Abstract
Recurrent excitatory connections in hippocampal regions CA3 and CA2 are thought to play a key role in the generation of sharp-wave ripples (SWRs), electrophysiological oscillations tightly linked with learning and memory consolidation. However, it remains unknown how defined populations of inhibitory interneurons regulate these events during behavior. Here, we use large-scale, three-dimensional calcium imaging and retrospective molecular identification in the mouse hippocampus to characterize molecularly identified CA3 and CA2 interneuron activity during SWR-associated memory consolidation and spatial navigation. We describe subtype- and region-specific responses during behaviorally distinct brain states and find that SWRs are preceded by decreased cholecystokinin-expressing interneuron activity and followed by increased parvalbumin-expressing basket cell activity. The magnitude of these dynamics correlates with both SWR duration and behavior during hippocampal-dependent learning. Together these results assign subtype- and region-specific roles for inhibitory circuits in coordinating operations and learning-related plasticity in hippocampal recurrent circuits.
Episodic memories formed from single experiences can be used to guide behavior throughout the lifetime of an organism1. For this to occur, continuous streams of sensory information must be discretized into snapshots, which can then be reactivated and consolidated into long-term memory2. In the mammalian brain, both the rapid encoding of episodic memories and their subsequent consolidation rely critically on the CA3 recurrent network in the hippocampus3–5. Furthermore, the adjacent CA2 region has recently emerged as another key recurrent network for hippocampal activity and hippocampal-dependent memory6–8. The initial encoding of hippocampal memories is thought to be implemented via plasticity of feedforward mossy fiber synapses onto CA3 pyramidal cells and at recurrent pyramidal cell synapses9,10. These nascent representations are subsequently consolidated during sharp-wave ripples (SWRs), synchronous population events generated within the CA3 and CA2 recurrent networks during which a compressed version of the memory traces is thought to be reactivated and transferred to the neocortex for long-term storage2,8,11,12. Accordingly, the main body of work so far has primarily focused on investigating the temporally structured recruitment of pyramidal cells during memory formation and consolidation13,14, leaving a major question mark regarding the role played by inhibition in supporting and regulating these memory processes. Indeed, experimental and computational studies strongly implicate inhibitory motifs in stabilizing recurrent networks and supporting efficient cortical computations15–17. However, as the CA1 output region of the hippocampus has traditionally served as a prototype circuit for the study of interneurons10,18–27, strikingly, little is known about inhibitory microcircuits in the upstream CA3 and CA2 networks and their role in structuring temporally ordered neuronal firing during behaviorally relevant network states associated with memory encoding and consolidation. Based on investigations in the CA1, inhibition is predominantly viewed as an immutable pacemaker of principal cell firing without itself being plastic, a concept reflected in the term ‘chronocircuit’28. Extending this static view of inhibition, in vivo structural and molecular studies have revealed plastic changes in CA3 and CA2 inhibitory circuits in response to behavioral manipulations29,30, raising the possibility that CA3/CA2 inhibitory dynamics are modifiable in an experience-dependent or learning-dependent manner10,31. Critically, major challenges associated with obtaining population-level recordings of molecularly defined cell types in deep brain regions have impeded addressing these major knowledge gaps concerning operations and plasticity of local inhibition in CA3/CA2 circuits of the hippocampus.
Here, we use a two-step method to simultaneously record large numbers of interneurons with fast, targeted three-dimensional (3D) calcium imaging during behavior and retrospectively identify their molecular profiles with post hoc immunohistochemistry in hippocampal areas CA3 and CA2. We provide a comprehensive description of molecularly identified interneuron dynamics during both active spatial navigation and awake SWRs associated with memory consolidation. Further, our study demonstrates subtype-specific, region-specific and learning-related changes in interneuron dynamics around SWRs that correlate with both SWR duration and behavior, suggesting a central role for local inhibition in hippocampal recurrent circuits in supporting memory consolidation.
Results
Large-scale imaging of molecularly identified interneurons
To obtain large-scale recordings from CA3/CA2 inhibitory circuits, we injected vesicular GABA transporter (VGAT)-Cre mice with a recombinant adeno-associated virus (rAAV) encoding Cre-dependent GCaMP7f into CA3/CA2 and implanted an imaging window over the alveus of the anterior hippocampus, providing unbiased access to GABAergic interneurons across all CA3/CA2 sublayers (Fig. 1a). We used acousto-optic deflection (AOD)-based two-photon (2p) imaging22 to record from hundreds of interneurons distributed in three dimensions while head-fixed mice were engaged on a treadmill32 in a spatial foraging task (total of 2,079 interneurons in 22 mice; 94.5 ± 23.3 per mouse (mean ± s.d.); Fig. 1b,c). After the conclusion of imaging experiments, multiplexed post hoc immunohistochemistry was performed on fixed brain slices that were registered to in vivo structural scans, revealing the molecular identity of imaged cells (Fig. 1d). The precise location of all imaged cells was determined with calbindin (CB) immunohistochemistry, which allows visualization of granule cell mossy fibers (Extended Data Fig. 1) within the stratum lucidum of CA3/CA2 (ref. 6). We used a combination of five molecular markers (parvalbumin (PV), somatostatin (SOM), SATB1, cholecystokinin (CCK) and CB) to identify five subtypes of CA3/CA2 pyramidal cell-targeting interneurons: PV+ basket cells (PVBCs), axoaxonic cells (AACs), somatostatin+ cells (SOMs), CCK+ cells (CCKs) and CB+ cells (CBs; Fig. 1e). These five markers were chosen to label some of the most abundant interneuron subtypes in the hippocampus23, including those not readily accessible via traditional transgenic driver lines (Fig. 1f). Finally, CA2 interneurons were separated from their CA3 counterparts post hoc via their proximity to pyramidal cells expressing STEP, a protein phosphatase specific for CA2 pyramidal neurons33 (Extended Data Fig. 1).
Fig. 1 |. Large-scale imaging of molecularly identified GABAergic interneurons in CA3 and CA2.
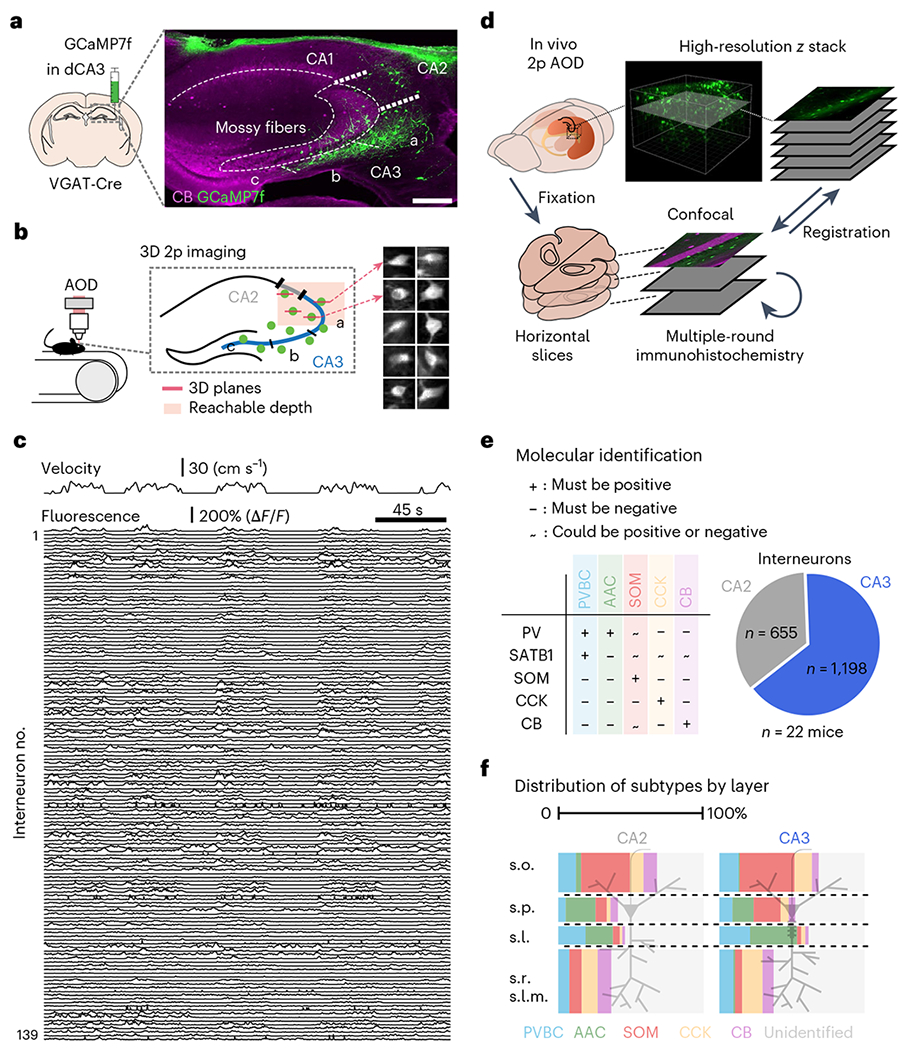
a, Experimental design. VGAT-Cre mice were Injected in CA2/CA3 with a Cre-dependent GCaMP7f virus to record all inhibitory interneurons with 2p imaging (n = 22 mice); scale bar, 250 μm; dCA3, dorsal CA3. b, Hundreds of interneurons can be recorded simultaneously at 5–10 Hz in three dimensions during behavior. Right, time average examples of ten interneurons. The images are 50 × 50 μm. c, Example fluorescence traces from 139 simultaneously recorded interneurons during several minutes of behavior (animal velocity is plotted above). d, Schematic of the experimental pipeline used to determine the molecular identity of imaged cells. Multiple rounds of immunohistochemistry were performed on fixed, horizontal slices that were registered to high-resolution in vivo Z-stacks. e, Left, combinatorial expression patterns of the five markers (PV, SOM, SATB1, CCK and CB) used to separate imaged cells into subtypes. Right, pie chart summary of the dataset. f, Layer distributions of all imaged and post hoc identified interneurons split by both region and subtype. s.o., stratum oriens; s.p., stratum pyramidale; s.l., stratum lucidum; s.r., stratum radiatum; s.l.m., stratum lacunosum-moleculare.
Online locomotion-related inhibitory dynamics during spatial navigation
Locomotory movements are the behavioral correlates of an online and actively engaged brain state, characterized in the hippocampus by a location-specific rate code in pyramidal cells. Thus, locomotion has been shown to strongly influence the recruitment or disengagement of distinct types of interneurons in CA1 (refs. 22,34,35), but the paucity of data in CA3/CA2 to date has hindered efforts in testing whether these interneuron dynamics are characteristic of the entire hippocampus. We trained and imaged water-restricted mice during a random foraging task on a 2-m-long cue-rich belt, during which several water rewards were delivered at random locations on each lap (Fig. 2a). We observed a tight correlation between the activity of most cells and the ambulatory state of the animal (locomotion versus immobility; Fig. 2b). To systematically characterize this relationship, we calculated the Pearson correlation coefficient between the activity of each cell and the animal’s velocity. At the population level, the average correlation was shifted toward positive values (Fig. 2c) among both CA3 and CA2 interneurons, although CA3 interneurons as a whole were significantly more positively correlated with velocity than CA2 interneurons (Fig. 2c). We then compared the coefficients across subtypes and regions and observed AACs to be overall more tightly correlated with velocity (Fig. 2d) in both CA3 and CA2. In addition, CB cells had overall lower coefficients, and many CB cells were more active during immobility than during locomotion (Fig. 2d). Closer inspection of the immobility-active CB cells revealed that the vast majority lacked SATB1 expression (CB+SATB1−), while SATB1+ CB cells (CB+SATB1+) displayed high correlations with locomotion (Fig. 2e), both in the CA3 and CA2. These trends were also reflected in the average run–start and run–stop responses for each subtype (Fig. 2f). As these functionally unique CB+SATB1− cells may represent a previously unrecognized interneuron subtype and were negative for the other tested markers (PV, SOM and CCK), we next determined what other molecules they may express and found that the majority of CB cells were also negative for most other hippocampal interneuron markers (VIP, CR, NPY and M2R) and expressed the transcription factor COUP-TFII (Extended Data Fig. 2), although no further differences were seen between SATB1+ and SATB1− cells.
Fig. 2 |. Locomotion-related dynamics during spatial navigation.
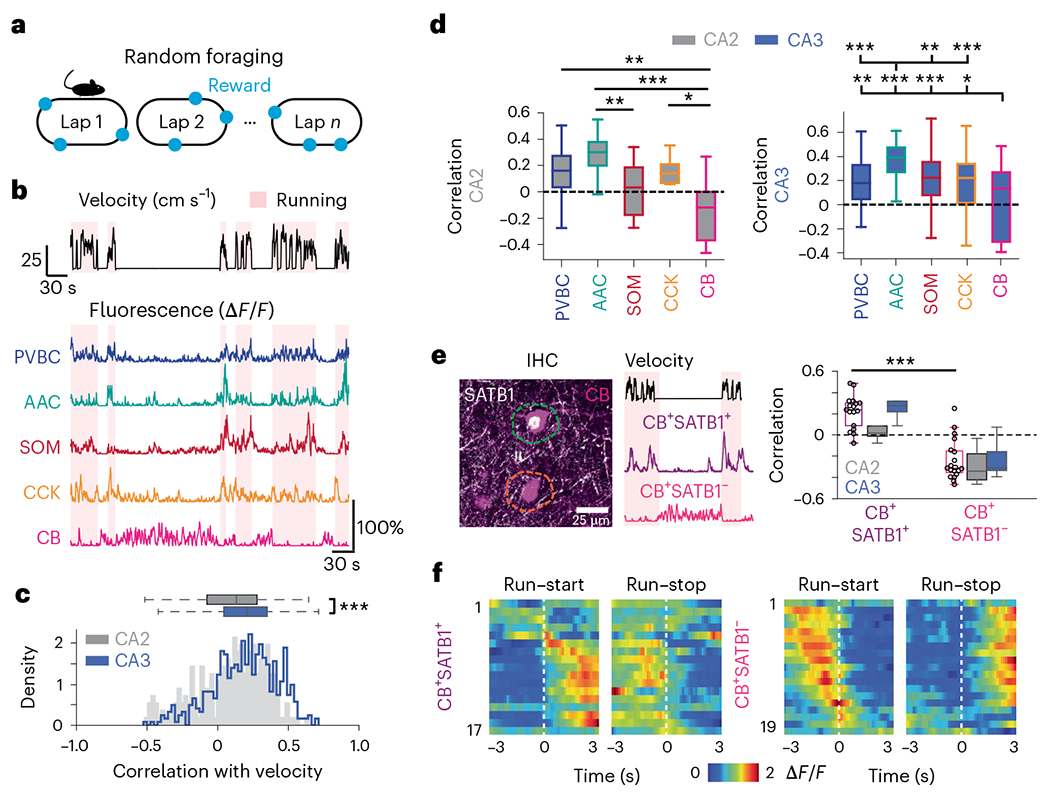
a, Mice were trained to run for randomly delivered water rewards on each lap. b, Representative fluorescence traces from different interneuron subtypes during running (red-shaded area) and immobility (non-shaded) bouts. c, Distribution of Pearson correlation coefficients between fluorescence and velocity for all recorded interneurons broken down by CA3 and CA2 regions. The activity of CA3 cells was significantly more correlated with velocity than the activity of CA2 cells (CA2 correlation: 0.092 ± 0.247; CA3 correlation: 0.191 ± 0.228; two-sided Mann–Whitney U-test, P = 9.73 × 10−7). Data are from n = 242 CA2 interneurons and 491 CA3 interneurons from n = 9 mice and are reported as mean ± s.d. d, Left, same velocity–activity correlation as in c but broken down by subtype for all CA2 cells (PVBC correlation: 0.141 ± 0.220; AAC correlation: 0.271 ± 0.175; SOM correlation: 0.026 ± 0.194; CCK correlation: 0.111 ± 0.282; CB correlation: −0.155 ± 0.225). Significant differences between CA2 subtypes are indicated. Data were analyzed by one-way ANOVA with post hoc multiple-testing correction: PPVBC–CB = 0.0019, PAAC–CB = 0.001, PAAC–SOM = 0.006 and PCCK–CB = 0.043. Data are from n = 45 PVBCs, 26 AACs, 16 SOMs, 12 CCKs and 10 CBs from n = 9 mice and are reported as mean ± s.d. Right, same velocity–activity correlation as in c but broken down by subtype for all CA3 cells (PVBC correlation: 0.196 ± 0.187; AAC: 0.359 ± 0.139; SOM: 0.229 ± 0.212; CCK: 0.179 ± 0.236; CB: 0.032 ± 0.289). Significant differences between CA3 subtypes are indicated. Data were analyzed by one-way ANOVA with post hoc multiple-testing correction: PAAC–CB = 0.001, PAAC–CCK = 0.001, PAAC–PVBC = 0.001, PAAC–SOM = 0.0031, PCB–CCK = 0.0259, PCB–PVBC = 0.0051 and PCB–SOM = 0.001. Data are from n = 76 PVBCs, 63 AACs, 70 SOMs, 55 CCKs and 26 CBs from n = 9 mice and are reported as mean ± s.d. e, Left, confocal image of a CB+SATB1+ cell (magenta/white) and a CB+SATB1− cell (magenta only). Example fluorescence traces from these cells during locomotion and immobility are shown. Right, same correlation as in d but for CB+SATB1+ and CB+SATB1− subtypes split by region. CB+SATB1+ cells were significantly more correlated with velocity than CB+SATB1− cells (CA2 and CA3 were considered together; CB+SATB1+ correlation: 0.219 ± 0.151; CB+SATB1−: −0.237 ± 0.186; two-sided Mann–Whitney U-test, P = 2.17 × 10−6). Data are from n = 17 CB+SATB1+ neurons and 19 CB+SATB1− neurons from n = 9 mice and are reported as mean ± s.d. f, Heat maps of average activity around run–start (left) and run–stop (right) events for all CB+SATB1+ (top) and CB+SATB1− (bottom) interneurons sorted by the location of their peak activity around run–start events (the same row on the left and right heat maps represents the same cell). Data are from n = 17 CB+SATB1+ neurons and 19 CB+SATB1− neurons from n = 9 mice; CA2 and CA3 data were considered together; *P < 0.05, **P <0.01, ***P <0.001.
