Highlights
-
•
In this study, we demonstrated that NSCLC was primarily involved in BCAA degradation.
-
•
We observed a significant increase in BCAA levels, downregulation of BCKDHA expression, and upregulation of BCKDK expression in NSCLC cells.
-
•
BCKDK promotes proliferation and inhibits apoptosis in NSCLC cells, and we observed that BCKDK affected Rab1A and p-S6 in A549 and H1299 cells via BCAA modulation.
-
•
Leucine affected Rab1A and p-S6 in A549 and H1299 cells and affected the apoptosis rate of H1299 cells.
-
•
BCKDK enhances Rab1A-mTORC1 signaling and promotes tumor proliferation by suppressing BCAA catabolism in NSCLC, suggesting a new biomarker for the early diagnosis and identification of metabolism-based targeted approaches for patients with NSCLC.
-
•
We provide new findings on the molecular mechanism of NSCLC that may suggest a new biomarker for early diagnosis or a way to identify metabolism-based targeted approaches for patients with NSCLC, resulting in more favorable outcomes.
Keywords: BCAA catabolism, BCKDK, Rab1A, mTORC1, NSCLC
Abstract
Background
Non-small cell lung cancer (NSCLC) is a leading cause of cancer death. Branched-chain amino acid (BCAA) homeostasis is important for normal physiological metabolism. Branched-chain keto acid dehydrogenase kinase (BCKDK) is a rate-limiting enzyme involved in BCAA degradation. BCAA metabolism has been highlighted in human cancers. The aberrant activation of mTORC1 has been implicated in tumor progression. Rab1A is a small GTPase, an activator of mTORC1, and an oncogene. This study aimed to reveal the specific role of BCKDK-BCAA-Rab1A-mTORC1 signaling in NSCLC.
Methods
We analyzed a cohort of 79 patients with NSCLC and 79 healthy controls. Plasma BCAA assays, immunohistochemistry, and network and pathway analyses were performed. The stable cell lines BCKDK-KD, BCKDK-OV A549, and H1299 were constructed. BCKDK, Rab1A, p-S6 and S6 were detected using western blotting to explore their molecular mechanisms of action in NSCLC. The effects of BCAA and BCKDK on the apoptosis and proliferation of H1299 cells were detected by cell function assays.
Results
We demonstrated that NSCLC was primarily involved in BCAA degradation. Therefore, combining BCAA, CEA, and Cyfra21-1 is clinically useful for treating NSCLC. We observed a significant increase in BCAA levels, downregulation of BCKDHA expression, and upregulation of BCKDK expression in NSCLC cells. BCKDK promotes proliferation and inhibits apoptosis in NSCLC cells, and we observed that BCKDK affected Rab1A and p-S6 in A549 and H1299 cells via BCAA modulation. Leucine affected Rab1A and p-S6 in A549 and H1299 cells and affected the apoptosis rate of H1299 cells.
In conclusion, BCKDK enhances Rab1A-mTORC1 signaling and promotes tumor proliferation by suppressing BCAA catabolism in NSCLC, suggesting a new biomarker for the early diagnosis and identification of metabolism-based targeted approaches for patients with NSCLC.
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide because of its high incidence and prevalence. Non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung cancers [1,2]. Nevertheless, it is gratifying that improved treatment has accelerated progress against lung cancer and driven a record drop in overall cancer mortality in recent years [3]. Thus, understanding the molecular mechanisms underlying NSCLC for further knowledge and advanced treatment options is urgently needed and accelerates progress against cancer.
Branched-chain amino acids (BCAAs), such as valine, leucine, and isoleucine, are essential amino acids for mammals. BCAA homeostasis plays a pivotal role in normal physiological metabolism [4]. In the BCAA catabolic pathway, BCAAs are reversibly metabolized by branched-chain aminotransferase (BCAT) into branched-chain alpha-ketoacids (BCKAs), followed by decarboxylation by the branched-chain alpha-ketoacid dehydrogenase (BCKDH) complex in an irreversible reaction, and metabolized into acetyl-CoA or succinyl-CoA. BCKDH, the flux-generating step in BCAA degradation, is controlled by a pair of enzymes, branched-chain keto acid dehydrogenase kinase (BCKDK) and Mg2+/Mn2+-dependent 1 K protein phosphatase (PPM1K) through phosphorylation status modulation [5], [6], [7].
