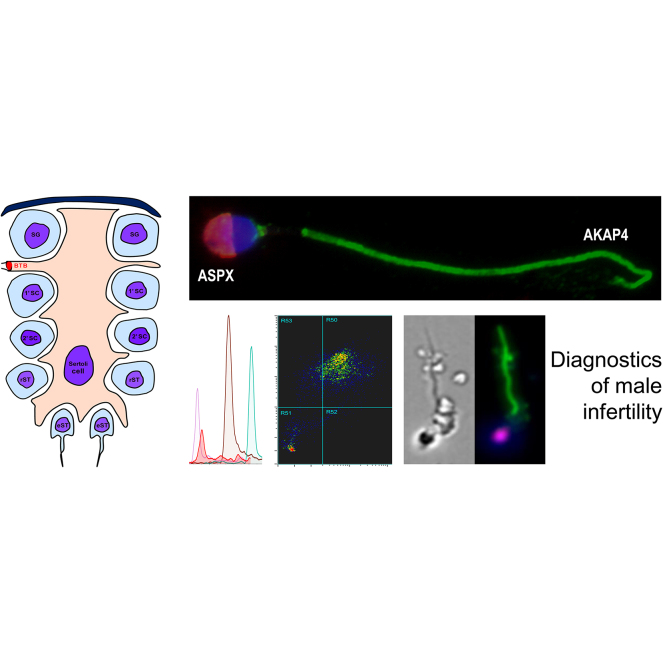
Abstract
Non-obstructive azoospermia (NOA), the most severe form of male infertility, could be treated with intracytoplasmic sperm injection, providing spermatozoa were retrieved with the microdissection testicular sperm extraction (mTESE). We hypothesized that testis-specific and germ cell–specific proteins would facilitate flow cytometry-assisted identification of rare spermatozoa in semen cell pellets of NOA patients, thus enabling non-invasive diagnostics prior to mTESE. Data mining, targeted proteomics, and immunofluorescent microscopy identified and verified a panel of highly testis-specific proteins expressed at the continuum of germ cell differentiation. Late germ cell–specific proteins AKAP4_HUMAN and ASPX_HUMAN (ACRV1 gene) revealed exclusive localization in spermatozoa tails and acrosomes, respectively. A multiplex imaging flow cytometry assay facilitated fast and unambiguous identification of rare but morphologically intact AKAP4+/ASPX+/Hoechst+ spermatozoa within debris-laden semen pellets of NOA patients. While the previously suggested markers for spermatozoa retrieval suffered from low diagnostic specificity, the multistep gating strategy and visualization of AKAP4+/ASPX+/Hoechst+ cells with elongated tails and acrosome-capped nuclei facilitated fast and unambiguous identification of the mature intact spermatozoa. AKAP4+/ASPX+/Hoechst+ assay may emerge as a noninvasive test to predict retrieval of morphologically intact spermatozoa by mTESE, thus improving diagnostics and treatment of severe forms of male infertility.
Keywords: ACRV1, AKAP4_HUMAN, AKAP4, ASPX_HUMAN, biomarkers, imaging flow cytometry, male infertility, non-obstructive azoospermia, testis-specific proteins, targeted proteomics
Graphical Abstract
Highlights
-
•
45 germ cell proteins were measured in human spermatozoa and seminal plasma.
-
•
Novel markers of obstructive and non-obstructive azoospermia were verified.
-
•
ASPX and AKAP4 proteins were exclusively localized to sperm acrosomes and tails.
-
•
Imaging flow cytometry identified rare intact AKAP4+/ASPX+/Hoechst+ sperm in NOA.
In Brief
Testis-specific proteins expressed at the continuum of germ cell differentiation were selected and evaluated by targeted proteomics, fluorescent microscopy, and flow cytometry. Novel markers of obstructive and non-obstructive azoospermia were identified. Late germ cell–specific proteins ASPX_HUMAN and AKAP4_HUMAN were exclusively localized to spermatozoa acrosomes and tails, respectively. Imaging flow cytometry identified rare but morphologically intact AKAP4+/ASPX+/Hoechst+ spermatozoa in semen pellets of patients with non-obstructive azoospermia. AKAP4+/ASPX+/Hoechst+ may improve diagnostics and treatment of severe male infertility.
Infertility affects 15% of couples globally, with a male factor contributing to infertility in approximately 50% of the cases (1). The routine diagnostics of male factor infertility relies on semen analysis for the spermatozoa count, motility, and morphology and on infertility in the couple (2). Common categories of male infertility based on abnormal sperm parameters include oligozoospermia, asthenozoospermia, teratozoospermia, azoospermia, and a combination of morphological abnormalities with low sperm count and diminished motility diagnosed as oligoasthenoteratozoospermia. Azoospermia is further classified as obstructive azoospermia (OA) with no sperm in ejaculate due to obstruction despite normal spermatogenesis and non-obstructive azoospermia (NOA) with no sperm in ejaculate due to testicular failure (2). Men with elevated follicle-stimulating hormone (FSH) in blood serum and smaller testicles are more likely to have testicular failure resulting in NOA. However, men with normal FSH levels could have either NOA or OA (3), and a testicular biopsy is typically required to determine the presence and degree of spermatogenesis and distinguish between OA and NOA.
Breakthroughs in assisted reproductive technologies allowed fertility clinics to offer successful treatments to couples with male factor infertility, including its severe forms (1). Patients with mild oligozoospermia or idiopathic male infertility have sufficient numbers of spermatozoa in semen to be offered either intrauterine insemination, conventional in vitro fertilization (IVF), or intracytoplasmic sperm injection (ICSI). Patients with moderate oligoasthenoteratozoospermia could be managed with IVF or ICSI depending on sperm count and spermatozoa quality. NOA, the most severe form of male infertility, could only be treated with ICSI, providing spermatozoa were retrieved through microdissection testicular sperm extraction (mTESE). The mTESE procedure has a ∼50% success rate and can identify spermatozoa even in those cases when an initial diagnostic testicular biopsy revealed no spermatozoa (4, 5). It should be noted that testicular needle biopsies, partly due to the random sampling, fail to identify spermatozoa in ∼30% of NOA patients who had spermatozoa following mTESE (6). Thus, extensive microsurgical dissection of seminiferous tubules and their examination with an operating microscope is the most widely used technique to identify pockets of spermatozoa. Seminiferous tubules with spermatogenesis are excised, and the mTESE spermatozoa are used for ICSI (7). The mTESE procedure is laborious, and patients often spend more than 3 hours under general anesthesia. The heterogeneous etiology of male infertility, the low diagnostic sensitivity of FSH, the inaccuracy of the initial testicular biopsy diagnostics, and the laborious surgical retrieval of spermatozoa may result in different and unpredictable mTESE outcomes at different fertility clinics. A noninvasive diagnostic test to predict mTESE outcome constitutes a recognized clinical need. Likewise, a viable approach for the rapid identification of rare mature spermatozoa within dissected seminiferous tubules of NOA patients, or within semen cell debris of patients with severe oligoasthenoteratozoospermia, would be highly beneficial to IVF programs.
Numerous studies provided potential biomarkers of male infertility, including genomic (8, 9, 10) and epigenetic markers (11), RNA (12), metabolites (13), hormones (14), and proteins (15, 16, 17, 18, 19). Numerous parameters, including testis size, serum testosterone, FSH, inhibin B, and seminal proteins, were evaluated to predict mTESE sperm retrieval outcome (20, 21). Likewise, we recently developed a noninvasive test to differentiate between NOA and OA (22, 23). While those approaches revealed diagnostic sensitivity of 60 to 80%, they suffered from low specificity (24, 25, 26). Interestingly, low diagnostic specificity for mTESE retrieval of spermatozoa was determined even for some highly promising testis-specific proteins, likely due to their presence at the continuum of germ cell differentiation and abundance at the early and late stages of spermatogenesis (25).
In this study, we hypothesized that testis-specific proteins further categorized as germ cell–specific proteins and measured in seminal plasma (SP) could facilitate noninvasive and more specific prediction of mTESE outcomes. In addition, we suggested that testis-, germ cell–, and organelle-specific proteins would enable noninvasive and more specific identification of rare morphologically intact spermatozoa in the NOA semen cell pellets using multiplex imaging flow cytometry. The unique morphology of spermatozoa including acrosome-capped nuclei and elongated tails could be visualized with the aid of some testis-, germ cell– and organelle-specific markers and could provide unambiguous identification of spermatozoa with nearly no false-positives and high diagnostic specificity probably unachievable with soluble biomarkers in SP.
Our pipeline to identify the most promising candidate markers relied on data mining for the testis and germ cell–specific proteins, in-depth identification and quantification of the spermatozoa proteome, extensive validation of markers by the targeted proteomic assays, and selection of markers which could be visualized with the multiplex immunofluorescent microscopy and imaging flow cytometry assays.
Experimental Procedures
Experimental Design and Statistical Rationale
We hypothesized that testis- and germ cell–specific proteins measured in SP or semen pellet could facilitate noninvasive and more specific prediction of mTESE outcomes. Our study was designed as a biomarker discovery pipeline to evaluate numerous testis- and germ cell–specific proteins as novel biomarkers of NOA. Our specific goals included (i) selection of the most promising testis- and germ cell–specific proteins; (ii) development of targeted selected reaction monitoring (SRM) assays; (iii) retrospective evaluation of dozens of candidate proteins by SRM assays in large sets of patient samples, (iv) extensive validation of few top candidates with alternative assays, such as immunofluorescence and flow cytometry, and (v) development of a multiplex imaging flow cytometry assay for noninvasive identification of rare spermatozoa in semen of NOA patients. According to power calculations (supplemental Table S1; G∗Power v3.1.9.6, Heinrich Heine University Dusseldorf), 80% power to detect highly testis-specific proteins, such as TX101_HUMAN (22), could be achieved with 16 independent prevasectomy versus 16 postvasectomy, or OA, or NOA samples (α = 0.05, two-tailed t test; TX101_HUMAN levels and standard deviations from our previous study (22)). To be able to establish sensitivities and specificities of each protein with higher accuracy, we evaluated our panel of testis-specific proteins in larger sets of SP samples already available for our studies on male infertility (36 pre-vasectomy, 36 post-vasectomy, 19 OA, 38 NOA with histological subtypes, and 84 NOA with the known mTESE outcomes). A single analytical (full process digestion, sample preparation, and SRM analysis) and two technical (LC-SRM injections) replicates were analyzed by SRM for each SP sample. Single analytical replicates were justified by (i) the enormous decrease of concentrations of testis-specific proteins in post-vasectomy, OA, and NOA samples, as compared to pre-vasectomy (22); (ii) median analytical and day-to-day CVs of 10% and 18%, respectively, as we previously demonstrated for a panel of 25 proteins quantified in SP by SRM with the internal standards and the identical proteomic sample preparation protocols (36). GraphPad Prism (v5.03) was used to generate scatter plots, perform statistical analysis, and calculate areas under the receiver operating characteristic curve (AUCs), sensitivity, and specificity. A one-tailed Mann-Whitney U (MWU) test was used for pairwise comparisons between groups, and p-values <0.05 were considered significant.
