Abstract
The X-box element present within the promoter region of genes belonging to the major histocompatibility complex (MHC) plays a pivotal role in the expression of class II molecules, since it contains the binding sites for several well-characterized transcription factors. We have analyzed a randomly selected compilation of viral genomes for the presence of elements homologous to the X box of the HLA-DRA gene. We found that human immunodeficiency virus type 1 (HIV-1) shows the highest frequency of X-like box elements per 1,000 bases of genome. Within the HIV-1 genome, we found an X-like motif in the TAR region of the HIV-1 long terminal repeat (LTR), a regulative region playing a pivotal role in Tat-induced HIV-1 transcription. The use of a decoy approach for nuclear proteins binding to this element, namely, XMAS (X-like motif activator sequence), performed by transfection of multiple copies of this sequence into cells carrying an integrated LTR-chloramphenicol acetyltransferase construct, suggests that this element binds to nuclear proteins that enhance Tat-induced transcription. In this report we have characterized two proteins, one binding to the XMAS motif and the other to the flanking regions of XMAS. Mobility shift assays performed on crude nuclear extracts or enriched fractions suggest that similar proteins bind to XMAS from HIV-1 and the X box of the HLA-DRA gene. Furthermore, a UV cross-linking assay suggests that one protein of 47 kDa, termed FAX (factor associated with XMAS)-1, binds to the XMAS of HIV-1. The other protein of 56 kDa was termed FAX-2. In a decoy ex vivo experiment, it was found that sequences recognizing both proteins are required to inhibit Tat-induced HIV-1 LTR-driven transcription. Taken together, the data reported in this paper suggest that XMAS and nearby sequences modulate Tat-induced HIV-1 transcription by binding to the X-box-binding proteins FAX-1 and FAX-2. The sequence homology between XMAS and X box is reflected in binding of a common protein, FAX-1, and similar functional roles in gene expression. To our knowledge, this is the first report showing that transcription factors binding to the X box of the MHC class II genes enhance the transcription of HIV-1.
The class II molecules of the major histocompatibility complex (MHC), a class of transmembrane glycoproteins expressed on the cell surface as heterodimers of A and B chains, are very important for immune function (4, 29). These molecules, encoded by the MHC class II genes HLA-DR, -DP, and -DQ, are generally expressed only on a small number of specialized cells, and expression is constitutive for antigen-presenting cells, including B lymphocytes, Langerhans cells, and monocyte-macrophage lineage cells, or can be induced or enhanced by different stimuli, such as gamma interferon (IFN-γ) treatment, in endothelial cells, epithelial cells, fibroblasts, muscle cells, and T lymphocytes (14, 18). MHC class II molecules play a pivotal role in the immune response, as demonstrated by the almost complete absence of circulating CD4+ T cells in MHC class II knockout mice (6, 17). These molecules are involved in acquisition of the mature T-cell repertoire through positive and negative selection in the thymus, in the induction of lymphocyte and macrophage-monocyte activation and differentiation, and in the presentation of processed antigens to CD4+ T lymphocytes (19).
Expression of MHC class II genes is primarily regulated at the level of transcription, via a highly conserved proximal promoter located upstream of the transcription start site that is sufficient to confer both constitutive and inducible expression in transient-transfection experiments (1, 14, 41). Two highly conserved cis sequences, the X and Y boxes, present in all murine and human class II proximal promoters (1, 25) are positive regulators interacting with trans-acting factors required for expression of MHC class II genes (3, 14, 41, 42). The X-box motif interacts with several well-characterized transcription factors, including RFX, RFX1, NF-X, NFXc, hXBP-1, X2BP, Jun/Fos, NF-S, and NF-X3/BCF-1 (29), and for at least one of them, the absence correlates with the class II-negative phenotype in a hereditary immunodeficiency disease termed bare lymphocyte syndrome (10, 33).
The level of transcription of the human immunodeficiency virus type 1 (HIV-1) genes is under control of the long terminal repeat (LTR), and within this region, a trans-activating region (TAR, spanning nucleotides +1 to +80 with respect to the transcription start site at +1) is present and plays a pivotal role in viral genes transcription. In fact, during the earliest phase of infection, the level of viral gene transcription is quite low, but increases sharply in the presence of the Tat trans-activating viral protein (5, 12, 21). The Tat protein binds to a structured TAR RNA element present in all viral transcripts. It has been suggested that Tat overcomes the premature termination of transcription by increasing the rate of transcription initiation and by stabilizing transcription elongation, or through a combination of the two (7, 15, 16, 24, 26, 27, 28, 30, 45). In this process, the TAR element acts as an enhancer of transcription that recruits viral Tat and cellular protein cofactors (20, 21, 23, 39). Although TAR RNA is critical for Tat activation, the role played by TAR DNA in regulating HIV-1 gene expression is far from being completely understood. It has been demonstrated that several nuclear proteins bind to the TAR DNA (13, 32) and also that TAR DNA-binding proteins purified from HeLa cells and T-lymphocyte nuclear extracts by conventional and DNA affinity chromatography are transcription factors playing both activating and repressing roles (13, 32).
In this study we have analyzed a randomly selected compilation of viral genomes for the presence of elements homologous to the X box of the HLA-DRA gene. We have focused attention on the HIV-1 genome, which shows a high frequency of X-box-like elements per 1,000 bases of genome. Within the HIV-1 genome we found an X-like motif in the TAR region of the HIV-1 LTR, a regulatory region playing a pivotal role in Tat-induced viral transcription. Herein we report data demonstrating that this sequence, termed XMAS for X-like motif activator sequence, binds to X-box-binding proteins and participates to the enhancement of Tat-induced HIV-1 gene transcription. A decoy strategy was used in order to demonstrate that XMAS-binding proteins act as transcription factors enhancing Tat-induced viral transcription. This is the first evidence suggesting the involvement of X-box-binding proteins in the modulation of HIV-1 transcription.
MATERIALS AND METHODS
Cell culture conditions and nuclear extract preparation.
Cells were grown in RPMI 1640 medium supplemented with 10% fetal calf serum at 37°C in a humidified atmosphere containing 5% CO2. Nuclear extracts were prepared by the standard procedure (8).
Plasmid constructions and preparation of X-HIV DNA concatamer resin.
Complementary synthetic HIV containing the XMAS motif of HIV-1 (X-HIV) oligonucleotides (sense strand, 5′-CTGGTTAGACCAGATCTGAGCGT-3′, sequence spanning +8 to +31) were combined in phosphate-buffered saline (PBS), heated to 100°C, and annealed at room temperature for 2 h. Concatamers of annealed X-HIV oligonucleotides were prepared by single-step kinase-ligase reaction at 25°C for 16 h, using T4 kinase and T4 ligase (MBI Fermentas, Vilnius, Lithuania). The product of the reaction was blunt-end ligated into the SmaI site of plasmid pUC18, and recombinant plasmids were sequenced. A clone containing 10 annealed X-HIV oligonucleotides was selected and termed pUC-XHIV. Biotinylated X-HIV concatamers were then obtained by PCR using the pUC-XHIV plasmid as the template and sense (5′-end biotinylated) and antisense X-HIV oligonucleotides as the primers. The DNA affinity resin was prepared by extensively coupling the 5′-end biotinylated X-HIV concatamers to 200 μl of streptavidin-conjugated agarose beads in TE buffer (10 mM Tris-Cl [pH 7.6], 1 mM EDTA) for 16 h at 4°C. Then, the DNA affinity resin was equilibrated in binding buffer (20 mM HEPES [pH 7.9], 20% glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 mM dithiothreitol [DTT], 1 μg of leupeptin per ml, 1 μg of aprotinin per ml).
Purification of X-HIV binding proteins.
T-lymphoid Jurkat cells (1010) were collected during the logarithmic phase of growth by centrifugation, washed in PBS, and subjected to the Dignam procedure (8) for nuclear extract preparation. In our hands, 13 mg of protein was obtained. Protein quantification was performed by the standard Bradford method (2). The X-HIV DNA-binding activity was determined by band shift. The nuclear extract was precipitated with ammonium sulfate at 40% saturation and centrifuged for 20 min at 100,000 × g. The supernatant was precipitated with ammonium sulfate at 60% saturation and centrifuged as above. The pellet was dissolved in Dignam's buffer D (20 mM HEPES [pH 7.9], 20% glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM PMSF, 0.5 mM DTT, 1 μg of leupeptin per ml, 1 μg of aprotinin per ml) to a final concentration of about 10 mg/ml and then dialyzed against two changes of buffer D at 4°C and loaded onto a heparin-Sepharose column (HiTrap; Pharmacia) preequilibrated in buffer D. After a washing step with buffer D containing 0.2 M KCl, the proteins bound were stepwise eluted in buffer D containing 0.3, 0.4, 0.5, 0.6, 0.7, or 1 M KCl. Under these conditions, X-HIV DNA-binding activity was eluted in the 0.3 to 0.5 M KCl fractions. These fractions were pooled, concentrated on a Microcon 10 device, and dialyzed against two changes of binding buffer (20 mM Tris-HCl [pH 7.5], 50 mM KCl, 1 mM MgCl2, 0.01% Triton X-100, 1 mM DTT, 0.2 mM EDTA, 5% glycerol, 0.5 mM PMSF, 1 μg of leupeptin per ml, 1 μg of aprotinin per ml) at 4°C. After addition of bovine serum albumin, poly(dI-dC) · poly(dI-dC) and 200 μl (packed volume) of X-HIV DNA concatamer resin, the mixture was incubated for 20 min at room temperature, and the resin was packed by gravity on a chromatographic support. The DNA affinity column was washed twice with 5 volumes of binding buffer, and stepwise elution was performed with buffer D containing, successively, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, and 1 M KCl. Under these conditions, X-HIV DNA-binding activity was eluted between 0.3 and 0.5 M KCl.