Spatial tuning of CA3 and CA2 inhibitory circuits during spatial navigation
Akin to pyramidal cells selective for particular regions of an environment36, interneurons have also been reported to display some degree of spatial selectivity in CA1 (refs. 22,37). Thus, we next sought to determine the nature and extent of the spatial tuning of interneurons during random foraging (Fig. 3a). We found that significant subsets of cells displayed positive (32.8% ± 23.7% (mean ± s.d.)) or negative (39.6% ± 21.8% (mean ± s.d.)) spatial selectivity and that while the activity of these tuned cells contained significant spatial information, we did not observe differences among CA3 subtypes in either positive or negative tuning (Fig. 3b,c and Extended Data Fig. 3a). Similarly, we found only subtle differences between CA2 subtypes, especially regarding their negative tuning (Extended Data Fig. 3d). The spatial representations of all interneurons were relatively stable both within and across days, although some differences were observed across subtypes and regions (Extended Data Fig. 3b,c). Under identical experimental conditions, CA3 pyramidal cell recordings (Fig. 3d) performed with the Grik4-Cre transgenic line (Extended Data Fig. 4) revealed significant fractions of spatially tuned place cells (33.7% ± 12.5% (mean ± s.d.)) whose representations collectively spanned the entirety of the belt (Fig. 3e,f). Finally, to more precisely quantify the relative contributions of various behavioral and spatial variables to interneuron activity, we constructed a generalized linear model to predict each cell’s fluorescence trace (Extended Data Fig. 3e–i) from a linear combination of velocity, position, reward and licking. We found that velocity displayed the highest predictive coefficient among the behavioral variables, while few subtype and region differences were observed in the other predictors, including position (Extended Data Fig. 3f–i). Taken together, our results suggest prominent spatial selectivity among many constituents of the CA3/CA2 microcircuit but generally do not support a prominent role for subtype or region specificity in these spatial representations.
Fig. 3 |. Spatial tuning of CA3 and CA2 circuit elements during spatial navigation.
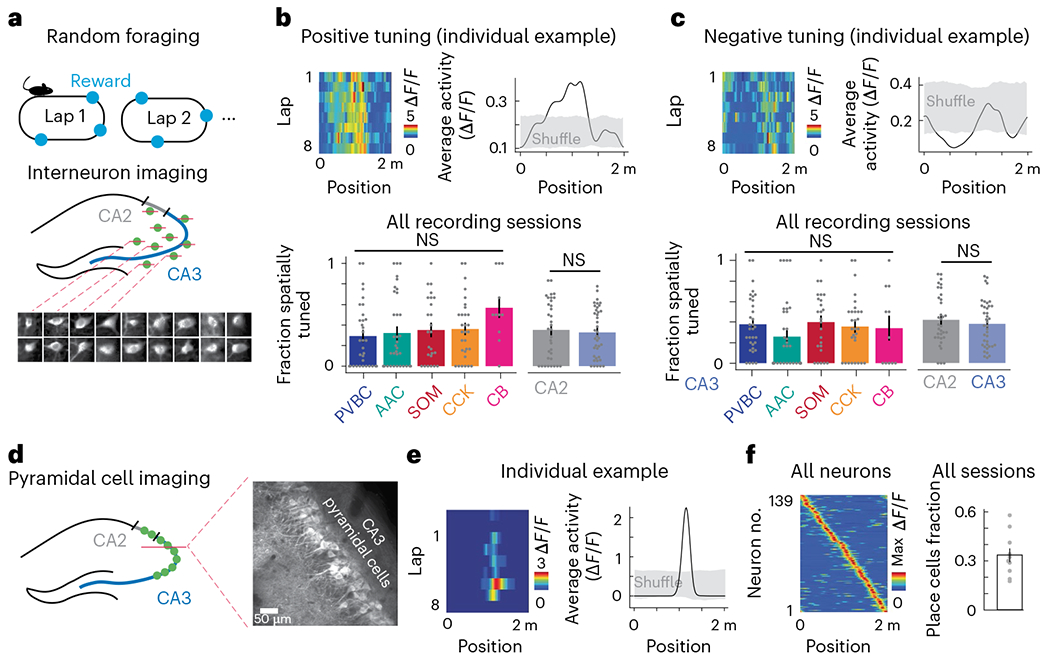
a, Schematic of the experimental paradigm. Top, mice were trained to run for water rewards that were randomly delivered on each lap. Bottom, CA3/CA2 interneurons were simultaneously recorded with 3D 2p imaging. b, Top left, representative example heat map of the lap-by-lap activity for an interneuron with positive spatial tuning. Top right, the same interneuron’s average tuning curve over all laps compared to a shuffle distribution (Methods). Bottom, summary of the fraction of positively tuned interneurons broken down by both region and CA3 subtypes (PVBC: 0.294 ± 0.304; AAC: 0.323 ± 0.348; SOM: 0.352 ± 0.309; CCK: 0.362 ± 0.291; CB: 0.568 ± 0.311; CA2 average: 0 353 ± 0.283; CA3 average: 0.328 ± 0.237). No significant differences in the fraction of spatially tuned cells were found between CA3 subtypes (one-way ANOVA, P = 0.18) or between the CA2 and CA3 regions (two-sided Mann–Whitney U-test, P = 0.37). The CB subtype represents only CB+SATB1+ neurons; immobility-active CB+SATB1− neurons were silenced during locomotion and were thus not considered in this analysis. Each data point represents an imaging session. PVBC data from 33 imaging sessions, AAC data from 35 sessions, SOM data from 25 sessions, CCK data from 30 sessions and CB+SATB1+ data from 11 sessions were included. CA2 data are from 34 sessions, and CA3 data are from 39 sessions (n = 9 mice). Data are reported as mean ± s.d.; NS, not significant. C, Top left, representative example heat map of the lap-by-lap activity for an interneuron with negative spatial tuning. Top right, the same interneuron’s average tuning curve over all laps compared to a shuffle distribution (Methods). Bottom, summary of the fraction of negatively tuned interneurons broken down by both region and CA3 subtypes (PVBC: 0.379 ± 0.273; AAC: 0.258 ± 0.331; SOM: 0.400 ± 0.290; CCK: 0.357 ± 0.286; CB: 0.341 ± 0.358; CA2 average: 0.423 ± 0.249; CA3 average: 0.384 ± 0.226). No significant differences in the fraction of spatially tuned cells were found between CA3 subtypes (one-way ANOVA, P = 0.41) or between the CA2 and CA3 regions (two-sided Mann–Whitney U-test, P = 0.24). The CB subtype represents only CB+SATB1+ neurons; immobility-active CB+SATB1− neurons were silenced during locomotion and were thus not considered in this analysis. Each data point represents an imaging session. Data are from the same number of sessions and the same number of mice as described earlier and are reported as mean ± s.d. d, Schematic of the imaging paradigm for CA3 pyramidal cells (left) and representative in vivo time average of the CA3 pyramidal cell field of view (right) using the Grik4-Cre transgenic line. CA3 pyramidal cells were imaged during the same random foraging paradigm. e, Left, representative example heat map of the lap-by-lap activity of a spatially tuned CA3 pyramidal cell (from a total of 139 cells from n = 5 mice). Right, the same pyramidal cell’s average spatial tuning curve over all laps compared to a shuffle distribution (Methods). f, Summary data of CA3PC spatial tuning. Left, heat map of the spatial tuning curves of all cells sorted by the location of each cell’s maximum activity (139 cells from n = 5 mice). Right, the fraction of spatially tuned cells during each imaging session; 33.7% ± 12.5% neurons were spatially selective during each imaging session (data are from ten sessions over n = 5 mice, with two sessions per mouse). Each dot represents an imaging session. Data are reported as mean ± s.e.m.; *P < 0.05, **P < 0.01, ***P < 0.001.
Inhibitory dynamics around offline memory consolidation events
The precise circuit mechanisms regulating SWR initiation in CA3/CA2 and the role played by the distinct populations of interneurons in these processes remain unknown. To address this knowledge gap, we used our imaging approach and combined it with contralateral local field potential (LFP) recordings (Fig. 4a) to detect SWR events during periods of immobility. We first confirmed that we could measure changes in CA3/CA2 circuits around detected SWR events38–40 by imaging pyramidal cell activity using the Grik4-Cre line (Methods). We found that their activity was indeed elevated around SWRs, allowing us to next consider the local interneurons (Extended Data Fig. 5a). We constructed average peri-SWR traces for each cell and subtype average traces (Extended Data Fig. 6a,b). To fully characterize the peri-SWR responses at the individual cell level and better appreciate potential within-subtype diversity, we calculated an SWR activity index for each cell as a measure of whether the cell was more activated or inhibited around SWRs (Fig. 4b and Methods). Using this index, we found that most PVBCs were strongly activated, and the vast majority of AACs were inhibited (Fig. 4b). SOM cells exhibited a long-tail distribution, as the majority of cells were either suppressed or only weakly activated, but a minority of strongly activated cells were also present (Fig. 4b), likely reflecting the presence of distinct SOM-expressing subtypes not captured by our molecular identification41,42. Similarly, the distribution of the activity index for CCKs and CBs was bimodal, as the majority of these cells showed a net inhibition, but prominent subpopulations of activated cells were also present (Fig. 4b), likely reflecting distinct cell types with unique molecular and axonal-targeting profiles43. In support of this hypothesis, the robustly activated CBs belonged to the CB+SATB1− subtype (Fig. 4b). The distribution of activated and inhibited cells in each subtype and their corresponding peri-SWR waveforms supported these subtype-specific dynamics (Fig. 4c,d). Overall, CA2 interneuron responses mirrored the dynamics of their CA3 counterparts, with relatively minor differences in the shape of the peri-SWR traces, especially for AACs, SOMs and CCKs (Extended Data Fig. 6c).
Fig. 4 |. Subtype-specific offline dynamics during SWR events.
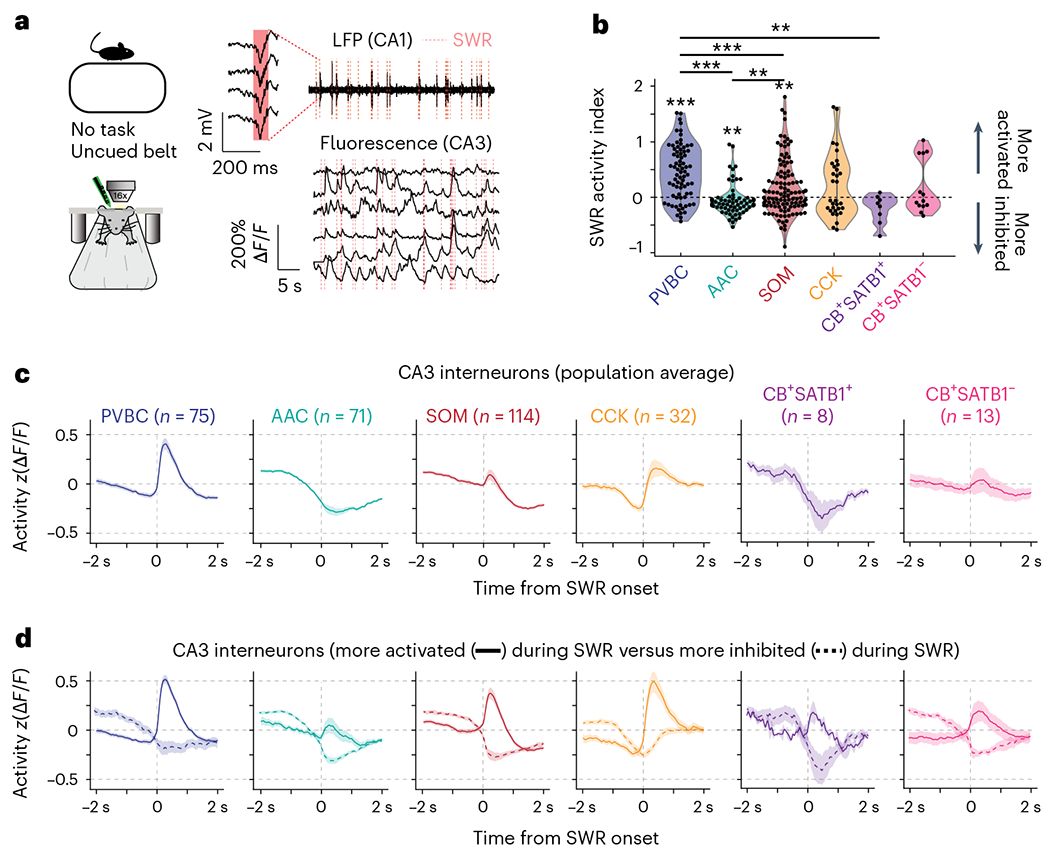
a, Experimental setup for simultaneous 2p AOD imaging and LFP recordings. SWRs were recorded on a four-channel silicon probe implanted in the contralateral CA1. Calcium traces are shown on the same timescale as the recorded LFP, with identified SWRs denoted with dashed red lines. b, Average SWR activity index for all CA3 interneurons grouped by subtype. Two-sided Wilcoxon signed-rank tests against a median of 0 were performed for each subtype (PVBC: 0.45 ± 0.48, P = 1.1 × 10−9; AAC: −0.037 ± 0.27, P = 0.004; SOM: 0.17 ± 0.47, P = 0.006; CCK: 0.20 ± 0.56, P = 0.11; CB+SATB1+: −0.19 ± 0.24, P = 0.07; CB+SATB1−: 0.24 ± 0.45, P = 0.31). Between-subtype statistical comparisons were performed using the Kruskal–Wallis test (P < 10−8) with post hoc Wilcoxon rank-sum tests with P values adjusted using the Bonferroni correction (significant adjusted P values: PVBC–AAC: <10−5; PVBC–SOM: 0.0006; PVBC–CB+SATB1+: 0.0047; AAC–SOM: 0.005). Data are from n = 75 PVBCs, 71 AACs, 114 SOMs, 32 CCKs, 8 CB+SATB1+ cells and 13 CB+SATB1− cells from n = 13 mice. Data are reported as mean ± s.d. c, Average peri-SWR traces for all CA3 interneuron subtypes. Cell numbers for each subtype are indicated; data are from n = 13 mice. Traces for each subtype represent mean ± s.e.m. The scale of the y axis is shared across all subtypes. d, Same as c, but the data are split by activated (solid lines) and inhibited (dashed lines) neurons. CA3 activated neurons: PVBCs = 59; AACs = 21; SOMs = 65; CCKs = 16; CB+SATB1+ cells = 2; CB+SATB1− cells = 7. Inhibited neurons: PVBCs = 16; AACs = 50; SOMs = 49; CCKs = 16; CB+SATB1+ cells = 6; CB+SATB1− cells = 6. Traces for each subtype represent mean ± s.e.m. The scale of the y axis is shared across all subtypes; *P <0.05, **P <0.01, ***P <0.001.
Predictive and reflective inhibitory dynamics around SWRs
SWR duration recorded in vivo exhibits marked event-to-event variability, which is thought to reflect differences in the size of recruited pyramidal cell ensembles (Extended Data Fig. 5b) in the hippocampal network2,44. If CA3/CA2 inhibitory interneurons influence SWR initiation or termination, we expect that the activity profile of those cells would be modulated by different SWR durations. We found that some interneuron subtypes indeed displayed responses largely correlated with the duration of the SWR they follow (PVBCs) or precede (CCKs; Fig. 5a). We measured these responses by analyzing the shortest (0–20th percentile of all durations) and the longest (80–100th percentile) SWR events and found that PVBCs and CCKs were differently recruited by the two conditions. PVBCs were more activated after long-duration SWR events, while CCKs were more inhibited around and before long-duration events (Fig. 5b). The timing of the most extreme difference between the average short- and long-duration SWR responses was, for CCKs, tens of milliseconds before the SWR onset, while PVBCs had differential responses after SWR onset (Fig. 5b). To more accurately quantify these responses, we averaged the activity of each interneuron from −500 to 0 ms preceding SWR events (pre-SWR) and from 0 to 500 ms after (post-SWR). We found that the negative response of CCKs pre-SWR was indeed strongly correlated with the duration of the subsequent SWR across all duration percentiles. Similarly, for PVBCs, the magnitude of the post-SWR response was highly correlated with the length of the SWR (Fig. 5c). Finally, when taking the difference of activity between the longest and shortest SWRs, we found that PVBCs and CCKs most significantly changed their response profiles from all subtypes, and these changes occurred after and before SWR onset, respectively (Fig. 5d). By comparison, while CA2 PVBCs also showed increased activation around long-duration SWR events, CA2 CCKs did not show a corresponding increased inhibition around long-duration SWRs (Extended Data Fig. 7). Overall, these results suggest a role for CA3 CCK interneurons in sculpting the inhibitory tone preceding SWRs and regulating their duration and an important role for PVBCs in potentially terminating the generated population activity.
Fig. 5 |. Peri-SWR dynamics are both predictive and reflective of SWR duration in a subtype-specific manner.