Patients
Semen, spermatozoa, SP, and orchiectomy testicular tissues (supplemental Table S2) with relevant clinical information were collected with institutional REB-approved consent at Murray Koffler Urologic Wellness Centre at Sinai Health System (REB# 15–0149-A) and CReATe Fertility Centre (University of Toronto REB# 09–0156-E30252 and Sinai Health System REB# 14–0032-E). Men with normal spermatogenesis were confirmed fertile men with normal sperm count (>15 million per mL). These men were referred for vasectomy, and SP samples were obtained prior to vasectomy. The post-vasectomy group included SP samples obtained from fertile men 3 to 6 months after vasectomy, and zero sperm count was confirmed by at least two semen analyses. The OA group included men with biopsy-confirmed OA, obstruction at the epididymis, normal testicular volume, and normal FSH (1–18 IU/L). The NOA group included men with azoospermia by semen analysis and elevated FSH (>18 IU/L) or NOA confirmed by testicular biopsy. Y chromosome deletion status was not used as an independent parameter for NOA cases. Serial SP samples were obtained at a baseline and 1 to 3 months and prior to mTESE. Patients were included if they had medical indications for the mTESE and IVF/ICSI and had no contraindications. Female infertility factors were excluded. The presented study was based on the retrospectively collected semen and SP samples, followed the Standards for Reporting Diagnostic Accuracy Studies (STARD 2015) recommendations (supplemental Table S3), and abided by the Declaration of Helsinki principles.
Processing of Clinical Specimens
Semen samples from men with normal spermatogenesis prior vasectomy and post vasectomy, as well as men with azoospermia, were collected after a minimum of 3 days of sexual abstinence. For proteomic measurements, semen was left to liquefy at room temperature for 1 h, then aliquoted in 1 ml portions, centrifuged once at 800 g to pellet spermatozoa, and then centrifuged twice at 13,000 g to remove cellular debris and obtain SP devoid of cells and cellular components. Spermatozoa pellet and SP were stored in internal biobanks at −80 °C. To obtain motile spermatozoa, semen was incubated at 37 ºC for 15 min, followed by one-layer density-gradient centrifugation. The spermatozoa pellet was washed twice with PBS and either cryopreserved or fixed for immunofluorescence microscopy and flow cytometry. The samples were anonymous and labeled with biobank identification numbers. The research laboratory did not have any identifying patient information but was provided with the results of the standard semen analysis. Testicular sperm was isolated from testicular tissues following mechanical dissociation and digestion by collagenase (1.4 mg/ml; #GM501, Gynemed) diluted with HTF Hepes media (#2002, InVitroCare).
Development and Analytical Validation of SRM Assays
SRM assays were developed for the TSQ Quantiva mass spectrometer (Thermo Scientific), as previously reported (27, 28, 29, 30, 31, 32, 33, 34). Briefly, peptides were desalted with OMIX C18 pipette tip microextraction (10 μl; Agilent; A5700310). The EASY-nLC 1000 liquid chromatography (Thermo Scientific Inc.) was coupled to TSQ Quantiva triple-quadrupole mass spectrometer and a Nanospray Flex ion source (Thermo Scientific Inc.). Peptides (18 μl) were loaded at 5 μl/min onto a 3 cm trap column (150 μm ID; 5 μm C18, New Objective) and separated at 300 nl/min using a 15 cm analytical column (3 μm C18, 75 μm ID, 8 μm tip ID) and a 16 min gradient with buffers A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile): 0% to 40% of buffer B for 8 min, 40% to 65% for 2 min, 65% to 100% for 2 min, and 100% for 4 min. The SRM parameters included positive polarity, 150 V declustering, and 10 V entrance potentials, 300°C ion transfer tube temperature, optimized collision energy (CE) values, 5 to 60 ms scan time, 0.4 Q1 and 0.7 Q3 FWHM resolution, and 1.5 mTorr Q2 collision gas pressure.
Peptide Atlas (www.peptideatlas.org) was used to select the top 5 to 7 peptides (charge +2 and +3) for each candidate protein. Fully tryptic peptides with 7 to 20 amino acids were chosen, and peptides with methionine and N-terminal cysteine residues were avoided, if possible. Peptide uniqueness was confirmed with the Protein BLAST search against the human UniProt database (https://blast.ncbi.nlm.nih.gov). A list of peptides and the top 10 transitions were downloaded. Previous shotgun MS data for spermatozoa and SP proteomes was used to confirm the most abundant peptides (35, 36). Following that, 58 synthetic heavy isotope-labeled SpikeTides_L peptides (JPT Peptide Technologies GmbH) were obtained (1–3 peptides per protein) and used as internal standards for the Tier 2 SRM assay development, selection of the best precursor-to-fragment ion transitions, and protein quantification. The amounts of synthetic heavy isotope-labeled peptides were provided by the manufacturer. Lyophilized peptides were reconstituted in 15% acetonitrile, diluted in water with 0.4 M L-methionine, aliquoted, and frozen. Substantial excess of L-methionine prevented oxidation of methionine-containing peptides during storage and analysis, as we previously demonstrated (37). An aliquot of each peptide was used to prepare a master mix of heavy peptide standards. Additional precautions were taken to ensure peptide stability during storage and analysis: (i) supplementation of the protein digests with 0.4 M methionine, (ii) storage of tryptic peptides at −20 °C until use; and (iii) sealing of 96-well plates with silicone rubber mats and maintaining 7 °C in autosampler during SRM analysis.
Survey multiplex SRM methods with 10 to 15 peptides, +2 and/or +3 charge states, up to 10 transitions per peptide, and 5 ms scan times were designed in Skyline Targeted Proteomics Environment v20.1.0 (MacCoss Lab) and tested with heavy peptides. Raw files were uploaded to Skyline, and peaks were analyzed manually. Low-intensity transitions, transitions with interferences in SP, and poorly performing peptides were removed. SRM methods were redesigned, and the top 3 to 6 light and heavy transitions were retested in the digest of prevasectomy SP and spermatozoa lysate. High-abundance peptides with clear peaks, high signal-to-noise intensities, and multiple overlapping transitions were selected, while medium- and low-abundance peptides were retested with higher sensitivity assays (30–40 ms scans per transition). CE for each precursor peptide was retested (±2 V from the predicted CE). Optimized CE for each peptide was selected based on the highest peak area observed. The final multiplex SRM assay for 18 proteins included one peptide per protein, three unique transitions, 2.4 min scheduling intervals, and 10 to 60 ms scan times (∼15 points per peak).
SRM assay limits of detection (LODs) were determined as we previously described (36). Briefly, serial dilutions of heavy peptides were spiked into a digest of a pre-vasectomy SP sample with high and constant levels of the endogenous “light” peptides. LODs (Table 1) were calculated using the SP volume, UniProt molecular mass, and the lowest amounts of each heavy peptide detectable with the signal-to-noise ratio ≥3 and light-to-heavy ratio within the linear range (supplemental Fig. S1). The nanoLC-MS system suitability for analysis was assessed every 10 runs with the BSA digest (10 μl of 10 fmol/μl). Well-to-well carryover was estimated in the range 0.05 to 0.2%.
Table 1.
