Abstract
Recent studies have revealed that the gut microbiome affects various health conditions via its metabolites, including short-chain fatty acids (SCFAs) and bile acids (BAs). In the analysis of these, appropriate collection, handling, and storage of fecal specimens are required, and convenient specimen handling processes will facilitate their investigation. Here, we developed a novel preservation solution, “Metabolokeeper®”, to stabilize fecal microbiota, organic acids including SCFAs, and BAs at room temperature. In the present study, we collected fecal samples from 20 healthy adult volunteers and stored them at room temperature with Metabolokeeper® and at −80°C without preservatives for up to four weeks to evaluate the usefulness of the novel preservative solution. We found that microbiome profiles and short chain fatty acid contents were stably maintained at room temperature with Metabolokeeper® for 28 days, while the bile acids were stably maintained for 7 days under the same conditions. We conclude that this convenient procedure to obtain a fecal sample for collecting the gut microbiome and gut metabolites can contribute to a better understanding of the health effects of fecal metabolites produced by the gut microbiome.
Keywords: preservation solution, intestinal microbiota, short-chain fatty acids, bile acids, fecal sampling
Introduction
Accumulating research of the gut microbiome has revealed that the composition of human gut microbiota is linked to host health and disease.(1–4) Although the analysis of gut microbiota in feces has been focused on evaluating the health or pathological conditions of humans and animals, more emphasis has recently been placed on the analysis of metabolites produced by bacteria to evaluate the intestinal environment.(5) In particular, short-chain fatty acids (SCFAs) and bile acids (BAs) are affected by alterations in the gut microbiota.(6,7)
SCFAs are metabolites of dietary fiber and carbohydrates produced by the gut microbiota, also increased during the digestion of proteins and peptides.(8) BAs are known to affect the gut microbiota owing to their high antibacterial activity. Primary BAs biosynthesized in the liver are converted to secondary BAs by intestinal bacteria.(9) Therefore, to measure fecal metabolites, it is very important to immediately store collected fecal samples in frozen conditions to prevent metabolism by intestinal bacteria. The analytical values of metabolites from intestinal bacteria and the structure of gut microbiota depend on the length of time between the collection of the fecal sample and analysis, the temperature during storage, and the storage method, including the preservation solution used.(10,11) Therefore, it is important to develop a preservation solution that does not affect fecal metabolites or gut bacterial composition. In particular, a preservative solution that can be stored at room temperature for a long period could greatly contribute to research on the intestinal environment.
Regarding the storage method of fecal samples for the measurement of gut metabolites, some researchers have used the OMNIgene-GUT solution and RNAlater, storage solutions for bacterial flora analysis.(12,13) In these studies, derivatized SCFAs were analyzed using GC-MS and LC-MS after storage at room temperature, and satisfactory results were not obtained. Here, we have already developed a preservation solution without bromothymol blue based on an existing guanidine thiocyanate solution for the preservation of fecal specimens,(14) which consists mainly of a protein denaturant and can analyze SCFAs without derivatization. In the present study, we evaluated the effect of a newly developed preservative solution (Metabolokeeper®; TechnoSuruga Laboratory, Shizuoka, Japan) on the stabilization of fecal microbiota and organic acids, including SCFAs, and BAs, at room temperature.
Materials and Methods
Fecal sample collection
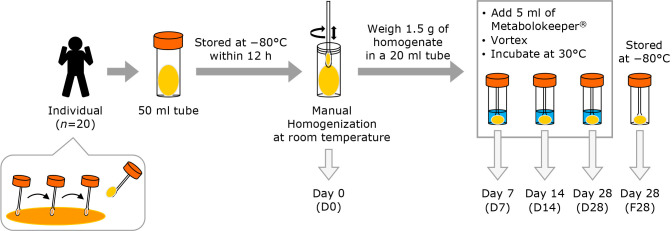
Feces were sampled from twenty healthy adult volunteers considered to be in good health (10 males: 39.7 ± 10.9 years old; 10 females: 35.2 ± 6.0 years old) at Kyoto Prefectural University. Fecal samples were collected without preservatives and stored at −80°C within one hour after defecation. After thawing to room temperature, 1.5 g of fecal material was weighed using a sterile swab and transferred into five tubes at room temperature, as shown in Fig. 1. One fecal sample out of a total of 20 samples was excluded due to deficiency of the necessary quantity; as a result, 19 samples were prepared for handling. One tube was supplied without intervention for fecal microbiota and organic acid analyses (D0), and one tube was frozen and stored at −80°C for 28 days (F28) for later analysis. Five ml of the newly developed preservative solution (Metabolokeeper®), containing guanidine thiocyanate and detergent, was added to the other three tubes. After vigorous mixing, the samples were stored at 30°C for 7 (D7), 14 (D14), or 28 (D28) days before analysis of SCFAs and BAs, and DNA extraction.
