Abstract
We have developed a cosmid and plasmid system to generate mutant strains of murine cytomegalovirus (MCMV). The system is based on a series of seven overlapping cosmid clones that regenerate MCMV when cotransfected into mouse cells. The unaltered cosmids produce MCMV that is indistinguishable from wild-type MCMV based on restriction enzyme digest patterns of virus DNA and growth rates both in vitro and in vivo. Analysis of viral DNA from plaque-purified recombinant isolates taken from in vitro and in vivo stocks indicated that regeneration did not introduce novel mutations in the recombinant viral genomes. Isolation of specific genes and subsequent generation of specific mutant MCMVs was accomplished by replacement of cosmids with overlapping plasmid subclones. A new vector, PmeSUB, featuring a multiple cloning site and a stringent origin of replication, was constructed to make large subclones for use with smaller subclones containing the gene of interest. The utility of this system was demonstrated by the generation of two different mutant MCMVs from different combinations of overlapping plasmid subclones of one cosmid. The advantages of this system are that (i) target genes are maintained as small clones making them amenable to standard in vitro mutagenesis manipulations and that (ii) no reporter or selection genes are necessary to identify mutants.
Murine cytomegalovirus (MCMV) is widely used as a model virus system for the study of human cytomegalovirus (HCMV) pathogenesis. MCMV and HCMV share an essentially collinear arrangement of genes (7, 31) and similar pathogenesis in their respective hosts (18). Our understanding of MCMV and HCMV biology has been greatly facilitated by the construction of recombinant viruses bearing specific mutations in genes of interest. The most widely used approach to construct recombinant MCMV involves the introduction of viral DNA and a plasmid bearing the target gene adjacent to a selectable marker and/or reporter gene into permissive cells (reviewed in reference 28). Mutant viruses arise by homologous recombination between the viral DNA and homologous sequences flanking the target gene and are isolated by multiple rounds of plaque purification. In addition, CMV genomes have been isolated as infectious bacterial artificial chromosomes (BACs) and, from these, specific mutants were constructed (2, 4, 27). A recent version of the MCMV BAC regenerates an intermediate virus that can delete its BAC-specific sequences when passaged in vitro, yielding progeny virus that replicates at wild-type levels in vivo (38). The CMV BACs have proven to be useful in the identification of open reading frames (ORFs) necessary for MCMV replication (5). However, the construction of BAC mutants requires challenging genetic manipulations to enable allelic exchange in Escherichia coli (30).
Alternatively, sets of overlapping cosmid and plasmid clones have been used in the construction and direct recovery of herpesvirus mutants that do not contain extra genetic material (10, 11, 22, 32, 36, 37). The utility of cosmids and plasmids for the rapid generation of mutant herpesviruses was demonstrated by the isolation of 17 different HCMV UL54 mutants (9). Here we describe the isolation of an infectious set of cosmid clones for MCMV and its development into a system for generating specific MCMV mutants. The utility of this system was demonstrated by the generation of two mutant MCMV strains by replacement of one cosmid with two different series of overlapping subclones. These combinations of cosmids and plasmids generated mutant strains of MCMV without the need for plaque purification.
MATERIALS AND METHODS
Cell culture and viruses.
Murine fibroblasts (NIH 3T3; ATCC CRL-1658) were maintained on Dulbecco modified Eagle medium (DMEM) supplemented with 5% calf serum and 2 mM glutamine at 37°C in 6% CO2 in a humidified incubator. MCMV (Smith strain, ATCC VR-1399) DNA was used for cosmid libraries constructed.
Vector constructions.
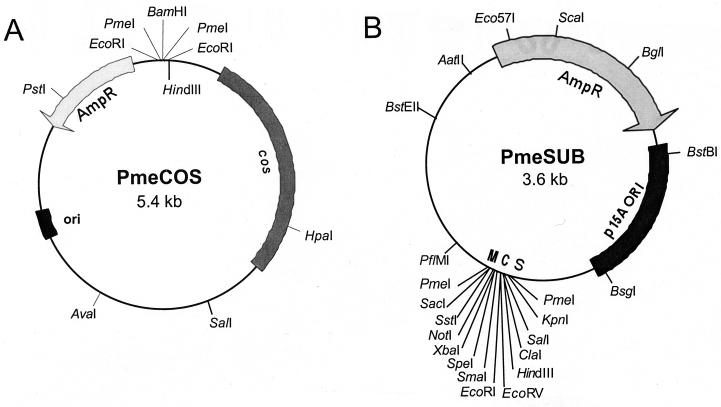
The cosmid vector, PmeCOS (pBH021; Fig. 1A), used to make libraries was derived from pJB8 (19). Complimentary oligonucleotides, 5′-AATTCCGTTTAAACAG-3′ and 5′-GATCCTGTTTAAACGG-3′, were phosphorylated using T4 polynucleotide kinase (33) and allowed to anneal forming an adaptomer. This adaptomer featured a cut EcoRI site on one end, with a PmeI site in the middle and a cut BamHI site on the other end. The adaptomer was ligated into EcoRI-cut pJB8 to yield PmeCOS, which has the cloning site configuration of EcoRI-PmeI-BamHI-PmeI-EcoRI, as confirmed by DNA sequence analysis.
FIG. 1.
Cosmid and plasmid vectors for making infectious MCMV clone sets. (A) The PmeCOS cosmid vector was constructed by adding a pair of PmeI sites surrounding the BamHI cloning site of pJB8 (19) so that any cloned MCMV sequences can be removed intact. (B) The PmeSUB vector was derived from pACYC177 by replacement of the Kanr gene with the MCS from pBluescript. The MCS was surrounded by PmeI sites to allow excision of MCMV sequences.
The plasmid vector PmeSUB (pBH060; Fig. 1B) was constructed for subcloning MCMV sequences from cosmid clones. The multiple cloning site (MCS) from pBluescript KS(−) (1, 34) was amplified by PCR using the primers 5′-ATTGACCACGTGTTTAAACGCCAGTGAATTGTAATACGAC-3′ and 5′-ACTGCTCACGTGGTTTAAACACTAAAGGGAACAAAAGC-3′. The amplified product contained an MCS surrounded by PmeI restriction sites with PmlI sites at its ends. The PCR fragment was blunt ended by digestion with PmlI, followed by ligation into the T4 DNA polymerase-blunted HindIII and XhoI sites within the kanamycin resistance gene of pACYC177 (6), yielding PmeSUB.
