Abstract
Background
miR-491-5p has been reported to regulate the expression of FGFR4 and promote gastric cancer metastasis. Hsa_circ_0001361 was demonstrated to play an oncogenic role in bladder cancer invasion and metastasis by sponging the expression of miR-491-5p. This work aimed to study the molecular mechanism of the effect of hsa_circ_0001361 on axillary response in the treatment of breast cancer.
Methods
Ultrasound examinations was performed to evaluate the response of breast cancer patients receiving NAC treatment. Quantitative real-time PCR, IHC assay, luciferase assay and Western blot were performed to analyze the molecular interaction between miR-491, circRNA_0001631 and FGFR4.
Results
Patients with low circRNA_0001631 expression had a better outcome after NAC treatment. The expression of miR-491 was remarkably higher in the tissue sample and serum collected from patients with lower circRNA_0001631 expression. On the contrary, the FGFR4 expression was notably suppressed in the tissue sample and serum collected from patients with lower circRNA_0001631 expression when compared with patients with high circRNA_0001631 expression. The luciferase activities of circRNA_0001631 and FGFR4 were effectively suppressed by miR-491 in MCF-7 and MDA-MB-231 cells. Moreover, inhibition of circRNA_0001631 expression using circRNA_0001361 shRNA effectively suppressed the expression of FGFR4 protein in MCF-7 and MDA-MB-231 cells. Up-regulation of circRNA_0001631 expression remarkably enhanced the expression of FGFR4 protein in MCF-7 and MDA-MB-231 cells.
Conclusion
Our study suggested that the up-regulation of hsa_circRNA-0001361 could up-regulate the expression of FGFR4 via sponging the expression of miR-491-5p, resulting in the alleviated axillary response after neoadjuvant chemotherapy (NAC) in breast cancer.
Keywords: circRNA-0001361, Axillary response, Ultrasound, Neoadjuvant chemotherapy, Breast cancer
Highlights
-
•
The therapeutic outcome of NAC treatment is remarkably better in patients with low circRNA_0001631 expression.
-
•
Upregulation of miR-491 and downregulation of FGFR4 are observed in patients with low circRNA_0001631 expression.
-
•
miR-491 is sponged by circRNA_0001361 and could target FGFR4.
-
•
Regulation of circRNA_0001631 remarkably altered the expression of FGFR4.
1. Introduction
Breast cancer (BC) is the most common cancer in women [1,2]. Neoadjuvant chemotherapy (NAC) has been developed as a common treatment for BC [[3], [4], [5]]. Many studies showed that people who accomplished a complete response after NAC was anticipated to show a significantly more advantageous outcome as compared to people without chemotherapy [6,7]. As just < 30% of BC patients achieve CR after therapy, most of BC patients still have a higher risk of relapse [8]. Some data has shown that the nodal phase of BC predicts prognosis much more efficiently than the initial axillary condition [9]. Thus, BC patients can manage to avoid postoperative morbidity, including upper arm pain [10,11]. People who obtain pathological complete responses may have considerably better survival as contrasted to those with a recurring ailment. Pathological complete response is defined by the loss of all intrusive cancer cells after therapy and is highly associated with boosted survival [12,13]. For individuals with BC who undergo therapy, the evaluation of axillary lymph nodes is a crucial factor in predicting the possibility of disease recurrence and the likelihood of disease-free survival [9,14,15]. These lymph nodes are the primary ones that breast cancer cells tend to spread to. As cancer cells break away from the primary tumor site, they can migrate to the axillary lymph nodes via the lymphatic system. Therefore, the presence of cancer cells in axillary lymph nodes indicates that cancer has metastasized from the breast to other parts of the body [16].
As a type of non-coding RNA, circRNAs are RNAs without a 5′ or 3′ ends [17]. Compared with regular RNAs, circRNAs are more stable in serum [18]. First uncovered via electron microscopy in the 1970s, circRNAs at first are thought of as the outcome of mRNA splicing error [19]. Lately, along with the development of the next-generation RNA sequencing techniques, circRNAs have been found to play numerous roles in managing gene expression [20]. Essentially, proofs have displayed that circRNAs are related to different human illnesses, such as atherosclerosis, heart failure, nerve ailments, and tumorigenesis [21,22]. It was also found that circ000136 is upregulated in BC cells, while high expression of circRNA_0001361 is positively correlated with the grade and invasion of BC. Significantly, it was found that circRNA_0001361 can inhibit miR-491-5p to upregulate the expression of MMP9 and subsequently boost the metastasis of BC.
Fibroblast growth factor (FGF)/FGF receptor (FGFR) signaling is involved in many biological events, such as cell differentiation, cell proliferation, angiogenesis, and cell motility [23]. The members of FGFR tyrosine kinase family are four related genes (FGFR1-FGFR4) [23]. The dysregulation, especially the overexpression of FGFR4 has been reported in various tumors, and the FGF/FGFR signaling has been implicated in cancer onset and tumor growth [23]. Also, FGFR4 is particularly high in breast cancer tissues, reaching almost as high as more than 2.5-fold of FGFR4 expression compared with normal tissues [24]. And FGFR4 has been recognized for its overexpression in approximately 30% of breast cancer cases compared to normal tissues [25]. Thussbas et al. revealed that FGFR4 Arg388 is an indicator of progression in BC patients receiving adjuvant therapy [26]. In a retrospective study, nonetheless, 85% of people received chemotherapy of cyclophosphamide + methotrexate + 5-fluorouracil but showed no response. Another research evaluated the possibility of using FGFR4 genotypes to predict the reaction to anthracycline + taxane chemotherapy carried out in a phase II trial of T2-4N0-2M0 BC [27].
NAC has been increasingly used for patients with operable breast cancer, while tumor size and cortical thickness were reported to be associated with axillary response [28,29]. miR-491-5p has been reported to regulate the expression of FGFR4 and promote gastric cancer metastasis [30]. Also, hsa_circ_0001361 was demonstrated to play an oncogenic role in bladder cancer invasion and metastasis by sponging the expression of miR-491-5p [31]. In this study, we enrolled breast cancer patients who received NAC and grouped them according to the expression level of hsa_circ_0001361, with the aim to study the molecular mechanism of the effect of hsa_circ_0001361 on axillary response in the treatment of breast cancer.