Fig. 1.
Schematic of the study design including the workflow. Fecal samples were collected from 20 healthy adults and each fecal sample was divided into five aliquots. One aliquot was supplied as it was (D0), and another was frozen and stored at −80°C for 28 days (F28). Another three aliquots were stored in Metabolokeeper® at 30°C for 7 (D7), 14 (D14), and 28 (D28) days. The microbiota and metabolites in each specimen were analyzed.
This study conformed to the ethical code defined in the Declaration of Helsinki. The Ethics Committee of the Kyoto Prefectural University of Medicine approved the research protocol (approval number: ERB-C-1277-1), and all participants provided written informed consent before enrollment.
Analysis of SCFAs
Frozen fecal samples (F28) were thawed on ice. 100 mg of fecal samples (D0 and F28) or 300 μl of samples stored in Metabolokeeper® (D7, D14, and D28) were placed in 2.0 ml tubes with zirconia beads, and MilliQ water was added to the tubes up to 1 ml. The tubes were heated at 80°C for 15 min, vortexed at 5 m/s for 45 s using a FastPrep 24 5G (MP Biomedicals, Irvine, CA), and centrifuged at 18,400 × g for 10 min. The supernatant was filtrated through a 0.2 μm filter (Ultrafree-MC; Merck Millipore, Billerica, MA). SCFAs (acetic acid, propionic acid, butyric acid, iso-butyric acid, succinic acid, lactic acid, formic acid, valeric acid, and iso-valeric acid) in feces were measured by high-performance liquid chromatography (Prominence; Shimadzu, Kyoto, Japan) using a post-column reaction with a detector (CDD-10A, Shimadzu), two tandem columns [Shim-pack SCR-102(H), 300 mm × 8 mm ID; Shimadzu], and a guard column [Shim-pack SCR-102(H), 50 mm × 6 mm ID; Shimadzu]. The LC system was used with a mobile phase (5 mM p-toluenesulfonic acid) and reaction solution (5 mM p-toluenesulfonic acid, 100 μM EDTA, and 20 mM Bis-Tris). The flow rate and oven temperature were 0.8 ml/min and 45°C, respectively. The detector cell temperature was maintained at 48°C. Measurements were performed using an absolute calibration curve (ranging from 5 to 1,000 mg/L). All analyses were performed using three aliquots from each tube. The data are presented as the average of triplicate analyses.
Analysis of BAs
BAs were extracted from fecal samples using a previously described method with minor modifications.(15) 100 mg of fecal samples (D0 and F28) or 100 μl of samples stored in Metabolokeeper® (D7, D14, and D28) were placed in 2.0 ml-tubes containing zirconia beads and suspended in 900 μl of 50 mM cold sodium acetate buffer (pH 5.6)/ethanol mixture (1:3, v/v). The tubes were vortexed (5 m/s, 45 s) using FastPrep 24 5G (MP Biomedicals) and heated at 85°C for 30 min. After centrifugation (18,400 × g, 10 min), the supernatant was diluted four times with MilliQ water and applied to a Bond Elute C18 cartridge (500 mg/6 ml, Agilent Technologies, Palo Alto CA). The cartridge was washed with 10% ethanol (5 ml), and BAs were eluted with ethanol (5 ml). The solvent was evaporated and the residue dissolved in 50% ethanol (1 ml). The extracted solution was diluted with 50% ethanol, including an internal standard, and transferred to a vial after filtration using a 0.2 μm filter (Ultrafree-MC; Merck Millipore).
