 2e). Remarkably, the addition of nCe didn't much improve the mechanical characteristics of GelMA, while the addition of G3@nCe resulted in an improved compressive modulus compared to pristine GelMA. It is expected that the supramolecular interactions that arise amongst the carboxyl groups in GelMA and abundant amine groups in G3@nCe are responsible for the better integration and mechanical stability of hydrogels [[50], [51], [52], [53]]. A. El-Fiqi et al. and X. Ding et al. also observed similar interactions after incorporating surface-aminated nanomaterials in biopolymers [54,55]. Following this, we evaluated the swelling kinetics of hydrogels using 3 different representative solutions (DW, PBS and Growth Media). An ideal tissue engineering scaffold should maintain a suitable amount of water to bear a resemblance to the native microenvironment, which benefits normal cell functions and tissue metabolism after implantation of the hydrogel [56]. There were no significant differences in the swelling kinetics between the GelMA and nCe/GelMA, but there was a decrease in the swelling behaviour of G3@nCe/GelMA hydrogels in all three solutions analyzed (Fig. 2f). For implantable hydrogels, this type of controlled swelling is particularly desired because it prevents the hydrogel from expanding from the boundaries of the trauma and separating from the implantation site in clinical applications [51].
Next, we analyzed the interior morphology of freeze-dried G3@nCe/GelMA hydrogels using SEM and elemental composition by EDS. The porous nature of hydrogels is also known to influence cell fate. Hydrogels exhibiting a porous microstructure facilitate efficient nutrient and fluid exchange that encourages stem cell adhesion and proliferation [49]. The uniformly distributed microsized pores in G3@nCe/GelMA suggest the creation of a porous, interconnected network without any observable aggregation of nanoparticles (Fig. 2g). However, the pore size distribution among the hydrogel groups showed no discernible changes. The EDS analysis of the freeze-dried G3@nCe/GelMA hydrogels confirmed the successful loading of G3@nCe in the GelMA matrix (Fig. 2h). Together with the morphological analysis and hygroscopic behaviour of hydrogels, it is expected that GelMA engineered using G3@nCe could result in bioactive hydrogels that serve as a promising biomaterial for engineering bones [57,58]. Biomatrices that support bone regeneration must essentially mimic the composition of bones for better regenerative efficiency [59]. The hydroxyapatite formation ability of the hydrogels was then evaluated by incubating hydrogels in SBF for 3 days. SEM images revealed the formation of spherical nodules of apatite deposited on hydrogel surfaces (Fig. S5). G3@n Ce/GelMA hydrogels showed relatively higher deposition of hydroxyapatite among other groups due to the presence of abundant –NH2 groups that exert strong electrostatic interactions with phosphate and calcium ions in SBF and could behave as nucleation regions for the mineralization of hydroxyapatite nodules [60,61].
3.3. G3@nCe/GelMA facilitates rMSC adhesion, proliferation, and migration
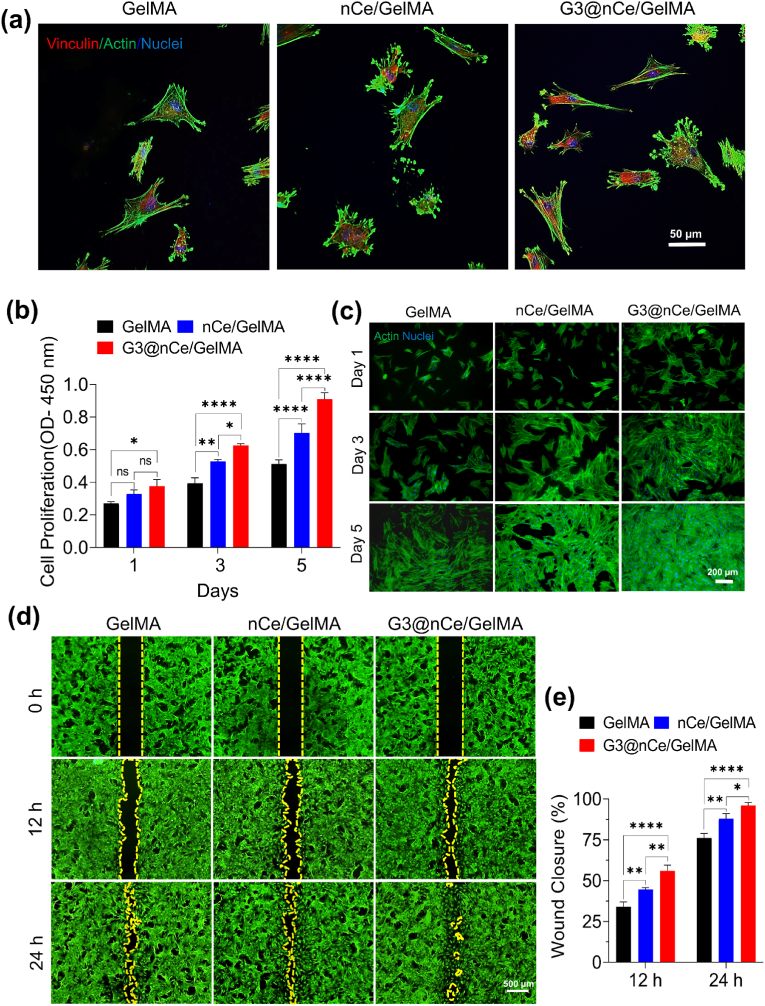
Cell adhesion is the initial step in cell-hydrogel interaction, where the cells attach to the substrate through specific binding interactions between cell-surface receptors and ligands present on the hydrogel surface [62]. The capability to enhance early cell attachment and their consequent growth is considered an important prerequisite while engineering various biomaterials for bone tissue regeneration [63]. So, we analyzed the initial response of cells on hydrogels and their focal adhesion was confirmed using immunofluorescence of vinculin protein at 6 h (Fig. 3a). Hydrogels integrated with amine-terminated nanomaterials are known to influence initial cell adhesion and proliferation by interacting with the negatively charged cell membrane. All such cationic surfaces also functioned as protein immobilization anchor points, which is also advantageous for cell migration [64]. After the rMSC has firmly attached to hydrogel, the proliferation rate was examined by CCK-8 assay for up to 5 days (Fig. 3b). Subsequently, the morphology of proliferating cells was observed by fluorescence staining. G3@nCe/GelMA showed a significantly higher number of cells almost confluent over the hydrogels on the 5th day indicating their cell proliferative effects (Fig. 3c). This was further confirmed using the immunostaining of the Ki67 proliferation marker. The results indicated that more Ki67-positive nuclei were observed in the case of G3@nCe/GelMA followed by nCe/GelMA and GelMA (Fig. S6). It is anticipated that G3@nCe/GelMA hydrogels deliver the nutrients and growth factors required for cells by sequestration and sustained release, which stimulates the ECM remodelling process, leading to the formation of a more favourable microenvironment for cell proliferation and migration [65].
Fig. 3.
Adhesion, proliferation, and migration of rMSCs promoted by G3@nCe/GelMA hydrogel. (a) Immunofluorescence images of vinculin expression in rMSCs at 6 h. Scale bar: 50 μm. (b) The proliferation of rMSCs evaluated up to 5 days of culture using CCK-8 assay. (c) Indicative fluorescent images of actin and nuclei of rMSCs after culturing up to 5 days. Scale bar: 200 μm. (d) Representative images from the migration assay using rMSCs up to 24 h. Scale bar: 500 μm. (e) The quantitative analysis of wound closure rate using ImageJ. All data expressed as mean ± SD (n = 3; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001).
The MSC migration in the initial stage of bone formation is very crucial because MSCs must first migrate to the bone surface before they can take part in bone formation. Hence cell migration is another key event involved in bone formation and bone disease treatment. So, we experimented with the directional behaviour or migration ability of rMSCs under the influence of hydrogels using a wound healing assay (Fig. 3d). All hydrogel groups exhibited cell migration over time. G3@nCe/GelMA hydrogel enhanced rMSC migration profoundly compared to pristine GelMA and nCe/GelMA at both 12 h and 24 h. We expect that in addition to the effects of various signalling molecules and ECM components, the physicochemical cues provided might have also triggered the stimulation of cell migration by G3@nCe/GelMA. It was also observed that the migration of cells between nCe/GelMA and pristine GelMA were also significantly different (Fig. 3e). We believe that because both nCe and G3@nCe are ROS-responsive, alternate ROS-related signalling pathways also played a role in the case of nCe/GelMA at 24 h [66]. The differences in hydrogel composition, stiffness, and the presence of signalling molecules can also play a role in activating alternative signalling pathways suggesting that the signalling pathways regulating various cellular processes may be differentially activated [67]. Overall, our findings suggest that the G3@nCe/GelMA hydrogels may provide an adequate microenvironment for cell adhesion, proliferation, and migration, which is beneficial for bone therapeutics.