2. Materials and methods
2.1. RNA isolation and real-time PCR
Breast cancer patients receiving NAC were recruited for the collection of BC samples. Depending on to the guidelines of an RNeasy Mini assay kit (Qiagen, Hilden, Germany) and TRIzol (Invitrogen, Carlsbad, CA), sample RNAs were removed from BC tissues or cells, and then subject to reverse transcription using primers designed by Primer 5.0 and made by GenePharma (Shanghai, China). The reverse transcription was done with the OneStep PrimeScript cDNA Synthesis reagent (Takara, Shiga, Japan). The real-time PCR was then executed per the instruction of the SYBR PrimeScript assay kit (Takara, Shiga, Japan), and the levels of circRNA_0001361 (sense: GAGATGCAGCTCAGCAGGTTA; anti-sense: AATGGTGGCAGTTCCAGAGG), miR-491 (sense: GGAGTGGGGAACCCTTCC; anti-sense: GTGCAGGGTCCGAGGT), and FGFR4 mRNA (sense: GAGGGGCCGCCTAGAGATT; anti-sense: CAGGACGATCATGGAGCCT) were computed via the 2-△△Ct method he possibility of, using U6 (sense: GCTTCGGCAGCACATATACTAAAAT; anti-sense: CGCTTCACGAATTTGCGTGTCAT) and GAPDH (sense primer: ACAGTCAGCCGCATCTTC; anti-sense: CTCCGACCTTCACCTTCC) as internal controls.
2.2. Human subjects
We recruited 134 breast cancer patients for our study and divided the patients into two groups according to the circRNA_0001361 expression level: High circRNA_0001361 group (N = 62) and Low circRNA_0001361 group (N = 62). The basic characteristics including age, clinical stage, tumor histologic type, ER status, PR status and HER2 status were compared between the two groups. The patients received therapy and were checked by US or MRI imaging in the course of therapy. The patients with the following conditions were excluded [1]: MRI images obtained with the 1.5 MRI system [2], MRI was not obtained at our medical center [3], insufficient size of axillary region. The enrolled patients had a mean age of 48 ± 5 years with positive US or MRI imaging of cortical thickening of LN by > 3 mm. The therapy given to all patients was a standard therapy of adriamycin+docetaxel/cyclophosphamide or adriamycin + cyclophosphamide + docetaxel/HER2 monoclonal antibody. After the therapy, all patients underwent a mastectomy to remove axillary LNs surgically. The institutional ethics committee has approved this study. Consent forms were signed by all participants before the initiation of this study.
2.3. Cell culture and transfection
MCF-7 as well as MDA-MB-231 cells were divided into 4 groups: 1. Mock group; 2. negative control group; 3. circRNA_0001361 shRNA1 group; 4. circRNA_0001361 shRNA2 group. The BC cell lines were got from the American Type Culture Collection and cultured in RPMI-1640 (Gibco, Thermo Fisher Scientific, Waltham, WA) with 10% FBS (Hyclone, GE Health Care, Baltimore, MD) and 1% streptomycin, with 5% CO2 and at 37 °C. The mycoplasma result was negative in all cells. When the cell confluency was 70%, the medium was switched to serum-free RPMI-1640, while the transfection of cells was done by making use of Lipofectamine 3000 (Invitrogen, Thermo Fisherman Scientific, Waltham, WA) per the manufacturer's method. The cells were harvested at 48 h after transfection for analysis.
2.4. Luciferase assay
Sequence analysis indicated that miR-491 could bind to circRNA_0001361. The luciferase vectors containing wild-type and mutant circRNA_0001361 were established and transfected into MCF-7 and MDA-MB-231 cells with miR-491. Similarly, sequence analysis indicated that miR-491 could bind to FGFR4, and the luciferase vectors containing wild-type and mutant FGFR4 were established and transfected into MCF-7 and MDA-MB-231 cells with miR-491. The plasmids were made by RiboBio (Guangzhou, China) and transfected into cells using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, Waltham, WA) per the manufacturer's method. The cells were harvested at 48 h after transfection for luciferase activity analysis.
2.5. Western blot analysis
The cells were homogenized through pulverization and lysed with a lysis buffer for 15 min. The protein in the supernatant was separated with 12% SDS-PAGE and transferred onto a PVDF membrane, which was blocked with 5% non-fat milk in TBST. Afterward, the PVDF membrane was incubated with anti-FGFR4 primary and secondary antibodies (Abcam, Cambridge, MA). After the membrane was thoroughly washed with TBST, protein bands were imagined with an enhanced chemiluminescence reagent (Millipore, New York, NY) to determine the expression of FGFR4 protein.
2.6. Immunohistochemistry
The wound tissues were fixed, deparaffinized, and rehydrated. Then, endogenous peroxidase activity was blocked using 3% hydrogen peroxide. The non-specific activity was blocked along with goat serum, and the expression of FGFR4 protein was determined using anti-FGFR4 primary antibodies (Abcam, Cambridge, MA) and HRP-labeled secondary antibodies (Abcam, Cambridge, MA). Counterstaining was done with hematoxylin.
2.7. Statistical analysis
All results were analyzed via SPSS 18.0 software (SPSS, Chicago, IL) and shown as mean ± standard deviation. The t-test was utilized for inter-group comparisons. Statistical significance was taken at P < 0.05.
3. Results
3.1. The therapeutic outcome of NAC treatment was remarkably better in breast cancer patients with low circRNA_0001631 expression
Breast cancer patients receiving NAC were recruited and subjected to circRNA_0001361 expression analysis. These patients were divided into two groups according to the circRNA_0001361 expression level: High circRNA_0001361 and Low circRNA_0001361 groups. The basic characteristics including age, clinical stage, tumor histologic type, ER status, PR status and HER2 status were compared between the two groups. No significant difference was observed (Table 1). Ultrasound screening was performed to measure the tumor size, reduction in tumor size, number of nodes and longitudinal diameter difference before and after NAC treatment. There was no remarkable difference in the tumor size between high circRNA_0001361 and low circRNA_0001361 groups before NAC treatment. However, the tumor size was significantly decreased in the low circRNA_0001361 group when compared with the high circRNA_0001361 group (Fig. 1A). Therefore, the reduction of tumor size was notably increased for low circRNA_0001361 group when compared with high circRNA_0001361 group (Fig. 1B). Besides, the number of nodules was apparently decreased in the low circRNA_0001361 group when compared with the high circRNA_0001361 group after NAC treatment (Fig. 1C). The longitudinal diameter of the nodules was also decreased in the low circRNA_0001361 group when compared with the high circRNA_0001361 group after NAC treatment (Fig. 1D). Moreover, the cortical thickness was notably decreased in the low circRNA_0001361 group when compared with the high circRNA_0001361 group after NAC treatment (Fig. 1E).
Table 1.