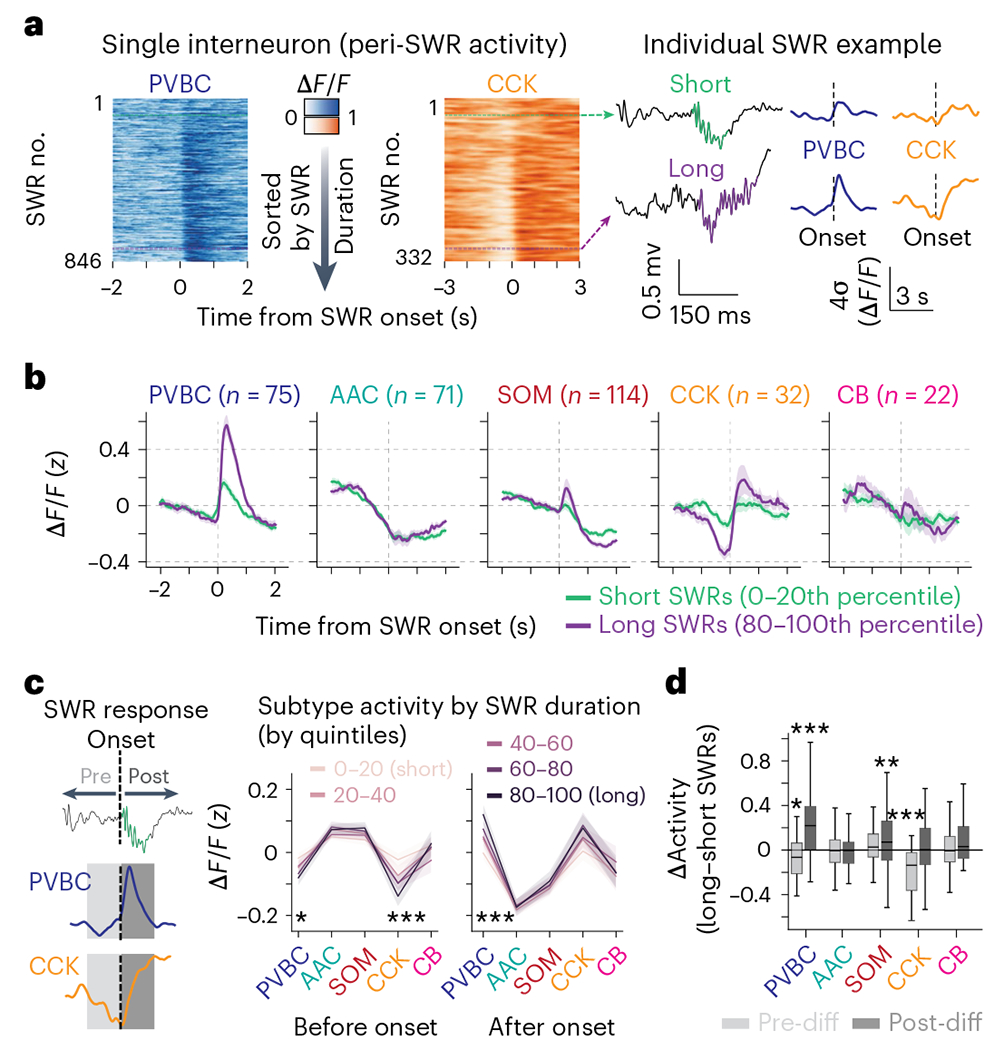
a, Example responses of a CA3 PVBC and a CA3 CCK cell to long- and short-duration SWRs. Left, heat maps illustrating each cell’s responses around SWR events ordered by their duration. Right, both activated and inhibited responses were strongly modulated by SWR duration. b, Average z-scored peri-SWR traces for both short-duration (0–20th percentile) and long-duration (80–100th percentile) SWRs for CA3 interneurons of each subtype (n = 75 PVBCs, 71 AACs, 114 SOMs, 32 CCKs and 22 CBs from n = 13 mice). CB+SATB1+ and CB+SATB1− neurons are considered together in the CB subtype for this analysis (see Extended Data Fig. 7a–c for the separated data). Traces for each subtype and condition represent mean ± s.e.m. The scale of the y axis is shared across all subtypes. c, Left, schematic illustrating the activity before and after each SWR that was considered for analysis. A 500-ms window before and after each SWR event was used. Right, average value of the activity of CA3 interneurons of each subtype before and after SWRs considered separately for SWRs falling into five groups based on their duration (0–20th, 20–40th, 40–60th, 60–80th and 80–100th percentiles). Statistics indicate the two-sided Spearman correlation between each subtype’s average activity in pre- and post-SWR epochs with SWR durations falling into different quantiles. Only PVBCs and CCKs have their activity significantly correlated with SWR duration (PVBC pre-SWR, P = 0.012; PVBC post-SWR, P = 0.0002; CCK pre-SWR, P = 0.001). Data are from n = 75 PVBCs, 71 AACs, 114 SOMs, 32 CCKs and 22 CBs from n = 13 mice and are presented as mean ± s.e.m. d, Average value of the difference (Δ or diff) in activity between long-duration (80–100th percentile) and short-duration (0–20th percentile) SWRs for each CA3 interneuron considered separately for pre-SWR and post-SWR activity and grouped by subtype. PVBCs were slightly more inhibited before long-duration SWRs and significantly more activated after long-duration SWRs, while CCKs were significantly more inhibited before long-duration SWRs. SOMs were more activated after long-duration SWRs. Only significant differences are indicated; the remaining subtypes did not show considerably different activity before or after long- duration SWRs compared to short-duration SWRs. Data were analyzed by two-way, one-sample t-tests against 0 for each subtype with a Bonferroni correction for multiple testing: PVBC pre: −0.067 ± 0.180, P = 0.020; PVBC post: 0.242 ± 0.347, P = 7.22 × 10−7; AAC pre: −0.0069 ± 0.155, P = 1.0; AAC post: −0.0025 ± 0.208, P = 1.0; SOM pre: 0.040 ± 0.198, P = 0.34; SOM post: 0.112 ± 0.310, P = 0.0020; CCK pre: −0.195 ± 0.209, P = 1.19 × 10−4; CCK post: 0.057 ± 0.339, P = 1.0; CB pre: 0.0023 ± 0.19, P = 1.0; CB post: 0.093 ± 0.245, P = 0.97). Data are from n = 75 PVBCs, 71 AACs, 114 SOMs, 32 CCKs and 22 CBs from n = 13 mice. Data are reported as mean ± s.d.; *P < 0.05, **P < 0.01, ***P < 0.001.
Learning-related changes in inhibitory activity around SWR events
Although individual SWR events show marked variation in their duration, hippocampal-dependent learning tasks have been shown to increase the average SWR duration in the rest periods immediately after the learning epoch38,44,45. This increase in SWR duration after learning suggests that hippocampal-dependent learning tasks induce changes in the recurrent circuits that generate SWRs such that post-learning SWRs are longer and support memory consolidation. To assess whether we could also observe learning-related changes in CA3/CA2 inhibitory circuit dynamics around SWR events, we trained water-restricted mice to solve a goal-oriented learning (GOL) paradigm in which they had to learn and remember the location of a water reward on the belt that contained various tactile cues (Fig. 6a and Methods). Before (PRE) and after (POST) the task, we performed simultaneous CA3/CA2 interneuron imaging and contralateral LFP recordings on a burlap belt devoid of cues while animals were immobile (Fig. 6a). Consistent with the previous literature, we observed a robust increase in SWR occurrence and duration between the PRE and POST sessions (occurrence PRE: 0.146 ± 0.088 s−1; occurrence POST: 0.456 ± 0.100 s−1; duration PRE: 74.5 ± 12.7 ms; duration POST: 80.4 ± 8.9 ms (mean ± s.d.); Fig. 6b). Because the reward location was changed from day to day, we measured the mouse’s behavioral performance, or learning, on each day by calculating the difference in the fraction of anticipatory licks between the second and first halves of the session (Fig. 6c). Using this metric, we observed that the increased SWR duration in the POST session compared to the PRE session followed the behavioral performance of the animal on the day (Fig. 6d,e). We thus divided the data into learning days (days when the difference in the fraction of anticipatory licks between the second and first halves of the imaging session was greater than 0) and non-learning days (days when this difference was not greater than 0). The significant increase in SWR duration was specific to learning days as opposed to non-learning days, while the SWR occurrence increased nonspecifically for both conditions, suggesting specific learning- versus task-related changes in SWR properties (Fig. 6e). We next calculated the interneuron activity profiles around SWR events during the PRE and POST sessions of both learning and non-learning days (Extended Data Fig. 8) and found that the responses of PVBCs and CCKs were different compared to other subtypes (Fig. 6f). More specifically, we observed a robust increase in PVBC activation in POST sessions on learning days but not on non-learning days (Fig. 6g). Similarly, we observed a more pronounced inhibition of CA3 CCKs around SWR events in POST sessions only on learning days (Fig. 6g and Extended Data Fig. 8). Interestingly, these PRE-to-POST changes in peri-SWR interneuron dynamics were not observed in CA2, as CA2 CCKs showed no change in peri-SWR inhibition between PRE and POST sessions on learning days (Extended Data Fig. 8), and CA2 PVBCs actually showed a small but statistically significant decrease in activation around SWR events between the PRE and POST sessions of learning days (Fig. 6h). Together, these results demonstrate clear subtype- and region-specific changes in SWR-associated inhibitory responses following spatial learning.
Fig. 6 |. Learning-related changes in interneuron dynamics around SWRs are both region and subtype specific.
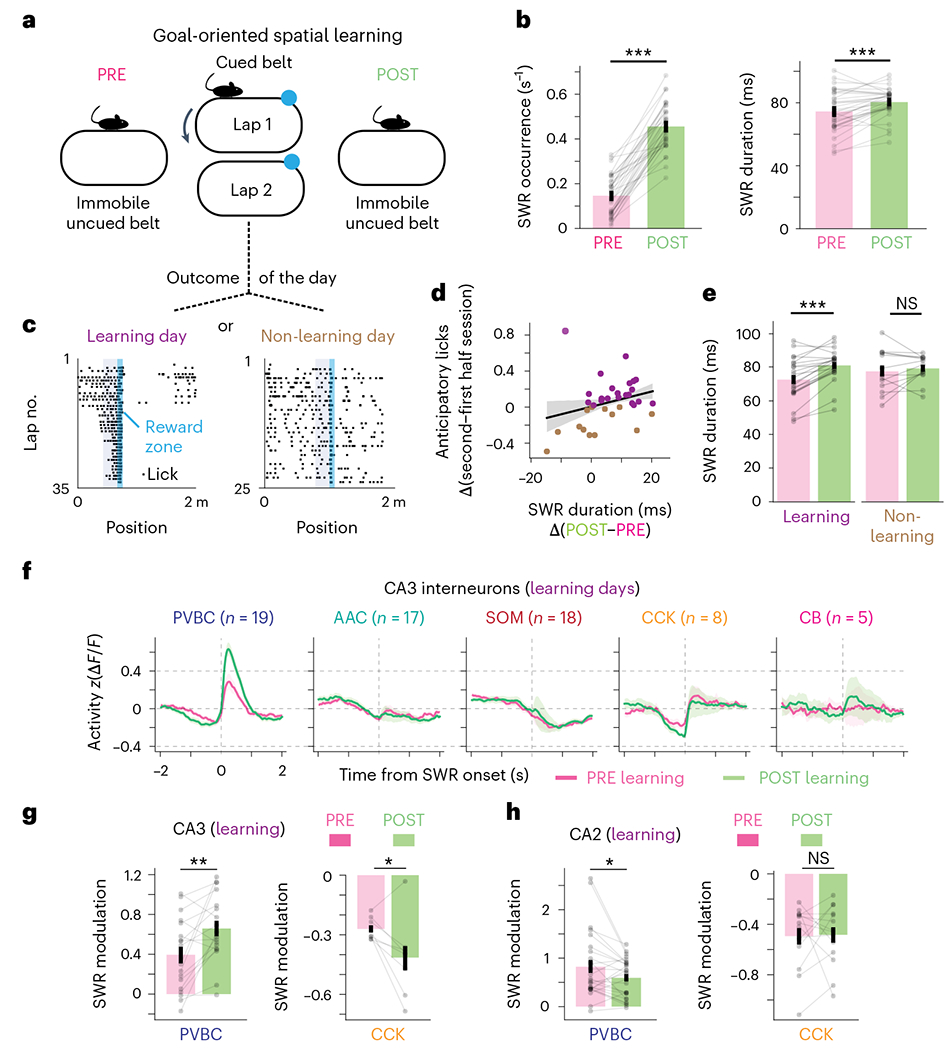
a, Schematic of the GOL task. CA3/CA2 interneurons were imaged during SWRs when mice rested on a cueless burlap belt during the PRE and POST sessions. During the task, water-restricted mice ran on a cued belt to receive a water reward that remained in the same location from lap to lap. b, Quantification of the average SWR occurrence rate (left) and average SWR duration (right) for PRE and POST sessions. Both the occurrence rate and the average duration increased significantly from PRE to POST (occurrence PRE: 0.146 ± 0.088 s−1; occurrence POST: 0.456 ± 0.100 s−1; two-sided Wilcoxon signed-rank test, P = 2.48 × 10−7; duration PRE: 74.5 ± 12.7 ms; duration POST: 80.4 ± 8.9 ms; two-sided Wilcoxon signed-rank test, P = 6.19 × 10−4; data are from n = 35 PRE sessions and 35 POST sessions from n = 8 mice). Data are reported as mean ± s.d. c, Example mouse behavior from both learning and non-learning days. On learning days, the fraction of anticipatory licks during the second half of the session was greater than in the first half. Dividing the dataset in this manner resulted in 22 learning days and 13 non-learning days. The blue shaded area represents the reward zone, and the gray shaded area represents the area where anticipatory licks were considered. d, Relationship between the change in SWR duration and the change in anticipatory licks between the second and first halves of the session on each imaging day. The linear regression fit is shown, with shaded bands representing the 95% confidence interval. Days on which the mouse learned were associated with greater changes in SWR duration from PRE to POST. e, Quantification of the difference in SWR duration from PRE to POST sessions on learning and non-learning days. On learning days, the SWR duration increased significantly from PRE to POST (PRE: 72.7 ± 12.7 ms; POST: 81.1 ± 9.5 ms; two-sided Wilcoxon signed-rank test, P = 2.60 × 10−4; data are from n = 22 imaging days in n = 8 mice). On non-learning days, the average SWR duration did not change significantly from PRE to POST sessions (PRE: 77.6 ± 12.2 ms; POST: 79.3 ± 7.6 ms; two-sided Wilcoxon signed-rank test, P = 0.600; data are from n = 13 imaging days in n = 8 mice). Data are reported as mean ± s.d. f, Average z-scored peri-SWR traces for each CA3 interneuron subtype during both PRE and POST sessions on learning days. PVBCs became more activated around SWRs after learning, while CCKs became more inhibited around SWRs after learning (n = 19 PVBCs, 17 AACs, 18 SOMs, 8 CCKs and 5 CBs from n = 8 mice). CB+SATB1+ and CB+SATB1− neurons are considered together in the CB subtype. Traces for each subtype and condition represent mean ± s.e.m. The scale of the y axis is shared across all subtypes. g, Quantification of the change in peri-SWR modulation for CA3 PVBCs and CCKs between PRE and POST sessions on learning days. PVBCs became more activated around SWRs after learning, while CCKs became more inhibited around SWRs after learning (PVBC PRE: 0.39 ± 0.36 and PVBC POST: 0.66 ± 0.33, two-sided Wilcoxon signed-rank test, P = 0.0057; CCK PRE: −0.27 ± 0.05 and CCK POST: −0.41 ± 0.18, two-sided Wilcoxon signed-rank test, P = 0.049; data are from n = 18 PVBCs and 8 CCKs from n = 8 mice). Data are reported as mean ± s.d. h, Same quantification as in g but now for CA2 PVBCs and CCKs. PVBCs became slightly less activated around SWRs after learning, while CCKs did not significantly change their activity (PVBC PRE: 0.82 ±0.68 and PVBC POST: 0.59 ± 0.39, two-sided Wilcoxon signed-rank test, P = 0.043; CCK PRE: −0.49 ± 0.24 and CCK POST: −0.48 ± 0.23, two-sided Wilcoxon signed-rank test, P = 0.68; data are from n = 26 PVBCs and 14 CCKs from n = 8 mice). Data are reported as mean ± s.d.; *P < 0.05, **P < 0.01, ***P < 0.001.
To determine whether similar changes in peri-SWR dynamics occur in response to passive sensory experience without explicit learning demand, we exposed mice to randomly delivered water, light or odor stimuli while head-fixed on a cueless burlap belt40. Additionally, we recorded interneuron activity during SWR events that occurred before (PRE) and after (POST) the presentation of a set of stimuli (Extended Data Fig. 9). CA3 CCKs were selectively activated in response to the sensory stimuli, while CA3 SOMs were inhibited, and the remaining CA3 subtypes showed no significant change in activity (Extended Data Fig. 9). Although this task resulted in a greater SWR occurrence rate from PRE to POST sessions, we did not detect a change in SWR duration (Extended Data Fig. 9). Correspondingly, we observed no significant changes from PRE to POST in the activation and inhibition of PVBCs and CCKs, respectively, around SWR events as we did in the spatial learning paradigm (Extended Data Fig. 9). Although not statistically significant, we observed a trend of increased CCK activation around SWR events in response to sensory stimulation (Extended Data Fig. 9), suggesting possibly different plasticity mechanisms in CA3 circuits in response to passive sensory experience and explicit learning.
Discussion
To date, the electrophysiological properties of CA3/CA2 interneurons have been mainly studied in vitro46–49, especially as they relate to plasticity of their mossy fiber inputs10,50. Additionally, previous work characterized interneuron activity in vivo, but these experiments were performed mostly under anesthesia43,51 and yielded only small numbers of neurons. The sparsity, diversity and depth of CA3/CA2 inhibitory circuits in the intact brain have continuously prevented the deployment of traditional population recording techniques during behavior. Our approach combining large-scale, unbiased sampling of inhibitory neurons, simultaneous LFP recordings and multiplexed molecular characterization post hoc allowed us to overcome these limitations and provide new insights into CA3 and CA2 interneuron activity dynamics during spatial navigation and SWR-associated memory reactivation.
We find that the in vivo dynamics of CA3/CA2 inhibitory circuits exhibit similarities to their CA1 counterparts with respect to their overall positive velocity modulation and spatially modulated activity in a subset of interneurons. One notable difference is the CCKs that are largely active during immobility in CA1 (refs. 22,35) but show more heterogeneity in CA3 and are active during locomotion in CA2. The differences between CA3 and CA2 that we observe with respect to the overall velocity modulation of interneuron subtypes may be related to the previously reported increased immobility-related activity of CA2 pyramidal cells14. Our finding that CA3/CA2 CBs comprise two functionally distinct subpopulations regarding their activity during locomotion strongly correlates with the expression of the transcription factor SATB1 in these cells. The immobility-active CBs we identify are potentially linked to a rare subtype recorded from in CA1 under anesthesia52. As SATB1 acts downstream of other transcription factors that specify an origin from the medial ganglionic eminence (MGE) and is thus enriched in MGE-derived interneurons, our finding suggests that the functional dichotomy we observe in CB-expressing interneurons may represent differences between MGE- and caudal ganglionic eminence(CGE)-derived CBs. We hypothesize that differences in ‘hard-wired’ input connectivity onto these two cell types might account for their complementary activity profiles during locomotion and immobility and that these connectivity differences likely apply to MGE- and CGE-derived interneurons more generally. It will be critical in future experiments to understand whether immobility-activated interneurons throughout the hippocampus share common inputs, if locomotion/immobility signals are passed from one region to the other and how this locomotion state-dependent inhibition from specific interneuron subtypes regulates information processing in pyramidal cell networks. Further, in contrast to the prominent subtype- and region-specific velocity modulation of activity we observe in CA3/CA2 interneurons, we also show robust and stable spatial modulation that differs little between subtypes and between the CA3 and CA2 regions.
Our study offers important new directions regarding the currently unknown mechanisms of SWR initiation and termination associated with memory consolidation by providing a large-scale report of multiple interneuron activity profiles during these events. It was originally proposed that SWRs are generated by transient disinhibition of recurrently connected CA3 pyramidal cells2,53, but experimental evidence for this hypothesis has been limited to neural recordings of only a handful of AACs showing inactivity during SWRs51. Here, we confirm that such a response profile is shared across the large majority of AACs. Additionally, we show that CCKs are also inhibited during SWRs, starting hundreds of milliseconds before the onset of each event (Fig. 4). Together, these interneuron populations may potentially provide the disinhibition necessary to trigger the synchronous activation of pyramidal cells. Moreover, the extent to which CA3 CCK activity is reduced preceding SWR events is specifically predictive of the resulting SWR duration (Fig. 5). Thus, while AAC inhibition and the resulting withdrawal of inhibition from the axon initial segment of pyramidal cells may be a necessary condition for SWRs to occur, our data also suggest a critical role for CCK-expressing interneurons in that process.
Furthermore, we found that the magnitude of CA3/CA2 PVBC responses is correlated with SWR duration (Fig. 5), which implicates these cells in providing feedback inhibition that regulates population activity, consistent with previous in vitro observations in CA1 (ref. 54). Relatedly, recent experiments using in vivo intracellular recordings from CA3 pyramidal cells in awake animals have found that larger SWR events evoke a more pronounced hyperpolarization just after SWR onset; our finding potentially provides the cellular mechanism for this increased inhibition55. Together, our results support a complementary role of CCKs and PVBCs in organizing pyramidal cell assembly recruitment in a predictive and reflective manner, respectively. Future experiments will require manipulations of specific interneuron populations in CA3 to determine the necessity and sufficiency of the various subtypes in generating SWRs.