Quantification of bile acids was performed using a Waters Acquity UPLC system with an Acquity UPLC BEH C18 column (2.1 × 150 mm, pore size 1.7 μm; Waters Co., Milford, MA) coupled with a Waters Xevo G2-S QTOF mass spectrometer with an electrospray ionization probe. The injection volume was 4 μl. Mobile phase A was water and mobile phase B was acetonitrile, both containing 0.1% formic acid. The flow rate was 0.5 ml/min. The mobile phase was increased to 35% B from 0.5 to 1.0 min, increased to 40% B from 1.0 to 7.0 min, and increased to 50 from 7.0 to 10.0. The mobile phase was then changed to 95% B over 0.5 min and maintained for 1.5 min. The proportion was adjusted to the initial ratio of 30% B, and this was maintained for 4 min to equilibrate the column. The column and autosampler temperatures were maintained at 65°C and 10°C, respectively. The Waters Xevo G2-S QTOF was run in negative mode (scan 50–850 amu at a rate of 0.3 scans per second). The following instrument conditions were used: capillary, 0.5 kV; source temperature, 150°C; sampling cone, 20 V; cone gas, 100 L/h; desolvation gas flow, 1,000 L/h at 450°C. Leucine enkephalin was used as the reference lock mass (m/z 554.2615) with a lock-mass spray to ensure mass accuracy and reproducibility. Data analysis was performed using TargetLynx software (Waters). All analyses were performed using three aliquots from each tube. The data are presented as the average of triplicate analyses.
DNA extraction
Fecal samples (100 mg; D0 and F28) or 100 μl of samples stored in Metabolokeeper® (D7, D14, and D28) were suspended in 900 μl GTC buffer [4 M guanidium thiocyanate, 100 mM Tris-HCl (pH 9.0), and 40 mM EDTA]. These samples were then beaten with zirconia beads using FastPrep 24 5G (MP Biomedicals). DNA was purified from bead-treated suspensions using GENE PREP STAR PI-480 (Kurabo Industries Ltd., Osaka, Japan). DNA concentrations were estimated by spectrophotometry using a NanoDrop 8000 (Thermo Fisher Scientific, Waltham, MA), and the final concentration of the DNA sample was adjusted to 10 ng/μl.
Illumina library preparation, amplicon sequencing of 16S rRNA genes, and data analysis
Library preparation, including PCR amplification of the V3–V4 region of the 16S rRNA gene, was carried out as previously described.(16) The forward and reverse primers also contained an 8-bp indexing sequence to allow multiplexing.(17) Sequencing was conducted using the Illumina MiSeq sequencing system and the MiSeq Reagent Kit v3 (600 cycle) chemistry (Illumina Inc., San Diego, CA). Paired-end sequencing reads were merged using the fastq-join program with default settings.(18) The joined amplicon sequence reads were processed using the QIIME2 software.(19) Quality filtering and deletion of chimeric sequences were performed, and representative sequences were created using DADA2 (Divisive Amplicon Denoising Algorithm 2) denoise-single plugin ver. 2017.6.0, with default settings.(20) The taxonomy of representative sequences was assigned using Greengenes database ver. 13.8 by training a naive Bayes classifier using the q2-feature-classifier plugin.(21)
Statistical analysis
The Shannon index for alpha diversity was calculated using the QIIME2 diversity alpha-rarefaction plugin. Bonferroni-corrected pairwise Wilcox tests were executed to compare the Shannon index among the three groups (D0, D28, and F28) using R.(22) For beta diversity, principal coordinate analysis (PCoA) was performed based on Bray–Curtis distances using the QIIME2 diversity core-metrics-phylogenetic plugin. To compare the Bray–Curtis distances from the D0 group to the other two groups, the Kruskal Wallis test was performed using R.(22)
PCoA plots as 2D graphics with alpha diversity indices were shown using tidyverse ver. 1.2.,(23) and qiime2R ver. 0.99.13 packages of R software.(22,23) The statistical significance of the similarity of bacterial communities among groups was assessed with the ANOSIM test using the beta-group-significance plugin.
Data availability
The nucleotide sequence dataset was deposited in the Sequence Read Archive of the DNA Data Bank of Japan under accession number DRADRR302116.
