Abstract
Residual antibiotics may affect human health by increasing challenges related to infection treatment due to antibiotic resistance development. Hence, determining whether residual antibiotics in the body can lead to antibiotic resistance is important. We developed a model to predict possible antibiotic resistance caused by residual antibiotics by simulating human digestion in vitro. Increased antibiotic resistance was found to be dependent on the digestion process. Ethical prediction of antibiotic resistance using fewer animals and no humans was possible by simulating the internal environment. Thus, preliminary studies to monitor antibiotic resistance that can affect human health may be safely conducted using this model.
Keywords: In vitro human digestion, Gut microbiota, Antibiotic resistance
1. Introduction
Antibiotics are widely used to prevent infection and treat infectious diseases in livestock and agriculture to maintain and improve the quality of industrially important animals and plants [[1], [2], [3]]. Residual antibiotics in food are gradually attracting interest worldwide [4,5]. Antibiotic overuse and misuse during the treatment of farm animals for bacterial diseases can lead to an increased risk of antibiotic resistance in humans due to the accumulation of residual antibiotics from animal products.
Antibiotic resistance that occurs when antibiotics are abused in livestock and crops may be transferred to humans by exposure to contaminated animal products [6]. Therefore, the defined daily dose of drugs is strictly restricted to limit human exposure to residual antibiotics from animal products in many countries [7,8]. Antibiotics are used during meat production for disease prevention and growth promotion [9,10]. More than 90% of all antibiotics used in the United States are administered to cattle, turkey, and pigs [11]. Additionally, a recent study showed that the use of these preventive antibiotics in livestock is increasing [[12], [13], [14]]. The consistent use of antibiotics could contribute to the accumulation of residual antibiotics in the muscles and organs of animals. Previous studies demonstrated that residual antibiotics in animal products can inhibit normal sausage fermentation and raise infection risk in humans by increasing the proliferation rate of pathogenic bacteria, such as Escherichia coli O157:H7 and Salmonella [15,16]. Furthermore, continuous consumption of animal products containing residual antibiotics may make disease treatment challenging due to the development of antibiotic resistance.
In vitro human digestion models that are good alternatives for human models are ethical and economical [17,18]. Moreover, various regulations have been implemented to safeguard the welfare and rights of laboratory animals [19]. Digestion models can be used in preliminary studies for rapid screening of the bioavailability of certain compounds and provide substantial information while preventing the overuse of experimental animals. Considering the ethical difficulties related to the assessment of antibiotic resistance in the human body, a research model that simulates the human body's environment is required. However, an in vitro human digestion model to analyze antibiotic resistance and thereby, confirm the effect of residual antibiotics in the human body has not yet been developed. Therefore, this study was aimed to develop such a model to identify the role of residual antibiotics in the development of antibiotic resistance in the human body due to the consumption of contaminated food.
2. Materials and methods
2.1. Chemicals
Digestive enzymes for the in vitro human digestion model were prepared using various reagents. Sodium chloride, uric acid, urea, hydrochloric acid (HCl), calcium chloride dihydrate, α-amylase, mucin, bovine serum albumin, pepsin, potassium chloride (KCl), potassium hydroxide (KOH), calcium chloride dihydrate (CaCl2·2H2O), pancreatin, lipase, sodium bicarbonate, and bile extract porcine were purchased from Sigma-Aldrich (St. Louis, MO, USA). Buffer solution (pH 7.0 ± 0.02) was purchased from Samchun Pure Chemicals, Co., Ltd., Korea. For cultivation of gut microbiota used in large intestine digestion, Luria-Bertani (LB) agar, lactobacilli DeMan, Rogosa, and Sharpe (MRS) broth, and lactobacilli MRS agar were purchased from Difco Laboratories (Difco, Sparks, MD, USA). Antibiotics, including tetracycline, ofloxacin, and penicillin, were purchased from Sigma-Aldrich (St. Louis, MO, USA). Tryptic soy broth (TSB), required for the cultivation of bacterial strains, was purchased from Difco Laboratories (Difco, Sparks, MD, USA).
2.2. Preparation of digestive enzymes and gut microbiota for digestion simulation
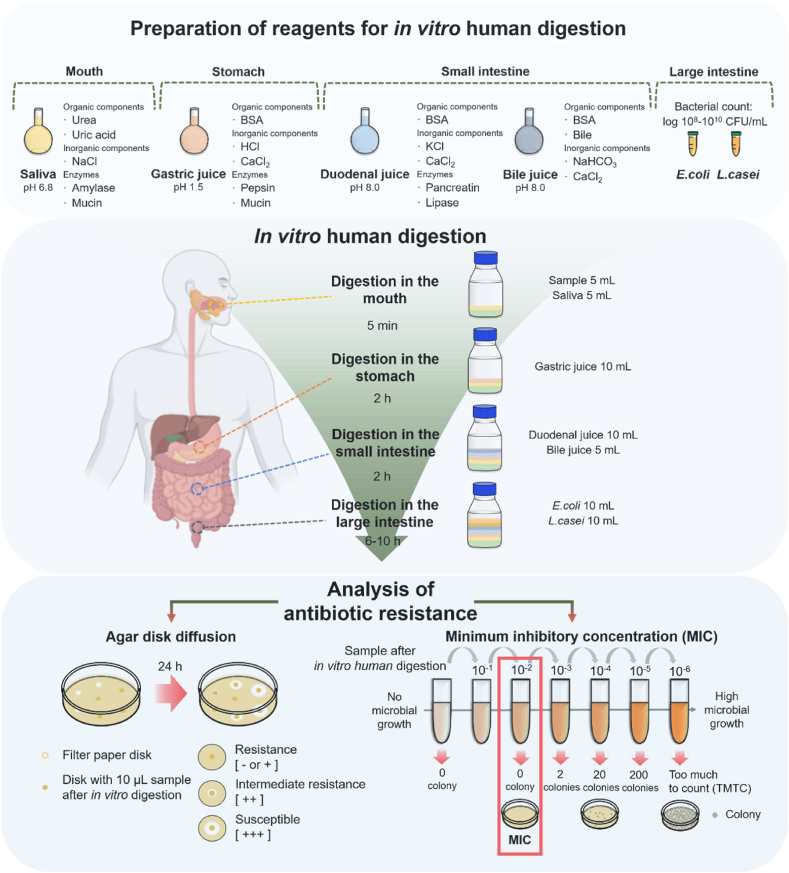
The study protocol comprising three steps is schematically presented in Fig. 1. The first step involves preparation of the digestive enzymes (saliva, gastric juice, duodenal juice, and bile juice) (Table 1) and gut microbiota; the second step, establishment of the in vitro model that simulates the human digestive tract (mouth, stomach, small intestine, and large intestine), and the third step, antibiotic resistance assessment using disk diffusion and minimum inhibitory concentration (MIC) methods.
Fig. 1.
Overview of the method employed to assess antibiotic resistance using the in vitro human digestion model. NaCl, sodium chloride; BSA, bovine serum albumin; HCl, hydrochloric acid; CaCl2, calcium chloride; KCl, potassium chloride; NaHCO3, sodium bicarbonate, E. coli: Escherichia coli; L. casei: Lactobacillus casei.