Our results describing changes in the activity of CA3/CA2 interneuron subtypes after spatial learning represent an assessment of learning-related and subtype-specific changes in inhibitory activity around SWR events. Early studies describing plastic changes in hippocampal inhibitory network activity were conducted after pharmacologic manipulations in CA3 in vitro56, under anesthesia after the electrical stimulation of CA1 afferents in vivo57 or during behavior in CA1 but without interneuron subtype identification24,58. Here, we demonstrate that explicit spatial learning induces changes in CA3 inhibitory network dynamics such that PVBCs and CCKs become more activated and inhibited around SWRs after learning, respectively. Notably, these subtype-specific dynamics are also characteristic of long-duration SWRs at baseline (Fig. 5), and our spatial reward learning task induced a significant increase in average SWR duration. Because SWRs are thought to be generated in the strongly recurrent CA3a/CA3b2 and CA2 (ref. 8), one possibility is that the changes we observe, namely the increased activation of local PVBCs and the increased inhibition of CCKs, may play a critical role in generating the long-duration SWRs that appear to be specifically associated with learning38,44,45. Under this scenario, we can speculate that the increased excitation onto CA3 PVBCs may come from recurrently connected pyramidal cells or from highly plastic mossy fiber synapses10, while the increased inhibition onto CCKs may originate from other local interneurons59, long-range inhibitory projections or other subcortical neuromodulatory projections that may undergo plastic reconfiguration after learning25. Support for this hypothesized subtype-specific synaptic plasticity onto interneurons comes from recent in vitro work, which has demonstrated increased excitatory and inhibitory inputs to CA3 PVBCs and CCKs, respectively, after pharmacologically induced gamma oscillations56. Alternatively, it is possible that the changes we observe in CA3 interneuron activity after learning are primarily a consequence of plastic changes within the CA3 excitatory recurrent circuitry. Of note, the changes we observe after learning are not only subtype specific but also region specific, as they are not observed in CA2. The lack of changes in SWR-associated recruitment of CA2 inhibitory circuits after learning could be related to the reduced capacity and unique regulation of canonical long-term synaptic plasticity in CA2 (ref. 6). It is also possible that our spatial learning task is more specific to CA3 functions and that CA2 circuits would be more engaged in social memory tasks60.
Our approach taken in this work also has limitations. First, calcium imaging of interneurons does not allow for single-action-potential resolution, and interneurons may have different spike–fluorescence relationships depending on their electrophysiological and morphological properties. Second, contralateral LFP recordings do not allow for the detection of local SWR events that occur in the hemisphere ipsilateral to the imaging. Third, our recording sessions were only performed during awake states, and dynamics during sleep SWRs remain unexplored. Finally, because our imaging depth is limited to hundreds of microns below the imaging window with 2p excitation, the results we report here are restricted to CA3a, the most recurrent CA3 subregion and the hypothesized primary locus of SWR initiation.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41593-023-01306-7.
Methods
Animals
All experiments were conducted in accordance with NIH guidelines and with the approval of the Columbia University Institutional Animal Care and Use Committee. Interneuron imaging experiments were performed with healthy, 3- to 5-month-old, heterozygous adult male and female VGAT-IRES-Cre mice (Jackson Laboratory, 016962) on a C57BL/6J background. Pyramidal cell imaging experiments were performed with healthy, 3- to 5-month-old, heterozygous adult male and female Grik4-cre mice (Jackson Laboratory, 006474) on a C57BL/6J background. Mice were housed in the vivarium at ambient temperature and humidity on a reversed 12-h light/12-h dark cycle with three to five mice in each cage. Mice with implanted silicon probes were housed individually. Experiments were performed during the dark portion of the cycle.
Viruses
Cre-dependent rAAV expressing GCaMP7f under the control of the synapsin promoter (rAAV1-Syn-FLEX-jGCaMP7f-WPRE-Sv40; Addgene, 104492; titer, 1 × 1013 viral genomes per ml) was used to express GCaMP7f in interneurons. For pyramidal cell imaging, a Cre-dependent GCaMP7f (described above) or GCaMP8s (AAV9-Syn-FLEX-jGCaMP8s-WPRE; Addgene, 162377; titer, 1 × 1013 viral genomes per ml) virus was used.
Virus injections and hippocampal window/headpost implant for in vivo imaging
For viral injections, 2- to 4-month-old mice were anesthetized with isoflurane and placed into a stereotaxic apparatus. Meloxicam and bupivacaine were administered subcutaneously to minimize discomfort. After the skin was cut in the midline to expose the skull, the skull was leveled, and a craniotomy was made over the right hippocampus using a drill. A sterile glass capillary loaded with rAAV was attached to a Nanoject syringe (Drummond Scientific) and slowly lowered into the right hippocampus. Dorsal CA3/CA2 was targeted with two x–y coordinates, each one consisting of two different injection sites separated in z: anterio–posterior −1.35, medio–lateral −1.6, dorso–ventral −2.1, −1.9 and anterio–posterior −1.6, medio–lateral −1.9, dorso–ventral −2.2, −2.0 relative to bregma, with 50–64 nl of virus injected at each dorsal–ventral location. After injection, the pipette was left in place for 5–10 min and slowly retracted from the brain. The skin was closed with several sutures, and the mice were allowed to recover for 4 d before the window/headpost implant.
The surgical procedure for the CA3/CA2 window/headpost implant was similar to the one implemented for CA1 imaging22. Briefly, the injected mice were anesthetized with isoflurane and placed into the stereotaxic apparatus. After subcutaneous administration of meloxicam and bupivacaine, the skull was exposed and leveled, and a 3-mm craniotomy was made over the right anterior hippocampus, centered between the two injection coordinates. The dura overlying the cortex was removed, and the cortex overlying the hippocampus was slowly removed with negative pressure while ice-cold cortex buffer was simultaneously applied. Care was taken to not damage the lateral ventricle. This process was performed until the white, anterior–posterior fibers overlying the hippocampus became visible and any bleeding subsided. A stainless steel, 3-mm wide and 2-mm long circular cannula fitted with a glass window was inserted into the craniotomy and pushed down to sufficiently flatten the natural curvature of the anterior hippocampus. The cannula was secured in place with Vetbond applied on the skull. Subsequently, dental cement was applied to the entire skull, and a headpost was affixed to the posterior skull with dental cement. The mice received a 1.0-ml subcutaneous injection of saline and recovered in their home cage while heat was applied. The mice were monitored for 3 d after the operation until behavioral training began.
AOD imaging
Before the random foraging experiments, mice first underwent a single imaging session consisting of a high-resolution structural scan. This step was necessary to obtain a reference z stack and derive the x–y–z positions of GCaMP-expressing neurons. The mice were head-fixed under a custom-modified AOD microscope (Femto3D-ATLAS, Femtonics) and anesthetized with ketamine/xylazine to reduce motion artifacts during the stack. To provide stable transmission parameters during chronic imaging in the entire 3D scanning volume, the AOD microscope was extended with a high-speed and precision beam stabilization unit, which was directly attached to the AOD scan head, sensitive to input beam misalignment. The beam stabilization unit consisted of two quadrant detectors (PDQ80A and TPA101, Thorlabs) and two broadband dielectric mirrors (Thorlabs) mounted on motorized mirror mounts (Femtonics). Beam alignment was performed by the LaserControl software (Femtonics). A water immersion objective (×16 Nikon CFI75) was placed above the glass window and lowered until the CA3 pyramidal cell layer was in focus. At this stage, the objective was fixed in position, and focus was subsequently adjusted using AO crystals61. The laser (Coherent Ultra II) was tuned to λ = 920 nm. The reference z stack was taken starting from the approximate location of CA2 (~200–300 μm below the glass window) and extending as deep into CA3 as possible (~500–600 μm below the glass window while still being able to visualize individual interneurons); 800 × 800 pixel images (x–y resolution of 1.25 μm per pixel) were taken every 4 μm. Laser power and photomultiplier tube (PMT) detectors (GaAsP, H10770PA-40, Hamamatsu) were compensated appropriately in z throughout the stack (power at 20–40 mW and detector gain at 80% at the top of the stack and power at 120–150 mW and detector gain at 90% at the bottom). After completion, the mice were returned to their home cage and allowed to recover for 24 h until the start of functional imaging.
Before simultaneous imaging and LFP experiments, the mice were not anesthetized, and the reference z stacks were taken on the same day that functional imaging was performed. Small z stacks of ~ 40 μm were taken while the mouse was immobile on the belt.
To determine x–y–z positions of GCaMP-expressing neurons, the z stack was scrolled through, and each visible interneuron was manually selected using the integrated software (MES, Femtonics) to generate a list of ~100 x–y–z coordinates defined as the center of each cell (~20 cells for simultaneous imaging/LFP experiments). These points constituted the centers of regions of interest (ROI) used on subsequent days for functional imaging. Each ROI was defined as a square of 40–50 μm2 (chessboard scan) with a resolution of 1–1.5 μm per pixel. The advantage of the chessboard scanning method is that only neurons and small areas around the preselected cells are recorded. Therefore, a high ratio of the total recording time (~20–50%) is spent reading out information from the selected neurons. By contrast, volumetric imaging with the same 2p excitation provides an orders-of-magnitude worse ratio for measurement time utilization, as the somata of interneurons occupy a relatively small ratio of the total scanning volume.
On each day of functional imaging, the same field of view was found using the reference z stack, and x–y–z coordinates were loaded into the software to perform 3D imaging. Once all cells were in focus, 10-min functional imaging sessions were conducted at a frame rate of 5–10 Hz for most experiments (frame rate was dependent on ROI size and resolution). For experiments involving contralateral LFP recordings, imaging was conducted at a higher rate (~40 Hz), which restricted imaging to only 10–20 cells simultaneously. During functional imaging, the laser power and detector gain were compensated based on the reference z stack parameters.
Two-photon resonant scanner imaging of CA3 pyramidal cells (CA3PCs)
For experiments involving imaging of CA3PCs, single-plane resonant scanning 2p imaging was performed as described elsewhere15. Briefly, imaging was conducted using a 2p 8-kHz resonant scanner (Bruker) and a 16X water immersion objective (Nikon). A 920-nm laser (50–100 mW; Coherent) was used for excitation, and fluorescence signals were collected with a GaAsP PMT (Hamamatsu). A custom dual-stage preamp (Bruker) was used to amplify signals before digitization.
Silicon probe implantation, LFP recordings and SWR identification
For experiments requiring simultaneous 2p calcium imaging and LFP recordings, mice were implanted with a glass window over the hippocampus as described above, and a chronic, four-channel silicon probe (Qtrode, Neuronexus) was inserted into the contralateral CA1 at a 45° angle. The probe was secured in place with dental acrylic, and the mouse was allowed to recover for several days, as described above. LFP signals were recorded with a multichannel recording system (Intan Technologies) synchronized with the AOD imaging system. The correct position of the silicon probe was confirmed by the presence of SWRs in the data. LFP signals were recorded at 20 kHz. To identify putative SWR events, the raw LFP signal was band-pass filtered from 150 to 300 Hz and thresholded at 2.5 s.d. above the mean value within the passband. All putative SWR events were then manually inspected, and false positives were discarded to obtain the final set of SWR events used for analysis.
Behavioral paradigm
After recovery from surgery, mice were handled for several days and habituated to head fixation. Mice were subsequently water restricted to 85–90% of their original weight and trained to run on a single-fabric, cue-free belt. Mice were trained to operantly lick and receive water rewards (water was delivered in response to tongue contact with a capacitive sensor) at random locations along the belt. As performance improved, the number of rewards delivered on each lap decreased. After several days of training on this cue-free belt, the mice were trained for ~1 week on a 2-m-long, cue-rich belt for randomly delivered water rewards. For random foraging experiments, imaging was started after mice could run approximately ten laps in 10 min (usually after 7–10 d of total training). For combined imaging and LFP experiments, data acquisition was started once GCaMP7f expression was optimal, hippocampal windows were clear and the mice were habituated to head fixation; these mice did not undergo additional behavioral training.
For the GOL paradigm, mice were initially trained on the random foraging paradigm as described above. Once they could run approximately ten laps in 10 min, a single reward zone was fixed on the cued belt, and mice were trained for several days with one reward zone that remained fixed throughout the session but changed locations from day to day. After several days of training on this GOL paradigm, imaging experiments were started. On imaging days, mice first rested on an uncued burlap belt for the PRE session, during which imaging and LFP recordings were performed. Mice were then placed back into their home cage while the belt was changed to the cued belt that they were trained on. Mice were subsequently imaged during the 10-min GOL task on the cued belt. Afterward, mice were placed back into their home cage, and the belt was again changed to the uncued burlap belt for the POST session, during which imaging and LFP recordings were again performed38. To control for the effect of elapsed time on the impact of potential learning during the GOL session on the POST session, data from the POST session were only included in the subsequent analysis if the POST data collection was started 30 min or less after the end of the GOL session.
The sensory stimulation experiments (random cue task) were performed as described previously40 on a burlap belt. Briefly, three sensory cues (odor, light and a non-operant water reward) were presented randomly at 15 trials per cue independently of the mouse’s position on the treadmill, and each cue presentation was separated by a random interstimulus interval of 10–15 s.
Behavior data (licking, velocity, position and reward) were collected using custom software implemented in Java via a microcontroller (Arduino DUE) on the treadmill.
Perfusion and tissue processing
After the completion of imaging experiments, mice were transcardially perfused with 40 ml of ice-cold PBS (Thermo Fisher), followed by 40 ml of ice-cold 4% paraformaldehyde (PFA; Electron Microscopy Sciences). Brains were stored overnight in 4% PFA at 4 °C. The next day, the 4% PFA was removed, and the brains were rinsed for 5 min three times in PBS. Horizontal sections (75 μm) of the imaged hippocampus were cut on a vibrating microtome (Leica VT1200S) and washed for 15 min three times in PBS. Subsequently, sections were permeabilized for 20 min twice in PBS with 0.3% Triton X-100 (Sigma-Aldrich). Blocking was then performed with 10% normal donkey serum (Jackson ImmunoResearch, 017-000-121) in PBS with 0.3% Triton X-100 for 45 min. The sections were then incubated in a PBS solution containing three primary antibodies (see below for antibody information and dilutions) for 1 h at room temperature, followed by 2 d at 4 °C. After 2 d, the primary antibody solution was removed from the slices, and the slices were washed three times for 15 min each in PBS to remove unbound primary antibodies. The slices were subsequently incubated in a PBS solution containing a mixture of appropriate secondary antibodies conjugated to fluorescent labels (see below for antibody information and dilutions) for 2 h at room temperature. The sections were then washed five times for 15 min each in PBS at room temperature. Finally, sections were mounted on glass slides in Fluoromount-G aqueous mounting medium (Thermo Fisher Scientific) and coverslipped. The slides were allowed to dry at 4 °C for at least 1 h before confocal imaging (see below). After confocal imaging, the slides were submerged in PBS to remove the coverslip, and the sections were removed from the slides with gentle rocking. After washing three times for 15 min each in PBS and blocking with 10% normal donkey serum in PBS with 0.3% Triton X-100 for 45 min, the sections were incubated in an additional two to three primary antibodies. Regarding the sensitivity and specificity of the multiple labeling and the potential cross-talk between the immunosignals, several steps were taken to ensure minimum cross-contamination and maximum accuracy of our immunolabels. First, we only used antibodies that were validated in previous studies. Second, we only analyzed cells that were clearly positive for one of the immunomarkers, and we also required that the staining pattern of the imaged cell reproduced the known staining pattern for that particular antigen. For example, to report a cell as PV+ or CB+, we required that the PV or CB immunolabel extended significantly throughout the cell (as opposed to just nuclear or perinuclear labeling), which is the known labeling pattern of both PV+ and CB+ interneurons. Similarly, for us to consider a cell to be SOM+ or CCK+, we required that the SOM or CCK immunolabel was restricted to the perinuclear region of the cell. For us to consider a cell to be SATB1+, we similarly required the immunolabel to be restricted to the nucleus, as SATB1 is known to be a nuclear antigen. Because we only analyzed cells that were clearly immunopositive, we acknowledge that not all imaged cells of a given subtype were included in our analysis, as they may have had only weak or unclear immunosignals and were therefore discarded. However, this ensured that our dataset was as accurate as possible.
Third, we did not remove the first round of labeling from the tissue. Instead, we simply added the second round of staining on top of the first round and so forth. We were able to do this because we carefully chose an immunolabeling strategy that allowed clear identification of two different antigens in the same confocal channel. The sections were subsequently washed, incubated in secondary antibodies, washed again, mounted and imaged, as in the first round of staining. This process of unmounting, staining, mounting and imaging was again repeated for a third round of staining.
Immunohistochemistry
Antibodies used for random foraging mice (data in Figs. 2 and 3) were as follows (see Reporting Summary for all antibody catalog numbers): First-round primary antibodies: rabbit anti-proCCK (1:500), rat anti-SOM (1:500) and goat anti-CB (1:500).
First-round secondary antibodies: donkey anti-rabbit DyLight 405 (1:300), donkey anti-rat Rhodamine Red (1:300) and donkey anti-goat Alexa 647 (1:300).
Second-round primary antibodies: chicken anti-PV (1:5,000) and rabbit anti-SATB1 (1:1,000).
Second-round secondary antibodies: donkey anti-chicken DyLight 405 (1:300) and donkey anti-rabbit Rhodamine Red (1:300).
Third-round primary antibody: mouse anti-STEP (1:500).
Third-round secondary antibody: goat anti-mouse IgG1 Alexa 488 (1:500).
Antibodies used for SWR mice (data in Figs. 4–6) were as follows:
First-round primary antibodies: chicken anti-PV (1:5,000), rat anti-SOM (1:500) and rabbit anti-SATB1 (1:1,000).
First-round secondary antibodies: donkey anti-chicken DyLight 405 (1:300), donkey anti-rat Rhodamine Red (1:300) and donkey anti-rabbit Alexa 647 (1:300).
Second-round primary antibodies: rabbit anti-proCCK (1:500) and goat anti-CB (1:500).
Second-round secondary antibodies: donkey anti-rabbit Rhodamine Red (1:300) and donkey anti-goat Alexa 647 (1:300).
Third-round primary antibody: mouse anti-STEP (1:500).
Third-round secondary antibody: goat anti-mouse IgG1 Alexa 488 (1:500).
Additional immunolabeling experiments.
To determine colocalization between CB, SATB1 and other interneuron markers not assessed in the main imaging experiments of the paper (including calretinin, M2R, NPY, COUP-TFII and VIP; Extended Data Figs. 2 and 4), VGAT-Cre mice were perfused, and fixed slices including the hippocampus were prepared as described above. In triple-immunolabeling experiments, these slices were each stained with one of the following labeling strategies at the specified antibody dilutions to determine colocalization.