Results
Effect of Metabolokeeper® on the preservation of SCFAs in feces
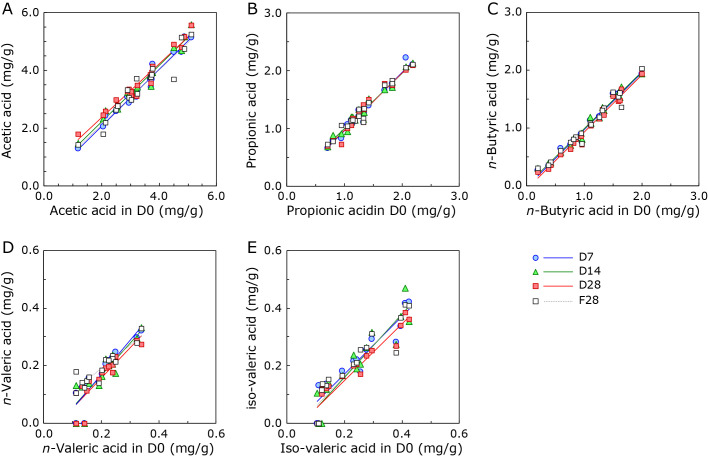
To evaluate the usefulness of Metabolokeeper®, feces from 19 healthy subjects were suspended in a preservative solution and stored at 30°C. Aliquots of the suspension were removed for the analysis of SCFAs on days 0, 7, 14, and 28 (D0, D7, D14, and D28, respectively). As a reference, an aliquot from the suspension stored at −80°C for 28 days (F28) was analyzed. Analysis data of the major SCFAs (acetic acid, propionic acid, n-butyric acid, iso-butyric acid, n-valeric acid, iso-valeric acid, and lactic acid) are presented in Supplemental Table 1*. Scatter plot analysis followed by the construction of a linear equation was performed to clarify the relationship between different groups (D0 and D7, D14, D28, and F28) and the major SCFAs, except for iso-valeric and lactic acid (Fig. 2). As shown in Table 1, the slopes (m) of equations for D0 vs D7, D14, and D28 were almost one (m = 0.9306 to 1.1889), which indicates that the SCFAs in Metabolokeeper® are very well preserved for 28 days at 30°C as well as at −80°C. No significant difference in the profile of SCFAs in each fecal specimen was observed between D0, D7, D14, and D28 (Friedman’s test, p>0.05, data not shown).
Fig. 2.
Scatter plot of short-chain fatty acids from D0 and D7, D14, D28, or F28. The correlation between fecal short-chain fatty acids from D0 and D7, D14, D28, and F28 was evaluated. (A) acetic acid, (B) propionic acid, (C) n-butyric acid, (D) n-valeric acid, and (E) iso-valeric acid.
Table 1.
Correlation between fresh (D0) and stored (D7, D14, D28 or F28) in short-chain fatty acid
| Linear equation (y = mx + b) |
r 2 | n | Linear equation (y = mx + b) |
r 2 | n | |||
|---|---|---|---|---|---|---|---|---|
| Acetic acid | n-Valeric acid | |||||||
| D0 vs D7 | y = 0.9858x + 0.0864 | 0.973 | 19 | D0 vs D7 | y = 1.1889x − 0.0689 | 0.817 | 13 | |
| D0 vs D14 | y = 0.9763x + 0.2549 | 0.973 | 19 | D0 vs D14 | y = 1.1235x − 0.0599 | 0.756 | 13 | |
| D0 vs D28 | y = 0.9306x + 0.4877 | 0.964 | 19 | D0 vs D28 | y = 1.0517x − 0.0538 | 0.786 | 13 | |
| D0 vs F28 | y = 0.9501x + 0.2203 | 0.924 | 19 | D0 vs F28 | y = 0.8016x + 0.0313 | 0.849 | 15 | |
| Propionic acid | Iso-valeric acid | |||||||
| D0 vs D7 | y = 1.012x − 0.0581 | 0.966 | 19 | D0 vs D7 | y = 0.9977x − 0.0288 | 0.881 | 15 | |
| D0 vs D14 | y = 0.9578x + 0.0372 | 0.968 | 19 | D0 vs D14 | y = 1.0924x − 0.0573 | 0.859 | 14 | |
| D0 vs D28 | y = 0.9902x − 0.0227 | 0.954 | 19 | D0 vs D28 | y = 0.9851x − 0.0486 | 0.898 | 14 | |
| D0 vs F28 | y = 0.9678x + 0.0203 | 0.965 | 19 | D0 vs F28 | y = 1.0628x − 0.048 | 0.866 | 14 | |
| n-Butyric acid | ||||||||
| D0 vs D7 | y = 0.987x − 0.0197 | 0.983 | 19 | |||||
| D0 vs D14 | y = 0.9798x + 0.0037 | 0.979 | 19 | |||||
| D0 vs D28 | y = 0.9894x − 0.0668 | 0.979 | 19 | |||||
| D0 vs F28 | y = 0.9544x + 0.0115 | 0.962 | 19 |
Numerical datasets for iso-butyric acid and lactic acid are not presented.
