Abstract
Open reading frame (ORF) O and ORF P partially overlap and are located antisense to the γ134.5 gene within the domain transcribed during latency. In wild-type virus-infected cells, ORF O and ORF P are completely repressed during productive infection by ICP4, the major viral transcriptional activator/repressor. In cells infected with a mutant in which ORF P was derepressed there was a significant delay in the appearance of the viral α-regulatory proteins ICP0 and ICP22. The ORF O protein binds to and inhibits ICP4 binding to its cognate DNA site in vitro. These characteristics suggested a role for ORF O and ORF P in the establishment of latency. To test this hypothesis, two recombinant viruses were constructed. In the first, R7538(P−/O−), the ORF P initiator methionine codon, which also serves as the initiator methionine codon for ORF O, was replaced and a diagnostic restriction endonuclease was introduced upstream. In the second, R7543(P−/O−)R, the mutations were repaired to restore the wild-type virus sequences. We report the following. (i) The R7538(P−/O−) mutant failed to express ORF O and ORF P proteins but expressed a wild-type γ134.5 protein. (ii) R7538(P−/O−) yields were similar to that of the wild type following infection of cell culture or following infection of mice by intracerebral or ocular routes. (iii) R7538(P−/O−) virus reactivated from latency following explanation and cocultivation of murine trigeminal ganglia with Vero cells at a frequency similar to that of the wild type, herpes simplex virus 1(F). (iv) The amount of latent R7538(P−/O−) virus as assayed by quantitative PCR is eightfold less than that of the repair virus. The repaired virus could not be differentiated from the wild-type parent in any of the assays done in this study. We conclude that ORF O and ORF P are not essential for the establishment of latency in mice but may play a role in determining the quantity of latent virus maintained in sensory neurons.
Herpes simplex virus 1 (HSV-1) and HSV-2 cause two types of infections in humans and experimental animals, productive and latent. Productive infection at the portal of entry involves the coordinated expression of >80 open reading frames (ORFs), replication, assembly of infectious progeny, and destruction of the cell (reviewed in references 49 and 50) (20, 21). More than half of the genes are dispensable for growth in cell culture and appear to have auxiliary functions that optimize viral replication and spread within its host. From the portal of entry, HSV infects innervating sensory neurons and is transported retrograde to the nucleus. The precise sequence of events that follows is unclear; it seems that in some neurons the virus replicates and destroys the neurons whereas in others the virus establishes a latent infection.
It is convenient to differentiate three stages of infection of neurons: the establishment phase, the maintenance phase, and the reactivation phase. Little is known of the establishment phase since the neurons which replicate the virus during the first several days after infection obscure the events taking place in the neurons committed to maintaining the virus in a latent state. In the maintenance state, signaled by the disappearance of all traces of replicating virus, viral DNA is maintained in an episomal form and only a small region of the genome is transcribed, the latency-associated transcripts (LATs) (55). The last phase, reactivation, is induced spontaneously in some experimental animal systems (reviewed in reference 15). It can be induced artificially by explanation and cocultivation of sensory ganglia harboring latent virus. In essence, our knowledge of the requirement for the establishment of latency has been based on whether latent virus can be detected during the maintenance stage or as a consequence of induced reactivation. Over the past decade several genes have been “identified” as playing a role in the establishment of latency (23, 32, 37, 38, 53, 56, 59). The list includes a large number of ORFs and also the LATs. In most instances where thorough investigations have been carried out, it has become apparent that these genes play a key role in viral replication. Consequently, recombinant viruses mutated in or lacking these genes replicate poorly at the portal of entry and during reactivation from latent phase.
The major focus of investigations into genes controlling the establishment or maintenance of latency has been the LATs, a family of transcripts arising from the inverted repeats flanking the unique long (UL) sequence. The full-length 8.3-kb transcript accumulates at low levels in latently infected neurons, while 2.0- and 1.5-kb introns processed from the full-length transcript are abundant (10, 25, 36, 47, 52, 55, 57, 58, 63). These introns are highly stable and appear to be lariat structures (14, 48, 60, 62). Viruses with LATs deleted have been reported to establish latency at levels within a threefold range of the wild type (3, 56). Deletion of LATs reduces the capacity of the virus to cause productive infections in the mouse and reduces the capacity of the virus to replicate following explanation of the neurons (2, 18, 22, 31, 50, 51, 54). The region of LAT associated with decreased reactivation has been mapped to a 348-bp sequence in the 5′ end (3, 19). LAT has not been shown to express ORFs. A recent report indicated that sequences containing the LAT introns can protect neurons from apoptosis and that a virus with LAT deleted induces apoptosis in rabbit trigeminal ganglia at higher levels than the wild-type virus (39). Thus, at least one function of LAT may be to promote neuronal survival during the maintenance of latent infection. Other studies have suggested that viral functions that repress lytic gene expression in vivo reside within the LAT domain (6, 16). The effectors of these functions are not identified. Irrespective of the final determination of the functions of LATs, the necessary conclusion is that LATs play a role in the maintenance of the latent state rather than in its establishment.
In earlier studies, we reported that the domain of the inverted repeats represented in LATs contains 16 ORFs of greater than 50 codons and that at least two of these, ORF O and ORF P, are expressed (27, 43). ORF O and ORF P are located in the 3′ domain of the LAT domain, almost entirely antisense to the γ134.5 gene. They are expressed from a promoter and associated RNA internal to and 3′ coterminal with LAT (4, 61). This transcript is completely repressed during productive infection by the binding of ICP4, the major viral transactivator/repressor, to a consensus ICP4 binding site that straddles the ORF P transcriptional initiation site (13, 26, 28, 29, 33–35, 42, 45, 46, 61). ORF O and ORF P are expressed only under conditions in which ICP4 repression is nonfunctional, i.e., in a virus containing a mutated ICP4 binding site or during infection and maintenance at 39.5°C, the nonpermissive temperature for ICP4, in HSV-1(F) and other limited-passage clinical isolates (11, 12). The repression of ORF O by the ICP4 binding site was surprising since ORF O was predicted to begin upstream of the ORF P transcript. Analyses of the products encoded by the ORFs showed that the translation of ORF O begins at the ORF P initiator methionine codon and then shifts into the ORF O reading frame before the amino acid 35 codon of ORF P (43). Thus, ORF O and ORF P are expressed under identical conditions and have not been detected during productive infection.
Investigations into the functions of ORF O and ORF P have revealed the following. (i) ORF P transcription is sufficient to preclude expression of the antisense γ134.5 gene and attenuates virulence (29, 42). (ii) ORF P protein inhibits the expression of ICP0 and ICP22. This correlates with an interference in the splicing of mRNAs inasmuch as (a) ORF P interacts and colocalizes with splicing factors (5); (b) a virus with ORF O and ORF P expression derepressed accumulates significantly less ICP0 and ICP22, which are translated from spliced mRNAs, while the levels of two proteins synthesized from intronless mRNAs, ICP4 and ICP27, are unchanged (5, 42); (c) ORF P protein expression is required for the inhibition of ICP0 and ICP22 expression (42); and (d) ORF P derepression alters the accumulation of the spliced LAT (30). (iii) ORF O protein specifically binds to and inhibits in vitro binding of ICP4 to its cognate site (43). ICP0 is a promiscuous transactivator required for efficient expression of viral genes. ICP4, the major viral transcriptional regulator, is required for the expression of β and γ genes. ICP22 positively regulates the expression of a subset of α and γ genes (reviewed in reference 50). Accordingly, ORF O and ORF P interfere with the expression of multiple α-regulatory genes which promote lytic gene expression.
