Abstract
Reactive oxygen species (ROS)-associated oxidative stress, inflammation storm, and massive hepatocyte necrosis are the typical manifestations of acute liver failure (ALF), therefore specific therapeutic interventions are essential for the devastating disease. Here, we developed a platform consisting of versatile biomimetic copper oxide nanozymes (Cu NZs)-loaded PLGA nanofibers (Cu NZs@PLGA nanofibers) and decellularized extracellular matrix (dECM) hydrogels for delivery of human adipose-derived mesenchymal stem/stromal cells-derived hepatocyte-like cells (hADMSCs-derived HLCs) (HLCs/Cu NZs@fiber/dECM). Cu NZs@PLGA nanofibers could conspicuously scavenge excessive ROS at the early stage of ALF, and reduce the massive accumulation of pro-inflammatory cytokines, herein efficiently preventing the deterioration of hepatocytes necrosis. Moreover, Cu NZs@PLGA nanofibers also exhibited a cytoprotection effect on the transplanted HLCs. Meanwhile, HLCs with hepatic-specific biofunctions and anti-inflammatory activity acted as a promising alternative cell source for ALF therapy. The dECM hydrogels further provided the desirable 3D environment and favorably improved the hepatic functions of HLCs. In addition, the pro-angiogenesis activity of Cu NZs@PLGA nanofibers also facilitated the integration of the whole implant with the host liver. Hence, HLCs/Cu NZs@fiber/dECM performed excellent synergistic therapeutic efficacy on ALF mice. This strategy using Cu NZs@PLGA nanofiber-reinforced dECM hydrogels for HLCs in situ delivery is a promising approach for ALF therapy and shows great potential for clinical translation.
Keywords: Acute liver failure, Nanozyme, Hepatocyte-like cells, Human adipose-derived mesenchymal stem/stromal cells, Electrospun nanofiber
Graphical abstract
Highlights
-
•
A synergistic composite consisting of antioxidant nanozyme-loaded nanofiber and dECM hydrogel for HLCs delivery were fabricated.
-
•
Cu NZs@PLGA nanofibers possessed ROS scavenging and pro-angiogenesis capabilities.
-
•
HLCs showed hepatic function compensation and anti-inflammatory activity. dECM hydrogels enhanced the functions of HLCs.
-
•
HLCs/Cu NZs@fiber/dECM exhibited synergistic effects in acute liver failure treatment.
1. Introduction
Acute liver failure (ALF), a life-threatening disease with widespread hepatocellular necrosis and acute deterioration of liver function, can rapidly evolve to a lethal outcome [1,2]. However, there is still no satisfactory treatment for curing ALF, other than liver transplantation. It is reported that overproduction and excessive accumulation of reactive oxygen species (ROS) are the early pathological hallmarks of ALF and closely correlated with the disease progression owing to the induced oxidative stress, inflammation storm, and massive hepatocyte necrosis [[3], [4], [5]]. Therefore, effective elimination of ROS at the early stages to avert irreversible damage of hepatocytes and replenishing functional hepatocytes are critical in the treatment of ALF.
With the rapid development of biotechnology and nanoscience, a variety of nanomaterials with enzymatic mimicking activities, namely nanozymes, have attracted tremendous interest [[6], [7], [8], [9], [10]]. The low cost, good stability, and high production make nanozymes widely applied in various biomedical scenarios [6,7,11]. Especially, nanozymes exhibiting antioxidant properties are beneficial for oxidative stress-related disease therapy [3,[8], [9], [10],12,13]. These nanozymes usually possessed similar enzyme catalytic properties of catalase (CAT), superoxide dismutase (SOD), and/or glutathione peroxidase (GPx). CAT mimics can decompose H2O2 into O2 and H2O. SOD mimics can scavenge O2·− through the disproportionation of O2·− to H2O2 and O2. GPx mimics can eliminate H2O2 into H2O and catalyze glutathione (GSH) to form glutathione disulfide (GSSG). Copper oxide nanozymes (Cu NZs) can protect cells from oxidative damage [[14], [15], [16]]. Cu NZs are one type of nanomaterials with typical properties of multiple antioxidant enzymes, including CAT, SOD, and GPx, which show enormous potential as biomimetic antioxidants to modulate ROS homeostasis [3,14,15,[17], [18], [19], [20], [21], [22]]. However, conventional intravenous administration of nanozymes often leads to undesired organ distribution, which may not only affect the therapeutic efficacy, but also evoke potential side effects [19,20,23,24]. Electrospun nanofibers, with the analogous structure of naïve extracellular matrix (ECM), have been used for the delivery of diverse drugs [25]. Poly(lactic-co-glycolic acid) (PLGA) is an FDA-approved biodegradable polymer compound with good biocompatibility and good processability [26]. Moreover, PLGA electrospun nanofibers can be easily functionalized by nanozymes, and electrospinning process does not affect the ROS scavenging activity of nanozymes [[27], [28], [29], [30]]. Thus, in situ delivery of Cu NZs using electrospun nanofibers can be an alternative solution for effectively scavenging the excessive ROS in the ALF liver.
Of note, although the effective clearance of ROS can alleviate the local harsh microenvironment, massive hepatocyte necrosis and liver function loss in ALF still call for the necessary compensation of functional hepatocytes. However, limited donors, poor cell proliferation, and difficulty in maintaining hepatocyte functionality in vitro impede their therapeutic applications [1,31]. Mesenchymal stem/stromal cells (MSCs) with multi-lineage differentiation potential, self-renewal ability, as well as anti-oxidative and anti-inflammatory functions, can be conveniently isolated and expanded in vitro. Moreover, MSCs also exhibit low immune responses without ethical issues in clinical translation [[32], [33], [34]]. Studies have evidenced that MSCs can effectively differentiate into functional hepatocyte-like cells (HLCs) with customized cytokine cocktails [35,36]. MSC-derived HLCs not only retain anti-inflammatory activity, but also possess hepatic-specific functions [[37], [38], [39]]. Therefore, MSC-derived HLCs show great potential as the alternative cell source of hepatocyte therapy [31,[40], [41], [42]]. However, studies have shown that most of the implanted cells are obstructed in the lungs after intravenous injection or cleared by macrophages/phagocytes after intrasplenic transplantation [41,43]. In addition, the microenvironment of ALF liver is a key consideration for cellular therapy. The inflammatory environment in which cells are implanted can cause necrosis and apoptosis of transplanted cells [31,44]. Alternatively, HLCs-laden hydrogels acting as engineered hepatic tissues for in situ hepatocyte transplantation have gained rapid development in recent years [45,46]. Among them, the liver decellularized extracellular matrix (dECM) hydrogels that provide a similar microenvironment of naïve liver for encapsulated cells without adverse immune responses are of high interest [34,47,48]. Nevertheless, the weak mechanical strength of dECM hydrogels and the harsh microenvironment at the injured site make it challenging for the proper functioning of implanted HLCs.
Here, we developed a synergistic therapeutic platform consisting of Cu NZs-loaded PLGA nanofibers (Cu NZs@PLGA nanofibers) and dECM hydrogels to deliver human adipose-derived mesenchymal stem/stromal cells-derived HLCs (hADMSCs-derived HLCs) for efficient treatment of ALF. Cu NZs@PLGA nanofibers were fabricated by electrospinning and showed distinct excessive ROS elimination ability, good biocompatibility, as well as proangiogenic effect. In addition, the hADMSCs-derived HLCs showed enhanced hepatic-functional expressions when cultured in the dECM hydrogels. After being implanted into ALF-challenged mice, mechanical support of nanofibers and the protective Cu NZs permitted the functioning of the implanted hADMSCs-derived HLCs in time and the relief of oxidative stress and inflammation at the early stage. The multifunctional therapeutic biohybrid platform exhibited a great and highly efficient therapeutic effect on ALF, which was evidenced by reduced cell apoptosis and tissue necrosis but increased cell proliferation and vascularization, as well as restored liver functionality (Scheme 1). These results indicate that in situ delivery of HLCs using Cu NZs@PLGA nanofiber-reinforced dECM hydrogels is a promising approach for the treatment of ALF and herein holds great potential for clinical translation.
Scheme 1.
Schematic diagram of the composite platform consisting of Cu NZs@PLGA nanofibers and dECM hydrogels to deliver hADMSCs-derived HLCs for the treatment of CCl4-induced ALF. (A, B) The synthesis of Cu NZs@PLGA nanofibers, dECM hydrogels, and the composite HLCs/Cu NZs@fiber/dECM. (C) Schematic illustration of the HLCs/Cu NZs@fiber/dECM for CCl4-induced ALF therapy through the effects of ROS elimination, angiogenesis promotion, anti-inflammation, hepatocyte-related functions, and facilitating liver regeneration.
2. Materials and methods
2.1. Synthesis and characterization of Cu NZs
Cu NZs were synthesized according to the previously reported procedures with slight modifications [14]. Briefly, CuCl2 (7447-39-4, Aladdin, China) was dispersed in 50 mL of deionized water at a final concentration of 10 mM. After stirring for 10 min at 80 °C, 50 mL of 100 mM l-ascorbic acid (50-81-7, Alfa Aesar, USA) aqueous solution was slowly added into the CuCl2 solution. The mixture pH was adjusted to 8.5 ± 0.5 using 1 M NaOH (1310-73-2, Macklin, China) and kept stirred for 12 h at 80 °C. After the reaction, the large spun precipitations were discarded while supernatants were further dialyzed against deionized water for 48 h using a 10 kDa dialysis tube (TX132, Viskase, China). The final Cu NZs were obtained by centrifugation. The morphology and size of Cu NZs were characterized by a transmission electron microscope (TEM, FEI Tecnai G2 Spirit) at a voltage of 120 kV. The crystal structure and oxidation state of Cu NZs were characterized using an X-ray diffractometer (XRD, Empyrean) at a voltage of 45 kV and a current of 40 mA.
2.2. Electrospinning of Cu NZs@PLGA nanofibers
Cu NZs (600 μg mL−1) and 10 wt/v% poly(lactic-co-glycolic acid) (PLGA, LA/GA = 50/50, Mw ∼100,000, Shenzhen Polymtek Biomaterial) were homogeneously dispersed in hexafluoroisopropanol (HFIP, ≥99.8%, Aladdin, China) and electrospun into nanofibers at a voltage of 13 kV with a feeding rate of 0.6 mL h−1. The electrospun nanofibers were collected and placed in vacuum at room temperature (RT) overnight to remove the residual solvent. The plain PLGA nanofibers were prepared following the same procedure without the addition of Cu NZs. The morphology characterization and energy dispersive spectroscopy (EDS) analysis for element mapping of the nanofibers were performed using a scanning electron microscope (SEM, Quanta 400F) at a voltage of 20 kV. The stretchability of plain PLGA fibers and Cu NZs@PLGA fibers were performed using an electronic universal material testing machine (CMT4202).
To investigate the releasing profile of Cu NZs, 25 mg Cu NZs@PLGA fibers were incubated with 1 mL ddH2O at 37 °C. After incubation for 0, 2, 4, 8, and 10 h. The released Cu NZs were detected by sodium diethyldithiocarbamate trihydrate (20624-25-3, Aladdin, China) colorimetry method. The releasing profile of Cu NZs from dECM hydrogels was used as the control.
2.3. H2O2 scavenging ability of Cu NZs and Cu NZs@PLGA nanofibers
Different concentrations of Cu NZs in 1 × phosphate-buffered saline (1 × PBS) ranging from 0 to 200 ng mL−1 were incubated with 1 mM H2O2 (CS7730, G-Clone, China) at 37 °C for 2 h, respectively. The remaining H2O2 was detected by Amplex Red (Meilunbio, China) and horseradish peroxidase (Meilunbio, China) following the manufacturer's protocol. Amplex Red reacted with H2O2 to form the resorufin products with an absorption peak at 571 nm. Resorufin was quantified using a UV-2600 ultraviolet–visible (UV–Vis) spectrophotometer (SHIMADZU, China). 1 × PBS without Cu NZs was used as the control.
Cu NZs@PLGA nanofibers containing different amounts of Cu NZs (0–200 ng mL−1) were immersed in 1 × PBS and incubated with 1 mM H2O2 at 37 °C for 0–10 h, respectively. The remaining H2O2 in groups was detected at 2, 4, 6, 8, and 10 h. PLGA nanofibers without Cu NZs were used as the control. The H2O2 clearance rate was calculated with the formula: clearance rate (%) = (Ac-Ai)/Ac × 100%. Ac is the absorbance of the control and Ai is the absorbance of the samples.
2.4. ·OH scavenging ability of Cu NZs@PLGA nanofibers
To study the scavenging ability of Cu NZs@PLGA nanofibers for other ROS, a typical 3,3′,5,5′-tetramethylbenzidine (TMB) (54827-17-7, Aladdin, China) chromogenic method was applied to monitor ·OH clearance rate at different time points. Cu NZs@PLGA nanofibers were incubated in acetic acid/sodium acetate buffer (HAc/NaAc, 0.5 M, pH 4.5) (Aladdin, China) with a final Cu NZs concentration of 150 ng mL−1 for 0, 2, 6, and 12 h, respectively. The supernatants were harvested and incubated with the working solution containing 1 mM FeSO4 (7782-63-0, Sangon, China), 1 mM H2O2, and 250 μM TMB for 7 min in dark. The HAc/NaAc buffer without pretreatment using nanofibers was used as the control. The oxidized TMB (oxTMB) by ·OH showed blue color and could be measured by a UV–Vis spectrophotometer at a wavelength of 652 nm. Accordingly, the ·OH clearance rate was calculated using the same formula of H2O2 clearance rate.
2.5. O2·− scavenging ability of Cu NZs@PLGA nanofibers
The O2·− depletion activity of Cu NZs@PLGA nanofibers was investigated via the nitroblue tetrazolium (NBT) chromogenic method. Cu NZs@PLGA nanofibers were immersed in Tris-HCl buffer (10 mM, pH 7.4) (Biosharp, China) with a final Cu NZs concentration of 150 ng mL−1. After 0, 2, 6, or 12 h preincubation, the working solution containing 6.5 mM l-methionine (C11385354, Macklin, China), 37.5 μM NBT (C11167116, Macklin, China), and 10 μM riboflavin (C11507808, Macklin, China) was homogeneously mixed with the pre-treated Tris-HCl solutions, respectively, and irradiated using a SCIENTZ03-II UV radiation instrument (Scientz, China) with a wavelength of 254 nm (60 W, 150 mm) for 3 min at RT. UV irradiation resulted in the generation of O2·− from riboflavin and l-methionine, which further converted NBT to blue formazan. After illumination, the absorbances of the mixtures were measured by a UV–Vis spectrophotometer at a wavelength of 560 nm. The clearance rate of O2·− was calculated according to the same formula of H2O2 clearance rate.
