Abstract
All positive-strand RNA viruses of eukaryotes studied assemble RNA replication complexes on the surfaces of cytoplasmic membranes. Infection of mammalian cells with poliovirus and other picornaviruses results in the accumulation of dramatically rearranged and vesiculated membranes. Poliovirus-induced membranes did not cofractionate with endoplasmic reticulum (ER), lysosomes, mitochondria, or the majority of Golgi-derived or endosomal membranes in buoyant density gradients, although changes in ionic strength affected ER and virus-induced vesicles, but not other cellular organelles, similarly. When expressed in isolation, two viral proteins of the poliovirus RNA replication complex, 3A and 2C, cofractionated with ER membranes. However, in cells that expressed 2BC, a proteolytic precursor of the 2B and 2C proteins, membranes identical in buoyant density to those observed during poliovirus infection were formed. When coexpressed with 2BC, viral protein 3A was quantitatively incorporated into these fractions, and the membranes formed were ultrastructurally similar to those in poliovirus-infected cells. These data argue that poliovirus-induced vesicles derive from the ER by the action of viral proteins 2BC and 3A by a mechanism that excludes resident host proteins. The double-membraned morphology, cytosolic content, and apparent ER origin of poliovirus-induced membranes are all consistent with an autophagic origin for these membranes.
Infection with positive-strand RNA viruses results in a range of membrane morphologies, many of which involve complex membrane rearrangements. Cells infected with poliovirus and other picornaviruses, for example, accumulate large quantities of membranous vesicles 150 to 400 nm in diameter (Fig. 1) (5, 12). Most of these vesicles are surrounded by double lipid bilayers (Fig. 1B and C) (41), precluding a simple budding mechanism. Instead, the presence of a double membrane suggests a wrapping mechanism for vesicle formation akin to the process of cellular autophagy, as suggested previously (12, 41). For all positive-strand RNA viruses studied to date, the RNA synthesis machinery is associated with the cytoplasmic surface of these cytoplasmic membranes, and many of the proteins required for viral RNA synthesis are membrane associated when expressed in isolation.
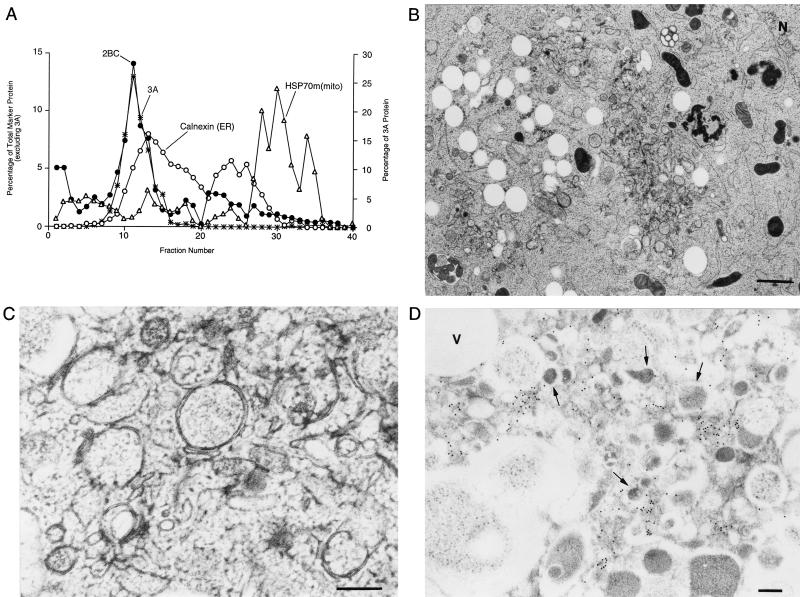
FIG. 1.
Electron micrographs of COS-1 cells preserved by high-pressure freezing show the ultrastructure of uninfected cells and cells infected with poliovirus for 4 h at 37°C. (A) Uninfected cell. N, nucleus; G, Golgi. Bar = 1 μm. (B) Infected cell; bar = 1 μm. (C) Infected cell; bar = 0.2 μm. (D) Immunostaining of infected cell to identify poliovirus 2C epitopes using 15-nm gold particles conjugated to secondary antibodies. Arrows indicate double-membraned vesicles. Bar = 0.3 μm.
Studies of several different positive-strand RNA viruses, including costaining and coisolation of viral and cellular proteins known to be residents of individual organelles, have diversely implicated the endoplasmic reticulum (ER), trans-Golgi endosomes, or lysosomes as likely sources for virally induced membranes. For example, Semliki Forest virus has long been thought to replicate on cellular endosomes modified during viral infection (19), and cowpea mosaic virus replication complexes colocalize with ER marker proteins (9). For poliovirus-infected cells, although images consistent with the budding of these vesicles from ER membranes have been reported (5), immunoisolation of the vesicles showed the presence of protein markers from the entire secretory apparatus, including ER, trans-Golgi, and lysosomal markers (41). It was concluded that the poliovirus-induced vesicles either derive from a pooled compartment or acquire many of their cellular markers after their formation (41).
To identify viral proteins which can, in isolation, cause membrane rearrangements, individual proteins from several positive-strand RNA viruses have been expressed in a variety of cell types. For poliovirus, the viral 2C protein, a 329-amino-acid membrane-associated protein with RNA-dependent ATPase activity (31, 38), is required for RNA replication and was shown to cause both membrane vesiculation and the formation of multilamellar structures when expressed in isolation (1, 10). When the 426-amino-acid precursor protein, 2BC, was expressed in human cells, a greater amount of vesiculation was observed, which was morphologically more consistent with the pattern seen in poliovirus-infected cells. The more authentic ultrastructural changes were seen only when 2B and 2C were covalently linked (1, 10, 48).
An 87-amino-acid protein, 3A, when expressed in isolation, associates with membranes of the ER, as shown by immunoelectron microscopy (13) and by coimmunofluorescence (14). Near the C terminus of the 3A coding region is a hydrophobic region whose integrity is required for membrane association in vitro (49) and is presumed to anchor 3A and 3AB to cellular membranes during poliovirus RNA synthesis. Membrane-associated 3AB binds directly to the poliovirus RNA-dependent RNA polymerase (22), stimulating protease activity of the polymerase precursor 3CD (25), and thus is thought to anchor 3D polymerase in the RNA replication complexes. Either the 22-amino-acid 3B or its precursor 3AB serves as the primer for RNA synthesis (33). Although 3A protein expression causes the inhibition of ER-to-Golgi protein traffic (14) and dramatically alters ER ultrastructure (13), these morphological changes do not in themselves resemble those observed in poliovirus-infected cells.
Autophagic vacuoles form in normal cells in response to nitrogen or amino acid starvation. Membranes thought to be derived from the ER (16) wrap around small pools of cytoplasm, forming immature autophagic vacuoles. The lumen of these double-membraned, immature autophagic vacuoles contains material morphologically indistinguishable from cytoplasm, sometimes including intact mitochondria or other small organelles. The evidence that autophagic vacuoles form from the ER has been predominately ultrastructural (16). Biochemical identification of the origin of the limiting membranes in these structures has proven difficult, possibly because these membranes are relatively depleted for protein markers (37, 45). However, those proteins present in immature autophagic vacuoles include the ER lumenal proteins calreticulin, protein disulfide isomerase (PDI), ER60 protease, and BiP (51). As the double-membraned autophagic vacuoles mature, they acquire from vesicles in the endosomal and lysosomal pathway protein markers and lumenal contents such as β-hexaminidase (17). The degradation of the inner lipid bilayer marks the transition to single-membraned, mature autophagic vacuoles (17). Poliovirus-induced vesicles resemble immature autophagic vacuoles in that they are frequently bounded by double membranes, bear markers from the late as well as early secretory pathway, and contain lumenal material indistinguishable from cytoplasm, sometimes including cytoplasmic organelles such as mitochondria (12, 41).
