Abstract
Expression of cytoplasmic mRNA from most adenovirus transcription units is subjected to a temporal regulation at the level of alternative pre-mRNA splicing. The general tendency is that splice site selection changes from proximal to distal late after infection. Interestingly, ASF/SF2, which is a prototypical member of the SR family of splicing factors, has the opposite effect on splice site selection, inducing an increase in proximal splice site usage. We have previously shown that SR proteins late during an adenovirus infection become partially inactivated as splicing regulatory proteins. A prediction from these results is that overexpression of an SR protein, such as ASF/SF2, during virus growth will interfere with virus replication by disturbing the balance of functional and nonfunctional ASF/SF2 in the infected cell. To test this hypothesis, we reconstructed a recombinant adenovirus expressing ASF/SF2 under the transcriptional control of a regulated promoter. The results show that, as predicted, induction of ASF/SF2 during lytic virus growth prevents the early to late shift in mRNA expression from both early (E1A and E1B) and late (L1) transcription units. Furthermore, ASF/SF2 overexpression blocks viral DNA replication and reduces selectively cytoplasmic accumulation of major late mRNA, resulting in a lower virus yield. Collectively, our results provide additional support for the hypothesis that viral control of SR protein function is important for the proper expression of viral proteins during lytic virus growth.
Adenovirus gene expression is to a large extent regulated at the level of pre-mRNA alternative splicing (reviewed in reference 21). Thus, the majority of adenovirus transcription units produce multiple differently spliced cytoplasmic mRNAs. The production of several mRNAs from each viral transcription unit increases the coding potential of the viral genome and, furthermore, enables the virus to control viral gene expression by regulating the specificity of RNA splicing. Importantly, the accumulation of alternatively spliced mRNAs is subjected to a temporal regulation, with distinct mRNA species accumulating at different time points of infection (reviewed in reference 21).
Although the mechanism(s) responsible for this shift in RNA splice site choice is far from clear, a number of studies have suggested that the cellular SR family of splicing factors plays a critical role in this regulation. SR proteins constitute a family of splicing factors that are essential for generic pre-mRNA splicing (reviewed in references 12 and 28). They contain one or two amino-terminal RNA binding domains and a characteristic carboxy-terminal RS domain consisting of repeats of serine/arginine dipeptides of variable length, hence the name SR proteins. The RNA recognition motif determines the RNA binding specificity, whereas the RS domain functions as an effector domain mediating interaction with other splicing factors (reviewed in references 12 and 28). SR proteins participate at multiple steps during the initial stages of splice site recognition. Most SR proteins interact with simple RNA elements containing a high content of purines (reviewed in reference 42) and have the capacity to enhance or repress splicing, depending on the position of the RNA recognition motif in the pre-mRNA (22). SR proteins appear to serve the same function in spliceosome assembly as regulatory transcription factors in preinitiation complex formation at a promoter (reviewed in reference 17). Importantly, SR proteins are highly phosphorylated, primarily within the RS domain (9), a modification that is required for their function as initiators of spliceosome assembly (7, 47) and which controls their activity in alternative splicing (23). During splicing catalysis, SR proteins become dephosphorylated. Thus, cycles of SR protein phosphorylation/dephosphorylation occur during spliceosome assembly and pre-mRNA splicing in vitro (7, 31).
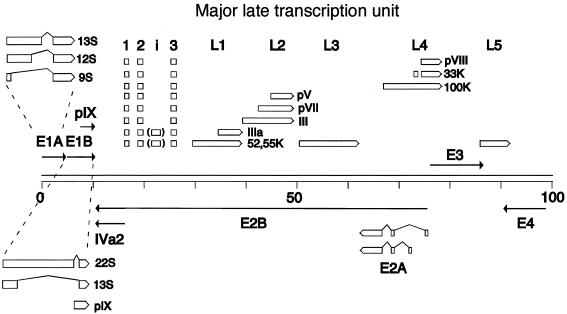
The adenovirus E1A and L1 pre-mRNAs have been subjected to the most thorough investigation (reviewed in reference 21). The E1A pre-mRNA gives rise to three major mRNAs, the 13S, 12S, and 9S mRNAs, by use of three alternative 5′ splice sites which are spliced to a common 3′ splice site (Fig. 1). The 13S mRNA is the most abundant E1A mRNA expressed at early times of infection, whereas the 9S mRNA becomes the predominant E1A mRNA expressed late after infection. Different members of the SR protein family have been shown to have distinct effects on E1A pre-mRNA splicing. Thus, ASF/SF2 and SC35 activate 13S splicing, both in vitro (11, 16, 48) and in transient transfection assays (6, 45). In contrast, SR protein p54 promotes selectively 9S splicing (49). Other SR proteins, e.g., SRp20, 9G8, SRp55, and SRp75, promote 12S splicing (18, 37, 48). The early to late shift from 13S to 9S mRNA splicing has been proposed to be triggered by a titration of functionally active SR proteins by high-molecular-weight adenovirus major late transcript, produced late after infection (13, 18, 25).
FIG. 1.
Schematic illustration showing the spliced structure of mRNAs expressed from regions E1A, E1B, E2A, L1, L2, and L4. For simplicity, the exon structures of mRNAs expressed from the major late transcription unit are shown as open boxes.
The adenovirus major late region 1 (L1) pre-mRNA produces two major mRNAs by alternative 3′ splice site selection (reviewed in reference 21). Thus, a common 5′ splice site is joined to either of two alternative 3′ splice sites (Fig. 1), resulting in the formation of the 52/55K mRNA (mRNA with an Mr of 52,000 to 55,000) (proximal 3′ splice site) or the IIIa mRNA (distal 3′ splice site). At early times of infection, the 52,55K mRNA is the exclusive L1 mRNA produced, whereas the IIIa mRNA becomes the predominant L1 mRNA expressed at late times of infection. In contrast to E1A pre-mRNA splicing, L1 IIIa mRNA splicing is suppressed by SR proteins. Thus, all SR proteins tested except SRp20 (22, 36) inhibit IIIa splicing in vitro, with the SRp30 fraction, which include ASF/SF2, being most effective. HeLa SR proteins block IIIa 3′ splice site usage by binding to the intronic IIIa repressor element (3RE), located immediately upstream of the IIIa branch site (22). SR proteins binding to the 3RE suppress IIIa splicing by preventing U2 snRNP recruitment to the spliceosome. Activation of IIIa splicing, late after infection, has been shown to be accompanied by a virus-induced dephosphorylation of HeLa SR proteins (23). This modification reduces the RNA binding capacity of SR proteins and as a consequence relieves their repression of IIIa 3′ splice site usage, resulting in a shift toward an increase in IIIa mRNA production.