2.6. Cell culture
hADMSCs were purchased from Saliai Stem Cell Technology, Inc. (China). hADMSCs were cultured in a serum-free medium containing 5% serum substitute (G03010, Saliai, China). hADMSCs between the passages 5 and 9 were used for the experiments. AML12 cells were purchased from the National Collection of Authenticated Cell Cultures (China) and cultured with Dulbecco's modified Eagle's medium/nutrient mixture F-12 1:1 (DMEM/F-12, 11330-032, Gibco, USA) containing 10% fetal bovine serum (FBS) (Gibco, USA), 1 × insulin-transferrin-selenium supplement (ITS) (ITSS-10201, Cyagen, China) and 40 ng mL−1 dexamethasone (D4902, Sigma-Aldrich, USA). Human umbilical vein endothelial cells (HUVECs) were cultured in DMEM/F-12 containing 10% FBS, 1% penicillin/streptomycin. Human monocytic cell line THP-1 was cultured with RPMI 1640 medium (Gibco, USA), 10% heated-inactivated FBS (Gibco, USA), 1% penicillin/streptomycin (Gibco, USA), and 50 μM beta-mercaptoethanol (Sigma-Aldrich, USA). RAW264.7 cells were cultured with Dulbecco's Modified Eagle's Medium (DMEM) (Gibco, USA), 10% heated-inactivated FBS, 1% penicillin/streptomycin. All cells were cultured in an incubator supplemented with 5% CO2 at 37 °C.
2.7. Biocompatibility evaluation of Cu NZs@PLGA nanofibers
AML12 cells, hADMSCs, and HLCs were seeded in 96-well plates at a density of 1 × 104 cells per well, respectively. After culture overnight, the supernatant media were replaced by fresh media containing various amounts of Cu NZs@PLGA nanofibers (equivalent Cu NZs concentrations ranging from 0 ng mL−1 to 200 ng mL−1). The media and nanofibers were discarded at 24 h and cell viability was assessed using a cell counting kit-8 (CCK-8, K1018, APExBIO, USA) following the manufacturer's instructions. The cell viability was quantified by the absorbance at 450 nm using a Synergy H1 microplate reader (BioTek).
2.8. ROS scavenging capability of Cu NZs@PLGA nanofibers in AML12 cells
AML12 cells seeded on 48-well plates at a density of 2 × 104 cells per well were pre-incubated with various amounts of Cu NZs@PLGA nanofibers (equivalent Cu NZs concentrations ranging from 0 ng mL−1 to 200 ng mL−1) overnight. The electrospun nanofibers were placed on the top surface of the seeded AML-12 cells. The cells were challenged with 500 μM H2O2 in the cell cultured medium for 24 h, after which the cell viability was assessed using CCK-8, and the intracellular ROS level was stained using 2′,7′-dichlorofluorescin diacetate (DCFH-DA, S0033S, Beyotime, China) in accordance with the manufacturer's instructions. Briefly, AML12 cells were gently washed with 1 × PBS and incubated with 10 μM DCFH-DA/1 × PBS staining solution at 37 °C in dark for 30 min. The staining solution was removed before the addition of 4% paraformaldehyde (PFA, Biosharp, China) for cell fixation. Cell nuclei were stained using 0.2 μg mL−1 4,6-diamidino-2-phenylindole (DAPI) at RT for 15 min. The intracellular ROS were imaged with an inverted fluorescence microscope (Nikon, Japan) and quantified by the mean fluorescent intensity via flow cytometry (Beckman, CytoFLEX, USA).
2.9. Effects of Cu NZs@PLGA nanofibers on migration and proliferation of HUVECs
The effect of Cu NZs@PLGA nanofibers on HUVECs migration was investigated via scratch assay. HUVECs were seeded on the 24-well plate at a density of 5 × 104 per well and cultured overnight. The cells were starved in serum-free medium for 24 h before a central straight line was scratched on the bottom of the multiple wells using a sterile 1 mL-sized pipette tip. The cells were gently washed with 1 × PBS to remove debris twice and fed with the complete medium containing 1% FBS and various amounts of Cu NZs@PLGA nanofibers (equivalent Cu NZs concentration ranging from 0 ng mL−1 to 200 ng mL−1). The remaining scratch areas were imaged and measured by Image J software (NIH, USA) at 24 and 48 h. Cell migration was characterized using the formula: Cell migration (%) = ((S0-St)/S0) × 100%, where S0 is the initial scratch area and St is the scratch area at 24 or 48 h.
EdU imaging kits (K1076, APExBIO, USA) were used to assess the effect of Cu NZs@PLGA nanofibers on HUVECs proliferation. The cell treatment procedures were the same as that used in the scratch assay except for a lower cell seeding density of 1 × 104 per well. The HUVECs were stained by EdU imaging kits according to the manufacturer's instructions and imaged by an inverted fluorescence microscope with an excitation wavelength of 646 nm and a maximum emission wavelength of 662 nm at 48 h. The relative proliferating cells with positive signals were measured by Image J. HUVECs proliferation rate (%) = EdU-positive cells/total cells × 100%. EdU-positive cells were quantified in three randomly selected areas of each well.
2.10. Hepatic differentiation of hADMSCs in vitro
hADMSCs were induced differentiation toward the hepatic lineage with cytokine cocktails as previously described [35,49,50]. Briefly, hADMSCs were first serum-deprived in the low glucose DMEM (Gibco, USA) with supplements of 20 ng mL−1 epidermal growth factor (EGF, PeproTech, USA) and 10 ng mL−1 basic fibroblast growth factor (bFGF, PeproTech, USA) for two days. The cells were subsequently differentiated in the low glucose DMEM supplemented with 20 ng mL−1 hepatocyte growth factor (HGF, PeproTech, USA), 10 ng mL−1 bFGF, and 0.61 mg mL−1 nicotinamide (Sigma-Aldrich, USA) for one week and maintained in the mature medium containing low glucose DMEM with supplements of 20 ng mL−1 oncostatin M (OSM, PeproTech, USA), 1 mM dexamethasone, and 1 × ITS for another two weeks. The media were changed twice per week.
The produced albumin (ALB) and urea in the supernatant media were measured by an ALB assay kit (CH0101002, Maccura, China) and a urea assay kit (CH0101051, Maccura, China) on day 7 and 21, respectively. After cells were fixed in 4% PFA for 15 min and washed with 1 × PBS twice, the accumulated lipids and glycogen were respectively stained by Oil Red O (Sigma-Aldrich, USA) and periodic acid-Schiff (PAS, Sigma-Aldrich, USA) following the manufacturer's instructions. The supernatant media of hADMSCs and HLCs were collected to measure the secreted HGF by an ELISA kit (EK1H01-48, MultiSciences, China) following the manufacturer's protocols.
2.11. Quantitative real time PCR
Total RNA was extracted from cells or mouse liver tissues by a TRIzon reagent (CW0580S, Cwbiotech, China) and used for reverse transcription with a HiFiScript cDNA synthesis kit (CW2569 M, Cwbiotech, China) following the manufacturer's protocols. Real-time fluorescent quantitative PCR was performed using UltraSYBR mixture (CW2601, Cwbiotech, China) by a QuantStudio™ 5 system (Applied Biosystems) with the primer pairs listed in Supplementary Table S1. All data were normalized to the values of ACTB or GAPDH and relative expression was calculated using the delta-delta Ct method.
2.12. Immunofluorescence staining
Cells were fixed in 4% PFA at RT for 15 min and washed with 1 × PBS twice, before blocking with 5% goat serum for 1 h. Thereafter, cells were incubated with primary antibody overnight at 4 °C. The rabbit-anti-human primary antibodies were diluted in 1 × PBS/0.3% Triton X-100 (Biosharp, China) as follows: ALB (16475-1-AP, Proteintech, China) (1 : 500), E-cadherin (710161, Invitrogen, USA) (1 : 200), dipeptidyl peptidase 4 (DPP4, PAB32046, Bioswamp, China) (1 : 200) and hepatocyte nuclear receptor 4 α (human) (HNF-4α, 61189, Active Motif, USA) (1 : 2000), ATP-binding cassette sub-family C member 2 (ABCC2, PAB35870, Bioswamp, China) (1 : 200). The rabbit-anti-mouse primary antibodies were diluted in 1 × PBS/0.3% Triton X-100 (Biosharp, China) as follows: tumor necrosis factor alpha (TNF-α) (EPR19147, Abcam, USA) (1 : 5000), interleukin 1 beta (IL-1β) (16806-1-AP, Proteintech, China) (1 : 500), inducible nitric oxide synthases (iNOS) (18985-1-AP, Proteintech, China) (1 : 500), interleukin 10 (IL-10) (20850-1-AP, Proteintech, China) (1 : 1000). The cells were subsequently incubated with the secondary antibody conjugated to Alexa Fluor 488 goat-anti-rabbit with a dilution rate of 1 : 500 in 1 × PBS/0.3% Triton X-100 at RT for 2 h in dark and washed by 1 × PBS twice. After staining of cell nuclei by 0.2 μg mL−1 DAPI at RT for 15 min, cells were imaged with an inverted fluorescence microscope.
2.13. Anti-inflammatory ability of HLCs
THP-1 cells were seeded on the 6-well plate at a density of 8 × 105 per well and treated with culture medium containing 100 ng mL−1 12-myristate 13-acetate (PMA) (Sigma-Aldrich, USA) for 48 h to induce activation of THP-1 cells (M0). Then, PMA-containing medium was removed. THP-1 cells (M0) were cultured with fresh medium and HLCs supernatant (1:1) for 24 or 48 h.
Activated THP-1 (M0) and Raw264.7 cells were stimulated with lipopolysaccharide (LPS, 100 ng mL−1) (Sigma-Aldrich, USA) for 24 h to induce M1 polarization. Then, the LPS-containing medium was removed. Polarized THP-1 and Raw264.7 cells were treated with fresh medium and HLCs supernatant (1 : 1) for 24 h.
After incubation in HLC supernatant media for 24 h, RAW264.7 cells were harvested to detect the expression level of the M2 macrophage marker (CD206, Rat monoclonal, 141704) (1 : 500) by flow cytometry following the manufacturer's instructions. The expressions of TNF-α and iNOS in RAW264.7 cells were also detected by Western blot. The rabbit-anti-mouse primary antibodies were diluted as follows: TNF-α (EPR19147, Abcam, USA) (1 : 1000), iNOS (18985-1-AP, Proteintech, China) (1 : 1000).
The expression of pro-inflammatory cytokines IL-1β, TNF-α, iNOS, and transforming growth factor beta 1 (TGF-β1) [20,51,52] as well as the anti-inflammatory cytokine and M2 macrophage marker IL-10 [52,53] and mannose receptor C type 1 (MRC1) [51,54], respectively, in THP-1 and RAW264.7 cells were detected by RT-qPCR.
2.14. Decellularization of liver tissues
Approximately 200 g of porcine liver from a market was gently minced after removal of connected tissues. The liver tissues were frozen at −80 °C and thawed at RT for 3 cycles, during the intervals deionized water wash was performed. The liver tissues were decellularized in the deionized water containing 1% Triton X-100, 0.1% NH3·H2O, 1% penicillin/streptomycin, and 0.01 mM phenylmethylsulfonyl fluoride (PMSF) for 3 days with solution change 3 times per day. Before freeze-drying, the decellularized liver tissues were washed in sterile water for 1 h. All procedures were performed in agitation at 150 rpm. To prepare dECM stock solution, the decellularized liver tissues were further digested by 10% pepsin (Sigma-Aldrich, USA) with a dECM: pepsin weight ratio of 10 : 1 in 0.1 M HCl and agitated at RT for 48 h. The morphology of dECM hydrogels was characterized by SEM.
To investigate the stability of dECM hydrogels in vitro, 500 μL 5 mg mL−1 dECM hydrogels were incubated with 500 μL 1 × PBS that contained 20 μg mL−1 collagenase type II at 37 °C. After incubation for 0, 24, 48, 72, and 96 h, the supernatant liquids were discarded, and the remaining hydrogels were lyophilized for measurement of weight change. The degradation was calculated using the formula: degradation rate (%) = (W0-Wi)/W0 × 100%, where W0 is the initial weight of dECM hydrogels and Wi is the weight of remaining hydrogels.
2.15. Biocompatibility of dECM hydrogel and its effects on hepatic functions
The dECM stock solution was diluted by basic medium to a final concentration of 5 mg mL−1 and adjusted pH to 7.4 by 1 M NaOH. HLCs or hADMSCs were homogeneously dispersed in the as-prepared dECM solution (HLCs/hADMSCs-laden dECM) at a density of 1 × 106 cells mL−1 in 96-well plates and incubated at 37 °C for 1 h to induce gelation. After the formation of stable dECM hydrogels, complete cell media were added. The cell viability was characterized by Live/Dead assay with 1 μM calcein AM (Beyotime, China) and 1 μM propidium iodide (PI, MP Biomedicals, France) in 24 h. The supernatant media were harvested on day 3 to measure the produced ALB and urea by HLCs, while the cell-laden dECM hydrogels were prepared for frozen section and immunofluorescence staining of hepatocyte-related markers, including ALB, HNF-4α, ABCC2, and E-cadherin. hADMSCs cultured in dECM hydrogels were used as the control.
2.16. ROS scavenging ability of Cu NZs@PLGA nanofiber-reinforced dECM hydrogels
The cytoprotection of Cu NZs@PLGA nanofibers from oxidative stress was further assessed for HLCs cultured in dECM hydrogels. Briefly, HLCs were embedded in the dECM hydrogels at a density of 1 × 106 cells mL−1 in 96-well plates and pre-incubated with Cu NZs@PLGA nanofibers (equivalent Cu NZs concentration of 150 ng mL−1) at 37 °C overnight. The cells were subsequently challenged with 500 μM H2O2 for 24 h, after which the cell viability of HLCs was assessed by Live/Dead assay, and the intracellular ROS were stained using DCFH-DA. The mean fluorescent intensities were measured by a microplate reader for ROS quantification.
2.17. Delivery of HLCs using Cu NZs@PLGA nanofiber-reinforced dECM hydrogels to ALF mice
All the animal studies were performed following the protocol approved by the Institutional Animal Care and Use Committee of Sun Yat-Sen University (SYSU-IACUC-2022-001799). The ALF mouse model was established by intraperitoneal infusion of 30 (v/v) % CCl4 in corn oil at a dose of 5 μL g−1 for 24 h before treatment. Male C57BL/6 mice (6-8 week-old, 18–22 g) were randomly divided into six groups, including (1) Blank (n = 5): health mice; (2) Control (n = 5): ALF mice with sham operation; (3) fiber/dECM (n = 5): ALF mice implanted with PLGA nanofiber-reinforced dECM hydrogels; (4) Cu NZs@fiber/dECM (n = 5): ALF mice implanted with Cu NZs@PLGA nanofiber-reinforced dECM hydrogels; (5) hADMSCs/Cu NZs@fiber/dECM (n = 5): ALF mice implanted with hADMSCs in the Cu NZs@PLGA nanofiber-reinforced dECM hydrogels; (6) HLCs/Cu NZs@fiber/dECM (n = 5): ALF mice implanted with HLCs in Cu NZs@PLGA nanofiber-reinforced dECM hydrogels. The materials with/without cells were implanted in situ on the liver surface.