Basic characters of participants enrolled in this study.
| Characteristics | High group (N = 62) | Low group (N = 62) | P value |
|---|---|---|---|
| Age, years | 46.3 ± 5.6 | 48.2 ± 6.2 | 0.673 |
| Clinical stage | 0.331 | ||
| II | 38 (61.3) | 35 (56.5) | |
| III | 24 (38.7) | 27 (43.5) | |
| Tumour histologic type | 0.342 | ||
| Ductal | 56 (90.3) | 58 (93.5) | |
| Lobular | 4 (6.5) | 1 (1.6) | |
| Others | 2 (3.23) | 3 (4.8) | |
| ER status | 0.241 | ||
| Negative | 41 (66.1) | 37 (59.7) | |
| Positive | 21 (33.9) | 25 (40.3) | |
| PR status | 0.306 | ||
| Negative | 44 (71.0) | 41 (66.1) | |
| Positive | 18 (29.0) | 21 (33.9) | |
| HER2 status | 0.313 | ||
| Negative | 38 (61.3) | 37 (59.7) | |
| Positive | 24 (38.7) | 25 (40.3) |
Fig. 1.
The therapeutic outcome of NAC treatment was remarkably better in breast cancer patients with low circRNA_0001631 expression (*P < 0.05 vs. High group).
A: Ultrasound examination indicated that the tumor size was remarkably decreased in patients with low circRNA_0001631 expression. B: Ultrasound examination indicated that reduction in tumor size was remarkably increased in patients with low circRNA_0001631 expression. C: Ultrasound examination indicated that the number of nodules was remarkably decreased in patients with low circRNA_0001631 expression. D: Ultrasound examination indicated that the longitudinal diameter was remarkably decreased in patients with low circRNA_0001631 expression. E: Ultrasound examination indicated that the cortical thickness was remarkably decreased in patients with low circRNA_0001631 expression.
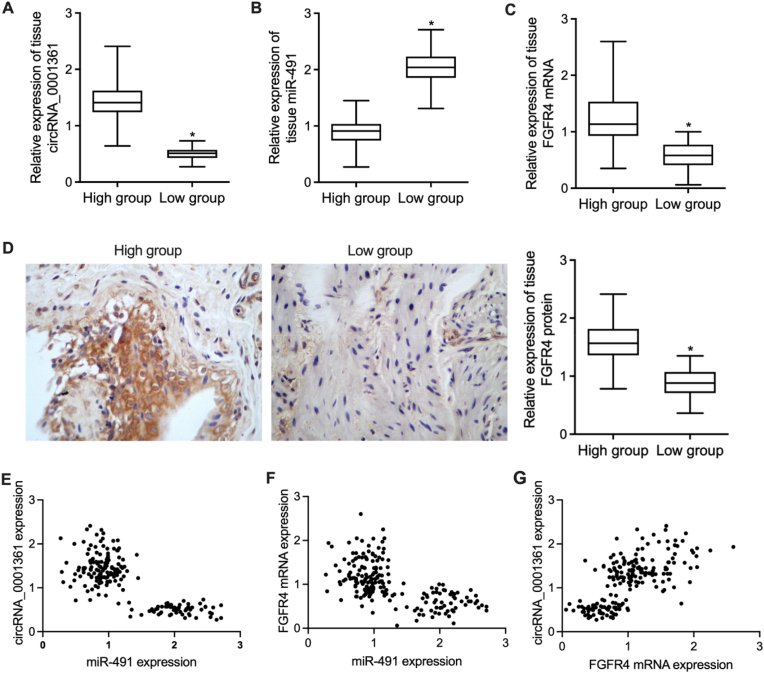
3.2. Upregulation of miR-491 and downregulation of FGFR4 in the tissue samples from patients with low circRNA_0001631 expression
The tumor tissue samples were collected after surgery. Quantitative real-time PCR was performed to analyze the expression of circRNA_0001361, miR-491 and FGFR4 mRNA in the tissue samples with differential expression of circRNA_0001631 (Fig. 2A). The expression of miR-491 was notably elevated in the tissue samples collected from patients with low circRNA_0001631 expression when compared with patients with high circRNA_0001631 expression (Fig. 2B). While the expression of FGFR4 mRNA was significantly suppressed in the tissue samples collected from patients with low circRNA_0001631 expression (Fig. 2C). IHC analysis and quantification also indicated that the expression of FGFR4 protein was significantly suppressed in the tissue samples collected from patients with low circRNA_0001631 expression (Fig. 2D). Moreover, the correlation analysis between circRNA_0001631, miR-491 and FGFR4 mRNA suggested possible correlation between these genes (Fig. 2E–G).
Fig. 2.
Upregulation of miR-491 and downregulation of FGFR4 in the tissue samples from patients with low circRNA_0001631 expression (*P < 0.05 vs. High group).
A: Differential expression of circRNA_0001361 in the tissue samples from breast cancer. B: The expression of miR-491 was remarkably higher in the tissue samples with circRNA_0001631 downregulation. C: The expression of FGFR4 mRNA was apparently suppressed in the tissue samples with circRNA_0001631 downregulation. D: The expression of FGFR4 protein was apparently suppressed in the tissue samples with circRNA_0001631 downregulation. E: Correlation analysis between circRNA_0001361 and miR-491 expression. F: Correlation analysis between FGFR4 mRNA and miR-491 expression. G: Correlation analysis between circRNA_0001361 and FGFR4 mRNA expression.
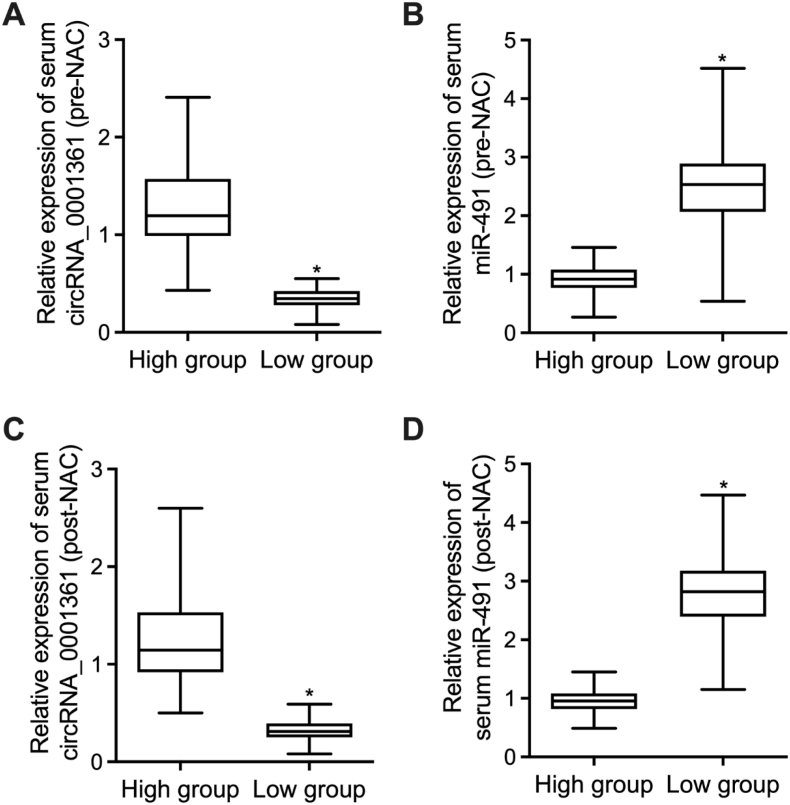
3.3. Activated expression of miR-491 in the serum collected from patients with low circRNA_0001631 expression
Moreover, the peripheral blood samples were collected from patients with distinct circRNA_0001631 expression before and after NAC treatment. The expression of circRNA_0001631 in the serum was significantly higher before NAC treatment for patients with high circRNA_0001631 expression (Fig. 3A). The expression of miR-491 in the serum was remarkably lower before NAC treatment for patients with high circRNA_0001631 expression (Fig. 3B). The expression of circRNA_0001631 in the serum was significantly higher after NAC treatment for patients with high circRNA_0001631 expression (Fig. 3C). The expression of miR-491 in the serum was remarkably lower after NAC treatment for patients with high circRNA_0001631 expression (Fig. 3D).