Collectively, the work on E1A and L1 alternative splicing has suggested that SR proteins play a critical role in the early to late shift in adenovirus pre-mRNA splicing. However, the conclusions reached so far are based solely on in vitro splicing experiments or transient transfection studies. Thus, the significance of SR proteins as regulators of viral pre-mRNA splicing during lytic virus growth has not been tested.
Assuming that SR proteins are important regulatory factors controlling viral gene expression, one would predict that overexpression of an SR protein during a lytic infection would have a significant impact on viral mRNA expression by shifting the balance of functional and nonfunctional SR proteins toward the active form and therefore interfere with viral multiplication. Here we test this hypothesis by constructing a recombinant adenovirus expressing the prototypical SR protein ASF/SF2 under the inducible control of an RU 486-regulated gene cassette (32). The results show that ASF/SF2 overexpression blocks the early to late shift in viral mRNA expression. Thus, the early splice patterns of the E1A, E1B, and L1 mRNAs were retained also at late time points of infection. Furthermore, ASF/SF2 overexpression reduced the efficiency of viral DNA replication, major late mRNA accumulation, and new virus particle formation. Collectively, our data emphasize the significance of viral control of alternative RNA splicing as a mean to regulate adenovirus gene expression during lytic virus growth.
MATERIALS AND METHODS
Construction of a recombinant adenovirus expressing ASF/SF2.
Plasmid maps and sequences are available on request or online (http://www.bmc.uu.se/IMIM/res/GA.html). Transfer plasmid pAdG5Trip(His)-ASF/SF2 was constructed by replacing the chloramphenicol acetyltransferase (CAT) gene in plasmid pAdG5Trip-CAT (32) with a synthetic double-stranded oligonucleotide encoding six histidines followed by a cDNA encoding the human ASF/SF2 protein (taken from Prey-ASF [46]). Plasmid pAdG5Trip(His)-ASF/SF2 was reconstructed into a recombinant adenovirus essentially as described by Stow (39). Briefly, a vector arm was produced by double digestion of genomic adenovirus type 5 (Ad5) dl309 DNA with XbaI and ClaI, followed by sucrose gradient purification of the long right-hand genomic fragment (from 3.71 to 100 map units [m.u.]). Recombinant viruses were generated by in vivo recombination. Thus, 5 μg of pAdG5Trip(His)-ASF/SF2 DNA was mixed with 1 μg of Ad5 dl309 vector arm and cotransfected by the calcium phosphate coprecipitation technique to 293 cells (14). Plaques typically appeared 5 to 6 days posttransfection. Recombinant viruses were verified by restriction enzyme cleavage of Hirt-extracted viral DNA (19). One plaque expressing His-ASF/SF2 was selected and purified by a second round of plaque assay. The final reporter virus was named AdG5(His)-ASF. The activator virus AdCMVProg has previously been described (32).
Purification of recombinant adenovirus.
High-titer stock of recombinant virus was produced essentially as described elsewhere (20). Briefly, six 15-cm plates of 293 cells were infected with recombinant virus. Three days postinfection, when a clear cytopathic effect was visible, cells were harvested by low-speed centrifugation. The cell pellet was freeze-thawed once and resuspended in 2 ml of 0.1 M Tris-HCl (pH 8.0) followed by lysis with 0.1 volume of 5% sodium deoxycholate on ice for 30 min. Subsequently, the cell lysate was sonicated on ice. Virus was purified by CsCl centrifugation. The virus band was collected and dialyzed against 100 volumes of phosphate-buffered saline (PBS) containing 1 mM CaCl2, 1 mM MgCl2, and 10% glycerol, using a Slide-A-Lyzer cassette (Pierce). Virus titer was determined by plaque assay. Typical virus titers exceeded 1010 PFU/ml.
Infections.
Virus stocks were diluted in 1 ml of Dulbecco modified Eagle medium (DMEM) supplemented with 2% newborn calf serum and used to infect subconfluent 6-cm-diameter plates of 293 or HeLa cells (final concentration, 10 PFU/cell). In all experiments, 5 PFU of AdCMVProg and 5 PFU of AdG5(His)-ASF per cell were used. In Fig. 6, 10 PFU of dl309 per cell was added to the activator-reporter mix. Inoculum was removed after a 1-h incubation at 37°C, cells were washed two times with fresh DMEM, and then 4 ml of DMEM supplemented with 10% newborn calf serum with or without 0.5 μM RU 486 was added. Infected cells were harvested at different time points and used to prepare RNA, DNA, or protein extracts.
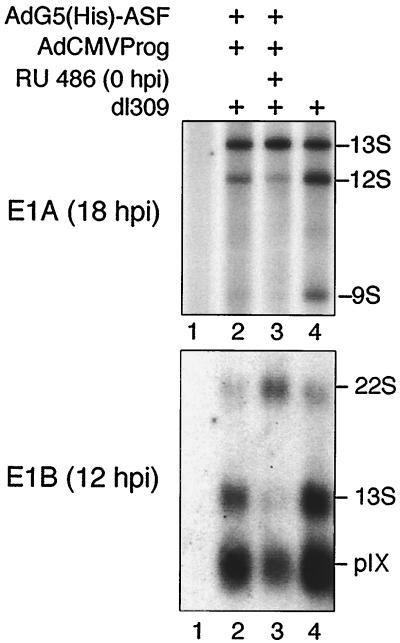
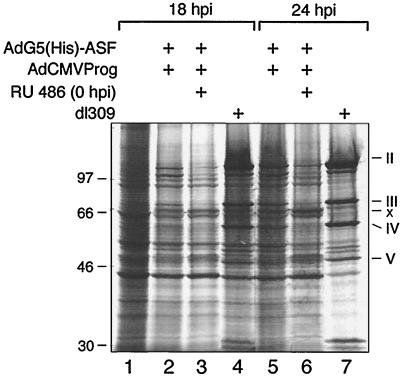
FIG. 6.
ASF/SF2 blocks the temporal shift in E1A and E1B mRNA accumulation. Total cytoplasmic RNA prepared from AdG5(His)-ASF/AdCMVProg/dl309-infected HeLa cells were subjected to neutral S1 analysis to detect individual E1A mRNAs (A) or Northern blot hybridization to detect E1B mRNA expression (B). dl309 was included in this experiment since the recombinant viruses lack the E1 region. Lane 1, mock-infected cells.
