Highlights
-
•
Dicarbonyl metabolism in the mitochondria remains unclear.
-
•
Knockout mice for mitochondrial protein ES1 were generated.
-
•
Metabolic activity of glyoxal decreased significantly in ES1-knockout mice.
-
•
ES1 was found to have glyoxalase III activity and modulate dicarbonyl metabolism.
-
•
Mouse recombinant ES1 protein exhibited metabolic activity for glyoxal.
Keywords: Mitochondria, ES1, Reactive dicarbonyl, Glyoxalase, Methylglyoxal degradation
Abstract
Glycation, caused by reactive dicarbonyls, plays a role in various diseases by forming advanced glycation end products. In live cells, reactive dicarbonyls such as glyoxal (GO) and methylglyoxal (MGO) are produced during cell metabolism, and these should be removed consistently. However, the dicarbonyl metabolic system in the mitochondria remains unclear. It has been speculated that the mammalian mitochondrial protein ES1 is a homolog of bacterial elbB possessing glyoxalase III (GLO3) activity. Therefore, in this study, to investigate ES1 functions and GLO3 activity, we generated ES1-knockout (KO) mice and recombinant mouse ES1 protein and investigated the biochemical and histological analyses. In the mitochondrial fraction obtained from ES1-KO mouse brains, the GO metabolism and cytochrome c oxidase activity were significantly lower than those in the mitochondrial fraction obtained from wildtype (WT) mouse brains. However, the morphological features of the mitochondria did not change noticeably in the ES1-KO mouse brains compared with those in the WT mouse brains. The mitochondrial proteome analysis showed that the MGO degradation III pathway and oxidative phosphorylation-related proteins were increased. These should be the response to the reduced GO metabolism caused by ES1 deletion to compensate for the dicarbonyl metabolism and damaged cytochrome c oxidase by elevated GO. Recombinant mouse ES1 protein exhibited catalytic activity of converting GO to glycolic acid. These results indicate that ES1 possesses GLO3 activity and modulates the metabolism of GO in the mitochondria. To our knowledge, this is the first study to show a novel metabolic pathway for reactive dicarbonyls in mitochondria.
1. Introduction
Glycation, the nonenzymatic glycosylation of biomolecules, leads to the formation of advanced glycation end products (AGEs). AGE formation is a common feature of diabetes [1,2] and aging [3] and is associated with various diseases, such as transthyretin amyloidosis, Alzheimer's disease, and Parkinson's disease [4], [5], [6], [7], [8], [9]. Reactive dicarbonyls, including glyoxal (GO) and methylglyoxal (MGO), are well characterized as the main causative agents of glycation. The reactivity of reactive dicarbonyls is 20,000 times higher than that of glucose, and they react with lysine and arginine residues in a nonenzymatic manner to form AGEs [10,11]. In particular, GO is involved in the formation of carboxymethyl lysine, a type of AGE that causes changes in protein molecular structure, enzyme activity, and receptor function [12]. Reactive dicarbonyls are primarily produced as glycolytic byproducts; lipid peroxidation and DNA oxidation are also sources of reactive dicarbonyls [13]. Hence, reactive dicarbonyls are present in all cells under both normal and pathological conditions. Dicarbonyl stress is caused by an imbalance between dicarbonyl formation and depletion; thus, the dicarbonyl metabolic system plays an essential role in live cells.
Dicarbonyls can diffuse across mitochondrial membranes and enter the mitochondria. Mitochondrial dicarbonyl stress can account, at least partially, for the mitochondrial dysfunction and oxidative damage observed with hyperglycemia and aging [14], [15], [16], [17]. Glycation damage in mitochondria can cause mitochondrial dysfunction in neurodegenerative diseases [18,19]. The presence of multiple dicarbonyl-metabolizing enzymes in the cytoplasm has been reported [20], [21], [22], but these enzymes have not been found in mitochondria. Therefore, the mechanism underlying the detoxification of mitochondrial dicarbonyls remains unclear.
We focused on the mitochondrial protein ES1, which is highly homologous to elbB in bacteria (approximately 43% identity of amino acid sequence). ES1 and elbB are members of the DJ-1/Pfpl family [23]. In mammals, ES1 expression has been confirmed in various species, including humans and mice; however, the function of mammalian ES1 remains unclear. elbB possesses glyoxalase (GLO) activity; GLO converts GO and MGO to glycolic acid and lactic acid, respectively [24,25]. The main GO-detoxification system is the glutathione-dependent pathway, which is mediated by GLO1 and GLO2 [26]. In contrast, elbB exhibits GLO3 activity, converting dicarbonyl to acid species in a one-step, glutathione-independent reaction [27,28]. Mammalian ES1, similar to other well-known proteins with GLO3 activity such as Hsp31, has Cys and Glu residues that are essential for catalytic activity [25,29]. Based on these facts, we speculated that mammalian ES1 also possesses GLO3 activity. When cells are exposed to dicarbonyls, the depletion of glutathione and NADPH leads to dysfunction of the cell detoxification systems that depend on these electron donors [30,31]. Therefore, GLO3 activity, which lacks this dependence, may play an essential role in protecting cells from dicarbonyl-induced cell death.
In the present study, ES1-knockout (KO) mice were generated to clarify the function of mammalian ES1 as human and mouse ES1 proteins are highly homologous and have common essential catalytic residues [29]. We analyzed mitochondrial morphological features and performed GLO and mitochondrial activity assays with the ES1-KO mice. We performed a comprehensive proteomic analysis of wildtype (WT) and ES1-KO mice using liquid chromatography-tandem mass spectrometry (LC-MS/MS). The semiquantitative proteomic data obtained were subjected to pathway and network analyses. A recombinant mouse ES1 protein was generated for detailed functional analysis. To our knowledge, this is the first study that aimed to evaluate the direct metabolic activity of dicarbonyl-metabolizing enzymes in the mitochondria and to identify any novel metabolic pathways involved.
2. Materials and methods
STAR★METHODS KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ES1 | This paper | N/A |
| Adenylate Kinase 2 (AK2) | Invitrogen | PA5-49482 |
| Porin | Sigma-Aldrich | 185-197 |
| COXIV | Abcam | Ab16056 |
| AIF | Invitrogen | MA5-15880 |
| ATP5A | Santa Cruz | sc-136178 |
| Beta-actin | Abcam | ab6276 |
| Chemicals and Recombinant Proteins | ||
| Protease-inhibitor cocktail tablets | Roche Diagnostics | 11836145001 |
| Mitochondrial isolation buffer | BioChain Institute | KC010100 |
| ECL Prime Western Blotting Detection Kit | Cytiva | RPN2236 |
| KOD OneTM PCR Master Mix | Toyobo | KMM-101 |
| Mitochondrial activity assay kit | BioChain Institute | KC310100 |
| Protease MAX surfactant | Promega | V2071 |
| TRI Reagent | Moleculer Research Center | TR118 |
| ReverTra Ace | Toyobo | TRT-100 |
| Wizard SV Gel and PCR Clean-up System | Promega | A9282 |
| In-Fusion HD Cloning Kit | Clontech Takara Cellartis | 639648 |
| TALON® Metal Affinity Resin | Clontech Laboratories, Inc. | 635502 |
| mES1 | This paper | N/A |
| Experimental Models: Animal | ||
| C57BL/6J mice | Japan SLC | C57BL/6JJmsSlc |
| ES1-KO mice | This paper | N/A |
| Software | ||
| Analyst TF 1.7.1 software program | AB Sciex | https://sciex.com |
| Peak View v2.2.0 | AB Sciex | https://sciex.com |
| ProteinPilot 5.0.1 software program | AB Sciex | https://sciex.com |
| Marker View software program v.1.3.0.1 | AB Sciex | https://sciex.com |
| SIMCA software program SIMCA15 | Infocom Corp. | https://www.infocom-science.jp |
| Ingenuity Pathway Analysis | Qiagen | http://ingenuity.com |
| Deposited Data | ||
| Proteomic data | E.W.Deutsch et al., 2017, S. Okuda et al., 2017 | PXD025954; JPST001170 |
2.1. Animals
All experiments were performed in accordance with the UK Animals (Scientific Procedures) Act, 1986; associated guidelines (EU Directive 2010/63/EU for animal experiments); and ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines. The study protocol was approved by the Committee for the Use of Live Animals at Iwate University (Morioka, Japan) (Approval Numbers: A201924, A202052, and A202222).
Adult male and female C57BL/6J mice were obtained from Kumagai-shigeyasu Co., Ltd. (Miyagi, Japan) and bred in our laboratory. After weaning, 2–5 pups were housed per cage. They were maintained under temperature-controlled conditions at 25°C with a 12/12-h light-dark cycle and provided food and water ad libitum. Eight to ten-weeks-old mice were used for GLO activity assay (2 male and 1 female mice), mitochondrial activity assay (2 male and 2 female mice), and proteomic analysis (2 male and 3 female mice). In the present study, there were no sex differences in any of the experiments.
2.2. Preparation of mouse tissue extracts for western blotting
Several C57BL/6J mice at 8–10 weeks of age were anesthetized via inhalation of isoflurane (Pfizer Inc., New York, NY, USA), perfused with radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) containing protease-inhibitor cocktail tablets (catalog number: 11836145001; Roche Diagnostics, Mannheim, Germany). The tissues of these mice, including the brain, heart, lung, liver, stomach, spleen, intestine, kidney, and muscle, were removed. Immediately after excision, each tissue was finely chopped and homogenized in two volumes of RIPA buffer containing protease-inhibitor cocktail tablets (Roche Diagnostics). Thereafter, the samples were centrifuged at 20,000 × g for 30 min. All procedures were conducted at 4°C. The supernatants were used for western blotting to determine ES1 tissue distribution.