3.4. G3@nCe/GelMA promotes osteogenic differentiation of rMSCs
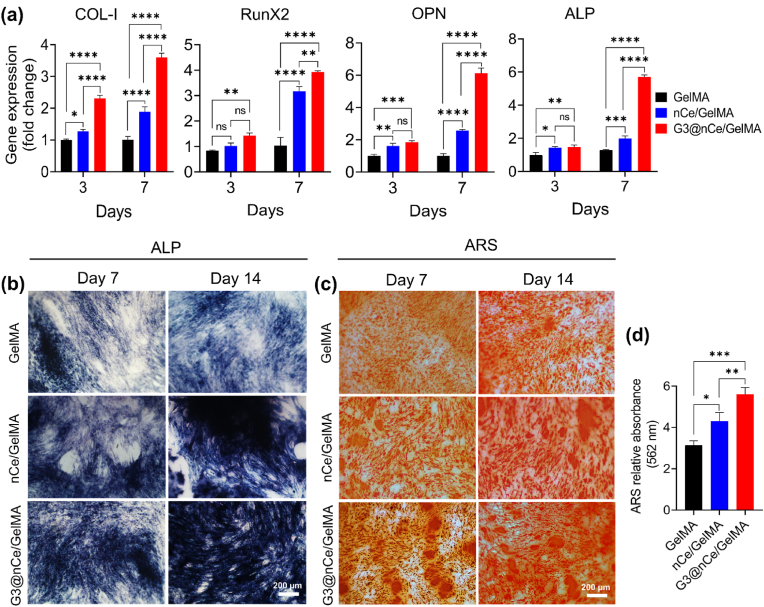
It is a fact that the intrinsic osteogenic function of the majority of hydrogels employed in tissue regeneration is insufficient, resulting in a limited therapeutic effect in clinical settings [68,69]. The successful implantation of hydrogels relies on their interaction with bone tissues. Understanding the molecular and cellular level events in the immediate microenvironment of bone progenitor cells is therefore essential. Hence to confirm the favourable osteogenic activity exhibited by G3@nCe/GelMA hydrogel on rMSCs, we performed osteogenic differentiation by supplementing DM to cells cultured on hydrogels. To confirm the differentiation of rMSCs to osteogenic lineage, the first PCR was performed on days 3 and 7 to estimate the relative expression of osteogenic genes. The analysis of bone-related genes such as Runx2, Col-I, ALP and OPN showed higher expression in G3@nCe/GelMA compared to pristine GelMA and nCe/GelMA. The Runx2 and Col-I being early markers of osteogenic differentiation, upregulated during the early timepoint of differentiation, while ALP and OPN expressed more during the late phase. (Fig. 4a). ALP and ARS staining, which are key osteogenic markers for osteogenesis of rMSCs were then carried out at 7 and 14 days of osteogenic differentiation to further ascertain the potential of G3@nCe/GelMA in promoting bone formation. G3@nCe/GelMA group possessed the highest ALP activity compared to nCe/GelMA and pristine GelMA at both 7 and 14 days of osteogenic differentiation (Fig. 4b). The same trend was observed in the case of ARS staining also. ARS staining showed mineralized nodules in all groups whereas denser bright red mineralized nodules were found in G3@nCe/GelMA (Fig. 4c). The relative quantification of the mineralized nodules on the 14th day of differentiation using CPC further confirmed that G3@nCe/GelMA possessed a higher rate of bio-mineralization suggesting an enhanced osteogenic potential of G3@nCe/GelMA (Fig. 4d).
Fig. 4.
Osteogenic differentiation of rMSCs promoted by G3@nCe/GelMA hydrogel (a) Relative gene expression by qRT-PCR for confirming osteogenic differentiation of rMSCs cultured on G3@nCe/GelMA hydrogels. (b) ALP activity of rMSCs at 7th and 14th day of differentiation. Scale bar: 200 μm and (c) Mineralization of rMSCs during differentiation observed by ARS. Scale bar: 200 μm. (d) Colorimetric estimation of mineralization at the 14th day of differentiation using CPC (562 nm). All data reported as mean ± SD (n = 3; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
To further confirm our observation, the expression of RunX2, a crucial transcription factor that is only expressed in mineralized tissues was done using immunofluorescence. As expected, the relative expression of RunX2 was observed to be higher in G3@nCe/GelMA compared to nCe/GelMA and pristine GelMA groups (Fig. S7). These findings indicate that G3@nCe/GelMA hydrogel matrix stimulated rMSCs by promoting the secretion of growth factors and other signalling molecules to differentiate towards the osteogenic lineage that involves a complex interplay of microenvironmental cues and signalling pathways. Although pristine GelMA already has some advantages for providing a suitable microenvironment for preosteoblast cells, the osteogenic microenvironment provided by G3@nCe/GelMA must be more effective and suitable for successful applications in bone therapeutics as they mimic the micro/nano composition of native bone as well as supports the dynamic mineralization process of the bone matrix.
3.5. G3@nCe/GelMA rescues rMSCs from H2O2-induced oxidative stress
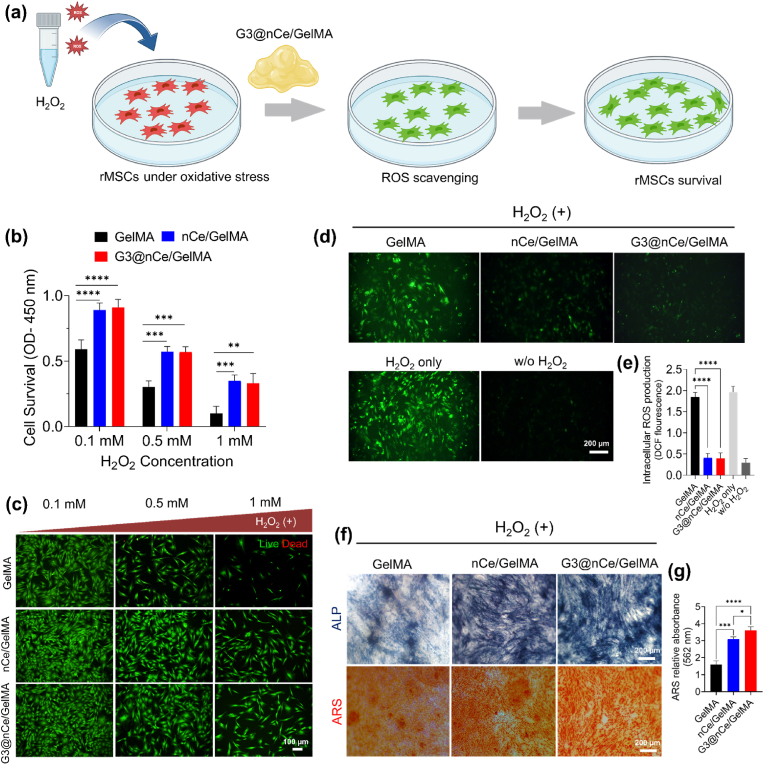
As there is an enhanced ROS level in the microenvironment of critical-sized bone defects, an efficient strategy for bone regeneration must comprise a fast and responsive elimination of extreme levels of ROS [70]. Inorganic nanomaterials like nCe are well known to be ROS-scavenging materials with supreme surface catalytic activity. The self-regenerative oxidation states of nCe allow them to behave like biological enzymes, making them one of the versatile ROS-responsive nanomaterials used in tissue engineering [22,23]. Fig. 5a illustrates a graphical outline of the experiment depicting the antioxidant nature of G3@nCe/GelMA under culture conditions. The in vitro ROS scavenging effects of G3@nCe/GelMA hydrogel was demonstrated using a cellular oxidative stress method by treating rMSCs at various concentration of H2O2 (0.1 mM, 0.5 mM, and 1 mM). G3@nCe/GelMA was then introduced along with several concentrations of H2O2, and the survival of cells was quantified by CCK-8 assay (Fig. 5b) and imaged by Live/Dead staining (Fig. 5c). When initially cells were treated with 0.1 mM H2O2, the number of live cells was significantly maintained in the case of G3@nCe/GelMA and nCe/GelMA while suppressed in the case of pristine GelMA. As the concentration was increased to 0.5 mM, cell viability was more repressed and finally at 1 mM, most of the cells were dead in pristine GelMA, while G3@nCe/GelMA and nCe/GelMA, still maintained a significant number of viable cells. Consequently, the intracellular ROS levels in rMSCs after treatment with hydrogels were detected by fluorescent signals and quantified by fluorescence measurement. The rMSCs cultured with G3@nCe/GelMA and nCe/GelMA significantly reduced the ROS levels by scavenging detrimental ROS induced by H2O2, hence exhibiting weak green fluorescence, while pristine GelMA lacking antioxidant activity showed strong green fluorescence signals indicating higher intracellular ROS levels (Fig. 5d). The intracellular ROS production in rMSCs quantified using DCF fluorescence also confirms the same (Fig. 5e). These findings also rule out the possibility that intracellular ROS levels in cells cultured with G3@nCe/GelMA and nCe/GelMA were comparable due to similar antioxidant properties.
Fig. 5.