Metabolic reprogramming occurs in various diseases, including cancer [8]. Altered BCAA metabolism is recently highlighted in many human cancer types, including NSCLC, pancreatic ductal adenocarcinoma (PDAC), leukemia, breast cancer, ovarian cancer, hepatocellular carcinoma (HCC), and other tumors [9], [10], [11], [12], [13], [14]. Many studies proved leucine binds to sestrin2 to activate rapamycin complex 1 (mTORC1) [15]. Aberrant activation of mTORC1 has been implicated in tumor progression and is considered a potential target for cancer treatment [16]. Altered BCAA metabolism in the aforementioned types of cancer cells is strongly associated with the activation of mTORC1 during tumor progression. Rab1A is a small GTPase [17] recently identified as a mTORC1 activator and oncogene [18]. Additionally, it was shown that Rab1A plays a pivotal role in amino acid signaling to regulate mTORC1 in normal and cancer cells [19]. In this study, we investigated the specific role of the BCKDK-BCAA-Rab1A-mTORC1 signaling pathway in NSCLC.
Materials and methods
Plasma and tissue samples
We analyzed a cohort of 79 patients with NSCLC and 79 healthy controls (HCs) at Tianjin Medical University Cancer Hospital. The clinicopathological data of patients with NSCLC (n = 79) and HCs (n = 79) are provided in Tables 1, 2. The mean age of the patients with NSCLC was 57 (standard deviation [SD], 10 years); 35 were males, and 44 were females. Classification according to the American Joint Committee on Cancer (AJCC) 8th edition revealed that 79 tumors were stage I. The HCs had a mean age of 51 (SD, 10 years); 35 were males, and 44 were females. This study was approved by the Ethics Committee of Tianjin Medical University. Samples were obtained from all patients with informed consent. Plasma samples were obtained from the Clinical Laboratory, Tianjin Medical University Cancer Hospital. The blood samples were centrifuged at 3500 rpm for 10 min, and then the supernatants were collected and stored at −80 °C until nuclear magnetic resonance (NMR) spectroscopy was conducted. Fresh lung cancer tissues and paired adjacent non-tumor lung tissues were obtained from patients who underwent surgery at the Tianjin Medical University Cancer Institute and Hospital.
Table 1.
Demographics and clinicopathologic characteristics of samples from patients with NSCLC.
| Characteristics | n = 79 |
|---|---|
| Age,M±SD(IQR), years | 57±10 (50-66) |
| Sex, n (%) | |
| Male | 35(44.3%) |
| Female | 44(55.7%) |
| Tumor diameter, n (%) | |
| ≤2 cm | 62(78.5%) |
| >2 cm | 17(21.5%) |
| TNM stage, n (%) | |
| IA | 56(70.9%) |
| IB | 23(29.1%) |
| Presence of lymph node metastasis, n (%) | |
| Metastasis | 6(7.6%) |
| No Metastasis | 73(92.4%) |
| CEA,M(IQR) | 2.58 (1.34-3) |
| Cyfra21.1,M(IQR) | 2.11(1.44-2.52) |
NSCLC, non-small cell lung cancer; TNM, tumor node metastasis; CEA, carcinoembryonic antigen.
Table 2.
Characteristics of samples from healthy controls (HC).
| Characteristics | n = 79 |
|---|---|
| Age,M±SD(IQR), years | 51±10(43-57) |
| Sex, n (%) | |
| Male | 35(44.3%) |
| Female | 44(55.7%) |
| CEA,M(IQR) | 1.84(1.3-2.2) |
| Cyfra21.1,M(IQR) | 1.36(0.91-1.7) |
CEA, carcinoembryonic antigen.