2.3. Mitochondrial isolation from mouse brain tissue for western blotting
The mitochondrial fraction was obtained using a mitochondrial isolation kit (catalog number: KC010100; BioChain Institute, Newark, CA, USA). The mice were euthanized via cervical dislocation and the brain tissues were removed. Each brain tissue sample was finely chopped and homogenized in two volumes of mitochondrial isolation buffer provided in the kit and then centrifuged at 600 × g for 10 min. The supernatant was collected and centrifuged at 12,000 × g for 15 min. The supernatant obtained was used as a lysosome- and microsome-containing cytosolic fraction. The mitochondrial fraction was obtained from the pellet of the second centrifugation and resuspended in two volumes of mitochondrial isolation buffer. The suspended mitochondria were sonicated (TOMY; Ultrasonic disruptor UD-100; output level: 40, 30 s × 2) and centrifuged at 105,000 × g for 30 min. The supernatant obtained was used as the mitochondrial soluble fraction, which contained the proteins in mitochondrial intermembrane space (IMS) and matrix (MAT). The pellet was resuspended in RIPA buffer and centrifuged at 20,000 × g for 30 min. The supernatant was used as the mitochondrial membrane fraction, which contained the mitochondrial outer membrane (OM) and inner membrane (IM). All procedures were conducted at 4°C. These samples were used for western blotting to determine ES1 subcellular localization.
2.4. Antibodies
The antibodies that were used in the present study included: anti-adenylate kinase 2 (Ak2; catalog number: PA5-49482; Invitrogen, Waltham, MA, USA), anti-porin (catalog number: 185-197; Sigma-Aldrich, St. Louis, MO, USA), anti-cytochrome c oxidase subunit IV (COX4; catalog number: ab16056; Abcam, Cambridge, UK), anti-apoptosis-inducing factor (AIF; catalog number: MA5-15880; Invitrogen, Carlsbad, CA, USA), anti-ATP5A (catalog number: sc-136178; Santa Cruz Biotechnology, Dallas, TX, USA), and anti-beta-actin (catalog number: ab6276; Abcam) antibodies. The anti-ES1 antibody was generated in our laboratory, as previously reported [29]. Keyhole limpet hemocyanin conjugated to the C-terminal 17 amino acid peptide of mouse ES1 (Cys-HDGIGAMVKNVLELTGK) was used as the antigen for antibody production.
2.5. Western blotting
Western blotting was conducted following the method of Ozaki et al. [32]. Briefly, each sample was separated using SDS-PAGE and transferred onto Immobilon-P polyvinylidene fluoride (PVDF) transfer membranes (catalog number: IPVH00010; EMD Millipore, Billerica, MA, USA). The membranes were blocked with blocking buffer (10 mM sodium phosphate buffer, pH 7.4, 0.14 M NaCl, and 0.05% Tween 20 [TW-PBS] containing 1% skim milk or TW-PBS containing 5% skim milk) for 4 h at 25°C and then incubated overnight with the primary antibodies at 4°C. After washing with TW-PBS, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies overnight at 4°C. Each antibody had been diluted with the blocking buffer as follows: anti-mouse ES1 (1:1000), anti-porin (1:1000), anti-Ak2 (1:2000 or 1:1000), anti-COX4 (1:2000), anti-AIF (1:500), anti-ATP5A (1:200), and anti-beta-actin (1:10,000). After incubation and washing, the immunoreactive signals were detected using the ECL Prime Western Blotting Detection Kit (catalog number: RPN2236; Cytiva, Tokyo, Japan). The images were captured using a luminescent image analyzer (ChemiDoc XRS Plus; catalog number: 1708265J1PC; Bio-Rad Laboratories, Hercules, CA, USA).
2.6. Generation of ES1-KO mice
The ES1-KO mouse line was generated using the technique for animal KO system via the electroporation method, a unique electroporation technique, using the CRISPR-Cas9 system. Adult female C57BL/6J mice were obtained from Japan SLC (Hamamatsu, Shizuoka, Japan). C57BL/6J mouse embryos were purchased from CLEA Japan (Tokyo, Japan). All the processes were conducted as described by Wake and Kaneko [33]. Cas9 nuclease V3, crRNA, and tracrRNA were obtained from Integrated DNA Technologies (Coralville, IA, USA). The crRNA was designed to target exon 1 of the mouse ES1 (Gatd3a). The nuclease solution was prepared by mixing crRNA (15 μM), tracrRNA (15 μM), and Cas9 protein (200 ng/μL) in Opti-MEM™ (Thermo Fisher Scientific, Waltham, MA, USA) immediately before electroporation. Next, the nuclease solution (5 μL) was added between the electrodes on a glass slide (catalog number: CUY501P1-1.5; Nepa Gene, Chiba, Japan), and the embryos were placed in line. The nucleases were then introduced into the embryo using the NEPA21 Super Electroporator (Nepa Gene). Each pulse was set as follows: poring pulse (voltage: 40 V; pulse length: 2 ms; pulse interval: 50 ms; number of pulses: 4; decay rate: 10%; polarity: +) followed by transfer pulse (voltage: 15 V; pulse length: 50 ms; pulse interval: 50 ms; number of pulses: 5; decay rate: 40%; polarity: +/–). Thirty embryos were then transferred into the oviducts of one pseudopregnant female mouse.
Two F0 mice obtained were genotyped using PCR-based analyses; genomic DNA extracted from mouse tails was used. PCR was performed with a 20-μL reaction mixture containing 100 μg of cDNA, 10 μL of KOD One™ PCR Master Mix (catalog number: KMM-101; Toyobo, Osaka, Japan), and 0.6 μL of 100 μM of each primer in water. The denaturation process at 94°C for 2 min was followed by 35 cycles at 98°C for 10 s, 60°C for 5 s, and 68°C for 10 s. DNA sequence analysis was outsourced to GENEWIZ Japan (Tokyo, Japan). The primer sequences were as follows: genotyping primers, 5′-TCCTACCGCTCTCCAGATACC-3′ and 5′-CTAGCAAGGGAAGGCACTCG-3′; and sequencing primers 5′-CGTTCTCCTGACACCACTGT-3′ and 5′-TGGCTGTGCTAACTTTGGCT-3′. To evaluate the decrease in ES1 level, we euthanized a subset of mice via cervical dislocation, removed their brains, collected samples as described in the section “Preparation of mouse tissue extracts for western blotting,” and performed western blotting.
2.7. GLO activity assay in mouse brain mitochondria
We assayed GLO enzyme activity in the mitochondrial soluble fractions obtained from WT and ES1-KO mouse brains using a procedure described by Lee et al. [31]. Mitochondrial soluble fractions were isolated using a mitochondrial isolation kit (KC010100; BioChain Institute) and 100 mM sodium phosphate buffer (pH 7.4) containing 5 mM dithiothreitol (DTT), as described in the section “Mitochondrial isolation from mouse brain tissue for western blotting,” and 50 µg of each fraction was incubated with 0.5 mM GO or 0.75 mM MGO for 4 h at 37°C in a total reaction volume of 82 μL. The reaction was stopped by adding 15 μL of 0.1% 2,4-dinitrophenylhydrazine (DNPH) in 2 N HCl and 34 μL of water. These samples were incubated for 15 min at 25°C, and then 69 μL of 10% NaOH was added. After subsequent incubation for 15 min at 25°C, the absorbance of the samples at 595 nm was measured.
2.8. Mitochondrial activity assay
Mitochondrial activity was assayed using a mitochondrial isolation kit (KC010100; BioChain Institute) and mitochondrial activity assay kit (KC310100; BioChain Institute) following the method established by Chukai et al. [34]. The isolated mitochondria were suspended in mitochondrial storage buffer. First, 850 μL of 1 × enzyme assay buffer was added to a cuvette. Next, the mitochondrial samples were dissolved in enzyme dilution buffer containing 1 × n-dodecyl-β-d-maltoside to 0.1–0.2 mg protein/mL and incubated on ice for 15 min. Two micrograms of mitochondrial sample and enzyme dilution buffer were mixed to a total volume of 100 μL, added to the cuvette, and mixed via inversion. Subsequently, 50 μL of ferrocytochrome c substrate solution was added to the cuvette and mixed via cuvette inversion. The amount of enzyme needed to oxidize 1 μmol of ferrocytochrome c per min was defined as one unit of activity. The absorption of samples at 550 nm was immediately measured and cytochrome c oxidase activity was calculated according to the manufacturer's instructions.
2.9. Transmission electron microscopy (TEM)
TEM was performed to compare the mitochondrial morphological features of WT and ES1-KO mice. The method of sampling was the same as that reported by Ozaki et al. [35]. The brains were obtained from WT and ES1-KO mice and stored in a fixative solution (2.5% glutaraldehyde, 2% paraformaldehyde, 0.1 M sodium phosphate buffer, pH 7.4) overnight at 4°C. The fixed brains were washed two times with 0.1 M sodium phosphate buffer (pH 7.4). The hippocampus and cerebral cortex were excised from the fixed brains, post-fixed with 1% osmium tetroxide for 2 h, dehydrated with serial dilutions of ethanol (50%, 70%, 80%, 90%, and 100%) for 15 min at each concentration, and then embedded in Epon 812 (TAAB Laboratory Equipment Ltd., Aldermaston, England). Ultrathin sections (100 nm) were prepared with an ultramicrotome (EM-UC6; Leica Microsystems, Wetzlar, Germany) and stained with 1% uranyl acetate and lead citrate. The images were captured using a transmission electron microscope (H-7650; Hitachi, Tokyo, Japan).