Survivability, intracellular ROS production, and osteogenic differentiation of rMSCs under H2O2-induced oxidative stress conditions. (a) A graphical illustration depicting the antioxidant nature G3@nCe/GelMA under culture conditions. (b) Cell survivability under the influence of hydrogels measured using CCK-8 assay after exposing rMSCs to various concentrations of H2O2. (c) Representative live/dead images of rMSCs after being exposed to various concentrations of H2O2. Scale bar: 100 μm. (d) Intracellular ROS production observed using fluorescence staining (e) Intracellular ROS production in rMSCs quantified using DCF fluorescence. Scale bar: 200 μm. (f) The influence of G3@nCe/GelMA in the osteogenic differentiation of rMSCs under oxidative stress was confirmed using ALP and ARS. Scale bar: 200 μm. (g) Colourimetric estimation of mineralization using CPC (562 nm). Data reported as mean ± SD (n = 3; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
It is also well-known that antioxidant biomaterials are also known for inducing the differentiation of osteoblast progenitor cells by modulating oxidative stress [47]. So further we evaluated the effect of G3@nCe/GelMA on the differentiation of rMSCs to osteogenic lineage by evaluating the ALP and ARS expression under the oxidative stress microenvironment. As shown in (Fig. 5f), G3@nCe/GelMA and nCe/GelMA maintained the osteogenic differentiation capacity of rMSCs under H2O2-induced oxidative stress conditions up to 14 days of differentiation, while pristine GelMA showed reduced differentiation capacity as evident from fewer ALP positive area and reduced calcium deposition. Fig. 5g indicates the colourimetric estimation of mineralization using CPC at 562 nm. Therefore, it is indicated that H2O2 reduced the differentiation capacity of rMSCs on GelMA while G3@nCe/GelMA and nCe/GelMA hydrogels improved the process of osteogenic differentiation by scavenging and neutralizing ROS which is beneficial for bone regeneration. Overall, the findings suggest that G3@nCe/GelMA protects preosteoblast cells from oxidative stress injury and is beneficial for bone formation under harsh conditions.
3.6. QuantSeq 3′ mRNA-sequencing to identify functions and signalling pathways activated by G3@nCe/GelMA
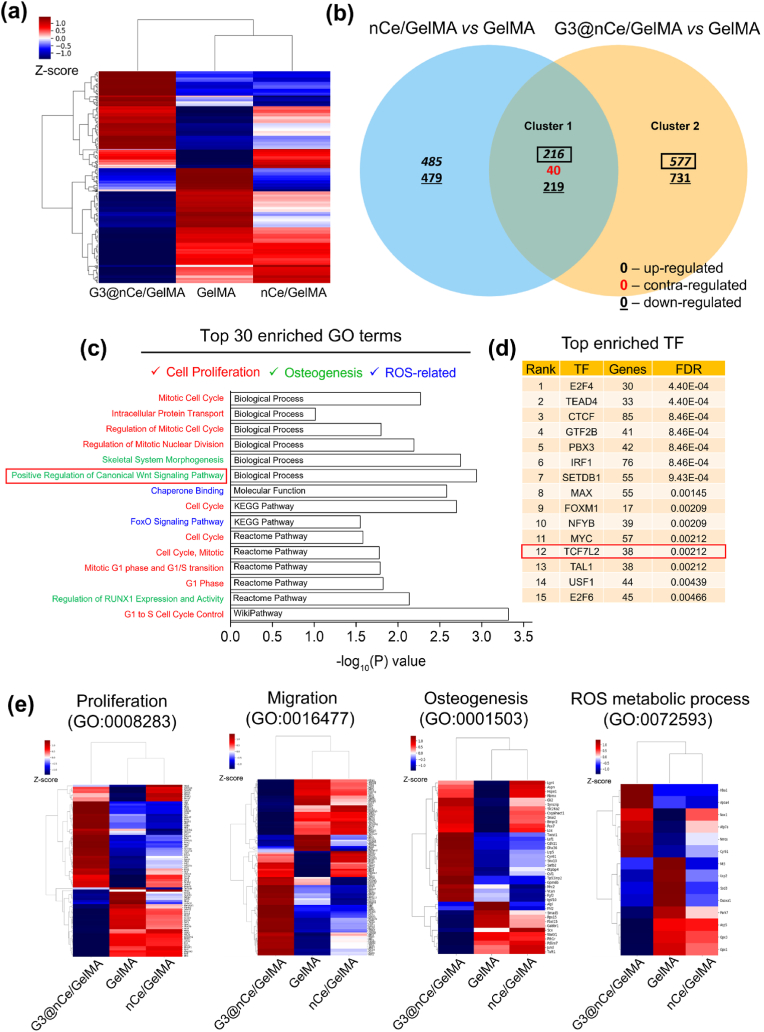
Transcriptome analysis (a total of 17,048 genes) was carried out to identify the important signalling pathways and biological or molecular processes through which G3@nCe/GelMA hydrogel controls cellular functions and promotes the formation of new bone. Fig. 6a indicates the distance-based clustering analysis from global transcriptional changes in GelMA, nCe/GelMA and G3@nCe/GelMA groups from the combinative comparison. The differentially expressed genes (DEGs) of rMSCs on different hydrogel groups were broadly detected between groups (577 upregulated genes in G3@nCe/GelMA vs GelMA, 485 upregulated genes in nCe/GelMA vs GelMA and 216 co-upregulated genes in G3@nCe/GelMA vs GelMA and nCe/GelMA vs GelMA), as shown in the Venn diagram (Fig. 6b). The cluster 1 and 2 gene sets were used for Gene Ontology (GO) and pathway enrichment analysis by DAVID. As an outcome, the top 30 enriched GO terms, including biological process (BP), molecular functions (MF), KEGG pathway, Reactome pathway, and Wikipathway, were displayed (Fig. S8), wherein DEGs of cluster 1, co-expressed in G3@nCe/GelMA and nCe/GelMA owing to the presence of ceria, were revealed to be primarily rich in cell proliferation (mitotic cell cycle, intracellular protein transport, regulation of mitotic cell cycle, regulation of mitotic nuclear division), osteogenesis (skeletal system morphogenesis, positive regulation of canonical Wnt signalling pathway), and ROS-metabolic process (chaperone binding) terms (Fig. 6c). Functional annotation clustering for cluster 1 using DAVID analysis were confirmed based on above key biological terms (Fig. S9). Canonical Wnt signalling is an important regulatory pathway in the bone formation process, regulating a variety of biological processes related to stem cell function such as proliferation, migration, differentiation, and so on [71,72]. The Wnt signalling-related genes like Aspm, Csnk1g1 were significantly upregulated in G3@nCe/GelMA. Additionally, the transcription factor enrichment analysis (TFEA) of cluster 1 DEGs using Chip-X enrichment analysis tool (ENCODE ChipSeq library) also disclosed possible involvement of Transcription factor 7-like 2 (TCF7L2), a key effector to promote osteogenesis through canonical Wnt-signalling pathway [73,74] (Fig. 6d). In-depth mechanistic studies accounting for the TCF7L2 related Wnt-signalling dependent bone regenerative potential of G3@nCe/GelMA would be thoroughly explored in future.
Fig. 6.
Transcriptome analysis reveals the key functions and signalling pathways activated by G3@nCe/GelMA. (a) The distance-based clustering analysis from global transcriptional changes in GelMA, nCe/GelMA and G3@nCe/GelMA groups with 1.5 fold change and over 2 log2 values from the combinative comparison. (b) Venn diagram showing the comparison between differential gene expression across 2 key comparisons (nCe/GelMA vs GelMA and G3@nCe/GelMA vs GelMA) based on co-up or –down and contra-regulated genes. (c) Among the top 30 enriched GO or pathway using up-regulated genes from cluster 1 (mainly due to the presence of nanoceria) using DAVID (BP, MF, KEGG, Reactome and Wikipathway), major terms related to cell proliferation, osteogenesis and ROS-related processes were disclosed. (d) Transcription factor enrichment (TFE) of cluster 1 was analyzed using ChEA3, a web-based tool indicating the enrichment of TCF7L2, a key transcription factor involved in Wnt signalling pathway. (e) Heat map showing DEGs in G3@nCe/GelMA, nCe/GelMA, and pristine GelMA from specific GO terms related to cell proliferation, migration, osteogenesis, and ROS-metabolic process.
The individual heat maps from specific GO terms also displayed differentially expressed genes in all groups; especially those key genes involved in cell proliferation (Hgf, Csf1, Cited2, Fzd6, Atf2 etc.), osteogenesis (Gli2, Dhx36, Bmpr2, Lef1, Vcan Gpm6b etc.) and migration (Ttbk2, Rock1, Aspm, Nck2, etc.) were significantly up-regulated in G3@nCe/GelMA as compared to nCe/GelMA and pristine GelMA. The ROS-metabolism-related genes such as Nox1, Atp7a were highly co-upregulated in G3@nCe/GelMA and nCe/GelMA while Mt3, Ucp2, Sod3, Duoxa1 genes got downregulated, indicating the role of ceria in regulating ROS that encourages bone regeneration (Fig. 6e). The entire gene list from heatmap and their respective fold change values are also given (Fig. S10). When the top 30 enriched GO terms from cluster 2 were analyzed by the DAVID database, cell proliferation, osteogenesis, and ROS-related terms were similarly detected owing to the presence of dendrimer on ceria (Fig. S11). The TFEA of cluster 2 DEGs using Chip-X also further revealed the possible involvement of TCF7L2 (Fig. S12). Taken all, G3@nCe/GelMA hydrogels would act as a therapeutic hydrogel that synergistically regulates stem cell function (proliferation and differentiation) and modulates ROS processes for efficient bone regeneration.