Table 1.
Constituents and concentration of digestive juices used in the in vitro human digestion model representing fed conditions.
| Saliva (digestion in mouth) | Gastric juice (digestion in stomach) | Duodenal juice (digestion in small intestine) | Bile juice (digestion in small intestine) | |
|---|---|---|---|---|
| Organic and inorganic components | 1.7 mL NaCl (3 M) | 6.5 mL HCl (1 M, 37%) | 6.3 mL KCl (1.2 M) | 68.3 mL NaHCO3 (1 M) |
| 8 mL urea (0.4 M) | 18 mL CaCl2.2H2O (0.15 M) | 9 mL CaCl2·2H2O (0.15 M) | 10 mL CaCl2·2H2O (0.15 M) | |
| 15 mg uric acid | 1 g bovine serum albumin | 1 g bovine serum albumin | 1.8 g bovine serum albumin 30 g bile acid | |
| Enzymes | 290 mg α-amylase (50 U/mg) | 2.5 g pepsin (250 U/mg) | 9 g pancreatin (8 × USP/g) | |
| 25 mg mucin | 3 g mucin | 1.5 g lipase (30–90 U/mg) | ||
| pH | 6.8 ± 0.2 | 1.50 ± 0.02 | 8.0 ± 0.2 | 7.0 ± 0.2 |
1) Values indicate concentrations used to make digestive juices.
2) Values in parentheses are the concentrations of inorganic or organic components per/1 L distilled water.
After mixing all ingredients (inorganic components, organic components, and enzymes), the volume was increased to 500 mL with distilled water. If necessary, the pH of the digestive juice was adjusted to the appropriate value.
NaCl, sodium chloride; HCl, hydrochloric acid; CaCl2·2H2O, calcium chloride dihydrate; KCl, potassium chloride; NaHCO3, sodium bicarbonate.
2.2.1. Digestive enzymes and inorganic and organic solutions
Digestive enzymes and other solutions were prepared according to previously reported methods [18,[20], [21], [22]]. The compositions of prepared saliva and gastric, duodenal, and bile juice are listed in Table 1.
2.2.2. Preparation of saliva for simulation of digestion in the mouth
-
1.
The organic components, namely 8 mL 400 mM urea and 0.015 g uric acid were first mixed; subsequently, the inorganic component, i.e., 1.7 mL 3 M sodium chloride was added.
-
2.
The enzymes were prepared by adding 0.29 g α-amylase (50 U/mg) and 0.025 g mucin to 800 mL distilled water (DW).
-
3.
The solution prepared in the first step was mixed with the enzyme solution, and pH was adjusted to 6.8 ± 0.2 with 1 M KOH.
-
4.
DW was added to the volumetric flask to obtain the required volume.
2.2.3. Preparation of gastric juice for simulation of digestion in the stomach
-
1.
The inorganic components, namely 6.5 mL 1 M HCl (purity 37%) and 18 mL 0.15 M CaCl2·2H2O were mixed with the organic component, i.e., 1 g bovine serum albumin.
-
2.
Next, 2.5 g pepsin (250 U/mg) and 3 g mucin was added to 0.8 L DW.
-
3.
The solution prepared in step 1 was mixed with the enzyme solution, and pH was adjusted to 1.50 ± 0.02 using 1 M HCl.
-
4.
DW was added to the volumetric flask until the required volume was obtained.
2.2.4. Preparation of duodenal juice for simulation of digestion in the small intestine
-
1.
The inorganic components, namely 6.3 mL 1.2 M KCl and 9 mL 0.15 M CaCl2.2H2O were mixed with the organic component, i.e., 1 g bovine serum albumin.
-
2.
Next, 9 g pancreatin and 1.5 g lipase (90 U/mg) were added to 800 mL DW.
-
3.
The solution prepared in step 1 was mixed with the enzyme solution, and pH was adjusted to 8.0 ± 0.2 using 1 M KOH.
-
4.
DW was added to the volumetric flask to obtain the required volume.
2.2.5. Preparation of bile juice for simulation of digestion in the small intestine
-
1.
The inorganic components, namely 68.3 mL 1 M sodium bicarbonate and 10 mL 0.15 M CaCl2·2H2O were mixed with the organic components, i.e., 1.8 g bovine serum albumin and 30 g bile extract porcine.
-
2.
The pH was adjusted to 8.0 ± 0.2 using 1 M KOH.
-
3.
DW was added to the volumetric flask until the required volume was obtained.
2.2.6. Preparation of gut microbiota for simulation of digestion in the large intestine
In a previous study regarding representative gut microbiota in the Korean population, it was identified that the dominant gut microbes contributing to digestion in the large intestine are E. coli and Lactobacillus casei [23]. Therefore, these organisms were used for the in vitro study of digestion in the large intestine.
-
1.Preparation of media for cultivation of E. coli to be used in the large intestinal digestion model:
-
1.1First, 250 g LB broth was added to 1 L deionized DW (DDW) in a sterile flask, and the mixture was stirred for 20 min at 180 rpm.
-
1.2Next, 40 g LB agar was added to 1 L DDW in a sterile flask, and the mixture was stirred for 20 min at 180 rpm.
-
1.1
-
2.Preparation of media for cultivation of L. casei (ATCC 393) used in large intestine digestion assays:
-
2.1First, 55 g lactobacilli MRS broth was mixed with 1 L DDW in a sterile flask, and the mixture was stirred for 20 min at 180 rpm.
-
2.2Next, 70 g lactobacilli MRS agar was mixed with 1 L DDW in a sterile flask, and the mixture was stirred for 20 min at 180 rpm.
-
2.1
-
3.
The mixture was dissolved completely. The prepared broth and agar were sterilized using an autoclave for 15 min at 121 °C.
-
4.
The sterilized broth was cooled to <40 °C and transferred to a new 15- or 50-mL tube. The sterilized agar was cooled to 50 ± 2 °C, and 20–25 mL was transferred to a 100 mm petri dish.
-
5.
The petri dish was left on a flat surface for a minimum of 30–40 min until completely dry.
-
6.
The broth and agar were maintained at 2–8 °C and stored in sealed containers to avoid moisture loss.
-
7.
The stock gut microbes (E. coli and L. casei) stored at −80 °C were thawed to ambient temperature, and 1% of each was inoculated into the appropriate broth.
-
8.
The inoculated micro-organisms were incubated at 37 °C for 12–24 h in a shaking incubator depending on the appropriate growth condition for each organism.
-
9.
Next, 0.1 mL of the activated gut micro-organisms was transferred to agar dishes and spread using a spreader.
-
10
The inoculated dishes were incubated at 37 °C for 12–24 h in an incubator. The total number of colonies was determined, and single colonies were collected and inoculated into the appropriate broth at a rate of approximately 108–1010 CFU/mL.