Fig. 3.
Activated expression of miR-491 in the serum collected from patients with low circRNA_0001631 expression (*P < 0.05 vs. High group).
A: The expression of circRNA_0001631 in the serum before NAC treatment was remarkably higher for patients with higher circRNA_0001631 expression. B: The expression of miR-491 in the serum before NAC treatment was remarkably lower for patients with higher circRNA_0001631 expression. C: The expression of circRNA_0001631 in the serum after NAC treatment was remarkably higher for patients with higher circRNA_0001631 expression. D: The expression of miR-491 in the serum after NAC treatment was remarkably lower for patients with higher circRNA_0001631 expression.
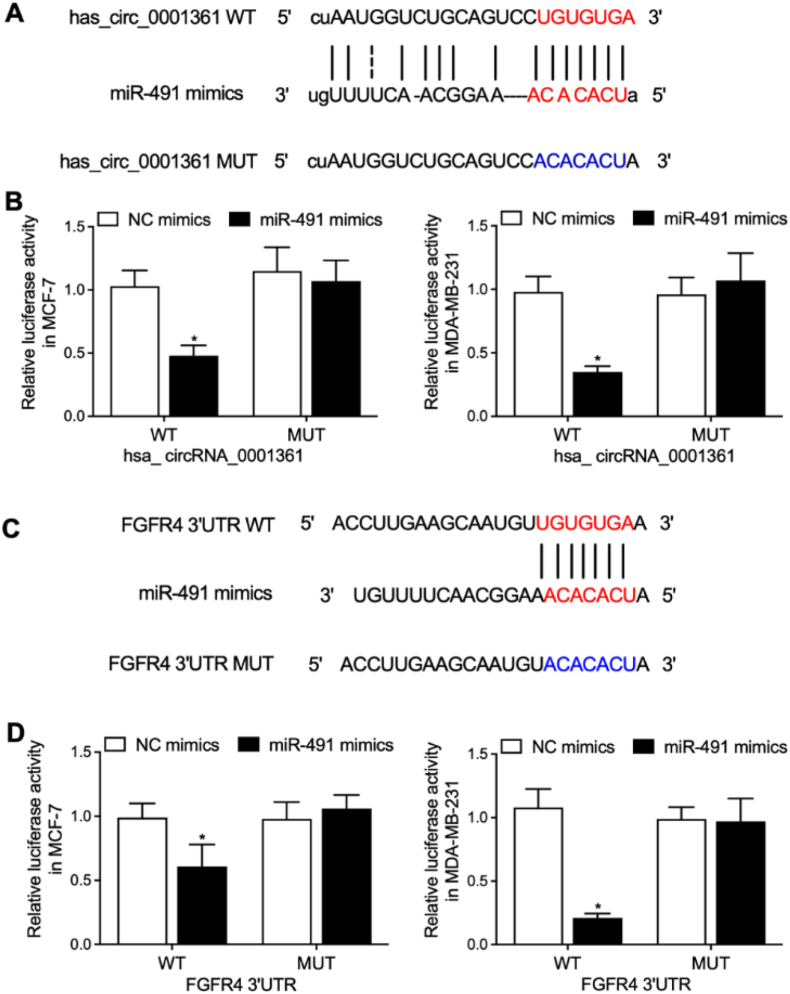
3.4. MiR-491 inhibited the luciferase activities of circRNA_0001361 and FGFR4 in MCF-7 and MDA-MB-231 cells
Sequence analysis indicated that miR-491 could bind to circRNA_0001361 (Fig. 4A). Luciferase vectors containing wild-type and mutant circRNA_0001361 were established and transfected into MCF-7 and MDA-MB-231 cells with miR-491. The luciferase activity of wild-type circRNA_0001631 was significantly suppressed by miR-491 in MCF-7 and MDA-MB-231 cells (Fig. 4B). Sequence analysis indicated that miR-491 could bind to FGFR4 (Fig. 4C). Luciferase vectors containing wild-type and mutant FGFR4 were established and transfected into MCF-7 and MDA-MB-231 cells with miR-491. The luciferase activity of wild-type FGFR4 was significantly suppressed by miR-491 in MCF-7 and MDA-MB-231 cells (Fig. 4D).
Fig. 4.
MiR-491 inhibited the luciferase activities of circRNA_0001361 and FGFR4 in MCF-7 and MDA-MB-231 cells (*P < 0.05 vs. NC mimics).
A: Sequence analysis indicated binding of miR-491 to circRNA_0001631. B: The luciferase activities of wild-type circRNA_0001631 were effectively suppressed by miR-491 in MCF-7 and MDA-MB-231 cells. C: Sequence analysis indicated binding of miR-491 to FGFR4. D: The luciferase activities of wild-type FGFR4 were effectively suppressed by miR-491 in MCF-7 and MDA-MB-231 cells.
3.5. Regulation of circRNA_0001631 expression remarkably altered the expression of FGFR4 in MCF-7 and MDA-MB-231 cells
We designed two circRNA_1361 shRNA vectors and transfected them into MCF-7 and MDA-MB-231 cells. Western blot was performed to analyze the expression of FGFR4 in MCF-7 and MDA-MB-231 cells under distinct conditions. The expression of FGFR4 was remarkably suppressed by both circRNA_1361 shRNA1 and circRNA_1361 shRNA2 in MCF-7 (Fig. 5A and B) and MDA-MB-231 cells (Fig. 5C and D). Moreover, circRNA_0001631 overexpression was carried out by transfecting circRNA_0001361 over-1 and circRNA_0001361 over-2 into MCF-7 and MDA-MB-231 cells. The expression of FGFR4 was remarkably activated by both circRNA_0001361 over-1 and circRNA_0001361 over-2 transfection in MCF-7 (Fig. 5E and F) and MDA-MB-231 cells (Fig. 5G and H).