Here, we provide biochemical and ultrastructural evidence for the effects of expression of individual viral proteins on host cytoplasmic membranes. Density gradient centrifugation was used to identify the host membranes with which the individual viral proteins 2C, 2BC, and 3A associate when expressed in isolation and during poliovirus infection. Corresponding ultrastructural studies of infected cells and cells transfected with individual viral proteins were performed using high-pressure freezing and cryosubstitution to preserve membrane morphology (reviewed in reference 11). The combined actions of 2BC and 3A were found to mimic both the biochemical and ultrastructural alterations in poliovirus-infected cells, forming structures consistent with an autophagic mechanism for the formation of the virus-induced vesicles from the ER.
MATERIALS AND METHODS
Cell culture and virus infection.
COS-1 cells were cultured as monolayers in Dulbecco's modified Eagle's medium supplemented with 10% calf serum, 5 mM glutamine, and 30,000 U of penicillin-streptomycin (Life Technologies, Inc., Gaithersburg, Md.) per liter at 37°C in a 5% CO2 incubator. Wild-type poliovirus type 1 Mahoney was used to infect cells as previously described (30). All infections were performed at a multiplicity of 50 PFU per cell. Virus titers were determined by plaque assays on COS-1 cells.
Plasmids and DNA transfections.
The pLINK-based DNA plasmids used in these experiments have been previously described (14). Briefly, these constructs, which are under the transcriptional control of the simian virus 40 late promoter, contain an internal ribosomal entry site followed by the coding sequence for the vesicular stomatitis G protein (VSV-G). In the parental pLINK plasmid, only VSV-G is produced. No effect of VSV-G expression from pLINK was observed in either ultrastructural or gradient fractionation experiments (data not shown). The dicistronic pLINK derivatives used here encode poliovirus protein 2B, 2C, 2BC, or 3A in the first cistron as well as VSV-G in the second cistron. DNA transfections were performed using Lipofectamine or Lipofectamine Plus reagents as described by the manufacturer (Life Technologies). Twelve micrograms of each plasmid DNA was used for transfection of each 150-mm-diameter plate of 4 × 106 COS-1 cells. Following a 48-h incubation period, cells were harvested and either used in biochemical fractionation experiments or prepared for electron microscopic analysis. The 48-h incubation time point yielded ultrastructural results similar to results of 24-h incubations for those proteins tested (data not shown), but expression levels of the viral proteins were higher at the later time point, which facilitated biochemical analysis.
Biochemical fractionation.
Cells were infected with poliovirus at 37°C for 4 h unless otherwise specified or transfected for 48 h at 37°C with the pLINK-based DNA plasmids. Each sample of 1.5 × 108 infected, transfected, or control cells was scraped into phosphate-buffered saline (PBS), collected by low-speed centrifugation, and resuspended in 1 ml of homogenization buffer (0.25 M sucrose, 5 mM morpholinepropanesulfonic acid [pH 7.4]). The plasma membranes were disrupted by pressure filtration through 14-μm-pore-size polycarbonate track-etch membranes (Osmonics, Livermore, Calif.), a technique useful in preserving the integrity of organelles during cell lysis (46). The low-salt conditions used were found to be optimal for the separation of cytoplasmic organelles from the poliovirus-induced membranes (Fig. 2 and 8) and did not lead to nonspecific aggregation, as evidenced by the successful separations achieved. The filtrate was centrifuged twice for 10 min at 4°C and 1,000 × g to sediment intact cells, nuclei, and large sheets of plasma membranes. The resultant supernatant was loaded onto 20 ml of self-forming Percoll density gradient medium (Pharmacia Biotech, Piscataway, N.J.); the Percoll concentrations indicated for each experiment were achieved by diluting stock isotonic Percoll (9 parts Percoll, 1 part 2.5 M sucrose [vol/vol]) with homogenization buffer. For some experiments, both the homogenization buffer and the Percoll suspension medium were adjusted to 60 mM KCl. The samples were centrifuged in a Beckman type 60 Ti rotor at 25,000 rpm for 23 min at 4°C. Density marker beads (Pharmacia Biotech) were used to calibrate the initial gradients (data not shown).
FIG. 2.
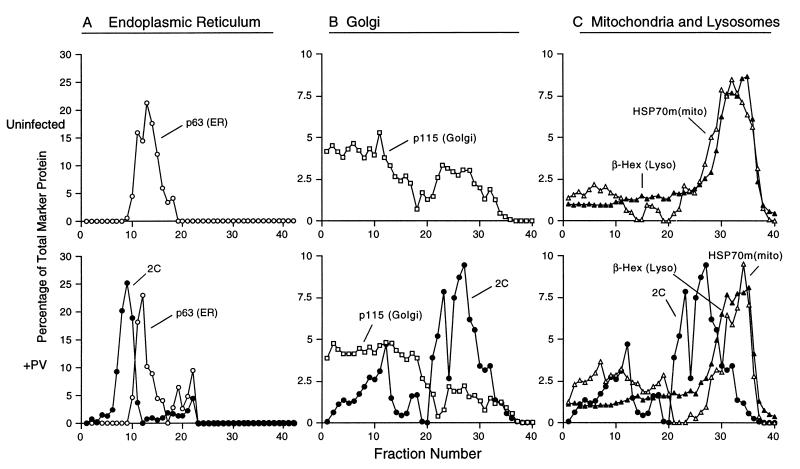
Density gradient analyses of the distributions of subcellular membranes of uninfected and poliovirus-infected cells. Following infection with poliovirus for 4 h, the plasma membranes of COS-1 cells were disrupted and the cytoplasmic organelles were separated on Percoll density gradients. Individual fractions (lightest fractions are at the left) were collected and either tested for enzymatic activity or analyzed by immunoblot assays to identify poliovirus 2C protein (A, B, and C), ER marker p63 (A), Golgi marker p115 (B), and mtHSP70 [HSP70m(mito)] (C). β-Hexosaminidase activity assays were used to identify fractions derived from lysosomes [β-Hex (Lyso)] (C). Stock isotonic Percoll concentrations were 20% (A) and 9.5% (B and C).
FIG. 8.
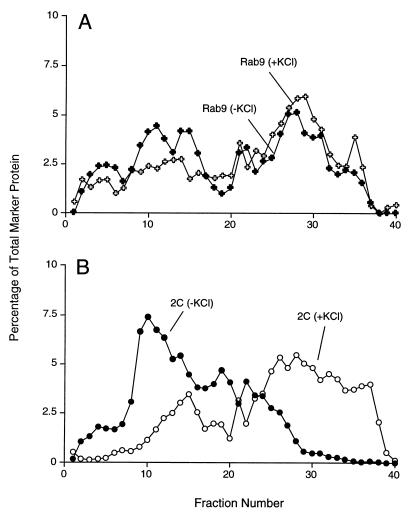
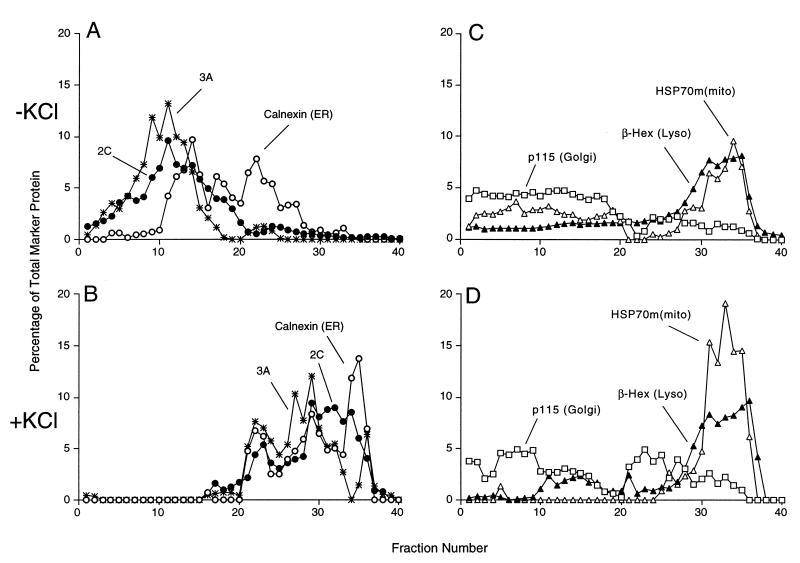
Effect of increased ionic strength on buoyant density distribution of endosomes and poliovirus-induced vesicles. Following infection with poliovirus for 4 h, the plasma membranes of COS-1 cells were disrupted in homogenization buffer in the presence and absence of 60 mM KCl, and the cellular organelles were separated on 12% stock isotonic Percoll density gradient that did or did not contain 60 mM KCl; lightest fractions are at the left. (A) Distribution of Rab9-containing membranes in the presence and absence of 60 mM KCl; (B) distribution of poliovirus 2C-containing membranes in the presence and absence of 60 mM KCl.