NMR
NMR analysis of the plasma samples and cell supernatants was conducted by ProteinT Biotechnology Co. Ltd. (Tianjin, China) following the Bruker Standard Operating Procedure for In Vitro Diagnostic Studies (IVDr SOP). In detail, 300 µL buffer (phosphate buffer [PBS] pH 7.4, containing TSP-D4; Bruker Corp., Billerica, MA, USA) was added to the 300 µL sample, then the mixture was transferred to a 5 mm NMR tube and analyzed in an NMR (Bruker 600 MHz). The data were analyzed using Bruker Topspin 3.6.0 software on NMRPC. BCAA was detected using NMR.
Immunohistochemistry (IHC)
Paraffin-embedded sections were used for IHC staining. Sections were incubated with the BCKDK antibody (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) overnight, followed by incubation with a secondary antibody conjugated with horseradish peroxidase (Abcam, Cambridge, UK). The nuclei were stained with hematoxylin. 3,3‐diaminobenzidine (BOSHIDE) showed immunoreactivity. Representative images were captured using an imaging microscope (Olympus).
Network and pathway analysis
Based on the SIMCA-P platform (Wu et al., 2010) (version 14.1, Umetrics, Sweden), we performed principal component analysis (PCA) and orthogonal partial least-squares discriminant analysis (PLS-DA) to identify metabolic alterations across the NSCLC and non-NSCLC groups. The variable importance in the projection (VIP) value was calculated for each covariate, and VIP> 1 indicated relevance to group discrimination. To explore the biological connections across potential metabolites, we conducted enrichment and pathway analyses of metabolomic data based on the Kyoto Encyclopedia of Genes and Genomes database using MetaboAnalyst software (version 5.0).
Immunofluorescence staining
A549 cells of equal density were inoculated in 6-well plates. When the cells reached 80% confluence, they were treated with MitoTracker Red CMXRos. The cells were fixed with 4% paraformaldehyde. After washing with PBS, the cells were blocked with 5% bovine serum albumin and incubated with an anti-BCKDK antibody (sc-374425; Santa Cruz Biotechnology Inc.), followed by incubation with the corresponding secondary antibody conjugated to fluorescein isothiocyanate (FITC; Invitrogen). Nuclei were stained with DAPI (4′, 6-diamino-2-phenylindole, Invitrogen). Images were captured using a fluorescence imaging module (Olympus).
Cell culture and treatment
The human NSCLC cell lines A549 and H1299 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). A549 and H1299 cells were cultured in RPMI-1640 (BI, 01-100-1ACS) supplemented with 10% fetal bovine serum (FBS, BI, 04-001-1ACS) at 37 °t in an atmosphere of 5% CO2. Leucine (7.5 mM) was prepared from leucine powder (Sigma-Aldrich).
Generation of stable cell lines
Lentiviruses purchased from GeneChem Corporation (Shanghai, China) were used to generate stable cell lines. A549 and H1299 cells were infected with lentivirus in the presence of polybrene (5 μg/mL, Sigma-Aldrich). At 48 h after infection, cells were selected using puromycin (1 mg/mL; Sigma-Aldrich).
Protein extraction and western blot analysis
A549 and H1299 cells were seeded in 6-well plates. When the cells reached 80% confluence, they were exposed to leucine (7.5 mM) for 15 min, 30 min, 1 h, 2 h, and 4 h, respectively. Stable A549 and H1299 cell lines were analyzed. The cells were lysed in radioimmunoprecipitation assay buffer (R0020, Solarbio) supplemented with a protease inhibitor cocktail and phosphatase inhibitor mixture. The total protein concentration was measured using a bicinchoninic acid protein assay kit. The proteins were heated at 97°C for 5 min, separated by 10–15% SDS-PAGE, and transferred onto polyvinylidene fluoride membranes (Millipore, USA). The membranes were then incubated with specific primary antibodies and corresponding secondary antibodies. Protein bands were visualized using an enhanced chemiluminescence reagent kit (Millipore). The following antibodies were used: anti-BCKDK (E-12) (sc-374425; Santa Cruz Biotechnology), anti-phospho-S6 (Ser235/236) (D57.2.2E; Cell Signaling Technology), anti-S6 (54D2; Cell Signaling Technology), and anti-Rab1A (G-10) (sc-377201; Santa Cruz Biotechnology). The mean optical densities of the bands were quantified using ImageJ acquisition and analysis software.