Hydrophilization of the electrospun PLGA nanofiber meshes, with a size of 1 cm × 1 cm, was performed by plasma treatment (plasma cleaner, CPC-A, CIF International Group Co., Ltd, China) before homogeneous distribution of 50 μL dECM precursor solution. The composites were left for 5–10 min before sandwiched implantation between the left liver lobe and the median liver lobe of ALF-challenged mice so that the internal pores of the electrospun nanofiber meshes could be fulfilled with the dECM hydrogel. Once gelation in situ at 37 °C, the surface tension of dECM hydrogel and the sandwich structure allowed the entire composite firmly attach to the liver tissues. The equivalent Cu NZs dosage in Cu NZs@PLGA fiber was 6 μg kg−1, while the number of implanted hADMSCs or HLCs in dECM hydrogels was approximately 1 × 106 cells.
1-day or 3-day post-transplantation, mice were intraperitoneally injected with 0.6% pentobarbital sodium solution (Aladdin, China) at a dose of 10 μL per gram of mouse weight and sacrificed to collect blood and organs. The ALB, aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels in blood serum were quantified by ALB assay kit (CH0101002, Maccura, China), AST assay kit (CH0101202, Maccura, China) and ALT assay kit (CH0101201, Maccura, China), respectively, following the manufacturer's protocols. The harvested hearts, livers, spleens, lungs, and kidneys were fixed in 4% PFA and prepared for hematoxylin and eosin staining (G1005, Servicebio, China) after the paraffin section. The left liver lobes were used for histology analysis, immunofluorescence staining, RT-qPCR, and RNA-sequencing unless specified. The terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) apoptosis assay kit (G1501, Servicebio, China) was used to determine cell apoptosis in the livers. Ki67 staining (GB11141, Servicebio, China) was performed to show proliferating cells in the livers. In addition, immunofluorescent staining of TNF-α (GB11188, Servicebio, China) and IL-6 (GB11141, Servicebio, China) in liver sections were performed to evaluate the degree of inflammation. Dihydroethidium (DHE, GDP1018, Servicebio, China) staining in frozen liver sections was applied to evaluate the ROS levels in the livers. Immunofluorescent stainings of iNOS (GB11119, Servicebio, China) and CD206 (60143-1-Ig, Proteintech, China) in liver sections were performed to evaluate the inflammatory modulating effects of HLCs. The mean fluorescence intensity was quantified using Image J.
CD31/HNF-4α immunofluorescence staining (anti-CD31 antibody, GB11063-1, Servicebio, China) of the liver tissues surrounding the composite transplantation areas was performed to illustrate the angiogenesis and vascularization of the implants.
2.18. RNA-Sequencing
Liver tissues from the control and the HLCs/Cu NZs@fiber/dECM groups were isolated (n = 2) and ground in the TRIzon reagent to extract mRNA. High-quality RNA samples were used to construct a sequencing library. The 2 × 150 bp paired-end sequencing (PE150) was conducted using Illumina Novaseq™ 6000 (Illumina, USA) according to the vendor's recommended protocol. All data were processed using R-packages provided by Lc-Bio Technology (Hangzhou, China).
2.19. Statistical analysis
The statistical software GraphPad Prism 9 (GraphPad Software, USA) was used for all statistical assessments. Data were presented as mean ± standard deviation (SD) and were compared by Student's t-test or one-way ANOVA analysis. The difference was determined as significant when *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, or #P < 0.05, ##P < 0.01, ###P < 0.001, ####P < 0.0001. ns means no significance.
3. Results
3.1. Characterization of Cu NZs@PLGA nanofibers
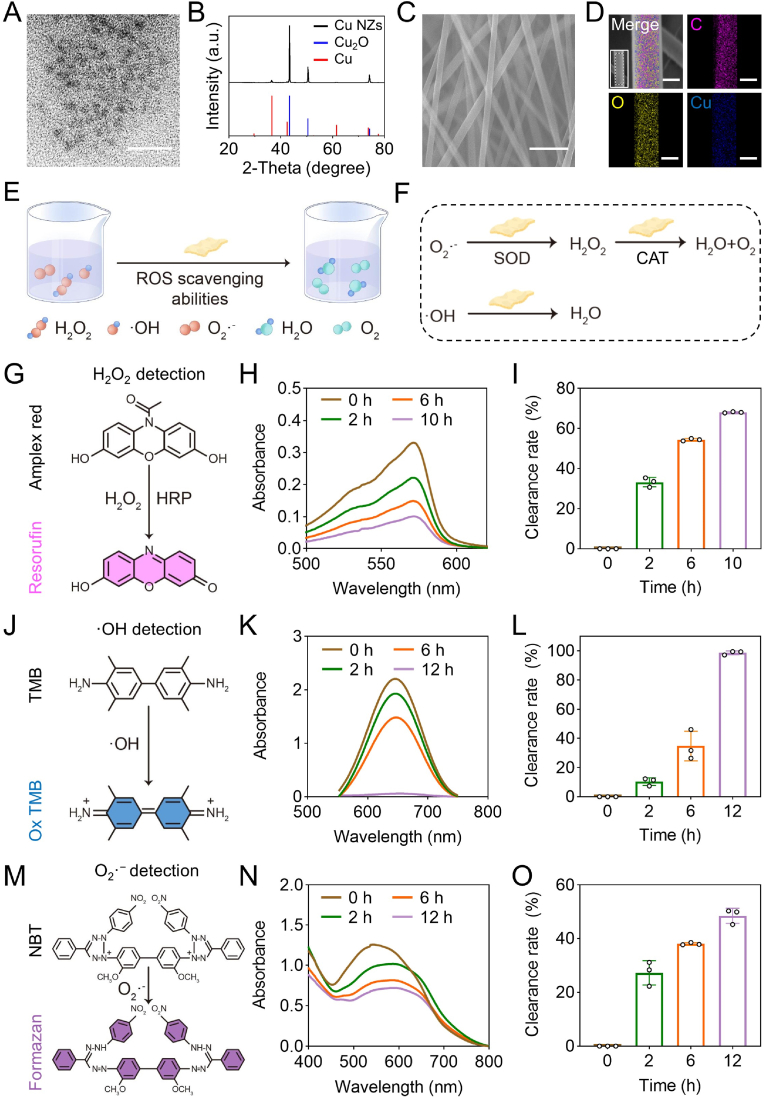
The devastating ALF is often accompanied by high oxidative stress and severe inflammation, which is challenging for effective treatment through conventional hepatocyte transplantation. Hence, we developed a Cu NZs@PLGA nanofiber-reinforced dECM hydrogel for regulation of the local harsh microenvironment. Cu NZs were first characterized by TEM and showed uniform distribution with an average particle size of approximately 4.0 nm in diameter (Fig. 1A, Fig. S1A). The zeta potential of Cu NZs was about −14.4 mV (Fig. S1B). Referred to the standard XRD patterns, Cu NZs were the compound of Cu2O and Cu, suggesting the successful synthesis of Cu NZs (Fig. 1B). Cu NZs were then loaded into implantable PLGA electrospun nanofibers. The SEM imaging indicated that Cu NZs did not significantly change the smooth surface and long fibrous morphology of PLGA nanofibers, with the average diameter of 382.5 ± 74.5 nm (Fig. 1C, Fig. S2). The EDS element mapping imaging identified that C, O, and Cu were homogeneously distributed throughout the PLGA nanofibers (Fig. 1D), further confirming the successful loading of Cu NZs in the nanofibers. The stretching tests of plain PLGA fibers and Cu NZs@PLGA fibers showed that electrospun PLGA fibers had a high stretching strength up to 6.5 MPa, indicating the flexibility of PLGA fibers as a liver patch (Fig. S2D). Meanwhile, the embedment of Cu NZs in PLGA fibers did not obviously affect the stretching ability of the scaffold. As shown in Fig. S3, Cu NZs were released from the PLGA fibers in a more sustained manner compared to that from dECM hydrogels, which was a premium to maintain the minimal working dosage in situ for the effective elimination of ROS and advancing liver regeneration.
Fig. 1.
The characterizations of Cu NZs and Cu NZs@PLGA nanofibers. (A) TEM image of Cu NZs (scale bar: 20 nm). (B) X-ray diffraction (XRD) pattern of the Cu NZs. (C) SEM image (scale bar: 2 μm) and (D) elemental mapping images of Cu NZs@PLGA nanofibers (scale bar: 200 nm). (E) Scheme of Cu NZs@PLGA nanofibers scavenging H2O2, ·OH and O2·−. (F) Multiple enzyme activities of Cu NZs@PLGA nanofibers. (G) H2O2 detection mechanism. (H) UV–Vis absorption spectrum of resorufin products after incubation with Cu NZs@PLGA nanofibers for 0–10 h. (I) H2O2 clearance rate of Cu NZs@PLGA nanofibers at 0, 2, 6, 10 h. (J) ·OH detection mechanism. (K) UV–Vis absorption spectrum of oxTMB after incubation with Cu NZs@PLGA nanofibers for 0–12 h. (L) ·OH clearance rate of Cu NZs@PLGA nanofibers at 0, 2, 6, 12 h. (M) O2·− detection mechanism. (N) UV–Vis absorption spectrum of formazan after incubation with Cu NZs@PLGA nanofibers for 0–12 h. (O) O2·− clearance rate of Cu NZs@PLGA nanofibers at 0, 2, 6, 12 h (Cu NZs@PLGA nanofibers containing equivalent Cu NZs concentration of 150 ng mL−1 were applied in G-O). All data are presented as mean ± SD, n = 3.
To evaluate multiple enzyme activities of Cu NZs and Cu NZs@PLGA nanofibers, their scavenging efficiencies against H2O2, ·OH, and O2·− were investigated (Fig. 1E and F). To investigate the H2O2 clearance rate, the remaining H2O2 was detected in the presence of horseradish peroxidase by employing Amplex Red as the probe, producing the red resorufin with the absorbance peak at 571 nm (Fig. 1G). The increased amount of Cu NZs addition could gradually induce the UV absorption intensity at 571 nm attenuation and colorimetric fading, suggesting that H2O2 clearance efficiency of Cu NZs was in a concentration-dependent manner (Fig. S4). Of note, the addition of Cu NZs at the ultralow concentration of 200 ng mL−1 could eliminate up to 83.4% of H2O2 (Fig. S4C). When Cu NZs were encapsulated in the PLGA nanofibers, the H2O2 scavenging efficiency was not compromised and showed a time and concentration-dependent manner (Fig. S3B). Cu NZs@PLGA nanofibers containing equivalent Cu NZs concentrations of 150 ng mL−1 and 200 ng mL−1 could clear about 68.1% and 75.0% of H2O2 at 10 h, respectively (Fig. 1H–I, Fig. S3B). In addition, during the removal of H2O2, air bubbles were generated as shown on the tube walls, indicating that H2O2 was converted into O2 by Cu NZs (Fig. S5). Moreover, taking the Cu NZs@PLGA nanofibers containing equivalent Cu NZs concentration of 150 ng mL−1 as an example, the ·OH and O2·− scavenging capabilities of Cu NZs@PLGA nanofibers were explored. Cu NZs@PLGA nanofibers were able to eliminate the ·OH generated by Fenton reagents. The UV absorption spectra of oxTMB gradually decreased as the incubation prolonged, indicating that ·OH generated from Fe2+ and H2O2 was effectively eliminated by Cu NZs in Cu NZs@PLGA nanofibers. In particular, the scavenging efficiency of ·OH by Cu NZs@PLGA nanofibers was near 100% after incubation for 12 h (Fig. 1J-L). Additionally, the descending UV absorbance of converted NBT at 560 nm also revealed the successful elimination of O2·− by Cu NZs@PLGA nanofibers, with about 48.5% of O2·− clearance rate at 12 h (Fig. 1M − O). Taken together, all these results strongly proved that Cu NZs@PLGA nanofibers enabled the effective elimination of H2O2, ·OH, and O2·−.
3.2. ROS scavenging activities of Cu NZs@PLGA nanofibers in vitro
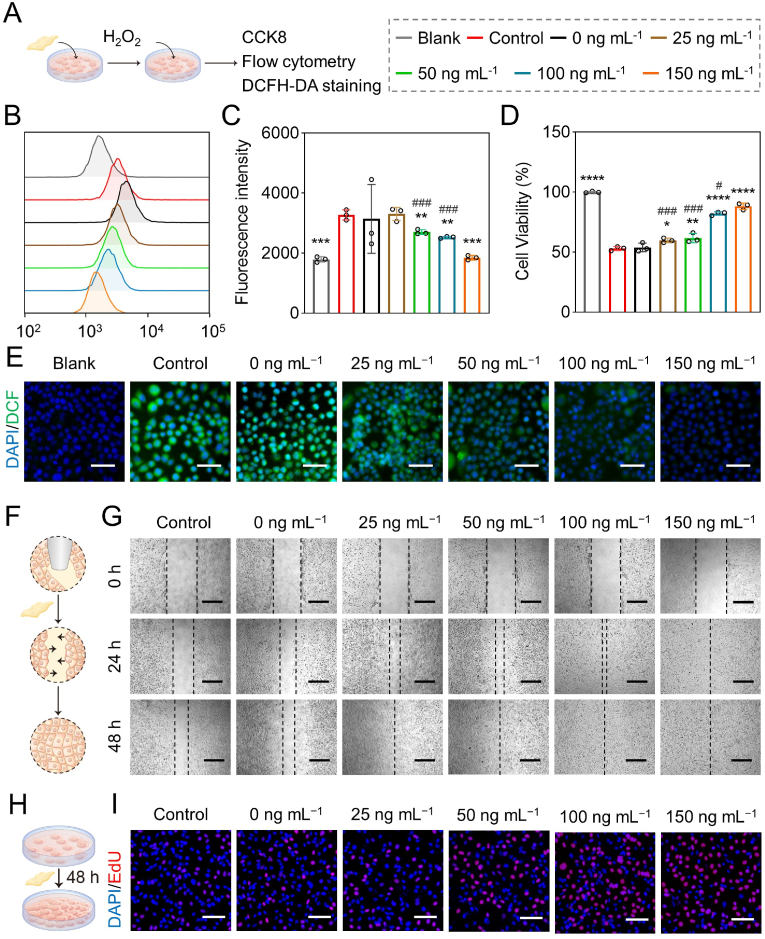
The biocompatibility of Cu NZs@PLGA nanofibers was firstly studied in multiple cell types, including AML12 cells, hADMSCs, and HLCs. The cells were incubated with Cu NZs@PLGA nanofibers containing equivalent Cu NZs concentrations ranging from 0 ng mL−1 to 200 ng mL−1 for 24 h. The CCK-8 assay showed that the cell viabilities varied from 95.0 to 107.9% compared with the blank group without treatment, suggesting the good biocompatibility of Cu NZs@PLGA nanofibers (Fig. S6). Afterwards, inspired by the excellent ROS scavenging ability of Cu NZs@PLGA nanofibers, we continued to investigate the cytoprotective capacity of Cu NZs@PLGA nanofibers against intracellular ROS. According to the flow cytometry analysis of intracellular ROS generation after DCFH-DA staining, when the equivalent concentration of Cu NZs in Cu NZs@PLGA nanofibers reached 150 ng mL−1, the fluorescence intensity decreased 1.8 times after the Cu NZs@PLGA nanofibers treatment compared with the control group. The results suggested that the intracellular ROS could be significantly eliminated (Fig. 2B and C), which were consistent with the DCFH-DA staining images (Fig. 2E). Likewise, the CCK-8 assay (Fig. 2D) also confirmed that in comparison with the control (cell viability 53.0%), Cu NZs@PLGA nanofibers distinctly enhanced the cell viability of H2O2-treated cells to 82.2% and 88.3%, respectively, at the equivalent Cu NZs concentrations of 100 ng mL−1 and 150 ng mL−1. Therefore, Cu NZs@PLGA nanofibers were able to evidently alleviate cell damage caused by oxidative stress, thereby promoting the survival of hepatocytes under the high ROS condition. Notably, Cu NZs@PLGA nanofibers with an equivalent concentration of 200 ng mL−1 Cu NZs did not distinctly exhibit increased ROS elimination efficiency and improved cell viability (Fig. S7A and B), which might be because that the increased fiber surface area affected the release of Cu NZs and herein compromise their ROS clearing efficiency (Fig. S3). Therefore, Cu NZs@PLGA nanofibers with equivalent 150 ng mL−1 Cu NZs were set as the maximum concentration for subsequent experiments.