DNase I footprinting assay.
For preparation of the footprinting probe, the 259-bp DNA fragment mimicking a region of the HIV-1 LTR was prepared by PCR using 10 ng of plasmid pTZIIICAT as the template DNA and 150 ng of HIV-1-F and HIV-1-R primers (see Fig. 2 for locations of the primers within the HIV-1 LTR) (11). PCR was performed using a 32P-labeled HIV-1-R PCR primer. PCR was performed in 25 μl of 50 mM KCl–10 mM Tris-HCl (pH 8.8)–2.5 mM MgCl2–0.1% Triton X-100 by using 2 U of Taq DNA polymerase (Dynazyme; Finnzymes, Espoo, Finland) per reaction. The cycles were as follows: denaturation for 1 min at 94°C; annealing for 1 min at 60°C; elongation for 1 min at 72°C. The 32P-labeled amplified fragment was analyzed by electrophoresis on a 2% agarose gel, purified by phenol-chloroform extraction, washed through a Microcon 30 (Amicon Inc.-Grace Company, Beverly, Mass.) with 400 μl of water, and dissolved in 100 μl of water.
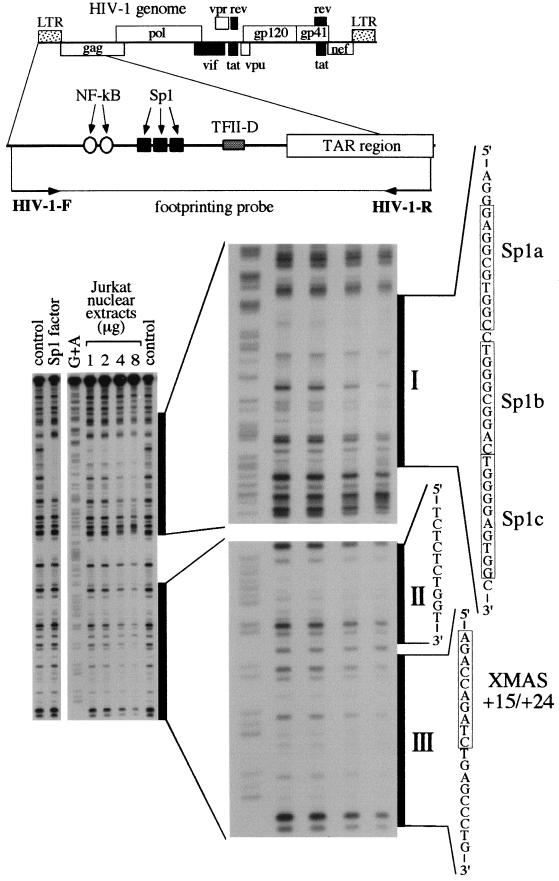
FIG. 2.
Analysis of nuclear protein binding to the HIV-1 LTR region carrying the XMAS motif, performed by DNase I footprinting. The footprinting probe was produced by PCR using the HIV-1-R 32P-labeled primer. The probe was incubated in the absence (control) or in the presence of either recombinant Sp1 protein (Sp1 factor) or increasing amounts of Jurkat cell nuclear extracts. G+A, G+A sequencing reaction performed on the footprinting probe. I, II, and III are three major footprints extending from −78 to −46, +4 to +13, and +15 to +32. The XMAS motif (nucleotides +15 to +24) is also shown.
The experimental conditions for the footprinting assay were as follows. Footprinting reactions were carried out in 50 μl containing 10,000 cpm of 32P-end-labeled DNA, 5% glycerol, 20 mM Tris-HCl (pH 7.5), 50 mM KCl, 1 mM MgCl2, 1 mM DTT, and 0.01% Triton X-100. Fifty microliters of 10 mM MgCl2–5 mM CaCl2 solution was added 1 min before the addition of DNase I. The footprinting reaction was blocked at room temperature by adding 90 μl of 200 mM NaCl–30 mM EDTA–1% sodium dodecyl sulfate (SDS)–100 μg of yeast RNA per ml. Reactions were phenol extracted and precipitated by adding 2.5 volumes of ethanol. The pellets were dissolved in 3 μl of loading dye, denatured for 2 min at 90°C, ice cooled, and layered onto an 8% polyacrylamide–7 M urea sequencing gel. After electrophoresis, gels were vacuum dried and exposed to Kodak X-OMAT films. Maxam-Gilbert G+A sequencing reactions were performed in 10 μl of TE buffer, using 3.6 ng of 32P-end-labeled DNA and 1 μg of calf thymus DNA. One microliter of 4% formic acid (pH 2) was added, and the reaction mixture was incubated for 25 min at 37°C. After the addition of 150 μl of 1 M piperidine and further incubation for 30 min at 90°C, the reaction mixtures were extracted with 1 ml of butanol. The pellets were washed with 150 μl of 1% SDS and 1 ml of butanol. After two additional washes with butanol, the pellets were dried, dissolved in loading dye, and analyzed by electrophoresis on the 8% polyacrylamide–7 M urea sequencing gel.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed by using double-stranded synthetic oligonucleotides mimicking the X-HIV motif present within the TAR region of the HIV-1 LTR (X-HIV mer) or the X box sequence of the HLA-DRA promoter (X-DRA mer). The sense strand sequence of X-DRA mer was 5′-ACCCTTCCCCTAGCAACAGATGCGTCATCT-3′. The synthetic oligonucleotides were 5′-end labeled using [γ-32P]ATP and T4 polynucleotide kinase. Binding was performed in a total volume of 25 μl containing 20 mM Tris-HCl (pH 7.5), 50 mM KCl, 1 mM MgCl2, 0.01% Triton X-100, 1 mM DTT, 0.2 mM EDTA, 5% glycerol, 1 μg of poly(dI-dC) · poly (dI-dC), crude nuclear extract or enriched fractions, and 10,000 cpm of labeled double-stranded oligonucleotides. After 30 min at room temperature, the samples were electrophoresed at constant voltage (200 V for 2 h) on a 6% polyacrylamide gel in 0.25× TBE. The gel was dried and autoradiographed.
UV cross-linking assay.
The double-stranded 32P-labeled X-HIV mer probe was prepared by using [γ-32P]ATP and T4 polynucleotide kinase. Binding conditions were the same as described for the EMSA. Protein samples from crude nuclear extracts or fractions eluted from the DNA affinity column in buffer plus 0.3 to 0.5 M were incubated with 2 ng of the labeled X-HIV DNA (200,000 cpm) probe in a final volume of 25 μl for 30 min at room temperature. The mixture was irradiated for 30 min using a UV transilluminator (254 nm; 7,000 mW/cm2) at a distance of 5 cm from the UV source. The reaction was blocked by addition of 5 μl of 6× Laemmli gel loading buffer and electrophoresed at constant voltage (150 V) on a 15% polyacrylamide–SDS gel. After electrophoresis, the gel was fixed, silver stained when appropriate, and dried under vacuum, and the DNA-binding proteins were identified by autoradiography.
Transfection and CAT assay.
HL3T1 cells, containing integrated copies of the LTR-chloramphenicol acetyltransferase (CAT) retroviral construct, were maintained on complete RPMI 1640 containing penicillin and streptomycin. Cells were split on the day prior to transfection so that each well of a six-well plate was 50 to 70% confluent at the time of transfection. Using 10 μl of Lipofectin reagent (Gibco-BRL-Life Technologies, Milan, Italy) per well, 2 μg of pUC-XHIV or 2 μg of plasmid pUC18 DNA (with or without 1 μg of HIV-1 Tat) was transfected onto identically prepared wells in Optimem (Gibco-BRL-Life Technologies). As a control for transfection efficiency, 1 μg of pCMV-SPORT-β-gal (Gibro-BRL-Life Technologies) was cotransfected in all experiments. Four hours after transfection, the medium was replaced with RPMI 1640 plus 10% fetal calf serum and 0.5 μg of HIV-1 Tat, and cells were harvested 72 h later. Cytoplasmic extracts were prepared and assayed for β-galactosidase expression, and protein amounts containing the same level of β-galactosidase activity were assayed for CAT activity (37).
RT-PCR assay.
Cell samples were collected for RNA extraction using the TRIzol reagent from Gibco-BRL-Life Technologies. Total RNA (2 μg) was used for each cDNA synthesis with oligo(dT)15 primer and the cDNA cycle kit from Invitrogen. Aliquots of the cDNA samples were used as a template for amplifying specific gene fragments by PCR. The primers used in this study were: for HLA-DRA mRNA amplification, 5′-CCTGTCACCACAGGAGTGTCAGAG-3′ (forward) and 5′-CAGAGGCCCCCTGC GTTCTGCTG-3′ (reverse); and for β-actin amplification, 5′-GTGGGGCGCCCCAGG CACCA-3′ (forward) and 5′-CTCCTTAATGTCACGCACGATTTC-3′ (reverse). β-Actin mRNA was used as an internal control for the amount of cDNA synthesized. To ensure the specificity of mRNA detection, all primers were designed to cover at least two exons, and parallel samples without reverse transcription (RT) were run as negative controls. The amplified DNA products were run on an agarose gel and visualized with ethidium bromide staining (37).
RESULTS
Distributions of X-box-like sequences within the HIV-1 genome.