Cell proliferation assay
Cell proliferation was detected using the CCK-8 and colony formation assays. CCK-8 assay: BCKDK KD and BCKDK OV H1299 cells were seeded in 6-well plates and incubated for 1–5 days. CCK-8 was added to the cells, and after 1 h of incubation, absorbance was detected at 570 nm on a microplate reader. The absorbance values on days 1, 2, 3, 4, and 5 were normalized to the data from day 0 to create a proliferation curve.
For colony formation assay, BCKDK KD and BCKDK OV H1299 cells were seeded in 6-well plates. The cells were cultured until the number of cells in most single clones was greater than 50. After cloning, the cells were washed once with PBS, and 1 mL of 4% paraformaldehyde was added to each well and fixed for 30–60 min. Next, 1 mL of crystal violet dye solution was added to each well, and the cells were incubated for 10–20 min, washed several times with PBS, dried, and photographed using a digital camera.
Cell apoptosis assay
H1299 cells were seeded into 6-well plates. When the cells reached 80% confluence, they were exposed to leucine (7.5 mM) for 15 min, 30 min, 1 h, 2 h, and 4 h, respectively. Apoptosis of BCKDK KD and BCKDK OV H1299 cells was measured. An annexin V–FITC/PI apoptosis assay kit (ZETA LIFE, San Francisco, CA, USA) was used to assess the apoptotic rates according to the manufacturer's instructions. Apoptotic cells were detected by flow cytometry (Bricyte E6, Mindray, Inc., Shenzhen, China).
Statistical analysis
All data are expressed as the means ± standard deviation from at least three independent experiments. Comparisons were assessed by two-way Student's t-test using GraphPad Prism software (version 7). The level of statistical significance was set at 0.05. Receiver operating characteristic (ROC) curves were drawn using the pROC package in the R software, and diagnostic efficacy was assessed using ROC curves. The optimal cutoff value for each biomarker was determined when the specificity was 95% in the training set to find a cutoff that is helpful for a definitive diagnosis. Kaplan–Meier survival plots and hazard ratios with 95% confidence intervals and log-rank P values were calculated and plotted in R using the Bioconductor packages.
Results
The metabolic profile of NSCLC patients differs from that of HCs
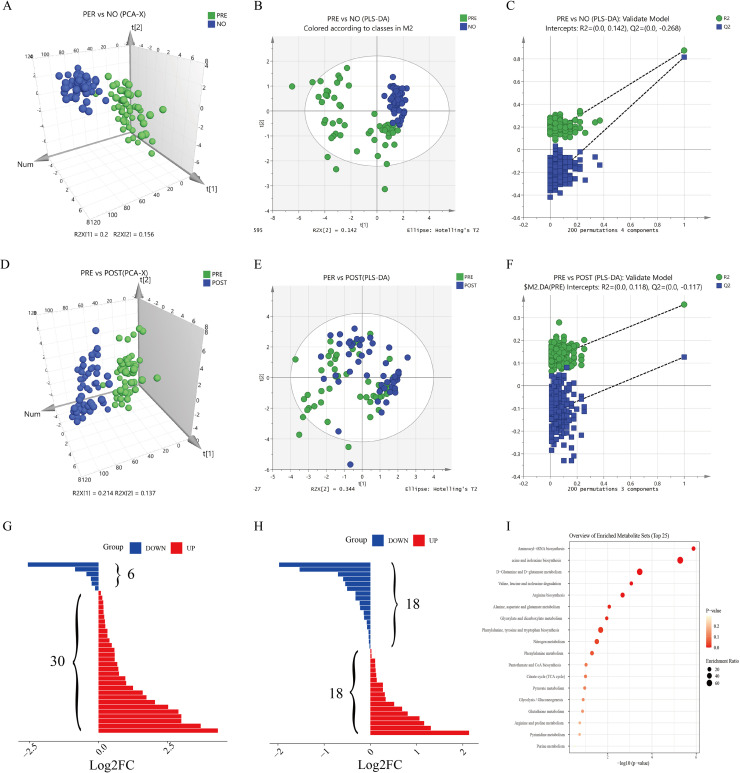
We analyzed a cohort of patients with NSCLC (pre- and post-operative) and healthy controls. For a comprehensive observation of the metabolic profiles, PCA and PLS-DA were performed on the respective NMR data of the sera. The PCA and PLS-DA results are shown in Fig. 1. The metabolic profiles of the pre-operative patients could be distinguished from those of the healthy control in the PCA score plot (Fig. 1A). In the PCA models, the metabolic profiles of the pre-operative group could be differentiated from those of the post-operative (Fig. 1D). For a deeper metabolomic study of NSCLC, PLS-DA was conducted to explore metabolic alterations across the NSCLC and non-NSCLC groups. Multivariate analysis in the PLS-DA model