Fig. 2.
Intracellular ROS scavenging capability of Cu NZs@PLGA nanofibers. (A) Schematic illustration of investigation about ROS scavenging activity of Cu NZs@PLGA nanofibers in vitro. (B, C) Flow cytometry analysis and fluorescence intensity quantification of DCFH-DA staining in AML12 cells treated with Cu NZs@PLGA nanofibers containing equivalent Cu NZs concentration ranging from 0 ng mL−1 to 150 ng mL−1 after incubation with H2O2. (D) Cell viability of AML12 cells (CCK-8). (E) Representative fluorescence images of AML12 cells (DCFH-DA staining). Scale bar: 50 μm. (B–E: Blank: AML12 cells without any treatment, Control: H2O2-challenged cells without incubation with nanofibers, 0 ng mL−1: The cells challenged by H2O2 and incubated with plain PLGA nanofibers in the absence of Cu NZs). (F) Schematic illustration of Cu NZs@PLGA nanofibers promoting HUVECs migration. (G) HUVECs migration images after Cu NZs@PLGA nanofibers incubation at 24 h and 48 h. Scale bar: 400 μm. (H) Schematic illustration of Cu NZs@PLGA nanofibers promoting HUVECs proliferation. (I) EdU staining images of HUVECs at 48 h. Scale bar: 100 μm. (F–I: Control: HUVECs without any treatment, 0 ng mL−1: HUVECs incubated with plain PLGA nanofibers in the absence of Cu NZs). All data are presented as mean ± SD, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 versus control, ns means no significance versus control. #P < 0.05, ##P < 0.01, ###P < 0.001, ####P < 0.0001 for 25 ng mL−1, 50 ng mL−1, and 100 ng mL−1 versus 150 ng mL−1.
3.3. Pro-angiogenic effects of Cu NZs@PLGA nanofibers
Studies suggested that biomaterials containing copper promoted cell proliferation, playing an important role in angiogenesis and tissue regeneration [15,16,55]. Therefore, the effects of Cu NZs@PLGA nanofibers on HUVECs migration and proliferation were further explored by scratch assay and EdU imaging assay, respectively. As shown in Fig. 2F and G, after incubation with Cu NZs@PLGA nanofibers, more cells migrated towards the central scratch in a concentration-dependent manner compared to the control without treatment. Particularly, full recovery of the scratch was achieved after the addition of Cu NZs@PLGA nanofibers with equivalent Cu NZs concentration of 150 ng mL−1 at 24 h and other concentrations at 48 h (Fig. 2F–G, S7C-D). Furthermore, there were more EdU-positive cells after treatment by Cu NZs@PLGA nanofibers for 48 h (Fig. 2H and I). The number of proliferating HUVECs treated with the Cu NZs@PLGA nanofibers containing an equivalent Cu NZs concentration of 150 ng mL−1 increased 21.1% compared to the control group without treatment (Fig. S7E). The pro-angiogenic effects of Cu NZs@PLGA nanofibers might accelerate the integration and functioning of the transplanted HLCs and herein promote liver regeneration.
3.4. Induction and characteristics of HLCs
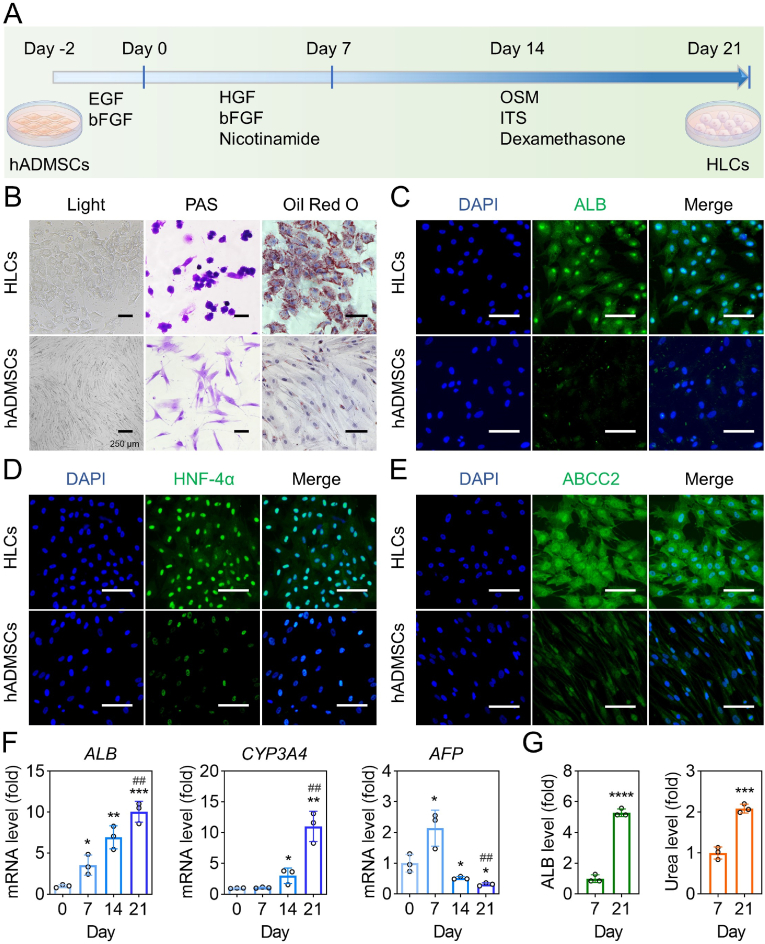
The multipotent MSCs can be induced towards hepatic lineage by different methods [35,36,49,56,57]. Here, a two-step differentiation protocol using cytokine cocktails was applied (Fig. 3A). After differentiation, spindle-like hADMSCs changed to polygonal shapes (Fig. 3B, Fig. S8). Compared to hADMSCs, the induced HLCs showed distinct glycogen production and lipid accumulation as evidenced by PAS staining and Oil Red O staining, respectively (Fig. 3B). Furthermore, the hepatocyte-related proteins, such as ALB, HNF-4α, ABCC2, E-cadherin and DPP4, were also highly expressed in HLCs (Fig. 3C–E, Fig. S9). The mRNA expression of hepatocyte-specific genes was tracked from the beginning of cell differentiation to the end. The ALB and CYP3A4 genes in HLCs showed over 10.0 times higher mRNA expression on day 21 than that on day 0 (Fig. 3F). In contrast, a much lower level of immature hepatocytes marker AFP was observed on day 21 as compared to day 7 (Fig. 3F). In addition, the secretion levels of ALB and urea in the medium supernatants of HLCs at the mature stage (day 21) were 5.3-fold and 2.1-fold higher than those in the first induction phase (day 7), respectively (Fig. 3G).
Fig. 3.
The characterization of hADMSCs-derived HLCs. (A) Schematic diagram of hepatic differentiation schedule. (B) The light field image, PAS staining, and Oil red O staining of HLCs and hADMSCs. (C–E) The ALB, HNF-4α, and ABCC2 immunofluorescence staining of HLCs and hADMSCs. Blue: DAPI-labelled cell nuclei; Green: Alexa Fluor 488-labelled ALB, HNF-4α, and ABCC2. (F) The hepatic gene expression levels (fold) of ALB, CYP3A4, and AFP (all data are normalized to the value of day 0) (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 versus day 0. ##P < 0.01 versus group of day 7). (G) The ALB and urea levels of culture supernatants in HLCs on days 7 and 21 (all data are normalized to the value of day 7) (***P < 0.001, ****P < 0.0001 versus day 7). All data are presented as mean ± SD, n = 3. Scale bar: 100 μm.
The HGF level in the supernatant media of HLCs was also significantly higher than that of hADMSCs, which demonstrated that the superior therapeutic efficacy of HLCs/Cu NZs@fiber/dECM than hADMSCs/Cu NZs@fiber/dECM might be attributed to the enhanced HGF secretion from HLCs in addition to their hepatocyte-related functions (Fig. S10). Taken together, these results suggested that the hADMSCs were successfully induced into HLCs with typical hepatocyte-related morphology and functions, which was favorable to improving liver biofunctions in ALF.
In addition to possessing hepatocyte-related functions, HLCs have been reported to exert inflammatory regulatory effects [37]. Therefore, we investigated the impact of HLCs supernatant on macrophage polarization. First, HLCs supernatant was incubated with THP-1 (M0) for 24 or 48 h. Gene expressions were analyzed by qPCR to assess the polarization of macrophages. Compared with THP-1 (M0), the expression of IL 1β, iNOS, and TGF β1 obviously decreased in THP-1 treated with HLCs supernatant with increasing treatment time, while the levels of IL 10 and MRC 1 increased 3.3-fold and 33.9-fold at 48 h, respectively (Fig. 4A and B). These suggested that HLCs supernatant might predispose macrophages to M2 polarization (anti-inflammatory phenotype). Next, whether HLCs supernatant reversed macrophage polarization in the inflammatory state was further explored. Results showed THP-1 exhibited the up-regulated expressions of IL 1β, TNFA, and TGF β1 after LPS treatment, indicating that THP-1 (M0) was successfully induced to M1 phenotype (pro-inflammatory phenotype). Notably, in comparison with the LPS-treated group, the expressions of IL 1β, TNFA, and TGF β1 significantly declined in HLCs supernatant-treated THP-1, approaching the level of THP-1 (M0). In contrast, the levels of IL 10 and MRC 1 markedly upregulated to 23.6-fold and 21.1-fold after the HLCs supernatant treatment (Fig. 4C and D). Meanwhile, we also obtained similar results in RAW264.7 cells (Fig. S11). Furthermore, we measured the expression levels of TNF-α, IL-1β, iNOS, and IL-10 in RAW264.7 cells after different treatments by immunofluorescence staining or Western blot. Fig. S12 illustrated that RAW264.7 cells treated with LPS alone showed remarkable high expression of pro-inflammation-related markers (TNF-α, IL-1β, and iNOS). After incubation in HLCs supernatant media for 24 h, the level of anti-inflammatory IL-10 was distinctly increased. Western blot analysis also revealed that HLCs supernatant media downregulated the protein expression of TNF-α and iNOS in LPS-stimulated RAW264.7 cells. According to the flow cytometry analysis (Fig. S12 E-F), the percentage of CD206-positive cells in HLCs supernatant treated group was 80.1%, while only 18.1% in LPS treated group. These results demonstrated the paracrine effects of HLCs on macrophage polarization. Additionally, we also investigated whether HLCs possessed the protective effect on hepatocytes under high oxidative stress. After H2O2 challenging, a large population of floating dead cells and debris were observed for the monoculture of AML12 cells (control), but not for the cells co-cultured with HLCs or hADMSCs (Fig. S13A). The AML12 cells cocultured with HLCs showed superior cell viability compared with monocultured AML12 cells and cocultured AML12 with hADMSCs (Fig. S13B). By characterization of antioxidant genes expression in the AML12 cells, we found that the mRNA expressions of SOD2, NQO1, and KEAP1 genes were significantly up-regulated by 2.2–3.8 times when the cells were cocultured with HLCs (Fig. S13C). Hence, the induced HLCs might advance liver regeneration by improving antioxidation and cell viability of host hepatocytes in ALF therapy.
Fig. 4.
The anti-inflammatory effect of HLCs. (A) Schematic representation of HLCs supernatant predisposing macrophages to M2 polarization. (B) IL 1β, INOS, TGF β1, IL 10, and MRC 1 expression levels of THP-1 after HLCs supernatants cultured for 24 or 48 h (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 versus THP-1 (M0), ##P < 0.01, ###P < 0.001 versus HLCs supernatant 24 h). (C) Schematic representation of HLCs supernatant promoting conversion of macrophages phenotype in inflammatory status. (D) IL 1β, TNFA, TGF β1, IL 10, and MRC 1 expression levels of LPS-stimulated THP-1 (**P < 0.01, ***P < 0.001, ****P < 0.0001 versus THP-1 (M0), ns means no significance versus THP-1 (M0), ###P < 0.001, ####P < 0.0001 versus 100 ng mL−1 LPS). All data are normalized to the value of THP-1 (M0) and presented as mean ± SD, n = 3.
3.5. Characterization of the Cu NZs@PLGA nanofiber-reinforced dECM hydrogel
For efficient delivery of HLCs in situ, liver dECM hydrogels were used in this study (Fig. 5A) due to the retaining of hepatic bioactive components and the 3D culture environment that promote the maturity of HLCs [[58], [59], [60]]. After incubation at 37 °C for 1 h, the dECM hydrogel was formed with a relatively larger storage modulus (G′) than loss modulus (G′′) (Fig. 5B, Fig. S14A). The SEM image illustrated that there were no residual cells in the porous dECM hydrogel after decellularization (Fig. 5C). To investigate the stability of dECM hydrogels in vitro, we performed a degradation experiment by immersion of the hydrogels in 1 × PBS containing 20 μg mL−1 collagenase type II at 37 °C. With the extension of incubation time in the digesting collagenase type II solution, dECM hydrogels gradually degraded (Fig. S15A). The degradation rate was up to 91.5% in 96 h (Fig. S15B). Only minor dECM hydrogel was seen remaining on the left liver lobe 3-day post-implantation (Fig. S15C). However, the weak hydrogel, with an elastic modulus of less than 100.0 Pa, might not be convenient for transplantation operation. Therefore, we used Cu NZs@PLGA nanofibers as the mechanical support of the dECM hydrogel (Fig. S14B). After cell embedment, HLCs were homogenously distributed within the Cu NZs@PLGA nanofiber-reinforced dECM hydrogel (Fig. 5D).