It has been predicted that genes that are involved in the establishment of latency (i) are located within the domain transcribed during latency, (ii) encode functions which inhibit the lytic gene expression program, and (iii) are repressed during productive expression, as they encode genes detrimental to viral replication. ORF O and ORF P fulfill these criteria and as such are excellent candidates for genes which regulate the establishment of latency (5, 27–29, 42, 43). Multiple studies have implicated the ORF O/ORF P/γ134.5 domain as important in the latent life cycle. Deletions in this region result in decreased establishment of and reactivation from latency (37, 38, 53, 59). However, since γ134.5 is required for virulence, these deletions also result in reduced viral replication (7–9, 17, 59). Fewer viruses reach the trigeminal neurons from peripheral sites, and the amount of latent virus is consequently reduced. A recombinant virus expressing a truncated ORF P protein and an ORF O protein containing a one amino acid substitution was reported to reactivate from latency with frequencies similar to that of the wild type (30). The amount of established latent virus was not examined. The contribution of ORF O and ORF P to the establishment of latency remains unknown.
The goal of this study was to address the significance of ORF O and ORF P in the virus life cycle without interfering with the expression of the antisense γ134.5 gene. ORF P encodes only two methionines, the initiator and one located eight amino acids from the C terminus. Previous studies have shown that mutation of the initiator methionine in the context of the ORF P-derepressed virus is sufficient to prevent ORF P protein translation (42). We have previously shown that ORF O may begin at the ORF P initiator methionine codon, such that mutation of this codon would also prevent ORF O translation (43). Two recombinant viruses were constructed, (i) R7538(P−/O−), which contains the initiator methionine codon mutation, and (ii) R7543(P−/O−)R, in which the mutation was repaired to wild type. R7538(P−/O−) does not express ORF O and ORF P proteins but does express γ134.5 at wild-type levels. We report that mutants lacking the capacity to synthesize ORF O and ORF P proteins were not affected in their ability to replicate in cell culture, mouse central nervous system, mouse eye, or mouse trigeminal ganglia. In the mouse, they established latency but at a reduced copy number per cell.
MATERIALS AND METHODS
Cells and viruses.
Rabbit skin and 143tk− cells were originally obtained from J. McClaren and Carlo Croce, respectively. Vero cells were from the American Type Culture Collection. HSV-1(F) is the prototype HSV-1 strain used in this laboratory; as is the case with fresh HSV-1 isolates with limited history of replication outside the human host, the α4 gene of HSV-1(F) is temperature sensitive and does not repress itself or ORF P at 39.5°C (12, 27). The recombinant virus R3659 has been previously described (28). It lacks the SacI-BglII sequence of the BamHI Q fragment encoding the thymidine kinase (TK) and UL24 genes. A sequence consisting of the coding domain of the TK gene under the control of the α27 promoter (40) replaced the BstEII-StuI sequence of the BamHI S fragment containing the γ134.5 and ORF P genes (Fig. 1, line 4).
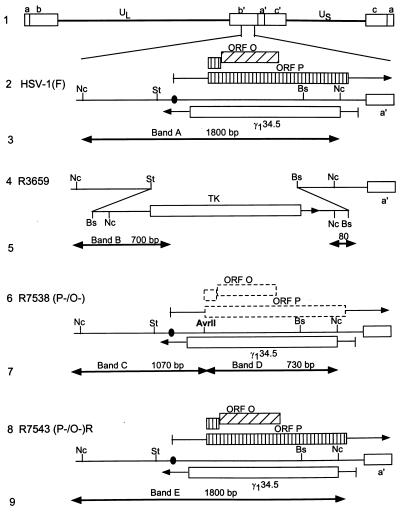
FIG. 1.
Schematic representations of sequence arrangements in the recombinant viruses used in these studies. (Line 1) Representation of the HSV-1(F) genome. Shown are the UL and unique short (US) sequences, which are flanked by inverted repeats c, a, and b and b′, a′, and c′. (Line 2) Domains of the ORF O, ORF P, and γ134.5 genes in the inverted repeat sequence b′ a′ flanking the UL sequences. The coding domains (boxes) and transcripts (lines with arrows denoting transcription direction) of the ORF O, ORF P, and γ134.5 genes are shown. Solid circle, wild-type ICP4 binding site. (Line 4) Sequence arrangement of the relevant domains of R3659. The StuI-BstEII sequences containing ORF P and γ134.5 were replaced in both repeats by the chimeric α27-tk gene. (Line 6) Sequence arrangement of the relevant domains of recombinant R7538(P−/O−). The α27-tk gene of R3659 was replaced with sequences containing a mutated ORF P initiation methionine codon introducing a diagnostic AvrII endonuclease site. (Line 8) Sequence arrangement of the relevant region of recombinant R7543(P−/O−)R. The mutations in the ORF O/P domain of R7538(P−/O−) were repaired by transfection with the NcoI fragment from the HSV-1(F) BamHI S fragment. (Lines 3, 5, 7, and 9) Expected sizes of fragments detected by hybridization of the 1,800-bp NcoI fragment with electrophoretically separated digests of viral DNAs with NcoI-AvrII, diagnostic of the replacement of the initiator methionine. Arrows, restriction cleavage sites present in the respective viruses and therefore fragment boundaries. HSV-1(F) DNA would be expected to yield band A, R3659 DNA would be expected to yield band B, R7538(P−/O−) DNA would be expected to yield bands C and D, and R7543(P−/O−)R DNA would be expected to yield band E. Abbreviations: St, StuI; Nc, NcoI; Bs, BstEII.
Antibodies.
Rabbit polyclonal antisera specific for γ134.5, BR4 (1), and rabbit polyclonal antisera specific for ORF O (43) have been described previously.
Plasmids.
pRB4930 contains the 830-bp NotI fragment of BamHI S cloned into the NotI site of pUC19. Plasmid pBR4929 contains a mutant ORF P initiator methionine codon and was made by insertion of a 160-bp PCR product into the SrfI-DraIII sites of pRB4930. The PCR primers were 5′-ACGGGCCTCGGGCCCTAGGCACGGCCCGATAACCGCCTCGGCCTC and 5′-GAGGCCGAGGCGGTTATCGGGCCGTGCCTAGGGCCCGAGGCCCGT. Underlined bases represent mutations that replace the ORF P initiator methionine codon (ATG) with an isoleucine codon (ATA) and create a diagnostic AvrII restriction site 15 bp upstream. Plasmid pRB4929 was used to construct the recombinant virus R7537. Plasmid pRB103, containing the BamHI Q fragment, was used to repair the deletion in the TK gene of R7537, resulting in the recombinant virus R7538. pRB4794 has been described elsewhere (28). It contains the 1,800-bp NcoI fragment of BamHI S, spanning the region between the start codons of the α0 and γ134.5 genes. It was used as a probe for analyses of recombinant viral DNA and was used to repair the mutations in the ORF P domain of R7538 resulting in the recombinant virus R7543.
Construction of recombinant viruses.
Viral stocks and titrations of viruses were done in Vero cells (American Type Culture Collection). R7537 was constructed by cotransfection of rabbit skin cells (originally obtained from J. McClaren) with pRB4929, which contains the ORF P initiator methionine codon mutation, with R3659 viral DNA (Fig. 1, lane 4) which contains a deletion in the TK gene, and with an α27 promoter-driven TK gene replacement of the 1,100-bp StuI-NcoI fragment encoding the ORF P and γ134.5 genes. TK− viruses were selected by plating the progeny of the cotransfection on 143TK− cells overlaid with Dulbecco modified Eagle medium containing 5% newborn calf serum and 40 μg of bromodeoxyuridine per ml of medium. Plaque-purified stocks were prepared as described elsewhere (41). Viral DNA was isolated from infected cells and purified on a 5 to 20% potassium acetate gradient as described elsewhere (27). Viral DNAs from single plaques were analyzed for the presence of novel AvrII endonuclease restriction sites diagnostic of the initiator methionine codon mutation. The TK gene of R7537 was repaired by plating the progeny of the cotransfection of rabbit skin cells with R7537 DNA and pBR103, which contains the BamHI Q fragment, on 143TK− cells overlaid in Dulbecco modified Eagle medium containing 5% fetal bovine serum, hypoxanthine, aminopterin, and thymidine. Virus was isolated, purified, and analyzed as described above. This process resulted in the isolation of R7538, which contains a wild-type BamHI Q fragment and the ORF P initiator methionine codon mutation. The mutations in the ORF P gene of R7538 were repaired by cotransfection of rabbit skin cells with R7538 viral DNA and pRB4794, which contains the 1,800-bp NcoI fragment from BamHI S of HSV-1(F). Plaques were isolated, and viral DNA was analyzed for a wild-type restriction endonuclease pattern indicating the absence of the introduced EcoRI and AvrII sites within BamHI S. R7543 has a wild-type restriction endonuclease pattern and therefore has a wild-type genotype.