-
11
The broth seeded with the gut microbiota (E. coli and L. casei) was incubated at 37 °C for 12 h. This incubation was repeated twice to simulate the conditions for activation. The activated gut microbiota with a cell density of 108–1010 CFU/mL was used as the reagent supporting digestion in the large intestine.
2.3. Simulation of human digestion using in vitro model comprising mouth, stomach, small intestine, and large intestine
2.3.1. Conditions for in vitro human digestion
The in vitro human digestion models simulating the human gastrointestinal tract, including the mouth, stomach, small intestine, and large intestine, were developed based on digestion temperatures, digestion times, digestive enzymes, and gut microbiota [20,23] in our previous studies, which aimed to determine the changes in structure, digestibility, and biological activity of pharmaceuticals, foods, and compounds [20,[24], [25], [26], [27], [28], [29]].
In our previous review [18,20], the study was determined that the optimal temperature for the in vitro human digestion model is 37 ± 0.5 °C. Based on previous physiological studies, transit times were established [12,20] and divided into four groups according to digestion location as follows: 1) mouth, 5 min; 2) stomach, 2 h; 3) small intestine, 2 h; and 4) large intestine, 6–10 h. According to previous physiological measurements, the in vitro human digestion model was maintained at 150 rpm using a shaking water bath to mimic gastric intestinal motility and facilitate digestion [20]. The pH of the digestive environment aids enzyme activity and influences the digestibility and absorption and/or the degradation of nutrients in the human body [30,31]. Therefore, pH levels in the in vitro human digestion model must be established according to physiological conditions [20,32]. In our model, the following pH levels were maintained: 1) mouth, pH 6.8 ± 0.2; 2) stomach, pH 1.5 ± 0.2; 3) small intestine, pH 8.0 ± 0.2; and 4) large intestine, pH 7.0 ± 0.2.
2.3.2. Preparations before beginning in vitro human digestion
-
1.
The digestive enzymes were prepared and stored at −80 °C (Table 1).
-
2.
The gut microbes were prepared and incubated at 37 °C.
-
3.
The temperature of the shaking water bath was set to 37 °C.
-
4.
The frozen digestive enzymes were thawed in a water bath at 37 °C.
-
5.
The required quantities of samples and digestive enzymes were calculated and weighed in advance.
2.3.3. In vitro human digestion
-
1.
Samples: First, 5 g of each sample was weighed. If the sample amount was low, the quantities of all reagents were reduced proportionally.
-
2.
Simulation of mouth digestion: First, 5 g sample was transferred to a 50-mL screw-capped tube containing 5 mL prepared saliva solution (pH 6.8), and the mixture was stirred at 150 rpm for 5 min at 37 °C in a shaking water bath.
-
3.
Simulation of stomach digestion: First, 10 mL prepared gastric juice (pH 1.5) was transferred to a 50-mL screw-capped tube, and the mixture was stirred at 150 rpm for 5 min at 37 °C in a shaking water bath.
-
4.
The dialysis tube (molecular weight cutoff, 50 kDa; flat width, 34 mm; thickness, 18 μm [Membrane Filtration Products, Inc., Seguin, TX, USA]) was prepared by cutting to proper size (2.5 × 10 cm/5 g sample) and soaking in DW until soft. The excess water was removed, and the softened dialysis tube was tied at one end. Next, 5 mL phosphate buffer (pH 7) was added to the prepared dialysis tube, and the other end was tied. It was then transferred to the 50-mL screw-capped tube to simulate digestion in the small intestine. The tube contents were stirred in a shaking water bath at 150 rpm for 2 h at 37 °C.
-
5
Simulation of small intestine digestion: First, 10 mL prepared duodenal juice and 5 mL bile juice were added to the 50-mL screw-capped tube which contained the mixture after mouth and stomach digestion. The mixture was stirred in a shaking water bath at 150 rpm for 2 h at 37 °C.
-
6
Simulation of large intestine digestion: First, 10 mL of each of the prepared gut microbes (E. coli and L. casei) was inoculated into the mixture obtained by performing the steps described above. The samples were then incubated for 4–6 h in a shaking water bath maintained at 37 °C and 150 rpm (Fig. 1, Fig. 2).
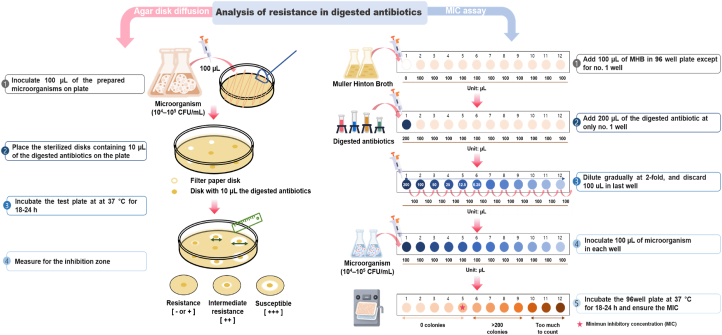
Fig. 2.
Procedure for analyzing resistance to digested antibiotics. MHB, Muller–Hinton broth; MIC, minimum inhibitory concentration.
The products obtained at each digestion step were used to determine antibiotic resistance in the human body.
2.4. Microbial strains for analysis
The following microbial strains were used: 1) SA RN4220, a fluoroquinolone-susceptible restriction-deficient mutant of Staphylococcus aureus, and 2) sequence type 72 (ST-72)-staphylococcal cassette chromosome mec (SSCmec) type IV methicillin-resistant S. aureus (MRSA) that was isolated from human blood. The microbial strains used in this study were provided by the Microbiology Laboratory of Animal Science and Technology of Chung-Ang University (Anseong, South Korea). The two microbial strains were cultured in TSB to obtain 106–107 CFU/mL according to a previous method [28].
2.5. Antibiotic resistance analysis
This study was analyzed resistance to residual antibiotics in the human body using the in vitro human digestion model. The following antibiotics were used: tetracycline, ofloxacin, and penicillin. Tetracycline and penicillin were dissolved in DDW and ofloxacin in a specific buffer (pH 3, w/v). Specific procedures for assessing antibiotic resistance, disk diffusion, and MIC were employed (Fig. 2). The large intestine digestion step was excluded from MIC assaying to eliminate the impact of gut microbiota. Antibiotic susceptibility or resistance testing was performed in accordance with the Clinical and Laboratory Standard Institute (CLSI) guidelines. The troubleshooting guide is presented in Table 2.
Table 2.