Following centrifugation, 40 individual 0.5-ml fractions were collected from each sample using a density gradient fractionator (Brandel, Gaithersburg, Md.); each fraction was adjusted to 1 mM phenylmethylsulfonyl fluoride and 0.1% Triton X-100. For direct analysis of lysosome-derived material, equivalent volumes of each fraction were measured for β-hexosaminidase activity (36). Briefly, 200 μl of substrate buffer (50 mM sodium citrate [pH 4.8], 0.1% Triton X-100, 1.7 mg of p-nitrophenol-β-N-acetylglucosamide [Sigma, St. Louis, Mo.] per ml) was added to 100 μl of each sample. After incubation for 60 min at 37°C, the reaction was developed by adding 0.9 ml of 0.15 M glycine (pH 10.8). Release of the p-nitrophenol product was measured by increased absorbance at 412 nm.
For immunoblot analysis, equivalent volumes of each sample were subjected to centrifugation at 100,000 × g at 4°C for 45 min in a Beckman TLA-100 rotor to pellet the Percoll resin. The proteins in the resultant supernatants were precipitated (55), resuspended in a minimal volume, and displayed by glycine-sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) or tricine-SDS-PAGE (40). The proteins were then transferred to Immobilon-P polyvinylidene fluoride membranes (Millipore, Bedford, Mass.), which were probed directly using antibodies generated against specific viral proteins or to resident marker proteins of cellular organelles. To identify the distribution of poliovirus proteins, anti-2C monoclonal antibodies, kindly provided by K. Bienz and D. Egger (University of Basel, Basel, Switzerland), were used at a dilution of 1:1,500, and anti-3A monoclonal antibodies (13) were used at a dilution of 1:50. Fractions containing proteins from the ER were identified either by an anticalnexin polyclonal serum (Stressgen, Victoria, British Columbia, Canada) at a dilution of 1:4,000 or by anti-p63 monoclonal antibodies, kindly provided by H.-P. Hauri (University of Basel). Anti-p115 (7DI) monoclonal antibodies, kindly provided by G. Waters (Princeton University) and used at a dilution of 1:20,000, and anti-1,4-galactosyltransferase polyclonal serum, kindly provided by E. Berger (Universität Zurich, Zurich, Switzerland) and used at a dilution of 1:250, were used to identify Golgi-containing fractions. Mitochondrial fractions were tracked using anti-mitochondrial HSP70 (mtHSP70) monoclonal antibodies, kindly provided by S. Pierce (Northwestern University), at a dilution of 1:500. Finally, endosomes were traced with anti-Rab9 monoclonal antibodies, kindly provided by S. Pfeffer (Stanford University), at a dilution of 1:100. Anti-mouse and anti-rabbit secondary antibodies, conjugated to alkaline phosphate (Zymed, South San Francisco, Calif.), were used at 1:4,000. Enhanced chemifluorescence (Amersham, Arlington Heights, Ill.) coupled with PhosphorImager analyses utilizing ImageQuant (version 4.0) software (Molecular Dynamics, Sunnyvale, Calif.) was used to generate signals that were linearly responsive to protein concentrations and to quantify the relative level of each protein marker. The results are presented as the percentage of the specific protein found within each fraction; the total amount of the protein of interest was determined by summing the enhanced chemifluorescence signals from the 40 gradient fractions.
High-pressure freezing and freeze-substitution.
For cryofixation and electron microscopy, transfected, infected, or untreated COS-1 cells from one 100-mm-diameter plate were washed twice with PBS, removed from the plates by treatment with trypsin, and collected by centrifugation. The cell pellet was resuspended in a minimal volume of 0.15 M sucrose in PBS. Aliquots of the cell slurry were frozen in a Balzers HPM 010 high-pressure freezing apparatus as described previously (41) and stored in liquid nitrogen. For observation of cellular morphology, samples were freeze-substituted in 0.1% tannic acid in acetone at −80°C, rinsed in acetone, then warmed to −20°C in the presence of 2% osmium tetroxide in acetone for 16 h, and incubated at 4°C for 4 h. After being rinsed in acetone at 4°C, samples were embedded in Epon-Araldite resin. Thin sections were stained with 2% uranyl acetate and lead citrate and then imaged at 80 kV in a JEOL 100C or Philips CM10 electron microscope.
For immunostaining, high-pressure-frozen samples were freeze-substituted in 0.1% glutaraldehyde–0.05% uranyl acetate in acetone and then embedded in Lowicryl K4M. Sections were mounted on Formvar-coated nickel grids and immunolabeled with either anti-2C or anti-3A monoclonal antibodies. Grids were floated on a drop of blocking solution in PBS that contained primary antibody as described previously (13, 41). Mouse monoclonal primary antibodies were detected with goat anti-mouse secondary antibodies coupled to gold particles 15 nm in diameter (Ted Pella, Inc., Redding, Calif.).
RESULTS
Poliovirus-induced vesicles do not cofractionate with ER, Golgi, lysosomal, or mitochondrial protein markers.
Buoyant density gradients were used to determine the proportion of protein markers from ER, Golgi, lysosomes, and mitochondria found in poliovirus-induced vesicles and in subcellular organelles during poliovirus infection. Separation of cellular components by centrifugation in polydisperse solutes such as Percoll is based on a combination of sedimentation rate and isopycnic effects, with the density of the organelles being the most critical parameter for separation (43). To fractionate the cytoplasmic organelles in uninfected and in poliovirus-infected COS-1 cells, plasma membranes were disrupted by pressure filtration homogenization (46); nuclei and intact cells were removed by low-speed centrifugation, and the postnuclear supernatants were fractionated on Percoll gradients. Fractions were collected and assayed either enzymatically or by SDS-PAGE followed by transfer to membranes for Western blot analysis. Membranes associated with poliovirus RNA replication complexes were identified using antibody to poliovirus protein 2C, known to be associated with the virus-induced vesicles and with active RNA replication complexes (7). The poliovirus 2C-containing fractions were narrowly distributed upon fractionation in 20% stock isotonic Percoll (Fig. 2A), although the fractions that contained the poliovirus RNA replication complexes became more disperse at lower Percoll concentrations (Fig. 2B and C). Thus, the range of buoyant densities displayed by the poliovirus-induced vesicles is narrow and Percoll gradients can be efficiently used in their purification.
To determine the location of ER membranes in Percoll gradients, the relative amount of the ER resident protein p63 (42) in each fraction was determined by quantitative immunoblot analysis (Materials and Methods). Although some initial controversy surrounded the intracellular location of p63, recent data have confirmed its ER localization (42). The distribution of p63 was similar in mock-infected and poliovirus-infected cells (Fig. 2A). In the 20% stock isotonic Percoll gradient shown in Fig. 2A, the peak of poliovirus-induced vesicles displayed a lower buoyant density than the p63-containing fractions. Therefore, the majority of the virus-induced vesicles do not cofractionate with ER membranes.