Diagnostic performance of 18 testis germ cell–specific proteins measured by SRM in SP
| Protein | Gene | The continuum of germ cell differentiation | LOD, μg/ml | PreV, μg/ml | PreV CV, % | PreV/postV FC | PreV/postV p-value | PreV/postV AUC | PreV/OA AUC | PreV/NOA AUC |
|---|---|---|---|---|---|---|---|---|---|---|
| ASPX_HUMAN | ACRV1 | rST → eST | 0.4 | 34 | 1.9 | 61 | 2 × 10−10 | 1.000 | 1.000 | 1.000 |
| ACRBP_HUMAN | ACRBP | SG → eST | 21 | 507 | 3.3 | 17 | 2 × 10−8 | 0.972 | 0.972 | 0.970 |
| DPEP3_HUMAN | DPEP3 | 1′SC→ eST | 1.1 | 24 | 2.3 | 16 | 2 × 10−10 | 1.000 | 1.000 | 0.955 |
| TX101_HUMAN | TEX101 | 2′SC→ eST | 1.1 | 20 | 2.4 | 13 | 2 × 10−10 | 1.000 | 1.000 | 0.954 |
| ACRO_HUMAN | ACR | 2′SC→ eST | 0.6 | 10 | 3.5 | 12 | 2 × 10−8 | 0.972 | 0.972 | 0.972 |
| LYZL5_HUMAN | SPACA5 | rST → eST | 0.2 | 2.0 | 3.5 | 7.6 | 4 × 10−7 | 0.944 | 0.944 | 0.944 |
| CRIS2_HUMAN | CRISP2 | SG → eST | 0.1 | 5.6 | 1.5 | 4.9 | 8 × 10−7 | 0.935 | 0.953 | 0.946 |
| SACA3_HUMAN | SPACA3 | rST → eST | 0.3 | 1.5 | 3.4 | 4.2 | 3 × 10−6 | 0.917 | 0.917 | 0.917 |
| LYZL4_HUMAN | LYZL4 | rST → eST | 0.8 | 3.5 | 2.4 | 3.3 | 2 × 10−5 | 0.889 | 0.889 | 0.882 |
| LY6K_HUMAN | LY6K | 2′SC→ eST | 0.2 | 1.1 | 13 | 3.2 | 4 × 10−7 | 0.944 | 0.944 | 0.924 |
| GTR14_HUMAN | SLC2A14 | 2′SC→ eST | 1.5 | 4.4 | 11 | 2.2 | 4 × 10−4 | 0.833 | 0.833 | 0.833 |
| MENT_HUMAN | MENT | rST → eST | 0.2 | 3.4 | 2.7 | 1.7 | 4 × 10−2 | 0.701 | 0.836 | 0.803 |
| ZPBP1_HUMAN | ZPBP | rST → eST | 0.8 | 1.9 | 1.8 | 1.7 | 1 × 10−3 | 0.806 | 0.806 | 0.806 |
| ADA30_HUMAN | ADAM30 | 2′SC→ eST | 0.3 | 0.47 | 9.5 | 1.3 | 2 × 10−3 | 0.806 | 0.806 | 0.806 |
| SACA4_HUMAN | SPACA4 | rST → eST | 0.1 | 0.1 | 12 | 1.3 | 1 × 10−3 | 0.806 | 0.806 | 0.806 |
| PRS55_HUMAN | PRSS55 | rST → eST | 0.4 | 0.65 | 15 | 1.3 | 1 × 10−3 | 0.750 | 0.750 | 0.750 |
| PRND_HUMAN | PRND | SG → eST | 0.7 | 1.1 | 5.8 | 1.1 | 4 × 10−3 | 0.778 | 0.778 | 0.778 |
| ADA18_HUMAN | ADAM18 | 1′SC→ eST | 0.2 | 0.21 | 16 | 1.0 | 5 × 10−2 | 0.694 | 0.694 | 0.694 |
| ESPB1_HUMAN | ELSPBP1 | Epididymis | 0.1 | 69 | 1.3 | 52 | 1 × 10−8 | 0.978 | 0.927 | 0.761 |
| CRIS1_HUMAN | CRISP1 | Epididymis | 0.4 | 231 | 0.8 | 9.0 | 5 × 10−7 | 0.941 | 0.871 | 0.714 |
The Human Protein Atlas immunohistochemistry data provided the continuum of germ cell specificity (spermatogonia to elongated spermatids, excluding mature spermatozoa). Epididymis-specific proteins were measured as controls. SP cohorts included normozoospermic pre-vasectomy (PreV; N = 18), unmatched post-vasectomy (PostV; N = 18), OA (N = 19), and NOA (N = 38) samples. SRM assay LODs, median levels of proteins in PreV, and median technical CVs in PreV samples are provided. The levels below LOD were adjusted to LOD to enable the calculation of the fold changes of median concentrations (FC), MWU p-values, and ROC AUCs.
2′SC, pachytene spermatocytes; 1′SC, preleptotene spermatocytess; eST, elongated spermatids; rST, round spermatid; SG, spermatogonia.
SRM Quantification of Testis-Specific Proteins in Clinical Specimens
Semen, spermatozoa, and SP were lysed with Rapigest SF surfactant (35, 38). Proteins were denatured at 60 °C, reduced, alkylated, and digested with sequencing-grade modified trypsin (Promega). TX101_HUMAN and DPEP3_HUMAN internal standards with trypsin-cleavable tags were added before trypsin digestion, while the rest of the heavy isotope-labeled peptides were spiked after digestion. Each digest was subjected to microextraction with 10 μl OMIX C18 pipette tips (Agilent) and analyzed with TSQ Quantiva triple-quadrupole mass spectrometer as described above. A single analytical (full process digestion and sample preparation) and two technical replicates were analyzed for each SP sample. Raw SRM files were analyzed with Skyline. SRM area was used to calculate L/H ratios, protein concentrations, and standard deviations. Peak integration boundaries were adjusted manually, and the integrated areas of all transitions were extracted. To mitigate the well-to-well carry-over, a restricted randomized block design was used for sample randomization. SP samples were split into pre-vasectomy, post-vasectomy, OA, or NOA blocks of 4 × 2 samples, and blocks were randomized on 96-well plates for sample preparation and SRM. Data analysis of patient samples was retrospective and nonblinded for the clinical outcomes.
Immunofluorescence Microscopy
Approximately 100 spermatozoa per slide were fixed with cytology fixative (Andwin Scientific; 930010C12), permeabilized with 70% ethanol, and mounted onto microscope slides for immunofluorescence staining. Mouse monoclonal antibodies included anti-ACRV1 (1 μg/ml final; Sinobiological; 11789-MM01), anti-ADAM20 (10 μg/ml final; Abnova; H00008748-M05), and anti-ADAM29 (10 μg/ml final; Abnova; H00011086-M09). Rabbit polyclonal and the corresponding secondary antibodies included anti-AKAP4 (5 μg/ml final; Thermo Fisher; PA5-38015), Alexa Fluor 594 goat-anti-mouse (2 μg/ml final; Thermo Fisher; A-21125), and Alexa Fluor 488 goat-anti-rabbit (1 μg/ml final; Thermo Fisher; A-11034). High-magnification fluorescence microscopy (100 × ; Olympus BX61) was used to visualize protein markers and DAPI staining of nuclei (SlowFade mountant S36968, Thermo Fisher), as previously described (35).
Flow Cytometry
Whole semen and washed motile spermatozoa were analyzed for ASPX_HUMAN and AKAP4_HUMAN levels using flow cytometry (MACSQuant Analyzer 10; Miltenyi Biotec) and FlowJo v10 data analysis software (BD Life Sciences).
Imaging Flow Cytometry
Imaging flow cytometry (Amnis ImageStreamX Mark II, Luminex) was used to evaluate the performance of tail- and acrosome-specific proteins and detect rare spermatozoa in semen pellets of azoospermic and oligozoospermic patients. Semen cell pellets were washed with PBS and stained with Hoechst 33342 and primary antibodies (mouse anti-ASPX_HUMAN #11789-MM01 and rabbit anti-AKAP4 #PA5-38015), followed by the secondary antibodies labeled with Alexa Fluor 594 and 488, respectively. ImageStream imaging flow cytometry (brightfield and side-scatter imaging, and fluorescence excitation with 405, 488, and 592 nm lasers) provided up to 100,000 images per run, with up to 14 individual runs per NOA semen pellets. Image analysis was performed with IDEAS v6.2 software (Luminex). Analysis of normozoospermic and oligozoospermic samples facilitated the development of a sequential gating template based on the brightfield area and aspect ratio, AKAP4 and Hoechst fluorescence intensity, followed by AKAP4 aspect ratio intensity sorting and visual inspection of the combined AKAP4+/ASPX+/Hoechst+ images. Such a multistep gating strategy facilitated the manual inspection of thousands of images and rapid identification of the morphologically intact AKAP4+/ASPX+/Hoechst+ spermatozoa.
Results
Data Mining and Identification of Testis- and Germ Cell–Specific Proteins
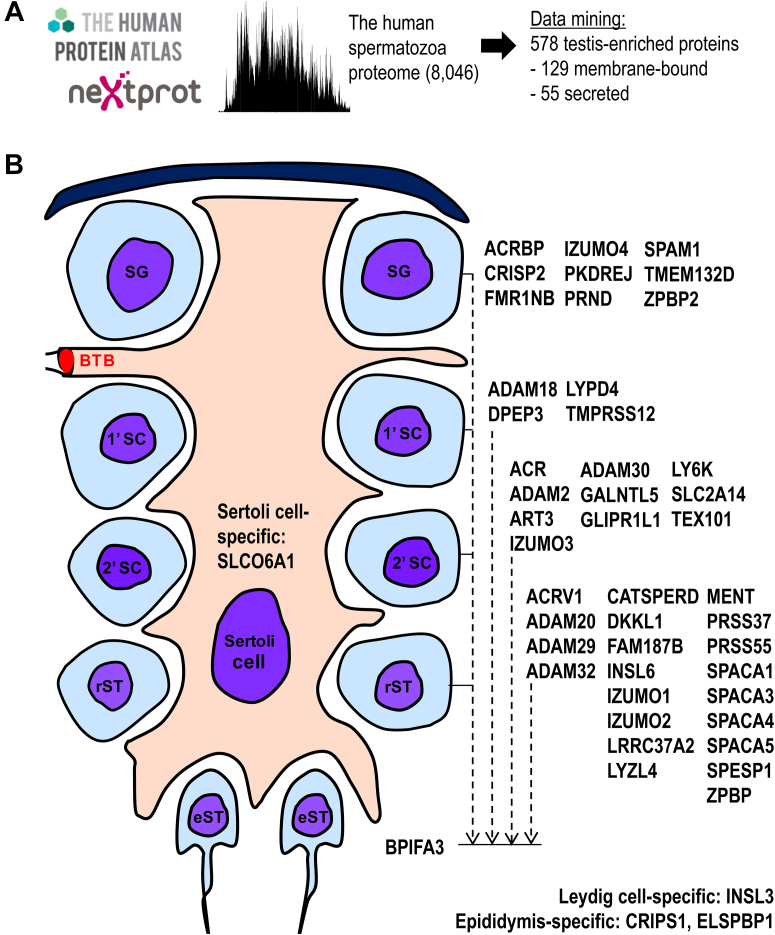
We previously identified one of the largest sets of proteins detectable by mass spectrometry in the human spermatozoa (8,046 proteins) (35) and SP (3,141 proteins) (36, 39, 40). Here, the spermatozoa proteome was searched across the Human Protein Atlas data (41) and revealed 578 testis-specific proteins (Fig. 1A and supplemental Table S4). NextProt data (42) further prioritized 129 proteins with at least one extracellular membrane-bound isoform and 55 proteins with at least one secreted isoform. We assumed that some proteins predicted as membrane-bound could have noncanonical secreted or membrane-shed isoforms, and vice versa. For instance, a secreted acrosomal protein ASPX_HUMAN, the product of ACRV1 gene with 11 different transcript isoforms, was previously found as a soluble protein in the acrosomal matrix and as a protein bound to the acrosomal membrane (43). Human Protein Atlas immunohistochemistry data for the membrane-bound and secreted proteins were manually examined to select 45 proteins expressed at the different stages of spermatogenesis (Fig. 1B and supplemental Table S5). Finally, the Antibodypedia portal (44) was searched for immunoassays and antibodies suitable for the development of validation assays but revealed that high-quality antibodies were not available for the majority of our candidate proteins. Thus, we proposed targeted proteomic assays as tools to experimentally validate 45 proteins in human spermatozoa, vas deferens fluid, and SP, and select the most promising candidates.