Fig. 5.
Regulation of circRNA_0001631 expression remarkably altered the expression of FGFR4 in MCF-7 and MDA-MB-231 cells (*P < 0.05 vs. negative control group).
A: Western blot analysis of FGFR4 protein expression in MCF-7 cells transfected with circRNA_0001361 shRNA under distinct conditions. B: Quantitative analysis of Western blot results indicated that circRNA_0001361 shRNA effectively suppressed the expression of FGFR4 protein in MCF-7 cells. C: Western blot analysis of FGFR4 protein expression in MDA-MB-231 cells transfected with circRNA_0001361 shRNA under distinct conditions. D: Quantitative analysis of Western blot results indicated that circRNA_0001361 shRNA effectively suppressed the expression of FGFR4 protein in MDA-MB-231 cells. E: Western blot analysis of FGFR4 protein expression in circRNA_0001361-overexpressed MCF-7 cells under distinct conditions. F: Quantitative analysis of Western blot results indicated that circRNA_0001361 overexpression effectively activated the expression of FGFR4 protein in MCF-7 cells. G: Western blot analysis of FGFR4 protein expression in circRNA_0001361-overexpressed MDA-MB-231 cells under distinct conditions. H: Quantitative analysis of Western blot results indicated that circRNA_0001361 overexpression effectively activated the expression of FGFR4 protein in MDA-MB-231 cells.
4. Discussion
NAC is standard for patients of locally advanced BC. The main objective of NAC is to boost disease-free survival [32]. In fact, > 70% BC patients might achieve a response after the treatment, with the rest presenting different levels of resistance [33]. Pre-NAC and post-NAC MRI values of tumor size have been linked to the level of axillary response, with the greatest diagnostic performance of an AUC of 0.760 [34]. Regarding the features of axillary LNs, the cortical thickness during and after NAC therapy are predictors for a higher OR [35]. When the tumor size is decreased by NAC therapy, the cortical thickness can also reflect the tumor burden inside the LN [35,36]. Not much information is available on the axillary images of LNs after the NAC therapy of breast cancer. On top of that, not all people reviewed in previous research underwent nodal staging by pre-operative needle biopsy, so some patients were staged by imaging as cN1 but some patients were staged as pre-NAC SLNB, additionally complicating the evaluation of images [37,38]. In this study, we recruited breast cancer patients receiving NAC treatment and explored the effect of circRNA_0001631 expression on NAC therapeutic outcome. Patients with low circRNA_0001631 expression had a better therapeutic outcomes of NAC therapy. In addition, we performed qPCR to analyze the expression of miR-491 and FGFR4 in the tissue samples as well as the serum collected from patients with differential circRNA_0001631 expression. The expression of miR-491 was significantly elevated in patients with low circRNA_0001631 expression and the expression of FGFR4 was notably suppressed in patients with low circRNA_0001631 expression.
CircRNAs are non-coding RNAs in a form of closed-loop structures [39]. Being highly conserved in eukaryotic cells, they have been shown to work as miRNA sponges via either being derived from exons encoding proteins or being distributed mainly in the cytoplasm [40]. Recently, research suggested that circRNAs serve as miRNA sponges and require numerous binding sites on the same miRNA, like Sry circRNA, ciRS-7, and HRCR [17,21]. The interactions between circRNAs and miRNAs are implicated in various pathophysiological functions, especially in tumor onset and progression [41]. For instance, Gao G et al. identified circ_0001946 as a potential target for BC therapy to overcome tamoxifen resistance. Their research showed that circ_0001946 is up-regulated in breast cancer tissues and can promote the growth and malignant invasion of tamoxifen-resistant BC cells by regulating the expression of miR-671-5p [42]. Also, Liang H et al. reported that circ-ABCB10 was highly expressed in breast cancer tissues, and circ-ABCB10 was demonstrated to function as a miRNA sponge for miR-1271 [43]. Other circRNAs such as circ_0052112 [44] and circ_ 0000520 [45] were also reported in the pathogenesis of BC, which each functions as miRNA sponge for different miRNAs. Especially, the suppressed expression of circ_0052112 not only inhibits BC cell migration and invasion, but also elevates the expression of miR-125-5p [44]. And circ_0000520 which directly regulates the expression of miR-1296 has been found to be significantly upregulated in BC tumors [45]. In this study, we aimed to unveil the relationship between axillary response in BC treatment and circ_0001361. As circ_0001361 is derived from two FNDC3B exons, it was actually shown that circ_0001361 serves as a miRNA sponge to control the levels of genes downstream FNDC3B [31]. The upregulation of circ_0001361 was found to be associated with pathologic grade and muscle invasion in bladder cancer tissues [31]. Similar to the above-mentioned circRNAs, circ_0001361 also promotes bladder cancer cell invasion and metastasis via sponging miR-491-5p and regulate the expression of MMP-9. It is noteworthy that circ_0001361 has two binding sites of miR-491-5p, although only one binding site is effective. Similarly, recent research additionally showed that many of circRNAs only have one or two sites for some miRNAs, and can serve as efficient sponges for miRNAs like circIRAK3, circ101555, circFNDC3B, as well as circTP63 [46,47]. Also, in lung adenocarcinoma tissues and cells, the expression of circ_0001361 was increased while that of miR-525-5p was decreased. And the functional experiments not only confirmed the inhibitory effect on tumor metastasis of circ_0001362, but also revealed the circRNA's inhibitory effect on lung adenocarcinoma cell growth [48]. Since the oncogenic role of circ_0001361 has been reported in various types of cancer, we suspect that circ_0001361 may also participate in the pathogenesis procedures in BC. In this study, we overexpressed and suppressed the expression of circ_0001361 in MCF-7 and MDA-MB-231 cells. Accordingly, we found that the expression of circ_0001361 was negatively correlated with the expression of miR-491 and positively correlated with the expression of FGFR4 in MCF-7 and MDA-MB-231 cells.
miR-491-5p is recognized as a tumor suppressor gene in many kinds of cancers to control cell proliferation, cell apoptosis, cell movement, and chemoresistance [49]. However, the activity of miR-491-5p in GC is unclear. Sun et al. 6 showed that miR-491-5p was actually downregulated in GC cells, but miR-491-5p restoration hampered tumor development with the targeting of Wnt3a/beta-catenin pathway. Having said that, the comprehensive functions of miR-491-5p in GC are unclear. In this study, we carried out luciferase assay to explore the inhibitory effect of miR_491 on circRNA_0001631 and FGFR4. The luciferase activities of circRNA_0001631 and FGFR4 were apparently inhibited by miR_491 in MCF-7 and MDA-MB-231 cells.