MCMV DNA isolation.
Viral DNA was isolated from purified virus as follows. Ten T-150 flasks of MCMV-infected cells, including media, were frozen at −80°C and thawed to lyse the cells. The lysate was clarified by two rounds of centrifugation (Beckman JA-14 rotor; 7,500 × g, 20°C, 20 min), and the supernatant was further centrifuged (Beckman JA-14 rotor; 30,000 × g, 20°C, 3 h) to pellet the virus. The crude virus pellet was resuspended in 0.5 ml of 10 mM Tris-HCl buffer (pH 8.0) containing 10 mM MgCl2. DNase (bovine pancreatic DNase I; Sigma) was added to a final concentration of 200 μg/ml, and the mixture incubated at 37°C for 1 h. EDTA was added to a final concentration of 50 mM, and the viral suspension was centrifuged through a sucrose cushion (20% sucrose in 10 mM Tris-HCl buffer [pH 8.0], 150 mM NaCl, and 1 mM EDTA) in a Beckman SW28 rotor (113,000 × g, 20°C, 1 h). The resulting pellet was resuspended in 200 μl of buffer (10 mM Tris-HCl buffer, pH 8.0; 10 mM EDTA) and incubated overnight at 37°C in the presence of sodium sarkosinate (1%), sodium dodecyl sulfate (SDS; 0.5%), and proteinase K (250 μg/ml). The lysate was extracted twice with phenol-chloroform-isoamyl alcohol, and purified MCMV DNA was recovered by precipitation with ethanol.
Infected-cell DNA was isolated as follows: MCMV-infected cells were scraped from tissue culture flasks (100% cytopathic effect) and washed twice with phosphate-buffered saline (PBS). The cells were resuspended in 0.5 ml of 10 mM Tris-HCl buffer (pH 8.0) containing 100 mM NaCl, 0.6% SDS, and 0.1 mg of proteinase K per ml and incubated at 37°C overnight. This mixture was extracted twice with phenol-chloroform-isoamyl alcohol, and the DNA was recovered by precipitation with ethanol.
Library construction.
Cosmid libraries were constructed from both purified virus DNA and MCMV-infected-cell DNA according to methods developed for library construction in pJB8 (19). DNA from either purified virions or virus-infected cells was partially digested with restriction endonuclease Sau3AI to an average length of 40 to 45 kb. Purified viral DNA digested with Sau3AI was dephosphorylated with shrimp alkaline phosphatase (Boehringer Mannheim), followed by ligation with BamHI-digested PmeCOS arms. In contrast, infected-cell MCMV DNA digested with Sau3AI was size fractionated by sucrose density gradient centrifugation (33), but it was not dephosphorylated prior to ligation. The left cosmid arm was the 5.4-kb HindIII-BamHI fragment of PmeCOS with the HindIII end dephosphorylated prior to cutting with BamHI, whereas the right arm was the 2.4-kb BamHI-SalI fragment with the SalI end dephosphorylated. Ligation reactions, containing Sau3AI-digested MCMV DNA and a molar excess of PmeCOS arms, were catalyzed by T4 DNA ligase at 16°C for 16 h. The ligation products were packaged into bacteriophage lambda using commercially available packaging mixes (Gigapack III XL and Gigapack III Plus; Stratagene). Escherichia coli DH10B (recA1) grown in the presence of 10 mM MgCl2 (33) was transduced with the packaged libraries followed by plating and growth on Luria-Bertani (LB) agar media plates containing 100 μg of ampicillin per ml.
Selection of overlapping cosmid clones.
Cosmid DNA from randomly picked colonies was digested with EcoRI to map cloned inserts, whereas DNA sequencing precisely identified the boundaries of selected clones. Colony hybridization with radioactive DNA probes against replicate filters (33) was used to find clones covering (i) MCMV nucleotides (nt) 83,000 to 85,000 and (ii) the fused genome ends formed during viral replication (20, 21). Clones overlapping the 83,000-nt region were identified with a 1.1-kb EcoRI fragment covering MCMV nt 83861 to 84978, whereas replicative intermediate clones were found using a 1.8-kb EcoRI-HindIII fragment from MCMV nt 888 to 2718.
Cosmid subclones.
The M78 ORF (31) was isolated from cosmid pBH551 on a 6.2-kb KpnI fragment in pUC18 (40), yielding pBH042. The remainder of the cosmid was isolated as a 5.8-kb PmlI fragment in pUC18, pBH045; a 13.4-kb NheI fragment in PmeSUB, pBH067; and a 22-kb PmeI-HindIII fragment in PmeSUB, pBH065. pBH042 was digested with NsiI, and the NsiI-ends were blunted with T4 DNA polymerase plus deoxynucleoside triphosphates, followed by intramolecular ligation yielding pBH057.
The M80 ORF was isolated as pBH058 by ligating the 3.3-kb XhoI fragment from pBH027 and the 3.5-kb XhoI-HindIII fragment of pBH045 into HindIII-plus-SalI-cut pUC18. pBH027 was derived from pJML28 (25) through ablation of the SspI site at MCMV nt 113,356 by oligonucleotide-directed mutagenesis (41, 42) using the oligonucleotide 5′-CGTGATGTGTAAGATTAGATGATTG-3′ and a commercially available kit (GeneEditor; Promega). pBH064 was constructed by ligating the 13-kb PmlI fragment from pBH551 into the SmaI site of PmeSUB.
Cotransfection.
Individual colonies from each cosmid set member or plasmid subclone were isolated on solid medium and used to inoculate 100-ml liquid cultures (LB medium containing 100 μg of ampicillin per ml) for DNA purification using the Qiagen Plasmid Midi Kit (Qiagen). Occasionally, some cosmid clones were unstable and converted into small, undefined plasmids when grown under these conditions. For these, a 1- to 2-ml liquid culture grown overnight at 37°C was used to inoculate 500-ml liquid cultures that were grown at 37°C to middle logarithmic phase (0.4 to 0.6 optical density at 600 nm) before isolation of cosmid DNA using the Qiagen Plasmid Midi Kit. The integrity of each cosmid DNA preparation was checked by digestion with EcoRI, followed by agarose gel electrophoresis. The MCMV sequences were released from the cosmid and plasmid vectors by digestion with either PmeI or the appropriate restriction enzymes, followed by a single phenol-chloroform-isoamyl alcohol extraction and precipitation and washing with ethanol. The dried DNA pellet was redissolved in sterile water in preparation for cotransfection.