Fig. 1.
Data mining to select testis germ cell–specific proteins.A, the deep proteome of the human spermatozoa (35), Human Protein Atlas, and NextProt data were used to identify and classify testis-enriched, membrane-bound, and secreted genes and proteins. B, Human Protein Atlas immunohistochemistry data were manually examined to select genes and proteins expressed at the continuum of germ cell differentiation. Human gene names are presented as identifiers. 1′SC, preleptotene spermatocytes; 2′SC, pachytene spermatocytes; BTB, blood–testis barrier; eST, elongated spermatids; rST, round spermatids; SG, spermatogonia.
Development of Targeted Proteomic Assays
We previously demonstrated applications of targeted proteomic assays for the quantification of proteins in various biological fluids and clinical samples (45, 46, 47, 48). To develop SRM assays, we re-searched our shotgun mass spectrometry and label-free quantification data with protein abundance in human spermatozoa and SP (35, 36) and selected the unique and high-intensity proteotypic peptides. The most promising proteotypic peptides (1–3 per protein) were synthesized as heavy isotope-labeled peptides and used as internal standards for the development of SRM assays (supplemental Tables S6 and S7).
Quantification of Testis-Specific Proteins in Spermatozoa, Vas Deferens Fluid, and SP
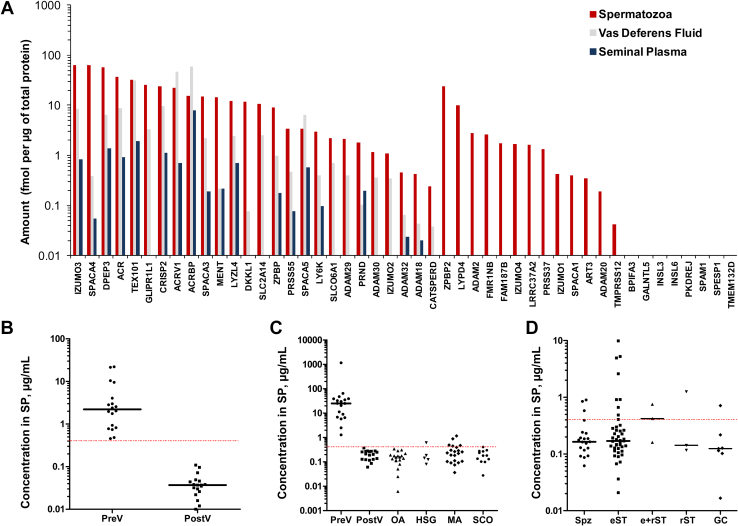
We hypothesized that the most promising markers to predict mTESE outcome would be the most abundant testis-specific proteins reproducibly measured in SP, vas deferens fluid, and spermatozoa lysate. We quantified by SRM the abundance of 45 germ cell–specific proteins in normozoospermic spermatozoa, SP and vas deferens fluid samples (Fig. 2A) and selected 18 most promising proteins (Table 1). We further optimized a multiplex SRM assay (supplemental Table S8), determined protein LODs (Table 1 and supplemental Fig. S1), and measured the levels of 18 testis-specific proteins in matched prevasectomy and postvasectomy SP samples (supplemental Fig. S2). As a result, levels of 12 proteins decreased below the SRM assay LOD, suggesting their high testis/epididymis tissue specificity, and excluding seminal vesicle- and prostate-expressed proteins. Finally, 18 proteins were remeasured in a larger independent set of SP samples including unmatched pre-vasectomy and post-vasectomy, OA, and NOA with the known histological subtypes of hypospermatogenesis, maturation arrest, and Sertoli cell-only syndrome (supplemental Fig. S3). Interestingly, we rediscovered TX101_HUMAN and DPEP3_HUMAN proteins reported in our previous studies (22, 49) and reconfirmed their high diagnostic performance to noninvasively detect post-vasectomy and OA with AUC = 1.0. In addition, we identified ASPX_HUMAN (acrosomal protein SP-10; ACRV1 gene) whose levels decreased 61-fold in post-vasectomy SP and provided AUC = 1.0 at its LOD of 0.4 μg/ml. ASPX_HUMAN was the most promising soluble marker for the noninvasive diagnosis of OA and NOA (Table 1 and Fig. 2, B and C). It should also be noted that two epididymis-specific proteins ESPB1_HUMAN and CRIS1_HUMAN emerged as novel markers to differentiate between NOA and OA/PV. ESPB1_HUMAN (AUC = 0.72, MWU p = 8 × 10−4) and CRIS1_HUMAN (AUC = 0.73, MWU p = 6 × 10−4) could be useful to complement ECM1_HUMAN, our previously discovered marker for the differential diagnosis of azoospermia (22, 25).
Fig. 2.
Levels of testis germ cell–specific proteins in human spermatozoa, vas deferens fluid, and SP measured by SRM assays.A, 45 germ cell–specific proteins were measured in a pool of spermatozoa lysates and matched SP from the fertile normozoospermic men, as well as unmatched vas deferens fluid samples collected during vasectomy. A Sertoli cell–specific SO6A1_HUMAN protein (SLCO6A1 gene) and Leydig cell–specific INSL3_HUMAN protein (INSL3 gene) were measured as controls. Human gene names are presented as identifiers. B, levels of ASPX_HUMAN protein (ACRV1 gene) decreased below the SRM assay LOD (dashed line) in matched post-vasectomy SP (PostV; N = 18), as compared to pre-vasectomy SP (PreV; N = 18) and suggested the high testis specificity of ASPX_HUMAN. C, ASPX_HUMAN levels measured in the independent SP sets of unmatched pre-vasectomy (N = 18), unmatched post-vasectomy (N = 18), OA (N = 19), as well as NOA samples with the histological subtypes of hypospermatogenesis (HGS; N = 5), maturation arrest (MA; N = 21) and Sertoli cell-only (SCO; N = 12). Interestingly, ASPX_HUMAN was detected in some HSG and MA samples. D, ASPX_HUMAN levels measured in the independent SP set of NOA patients (N = 81) with the confirmed mTESE outcomes. ASPX_HUMAN was detected in some mTESE spermatozoa (Spz)- and elongated spermatid (eST)-positive SP samples. The performance of other germ cell–specific proteins is presented in supplemental Figs. S2–S4. e+rST, elongated+round spermatids; GC, germ cells; rST, round spermatids.
Prediction of mTESE Outcomes Based On the SP Levels of 18 Proteins
We measured 18 proteins in 84 serial SP samples (obtained at baseline and at 1–3 months) of 27 NOA patients with the known mTESE outcomes (Figs. 2D and S4). Extensive data analysis revealed insufficient analytical sensitivity of SRM assays in NOA SP samples and poor diagnostic sensitivity to predict spermatozoa or elongated spermatids on mTESE. At assay LOD levels, only a few proteins (ASPX_HUMAN, ADA18_HUMAN, DPEP3_HUMAN, PRND_HUMAN, and TX101_HUMAN) revealed sensitivity of ∼10 to 40%. Even though the more sensitive SRM or immunoprecipitation-SRM assays could be a viable approach to validate this panel of soluble testis-specific proteins, as we previously demonstrated for TX101_HUMAN (22), we decided to explore alternative approaches.
Selection of Organelle-Specific Proteins
We previously evaluated the abundance levels and localization of testis-specific proteins TX101_HUMAN, DPEP3_HUMAN, and LY6K_HUMAN in mature spermatozoa (35, 49) and revealed their cell surface localization within the postacrosomal region. In this study, we selected two additional proteins ADA20_HUMAN (ADAM20 gene) and ADA29_HUMAN (ADAM29 gene) due to their elevated levels at the late-stage germ cells (supplemental Table S5), their similarity to the sperm-egg binding fertilin beta ADAM2_HUMAN (50), and availability of antibodies. Immunofluorescent microscopy revealed cell surface localization of ADA20_HUMAN and ADA29_HUMAN within the postacrosomal region of spermatozoa (supplemental Fig. S5). Finally, we realized that proteins localized within the postacrosomal region could hardly distinguish spermatozoa heads from the round spermatids within the complex cell mixtures.
We then suggested that the best markers to identify morphologically intact spermatozoa in debris-laden NOA semen pellets would include acrosome- and tail-specific proteins. While some markers, such as ASPX_HUMAN, were the acrosome-localized proteins, there was no indication that any of the selected 45 proteins (supplemental Table S5) could be localized within the spermatozoa tails. Thus, we researched our initial list of testis-specific proteins (supplemental Table S4), the Human Protein Atlas, and shotgun mass spectrometry data to review the previously excluded intracellular proteins. Interestingly, a testis- and spermatid-specific intracellular protein AKAP4_HUMAN (A-kinase anchoring protein 4) emerged as one of the most abundant human spermatozoa proteins. AKAP4 has previously been identified as a tail-localized protein in mouse spermatozoa (51).
As a result, we suggested that simultaneous detection of spermatozoa acrosomes with ASPX_HUMAN, tails with AKAP4_HUMAN, and nuclei with the DNA-staining DAPI or Hoechst dyes could reveal morphologically intact and mature spermatozoa within the highly heterogenous debris-laden NOA semen pellets and homogenized testicular tissues. Likewise, the identification of triple-positive AKAP4+/ASPX+/Hoechst+ cells of elongated shapes would distinguish mature spermatozoa from germ cells and spermatids.