Fig. 5.
The characterization and function of HLCs/dECM hydrogel. (A) Schematic illustration of dECM preparation and HLCs embedment. (B) Rheological behaviors of dECM (storage modulus (G′), loss modulus (G″)). (C) SEM of dECM hydrogels. Scale bar: 50 μm. (D) SEM of cells/Cu NZs@fiber/dECM. Yellow arrows point to cells in dECM. Scale bar: 30 μm. (E–H) The ALB, HNF-4α, E-Cadherin, and ABCC2 immunofluorescence staining images of HLCs or hADMSCs cultured in the dECM hydrogel for 3 days. Scale bar: 100 μm. (I) The hepatic gene expression levels of ALB, CYP3A4, and CK18. (J) The ALB and urea levels in the culture supernatant of HLCs cultured on 2D or embedded in 3D on day 3. Blue: DAPI-labelled cell nuclei; Green: Alexa Fluor 488-labelled ALB, HNF-4α, E-Cadherin, and ABCC2. All data are presented as mean ± SD, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 versus 2D.
The dECM hydrogel possesses the excellent biocompatibility. When cultured in the dECM hydrogel for up to three days, HLCs showed good cell viability (Fig. S14C). In addition, after 3D culture of HLCs in the dECM hydrogel, hepatocyte-related markers were maintained, including ALB, HNF-4α, E-cadherin, and ABCC2 (Fig. 5E–H). The mRNA expressions of ALB and CK18 genes were over 8.0 times higher for HLCs cultured in the 3D dECM hydrogel than those on 2D plates. Meanwhile, the drug metabolism-related gene CYP3A4 was also significantly upregulated for the HLCs embedded in the dECM hydrogel (Fig. 5I). Moreover, the secreted ALB and urea in the medium supernatants of HLCs were improved as well (Fig. 5J). Particularly, the urea derived from 3D-cultured HLCs showed a 2.3-fold increase compared to that from 2D-cultured HLCs. Therefore, dECM hydrogel is beneficial for the maintenance of HLC phenotypes and the enhancement of hepatocyte-related functions.
In addition to mechanical support, the cytoprotective effect of the Cu NZs@PLGA nanofibers for the embedded HLCs in the dECM hydrogel was further explored. After H2O2 challenging, there were more dead HLCs in the system without Cu NZs@PLGA nanofibers (control) than that with an equivalent Cu NZs concentration of 150 ng mL−1 (Fig. S16A). This was because the high intracellular ROS level was distinctly mitigated by the multifaceted Cu NZs (Fig. S16B).
3.6. Delivery of HLCs by Cu NZs@PLGA nanofiber-reinforced dECM hydrogel for ALF treatment
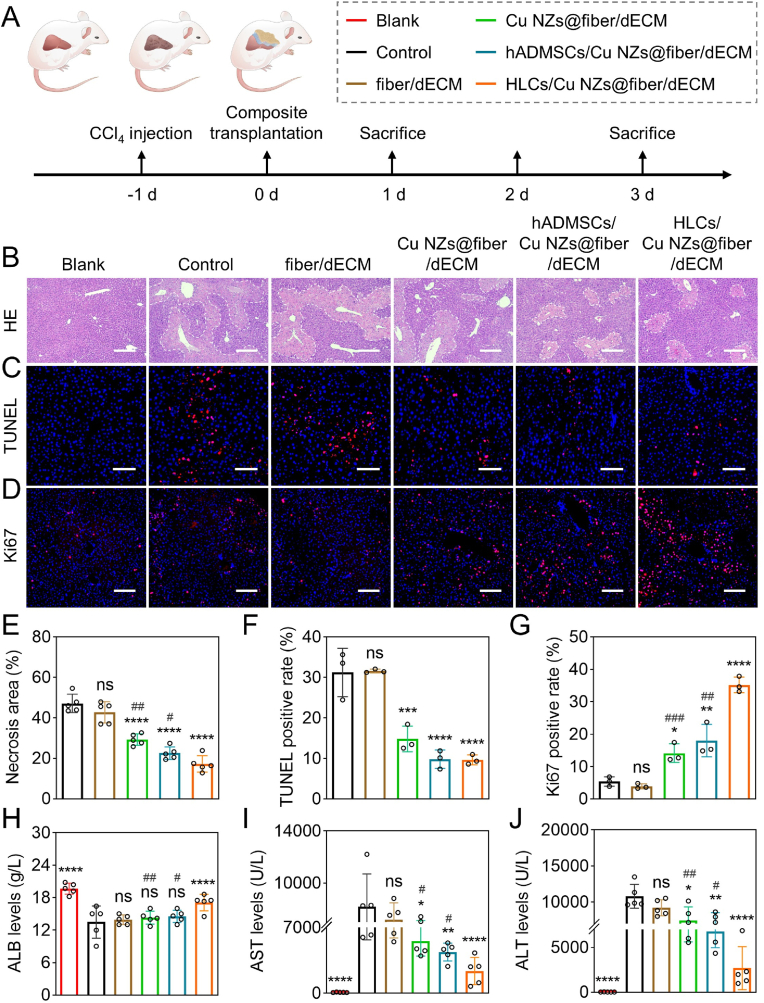
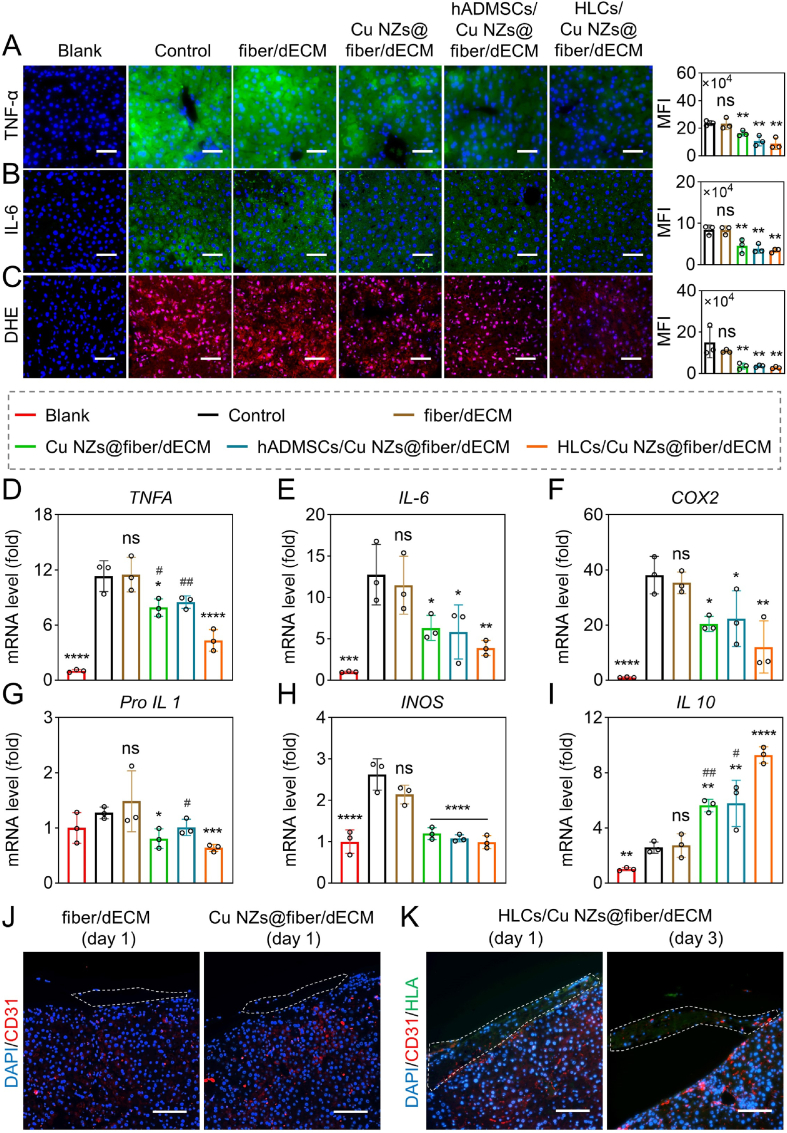
Inspired by the powerful ROS scavenging efficiency and pro-angiogenic effects of Cu NZs@PLGA nanofibers, enhanced hepatic functions of HLCs by dECM hydrogels, as well as convenient operation of PLGA nanofibers, we further investigated the therapeutic potential of the synergic effects brought by HLCs/Cu NZs@fiber/dECM in the treatment of CCl4-induced ALF (Fig. 6A). After administration of CCl4, livers became pale with hard texture and a large amount of necrosis near the portal area (Fig. S17). The implantation of Cu NZs@PLGA nanofibers significantly reduced necrotic areas by 17.9%–29.9% on day 1 according to the HE-stained images (Fig. 6B, E). Similarly, TUNEL and Ki67 staining also showed that fewer apoptotic cells and more proliferating cells in the livers of ALF-challenged mice from the “Cu NZs@fiber/dECM” group were found as compared to the “control” and “fiber/dECM” groups, suggesting the cytoprotective effects of Cu NZs against oxidative stress in vivo at the initial stage of ALF. Of note, mice implanted with HLCs/Cu NZs@fiber/dECM showed less tissue necrosis and more obvious cell proliferation compared with that treated with hADMSCs/Cu NZs@fiber/dECM, which indicated the importance of the implanted HLCs with alternative hepatic functions and premium protective effects on the host hepatocytes (Fig. 6C–D, F-G). These acquired results were consistent with the in vitro findings [38,[61], [62], [63]]. The superior therapeutic effects of HLCs were also evidenced by the reduced serum levels of AST and ALT and the recovery of the serum ALB level in the mice treated with HLCs/Cu NZs@fiber/dECM (Fig. 6H–J). In terms of inflammation and oxidative stress, the implantation of Cu NZs@PLGA nanofibers distinctly reduced the accumulation of inflammatory factors, such as TNF-α and IL-6, and intracellular ROS compared with the control without treatment on day 1 (Fig. 7A–C). In addition, the mRNA expression of TNFA, IL-6, and COX2 genes in the liver cells of the control group without treatment was over 10.0 times higher than that in normal mice, which largely decreased after treatment by Cu NZs@PLGA nanofibers (Fig. 7D–F). In particular, the mRNA expression of IL1 and INOS genes was in the normal range (Fig. 7G–I), while the anti-inflammatory IL10 gene highly increased by more than 5.0 times for the “Cu NZs@fiber” group compared to the control without treatment (Fig. 7I). Furthermore, the implantation of HLCs further ameliorated the mRNA expression of pro-inflammatory TNFA and IL1 genes and upregulated anti-inflammatory IL10 gene in the ALF-challenged liver cells. The TNFA and IL1 gene expressions of the HLCs/Cu NZs@fiber/dECM group significantly decreased by 2.6 and 2.0 times, respectively, compared with the control group, and the IL10 expressions markedly upregulated to 9.3-fold after the HLCs/Cu NZs@fiber/dECM treatment, suggesting the premium benefits of HLCs/Cu NZs@fiber/dECM for the regulation of local harsh microenvironment. Meanwhile, to track macrophage polarization in vivo, immunofluorescence staining of iNOS (M1 macrophage marker) and CD206 (M2 macrophage marker) in the liver tissue sections was performed. As shown in Fig. S18, the iNOS-positive cells in “HLCs/Cu NZs@fiber/dECM” group were the fewest after 1 day of treatment. In contrast, CD206-positive cells were significantly more than that in the other groups on day 1 and 3, indicating the inflammatory modulation of HLCs. We also evaluated the pro-angiogenic effects of Cu NZs in vivo. As shown in Fig. 7J, there was no CD31-positive signal in the implant of “fiber/dECM” group 1-day post-implantation. However, CD31-positive cells were observed in the grafts of “Cu NZs@fiber/dECM” and “HLCs/Cu NZs@fiber/dECM” groups. In addition, the number of CD31-positive cells in the “HLCs/Cu NZs@fiber/dECM” group further increased on day 3 (Fig. 7K). These results demonstrated the pro-angiogenic effects of Cu NZs in vivo.
Fig. 6.
Therapeutic efficacy of HLCs/Cu NZs@fiber/dECM on CCl4-induced ALF after 1 day of treatment. (A) Schematic illustration of treatment schedule for in vivo CCl4-induced ALF therapy. (B) The HE staining images of the livers. Scale bar: 400 μm. (C) The TUNEL staining images of the livers. Scale bar: 200 μm. (D) The Ki67 staining images of the livers. Scale bar: 200 μm. (E) Quantitative analysis of the necrosis area according to HE staining images (n = 5). (F–G) Quantitative analysis of the TUNEL and Ki67 staining images (n = 3). (H–J) The serum levels of ALB, AST, and ALT. Blue: DAPI-labelled cell nuclei; Red: TUNEL-positive dead cells, Ki67-positive proliferating cells. All data are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 versus control, ns means no significance versus control. #P < 0.05, ##P < 0.01, ###P < 0.001 for Cu NZs@fiber/dECM and hADMSCs/Cu NZs@fiber/dECM versus HLCs/Cu NZs@fiber/dECM.
Fig. 7.
The inflammation level of liver in CCl4-induced ALF mice and the angiogenesis in grafts. (A–C) The TNF-α, IL-6, and DHE staining images of liver tissues and quantitative analysis of mean fluorescence intensity (MFI). Scale bar: 200 μm. (D–I) The mRNA expression levels of TNFA, IL-6, COX2, Pro-IL 1, INOS, and IL 10. (J) Immunofluorescence staining images of CD31 in the liver tissue sections of “fiber/dECM” and “Cu NZs@fiber/dECM” groups on day 1. (K) The immunofluorescence staining images of CD31 and HLA in the liver tissue sections of “HLCs/Cu NZs@fiber/dECM” group on day 1 and 3. Blue: DAPI-labelled cell nuclei; Red: Cy3-labelled CD31, ROS-reacted DHE; Green: Alexa Fluor 488-labelled TNF-α, IL-6, HLA. Scale bar: 100 μm. All data are presented as mean ± SD, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 versus control, ns means no significance versus control, #P < 0.05, ##P < 0.01 for Cu NZs@fiber/dECM and hADMSCs/Cu NZs@fiber/dECM versus HLCs/Cu NZs@fiber/dECM.
As shown in Fig. S19 and Fig. S20, the livers of “HLCs/fiber/dECM” group had similar size of necrotic area, inflammation, and ROS level compared to that of Cu NZs@fiber/dECM group. Notably, the number of proliferating cells in the “HLCs/fiber/dECM” group was obviously larger than that of “Cu NZs@fiber/dECM” group, which might be due to the paracrine effects of HLCs as evidenced in vitro (Fig. S10). The ALB level was much higher in “HLCs/fiber/dECM” group compared to “Cu NZs@fiber/dECM” group, indicating the functioning of HLCs. However, inflammation and oxidative stress (Fig. S19 D-F) were more serious in the “HLCs/fiber/dECM” group than that in the “HLCs/Cu NZs@fiber/dECM” group, revealing the important antioxidative role of Cu NZs.