Western blot analysis.
Protein extracts were prepared at 12 to 24 h postinfection (hpi) by lysis of infected cells with radioimmunoprecipitation assay buffer (10 mM Tris buffer [pH 7.4], 150 mM NaCl, 1% NP-40, 1% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 μg of aprotonin/ml, 50 μg of phenylmethylsulfonyl fluoride/ml, 10 mM β-glycerophosphate). Extracts were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on a 15% gel and transferred to a nitrocellulose membrane. Filters were treated as previously described (36), and proteins were visualized by chemiluminescence as instructed by the manufacturer (Amersham). His-ASF/SF2 was detected using a His6 monoclonal antibody (Clontech), total ASF/SF2 was detected using a peptide antiserum (kind gift from J. Stevenin), and E2A-72K was detected with monoclonal antibody B610 (kindly provided by E. Bridge). In Fig. 2C, 2 μg of 293 cell extract, prepared at 18 hpi from AdG5(His)-ASF/AdCMVProg-infected cells treated with RU 486, was incubated with 5 U of calf intestinal alkaline phosphatase (Fermentas) for 30 min at 30°C before separation by SDS-PAGE on a long 15% gel. His-ASF/SF2 was visualized by chemiluminescence using a His6 monoclonal antibody (Clontech).
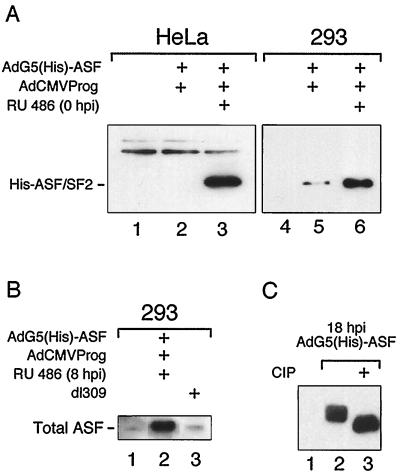
FIG. 2.
Western blot analysis of His-ASF/SF2 expression in AdG5(His)-ASF/AdCMVProg-infected HeLa and 293 cells. Fifty micrograms of protein from HeLa cells and 2 μg of protein from 293 cells were separated by SDS-PAGE on a 15% gel and probed with an anti-His monoclonal antibody (A) or ASF/SF2 peptide antiserum (B). His-ASF/SF2 was induced at the start of infection (A) or at 8 hpi (B), and cells were harvested at 18 (A) or 24 (B) hpi. Panel A shows a short exposure of His-ASF/SF2 expression in 293 cells, to enable comparison of the background expression of His-ASF/SF2 in the absence of added inducer in HeLa and 293 cells. (C) 293 cell protein extracts prepared from AdG5(His)- and ASF/AdCMVProg-infected cells at 18 hpi were treated with calf intestinal alkaline phosphatase (CIP), separated on a long SDS-polyacrylamide gel, and probed with an anti-His monoclonal antibody. Lanes 1 and 4, mock-infected cells.
Northern blot analysis.
Total cytoplasmic RNA was prepared by lysis with IsoB–NP-40 (10 mM Tris-HCl [pH 7.9], 150 mM NaCl, 1.5 mM MgCl2, 1% NP-40), followed by two rounds of phenol-chloroform-isoamyl alcohol extraction and one extraction with chloroform-isoamyl alcohol (3). Two micrograms of cytoplasmic RNA was separated on a 1% agarose gel containing 2.2 M formaldehyde, transferred to a nitrocellulose filter, and hybridized with probe DNA 32P labeled by random priming as previously described (3). The DNA probes used were as follows: L1, SmaI-HindIII (36.3 to 37.9 m.u.); L2, KpnI-SphI (42.8 to 49.5 m.u.); L4, SmaI-L (76.1 to 76.7 m.u.); and E1B, BglII-SmaI (9.2 to 10.9 m.u.).
S1 protection assay.
Ten micrograms of total cytoplasmic RNA prepared as described above was hybridized with an EcoRI/XbaI fragment 5′-end labeled at the XbaI site (41). Single-stranded RNA was digested with S1 nuclease, and RNA-DNA duplexes were separated on a nondenaturing 1% agarose gel as previously described (41).
Dot blot analysis of viral DNA replication.
Viral DNA was prepared from infected 293 cells by Hirt extraction (19). RNase-treated viral DNA was adjusted to 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and denatured by boiling for 10 min. DNA was transferred to a nitrocellulose filter using a vacuum device. The filter was subsequently incubated for 10 min on Whatman paper wetted with 1.5 M NaCl–0.5 M NaOH. The pH was neutralized by placing the filter on Whatman paper wetted with 1 M NaCl–0.5 M Tris (pH 7.0). The DNA was UV cross-linked to the filter and hybridized with the Ad5 XhoI G fragment (27.0 to 28.6 m.u.) 32P labeled by random priming (3).
Titration of virus production.
Infected 293 cells were collected 40 hpi by low-speed centrifugation, washed once with PBS, and then resuspended in PBS containing 1 mM CaCl2, 1 mM MgCl2 and 10% glycerol. Cell suspensions were freeze (−70°C) thawed (37°C) three times. Serial dilutions of freeze-thawed extracts were used for infections of 293 cells. Plaques were counted 13 days postinfection.
35S labeling of infected cells.
Infected 293 cells were pulse-labeled with 100 μCi of 35S-Promix (Amersham) for 2 h beginning at 16 or 22 hpi. Protein extracts were prepared by IsoB–1% NP-40 extraction, and 70-μg aliquots of total proteins were separated by SDS-PAGE on a 10% gel, which was then subjected to autoradiography.
RESULTS
High ASF/SF2 expression using a recombinant adenovirus.
To determine the effect of SR protein overexpression on virus multiplication, we constructed a recombinant adenovirus expressing a His-tagged ASF/SF2 protein under the transcriptional control of a progesterone antagonist-induced promoter (32). The experimental approach is based on a double-infection strategy, where equal numbers of PFU of an activator and a reporter virus are mixed and used for infection. Thus, activator virus, AdCMVProg (32), encoding a chimeric Gal4-VP16-progesterone transactivator protein was mixed with reporter virus, AdG5(His)-ASF, expressing His-ASF/SF2 from a Gal4-regulated promoter. Transcription of the His-ASF/SF2 gene was activated by addition of the progesterone antagonist RU 486 to the culture medium.