CB/SATB1/calretinin.
Primary antibodies: goat anti-CB (1:500), guinea pig anti-calretinin (1:1,000) and rabbit anti-SATB1 (1:1,000).
Secondary antibodies: donkey anti-goat 488 F(ab′)2 (1:300), donkey anti-guinea pig Rhodamine Red (1:300) and donkey anti-rabbit 647 (1:300).
CB/SATB1/M2R.
Primary antibodies: goat anti-CB (1:500), rabbit anti-SATB1 (1:1,000) and rat anti-M2R (1:2,000).
Secondary antibodies: donkey anti-goat 488 F(ab’)2 (1:300), donkey anti-rabbit Rhodamine Red (1:300) and donkey anti-rat 647 (1:300).
CB/SATB1/NPY.
Primary antibodies: guinea pig anti-CB (1:500), rabbit anti-SATB1 (1:1,000) and sheep anti-NPY (1:500).
Secondary antibodies: donkey anti-guinea pig 488 (1:300), donkey anti-rabbit Rhodamine Red (1:300) and donkey anti-sheep F(ab′)2 647 (1:300).
CB/SATB1/COUP-TFII.
Primary antibodies: guinea pig anti-CB (1:500), rabbit anti-SATB1 (1:1,000) and mouse anti-COUP-TFII (1:1,000).
Secondary antibodies: donkey anti-guinea pig 488 (1:300), donkey anti-rabbit Rhodamine Red (1:300) and goat anti-mouse IgG2a 647 (1:300).
CB/SATB1/VIP.
To determine colocalization between CB, SATB1 and VIP, VIP-Cre mice were injected into CA3/CA2 with an AAV expressing Cre-dependent green fluorescent protein. After ~2 weeks to allow for virus expression, the mice were perfused, and slices of the hippocampus were prepared, as described above. These slices were then stained with primary goat anti-CB and rabbit anti-SATB1 described above together with secondary donkey anti-rabbit Rhodamine Red (1:300) and donkey anti-goat 647 (1:300) to determine colocalization between these three markers.
Verification of the Grik4-Cre line.
For immunohistochemical verification of the Grik4-Cre line, a FLEX-GCaMP virus was injected into CA3/CA2 as described above. After 2–3 weeks, mice were perfused, and brains were extracted, fixed and sliced as described above. Slices including the hippocampus were stained for the CA2-specific marker PCP4 (ref. 33) with rabbit anti-PCP4 (1:500 dilution). Secondary antibody staining was performed with donkey anti-rabbit Alexa 647 (1:300 dilution) as described above.
Confocal imaging
A Nikon A1 confocal microscope was used to acquire multichannel fluorescence images of the immunolabeled tissue sections; 405-nm, 488-nm, 561-nm and 640-nm laser lines were used for excitation. Each channel was acquired sequentially with a ×10/0.45-NA Plan Apo objective (Nikon) at ×1.2–1.3 zoom; 2,048 × 2,048 pixel images were acquired every ~3 μm through the entire depth of the tissue sections, with the pinhole size set to ~1 Airy unit. Fluorescence was collected with two GaAsP PMTs (488-nm and 561-nm channels) and two multialkali PMTs (405-nm and 640-nm channels). The resulting four-channel z stacks were viewed in ImageJ (NIH).
Registration of confocal images to in vivo z stacks and identification of immunopositivity/immunonegativity
The following steps were performed by an experimenter without the use of any automated methods. First, the confocal stacks were rotated and translated until the cells in the green channel (GCaMP labeled) matched the cells seen in the in vivo z stack. Second, each imaged cell was found in the confocal stacks, and it was evaluated for immunopositivity or immunonegativity for the tested molecule. For a cell to be considered positive, the fluorescence intensity inside the cell had to be significantly greater than the background intensity level. A cell was considered positive for a given marker only if clear examples of immunonegative cells could be found on the same tissue section. Similarly, a cell was considered negative for a given marker only if clear examples of immunopositive cells could be found on the same tissue section. In the case of ambiguous immunolabeling, cells were discarded and not grouped into a subtype for further analysis. Overall, all efforts were made to use the most stringent criteria for cell classification before analysis.
For region assignment, cells were classified as CA2 interneurons if they were on the same 75-μm slice as CA2 pyramidal cells. For pyramidal cells on a given slice to be considered CA2, the vast majority (>90%) of the pyramidal cells within the imaging FOV had to be STEP+33. If this criterion was not met (no majority of STEP+ pyramidal cells) but the CB+ mossy fibers were visible on the slice, then the interneurons on that slice were considered to be in CA3.
Subtype assignment
Subtypes were assigned based on the immunoreactivity of cells to the five tested markers and the association between these markers and defined interneuron subtypes based on the previous literature. All imaged cells not within the region innervated by CB+ mossy fibers were assumed to be within CA1 and were excluded from all analyses.
PVBCs.
PVBCs were positive for PV and SATB1 and negative for the other three tested markers (SOM, CCK and CB).
AACs.
AACs were positive for PV but negative for SATB1 and negative for the other three tested markers (SOM, CCK and CB).
SOMs.
All cells positive for SOM were categorized as SOMs. While all cells within this category were negative for CCK, some were also positive for PV, SATB1 or CB. Notably, SOM/CB-coexpressing cells were included in this category because they represent long-range projecting interneurons located almost exclusively in the stratum oriens of CA1–CA3 (refs. 41,62). Thus, the SOM subtype here represents putative dendrite-targeting and some long-range projecting interneurons.
CCKs.
CCKs were necessarily positive for CCK, and all of these cells were always negative for PV and SOM. Cells within this category could express SATB1, although the vast majority of them were SATB1−. Although some cells within the dataset coexpressed CCK and CB, the vast majority of CCK- and/or CB-expressing cells expressed only one of the two markers. Thus, CCK and CB double-positive neurons were excluded from further analysis, and this category represents those cells positive for CCK only.
CBs.
CBs were necessarily positive for CB, and all of these cells were always negative for PV. Although some cells coexpressed SOM and CB, these cells were included in the SOM subtype (see above). Similarly, while some cells coexpressed CCK and CB, only the CB+ and CCK− cells were included in this subtype (see above). CB cells could be either positive or negative for SATB1. Thus, this category represents putative dendrite-targeting, CB-expressing interneurons63.
Calcium imaging data preprocessing
The raw movies containing each cell were motion corrected independently using a whole-frame cross-correlation algorithm, as implemented in the SIMA software64. The time average of each imaged cell was manually inspected, and an ROI was hand drawn over each cell. Fluorescence was extracted from each ROI using the FISSA software65 package to correct for neuropil contamination using six patches of 50% the size of the original ROI. For each resulting raw fluorescence trace, a baseline F was calculated by taking the first percentile in a rolling window of 30 s, and a ΔF/F trace was calculated. The ΔF/F trace for each cell was smoothed using an exponential filter, and all further analyses were performed on the resulting ΔF/F traces. All further analyses were implemented in Python 2.7 and are detailed below.
Locomotion and immobility modulation
To calculate the correlation between each cell’s activity and the animal’s velocity, the Pearson correlation coefficient was calculated between each cell’s ΔF/F trace and the smoothed velocity trace. The velocity trace was smoothed with a Hanning filter with a five-point window length.
Run–start and run–stop responses
Run–start and run–stop events were identified in the imaging data as frames during which the animal’s velocity increased above 0.2 cm s−1 (run–start event) or decreased below 0.2 cm s−1 (run–stop event). In addition, each run–start/run–stop event had to be separated from the previous run–start/run–stop event by at least several seconds to be considered as a separate event. For each event, the mean of the pre-event ΔF/F was subtracted from the mean of the post-event ΔF/F in a −3-s to +3-s window to calculate a response magnitude. For each cell, the run–start and run–stop response magnitudes were averaged over all run–start and run–stop events in the given imaging experiment. If a cell was imaged across more than one imaging experiment, the average run–start and run–stop responses from each experiment were averaged over all experiments.
Interneuron spatial tuning curves
To calculate a spatial tuning curve for each imaged cell in a given experiment, the 2-m treadmill was divided into 100 2-cm-long bins. For each bin, we calculated the average ΔF/F from frames where the animal was in locomotion (velocity of >5 cm s−1). To determine whether a cell was spatially tuned during an imaging session, we generated 1,000 shuffled tuning curves by circularly rotating positions in relation to ΔF/F traces (restricted to frames during locomotion). A cell was determined as spatially selective if it had ten consecutive bins (20 cm) exceeding the 95th percentile of the shuffle distribution (or lower than the 5th percentile for negative fields).
Pyramidal cell spatial tuning curves
Detailed methods for determining statistically significant place cells and their place fields are described previously66. Briefly, for each pyramidal cell, calcium transients with onsets during running bouts of at least 1 s in duration were used to calculate the spatial information of the cell. Transients were randomly shuffled to different times during the running events, and the spatial information was recalculated. One thousand iterations were performed to create a null distribution for spatial information, and the cell was considered to be a place cell if its spatial information was above the 95th percentile of the null distribution. The belt was evenly divided into 100 spatial bins, and the place field was calculated from its transient rate map over these bins. The rate map was the number of transients in a given spatial bin normalized by the animal’s occupancy in that spatial bin, which was then smoothed with a Gaussian kernel (s = 3 spatial bins). To detect individual place fields, each local maximum of the smoothed rate map was fitted with a Gaussian curve centered at that location. For each smoothed rate map, the place fields where the associated Gaussian was smaller than 50% of the largest Gaussian (by measuring the total area under the curve) were discarded. The remaining Gaussians were considered place fields. The centroid of each place field was determined by the location of the peak of the Gaussian, and the location of the centroid of the largest place field was used for enrichment analysis.
Generalized linear model
To more explicitly dissociate the effects of the various behavioral variables on each cell’s activity during navigation, we developed a multivariate linear regression model to predict each cell’s fluorescence activity (ΔF/F) from the following behavioral variables: (1) the animal’s velocity, (2) position, (3) reward and (4) licking. Velocity was encoded as a continuous variable, reward and licking were encoded as binary variables corresponding to each calcium-imaging frame, and the position variable was modeled using binary ‘one-hot-encoding’ consisting of ten evenly spaced 20-cm bins. For each predictor, additional time-shifted versions corresponding to ±3 s (in two-frame steps) were also included. The model used Ridge regression to minimize the effects of potential relationships between the independent variables. Following a previously established method67, each of these four predictors was fitted independently using a cross-validated paradigm in which one lap was excluded from the training set and used for model testing. For each cell for each predictor, the cross-validated fit values were assessed as the correlation coefficient between the observed and predicted ΔF/F on the left-out lap. The significance of each predictor was assessed based on a one-way, one-sided t-test of its cross-validated fit values across laps. If for a given cell no predictors were found to significantly predict ΔF/F, then the procedure stopped at this step. If one predictor was found to be predictive, then that predictor was retained. If more than one predictor was significant, the one with the best average fit value across laps was retained. Next, new models were constructed including the retained predictor and each of the remaining predictors. The fit of these multipredictor models was compared to the fit of the single-predictor model as described above. If at least one of these joint-predictor models was significantly more predictive than the single-predictor model, the additional predictor variables were retained, and the procedure above was repeated with the remaining predictors.
Spatial information and stability analysis
Spatial information was calculated from the position bin-averaged ΔF/F vectors as described previously68. Spatial information was calculated for each cell’s observed and shuffled by-position mean ΔF/F vectors; for each cell, the observed spatial information was normalized by z scoring, by subtracting the mean and dividing by the standard deviation of that cell’s shuffled spatial information distribution. Spatial stability was calculated as the correlation coefficient across spatial bins between the by-position mean ΔF/F vectors of a given cell imaged over multiple sessions. For each cell, the mean stability value was taken either for pairs of sessions occurring on the same belt 1 h apart on the same day or for pairs of sessions occurring on the same belt but on different imaging days.
Peri-SWR time histogram
To construct the average peri-SWR time histogram for a given cell, the cell’s ΔF/F trace was z scored in a −3-s to +3-s window around each SWR event, and the resulting peri-SWR traces were averaged together across all SWR events to obtain one trace. All SWR events were considered, none were excluded. In certain figures, the resulting peri-SWR traces are shown only from −2 s to +2 s.
SWR activity index
For each cell, a baseline activity was calculated as the mean fluorescence activity 3 s to 2 s before a given SWR (−3 to −2 s from onset) from the cell’s average peri-stimulus time histogram (PSTH). Then, both negative and positive modulation values were computed by respectively subtracting the baseline from the absolute minimum and maximum activity value in the window −1 s to +1 s from onset. The largest value of the two was kept. If the modulation value was derived from the maximum, the cell was considered activated; otherwise, the cell was considered inhibited.
SWR duration
To obtain the duration of each SWR event, the detected start time of the SWR was simply subtracted from the detected end time. To calculate PSTHs for each subtype around short- and long-duration SWRs, we first calculated PSTHs around short-duration (0–20th percentile) and long-duration (80–100th percentile) SWRs for each cell. The PSTHs for all cells within a subtype were then averaged to produce the subtype averages (Fig. 5b and Extended Data Fig. 7). To compare the activity of each cell across the different percentile groups (0–20th percentile, 20–40th percentile and so on), an average peri-SWR trace was first calculated for each percentile group. The average value of this trace in the −500-ms to 0 (SWR onset) interval was taken as the average PRE. The average value of this trace in the 0 (SWR onset) to +500-ms interval was taken as the average POST. Average PRE and POST values were subsequently compared across the different SWR duration percentile groups (Fig. 5c). To calculate the difference in activity between long- and short-duration SWRs for each cell, the average short-duration trace (0–20th percentile) was first subtracted from the average long-duration trace (80–100th) to produce a difference trace. The average of this difference trace in the interval −500 ms to 0 (SWR onset) was taken as the PRE difference in activity, and the average of this difference trace in the interval 0 (SWR onset) to 500 ms was taken as the POST difference in activity (Fig. 5d and Extended Data Fig. 7).
SWR power
To obtain the amplitude of each SWR event, the broadband LFP trace between the start and end of each detected SWR event was band-pass filtered between 150 and 300 Hz, and the maximum of the absolute value of the filtered trace was taken as the amplitude. To compare a given cell’s activity across SWR events of varying power, all SWRs that occurred during the time that cell was imaged were z scored. Thus, while SWR power is highly dependent on the electrode position within CA1, all comparisons were made only between SWR events that occurred on the same day in a given mouse. Additionally, because we consider only z-scored SWR amplitudes and not absolute figures, we use the terms ‘SWR amplitude’ and ‘SWR power’ interchangeably.
Transient detection of CA3 pyramidal cells
Transient intervals were detected from the CA3PC data as intervals where the z-scored activity for a given cell exceeded 2 and stayed above 0.5 for at least 0.5 s. The first frame of this interval was taken as the transient onset and was used for analysis.
Separation of GOL data into learning days and non-learning days
Learning days were defined as imaging sessions when the difference in the fraction of anticipatory licks between the second and first halves of the session was greater than 0. The fraction of anticipatory licks in either half of the session was defined as the number of licks in the anticipatory zone, which was the 25-cm portion of the belt immediately preceding the reward zone (total belt length is 2 m) divided by the total number of licks on the whole belt. Licks that occurred after reward delivery in the reward zone were excluded from consideration. Non-learning days were defined as days when this difference in the fraction of anticipatory licks between the second and first halves of the session was either negative or 0. Dividing the dataset in this manner resulted in 22 learning days and 13 non-learning days.
SWR modulation
To calculate the SWR modulation for a given cell, the average peri-SWR time histogram was first computed, as described above. For predominantly activated subtypes (PVBCs), we defined the SWR modulation as the maximum of this average trace in the −1-s to +1-s interval around the SWR event minus the average of the pre-SWR baseline (defined as the −3-s to −1-s interval preceding the SWR). Similarly, for the predominantly inhibited subtypes (AACs, SOMs, CCKs and CBs), we defined the SWR modulation as the minimum of this average trace in the −1-s to +1-s interval around the SWR event minus the average of the pre-SWR baseline. Thus, the SWR modulation measures the extent of peri-SWR PVBC activation and the extent of peri-SWR inhibition of the other subtypes.
Cue responses
The cue response for each interneuron was calculated as the difference between the mean activity from 0 to 1 s after cue presentation and the baseline (−1 to 0 s before), regardless of cue identity.
Statistics and reproducibility
Statistical details of comparisons are specified in either the main text or figure legends. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous studies15,22,38. Mice showing poor imaging windows (low-quality viral expression or scarring), animals that did not display clear SWRs in their LFP signals and those that did not progress along the behavioral paradigms described above were excluded before commencing experiments. This study contained a single experimental group; therefore, no blinding was involved in this study. Box plots represent median and interquartile range, while whiskers extend to cover the distribution without outliers (defined as points above 1.5× the interquartile range below or above the box edges). Bar plots represent the mean, with error bars indicating the 95% confidence interval. Between-subtype comparisons were tested using a one-way analysis of variance (ANOVA) followed by a Tukey’s range test with correction for multiple testing if appropriate. For comparisons between two populations, a paired sample or unpaired t-test was applied if the data points followed a normal distribution. In the instances where parametric tests were used, distributions were assumed to be normal, but this was not formally tested. To analyze data that were not normally distributed, a Mann–Whitney U-test was used. All tests were two sided unless otherwise indicated in the figure legends; *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data analysis was performed and figures were generated using custom-made software in Python 2.7.15.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 |. Immunohistochemical identification of interneuron subtypes and separation of CA2 and CA3 interneurons with anti-STEP immunohistochemistry.