3.7. Biocompatibility implies the tissue healing capacity of G3@nCe/GelMA
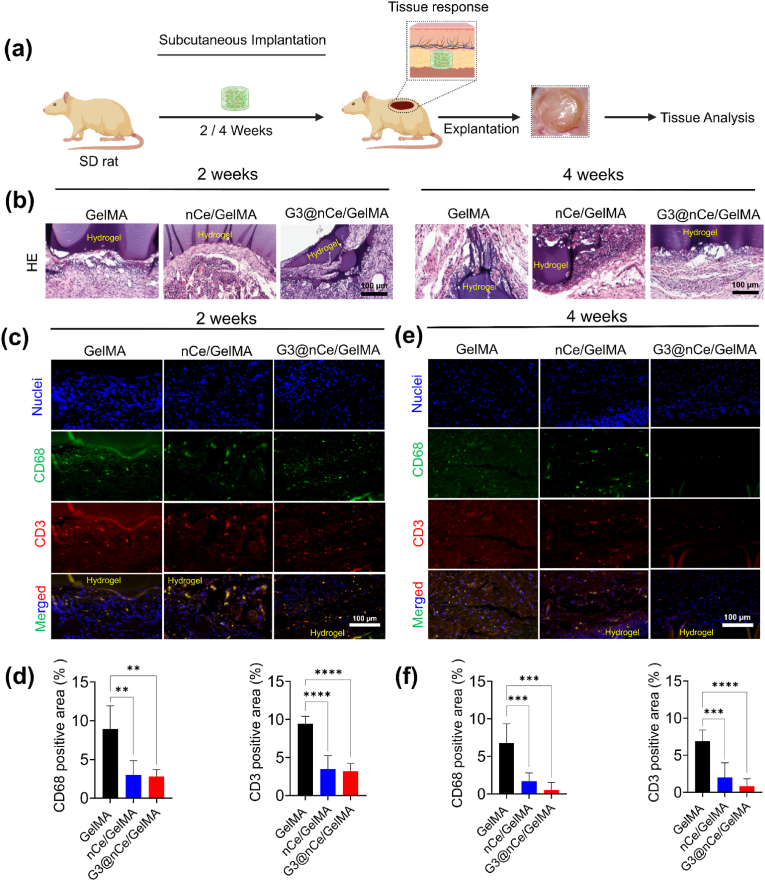
To ensure the safety, biocompatibility, and functionality of an implantable hydrogel, it may be essential to have a thorough grasp of how the immune system responds to it. The clinical applications using conventional hydrogel-based materials are generally limited due to the reason that they are often not properly integrated into in vivo, which leads to insistent inflammatory responses in the organism [6,75]. First, we used an in vitro hemolysis assay to assess hydrogel biocompatibility. The haemolysis ratio of all hydrogel samples falls below the critical safe haemolysis ratio for biomaterials, allowing them to be used in clinical studies (Fig. S13). Further, we implanted hydrogel constructs in rat subcutaneous sites and examined the resultant immune responses evoked by them with the local tissues (Fig. 7a). The results revealed that G3@nCe/GelMA hydrogels displayed tissue compatibility with observance of some residual hydrogels (Fig. S14). Being highly bioactive and biocompatible we expect that the residual hydrogels would minimize any potential negative effects. Histology analyses using high-magnification images revealed that neither implanted hydrogel elicited significant inflammatory responses and had less deposition of a fibrous collagenous capsule, indicating its tissue biocompatibility (Fig. 7b).
Fig. 7.
Subcutaneous implantation of G3@nCe/GelMA hydrogels in vivo. (a) Schematic illustration showing the steps involved in subcutaneous implantation of G3@nCe/GelMA hydrogel. (b) Histological analysis of explanted tissue samples by H&E staining. Scale bar: 100 μm. (c) Expression of CD68; pan macrophage marker (green) and CD3; pan T-cell marker (red) in the interface of hydrogel and tissue after 2 weeks of implantation by immunofluorescence. Scale bar: 100 μm. (d) Quantitative analysis of CD68 and CD3 expression at 2 weeks. (e) Relative expression of CD68 (green) and CD3 (red) inflammatory markers in the interface of hydrogel and tissue after 4 weeks of implantation. Scale bar: 100 μm. (f) Quantitative analysis of CD68 and CD3 expression at 4 weeks.. All data expressed as mean ± SD (n = 5; ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
It was also observed that G3@nCe/GelMA exhibited a faster partial degradation at week 2 but did not show any signs of degradation at week 4. This observation may have various underlying reasons [76,77]. For example, one possibility is that the hydrogel had a shorter degradation profile, which means that it quickly degraded in the initial stages but then stabilized or slowed down in the later stages. Another possibility is that the microenvironmental conditions (the presence of enzymes or other degradation-promoting factors) of the hydrogel may have changed between weeks 2 and 4. Alternatively, the hydrogel may have been exposed to different pH values or mechanical stresses that could have accelerated the degradation rate. Despite differences in degradation rates among the hydrogel samples tested, all of them exhibited evidence of degradation over a period. Furthermore, the explanted tissues did not show any apparent necrosis, indicating that the hydrogels were well-accepted by the host animals. Some studies also reported the use of an enzyme (collagenase) and/or chelating agents (EDTA) to disrupt the crosslinking to accelerate the degradation of the hydrogels after implantation for enhancing the integration of the implanted hydrogel with the surrounding tissues [78]. However, none of these methods were employed in our investigation.
It is well known that under clinical conditions, the immune reactions that arise from newly developed biomaterials are a critical factor influencing its efficiency [75]. Hence, furthermore, we evaluated the immunogenicity of hydrogels using surface marker immunofluorescence for T-lymphocytes (CD3) and macrophages (CD68) at 2 weeks and 4 weeks and semi-quantifications were made using ImageJ. Like our in vitro results, nanomaterial incorporation doesn't cause any observable toxicity. All implanted samples elicited moderate inflammatory responses at 2 weeks (Fig. 7c–d), but their expression gradually decreased by the 4th week (Fig. 7e–f). It is well known that nCe-based biomaterials exhibit anti-inflammatory properties in subcutaneous models, which may be attributed to the antioxidant properties of nCe [79]. These materials scavenge ROS and can prevent the activation of macrophages and T cells, which produce ROS as part of their inflammatory response. Both nCe/GelMA and G3@nCe/GelMA reduced macrophage and T-cell activation and infiltration in the tissue surrounding the hydrogels by lowering ROS levels. However, pristine GelMA, which lacks ROS responsiveness, may still cause tissue reactions. As a result, it has been demonstrated that G3@nCe/GelMA hydrogels cause minimal inflammatory responses in an organism and provide functional benefits for use as a nanobiomaterial in a variety of biomedical applications, particularly bone regeneration.
3.8. Tissue regenerative property of G3@nCe/GelMA in critical-sized bone defects
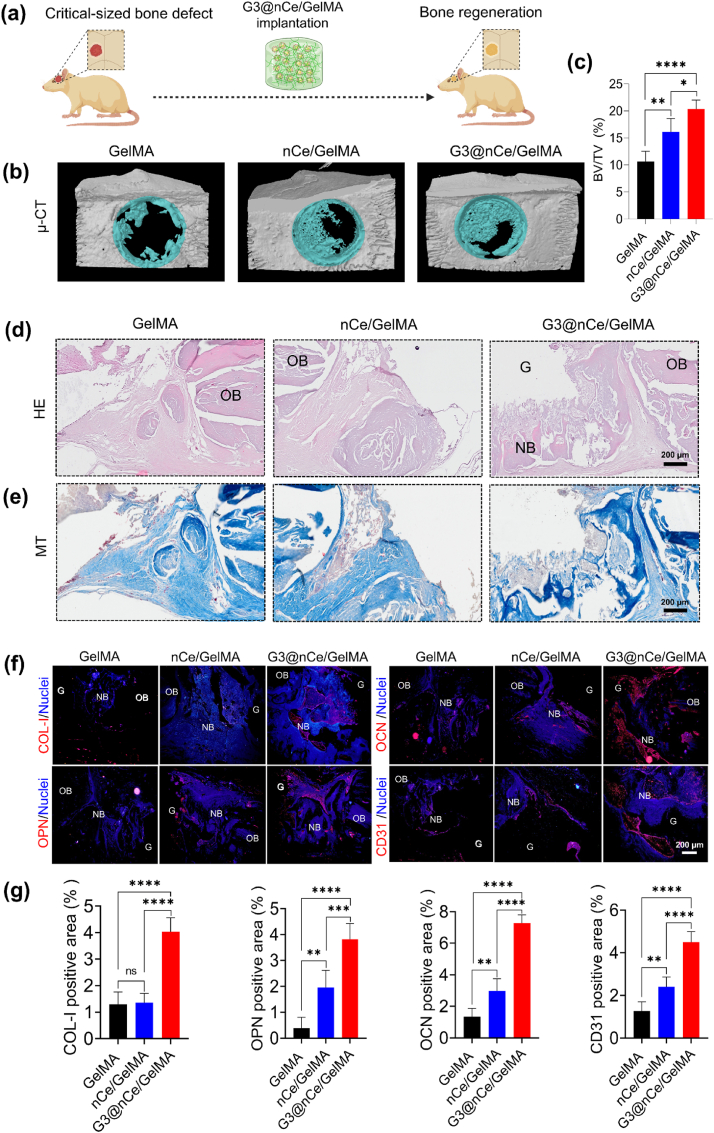
Based on the promising results from subcutaneous implantation, we next sought to demonstrate the function of G3@nCe/GelMA hydrogels in fostering bone tissue regeneration. Inspired by previous studies, we made a critical-sized calvaria bone defect model which demands physiological conditions demanding both ROS-modulation and enhanced cellular and osteogenic processes for bone remodelling. Fig. 8a shows a graphical illustration of the implantation of G3@nCe/GelMA hydrogel constructs in rat critical-sized bone defects. As an implant, we used hydrogel constructs that were 5 mm in size (Fig. S15). The local tissue simply accepted the hydrogels when they were implanted at the rat cranial defects, demonstrating osteointegration, and they also gradually promoted neo-bone formation. Following 12 weeks of hydrogel implantation, new autologous bone formation occurred at the margin of the calvaria defects growing towards the centre in G3@nCe/GelMA groups and nCe/GelMA groups while the pristine GelMA group exhibited inadequate neo-bone formation. The examination of cranial defects using μCT analysis revealed bone structure parameters such as BV/TV (Fig. 8c) and BSD (Fig. S16), confirming that the G3@nCe/GelMA groups displayed a comparatively higher extent of regenerated bone, implying that the presence of G3@nCe in GelMA might facilitate the effective repair of bone deformities. Additionally, it was noted that under the impact of G3@nCe/GelMA, the density of bones at defects continued to harden and remodel.