As shown in Fig. 2A, His-ASF/SF2 expression is tightly regulated in HeLa cells with a high level of induced expression (lane 3) and no detectable background (lane 2). Infection of 293 cells resulted in an approximately 100-fold increase in His-ASF/SF2 expression compared to HeLa cells (data not shown). The higher expression level results from the fact that the E1-deleted viral vectors replicate in 293 cells. Importantly, infection of 293 cells also results in a detectable background of His-ASF/SF2 in the absence of inducer (Fig. 2A, lane 5). Thus, the fold induction of His-ASF/SF2 is substantially lower in 293 cells than in HeLa cells. Note that much less extract was analyzed in lanes 4 to 6 (293 cells) than in lanes 1 to 3 (HeLa cells). This was done in order to show that the background expression of His-ASF/SF2 in 293 cells was not due to the higher levels of expression obtained in this cell line. The troublesome background expression of His-ASF/SF2 in 293 cells appears to result, in part, from aberrant transcription activation by the promiscuous E1A transactivator protein expressed in the 293 cell line (data not shown).
Interestingly, probing a filter with a peptide antibody that recognizes both endogenously and exogenously expressed ASF/SF2 shows that AdG5(His)-ASF overproduces His-ASF/SF2 more than 20-fold compared to endogenous ASF/SF2 in 293 cells (Fig. 2B). In fact, at 24 hpi the His-ASF/SF2 protein constitutes as much as 2% of total proteins.
We have previously shown that SR proteins late during an adenovirus infection are hypophosphorylated (23). As shown in Fig. 2C, separation of extracts prepared at 18 hpi on a long gel revealed that His-ASF/SF2 overexpression resulted in the accumulation of multiple protein species (lane 2). Treatment of the sample with calf intestinal alkaline phosphatase generated a single faster-migrating protein species (lane 3), indicating that the slower-migrating His-ASF/SF2 species vary in phosphorylated status.
IIIa mRNA splicing is inhibited by ASF/SF2 overexpression.
Since addition of an excess of ASF/SF2 blocks IIIa mRNA splicing in vitro (22), we predicted that His-ASF/SF2 overexpression during a lytic infection would block the early to late shift in L1 alternative splicing. As shown in Fig. 3A, addition of inducer from the start of infection resulted in a dramatic reduction in L1 mRNA accumulation compared to wild type (compare lanes 8 and 9). In contrast, addition of inducer at the start of the late phase of infection (8 hpi) resulted in a less severe effect on total L1 mRNA accumulation (Fig. 3B, lanes 2 and 3). We interpret these results to indicate that His-ASF/SF2 perturbation of early viral pre-mRNA splicing and protein synthesis causes a block in L1 mRNA accumulation (see below).
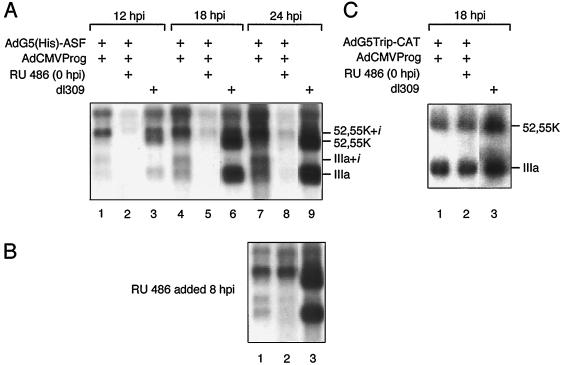
FIG. 3.
ASF/SF2 overexpression inhibits L1 mRNA production. Cytoplasmic RNA was prepared from AdG5(His)-ASF/AdCMVProg (A and B)- or AdG5Trip-CAT/AdCMVProg (C)-infected 293 cells at the indicated time points and separated on a 1% agarose gel containing 2.2 M formaldehyde. RNA was transferred to a nitrocellulose filter and hybridized with a 32P-labeled probe specific for region L1. His-ASF/SF2 was induced from the start of infection (A and C) or at 8 hpi (B).
However, His-ASF/SF2 overexpression also blocked the temporal shift in L1 alternative splicing. Noteworthy, because of the leakiness of the viral vector system (see above), a substantial production of His-ASF/SF2 is also detectable in the absence of inducer (Fig. 2A, lane 5). The effects on alternative RNA splicing are, in fact, best seen in the uninduced samples (Fig. 3A, lanes 1, 4, and 7), with a stronger trend observed after induction (Fig. 3A, lanes 2, 5, and 8). As predicted from our in vitro experiments (22, 23, 36), the relative expression of the IIIa mRNA compared to the 52,55K mRNA was dramatically reduced in AdG5(His)-ASF-infected cells compared to the wild type. Also, the i-leader, which is selectively retained on the 52,55K mRNA population expressed at early and intermediate times of a wild-type infection (Fig. 3A, lane 3), is not efficiently spliced out in AdG5(His)-ASF-infected cells. This results in an almost exclusive accumulation of the 52,55K+i mRNA also at late times of infection (Fig. 3A, lanes 7 and 8). Similarly, the small amounts of IIIa mRNA produced in the recombinant virus-infected cells accumulate with approximately half of the population containing the i-leader (Fig. 3A, lanes 4 and 7). The IIIa+i mRNA has previously been detected only under experimental conditions where late viral protein synthesis was stringently blocked with protein synthesis inhibitors (25).
Collectively, the observed effects of His-ASF/SF2 on the L1 mRNA profile are compatible with the known effects of ASF/SF2 on splicing. Thus, ASF/SF2 has previously been shown to stimulate proximal splice site usage both in vitro and in transiently transfected cells (4, 15, 24, 30, 38), here resulting in the formation of the 52,55K mRNA. ASF/SF2 also stimulates exon inclusion (6, 27, 29), here resulting in the preferential accumulation of L1 mRNAs containing the i-leader.
The effect of His-ASF/SF2 on L1 mRNA accumulation is specific and not a consequence of protein overexpression since AdG5TripCAT (32), overproducing the enzyme CAT, has an L1 phenotype almost identical to that of the parental virus, dl309 (Fig. 3C).