Analyses of viral DNAs.
Viral DNAs were digested with appropriate restriction enzymes as detailed in the legend to Fig. 1. They were then subjected to electrophoresis on a 28-cm-long, 0.85% agarose gel and transferred to a Zeta probe (Bio-Rad, Richmond, Calif.) by capillary action in 0.5 M NaOH. The membrane was rinsed in 2× SSC (0.3 M NaCl plus 0.015 M sodium citrate) and prehybridized in 30% formamide–6× SSC–1% milk–1% sodium dodecyl sulfate (SDS)–100 μg of single-stranded calf thymus DNA per ml for 30 min at 68°C. Denatured, 32P-labeled pRB4794 (106 cpm) was then added overnight, and the blot was rinsed as recommended by the manufacturer. Autoradiographic images were obtained on Kodak XAR5 film.
Immunoblots.
Immunoblots were done as previously described (27). Briefly, infected cells were scraped into phosphate-buffered saline (PBS), pelleted under low-speed centrifugation, resuspended in disruption buffer containing 0.7 M β-mercaptoethanol, 2% SDS, 50 mM Tris, and 2.75% sucrose, sonicated briefly, boiled, and electrophoretically separated on a denaturing polyacrylamide gel cross-linked with N,N′-diallyltartardiamide (Bio-Rad). The electrophoretically separated, denatured proteins were electrically transferred to a nitrocellulose sheet, blocked, reacted with the appropriate antiserum, rinsed, and reacted with either goat anti-rabbit immunoglobulin G (IgG) conjugated to alkaline phosphatase for rabbit polyclonal antisera or goat anti-mouse IgG conjugated to alkaline phosphatase for mouse monoclonal antisera. Proteins were then rinsed again and developed as recommended by the manufacturer (Bio-Rad).
Viral replication in cell culture.
Triplicate 25-cm2 plaque dishes containing Vero or SK-N-SH cells were infected with 5 PFU of HSV-1(F), R7538(P−/O−), or R7543(P−/O−)R. Cells were harvested at 6, 12, or 24 h, washed in PBS, resuspended in 1 ml of sterile milk, taken through three freeze-thaw cycles, sonicated, and used to infect Vero cells in 25-cm2 plaque dishes at 10-fold dilutions. Cells were maintained in 199“O” medium, and plaques were counted 2 days after infection.
Intracranial inoculation of mice.
CBA/J mice (3.5 weeks old) from Jackson Laboratory were anesthetized with pentobarbital sodium (Nembutal) and injected intracerebrally with 10-fold serial dilutions of virus, seven mice per dilution. Mice were monitored daily; mortality from 2 to 21 days after infection was attributed to the inoculated virus. The 50% lethal dose (LD50) ratios were calculated by the method of Reed and Muench (44).
Assays of viral replication in murine eye and trigeminal ganglia.
CBA/J mice from Jackson Laboratory, 4.5 weeks of age, were anesthetized and inoculated with 10 μl (5 × 105 PFU) of virus on scarified corneas as previously described (29). Mice were sacrificed 1, 3, 5, and 7 days after infection; the eyes or trigeminal ganglia were removed, placed in 1 ml of 199V containing nystatin, homogenized in a mechanical tissue grinder, and plated on Vero cells (6 eyes or 20 ganglia per virus for each time point). Plaques were counted 2 days after being seeded on Vero cells.
Viral reactivation in murine trigeminal ganglia.
CBA/J mice from Jackson Laboratory, 4.5 weeks of age, were anesthetized and inoculated with 10 μl of virus on scarified corneas as described above. Mice were sacrificed at 30 days after infection, and the trigeminal ganglia were removed, placed in 1 ml of 199V containing nystatin, and incubated for 5 days. The ganglia were then homogenized in a mechanical tissue grinder and plated on Vero cells (28 ganglia per virus). Cytopathic effect (CPE) was monitored daily for 8 days after cocultivation.
Quantitation of latent virus.
CBA/J mice (4.5 weeks old) were ocularly infected as described above. At 30 days after infection, 26 trigeminal ganglia per virus were removed, flash frozen, and stored at −80°C. Quantitative PCR was performed as previously described (23, 24). Briefly, ganglia were homogenized in 1 ml of solubilization solution (5 M guanidine thiocyanate, 50 mM Tris [pH 7.5], 10 mM EDTA, 5% β-mercaptoethanol) using a Dounce homogenizer mechanical tissue grinder. DNA was precipitated, resuspended in PCR buffer, and digested with 0.2 μg of proteinase K/ml at 55°C for 2 h, 80°C for 20 min, and 95°C for 5 min. One hundred nanograms of DNA was used for PCR with the following primers specific for the viral TK gene or the cellular β-actin gene: TK-1, 5′-CTTAACAGCGTCAACAGCGTGCCG; TK-2, 5′-CAAAGAGGTGCGGGAGT; Act-1, 5′-AACCCTAAGGCCAACCGTGAAAAGATGACC; Act-2, 5′-CCAGGGAGGAAGAGGATGCGGC. PCR was performed under previously described conditions (24). TK products were amplified for 30 cycles at 95°C for 1 min, 55°C for 1 min, and 72°C for 2 min, with a final extension of 5 min at 72°C. Actin PCR conditions were as above except that the annealing temperature was 60°C. Aliquots of the PCR mixture were separated by nondenaturing polyacrylamide gel electrophoresis, electroblotted to a Zeta probe nylon membrane, denatured, and probed with the following 32P-labeled oligonucleotides internal to the PCR primers: TK-3, 5′-CAGATCTTGGTGGCGTG; Act-3, 5′-GCTCTAGACTTCGAGCAGGAGATGGCCACT. The labeled probes were hybridized overnight at 50°C in 5× SSC–7% SDS–1× Denhardt's solution–25 mM sodium phosphate, pH 7.2, and washed as recommended by the manufacturer. The levels of amplified TK product were quantitated with a Storm phosphorimager and compared with a linear standard curve generated by PCR of 100 ng of uninfected murine trigeminal ganglion DNA spiked with 10-fold dilutions of purified HSV-1(F) DNA. The values were normalized for DNA content by comparison of the amplified β-actin product.
RESULTS
Construction and characterization of the recombinant virus R7538(P−/O−), containing a mutated ORF P initiator methionine codon, and the repaired recombinant virus, R7543(P−/O−)R.