Fig. 8.
In vivo bone regeneration promoted by G3@nCe/GelMA hydrogel evaluated at the 12th week of implantation. (a) Graphical illustration showing the implantation of G3@nCe/GelMA hydrogel constructs in rat critical-sized bone defects. (b) Representative 3D reconstructed μCT images and (c) Quantification of the neo-bone formation using BV/TV in specific regions of interest. (d) Representative images of HE-stained tissues. Scale bar: 200 μm. (e) Representative images from MT staining. Scale bar: 200 μm. (f) Immunofluorescence of tissue samples showing the expression of various bone-related proteins (Col-I, OPN, OCN, & CD31). Scale bar: 200 μm. The annotation G stands for the residual hydrogel, OB for old bone and NB for new bone. (g) The quantitative analysis of various bone-related proteins (Col-I, OPN, OCN, & CD31 using ImageJ. All data expressed as mean ± SD (n = 5; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
Following the confirmations using μCT, we performed the histological studies (Fig. S17). The HE and MT staining were performed to verify the neo-bone formation and collagen deposition. From the high-magnification histology images (Fig. 8d), it was evident that after 12 weeks, the formation of mature bone-like tissues along the border of the defects was more observable in the case of G3@nCe/GelMA. Additionally, a calcified structure that is typical of newly formed bone was evident from the staining. MT staining also confirmed the dense collagen deposition in G3@nCe/GelMA while less dense collagen was observed in pristine GelMA and nCe/GelMA groups (Fig. 8e). Hydrogel residues were also observed during tissue examination by histology. Although complete degradation of the hydrogel component is ideal for optimal tissue integration, G3@nCe/GelMA may not necessarily restrict new bone formation because of its exceptional bone-supporting qualities which surpass those of pristine GelMA [80,81].
Next, the relative expression of important proteins associated with the development of new bone was examined using immunohistochemistry. COl-1, OPN, OCN, and the angiogenic protein CD31 were used as representative markers (Fig. 8f) and semi-quantifications were made using ImageJ (Fig. 8g). The findings show that G3@nCe/GelMA groups showed increased protein expression, whereas the pristine GelMA and nCe/GelMA group showed relatively reduced expression for all the bone-related proteins. We anticipate that the increased osteogenic activities of G3@nCe/GelMA in the in vivo microenvironment may have contributed to these elevated bone-related protein expressions. Despite our encouraging results, further research is ongoing to better understand the processes underlying effective bone regeneration to improve the material's applicability for future applications. The G3@nCe/GelMA hydrogels were shown to have excellent tissue compatibility with lesser inflammatory responses in a subcutaneous model and enhanced neo-bone formation in a critical-sized bone defect model, which collectively suggests the effectiveness of the proposed G3@nCe/GelMA as a promising cell-free biomaterial to enhance the bone regeneration in critical-sized bone defects.
4. Discussion
A popular approach in BTE involves the development of biomaterials that can accelerate natural healing mechanisms by attracting stem cells to injured sites. Unlike conventional cell-based therapies, which have limited clinical applications, strategies utilizing biomaterials to facilitate tissue regeneration without external cell sources are preferred [82]. By utilizing engineered biomaterials, it is possible to regulate the biological cues, which can mimic the intricate signalling patterns of endogenous tissue regeneration, thereby offering a feasible solution. Over the years, the formulation of nCe-engineered GelMA hydrogels has been extensively studied for their potential applications in tissue engineering and regenerative medicine because of the unique cell-friendly characteristics of GelMA and the intrinsic antioxidant properties of nCe. However, most of the studies have focused on the engineering of soft tissues such as skin, because the incorporation of nCe alone may not provide the necessary microenvironmental cues required for BTE. To address these limitations, and to engineer GelMA as a biofunctional matrix, we introduced an approach to develop hybrid nanomaterials based on nCe, which is surface functionalized with G3 dendrimers. The resulting G3@nCe harnesses the intrinsic antioxidant and bioactivity properties of nCe while also integrating the additional bioactivity offered by the dendrimer.
When incorporated into the GelMA matrix by photogelation, G3@nCe/GelMA offers suitable physicochemical, and biological cues for activating stem cell functions. Due to the protein or growth factor immobilization capacity of dendrimers, G3@nCe/GelMA hydrogels could act as a “nano-reservoir” that immobilizes and deliver various bioactive molecules (growth factors, ECM proteins etc.) to cells in a sustained manner thus allowing their proliferation, migration, and differentiation. Besides, the biological cues provided by G3@nCe/GelMA (cell-to-cell and cell-to-matrix interactions) can also stimulate cell proliferation and differentiation by promoting the secretion of growth factors and other signalling molecules, compared to nCe/GelMA and pristine GelMA. Furthermore, G3@nCe/GelMA mimicked the ECM of bone tissues by offering superior microenvironmental cues that support the differentiation of preosteoblast cells. These processes are tightly regulated by the complex interplay between microenvironmental cues and signalling pathways. Additionally, G3@nCe/GelMA protected preosteoblast cells from H2O2-induced oxidative stress injury and was beneficial for osteogenesis under harsh conditions. The transcriptome analysis identified the important signalling pathways and biological or molecular processes through which the G3@nCe/GelMA hydrogel controlled cellular function and promoted the formation of new bone. It was revealed that G3@nCe/GelMA hydrogels would synergistically regulate stem cell function (proliferation and differentiation) and modulate ROS processes for efficient bone regeneration through positive regulation of the canonical Wnt signalling pathway. Blood and tissue compatibility analysis of G3@nCe/GelMA by haemocompatibility assay and subcutaneous implantation also assured that this material has a critically safe haemolysis rate and exhibits low immunogenicity. When implanted at rat cranial defects, G3@nCe/GelMA demonstrated osteointegration and gradually accelerated the neo-bone formation. In short, this work details the multifunctional design of hydrogels by introducing a hybrid nanostructure, highlighting the design of materials according to bone tissue biology, and providing the rationale for designing next-generation nanoengineered hydrogels that support natural bone healing and regeneration.
The proposed hydrogel platform also sets the stage for future research to employ the dendritic functionalization approach to create new functional nanobiomaterials with the potential to extend this methodology to other types of nanomaterials. Recent studies have also shown that dendrimer functionalized nanomaterials can act as cationic nanomaterials to have a therapeutic effect on many inflammatory diseases in animal models by scavenging the negatively charged cell-free DNA (cfDNA) through Toll-like receptor-9 (TLR9) inhibitory activity, which includes periodontitis [83], sepsis [84], obesity-related chronic inflammation [85] etc. In the future, G3@nCe/GelMA hydrogel formulations could also be used as “dual scavengers” (ROS and cfDNA) for targeting several inflammatory diseases. Since the current study already explored the potential effects of G3@nCe/GelMA in bone tissue regeneration, future research can explore the use of these formulation in the treatment of inflammatory bone disorders, such as rheumatoid arthritis, which involves the role of both ROS and cfDNA in disease progression [86]. By carefully optimizing the hydrogel formulation, we also believe that the currently explored hydrogel platforms will have promising outcomes when extended to soft tissue regeneration, such as diabetic wound healing. Although we have successfully engineered GelMA hydrogel using G3@nCe, which activates stem cells and promotes bone regeneration by orchestrating cellular responses, we did not examine the potential effect of G3@nCe/GelMA-mediated macrophage polarization for in vivo bone regeneration. Moreover, recent reports have confirmed the role of dendrimeric materials in modulating macrophage response [87]. This is another promising area to explore; hence, further research is needed to evaluate the immunomodulatory role of G3@nCe/GelMA in bone defect repair to confirm its effectiveness. In essence, our report on engineering GelMA using G3@nCe indicates that it has the potential to be an effective method for creating hydrogels that mimic the properties of natural bone tissues. This could have significant translational applications, especially in promoting bone remodelling and tissue regeneration. Overall, our findings suggest that this approach holds great promise for the development of new biomaterials with therapeutic potential for various tissue regeneration applications in medical settings.