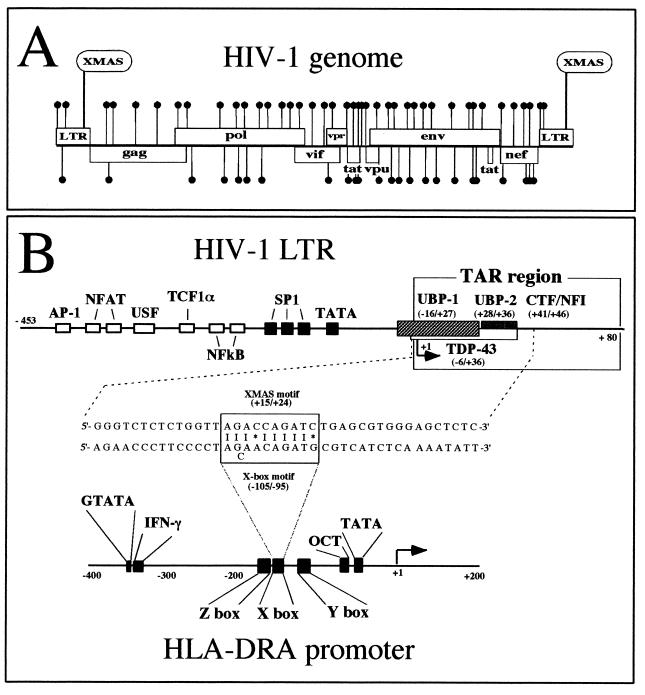
We have studied the presence of X-box-like sequences in a randomly selected group of viral genomes by screening at 90, 80, and 70% homology for the X box of the HLA-DRA gene. This search was performed by using the MacVector Software (IBI Kodak) for Macintosh computers. The results, summarized in Table 1, show that X-like motifs (sequences 70% homologous to the X box) are present in many viral genomes, and the HIV-1 genome (GenBank entry HIVHXB2CG) shows a high frequency of these X-like elements per 1,000 bp. When the search was performed on other human viral genomes, we found that X-like elements were present at a lower frequency within most of them (for instance, HS11CG and HS4 in Table 1). We found 68 X-like elements within the 9.7 kb of the HIV-1 genome (Fig. 1A), and interestingly, one of these was found within the HIV-1 TAR region (named XMAS, for X-like motif activator sequence; see Fig. 1A and B). A noteworthy, low level of homology was observed between the 5′- and 3′-flanking sequences of the XMAS motif and the flanking sequences of the X box of the HLA-DRA gene (Fig. 1B). In addition, we performed a comparative analysis of different HIV-1 genomes, demonstrating that the XMAS motif is conserved within the TAR regions of different HIV-1 isolates (Table 2).
TABLE 1.
Distribution of sequences homologous to the X box of MHC class II genes within the genomes of randomly selected human viruses
| GenBank entry | Virus | Genome length (bp) | No. of elements homologous to X box
|
Frequency of X-like motifsa | ||
|---|---|---|---|---|---|---|
| 90% | 80% | 70% | ||||
| HIVHXB2CG | HIV-1 | 9,718 | 2 | 11 | 68 | 7 |
| HPA | Hepatitis A virus | 7,478 | 1 | 7 | 45 | 6 |
| HIV2BEN | HIV-2 | 10,359 | 1 | 8 | 49 | 4.7 |
| HRV | Rhinovirus | 7,212 | 0 | 3 | 33 | 4.6 |
| HPBADR | Hepatitis B virus | 3,188 | 0 | 0 | 11 | 3.4 |
| HS5HCMVCG | Cytomegalovirus | 229,354 | 8 | 85 | 744 | 3.2 |
| PPL | Papovavirus | 5,270 | 0 | 1 | 15 | 2.8 |
| POL1SAB | Poliovirus | 7,441 | 0 | 1 | 19 | 2.5 |
| VA2CG | Varicella-zoster virus | 124,884 | 3 | 33 | 308 | 2.5 |
| HS4 | Epstein-Barr virus | 172,282 | 6 | 50 | 380 | 2.2 |
| HS11CG | Herpes simplex virus type 1 | 152,260 | 0 | 14 | 240 | 1.6 |
The number of X-like motifs at least 70% homologous to the X box of the HLA-DRA gene per 1,000 bp of genome.
FIG. 1.
(A) Distribution of sequences 70% homologous to the X box of MHC class II genes within the HIV-1 genome (GenBank entry HIVHXB2CG). The search was performed with MacVector Software. The 68 X-like elements found within the 9.7 kb of the HIV-1 genome are marked with solid circles. (B) Structure of the HIV-1 and HLA-DRA promoter regions. The binding sites for transcription factors binding to the promoter regions have been boxed. +1, transcription start site. HIV-1 LTR and HLA-DRA genomic regions carrying XMAS and X-box motifs have been aligned in the middle of the figure. XMAS and X-box motif homologies are marked by vertical lines. The asterisk represents mismatched nucleotides.
TABLE 2.
Sequences homologous to the X box of class II MHC genes are conserved in the LTRs of different HIV-1 isolatesa
| GenBank entry | Sequence of X-like element |
|---|---|
| HIVBH101 | TGGTTAGACCAGATCTGAGC |
| HIVBH102 | TGGTTAGACCAGATCTGAGC |
| HIVBRUCG | TGGTTAGACCAGATTTGAGC |
| HIVCDC41 | TGGTTAGACCAGATCTGAGT |
| HIVHXB2CG | TGGTTAGACCAGATCTGAGC |
| HIVPV22 | TGGTTAGACCAGATCTGAGC |
| HIVSF2CG | TGGTTAGACCAGATCTGAGC |
The X box sequence of the HLA-DRA gene is AGCAACAGATG. The X box sequences in the table are shown in boldface.
Binding of nuclear proteins to the XMAS motif present within the HIV-1 TAR region.
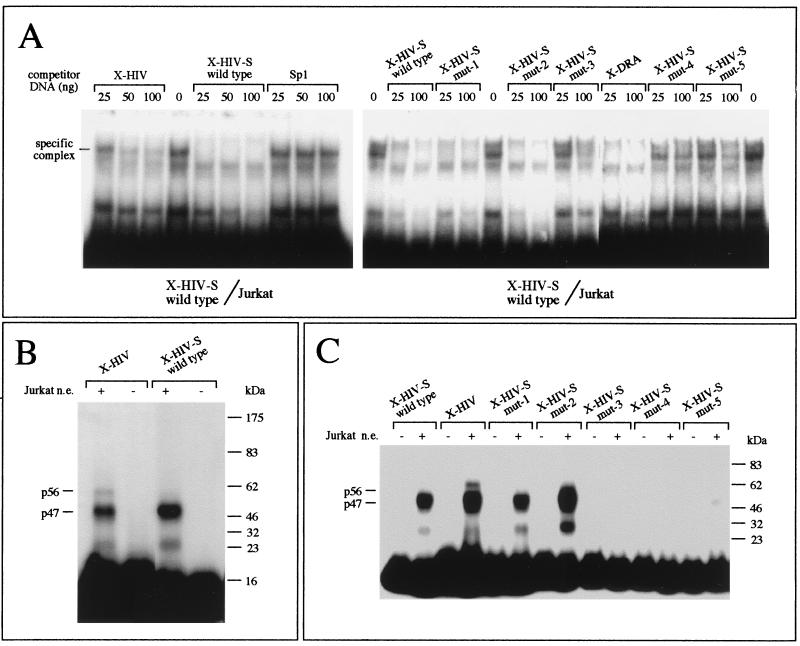
First, we analyzed the interaction between nuclear factors present in T-lymphoid Jurkat cells and the HIV-1 LTR by a DNase I footprinting assay. In order to focus the study on binding of nuclear proteins to the TAR region, the probe was labeled on the noncoding strand. Binding conditions were verified by incubation of the footprinting probe with recombinant Sp1 protein (Fig. 2), which generates a large footprint at the level of the LTR Sp1 binding sites. Then, the probe was incubated in the presence of increasing amounts of nuclear extracts (from 1 to 8 μg/reaction) and digested with DNase I, and the generated fragments were resolved by electrophoresis on a sequencing gel (Fig. 2). In our experimental conditions, three major footprints were observed: one extending from −78 to −46 and covering the Sp1 binding sites; the other two footprints were observed within the TAR region from +4 to +13 and from +15 to +32 (the last covering the XMAS motif and its 3′-flanking region). It should be mentioned that the XMAS motif (nucleotides +15 to +24; Fig. 1B) is localized in a region that could be bound by several nuclear proteins (13, 32). For this reason, further investigations were undertaken in order to characterize the protein(s) binding the XMAS element.
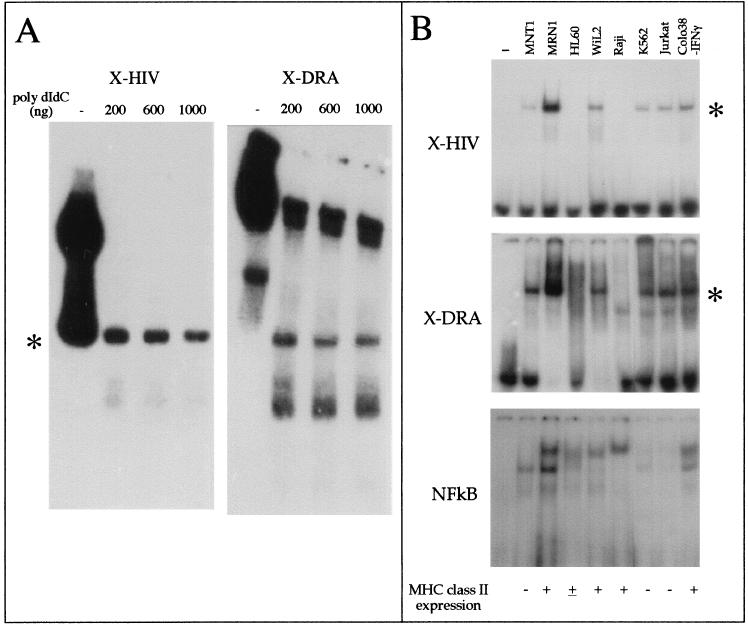
In order to determine whether the XMAS-containing region is recognized by nuclear proteins, we performed an EMSA using an oligonucleotide mimicking the LTR region (X-HIV, spanning from +8 to +31). In a preliminary experiment, we inhibited the binding of Jurkat cell nuclear extracts to X-HIV (carrying the XMAS motif) or X-DRA (carrying the X-box motif of the HLA-DRA gene) double-stranded oligonucleotides by adding increasing amounts of poly(dI-dC) · poly (dI-dC) in the binding reaction. DNA-protein complexes generated in the presence of 1 μg of poly(dI-dC) · poly(dI-dC) were considered specific. Although the X-HIV and X-DRA probes generate distinct binding patterns, as clearly evident in the experiment reported in Fig. 3A, one specific protein-DNA complex showing the same electrophoretic mobility was generated by both probes (asterisk, Fig. 3A).