Effect of Metabolokeeper® on the preservation of BAs in feces
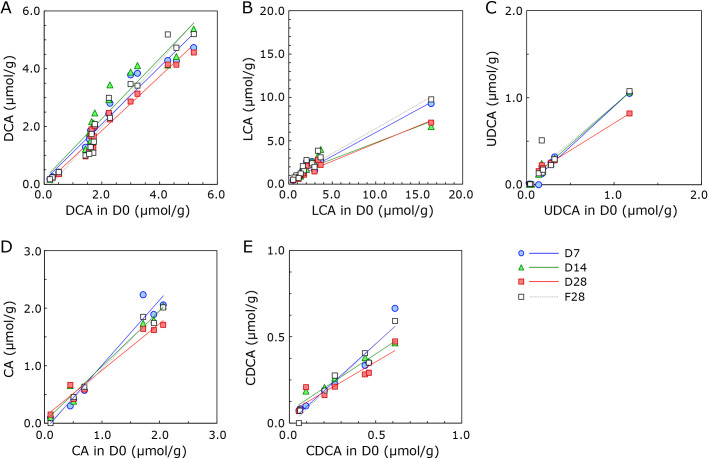
The test for preservation of BAs in Metabolokeeper® was performed as for SCFAs. Analysis data of the major BAs [deoxycholic acid (DCA), lithocholic acid (LCA), ursodeoxycholic acid (UDCA), cholic acid (CA), chenodeoxycholic acid (CDCA), glycocholic acid (GCA), and 7-oxo-LCA] are presented in Supplemental Table 1*. As shown in Fig. 3, the amount of BAs, except DCA, tended to decrease during storage. However, as shown in Table 2, the amount of each BA was well preserved during the period of 28 days for DCA (m = 0.9347), 14 days for UDCA (m = 0.8901) and CA (m = 0.9631), and 7 days for CDCA (m = 0.9080). LCA was not well preserved even in the 7 day-storage period (m = 0.5458), but a high correlation was observed between D0 and D7, D14, and D28 (r2 = 0.830–0.967). When subject No. 12, which had outlying LCA levels, was removed from calculations [represented as LCA(−No. 12)], the slope (m) of the linear equation increased (m = 0.7028). The amount of LCA also decreased in F28 (m = 0.5756). The correlation between D0 and D7, D14, and D28 in GCA and 7-oxo-LCA was considerably lower than that of the other BAs (Table 2), indicating that the quantified values of GCA and 7-oxo-LCA stored in Metabolokeeper® are unreliable. No significant difference in the BA profile of each fecal specimen was observed between D0, D7, D14, and D28 (Friedman’s test, p>0.05, data not shown). Consequently, it is recommended that the storage period in Metabolokeeper® for BA analysis be less than or equal to seven days.
Fig. 3.
Scatter plot of bile acids from D0 and D7, D14, D28, or F28. The correlation between bile acids from D0 and D7, D14, D28, and F28 was evaluated. (A) DCA, deoxycholic acid; (B) LCA, lithocholic acid; (C) UDCA, ursodeoxycholic acid; (D) CA, cholic acid; (E) CDCA, chenodeoxycholic acid. A scatter plot for glycocholic acid (GCA) or 7-oxo-lithocholic acid (LCA) is not shown.
Table 2.