The CAT enzyme serves as a good control for unspecific effects of protein overexpression on viral mRNA accumulation since CAT is of bacterial origin and therefore is not expected to interfere with regulatory circuits in human cells. Furthermore, CAT accumulates predominantly in the nucleus in AdG5TripCAT-infected cells (data not shown). A careful quantitation of His-ASF/SF2 and CAT expression in recombinant virus-infected cells demonstrates that at 18 hpi, His-ASF/SF2 is expressed at approximately fivefold-higher levels compared to CAT. The negative effects of His-ASF/SF2 expression on L1 mRNA accumulation (Fig. 3A) could therefore result from unspecific effects of the high levels of His-ASF/SF2 expression. However, the quantity of His-ASF/SF2 expression at 12 hpi is similar to the CAT expression level at 18 hpi (approximately 0.3 to 0.5% of total protein). Nevertheless, the L1 mRNA profile in CAT-expressing cells is essentially wild type (Fig. 3C), whereas L1 mRNA expression is essentially abolished in His-ASF/SF2-expressing cells (Fig. 3A, lane 2). Collectively, these results suggest that the negative effects of His-ASF/SF2 on major late mRNA accumulation are not unspecific.
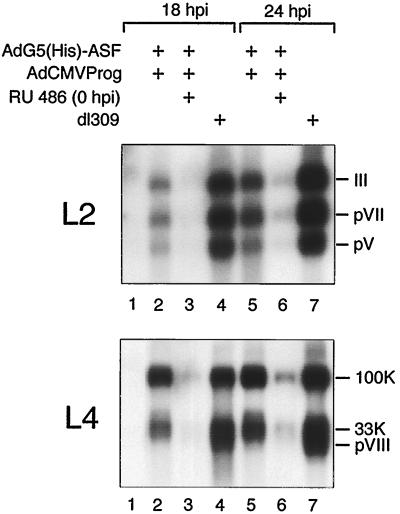
Different effects of ASF/SF2 overproduction on major late region 2 and 4 mRNA accumulation.
In our previous work we have used the L1 region as a model system to study the significance of SR proteins as regulators of alternative RNA splicing. To determine if ASF/SF2 also affects mRNA accumulation from other late mRNA families, we probed Northern blot filters with 32P-labeled probes specific for the L2 and L4 families of mRNAs. As shown in Fig. 4, inducing His-ASF/SF2 expression from the start of infection resulted in dramatic decrease in the steady-state levels of L2 and L4 mRNAs. Unlike L1 splicing, the spliced structure of the individual L2 mRNA species did not change significantly, suggesting that ASF/SF2 is not a critical factor regulating L2 alternative splicing. The mRNA profile from region L4 was intermediate, compared to L1 and L2, and showed a slight shift toward an increase in 100K mRNA production. Thus, the 33K and particularly the pVIII mRNA species were reduced.
FIG. 4.
Effect of ASF/SF2 overexpression on L2 and L4 mRNA expression. Cytoplasmic RNA was prepared from AdG5(His)-ASF/AdCMVProg-infected 293 cells at the indicated time points and separated on a 1% agarose gel containing 2.2 M formaldehyde. RNA was transferred to a nitrocellulose filter and hybridized with 32P-labeled probes specific for region L2 or L4. Lanes 1, mock-infected cells.
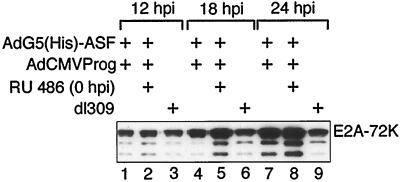
ASF/SF2 overexpression does not block E2A-72K protein expression.
The observation that His-ASF/SF2 almost completely eliminated major late mRNA expression could indicate that ASF/SF2 unspecifically blocks viral gene expression. To test this, we analyzed E2A-72K protein levels in AdG5(His)-ASF-infected cells. In contrast to most adenovirus units, E2A mRNA expression is not regulated at the level of alternative splicing (reviewed in reference 21). Instead, transcription initiation occur at two alternative promoters (Fig. 1); the distal promoter is used early and the proximal promoter is used late after infection. As shown in Fig. 5, E2A-72K protein expression and E2A mRNA accumulation (data not shown) were enhanced approximately threefold at 18 and 24 hpi in His-ASF/SF2-expressing cells compared to the parental virus, dl309. Similarly, the steady-state levels of E1A and E1B mRNA accumulation are not significantly reduced by His-ASF/SF2 overexpression (see below). Taken together, these results suggest that the negative effects of ASF/SF2 overproduction primarily affect mRNA accumulation from the major late transcription unit (MLTU).
FIG. 5.
ASF/SF2 does not block E2A-72K expression. 293 cells infected with AdG5(His)-ASF/AdCMVProg were harvested at the indicated time points, and E2A-72K protein expression was analyzed by a Western blot assay. The faster-migrating species most likely represent degradation products of the E2A-72K protein.
ASF/SF2 overexpression inhibits the temporal shift in E1A and E1B alternative splicing.
The E1A and E1B transcription units represent examples of genes where a common 3′ splice site is joined to alternative 5′ splice sites (Fig. 1). During a lytic infection, the relative ratios of individual E1A and E1B mRNAs change. Thus, the E1A 13S and the E1B 22S mRNAs are the predominant mRNAs expressed at early times of infection, whereas the E1A 9S and E1B 13S mRNAs become the most abundant mRNAs expressed late after infection.
Since the progesterone-regulated gene cassette replaces the E1 region in the recombinant viruses, AdCMVProg and AdG5(His)-ASF do not express E1A and E1B mRNAs. To investigate the effect of ASF/SF2 overexpression on the accumulation of region E1 mRNAs, we therefore included the parental virus dl309 in the infection cocktail. Thus, a mixture of AdCMVProg, AdG5(His)-ASF, and dl309 was used to infect monolayer cultures of HeLa cells. His-ASF/SF2 expression was induced at the start of infection, and cytoplasmic RNAwas isolated at 12 and 18 hpi. The structure of E1A and E1B mRNAs expressed from the dl309 genome was determined by S1 protection (E1A) and Northern blot (E1B) analysis.
As shown in Fig. 6, His-ASF/SF2 induction resulted in an increase in E1A 13S mRNA accumulation, at the expense of E1A 12S and 9S mRNA accumulation. Similarly, His-ASF/SF2 overexpression resulted in a substantial shift toward E1B 22S mRNA accumulation accompanied by a dramatic reduction in E1B 13S mRNA accumulation. The steady-state levels of the pIX mRNA, which is an unspliced mRNA (2) encoded by a separate transcription unit located within E1B (Fig. 1), was slightly reduced.
Taken together, the results extend previous data by showing that ASF/SF2 promotes proximal splice site usage also during an adenovirus infection. This general activity of ASF/SF2 maintains E1A and E1B mRNA expression in a prolonged early phase of mRNA expression.