Immunoblot analysis using an antibody generated against p115 (53) was used to determine the distribution of Golgi membranes in buoyant density gradients. In both poliovirus-infected and uninfected cells, p115 was widely scattered across 9.5% stock isotonic Percoll gradients (Fig. 2B). To address the concern that this diffuse distribution might have resulted from the removal of p115, a peripheral membrane protein, from the Golgi membranes during extraction, the fractionation of another Golgi marker, 1,4-galactosyltransferase (54), was tested and found to display an identical distribution in this gradient (data not shown). Despite the diffuse distribution of the Golgi membranes in these Percoll gradients, it is clear that these membranes were not consumed by the formation of the poliovirus-induced membranes, and the poliovirus-induced vesicles did not cofractionate with the majority of the Golgi membranes.
A lysosomal origin for poliovirus-induced vesicles would be consistent with immunoelectron microscopic analysis that showed staining of poliovirus-induced vesicles with LAMP-1, a marker of endosomes and lysosomes (41), and with the documented increases in lysosomal enzymes in the cytoplasm of poliovirus-infected cells (21). All fractions from uninfected and virus-infected cells were assayed for β-hexosaminidase activity (Fig. 2C) to identify the lysosome-containing fractions in 9.5% stock isotonic Percoll gradients. The discrete peak of β-hexosaminidase activity observed in uninfected cells was not significantly altered after 4 h of poliovirus infection, and very little overlap was seen between the β-hexosaminidase- and poliovirus 2C-containing fractions.
Mitochondria form an unlikely source for the poliovirus-induced vesicles, and no disruption of mitochondria has been reported during poliovirus infection. Therefore, as a representative organelle that should not be disrupted during poliovirus infection, the fractionation of mitochondria in 9.5% stock isotonic Percoll gradients was determined by immunoblot analysis using antibodies generated against the mtHSP70 (20). In mock- and poliovirus-infected cells, mitochondria formed a discrete peak that cofractionated with lysosomes (Fig. 2C). The mitochondrial peak was not disrupted by poliovirus infection, and the majority of the poliovirus 2C-containing membranes did not contain the GRP75 mitochondrial marker.
Thus, the poliovirus-induced vesicles are either generated de novo, formed from a subcellular compartment whose resident proteins were not tested, or generated from one of the tested organelles (ER, Golgi, lysosomes, or mitochondria) by a mechanism that excludes at least some of the resident proteins. To test the latter possibility, we sought to determine whether the characteristic biochemistry and intracellular morphology of poliovirus-infected cells could be mimicked by the expression of a subset of the viral proteins and, if so, to determine their intracellular localization.
Viral proteins 2C and 3A fractionate as resident ER proteins and alter ER morphology when expressed in isolation.
Previous studies have shown that the ER membranes of cells that expressed the 3A protein displayed a characteristic swollen and distended morphology by electron microscopy (13). Immunoelectron microscopy demonstrated that 3A protein localizes to these swollen membranes (13). Coimmunofluorescence of the ER marker PDI secretory cargo (α-1 protease inhibitor) whose transport was disrupted by 3A function, and 3A protein showed that the swollen membranes that contain both 3A and arrested secretory cargo are ER (13, 14). To examine the distribution of 3A protein in Percoll gradients, COS-1 cells were transfected with a plasmid that expressed 3A protein from a dicistronic RNA that also encoded VSV-G (14). Cell extracts were prepared and fractionated on 12% stock isotonic Percoll gradients, the concentration found to be optimal for separation of the organelles of interest. The fractions that contained 3A protein were identified by immunoblot analysis. The positions of ER, Golgi, lysosomal, and mitochondrial membranes were also determined, although only a subset of these distributions is shown for simplicity. In both transfected and untransfected COS-1 cells, the ER marker calnexin was dispersed over many fractions; 3A protein in the transfected cells cofractionated with the ER fractions of the lightest buoyant density (Fig. 3A). The expression of 3A protein did not perturb the distribution of ER membranes in Percoll gradients compared to untransfected cells (data not shown); because only 30 to 40% of the cells were transfected, a subtle disturbance in the transfected cells may not have been detectable. Electron microscopy was performed on populations of cells that were transfected with 3A-expressing plasmids. Approximately 40% of the cells displayed the characteristic morphology reported previously to be caused by 3A expression (Fig. 3B) (13).
FIG. 3.
Density gradient and ultrastructural analysis of COS-1 cells that expressed poliovirus 3A protein. (A) Cytoplasmic membranes from cells expressing 3A protein were separated on a 12% stock isotonic Percoll; proteins in each fraction were detected by immunoblotting using antibodies directed against the poliovirus 3A protein, calnexin (ER), and mtHSP70 [HSP70m(mito); mitochondria]; lightest fractions are at the left. The distribution profiles of Golgi and lysosomal proteins did not vary significantly from the patterns displayed in Fig. 2 and were not included for simplicity. (B) Ultrastructure of COS-1 cell expressing poliovirus 3A protein. N, nucleus. Bar = 1 μm.
To test whether the expression of poliovirus 2C protein altered the ultrastructure of 3A-expressing cells, and to localize 2C protein expression in the presence of 3A protein, COS cells were cotransfected with plasmids that expressed 2C and 3A proteins. When cytoplasmic extracts of these transfected cells were separated on 12% stock isotonic Percoll gradients, 2C and 3A cofractionated with each other as well as with the ER-containing fractions of the lightest buoyant density (Fig. 4A).
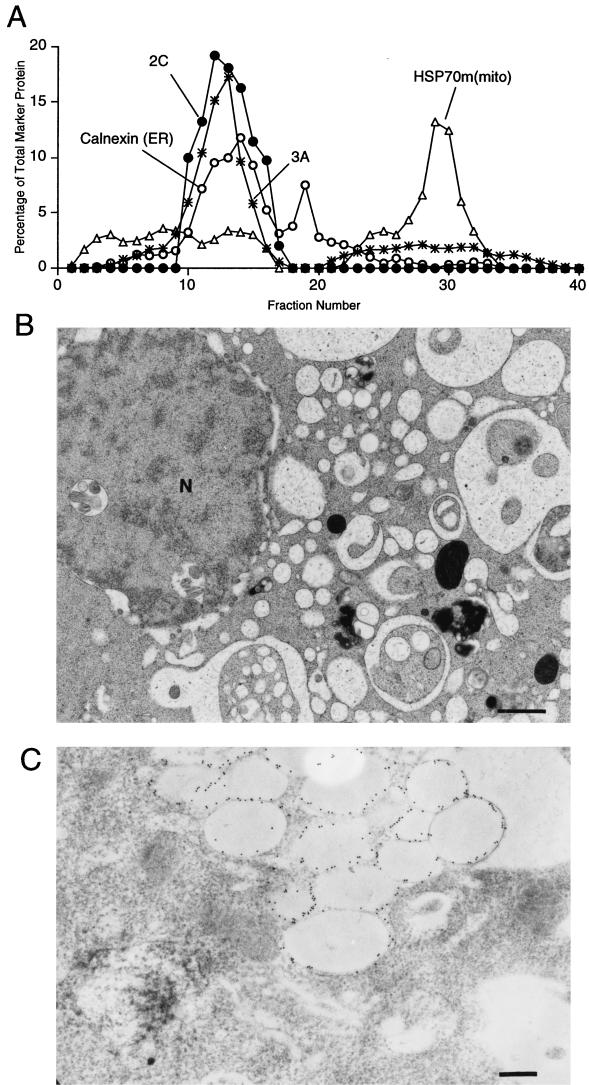
FIG. 4.
Density gradient and ultrastructural analysis of COS-1 cells that expressed poliovirus proteins 2C and 3A. (A) Cytoplasmic membranes from cells expressing 2C and 3A proteins were separated on a 12% stock isotonic Percoll gradient; proteins in each fraction were detected by immunoblotting using antibodies directed against poliovirus 2C, poliovirus 3A, calnexin (ER), and mtHSP70 [HSP70m(mito) mitochondria]; lightest fractions are at the left. (B) Electron microscopy reveals the effects of 2C and 3A expression on the ultrastructure of COS-1 cells. N, nucleus. Bar = 1 μm. (C) Section labeled with 15-nm gold particles coupled to secondary antibodies. The primary antibody was directed against poliovirus protein 2C. Bar = 0.5 μm.