Correlations between fresh (D0) and stored (D7, D14, D28 or F28) in bile acids
| Linear equation (y = mx + b) |
r 2 | n | Linear equation (y = mx + b) |
r 2 | n | |||
|---|---|---|---|---|---|---|---|---|
| DCA | CDCA | |||||||
| D0 vs D7 | y = 0.9817x + 0.1280 | 0.949 | 18 | D0 vs D7 | y = 0.9080x + 0.0089 | 0.920 | 10 | |
| D0 vs D14 | y = 1.0394x + 0.1870 | 0.919 | 18 | D0 vs D14 | y = 0.6606x + 0.1417 | 0.960 | 9 | |
| D0 vs D28 | y = 0.9347x − 0.0131 | 0.963 | 18 | D0 vs D28 | y = 0.5860x + 0.1225 | 0.873 | 10 | |
| D0 vs F28 | y = 1.1305x − 0.2633 | 0.950 | 18 | D0 vs F28 | y = 0.9375x − 0.0222 | 0.960 | 6 | |
| LCA | GCA | |||||||
| D0 vs D7 | y = 0.5458x + 0.5000 | 0.967 | 15 | D0 vs D7 | y = 0.7269x + 0.1389 | 0.6169 | 19 | |
| D0 vs D14 | y = 0.3826x + 0.9571 | 0.830 | 15 | D0 vs D14 | y = 0.6311x + 0.1193 | 0.6223 | 19 | |
| D0 vs D28 | y = 0.4119x + 0.5795 | 0.938 | 15 | D0 vs D28 | y = 0.7978x + 0.1717 | 0.5596 | 19 | |
| D0 vs F28 | y = 0.5756x + 0.6568 | 0.944 | 15 | D0 vs F28 | y = 0.5980x + 0.1216 | 0.5871 | 19 | |
| UDCA | 7-oxo-LCA | |||||||
| D0 vs D7 | y = 0.9253x − 0.0268 | 0.979 | 6 | D0 vs D7 | y = 6.0248x + 2.3714 | 0.3698 | 13 | |
| D0 vs D14 | y = 0.8901x + 0.0167 | 0.989 | 7 | D0 vs D14 | y = 4.6485x + 2.3714 | 0.2254 | 13 | |
| D0 vs D28 | y = 0.6260x + 0.0808 | 0.996 | 7 | D0 vs D28 | y = 2.7359x + 3.1154 | 0.0655 | 13 | |
| D0 vs F28 | y = 0.8417x + 0.0688 | 0.844 | 7 | D0 vs F28 | y = 0.5414x + 0.1277 | 0.6206 | 13 | |
| CA | LCA(−No. 12)* | |||||||
| D0 vs D7 | y = 1.1313x − 0.1236 | 0.953 | 8 | D0 vs D7 | y = 0.7028x + 0.1069 | 0.7826 | 14 | |
| D0 vs D14 | y = 0.9631x + 0.0245 | 0.982 | 7 | D0 vs D14 | y = 1.0079x + 0.0721 | 0.9422 | 14 | |
| D0 vs D28 | y = 0.8104x + 0.1084 | 0.969 | 9 | D0 vs D28 | y = 0.6679x + 0.0958 | 0.8021 | 14 | |
| D0 vs F28 | y = 1.0222x − 0.0772 | 0.987 | 6 | D0 vs F28 | y = 0.9178x + 0.1257 | 0.8162 | 14 |
*The subject No. 12 was removed from the calculation, because it had outlier in LCA. DCA, deoxycholic acid; LCA, lithocholic acid; UDCA, ursodeoxycholic acid; CA, cholic acid; CDCA, chenodeoxycholic acid; GCA, glycocholic acid; 7-oxo-LCA, 7-oxo-lithocholic acid.
Effect of Metabolokeeper® on the preservation of structure and diversity of microbiota
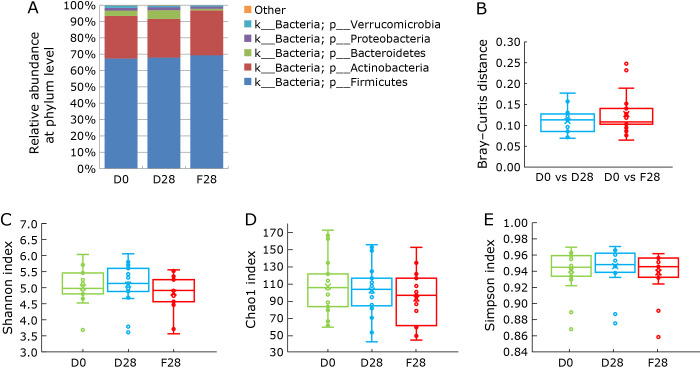
In this study, we evaluated the structure and diversity of fecal microbiota. As shown in Fig. 4A, there was no obvious change in gut microbiota composition at the phylum level between D0, D28, and F28. In addition, there was no obvious alteration in the gut microbiota composition at the genus level between D0, D28, and F28 (Supplemental Fig. 1*). In the analysis of microbial community structure based on Bray–Curtis distance, no significant differences in the structure of fecal microbiota were detected between the storage conditions (Fig. 4B, Supplemental Fig. 2*). In addition, no significant differences in the diversity of fecal microbiota were detected between storage conditions using α-diversity indices (Shannon, Chao1, and Simpson indices; Fig. 4C–E). These results suggest that Metabolokeeper® well preserved the fecal microbiota at 30°C and −80°C for 28 days.