2.10. Proteomic analysis
Mitochondrial and cytosolic fractions were isolated from 5 WT and 5 ES1-KO mouse brains in 1% Triton X-100, 20 mM Tris-HCl, pH 7.4, as described in the section “Mitochondrial isolation from mouse brain tissue for western blotting.” The samples were concentrated using SDS-PAGE, stained with Coomassie Blue, and then excised from the gels. The gel sections were finely chopped and washed with water. The gel pieces were then destained with 50% methanol and vortexed for 5 min in 25 mM ammonium bicarbonate containing 50% acetonitrile. The supernatant was removed, and the gel pieces were soaked in 100% acetonitrile at 25°C for 1 min. After removing the solution, the gel pieces were dried under vacuum, rehydrated with a reducing solution (50 mM ammonium bicarbonate and 25 mM DTT), and incubated for 40 min at 56°C. After removing the reducing buffer, the gel pieces were immersed in an alkylating solution (50 mM ammonium bicarbonate containing 55 mM iodoacetamide) for 30 min at 25°C in the dark. The alkylating solution was then removed, and the gel pieces were washed twice with water, vortexed for 5 min in 25 mM ammonium bicarbonate containing 50% acetonitrile, and soaked in 100% acetonitrile for 1 min. After removing the solution, the gel pieces were evaporated to dryness under vacuum and rehydrated using 50 mM ammonium bicarbonate containing 4 ng/μL sequencing-grade trypsin and 0.01% ProteaseMAX surfactant (catalog number: V2071; Promega, Madison, WI, USA). An equivalent volume of 50 mM ammonium bicarbonate containing 0.01% ProteaseMAX surfactant was then added to the mixture, which was then incubated for 3 h at 37°C to digest the proteins in the gels. An equivalent volume of 1% trifluoroacetic acid was added, and the mixture was vortexed for 10 min to extract the tryptic fragments. After centrifuging the mixture at 12,000 × g for 15 min at 4°C, the supernatant containing the peptides was collected. The peptide samples were desalted using C18 ZipTips (Millipore). The ZipTip eluates were dried by vacuum. The dried samples were rehydrated in 0.1% formic acid and analyzed using LC-MS/MS with a nanoLC Eksigent 400 system (Eksigent, AB Sciex, Tokyo, Japan) coupled online to a TripleTOF 6600 mass spectrometer (AB Sciex), as described previously [36].
Spectra acquired with data-dependent acquisition were searched against the UniProt reviewed database using the Paragon Algorithm built into the ProteinPilot 5.0.1 software program (AB Sciex). Proteins and peptides that achieved 1% local false discovery rate (FDR) for the identified proteins and peptides were considered positive identifications. The obtained group file was loaded onto PeakView (v2.2.0; AB Sciex). The files were then exported to the MarkerView software program (version 1.3.0.1; AB Sciex), and the peak areas of individual peptides were normalized to the sum of the peak areas of all detected peptides. Principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) were used for multivariate analyses; they were performed using the SIMCA software program (SIMCA 15; Infocom Corp., Tokyo, Japan). Pareto scaling was applied to the normalized peak area values. Canonical pathways and networks of significantly regulated proteins (p < 0.05) were generated using Ingenuity Pathway Analysis (IPA; Qiagen, Hilden, Germany). Mitochondrial proteins were selected using Gene Ontology terms classified by the Gene Ontology Consortium (http://www.geneontology.org/), and their UniProt IDs and fold changes were input into IPA.
2.11. Expression of recombinant mouse ES1 protein
To construct the plasmid, ES1 cDNA without the mitochondrial localization signal was isolated from total mouse brain RNA via RT-PCR. The total RNA was extracted from mouse brains using TRI Reagent (catalog number: TR118; Molecular Research Center, Inc., OH, USA). The mouse brains were homogenized in TRI Reagent (1 mL/100 mg brains) using a polytron homogenizer. The homogenate was treated with chloroform (0.2 mL/1 mL TRI Reagent) and centrifuged at 10,000 × g for 15 min at 4°C. The aqueous phase was transferred to a fresh tube and mixed with isopropanol (0.5 mL/1 mL TRI Reagent). The mixture was stored at 25°C for 10 min and centrifuged at 10,000 × g for 20 min at 4°C. The supernatant was removed, and the RNA pellet was washed with 70% ethanol and subjected to subsequent centrifugation at 10,000 × g for 5 min at 4°C. The ethanol was removed, and the RNA pellet was air-dried and dissolved in RNase-free water. The purified RNAs were reverse-transcribed using ReverTra Ace™ (catalog number: TRT-101; Toyobo, Osaka, Japan). The primer set was designed using Online In-Fusion Tools (Takara Bio Inc.; https://www.takara-bio.co.jp/research/infusion_primer/infusion_primer_form.php) to amplify the mES1 cDNA without the mitochondrial localization signal (1–117 bp). The primer sequences were as follows: 5′-GTGCGGCCGCAAGCTTTACTTTCCCGTGAGTTCC-3′ (forward primer) / 5′-CGCGCGGCAGCCATATTGGGGCCAGGGTGGCGT-3′ (reverse primer). PCR was performed with a 30-µL mixture containing 1 µL of cDNA, 15 µL of KOD One™ PCR Master Mix (KMM-101; Toyobo), 0.9 µL of 100 µM of each primer, and 12.2 µL of Milli-Q water. The initial denaturation step at 94°C for 2 min was followed by 30 cycles at 98°C for 10 s, 60°C for 5 s, and 68°C for 10 s.
The PCR product was purified using the Wizard SV Gel and PCR Clean-Up System (catalog number: A9282; Promega, Madison, WI, USA). The purified PCR product was inserted into the HindIII and NdeI-linearized pET-28a(+) vector (Novagen) using the In-Fusion HD Cloning Kit (catalog number 639648; Clontech Takara Cellartis, CA, USA). The plasmid construct was transformed into Escherichia coli strain BL21 (DE3) using the heat shock method. The presence of the insert was confirmed via restriction enzyme digestion.
2.12. Purification of recombinant mouse ES1 protein
The N-terminal 6 His-tagged protein was expressed in BL21 (DE3). BL21 (DE3) containing the constructed plasmid was incubated in 400 mL of LB broth (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) containing 0.03 mg/mL kanamycin at 200 rpm until the OD600 reached 0.7 in a shaker at 37°C. Next, isopropyl-β-D-thiogalactoside (IPTG, 1 mM) was added, and the mixture was further incubated at 200 rpm for 4 h at 37°C. After centrifuging at 5,800 × g for 3 min at 4 °C, the cells were sonicated (TOMY; Ultrasonic disruptor UD-100; output level: 40, 10 s × 15) in lysis buffer (100 mM sodium phosphate, 500 mM NaCl, pH 7.4) and centrifuged at 20,600 × g for 15 min at 4°C. The supernatant was loaded onto a TALON® Metal Affinity Resin (catalog number: 635502; Clontech Laboratories, Inc., CA, USA). The column containing the N-terminal 6 His-tagged protein was eluted with elution buffer (100 mM sodium phosphate, 500 mM NaCl, 10-200 mM imidazole, pH 7.4). The purity of recombinant mouse ES1 protein was confirmed using Coomassie Brilliant Blue (CBB) staining. The purified proteins were dialyzed using 100 mM sodium phosphate buffer (pH 7.4).
2.13. GLO activity assay for recombinant mouse ES1 protein
We measured the GLO activity of the recombinant mouse ES1 protein using the procedure described by Lee et al. [31] as well as the procedure described in the section "GLO activity assay for mouse brain." Initially, 100 µg of purified recombinant mouse ES1 protein was incubated with 0.5 mM GO or 0.75 mM MGO for 1 h at 42°C in a total reaction volume of 500 μL using 100 mM sodium phosphate buffer (pH 7.4). Subsequently, 90 µL of 0.1% 2,4-dinitrophenylhydrazine (DNPH) in 2 N HCl and 210 µL of water were added to stop the reaction. These samples were incubated at 25°C for 15 min and diluted 10-fold to a total of 800 µL per sample with a diluent (500 µL of sodium phosphate buffer + 90 µL of 0.1% DNPH + 210 µL of water). Thereafter, 420 μL of 10% NaOH was added. The samples were incubated at 25°C for 15 min and their absorbance at 595 nm was measured.
2.14. High-performance liquid chromatography (HPLC)
To determine the reaction products from ES1 activity, HPLC was performed according to the method of Yamada et al. [37]. Briefly, recombinant mouse ES1 protein was reacted with GO following the protocol described in the section "GLO activity assay for recombinant mouse ES1 protein." After incubation at 42°C for 1 h, glycolic acid in the reaction mixture was determined using HPLC with an ULTRON PS-80H column (Shinwa Chemical Industries, Tokyo, Japan). The elution was performed with perchloric acid solution (pH 2.1) at a flow rate of 1.0 mL/min at 60°C for 20 min. The elution peak of glycolic acid was at 9.8 min.
2.15. Statistical analysis
The results of cytochrome c oxidase and GLO activity assays were compared using Student's t-test. A p-value < 0.05 or 0.01 was used to indicate statistical significance. All data were analyzed using Statistical Package for the Social Sciences (SPSS) 28.0 (IBM Corp., Armonk, NY, USA).