PmeI-digested cosmids or cosmids plus plasmids were used to cotransfect mouse cells pretreated with DEAE-dextran (23, 29) in T-25 flasks using the calcium phosphate coprecipitation method (16), followed by glycerol shock at 6 h after cotransfection (15). At 36 to 48 h following cotransfection, the medium was replaced with fresh medium containing penicillin and streptomycin. A cytopathic effect was visible 5 to 7 days after cotransfection and was allowed to progress until 100% cytopathic effect, at which point the flasks were frozen at −80°C. A portion of the thawed lysate was used to infect cells in T-150 flasks to establish virus stocks for further study.
In vitro growth of cosmid-derived MCMV.
Multistep growth curves were determined for wild-type (Smith strain) and cosmid-derived virus in NIH 3T3 cells. Cells were infected at an MOI of 0.01 for 1.5 h at 37°C, washed once with medium, and incubated for various times in a six-well plate. At each time point cells, including media, were frozen at −80°C until titers were determined by standard plaque assay on NIH 3T3 cells.
Analysis of DNA from plaque-purified virus isolates.
Plaque-purified strains (“isolates”) were established by four sequential rounds of plaque isolation using medium containing 0.5% agarose as an overlay. DNA was isolated from purified virus and further refined using GeneClean (Bio 101) prior to restriction enzyme digestion or amplification by PCR. DNA was exhaustively digested with HinP1I, followed by radioactive end labeling using T4 polynucleotide kinase and [33P]ATP (33). The fragments were resolved by polyacrylamide gel electrophoresis on a gel composed of 10% acrylamide–0.11% bisacrylamide over a 1×-to-5× TAE buffer gradient (1× TAE is 40 mM Tris-acetate buffer [pH 8.0] containing 1 mM EDTA) using a standard sequencing gel apparatus with the dried gel used to make an autoradiogram. Selected regions of the viral genome were amplified using PCR with three different pairs of primers as follows: (i) 5′-GACCTAAACCTCCACCAAGACG and 5′-CGGAACTAGTCCCAAATCCATAC to amplify MCMV nt 6936 to 8827, (ii) 5′-AAACACAGGCTGACGAAGGT and 5′-CACGGAGTTTTCGCAAAGATA to amplify MCMV nt 46119 to 47529, and (iii) 5′-AGCATACTGCACGGCGGTCT and 5′-TGGCAGGGATTGAAAGCGAA to amplify MCMV nt 83321 to 84466. These PCR products were cloned into pCR2.1 TOPO (Invitrogen), followed by DNA sequence analysis of the inserts.
Southern hybridization.
DNA (total infected cell, 1 μg; viral DNA, 0.25 μg) was digested overnight with restriction endonuclease before fractionation by agarose gel electrophoresis and blotting to a nitrocellulose membrane using standard methods (33). DNA probes were labeled with [32P]dCTP using the random priming method (14) with a commercially available kit (Boehringer Mannheim) and hybridized overnight in 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–1× Denhardt's solution–0.1% SDS at 65°C. Following hybridization, the membrane was washed three times for 20 min in 0.1× SSPE plus 0.1% SDS at 68°C before exposure to X-ray film.
In vivo growth of cosmid-derived MCMV.
BALB/c mice were infected intraperitoneally with 5.0 × 105 PFU of tissue culture-grown wild-type (Smith strain) or regenerated MCMV. At 10 days postinfection, the animals were euthanized and the salivary glands were removed and homogenized in DMEM containing 5% calf serum, penicillin (100 IU/ml), streptomycin (100 μg/ml), and gentamicin sulfate (50 μg/ml). Virus titers for each animal were determined by standard plaque assay on NIH 3T3 cells.
RESULTS
These results describe the construction of a set of infectious cosmid clones and its development into a system of cosmids and plasmids that generate site-specific MCMV mutants.
Cosmid libraries were constructed using a new cosmid vector, PmeCOS, which was derived from the cosmid vector, pJB8 (19), by surrounding the BamHI cloning site with PmeI restriction sites (Fig. 1A). PmeI sites are not found in the genome of MCMV (31), and therefore any cloned MCMV sequence could be removed by digestion with this enzyme. PmeCOS is 5.4 kb and will accept cloned inserts of up to 45 kb.
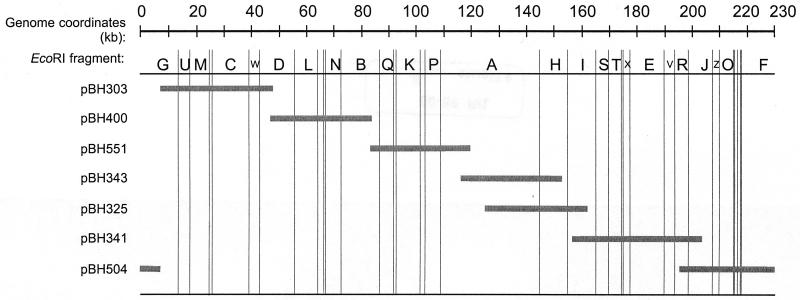
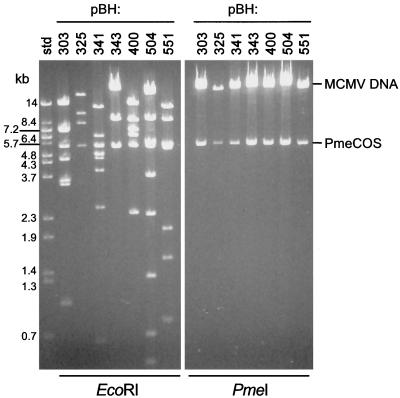
Establishing an infectious set of cosmid clones required isolating recombinants containing inserts that covered the entire MCMV sequence, including the fused ends of the replicative intermediate genome (20, 21). Our initial selection of cosmid clones was deficient in those that covered the region around nt 83,000 and the replicative intermediate. These were isolated from infected-cell DNA-derived libraries using hybridization probes specific to the respective regions of the genome (see Materials and Methods). The genome locations of the cosmid set are illustrated diagrammatically in Fig. 2 with the precise insert coordinates given in Table 1. The inserts ranged from 35 to 46 kb, with sequence overlaps ranging from 884 nt between pBH400 and pBH551 to 28,644 nt between pBH343 and pBH325 (Table 1). Although these latter cosmid clones overlap by approximately 30 kb, they were included in the cosmid set analyzed here since our immediate goal was to cover the entire genome and then demonstrate the feasibility of using these clones to regenerate MCMV. We have mapped approximately 200 other cosmid clones from which others having different regions and extents of overlap may be selected to isolate different genes of interest. The restriction digests shown in Fig. 3 demonstrate the utility of using EcoRI to identify the approximate genomic location of each cosmid clone. EcoRI has 34 recognition sites in the Smith strain sequence (31) and, as a consequence, produces readily interpretable restriction patterns when used to digest the cosmid clones (see references 13 and 26). As expected, digestion of the cosmid clones with PmeI releases the high-molecular-weight MCMV sequences from the 5.4-kb PmeCOS vector (Fig. 3).