Fig. 4.
Taxonomic composition and diversity of the fecal microbiota in fresh (D0) and stored (D28 or F28) fecal samples. (A) Each component of the cumulative bar chart indicates a phylum. Taxonomic composition of the fecal microbiota at phylum level was evaluated among D0, F28, and D28. (B) Microbial community structure was evaluated by Bray–Curtis distance (p>0.05, Kruskal–Wallis test). The α-diversity was evaluated using Shannon (C), Chao1 (D), and Simpson (E) indices of the D0 and stored samples (D28 or F28) (p>0.05, Bonferroni–Dunn test).
Discussion
In the present study, we evaluated the usefulness of preservation using our newly developed preservation solution, “Metabolokeeper®”, at 30°C for 28 days, compared to the gold standard method of immediate freezing at −80°C, on the observed microbial community structure and the metabolite contents of fecal samples. Our results show that the new preservative successfully preserved the fecal structure of gut microbiota and fecal concentration of SCFAs, even when samples were stored at 30°C for 28 days, as well as when samples were frozen at −80°C for 28 days. BAs were successfully preserved at 30°C for 7 days only.
As the integrated analysis of gut microbiota and metabolites holds the potential to reveal interactions between the host and microbiota in relation to disease risks, the convenient and accurate simultaneous measurement of gut microbiota and metabolites in fecal samples is deeply important. Although several reports have demonstrated the stability of fecal microbiome composition,(14,24,25) there is limited knowledge on the stability of fecal metabolites. Wang et al.(12) have demonstrated that five fecal collection methods [immediate freezing at −20°C without preservative, OMNIgene GUT, 95% ethanol, RNAlater, and Flinders Technology Associates (FTA) cards] exhibit differences in certain measures of the gut microbiome and fecal metabolome, suggesting the importance of developing a preservative solution to stabilize fecal microbiota and organic acids such as SCFAs and BAs.
Lim et al.(13) have described the usefulness of using OMNIgene-GUT solution to store fecal samples for the measurement of gut metabolites; however, satisfactory results were not obtained at room temperature storage. We developed a new preservative solution containing a detergent and guanidine thiocyanate. The detergent makes it possible to promptly dissolve fecal samples in the preservation solution so that more stabilization of fecal microbiota, SCFAs, and BAs at room temperature can be achieved. As the immediate deep-freezing of fecal samples is often inconvenient in routine clinical practice, this simple storage method using a preservative solution, which has an equivalent efficacy to deep-freezing, may act to stabilize fecal samples in a convenient manner.
In conclusion, this study demonstrated the usefulness of our newly developed preservation solution, “Metabolokeeper®”, which does not change the structure of the gut microbiota or its metabolite levels, even when samples are left at room temperature for a long period. Although the evaluation of metabolites other than SCFAs and BAs, and the potential for longer-term preservation, with this novel solution remain unknown and further studies are warranted, this preservation solution is expected to make a great contribution to fecal collection in clinical settings, and to further our understanding of the health effects of fecal metabolites produced by the gut microbiome.
Author Contributions
Study concept and design, TT and YN; acquisition of data, TK, ST, TH, and KM; analysis and interpretation of data, TT, TK, ST, TH, and JM; drafting of the manuscript, TT and TK; critical revision of the manuscript for important intellectual content, KN; final approval, all authors; study supervision, YN.
Acknowledgments
We would like to thank Editage (www.editage.jp) for assistance with English language editing.
Abbreviations
- BA
bile acid
- CA
cholic acid
- CDCA
chenodeoxycholic acid
- DCA
deoxycholic acid
- GCA
glycocholic acid
- LCA
lithocholic acid
- 7-oxo-LCA
7-oxo-lithocholic acid
- PCoA
principal coordinate analysis
- SCFA
short-chain fatty acid
- UDCA
ursodeoxycholic acid
Conflict of Interest
TT received collaborative research funds from Fujifilm Medical Co., Ltd. and lecture fees from Mochida Pharma. Co. Ltd., and Yanssen Pharmaceutical K.K. YN received scholarship funds from EA Pharma Co. Ltd., collaboration research funds from Taiyo Kagaku Co., Ltd., and lecture fees from Mylan EPD Co., Takeda Pharma. Co., Ltd. and Mochida Pharma. Co. Ltd., EA Pharma Co., Ltd., Otsuka Pharma. Co. Ltd. and Miyarisan Pharma. Co. Ltd. This research was not supported by any of these funds. In addition, neither the funding agencies nor any outside organizations participated in the study design nor had any competing interests.