5. Conclusion
In summary, a bioactive G3@nCe/GelMA hydrogel was prepared by photo-gelation combining the benefits of GelMA and G3@nCe. The hydrogels provided suitable physicochemical cues supporting bone formation. Further, G3@nCe/GelMA provided biological cues by performing as a multifunctional platform that supports cell adhesion, proliferation, migration, and differentiation. Additionally, G3@nCe demonstrated ROS scavenging activity like nCe, which rendered G3@nCe/GelMA antioxidant properties and reduced oxidative stress in the cellular microenvironment. Transcriptome analysis showed that G3@nCe/GelMA upregulated various genes related to orchestrated cellular activities that coordinate bone regeneration and revealed the involvement of the canonical Wnt signalling pathway. Furthermore, G3@nCe/GelMA exhibited low immunogenicity under in vivo conditions, like nCe/GelMA, indicating its excellent biocompatibility compared to pristine GelMA. Implantation of G3@nCe/GelMA in critical-sized rat cranial defects revealed that the G3@nCe/GelMA hydrogel accelerated new bone formation in vivo compared to the pristine GelMA and nCe/GelMA. Altogether, our report on engineering GelMA using G3@nCe could be considered a promising strategy for designing bioactive materials with the translational potential to endorse the bone remodelling process and for other tissue regeneration applications.
Author contributions
Amal George Kurian: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing an original draft. Nandin Mandakhbayar: Methodology, Formal analysis, Validation. Rajendra K. Singh: Conceptualization, Methodology, Validation, Visualization. Jung-Hwan Lee: Supervision, Methodology, Validation, Project administration. Gangshi Jin: Formal analysis, Data curation. Hae-Won Kim: Conceptualization, Supervision, Funding acquisition, Project administration, Writing review & editing. All authors have approved the final version of this manuscript.
Declaration of competing interest
The authors confirm that they have no known financial or personal conflicts that would have appeared to influence the findings presented in this study.
Acknowledgements
This work was supported by the grants (2018R1A2B3003446, 2018K1A4A3A01064257, 2019R1C1C1002490, 2019R1A6A1A11034536, 2020R1I1A1A01071828, 2021R1A5A2022318), National Research Foundation (NRF), Republic of Korea.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2023.100664.
Contributor Information
Rajendra K. Singh, Email: singhiitgphd@gmail.com.
Jung-Hwan Lee, Email: ducious@gmail.com.
Hae-Won Kim, Email: kimhw@dku.edu.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Petite H., Viateau V., Bensaid W., Meunier A., de Pollak C., Bourguignon M., Oudina K., Sedel L., Guillemin G. Tissue-engineered bone regeneration. Nat. Biotechnol. 2000;18(9):959–963. doi: 10.1038/79449. [DOI] [PubMed] [Google Scholar]
- 2.Dimitriou R., Jones E., McGonagle D., Giannoudis P.V. Bone regeneration: current concepts and future directions. BMC Med. 2011;9(1):1–10. doi: 10.1186/1741-7015-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao C., Peng S., Feng P., Shuai C. Bone biomaterials and interactions with stem cells. Bone research. 2017;5(1):1–33. doi: 10.1038/boneres.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billström G.H., Blom A.W., Larsson S., Beswick A.D. Application of scaffolds for bone regeneration strategies: current trends and future directions. Injury. 2013;44:S28–S33. doi: 10.1016/S0020-1383(13)70007-X. [DOI] [PubMed] [Google Scholar]
- 5.Stevens M.M. Biomaterials for bone tissue engineering. Mater. Today. 2008;11(5):18–25. [Google Scholar]
- 6.Lee K.Y., Mooney D.J. Hydrogels for tissue engineering. Chem. Rev. 2001;101(7):1869–1880. doi: 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen K.T., West J.L. Photopolymerizable hydrogels for tissue engineering applications. Biomaterials. 2002;23(22):4307–4314. doi: 10.1016/s0142-9612(02)00175-8. [DOI] [PubMed] [Google Scholar]
- 8.Hunt J.A., Chen R., van Veen T., Bryan N. Hydrogels for tissue engineering and regenerative medicine. J. Mater. Chem. B. 2014;2(33):5319–5338. doi: 10.1039/c4tb00775a. [DOI] [PubMed] [Google Scholar]
- 9.Yue K., Trujillo-de Santiago G., Alvarez M.M., Tamayol A., Annabi N., Khademhosseini A. Synthesis, properties, and biomedical applications of gelatin methacryloyl (GelMA) hydrogels. Biomaterials. 2015;73:254–271. doi: 10.1016/j.biomaterials.2015.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao S., Zhao T., Wang J., Wang C., Du J., Ying L., Lin J., Zhang C., Hu W., Wang L. Gelatin methacrylate (GelMA)-based hydrogels for cell transplantation: an effective strategy for tissue engineering. Stem cell reviews and reports. 2019;15:664–679. doi: 10.1007/s12015-019-09893-4. [DOI] [PubMed] [Google Scholar]
- 11.Klotz B.J., Gawlitta D., Rosenberg A.J., Malda J., Melchels F.P. Gelatin-methacryloyl hydrogels: towards biofabrication-based tissue repair. Trends Biotechnol. 2016;34(5):394–407. doi: 10.1016/j.tibtech.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurian A.G., Singh R.K., Patel K.D., Lee J.-H., Kim H.-W. Multifunctional GelMA platforms with nanomaterials for advanced tissue therapeutics. Bioact. Mater. 2022;8:267–295. doi: 10.1016/j.bioactmat.2021.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakr M.A., Sakthivel K., Hossain T., Shin S.R., Siddiqua S., Kim J., Kim K. Recent trends in gelatin methacryloyl nanocomposite hydrogels for tissue engineering. J. Biomed. Mater. Res. 2022;110(3):708–724. doi: 10.1002/jbm.a.37310. [DOI] [PubMed] [Google Scholar]
- 14.Wu Z., Bai J., Ge G., Wang T., Feng S., Ma Q., Liang X., Li W., Zhang W., Xu Y. Regulating macrophage polarization in high glucose microenvironment using lithium-modified bioglass-hydrogel for diabetic bone regeneration. Advanced Healthcare Materials. 2022;11(13) doi: 10.1002/adhm.202200298. [DOI] [PubMed] [Google Scholar]
- 15.Huang K., Ou Q., Xie Y., Chen X., Fang Y., Huang C., Wang Y., Gu Z., Wu J. Halloysite nanotube based scaffold for enhanced bone regeneration. ACS Biomater. Sci. Eng. 2019;5(8):4037–4047. doi: 10.1021/acsbiomaterials.9b00277. [DOI] [PubMed] [Google Scholar]
- 16.Ou Q., Huang K., Fu C., Huang C., Fang Y., Gu Z., Wu J., Wang Y. Nanosilver-incorporated halloysite nanotubes/gelatin methacrylate hybrid hydrogel with osteoimmunomodulatory and antibacterial activity for bone regeneration. Chem. Eng. J. 2020;382 [Google Scholar]
- 17.Xavier J.R., Thakur T., Desai P., Jaiswal M.K., Sears N., Cosgriff-Hernandez E., Kaunas R., Gaharwar A.K. Bioactive nanoengineered hydrogels for bone tissue engineering: a growth-factor-free approach. ACS Nano. 2015;9(3):3109–3118. doi: 10.1021/nn507488s. [DOI] [PubMed] [Google Scholar]
- 18.Liu L., Li X., Shi X., Wang Y. Injectable alendronate-functionalized GelMA hydrogels for mineralization and osteogenesis. RSC Adv. 2018;8(40):22764–22776. doi: 10.1039/c8ra03550d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J., Han F., Ma J., Wang H., Pan J., Yang G., Zhao H., Zhao J., Liu J., Liu Z. Targeting endogenous hydrogen peroxide at bone defects promotes bone repair. Adv. Funct. Mater. 2022;32(10) [Google Scholar]
- 20.Rajeshkumar S., Naik P. Synthesis and biomedical applications of cerium oxide nanoparticles–a review. Biotechnology Reports. 2018;17:1–5. doi: 10.1016/j.btre.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thakur N., Manna P., Das J. Synthesis and biomedical applications of nanoceria, a redox active nanoparticle. J. Nanobiotechnol. 2019;17(1):1–27. doi: 10.1186/s12951-019-0516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dashnyam K., Lee J.-H., Singh R.K., Yoon J.-Y., Lee J.H., Jin G.-Z., Kim H.-W. Optimally dosed nanoceria attenuates osteoarthritic degeneration of joint cartilage and subchondral bone. Chem. Eng. J. 2021;422 [Google Scholar]