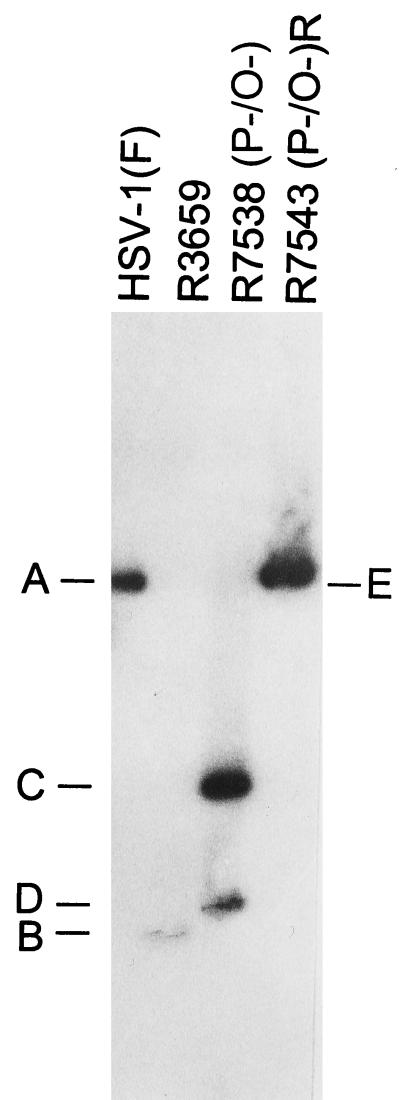
The procedures for the construction of recombinant viruses R7538(P−/O−) and R7543(P−/O−)R are described in Materials and Methods. R7538(P−/O−) contains two nucleotide substitutions, one at the initiator methionine codon (ATG→ATA) and one 15 bp upstream creating a unique AvrII restriction endonuclease site (CCCAGG→CCTAGG). The mutation creating the novel AvrII restriction endonuclease site also introduced a TAG stop codon into the predicted ORF O gene, upstream of the ORF P initiator methionine codon. Both mutations are in wobble codons of γ134.5, and as such they do not alter the amino acid sequence of the γ134.5 protein. The genotype of R7538(P−/O−) was verified by the presence of the unique AvrII restriction endonuclease site diagnostic of the ORF P initiator methionine codon mutation, the location of which is shown in Fig. 1, line 6. R7538(P−/O−) DNA was purified and incubated with restriction endonucleases NcoI and AvrII, electrophoretically separated on 0.85% agarose gels, transferred to Zeta probe membranes, and hybridized with 32P-labeled pRB4794, which contains the 1,800-bp NcoI fragment of BamHI S (Fig. 2, lane 3). Restriction endonuclease cleavage of R7538(P−/O−) with NcoI and AvrII resulted in the predicted 1,070- and 730-bp DNA fragments (Fig. 1, line 7, and Fig. 2, bands C and D), verifying the presence of the ORF P initiator methionine codon mutation. This pattern is distinct from those of the 1,800-bp fragment of HSV-1(F) (Fig. 1, line 3, and Fig. 2, band A) and the 700-bp DNA fragment of the parental virus R3659 (Fig. 1, line 5, and Fig. 2, band B).
FIG. 2.
Autoradiographic image of electrophoretically separated viral DNA fragments containing sequences in the domain of the ORF O, ORF P, and γ134.5 genes. Viral DNAs were digested with NcoI and AvrII, electrophoretically separated on a 0.85% agarose gel, transferred to a Zeta probe membrane, hybridized to radiolabeled DNA of plasmid pRB4794 containing the 1,800-bp NcoI fragment of the BamHI S fragment (28), and exposed to Kodak XAR5 film. The expected sizes of the fragments generated by cleavages (bands A through E) are shown in Fig. 1.
As is necessary in all cases in which the phenotypes of recombinant viruses are tested, the mutations were repaired by cotransfection of rabbit skin cells with R7538(P−/O−) and pRB4794 (described above). Viral DNAs from plaque isolates of progeny virus were screened for the wild-type restriction pattern. Incubation of R7543(P−/O−)R with NcoI and AvrII resulted in the wild-type 1,800-bp band (Fig. 1, line 9, and Fig. 2, band E), indicating that the introduced mutations were replaced with wild-type sequences.
Expression of ORF O and γ134.5 proteins in cells infected with HSV-1(F), R7538(P−/O−), and R7543(P−/O−)R recombinant viruses.
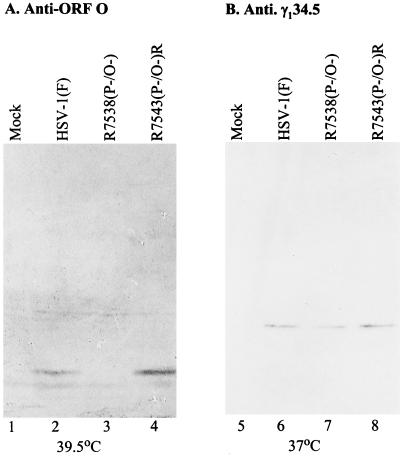
The ORF P initiator methionine codon mutation previously was shown to prevent ORF P translation from a derepressed ORF P transcript (42). The effect of the mutations on ORF O expression was unknown. Therefore, cells in replicate 25-cm2 flasks were mock infected or infected (10 PFU/cell) with HSV-1(F), R7538(P−/O−), or R7543(P−/O−)R at 4°C and maintained at 39.5°C, the nonpermissive temperature for ICP4. After 20 h the cells were harvested, lysed by sonication in disruption buffer, boiled for 5 min, subjected to electrophoresis in denaturing polyacrylamide gels, transferred to a nitrocellulose sheet, and reacted with polyclonal antiserum specific for ORF O. As shown in Fig. 3A, ORF O accumulated in cells infected with either HSV-1(F) or the repair virus R7543(P−/O−)R and maintained at the nonpermissive temperature for ICP4. ORF O was not detected in R7538(P−/O−), indicating that the introduced mutations are sufficient to prevent ORF O expression.
FIG. 3.
Photograph of infected-cell proteins electrophoretically separated on a denaturing polyacrylamide gel and reacted with polyclonal rabbit antisera specific for ORF O (A) or γ134.5 (B). Replicate Vero cell cultures grown in 25-cm2 flasks were infected with 10 PFU of HSV-1(F), R7538(P−/O−), or R7543(P−/O−)R per cell; maintained at either 39.5 (A) or 37°C (B) for 18 h; harvested and solubilized; electrophoretically separated on 12.5% polyacrylamide denaturing gels; and transferred to a nitrocellulose sheet. The sheets were reacted with rabbit polyclonal antiserum anti-ORF O (43) (A) or BR4 (1) or anti-γ134.5 (B) and then with goat anti-rabbit IgG conjugated to alkaline phosphatase and alkaline phosphatase substrate.
Since the mutations introduced into ORF O and ORF P are also in the γ134.5 gene, we verified that γ134.5 expression in the recombinant viruses was comparable to that in the wild-type virus. Cells in replicate 25-cm2 flasks were mock infected or infected with 10 PFU of HSV-1(F), R7538(P−/O−), and R7543(P−/O−)R and maintained at 37°C. Infected-cell lysates were processed as described above and reacted with polyclonal antiserum BR4, specific for γ134.5. As shown in Fig. 3B, the γ134.5 protein was present in all infected lysates at comparable levels. The results of Fig. 3 show that, in addition to precluding ORF P protein translation, the mutations introduced into R7538(P−/O−) prevent ORF O synthesis without affecting the expression of the antisense γ134.5 gene.
Replication of wild-type and mutant viruses in cell culture and in vivo.
The replication competence of recombinant viruses was tested in four series of experiments involving (i) the production of infectious progeny in cell culture, (ii) determination of the neurovirulence of the recombinant viruses compared to that of the wild type, (iii) the isolation of infectious virus from murine eyes, and (iv) the isolation of infectious virus from murine trigeminal ganglia. In the first series of experiments, replicate 25-cm2 cultures of Vero or SK-N-SH cells were infected with 5 PFU of HSV-1(F) or R7538(P−/O−) per cell. Infected cells were harvested at 6, 12, and 24 h after infection. As shown in Table 1, the yields of HSV-1(F) and R7538(P−/O−) viruses were comparable in Vero cells and in SK-N-SH cells. Thus, the absence of ORF O and ORF P protein synthesis did not affect viral growth in cells of neuronal and nonneuronal origin.
TABLE 1.
Replication of HSV-1(F) and R7538(P−/O−) in Vero and SK-N-SH cells
| Virus | Virus yielda (log10 PFU) from:
|
|||||
|---|---|---|---|---|---|---|
| Vero cells at:
|
SK-N-SH cells at:
|
|||||
| 6 h | 12 h | 24 h | 6 h | 12 h | 24 h | |
| HSV-1(F) | 4.9 | 6.8 | 7.8 | 4.8 | 7.4 | 8.0 |
| R7538(P−/O−) | 4.9 | 6.9 | 7.9 | 4.8 | 7.5 | 8.1 |
Virus yields from Vero and SK-N-SH cells infected with 5 PFU of HSV-1(F) or R7538(P−/O−) per cell and harvested at 6, 12, and 24 h after infection.