Troubleshooting guide for the method developed to assess antibiotic resistance using the in vitro human digestion model.
| Procedure | Problem | Cause | Solution |
|---|---|---|---|
| Preparation of gut microbiota for digestion | Incorrect gut microbial population | Insufficient gut microbial population (108–1010 CFU/mL) for use in the in vitro human digestion model | Adjust the experiment timing. If the gut microbial population does not reach the required count, cultivate it for few more hours |
| Enzyme activity (amylase, pepsin, lipase, trypsin) | Assay not working | · Data read at wrong wavelength | · Check the wavelength in the data and filter settings of the microplate reader |
| · Using for different type plate | · In case of colorimetric method, use clear 96-well plates. (Fluorescence, black plates with clear bottoms; luminescence, white plates) | ||
| Irregular values in samples | · Samples made in an incorrect buffer | · Use assay buffer in the specific kit or refer to instructions | |
| · Poorly dissolved sample and reagents | · Dissolve all samples and reagents completely and mix gently before testing | ||
| Assay failure during measurement of linear pattern for standard curve | · Incompletely dissolved standards | · Completely mix all components before making standard stock | |
| · Incorrect pipetting of standard or reagents | · Avoid pipetting small amounts to prevent bubble generation | ||
| · Possible measurement of well plate in the presence of bubbles | · Inject all mixtures gently against the wall of the well plates | ||
| · Calculation errors | · Check calculations and values by referring to the data sheet | ||
| In vitro human digestion steps | Preparation of samples for analyzing changes at each digestion step | To analyze changes (quantitative or structural changes) in the samples at each digestion step (mouth, stomach, small intestine, and large intestine), the number of samples prepared must be more than the number of treatments | For analysis at each digestion step, the sample must be collected and analyzed at the end of each step (mouth, stomach, small intestine, and large intestine); thus, the number of samples prepared should be four times greater than the number of treatment groups |
| E.g., a total of 36 samples should be prepared for 3 treatments × 3 replication experiments (3 treatments × 3 replications × 4 digestion steps) | |||
| Short digestion time in mouth | Short digestion time (<5 min) in the mouth may be a result of considerable time spent adding the digestive enzyme | Do not experiment with too many samples at the same time; add the samples first, followed by enzymes randomly and quickly | |
| Difficulty in handling dialysis tube | It is not easy to tie the dialysis tube, and there may be many digested samples (including digestive juices) on the outer surface of the tube after the step involving digestion in the small intestine | The dialysis tube should be tied with a clip (not straps). After digestion in the small intestine, it should be cleaned with a phosphate buffer solution (pH 7) and then used for the experiments | |
| Effects of dilution | Increasing the sample volume by adding digestive juices | Before analysis, use a buffer that does not influence the data to adjust sample volume | |
| Agar disk diffusion | Difficulty in measuring inhibition zone | · Overlap with inhibition zone disks | · Use no more than 5 disks on a 100-mm agar plate or no more than 12 disks on a 150-mm agar plate |
| · No inhibition zone possessing a fully round shape | · Do not place disks on the edge of the agar plate | ||
| · Irregular inhibition zone shape | · Each disk must be pushed down using forceps to confirm complete contact with the agar surface | ||
| Disappearance of microorganism | · No growth of the test microorganism | · Check the optimal incubation conditions (e.g., temperature, time) and dilute appropriately to the optimal microorganism concentration | |
| MIC assay | Failure in determining MIC | Simply ensuring that the appropriate antibiotic has been selected | Ensure that the appropriate dose according to sensitivity testing is used |
| Inappropriate MIC | MICs are lower or higher than expected | Repeat testing using McFarland 0.5 turbidity or values at 600 nm. Check inoculation procedure and inoculum preparation steps |
MIC, minimum inhibitory concentration.
2.5.1. Agar disk diffusion
The disk diffusion assay is widely used to test antimicrobial activity according to the size of microbial growth inhibition zone on media containing antibiotics. Disks containing antibiotics are placed in a dish containing microorganism cultures, and sizes of growth inhibition zones are measured. Results are presented in terms of full susceptibility, intermediate susceptibility, and resistance based on CLSI guidelines.
-
1.Preparation of media for determination of antibiotic resistance:
-
1.1.First, 38 g Muller–Hinton agar (MHA) was mixed with 1 L DDW in a sterile flask; the mixture was stirred for 20 min at 180 rpm.
-
1.2.Next, 21 g Muller–Hinton broth (MHB) was mixed with 1 L DDW in a sterile flask; the mixture was stirred for 20 min at 180 rpm.
-
1.3.The mixture was completely dissolved, and the prepared agar was sterilized in an autoclave for 15 min at 121 °C.
-
1.4.The sterilized broth was cooled to <40 °C and transferred to a new 15- or 50-mL tube, which was then cooled to 50 ± 2 °C. Subsequently, 20–25 mL aliquots were transferred to 100-mm petri dishes.
-
1.5.Before use, the agar plates were dried for no more than 30 min or until the excess surface moisture evaporated.
-
1.6.The prepared agar plates were stored using cover wraps and plastic bags at 2–8 °C for up to 1 month.
-
1.1.
-
2.Inoculation of micro-organisms for agar disk diffusion:
-
2.1.In the direct colony suspension method, four or five colonies were sampled using a loop.
-
2.2.The colonies were suspended in 5 mL MHB and incubated at 37 °C until turbidity equivalent to that of 0.5 McFarland standard or absorbance of 0.07–0.1 (approximately, 107–108 CFU/mL) at 600 nm was reached. The cells were then diluted to 104–105 CFU/mL using MHB.
-
2.1.
-
3.
Subsequently, 100 μL test microorganism was added to an MHA plate and spread using a spreader.
-
4.
The plate was rotated by 60°, and the procedure was repeated twice. Inoculation distribution was observed, and the medium surface was allowed to dry for 3–5 min.
-
5.
The sterilized disks containing 10 μL digested antibiotics were placed on the plate surface within 15 min.
-
6.
The test plate was incubated at 37 °C or the appropriate optimum growth temperature.
-
7.
The inhibition zone was measured using a ruler graduated to 0.5 mm after 16–18 h.
2.5.2. MIC assay
The MIC assay is a type of dilution method that is widely used to determine the lowest concentration (mg/mL or μg/mL) of antimicrobial agent that inhibits microorganism growth by measuring broth turbidity [33,34]. The MIC assay steps described below adheres to guidelines of the CLSI and European Committee on Antimicrobial Susceptibility Testing (EUCAST).
-
1.
First, 100 μL MHB was added into all wells except the first one of a 96-well plate. An equal amount of digested antibiotics was added only to the first well.
-
2.
Dilution was initiated in the first well, and after dilution until the twelfth one, 100 μL solution was discarded from the last well so that each well contained equal amount of solution. Next, MHB and antibiotic or digested antibiotic solutions were mixed by pipetting.
-
3.
Subsequently, 100 μL microbial cultures (approximately 104–105 CFU/mL) were inoculated into each well.
-
4.
The 96-well plate was incubated at 37 °C or at temperatures corresponding to the optimum growth temperature of each organism.
-
5.
After incubation, the growth end point (time at which no bacterial growth was detected) was determined.
2.6. Statistical analysis
Five replicates of the experiments were performed. The results were expressed as mean ± standard deviation (SD). All statistical analyses were conducted by performing one-way analysis of variance using IBM SPSS Statistics for Windows, version 26.0 (IBM Corp., Armonk, N.Y., USA). Tukey's multiple comparison test was used to determine significant differences between mean values; evaluations were based on a significance level of p < 0.05.