FGFR4 is an EMT inducer and can promote metastasis [50]. In GC, Ye et al. showed that FGFR4 is strongly expressed in GC tissues to promote GC proliferation and decrease apoptosis [51]. The FGFR4 expression level is also positively correlated with the expression of SNAIL and a poor prognosis. In cancers, FGFR4 aberrations include excessive FGFR4 amplification, FGFR4 mutation activation, as well as overexpression of FGFR4, which can trigger cell proliferation and tumor growth [52]. FGFR4 dysregulation was observed in epithelial cancers including hepatocellular and prostate tumors [26]. A research presented that during the course of doxorubicin therapy, the aberrant FGFR4 expression in cancer cells lowered the level of apoptosis, and FGFR4 overexpression is substantially associated with a high tumor grade in prostate cancer and chemotherapy resistance in patients with BC [53].
5. Conclusion
In conclusion, our study suggested that the up-regulation of hsa_circRNA-0001361 could up-regulate the expression of FGFR4 via sponging the expression of miR-491-5p, and the signaling of the circRNA-0001361/miR-491/FGFR4 axis can therefore alleviate axillary response after neoadjuvant chemotherapy in breast cancer.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Funding statement
No funding was received.
Ethical approval and study consent
Institutional ethics committee has approved this study. Consent forms were signed by all participants before the initiation of this study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Ward E.M., DeSantis C.E., Lin C.C., Kramer J.L., Jemal A., Kohler B., et al. Cancer statistics: breast cancer in situ. CA A Cancer J. Clin. 2015;65(6):481–495. doi: 10.3322/caac.21321. PubMed PMID: 26431342. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis C., Ma J., Bryan L., Jemal A. Breast cancer statistics. CA A Cancer J. Clin. 2014;64(1):52–62. doi: 10.3322/caac.21203. 2013. PubMed PMID: 24114568. [DOI] [PubMed] [Google Scholar]
- 3.Mamounas E.P. Impact of neoadjuvant chemotherapy on locoregional surgical treatment of breast cancer. Ann. Surg Oncol. 2015;22(5):1425–1433. doi: 10.1245/s10434-015-4406-6. PubMed PMID: 25727558. [DOI] [PubMed] [Google Scholar]
- 4.Iqbal J., Shafi A.A., Alharthi B.N. Neoadjuvant chemotherapy in locally advanced breast cancer. J. Coll. Physicians Surg. Pak. 2014;24(11):845–848. 11.2014/JCPSP.845848. PubMed PMID: 25404445. [PubMed] [Google Scholar]
- 5.Golshan M., Cirrincione C.T., Sikov W.M., Berry D.A., Jasinski S., Weisberg T.F., et al. Impact of neoadjuvant chemotherapy in stage II-III triple negative breast cancer on eligibility for breast-conserving surgery and breast conservation rates: surgical results from CALGB 40603 (Alliance) Ann. Surg. 2015;262(3):434–439. doi: 10.1097/SLA.0000000000001417. discussion 8-9. PubMed PMID: 26222764; PubMed Central PMCID: PMC4710511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seo A.N., Lee H.J., Kim E.J., Kim H.J., Jang M.H., Lee H.E., et al. Tumour-infiltrating CD8+ lymphocytes as an independent predictive factor for pathological complete response to primary systemic therapy in breast cancer. Br. J. Cancer. 2013;109(10):2705–2713. doi: 10.1038/bjc.2013.634. PubMed PMID: 24129232; PubMed Central PMCID: PMC3833219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaudry M., Lei X., Gonzalez-Angulo A.M., Mittendorf E.A., Valero V., Tripathy D., et al. Recurrence and survival among breast cancer patients achieving a pathological complete response to neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2015;153(2):417–423. doi: 10.1007/s10549-015-3533-x. PubMed PMID: 26272743. [DOI] [PubMed] [Google Scholar]
- 8.Redden M.H., Fuhrman G.M. Neoadjuvant chemotherapy in the treatment of breast cancer. Surg. Clin. 2013;93(2):493–499. doi: 10.1016/j.suc.2013.01.006. PubMed PMID: 23464698. [DOI] [PubMed] [Google Scholar]
- 9.Rouzier R., Extra J.M., Klijanienko J., Falcou M.C., Asselain B., Vincent-Salomon A., et al. Incidence and prognostic significance of complete axillary downstaging after primary chemotherapy in breast cancer patients with T1 to T3 tumors and cytologically proven axillary metastatic lymph nodes. J. Clin. Oncol. 2002;20(5):1304–1310. doi: 10.1200/JCO.2002.20.5.1304. PubMed PMID: 11870173. [DOI] [PubMed] [Google Scholar]
- 10.Mansel R.E., Fallowfield L., Kissin M., Goyal A., Newcombe R.G., Dixon J.M., et al. Randomized multicenter trial of sentinel node biopsy versus standard axillary treatment in operable breast cancer: the ALMANAC Trial. J. Natl. Cancer Inst. 2006;98(9):599–609. doi: 10.1093/jnci/djj158. PubMed PMID: 16670385. [DOI] [PubMed] [Google Scholar]
- 11.Lucci A., McCall L.M., Beitsch P.D., Whitworth P.W., Reintgen D.S., Blumencranz P.W., et al. Surgical complications associated with sentinel lymph node dissection (SLND) plus axillary lymph node dissection compared with SLND alone in the American College of Surgeons Oncology Group Trial Z0011. J. Clin. Oncol. 2007;25(24):3657–3663. doi: 10.1200/JCO.2006.07.4062. PubMed PMID: 17485711. [DOI] [PubMed] [Google Scholar]
- 12.Rastogi P., Anderson S.J., Bear H.D., Geyer C.E., Kahlenberg M.S., Robidoux A., et al. Preoperative chemotherapy: updates of national surgical adjuvant breast and bowel project protocols B-18 and B-27. J. Clin. Oncol. 2008;26(5):778–785. doi: 10.1200/JCO.2007.15.0235. PubMed PMID: 18258986. [DOI] [PubMed] [Google Scholar]
- 13.Liedtke C., Mazouni C., Hess K.R., Andre F., Tordai A., Mejia J.A., et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 2008;26(8):1275–1281. doi: 10.1200/JCO.2007.14.4147. PubMed PMID: 18250347. [DOI] [PubMed] [Google Scholar]