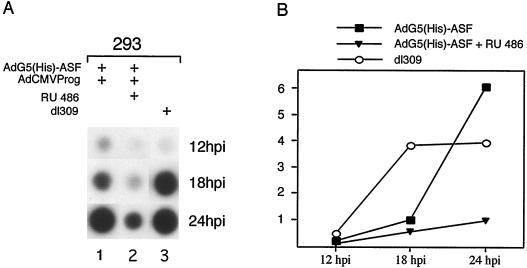
Viral DNA replication is inhibited by ASF/SF2 overexpression.
It has previously been shown that viral DNA replication is coupled to efficient mRNA expression from the MLTU (reviewed in reference 21). Thus, the observation that high levels of His-ASF/SF2 expression severely reduced cytoplasmic L1 mRNA accumulation (Fig. 3A) suggested that ASF/SF2, directly or indirectly, may block viral DNA replication. To test this hypothesis, the efficiency of viral DNA replication in AdG5(His)-ASF-infected cells was measured at different time points of infection by a dot blot assay. As shown in Fig. 7, inducing His-ASF/SF2 expression from the start of infection indeed caused a severe reduction in viral DNA replication compared to dl309. Interestingly, the background expression of His-ASF/SF2, in the absence of inducer, was sufficient to cause a delay in viral DNA replication. Thus, viral DNA accumulation was severely reduced at 18 hpi but back to normal at 24 hpi.
FIG. 7.
Effect of ASF/SF2 on adenovirus DNA replication. Viral DNA was prepared from AdG5(His)-ASF/AdCMVProg-infected 293 cells by Hirt extraction (19) at the indicated time points and analyzed by a dot blot assay (A). The result was quantitated by PhosphorImager scanning (B). The graph represents the mean value of three separate experiments.
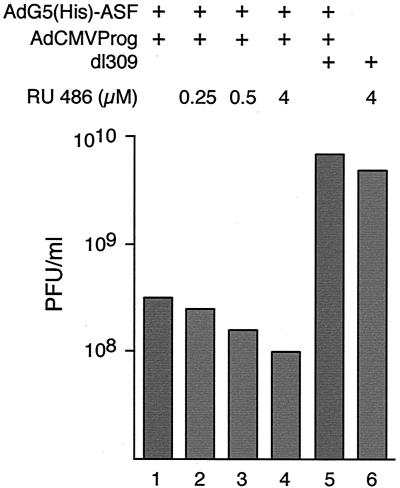
ASF/SF2 expression blocks formation of new virus particles.
A consequence of the negative effects of ASF/SF2 on the expression of major late mRNA would be less efficient growth of AdG5(His)-ASF. To test this hypothesis, we measured the effect of ASF/SF2 overexpression on late viral protein synthesis and new virus particle formation. AdG5(His)-ASF was grown in the presence of increasing amounts of inducer, added at the start of infection. Cells were harvested 40 hpi, and virus production was quantitated by plaque assay. As shown in Fig. 8, growth of AdG5(His)-ASF in the presence of increasing amounts of inducer resulted in a more than 60-fold reduction in virus yield. Noteworthy, growth of the recombinant virus in the absence of inducer resulted in a 20-fold reduction in virus yield. At first glance, this may seem surprising. However, as shown in Fig. 2A, a high background expression of His-ASF/SF2 is detectable also in the absence of inducer. This spurious His-ASF/SF2 expression is sufficient to significantly reduce expression of late viral proteins needed for virion assembly (Fig. 9). Collectively, the results strengthen our hypothesis that viral control of SR protein function is essential for efficient virus multiplication.
FIG. 8.
ASF/SF2 decreases the formation of infectious virus particles. 293 cells were infected with AdG5(His)-ASF/AdCMVProg in the presence of different concentrations of RU 486. Virus was prepared 40 hpi by freeze-thawing and quantitated by plaque titration.
FIG. 9.
ASF/SF2 overexpression inhibits late viral protein synthesis. 293 cells infected with AdG5(His)-ASF/AdCMVProg were pulse-labeled with [35S]cysteine/methionine for 2 h prior to harvest. Protein extracts prepared at the indicated time points were separated by SDS-PAGE on a 10% gel, which was then subjected to autoradiography. Lane 1, mock-infected cells.
DISCUSSION
During a lytic adenovirus infection, the production of viral proteins is regulated, to a large extent, at the level of alternative RNA splicing, i.e., production of multiple mRNAs with different coding capacities from a single transcription unit. The spliced structure of mRNAs expressed from most transcription units changes during the infectious cycle. The general tendency is that shorter mRNAs, produced by splicing out larger introns, dominate at later time points of infection. Interestingly, the prototypical SR protein ASF/SF2 has been shown to have the opposite effect on RNA splice site choice. Thus, addition of an excess of ASF/SF2 in vitro or transfection of a plasmid expressing ASF/SF2 results in a shift toward production of long mRNAs, i.e., proximal splice site usage (4, 6, 16, 24, 30, 38). We have previously shown that SR proteins purified from late adenovirus-infected cells are partially inactivated as splicing enhancer or splicing repressor proteins by a virus-induced dephosphorylation (23). Based on this finding, we speculated that the temporal shift from proximal to distal splice site usage in adenovirus alternative splicing was coupled to this SR protein inactivation. A prediction of this model is that overexpression of an SR protein during a productive infection should cause a block in the temporal shift in adenovirus alternative splicing, by disturbing the balance of functional and nonfunctional SR proteins in infected cell.
Here we test this prediction by analyzing the phenotype of a recombinant adenovirus expressing the prototypical SR protein, ASF/SF2. We show that overexpression of ASF/SF2 has negative effects, particularly on major late gene expression (Fig. 3 and 4), with no (E2A-72K [Fig. 5]) or small (E1A and E1B [Fig. 6]) inhibitory effects on early viral mRNA accumulation. As the model also predicts, ASF/SF2 overexpression blocks the early to late shift in E1A, E1B, and L1 alternative splicing (Fig. 3 and 5). Furthermore, high ASF/SF2 expression also caused a severe inhibition of viral DNA replication (Fig. 7), late protein synthesis (Fig. 9), and virus production (Fig. 8). Collectively, our results support the hypothesis that viral control of SR protein activity is important for the correct temporal expression of early and late viral mRNAs during infection, and as a consequence effective virus multiplication.