3. Results and discussion
This study was performed to ethically determine and assess resistance to residual antibiotics in the human body using an in vitro human digestion model. Antibiotic resistance was assessed using assays that confirmed variation in resistance to different antibiotics. Our previous studies examined changes in antimicrobial activity and resistance to antibiotics that were digested during in vitro human digestion using the disk diffusion method [28,29]. In addition, health risks due to antibiotic-resistant bacteria and antibiotic resistance genes in humans [35] as well as the presence of antibiotic-resistant enterococci in seafood [36] were determined using an in vitro digestion model. The MIC method enables rapid screening of microbial resistance to antibiotics [[37], [38], [39], [40]] and pharmaceuticals [[41], [42], [43], [44]]. As opposed to in vivo studies involving human trials and animal models, in vitro human digestion models provide rapid results and are inexpensive, resource-efficient, and ethical [45].
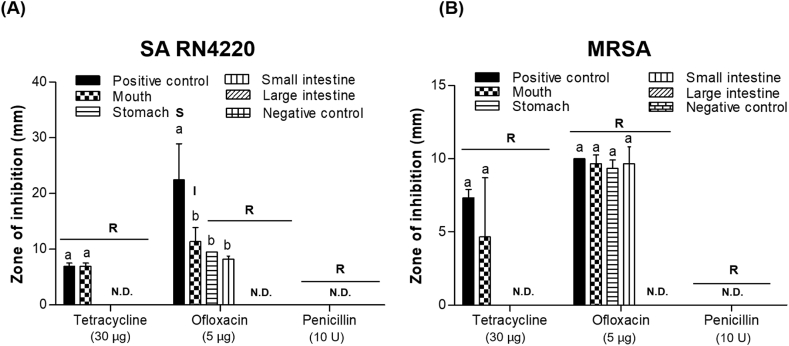
Agar disk diffusion experiments revealed differences in resistance that were dependent on the antibiotic used and specific digestion steps (Fig. 3). The positive control was to be the antibiotics before digestion, and the negative control was to be the dissolved buffer such as DDW and a specific buffer (pH 3). All negative control did not show inhibition zones regardless of microbial strains. Before digestion, treatment with tetracycline and ofloxacin resulted in inhibition zones indicative of antimicrobial activity against SA RN4220 (6.96 ± 0.55 mm and 22.49 ± 6.47 mm, respectively); however, inhibition by penicillin was not detected. Similar results were observed for MRSA pertaining to antibiotics. These results, which were observed before digestion, indicate that only SA RN4220 is susceptible to ofloxacin and other strains are all resistant.
Fig. 3.
Analysis of resistance to antibiotics by (A) Staphylococcus aureus RN4220 (SA RN4220) and (B) methicillin-resistant S. aureus (MRSA) during in vitro human digestion by agar disk diffusion. Antibiotics before digestion and dissolved buffer of antibiotics were taken as positive and negative control, respectively. R, resistant; I, intermediately resistant; S, susceptible; N.D., not detected. Antimicrobial susceptibility testing was performed using the disk diffusion method according to Clinical and Laboratory Standards Institute (CLSI) guidelines. a,b Values marked with different letters differed significantly according to the inhibition zones of the antibiotics used during in vitro human digestion (P < 0.05).
After digestion in the mouth, antimicrobial activity against SA RN4220 and MRSA was higher in the ofloxacin than in the tetracycline and penicillin (not detectable) groups, indicating that SA RN4220 exhibits intermediate resistance to ofloxacin but is resistant to tetracycline and penicillin. Although tetracycline and ofloxacin produced inhibition zones, MRSA was resistant to all antibiotics after digestion in the mouth. After digestion in the stomach and small intestine, only ofloxacin produced an inhibition zone against SA RN4220 and MRSA, while tetracycline and penicillin did not. However, some resistance was observed for all antibiotics, regardless of the microbial strain.
After simulated digestion in the large intestine, none of the antibiotics demonstrated antimicrobial activity, with SA RN4220 and MRSA showing resistance to all of them. Moreover, penicillin did not exhibit antimicrobial activity against any of the strains. This may be attributed to the structural degradation of antibiotics by pH alteration and gut microbiota used to simulate digestion in the large intestine, thus suggesting that increased antibiotic resistance is related to the digestion process.
Increased MIC may be attributed to reduced antimicrobial activity or increased resistance. Furthermore, the difference in antibiotic resistance depends on specific digestion steps, as demonstrated by the results in Table 3. Resistance to the three antibiotics increased as digestion progressed, corresponding to each MIC interpretive criterion.
Table 3.
Changes in MIC of antibiotics during in vitro human digestion.
| Bacterial strain | Antibiotic | MIC (μg/mL) |
|||
|---|---|---|---|---|---|
| Before digestion |
In vitro human digestion phase |
||||
| Mouth | Stomach | Small intestine | |||
| SA RN4220 | Tetracycline | 2 (S) | 4 (S) | 8 (I) | 16 (R) |
| Ofloxacin | 1 (S) | 2 (I) | 2 (I) | 8 (R) | |
| Penicillin | >2 (R) | >2 (R) | >2 (R) | >2 (R) | |
| MRSA | Tetracycline | 8 (I) | 16 (R) | 16 (R) | 16 (R) |
| Ofloxacin | 4 (R) | 4 (R) | 4 (R) | 8 (R) | |
| Penicillin | >2 (R) | >2 (R) | >2 (R) | >2 (R) | |
SA RN4220, fluoroquinolone-susceptible restriction-deficient mutant of Staphylococcus aureus, MRSA, methicillin-resistant S. aureus, R, resistant; I, intermediately resistant; S, susceptible; MIC, minimum inhibitory concentration.
MIC was determined according to the Clinical and Laboratory Standards Institute (CLSI) guidelines for Staphylococcus spp.
Before digestion, the MICs of tetracycline, ofloxacin, and penicillin against SA RN4220 were 2, 1, and >2 μg/mL, respectively. After digestion in the mouth, the MIC for tetracycline was 4 μg/mL, indicating susceptibility, while that for ofloxacin was 2 μg/mL, indicating intermediate resistance against SA RN4220. After digestion in the stomach, the MICs for tetracycline and ofloxacin were 8 and 2 μg/mL, respectively, indicating intermediate resistance against SA RN4220. However, SA RN4220 showed resistance to all the antibiotics after digestion in the small intestine. Both strains showed the following the trend of resistance to antibiotics: penicillin > ofloxacin ≥ tetracycline. Furthermore, MRSA were resistant to all antibiotics, and the MICs of the three antibiotics were higher for MRSA than they were for SA RN4220. All the antibiotics’ MICs after digestion were higher than those before digestion, indicating resistance by both strains, which might have been because of change in resistance to tetracycline and ofloxacin due to the simulated intestinal tract conditions.
Increasing resistance or decreasing antimicrobial activity of antibiotics was a prominent phenomenon observed during the in vitro digestion process in this study; however, the underlying mechanisms remain unclear. This study can suggest some possible ones.