FIG. 2.
Genome coordinates of the cosmid set that regenerates MCMV. The gray bars indicate the extent and location of each cosmid insert. The MCMV genome nucleotide coordinates are indicated at the top in kilobases. The alphabetic designations for the larger EcoRI fragments of Smith strain (13, 26) are indicated below the genome coordinates. The vertical lines indicate the location of EcoRI sites.
TABLE 1.
MCMV genome coordinates of infectious cosmid and plasmid inserts
| Insert | MCMV genome coordinatesb (nt) | Vector |
|---|---|---|
| Cosmidsa | ||
| pBH303 | 7178–47328 | |
| pBH400 | 46286–84313 | |
| pBH551 | 83429–119509 | |
| pBH343 | 117180–152764 | |
| pBH325 | 124120–162352 | |
| pBH341 | 158972–204632 | |
| pBH504 | 194718–8740 | |
| Plasmids | ||
| pBH042 | 107769–113962 | pUC18 |
| pBH045 | 113131–118938 | pUC18 |
| pBH057 | 107769–113962 | pUC18 |
| pBH058 | 112106–118938 | pUC18 |
| pBH065 | 83429–105054 | PmeSUB |
| pBH064 | 100543–113131 | PmeSUB |
| pBH067 | 95767–109280 | PmeSUB |
Cosmid clones were maintained in PmeCOS.
MCMV genome coordinates are from the sequence of the Smith strain (31).
FIG. 3.
Restriction digests of the infectious cosmid set. The complete set of infectious cosmids for reconstituting MCMV was digested with either EcoRI or PmeI, fractionated by agarose gel electrophoresis and stained with ethidium bromide. The released MCMV sequences and cosmid vector, PmeCOS, are indicated on the right. The molecular size standards (std) are BstEII-digested λ DNA.
The set of cosmid clones was digested exhaustively with PmeI, mixed in equal proportions, and used to cotransfect mouse fibroblasts. After 5 to 6 days of incubation, the monolayer showed areas of cytopathic effect that were similar in appearance to those in MCMV-infected cells (not shown). This result was achieved using a range of from 1 to 3 μg of each cosmid, with the larger amounts causing more advanced infection at somewhat earlier times after cotransfection (not shown). Three cosmid-derived MCMV lines, cM4, cM5, and cM6 (“cM” derived from cosmid MCMV) were established from cotransfections of 1, 2, or 3 μg of each cosmid, respectively, and these were analyzed as described below.
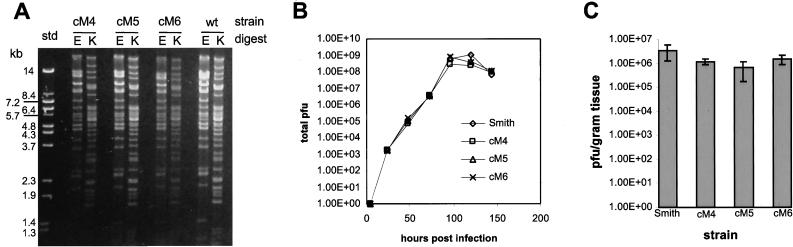
Virus DNA prepared from stocks of Smith and regenerated lines of MCMV and was compared by visual examination of restriction enzyme digests separated on electrophoretic gels (Fig. 4A). The EcoRI and KpnI digests of cosmid-derived virus DNAs were indistinguishable from Smith MCMV DNA, indicating that large genomic rearrangements did not take place when the virus was regenerated. The in vitro growth characteristics of the regenerated lines were also indistinguishable from wild-type MCMV, as evidenced by the equivalence of viral yields at each time postinfection (Fig. 4B). Our long-term interest in developing a recombinant virus generation system was to eventually construct MCMV mutants for pathogenesis studies. Thus, it was necessary to show that cosmid-derived MCMVs demonstrated unaltered growth in vivo. Mice were infected with tissue culture-grown stocks of either Smith strain or cosmid-derived MCMVs, and salivary gland titers were determined at 10 days postinfection (Fig. 4C). The organ titers developed by each recombinant strain resembled those from the wild-type Smith strain, indicating that cosmid-derived MCMVs are unaltered in their ability to replicate in vivo. Thus, comparison of in vitro and in vivo growth indicates that cosmid-derived MCMVs are similar to wild-type MCMV.
FIG. 4.
Analysis of regenerated MCMV. (A) Analysis of viral DNA from cosmid-generated MCMVs. DNA from purified virions of three independent cosmid-generated MCMV strains (cM4, cM5, and cM6) was digested with EcoRI (E) or KpnI (K), separated on a 0.8% agarose electrophoretic gel, and stained with ethidium bromide. The wild-type MCMV DNA (wt) is the Smith strain, and the size standards (std) are λ DNA digested with BstEII. (B) In vitro multistep growth of cosmid-generated MCMV. Three independently generated strains of MCMV, cM4, cM5, and cM6, were used to infect NIH 3T3 cells at an MOI of 0.01. Virus was harvested at the times indicated postinfection, and titers were measured by a standard plaque assay. (C) In vivo growth of cosmid-generated MCMV. BALB/c mice were infected with 5.0 × 105 PFU of the indicated strain of tissue culture-grown MCMV. At 10 days postinfection salivary glands were removed, and the titers for the individual mice were determined. The results are the means of three to five mice, with error bars showing the standard deviation for each group.