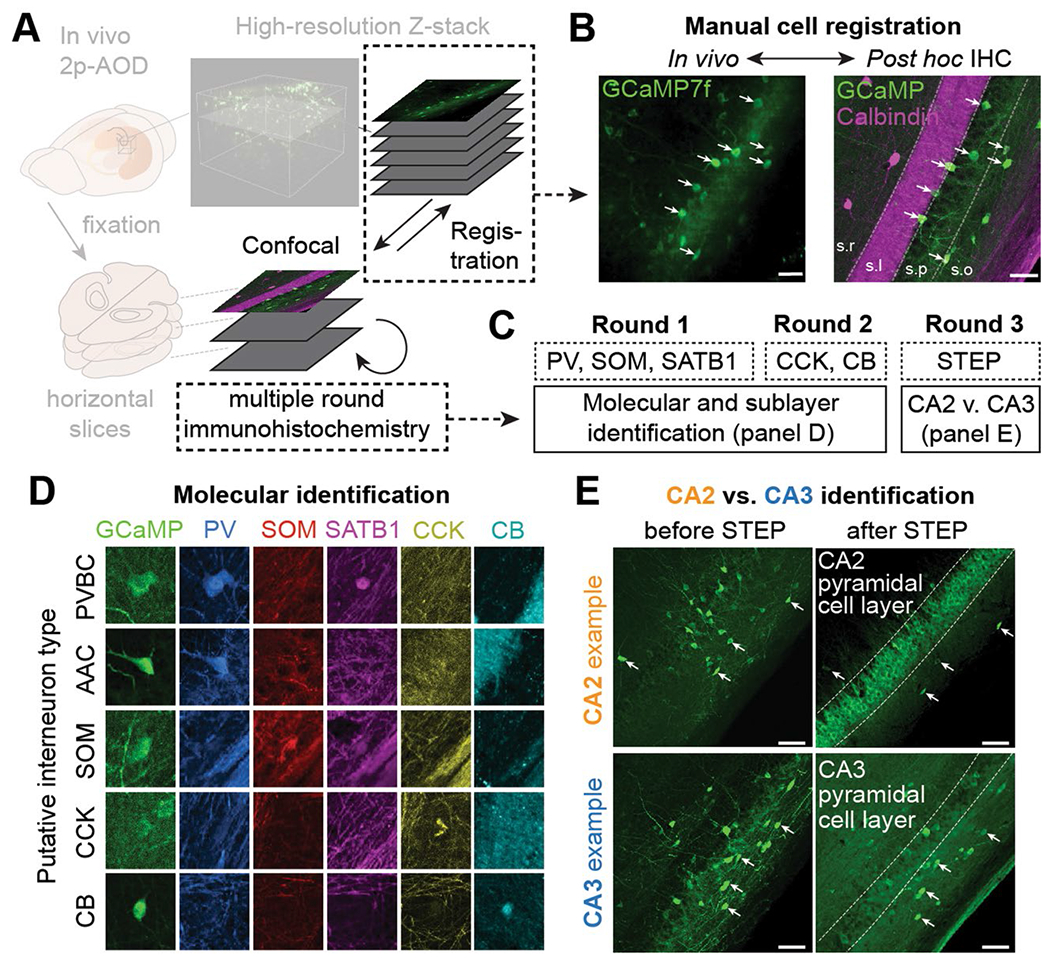
(a) Schematic of the experimental pipeline used to determine the molecular identity of imaged cells. Multiple rounds of immunohistochemistry were performed on fixed, horizontal slices that were registered to high-resolution in vivo Z-stacks. (b) Example in vivo 2p-AOD image (left) and confocal image (right) of the registered FOV. White arrows indicate the registered cells. Calbindin immunohistochemistry was used to label the mossy fibers of stratum lucidum of CA3/CA2. This procedure was repeated in n = 22 imaged mice. Scale bars on the left and right images represent 50 and 100 μm, respectively. (c) Example labeling strategy used to determine the subtype and region identity of imaged cells. Immunohistochemical labels were not removed between the different rounds (see Methods). (d) Example immunohistochemical labeling and combinatorial expression patterns of the 5 markers (PV, SOM, SATB1, CCK, CB) used to separate imaged cells into subtypes. This procedure was repeated in n = 22 imaged mice. All images are approximately 60 × 60 μm. (e) CA2 interneurons were identified by their proximity to STEP-expressing CA2 pyramidal cells (top row). In comparison, CA3 interneurons occupied slices where Calbindin-positive mossy fibers were present but where the majority of pyramidal cells were not STEP-expressing (bottom row). This procedure was repeated in n = 22 imaged mice. Scale bars on all four images represent approximately 100 μm.
Extended Data Fig. 2 |. Molecular profiling of calbindin-positive SATB1-negative immobility-active interneurons.
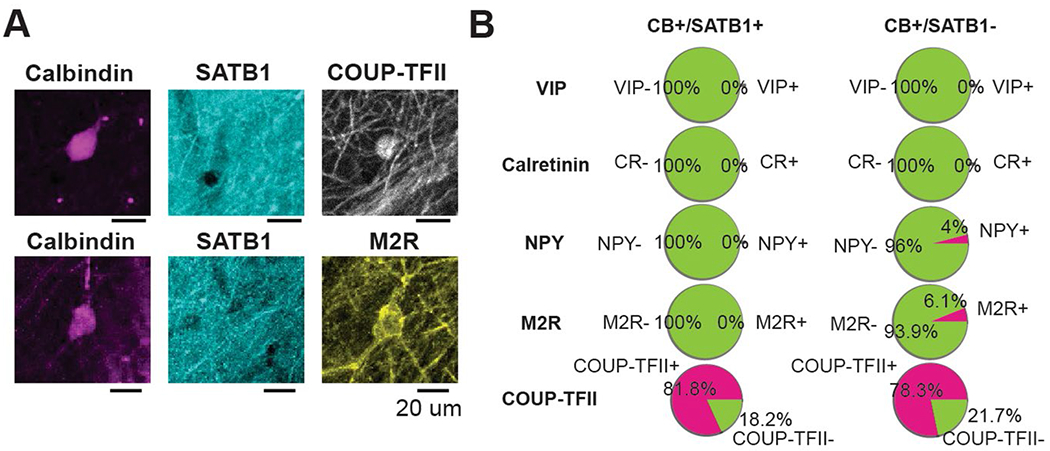
(a) Confocal micrograph of CB-expressing interneurons, negative for SATB1 but positive for COUP-TFII (top) and M2R (bottom). This staining was repeated in n = 2 mice. Scale bars represent 20 μm. (b) Quantification of the overlap of CB-expressing interneurons split by immunoreactivity to SATB1 with other markers.
Extended Data Fig. 3 |. Additional data on interneuron spatial selectivity and generalized-linear model of interneuron activity during spatial navigation.
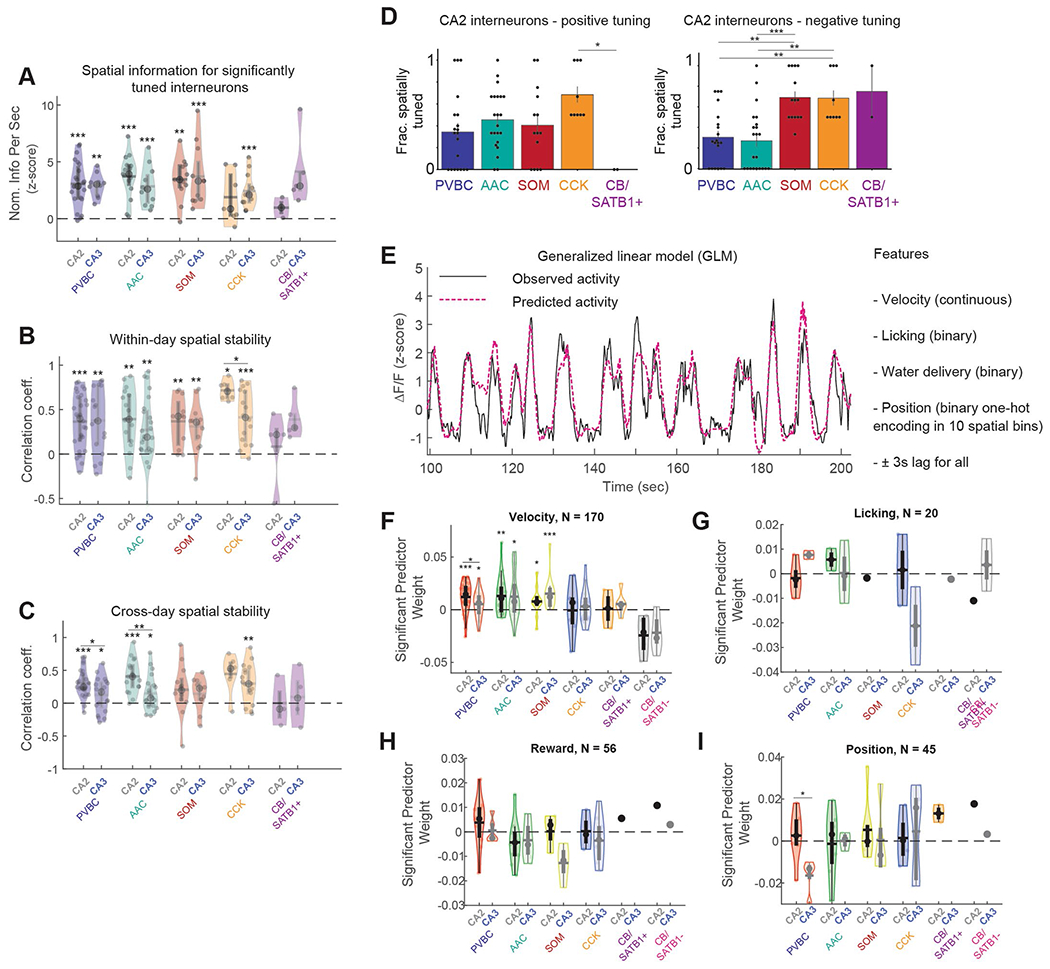
(a) Spatial information for significantly tuned Interneurons, broken down by both region and subtype. Immobility-active CB/SATB1− neurons were silenced during locomotion and were thus not considered in this analysis. Data from 188 cells in n = 9 mice. Significance values over individual violin plots show the results of (one-way) signed-rank tests. (b) Within-day spatial stability of all interneurons, broken down by both region and subtype. Plotted as in A. Significance values over individual violin plots show the results of (one-way) signed-rank tests. Significance values over pairs of violin plots show results from (two-way) ranked-sum tests (only significant differences are shown). Data from 152 cells in n = 9 mice. (c) Across-day spatial stability of all interneurons, broken down by both region and subtype. Plotted as in A. Significance values over individual violin plots show the results of (one-way) signed-rank tests. Significance values over pairs of violin plots show results from (two-way) ranked-sum tests (only significant differences are shown). Data from 142 cells in n = 9 mice. (d) Left: Summary of the fraction of positively tuned CA2 interneurons, broken down by subtype (PVBC: 0.343 ± 0.341, AAC: 0.455 ± 0.267, SOM: 0.405 ± 0.339, CCK: 0.685 ± 0.228, CB/SATB1 + : 0.0 ± 0.0). CA2 CCK cells were more likely to be positively spatially tuned cells than CA2 CB/SATB1+ cells (one-way ANOVA with post-hoc multiple testing correction, p = 0.048). Immobility-active CB/SATB1− neurons were silenced during locomotion and were thus not considered in this analysis. Each data point represents an imaging session. PVBC data from 20 imaging sessions, AAC data from 22 sessions, SOM data from 14 sessions, CCK data from 9 sessions, and CB/SATB1 + data from 2 sessions; data from n = 9 mice. Data reported as mean ± s.d. Right: Same data as on the left, but for negatively tuned CA2 interneurons (PVBC: 0.304 ± 0.261, AAC: 0.270 ± 0.293, SOM: 0.690 ± 0.226, CCK: 0.685 ± 0.228, CB/SATB1 + : 0.75 ± 0.25). Significant differences in the fraction of negatively tuned CA2 interneurons by subtype are indicated (one-way ANOVA with post-hoc multiple testing correction: p(CCK-PVBC) = 0.0075, p(CCK-AAC) = 0.0024, p(SOM-PVBC) = 0.0012, p(SOM-AAC) = 0.001). Immobility-active CB/SATB1− neurons were silenced during locomotion and were thus not considered in this analysis. Each data point represents an imaging session. PVBC data from 20 imaging sessions, AAC data from 22 sessions, SOM data from 14 sessions, CCK data from 9 sessions, and CB/SATB1 + data from 2 sessions; data from n = 9 mice. Data reported as mean ± s.d. (e) Example of 100 seconds of real interneuron activity during locomotion and the predicted activity from a GLM. The predicted activity for each cell was calculated based on 4 predictor behavioral variables: velocity, position, licking, and water delivery (see Methods). (f) Comparison of the GLM weights for each cell for the velocity predictor, separated by both subtype and region. Only cells for which the velocity predictor in the model was a significant predictor are shown. Significance values over individual violin plots show the results of (oneway) signed-rank tests. Significance values over pairs of violin plots show results from (two-way) ranked-sum tests. Only significant differences are shown. Data from 170 cells from n = 9 mice. (g) Same data as shown in F, but now for the licking predictor. Only cells for which the licking predictor in the model was significant are shown. No significance at the subtype or region level was found. Data from 20 cells in n = 9 mice. (h) Same data as shown in F, but now for the reward predictor. Only cells for which the reward predictor in the model was significant are shown. No significance at the subtype or region level was found. Data from 56 cells from n = 9 mice. (i) Same data as shown in F, now for the position predictor. Only cells for which the position predictor in the model was significant are shown. Significance values over pairs of violin plots show results from (two-way) ranked-sum tests. Only significant differences are shown. Data from 45 cells from n = 9 mice.
Extended Data Fig. 4 |. Immunohistochemical verification of Grik4-Cre transgenic line.
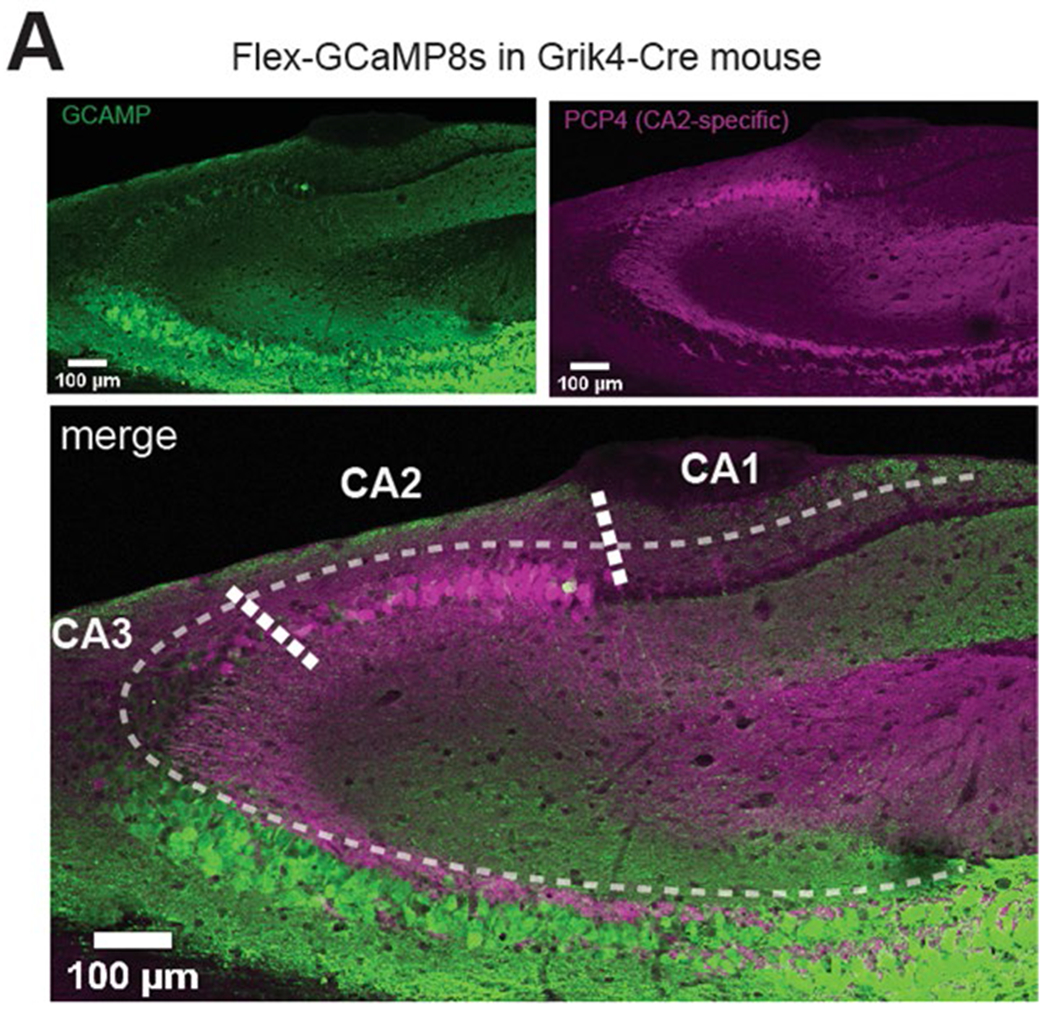
(a) Top left: Confocal image of FLEX-GCaMP8s expression when injected into CA3/CA2 of the Grik4-Cre transgenic line. Top right: CA2 pyramidal cells identified by their PCP4 immunosignal. Bottom: Merge of the above images. Note that GCaMP expression is largely confined to CA3 in the Grik4-Cre line, although some GCaMP8s-positive pyramidal cells in CA2 can be seen as well. This staining was repeated in n = 3 mice.
Extended Data Fig. 5 |. CA3 pyramidal cell dynamics around SWRs and correlations between SWR properties.
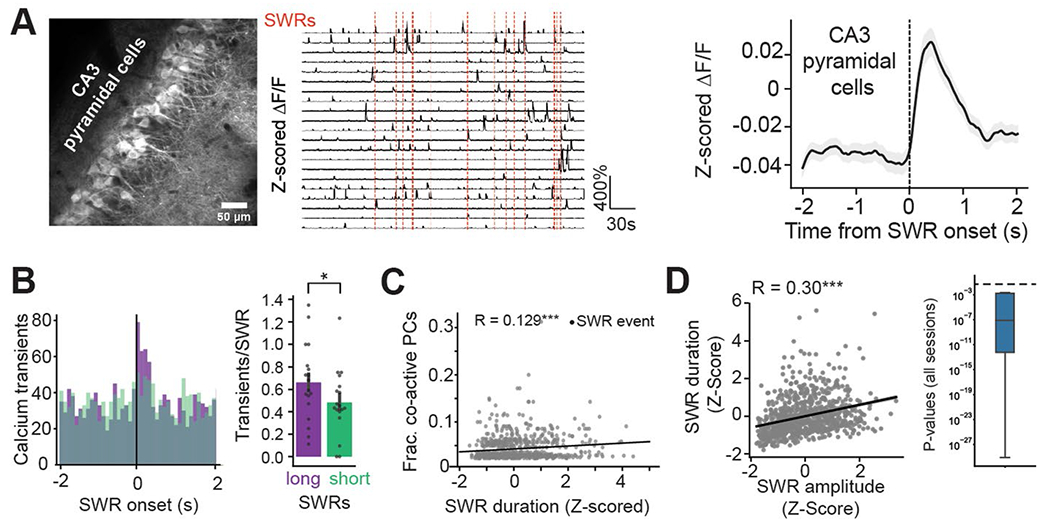
(a) Left: Representative in vivo two-photon time-average image of a CA3PC FOV. Center: Example CA3PC ΔF/F traces with detected SWRs depicted as vertical red lines. Right: Peri-SWR fluorescence for the entire CA3PC population, averaged over all SWR events. Data from n = 3 mice. Trace represents mean ± s.e.m. (b) Left: Distribution of peri-SWR CA3PC calcium transients for SWRs with short duration (green, taken as SWRs with duration falling between 0–20th percentile of all SWRs for a given mouse) and long duration (purple, for SWRs falling between the 80–100th duration percentile). Right: Quantification of the population transient rate for long- and short-duration SWRs. CA3PCs emitted significantly more transients during long-duration SWRs than during short-duration SWRs (Short duration SWRs: 0.483 ± 0.285 transients/SWR, Long duration SWRs: 0.661 ± 0.331 transients/SWR, two-sided Wilcoxon signed-rank test: p = 0.011). Data from 18 sessions from n = 3 mice. Data reported as mean ± s.d. (c) Correlation between SWR duration and the number of co-active pyramidal cells around the SWR. Each dot represents a SWR event. Long-duration SWRs were associated with greater fractions of co-active CA3PCs around the SWR event (linear regression, r = 0.129, p = 2.50 × 10–4). Only SWRs associated with at least 1 transient in the CA3PC FOV are considered. Data from n = 3 mice. (d) Correlation between amplitude and duration for individual SWRs. Left: Example scatter plot and linear regression line depicting the relationship between amplitude and duration for all SWRs recorded during the imaging of one example interneuron. Right: Distribution of p-values for the two-sided regression between amplitude and duration, calculated over all imaging sessions. The horizontal dashed line corresponds to a p-value of 0.05. A strong relationship between SWR amplitude and duration was present in all imaging sessions. Data from n = 13 mice. * p < 0.05, ** p <0.01, *** p < 0.001.