3-day post-treatment, livers were shown with obviously reduced necrosis, whereas infiltration of inflammatory cells predominated (Fig. S21A and B). There were few TUNEL-positive cells, while Ki67-positive cells were widely distributed throughout the liver tissues in all the groups (Fig. S21C and D). The serum levels of AST and ALT in the HLCs/Cu NZs@fiber/dECM group were similar to normal values but remarkably lower than the other counterparts (Fig. S21E and F). In the meantime, the mice treated with HLCs/Cu NZs@fiber/dECM showed full recovery of serum ALB (Fig. S21G). The HLCs/Cu NZs@fiber/dECM also slowed the weight loss of ALF-challenged mice at the initial stage and accelerated weight recovery from day 3 onward (Fig. S21H). Moreover, the relief of inflammation storm and excessive intracellular ROS by Cu NZs@PLGA nanofibers were still evidenced with TNF-α, IL-6, and DHE staining on day 3 (Fig. S22A–C). Compared with the hADMSCs/Cu NZs@fiber/dECM group, liver cells of the mice in the HLCs/Cu NZs@fiber/dECM group showed significantly lower mRNA expression of TNFA, COX2, and NLRP3 genes (Fig. S22D–F). Therefore, HLCs embedded in Cu NZs@PLGA nanofiber-reinforced dECM hydrogels could consistently and effectively alleviate inflammation storm and suppress oxidative stress, establishing a desired microenvironment for the host liver cells, herein advancing liver regeneration. In addition, the pro-angiogenic Cu NZs also enhanced migration of host CD31-positive endothelial cells towards the HNF-4α-positive cell-laden dECM hydrogels and formation of blood vessels throughout the implants, indicating the promising therapeutic efficacy of HLCs/Cu NZs@fiber/dECM might be partially attributed to the sufficient vascularization for the functioning of HLCs (Fig. S23). The systemic toxicity of HLCs/Cu NZs@fiber/dECM was also evaluated by H&E staining of major organs. There was no obvious change in heart, spleen, lung, and kidney in mice at day 3 after implantation, indicating the excellent biocompatibility of HLCs/Cu NZs@fiber/dECM in vivo (Fig. S24). Taken together, all these results of anti-inflammatory, hepatic functions, proliferation, and apoptotic analyses based on blood and liver tissue confirmed the excellent synergistic therapeutic performance of HLCs/Cu NZs@fiber/dECM against ALF by combining the anti-oxidation and pro-angiogenesis activities of Cu NZs, as well as the anti-inflammation and hepatic function enhancement of HLCs.
Additionally, the therapeutic efficacy of HLCs/Cu NZs@fiber/dECM composite on ALF was also studied 1-week post-implantation. After treatment by the HLCs/Cu NZs@fiber/dECM composite for one week, the serum levels of ALB, AST, and ALT recovered to the normal range (Fig. S25A). The liver of “HLCs/Cu NZs@fiber/dECM” group was similar to that of normal mice in the absence of tissue necrosis, cell apoptosis, inflammation, and oxidative stress, but more proliferating cells (Fig. S25B). In addition, as shown in Fig. S25C, Ki67/human leukocyte antigen (HLA)-positive cells distributed within the graft and integrated into the host liver, indicating survival of the implanted HLCs and their migration.
3.7. Transcriptome analysis to reveal the therapeutic mechanism of HLCs/Cu NZs@fiber/dECM
To further investigate the therapeutic mechanism, the synergic effects of HLCs/Cu NZs@fiber/dECM in the ALF-challenged mice were investigated by transcriptome analysis of the attached liver tissues in the treated group (HLCs/Cu NZs@fiber/dECM: HLC-1 and HLC-2) in comparison with that of the control group without treatment (Control: Ctrl-1 and Ctrl-2) on day 1. The Volcano plot illustrated 833 upregulated genes and 1184 downregulated genes were differentially expressed in the livers of HLCs/Cu NZs@fiber/dECM-treated group (fold change (FC) > 2; p < 0.05) (Fig. 8A). Moreover, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that the top 20 enriched pathways of differential genes were involved with cell cycle, liver biosynthesis, drug metabolism, apoptosis, and liver regeneration (Fig. 8B). Further, system screening of the highly expressed drug metabolism-related genes in the HLCs/Cu NZs@fiber/dECM-treated group and their gene set enrichment analysis (GSEA) indicated the important role of cytochrome P450-involved drug metabolism in the treatment of ALF (Fig. 8C, Fig. S26). In addition to the important Hippo signaling pathway that participated in liver regeneration, the apoptosis-related p53 signaling pathway was of interest. The GSEA showed that 88% of the key genes in the p53 signalling pathway were differentially downregulated after implantation of HLCs/Cu NZs@fiber/dECM (Fig. 8C, Fig. S27). Notably, a part of genes related to Wnt signaling pathway, a crucial pathway mediated liver regeneration, were significantly upregulated (Fig. S28), in which AXIN2 gene as the marker of the hepatocyte proliferation/self-renewal was highly expressed in the HLCs/Cu NZs@fiber/dECM group [64,65]. These observations were consistent with the TUNEL and Ki67 staining results (Fig. 6C and 6D). Moreover, transcriptome analysis showed that genes associated with inflammation or oxidative stress such as IL33 and HIF1A were significantly downregulated in the liver of mice implanted with HLCs/Cu NZs@fiber/dECM, while antioxidative genes, such as SOD1, SOD2, GPX1, and CAT, as well as angiogenesis-related VEGFA and ANG genes were highly expressed (Fig. 8D) [[66], [67], [68]]. More importantly, in the liver tissues of mice from the HLCs/Cu NZs@fiber/dECM group, the expression of liver regeneration and hepatocyte-related genes, such as ALB, HNF4A, DPP4, ABCC2, TTR, UGT1A1, GSTM1, and SULT1A1, were distinctly increased (Fig. 8D) [69,70].
Fig. 8.
Transcriptome analysis of liver tissues for in vivo experiment. (A) Volcano plot of the differentially expressed genes. (≥2-fold difference. red: upregulated genes; blue: downregulated genes). (B) The gene enrichment KEGG pathway analysis (The top 20 terms are displayed). (C) The GSEA plot of drug metabolism cytochrome P450 (Enrichment Score (ES) = 0.61718786, Normalized Enrichment Score (NES) = 2.2412274; nominal p value < 0.001) and p53 signaling pathway (ES = −0.49999398, NES = −1.620217; nominal p value < 0.05). (D) The heat maps of differentially expressed genes related to oxidative stress, inflammation, antioxidation, angiogenesis, hepatocyte marker, and liver regeneration.
4. Discussion
Excessive ROS, severe inflammation storm, and massive liver necrosis are the main pathological features of ALF [18,71]. Therefore, scavenging ROS to reduce hepatocyte necrosis and supplementing hepatocytes to achieve hepatic function compensation are essential for liver regeneration in ALF therapy. In this study, a synergistic therapeutic platform consisting of antioxidant nanozyme-loaded nanofibers and functional HLCs (HLCs/Cu NZs@fiber/dECM) was fabricated and used for ameliorating CCl4-induced ALF.
Nanozymes that resemble natural enzymes, such as CAT, GPx, and SOD, can protect cells from oxidative stress [3,14]. Recent studies reported that copper-based nanoenzyme materials showed considerable efficacy in the treatment of wound healing, acute kidney injury, acute lung injury, rheumatoid arthritis, and other inflammation-related diseases [14,15,53,72]. Here, Cu NZs with good biocompatibility and high ROS scavenging efficiency were successfully synthesized and characterized. Our (Fig. 1G-O, Fig. 2B–E, Fig. S3, and Fig. S5) and other similar studies have shown that electrospinning process did not affect ROS scavenging activity of nanozymes [[27], [28], [29], [30]]. When embedment of Cu NZs in the electrospun fibers, the H2O2 clearance rate gradually increased with the extension of incubation time (Fig. S3B), which was in line with the releasing profile of Cu NZs (Fig. S3A). Therefore, we believe that the incorporation of Cu NZs in the electrospun fibers could effectively prolong the antioxidative effects of the nanoenzyme in vitro and in vivo. Besides, Cu NZs@PLGA nanofibers also significantly enhanced HUVECs migration and proliferation, which was conducive to rapid vascularization of the implants in vivo.
Hepatocyte transplantation is one of the effective methods to compensate for the loss of hepatic function in ALF patients [31,41]. However, limited donor sources, poor cell proliferation in vitro, and potential allogenic immune rejection impede the popularity of the therapeutic approach [1,31]. Stem cell-derived HLCs are readily available with low immunogenicity. HLCs exhibit similar hepatic functions as primary hepatocytes and possess anti-inflammatory and regenerative effects [[37], [38], [39],41,47]. Studies indicated that after hepatocyte differentiation, MSCs-derived HLCs gained hepatic functions including ALB formation, cytochrome P450 enzyme activity, urea secretion, and so on. HLCs were able to recover the ammonia and purine metabolism of liver, decrease aminotransferase and ammonia levels, and improve serum ALB levels, thereby showing more efficient than undifferentiated MSCs in liver diseases treatment [38,[61], [62], [63]]. HLCs also reduced the production of inflammatory factors IL-1β, IL-6, and TNF-α to alleviate inflammation in CCl4-induced ALF mice [39]. Therefore, hADMSCs were induced differentiation towards hepatic lineage by cytokine cocktails to obtain HLCs for the treatment of ALF. As most cells can be removed by macrophages or phagocytes after intrasplenic transplantation [41,43], and an inflammatory environment in the liver also impairs the viability of transplanted cells [31,44], Cu NZs@PLGA nanofibers integrated with dECM hydrogels were applied for in situ delivery of HLCs. Results indicated that as the vehicle of HLCs, dECM provided a desirable 3D culture environment, thereby conspicuously enhancing hepatic functions of HLCs, due to its porous structure and natural ingredients (Fig. 5). These would be conducive to HLCs achieving hepatic function compensation in the injured liver [16,73,74]. Furthermore, Fig. S16 proved that Cu NZs@PLGA nanofibers scavenged H2O2 and exhibited a cytoprotection effect on the embedded HLCs in dECM hydrogel. Besides, the low mechanical strength of dECM hydrogels made it challenging to transplant in situ without disruption. Cu NZs@PLGA nanofiber-reinforced dECM hydrogels improved the operability during in vivo transplantation. The nanofibers also could minimize the implants stuck to neighboring organs or tissues, facilitating migration and integration of HLCs with the host liver (Fig. S23) [46,[75], [76], [77]]. In this study, we fabricated a multifunctional and synergistic therapeutic platform (HLCs/Cu NZs@fiber/dECM), in which each component is equipped with particular functions.
The synergistic therapeutic efficacy of HLCs/Cu NZs@fiber/dECM was evidenced in vivo. On the one hand, the multifaceted antioxidant nanozyme Cu NZs scavenged the excessive ROS with high efficiency, preventing the deterioration of liver tissue necrosis as shown in Fig. 6 and Fig. S17. The decreased cell apoptosis resulted in less accumulation of inflammatory factors, thereby avoiding severe inflammation storms in the liver (Fig. 7, Fig. S18). Meanwhile, the regulated microenvironment also could promote the survival of HLCs. Additionally, as indicated in Fig. 7J–K and Fig. S23, the pro-angiogenic Cu NZs further improved the angiogenesis and vascularization of the implants. On the other hand, HLCs possessed the hepatocyte-related features, which were further elevated in the dECM hydrogel. Fig. 6 and Fig. S21 revealed that HLCs prominently improved the ALB levels of ALF mice. Besides, HLCs possessed inherent anti-inflammatory activity [[37], [38], [39],78], thereby synergistically protecting the host liver cells from oxidative stress and accelerating liver regeneration. Compared with the HLCs/Cu NZs@fiber/dECM group, the serological measurement showed that there was no significant increase in the ALB level of HLCs/fiber/dECM group (Fig. S19 and Fig. S20). That's probably because the excess ROS in the liver cannot be effectively cleared, which influenced the survival and function of implanted HLCs. Likewise, the serum ALB level of HLCs/Cu NZs@fiber group was similar to that of Cu NZs@fiber/dECM group due to the absence of dECM, which likely led to the loss of hepatocyte-related functions in HLCs (Fig. S29 and Fig. S30). As expected, HLCs/Cu NZs@fiber/dECM showed superior therapeutic efficacy on CCl4-induced ALF. Taken together, the synergic effects of HLCs/Cu NZs@fiber/dECM contributed to the effective liver regeneration in ALF-challenged mice, and facilitated long-term hepatic function integration in vivo.
The no-contact liver lobes in the “HLCs/Cu NZs@fiber/dECM” group were also analyzed. There was no significant difference in the necrotic areas of livers between different lobes either on day 1 or day 3 (Fig. S31). Moreover, TUNEL and Ki67 staining images showed similar number of apoptotic cells and proliferating cells in all liver lobes. The ROS level and pro-inflammatory factors (TNF-α and IL-6) were also similar. These results revealed that the HLCs/Cu NZs@fiber/dECM composite had considerable therapeutic efficacy for the whole liver. This might be because of the released Cu NZs in vivo that could diffuse throughout the liver and herein work on other no-contact liver lobes. Furthermore, a recent study also showed that cell migration could be observed in other liver lobes after cell patch grafting in livers [46].
Transcriptome analysis was further conducted to detect the underlying mechanism in vivo. From the heatmap of differential gene expressions, the differential downregulated genes related to oxidative stress and inflammation were displayed in Fig. 8D, which corresponded to the ROS elimination and anti-inflammatory ability of HLCs/Cu NZs@fiber/dECM. The highly expressed angiogenesis-related genes implied the pro-angiogenesis effect of Cu NZs. Similarly, previous research confirmed that Cu NZs substantially contributed to the tissue repairment in wound healing [15]. The KEGG and GSEA analysis were performed to further investigate the pathways influenced by HLCs/Cu NZs@fiber/dECM. Studies suggested that massive hepatocyte senescence in acute liver injury may break the balance between liver regeneration and liver failure [64]. Our results showed that the genes enriched in the p53 signaling pathway were differentially downregulated, and AXIN2 was significantly upregulated after HLCs/Cu NZs@fiber/dECM treatment. Upregulation of AXIN2 in hepatocytes has been identified as a surface marker of hepatocyte self-renew, cell cycle reentrance, and regeneration [64,79]. These results implied that HLCs/Cu NZs@fiber/dECM could prevent hepatocyte senescence and might facilitate liver regeneration by influencing the Wnt signaling pathway. Moreover, differential genes are also enriched in pathways concerning liver biosynthesis, drugs, and substances metabolism. It's worth noting that cytochrome P450-involved drug metabolism pathway differentially upregulated in the liver of HLCs/Cu NZs@fiber/dECM group. In addition, hepatocyte-related genes were also highly expressed. These observations suggested the potential mechanism of hepatic function compensation of HLCs could mainly involve substances biosynthesis and drug metabolism. Collectively, the transcriptome analysis also confirmed the synergistic effect of anti-oxidation, angiogenesis promotion, anti-inflammation, and hepatocyte-related functions in HLCs/Cu NZs@fiber/dECM-based ALF therapy.