showed a specific separation of metabolomic features between the plasma of patients with NSCLC and non-NSCLC (Q2 = 0.58, R2 = 0.52; Fig. 1B). The specificity and reliability of the patient classification were revealed in the validation model using a permutation test (R2 = 0.142, Q2 = −0.268; Fig. 1C). Multivariate analysis in the PLS-DA model showed a specific separation of metabolomic features between the plasma of patients with NSCLC and the post-operative NSCLC group (Q2 = 0.358, R2 = 0.127, Fig. 1E). The specificity and reliability of patient classification were revealed in the validation model using a permutation test (R2 = 0.118, Q2 = −0.117, Fig. 1F). These results indicated that the metabolic profiles of patients with NSCLC differed from those of healthy controls.
Fig. 1.
PCA score plots, PLS-DA 3D score plots, validation plots of metabolic analysis results between LUAD and LUAD post-operative and between LUAD and normal groups, and enrichment of pathways based on relevant metabolites. PCA score plots compared (A) LUAD and normal groups (D) LUAD and LUAD post-operative groups. (B) PLS-DA score plot (two latent variables, R2Y = 0.846, Q2 = 0.817) and (C) the 200-permutation plot validated LUAD versus normal groups. All permuted R2 and Q2 values on the left were lower than the point on the right (R2 = 0.133, Q2 = −0.27). (E) PLS-DA score plot (two latent variables, R2Y = 0.358, Q2 = 0.127), and (F) the 200-permutation plot validated LUAD versus LUAD post-operative groups. All permuted R2 and Q2 values on the left were lower than the point on the right (R2 = 0.118, Q2 = −0.117). (G) Bar chart discriminating the components with significant increments (red bar, n = 30) or decrement (blue bar, n = 6) in LUAD versus normal groups. (H) Bar chart discriminating the components with significant increments (red bar, n = 17) or decrements (blue bar, n = 18) in LUAD versus LUAD post-operative groups. (I) A total of 18 metabolic pathways were detected by MetaboAnalyst using pathway enrichment with differential substances common to both groups (LUAD versus normal and LUAD versus post-LUAD surgery).
NSCLC is primarily involved in BCAA degradation
Through univariate ANOVA analysis, we identified significant variation across the plasma of patients with NSCLC and the non-NSCLC group in n metabolites (higher in 30 but lower in six features for the plasma of patients with NSCLC) (Fig. 1G). In addition, significant variation in n metabolites was observed across the plasma of patients with NSCLC and the post-operative NSCLC group through univariate ANOVA analysis (higher in 18 but lower in 18 features for the plasma of patients with NSCLC) (Fig. 1H). A set of differential substances (glutamine, phenylalanine, citric acid, lactic acid, ornithine, and BCAAs) was obtained by taking the intersection of these two sets of differential substances; their trends were the same. Further functional pathway analysis revealed that the differentially expressed metabolites in NSCLC were primarily involved in BCAA degradation (Fig. 1I).