Subcellular Localization of AKAP4_HUMAN and ASPX_HUMAN Proteins in Spermatozoa
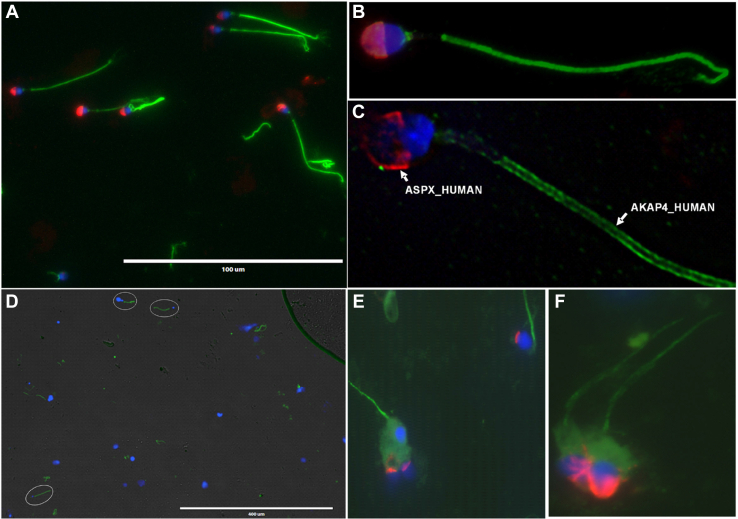
To develop multiplex immunofluorescence microscopy assays, we selected anti-ASPX_HUMAN mouse monoclonal and anti-AKAP4_HUMAN rabbit polyclonal antibodies, as well as secondary antibodies labeled with Alexa 594 and Alexa 488, respectively. We confirmed the localization of ASPX_HUMAN within acrosomes and ASPX_HUMAN within tails of the motile normozoospermic spermatozoa (Fig. 3, A–C), as well as spermatozoa present in the homogenized orchiectomy testicular tissues (Fig. 3, D–F).
Fig. 3.
Immunofluorescent microscopy analysis of ASPX_HUMAN and AKAP4_HUMAN proteins in motile spermatozoa and testicular tissues.A–C, fluorescence imaging of AKAP4_HUMAN (green), ASPX_HUMAN (red), and cell nuclei (blue) in motile normozoospermic spermatozoa at 400× (A) and 1000× magnification (B). AKAP4_HUMAN and ASPX_HUMAN protein localization in motile spermatozoa visualized and deconvoluted with Huygens software (C). D–F, testicular spermatozoa visualized in orchiectomy testicular tissue homogenate at 100× (D) and 1000× (E and F) magnification.
Flow Cytometry Analysis of AKAP4_HUMAN and ASPX_HUMAN in Spermatozoa
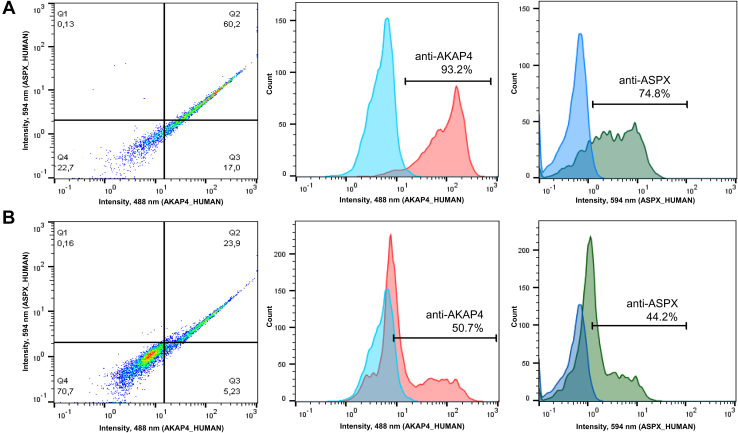
Following immunofluorescence microscopy analysis, we developed and optimized flow cytometry and imaging flow cytometry assays for ASPX_HUMAN and AKAP4_HUMAN proteins. We discovered that the permeabilization of spermatozoa with 70% ethanol substantially improved flow cytometry analysis. Flow cytometry (Fig. 4) and imaging flow cytometry (Figs. 5 and S6) revealed the simultaneous presence of high levels of ASPX_HUMAN and AKAP4_HUMAN proteins in >70% of motile normozoospermic spermatozoa.
Fig. 4.
Flow cytometry analysis of AKAP4+and ASPX+spermatozoa.A and B, normozoospermic whole semen pellets were washed, and spermatozoa were treated with 70% ethanol (A) or PBS (B) prior to staining with anti-AKAP4 and anti-ASPX primary antibodies and the corresponding secondary antibodies. Ethanol permeabilization significantly increased the fraction of AKAP4+/ASPX+ spermatozoa.
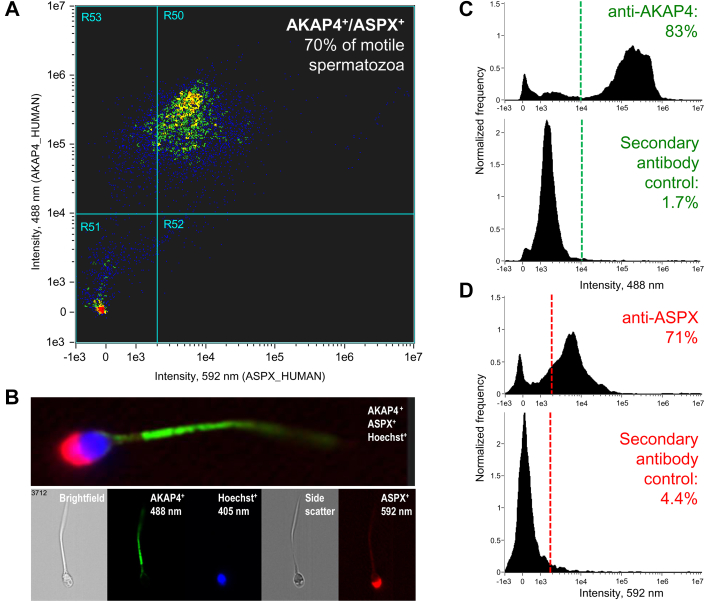
Fig. 5.
Imaging flow cytometry analysis of AKAP4+/ASPX+/Hoechst+spermatozoa.A, 70% of motile spermatozoa permeabilized with ethanol were AKAP4+/ASPX+ positive. B, fluorescence imaging of motile spermatozoa with AKAP4 (green; Alexa 488), ASPX (red; Alexa 594), and nuclear (blue; Hoechst) staining. C and D, staining for AKAP4 (83% positive) and ASPX (71% positive) in motile spermatozoa versus secondary antibody-only controls.
Identification of Rare Intact AKAP4+/ASPX+/Hoechst+ Spermatozoa in NOA Semen Pellet
Analysis of normozoospermic and oligozoospermic semen pellet samples by imaging flow cytometry (supplemental Figs. S6-7) facilitated the development of a multistep gating strategy based on spermatozoa brightfield area and aspect ratio, AKAP4, and Hoechst fluorescence intensity, AKAP4 aspect ratio intensity sorting, and rapid visual inspection of the combined AKAP4+/ASPX+/Hoechst+ images. The combined approach facilitated the analysis of millions of single-cell images, straightforward visualization, and rapid identification of the rare morphologically intact triple-positive AKAP4+/ASPX+/Hoechst+ spermatozoa (as few as 1 spermatozoon in ∼100,000 ImageStream images) even within the debris-laden NOA semen pellets (supplemental Fig. S8).
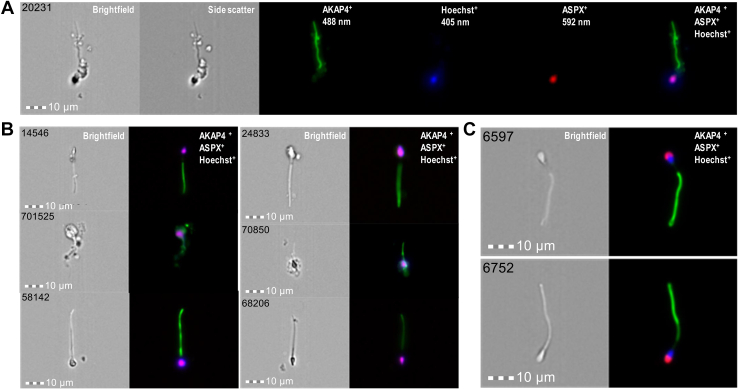
Semen pellets of 30 patients (Table 2) were subjected to the exhaustive imaging flow cytometry analysis for AKAP4+/ASPX+/Hoechst+ cells of elongated shapes. As a result, numerous AKAP4+/ASPX+/Hoechst+ spermatozoa were identified in semen pellets of normozoospermic and oligozoospermic samples (supplemental Figs. S6-7). In addition, we identified several AKAP4+/ASPX+/Hoechst+ triple-positive morphologically intact spermatozoa in semen pellets of NOA patients with no spermatozoa identified at the routine laboratory semen analysis (Figs. 6 and S8). Our NOA-mTESE reference set of seven patients with the known mTESE outcomes provided 83% diagnostic sensitivity to noninvasively detect intact spermatozoa in NOA semen pellets. Finally, we evaluated the performance of AKAP4/ASPX/Hoechst imaging flow cytometry with an independent set of azoospermia samples with unknown mTESE outcomes. As a result, we identified numerous morphologically intact spermatozoa in the semen pellets of three patients (Table 2). Collectively, these results revealed that our multistep gating strategy and imaging flow cytometry visualization of AKAP4+/ASPX+/Hoechst+ cells with elongated tails and acrosome-capped nuclei could enable fast and unambiguous identification of rare morphologically intact spermatozoa in NOA semen pellet samples.
Table 2.
Imaging flow cytometry identification of morphologically intact AKAP4+/ASPX+/Hoechst+ spermatozoa in NOA semen pellet samples
| Reference NOA-mTESE set (Murray Koffler Urologic Wellness Centre) | |||
|---|---|---|---|
| Patient ID | mTESE outcome | TX101_HUMAN in SP, ng/ml (26) | AKAP4+/ASPX+/Hoechst+ count |
| Patient #88 | Spermatozoa | 0.5 | 11 spermatozoa |
| Patient #117 | Spermatozoa | 14.2 | 2 spermatozoa |
| Patient #55 | Spermatozoa | 0.5 | 1 spermatozoon |
| Patient #85 | Spermatozoa | <0.2 | 1 spermatozoon |
| Patient #93 | Spermatozoa | 504 | 1 spermatozoon |
| Patient #60 | Spermatozoa | <0.2 | No spermatozoa |
| Patient #29 | No spermatozoa | <0.2 | No spermatozoa |
|
Additional NOA set (CReATe Fertility Centre) | |||
|---|---|---|---|
| Patient ID | Patient group | Andrology lab count | AKAP4+/ASPX+/Hoechst+count |
| Patient #1 | NOA | No spermatozoa | 77 spermatozoa |
| Patient #2 | NOA | No spermatozoa | 15 spermatozoa |
| Patient #3 | NOA | No spermatozoa | 10 spermatozoa |
| Patient #4 | NOA | No spermatozoa | 2 spermatozoa |
| Patients #5–13 | NOA | No spermatozoa | No spermatozoa |
| Patients #14–20 | Azoospermiaa | No spermatozoa | No spermatozoa |
| Patient #21 | OA | No spermatozoa | No spermatozoa |
| Patient #22 | Oligozoospermia | 1.6 million/mL | >3000 spermatozoa |
| Patient #23 | Normozoospermia | 15 million/mL | Numerous spermatozoa |
The reference NOA-mTESE set provided 83% sensitivity to noninvasively detect spermatozoa in NOA patients.