- 14.Hennessy B.T., Hortobagyi G.N., Rouzier R., Kuerer H., Sneige N., Buzdar A.U., et al. Outcome after pathologic complete eradication of cytologically proven breast cancer axillary node metastases following primary chemotherapy. J. Clin. Oncol. 2005;23(36):9304–9311. doi: 10.1200/JCO.2005.02.5023. PubMed PMID: 16361629. [DOI] [PubMed] [Google Scholar]
- 15.von Minckwitz G., Untch M., Ju Blohmer, Costa S.D., Eidtmann H., Fasching P.A., et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J. Clin. Oncol. 2012;30(15):1796–1804. doi: 10.1200/JCO.2011.38.8595. PubMed PMID: 22508812. [DOI] [PubMed] [Google Scholar]
- 16.Rahman M., Mohammed S. Breast cancer metastasis and the lymphatic system. Oncol. Lett. 2015;10(3):1233–1239. doi: 10.3892/ol.2015.3486. Epub 20150713. PubMed PMID: 26622656; PubMed Central PMCID: PMC4533217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. PubMed PMID: 23446346. [DOI] [PubMed] [Google Scholar]
- 18.Barrett S.P., Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143(11):1838–1847. doi: 10.1242/dev.128074. PubMed PMID: 27246710; PubMed Central PMCID: PMC4920157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu M.T., Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–340. doi: 10.1038/280339a0. PubMed PMID: 460409. [DOI] [PubMed] [Google Scholar]
- 20.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. PubMed PMID: 23446348. [DOI] [PubMed] [Google Scholar]
- 21.Wang K., Long B., Liu F., Wang J.X., Liu C.Y., Zhao B., et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37(33):2602–2611. doi: 10.1093/eurheartj/ehv713. PubMed PMID: 26802132. [DOI] [PubMed] [Google Scholar]
- 22.Burd C.E., Jeck W.R., Liu Y., Sanoff H.K., Wang Z., Sharpless N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12) doi: 10.1371/journal.pgen.1001233. PubMed PMID: 21151960; PubMed Central PMCID: PMC2996334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powers C.J., McLeskey S.W., Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocr. Relat. Cancer. 2000;7(3):165–197. doi: 10.1677/erc.0.0070165. PubMed PMID: 11021964. [DOI] [PubMed] [Google Scholar]
- 24.Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., et al. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. PubMed PMID: 24071849; PubMed Central PMCID: PMC3919969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penault-Llorca F., Bertucci F., Adélaïde J., Parc P., Coulier F., Jacquemier J., et al. Expression of FGF and FGF receptor genes in human breast cancer. Int. J. Cancer. 1995;61(2):170–176. doi: 10.1002/ijc.2910610205. PubMed PMID: 7705943. [DOI] [PubMed] [Google Scholar]
- 26.Thussbas C., Nahrig J., Streit S., Bange J., Kriner M., Kates R., et al. FGFR4 Arg388 allele is associated with resistance to adjuvant therapy in primary breast cancer. J. Clin. Oncol. 2006;24(23):3747–3755. doi: 10.1200/JCO.2005.04.8587. PubMed PMID: 16822847. [DOI] [PubMed] [Google Scholar]
- 27.Marme F., Werft W., Benner A., Burwinkel B., Sinn P., Sohn C., et al. FGFR4 Arg388 genotype is associated with pathological complete response to neoadjuvant chemotherapy for primary breast cancer. Ann. Oncol. 2010;21(8):1636–1642. doi: 10.1093/annonc/mdq017. PubMed PMID: 20147743. [DOI] [PubMed] [Google Scholar]
- 28.Eun N.L., Son E.J., Gweon H.M., Kim J.A., Youk J.H. Prediction of axillary response by monitoring with ultrasound and MRI during and after neoadjuvant chemotherapy in breast cancer patients. Eur. Radiol. 2020;30(3):1460–1469. doi: 10.1007/s00330-019-06539-4. PubMed PMID: 31802216. [DOI] [PubMed] [Google Scholar]
- 29.Choi H.J., Ryu J.M., Kim I., Nam S.J., Kim S.W., Yu J., et al. Prediction of axillary pathologic response with breast pathologic complete response after neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2019;176(3):591–596. doi: 10.1007/s10549-019-05214-y. PubMed PMID: 31065874. [DOI] [PubMed] [Google Scholar]
- 30.Yu T., Wang L.N., Li W., Zuo Q.F., Li M.M., Zou Q.M., et al. Downregulation of miR-491-5p promotes gastric cancer metastasis by regulating SNAIL and FGFR4. Cancer Sci. 2018;109(5):1393–1403. doi: 10.1111/cas.13583. PubMed PMID: 29569792; PubMed Central PMCID: PMC5980274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu F., Zhang H., Xie F., Tao D., Xiao X., Huang C., et al. Hsa_circ_0001361 promotes bladder cancer invasion and metastasis through miR-491-5p/MMP9 axis. Oncogene. 2020;39(8):1696–1709. doi: 10.1038/s41388-019-1092-z. PubMed PMID: 31705065. [DOI] [PubMed] [Google Scholar]
- 32.Estevez L.G., Gradishar W.J. Evidence-based use of neoadjuvant taxane in operable and inoperable breast cancer. Clin. Cancer Res. 2004;10(10):3249–3261. doi: 10.1158/1078-0432.CCR-03-0133. PubMed PMID: 15161677. [DOI] [PubMed] [Google Scholar]
- 33.Zhang F., Yang Y., Smith T., Kau S.W., McConathy J.M., Esteva F.J., et al. Correlation between HER-2 expression and response to neoadjuvant chemotherapy with 5-fluorouracil, doxorubicin, and cyclophosphamide in patients with breast carcinoma. Cancer. 2003;97(7):1758–1765. doi: 10.1002/cncr.11245. PubMed PMID: 12655533. [DOI] [PubMed] [Google Scholar]
- 34.Ye B.B., Zhao H.M., Yu Y., Ge J., Wang X., Cao X.C. Accuracy of axillary ultrasound after different neoadjuvant chemotherapy cycles in breast cancer patients. Oncotarget. 2017;8(22):36696–36706. doi: 10.18632/oncotarget.13313. PubMed PMID: 27852041; PubMed Central PMCID: PMC5482689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le-Petross H.T., McCall L.M., Hunt K.K., Mittendorf E.A., Ahrendt G.M., Wilke L.G., et al. Axillary ultrasound identifies residual nodal disease after chemotherapy: results from the American college of surgeons oncology group Z1071 trial (alliance) AJR Am. J. Roentgenol. 2018;210(3):669–676. doi: 10.2214/AJR.17.18295. PubMed PMID: 29381381; PubMed Central PMCID: PMC5884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim W.H., Kim H.J., Park H.Y., Park J.Y., Chae Y.S., Lee S.M., et al. Axillary pathologic complete response to neoadjuvant chemotherapy in clinically node-positive breast cancer patients: a predictive model integrating the imaging characteristics of ultrasound restaging with known clinicopathologic characteristics. Ultrasound Med. Biol. 2019;45(3):702–709. doi: 10.1016/j.ultrasmedbio.2018.10.026. PubMed PMID: 30567630. [DOI] [PubMed] [Google Scholar]