3. Results
3.1. Presence of ES1 in the mitochondrial soluble fraction of mouse tissues
To investigate ES1 tissue distribution, western blotting of mouse tissue extracts was performed with an ES1-specific antibody. The ES1 band was detected in the brain, heart, lung, liver, stomach, spleen, intestine, kidney, and muscle samples, implying that ES1 is widely expressed in mouse bodies (Fig. 1a). ES1 antibody specificity was confirmed by blocking antibody binding with the antigen peptide (10 μg/mL) (Fig. 1b). As we found that ES1 was expressed in the brain, subsequent experiments were performed using brain tissue.
Fig. 1.
ES1 tissue distribution and subcellular localization in mice. (a) ES1 -expressed in the extract of each mouse tissue was analyzed (40 μg proteins/lane). (b) Verification of antibody specificity. The ES1 band disappeared by competitively inhibiting antibody binding with the antigen peptide. COX4 was used as the loading control for mitochondria. (c) Subcellular localization of ES1 in the mouse brain. The cytosolic fraction (Cytosol) contained lysosomes and microsomes in addition to soluble component in the cytosol. Mitochondria were separated into the following two fractions: OM + IM and intermembrane space (IMS) + matrix (MAT). The purity of the mitochondrial membrane (OM + IM) and soluble (IMS + MAT) fractions was determined using marker proteins (porin, mitochondrial membrane; Ak2, mitochondrial soluble fraction [10 μg proteins/lane]). ES1 was mainly localized in the mitochondrial soluble fractions. M, molecular weight marker; OM, outer membrane; IM, inner membrane.
To investigate ES1 subcellular localization, we performed western blotting of ES1 on mitochondria isolated from the mouse brain. The mitochondria were separated into a soluble fraction, which consisted of the IMS and MAT, and membrane fraction, which consisted of the OM and IM. Fraction purity was confirmed using antibodies against marker proteins. The western blots revealed that ES1 was mainly localized in the mitochondrial soluble fraction (Fig. 1c, IMS+MAT lane).
3.2. Evaluation of ES1-KO mice
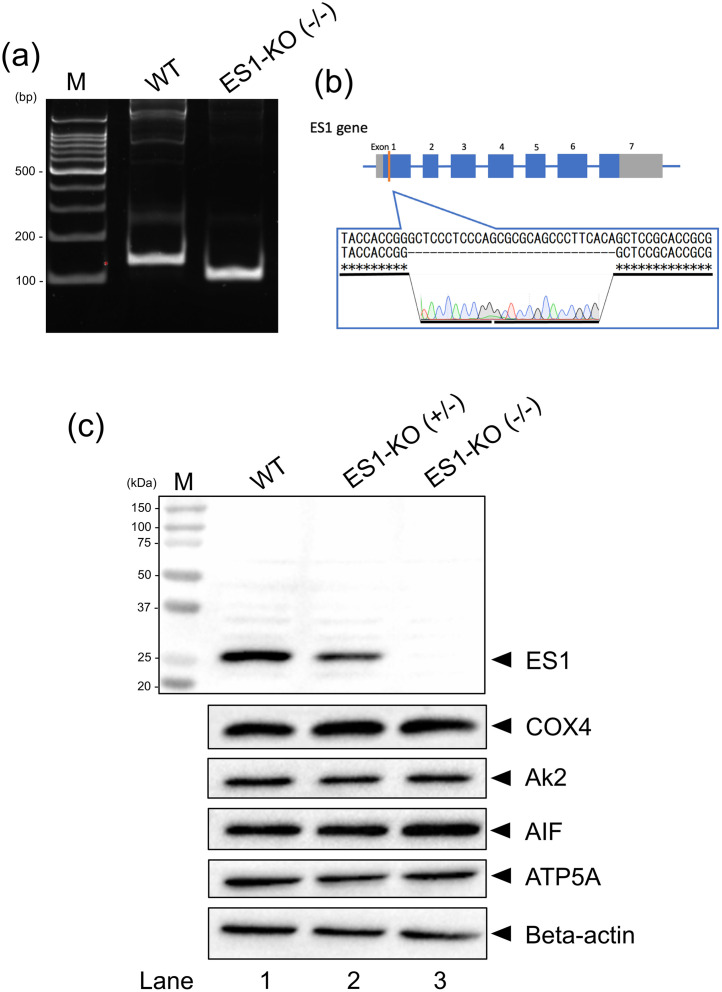
We generated ES1-KO mice to investigate the function of ES1. To ensure the loss of ES1 expression and function, exon 1 of the ES1 (Gatd3a) was targeted to maximize the effect of the frameshift mutation. Two CRISPR/Cas9-edited F0 mice were obtained from the 30 transferred embryos. F1 mice were obtained by mating the F0 mice with WT mice. Next, homozygous ES1-KO mice were established by backcrossing with the F0 mice. Both male and female homozygous ES1-KO mice showed normal growth and reproductive behaviors and normal phenotypes. The ES1-KO mice obtained were genotyped using PCR (Fig. 2a), and the PCR products were analyzed via DNA sequencing (Fig. 2b). Western blotting confirmed the absence of ES1 in the brains of the KO mice (Fig. 2c). The mitochondrial and cytosolic marker proteins did not show any significant changes and were used as loading controls.
Fig. 2.
ES1 deficiency in knockout mice. (a) Identification of the mouse genotype using PCR analysis. The PCR products approximately 140 and 110 bp in size indicate the presence of the WT+/+ and KO−/− alleles, respectively. (b) DNA sequencing of PCR products. A 28-bp fragment deletion was confirmed in ES1-KO mice. (c) ES1 protein level in the brains of WT, heterozygote, and KO mice. ES1 deficiency was confirmed in ES1-KO−/− mice (40 µg proteins/lane). Anti-COX4, anti-Ak2, anti-AIF, and anti-ATP5A antibodies were used as mitochondrial markers. Anti-beta-actin antibody was used as the cytosolic marker. M, molecular weight marker; KO, knockout; WT, wildtype.
3.3. Mitochondrial GLO activity in the ES1-KO mouse brain
To analyze the GLO3 activity of ES1, we compared the GLO activity in ES1-KO mice with that in WT mice by using the DNPH assay. GO and MGO were used as substrates. To avoid the contribution of glutathione (GSH)-requiring GLO1, GLO activity was measured in the mitochondrial soluble fraction that had been separated in sodium phosphate buffer containing 5 mM DTT. The metabolic activity toward GO in the mitochondria of ES1-KO mice significantly decreased compared with that in the mitochondria of WT mice (n = 3; WT, 3.03 ± 0.18 × 10−3 unit/mg protein; ES1-KO, 2.54 ± 0.18 × 10−3 unit/mg protein) (Fig. 3a). The metabolic activity toward MGO in the mitochondria of ES1-KO mice slightly increased compared with that in the mitochondria of WT mice (n = 3; WT, 3.12 ± 0.11 × 10−3 unit/mg protein; ES1-KO, 3.38 ± 0.03 × 10−3 unit/mg protein) (Fig. 3b).
Fig. 3.
Glyoxalase activity of brain mitochondria from WT and ES1-KO mice. (a) Enzyme activity assay results for WT and ES1-KO mice with glyoxal (GO) as the substrate, obtained using the DNPH assay. The metabolic activity of ES1-KO mouse samples was significantly lower than that of the WT mouse samples. Values are presented as mean ± SD (n = 3 for each sample). *p < 0.05, Student's t-test (p = 0.03, WT vs. ES1-KO). (b) Enzyme activity assay results for WT and ES1-KO mice with methylglyoxal (MGO) as the substrate, obtained using the DNPH assay. There was a slightly higher metabolic activity in the ES1-KO mouse samples than in the WT mouse samples. Values are presented as mean ± SD (n = 3 for each sample). *p < 0.05, Student's t-test (p = 0.017, WT vs. ES1-KO). One unit of activity was defined as the conversion of 1 µmol substrate per min. Glyoxalase activity is expressed as units per milligram of protein. (c) Cytochrome c oxidase activity of the brain mitochondria from WT and ES1-KO mice. ES1-KO mouse samples showed a significant decrease in cytochrome c oxidase activity. Values are presented as mean ± SD (n = 4 for each sample). *p < 0.05, Student's t-test (p = 0.015, WT vs. ES1-KO). One unit of activity was defined as the oxidation of 1 µmol ferrocytochrome c per min. Cytochrome c oxidase activity is expressed as units per milligram of protein. KO, knockout; WT, wildtype.
3.4. Mitochondrial activity in ES1-KO mouse brain
We compared mitochondrial activity in WT and ES1-KO mouse brains by measuring total cytochrome c oxidase activity. This total activity was measured in the mitochondria after solubilizing the membrane. The cytochrome c oxidase activity was significantly decreased by ES1-KO (n = 4; WT, 58.09 ± 10.32 unit/mg; ES1-KO, 37.81 ± 6.22 unit/mg) (Fig. 3c).
3.5. Mitochondrial morphological features in ES1-KO mouse cerebral cortex and hippocampus
To examine the effect of ES1-KO on mitochondrial morphological features, we examined the brain mitochondria of WT and ES1-KO mice using TEM. Mitochondrial fragmentation and swelling, which are characteristic of mitochondrial dysfunction [38,39], were not observed. “Normal” mitochondria with intact cristae organized vertically to the long axis of the mitochondrion and with intact OMs were observed in the cerebral cortex (Fig. 4a–4d) and hippocampus of ES1-KO mice (Fig. 4e–4h).