Unknown NOA or OA subtype.
Fig. 6.
Imaging flow cytometry identification and visualization of morphologically intact AKAP4+/ASPX+/Hoechst+spermatozoa in semen pellets.A and B, a total of 1,386,670 ImageStream images were recorded during exhaustive 14 runs of a semen pellet sample of NOA patient #88, and 11 morphologically intact AKAP4+/ASPX+/Hoechst+ spermatozoa were identified. C, a single run of 21,375 images of a semen pellet sample of oligozoospermic patient #22 revealed numerous AKAP4+/ASPX+/Hoechst+ spermatozoa. Additional images are presented in supplemental Figs. S6–S8. NOA, nonobstructive azoospermia.
Discussion
The Human Protein Atlas reported that nearly 4.5% of all human protein-coding genes were testis-enriched, with at least four-fold higher mRNA level in testis compared to any other human tissue (41). The unique and highly specialized roles of testis-specific proteins in spermatogenesis, such as remodeling of the spermatozoa cell surface, sperm transit, sperm-oocyte interaction, and fusion were discovered (52). Prominent examples included metalloprotease-disintegrin ADAM2 (53), the cell adhesion tetraspanin CD9 (54), and the sperm-egg fusion protein IZUMO1 (55). The discovery of the cell surface recognition complex of IZUMO1 protein and the sperm–egg fusion protein JUNO provided detailed insights into gamete recognition and sperm-oocyte fusion (56, 57).
In this study, we hypothesized that some testis- and germ cell–specific proteins visualized with fluorescent microscopy could emerge as markers for the noninvasive and highly specific identification of rare spermatozoa within the debris-laden semen pellets of NOA patients. Previous studies on the identification of rare intact spermatozoa in NOA semen pellets were often inconsistent due to difficulties with the correct differentiation of intact spermatozoa from debris (2). Our approach to the selection of the most promising testis-, germ cell– and organelle-specific proteins relied on reanalysis of our previously identified spermatozoa (35) and SP proteomes (36), and on mining of the well-established proteomic databases. Since high-quality protein assays are essential for biomarker discovery and development (58, 59, 60), we suggested that targeted proteomic assays due to their rapid design, development, and execution could be indispensable tools to prioritize candidate proteins and evaluate the clinical utility of proteins lacking high-quality immunoassays or antibodies. We previously demonstrated that targeted proteomic assays provided robust quantification of proteins in human cell lines (37, 38), primary cells (61, 62), tissues (45), biological fluids (27, 28, 29, 34, 63), and serum (64, 65, 66). Immunofluorescence microscopy and flow cytometry assays would independently confirm subcellular localization and abundance levels of the most promising candidates.
We previously identified and validated numerous testis-specific proteins as biomarkers for the differential diagnosis of azoospermia (22, 23). The combination of testis- and germ cell–specific protein TX101_HUMAN and an epididymis-specific protein ECM1_HUMAN provided 81% sensitivity at 100% specificity to differentiate between nonobstructive and obstructive azoospermia (25). Our initial study (25) revealed the moderate performance of TX101_HUMAN to predict spermatozoa retrieval by mTESE (73% sensitivity and 64% specificity at the assay limit of detection ≥0.6 ng/ml). A follow-up study (26) with an improved immunoassay revealed 87% sensitivity and 53% specificity at ≥0.2 ng/ml to extract mature spermatozoa.
Our success with azoospermia biomarkers motivated us to extend our search for additional testis- and germ cell–specific proteins and to identify more specific and sensitive biomarkers for the prediction of mTESE outcomes. Integration of our experimental proteomic data with mining of the Human Protein Atlas (41) and NextProt (42) data revealed that spermatozoa expressed 578 testis-specific proteins, of which 129 and 55 proteins had at least a single membrane-bound or secreted isoform, respectively. Our shotgun mass spectrometry data on protein abundance and tryptic peptide fragmentation enabled the prioritization of candidate proteins and the selection of proteotypic peptides suitable for the development of targeted proteomic assays.
Experimental verification with SRM assays excluded poor markers, confirmed our previous findings for TX101_HUMAN and DPEP3_HUMAN, and revealed ASPX_HUMAN as an additional marker with AUC = 1.0 to detect post-vasectomy or OA. ASPX_HUMAN emerged as the most promising marker for the subsequent validation due to (i) late germ cell-specificity and subcellular localization in the acrosomal versus postacrosomal regions of spermatozoa; (ii) detection in some SP samples of NOA with maturation arrest (Fig. 2C), and SP samples of an mTESE spermatozoa/elongated spermatid group (Fig. 2D); (iii) availability of high-quality antibodies to detect the native nondenatured ASPX_HUMAN. Likewise, a tail-specific AKAP4_HUMAN was readily detected by immunofluorescence microscopy and flow cytometry assays.
It should be emphasized that previous studies on the soluble mTESE biomarkers in SP, including our studies (25, 26), revealed relatively high diagnostic sensitivity but suffered from low diagnostic specificity to predict spermatozoa retrieval. Low diagnostic specificity was found even for the highly testis-specific proteins measured in SP (25), likely due to their presence at the continuum of germ cell differentiation, including early and late germ cells (Table 1). Unlike measurements of soluble markers in SP, AKAP4+/ASPX+/Hoechst+ imaging flow cytometry provided additional visualization of the unique morphology of spermatozoa including acrosome-capped nuclei and elongated tails. Manual inspection of AKAP4+/ASPX+/Hoechst+ cells provided nearly unambiguous identification of mature spermatozoa suggesting nearly no false positives and high diagnostic specificity previously unachievable with soluble markers in SP.
In the future, a combination of our less expensive TX101_HUMAN immunoassay in SP as an initial screening approach with 87% sensitivity (25, 26) and the more expensive AKAP4+/ASPX+/Hoechst+ assay with high specificity to follow-up on TX101_HUMAN positive result could emerge as a clinical strategy for noninvasive prediction of mTESE outcomes. However, independent prospective validation of such a combination and corresponding diagnostic cutoffs in large sets of samples will be required. As potential limitations, it should be noted that rare AKAP4+/ASPX+/Hoechst+ spermatozoa selected by imaging flow sorting approaches may not be suitable for ICSI procedures due to the use of ethanol permeabilization and due to the residual anti-AKAP4 and anti-ASPX antibodies bound to spermatozoa following imaging flow cytometry analysis. The presented approach for flow cytometry identification of the rare intact AKAP4+/ASPX+/Hoechst+ spermatozoa in the debris-laden testicular tissue homogenates may facilitate sorting of the testicular spermatozoa to enable their proteome profiling. Comparison of the testicular versus ejaculate spermatozoa may identify proteins contributed during sperm maturation in rete testis and epididymis.
Our work may represent one of the most comprehensive studies on testis- and germ cell–specific proteins. Since no immunoassays are currently available for most germ cell–specific proteins evaluated in our study, our SRM assays are first-of-its-kind analytical tools to measure 18 testis-specific proteins in semen, SP, vas deferens fluid, and spermatozoa. Some of these proteins could be essential for spermatozoa-oocyte interaction (67) and emerge as targets to develop nonhormonal male contraceptives (68) or emerge as noninvasive compound biomarkers to evaluate male contraceptives.
Conclusions
AKAP4_HUMAN and ASPX_HUMAN proteins emerged as the best combination of markers for the highly specific detection and visualization of the morphologically intact spermatozoa in NOA semen pellets using the multiplex imaging flow cytometry. Pending further validation, our assay may emerge as a noninvasive test to predict the retrieval of spermatozoa by mTESE, thus improving diagnostics and treatment of the severe forms of male infertility. The reported panel of testis- and germ cell–specific proteins will facilitate the evaluation of spermatozoa and acrosome integrity and enable future functional and translational studies on human reproduction and fertility.
Data Availability
Raw SRM data and processed Skyline files were deposited to Peptide Atlas (www.peptideatlas.org/PASS/PASS01755 or ftp://PASS01755:QE3725r@ftp.peptideatlas.org) and can be downloaded with FTP software (https://filezilla-project.org or etc.) using the host server name ftp.peptideatlas.org, password QE3725r, and username PASS01755.
Supplemental data
This article contains supplemental data (22, 35, 36, 49).
Conflict of interest
The authors declare no competing interest.
Acknowledgments
We thank Susan Lau (Sinai Health System) and CReATe andrology lab personnel for assistance with the clinical specimens. We thank the Advanced Center for Detection of Cancer mass spectrometry facility (Sinai Health System) and the Lunenfeld-Tanenbaum Research Institute Flow Cytometry facility for the cost-recovery mass spectrometry and imaging flow cytometry services.
Author contributions
A. P. D. methodology; A. P. D., J. Z., and M. K. investigation; A. P. D. and J. Z. formal analysis; K. J., A. G-F., S. I. M., and C. L. resources; A. P. D. writing-original draft; A. P. D., J. Z., M. K., K. J., A. G-F., S. I. M., and C. L. writing-review and editing; J. Z., S. I. M., A. G-F., and A. P. D. data curation; K. J. and A. P. D. funding acquisition; A. P. D. conceptualization; A. P. D. validation; A. P. D. visualization; A. P. D. supervision; A. P. D. project administration.
Funding and additional information
This work was supported by the Canadian Institutes of Health Research Proof of Principle Program – PoP (#303100) and Phase I (#355146) grants to K. J. and A. P. D., Physicians Services Incorporated (PSI) Foundation Health Research grant to K. J., Prostate Cancer Canada Movember Rising Star to A. P. D., and the University of Alberta startup funds to A. P. D.