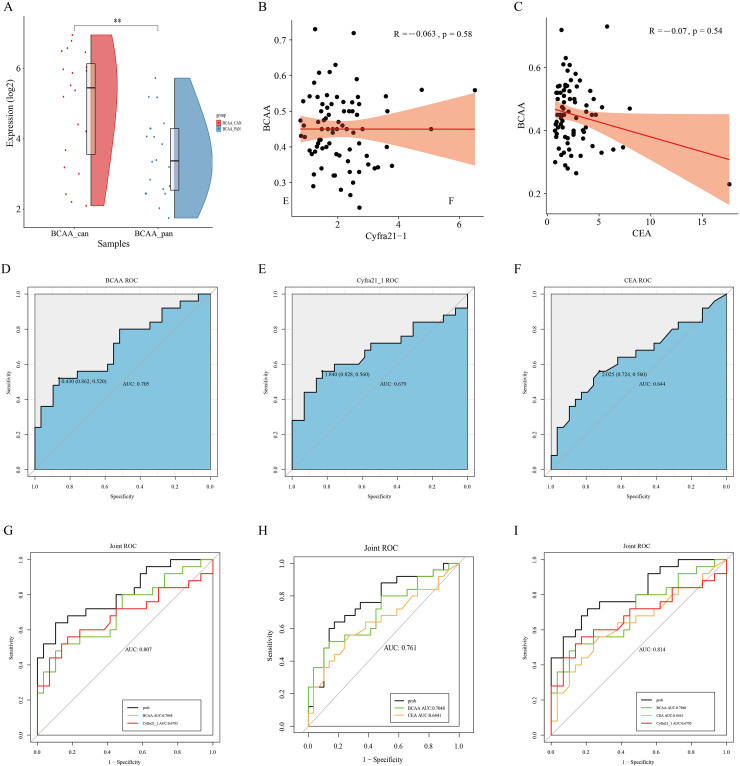
Combination of BCAA, CEA, and Cyfra21-1 is clinically useful in NSCLC
BCAA levels were higher in the cancerous tissues of patients with NSCL compared to the cancerous and paraneoplastic tissues of patients with NSCLC (Fig. 2A). To further evaluate the diagnostic performance of BCAA, first, according to the analyses comparing BCAA with Cyfra21-1 expression in NSCLC (R: −0.07, p = 0.54, Fig. 2B) and BCAA with CEA expression in NSCLC (R: −0.07, p = 0.54, Fig. 2C), BCAA was not correlated with CEA and Cyfra21-1 after an analysis was performed using ROC curves to assess the accuracy of the model. Tumor markers (CEA, CYFRA 21-1) were measured in the sera of patients with validated NSCLC and healthy subjects. The results of the validation group were statistically analyzed according to whether they were diseased. ROC analysis showed area under the curve (AUC) values (AUC = 0.705 for BCAA and AUC = 0.644 for CEA) for single metabolic markers (BCAA, sensitivity 52%, specificity 86.2%) and tumor markers (CEA, sensitivity 56%, specificity 72.4%; Cyfra21-1, sensitivity 56%, specificity 82.8%) and AUC = 0.679 for Cyfra21-1, (Fig. 2D–F) were lower than the combinations of these markers (AUC = 0.807 for BCAA+Cyfra21-1, AUC = 0.761 for BCAA + CEA, AUC = 0.814 for BCAA + CEA + Cyfra21-1, Fig. 2G–I), and the three combinations (BCAA + CEA + Cyfra21-1) had the highest AUC values. Thus, there is a link between the metabolic markers of tumor microenvironmental changes and tumor markers, and combinations of these markers (BCAA, CEA, and Cyfra21-1) are clinically useful for the differential diagnosis of NSCLC.
Fig. 2.
(A) Comparative results of BCAA content in 20 pairs of cancer and paracancerous tissues of patients with NSCLC using NMR experimental data. (B) Comparison of BCAA vs. CEA expression in NSCLC by analysis (R: −0.07, p = 0.54). (C) Comparison of BCAA vs. Cyfra21-1 expression in NSCLC by analysis (R: −0.07, p = 0.54). (D-I) Receiver operating characteristic (ROC) curves that distinguish the ability of patients with non-small cell lung cancer from that of normal subjects. (D) Scores of ROC curves for BCAA. (E) Scores of the ROC curve of Cyfra21_1. (F) Scores of the ROC curve of the CEA. (G) Scores of the ROC curve of BCAA combined with Cyfra21_1. (H) Scores of ROC curves of BCAA combined with CEA. (I) Scores of ROC curves of BCAA combined with Cyfra21_1 and CEA.