FIG. 3.
Comparison of EMSA patterns obtained by using oligonucleotides carrying the XMAS and X-box motifs. (A) Competition of nuclear factor binding to oligonucleotides X-HIV, mimicking the LTR region and spanning from +8 to +31, and X-DRA, carrying the X-box motif of the HLA-DRA gene, by increasing amounts of poly(dI-dC) · poly(dI-dC). Asterisk, protein-DNA complex generated by both X-HIV and X-DRA oligonucleotides. (B) Screening for the presence of XMAS and X-box motif-binding activity in different cell lines expressing or not MHC class II molecules (upper and middle panels), performed by EMSA. In the lower panel, the EMSA-grade quality of the nuclear extracts used with the X-HIV and X-DRA probes has been tested by detecting the binding of NF-κB factor to its target DNA sequence. Lane —, free probe.
By virtue of the sequence homology of XMAS and the X-box motif, it should be expected that common proteins bind to both motifs. By EMSA, we tested the simultaneous presence of XMAS motif- and X-box motif-binding activity in different cell lines expressing or not MHC class II molecules, including melanoma (MNT1, MRN1, and Colo38), T-lymphoid (Jurkat), B-lymphoid (Raji and WIL2), erythroleukemic (K562), and promyelocytic (HL60) cell lines. As a control, we tested the EMSA-grade quality of the nuclear extracts by probing the binding of NF-κB to its target DNA sequence (Fig. 3B, lower panel). Interestingly, the results (Fig. 3B, upper and middle panels) revealed the simultaneous presence (in MNT1, MRN1, WIL2, K562, Jurkat cells, and IFN-γ-treated Colo38 melanoma cells) and absence (in HL60 and Raji cells) of a DNA-protein complex evidenced by the asterisk, suggesting that common nuclear proteins bind to both the X-HIV and X-DRA probes. Noteworthy, although Raji cell nuclear extracts do not give retarded bands in the presence of X-HIV and X-DRA, the same nuclear extracts generate a specific band when tested in the presence of the double-stranded oligonucleotide carrying the consensus NF-κB binding sequence. Taken together, the data support the hypothesis that the HIV-1 XMAS motif and HLA-DRA gene X-box motif bind to similar nuclear proteins.
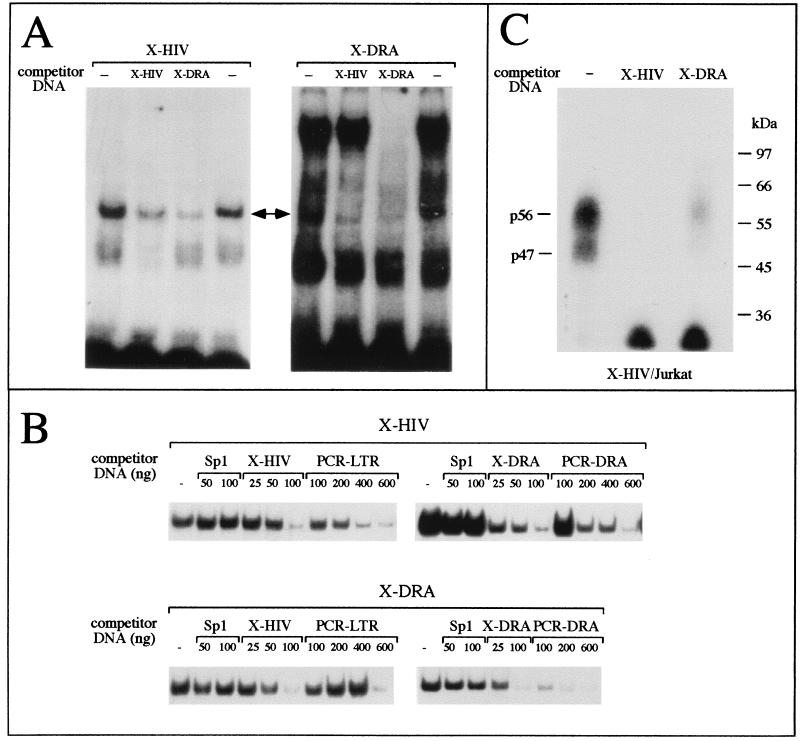
In order to verify the hypothesis that similar proteins bind to the X-HIV and X-DRA oligonucleotides, the competitive EMSAs shown in Fig. 4 were performed. We used nuclear extracts from T-lymphoid Jurkat cells. The binding of nuclear proteins to X-HIV (Fig. 4A, left panel) or to X-DRA (Fig. 4A, right panel) was inhibited by addition of a 100-fold molar excess of unlabeled X-HIV or X-DRA double-stranded oligonucleotides. Noteworthy, the competitor X-DRA inhibits the assembly of the X-HIV-protein complex (Fig. 4A, left panel, X-DRA lane, double arrow), whereas a molar excess of unlabeled X-HIV inhibits only the formation of one specific X-DRA-protein complex (Fig. 4A, right panel, X-HIV lane, double arrow). On the contrary, the competitor X-DRA inhibits the formation of both arrowhead (Fig. 4A, right panel, X-DRA lane) and low-speed-migrating X-DRA-protein complexes. These data suggest that, among the X-box-binding proteins, only some of them bind to the HIV-1 XMAS motif.
FIG. 4.
(A) Competitive band shift assay. The binding of proteins from Jurkat nuclear extracts to X-HIV (left panel) or X-DRA (right panel) double-stranded oligonucleotides was inhibited by addition of a 100-fold molar excess of unlabeled X-HIV or X-DRA double-stranded oligonucleotide. The double arrow indicates the protein-DNA complex generated by both probes. (B) Binding of Jurkat nuclear proteins to X-HIV (upper panel) or X-DRA (lower panel) double-stranded oligonucleotides was inhibited by addition of increasing amounts of unlabeled X-HIV, X-DRA, or HIV-1 Sp1-binding site-containing (Sp1) double-stranded oligonucleotides or by addition of increasing amounts of PCR products mimicking the HIV-1 (PCR-LTR) and HLA-DRA (PCR-DRA) promoters. Only the band corresponding to the complex marked by a double arrow in panel A is shown. (C) Competitive UV cross-linking assay. Crude nuclear extracts from Jurkat cells were incubated with the X-HIV radiolabeled probe in the absence (—) or in the presence of a 100-fold molar excess of unlabeled X-HIV or X-DRA double-stranded oligonucleotide. After the binding reaction, the assembled protein-DNA complexes were cross-linked by UV light exposure. The migration of protein size markers is shown on the right side of the autoradiograph. p47 and p57 are the proteins binding to the X-HIV probe.
In Fig. 4B, the X-HIV probe was incubated with Jurkat cell nuclear extracts in the presence of X-DRA or in the presence of a PCR product mimicking the entire HIV-1 LTR region or in the presence of a PCR product mimicking the promoter region of the HLA-DRA gene containing X, Y, and Z boxes. Control experiments were performed by incubating X-HIV with nuclear extracts in the presence of a molar excess of X-HIV or double-stranded oligonucleotide carrying the binding site for HIV-1 Sp1 (Sp1 mer). In these experiments, we focused attention on the retarded DNA-protein complex marked with an asterisk in Fig. 3, since this complex contains both X-box- and XMAS-binding proteins. The complex generated by the binding of nuclear proteins to labeled X-HIV was not inhibited by Sp1 mer, but was inhibited by X-HIV, X-DRA, and PCR products mimicking the HIV-1 LTR or the HLA-DRA gene promoter. Fully in agreement with these results, the complex generated by the binding of nuclear proteins to labeled X-DRA was not inhibited by Sp1 mer, but was inhibited by X-DRA, X-HIV, and PCR products mimicking the HIV-1 LTR or the HLA-DRA gene promoter. These data confirm the specificity of the DNA-protein complex assembled by both X-HIV and X-DRA mers and support the hypothesis that common proteins bind to both XMAS and X-box motifs.
In addition, we characterized nuclear protein binding to XMAS by a UV cross-linking assay. Since the covalent linkage of protein to an oligonucleotide up to 35 bp in length has little effect on the mobility of proteins in SDS-PAGE, allowing us to establish the molecular weight of DNA-binding proteins (36), we used labeled X-HIV double-stranded oligonucleotide and crude nuclear extracts from Jurkat cells. As shown in Fig. 4C, two proteins of 56 and 47 kDa bind to the X-HIV mer. Since the assembly of radiolabeled UV cross-linked complexes was inhibited in the presence of unlabeled X-HIV or X-DRA mers, the data suggest that both p56 and p47 Jurkat cell nuclear proteins are X-HIV- and X-box-binding proteins.
Purification of XMAS-binding proteins.