The first mechanism considers the impact of pH during the digestion process. Gastrointestinal digestion leads to pH fluctuations that are required for the activation of digestive enzymes. Tetracycline possesses a basic tetracycline class structure composed of a tetracyclic naphthacene carboxamide ring system, wherein the dimethylamine group at C4 in ring A is necessary to facilitate antimicrobial activity [46]. Additionally, the amino group at the C4 position and ketoenolic tautomers at the C1 and C3 positions in the ring group all contribute to antibacterial activities [46]. Previous studies that investigated factors causing tetracycline degradation, such as pH, photodegradation, and chelation have reported that hydrolysis of tetracycline is affected by pH and temperature [47,48]. Tetracycline can induce reversible epimerization at C4 under low pH conditions and can lead to the formation of 4-epi-tetracycline in the presence of hydrogen ions from water under acidic conditions [49]. Additionally, tetracycline can be converted to iso-tetracyclines such that the hydroxyl group at C6 may attack the C11 carbonyl group at alkaline pH, which in turn decreases the tetracycline amount during hydrolysis [47,50]. Ofloxacin is a quinolone antibiotic that is widely used to inhibit gram-positive bacteria such as S. aureus, Streptococcus pyogenes, and Streptococcus pneumoniae [51]. A previous study showed that initial pH of the liquid solution affects the outbreak of active species, and surface charge characteristics exert a significant impact on photocatalytic performance [52]. Another study regarding the effect of initial pH values ranging from 3.0 to 11.0 revealed the degradation characteristics of ofloxacin [53]. Tian et al. also demonstrated that ofloxacin is efficiently degraded over a pH range of 3.6–10.0 [54]. The carboxylic acid residue in ofloxacin is associated with antimicrobial activity [55] and contributes to the binding between quinolones and DNA gyrase. Thus, decarboxylation at the quinolone moiety may be accompanied by reduced antimicrobial activity. Furthermore, pH may affect ofloxacin activity by Fenton oxidation, resulting in poor performance and increased resistance. Thus, resistance to tetracycline and ofloxacin during in vitro human digestion may be due to structural degradation caused by pH fluctuations.
The second possible mechanism concerns increased antibiotic consumption that activates intestinal microbes. Antibiotics predominantly alter the metabolic state of bacteria [56]. Specifically, the metabolic state of bacteria influences the effects of antibiotics. Gut microbial diversity is influenced by oral administration of antibiotics, which greatly diminishes the total bacterial genomic DNA amount [57]. A previous study has reported increased resistance of fecal E. coli to tetracycline [58]. Moreover, commensal gut microbiota may be responsible for antibiotic resistance. Previously, potential antibiotic resistance genes against 53 antibiotics were found in the human gut microbiota; furthermore, four distinct clusters of gut microbiomes exhibiting similarities in antibiotic resistance, known as resistotypes, were identified [59]. In a study evaluating the resistome, which is the repository of antibiotic resistance genes, of Bifidobacterium, to evaluate its resistance in the gut metagenome, the tetracycline-resistant gene, namely tet (W) was predicted to be transported via conjugate transposons in adults and infants [60]. In our study, none of the antibiotics showed antimicrobial activity against SARN4220 and MRSA. These findings were similar to those regarding antibiotic resistance. Therefore, reduced antimicrobial activity or increased resistance to antibiotics during the simulated large intestinal digestion stage might have occurred because of decreased antibiotic concentration due to reactions with the gut microbiota that were introduced for simulation of digestion in the large intestine.
Although certain factors, such as structural degradation of antibiotics, require validation, our study suggests that the in vitro human digestion model is a promising tool for the detection of antibiotic resistance. Further research that considers changes in the molecular structure of antibiotics is necessary to improve our understanding regarding antimicrobial activity and development of antibiotic resistance depending on the digestion process.
4. Conclusion
This study demonstrated variations in antibiotic resistance during in vitro human digestion using disk diffusion and MIC assays. Our results suggest that increased resistance or reduced antimicrobial activity of antibiotics during digestion may be associated with decreased residual antibiotics in the body. Additionally, alterations in these antibiotics are likely related to changes in their structural stability and activity due to certain factors, such as pH fluctuation and gut microbiota, related to in vitro human digestion. It was also found that resistance to residual antibiotics was affected by the digestion process. Thus, the in vitro human digestion model may be used to determine resistance to residual antibiotics. Further research is required to verify the exact factors that contribute to changes in resistance and antimicrobial activity of antibiotics during human digestion.
Author contribution statement
Seung Yun Lee: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Da Young Lee: Hea Jin Kang: Seung Hyeon Yun: Ermie Jr. Mariano: Juhyun Lee: Performed the experiments.
Jong Hyuk Kim: Analyzed and interpreted the data.
Sun Jin Hur: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Data availability statement
Data will be made available on request.
Additional information
No additional information is available for this paper.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was carried out with the support of "Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ016201)" Rural Development Administration, Republic of Korea. This research was also supported by the Chung-Ang University Young Scientist Scholarship (CAYSS) in 2022.
References
- 1.Chiesa L.M., DeCastelli L., Nobile M., Martucci F., Mosconi G., Fontana M., et al. Analysis of antibiotic residues in raw bovine milk and their impact toward food safety and on milk starter cultures in cheese-making process. LWT. 2020;131 [Google Scholar]
- 2.Dibner J.J., Richards J.D. Antibiotic growth promoters in agriculture: history and mode of action. Poultry Sci. 2005;84(4):634–643. doi: 10.1093/ps/84.4.634. [DOI] [PubMed] [Google Scholar]