In the second series of experiments, the neurovirulence of HSV-1(F), R7538(P−/O−), and R7543(P−/O−)R was tested. Neurovirulence represents the capacity of HSV to replicate in and destroy the central nervous system and, as such, is frequently used to assess the replication competence of recombinant viruses in vivo. Four-week-old CBA/J mice were inoculated intracranially with 10-fold dilutions of HSV-1(F), R7538(P−/O−), or R7543(P−/O−)R, seven mice per dilution, as described in Materials and Methods. Mice were monitored daily, and mortality was recorded from days 2 to 21 after infection. The PFU/LD50 ratios for all viruses tested were within a 2.5-fold range. HSV-1(F) and R7543(P−/O−) had PFU/LD50 ratios of 63 and 60, respectively, while the value for R7538(P−/O−) was 164. Thus, the absence of ORF O and ORF P protein synthesis in cells infected with R7538(P−/O−) did not significantly affect the ability of the virus to replicate in cell culture or the murine central nervous system.
In the third series of experiments, 4.5-week-old CBA/J mice were infected with 5 × 105 PFU of HSV-1(F), R7538(P−/O−), or R7543(P−/O−)R per eye. At days 1, 3, 5, and 7 after infection, three mice per group were anesthetized and sacrificed and the eyes were removed, washed, homogenized in 1 ml of 199V with nystatin using a mechanical tissue grinder, and frozen. Vero cells in replicate 25-cm2 flasks were infected with 10-fold dilutions of the respective virus, maintained in the presence of HSV-1 neutralizing antibody, and plaque titers were determined 2 days after infection. As shown in Table 2, the amounts of infectious R7538(P−/O−), HSV-1(F), and R7543(P−/O−)R isolated from eyes following ocular infection generally fell within a threefold range. The amounts of infectious R7538(P−/O−) were not statistically different from those of HSV-1(F) or R7543(P−/O−)R at any of the times tested (P > 0.05). Thus, the relative amounts of infectious wild-type and recombinant viruses isolated from the site of inoculation were indistinguishable.
TABLE 2.
Isolation of infectious HSV-1(F) or of recombinant viruses R7538(P−/O−) and R7543(P−/O−)R from murine eye
| Virus | PFU/eye (100)a on day:
|
|||
|---|---|---|---|---|
| 1 | 3 | 5 | 7 | |
| HSV-1(F) | 880 ± 230 | 16 ± 6.6 | 3.3 ± 1.5 | 2.1 ± 1.2 |
| R7538(P−/O−) | 1,400 ± 490 | 11 ± 3.1 | 2.6 ± 0.8 | 0.4 ± 0.2 |
| R7543(P−/O−)R | 1,600 ± 770 | 28 ± 22 | 2.3 ± 1.8 | 1.6 ± 1.3 |
Average counts of plaques ± standard error appearing in flasks containing Vero cells, which were infected with 10-fold dilutions of eye homogenates (12 samples total per virus), harvested at the indicated days after ocular infection.
In the fourth series of experiments, 4.5-week-old CBA/J mice were infected with 5 × 105 PFU of HSV-1(F), R7538(P−/O−), or R7543(P−/O−)R per eye. At days 3 and 5 after infection, the trigeminal ganglia were removed and virus was titered as described above. As shown in Table 3, the amounts of infectious R7538(P−/O−), HSV-1(F), and R7543(P−/O−)R isolated from ganglia 3 days after ocular infection fell within a threefold range (9.0 × 103, 2.7 × 104, and 2.9 × 104 PFU per ganglion, respectively). At day 5, the level of infectious R7538(P−/O−) was 2 to 3 log units less than that of HSV-1(F) or the repair virus (3.2 × 102, 1.4 × 105, and 7.7 × 104 PFU per ganglion, respectively). It is unclear whether the decrease in infectious R7538(P−/O−) virus at day 5 reflects an alteration in the efficiency of the establishment of latency. We conclude that at day 3, a time at which acute infection and the establishment of latency. We conclude that at day 3, a time at which acute infection and the establishment of latency are occurring in mice, infectious R7538(P−/O−) was present in trigeminal ganglia at levels comparable to those of HSV-1(F) and the repair virus.
TABLE 3.
Isolation of infectious HSV-1(F) or of recombinant viruses R7538(P−/O−) and R7543(P−/O−)R from murine trigeminal ganglia
| Virus | PFU/ganglion (1,000)a on day:
|
|
|---|---|---|
| 3 | 5 | |
| HSV-1(F) | 27 ± 4.2 | 138 ± 23 |
| R7538(P−/O−) | 9.0 ± 2.3 | 0.32 ± 0.12 |
| R7543(P−/O−)R | 29 ± 9.3 | 77 ± 16 |
Average counts of plaques ± standard error appearing in flasks containing Vero cells, which were infected with 10-fold dilutions of trigeminal ganglion homogenates (20 ganglia total per virus), harvested at the indicated days after ocular infection.
Reactivation from latency in mice infected with HSV-1(F), R7538(P−/O−), and R7543(P−/O−)R.
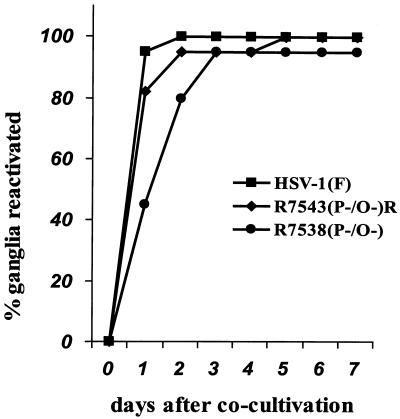
The significance of ORF O and ORF P in the latent life cycle was assessed by measuring the reactivation of wild-type and recombinant viruses. CBA/J mice (4.5 weeks old) were infected by ocular scarification with 5 × 105 PFU of HSV-1(F), R7538(P−/O−), or R7543(P−/O−)R per eye. After 30 days, a time at which replicating HSV-1 or R7538(P−/O−) could not be isolated in this animal model (reviewed in reference 15) (data not shown), mice were anesthetized; the trigeminal ganglia were removed, incubated for 5 days in 199V plus nystatin, homogenized, and cocultivated in replicate 25-cm2 flasks containing Vero cells; and CPE was scored for each ganglion. Figure 4 shows the percentage of ganglia that produced reactivated virus versus the day after cocultivation. Both HSV-1(F)- and R7543(P−/O−)R-infected ganglia reactivated virus in 100% of the samples, with 90% showing obvious CPE 1 day after cocultivation. R7538(P−/O−)-infected ganglia reactivated virus in 95% of samples; however, CPE in 90% of the ganglia was not achieved until 3 days after cocultivation. This likely reflects smaller amounts of reactivated virus. The data indicate that ORF O and ORF P proteins are not required for reactivation from latency, but a virus precluded from synthesizing ORF O and ORF P proteins reactivated with reduced kinetics.
FIG. 4.
Graph plotting the percentage of latently infected murine ganglia reactivating HSV-1(F), R7538(P−/O−), or R7543(P−/O−)R versus days after cocultivation. CBA/J mice (4.5 weeks old) were anesthetized, ocularly scarified, and infected with 106 PFU of HSV-1(F), R7538(P−/O−), or R7543(P−/O−)R. After 30 days, mice were sacrificed, and the trigeminal ganglia were removed, incubated in 1 ml of 199V medium plus nystatin for 5 days at 37°C, homogenized, and used to infect 25-cm2 plaque dishes containing Vero cells. CPE was monitored daily for 8 days. The graph represents the percentage of ganglia producing infectious virus versus days after cocultivation.