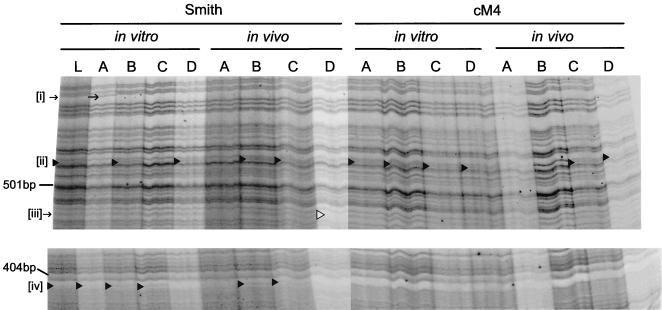
To further analyze regenerated virus, we prepared plaque-purified isolates from one of the regenerated lines, cM4, and compared these to plaque-purified isolates from Smith MCMV. Specifically, 16 isolates were prepared and included 4 isolates from the cM4 in vitro-passaged stock, 4 isolates from cM4 in vivo-passaged (salivary gland) stock, 4 isolates from in vitro-passaged Smith stock, and 4 isolates from in vivo-passaged (salivary gland) Smith stock. Virus DNA from each isolate was purified and digested with HinP1I, and the resulting fragments were radioactively end labeled before separation by gel electrophoresis and autoradiography (Fig. 5). HinP1I has 1,826 recognition sites in the MCMV genome sequence (31), and thus digestion with this restriction enzyme might reveal more-subtle differences between the strains. Our electrophoresis system allowed us to resolve more than 200 distinct fragments for each isolate, and the majority of these were identical between all the isolates. However, there were a small number of differences between the isolates. In one case, an additional fragment at approximately 510 bp (Fig. 5, indicated as “[ii]”) was found in two of the Smith in vitro and in vivo isolates, all four of the cM4 in vitro isolates, and two of the cM4 in vivo isolates. Additionally, one of the Smith in vitro isolates possessed a unique fragment shift (Fig. 5, [i]), whereas another of the Smith in vivo isolates was missing a fragment (Fig. 5, [iii]). Lastly, an additional ∼400-bp fragment (Fig. 5, [iv]) was found in three of the Smith in vitro isolates and two of Smith in vivo isolates but was not found in any of the cM4 in vitro or in vivo isolates. A HinP1I digest of DNA from a non-plaque-purified stock of Smith MCMV was included in this analysis (Fig. 5, lane L). This sample possessed the additional fragments [ii] and [iv], but not the variations [i] or [iii] described above.
FIG. 5.
Analysis of plaque-purified isolates of MCMV. Radioactively labeled HinP1I restriction fragments separated by gel electrophoresis were used to produce the autoradiogram sections illustrated. Plaque-purified isolates, designated A through D at the top of the lanes, were derived from stocks of in vitro- and in vivo-propagated Smith stocks (wild-type) and in vitro- and in vivo-propagated cM4 stocks. DNA from non-plaque-purified Smith stock, designated L, was included for comparison. Only portions of the autoradiogram showing differences between the isolates are shown, with the location of size standards (MspI-digested pUC19 fragments) indicated on the left. Fragment variations, [i] through [iv], are indicated where they occur by symbols (▸, ▹, and →).
One of the cM4 isolates, in vitro isolate B (see Fig. 5) was further examined by DNA sequence analysis through three regions, where its component cosmids overlap. Specifically, the overlap regions between cosmids pBH504 and pBH303 (MCMV nt 6936 to 8827), pBH303 and pBH400 (MCMV nt 467119 to 47529), and pBH400 and pBH551 (MCMV nt 83321 to 84466) were amplified by PCR, cloned, and sequenced. As a control, the same regions of viral DNA from a non-plaque-purified Smith stock were also amplified, cloned, and sequenced. The DNA sequence from each region of the cM4 in vitro isolate B was identical to the Smith MCMV sequence (data not shown and reference 33). This result indicates that regeneration with cosmids did not introduce any of the cosmid vector sequences in the recombinant viral genome. In contrast, one PCR product from Smith MCMV DNA possessed a T-to-C point mutation at MCMV nt 46272. We cannot rule out the possibility that this mutation was an artifact of PCR amplification, but if it exists in the MCMV genome it would result in a Ser-to-Pro mutation in the M25 gene product (31). Most importantly, the HinP1I restriction patterns and DNA sequence analysis of plaque-purified regenerated virus isolates, propagated either in vitro and in vivo, indicates that they do not possess any novel or excessive numbers of mutations when compared to wild-type MCMV.
The development of the infectious cosmids into a system for producing MCMV mutants required an approach to alter specific genes since cosmids are generally too large for routine molecular genetic manipulations. The route taken was to isolate the target gene on a smaller plasmid clone and include this in a series of overlapping subclones to replace one of the infectious cosmids. This approach was taken to demonstrate the generation of two strains of MCMV with mutations in the M78 and M80 ORFs, respectively.
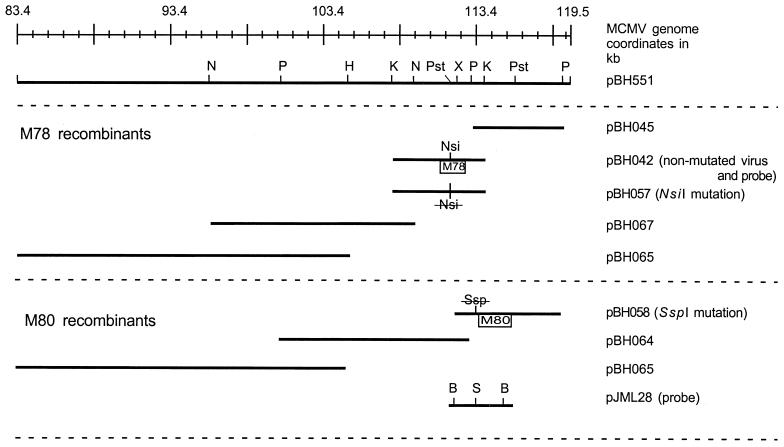
The M78 gene encodes a putative G-protein coupled receptor homologue (31) and is likely involved in pathogenesis (3, 8, 12). Hence, a mutation in this gene was not likely to cause serious impairment of in vitro replication, thus allowing us to use this as an uncomplicated test of our mutagenesis system. The M78 ORF is contained on cosmid pBH551 and was subcloned as a 6.2-kb KpnI fragment in pUC18, yielding pBH042 (Fig. 6). A mutation was introduced into the M78 gene so that it could be identified in the recombinant virus. Specifically, an NsiI restriction site within the translated region was removed yielding pBH057 (Fig. 6). Another subclone, pBH045, consisting of a 5.8-kb PmlI fragment from pBH551 maintained in pUC18, was constructed to overlap with pBH042 and the adjacent cosmid pBH343 (Fig. 6). To facilitate subcloning, the remainder of the cosmid a new vector, PmeSUB, was constructed (Fig. 1B). PmeSUB was derived from pACYC177 by the addition of an MCS surrounded by PmeI sites. The significant features of this vector are that it can maintain large cloned inserts because of its stringent origin of replication (6) and that any cloned MCMV sequence can be released by digestion with PmeI. PmeSUB was used to maintain a 13.4-kb NheI fragment, pBH067, and a 22-kb PmeI-HindIII fragment, pBH065, to complete the overlapping series of subclones from cosmid pBH551 (Fig. 6).