Using a combination of conventional and DNA affinity chromatography, cellular nuclear proteins binding to XMAS (spanning from +15 to +24) were purified. A scheme of the purification protocol is shown in Fig. 5A. We evaluated the purification process by probing the presence of both X-HIV- and X-box-binding activities by EMSA. We started the purification procedure using 1010 T-lymphoid Jurkat cells. Thirteen milligrams of nuclear extract was prepared by the standard Dignam procedure (8). In the preliminary experiment, we determined the concentration of ammonium sulfate that precipitates the low-migrating X-DRA-protein complex but not the complex identified by the asterisk in Fig. 5B. Forty percent saturation with ammonium sulfate was sufficient to separate the X-DRA-binding proteins involved in the formation of the low-speed-migrating complex in the supernatant from those forming the high-speed-migrating complex in the pellet. Therefore, in the preparative ammonium sulfate precipitation, we first saturated at 40%, and the supernatant was then saturated at 60%. The pellet, containing proteins forming the complex asterisked in Fig. 5B and in Fig. 5C, lane L, was fractionated by heparin-Sepharose chromatography, using stepwise elution with buffer D (see Materials and Methods) plus increasing concentrations of salt. Under these conditions, X-HIV-binding activity was eluted with buffers containing 0.3 to 0.5 M KCl (Fig. 5C, lanes c to e). The X-DRA-binding activity was eluted into the same fractions that were positive for X-HIV-binding activity. Fractions containing the X-HIV-binding proteins were pooled and applied to the DNA affinity column carrying X-HIV concatamers, and the on-column retained proteins were collected by stepwise elution with buffer D carrying increasing concentrations of salt. Under these conditions, X-HIV- and X-DRA-binding activities were eluted in the fractions at 0.3 to 0.5 M KCl (Fig. 5D, lanes c to e). As shown in Fig. 5D, the proteins eluted in these fractions generate three distinct DNA-protein complexes in the EMSA (complexes A, B, and C), with either the X-HIV or X-DRA probe. The final enrichment of X-HIV-binding proteins was 950-fold with respect to the crude nuclear extract (Table 3). A similar amount of protein fractionated after each purification step was loaded onto SDS–10% PAGE, and the gel was stained by the Coomassie R250 procedure in order to display the enrichment process (Fig. 5E, left panel). The reduction in complexity and protein amount observed during the purification steps correlates inversely to the increase in X-HIV-binding activity (see also Table 3). In Fig. 5E, the fractions eluted from the DNA affinity column were UV cross-linked to the radiolabeled X-HIV mer, and the gel was silver stained and then autoradiographed. Although three major proteins were evidenced by silver stain, 56, 50, and 47 kDa (Fig. 5E, middle panel), autoradiography of the gel showed that only p56 and p47 directly contact the X-HIV probe (Fig. 5E, right panel). Interestingly, these proteins are the same sizes as the X-HIV-binding proteins detected in crude nuclear extracts.
FIG. 5.
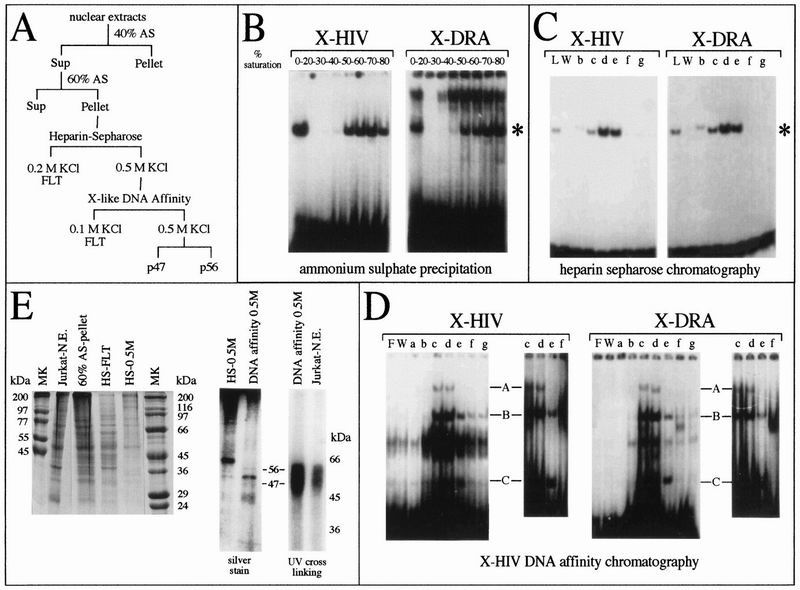
Enrichment of XMAS-binding proteins by conventional and DNA affinity chromatography. (A) Scheme of the protocol used for enrichment of cellular nuclear proteins binding to HIV-1 XMAS. Sup, supernatant; FLT, flowthrough; AS, ammonium sulfate. (B, C, and D) Similar volumes of fractions obtained from ammonium sulfate precipitation (B), heparin-Sepharose chromatography (C) and X-HIV DNA affinity chromatography (D) were assayed for the presence of X-HIV- and X-DRA-binding activities by EMSA. (E) In the left panel, similar amounts of total proteins for the selected fractions were loaded onto SDS–10% PAGE and the gel was stained with Coomassie R250 stain. In the middle panel, proteins from the fraction loaded onto DNA affinity chromatography (HS-0.5M) or eluted from DNA affinity chromatography in buffer plus 0.5 M KCl (DNA affinity 0.5 M) were fractionated by SDS-PAGE and the gel was silver stained. In the right panel, proteins eluted from X-HIV DNA affinity chromatography in buffer plus 0.5 M KCl (DNA affinity 0.5M) and proteins from Jurkat cell crude nuclear extracts (Jurkat-N.E.) were UV cross-linked to the X-HIV probe; the radiolabeled protein-DNA complexes have been evidenced by SDS-PAGE and autoradiography. Jurkat N.E., crude nuclear extracts; 60% AS-pellet, proteins present in the pellet obtained after 60% saturation with ammonium sulfate; HS-FLT, proteins not retained on the heparin-Sepharose column; HS-0.5M, proteins eluted from the heparin-Sepharose column with buffer containing 0.5 M KCl; MK, protein size markers; L, proteins loaded onto heparin-Sepharose column; F, free probe; W, proteins washed from chromatographic column; a, b, c, d, e, f, and g, fractions eluted from chromatographic matrix using buffer containing, respectively, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, and 0.7 M KCl.
TABLE 3.
Purification of XMAS-binding nuclear proteins
| Fraction | Concn (μg/μl) | Total protein (μg) | Binding activitya (BSU) | Sp act (BSU/μg) | Purification (fold) |
|---|---|---|---|---|---|
| Crude nuclear extract | 2 | 13,000 | 5,500 | 0.42 | 1 |
| Ammonium sulfate precipitation | 16.5 | 3,300 | 2,600 | 0.79 | 1.9 |
| Heparin-Sepharose | 1.9 | 570 | 370 | 0.65 | 1.5 |
| X-HIV DNA affinity | 0.01 | 0.5 | 200 | 400 | 950 |
A band shift unit (BSU) is the amount of probe retarded in the presence of 2 μg of Jurkat nuclear extract and a large excess of probe.
Modulation of Tat-induced HIV-1 transcription.
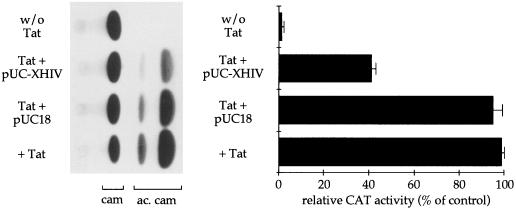
The experiments reported in this section were performed in order to determine whether the region containing the XMAS motif enhances Tat-induced HIV-1 transcription. The pivotal role played by TAR in the regulation of Tat-induced HIV-1 transcription is well known. Since constructs carrying deletions or mutations in the TAR sequence could be reflected in unstructured TAR RNA, a decoy approach was used in order to study the effect of protein binding to the region containing the XMAS motif on Tat-induced HIV-1 transcription. The double-stranded oligonucleotide X-HIV, mimicking the region from +8 to +31 and containing the XMAS motif, was used in order to produce concatamers of the XMAS motifs. These concatamers were ligated into SmaI-linearized plasmid pUC18 DNA, and recombinant clones were selected. One of them, pUC-XHIV, carrying 10 repeated XMAS motifs, was used in the decoy experiments diagrammed in Fig. 6. As a cellular system mimicking HIV-1 infection, we used HL3T1 cells, which contain integrated copies of the LTR-CAT construct. The cells were transfected with plasmid pUC-XHIV or pUC18 DNA in the presence or absence of HIV-1 Tat. Transfections were performed in triplicate, and cells were harvested 72 h later. Extracts were assayed for CAT activity. Basal CAT activity was also measured. The CAT activity present in cells treated with Tat alone was assumed to be 100%. Interestingly, a significant decrease in CAT activity (up to 40% ± 2% of the control value) was obtained in cells transfected with plasmid pUC-XHIV, whereas transfection with the backbone vector had only minor effects (up to 92% ± 4% of control). These data suggest that the X-HIV sequence binds transcription factors enhancing Tat-induced HIV-1 transcription.
FIG. 6.
Effect of decoy of proteins binding to the X-HIV element on Tat-induced HIV-1 LTR-driven transcription. HL3T1 cells carrying the LTR-CAT construct were transfected with plasmid pUC-XHIV (containing 10 copies of the X-HIV element) or with pUC18 (plasmid backbone of pUC-XHIV) in the presence of two-exon HIV-1 Tat. Basal CAT activity was also measured (w/o Tat). A representative result obtained by thin-layer chromatography (TLC) is shown in the left panel. In the right panel, acetylated chloramphenicol (ac. cam) spots were scraped from the TLC support, and radioactivity was measured. The CAT activity of cells transfected with pUC18 was set at 100%. The data reported are the averages of three independent experiments. cam, chloramphenicol.
Mutational analysis of the XMAS motif.