Electron micrographs of cells that express both 2C and 3A (Fig. 4B) showed that, like in 3A-expressing cells, the ER membranes were distended and the lumen was highly enlarged. Unlike the majority of the ER membranes in 3A-expressing cells, the ER in cells that expressed both 2C and 3A showed apparent invaginations, so that the swollen membranes encircled cellular material similar in morphology and electron density to cytosol. Immunostaining of sections prepared for electron microscopy showed that the swollen ER membranes contained 2C protein (Fig. 4C). Often, these cells also contained clusters of large, clear, single-membraned vesicles. No small double-membraned vesicles such as those seen in poliovirus-infected cells were observed.
Viral protein 2BC in isolation forms membranes of the buoyant density observed in poliovirus-infected cells.
Previous electron microscopic studies of cells that express individual poliovirus proteins have suggested that the expression of poliovirus 2BC protein mimics the morphology of poliovirus-infected cells (1, 10, 48). To test the effect of 2BC protein expression on the biochemical distribution of cytoplasmic membranes in Percoll gradients, COS-1 cells were transfected with a DNA plasmid that expressed 2BC and cytoplasmic extracts were fractionated on a 12% stock isotonic Percoll gradient (Fig. 5A). The virally encoded protease responsible for cleavage of the 2BC protein was not expressed in the transfected cells; therefore, only the 48 kDa 2BC protein, and not its final processing products, was detected by immunoblot analysis (data not shown). Like the membranes from poliovirus-infected cells, but unlike cells expressing 3A and 2C, the peak of the 2BC-containing membranes displayed a lighter buoyant density, centered at fraction 11, than even the lightest of the ER-containing fractions, centered at fraction 13 (Fig. 5A). Therefore, 2BC expression in isolation was sufficient to generate membranes of buoyant densities similar to those observed in poliovirus-infected cells.
FIG. 5.
Density gradient and ultrastructural analysis of COS-1 cells that expressed poliovirus 2BC protein. (A) Cytoplasmic membranes from cells expressing 2BC protein were separated on a 12% stock isotonic Percoll gradient; proteins in each fraction were detected by immunoblotting using antibodies directed against poliovirus 2C, calnexin (ER), and mtHSP70 [HSP70m(mito) mitochondria]; lightest fractions are at the left. (B) Ultrastructure of COS-1 cells transfected with a 2BC-expressing plasmid. G, Golgi. Bar = 2 μm. (C) An immunostained section using anti-2C antibodies and secondary antibodies coupled to 15-nm gold particles. Bar = 0.5 μm.
Expression of 2BC in COS-1 cells induced the formation of at least two distinct membrane morphologies, as visualized by high-pressure freezing and freeze-substitution followed by electron microscopy. The predominant consequence of 2BC expression was the formation of clusters of empty vacuoles limited by a single membrane, usually in peripheral regions of the cell (Fig. 5B and C). These membranes contained a high concentration of the 2C epitope. A few transfected cells showed small numbers of a second class of vesicle, morphologically more reminiscent of those formed during poliovirus infection (Fig. 5C). As in poliovirus-infected cells, these vesicles were limited by double membranes, had more intensely stained lumen, and were clustered. However, even in these cells, the vast majority of the 2BC protein was associated with the empty vacuoles. Expression of 2BC in the absence of other poliovirus proteins resembled poliovirus infection in that it caused the formation of membranes of lighter buoyant density than ER but induced the formation of morphologically similar membranes to only a very limited extent.
2BC expression causes 3A protein to change its localization from the ER to a fraction with a lower buoyant density.
The effect of coexpression of 2BC and 3A was tested in an attempt to mimic more closely the biochemistry and ultrastructure of poliovirus-infected cells. COS-1 cells were cotransfected with plasmids that expressed 2BC and 3A, and cytoplasmic extracts were displayed on 12% stock isotonic Percoll gradients (Fig. 6A). As in Fig. 5A with 2BC alone, 2BC and 3A coexpression led to the formation of a peak of lighter buoyant density than the calnexin-containing ER membranes. In addition, the vast majority of the 3A protein in these transfected cells was also detected in this light fraction. Therefore, 2BC expression was sufficient to recruit the 3A protein quantitatively into membranes similar in buoyant density to those induced during poliovirus infection, even though 3A colocalized to ER membranes when expressed in isolation (Fig. 3) (13).
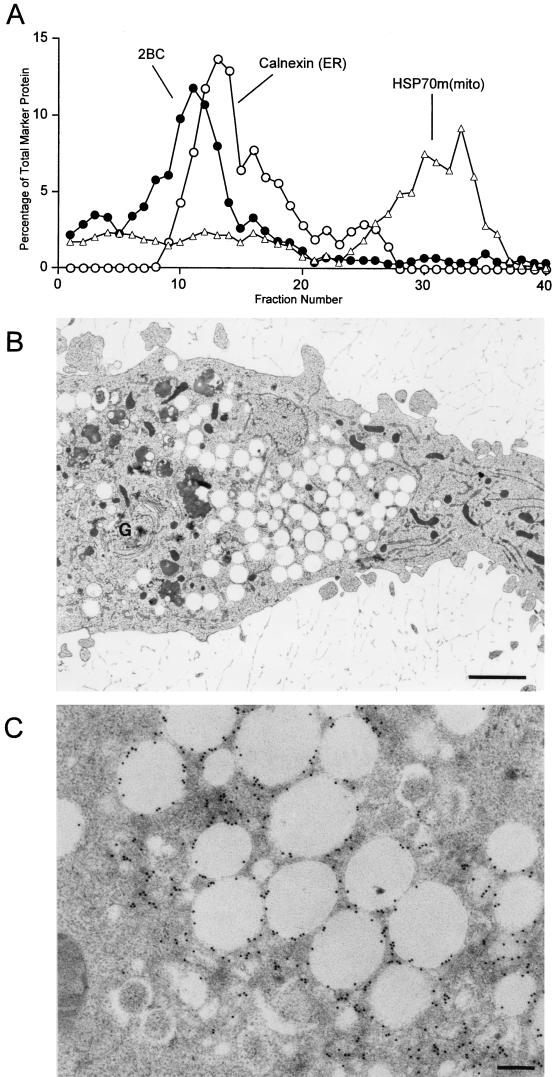
FIG. 6.
Density gradient and ultrastructural analysis of COS-1 cells cotransfected with plasmids that encode the poliovirus 2BC and 3A proteins. (A) Cytoplasmic membranes from cells expressing 2BC and 3A proteins were separated on a 12% stock isotonic Percoll gradient; proteins in each fraction were detected by immunoblotting using antibodies directed against poliovirus 2C, poliovirus 3A, calnexin (ER), and mtHSP70 [HSP70m(mito) mitochondria]; lightest fractions are at the left. (B) Ultrastructure of COS-1 cells transfected with 2BC- and 3A-expressing plasmids. N, nucleus. Bar = 1 μm. (C) Higher-resolution image of a section of a 2BC- and 3A-transfected cell. Bar = 0.2 μm. (D) A 2BC- and 3A-transfected cell that was immunostained with antibodies directed against the 2C protein. Secondary antibodies were coupled to 15-nm gold particles. Arrows indicate double-membraned vesicles. Bar = 0.5 μm.
The morphology of cells that express 2BC and 3A resembles that of poliovirus infection.
Ultrastructural analysis of COS-1 cells transfected with the 2BC and 3A expression plasmids revealed morphologies highly reminiscent of poliovirus infection, with both small clustered vesicles and large clear vacuoles formed (Fig. 6B). Higher magnifications revealed that many of the smaller membranous vesicles formed contained double membranes and cytoplasmic lumen (Fig. 6C), were similar in size and shape to the vesicles induced during poliovirus infection (Fig. 1B and C), and tended to be clustered in the centrosomal regions of the transfected cells. As in poliovirus infection (Fig. 1D), the 2C epitopes were found predominantly on or in the immediate vicinity of the double-membraned vesicles (Fig. 6D). Although empty vacuoles such as those found during expression of 2BC alone (Fig. 5B) were observed, they were not heavily labeled with the anti-2C antibody (Fig. 6D). Therefore, the vesicles induced by coexpression of 3A and 2BC resembled the vesicles induced during poliovirus infection more closely than the vesicles induced by expression of 2BC alone.