FIG. 6.
MCMV genome coordinates of plasmids replacing cosmid pBH551. Two series of overlapping plasmid clones were constructed to isolate the M78 and M80 ORFs on smaller replicons. Restriction sites: B, BamHI; H, HindIII; K, KpnI; N, NheI; Nsi, NsiI; P, PmlI; Pst, PstI; S and Ssp, SspI; X, XhoI. Nsi (crossed out) in pBH057 and Ssp (crossed out) in pBH058 indicate the location of mutations ablating the respective restriction sites.
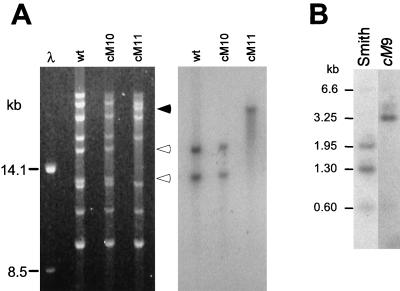
Two recombinant MCMVs were generated by cotransfection of NIH 3T3 cells using cosmids (pBH303, -400, -343, -325, -341, and -504, excluding pBH551) and plasmids (pBH045, -067, and -065) plus either the wild-type M78 gene on pBH042 or the mutated M78 gene on pBH057, yielding strains cM10 and cM11, respectively. Strain cM10 was constructed from nonmutated plasmids and should resemble wild-type MCMV. In contrast, strain cM11 was expected to have a mutation (NsiI site ablation) since it was generated by using the mutated M78 ORF from plasmid pBH057. Both cotransfections resulted in a cytopathic effect in the cells following 5 to 6 days of incubation, and stocks of cM10 and cM11 were established without complementation on NIH 3T3 cells. Viral DNA from cM10 and cM11 and wild-type MCMV was digested with NsiI and separated by gel electrophoresis, followed by blotting to a nitrocellulose membrane for hybridization with a probe for the M78 ORF (Fig. 7A). Ablation of the NsiI site in cM11 DNA yielded a new 28.6-kb NsiI fragment derived from the 15.9- and 12.7-kb fragments found in wild-type and cM10 MCMV DNA (Fig. 7A). A radioactive M78-specific probe, pBH042, hybridized only to the aforementioned DNA fragments, confirming that the NsiI mutation was incorporated into the M78 ORF of cM11 and not cM10 as expected (Fig. 7A). The recovery of these particular NsiI fragments in the respective strains indicates that regeneration of MCMV from a mixture of cosmids and plasmids produced only the desired viruses.
FIG. 7.
Analysis of recombinant MCMV bearing directed mutations. (A) Analysis of strains cM10 and cM11. DNA from purified virions was digested with NsiI and separated by agarose gel electrophoresis (left panel), followed by blotting to a nitrocellulose membrane. The membrane was hybridized to a radioactively labeled DNA probe, pBH042 (right panel). The closed arrowhead indicates the location of the 28.6-kb NsiI fragment unique to cM11, and the open arrowheads indicate the 15.9- and 12.7-kb NsiI fragments unique to wild-type (Smith strain) and cM10. (B) Analysis of strain cM9. Infected cell DNA was digested with BamHI plus SspI and separated by agarose gel electrophoresis, followed by blotting to a nitrocellulose membrane. The membrane was hybridized to a radioactively labeled DNA probe, pJML28 (see Fig. 6).
As a different example, the cosmid pBH551 was replaced with overlapping subclones so that mutations could be introduced into the M80 gene. The M80 gene, which encodes the protease and assembly protein (24, 39), was isolated from cosmid pBH551 on a 6.8-kb fragment subcloned into pUC18, yielding pBH058 (see Materials and Methods). This construct overlaps another plasmid, pBH064, and the cosmid pBH343 (Fig. 6). Additionally, pBH058 carries an ablated SspI site mutation located 57 nt proximal to the initiation codon of M80 and in the adjacent M79 ORF (25, 31). The SspI mutation was introduced to distinguish the regenerated virus, but it did not change the amino acid sequence of the predicted M79 gene product. The remainder of cosmid pBH551 was isolated in PmeSUB as a 22-kb PmeI-HindIII fragment, pBH065, and a 13-kb PmlI fragment, pBH064 (Fig. 6). Cotransfection of the three pBH551 subclones (pBH058, -064, and -065) and the remainder of the cosmids (pBH303, -400, -343, -325, -341, and -504, excluding pBH551) into mouse cells resulted in a cytopathic effect at 5 to 6 days posttransfection. This recombinant MCMV was designated cM9. Hybridization analysis of infected-cell DNA showed that cM9 MCMV DNA lacked the SspI site originally ablated in pBH058 (Fig. 7B). Specifically, the M80 probe, pJML28 (25; Fig. 6) hybridized to 1.95- and 1.3-kb fragments in Smith strain DNA in contrast to a single 3.25-kb fragment in cM9 DNA. The probe did not hybridize to 1.95- and 1.3-kb fragments in cM9 DNA even upon extended exposure, indicating that this was the only product of regeneration (data not shown). The probe also hybridized to short sequences outside the BamHI sites of pJML28, yielding weak signals at 6.6 and 0.7 kb for both Smith and cM9 DNA (Fig. 7B). Overall, the results of manipulating the M78 and M80 ORFs demonstrate that a combination of cosmids and plasmids could be used to generate different desired mutant strains of MCMV.