Supplementary Data
References
- 1.Jarvi K., Lo K., Grober E., Mak V., Fischer A., Grantmyre J., et al. The workup and management of azoospermic males. Can Urol. Assoc. J. 2015;9:229–235. doi: 10.5489/cuaj.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell M.J., Lotti F., Baldi E., Schlatt S., Festin M.P.R., Bjorndahl L., et al. Distribution of semen examination results 2020 - a follow up of data collated for the WHO semen analysis manual 2010. Andrology. 2021;9:817–822. doi: 10.1111/andr.12983. [DOI] [PubMed] [Google Scholar]
- 3.von Eckardstein S., Simoni M., Bergmann M., Weinbauer G.F., Gassner P., Schepers A.G., et al. Serum inhibin B in combination with serum follicle-stimulating hormone (FSH) is a more sensitive marker than serum FSH alone for impaired spermatogenesis in men, but cannot predict the presence of sperm in testicular tissue samples. J. Clin. Endocrinol. Metab. 1999;84:2496–2501. doi: 10.1210/jcem.84.7.5855. [DOI] [PubMed] [Google Scholar]
- 4.Schlegel P.N., Palermo G.D., Goldstein M., Menendez S., Zaninovic N., Veeck L.L., et al. Testicular sperm extraction with intracytoplasmic sperm injection for nonobstructive azoospermia. Urology. 1997;49:435–440. doi: 10.1016/S0090-4295(97)00032-0. [DOI] [PubMed] [Google Scholar]
- 5.Schlegel P.N. Testicular sperm extraction: microdissection improves sperm yield with minimal tissue excision. Hum. Reprod. 1999;14:131–135. doi: 10.1093/humrep/14.1.131. [DOI] [PubMed] [Google Scholar]
- 6.Ramasamy R., Schlegel P.N. Microdissection testicular sperm extraction: effect of prior biopsy on success of sperm retrieval. J. Urol. 2007;177:1447–1449. doi: 10.1016/j.juro.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 7.Aboukhshaba A., Punjani N., Doukakis S., Zaninovic N., Palermo G., Schlegel P.N. Testicular sperm characteristics in men with nonobstructive azoospermia and their impact on intracytoplasmic sperm injection outcome. Fertil. Steril. 2022;117:522–527. doi: 10.1016/j.fertnstert.2021.09.024. [DOI] [PubMed] [Google Scholar]
- 8.Yatsenko A.N., Georgiadis A.P., Ropke A., Berman A.J., Jaffe T., Olszewska M., et al. X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N. Engl. J. Med. 2015;372:2097–2107. doi: 10.1056/NEJMoa1406192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen Y., Zhang F., Li F., Jiang X., Yang Y., Li X., et al. Loss-of-function mutations in QRICH2 cause male infertility with multiple morphological abnormalities of the sperm flagella. Nat. Commun. 2019;10:433. doi: 10.1038/s41467-018-08182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coutton C., Vargas A.S., Amiri-Yekta A., Kherraf Z.E., Ben Mustapha S.F., Le Tanno P., et al. Mutations in CFAP43 and CFAP44 cause male infertility and flagellum defects in Trypanosoma and human. Nat. Commun. 2018;9:686. doi: 10.1038/s41467-017-02792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siklenka K., Erkek S., Godmann M., Lambrot R., McGraw S., Lafleur C., et al. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science. 2015;350 doi: 10.1126/science.aab2006. [DOI] [PubMed] [Google Scholar]
- 12.Jodar M., Sendler E., Moskovtsev S.I., Librach C.L., Goodrich R., Swanson S., et al. Absence of sperm RNA elements correlates with idiopathic male infertility. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.aab1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J., Mu X., Xia Y., Martin F.L., Hang W., Liu L., et al. Metabolomic analysis reveals a unique urinary pattern in normozoospermic infertile men. J. Proteome Res. 2014;13:3088–3099. doi: 10.1021/pr5003142. [DOI] [PubMed] [Google Scholar]
- 14.Yu W., Zheng H., Lin W., Tajima A., Zhang Y., Zhang X., et al. Estrogen promotes Leydig cell engulfment by macrophages in male infertility. J. Clin. Invest. 2014;124:2709–2721. doi: 10.1172/JCI59901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hetherington L., Schneider E.K., Scott C., DeKretser D., Muller C.H., Hondermarck H., et al. Deficiency in Outer Dense Fiber 1 is a marker and potential driver of idiopathic male infertility. Mol. Cell Proteomics. 2016;15:3685–3693. doi: 10.1074/mcp.M116.060343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diao R., Fok K.L., Chen H., Yu M.K., Duan Y., Chung C.M., et al. Deficient human beta-defensin 1 underlies male infertility associated with poor sperm motility and genital tract infection. Sci. Transl Med. 2014;6:249ra108. doi: 10.1126/scitranslmed.3009071. [DOI] [PubMed] [Google Scholar]
- 17.Bieniek J.M., Drabovich A.P., Lo K.C. Seminal biomarkers for the evaluation of male infertility. Asian J. Androl. 2016;18:426–433. doi: 10.4103/1008-682X.175781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drabovich A.P., Saraon P., Jarvi K., Diamandis E.P. Seminal plasma as a diagnostic fluid for male reproductive system disorders. Nat. Rev. Urol. 2014;11:278–288. doi: 10.1038/nrurol.2014.74. [DOI] [PubMed] [Google Scholar]
- 19.Schiza C.G., Jarv K., Diamandis E.P., Drabovich A.P. An emerging role of TEX101 protein as a male infertility biomarker. EJIFCC. 2014;25:9–26. [PMC free article] [PubMed] [Google Scholar]
- 20.Tsujimura A., Matsumiya K., Miyagawa Y., Takao T., Fujita K., Koga M., et al. Prediction of successful outcome of microdissection testicular sperm extraction in men with idiopathic nonobstructive azoospermia. J. Urol. 2004;172:1944–1947. doi: 10.1097/01.ju.0000142885.20116.60. [DOI] [PubMed] [Google Scholar]
- 21.Vernaeve V., Tournaye H., Schiettecatte J., Verheyen G., Van Steirteghem A., Devroey P. Serum inhibin B cannot predict testicular sperm retrieval in patients with non-obstructive azoospermia. Hum. Reprod. 2002;17:971–976. doi: 10.1093/humrep/17.4.971. [DOI] [PubMed] [Google Scholar]
- 22.Drabovich A.P., Dimitromanolakis A., Saraon P., Soosaipillai A., Batruch I., Mullen B., et al. Differential diagnosis of azoospermia with proteomic biomarkers ECM1 and TEX101 quantified in seminal plasma. Sci. Transl. Med. 2013;5:212ra160. doi: 10.1126/scitranslmed.3006260. [DOI] [PubMed] [Google Scholar]
- 23.Drabovich A.P., Jarvi K., Diamandis E.P. Verification of male infertility biomarkers in seminal plasma by multiplex selected reaction monitoring assay. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bohring C., Schroeder-Printzen I., Weidner W., Krause W. Serum levels of inhibin B and follicle-stimulating hormone may predict successful sperm retrieval in men with azoospermia who are undergoing testicular sperm extraction. Fertil. Steril. 2002;78:1195–1198. doi: 10.1016/s0015-0282(02)04259-0. [DOI] [PubMed] [Google Scholar]
- 25.Korbakis D., Schiza C., Brinc D., Soosaipillai A., Karakosta T.D., Legare C., et al. Preclinical evaluation of a TEX101 protein ELISA test for the differential diagnosis of male infertility. BMC Med. 2017;15:60. doi: 10.1186/s12916-017-0817-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarvi K.A., Schlegel P., Schiza C., Drabovich A., Lau S., Soosaipillai A., Korbakis D., et al. Semen biomarker TEX101 predicts sperm retrieval success for men with testicular failure, F1000Research. 2021;10:569. [Google Scholar]
- 27.Begcevic I., Brinc D., Drabovich A.P., Batruch I., Diamandis E.P. Identification of brain-enriched proteins in the cerebrospinal fluid proteome by LC-MS/MS profiling and mining of the Human Protein Atlas. Clin. Proteomics. 2016;13:11. doi: 10.1186/s12014-016-9111-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Begcevic I., Brinc D., Dukic L., Simundic A.M., Zavoreo I., Basic Kes V., et al. Targeted mass spectrometry-based assays for relative quantification of 30 brain-related proteins and their clinical applications. J. Proteome Res. 2018;17:2282–2292. doi: 10.1021/acs.jproteome.7b00768. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Morillo E., Cho C.K., Drabovich A.P., Shaw J.L., Soosaipillai A., Diamandis E.P. Development of a multiplex selected reaction monitoring assay for quantification of biochemical markers of down syndrome in amniotic fluid samples. J. Proteome Res. 2012;11:3880–3887. doi: 10.1021/pr300355a. [DOI] [PubMed] [Google Scholar]
- 30.Prakash A., Rezai T., Krastins B., Sarracino D., Athanas M., Russo P., et al. Platform for establishing interlaboratory reproducibility of selected reaction monitoring-based mass spectrometry peptide assays. J. Proteome Res. 2010;9:6678–6688. doi: 10.1021/pr100821m. [DOI] [PubMed] [Google Scholar]
- 31.Prakash A., Rezai T., Krastins B., Sarracino D., Athanas M., Russo P., et al. Interlaboratory reproducibility of selective reaction monitoring assays using multiple upfront analyte enrichment strategies. J. Proteome Res. 2012;11:3986–3995. doi: 10.1021/pr300014s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karakosta T.D., Soosaipillai A., Diamandis E.P., Batruch I., Drabovich A.P. Quantification of human kallikrein-related peptidases in biological fluids by multiplatform targeted mass spectrometry assays. Mol. Cell Proteomics. 2016;15:2863–2876. doi: 10.1074/mcp.M115.057695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korbakis D., Brinc D., Schiza C., Soosaipillai A., Jarvi K., Drabovich A.P., et al. Immunocapture-selected reaction monitoring screening facilitates the development of ELISA for the measurement of native TEX101 in biological fluids. Mol. Cell Proteomics. 2015;14:1517–1526. doi: 10.1074/mcp.M114.047571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho C.K., Drabovich A.P., Batruch I., Diamandis E.P. Verification of a biomarker discovery approach for detection of Down syndrome in amniotic fluid via multiplex selected reaction monitoring (SRM) assay. J. Proteomics. 2011;74:2052–2059. doi: 10.1016/j.jprot.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 35.Schiza C., Korbakis D., Jarvi K., Diamandis E.P., Drabovich A.P. Identification of TEX101-associated proteins through proteomic measurement of human spermatozoa homozygous for the missense variant rs35033974. Mol. Cell Proteomics. 