- 23.Kim J.W., Mahapatra C., Hong J.Y., Kim M.S., Leong K.W., Kim H.W., Hyun J.K. Functional recovery of contused spinal cord in rat with the injection of optimal-dosed cerium oxide nanoparticles. Adv. Sci. 2017;4(10) doi: 10.1002/advs.201700034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh R.K., Yoon D.S., Mandakhbayar N., Li C., Kurian A.G., Lee N.-H., Lee J.-H., Kim H.-W. Diabetic bone regeneration with nanoceria-tailored scaffolds by recapitulating cellular microenvironment: activating integrin/TGF-β co-signaling of MSCs while relieving oxidative stress. Biomaterials. 2022;288 doi: 10.1016/j.biomaterials.2022.121732. [DOI] [PubMed] [Google Scholar]
- 25.Kurian A.G., Singh R.K., Lee J.-H., Kim H.-W. Surface-engineered hybrid gelatin methacryloyl with nanoceria as reactive oxygen species responsive matrixes for bone therapeutics. ACS Appl. Bio Mater. 2022;5(3):1130–1138. doi: 10.1021/acsabm.1c01189. [DOI] [PubMed] [Google Scholar]
- 26.Ren S., Zhou Y., Zheng K., Xu X., Yang J., Wang X., Miao L., Wei H., Xu Y. Cerium oxide nanoparticles loaded nanofibrous membranes promote bone regeneration for periodontal tissue engineering. Bioact. Mater. 2022;7:242–253. doi: 10.1016/j.bioactmat.2021.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Augustine R., Zahid A.A., Hasan A., Dalvi Y.B., Jacob J. Cerium oxide nanoparticle-loaded gelatin methacryloyl hydrogel wound-healing patch with free radical scavenging activity. ACS Biomater. Sci. Eng. 2020;7(1):279–290. doi: 10.1021/acsbiomaterials.0c01138. [DOI] [PubMed] [Google Scholar]
- 28.Cheng H., Shi Z., Yue K., Huang X., Xu Y., Gao C., Yao Z., Zhang Y.S., Wang J. Sprayable hydrogel dressing accelerates wound healing with combined reactive oxygen species-scavenging and antibacterial abilities. Acta Biomater. 2021;124:219–232. doi: 10.1016/j.actbio.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Shaikh A., Kesharwani P., Gajbhiye V. Dendrimer as a momentous tool in tissue engineering and regenerative medicine. J. Contr. Release. 2022;346:328–354. doi: 10.1016/j.jconrel.2022.04.008. [DOI] [PubMed] [Google Scholar]
- 30.Oliveira J.M., Salgado A.J., Sousa N., Mano J.F., Reis R.L. Dendrimers and derivatives as a potential therapeutic tool in regenerative medicine strategies—a review. Prog. Polym. Sci. 2010;35(9):1163–1194. [Google Scholar]
- 31.Lee C.C., MacKay J.A., Fréchet J.M., Szoka F.C. Designing dendrimers for biological applications. Nat. Biotechnol. 2005;23(12):1517–1526. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- 32.Chen W., Li W., Xu K., Li M., Dai L., Shen X., Hu Y., Cai K. Functionalizing titanium surface with PAMAM dendrimer and human BMP2 gene via layer-by-layer assembly for enhanced osteogenesis. J. Biomed. Mater. Res. 2018;106(3):706–717. doi: 10.1002/jbm.a.36273. [DOI] [PubMed] [Google Scholar]
- 33.Staehlke S., Lehnfeld J., Schneider A., Nebe J.B., Müller R. Terminal chemical functions of polyamidoamine dendrimer surfaces and its impact on bone cell growth. Mater. Sci. Eng. C. 2019;101:190–203. doi: 10.1016/j.msec.2019.03.073. [DOI] [PubMed] [Google Scholar]
- 34.Pan B.-f., Gao F., Gu H.-c. Dendrimer modified magnetite nanoparticles for protein immobilization. J. Colloid Interface Sci. 2005;284(1):1–6. doi: 10.1016/j.jcis.2004.09.073. [DOI] [PubMed] [Google Scholar]
- 35.Sun W., Mignani S., Shen M., Shi X. Dendrimer-based magnetic iron oxide nanoparticles: their synthesis and biomedical applications. Drug Discov. Today. 2016;21(12):1873–1885. doi: 10.1016/j.drudis.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 36.Loessner D., Meinert C., Kaemmerer E., Martine L.C., Yue K., Levett P.A., Klein T.J., Melchels F.P., Khademhosseini A., Hutmacher D.W. Functionalization, preparation and use of cell-laden gelatin methacryloyl–based hydrogels as modular tissue culture platforms. Nat. Protoc. 2016;11(4):727–746. doi: 10.1038/nprot.2016.037. [DOI] [PubMed] [Google Scholar]
- 37.Mahapatra C., Singh R.K., Lee J.-H., Jung J., Hyun J.K., Kim H.-W. Nano-shape varied cerium oxide nanomaterials rescue human dental stem cells from oxidative insult through intracellular or extracellular actions. Acta Biomater. 2017;50:142–153. doi: 10.1016/j.actbio.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 38.Park I.-S., Mahapatra C., Park J.S., Dashnyam K., Kim J.-W., Ahn J.C., Chung P.-S., Yoon D.S., Mandakhbayar N., Singh R.K. Revascularization and limb salvage following critical limb ischemia by nanoceria-induced Ref-1/APE1-dependent angiogenesis. Biomaterials. 2020;242 doi: 10.1016/j.biomaterials.2020.119919. [DOI] [PubMed] [Google Scholar]
- 39.Hong S.M., Yoon J.Y., Cha J.R., Ahn J., Mandakhbayar N., Park J.H., Im J., Jin G., Kim M.Y., Knowles J.C. Bioengineering & Translational Medicine; 2022. Hyperelastic, Shape-memorable and Ultra-cell-adhesive Degradable Polycaprolactone-polyurethane Copolymer for Tissue Regeneration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):1–16. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 44.Keenan A.B., Torre D., Lachmann A., Leong A.K., Wojciechowicz M.L., Utti V., Jagodnik K.M., Kropiwnicki E., Wang Z., Ma’ayan A. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019;47(W1):W212–W224. doi: 10.1093/nar/gkz446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z., Wang Z., Lu W.W., Zhen W., Yang D., Peng S. Novel biomaterial strategies for controlled growth factor delivery for biomedical applications. NPG Asia Mater. 2017;9(10):e435. e435. [Google Scholar]
- 46.Agarwal R., García A.J. Biomaterial strategies for engineering implants for enhanced osseointegration and bone repair. Adv. Drug Deliv. Rev. 2015;94:53–62. doi: 10.1016/j.addr.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cerqueni G., Scalzone A., Licini C., Gentile P., Mattioli-Belmonte M. Insights into oxidative stress in bone tissue and novel challenges for biomaterials. Mater. Sci. Eng. C. 2021;130 doi: 10.1016/j.msec.2021.112433. [DOI] [PubMed] [Google Scholar]
- 48.Engler A.J., Sen S., Sweeney H.L., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 49.Tsou Y.-H., Khoneisser J., Huang P.-C., Xu X. Hydrogel as a bioactive material to regulate stem cell fate. Bioact. Mater. 2016;1(1):39–55. doi: 10.1016/j.bioactmat.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chakraborty A., Pacelli S., Alexander S., Huayamares S., Rosenkrans Z., Vergel F.E., Wu Y., Chakravorty A., Paul A. Nanoparticle-Reinforced tough hydrogel as a versatile platform for pharmaceutical drug delivery: preparation and in vitro characterization. Mol. Pharm. 2022;20:767–774. doi: 10.1021/acs.molpharmaceut.2c00564. [DOI] [PubMed] [Google Scholar]
- 51.Lu C.H., Yeh Y.C. Fabrication of multiresponsive magnetic nanocomposite double-network hydrogels for controlled release applications. Small. 2021;17(52) doi: 10.1002/smll.202105997. [DOI] [PubMed] [Google Scholar]
- 52.Ning Z., Tan B., Chen B., Lau D.S.A., Wong T.M., Sun T., Peng S., Li Z., Lu W.W. Precisely controlled delivery of abaloparatide through injectable hydrogel to promote bone regeneration. Macromol. Biosci. 2019;19(6) doi: 10.1002/mabi.201900020. [DOI] [PubMed] [Google Scholar]
- 53.Zhao X., Liu S., Yildirimer L., Zhao H., Ding R., Wang H., Cui W., Weitz D. Injectable stem cell-laden photocrosslinkable microspheres fabricated using microfluidics for rapid generation of osteogenic tissue constructs. Adv. Funct. Mater. 2016;26(17):2809–2819. [Google Scholar]
- 54.Ding X., Shi J., Wei J., Li Y., Wu X., Zhang Y., Jiang X., Zhang X., Lai H. A biopolymer hydrogel electrostatically reinforced by amino-functionalized bioactive glass for accelerated bone regeneration. Sci. Adv. 2021;7(50) doi: 10.1126/sciadv.abj7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.El-Fiqi A., Lee J.H., Lee E.-J., Kim H.-W. Collagen hydrogels incorporated with surface-aminated mesoporous nanobioactive glass: improvement of physicochemical stability and mechanical properties is effective for hard tissue engineering. Acta Biomater. 2013;9(12):9508–9521. doi: 10.1016/j.actbio.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 56.Drury J.L., Mooney D.J. Hydrogels for tissue engineering: scaffold design variables and applications. Biomaterials. 2003;24(24):4337–4351. doi: 10.1016/s0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]