Since the regulation and functions of ORF O and ORF P imply a role for these genes in the establishment of latency, it seemed likely that the decreased levels of reactivated virus correspond to a decrease in the amount of virus which has established latency. To test this hypothesis, a quantitative PCR analysis was employed to calculate the number of latent viral DNA copies, as previously described (23, 24). CBA/J mice (4.5 weeks old) were ocularly infected as described above, and ganglia were harvested after 30 days and flash frozen. Ganglia were homogenized in 1 ml of DNA extraction buffer, precipitated, washed, resuspended, and treated with proteinase K as described in Materials and Methods. DNA (100 ng) from each sample was subjected to PCR using primers specific for the viral TK gene or the murine β-actin gene (24). Aliquots of the PCR mixture were separated by nondenaturing polyacrylamide gel electrophoresis, electroblotted to a nylon membrane, denatured, and probed with 32P-labeled oligonucleotides internal to the PCR primers. The levels of amplified TK gene product were quantified with a Storm phosphorimager and compared with a linear standard curve generated by PCR of 100 ng of uninfected murine trigeminal ganglion DNA spiked with 10-fold dilutions of purified HSV-1(F) DNA. No amplified TK gene product was detected in uninfected murine trigeminal ganglia samples (data not shown). The values were normalized for DNA content by comparison of the amplified β-actin gene product. The results of these studies are shown in Table 4. Ganglia latently infected with R7538(P−/O−) contained 0.14 +/− 0.03 viral DNA copies per cell equivalent, whereas the HSV-1(F) latent DNA copy number was estimated to be sixfold higher, 0.84 copies per cell (P < 0.005). The amount of latent R7543(P−/O−)R was eightfold higher than the amount of R7538(P−/O−), 1.10 copies per cell (P < 0.0001). These results indicate that ORF O and ORF P proteins may play a role in the ultimate number of viral DNA copies maintained in the latent state.
TABLE 4.
Quantification of viral DNA in latently infected murine trigeminal ganglia
| Virus | No. of viral DNA copies/cella (no. of ganglia) | Pb |
|---|---|---|
| HSV-1(F) | 0.84 ± 0.17 (21) | <0.005 |
| R7538(P−/O−) | 0.14 ± 0.03 (26) | |
| R7543(P−/O−)R | 1.10 ± 0.21 (19) | <0.0001 |
Quantitation of PCR-amplified HSV-1 TK gene from 100 ng of purified trigeminal ganglion DNA. Values were extrapolated from a standard curve ranging from 0.0001 to 10 viral DNA copies per cell and normalized for DNA content by comparison to the PCR-amplified gene encoding murine β-actin. Values are averages ± standard error.
P values for R7538(P−/O−) latent DNA amounts compared with either HSV-1(F) or R7543(P−/O−)R DNA amounts.
DISCUSSION
A unique property of HSV is that it carries a large number of accessory genes designed in large part to control both the intracellular and extracellular environment in which it replicates. Latency is a significant mechanism for the perpetuation of HSV in human populations. Other herpesviruses, notably Epstein-Barr virus and other members of the gammaherpesvirus subfamily, have evolved elaborate mechanisms for the establishment of the latent state by virally encoded proteins. The presumption that HSV would also encode functions designed to facilitate the establishment of latency was the basis of the search that led to the identification of ORFs P and O.
ORF P and O and their products appear to be ideal candidates for the control of the latent state. Specifically, (i) ORF O and ORF P are located in the domain transcribed during latency, (ii) both ORFs are completely repressed during productive infection by ICP4, the major viral transcriptional transactivator/repressor, (iii) the ORF P protein inhibits the synthesis of the important α-regulatory proteins ICP0 and ICP22 in cells infected with ORF P-derepressed viruses, and (iv) the ORF O protein made under similar circumstances binds to and inhibits ICP4 binding to its cognate DNA site in vitro. The objective of the studies described in this report was to test the role of ORF O and ORF P in the establishment of latency in the mouse model in vivo.
To test the role of ORF O and ORF P proteins, we constructed the recombinant virus R7538(P−/O−), in which the initiator methionine codon of both coding sequences was mutated such that it would not affect the amino acid sequence of the product of the antisense γ134.5 gene. This virus contained two nucleotide substitutions, one mutating the ORF P initiator methionine codon and the other creating a unique restriction endonuclease site 15 bp upstream. These mutations were repaired in the recombinant virus R7543(P−/O−)R. The salient features of the results and the key conclusions derived from characterization of the recombinant viruses follow.
(i) The initiator methionine codon mutation was previously shown to preclude ORF P protein expression in the context of a virus with ORF P transcription derepressed (42). Characterization of R7538(P−/O−) showed that this mutation also prevents ORF O protein expression in cells infected and maintained at 39.5°C, the nonpermissive temperature for ICP4. The absence of ORF P and ORF O proteins is not surprising since earlier studies have shown that ORF O and ORF P proteins share the ORF P protein initiator methionine and then diverge within the first 34 amino acids of the ORF P protein (43). To preclude a potential low level of expression of ORF O from the single methionine codon in its own reading frame, the base substitution creating a unique restriction endonuclease site also introduced a TAG stop codon in the predicted ORF O frame upstream of the ORF P initiator methionine codon. γ134.5 protein expression and function in the mutant virus, as assayed by replication in neuronal cell lines and determination of neurovirulence, were the same as those in the wild type.
(ii) Consistent with their absence in productively infected cells, ORF O and ORF P proteins play no discernible role in viral replication in cell culture and in vivo. Specifically, R7538(P−/O−) replicated to wild-type levels in cell lines of neuronal and nonneuronal origin (SK-N-SH and Vero cells, respectively). Neurovirulence was similarly unaffected. HSV-1(F), R7538(P−/O−), and R7543(P−/O−)R all had PFU/LD50 ratios within a 2.5-fold range. Replication at a peripheral site was tested by the isolation of infectious virus from eyes following ocular infection. Differences in the amounts of infectious HSV-1(F), R7538(P−/O−), and R7543(P−/O−)R were not statistically significant at 1, 3, 5, and 7 days after infection. A fourth assay of replication competence was the recovery of infectious virus from ganglia at the peak of acute infection. After 3 days, the amounts of infectious wild-type and recombinant viruses fell within a threefold range. Thus at a time in which replication and the establishment of latency are occurring concurrently, similar levels of wild-type and recombinant viruses were present in the trigeminal ganglion. Interestingly, at 5 days after ocular infection, the level of infectious R7538(P−/O−) dropped 2 to 3 log units compared with the level of wild-type or repair virus. The mechanism responsible for this decrease is uncertain, but the observation itself does not support the hypothesis that ORF O and P proteins play a significant role in the establishment of latency.
(iii) R7538(P−/O−) virus appears to establish latent infections at reduced levels compared with those of the wild-type parent and repaired viruses. HSV-1(F) and R7543(P−/O−)R reactivated in 100% of infected ganglia with rapid kinetics. The virus with ORF O and ORF P mutated reactivated in 95% of infected ganglia; however, reactivation occurred with reduced kinetics. The decrease in the amount of virus recovered after explanation of trigeminal ganglia correlated with the six- to eightfold-lower numbers of copies of viral DNA/cell in trigeminal ganglia harboring latent R7538(P−/O−) than in those harboring wild-type or repaired viruses.
The results presented here suggest that ORF O and ORF P proteins may play a role but are not essential for the establishment of latency in the mouse model. One interpretation of our results is that the establishment of latency is a multifactorial event involving several gene products both inside and outside of the HSV LAT domain and that each of them contributes to the switching off of replicative functions in dorsal root neurons. An alternative hypothesis is that ORF P and ORF O are effective in the maintenance phase rather than the establishment phase of latency. For example, it is conceivable that the reduction in the DNA copy number reflects a loss of neurons harboring virus due to reactivation of the latent virus and consequent destruction of neurons harboring them rather than a reduction in the number of neurons in which latent infections had been established. Sorting out this role of ORF P and O would require construction of viral mutants constitutively expressing ORF P and ORF O proteins. This is not an easy task since the expression of ORFs P and O and of the γ134.5 gene is mutually exclusive. The γ134.5 gene plays a key role in breaching host defenses against infection; failure to express ICP34.5 would significantly impair productive infection and the ability of the virus to establish latent infections. The solution to this problem remains to be found.