3A protein colocalizes with 2C in fractions less dense than ER during poliovirus infection.
To determine whether, during poliovirus infection, viral 3A protein relocalized from the ER to lighter, 2BC-containing fractions as it did during coexpression with 2BC protein, the cytoplasmic organelles from poliovirus-infected COS-1 were separated on 12% stock isotonic Percoll gradients. The 3A-containing fractions completely colocalized with the 2C-containing fractions at a buoyant density lighter than that of the calnexin-containing ER fractions in extracts from infected cells (Fig. 7A). In these fractions, 2C, 3A, and their precursors 3AB and 2BC were all present (data not shown), as expected for poliovirus replication complexes (6, 18).
FIG. 7.
Effect of increased ionic strength on cellular membranes and poliovirus-induced vesicles. Following infection with wild-type poliovirus for 4 h, the plasma membranes of COS-1 cells were disrupted in homogenization buffer and the cellular organelles were separated on a 12% stock isotonic Percoll density gradient that did not (A and C) or did (B and D) contain 60 mM KCl; lightest fractions are at the left. (A and B) Distributions of ER membranes and poliovirus-induced vesicles identified by immunoblotting as indicated. (C and D) Distributions of Golgi, mitochondrial, and lysosomal (Lyso) membranes identified by immunoblotting against p115, immunoblotting against mtHSP70 [HSP70m(mito), and β-hexosaminidase (β-Hex) assays as indicated.
The buoyant density of both virally induced and ER membranes increases after KCl treatment.
To monitor physical attributes of the various membrane organelles other than protein composition, we tested the effect of increased KCl concentration on their fractionation in Percoll gradients. When organelles from poliovirus-infected and uninfected COS-1 cells were separated in the presence and absence of 60 mM KCl, the buoyant densities of Golgi, lysosomes, and mitochondria remained unchanged (Fig. 7C and D). However, both the calnexin-containing ER membranes and the 3A- and 2C-containing membranes showed large increases in buoyant density in the presence of 60 mM KCl (Fig. 7A and B). Thus, the membranes incorporated into the virus-induced vesicles and ER membranes responded similarly to ionic change.
Endosomal and poliovirus-induced membranes do not cofractionate.
Modified endosomal membranes have been suggested to support the RNA replication complexes of other positive-strand RNA viruses such as Semliki Forest virus and rubella virus (19, 29). The distribution of organelles containing Rab9, an endosomal marker, was determined for poliovirus-infected cells. The distribution of Rab9 was disperse, making it difficult to determine the extent of colocalization, or lack thereof, with the 2C-containing poliovirus-induced membranes. However, upon the addition of 60 mM KCl to the Percoll gradients, the 2C-containing and Rab9-containing membranes responded quite differently (Fig. 8). Specifically, the distribution of the Rab9-containing endosomal fractions obtained from the poliovirus-infected cells was not substantially altered by the addition of KCl (Fig. 8A), whereas the 2C-containing membranes displayed a higher apparent buoyant density in the presence of 60 mM KCl (Fig. 8B). Therefore, although there may be some endosomal membranes or proteins present in the poliovirus-induced membranes, the virally induced membranes did not display the osmotic properties of endosomes and the cellular endosomal population was not consumed during poliovirus infection.
DISCUSSION
Poliovirus-induced vesicles derive from the ER by the action of 2BC and 3A proteins.
Several hypotheses have been proposed to explain the origin of the large numbers of membranous vesicles that accumulate in poliovirus-infected cells. Ultrastructural evidence for the budding of poliovirus-induced vesicles from ER membranes has been reported (5). The disappearance of the Golgi apparatus in poliovirus-infected cells (8, 10, 12, 41, 48) is consistent with the idea that the virus-induced vesicles derive from dispersed Golgi membranes (39). The inhibition of poliovirus replication by brefeldin A (24, 30), which inhibits retrograde traffic from the Golgi to the ER and causes mixing of the Golgi and ER compartments, argues that either Golgi-to-ER traffic or the integrity of the ER and Golgi are critical in supporting poliovirus infection.
Although the buoyant densities of the poliovirus-induced vesicles were not identical to either ER, Golgi, lysosomes, endosomes, or mitochondria, they were closest to that of ER membranes (Fig. 2 and 8). Upon changes in ionic strength of the density gradient media, the poliovirus-induced vesicles behaved most like ER (Fig. 7).
Poliovirus 2C and 3A cofractionated with ER membranes in Percoll gradients when expressed together in the absence of other viral proteins (Fig. 4). In contrast, 3A protein coexpressed with 2BC or in the context of poliovirus infection quantitatively localized to membranes whose buoyant density was lighter than that of ER (Fig. 6 and 8). The simplest explanation of these data is that 2BC localizes to the ER and causes the formation of membranous vesicles into which viral 3A protein is recruited but most resident cellular ER proteins are excluded.
Further support for the derivation of the poliovirus-induced vesicles from the ER comes from studies in which host cells exhibited an increase in free cytosolic calcium concentrations as the infectious cycle of poliovirus progressed. The independent expression of 2BC, but not 2C, was sufficient to cause this increase in intracellular calcium concentration (23). The loosely bound pool of calcium utilized in second messenger signaling systems is stored within the ER (reviewed in reference 50), suggesting the possibility that the large increase in cytosolic calcium that occurs during poliovirus infection may result from the disruption of ER membranes by the action of 2BC.
It has been reported previously (1, 10, 48) that the expression of poliovirus 2BC protein mimics the morphology of poliovirus-infected cells. Consistent with this idea, the same decrease in buoyant density was observed for membranes isolated from poliovirus-infected and 2BC-transfected cells. Furthermore, we observed in the present study a small number of membranous vesicles similar in morphology to those observed during poliovirus infection. However, most of the membranous vesicles containing 2C epitopes (Fig. 6C) that formed during the expression of 2BC in isolation were much larger than those observed during poliovirus infection. The 2BC-induced membranes further differed from poliovirus-induced vesicles in that they displayed single membranes and lacked electron-dense material in the lumen (Fig. 6B and C).
In contrast, by both biochemical and ultrastructural criteria, the membrane rearrangements observed in cells that expressed both 3A and 2BC resembled those observed during poliovirus infection. Membrane vesicles that displayed double membranes, cytoplasmic lumenal contents, and substantial immunolabeling by anti-2C antibody were observed both in poliovirus-infected cells (Fig. 1B and C) (41) and in cells that coexpressed 3A and 2BC (Fig. 6B and C). Furthermore, 3A was recruited into the membranes that contain 2BC protein in both poliovirus-infected (Fig. 7A) and 3A- and 2BC-transfected (Fig. 6A) cells. Whether the interaction between 2BC and 3A proteins is direct or indirect, mediated by cellular proteins or lipids, remains to be determined.
Similarities between vesicles induced by positive-strand RNA viruses and autophagic vacuoles.
The appearance of double-membraned vesicles is not unique to cells infected with picornaviruses; double-membraned vesicles have been observed in cells infected with arteriviruses (34) and coronaviruses (15) as well. The observation that these virus-induced vesicles are bounded by double lipid bilayers argued that they might form by a mechanism similar to that of autophagic vacuoles (12, 41). Other observations consistent with an autophagy-like origin for the poliovirus-induced vesicles are (i) the apparent ER origin (reference 5 and this work), even though resident ER proteins are, for the most part, excluded (Fig. 2); (ii) the presence of lysosomal enzymes in the vesicles (41), even though they derive from the ER; and (iii) the ability of 2C protein to cause invaginations into ER membranes such that pools of cytoplasm are trapped within ER membranes that contain 3A protein (Fig. 4), providing a potential intermediate in the formation of double-membraned vesicles with cytoplasmic lumen (12, 41). Like the poliovirus-induced vesicles studied here, autophagic vacuoles isolated from primary rat hepatocytes migrate very similarly to ER membranes in Percoll gradients, at a lower buoyant density than lysosomes (32). For both autophagic vacuoles and poliovirus-induced vesicles, the apparent exclusion from the ER of protein markers such as calnexin and PDI could be a selective sorting process.