- 57.Liu F., Wang X., Li Y., Ren M., He P., Wang L., Xu J., Yang S., Ji P. Dendrimer-modified gelatin methacrylate hydrogels carrying adipose-derived stromal/stem cells promote cartilage regeneration. Stem Cell Res. Ther. 2022;13(1):1–15. doi: 10.1186/s13287-022-02705-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li B., Wang F., Hu F., Ding T., Huang P., Xu X., Liang J., Li C., Zhou Q., Lu M. Injectable “nano-micron” combined gene-hydrogel microspheres for local treatment of osteoarthritis. NPG Asia Mater. 2022;14(1):1–15. [Google Scholar]
- 59.Singh R.K., Jin G.-Z., Mahapatra C., Patel K.D., Chrzanowski W., Kim H.-W. Mesoporous silica-layered biopolymer hybrid nanofibrous scaffold: a novel nanobiomatrix platform for therapeutics delivery and bone regeneration. ACS Appl. Mater. Interfaces. 2015;7(15):8088–8098. doi: 10.1021/acsami.5b00692. [DOI] [PubMed] [Google Scholar]
- 60.Wang S., Yang Y., Wang R., Kong X., Wang X. Mineralization of calcium phosphate controlled by biomimetic self-assembled peptide monolayers via surface electrostatic potentials. Bioact. Mater. 2020;5(2):387–397. doi: 10.1016/j.bioactmat.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou L., Tan G., Tan Y., Wang H., Liao J., Ning C. Biomimetic mineralization of anionic gelatin hydrogels: effect of degree of methacrylation. RSC Adv. 2014;4(42):21997–22008. [Google Scholar]
- 62.Ahearne M. Introduction to cell–hydrogel mechanosensing. Interface focus. 2014;4(2) doi: 10.1098/rsfs.2013.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peppas N.A., Langer R. New challenges in biomaterials. Science. 1994;263(5154):1715–1720. doi: 10.1126/science.8134835. [DOI] [PubMed] [Google Scholar]
- 64.Serpooshan V., Mahmoudi M., Zhao M., Wei K., Sivanesan S., Motamedchaboki K., Malkovskiy A.V., Goldstone A.B., Cohen J.E., Yang P.C. Protein corona influences cell–biomaterial interactions in nanostructured tissue engineering scaffolds. Adv. Funct. Mater. 2015;25(28):4379–4389. doi: 10.1002/adfm.201500875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Teixeira S.P., Domingues R.M., Shevchuk M., Gomes M.E., Peppas N.A., Reis R.L. Biomaterials for sequestration of growth factors and modulation of cell behavior. Adv. Funct. Mater. 2020;30(44) [Google Scholar]
- 66.Hurd T.R., DeGennaro M., Lehmann R. Redox regulation of cell migration and adhesion. Trends Cell Biol. 2012;22(2):107–115. doi: 10.1016/j.tcb.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim S.-H., Turnbull J., Guimond S. Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. J. Endocrinol. 2011;209(2):139–151. doi: 10.1530/JOE-10-0377. [DOI] [PubMed] [Google Scholar]
- 68.Bai X., Gao M., Syed S., Zhuang J., Xu X., Zhang X.-Q. Bioactive hydrogels for bone regeneration. Bioact. Mater. 2018;3(4):401–417. doi: 10.1016/j.bioactmat.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yue S., He H., Li B., Hou T. Hydrogel as a biomaterial for bone tissue engineering: a review. Nanomaterials. 2020;10(8):1511. doi: 10.3390/nano10081511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Domazetovic V., Marcucci G., Iantomasi T., Brandi M.L., Vincenzini M.T. Oxidative stress in bone remodeling: role of antioxidants. Clinical Cases in Mineral and Bone Metabolism. 2017;14(2):209. doi: 10.11138/ccmbm/2017.14.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim J.H., Liu X., Wang J., Chen X., Zhang H., Kim S.H., Cui J., Li R., Zhang W., Kong Y. Wnt signaling in bone formation and its therapeutic potential for bone diseases. Therapeutic advances in musculoskeletal disease. 2013;5(1):13–31. doi: 10.1177/1759720X12466608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kobayashi Y., Uehara S., Udagawa N., Takahashi N. Regulation of bone metabolism by Wnt signals. J. Biochem. 2016;159(4):387–392. doi: 10.1093/jb/mvv124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yi F., Brubaker P.L., Jin T. TCF-4 mediates cell type-specific regulation of proglucagon gene expression by β-catenin and glycogen synthase kinase-3β. J. Biol. Chem. 2005;280(2):1457–1464. doi: 10.1074/jbc.M411487200. [DOI] [PubMed] [Google Scholar]
- 74.Li Z., Xu Z., Duan C., Liu W., Sun J., Han B. Role of TCF/LEF transcription factors in bone development and osteogenesis. Int. J. Med. Sci. 2018;15(12):1415. doi: 10.7150/ijms.26741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Franz S., Rammelt S., Scharnweber D., Simon J.C. Immune responses to implants–a review of the implications for the design of immunomodulatory biomaterials. Biomaterials. 2011;32(28):6692–6709. doi: 10.1016/j.biomaterials.2011.05.078. [DOI] [PubMed] [Google Scholar]
- 76.Lutolf M.P., Hubbell J. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol. 2005;23(1):47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 77.Paul A., Manoharan V., Krafft D., Assmann A., Uquillas J.A., Shin S.R., Hasan A., Hussain M.A., Memic A., Gaharwar A.K. Nanoengineered biomimetic hydrogels for guiding human stem cell osteogenesis in three dimensional microenvironments. J. Mater. Chem. B. 2016;4(20):3544–3554. doi: 10.1039/C5TB02745D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Deshayes S., Kasko A.M. Polymeric biomaterials with engineered degradation. J. Polym. Sci. Polym. Chem. 2013;51(17):3531–3566. [Google Scholar]
- 79.Augustine R., Dalvi Y.B., Dan P., George N., Helle D., Varghese R., Thomas S., Menu P., Sandhyarani N. Nanoceria can act as the cues for angiogenesis in tissue-engineering scaffolds: toward next-generation in situ tissue engineering. ACS Biomater. Sci. Eng. 2018;4(12):4338–4353. doi: 10.1021/acsbiomaterials.8b01102. [DOI] [PubMed] [Google Scholar]
- 80.El-Sherbiny I.M., Yacoub M.H. Hydrogel scaffolds for tissue engineering: progress and challenges. Global Cardiology Science and Practice. 2013;2013(3):38. doi: 10.5339/gcsp.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nguyen M.K., Jeon O., Dang P.N., Huynh C.T., Varghai D., Riazi H., McMillan A., Herberg S., Alsberg E. RNA interfering molecule delivery from in situ forming biodegradable hydrogels for enhancement of bone formation in rat calvarial bone defects. Acta Biomater. 2018;75:105–114. doi: 10.1016/j.actbio.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee C.-S., Singh R.K., Hwang H.S., Lee N.-H., Kurian A.G., Lee J.-H., Kim H.S., Lee M., Kim H.-W. Materials-based nanotherapeutics for injured and diseased bone. Prog. Mater. Sci. 2023 [Google Scholar]
- 83.Huang H., Pan W., Wang Y., Kim H.S., Shao D., Huang B., Ho T.-C., Lao Y.-H., Quek C.H., Shi J. Nanoparticulate cell-free DNA scavenger for treating inflammatory bone loss in periodontitis. Nat. Commun. 2022;13(1):5925. doi: 10.1038/s41467-022-33492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dawulieti J., Sun M., Zhao Y., Shao D., Yan H., Lao Y.-H., Hu H., Cui L., Lv X., Liu F. Treatment of severe sepsis with nanoparticulate cell-free DNA scavengers. Sci. Adv. 2020;6(22) doi: 10.1126/sciadv.aay7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang B., Wan Q., Li T., Yu L., Du W., Calhoun C., Leong K.W., Qiang L. Polycationic PAMAM ameliorates obesity-associated chronic inflammation and focal adiposity. Biomaterials. 2023;293 doi: 10.1016/j.biomaterials.2022.121850. [DOI] [PubMed] [Google Scholar]
- 86.Lee J., Sohn J.W., Zhang Y., Leong K.W., Pisetsky D., Sullenger B.A. Nucleic acid-binding polymers as anti-inflammatory agents. Proc. Natl. Acad. Sci. USA. 2011;108(34):14055–14060. doi: 10.1073/pnas.1105777108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Posadas I., Romero-Castillo L., El Brahmi N., Manzanares D., Mignani S., Majoral J.-P., Ceña V. Neutral high-generation phosphorus dendrimers inhibit macrophage-mediated inflammatory response in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 2017;114(37):E7660–E7669. doi: 10.1073/pnas.1704858114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.