Extended Data Fig. 6 |. Additional data on CA3 and CA2 interneuron dynamics around SWRs.
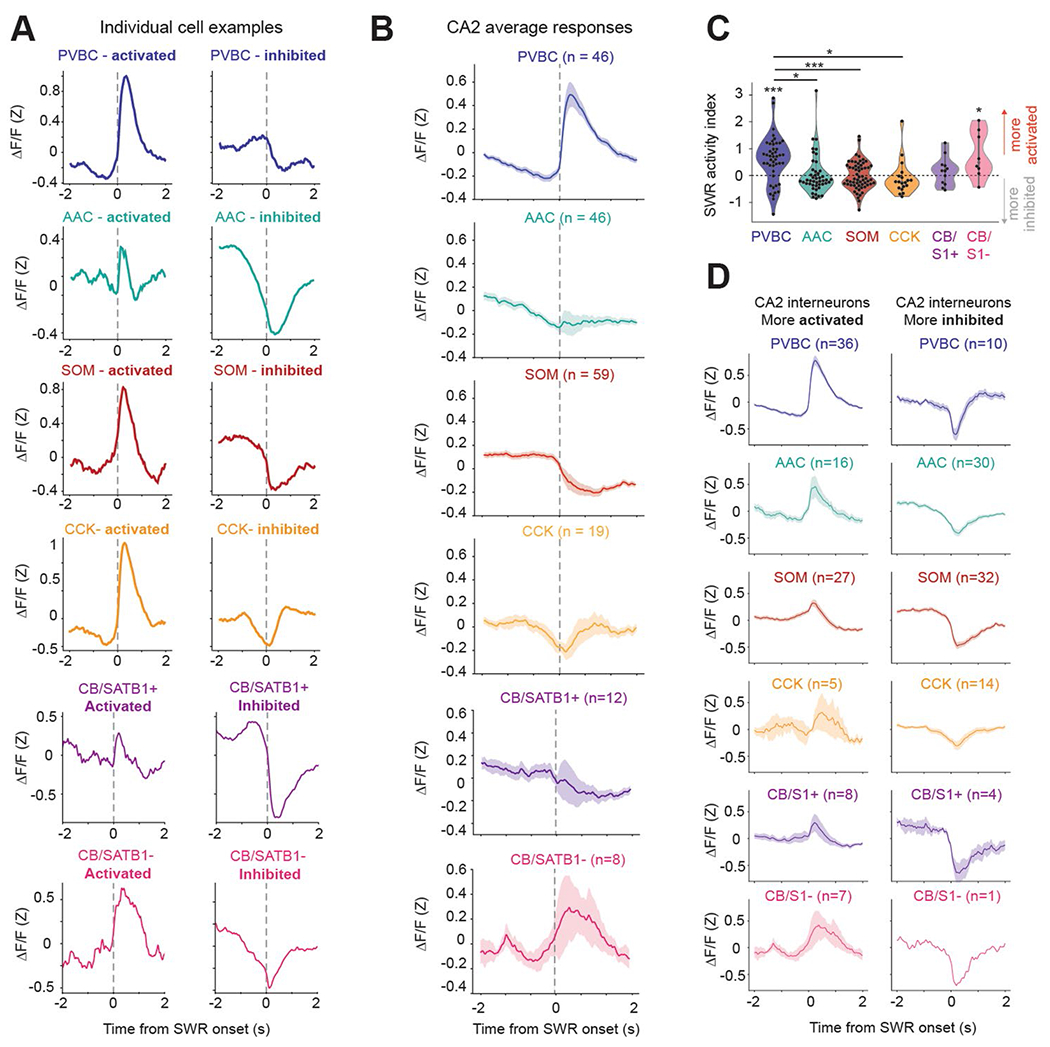
(a) Example Z-scored peri-SWR traces for both activated and inhibited cells of each subtype. (b) Average peri-SWR traces for all CA2 interneuron subtypes (n = 46 PVBCs, 46 AACs, 59 SOMs, 19 CCKs, 12 CB/SATB1 + neurons, and 8 CB/SATB1− neurons from n = 13 mice). Traces for each subtype represent mean ± s.e.m. (c) Average SWR activity index for all CA2 interneurons, grouped by subtype. Wilcoxon signed-rank tests against a median of 0 were performed for each subtype (PVBC: 0.56 ± 0.83, p = 1 × 10–5, AAC:0.03 ± 0.65, p = 0.18; SOM: −0.002 ± 0.5, p = 0.96; CCK: −0.08 ± 0.61, p = 0.12; CB/SATB1 + : 0.17 ± 0.5, p = 0.43; CB/SATB1−: 0.77 ± 0.82, p = 0.04). Between-subtype statistical comparisons were performed using the Kruskal-Wallis test (p = 9 × 10–5) with post-hoc Wilcoxon rank sum tests with p-values adjusted using the Bonferroni correction (significant adjusted p-values: PVBC-AAC = 0.038, PVBC-CCK = 0.016, PVBC-SOM = 0.0004). Data from n = 46 PVBCs, 46 AACs, 59 SOMs, 19 CCKs, 12 CB/SATB1 +, and 8 CB/SATB1− from n = 13 mice. Data reported as mean ± s.d. (d) Left: Average peri-SWR traces for all activated CA2 interneurons, grouped by subtype. Right: Same traces for all inhibited CA2 interneurons, grouped by subtype. Cell numbers for each subtype and condition indicated on the figure, data from n = 13 mice. Traces for each subtype represent mean ± s.e.m. * p < 0.05, ** p < 0.01, *** p < 0.001.
Extended Data Fig. 7 |. Dynamics of CA3 CB subtypes and all CA2 interneuron subtypes around short- and long-duration SWRs.
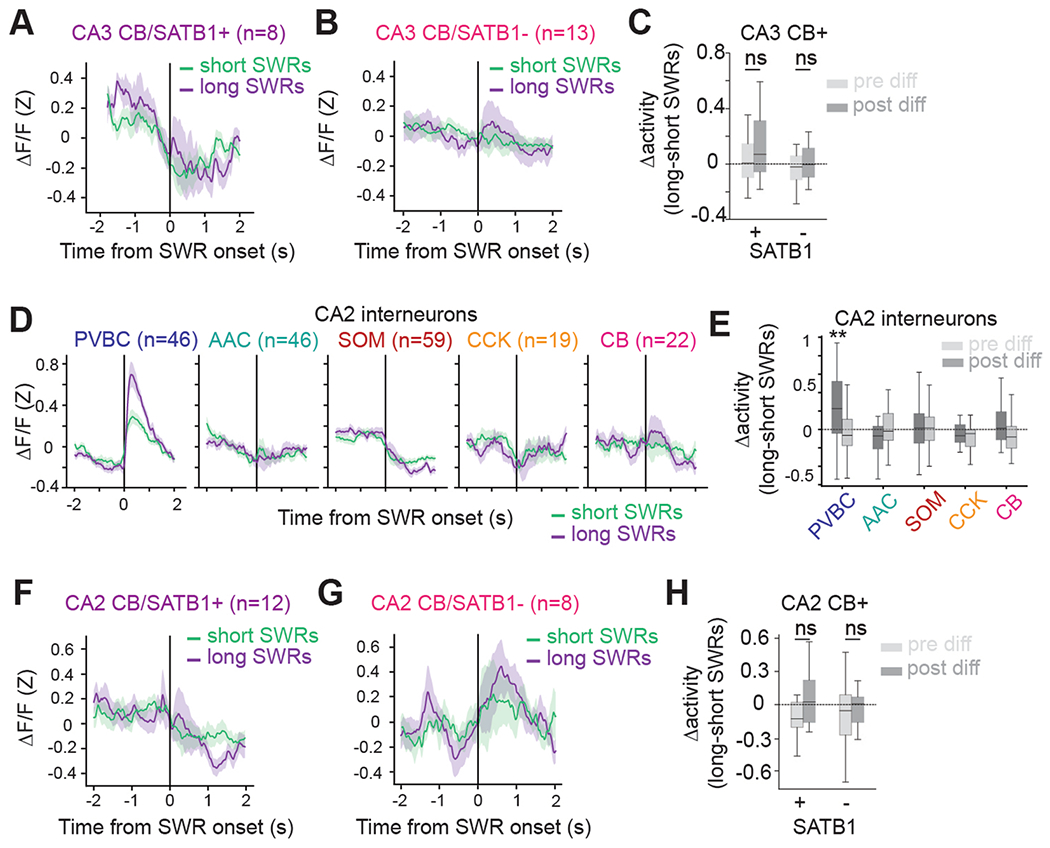
(a) Average Z-scored peri-SWR traces for both short- (0–20th percentile) and long- (80–100th percentile) duration SWRs for CA3 CB/SATB1 + interneurons (n = 8 CB/SATB1+ cells from n = 13 mice). Traces for each condition represent mean ± s.e.m. (b) Average Z-scored peri-SWR traces for both short- (0–20th percentile) and long- (80–100th percentile) duration SWRs for CA3 CB/SATB1− interneurons (n = 13 CB/SATB1− cells from n = 13 mice). Traces for each condition represent mean ± s.e.m. (c) Average value of the difference in activity between long- and short-duration SWRs for CA3 CB+/SATB1+ and CB+/SATB1− subtypes, considered separately for the pre-SWR and post-SWR activity. Neither CB+/SATB1+ nor CB+/SATB1− interneurons responded differently during long-duration compared to short-duration SWRs (two-way, one-sample t-tests against 0 for each subtype and condition: CB+/SATB1+ PRE: 0.028 ± 0.176, p = 0.69; CB+/SATB1+ POST: 0.124 ± 0.251, p = 0.23; CB+/SATB1−PRE: −0.047 ± 0.150, p = 0.30; CB+/SATB1−POST: 0.056 ± 0.239, p = 0.43). Data from 8 CB+/SATB1+ and 13 CB+/SATB1− cells from n = 13 mice. Data reported as mean ± s.d. (d) Average Z-scored peri-SWR traces for both short- (0–20th percentile) and long- (80–100th percentile) duration SWRs for CA2 interneurons of each subtype (n = 46 PVBCs, 46 AACs, 59 SOMs, 19 CCKs, and 22 CBs from n = 13 mice). CB+/SATB1+ and CB+/SATB1− neurons are considered together in the CB subtype. Traces for each subtype and condition represent mean ± s.e.m. (e) Average value of the difference in activity between long- and short-duration SWRs for each CA2 interneuron, considered separately for the pre-SWR and post-SWR activity, and grouped by subtype. PVBCs were significantly more activated after long-duration SWRs, while the other subtypes did not show different dynamics during short-compared to long-duration SWRs. Only significant differences are indicated (two-way, one-sample t-tests against 0 for each subtype with Bonferroni correction for multiple testing: PVBC PRE: −0.044 ± 0.264, p = 1.0; PVBC POST: 0.267 ± 0.464, p = 0.0037; AAC PRE: 0.024 ± 0.250, p = 1.0; AAC POST: −0.053 ± 0.322, p = 1.0; SOM PRE: 0.054 ± 0.389, p = 1.0; SOM POST: 0.042 ± 0.340, p = 1.0; CCK PRE: −0.116 ± 0.252, p = 0.70; CCK POST: −0.036 ± 0.360, p = 1.0; CB PRE: −0.050 ± 0.351, p = 1.0; CB POST: 0.103 ± 0.328, p = 1.0). Data from n = 46 PVBCs, 46 AACs, 59 SOMs, 19 CCKs, and 22 CBs from n = 13 mice. CB+/SATB1+ and CB+/SATB1− neurons are considered together in the CB subtype. Data reported as mean ± s.d. (f) Average Z-scored peri-SWR traces for both short- (0–20th percentile) and long- (80–100th percentile) duration SWRs for CA2 CB+/SATB1+ interneurons (n = 12 CB+/SATB1+ cells from n = 13 mice). Traces for each condition represent mean ± s.e.m. (g) Average Z-scored peri-SWR traces for both short- (0–20th percentile) and long- (80–100th percentile) duration SWRs for CA2 CB/SATB1− interneurons (n = 8 CB+/SATB1− cells from n = 13 mice). Traces for each condition represent mean ± s.e.m. (h) Average value of the difference in activity between long- and short-duration SWRs for CA2 CB+/SATB1+ and CB+/SATB1− subtypes, considered separately for the pre-SWR and post-SWR activity. Neither CB+/SATB1+ nor CB+/SATB1− interneurons responded differently during long-duration compared to short-duration SWRs (two-way, one-sample t-tests against 0 for each subtype and condition: CB+/SATB1+ PRE: −0.034 ± 0.373, p = 0.77; CB+/SATB1+ POST: 0.068 ± 0.263, p = 0.41; CB+/SATB1− PRE: −0.078 ± 0.333, p = 0.56; CB+/SATB1− POST: 0.058 ± 0.374, p = 0.69). Data from 12 CB+/SATB1+ and 8 CB+/SATB1− cells from n = 13 mice. Data reported as mean ± s.d. * p < 0.05, ** p < 0.01, *** p < 0.001.
Extended Data Fig. 8 |. Additional data on learning-related CA3/CA2 interneuron dynamics during the GOL task.
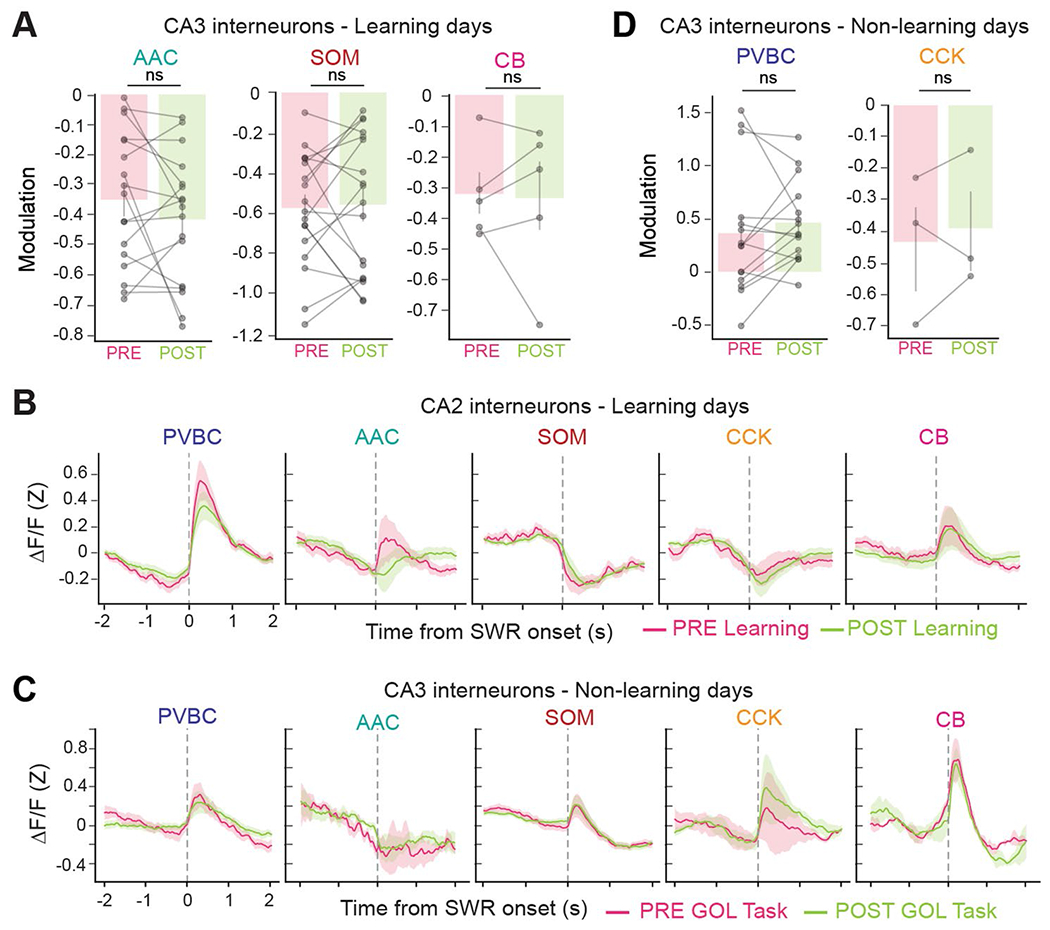
(a) Quantification of the peri-SWR modulation for CA3 AACs, SOMs, and CBs during PRE and POST sessions on Learning Days. None of these three subtypes significantly changed their activity around SWRs after learning (AAC PRE: −0.350 ± 0.217, AAC POST: −0.416 ± 0.211, two-sided Wilcoxon signed-rank test: p = 0.33; SOM PRE: −0.571 ± 0.275, SOM POST: −0.555 ± 0.334, two-sided Wilcoxon signed-rank test: p = 0.68; CB PRE: −0.322 ± 0.135, CB POST: −0.335 ± 0.227, two-sided Wilcoxon signed-rank test: p = 0.69). Data from n = 17 AACs, 18 SOMs, and 5 CBs from n = 8 mice. CB+/SATB1+ and CB/SATB1− neurons are considered together in the CB subtype. Data reported as mean ± s.d. (b) Average Z-scored peri-SWR traces for both PRE and POST sessions for all CA2 subtypes on Learning Days. Data from 27 PRE and 26 POST PVBCs, 21 PRE and 19 POST AACs, 27 PRE and 27 POST SOMs, 14 PRE and 14 POST CCKs, and 18 PRE and 16 POST CBs from n = 8 mice. CB/SATB1+ and CB/SATB1− neurons are considered together in the CB subtype. Traces for each subtype and condition represent mean ± s.e.m. (c) Average Z-scored peri-SWR traces for both PRE and POST sessions for all CA3 subtypes on Non-Learning Days. Data from 15 PRE and 16 POST PVBCs, 4 PRE and 4 POST AACs, 26 PRE and 25 POST SOMs, 4 PRE and 4 POST CCKs, and 2 PRE and 2 POST CBs from n = 8 mice. CB/SATB1+ and CB/SATB1− neurons are considered together in the CB subtype. Traces for each subtype and condition represent mean ± s.e.m. (d) Quantification of the change in peri-SWR modulation for CA3 PVBCs and CCKs between PRE and POST sessions on Non-Learning Days. Neither subtype changed its activity significantly after the GOL task (PVBC PRE: 0.360 ± 0.581, PVBC POST: 0.460 ± 0.370, two-sided Wilcoxon signed-rank test: p = 0.39; CCK PRE: −0.433 ± 0.195, CCK POST: −0.390 ± 0.177, two-sided Wilcoxon signed-rank test: p = 0.59; data from n = 15 PVBCs and 3 CCKs from n = 8 mice). Data reported as mean ± s.d. * p < 0.05, ** p <0.01, *** p < 0.001.