In order to define the nucleotides involved in the binding of proteins to the X-HIV mer, we first synthesized an oligonucleotide (X-HIV-S wild-type mer) in which the regions flanking the XMAS motif were deleted; second, we synthesized mutant oligonucleotides in which dinucleotide mutations were introduced in the XMAS motif (Table 4). The binding of nuclear proteins to these oligonucleotides was assayed by EMSA and UV cross-linking (Fig. 7). In a preliminary experiment, we inhibited the binding of Jurkat nuclear proteins to X-HIV-S wild type (Fig. 7A, left panel), carrying the XMAS motif alone, by addition of an increasing molar amount of unlabeled X-HIV, carrying both XMAS and flanking regions, or X-HIV-S wild type. As a control, unrelated cold Sp1 mer mimicking the Sp1-binding sites present in the HIV-1 LTR was used. In our experimental conditions, the competitor Sp1 mer did not inhibit the formation of the specific complex assembled on the XMAS motif. On the contrary, the competitor X-HIV was able to inhibit the assembly of the specific X-HIV-S wild-type/protein complex. These data suggest that X-HIV and X-HIV-S wild type double-stranded oligonucleotides bind similar proteins. Afterwards, the binding of Jurkat cell nuclear proteins to the X-HIV-S wild-type probe was performed in the absence and presence of double-stranded oligonucleotides carrying dinucleotide mutations within the XMAS motif (see sequences in Table 4) or in the presence of the double-stranded X-DRA mer (Fig. 7A, right panel). The specific complex generated by the binding of nuclear proteins to the 32P-labeled X-HIV-S wild type was not efficiently inhibited by X-HIV-S mutants 3, 4, and 5; on the contrary, the binding of nuclear proteins to labeled X-HIV-S wild type was efficiently inhibited by X-HIV-S mutants 1 and 2. These data suggest that nucleotides present within the sequence CAGATC (+19 to +24) are required for the binding of nuclear proteins to the XMAS motif.
TABLE 4.
Sequences of the oligodeoxyribonucleotides carrying wild-type and mutated XMAS motifs
| Oligonucleotide | Forward strand sequence (5′→3′)a |
|---|---|
| X-HIV | CTGGTTAGACCAGATCTGAGCGT |
| X-HIV-mut | CTGGTTAGACATCTGTTGAGCGT |
| X-HIV-S wild type | TTAGACCAGATCTG |
| X-HIV-S mut-1 | TGTGACCAGATCTG |
| X-HIV-S mut-2 | TTACTCCAGATCTG |
| X-HIV-S mut-3 | TTAGACATGATCTG |
| X-HIV-S mut-4 | TTAGACCACTTCTG |
| X-HIV-S mut-5 | TTAGACCAGAGTTG |
Mutations are underlined.
FIG. 7.
(A) Competitive band shift assay. The binding of nuclear proteins from Jurkat cells to short double-stranded oligonucleotides carrying the XMAS motif (without flanking regions [X-HIV-S wild type]) was inhibited by addition of a molar excess of unlabeled X-HIV, X-HIV-S wild type, or HIV-1 Sp1-binding site-containing (Sp1) double-stranded oligonucleotide (left panel); in the right panel, the binding was inhibited by addition of a molar excess of unlabeled X-HIV-S wild type or mutants (X-HIV-S mut-1 to mut-5) carrying dinucleotide substitution involving the HIV-1 XMAS motif. (B) The sizes of the DNA-binding proteins contacting the X-HIV or the X-HIV-S wild type double-stranded oligonucleotide were compared by a UV cross-linking assay. The radiolabeled probes were incubated in the presence (+) or in the absence (−) of crude nuclear extracts (n.e.) from Jurkat cells. After the binding reaction, the assembled protein-DNA complexes were cross-linked by UV light exposure. The migration of the protein size markers is shown on the right side of the autoradiograph. The sizes of the protein-DNA complexes detected are reported on the left side of the autoradiograph. (C) The sizes of the DNA-binding proteins contacting the X-HIV-S wild type or mutant double-stranded oligonucleotides were compared by a UV cross-linking assay, as described above.
In addition, we have characterized the binding of nuclear proteins to X-HIV and to X-HIV-S wild type by a UV cross-linking assay. In this experiment we used labeled X-HIV or X-HIV-S wild type double-stranded oligonucleotides and crude nuclear extracts from Jurkat cells. As shown in Fig. 7B, while both p56 and p47 bind to the X-HIV mer, only the protein termed FAX-1, for factor activating XMAS, binds to X-HIV-S wild type. Since the oligonucleotide X-HIV-S wild type lacks the regions flanking the XMAS motif, the results suggest that only FAX-1 binds to the XMAS motif, while p56 need the presence of the XMAS-flanking regions in order to obtain efficient interaction. In addition, when the UV cross-linking assay was performed as described above but using the X-HIV-S mutants as probes (Fig. 7C), only mutants X-HIV-S mut 1 and 2 directly bind to FAX-1, sustaining the hypothesis that nucleotides mutated in X-HIV-S mut 3 to 5 (involving the wild-type CAGATC, +19 to +24) are important for the binding of FAX-1 to the XMAS motif. On the contrary, the region of the XMAS motif spanning nucleotides +15 to +18 does not seem to be involved in the binding of FAX-1. Noteworthy, instead of the region of the X-box motif of the HLA-DRA gene spanning nucleotides −105 to −100, which contain an extra nucleotide with respect to the homologous region within the XMAS motif, the sequence −100 to −95 shows good homology to the XMAS motif (see Fig. 1). Thus, it is not surprising that regions spanning nucleotides −100 to −95 of the X-box motif and nucleotides +19 to +24 of the XMAS motif could bind the same factor.
Oligonucleotides carrying the XMAS motif inhibit IFN-γ-induced HLA-DRA gene expression.
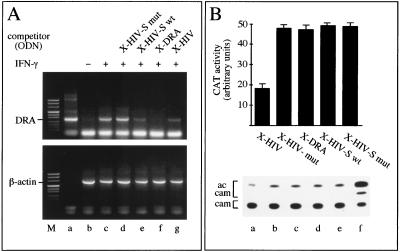
In order to determine whether the XMAS motif could modulate an X-box-dependent gene transcription, the decoy effect of double-stranded oligonucleotides carrying the XMAS motif on the HLA-DRA gene expression was analyzed. As cellular model system we used the Colo38 cell line, an MHC class II-negative cell line derived from a human melanoma. In these cells, IFN-γ dramatically induces the expression of HLA-DRA mRNA. To determine whether double-stranded oligonucleotides carrying the XMAS motif inhibit HLA-DRA gene transcription, HLA-DRA mRNA levels were evaluated by semiquantitative RT-PCR (Fig. 8A). Colo38 cells were treated in the absence or in the presence of the double-stranded oligonucleotides X-HIV and X-HIV-S wild type (carrying the XMAS motif), X-HIV-S mut-4 (carrying a dinucleotide mutation that abrogates the binding of FAX-1), and X-DRA (carrying the X box of the HLA-DRA gene). Afterwards, expression of the HLA-DRA gene was induced for 24 h by the addition of IFN-γ, and cells were collected for RNA extraction. Total RNA was used for each cDNA synthesis with the oligo(dT) primer, and aliquots of the cDNA samples were used as a template for amplifying the HLA-DRA and β-actin mRNAs. As a control for amplification, plasmid pIIDRα1 carrying the entire cDNA of the HLA-DRA gene was used (Fig. 8A, lane a). The induction of HLA-DRA expression is clearly evident by comparing lanes b and c in Fig. 8A. As expected, the expression induced by IFN-γ is inhibited by pretreating the cells with double-stranded oligonucleotide X-DRA, carrying the X box of the HLA-DRA gene (Fig. 8A, lane f). Also, pretreatment with double-stranded X-HIV and X-HIV-S oligonucleotides was found to inhibit HLA-DRA mRNA accumulation (Fig. 8A, lanes g and e, respectively). On the contrary, accumulation of HLA-DRA mRNA was not inhibited by pretreating the cells with the double-stranded oligonucleotide X-HIV-S mut-4, carrying a dinucleotide mutation abrogating the binding of FAX-1 to the XMAS motif (Fig. 8A, lane d). Taken together, the data demonstrate that the XMAS motif exhibits a decoy effect leading to inhibition of the X-box-dependent transcription of the HLA-DRA gene. In addition, these data suggest that FAX-1 might be important for HLA-DRA gene expression induced by IFN-γ.
FIG. 8.
(A) Inhibition of IFN-γ-inducible MHC class II expression by decoy oligodeoxyribonucleotides (ODN). Colo38 melanoma cells, treated with or without decoys (d to g) or not (b and c), were stimulated with IFN-γ (c to g) and collected for RT-PCR analysis 24 h later. Cells were stimulated with IFN-γ at 400 U/ml. Gene-specific primers were used to amplify the cDNA templates. Lane b, RT-PCR amplification of the RNA isolated from Colo38 cells not induced with IFN-γ. In lane a, plasmid pIIDRα1 carrying the entire HLA-DRA cDNA was used as the template in a control PCR amplification. Lane M, size markers. (B) Inhibition of Tat-induced HIV-1 LTR-driven transcription by decoys. HL3T1 cells were transfected with decoys (a to e) and induced by Tat. Cells were collected 48 h later and assayed for CAT activity. A representative result obtained by TLC is shown (bottom). In the top panel, acetylated chloramphenicol (ac cam) spots were quantified with a phosphor imager and values were plotted. The data are the averages of three independent experiments. cam, chloramphenicol. Lane f, control reaction performed by using purified CAT.
Effects of oligonucleotides carrying the XMAS motif on Tat-induced HIV-1 LTR-driven transcription.