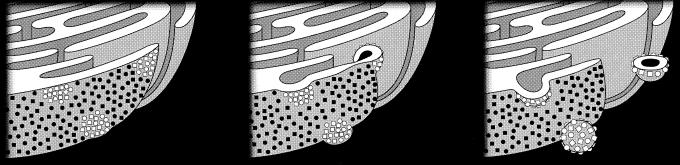
A model for the formation of poliovirus-induced membranes is shown in Fig. 9. In the presence of both 2BC and 3A, double-membraned vesicles that surround cytoplasm and contain both 2BC and 3A protein but exclude resident ER proteins are formed. The formation of predominately single-membraned vesicles by 2BC in isolation can be rationalized by two models. First, it is possible that in the absence of 3A protein, the engagement of the second membrane that changes a budding mechanism into a wrapping mechanism is not accomplished. A second possibility is that, by analogy to the maturation of autophagic vacuoles, 2BC induces the formation of double-membraned vesicles, but in the absence of 3A protein, they rapidly mature into degradative organelles that have lost their inner membranes and cytoplasmic lumen.
FIG. 9.
Model for the formation of poliovirus-induced vesicles. Proteins of the poliovirus replication complex, especially 2BC and 3A (open squares and circles), accumulate in patches on the ER. Double-membraned vesicles derive from these ER membranes by a mechanism that excludes cellular membrane proteins (closed squares and circles) but includes cytoplasmic material (black) in the lumen.
Formation of viral RNA replication complexes on intact or rearranged intracellular membranes.
There is disparity in the literature concerning the cellular origin of the membranes on which positive-strand RNA viruses assemble their RNA replication complexes. If, as we suggest, virus-induced membranes for many positive-strand RNA viruses are formed from the ER by a process that is similar to cellular autophagy, the presence of markers from later in the secretory pathway as well as endosomal contents can be readily explained. For autophagic vacuoles, delivery of extracellular materials such as colloidal gold has been reported to occur within 10 min of its addition to the extracellular milieu (28). Therefore, the presence of these markers does not necessary argue for an endosomal or lysosomal origin for the membranous vesicles induced by positive-strand RNA viruses and is just as consistent with an autophagic origin for the cytopathic vacuoles formed during alphavirus, rubella virus, and mouse hepatitis virus infection (19, 29, 35, 52).
The molecular biology of autophagy in mammalian cells is just beginning to be investigated but may play a significant role in host-pathogen interactions. Twelve genes in Saccharomyces cerevisiae are known to be required for autophagy in yeast (4); it has been demonstrated recently that the human homolog of one of them, beclin-1, is required for the induction of autophagy in human cells (27) and reduces Sindbis virus pathogenesis in mice (26). The maturation of vaccinia virus (44) and African swine fever virus (2), both DNA viruses, involves the addition of two membranes acquired by wrapping subviral particles by the intermediate compartment and the ER, respectively. The pathogenic bacterium Legionella pneumophila grows in modified membranous compartments within infected mammalian cells; these compartments are thought to form by an autophagic mechanism by virtue of their double lipid bilayers derived, at least in part, from the ER (47). Mutant bacteria that fail to thrive in mouse macrophages showed an increase in the accumulation of lysosomal markers and degradative enzymes in these compartments, suggesting that the wild-type bacterial proteins prevent normal maturation of the autophagic vacuoles into degradative compartments (3). Finally, as we have shown here, poliovirus proteins 2BC and 3A, which mimic the biochemical and morphological changes that occur during poliovirus infection, are likely to promote a process similar to cellular autophagy, potentially providing valuable tools to dissect this crucial cellular process.
ACKNOWLEDGMENTS
We thank our colleagues Peter Sarnow, Amy Clewell, Stephen Deitz, Dana Dodd, Stanley Falkow, Erin Gaynor, Richard Scheller, and Elizabeth Stillman for comments on the manuscript, and we are grateful to John Doedens and Dana Dodd for performing initial transfection experiments. We thank Kurt Bienz, Denise Egger, Gerry Waters, Sue Pierce, Hans-Peter Hauri, and Suzanne Pfeffer for antibodies used in this study.
This work was supported by the National Institutes of Health and by the Hutchison Program for Translational Medicine of Stanford University.
REFERENCES
- 1.Aldabe R, Carrasco L. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem Biophys Res Commun. 1995;206:64–76. doi: 10.1006/bbrc.1995.1010. [DOI] [PubMed] [Google Scholar]
- 2.Andres G, Garcia-Escudero R, Simon-Mateo C, Vinuela E. African swine fever virus is enveloped by a two-membraned collapsed cisterna derived from the endoplasmic reticulum. J Virol. 1998;72:8988–9001. doi: 10.1128/jvi.72.11.8988-9001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews H L, Vogel J P, Isberg R R. Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect Immun. 1998;66:950–958. doi: 10.1128/iai.66.3.950-958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba M, Osumi M, Scott S V, Klionsky D J, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bienz K, Egger D, Pasamontes L. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology. 1987;160:220–226. doi: 10.1016/0042-6822(87)90063-8. [DOI] [PubMed] [Google Scholar]
- 6.Bienz K, Egger D, Pfjister T, Troxler M. Structural and functional characterization of the poliovirus replication complex. J Virol. 1992;66:2740–2747. doi: 10.1128/jvi.66.5.2740-2747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bienz K, Egger D, Troxler M, Pasamontes L. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J Virol. 1990;64:1156–1163. doi: 10.1128/jvi.64.3.1156-1163.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bienz K, Egger D, Wolff D A. Virus replication, cytopathology, and lysosomal enzyme response of mitotic and interphase HEp-2 cells infected with poliovirus. J Virol. 1973;11:565–574. doi: 10.1128/jvi.11.4.565-574.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carette J E, Stuiver M, van Lent J, Wellink J, van Kammen A. Cowpea mosaic virus infection induces a massive proliferation of endoplasmic reticulum but not Golgi membranes and is dependent on de novo membrane synthesis. J Virol. 2000;74:6556–6563. doi: 10.1128/jvi.74.14.6556-6563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho M W, Teterina N, Egger D, Bienz K, Ehrenfeld E. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology. 1994;202:129–145. doi: 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- 11.Dahl R, Staehelin L A. High-pressure freezing for the preservation of biological structure: theory and practice. J Electron Microsc Tech. 1989;13:165–174. doi: 10.1002/jemt.1060130305. [DOI] [PubMed] [Google Scholar]
- 12.Dales S, Eggers H J, Tamm I, Palade G E. Electron microscopic study of the formation of poliovirus. Virology. 1965;26:379–389. doi: 10.1016/0042-6822(65)90001-2. [DOI] [PubMed] [Google Scholar]
- 13.Doedens J R, Giddings T H, Jr, Kirkegaard K. Inhibition of ER-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J Virol. 1997;71:9054–9064. doi: 10.1128/jvi.71.12.9054-9064.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doedens J R, Kirkegaard K. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 1995;14:894–907. doi: 10.1002/j.1460-2075.1995.tb07071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubois-Dalcq M, Holmes K V, Rentier B. Assembly of enveloped RNA viruses. Vienna, Austria: Springer-Verlag; 1984. [Google Scholar]