- 37.Klauber-Demore N., Kuzmiak C., Rager E.L., Ogunrinde O.B., Ollila D.W., Calvo B.F., et al. High-resolution axillary ultrasound is a poor prognostic test for determining pathologic lymph node status in patients undergoing neoadjuvant chemotherapy for locally advanced breast cancer. Am. J. Surg. 2004;188(4):386–389. doi: 10.1016/j.amjsurg.2004.06.022. PubMed PMID: 15474431. [DOI] [PubMed] [Google Scholar]
- 38.Alvarado R., Yi M., Le-Petross H., Gilcrease M., Mittendorf E.A., Bedrosian I., et al. The role for sentinel lymph node dissection after neoadjuvant chemotherapy in patients who present with node-positive breast cancer. Ann. Surg Oncol. 2012;19(10):3177–3184. doi: 10.1245/s10434-012-2484-2. PubMed PMID: 22772869. [DOI] [PubMed] [Google Scholar]
- 39.Panda A.C., Grammatikakis I., Munk R., Gorospe M., Abdelmohsen K. Emerging roles and context of circular RNAs. Wiley Interdiscip. Rev. RNA. 2017;8(2) doi: 10.1002/wrna.1386. Epub 20160909. PubMed PMID: 27612318; PubMed Central PMCID: PMC5315638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnaiz E., Sole C., Manterola L., Iparraguirre L., Otaegui D., Lawrie C.H. CircRNAs and cancer: biomarkers and master regulators. Semin. Cancer Biol. 2019;58:90–99. doi: 10.1016/j.semcancer.2018.12.002. PubMed PMID: 30550956. [DOI] [PubMed] [Google Scholar]
- 41.Patop I.L., Kadener S. circRNAs in Cancer. Curr. Opin. Genet. Dev. 2018;48:121–127. doi: 10.1016/j.gde.2017.11.007. Epub 20171212. PubMed PMID: 29245064; PubMed Central PMCID: PMC5877416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao G., Li X., Zhang J., Yu H. YY1 as a promoter regulating the circ_0001946/miR-671-5p/EGFR axis to promote chemotherapy resistance in breast cancer cells. Am. J. Transl. Res. 2022;14(4):2550–2566. Epub 20220415. PubMed PMID: 35559420; PubMed Central PMCID: PMC9091114. [PMC free article] [PubMed] [Google Scholar]
- 43.Liang H.F., Zhang X.Z., Liu B.G., Jia G.T., Li W.L. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am. J. Cancer Res. 2017;7(7):1566–1576. Epub 20170701. PubMed PMID: 28744405; PubMed Central PMCID: PMC5523036. [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H.D., Jiang L.H., Hou J.C., Zhong S.L., Zhou S.Y., Zhu L.P., et al. Circular RNA hsa_circ_0052112 promotes cell migration and invasion by acting as sponge for miR-125a-5p in breast cancer. Biomed. Pharmacother. 2018;107:1342–1353. doi: 10.1016/j.biopha.2018.08.030. Epub 20180829. PubMed PMID: 30257349. [DOI] [PubMed] [Google Scholar]
- 45.Zang H., Li Y., Zhang X., Huang G. Blocking circ_0000520 suppressed breast cancer cell growth, migration and invasion partially via miR-1296/SP1 Axis both in vitro and in vivo. Cancer Manag. Res. 2020;12:7783–7795. doi: 10.2147/cmar.S251666. Epub 20200825. PubMed PMID: 32922078; PubMed Central PMCID: PMC7457856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu J., Jiang Z., Chen C., Hu Q., Fu Z., Chen J., et al. CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Lett. 2018;430:179–192. doi: 10.1016/j.canlet.2018.05.033. PubMed PMID: 29803789. [DOI] [PubMed] [Google Scholar]
- 47.Chen Z., Ren R., Wan D., Wang Y., Xue X., Jiang M., et al. Hsa_circ_101555 functions as a competing endogenous RNA of miR-597-5p to promote colorectal cancer progression. Oncogene. 2019;38(32):6017–6034. doi: 10.1038/s41388-019-0857-8. PubMed PMID: 31300733. [DOI] [PubMed] [Google Scholar]
- 48.Shen H.Y., Shi L.X., Wang L., Fang L.P., Xu W., Xu J.Q., et al. Hsa_circ_0001361 facilitates the progress of lung adenocarcinoma cells via targeting miR-525-5p/VMA21 axis. J. Transl. Med. 2021;19(1):389. doi: 10.1186/s12967-021-03045-4. Epub 20210910. PubMed PMID: 34507559; PubMed Central PMCID: PMC8434718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X., Liu Y., Granberg K.J., Wang Q., Moore L.M., Ji P., et al. Two mature products of MIR-491 coordinate to suppress key cancer hallmarks in glioblastoma. Oncogene. 2015;34(13):1619–1628. doi: 10.1038/onc.2014.98. PubMed PMID: 24747968; PubMed Central PMCID: PMC4205227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu R., Li J., Xie K., Zhang T., Lei Y., Chen Y., et al. FGFR4 promotes stroma-induced epithelial-to-mesenchymal transition in colorectal cancer. Cancer Res. 2013;73(19):5926–5935. doi: 10.1158/0008-5472.CAN-12-4718. PubMed PMID: 23943801. [DOI] [PubMed] [Google Scholar]
- 51.Ye Y.W., Zhou Y., Yuan L., Wang C.M., Du C.Y., Zhou X.Y., et al. Fibroblast growth factor receptor 4 regulates proliferation and antiapoptosis during gastric cancer progression. Cancer. 2011;117(23):5304–5313. doi: 10.1002/cncr.26207. PubMed PMID: 21567388. [DOI] [PubMed] [Google Scholar]
- 52.Shimura T., Sasatani M., Kawai H., Kamiya K., Kobayashi J., Komatsu K., et al. Radiation-Induced myofibroblasts promote tumor growth via mitochondrial ROS-activated TGFbeta signaling. Mol. Cancer Res. 2018;16(11):1676–1686. doi: 10.1158/1541-7786.MCR-18-0321. PubMed PMID: 30042177. [DOI] [PubMed] [Google Scholar]
- 53.Roidl A., Berger H.J., Kumar S., Bange J., Knyazev P., Ullrich A. Resistance to chemotherapy is associated with fibroblast growth factor receptor 4 up-regulation. Clin. Cancer Res. 2009;15(6):2058–2066. doi: 10.1158/1078-0432.CCR-08-0890. PubMed PMID: 19240166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.