There are still some shortcomings in the current research. A long-term tracking of the implanted HLCs in vivo might be necessary in a future study. In addition, although Cu NZs have been evidenced with good biocompatibility both in vitro and in vivo, further investigation of Cu NZs metabolism and distribution might be a premium in the future, which can provide safe guidance for the clinical application of the nanoenzymes. Meanwhile, the effective dosage of HLCs might also need a comprehensive study.
5. Conclusion
In summary, we developed a Cu NZs@PLGA nanofiber-reinforced dECM hydrogel for the delivery of functional HLCs and evidenced the effectiveness of the strategy for treatment of CCl4-induced ALF in this study. Cu NZs possessed high efficiency of ROS scavenging, and herein avoided further progression of ALF. The cytoprotective role and pro-angiogenic effects of Cu NZs, mechanical support of PLGA nanofibers, as well as the functionally enhanced HLCs in the dECM hydrogel synergically contributed to the final promising therapeutic efficacy, including reduced tissue necrosis and inflammation, enhanced liver regeneration and function recovery. Hence, we believe our approach that delivers functional HLCs using Cu NZs@PLGA nanofiber-reinforced dECM hydrogels holds great potential for clinical translation in the treatment of ALF.
Ethics approval and consent to participate
All the animal studies were performed following the protocol approved by the Institutional Animal Care and Use Committee of Sun Yat-Sen University.
CRediT authorship contribution statement
Yuanyuan Jin: Methodology, Investigation, Formal analysis, Writing – original draft. Jiabin Zhang: Investigation, Methodology, Writing – review & editing. Yanteng Xu: Investigation, Methodology. Ke Yi: Methodology. Fenfang Li: Methodology. Huicong Zhou: Methodology, Resources. Haixia Wang: Software, Supervision. Hon Fai Chan: Resources, Writing – review & editing. Yeh-Hsing Lao: Visualization, Writing – review & editing. Shixian Lv: Visualization, Writing – review & editing. Yu Tao: Conceptualization, Methodology, Investigation, Validation, Visualization, Writing – review & editing, Funding acquisition, Project administration, Resources, Supervision. Mingqiang Li: Conceptualization, Methodology, Visualization, Writing – review & editing, Funding acquisition, Project administration, Resources, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work is supported by the National Key Research and Development Program of China (2019YFA0111300), the National Natural Science Foundation of China (22277155, 32001012), the Science and Technology Program of Guangzhou (202102010225, 202102010217), the Guangdong Provincial Pearl Talents Program (2019QN01Y131), Guangdong Basic and Applied Basic Research Foundation (2022A1515110685, 2023A1515011067), the China Postdoctoral Science Foundation (2020M683060), the Talent Introduction Program of Postdoctoral International Exchange Program (YJ20200313), the Thousand Talents Plan, and the China Primary Health Care Foundation (2022-003).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2023.05.001.
Contributor Information
Yu Tao, Email: taoy28@mail.sysu.edu.cn.
Mingqiang Li, Email: limq567@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Shi X., Gao Y., Yan Y., Ma H., Sun L., Huang P., Ni X., Zhang L., Zhao X., Ren H., Hu D., Zhou Y., Tian F., Ji Y., Cheng X., Pan G., Ding Y., Hui L. Improved survival of porcine acute liver failure by a bioartificial liver device implanted with induced human functional hepatocytes. Cell Res. 2016;26(2):206–216. doi: 10.1038/cr.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stravitz R.T., Lee W.M. Acute liver failure. Lancet. 2019;394(10201):869–881. doi: 10.1016/S0140-6736(19)31894-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu H., Xia F., Zhang L., Fang C., Lee J., Gong L., Gao J., Ling D., Li F. A ROS-sensitive nanozyme-augmented photoacoustic nanoprobe for early diagnosis and therapy of acute liver failure. Adv. Mater. 2022;34(7) doi: 10.1002/adma.202108348. [DOI] [PubMed] [Google Scholar]
- 4.Huang J., Li J., Lyu Y., Miao Q., Pu K. Molecular optical imaging probes for early diagnosis of drug-induced acute kidney injury. Nat. Mater. 2019;18(10):1133–1143. doi: 10.1038/s41563-019-0378-4. [DOI] [PubMed] [Google Scholar]
- 5.Woolbright B.L., Jaeschke H. Role of the inflammasome in acetaminophen-induced liver injury and acute liver failure. J. Hepatol. 2017;66(4):836–848. doi: 10.1016/j.jhep.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ai Y., Hu Z., Liang X., Sun H., Xin H., Liang Q. Recent advances in nanozymes: from matters to bioapplications. Adv. Funct. Mater. 2022;32(14) [Google Scholar]
- 7.Tu Z., Zhong Y., Hu H., Shao D., Haag R., Schirner M., Lee J., Sullenger B., Leong K.W. Design of therapeutic biomaterials to control inflammation. Nat. Rev. Mater. 2022;7(7):557–574. doi: 10.1038/s41578-022-00426-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mu J., Li C., Yu S., Liu G., Zou J., Zhang D., Jiang C., Wang X., He L., Huang P., Yin Y., Chen X. Protective effect of platinum nano-antioxidant and nitric oxide against hepatic ischemia-reperfusion injury. Nat. Commun. 2022;13(1):2513. doi: 10.1038/s41467-022-29772-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J., Wang Y., Wang W., Tian Y., Gan Z., Wang Y., He H., Chen W., Zhang X., Wu Y., Jia R., Shi M., Wei W., Ma G. In situ growth of nano-antioxidants on cellular vesicles for efficient reactive oxygen species elimination in acute inflammatory diseases. Nano Today. 2021;40 [Google Scholar]
- 10.Tao Y., Ju E., Ren J., Qu X. Bifunctionalized mesoporous silica-supported gold nanoparticles: intrinsic oxidase and peroxidase catalytic activities for antibacterial applications. Adv. Mater. 2015;27(6):1097–1104. doi: 10.1002/adma.201405105. [DOI] [PubMed] [Google Scholar]
- 11.Wu J., Wang X., Wang Q., Lou Z., Li S., Zhu Y., Qin L., Wei H. Nanomaterials with enzyme-like characteristics (nanozymes): next-generation artificial enzymes (II) Chem. Soc. Rev. 2019;48(4):1004–1076. doi: 10.1039/c8cs00457a. [DOI] [PubMed] [Google Scholar]
- 12.Xia F., Hu X., Zhang B., Wang X., Guan Y.A., Lin P.H., Ma Z.Y., Sheng J.P., Ling D.S., Li F.Y. Ultrasmall ruthenium nanoparticles with boosted antioxidant activity upregulate regulatory T cells for highly efficient liver injury therapy. Small. 2022;18(29) doi: 10.1002/smll.202201558. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y., Cheng Y., Zhang H., Zhou M., Yu Y., Lin S., Jiang B., Zhao X., Miao L., Wei C., Liu Q., Lin Y., Du Y., Butch C.J., Wei H. Integrated cascade nanozyme catalyzes in vivo ROS scavenging for anti-inflammatory therapy. Sci. Adv. 2020;6(29) doi: 10.1126/sciadv.abb2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu T., Xiao B., Xiang F., Tan J., Chen Z., Zhang X., Wu C., Mao Z., Luo G., Chen X., Deng J. Ultrasmall copper-based nanoparticles for reactive oxygen species scavenging and alleviation of inflammation related diseases. Nat. Commun. 2020;11(1):2788. doi: 10.1038/s41467-020-16544-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng Y., He D., Ge X., Lu Y., Chai Y., Zhang Y., Mao Z., Luo G., Deng J., Zhang Y. Construction of heparin-based hydrogel incorporated with Cu5.4O ultrasmall nanozymes for wound healing and inflammation inhibition. Bioact. Mater. 2021;6(10):3109–3124. doi: 10.1016/j.bioactmat.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao Z., Wang H., Chen J., Zhang Y., Mo Q., Zhang P., Wang M., Liu H., Bao X., Sun Y., Zhang W., Yao Q. Silk-based hydrogel incorporated with metal-organic framework nanozymes for enhanced osteochondral regeneration. Bioact. Mater. 2023;20:221–242. doi: 10.1016/j.bioactmat.2022.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahu A., Jeon J., Lee M.S., Yang H.S., Tae G. Nanozyme impregnated mesenchymal stem cells for hepatic ischemia-reperfusion injury alleviation. ACS Appl. Mater. Interfaces. 2021;13(22):25649–25662. doi: 10.1021/acsami.1c03027. [DOI] [PubMed] [Google Scholar]
- 18.Liu M., Huang Q., Zhu Y., Chen L., Li Y., Gong Z., Ai K. Harnessing reactive oxygen/nitrogen species and inflammation: nanodrugs for liver injury. Mater. Today Bio. 2022;13 doi: 10.1016/j.mtbio.2022.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang D.Y., Tu T., Younis M.R., Zhu K.S., Liu H., Lei S., Qu J., Lin J., Huang P. Clinically translatable gold nanozymes with broad spectrum antioxidant and anti-inflammatory activity for alleviating acute kidney injury. Theranostics. 2021;11(20):9904–9917. doi: 10.7150/thno.66518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ni D., Wei H., Chen W., Bao Q., Rosenkrans Z.T., Barnhart T.E., Ferreira C.A., Wang Y., Yao H., Sun T., Jiang D., Li S., Cao T., Liu Z., Engle J.W., Hu P., Lan X., Cai W. Ceria nanoparticles meet hepatic ischemia-reperfusion injury: the perfect imperfection. Adv. Mater. 2019;31(40) doi: 10.1002/adma.201902956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J., Hong G., Mazaleuskaya L., Hsu J.C., Rosario-Berrios D.N., Grosser T., Cho-Park P.F., Cormode D.P. Ultrasmall antioxidant cerium oxide nanoparticles for regulation of acute inflammation. ACS Appl. Mater. Interfaces. 2021;13(51):60852–60864. doi: 10.1021/acsami.1c16126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F., Qiu Y., Xia F., Sun H., Liao H., Xie A., Lee J., Lin P., Wei M., Shao Y., Yang B., Weng Q., Ling D. Dual detoxification and inflammatory regulation by ceria nanozymes for drug-induced liver injury therapy. Nano Today. 2020;35 [Google Scholar]
- 23.Zhang D.Y., Liu H., Li C., Younis M.R., Lei S., Yang C., Lin J., Li Z., Huang P. Ceria nanozymes with preferential renal uptake for acute kidney injury alleviation. ACS Appl. Mater. Interfaces. 2020;12(51):56830–56838. doi: 10.1021/acsami.0c17579. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X., Hu J., Becker K.V., Engle J.W., Ni D., Cai W., Wu D., Qu S. Antioxidant and C5a-blocking strategy for hepatic ischemia-reperfusion injury repair. J. Nanobiotechnol. 2021;19(1):107. doi: 10.1186/s12951-021-00858-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L., Cheng L., Wang Z., Zhang J., Mao X., Liu Z., Zhang Y., Cui W., Sun X. Conditioned medium-electrospun fiber biomaterials for skin regeneration. Bioact. Mater. 2021;6(2):361–374. doi: 10.1016/j.bioactmat.2020.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu M., Huang L. Nanomaterial manipulation of immune microenvironment in the diseased liver. Adv. Funct. Mater. 2019;29(7) [Google Scholar]
- 27.Jain A., Behera M., Mahapatra C., Sundaresan N.R., Chatterjee K. Nanostructured polymer scaffold decorated with cerium oxide nanoparticles toward engineering an antioxidant and anti-hypertrophic cardiac patch. Mater. Sci. Eng., C. 2021;118 doi: 10.1016/j.msec.2020.111416. [DOI] [PubMed] [Google Scholar]
- 28.Rather H.A., Thakore R., Singh R., Jhala D., Singh S., Vasita R. Antioxidative study of Cerium Oxide nanoparticle functionalised PCL-Gelatin electrospun fibers for wound healing application. Bioact. Mater. 2018;3(2):201–211. doi: 10.1016/j.bioactmat.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X., Yang J., Wang L., Ran B., Jia Y., Zhang L., Yang G., Shao H., Jiang X. Pharmaceutical intermediate-modified gold nanoparticles: against multidrug-resistant bacteria and wound-healing application via an electrospun scaffold. ACS Nano. 2017;11(6):5737–5745. doi: 10.1021/acsnano.7b01240. [DOI] [PubMed] [Google Scholar]
- 30.Hu Y.W., Xu Y.T., Mintz R.L., Luo X., Fang Y.Q., Lao Y.H., Chan H.F., Li K., Lv S.X., Chen G.J., Tao Y., Luo Y., Li M.Q. Self-intensified synergy of a versatile biomimetic nanozyme and doxorubicin on electrospun fibers to inhibit postsurgical tumor recurrence and metastasis. Biomaterials. 2023;293 doi: 10.1016/j.biomaterials.2022.121942. [DOI] [PubMed] [Google Scholar]
- 31.Dwyer B.J., Macmillan M.T., Brennan P.N., Forbes S.J. Cell therapy for advanced liver diseases: repair or rebuild. J. Hepatol. 2021;74(1):185–199. doi: 10.1016/j.jhep.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 32.Hazrati A., Malekpour K., Soudi S., Hashemi S.M. Mesenchymal stromal/stem cells and their extracellular vesicles application in acute and chronic inflammatory liver diseases: emphasizing on the anti-fibrotic and immunomodulatory mechanisms. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.865888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shokravi S., Borisov V., Zaman B.A., Niazvand F., Hazrati R., Khah M.M., Thangavelu L., Marzban S., Sohrabi A., Zamani A. Mesenchymal stromal cells (MSCs) and their exosome in acute liver failure (ALF): a comprehensive review. Stem Cell Res. Ther. 2022;13(1):192. doi: 10.1186/s13287-022-02825-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J., Chan H.F., Wang H., Shao D., Tao Y., Li M. Stem cell therapy and tissue engineering strategies using cell aggregates and decellularized scaffolds for the rescue of liver failure. J. Tissue Eng. 2021;12 doi: 10.1177/2041731420986711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee C.W., Chen Y.F., Wu H.H., Lee O.K. Historical perspectives and advances in mesenchymal stem cell research for the treatment of liver diseases. Gastroenterology. 2018;154(1):46–56. doi: 10.1053/j.gastro.2017.09.049. [DOI] [PubMed] [Google Scholar]