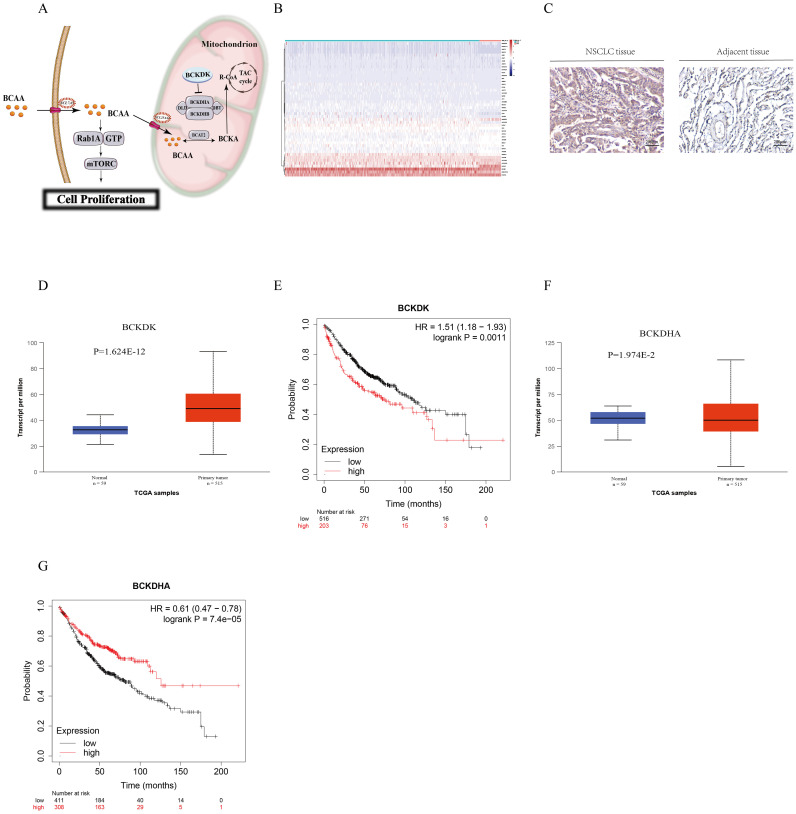
Downregulated BCKDH and upregulated BCKDK were shown in tissues of patients with NSCLC and were related to prognosis in NSCLC
As BCAA are clinically useful in NSCLC, we explored whether key enzymes in the BCAA catabolic pathway play a role in NSCLC. As shown in Fig. 3A, BCKDH is a key enzyme in the BCAA catabolic pathway, and BCKDK inhibits BCKDH activity. BCKDK inactivates the E1α (BCKDHA) subunit of this complex to suppress BCAA catabolism. A summary of the enzyme transcript levels in the NSCLC and non-NSCLC groups showed that BCKDH was downregulated in NSCLC (Fig. 3B). Immunohistochemical results showed that BCKDK was upregulated at the protein level in NSCLC tissues compared to that in adjacent tissues (Fig. 3C). We investigated BCKDK and BCKDHA expression levels in NSCLC tissues and normal adjacent tissues using the TCGA database, which consists of 515 NSCLC samples and 59 corresponding normal samples. We observed that BCKDK expression was higher in cancer tissues than in adjacent non-cancerous tissues (http://ualcan.path.uab.edu/cgi-bin/TCGAExResultNew2.pl?genenam=BCKDK&ctype=NSCLC, P <0.01, Fig. 3D) and lower in cancer tissues than in adjacent non-cancerous tissues (http://ualcan.path.uab.edu/cgi-bin/TCGAExResultNew2.pl?genenam=BCKDHA&ctype=NSCLC, P < 0.01, Fig. 3F). These results indicate that NSCLC downregulates BCKDHA expression and upregulates BCKDK expression. Furthermore, high expression of BCKDH and low expression of BCKDK were associated with significantly better patient outcomes by allowing the efficient catabolism of BCAA, indicating that BCKDK is related to the prognosis of NSCLC (Fig. 3E, G).