- 3.Rana M.S., Lee S.Y., Kang H.J., Hur S.J. Reducing veterinary drug residues in animal products: a review. Food Sci. Anim. Resour. 2019;39(5):687–703. doi: 10.5851/kosfa.2019.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laxminarayan R., Duse A., Wattal C., Zaidi A.K., Wertheim H.F., Sumpradit N., et al. Antibiotic resistance—the need for global solutions. Lancet Infect. Dis. 2013;13(12):1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention . National Center for Emerging and Zoonotic Infectious Diseases (NCEZID); Atlanta, GA: 2018. Antibiotic/antimicrobial Resistance (AR/AMR): Biggest Threats and Data. [Google Scholar]
- 6.Bartlett J.G., Gilbert D.N., Spellberg B. Seven ways to preserve the miracle of antibiotics. Clin. Infect. Dis. 2013;56(10):1445–1450. doi: 10.1093/cid/cit070. [DOI] [PubMed] [Google Scholar]
- 7.Singer R.S., Porter L. Mindwalk Consulting Group, LLC; Falcon Heights, MN, USA: 2019. Estimates of On-Farm Antimicrobial Usage in Broiler Chicken and turkey Production in the United States, 2013–2017. [Google Scholar]
- 8.Kirchhelle C. Pharming animals: a global history of antibiotics in food production (1935–2017) Palgrave Commun. 2018;4(1):1–13. [Google Scholar]
- 9.Aidara-Kane A., Angulo F.J., Conly J.M., Minato Y., Silbergeld E.K., McEwen S.A., et al. World Health Organization (WHO) guidelines on use of medically important antimicrobials in food-producing animals. Antimicrob. Resist. Infect. Control. 2018;7(1):7. doi: 10.1186/s13756-017-0294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keum G.B., Kim E.S., Cho J.H., Song M., Oh K.K., Cho J.H., et al. Analysis of antibiotic resistance genes in pig feces during the weaning transition using whole metagenome shotgun sequencing. J. Anim. Sci. Technol. 2022;65(1):175–182. doi: 10.5187/jast.2022.e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.U. S. Food and Drug Administration: Center for Veterinary Medicine . 2017. Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals. Tulsa, OK, USA. [Google Scholar]
- 12.Wallinga D., Roach S. Antibiotic consumption in US pork, beef, and Turkey industries vastly outstrips comparable industries in Europe, and the US chicken industry. Food Anim. Concerns Trust. 2018 [Google Scholar]
- 13.Yaqoob M.U., Wang G., Wang M. An updated review on probiotics as an alternative of antibiotics in poultry—a review. Animal Biosci. 2022;35(8):1109. doi: 10.5713/ab.21.0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khalid A., Khalid F., Mahreen N., Hussain S.M., Shahzad M.M., Khan S., Wang Z. Effect of spore-forming probiotics on the poultry production: a review. Food Sci. Anim. Resour. 2022;42(6):968–980. doi: 10.5851/kosfa.2022.e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kjeldgaard J., Cohn M.T., Casey P.G., Hill C., Ingmer H. Residual antibiotics disrupt meat fermentation and increase risk of infection. mBio. 2012;3(5) doi: 10.1128/mBio.00190-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holley R.A., Blaszyk M. Antibiotic challenge of meat starter cultures and effects upon fermentations. Food Res. Int. 1997;30(7):513–522. [Google Scholar]
- 17.Coles L., Moughan P., Darragh A.J. In vitro digestion and fermentation methods, including gas production techniques, as applied to nutritive evaluation of foods in the hindgut of humans and other simple-stomached animals. Anim. Feed Sci. Technol. 2005;123:421–444. [Google Scholar]
- 18.Lee S.J., Lee S.Y., Chung M.-S., Hur S.J. Development of novel in vitro human digestion systems for screening the bioavailability and digestibility of foods. J. Funct.Foods. 2016;22:113–121. [Google Scholar]
- 19.Guillén J., Steckler T. In: Good Research Practice in Non-clinical Pharmacology and Biomedicine. Bespalov A., Michel M.C., Steckler T., editors. Springer International Publishing; Cham: 2020. Good research practice: lessons from animal care and use; pp. 367–382. [DOI] [PubMed] [Google Scholar]
- 20.Hur S.J., Lim B.O., Decker E.A., McClements D.J. In vitro human digestion models for food applications. Food Chem. 2011;125(1):1–12. [Google Scholar]
- 21.Oomen A.G., Rompelberg C.J.M., Bruil M.A., Dobbe C.J.G., Pereboom D.P.K.H., Sips A.J.A.M. Development of an in vitro digestion model for estimating the bioaccessibility of soil contaminants. Arch. Environ. Contam. Toxicol. 2003;44:281–287. doi: 10.1007/s00244-002-1278-0. [DOI] [PubMed] [Google Scholar]
- 22.Versantvoort C.H., Oomen A.G., Van de Kamp E., Rompelberg C.J., Sips A.J. Applicability of an in vitro digestion model in assessing the bioaccessibility of mycotoxins from food. Food Chem. Toxicol. 2005;43(1):31–40. doi: 10.1016/j.fct.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Lee S.Y., Lee D.Y., Kang H.J., Kang J.H., Cho M.G., Jang H.W., et al. Differences in the gut microbiota between young and elderly persons in Korea. Nutr. Res. 2021;87:31–40. doi: 10.1016/j.nutres.2020.12.013. [DOI] [PubMed] [Google Scholar]
- 24.Hur S.J., Lee S.Y., Lee S.J. Effect of biopolymer encapsulation on the digestibility of lipid and cholesterol oxidation products in beef during in vitro human digestion. Food Chem. 2015;166:254–260. doi: 10.1016/j.foodchem.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 25.Kim H.S., Hur S.J. Changes of sodium nitrate, nitrite, and N-nitrosodiethylamine during in vitro human digestion. Food Chem. 2017;225:197–201. doi: 10.1016/j.foodchem.2017.01.036. [DOI] [PubMed] [Google Scholar]
- 26.Hur S.J., Lee S.J., Lee S.Y., Bahk Y.Y., Kim C.G. Effect of emulsifiers on microstructural changes and digestion of lipids in instant noodle during in vitro human digestion. LWT. 2015;60(1):630–636. [Google Scholar]
- 27.Lee S.Y., Kang H.J., Kang J.H., Jang H.W., Kim B.K., Hur S.J. Analysis of in vitro digestion using human gut microbiota in adult and elderly individuals. Food Chem. 2021;362 doi: 10.1016/j.foodchem.2021.130228. [DOI] [PubMed] [Google Scholar]
- 28.Lee S.Y., Kim O.Y., Yoon S.Y., Hur S.J. Changes in resistance to and antimicrobial activity of antibiotics during in vitro human digestion. J. Glob. Antimicrob. Resist. 2018;15:277–282. doi: 10.1016/j.jgar.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 29.Lee S.Y., Lee D.Y., Kang H.J., Kang J.H., Hur S.J. Changes in antimicrobial activity and resistance of antibiotics in meat patties during in vitro human digestion. LWT. 2021;137 [Google Scholar]
- 30.Hinsberger A., Sandhu B. Digestion and absorption. Curr. Pediatr. 2004;14(7):605–611. [Google Scholar]
- 31.Fallingborg J. Intraluminal pH of the human gastrointestinal tract. Dan. Med. Bull. 1999;46(3):183–196. [PubMed] [Google Scholar]
- 32.Brodkorb A., Egger L., Alminger M., Alvito P., Assunção R., Ballance S., et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019;14(4):991–1014. doi: 10.1038/s41596-018-0119-1. [DOI] [PubMed] [Google Scholar]
- 33.Ucak S., Sudagidan M., Borsa B.A., Mansuroglu B., Ozalp V.C. Inhibitory effects of aptamer targeted teicoplanin encapsulated PLGA nanoparticles for Staphylococcus aureus strains. World J. Microbiol. Biotechnol. 2020;36(5):69. doi: 10.1007/s11274-020-02845-y. [DOI] [PubMed] [Google Scholar]