Fig. 4.
Morphological features of brain mitochondria from WT and ES1-KO mice. (a) Transmission electron microscopy (TEM) micrograph of the cerebral cortex from WT mice. (b) Magnified view of the boxed area in panel a. (c) TEM micrograph of the cerebral cortex from ES1-KO mice. (d) Magnified view of the boxed area in panel c. (e) TEM micrograph of the hippocampus from WT mice. (f) Magnified view of the boxed area in panel e. (g) TEM micrograph of the hippocampus from ES1-KO mice. (h) Magnified view of the boxed area in panel g. Scale bar: 2 μm. KO, knockout; WT, wildtype.
3.6. Proteomic analysis of the brain of WT and ES1-KO mice
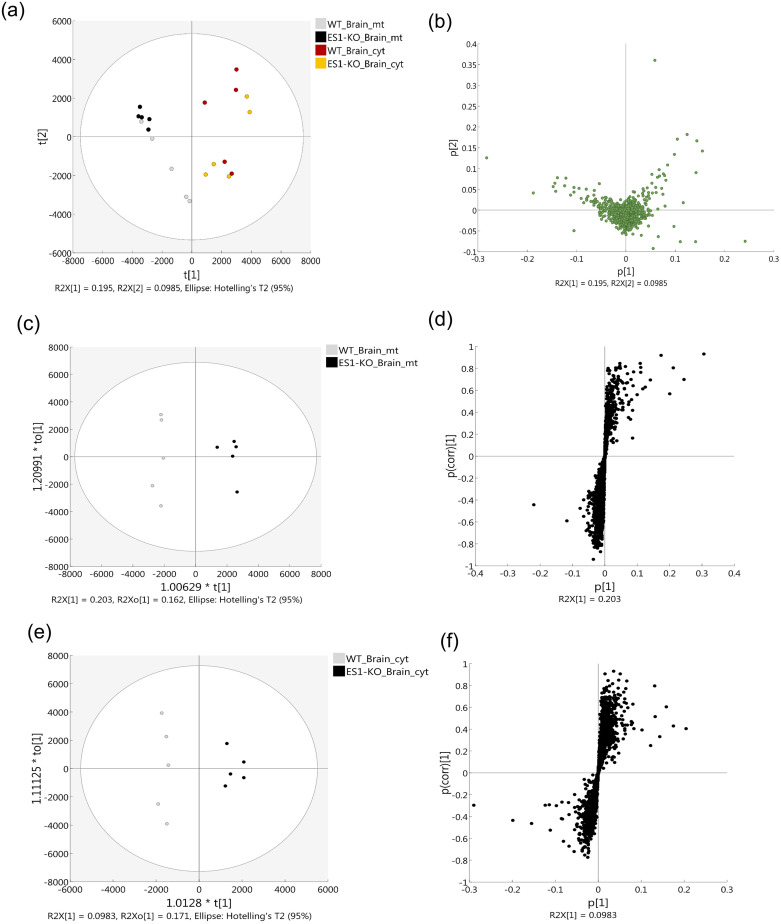
As the abundance of a protein is closely related to its intrinsic properties, such as evolution, structure, and function [40], changes in its abundance appear to reflect changes in metabolic and signaling pathways within the cells or intracellular organelles. Here, we performed a proteomic analysis of brain mitochondria and cytosol of WT and ES1-KO mice in order to elucidate the function of ES1 by capturing the metabolic and signaling pathway alterations that might occur within the cells as a result of ES1 deletion. To obtain an overview of the proteome data, the complete dataset from all mouse brain samples was subjected to PCA. A plot of the first two principal component scores, which accounted for 29.35% (PC1, 19.5%; PC2, 9.85%) of the original variation, is shown in Fig. 5a and 5b. There was some evidence of separation between the cellular component groups and the first principal component. However, the high degree of overlap among the genotype groups (WT and ES1-KO) indicated that the model did not have practical value and suggested that most of the spectral variation was independent of class differences. No large outliers were observed.
Fig. 5.
Score scatter plot (a) and loading plot (b) of all 20 mouse samples for principal component analysis overview. The sample groups are represented as follows: gray, mitochondrial fraction from WT mouse brain; black, mitochondrial fraction from ES1-KO mouse brain; red, cytosolic fraction from WT mouse brain; and yellow, cytosolic fraction from ES1-KO mouse brain. Uncharacterized samples are plotted at the center, and those with features are plotted at a distance from the center. Similar features are plotted close together. Owing to the positional relationship between the score plot and loading plot, the characteristic component of each sample is known. A plot of the first and second principal component scores, which accounted for 29.35% (PC1, 19.5%; PC2, 9.85%) of the original variation, is shown (ellipse, 95% tolerance range for the two scores based on Hotelling's T2). (c–f) Orthogonal partial least squares discriminant analysis (OPLS-DA) model for discriminating the ES1-KO from the WT mouse proteome. OPLS-DA results based on fractionated components from brain proteome data from WT mice vs. ES1-KO mice: (c, d) mitochondrial fraction from the brain and (e, f) cytosolic fraction from the brain. (c, e) Score scatter plots. (d, f) S-plots are presented for each analysis. The ellipse in each score scatter plot indicates the Hotelling's T2 (0.95) range for this model. KO, knockout; WT, wildtype.
The OPLS-DA was used to visualize class separation in the samples. The OPLS-DA model aggregates all identifying information into the first component [41]. The score scatter plots of the OPLS-DA model showed satisfactory separation between the WT and ES1-KO mouse groups while using 1 predictive component and 1 orthogonal component (Fig. 5c and 5e). These results indicate that cellular component proteome profiles could be used to distinguish the ES1-KO group from WT group. The resultant S-plot between the WT and ES1-KO mice in the OPLS-DA model that was developed helped identify highly correlated proteins in the separation of the groups (|p1(corr)| > 0.7); 77 and 33 proteins were identified in the mitochondrial and cytosolic fractions, respectively, from the brain (Fig. 5d, 5f, Tables S1, S2). The relative amount of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) to total protein was higher in the mitochondrial fraction than in the cytosolic fraction.
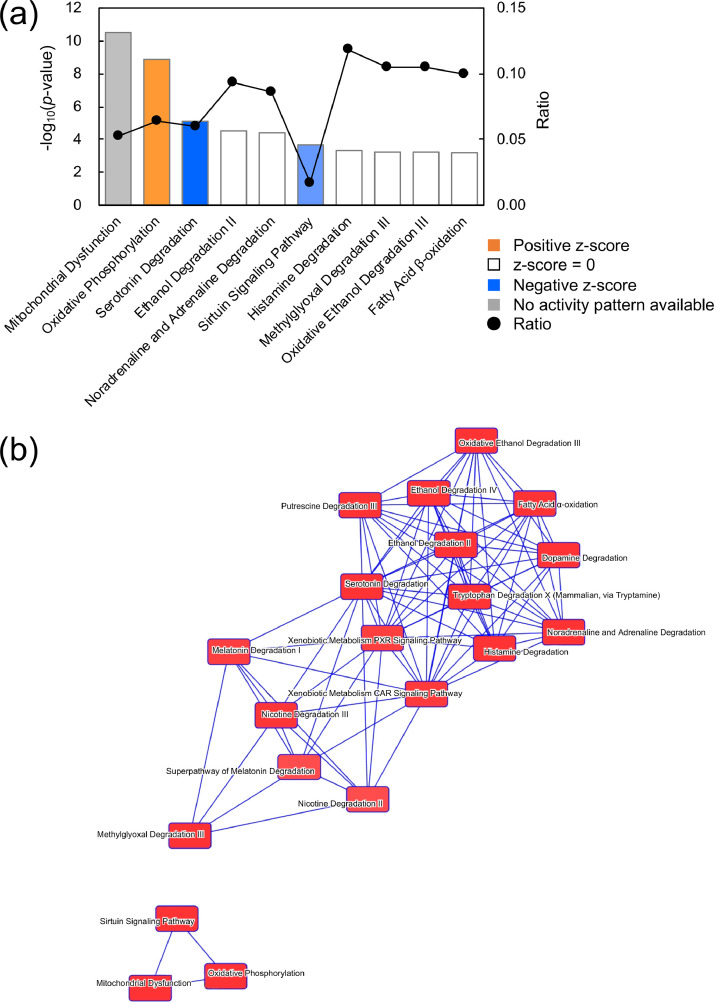
Cytosolic proteins detected in the mitochondrial fraction can be derived from cytosolic fraction contamination. Therefore, among the proteins that showed significant changes in the brain mitochondrial fractions, we extracted proteins that have been confirmed to be localized in mitochondria in the Gene Ontology database (http://www.geneontology.org/). The extracted proteins were subjected to pathway enrichment and network analyses using IPA software, which helped identify and rank the signaling pathways from the IPA library on the basis of IPA-based statistical analysis. The results of the mitochondrial fractions from WT and ES1-KO brains showed 50 pathways, the top 10 of which are shown in Fig. 6a. In particular, oxidative phosphorylation was characterized as being activated with ES1-KO (z-score, 1.89). Overlapping the enriched pathways showed that, in addition to the pathways related to mitochondrial dysfunction-forming clusters, there were overlapping pathways involving the MGO degradation III pathway and pathways sharing aldehyde dehydrogenases (Fig. 6b). The altered proteins were also superimposed on a global molecular network created from the curated information of the IPA Knowledge Base. The network of the proteins was algorithmically generated on the basis of their connectivity (Fig. 6c). This network contained proteins that were mainly involved in mitochondrial dysfunction. These results suggest that ES1-KO may cause mitochondrial dysfunction. The MGO degradation III pathway was also enriched in the canonical pathway (Fig. 6). The two main enzymes (AKR7A2 and CYP2E1) in the MGO degradation III pathway were upregulated in the ES1-KO mice, indicating that this pathway may mitigate the effects of ES1 deletion.