Supplementary Material
References
- 1.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med 2016; 375: 2369–2379. [DOI] [PubMed] [Google Scholar]
- 2.Naito Y, Takagi T, Inoue R, et al. Gut microbiota differences in elderly subjects between rural city Kyotango and urban city Kyoto: an age-gender-matched study. J Clin Biochem Nutr 2019; 65:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takagi T, Naito Y, Inoue R, et al. Differences in gut microbiota associated with age, sex, and stool consistency in healthy Japanese subjects. J Gastroenterol 2019; 54: 53–63. [DOI] [PubMed] [Google Scholar]
- 4.Takagi T, Naito Y, Kashiwagi S, et al. Changes in the gut microbiota are associated with hypertension, hyperlipidemia, and type 2 diabetes mellitus in Japanese subjects. Nutrients 2020; 12: 2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchesi JR, Adams DH, Fava F, et al. The gut microbiota and host health: a new clinical frontier. Gut 2016; 65: 330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staley C, Weingarden AR, Khoruts A, Sadowsky MJ. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl Microbiol Biotechnol 2017; 101: 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nyangale EP, Mottram DS, Gibson GR. Gut microbial activity, implications for health and disease: the potential role of metabolite analysis. J Proteome Res 2012; 11: 5573–5585. [DOI] [PubMed] [Google Scholar]
- 8.Markowiak-Kopeć P, Śliżewska K. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients 2020; 12: 1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol 2014; 30: 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salonen A, Nikkilä J, Jalanka-Tuovinen J, et al. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods 2010; 81: 127–134. [DOI] [PubMed] [Google Scholar]
- 11.Panek M, Čipčić Paljetak H, Barešić A, et al. Methodology challenges in studying human gut microbiota-effects of collection, storage, DNA extraction and next generation sequencing technologies. Sci Rep 2018; 8: 5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Zolnik CP, Qiu Y, et al. Comparison of fecal collection methods for microbiome and metabolomics studies. Front Cell Infect Microbiol 2018; 8: 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim MY, Hong S, Kim BM, Ahn Y, Kim HJ, Nam YD. Changes in microbiome and metabolomic profiles of fecal samples stored with stabilizing solution at room temperature: a pilot study. Sci Rep 2020; 10: 1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosomi K, Ohno H, Murakami H, et al. Method for preparing DNA from feces in guanidine thiocyanate solution affects 16S rRNA-based profiling of human microbiota diversity. Sci Rep 2017; 7: 4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kakiyama G, Muto A, Takei H, et al. A simple and accurate HPLC method for fecal bile acid profile in healthy and cirrhotic subjects: validation by GC-MS and LC-MS. J Lipid Res 2014; 55: 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi S, Tomita J, Nishioka K, Hisada T, Nishijima M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS One 2014; 9: e105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hisada T, Endoh K, Kuriki K. Inter- and intra-individual variations in seasonal and daily stabilities of the human gut microbiota in Japanese. Arch Microbiol 2015; 197: 919–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aronesty E. Comparison of sequencing utility programs. Open Bioinforma J 2013; 7: 1–8. [Google Scholar]
- 19.Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019; 37: 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016; 13: 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R: A language and environment for statistical computing. R Core Team, R Foundation for Statistical Computing. https://www.r-project.org/. Accessed 4 Jul 2022.
- 23.Wickham H, Averick M, Bryan J, et al. Welcome to the tidyverse. J Open Source Softw 2019; 4: 1686. [Google Scholar]
- 24.Kawada Y, Naito Y, Andoh A, Ozeki M, Inoue R. Effect of storage and DNA extraction method on 16S rRNA-profiled fecal microbiota in Japanese adults. J Clin Biochem Nutr 2019; 64: 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishimoto Y, Mizutani S, Nakajima T, et al. High stability of faecal microbiome composition in guanidine thiocyanate solution at room temperature and robustness during colonoscopy. Gut 2016; 65: 1574–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nucleotide sequence dataset was deposited in the Sequence Read Archive of the DNA Data Bank of Japan under accession number DRADRR302116.