- 36.Hu C.X., Zhao L.F., Li L.J. Current understanding of adipose-derived mesenchymal stem cell-based therapies in liver diseases. Stem Cell Res. Ther. 2019;10:199. doi: 10.1186/s13287-019-1310-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bogliotti Y., Vander Roest M., Mattis A.N., Gish R.G., Peltz G., Anwyl R., Kivlighn S., Schuur E.R. Clinical application of induced hepatocyte-like cells produced from mesenchymal stromal cells: a literature review. Cells. 2022;11(13):1998. doi: 10.3390/cells11131998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aurich H., Sgodda M., Kaltwasser P., Vetter M., Weise A., Liehr T., Brulport M., Hengstler J.G., Dollinger M.M., Fleig W.E., Christ B. Hepatocyte differentiation of mesenchymal stem cells from human adipose tissue in vitro promotes hepatic integration in vivo. Gut. 2009;58(4):570–581. doi: 10.1136/gut.2008.154880. [DOI] [PubMed] [Google Scholar]
- 39.Schuur E., Bogliotti Y., Vander Roest M. Effects of a novel cell therapy in mice with chemically induced acute liver injury using hepatocyte-like cells derived from adipose stromal cells reduces liver damage and inflammation. Hepatology. 2021;74(6):1412A–1413A. [Google Scholar]
- 40.Yuan J., Li W., Huang J., Guo X., Li X., Lu X., Huang X., Zhang H. Transplantation of human adipose stem cell-derived hepatocyte-like cells with restricted localization to liver using acellular amniotic membrane. Stem Cell Res. Ther. 2015;6:217. doi: 10.1186/s13287-015-0208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagamoto Y., Takayama K., Ohashi K., Okamoto R., Sakurai F., Tachibana M., Kawabata K., Mizuguchi H. Transplantation of a human iPSC-derived hepatocyte sheet increases survival in mice with acute liver failure. J. Hepatol. 2016;64(5):1068–1075. doi: 10.1016/j.jhep.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 42.Hwang Y., Goh M., Kim M., Tae G. Injectable and detachable heparin-based hydrogel micropatches for hepatic differentiation of hADSCs and their liver targeted delivery. Biomaterials. 2018;165:94–104. doi: 10.1016/j.biomaterials.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Español-Suñer R., Carpentier R., Van Hul N., Legry V., Achouri Y., Cordi S., Jacquemin P., Lemaigre F., Leclercq I.A. Liver progenitor cells yield functional hepatocytes in response to chronic liver injury in mice. Gastroenterology. 2012;143(6):1564–1575. doi: 10.1053/j.gastro.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 44.Bird T.G., Mueller M., Boulter L., Vincent D.F., Ridgway R.A., Lopez-Guadamillas E., Lu W.-Y., Jamieson T., Govaere O., Campbell A.D., Ferreira-Gonzalez S., Cole A.M., Hay T., Simpson K.J., Clark W., Hedley A., Clarke M., Gentaz P., Nixon C., Bryce S., Kiourtis C., Sprangers J., Nibbs R.J.B., Van Rooijen N., Bartholin L., McGreal S.R., Apte U., Barry S.T., Iredale J.P., Clarke A.R., Serrano M., Roskams T.A., Sansom O.J., Forbes S.J. TGF beta inhibition restores a regenerative response in acute liver injury by suppressing paracrine senescence. Sci. Transl. Med. 2018;10(454) doi: 10.1126/scitranslmed.aan1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim Y., Kim Y.W., Lee S.B., Kang K., Yoon S., Choi D., Park S., Jeong J. Hepatic patch by stacking patient-specific liver progenitor cell sheets formed on multiscale electrospun fibers promotes regenerative therapy for liver injury. Biomaterials. 2021;274 doi: 10.1016/j.biomaterials.2021.120899. [DOI] [PubMed] [Google Scholar]
- 46.Zhang W., Lanzoni G., Hani H., Overi D., Cardinale V., Simpson S., Pitman W., Allen A., Yi X., Wang X., Gerber D., Prestwich G., Lozoya O., Gaudio E., Alvaro D., Tokaz D., Dominguez-Bendala J., Adin C., Piedrahita J., Mathews K., Sethupathy P., Carpino G., He Z., Wauthier E., Reid L.M. Patch grafting, strategies for transplantation of organoids into solid organs such as liver. Biomaterials. 2021;277 doi: 10.1016/j.biomaterials.2021.121067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luce E., Messina A., Duclos-Vallee J.-C., Dubart-Kupperschmitt A. Advanced techniques and awaited clinical applications for human pluripotent stem cell differentiation into hepatocytes. Hepatology. 2021;74(2):1101–1116. doi: 10.1002/hep.31705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.da Silva Morais A., Vieira S., Zhao X., Mao Z., Gao C., Oliveira J.M., Reis R.L. Advanced biomaterials and processing methods for liver regeneration: state-of-the-art and future trends. Adv. Healthcare Mater. 2020;9(5) doi: 10.1002/adhm.201901435. [DOI] [PubMed] [Google Scholar]
- 49.Banas A., Teratani T., Yamamoto Y., Tokuhara M., Takeshita F., Quinn G., Okochi H., Ochiya T. Adipose tissue-derived mesenchymal stem cells as a source of human hepatocytes. Hepatology. 2007;46(1):219–228. doi: 10.1002/hep.21704. [DOI] [PubMed] [Google Scholar]
- 50.Xu L.J., Wang S.F., Wang D.Q., Ma L.J., Chen Z., Chen Q.Q., Wang J., Yan L. Adipose-derived stromal cells resemble bone marrow stromal cells in hepatocyte differentiation potential in vitro and in vivo. World J. Gastroenterol. 2017;23(38):6973–6982. doi: 10.3748/wjg.v23.i38.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perea L., Coll M., Sanjurjo L., Blaya D., Taghdouini A.E., Rodrigo-Torres D., Altamirano J., Graupera I., Aguilar-Bravo B., Llopis M., Vallverdu J., Caballeria J., van Grunsven L.A., Sarrias M.R., Gines P., Sancho-Bru P. Pentraxin-3 modulates lipopolysaccharide-induced inflammatory response and attenuates liver injury. Hepatology. 2017;66(3):953–968. doi: 10.1002/hep.29215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Triantafyllou E., Pop O.T., Possamai L.A., Wilhelm A., Liaskou E., Singanayagam A., Bernsmeier C., Khamri W., Petts G., Dargue R., Davies S.P., Tickle J., Yuksel M., Patel V.C., Abeles R.D., Stamataki Z., Curbishley S.M., Ma Y., Wilson I.D., Coen M., Woollard K.J., Quaglia A., Wendon J., Thursz M.R., Adams D.H., Weston C.J., Antoniades C.G. MerTK expressing hepatic macrophages promote the resolution of inflammation in acute liver failure. Gut. 2018;67(2):333–347. doi: 10.1136/gutjnl-2016-313615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ji H., Zhang C., Xu F., Mao Q., Xia R., Chen M., Wang W., Lv S., Li W., Shi X. Inhaled pro-efferocytic nanozymes promote resolution of acute lung injury. Adv. Sci. 2022;9(26) doi: 10.1002/advs.202201696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu S.Y., Zhao M., Zhou Y.J., Li L., Wang C.S., Yuan Y.J., Li L., Liao G.N., Bresette W., Chen Y.N., Cheng J.Q., Lu Y.R., Liu J.P. A self-assembling peptide hydrogel-based drug co-delivery platform to improve tissue repair after ischemia-reperfusion injury. Acta Biomater. 2020;103:102–114. doi: 10.1016/j.actbio.2019.12.011. [DOI] [PubMed] [Google Scholar]
- 55.Yang R.H., Li G., Zhuang C.Y., Yu P., Ye T.J., Zhang Y., Shang P.Y., Huang J.J., Cai M., Wang L., Cui W.G., Deng L.F. Gradient bimetallic ion-based hydrogels for tissue microstructure reconstruction of tendon-to-bone insertion. Sci. Adv. 2021;7(26) doi: 10.1126/sciadv.abg3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Afshari A., Shamdani S., Uzan G., Naserian S., Azarpira N. Different approaches for transformation of mesenchymal stem cells into hepatocyte-like cells. Stem Cell Res. Ther. 2020;11(1):54. doi: 10.1186/s13287-020-1555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stock P., Brückner S., Ebensing S., Hempel M., Dollinger M.M., Christ B. The generation of hepatocytes from mesenchymal stem cells and engraftment into murine liver. Nat. Protoc. 2010;5(4):617–627. doi: 10.1038/nprot.2010.7. [DOI] [PubMed] [Google Scholar]
- 58.Ardalani H., Sengupta S., Harms V., Vickerman V., Thomson J.A., Murphy W.L. 3-D culture and endothelial cells improve maturity of human pluripotent stem cell-derived hepatocytes. Acta Biomater. 2019;95:371–381. doi: 10.1016/j.actbio.2019.07.047. [DOI] [PubMed] [Google Scholar]
- 59.Asadi M., Lotfi H., Salehi R., Mehdipour A., Zarghami N., Akbarzadeh A., Alizadeh E. Hepatic cell-sheet fabrication of differentiated mesenchymal stem cells using decellularized extracellular matrix and thermoresponsive polymer. Biomed. Pharmacother. 2021;134 doi: 10.1016/j.biopha.2020.111096. [DOI] [PubMed] [Google Scholar]
- 60.Wang B., Li W., Dean D., Mishra M.K., Wekesa K.S. Enhanced hepatogenic differentiation of bone marrow derived mesenchymal stem cells on liver ECM hydrogel. J. Biomed. Mater. Res. 2018;106(3):829–838. doi: 10.1002/jbm.a.36278. [DOI] [PubMed] [Google Scholar]
- 61.Ramanathan R., Pettinato G., Beeston J.T., Lee D.D., Wen X., Mangino M.J., Fisher R.A. Transplantation of human stem cell-derived hepatocytes in an animal model of acute liver failure. Surgery. 2015;158(2):349–359. doi: 10.1016/j.surg.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 62.Xu D., Nishimura T., Zheng M., Wu M., Su H., Sato N., Lee G., Michie S., Glenn J., Peltz G. Enabling autologous human liver regeneration with differentiated adipocyte stem cells. Cell Transplant. 2014;23(12):1573–1584. doi: 10.3727/096368913X673432. [DOI] [PubMed] [Google Scholar]
- 63.Amer M.-E.M., El-Sayed S.Z., Abou El-Kheir W., Gabr H., Gomaa A.A., El-Noomani N., Hegazy M. Clinical and laboratory evaluation of patients with end-stage liver cell failure injected with bone marrow-derived hepatocyte-like cells. Eur. J. Gastroenterol. Hepatol. 2011;23(10):936–941. doi: 10.1097/MEG.0b013e3283488b00. [DOI] [PubMed] [Google Scholar]
- 64.Campana L., Esser H., Huch M., Forbes S. Liver regeneration and inflammation: from fundamental science to clinical applications. Nat. Rev. Mol. Cell Biol. 2021;22(9):608–624. doi: 10.1038/s41580-021-00373-7. [DOI] [PubMed] [Google Scholar]
- 65.Wang B., Zhao L., Fish M., Logan C.Y., Nusse R. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature. 2015;524(7564):180–185. doi: 10.1038/nature14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eming S.A., Murray P.J., Pearce E.J. Metabolic orchestration of the wound healing response. Cell Metabol. 2021;33(9):1726–1743. doi: 10.1016/j.cmet.2021.07.017. [DOI] [PubMed] [Google Scholar]
- 67.Cayrol C., Girard J.-P. Interleukin-33 (IL-33): a nuclear cytokine from the IL-1 family. Immunol. Rev. 2018;281(1):154–168. doi: 10.1111/imr.12619. [DOI] [PubMed] [Google Scholar]
- 68.Corcoran S.E., O'Neill L.A.J. HIF1 alpha and metabolic reprogramming in inflammation. J. Clin. Invest. 2016;126(10):3699–3707. doi: 10.1172/JCI84431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oliva-Vilarnau N., Vorrink S.U., Ingelman-Sundberg M., Lauschke V.M. A 3D cell culture model identifies wnt/beta-catenin mediated inhibition of p53 as a critical step during human hepatocyte regeneration. Adv. Sci. 2020;7(15) doi: 10.1002/advs.202000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thi V.L.D., Wu X.F., Belote R.L., Andreo U., Takacs C.N., Fernandez J.P., Vale-Silva L.A., Prallet S., Decker C.C., Fu R.M., Qu B.Q., Uryu K., Molina H., Saeed M., Steinmann E., Urban S., Singaraja R.R., Schneider W.M., Simon S.M., Rice C.M. Stem cell-derived polarized hepatocytes. Nat. Commun. 2020;11(1):1677. doi: 10.1038/s41467-020-15337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin Y., Wang H., Yi K., Lv S., Hu H., Li M., Tao Y. Applications of nanobiomaterials in the therapy and imaging of acute liver failure. Nano-Micro Lett. 2021;13(1):25. doi: 10.1007/s40820-020-00550-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu Y., Li Z., Li L.H., Chen J.L., Xu X.Y., Lin Z.F., Zhang T., Zhu Y., Ding C.H., Mao C.B. Highly effective rheumatoid arthritis therapy by peptide-promoted nanomodification of mesenchymal stem cells. Biomaterials. 2022;283 doi: 10.1016/j.biomaterials.2022.121474. [DOI] [PubMed] [Google Scholar]
- 73.Wu Q., Li Y., Yang Z., Li L., Yang J., Zhu X., Liu Y., Bao J., Bu H. Ectopic expansion and vascularization of engineered hepatic tissue based on heparinized acellular liver matrix and mesenchymal stromal cell spheroids. Acta Biomater. 2022;137:79–91. doi: 10.1016/j.actbio.2021.10.017. [DOI] [PubMed] [Google Scholar]
- 74.Dong Y.X., Sigen A., Rodrigues M., Li X.L., Kwon S.H., Kosaric N., Khong S., Gao Y.S., Wang W.X., Gurtner G.C. Injectable and tunable gelatin hydrogels enhance stem cell retention and improve cutaneous wound healing. Adv. Funct. Mater. 2017;27(24) [Google Scholar]
- 75.Liu C., Xu X., Cui W., Zhang H. Metal-organic framework (MOF)-based biomaterials in bone tissue engineering. Engineered Regeneration. 2021;2:105–108. [Google Scholar]
- 76.Cuidi Li W.C. 3D bioprinting of cell-laden constructs for regenerative medicine. Engineered Regeneration. 2021;2:195–205. [Google Scholar]
- 77.Cai L., Li N., Zhang Y., Gu H., Zhu Y. Microfluidics-derived microcarrier systems for oral delivery. Biomed. Technol. 2023;1:30–38. [Google Scholar]
- 78.Chen F., Jimenez R.J., Sharma K., Luu H.Y., Hsu B.Y., Ravindranathan A., Stohr B.A., Willenbring H. Broad distribution of hepatocyte proliferation in liver homeostasis and regeneration. Cell Stem Cell. 2020;26(1):27–33. doi: 10.1016/j.stem.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun T., Pikiolek M., Orsini V., Bergling S., Holwerda S., Morelli L., Hoppe P.S., Planas-Paz L., Yang Y., Ruffner H., Bouwmeester T., Lohmann F., Terracciano L.M., Roma G., Cong F., Tchorz J.S. AXIN2(+) pericentral hepatocytes have limited contributions to liver homeostasis and regeneration. Cell Stem Cell. 2020;26(1):97–107. doi: 10.1016/j.stem.2019.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.