Fig. 6.
Signaling pathways induced by ES1-KO in the mitochondrial fraction of mouse brain. (a) Top 10 canonical pathways enriched by the altered proteins between WT and ES1-KO mouse mitochondrial fraction of the brain. Bars and closed cycles correspond to –log10(p-value) and ratio, respectively. The ratio was calculated by dividing the number of proteins mapped to a pathway from the dataset by the total number of proteins mapped to the canonical pathway. Based on the ratio, the significance of the association between the dataset and the canonical IPA pathways was determined. Using Fisher's exact test, we calculated a p-value determining the probability that the association between the proteins in the dataset and the canonical pathway is not explained by chance alone. Determined pathways that were significantly altered from the cutoff of p < 0.05 are shown. If there was a consistent change with upregulation or downregulation of a pathway, IPA produced a z-score that predicted activation (positive z-score, orange bars) or inhibition (negative z-score, blue bars), which is a statistical measure of the correlation between relationship direction and protein level. Deeper orange shades indicate more upregulated pathways, and deeper blue shades indicate more downregulated pathways; the pale blue shade indicates that the pathway was weakly downregulated. (b) Overlapping of the enriched canonical pathways. Pathways that share proteins are interconnected by blue lines, and the more overlapping pathways are located closer together. We modified the text position and font in the figure generated from IPA to improve visibility. (c) Most significant protein network developed from the crated information of the IPA Knowledge Base. The intensity of the node colors (red and green) indicates the degree of upregulation and downregulation, respectively. Proteins in uncolored nodes were not identified as differentially expressed in our experiment and were merged into the computationally created networks based on the evidence stored in the IPA Knowledge Base. The node shapes denote enzymes (diamonds), subunits (trapezoids), complex/group/others (double circles) and others (circles). Labels indicate the name of the complex or the protein contained in the complex. Nodes classified as mitochondrial dysfunction are highlighted with a magenta frame. (d) Methylglyoxal degradation III pathway in the IPA Knowledge Base. The proteins denoted in pink are the two main enzymes of the MGO degradation III pathway, and their expression was upregulated in the mitochondria of ES1-KO mouse brain in our experiment. We modified the text position, font, and lines in the figure generated from IPA to improve visibility. (e, f) The box plots below the pathway diagram shows the normalized peak areas of AKR7A2 and CYP2E1 comparing ES1-KO with WT mice. The p-values and p1(corr) represent the analysis results of ES1-KO and WT mice using t-test and OPLS-DA, respectively. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; KO, knockout; WT, wildtype.
In summary, these results indicate that the MGO degradation III pathway was upregulated and oxidative phosphorylation-related protein expression was increased to compensate for the decrease in GO metabolism and cytochrome c oxidase activity caused by ES1 deficiency.
3.7. Purified recombinant mouse ES1 protein
To analyze the function of ES1, we synthesized a recombinant mouse ES1 protein (mES1). The N-terminal 6 His-tagged protein was expressed in BL21 (DE3). The His-tagged mES1 was purified using the Co-affinity column. CBB staining was performed, and a band that appeared to be mES1 was observed in the fraction eluted with 100 mM imidazole (Fig. 7a).
Fig. 7.
Expression and glyoxalase activity of recombinant mouse ES1 protein. (a) CBB staining of the recombinant protein. mES1 was detected in fractions of 100 mM imidazole. (b) The results of enzymatic activity of control and recombinant mouse ES1 protein (mES1) using GO as the substrate. mES1 metabolic activity was significantly higher than that of the Control. Values are presented as mean ± SD (n = 3 for each sample). *p < 0.01, Student's t-test (p = 0.005, Control vs. mES1). (c) Measurement results when MGO was used as the substrate. Comparison of Control and mES1 showed no significant differences. Values are presented as mean ± SD (n = 3 for each sample). n.s.: not significant, Student's t-test (p = 0.76, Control vs. mES1). One unit of activity was defined as the amount of enzyme used to convert 1 μmol substrate per min. (d) Purified mES1 (100 µg) was mixed with 0.5 mM GO, and mixture was incubated at 42°C for 1 h. The reaction product was characterized using HPLC. (e) Enlarged image of panel d. The peak of glycolic acid was detected only in the mES1 sample.
3.8. GLO activity in the recombinant mouse ES1 protein
To further elucidate that ES1 exhibits GLO activity, the recombinant protein was produced and GLO activity was measured. As a control, we used mES1 inactivated by heat denaturation. The results showed metabolic activity against GO (n = 3; Control, 4.32 ± 2.25 × 10−3 unit/mg protein; mES1, 15.03 ± 2.38 × 10−3 unit/mg protein) (Fig. 7b). The metabolic activity of MGO was not observed (n = 3; Control, 5.49 ± 1.21 × 10−3 unit/mg protein; mES1, 5.21 ± 0.81 × 10−3 unit/mg protein) (Fig. 7c). This specificity was similar to that of elbB. The HPLC analysis revealed that the reaction product of the metabolic activity of ES1 was glycolic acid (Fig. 7d and 7e). The ratio of GO metabolized to glycolic acid produced was approximately 2:1 (metabolism of GO, 128.45 ± 3.54 µM/100 µg proteins; production of glycolic acid, 55.94 ± 10.19 µM/100 µg proteins). The low amount of glycolic acid produced relative to the amount of GO metabolized suggests that another metabolite might be produced. This finding warrants further analysis.
4. Discussion
The current study revealed that ES1 possesses GLO3 activity and metabolizes GO specifically in the mitochondria. ES1-KO decreased the function of mitochondria, which are predicted to be dysfunctional upon glycation. Thus, we have demonstrated a novel metabolic pathway for reactive dicarbonyls in the mitochondria (Fig. 8).
Fig. 8.
Schematic diagram of a novel dicarbonyl metabolic pathway in the mitochondria. In this pathway, GLO1 and GLO2 convert GO to glycolic acid in a GSH-dependent manner in the cytoplasm. ES1, a mitochondrial soluble protein, metabolizes GO, which enters the mitochondria, in a GSH-independent manner. GO, glyoxal; GLO, glyoxalase; IMS, intermembrane space; OM, outer membrane.
ES1 was expressed in all tissues examined. These results are consistent with the information available in the National Center for Biotechnology Information (NCBI) database (Gene cluster; https://www.ncbi.nlm.nih.gov/gene). Western blotting showed that ES1 was localized in the mitochondrial soluble fraction. Previous studies have indicated that ES1 is a soluble mitochondrial protein in zebrafish and pigs [29,42]; the present study results are consistent with those of the previous studies.
Here, we found that in the mitochondria, the metabolic activity toward GO was significantly lower, but the metabolic activity toward MGO was slightly higher in ES1-KO mice than in WT mice. In the recombinant mouse ES1 protein, only metabolic activity toward GO was detected. A previous study showed that elbB can metabolize GO but not MGO, and that ES1 possesses properties similar to those of elbB [24]. Our proteomic analysis indicated the upregulation of cytochrome P450 2E1 (CYP2E1) and aflatoxin B1 aldehyde reductase member 2 (AKR7A2), which are involved in the MGO degradation III pathway, in ES1-KO mice (Table S1). The methylglyoxal degradation III pathway metabolizes only MGO, with aldo-keto reductase and aldose reductase as the major enzymes. We postulate that these proteins contributed to the increase in activity against MGO in ES1-KO mice.
Several studies have reported that glycation stress in mitochondria affects the electron transport chain and causes mitochondrial dysfunction [19,[43], [44], [45]]. In the current study, mitochondrial cytochrome c oxidase activity was found to decrease in ES1-KO mice, suggesting that the mitochondrial electron transport chain activity was decreased. In addition, the canonical pathway analysis showed that ES1-KO caused the upregulation of proteins involved in oxidative phosphorylation and that thioredoxin reductase 2 (TXNRD2), which is involved in mitochondrial dysfunction [46], was downregulated. On the basis of these results, ES1 protects mitochondria from dicarbonyls, and mitochondrial dysfunction is caused by glycation stress and the associated oxidative stress in the mitochondria of ES1-KO mice. However, TEM analysis of the mitochondria showed that ES1-KO did not significantly affect mitochondrial morphological features (Fig. 4). These results suggest that glycation stress in ES1-KO mice was mild and not sufficient to alter mitochondrial morphological features.
The proteomic data showed an increased relative amount of GAPDH in ES1-KO mouse mitochondria (Table S1, p1(corr) = 0.93 [OPLS-DA]). GAPDH is mainly localized in the cytosol, and GAPDH mitochondrial levels are low under basal conditions. However, a previous study indicated that GAPDH is imported into mitochondria when apoptosis-causing treatments/agents, such as serum deprivation, DNA-damaging agents, and mitochondriotoxic compounds, are used [47]. The translocation of GAPDH to mitochondria has been reported to cause a decrease in the inner transmembrane potential, matrix swelling, mitochondrial IM permeabilization, and the release of two proapoptotic proteins, namely cytochrome c and AIF [47,48]. Therefore, the increase in GAPDH in ES1-KO mouse mitochondria was considered to be because of the transfer of GAPDH to the mitochondria. Another study also indicated that GAPDH aggregates are formed in mitochondrial fractions under oxidative stress and induce disruption of the mitochondrial membrane potential and mitochondrial dysfunction [49]. On the basis of our results and those from previous studies, we postulate that ES1-KO causes a small increase in glycation stress and oxidative stress and leads to GAPDH translocation to mitochondria, decreased mitochondrial electron transport chain activity, and mitochondrial dysfunction.