In order to identify the DNA motifs required to inhibit Tat-induced HIV-1 LTR-driven transcription, an experiment similar to that in Fig. 8A was performed using transfection of synthetic oligonucleotides to Tat-induced HL3T1 cells as a model system. The results of the experiment are shown in Fig. 8B. As is clearly appreciable, the X-DRA, X-HIV-S wild type, and X-HIV-S-mut oligonucleotides do not inhibit Tat-induced HIV-1 LTR-driven transcription. By contrast, the X-HIV oligonucleotide, as expected (see also Fig. 6), leads to about 64% inhibition. This finding suggests that the XMAS motif is not sufficient to inhibit HIV-1 LTR-driven transcription in decoy experiments. In order to determine whether the XMAS motif was important for HIV-1 transcription, we compared the decoy effect of the X-HIV oligonucleotide with that of X-HIV-mut, which contains the scrambled sequence ATCTGT instead of CAGATC (Table 4). The results obtained (Fig. 8B) demonstrate that X-HIV-mut does not lead to any extent of inhibition of Tat-induced HIV-1 LTR-driven transcription. Taken together, these findings are in line with the hypothesis that, in decoy experiments, the XMAS sequence is required and sufficient to inhibit IFN-γ-induced expression of the human HLA-DRA gene (Fig. 8A), but the same sequence is required but not sufficient for inhibition of Tat-induced HIV-1 LTR-driven transcription (Fig. 8B). In order to obtain inhibition of Tat-induced HIV-1 LTR-driven transcription by using decoy oligonucleotides, both X-MAS and flanking regions are required, suggesting that both FAX-1 and FAX-2 are required to enhance HIV-1 transcription.
DISCUSSION
It has been well established that expression of HIV-1 genes is modulated by both viral and cellular factors (13). Accordingly, the identification of cellular transcription factors modulating Tat function is critical in order to understand the mechanisms regulating HIV-1 gene expression. Multiple regulatory elements direct HIV-1 LTR-driven transcription by serving as binding sites for a variety of cellular transcription factors (9, 22, 24, 32, 34, 35, 38) and influence the degree of transactivation by the Tat protein. Among these, YY1 (31), USF (9), TFII-I (34, 35), LBP family proteins (43), PRDII-BF1 (38), CTF/NF-1 (22), and TDP-43 (32) have been described to bind in the vicinity of the HIV-1 initiator spanning nucleotides −4 to +25 (40) and function as either repressors or activators of HIV-1 transcription.
In order to gain further insight on the mechanisms regulating HIV-1 gene expression, we screened the HIV-1 genome for the presence of additional binding sites for cellular transcription factors modulating Tat function. Since the X-box sequence present within the promoter region of MHC class II genes plays a pivotal role in transcriptional modulation, we searched for sequences homologous to this DNA motif in the HIV-1 genome. In our working hypothesis, a sequence homologous to this motif present within HIV-1 LTR could retain modulator activity. The screening showed that the HIV-1 genome is enriched in X-like elements (Table 1), and interestingly, we identified a sequence within the TAR region spanning nucleotides +15 to +24, herein termed XMAS for X-box motif activator sequence. Comparing the XMAS motifs present within the TAR region of different HIV-1 genomes (Table 2), we found that the sequence was highly conserved. This finding suggests that this sequence could have a regulatory function. However, conservation of nucleotide sequence within the TAR region is not surprising since, upon transcription, the XMAS motif is part of the stem-bulge TAR RNA.
Since XMAS was selected by sequence homology to the X-box element, we determined whether XMAS mediates interactions with cellular nuclear proteins, in particular with X-box-binding proteins, and if this sequence plays a role in modulating HIV-1 LTR-driven transcription.
Some inferences can be drawn from the binding data. The DNase I footprinting assay performed using T-lymphoid cell nuclear extracts and shown in Fig. 2 showed the presence of two footprints localized on +4 to +13 and +15 to +32, the last including the XMAS motif (nucleotides +15 to +24; Fig. 1B) and its 3′-flanking region. The UV cross-linking assay (Fig. 4C) suggests that two proteins of about 56 and 47 kDa bind to the X-HIV sequence in T-lymphoid Jurkat cells. These sizes do not match those of previously described ubiquitous transcription factors UBP-1/LBP-1 (63 kDa, binding the sequence from −16 to +27), UBP-2 (50 kDa, binding the sequence from +28 to +36), and TDP-43 (43 kDa, binding the sequence from −6 to +36) (13, 32) that bind in the region from −16 to +36. The two proteins p56 and p47 could represent previously uncharacterized nuclear proteins present in T-lymphoid cells.
Although competitive EMSAs (Fig. 3A and 4A and B) strongly suggest that X-box-binding proteins bind to the LTR region (+8 to +31), the differences in the binding patterns with probes X-HIV (+8 to +31 and carrying XMAS; see Fig. 1) and X-DRA (carrying the X-box motif and flanking sequences; see Fig. 1) suggest that sequences flanking the X box and XMAS contribute to generate different EMSA patterns. In addition, the data in Fig. 3B highlight a tight correlation between XMAS- and X-box-binding activities in different cell lines, including melanoma, T- and B-lymphoid, erythroleukemic, and promyelocytic cells, that could be explained by sequence homologies between the XMAS and X-box motifs. In addition, the competitive UV cross-linking assay (Fig. 4C) confirmed these data.
Furthermore, we performed a small-scale enrichment of cellular nuclear proteins binding to XMAS (Fig. 5) by a combination of conventional and DNA affinity chromatography in order to characterize the DNA-protein complexes assembled on XMAS and X-box motifs. It should be noted that during fractionation, the XMAS and X-box motif-binding activities always cofractionated (Fig. 5B, C, and D), confirming the hypothesis that common proteins bind to both motifs. In addition, fractions eluted from the DNA affinity matrix at 0.3 to 0.5 M KCl give more strongly labeled complexes in the UV cross-linking assay than the crude Jurkat nuclear extracts (Fig. 5E), confirming the enrichment for a XMAS-binding proteins.
The role played by XMAS and flanking regions on Tat-induced HIV-1 LTR-driven transcription was verified by a decoy approach using HL3T1 cells, which contain integrated copies of the LTR-CAT construct, as a cellular system mimicking HIV-1 infection (Fig. 6). We selected this approach because plasmid constructs carrying deletions or mutations in the TAR sequence could be reflected in an unstructured TAR RNA. In this approach, plasmid DNA carrying 10 repeated XMAS motifs was used as the decoy molecule. Interestingly, a significant decrease in CAT activity (up to 40% ± 2% of the control value) was obtained in cells transfected with the decoy plasmid, whereas transfection with the backbone vector has only a small effect (up to 92% ± 4% of the control). These data suggest that the X-HIV sequence binds transcription factors enhancing Tat-induced HIV-1 transcription. Furthermore, we performed a mutational analysis in order to define the nucleotides involved in the binding of proteins to the X-HIV sequence. Interestingly, deletion of nucleotides flanking the XMAS motif abrogates the binding of p56 but not the binding of p47, suggesting that p47 is a factor associated with XMAS (FAX-1). On the contrary, the binding of p56 requires nucleotides localized outside the XMAS motif (Fig. 7B and C). Mutational analysis of the XMAS motif revealed that sequence from +19 to +24 is necessary for the binding of FAX-1 (Fig. 7A and C).
In order to determine the effects of the XMAS motif on transcription, we first determined the effects of decoy oligonucleotides on X-box-dependent transcription of cellular genes. To this end, we analyzed the decoy effect of XMAS-containing double-stranded oligonucleotides on human HLA-DRA gene expression induced by IFN-γ. It should be underlined that recognition of infected CD4+ T cells requires IFN-γ induction of MHC class II expression and that suppression of IFN-γ-inducible MHC class II expression may represent an efficient immune evasion strategy used by intracellular pathogens to escape host defenses (44). At least in ex vivo experiments, we provide data demonstrating that the viral XMAS sequence (X-HIV-S wild type mer, Fig. 8, lane e) inhibits the transcription of HLA-DRA gene, a gene in which the X-box motif has been well characterized to enhance transcription. Thus, the protein binding to the XMAS motif, FAX-1, could be involved in coordinated activation of the HIV-1 genome and induction of the HLA-DRA gene.
Finally, in order to identify the DNA motifs required to inhibit Tat-induced HIV-1 LTR-driven transcription, a similar experiment was performed using as a model system Tat-induced HL3T1 cells. The results of the experiment show that the XMAS sequence is required but not sufficient for inhibition of Tat-induced HIV-1 LTR-driven transcription (Fig. 8B). In order to obtain inhibition of Tat-induced HIV-1 LTR-driven transcription by using decoy oligonucleotides, both X-MAS and flanking regions are required, suggesting that both FAX-1 and FAX-2 are required to enhance HIV-1 transcription.
In conclusion, our data suggest that one X-box-binding protein, FAX-1, binds to the XMAS motif present within the HIV-1 LTR TAR region, in proximity to the initiator sequence. This sequence, like the X-box motif in the context of the MHC class II gene promoters, plays an enhancing role in Tat-induced HIV-1 transcription and binds proteins involved in the regulation of IFN-γ-induced HLA-DRA gene transcription. This sequence is required but not sufficient to inhibit Tat-induced HIV-1 LTR-driven transcription. From the theoretical point of view, this finding suggests that coordinated expression of MHC class II and HIV-1 genes could be realized by the X-box-binding protein FAX-1. In this respect, we speculate that in latently infected T lymphocytes, the induction of MHC class II gene expression could be a signal provoking active viral replication and AIDS progression.
ACKNOWLEDGMENTS
This work was supported by the Istituto Superiore di Sanità (AIDS-1998), CNR-P.F. Biotecnologie, PRIN-98, and Finalized Research funds (year 1998) from the Italian Ministry of Health. C.M. was the recipient of an AIRC fellowship.