- 16.Dunn W A., Jr Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunn W A., Jr Studies on the mechanisms of autophagy: maturation of the autophagic vacuole. J Cell Biol. 1990;110:1935–1945. doi: 10.1083/jcb.110.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egger D, Pasamontes L, Bolten R, Boyko V, Bienz K. Reversible dissociation of the poliovirus replication complex: functions and interactions of its components in viral RNA synthesis. J Virol. 1996;70:8675–8683. doi: 10.1128/jvi.70.12.8675-8683.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Froshauer S, Kartenbeck J, Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green J M, Gu L, Ifkovits C, Kaumaya P T, Conrad S, Pierce S K. Generation and characterization of monoclonal antibodies specific for members of the mammalian 70-kDa heat shock protein family. Hybridoma. 1995;14:347–354. doi: 10.1089/hyb.1995.14.347. [DOI] [PubMed] [Google Scholar]
- 21.Guskey L E, Smith P C, Wolff D A. Patterns of cytopathology and lysosomal enzyme release in poliovirus-infected HEp-2 cells treated with either 2-(α-hydroxybenzyl)-benzimidazole or guanidine HCl. J Gen Virol. 1970;6:151–161. doi: 10.1099/0022-1317-6-1-151. [DOI] [PubMed] [Google Scholar]
- 22.Hope D A, Diamond S E, Kirkegaard K. Genetic dissection of interaction between poliovirus RNA-dependent RNA polymerase and viral protein 3AB. J Virol. 1997;71:9490–9498. doi: 10.1128/jvi.71.12.9490-9498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irurzun A, Arroyo J, Alvarez A, Carrasco L. Enhanced intracellular calcium concentration during poliovirus infection. J Virol. 1995;69:5142–5146. doi: 10.1128/jvi.69.8.5142-5146.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irurzun A, Perez L, Carrasco L. Involvement of membrane traffic in the replication of poliovirus genomes: effects of brefeldin A. Virology. 1992;191:166–175. doi: 10.1016/0042-6822(92)90178-r. [DOI] [PubMed] [Google Scholar]
- 25.Lama J, Paul A V, Harris S, Wimmer E. Properties of purified recombinant poliovirus protein 3AB as substrate for viral proteinases and as co-factor for RNA polymerase 3Dpol. J Biol Chem. 1994;269:66–70. [PubMed] [Google Scholar]
- 26.Liang X H, Kleeman L K, Jiang H H, Gordon G, Goldman J E, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel BCL-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang X H, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 28.Liou W, Geuze H J, Geelen M J H, Slot J W. The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J Cell Biol. 1997;136:61–70. doi: 10.1083/jcb.136.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magliano D, Marshall J A, Bowden D S, Vardaxis N, Meanger J, Lee J-Y. Rubella virus replication complexes are virus-modified lysosomes. Virology. 1998;240:57–63. doi: 10.1006/viro.1997.8906. [DOI] [PubMed] [Google Scholar]
- 30.Maynell L A, Kirkegaard K, Klymkowsky M W. Inhibition of poliovirus RNA synthesis by brefeldin A. J Virol. 1992;66:1985–1994. doi: 10.1128/jvi.66.4.1985-1994.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirzayan C, Wimmer E. Biochemical studies on poliovirus polypeptide 2C: evidence for ATPase activity. Virology. 1994;199:176–187. doi: 10.1006/viro.1994.1110. [DOI] [PubMed] [Google Scholar]
- 32.Niioka S, Goto M, Ishibashi T, Kadowaki M. Identification of autolysosomes directly associated with proteolysis on the density gradients in isolated rat hepatocytes. J Biochem. 1998;124:1086–1093. doi: 10.1093/oxfordjournals.jbchem.a022225. [DOI] [PubMed] [Google Scholar]
- 33.Paul A V, van Boom J H, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 34.Pedersen I W, van der Meer Y, Roos N, Snijder E J. ORF1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double membrane vesicles which carry the viral replication complex. J Virol. 1999;73:2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peranen J, Laakkonen P, Hyvonin M, Kaarianen L. The alphavirus replicase protein nsP1 is membrane-associated and has affinity to endocytic organelles. Virology. 1995;208:610–620. doi: 10.1006/viro.1995.1192. [DOI] [PubMed] [Google Scholar]
- 36.Peters T J, Muller M, De Duve C. Lysosomes of the arterial wall. I. Isolation and subcellular fractionation of cells from normal rabbit aorta. J Exp Med. 1972;136:1117–1139. doi: 10.1084/jem.136.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reunanen H, Punnonen E L, Hirsimaki P. Studies on vinblastine-induced autophagocytosis in mouse liver. V. A cytochemical study on the origin of membranes. Histochemistry. 1985;83:513–517. doi: 10.1007/BF00492453. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez P L, Carrasco L. Poliovirus protein 2C has ATPase and GTPase activities. J Biol Chem. 1993;268:8105–8110. [PubMed] [Google Scholar]
- 39.Sandoval I V, Carrasco L. Poliovirus infection and expression of the poliovirus protein 2B provoke the disassembly of the Golgi complex, the organelle target for the antipoliovirus drug Ro-090179. J Virol. 1997;71:4679–4693. doi: 10.1128/jvi.71.6.4679-4693.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 41.Schlegel A, Giddings T H, Ladinsky M S, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schweizer A, Rohrer J, Slot J W, Geuze H J, Kornfeld S. Reassessment of the subcellular localization of p63. J Cell Sci. 1995;108:2477–2485. doi: 10.1242/jcs.108.6.2477. [DOI] [PubMed] [Google Scholar]
- 43.Sheeler P. Density gradient centrifugation. In: Sheeler P, editor. Centrifugation in biology and medical science. New York, N.Y: John Wiley & Sons, Inc.; 1981. pp. 81–99. [Google Scholar]
- 44.Sodiek B, Doms R W, Ericsson M, Hiller G, Machamer C E, van't Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stromhaug P E, Berg T O, Fengsrud M, Seglen P O. Purification and characterization of autophagosomes from rat hepatocytes. Biochem J. 1998;335:217–224. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suhy D A, Simon K D, Linzer D I, O'Halloran T V. Metallothionein is part of a zinc-scavenging mechanism for cell survival under conditions of extreme zinc deprivation. J Biol Chem. 1999;274:9183–9192. doi: 10.1074/jbc.274.14.9183. [DOI] [PubMed] [Google Scholar]
- 47.Swanson M S, Isberg R R. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teterina N L, Gorbalenya A E, Egger D, Bienz K, Ehrenfeld E. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J Virol. 1997;71:8962–8972. doi: 10.1128/jvi.71.12.8962-8972.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Towner J, Semler B L. Determinants of membrane association on poliovirus protein 3AB. J Biol Chem. 1996;271:26810–26818. doi: 10.1074/jbc.271.43.26810. [DOI] [PubMed] [Google Scholar]
- 50.Tsien R W, Tsien R Y. Calcium channels, stores and oscillations. Annu Rev Cell Biol. 1990;6:715–760. doi: 10.1146/annurev.cb.06.110190.003435. [DOI] [PubMed] [Google Scholar]
- 51.Ueno T, Ishidoh K, Mineki R, Tanida I, Murayama K, Kadowaki M, Kominami E. Autolysosomal membrane-associated betaine homocysteine methyltransferase. Limited degradation fragment of a sequestered cytosolic enzyme monitoring autophagy. J Biol Chem. 1999;274:15222–15229. doi: 10.1074/jbc.274.21.15222. [DOI] [PubMed] [Google Scholar]
- 52.van der Meer Y, Snijder E J, Dobbe J C, Schleich S, Denison M R, Spaan W J M, Krijnse Locker J. Location of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J Virol. 1999;73:7641–7657. doi: 10.1128/jvi.73.9.7641-7657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waters M G, Clary D O, Rothman J E. A novel 115-kD peripheral membrane protein is required for intercisternal transport in the Golgi stack. J Cell Biol. 1992;118:1015–1026. doi: 10.1083/jcb.118.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watzele G, Bachofner R, Berger E G. Immunocytochemical localization of the Golgi apparatus using protein-specific antibodies to galactosyltransferase. Eur J Cell Biol. 1991;56:451–458. [PubMed] [Google Scholar]
- 55.Wessel D, Flugge U I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]