Our results suggest that ES1 possesses GLO3 activity and modulates the metabolism of reactive dicarbonyls in the mitochondria. ES1 could protect mitochondrial proteins against dicarbonyl stress. Thus, the current study reveals a novel metabolic pathway for reactive dicarbonyls in mitochondria. In our previous studies, we produced peptide compounds that modulate the activity of mitochondrial proteins [50], [51], [52]. In the future, we aim to develop compounds that promote the activity of ES1, leading to the development of new therapeutic methods for glycation-related diseases.
Funding
The work was partially supported by the Uehara Memorial Foundation [202020134 (for T.O.)].
Role of the sponsor
The funding organization played no role in the study design and conduction; data collection, management, analysis, and interpretation; and manuscript preparation or review. The funding organization played no role in the approval of the manuscript or in the decision to submit the manuscript for publication.
Data availability statement
The proteomic data are available online at the ProteomeXchange consortium under accession number PXD025954 [53] and the jPOST repository under accession number JPST001170 [54]. The other data are available from the corresponding author, Taku Ozaki, on reasonable request.
Supplementary material
This article contains supplementary material.
Declaration of Competing Interest
None.
Acknowledgments
We would like to thank Ms. Satomi Fujisaki for her assistance in creating the illustrations, and Mr. Shinto Utsumi for technical assistance. We are grateful to Ms. Miyu Miyazaki at the Center for Scientific Equipment Management, Hirosaki University Graduate School of Medicine, for her assistance with the LC-MS/MS analysis.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.bbadva.2023.100092.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- 1.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 2.Rabbani N., Thornalley P.J. Glyoxalase in diabetes, obesity and related disorders. Semin. Cell Dev. Biol. 2011;22:309–317. doi: 10.1016/j.semcdb.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Xue M., Rabbani N., Thornalley P.J. Glyoxalase in ageing. Semin. Cell Dev. Biol. 2011;22:293–301. doi: 10.1016/j.semcdb.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Brownlee M. Advanced protein glycosylation in diabetes and aging. Annu. Rev. Med. 1995;46:223–234. doi: 10.1146/annurev.med.46.1.223. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhuri J., Bains Y., Guha S., Kahn A., Hall D., Bose N., Gugliucci A., Kapahi P. The role of advanced glycation end products in aging and metabolic diseases: bridging association and causality. Cell Metab. 2018;28:337–352. doi: 10.1016/j.cmet.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lüth H.J., Ogunlade V., Kuhla B., Kientsch-Engel R., Stahl P., Webster J., Arendt T., Münch G. Age- and stage-dependent accumulation of advanced glycation end products in intracellular deposits in normal and Alzheimer's disease brains. Cereb. Cortex. 2005;15:211–220. doi: 10.1093/cercor/bhh123. [DOI] [PubMed] [Google Scholar]
- 7.Maessen D.E., Stehouwer C.D., Schalkwijk C.G. The role of methylglyoxal and the glyoxalase system in diabetes and other age-related diseases. Clin. Sci. (Lond.). 2015;128:839–861. doi: 10.1042/CS20140683. [DOI] [PubMed] [Google Scholar]
- 8.Münch G., Lüth H.J., Wong A., Arendt T., Hirsch E., Ravid R., Riederer P. Crosslinking of α-synuclein by advanced glycation endproducts — an early pathophysiological step in Lewy body formation? J. Chem. Neuroanat. 2000;20:253–257. doi: 10.1016/s0891-0618(00)00096-x. [DOI] [PubMed] [Google Scholar]
- 9.Rabbani N., Thornalley P.J. Dicarbonyl stress in cell and tissue dysfunction contributing to ageing and disease. Biochem. Biophys. Res. Commun. 2015;458:221–226. doi: 10.1016/j.bbrc.2015.01.140. [DOI] [PubMed] [Google Scholar]
- 10.Lin J.A., Wu C.H., Lu C.C., Hsia S.M., Yen G.C. Glycative stress from advanced glycation end products (AGEs) and dicarbonyls: an emerging biological factor in cancer onset and progression. Mol. Nutr. Food Res. 2016;60:1850–1864. doi: 10.1002/mnfr.201500759. [DOI] [PubMed] [Google Scholar]
- 11.Thornalley P.J. Dicarbonyl intermediates in the Maillard reaction. Ann. N. Y. Acad. Sci. 2005;1043:111–117. doi: 10.1196/annals.1333.014. [DOI] [PubMed] [Google Scholar]
- 12.Singh V.P., Bali A., Singh N., Jaggi A.S. Advanced glycation end products and diabetic complications. Korean J. Physiol. Pharmacol. 2014;18:1–14. doi: 10.4196/kjpp.2014.18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C., Park C. Bacterial responses to glyoxal and methylglyoxal: reactive electrophilic species. Int. J. Mol. Sci. 2017;18:169. doi: 10.3390/ijms18010169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newsholme P., Gaudel C., Krauce M. Mitochondria and diabetes. An intriguing pathogenetic role. Adv. Exp. Med. Biol. 2012;942:235–247. doi: 10.1007/978-94-007-2869-1_10. https://www.ncbi.nlm.nih.gov/pubmed/22399425 [DOI] [PubMed] [Google Scholar]
- 15.Rosca M.G., Mustata T.G., Kinter M.T., Ozdemir A.M., Kern T.S., Szweda L.I., Brownlee M., Monnier V.M., Weiss M.F. Glycation of mitochondrial proteins from diabetic rat kidney is associated with excess superoxide formation. Am. J. Physiol. Renal. Physiol. 2005;289:F420–F430. doi: 10.1152/ajprenal.00415.2004. https://www.ncbi.nlm.nih.gov/pubmed/15814529 [DOI] [PubMed] [Google Scholar]
- 16.Yoon Y., Galloway C.A., Jhun B.S., Yu T. Mitochondrial dynamics in diabetes. Antioxid. Redox Signal. 2011;14:439–457. doi: 10.1089/ars.2010.3286. https://www.ncbi.nlm.nih.gov/pubmed/20518704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green K., Brand M.D., Murphy M.P. Prevention of mitochondrial oxidative damage as a therapeutic strategy in diabetes. Diabetes. 2004;53:S110–S118. doi: 10.2337/diabetes.53.2007.s110. https://www.ncbi.nlm.nih.gov/pubmed/14749275 [DOI] [PubMed] [Google Scholar]
- 18.Goudarzi M., Kalantari H., Rezaei M. Glyoxal toxicity in isolated rat liver mitochondria. Hum. Exp. Toxicol. 2018;37:532–539. doi: 10.1177/0960327117715900. [DOI] [PubMed] [Google Scholar]
- 19.Pun P.B., Murphy M.P. Pathological significance of mitochondrial glycation. Int. J. Cell Biol. 2012;2012 doi: 10.1155/2012/843505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain M., Nagar P., Sharma A., Batth R., Aggarwal S., Kumari S., Mustafiz A. GLYI and D-LDH play key role in methylglyoxal detoxification and abiotic stress tolerance. Sci. Rep. 2018;8:5451. doi: 10.1038/s41598-018-23806-4. https://www.ncbi.nlm.nih.gov/pubmed/29615695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant A.W., Steel G., Waugh H., Ellis E.M. A novel aldo-keto reductase from Escherichia coli can increase resistance to methylglyoxal toxicity. FEMS Microbiol. Lett. 2003;218:93–99. doi: 10.1111/j.1574-6968.2003.tb11503.x. https://www.ncbi.nlm.nih.gov/pubmed/12583903 [DOI] [PubMed] [Google Scholar]
- 22.Chen C.N., Porubleva L., Shearer G., Svrakic M., Holden L.G., Dover J.L., Johnston M., Chitnis P.R., Kohl D.H. Associating protein activities with their genes: rapid identification of a gene encoding a methylglyoxal reductase in the yeast Saccharomyces cerevisiae. Yeast. 2003;20:545–554. doi: 10.1002/yea.979. https://www.ncbi.nlm.nih.gov/pubmed/12722185 [DOI] [PubMed] [Google Scholar]
- 23.Shin J.H., Weitzdoerfer R., Fountoulakis M., Lubec G. Expression of cystathionine beta-synthase, pyridoxal kinase, and ES1 protein homolog (mitochondrial precursor) in fetal Down syndrome brain. Neurochem. Int. 2004;45:73–79. doi: 10.1016/j.neuint.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Lee C., Lee J., Lee J.Y., Park C. Characterization of the Escherichia coli YajL, YhbO and ElbB glyoxalases. FEMS Microbiol. Lett. 2016:363. doi: 10.1093/femsle/fnv239. [DOI] [PubMed] [Google Scholar]
- 25.Subedi K.P., Choi D., Kim I., Min B., Park C. Hsp31 of Escherichia coli K-12 is glyoxalase III. Mol. Microbiol. 2011;81:926–936. doi: 10.1111/j.1365-2958.2011.07736.x. [DOI] [PubMed] [Google Scholar]
- 26.Thornalley P.J. The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem. J. 1990;269:1–11. doi: 10.1042/bj2690001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Misra K., Banerjee A.B., Ray S., Ray M. Glyoxalase III from Escherichia coli: a single novel enzyme for the conversion of methylglyoxal into D-lactate without reduced glutathione. Biochem. J. 1995;305:999–1003. doi: 10.1042/bj3050999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sousa Silva M., Gomes R.A., Ferreira A.E., Ponces Freire A., Cordeiro C. The glyoxalase pathway: the first hundred years... and beyond. Biochem. J. 2013;453:1–15. doi: 10.1042/BJ20121743. [DOI] [PubMed] [Google Scholar]