DISCUSSION
The aim of this work was to develop a facile system for constructing specific mutant strains of MCMV. Our results demonstrate that an appropriately designed set of cosmid and plasmid clones can be used to accomplish this goal. In one example, a recombinant virus, cM11, bearing a restriction site mutation in the M78 ORF was generated using an overlapping set of six cosmids and four plasmid clones. The mutation was introduced into one plasmid, pBH057, which was used to replace the nonmutated plasmid, pBH042, yielding the desired mutant virus. Use of the nonmutated cosmid and plasmid set produced a “wild-type” recombinant, cM10, which served as a control for regeneration efficiency. In a different example, we produced another recombinant virus, cM9, with a restriction site mutation in the M79 ORF using a set of six cosmids and three plasmids. Again, the mutation was engineered into one of the plasmids, pBH058, using standard molecular genetic methods.
These results suggest that it is feasible to generate specific mutant viruses for any gene provided the mutation does not incapacitate basic replication functions. However, it should be possible to generate viruses bearing lethal mutations by cotransfection into appropriately constructed complementing cell lines. We have constructed the mutants described here using a set of seven cosmid clones, but we have approximately 200 other partially characterized cosmid clones available. It is reasonable to assume that different cosmids can be used to generate mutants specific to genes located in the overlap regions of the cosmid set used here. The ability to vary the composition of the cosmids and plasmids immediately suggests that more than one mutation might be simultaneously introduced into a single recombinant virus. MCMVs carrying multiple specific lesions may become useful tools to gain a better understanding of viral replication and pathogenesis.
Cosmid and plasmid combinations have been used to regenerate specific mutants in other herpesviruses, including herpes simplex virus type 1 (11, 32), pseudorabies virus (37), varicella-zoster virus (10), Epstein-Barr virus (36), and HCMV (9, 22). In all cases, the regeneration of virus depended upon using a set of clones covering the entire sequence of the virus with sufficient overlap across adjacent clones to allow homologous recombination of the viral DNA sequences. MCMV regeneration from cosmids further shows that this method does not result in the production of unwanted mutations or rearrangements in the viral genome (see Fig. 5). The development of these systems followed the seminal observation by Graham et al. (17) showing that herpesvirus DNA is infectious. In addition to isolating infectious cosmids for MCMV, we have developed a general system for producing MCMV mutants by including the subcloning vector, PmeSUB. This vector was designed to make sets of overlapping plasmid subclones for the replacement of cosmids. Although most MCMV genes are small enough to be stably maintained in high-copy-number vectors such as pUC18, the subcloning of the remaining cosmid sequences is hindered by their large size. PmeSUB was derived from pACYC177 (6) in order to take advantage of its stringent origin of replication which can maintain large cloned inserts. The MCMV genome was originally restriction mapped using a series of clones maintained in pACYC177 (13, 26), thus proving the value of this replicon.
Most commonly, the construction of mutant MCMVs involved the introduction of viral DNA and a plasmid bearing the target gene plus an extraneous selection and/or reporter gene into permissive cells. Mutants arose by homologous recombination and were isolated from an excess of wild-type virus by multiple cycles of plaque purification. The successes from using this approach are underscored by the many different mutants it has produced (reviewed in reference 28). In some instances, however, the insertion of extraneous selection or reporter genes in the recombinant virus may interfere with the expression of adjacent genes preventing recovery of the desired mutant. The development of the MCMV cosmid and plasmid system was prompted because of this problem. The construction of specific mutants in the M80 gene is encumbered by the arrangement of adjacent genes. Specifically, the initiation codon of the M80 ORF overlaps with the initiation codon of the upstream adjacent M79 ORF, whereas the poly(A) signals of M80 overlap with those of the downstream M82 ORF (31). Although we placed a β-galactosidase expression cassette between duplicated poly(A) signals for M80 and M82, we were still unable to recover a legitimate recombinant virus (unpublished observations).
The construction of mutant herpesviruses has also been facilitated by the isolation of the respective genomes as infectious BACs (2, 4, 27, 35). A limitation of the first-described BAC MCMVs was their inability to replicate in vivo, but a later version deletes incorporated BAC sequences upon repeated passage in vitro, yielding virus that replicates normally in vivo (38). The BAC CMVs promise to be a great utility in the identification of genes necessary for virus replication (for example, see reference 5). However, construction of specific mutants requires the manipulation of E. coli recombination genetics to recover the desired product of allelic exchange between the wild-type BAC and a plasmid bearing the mutant gene of interest (30). Although elegant in design, manipulation of BACs is not readily mastered in most virology laboratories.
In comparison to other methods of constructing specific MCMV mutants, the cosmid and plasmid system has the major advantage that only the desired recombinant virus is produced. This allows the rapid isolation of different mutant strains using standard molecular techniques once a target gene is isolated and appropriately configured in a cosmid and plasmid set. This system is designed to complement existing methods of mutant virus construction and will be of use in ongoing studies of MCMV replication and pathogenesis.
ACKNOWLEDGMENTS
We are grateful to our colleagues Greg Chinchar, Carol Wu, Paul Hippenmeyer, and Ed Mocarski for critical reading of the manuscript.
This work was supported by Monsanto grant MON-99-018, University of Missouri Research Board grant RB-98-144, and the University of Missouri College of Veterinary Medicine, Committee on Research grant COR FY 98/99.