- 34.Andrews J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001;48(suppl_1):5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 35.Zhou M., Cai Q., Zhang C., Ouyang P., Yu L., Xu Y. Antibiotic resistance bacteria and antibiotic resistance genes survived from the extremely acidity posing a risk on intestinal bacteria in an in vitro digestion model by horizontal gene transfer. Ecotoxicol. Environ. Saf. 2022;247 doi: 10.1016/j.ecoenv.2022.114247. [DOI] [PubMed] [Google Scholar]
- 36.Citterio B., Mangiaterra G., Meli M.A., Cedraro N., Roselli C., Vignaroli C., et al. Gastrointestinal survival and adaptation of antibiotic-resistant enterococci subjected to an in vitro digestion model. Food Control. 2020;110 [Google Scholar]
- 37.Agarwal M., Rathore R.S., Chauhan A. A rapid and high throughput MIC determination method to screen uranium resistant microorganisms. Methods Protoc. 2020;3(1):21. doi: 10.3390/mps3010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Francesco V., Zullo A., Fiorini G., Saracino I.M., Pavoni M., Vaira D. Role of MIC levels of resistance to clarithromycin and metronidazole in Helicobacter pylori eradication. J. Antimicrob. Chemother. 2018;74(3):772–774. doi: 10.1093/jac/dky469. [DOI] [PubMed] [Google Scholar]
- 39.Sierra-Arguello Y.M., Quedi Furian T., Perdoncini G., Moraes H.L.S., Salle C.T.P., Rodrigues L.B., et al. Fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli from poultry and human samples assessed by PCR-restriction fragment length polymorphism assay. PLoS One. 2018;13(7) doi: 10.1371/journal.pone.0199974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ge B., Wang F., Sjölund-Karlsson M., McDermott P.F. Antimicrobial resistance in Campylobacter: susceptibility testing methods and resistance trends. J. Microbiol. Methods. 2013;95(1):57–67. doi: 10.1016/j.mimet.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 41.Fayad E., El-Sawalhi S., Azizi L., Beyrouthy M., Abdel-Massih R.M. Yerba Mate (Ilex paraguariensis) a potential food antibacterial agent and combination assays with different classes of antibiotics. LWT. 2020;125 [Google Scholar]
- 42.Klančnik A., Piskernik S., Jeršek B., Možina S.S. Evaluation of diffusion and dilution methods to determine the antibacterial activity of plant extracts. J. Microbiol. Methods. 2010;81(2):121–126. doi: 10.1016/j.mimet.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 43.Iwalokun B.A., Ogunledun A., Ogbolu D.O., Bamiro S.B., Jimi-Omojola J. In vitro antimicrobial properties of aqueous garlic extract against multidrug-resistant bacteria and candida species from Nigeria. J. Med. Food. 2004;7(3):327–333. doi: 10.1089/jmf.2004.7.327. [DOI] [PubMed] [Google Scholar]
- 44.Di Lodovico S., Menghini L., Ferrante C., Recchia E., Castro-Amorim J., Gameiro P., et al. Hop extract: an efficacious antimicrobial and anti-biofilm agent against multidrug-resistant Staphylococci strains and Cutibacterium acnes. Front. Microbiol. 2020;11(1852) doi: 10.3389/fmicb.2020.01852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S.Y., Kang J.H., Jeong J.W., Kim J.H., Kim H.W., Oh D.H., et al. Alternative experimental approaches to reduce animal use in biomedical studies. J. Drug Deliv. Sci. Technol. 2022 [Google Scholar]
- 46.Tariq S., Rizvi S.F.A., Anwar U. Tetracycline: classification, structure activity relationship and mechanism of action as a theranostic agent for infectious lesions-a mini review. Biomed. J. Sci. Tech. Res. 2018;7:5787–5796. [Google Scholar]
- 47.Halling-Sørensen B., Sengeløv G., Tjørnelund J. Toxicity of tetracyclines and tetracycline degradation products to environmentally relevant bacteria, including selected tetracycline-resistant bacteria. Arch. Environ. Contam. Toxicol. 2002;42(3):263–271. doi: 10.1007/s00244-001-0017-2. [DOI] [PubMed] [Google Scholar]
- 48.Loftin K.A., Adams C.D., Meyer M.T., Surampalli R. Effects oF ionic strength, temperature, and pH on degradation of selected antibiotics. J. Environ. Qual. 2008;37(2):378–386. doi: 10.2134/jeq2007.0230. [DOI] [PubMed] [Google Scholar]
- 49.Chen W.R., Huang C.H. Transformation kinetics and pathways of tetracycline antibiotics with manganese oxide. Environ. Pollut. 2011;159(5):1092–1100. doi: 10.1016/j.envpol.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 50.Waller M.D. Vibrations of free plates: line symmetry; corresponding modes. Proc. Math. Phys. Eng. Sci. 1952;211(1105):265–276. [Google Scholar]
- 51.Koulenti D., Xu E., Mok I.Y.S., Song A., Karageorgopoulos D.E., Armaganidis A., et al. Novel antibiotics for multidrug-resistant gram-positive microorganisms. Microorganisms. 2019;7(8):270. doi: 10.3390/microorganisms7080270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang R., Lu G., Yan Z., Wu D., Zhou R., Bao X. Insights into a CQD-SnNb2O6/BiOCl Z-scheme system for the degradation of benzocaine: influence factors, intermediate toxicity and photocatalytic mechanism. Chem. Eng. J. 2019;374:79–90. [Google Scholar]
- 53.Zhao G., Ding J., Zhou F., Chen X., Wei L., Gao Q., et al. Construction of a visible-light-driven magnetic dual Z-scheme BiVO4/g-C3N4/NiFe2O4 photocatalyst for effective removal of ofloxacin: mechanisms and degradation pathway. Chem. Eng. J. 2021;405 [Google Scholar]
- 54.Tian X., Jin H., Nie Y., Zhou Z., Yang C., Li Y., Wang Y. Heterogeneous Fenton-like degradation of ofloxacin over a wide pH range of 3.6–10.0 over modified mesoporous iron oxide. Chem. Eng. J. 2017;328:397–405. [Google Scholar]
- 55.Pi Y., Feng J., Song M., Sun J. Degradation potential of ofloxacin and its resulting transformation products during Fenton oxidation process. Chin. Sci. Bull. 2014;59(21):2618–2624. [Google Scholar]
- 56.Stokes J.M., Lopatkin A.J., Lobritz M.A., Collins J.J. Bacterial metabolism and antibiotic efficacy. Cell Metabol. 2019;30(2):251–259. doi: 10.1016/j.cmet.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu L., Surathu A., Raplee I., Chockalingam A., Stewart S., Walker L., et al. The effect of antibiotics on the gut microbiome: a metagenomics analysis of microbial shift and gut antibiotic resistance in antibiotic treated mice. BMC Genom. 2020;21(1):263. doi: 10.1186/s12864-020-6665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levy S.B. Factors impacting on the problem of antibiotic resistance. J. Antimicrob. Chemother. 2002;49(1):25–30. doi: 10.1093/jac/49.1.25. [DOI] [PubMed] [Google Scholar]
- 59.Ghosh T.S., Gupta S.S., Nair G.B., Mande S.S. In silico analysis of antibiotic resistance genes in the gut microflora of individuals from diverse geographies and age-groups. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0083823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duranti S., Lugli G.A., Mancabelli L., Turroni F., Milani C., Mangifesta M., et al. Prevalence of antibiotic resistance genes among human gut-derived bifidobacteria. Appl. Environ. Microbiol. 2017;83(3) doi: 10.1128/AEM.02894-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.