ACKNOWLEDGMENTS
We thank Lindsay Smith for expert technical assistance.
These studies were aided by grants from the National Cancer Institute (CA47451, CA71933, and CA78766) and the United States Public Health Service.
REFERENCES
- 1.Ackermann M, Chou J, Sarmiento M, Lerner R A, Roizman B. Identification by antibody to a synthetic peptide of a protein specified by a diploid gene located in the terminal repeats of the L component of herpes simplex virus genome. J Virol. 1986;58:843–850. doi: 10.1128/jvi.58.3.843-850.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Block T M, Deshmane S, Masonis J, Maggioncalda J, Valyi-Nagi T, Fraser N W. An HSV LAT null mutant reactivates slowly from latent infection and makes small plaques on CV-1 monolayers. Virology. 1993;192:618–630. doi: 10.1006/viro.1993.1078. [DOI] [PubMed] [Google Scholar]
- 3.Bloom D C, Devi-Rao G, Wagner E K, Feldman L T, Stevens J G. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus 1 reactivation. J Virol. 1996;70:2449–2459. doi: 10.1128/jvi.70.4.2449-2459.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohenzky R A, Papavassiliou A G, Gelman I H, Silverstein S. Identification of a promoter mapping within the reiterated sequences that flank the herpes simplex virus type 1 UL region. J Virol. 1993;67:632–642. doi: 10.1128/jvi.67.2.632-642.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruni R, Roizman B. Open reading frame P—a herpes simplex virus gene repressed during productive infection encodes a protein that binds a splicing factor and reduces synthesis of viral proteins made from spliced mRNA. Proc Natl Acad Sci USA. 1996;93:10423–10427. doi: 10.1073/pnas.93.19.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S H, Kramer M F, Schaffer P A, Coen D M. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J Virol. 1997;71:5578–5584. doi: 10.1128/jvi.71.8.5878-5884.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou J, Roizman B. The terminal a sequence of herpes simplex virus genome contains the promoter of a gene located in the repeat sequences of the L component. J Virol. 1986;57:629–637. doi: 10.1128/jvi.57.2.629-637.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou J, Roizman B. The γ134.5 gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programmed cell death in neuronal cells. Proc Natl Acad Sci USA. 1992;89:3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou J, Kern E R, Whitley R J, Roizman B. Mapping of herpes simplex virus-1 neurovirulence to γ134.5, a gene nonessential for growth in culture. Science. 1990;250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 10.Devi-Rao G B, Goodart S A, Hecht L M, Rochford R, Rice M K, Wagner E K. Relationship between polyadenylated and nonpolyadenylated herpes simplex virus type 1 latency-associated transcripts. J Virol. 1991;65:2179–2190. doi: 10.1128/jvi.65.5.2179-2190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon R A F, Schaffer P A. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate early protein VP175. J Virol. 1980;36:189–203. doi: 10.1128/jvi.36.1.189-203.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ejercito P M, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J Gen Virol. 1967;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 13.Faber S W, Wilcox K W. Association of the herpes simplex virus regulatory protein ICP4 with specific nucleotide sequences in DNA. Nucleic Acids Res. 1986;14:6067–6083. doi: 10.1093/nar/14.15.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrell M J, Dobson A T, Feldman L T. Herpes simplex virus latency-associated transcript is a stable intron. Proc Natl Acad Sci USA. 1991;88:790–794. doi: 10.1073/pnas.88.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fawl R L, Roizman B. The molecular basis of herpes simplex virus pathogenicity. Semin Virol. 1994;5:261–271. [Google Scholar]
- 16.Garber D A, Schaffer P A, Knipe D M. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J Virol. 1997;71:5885–5893. doi: 10.1128/jvi.71.8.5885-5893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He B, Gross M, Roizman B. The γ134.5 protein of herpes simplex virus I complexes with protein phosphatase 1α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill J M, Sedarati F, Javier R T, Wagner E K, Stevens J G. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology. 1990;174:117–125. doi: 10.1016/0042-6822(90)90060-5. [DOI] [PubMed] [Google Scholar]
- 19.Hill J M, Maggioncalda J B, Garza H H, Su Y H, Fraser N W, Block T M. In vivo epinephrine reactivation of ocular herpes simplex virus type 1 in the rabbit is correlated to a 370-base-pair region located between the promoter and the 5′ end of the 2.0-kilobase latency-associated transcript. J Virol. 1996;70:7270–7274. doi: 10.1128/jvi.70.10.7270-7274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc Natl Acad Sci USA. 1975;72:1276–1280. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Javier R T, Stevens J G, Dissette V B, Wagner E K. A herpes simplex virus transcript abundant in latently infected neurons is dispensable for establishment of the latent state. Virology. 1988;166:254–257. doi: 10.1016/0042-6822(88)90169-9. [DOI] [PubMed] [Google Scholar]
- 23.Katz J P, Bodin E T, Coen D M. Quantitative polymerase chain reaction analysis of herpes simplex virus DNA in ganglia infected with replication-incompetent mutants. J Virol. 1990;64:4288–4295. doi: 10.1128/jvi.64.9.4288-4295.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer M F, Coen D M. Quantification of transcripts from the ICP4 and thymidine kinase genes in mouse ganglia latently infected with herpes simplex virus. J Virol. 1995;69:1389–1399. doi: 10.1128/jvi.69.3.1389-1399.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krause P R, Croen K D, Straus S E, Ostrove J M. Detection and preliminary characterization of herpes simplex virus type 1 transcripts in latently infected human trigeminal ganglia. J Virol. 1988;62:4819–4823. doi: 10.1128/jvi.62.12.4819-4823.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kristie T, Roizman B. α4, the major regulatory protein of herpes simplex type 1, is stably and specifically associated with promoter-regulatory domains of α genes and of selected other viral genes. Proc Natl Acad Sci USA. 1986;83:3218–3222. doi: 10.1073/pnas.83.10.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagunoff M, Roizman B. Expression of a herpes simplex virus 1 open reading frame antisense to the γ134.5 gene and transcribed by an RNA 3′ coterminal with the unspliced latency-associated transcript. J Virol. 1994;68:6021–6028. doi: 10.1128/jvi.68.9.6021-6028.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagunoff M, Roizman B. The regulation of synthesis and properties of the protein product of open reading frame P of the herpes simplex virus 1 genome. J Virol. 1995;69:3615–3623. doi: 10.1128/jvi.69.6.3615-3623.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagunoff M, Randall G, Roizman B. Phenotypic properties of herpes simplex virus 1 containing a derepressed open reading frame P gene. J Virol. 1996;70:1810–1817. doi: 10.1128/jvi.70.3.1810-1817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee L Y, Schaffer P A. A virus with a mutation in the ICP4-binding site in the L/ST promoter of herpes simplex virus type 1, but not a virus with a mutation in open reading frame P, exhibits cell-type-specific expression of γ134.5 transcripts and latency-associated transcripts. J Virol. 1998;72:4250–4264. doi: 10.1128/jvi.72.5.4250-4264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leib D A, Bogard C L, Kosz-Vnenchak M, Hicks K A, Coen D M, Knipe D M, Schaffer P A. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J Virol. 1989;63:2893–2900. doi: 10.1128/jvi.63.7.2893-2900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leib D A, Coen D M, Bogard C L, Hicks K A, Yager O R, Knipe D M, Tyler K L, Schaffer P A. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759–768. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leopardi R, Michael N, Roizman B. Repression of the herpes simplex virus 1 α4 gene by its gene product (ICP4) within the context of the viral genome is conditioned by the distance and stereoaxial alignment of the ICP4 DNA binding site relative to the TATA box. J Virol. 1995;69:3042–3048. doi: 10.1128/jvi.69.5.3042-3048.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michael N, Roizman B. Repression of the herpes simplex virus 1 a4 gene by its gene product occurs within the context of the viral genome and is associated with all three identified cognate sites. Proc Natl Acad Sci USA. 1993;90:2286–2290. doi: 10.1073/pnas.90.6.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michael N, Spector D, Mavromara-Nazos P, Kristie T M, Roizman B. The DNA-binding properties of the major regulatory protein α4 of herpes simplex viruses. Science. 1988;239:1531–1534. doi: 10.1126/science.2832940. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell W J, Lirette R P, Fraser N W. Mapping of low abundance latency-associated RNA in the trigeminal ganglia a mice latently infected with herpes simplex virus type 1. J Gen Virol. 1990;71:125–132. doi: 10.1099/0022-1317-71-1-125. [DOI] [PubMed] [Google Scholar]