We thank Mauro Giacca for kindly providing Tat expression plasmid pGEX2T-Tat2E (ICGEB, Trieste, Italy). We thank P. G. Balboni for the generous gift of HL3T1 cells.
REFERENCES
- 1.Benoist C, Mathis D. Regulation of major histocompatibility complex class-II genes: X, Y and other letters of the alphabet. Annu Rev Immunol. 1990;8:681–715. doi: 10.1146/annurev.iy.08.040190.003341. [DOI] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Cogswell J P, Zeleznik-Le N, Ting J P-Y. Transcriptional regulation of the HLA-DRA gene. Crit Rev Immunol. 1991;11:87–112. [PubMed] [Google Scholar]
- 4.Cresswell P. Immunology: questions of presentation. Nature (London) 1990;343:593–594. doi: 10.1038/343593a0. [DOI] [PubMed] [Google Scholar]
- 5.Cullen B R. The HIV-1 Tat protein: an RNA sequence-specific processivity factor? Cell. 1990;63:655–657. doi: 10.1016/0092-8674(90)90129-3. [DOI] [PubMed] [Google Scholar]
- 6.Dardell S, Merkenschlager M, Bodmer H, Chan S, Cosgrove D, Benoist C, Mathis D. The immune system of mice lacking conventional MHC class II molecules. Adv Immunol. 1994;55:423–440. doi: 10.1016/s0065-2776(08)60515-5. [DOI] [PubMed] [Google Scholar]
- 7.Das A. Control of transcription termination by RNA-binding proteins. Annu Rev Biochem. 1993;62:893–930. doi: 10.1146/annurev.bi.62.070193.004333. [DOI] [PubMed] [Google Scholar]
- 8.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du H, Roy A L, Roeder R G. Human transcription factor USF stimulates transcription through the initiator elements of the HIV-1 and the Ad-ML promoters. EMBO J. 1993;12:501–511. doi: 10.1002/j.1460-2075.1993.tb05682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durand B, Kobr M, Reith W, Mach B. Functional complementation of major histocompatibility complex class II regulatory mutants by the purified X-box-binding protein RFX. Mol Cell Biol. 1994;14:6839–6847. doi: 10.1128/mcb.14.10.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feriotto G, Mischiati C, Gambari R. Sequence-specific recognition of the HIV-1 long terminal repeat by distamycin: a DNase I footprinting study. Biochem J. 1994;299:451–458. doi: 10.1042/bj2990451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frankel A D. Activation of HIV transcription by Tat. Curr Opin Genet Dev. 1992;2:293–298. doi: 10.1016/s0959-437x(05)80287-4. [DOI] [PubMed] [Google Scholar]
- 13.Gaynor G. Cellular transcription factors involved in the regulation of HIV-1 gene expression. AIDS. 1992;6:347–363. doi: 10.1097/00002030-199204000-00001. [DOI] [PubMed] [Google Scholar]
- 14.Glimcher L H, Kara C J. Sequences and factors: a guide to MHC class II transcription. Annu Rev Immunol. 1992;10:13–49. doi: 10.1146/annurev.iy.10.040192.000305. [DOI] [PubMed] [Google Scholar]
- 15.Graeble M A, Churcher M J, Lowe A D, Gait M J, Karn J. Human immunodeficiency virus type 1 trans-activator protein Tat stimulates transcriptional read-through of distal terminator sequences in vitro. Proc Natl Acad Sci USA. 1993;90:6184–6188. doi: 10.1073/pnas.90.13.6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenberg M E, Mathews M B. Effects of heterologous downstream sequences on the activity of the HIV-1 promoter and its response to Tat. Nucleic Acids Res. 1997;25:5017–5024. doi: 10.1093/nar/25.24.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grusby M J, Glimcher L H. Immune responses in MHC class II-deficient mice. Annu Rev Immunol. 1995;13:417–435. doi: 10.1146/annurev.iy.13.040195.002221. [DOI] [PubMed] [Google Scholar]
- 18.Guardiola J, Maffei A. Control of MHC class II gene expression in autoimmune, infectious, and neoplastic diseases. Crit Rev Immunol. 1993;13:247–268. [PubMed] [Google Scholar]
- 19.Itoh-Lindstrom Y, Peterlin B M, Ting J P-Y. Affinity enrichment and functional characterization of TRAX1, a novel transcription activator and X1-sequence-binding protein of HLA-DRA. Mol Cell Biol. 1995;15:282–289. doi: 10.1128/mcb.15.1.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeang K T, Xiao H, Rich E. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J Biol Chem. 1999;274:28837–28840. doi: 10.1074/jbc.274.41.28837. [DOI] [PubMed] [Google Scholar]
- 21.Jones K A, Peterlin B M. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 22.Jones K A, Luciw P A, Duchange N. Structural arrangements of transcription control domains within the 5′-untranslated leader regions of the HIV-1 and HIV-2 promoters. Genes Dev. 1988;2:1101–1114. doi: 10.1101/gad.2.9.1101. [DOI] [PubMed] [Google Scholar]
- 23.Karn J. Tackling Tat. J Mol Biol. 1999;293:235–254. doi: 10.1006/jmbi.1999.3060. [DOI] [PubMed] [Google Scholar]
- 24.Kato H, Sumimoto H, Pognonec P, Chen C-H, Rosen C A, Roeder R G. HIV-1 Tat acts as a processivity factor in vitro in conjunction with cellular elongation factors. Genes Dev. 1992;6:655–666. doi: 10.1101/gad.6.4.655. [DOI] [PubMed] [Google Scholar]
- 25.Kelly A, Trowsdale J. Complete nucleotide sequence of a functional HLA-DPβ gene and the region between the DPβ1 and DPα1 genes: comparison of the 5′ ends of HLA class II genes. Nucleic Acids Res. 1985;13:1607–1620. doi: 10.1093/nar/13.5.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laspia M F, Rice A P, Mathews M B. Synergy between HIV-1 Tat and adenovirus E1a is principally due to stabilization of transcriptional elongation. Genes Dev. 1990;4:2397–2408. doi: 10.1101/gad.4.12b.2397. [DOI] [PubMed] [Google Scholar]
- 27.Laspia M F, Rice A P, Mathews M B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989;59:283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- 28.Laspia M F, Wendel P, Mathews M B. HIV-1 Tat overcomes inefficient transcriptional elongation in vitro. J Mol Biol. 1993;232:732–746. doi: 10.1006/jmbi.1993.1427. [DOI] [PubMed] [Google Scholar]
- 29.Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: lessons from a disease. Annu Rev Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 30.Marciniak R A, Sharp P A. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Margolis D M, Somasundaran M, Green M R. Human transcription factor YY1 represses human immunodeficiency virus type 1 transcription and virion production. J Virol. 1994;68:905–910. doi: 10.1128/jvi.68.2.905-910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ou S-H I, Wu F, Harrich D, Garcia-Martinez L F, Gaynor R B. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reith W, Satola S, Herrero-Sanchez C, Amaldi I, Lisowska-Grospierre B, Griscelli C, Hadam M R, Mach B. Congenital immunodeficiency with a regulatory defect in MHC class II gene expression lacks a specific HLA-DR promoter binding protein, RF-X. Cell. 1988;53:897–906. doi: 10.1016/s0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 34.Roy A L, Malik S, Meisterernst M, Roeder R G. An alternative pathway for transcription initiation involving TFII-I. Nature (London) 1993;365:355–359. doi: 10.1038/365355a0. [DOI] [PubMed] [Google Scholar]
- 35.Roy A L, Meisterernst P, Pognonec P, Roeder R G. Cooperative interaction of an initiator-binding transcription initiator factor and the helix-loop-helix activator USF. Nature (London) 1991;354:245–248. doi: 10.1038/354245a0. [DOI] [PubMed] [Google Scholar]
- 36.Sagami I, Tsai S Y, Wang H, Tsai M-J, O'Malley B W. Identification of two factors required for transcription of the ovalbumin gene. Mol Cell Biol. 1986;6:4259–4267. doi: 10.1128/mcb.6.12.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Seeler J S, Muchardt C, Suessle A, Gaynor R B. Transcription factor PRDII-BF1 activates human immunodeficiency virus type 1 gene expression. J Virol. 1994;68:1002–1009. doi: 10.1128/jvi.68.2.1002-1009.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharp P A, Marciniak R A. HIV TAR: an RNA enhancer? Cell. 1989;59:229–230. doi: 10.1016/0092-8674(89)90279-1. [DOI] [PubMed] [Google Scholar]
- 40.Sheldon M, Ratnasabapathy R, Hernandez N. Characterization of the inducer of short transcripts, a human immunodeficiency virus type 1 transcriptional element that activates the synthesis of short RNAs. Mol Cell Biol. 1993;13:1251–1263. doi: 10.1128/mcb.13.2.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ting J P-Y, Baldwin A S. Regulation of MHC gene expression. Curr Opin Immunol. 1993;5:8–16. doi: 10.1016/0952-7915(93)90074-3. [DOI] [PubMed] [Google Scholar]
- 42.Tsang S Y, Nakanishi M, Peterlin B M. Mutational analysis of the DRA promoter: cis-acting sequences and trans-acting factors. Mol Cell Biol. 1990;10:711–719. doi: 10.1128/mcb.10.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon J B, Li G, Roeder R G. Characterization of a family of related cellular transcription factors which can modulate human immunodeficiency virus type 1 transcription in vitro. Mol Cell Biol. 1994;14:1776–1785. doi: 10.1128/mcb.14.3.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong G, Fan T, Liu L. Chlamydia inhibits interferon γ-inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor. J Exp Med. 1999;89:1931–1937. doi: 10.1084/jem.189.12.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Q, Sharp P A. Novel mechanism and factor for regulation by HIV-1 Tat. EMBO J. 1995;14:321–328. doi: 10.1002/j.1460-2075.1995.tb07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]