REFERENCES
- 1.Alting-Mees M A, Short J M. pBluescript II: gene mapping vectors. Nucleic Acids Res. 1989;17:9494. doi: 10.1093/nar/17.22.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angulo A, Messerle M, Koszinowski U H, Ghazal P. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J Virol. 1998;72:8502–8509. doi: 10.1128/jvi.72.11.8502-8509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billstrom M A, Johnson G L, Avdi N J, Worthen G S. Intracellular signaling by the chemokine receptor US28 during human cytomegalovirus infection. J Virol. 1998;72:5535–5544. doi: 10.1128/jvi.72.7.5535-5544.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst E M, Hahn G, Koszinowski U H, Messerle M. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J Virol. 1999;73:8320–8329. doi: 10.1128/jvi.73.10.8320-8329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brune W, Menard C, Hobom U, Odenbreit S, Messerle M, Koszinowski U H. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nat Biotechnol. 1999;17:360–364. doi: 10.1038/7914. [DOI] [PubMed] [Google Scholar]
- 6.Chang A C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchinson III C A, Kouzarides T, Matignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrel B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 8.Chee M S, Satchwell S C, Preddie E, Weston K M, Barrell B G. Human cytomegalovirus encodes three G protein-coupled receptor homologues. Nature. 1990;344:774–777. doi: 10.1038/344774a0. [DOI] [PubMed] [Google Scholar]
- 9.Cihlar T, Fuller M D, Cherrington J M. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J Virol. 1998;72:5927–5936. doi: 10.1128/jvi.72.7.5927-5936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen J I, Seidel K E. Generation of varicella-zoster virus (VZV) and viral mutants from cosmid DNAs: VZV thymidylate synthetase is not essential for replication in vitro. Proc Natl Acad Sci USA. 1993;90:7376–7380. doi: 10.1073/pnas.90.15.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham C, Davison A J. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology. 1993;197:116–124. doi: 10.1006/viro.1993.1572. [DOI] [PubMed] [Google Scholar]
- 12.Davis-Poynter N J, Lynch D M, Vally H, Shellam G R, Rawlinson W D, Barrell B G, Farrell H E. Identification and characterization of a G protein-coupled receptor homolog encoded by murine cytomegalovirus. J Virol. 1997;71:1521–1529. doi: 10.1128/jvi.71.2.1521-1529.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebeling A, Keil G M, Knust E, Koszinowski U H. Molecular cloning and physical mapping of murine cytomegalovirus DNA. J Virol. 1983;47:421–433. doi: 10.1128/jvi.47.3.421-433.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 15.Frost E, Williams J. Mapping temperature-sensitive and host-range mutations of adenovirus type 5 by marker rescue. Virology. 1978;91:39–50. doi: 10.1016/0042-6822(78)90353-7. [DOI] [PubMed] [Google Scholar]
- 16.Graham F L, Van der Eb A J. Transformation of rat cells by DNA of human adenovirus 5. Virology. 1973;54:536–539. doi: 10.1016/0042-6822(73)90163-3. [DOI] [PubMed] [Google Scholar]
- 17.Graham F L, Veldhuisen G, Wilkie N M. Infectious herpesvirus DNA. Nat New Biol. 1973;245:265–266. doi: 10.1038/newbio245265a0. [DOI] [PubMed] [Google Scholar]
- 18.Ho M. Cytomegalovirus: biology and infection. New York, N.Y: Plenum Medical Book Company; 1991. [Google Scholar]
- 19.Ish-Horowicz D, Burke J F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981;9:2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacob R J, Morse L S, Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J Virol. 1979;29:448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacob R J, Roizman B. Anatomy of herpes simplex virus DNA VIII. Properties of the replicating DNA. J Virol. 1977;23:394–411. doi: 10.1128/jvi.23.2.394-411.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kemble G, Duke G, Winter R, Spaete R. Defined large-scale alterations of the human cytomegalovirus genome constructed by cotransfection of overlapping cosmids. J Virol. 1996;70:2044–2048. doi: 10.1128/jvi.70.3.2044-2048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laithier M, Sheldrick P. Influence of DEAE-dextran treatment and cellular “age” on infection by herpes simplex virus DNA. In: Buccalossi P, Veronesi U, Coscinelli N, editors. Chemical and viral carcinogenesis 2. Amsterdam, The Netherlands: IARC Scientific Publications; 1975. pp. 85–90. [PubMed] [Google Scholar]
- 24.Liu F Y, Roizman B. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J Virol. 1991;65:5149–5156. doi: 10.1128/jvi.65.10.5149-5156.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loutsch J M, Galvin N J, Bryant M L, Holwerda B C. Cloning and sequence analysis of murine cytomegalovirus protease and capsid assembly protein genes. Biochem Biophys Res Commun. 1994;203:472–478. doi: 10.1006/bbrc.1994.2206. [DOI] [PubMed] [Google Scholar]
- 26.Mercer J A, Marks J R, Spector D H. Molecular cloning and restriction endonuclease mapping of the murine cytomegalovirus genome (Smith strain) Virology. 1983;129:94–106. doi: 10.1016/0042-6822(83)90398-7. [DOI] [PubMed] [Google Scholar]
- 27.Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski U H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mocarski E S, Jr, Kemble G W. Recombinant cytomegaloviruses for study of replication and pathogenesis. Intervirology. 1996;39:320–30. doi: 10.1159/000150503. [DOI] [PubMed] [Google Scholar]
- 29.Mocarski E S, Post L E, Roizman B. Molecular engineering of the herpes simplex virus genome: insertion of a second L-S junction into the genome causes additional genome inversions. Cell. 1980;22:243–55. doi: 10.1016/0092-8674(80)90172-5. [DOI] [PubMed] [Google Scholar]
- 30.O'Connor M, Peifer M, Bender W. Construction of large DNA segments in Escherichia coli. Science. 1989;244:1307–1312. doi: 10.1126/science.2660262. [DOI] [PubMed] [Google Scholar]
- 31.Rawlinson W D, Farrell H E, Barrell B G. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Register R B, Shafer J A. A facile system for construction of HSV-1 variants: site-directed mutation of the UL26 protease gene in HSV-1. J Virol Methods. 1996;57:181–193. doi: 10.1016/0166-0934(95)01984-7. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 34.Short J M, Fernandez J M, Sorge J A, Huse W D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988;16:7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith G A, Enquist L W. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J Virol. 1999;73:6405–6414. doi: 10.1128/jvi.73.8.6405-6414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomkinson B, Robertson E, Yalamanchili R, Longnecker R, Kieff E. Epstein-Barr virus recombinants from overlapping cosmid fragments. J Virol. 1993;67:7298–7306. doi: 10.1128/jvi.67.12.7298-7306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Zijl M, Quint W, Briaire J, de Rover T, Gielkens A, Berns A. Regeneration of herpesviruses from molecularly cloned subgenomic fragments. J Virol. 1988;62:2191–2195. doi: 10.1128/jvi.62.6.2191-2195.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wagner M, Jonjic S, Koszinowski U H, Messerle M. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J Virol. 1999;73:7056–7060. doi: 10.1128/jvi.73.8.7056-7060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welch A R, Woods A S, McNally L M, Cotter R J, Gibson W. A herpesvirus maturational proteinase, assemblin: identification of its gene, putative active site domain, and cleavage site. Proc Natl Acad Sci USA. 1991;88:10792–10796. doi: 10.1073/pnas.88.23.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 41.Zoller M J, Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]
- 42.Zoller M J, Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982;10:6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]