- 37.Perng G C, Thompson R L, Sawtell N M, Taylor W E, Slanina S M, Ghiasi H, Kaiwar R, Nesburn A B, Wechsler S L. An avirulent ICP34.5 deletion mutant of herpes simplex virus type 1 is capable of in vivo spontaneous reactivation. J Virol. 1995;69:3033–3041. doi: 10.1128/jvi.69.5.3033-3041.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perng G C, Ghiasi H, Slanina S M, Nesburn A B, Wechsler S L. High-dose ocular infection with a herpes simplex virus type 1 ICP34.5 deletion mutant produces no corneal disease or neurovirulence yet results in wild-type levels of spontaneous reactivation. J Virol. 1996;70:2883–2893. doi: 10.1128/jvi.70.5.2883-2893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perng G C, Jones C, Ciacci-Zanella J, Stone M, Henderson G, Yukht A, Slanina S M, Hofman F M, Ghiasi H, Nesburn A B, Wechsler S L. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science. 2000;287:1500–1503. doi: 10.1126/science.287.5457.1500. [DOI] [PubMed] [Google Scholar]
- 40.Post L E, Mackem S, Roizman B. Regulation of α genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with α gene promoters. Cell. 1981;24:555–565. doi: 10.1016/0092-8674(81)90346-9. [DOI] [PubMed] [Google Scholar]
- 41.Post L E, Roizman B. A generalized technique for deletion of specific genes in large genomes: α gene 22 of herpes simplex virus 1 is not essential for growth. Cell. 1981;25:227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 42.Randall G, Roizman B. Transcription of the derepressed open reading frame P of herpes simplex virus 1 precludes the expression of the antisense γ134.5 gene and may account for attenuation of the mutant virus. J Virol. 1997;71:7750–7757. doi: 10.1128/jvi.71.10.7750-7757.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Randall G, Lagunoff M, Roizman B. The product of ORF O located within the domain of herpes simplex virus 1 genome transcribed during latent infection binds to and inhibits in vitro binding of infected cell protein 4 to its cognate DNA site. Proc Natl Acad Sci USA. 1997;94:10379–10384. doi: 10.1073/pnas.94.19.10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reed L J, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 45.Resnick J, Boyd B A, Haffey M L. DNA binding by the herpes simplex virus type 1 ICP4 protein is necessary for efficient down regulation of the ICP0 promoter. J Virol. 1989;63:2497–2503. doi: 10.1128/jvi.63.6.2497-2503.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rivera-Gonzalez R, Imbalzano A N, Gu G, Deluca N A. The role of ICP4 repressor activity in temporal expression of the IE-3 and latency-associated transcript promoters during HSV-1 infection. Virology. 1994;202:550–564. doi: 10.1006/viro.1994.1377. [DOI] [PubMed] [Google Scholar]
- 47.Rock D L, Nesburn A B, Ghiasi H, Ong J, Lewis T L, Lokensgard J R, Wechsler S L. Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. J Virol. 1987;61:3820–3826. doi: 10.1128/jvi.61.12.3820-3826.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodahl E, Haarr L. Analysis of the 2-kilobase latency-associated transcript expressed in PC12 cells productively infected with herpes simplex virus type 1: evidence for a stable, nonlinear structure. J Virol. 1997;71:1703–1707. doi: 10.1128/jvi.71.2.1703-1707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roizman B. The function of herpes simplex virus genes: a primer for genetic engineering of novel vectors. Proc Natl Acad Sci USA. 1996;93:11307–11312. doi: 10.1073/pnas.93.21.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roizman B, Sears A E. In: Fields virology. 3rd ed. Fields B N, Knipe D M, Howley P M, editors. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 51.Sedarati F, Izumi K M, Wagner E K, Stevens J G. Herpes simplex virus type 1 latency-associated transcription plays no role in establishment or maintenance of a latent infection in murine sensory neurons. J Virol. 1989;63:4455–4458. doi: 10.1128/jvi.63.10.4455-4458.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spivack J G, Fraser N W. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J Virol. 1987;61:3841–3847. doi: 10.1128/jvi.61.12.3841-3847.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spivack J G, Fareed M U, Valyi-Nagy T, Nash T C, O'Keefe J S, Gesser R M, McKie E A, Maclean A R, Fraser N W, Brown S M. Replication, establishment of latent infection, expression of the latency-associated transcripts and explant reactivation of herpes simplex virus type 1 gamma 34.5 mutants in a mouse eye model. J Gen Virol. 1995;76:321–332. doi: 10.1099/0022-1317-76-2-321. [DOI] [PubMed] [Google Scholar]
- 54.Steiner I, Spivack J G, Lirette R P, Brown S M, MacLean A R, Subak-Sharpe J H, Fraser N W. Herpes simplex virus type 1 latency-associated transcripts are evidently not essential for latent infection. EMBO J. 1989;8:505–511. doi: 10.1002/j.1460-2075.1989.tb03404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stevens J G, Wagner E K, Devi-Rao G B, Cook M L, Feldman L T. RNA complementary to a herpesvirus α gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 56.Thompson R L, Sawtell N M. The herpes simplex virus type 1 latency-associated transcript gene regulates the establishment of latency. J Virol. 1997;71:5432–5440. doi: 10.1128/jvi.71.7.5432-5440.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner E K, Devi-Rao G, Feldman L T, Dobson A T, Zhang Y, Flanagan W M, Stevens J G. Physical characterization of the herpes simplex virus latency-associated transcript in neurons. J Virol. 1988;62:1194–1202. doi: 10.1128/jvi.62.4.1194-1202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagner E K, Flanagan W M, Devi-Rao G, Zhang Y, Hill J M, Anderson K P, Stevens J G. The herpes simplex virus latency-associated transcript is spliced during the latent phase of infection. J Virol. 1988;62:4577–4585. doi: 10.1128/jvi.62.12.4577-4585.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whitley R J, Kern E R, Chatterjee S, Chou J, Roizman B. Replication, establishment of latency, and induced reactivation of herpes simplex virus γ134.5 deletion mutants in rodent models. J Clin Investig. 1993;91:2837–2843. doi: 10.1172/JCI116527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu T T, Su Y H, Block T M, Taylor J M. Evidence that the latency-associated transcripts of herpes simplex virus type 1 are nonlinear. J Virol. 1996;70:5962–5967. doi: 10.1128/jvi.70.9.5962-5967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yeh L, Schaffer P A. A novel class of transcripts expressed with late kinetics in the absence of ICP4 spans the junction between the long and short segments of the herpes simplex virus type 1 genome. J Virol. 1993;67:7373–7382. doi: 10.1128/jvi.67.12.7373-7382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zabolotny J M, Krummenacher C, Fraser N W. The herpes simplex virus type 1 2.0-kilobase latency-associated transcript is a stable intron which branches at a guanosine. J Virol. 1997;71:4199–4208. doi: 10.1128/jvi.71.6.4199-4208.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zwaagstra J C, Ghiasi H, Slanina S M, Nesburn A B, Wheatley S C, Lillycrop K, Wood J, Latchman D S, Patel K, Wechsler S L. Activity of herpes simplex virus type 1 latency-associated transcript (LAT) promoter in neuron-derived cells: evidence for neuron specificity and for a large LAT transcript. J Virol. 1990;64:5019–5028. doi: 10.1128/jvi.64.10.5019-5028.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]