- 29.Utsumi S., Sakamoto K., Yamashita T., Tomita H., Sugano E., Ishida K., Ishiyama E., Ozaki T. Presence of ES1 homolog in the mitochondrial intermembrane space of porcine retinal cells. Biochem. Biophys. Res. Commun. 2020;524:542–548. doi: 10.1016/j.bbrc.2020.01.127. [DOI] [PubMed] [Google Scholar]
- 30.Morcos M., Du X., Pfisterer F., Hutter H., Sayed A.A., Thornalley P., Ahmed N., Baynes J., Thorpe S., Kukudov G., Schlotterer A., Bozorgmehr F., El Baki R.A., Stern D., Moehrlen F., Ibrahim Y., Oikonomou D., Hamann A., Becker C., Zeier M., Schwenger V., Miftari N., Humpert P., Hammes H.P., Buechler M., Bierhaus A., Brownlee M., Nawroth P.P. Glyoxalase-1 prevents mitochondrial protein modification and enhances lifespan in Caenorhabditis elegans. Aging Cell. 2008;7:260–269. doi: 10.1111/j.1474-9726.2008.00371.x. [DOI] [PubMed] [Google Scholar]
- 31.Lee J.Y., Song J., Kwon K., Jang S., Kim C., Baek K., Kim J., Park C. Human DJ-1 and its homologs are novel glyoxalases. Hum. Mol. Genet. 2012;21:3215–3225. doi: 10.1093/hmg/dds155. [DOI] [PubMed] [Google Scholar]
- 32.Ozaki T., Yamashita T., Ishiguro S. ERp57-associated mitochondrial μ-calpain truncates apoptosis-inducing factor. Biochim. Biophys. Acta. 2008;1783:1955–1963. doi: 10.1016/j.bbamcr.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 33.Wake Y., Kaneko T. Production of genome-edited mice by visualization of nucleases introduced into the embryos using electroporation. J. Reprod. Dev. 2020;66:469–473. doi: 10.1262/jrd.2020-068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chukai Y., Iwamoto T., Itoh K., Tomita H., Ozaki T. Characterization of mitochondrial calpain-5. Biochim. Biophys. Acta Mol. Cell Res. 2021;1868 doi: 10.1016/j.bbamcr.2021.118989. [DOI] [PubMed] [Google Scholar]
- 35.Ozaki T., Utsumi S., Iwamoto T., Tanaka M., Tomita H., Sugano E., Ishiyama E., Ishida K. Data on mitochondrial ultrastructure of photoreceptors in pig, rabbit, and mouse retinas. Data Brief. 2020;30 doi: 10.1016/j.dib.2020.105544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Almadanim M.C., Gonçalves N.M., Rosa M.T.G., Alexandre B.M., Cordeiro A.M., Rodrigues M., Saibo N.J.M., Soares C.M., Romão C.V., Oliveira M.M., Abreu I.A. The rice cold-responsive calcium-dependent protein kinase OsCPK17 is regulated by alternative splicing and post-translational modifications. Biochim. Biophys. Acta. Mol. Cell Res. 2018;1865:231–246. doi: 10.1016/j.bbamcr.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Yamada M., Higashiyama T., Kishino S., Kataoka M., Ogawa J., Shimizu S., Isobe K. Novel alcohol oxidase with glycolate oxidase activity from Ochrobactrum sp. AIU 033. J. Mol. Catal. B Enzym. 2014;105:41–48. doi: 10.1016/j.molcatb.2014.03.022. [DOI] [Google Scholar]
- 38.Heidari R. Brain mitochondria as potential therapeutic targets for managing hepatic encephalopathy. Life Sci. 2019;218:65–80. doi: 10.1016/j.lfs.2018.12.030. [DOI] [PubMed] [Google Scholar]
- 39.Kim D., Roy S. Effects of diabetes on mitochondrial morphology and its implications in diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 2020;61:10. doi: 10.1167/iovs.61.10.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong F., Yang D., Hao Y., Lin C., Jiang Y., Ying W., Wu S., Zhu Y., Liu S., Yang P., Qian X., He F. Regular patterns for proteome-wide distribution of protein abundance across species. PLoS One. 2012;7:e32423. doi: 10.1371/journal.pone.0032423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bylesjö M., Rantalainen M., Cloarec O., Nicholson J.K., Holmes E., Trygg J. OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J. Chemom. 2006;20:341–351. doi: 10.1002/cem.1006. [DOI] [Google Scholar]
- 42.Masuda T., Wada Y., Kawamura S. ES1 is a mitochondrial enlarging factor contributing to form mega-mitochondria in zebrafish cones. Sci. Rep. 2016;6:22360. doi: 10.1038/srep22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pamplona R., Requena J.R., Portero-Otín M., Prat J., Thorpe S.R., Bellmunt M.J. Carboxymethylated phosphatidylethanolamine in mitochondrial membranes of mammals—evidence for intracellular lipid glycoxidation. Eur. J. Biochem. 1998;255:685–689. doi: 10.1046/j.1432-1327.1998.2550685.x. [DOI] [PubMed] [Google Scholar]
- 44.Parker A.R. A single arginine residue is required for the interaction of the electron transferring flavoprotein (ETF) with three of its dehydrogenase partners. Mol. Cell. Biochem. 2003;254:91–100. doi: 10.1023/a:1027349303797. [DOI] [PubMed] [Google Scholar]
- 45.Rabbani N., Thornalley P.J. Dicarbonyls linked to damage in the powerhouse: glycation of mitochondrial proteins and oxidative stress. Biochem. Soc. Trans. 2008;36:1045–1050. doi: 10.1042/BST0361045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandez A., Meechan D.W., Karpinski B.A., Paronett E.M., Bryan C.A., Rutz H.L., Radin E.A., Lubin N., Bonner E.R., Popratiloff A., Rothblat L.A., Maynard T.M., LaMantia A.S. Mitochondrial dysfunction leads to cortical under-connectivity and cognitive impairment. Neuron. 2019;102:1127–1142. doi: 10.1016/j.neuron.2019.04.013. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tristan C., Shahani N., Sedlak T.W., Sawa A. The diverse functions of GAPDH: views from different subcellular compartments. Cell. Signal. 2011;23:317–323. doi: 10.1016/j.cellsig.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarze A., Deniaud A., Bras M.Le, Maillier E., Molle D., Larochette N., Zamzami N., Jan G., Kroemer G., Brenner C. GAPDH, a novel regulator of the pro-apoptotic mitochondrial membrane permeabilization. Oncogene. 2007;26:2606–2620. doi: 10.1038/sj.onc.1210074. [DOI] [PubMed] [Google Scholar]
- 49.Nakajima H., Itakura M., Kubo T., Kaneshige A., Harada N., Izawa T., Azuma Y.T., Kuwamura M., Yamaji R., Takeuchi T. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) aggregation causes mitochondrial dysfunction during oxidative stress-induced cell death. J. Biol. Chem. 2017;292:4727–4742. doi: 10.1074/jbc.M116.759084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozaki T., Yamashita T., Ishiguro S. Mitochondrial m-calpain plays a role in the release of truncated apoptosis-inducing factor from the mitochondria. Biochim. Biophys. Acta. 2009;1793:1848–1859. doi: 10.1016/j.bbamcr.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 51.Ozaki T., Nakazawa M., Yamashita T., Sorimachi H., Hata S., Tomita H., Isago H., Baba A., Ishiguro S. Intravitreal injection or topical eye-drop application of a µ-calpain C2L domain peptide protects against photoreceptor cell death in Royal College of Surgeons' rats, a model of retinitis pigmentosa. Biochim. Biophys. Acta. Mol. Basis Dis. 2012;1822:1783–1795. doi: 10.1016/j.bbadis.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 52.Sugawara M., Abe T., Kasai S., Itoh K., Ozaki T. Calpain-1 C2L domain peptide protects mouse hippocampus-derived neuronal HT22 cells against glutamate-induced oxytosis. Biochem. Biophys. Rep. 2021;27 doi: 10.1016/j.bbrep.2021.101101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deutsch E.W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., Campbell D.S., Bernal-Llinares M., Okuda S., Kawano S., Moritz R.L., Carver J.J., Wang M., Ishihama Y., Bandeira N., Hermjakob H., Vizcaíno J.A. The ProteomeXchange Consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017;45:D1100–D1106. doi: 10.1093/nar/gkw936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okuda S., Watanabe Y., Moriya Y., Kawano S., Yamamoto T., Matsumoto M., Takami T., Kobayashi D., Araki N., Yoshizawa A.C., Tabata T., Sugiyama N., Goto S., Ishihama Y. jPOSTrepo: an international standard data repository for proteomes. Nucleic Acids Res. 2017;45:D1107–D1111. doi: 10.1093/nar/gkw1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomic data are available online at the ProteomeXchange consortium under accession number PXD025954 [53] and the jPOST repository under accession number JPST001170 [54]. The other data are available from the corresponding author, Taku Ozaki, on reasonable request.
Data will be made available on request.