Expression of mRNAs from the MLTU is subjected to a temporal regulation of poly(A) site usage (reviewed in reference 21). Thus, the L1 region is the only part of the MLTU that is expressed at early times of infection (1, 8, 33). At a transient stage following initiation of viral DNA replication, mRNAs from regions L1 and L4 accumulate preferentially (26, 40). It has been proposed that efficient expression of mRNA from regions L1 through L5 requires both viral DNA replication and late protein synthesis (26). Previous results also have suggested that viral DNA replication per se is sufficient to induce accumulation of normal steady-state levels of L1 mRNAs (25). However, under such conditions the temporal shift in alternative splicing was shown to be incomplete, with a preferential accumulation of the 52,55K+i and IIIa+i mRNA species (25). Here we show that induction of ASF/SF2 expression from the start of infection blocks viral DNA replication (Fig. 7) and results in an almost complete loss of mRNA expression from the MLTU (Fig. 3 and 4). Also, we show that the small amounts of L1 mRNAs produced are incompletely spliced, with a preferential accumulation of 52,55K+i and IIIa+i mRNAs (Fig. 3A, lanes 7 to 9). The effects of ASF/SF2 on L1 mRNA expression were specific since overproduction of the CAT protein in the same type of experiment did not have an adverse effect on L1 mRNA expression (Fig. 3C).
The inhibitory effect of ASF/SF2 on viral DNA replication (Fig. 7) probably results from a block in proper expression of mRNAs from the E2B region (Fig. 1). In fact, preliminary experiments suggest that expression of the mRNA encoding for pTP, the adenovirus protein functioning as a primer protein for initiation of viral DNA replication (reviewed in reference 44), is reduced by ASF/SF2 overexpression (data not shown). However, pTP is expressed at low amounts during infection, and we were unable to detect pTP by Western blotting in most experiments.
Interestingly, the background expression of His-ASF/SF2 observed in 293 cells in the absence of inducer (Fig. 2A) was sufficient to disturb major late mRNA expression (Fig. 3A and 4), and late protein synthesis (Fig. 9), also under conditions where viral DNA replication was back to wild-type levels (24 hpi) (Fig. 7). This finding is significant and differs from previous results by suggesting that DNA replication per se is not enough to activate mRNA expression from the MLTU. However, all data can be reconciled in a model where it is postulated that an early viral protein(s) is required for efficient mRNA production from the L1 region. Thus, inducing ASF/SF2 expression from the start of infection interferes with the temporal shift in early viral mRNA expression (Fig. 6) and, as a consequence, synthesis of the complete repertoire of early viral proteins. Some of these proteins are likely to be required at various stages of gene expression, such as transcription, RNA processing, RNA transport, and RNA stability. For example, we have previously shown that several E4 proteins have the capacity to modulate RNA splicing. Thus, the E4-ORF4 protein inactivates SR proteins as IIIa splicing repressor proteins (23). The E4-ORF3 protein induces i-leader inclusion, while the E4-ORF6 protein causes i-leader skipping (35). However, we do not exclude that ASF/SF2 interferes at other levels of adenovirus gene expression. For example, high amounts of ASF/SF2 may block major late poly(A) site usage. It has been well documented that RNA splice site selection can affect the efficiency of polyadenylation, and vice versa (10, 34, 43). Alternatively, and perhaps more attractive, ASF/SF2 overexpression may directly interfere with transcription from the major late promoter. Recent data have indicated that RNA polymerase II transcription and RNA processing are coordinated events. Thus, many RNA processing factors, including ASF/SF2, have been shown to interact with the C-terminal domain tail of RNA polymerase II (reviewed in reference 5). Such an interaction may have, or induce, adverse effects on transcription initiation and/or elongation at the major late promoter. Clearly the approach described here should make it possible to further define the mechanistic details of ASF/SF2 inhibition of virus multiplication.
The experimental strategy used here provides a new in vivo method to study the significance of defined proteins on adenovirus multiplication. Thus, by reconstructing recombinant viruses expressing a gene of interest under the transcriptional control of an inducible promoter, it should be possible to use on/off switches of reporter gene expression to further define protein function. However, our experiments also illustrate an unexpected complexity of gene regulation with the current generation of our adenovirus vector system. Thus, the vector system functions extraordinarily well under nonreplicative conditions (HeLa cells [Fig. 2A]), where we reach induction levels exceeding 500-fold (32). In contrast, a significant background of reporter gene expression is observed in 293 cells, also in the absence of inducer (Fig. 2A and data not shown). This troublesome His-ASF/SF2 expression is, to a large extent, caused by the E1A transcriptional activator protein expressed in 293 cells, which appears to activate transcription from the TATA element present in the G5 minimal major late promoter driving reporter gene expression (data not shown). It is important for our future work to reduce this basal reporter gene expression in 293 cells. We are currently developing new strategies to silence the 293-cell specific background expression of the reporter gene (D. Edholm, M. Molin, and G. Akusjärvi, unpublished data).
ACKNOWLEDGMENTS
We are grateful to T. Maniatis for providing the Prey-ASF plasmid, J. Stevenin for the ASF/SF2 peptide antisera, and E. Bridge for the E2A-72K antibody. RU 486 was kindly provided by Roussel-Uclaf.
This work was supported by the Swedish Cancer Society and the Göran Gustafsson Foundation for Natural and Medical Research.