Extended Data Fig. 9 |. CA3 interneuron dynamics during the random cue task.
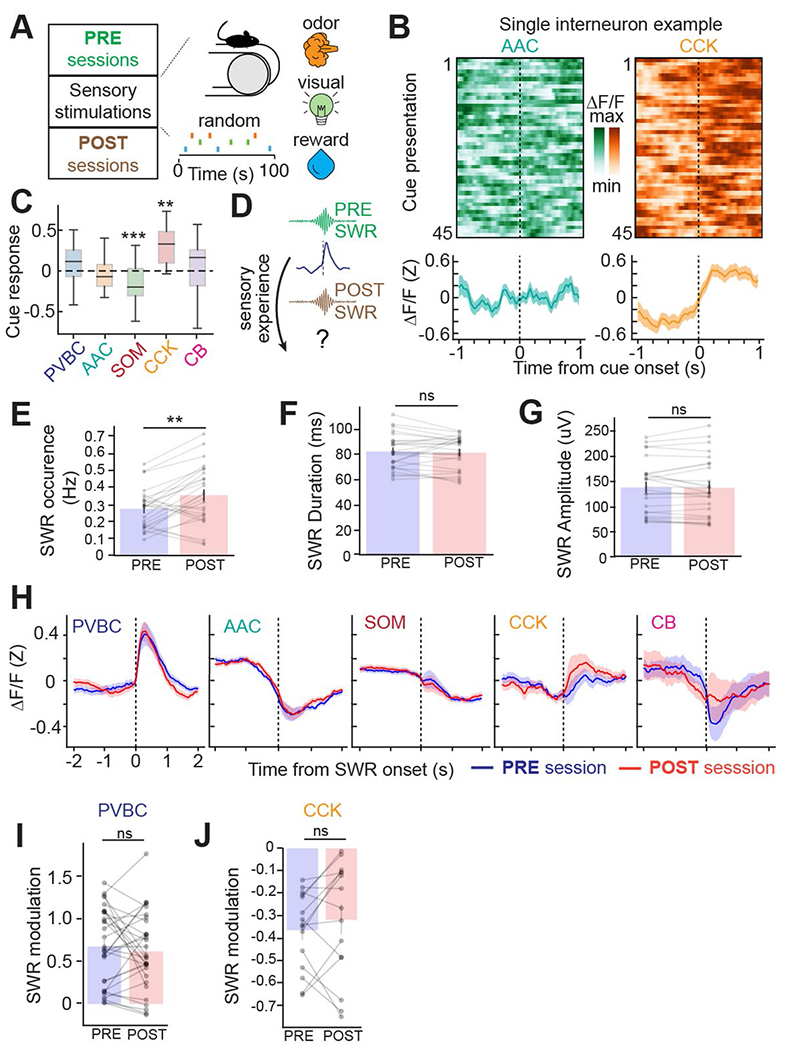
(a) Sensory stimulation paradigm. Water, light, and odor stimuli were presented pseudorandomly while the mouse remained head-fixed on a cue-less, burlap belt. Interneurons were imaged during SWRs in both the PRE and POST sessions as well as during stimulus presentations. (b) Representative example of an individual AAC (green) and CCK (orange) interneuron. Heatmaps represent the activity during all sensory stimulus presentations (45 in total) with the corresponding average response (bottom). The CCK neuron is consistently and significantly activated by cue presentations. The traces are plotted as the mean ± s.e.m. (c) Average sensory cue response for each cell, grouped by subtype (PVBC response: 0.078 ± 0.231, AAC: −0.032 ± 0.208, SOM: −0.145 ± 0.215, CCK: 0.328 ± 0.252, CB: 0.038 ± 0.349). CCK cells were significantly activated by cue presentation, while SOM cells were significantly inhibited (two-way, one-sample t-tests against 0 for each subtype: PVBC: p = 0.12; AAC: p = 0.41; SOM: p = 4.17 × 10–4; CCK: p = 0.0021; CB: p = 0.73). Data from n = 24 PVBCs, 31 AACs, 35 SOMs, 11 CCKs, and 12 CBs from n = 3 mice. CB+/SATB1+ and CB+/SATB1− neurons are considered together in the CB subtype. All reported cells are from CA3; CA2 data not reported. Data reported as mean ± s.d. (d) Sessions PRE and POST cue presentations are split to examine whether sensory cue presentations induced a change in interneuron dynamics around SWRs. (e) Quantification of the average SWR occurrence rate for PRE and POST sessions. The occurrence rate increased significantly from PRE to POST (occurrence PRE: 0.273 ± 0.115 s-1, occurrence POST: 0.354 ± 0.169 s-1, two-sided Wilcoxon signed-rank test: p = 0.0024). Data from n = 26 PRE sessions and 26 POST sessions from n = 5 mice. Data reported as mean ± s.d. (f) Quantification of the average SWR duration for PRE and POST sessions. The SWR duration did not change between PRE and POST sessions (duration PRE: 82.7 ± 13.6 ms, POST: 81.8 ± 13.0 ms, two-sided Wilcoxon signed-rank test: p = 0.28). Data from n = 26 PRE sessions and 26 POST sessions from n = 5 mice. Data reported as mean ± s.d. (g) Quantification of the average SWR maximum amplitude for PRE and POST sessions. The amplitude did not change between PRE and POST sessions (amplitude PRE: 137.3 ± 52.3 μV, POST: 136.6 ± 57.3 μV, two-sided Wilcoxon signed-rank test: p = 0.66). Data from n = 26 PRE sessions and 26 POST sessions from n = 5 mice. Data reported as mean ± s.d. (h) Average Z-scored peri-SWR traces for both PRE and POST sessions for all subtypes. Data from 31 PRE and 32 POST PVBCs, 37 PRE and 34 POST AACs, 49 PRE and 51 POST SOMs, 14 PRE and 15 POST CCKs, and 7 PRE and 7 POST CBs from n = 5 mice. CB+/SATB1+ and CB+/SATB1− neurons are considered together in the CB subtype. All reported cells are from CA3; CA2 data are not reported. Traces for each subtype and condition represent mean ± s.e.m. (i) Quantification of the change in SWR modulation for CA3 PVBCs between PRE and POST sessions. PVBCs did not become more activated around SWRs in response to the sensory stimulation task (PVBC PRE: 0.67 ± 0.44, PVBC POST: 0.62 ± 0.44, two-sided Wilcoxon signed-rank test: p = 0.64; data from n = 31 PVBCs from n = 5 mice). Data reported as mean ± s.d. (j) Quantification of the change in SWR modulation for CA3 CCKs between PRE and POST sessions. CCKs did not become more inhibited around SWRs in response to the sensory stimulation task (CCK PRE: −0.36 ± 0.17, CCK POST: −0.32 ± 0.25, two-sided Wilcoxon signed-rank test: p = 0.23). Data from n = 15 CCKs from n = 5 mice. Data reported as mean ± s.d. * p < 0.05, ** p <0.01, ***p < 0.001.
Acknowledgements
B.V. is supported by T32GM007367 (NIH) and F30MH125628 (NIMH). T.G. is supported by K99MH129565 (NIMH). A.L. is supported by 1R01MH124047 and 1R01MH124867 (NIMH), 1U19NS104590 and 1U01NS115530 (NINDS) and the Kavli Foundation. We thank members of the Losonczy lab for their invaluable comments on previous versions of the paper.
Footnotes
Code availability
The code used for this study is available at https://github.com/losonczylab/Vancura_Geiller_NatNeuro_2023.
Competing interests
The authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s41593-023-01306-7.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41593-023-01306-7.
Data availability
The data analyzed for this study are available at https://github.com/losonczylab/Vancura_Geiller_NatNeuro_2023. Source data are provided with this paper.
References
- 1.Tulving E Episodic memory: from mind to brain. Annu. Rev. Psychol 53, 1–25 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Buzsáki G Hippocampal sharp wave-ripple: a cognitive biomarker for episodic memory and planning. Hippocampus 25, 1073–1188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daumas S, Halley H, Francés B & Lassalle J-M Encoding, consolidation, and retrieval of contextual memory: differential involvement of dorsal CA3 and CA1 hippocampal subregions. Learn. Mem 12, 375–382 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kesner RP Behavioral functions of the CA3 subregion of the hippocampus. Learn. Mem 14, 771–781 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Nakashiba T, Buhl DL, McHugh TJ & Tonegawa S Hippocampal CA3 output is crucial for ripple-associated reactivation and consolidation of memory. Neuron 62, 781–787 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dudek SM, Alexander GM & Farris S Rediscovering area CA2: unique properties and functions. Nat. Rev. Neurosci 17, 89–102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexander GM et al. Social and novel contexts modify hippocampal CA2 representations of space. Nat. Commun 7, 10300 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliva A, Fernández-Ruiz A, Buzsáki G & Berényi A Role of hippocampal CA2 region in triggering sharp-wave ripples. Neuron 91, 1342–1355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mishra RK, Kim S, Guzman SJ & Jonas P Symmetric spike timing-dependent plasticity at CA3–CA3 synapses optimizes storage and recall in autoassociative networks. Nat. Commun 7, 11552 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rebola N, Carta M & Mulle C Operation and plasticity of hippocampal CA3 circuits: implications for memory encoding. Nat. Rev. Neurosci 18, 208–220 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Norimoto H et al. Hippocampal ripples down-regulate synapses. Science 359, 1524–1527 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Pfeiffer BE The content of hippocampal ‘replay’. Hippocampus 30, 6–18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunt DL, Linaro D, Si B, Romani S & Spruston N A novel pyramidal cell type promotes sharp-wave synchronization in the hippocampus. Nat. Neurosci 21, 985–995 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Kay K et al. A hippocampal network for spatial coding during immobility and sleep. Nature 531, 185–190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiller T et al. Local circuit amplification of spatial selectivity in the hippocampus. Nature 601, 105–109 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadeh S & Clopath C Inhibitory stabilization and cortical computation. Nat. Rev. Neurosci 22, 21–37 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Vogels TP, Sprekeler H, Zenke F, Clopath C & Gerstner W Inhibitory plasticity balances excitation and inhibition in sensory pathways and memory networks. Science 334, 1569–1573 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Arriaga M & Han EB Structured inhibitory activity dynamics in new virtual environments. eLife 8, e47611 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Booker SA & Vida I Morphological diversity and connectivity of hippocampal interneurons. Cell Tissue Res. 373, 619–641 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudok B et al. Recruitment and inhibitory action of hippocampal axo-axonic cells during behavior. Neuron 109, 3838–3850 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geiller T, Royer S, & Choi J-S. Segregated cell populations enable distinct parallel encoding within the radial axis of the CA1 pyramidal layer. Exp. Neurobiol 26, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geiller T et al. Large-scale 3D two-photon imaging of molecularly identified CA1 interneuron dynamics in behaving mice. Neuron 108, 968–983 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelkey KA et al. Hippocampal GABAergic inhibitory interneurons. Physiol. Rev 97, 1619–1747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dupret D, O’Neill J & Csicsvari J Dynamic reconfiguration of hippocampal interneuron circuits during spatial learning. Neuron 78, 166–180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaufman AM, Geiller T & Losonczy A A role for the locus coeruleus in hippocampal CA1 place cell reorganization during spatial reward learning. Neuron 105, 1018–1026 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Topolnik L, & Tamboli S, The role of inhibitory circuits in hippocampal memory processing. Nat. Rev. Neurosci 23, 476–492 (2022). [DOI] [PubMed] [Google Scholar]
- 27.Chamberland S, & Topolnik L, Inhibitory control of hippocampal inhibitory neurons. Front. Neurosci 6, 165 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klausberger T & Somogyi P Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science 321, 53–57 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donato F, Rompani SB, & Caroni P Parvalbumin-expressing basket-cell network plasticity induced by experience regulates adult learning. Nature 504, 272–276 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Ruediger S et al. Learning-related feedforward inhibitory connectivity growth required for memory precision. Nature 473, 514–518 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Sharma V, et al. eIF2a controls memory consolidation via excitatory and somatostatin neurons. Nature 586, 412–416 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geiller T, Fattahi M, Choi JS, & Royer S Place cells are more strongly tied to landmarks in deep than in superficial CA1. Nat. Commun 8, 14531 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hitti FL & Siegelbaum SA The hippocampal CA2 region is essential for social memory. Nature 508, 88–92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arriaga M, & Han EB, Dedicated hippocampal inhibitory networks for locomotion and immobility. J. Neurosci 37, 9222–9238 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dudok B et al. Alternating sources of perisomatic inhibition during behavior. Neuron 109, 997–1012 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Keefe J & Dostrovsky J The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 34, 171–175 (1971). [DOI] [PubMed] [Google Scholar]
- 37.Ego-Stengel V, & Wilson MA, Spatial selectivity and theta phase precession in CA1 interneurons. Hippocampus 17, 161–174 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Grosmark AD, Sparks FT, Davis MJ, & Losonczy A, Reactivation predicts the consolidation of unbiased long-term cognitive maps. Nat. Neurosci 24, 1574–1585 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Malvache A, Reichinnek S, Villette V, Haimerl C, & Cossart R, Awake hippocampal reactivations project onto orthogonal neuronal assemblies. Science 353, 1280–1283 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Terada S et al. Adaptive stimulus selection for consolidation in the hippocampus. Nature 601, 240–244 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jinno S et al. Neuronal diversity in GABAergic long-range projections from the hippocampus. J. Neurosci 27, 8790–8804 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katona L, et al. Sleep and movement differentiates actions of two types of somatostatin-expressing GABAergic interneuron in rat hippocampus. Neuron 82, 872–886 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lasztóczi B, Tukker JJ, Somogyi P, & Klausberger T, Terminal field and firing selectivity of cholecystokinin-expressing interneurons in the hippocampal CA3 area. J. Neurosci 31, 18073–18093 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernández-Ruiz A et al. Long-duration hippocampal sharp wave ripples improve memory. Science 364, 1082–1086 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grosmark AD & Buzsáki G Diversity in neural firing dynamics supports both rigid and learned hippocampal sequences. Science 351, 1440–1443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kohus Z et al. Properties and dynamics of inhibitory synaptic communication within the CA3 microcircuits of pyramidal cells and interneurons expressing parvalbumin or cholecystokinin. J. Physiol 594, 3745–3774 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mercer A, Trigg HL & Thomson AM Characterization of neurons in the CA2 subfield of the adult rat hippocampus. J. Neurosci 27, 7329–7338 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papp OI, Karlócai MR, Tóth IE, Freund TF & Hájos N Different input and output properties characterize parvalbumin-positive basket and Axo-axonic cells in the hippocampal CA3 subfield. Hippocampus 23, 903–918 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Vida I & Frotscher M A hippocampal interneuron associated with the mossy fiber system. Proc. Natl Acad. Sci. USA 97, 1275–1280 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maccaferri G, Tóth K & McBain CJ Target-specific expression of presynaptic mossy fiber plasticity. Science 279, 1368–1370 (1998). [DOI] [PubMed] [Google Scholar]
- 51.Viney TJ et al. Network state-dependent inhibition of identified hippocampal CA3 axo-axonic cells in vivo. Nat. Neurosci 16, 1802–1811 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuentealba P et al. Expression of COUP-TFII nuclear receptor in restricted GABAergic neuronal populations in the adult rat hippocampus. J. Neurosci 30, 1595–1609 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evangelista R et al. Generation of sharp wave-ripple events by disinhibition. J. Neurosci 40, 7811–7836 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mizunuma M et al. Unbalanced excitability underlies offline reactivation of behaviorally activated neurons. Nat. Neurosci 17, 503–505 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Kajikawa K, Hulse BK, Siapas AG & Lubenov EV UP–DOWN states and ripples differentially modulate membrane potential dynamics across DG, CA3, and CA1 in awake mice. eLife 11, e69596 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zarnadze S et al. Cell-specific synaptic plasticity induced by network oscillations. eLife 5, e14912 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lau PY-P et al. Long-term plasticity in identified hippocampal GABAergic interneurons in the CA1 area in vivo. Brain Struct. Funct 222, 1809–1827 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rolotti SV et al. Local feedback inhibition tightly controls rapid formation of hippocampal place fields. Neuron 110, 783–794 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Francavilla R et al. Connectivity and network state-dependent recruitment of long-range VIP–GABAergic neurons in the mouse hippocampus. Nat. Commun 9, 5043 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oliva A, Fernández-Ruiz A, Leroy F & Siegelbaum SA Hippocampal CA2 sharp-wave ripples reactivate and promote social memory. Nature 587, 264–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szalay G et al. Fast 3D imaging of spine, dendritic, and neuronal assemblies in behaving animals. Neuron 92, 723–738 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jinno S, Araki K, Matsumoto Y, Suh Y-H & Yamamoto T. Selective apoptosis induction in the hippocampal mossy fiber pathway by exposure to CT105, the C-terminal fragment of Alzheimer’s amyloid precursor protein. Brain Res. 1249, 68–78 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Gulyás AI, Hájos N & Freund TF Interneurons containing calretinin are specialized to control other interneurons in the rat hippocampus. J. Neurosci 16, 3397–3411 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaifosh P, Zaremba JD, Danielson NB & Losonczy A SIMA: Python software for analysis of dynamic fluorescence imaging data. Front. Neuroinform 8, 80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keemink SW et al. FISSA: a neuropil decontamination toolbox for calcium imaging signals. Sci. Rep 8, 3493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Danielson NB et al. Sublayer-specific coding dynamics during spatial navigation and learning in hippocampal area CA1. Neuron 91, 652–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hardcastle K, Maheswaranathan N, Ganguli S & Giocomo LM A multiplexed, heterogeneous, and adaptive code for navigation in medial entorhinal cortex. Neuron 94, 375–387 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hainmueller T & Bartos M Parallel emergence of stable and dynamic memory engrams in the hippocampus. Nature 558, 292–296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data analyzed for this study are available at https://github.com/losonczylab/Vancura_Geiller_NatNeuro_2023. Source data are provided with this paper.