2019;18:338–351. doi: 10.1074/mcp.RA118.001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drabovich A.P., Saraon P., Drabovich M., Karakosta T.D., Dimitromanolakis A., Hyndman M.E., et al. Multi-omics biomarker pipeline reveals elevated levels of Protein-glutamine Gamma-glutamyltransferase 4 in seminal plasma of prostate cancer patients. Mol. Cell Proteomics. 2019;18:1807–1823. doi: 10.1074/mcp.RA119.001612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drabovich A.P., Pavlou M.P., Schiza C., Diamandis E.P. Dynamics of protein expression reveals primary targets and secondary messengers of estrogen receptor alpha signaling in MCF-7 breast cancer cells. Mol. Cell Proteomics. 2016;15:2093–2107. doi: 10.1074/mcp.M115.057257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drabovich A.P., Pavlou M.P., Dimitromanolakis A., Diamandis E.P. Quantitative analysis of energy metabolic pathways in MCF-7 breast cancer cells by selected reaction monitoring assay. Mol. Cell Proteomics. 2012;11:422–434. doi: 10.1074/mcp.M111.015214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Batruch I., Lecker I., Kagedan D., Smith C.R., Mullen B.J., Grober E., et al. Proteomic analysis of seminal plasma from normal volunteers and post-vasectomy patients identifies over 2000 proteins and candidate biomarkers of the urogenital system. J. Proteome Res. 2011;10:941–953. doi: 10.1021/pr100745u. [DOI] [PubMed] [Google Scholar]
- 40.Batruch I., Smith C.R., Mullen B.J., Grober E., Lo K.C., Diamandis E.P., et al. Analysis of seminal plasma from patients with non-obstructive azoospermia and identification of candidate biomarkers of male infertility. J. Proteome Res. 2012;11:1503–1511. doi: 10.1021/pr200812p. [DOI] [PubMed] [Google Scholar]
- 41.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347 doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 42.Gaudet P., Michel P.-A., Zahn-Zabal M., Britan A., Cusin I., Domagalski M., et al. The neXtProt knowledgebase on human proteins: 2017 update. Nucl. Acids Res. 2017;45:D177–D182. doi: 10.1093/nar/gkw1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foster J.A., Klotz K.L., Flickinger C.J., Thomas T.S., Wright R.M., Castillo J.R., et al. Human SP-10: acrosomal distribution, processing, and fate after the acrosome reaction. Biol. Reprod. 1994;51:1222–1231. doi: 10.1095/biolreprod51.6.1222. [DOI] [PubMed] [Google Scholar]
- 44.Bjorling E., Uhlen M. Antibodypedia, a portal for sharing antibody and antigen validation data. Mol. Cell Proteomics. 2008;7:2028–2037. doi: 10.1074/mcp.M800264-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Fu Z., Rais Y., Bismar T.A., Hyndman M.E., Le X.C., Drabovich A.P. Mapping isoform abundance and interactome of the endogenous TMPRSS2-ERG fusion protein by orthogonal immunoprecipitation-mass spectrometry assays. Mol. Cell Proteomics. 2021;20 doi: 10.1016/j.mcpro.2021.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rais Y., Fu Z., Drabovich A.P. Mass spectrometry-based proteomics in basic and translational research of SARS-CoV-2 coronavirus and its emerging mutants. Clin. Proteomics. 2021;18:19. doi: 10.1186/s12014-021-09325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saraon P., Cretu D., Musrap N., Karagiannis G.S., Batruch I., Drabovich A.P., et al. Quantitative proteomics reveals that enzymes of the ketogenic pathway are associated with prostate cancer progression. Mol. Cell Proteomics. 2013;12:1589–1601. doi: 10.1074/mcp.M112.023887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saraon P., Musrap N., Cretu D., Karagiannis G.S., Batruch I., Smith C., et al. Proteomic profiling of androgen-independent prostate cancer cell lines reveals a role for protein S during the development of high grade and castration-resistant prostate cancer. J. Biol. Chem. 2012;287:34019–34031. doi: 10.1074/jbc.M112.384438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schiza C., Korbakis D., Panteleli E., Jarvi K., Drabovich A.P., Diamandis E.P. Discovery of a human testis-specific protein complex TEX101-DPEP3 and selection of its disrupting antibodies. Mol. Cell Proteomics. 2018;17:2480–2495. doi: 10.1074/mcp.RA118.000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho C., Bunch D.O., Faure J.E., Goulding E.H., Eddy E.M., Primakoff P., et al. Fertilization defects in sperm from mice lacking fertilin beta. Science. 1998;281:1857–1859. doi: 10.1126/science.281.5384.1857. [DOI] [PubMed] [Google Scholar]
- 51.Miki K., Willis W.D., Brown P.R., Goulding E.H., Fulcher K.D., Eddy E.M. Targeted disruption of the Akap4 gene causes defects in sperm flagellum and motility. Dev. Biol. 2002;248:331–342. doi: 10.1006/dbio.2002.0728. [DOI] [PubMed] [Google Scholar]
- 52.Ikawa M., Inoue N., Benham A.M., Okabe M. Fertilization: a sperm's journey to and interaction with the oocyte. J. Clin. Invest. 2010;120:984–994. doi: 10.1172/JCI41585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen M.S., Tung K.S., Coonrod S.A., Takahashi Y., Bigler D., Chang A., et al. Role of the integrin-associated protein CD9 in binding between sperm ADAM 2 and the egg integrin alpha6beta1: implications for murine fertilization. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11830–11835. doi: 10.1073/pnas.96.21.11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jegou A., Ziyyat A., Barraud-Lange V., Perez E., Wolf J.P., Pincet F., et al. CD9 tetraspanin generates fusion competent sites on the egg membrane for mammalian fertilization. Proc. Natl. Acad. Sci. U. S. A. 2011;108:10946–10951. doi: 10.1073/pnas.1017400108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inoue N., Ikawa M., Isotani A., Okabe M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature. 2005;434:234–238. doi: 10.1038/nature03362. [DOI] [PubMed] [Google Scholar]
- 56.Bianchi E., Doe B., Goulding D., Wright G.J. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature. 2014;508:483–487. doi: 10.1038/nature13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aydin H., Sultana A., Li S., Thavalingam A., Lee J.E. Molecular architecture of the human sperm IZUMO1 and egg JUNO fertilization complex. Nature. 2016;534:562–565. doi: 10.1038/nature18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Drabovich A.P., Martinez-Morillo E., Diamandis E.P. Toward an integrated pipeline for protein biomarker development. Biochim. Biophys. Acta. 2015;1854:677–686. doi: 10.1016/j.bbapap.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 59.Drabovich A.P., Pavlou M.P., Batruch I., Diamandis E.P. In: Proteomic and Metabolomic Approaches to Biomarker Discovery. Issaq H.J., Veenstra T.D., editors. Academic Press (Elsevier); Waltham, MA: 2013. Chapter 2 - Proteomic and mass spectrometry technologies for biomarker discovery; pp. 17–37. Number of 17-37. [Google Scholar]
- 60.Drabovich A.P., Martínez-Morillo E., Diamandis E.P. In: Proteomics for Biological Discovery. 2nd Edition. Veenstra T.D., Yates III J., editors. Wiley-Blackwell; Hoboken, NJ: 2019. Chapter 3 - Protein Biomarker Discovery: An Integrated Concept; pp. 63–88. [Google Scholar]
- 61.Konvalinka A., Zhou J., Dimitromanolakis A., Drabovich A.P., Fang F., Gurley S., et al. Determination of an angiotensin II-regulated proteome in primary human kidney cells by stable isotope labeling of amino acids in cell culture (SILAC) J. Biol. Chem. 2013;288:24834–24847. doi: 10.1074/jbc.M113.485326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cho C.K., Drabovich A.P., Karagiannis G.S., Martinez-Morillo E., Dason S., Dimitromanolakis A., et al. Quantitative proteomic analysis of amniocytes reveals potentially dysregulated molecular networks in Down syndrome. Clin. Proteomics. 2013;10:2. doi: 10.1186/1559-0275-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Konvalinka A., Batruch I., Tokar T., Dimitromanolakis A., Reid S., Song X., et al. Quantification of angiotensin II-regulated proteins in urine of patients with polycystic and other chronic kidney diseases by selected reaction monitoring. Clin. Proteomics. 2016;13:16. doi: 10.1186/s12014-016-9117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martinez-Morillo E., Nielsen H.M., Batruch I., Drabovich A.P., Begcevic I., Lopez M.F., et al. Assessment of peptide chemical modifications on the development of an accurate and precise multiplex selected reaction monitoring assay for apolipoprotein e isoforms. J. Proteome Res. 2014;13:1077–1087. doi: 10.1021/pr401060x. [DOI] [PubMed] [Google Scholar]
- 65.Drabovich A.P., Diamandis E.P. Combinatorial peptide libraries facilitate development of multiple reaction monitoring assays for low-abundance proteins. J. Proteome Res. 2010;9:1236–1245. doi: 10.1021/pr900729g. [DOI] [PubMed] [Google Scholar]
- 66.Fu Z., Rais Y., Dara D., Jackson D., Drabovich A.P. Rational design and development of SARS-CoV-2 serological diagnostics by immunoprecipitation-targeted proteomics. Anal. Chem. 2022;94:12990–12999.. doi: 10.1021/acs.analchem.2c01325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Inoue N., Hagihara Y., Wright D., Suzuki T., Wada I. Oocyte-triggered dimerization of sperm IZUMO1 promotes sperm-egg fusion in mice. Nat. Commun. 2015;6:8858. doi: 10.1038/ncomms9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aitken R.J., Baker M.A., Doncel G.F., Matzuk M.M., Mauck C.K., Harper M.J. As the world grows: contraception in the 21st century. J. Clin. Invest. 2008;118:1330–1343. doi: 10.1172/JCI33873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw SRM data and processed Skyline files were deposited to Peptide Atlas (www.peptideatlas.org/PASS/PASS01755 or ftp://PASS01755:QE3725r@ftp.peptideatlas.org) and can be downloaded with FTP software (https://filezilla-project.org or etc.) using the host server name ftp.peptideatlas.org, password QE3725r, and username PASS01755.