REFERENCES
- 1.Akusjärvi G, Persson H. Controls of RNA splicing and termination in the major late adenovirus transcription unit. Nature. 1981;292:420–426. doi: 10.1038/292420a0. [DOI] [PubMed] [Google Scholar]
- 2.Aleström P, Akusjärvi G, Perricaudet M, Mathews M B, Klessig D, Pettersson U. The gene for polypeptide IX of adenovirus type 2 and its unspliced messenger RNA. Cell. 1980;19:671–681. doi: 10.1016/s0092-8674(80)80044-4. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 4.Bai Y, Lee D, Yu T, Chasin L A. Control of 3′ splice site choice in vivo by ASF/SF2 and hnRNP A1. Nucleic Acids Res. 1999;27:1126–1134. doi: 10.1093/nar/27.4.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr Opin Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 6.Caceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 7.Cao W, Jamison S F, Garcia-Blanco M A. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;3:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- 8.Chow L T, Broker T R, Lewis J B. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J Mol Biol. 1979;134:265–303. doi: 10.1016/0022-2836(79)90036-6. [DOI] [PubMed] [Google Scholar]
- 9.Colwill K, Feng L L, Yeakley J M, Gish G D, Caceres J F, Pawson T, Fu X D. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J Biol Chem. 1996;271:24569–24575. doi: 10.1074/jbc.271.40.24569. [DOI] [PubMed] [Google Scholar]
- 10.Cooke C, Hans H, Alwine J C. Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol Cell Biol. 1999;19:4971–4979. doi: 10.1128/mcb.19.7.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eperon I C, Ireland D C, Smith R A, Mayeda A, Krainer A R. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu X D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 13.Gattoni R, Chebli K, Himmelspach M, Stevenin J. Modulation of alternative splicing of adenoviral E1A transcripts: factors involved in the early-to-late transition. Genes Dev. 1991;5:1847–1858. doi: 10.1101/gad.5.10.1847. [DOI] [PubMed] [Google Scholar]
- 14.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–574. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 15.Harper J E, Manley J L. A novel protein factor is required for use of distal alternative 5′ splice sites in vitro. Mol Cell Biol. 1991;11:5945–5953. doi: 10.1128/mcb.11.12.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harper J E, Manley J L. Multiple activities of the human splicing factor ASF. Gene Expr. 1992;2:19–29. [PMC free article] [PubMed] [Google Scholar]
- 17.Hertel K J, Lynch K W, Maniatis T. Common themes in the function of transcription and splicing enhancers. Curr Opin Cell Biol. 1997;9:350–357. doi: 10.1016/s0955-0674(97)80007-5. [DOI] [PubMed] [Google Scholar]
- 18.Himmelspach M, Cavaloc Y, Chebli K, Stevenin J, Gattoni R. Titration of serine/arginine (SR) splicing factors during adenoviral infection modulates E1A pre-mRNA alternative splicing. RNA. 1995;1:794–806. [PMC free article] [PubMed] [Google Scholar]
- 19.Hirt B. Selective extraction of polyoma DNA from infected mouse cells. J Mol Biol. 1967;14:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 20.Hitt M, Bett A J, Prevec L, Graham F L. Construction and propagation of human adenovirus vectors. In: Celis J E, editor. Cell biology: a laboratory handbook. Vol. 1. San Diego, Calif: Academic Press; 1994. pp. 479–490. [Google Scholar]
- 21.Imperiale M J, Akusjärvi G, Leppard K N. Post-transcriptional control of adenovirus gene expression. Curr Top Microbiol Immunol. 1995;199:139–171. doi: 10.1007/978-3-642-79499-5_6. [DOI] [PubMed] [Google Scholar]
- 22.Kanopka A, Mühlemann O, Akusjärvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- 23.Kanopka A, Mühlemann O, Petersen-Mahrt S, Estmer C, Öhrmalm C, Akusjärvi G. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature. 1998;393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- 24.Krainer A R, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 25.Larsson S, Kreivi J P, Akusjärvi G. Control of adenovirus alternative RNA splicing: effect of viral DNA replication on RNA splice site choice. Gene. 1991;107:219–227. doi: 10.1016/0378-1119(91)90322-3. [DOI] [PubMed] [Google Scholar]
- 26.Larsson S, Svensson C, Akusjärvi G. Control of adenovirus major late gene expression at multiple levels. J Mol Biol. 1992;225:287–298. doi: 10.1016/0022-2836(92)90922-7. [DOI] [PubMed] [Google Scholar]
- 27.Lim L P, Sharp P A. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol Cell Biol. 1998;18:3900–3906. doi: 10.1128/mcb.18.7.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 29.Mayeda A, Helfman D M, Krainer A R. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol Cell Biol. 1993;13:2993–3001. doi: 10.1128/mcb.13.5.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayeda A, Krainer A R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 31.Mermoud J E, Cohen P T, Lamond A I. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 1994;13:5679–5688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molin M, Shoshan M C, Öhman-Forslund K, Linder S, Akusjärvi G. Two novel adenovirus vector systems permitting regulated protein expression in gene transfer experiments. J Virol. 1998;72:8358–8361. doi: 10.1128/jvi.72.10.8358-8361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nevins J R, Wilson M C. Regulation of adenovirus-2 gene expression at the level of transcriptional termination and RNA processing. Nature. 1981;290:113–118. doi: 10.1038/290113a0. [DOI] [PubMed] [Google Scholar]
- 34.Niwa M, Rose S D, Berget S M. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 35.Nordqvist K, Öhman K, Akusjärvi G. Human adenovirus encodes two proteins which have opposite activities on accumulation of alternatively spliced mRNAs. Mol Cell Biol. 1994;14:437–443. doi: 10.1128/mcb.14.1.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petersen-Mahrt S K, Estmer C, Öhrmalm C, Matthews D A, Russell W C, Akusjärvi G. The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J. 1999;18:1014–1024. doi: 10.1093/emboj/18.4.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Screaton G R, Caceres J F, Mayeda A, Bell M V, Plebanski M, Jackson D G, Bell J I, Krainer A R. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shih S R, Krug R M. Novel exploitation of a nuclear function by influenza virus: the cellular SF2/ASF splicing factor controls the amount of the essential viral M2 ion channel protein in infected cells. EMBO J. 1996;15:5415–5427. [PMC free article] [PubMed] [Google Scholar]
- 39.Stow N D. Cloning of a DNA fragment from the left-hand terminus of the adenovirus type 2 genome and its use in site-directed mutagenesis. J Virol. 1981;37:171–180. doi: 10.1128/jvi.37.1.171-180.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svensson C, Akusjärvi G. Defective RNA splicing in the absence of adenovirus-associated RNAI. Proc Natl Acad Sci USA. 1986;83:4690–4694. doi: 10.1073/pnas.83.13.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svensson C, Pettersson U, Akusjärvi G. Splicing of adenovirus 2 early region 1A mRNAs is non-sequential. J Mol Biol. 1983;165:475–495. doi: 10.1016/s0022-2836(83)80214-9. [DOI] [PubMed] [Google Scholar]
- 42.Tacke R, Manley J L. Determinants of SR protein specificity. Curr Opin Cell Biol. 1999;11:358–362. doi: 10.1016/S0955-0674(99)80050-7. [DOI] [PubMed] [Google Scholar]
- 43.Vagner S, Vagner C, Mattaj I W. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 2000;14:403–413. [PMC free article] [PubMed] [Google Scholar]
- 44.van der Vliet P C. Adenovirus DNA replication. Curr Top Microbiol Immunol. 1995;199:1–30. doi: 10.1007/978-3-642-79499-5_1. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Manley J L. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- 46.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 47.Xiao S H, Manley J L. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 1998;17:6359–6367. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zahler A M, Neugebauer K M, Lane W S, Roth M B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 49.Zhang W J, Wu J Y. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol Cell Biol. 1996;16:5400–5408. doi: 10.1128/mcb.16.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]