Fig. 3.
(A) Graphical abstract of BCAA degradation (B) Heat map of differences between the lung cancer group and the paracancer group in the TCGA database. (C) IHC staining of BCKDK in NSCLC. Representative images are shown. (200 ×, scale bar = 40 µm). (D) Boxplot showing the relative expression of BCKDK in normal and non-small cell lung cancer samples. (E) Kaplan–Meier analysis of non-small cell lung cancer patients with low (blue curve) and high (red curve) expression of BCKDK in the TCGA datasets. p values were determined by the log-rank test. (F) Boxplot showing the relative expression of BCKDHA in normal and non-small cell lung cancer samples. (G) Kaplan–Meier analysis of non-small cell lung cancer patients with low (blue curve) and high (red curve) expression of BCKDHA in the TCGA datasets. p values were determined by the log-rank test.
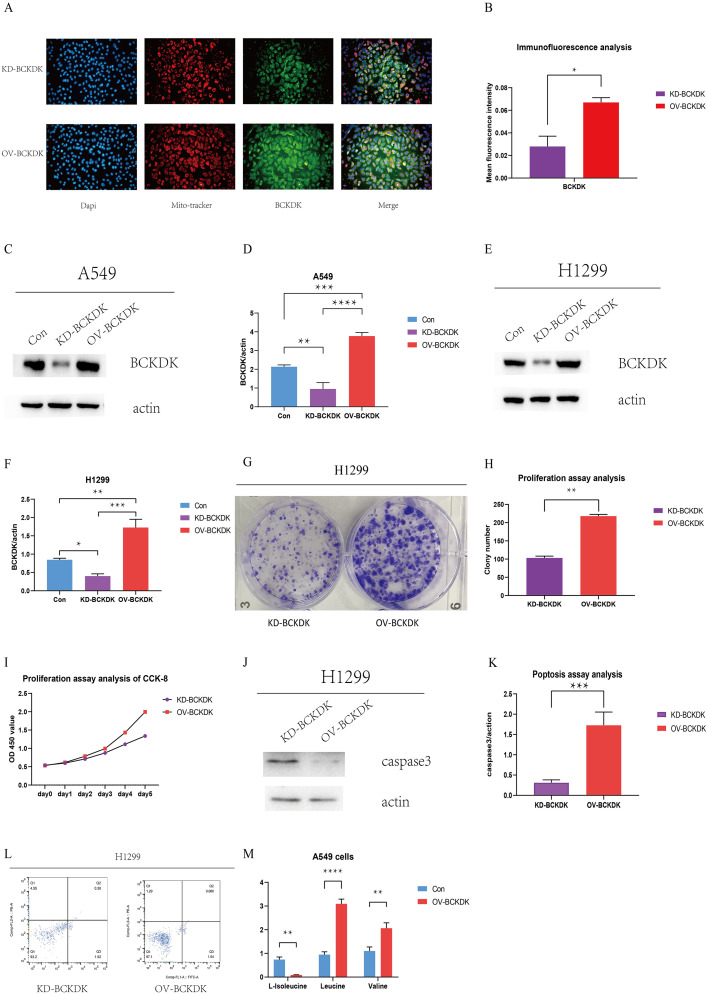
BCKDK promotes cell proliferation and inhibits apoptosis in NSCLC cells
We examined the role of BCKDK in NSCLC. Immunofluorescence staining revealed that BCKDK was localized in the nucleus, mitochondria, and cytoplasm of A549 cells, providing a basis for follow-up studies (Fig. 4A). To ascertain the downstream signaling pathways of BCAA in NSCLC, we selected A549 and H1299 cells with high and low BCKDK expression, respectively. BCKDK was knocked down and overexpressed in A549 and H1299 cell lines (Fig. 4C–F), indicating that stable cell lines were successfully constructed. We explored the effect of BCKDK on NSCLC cell function, and cell proliferation was detected using colony formation and CCK-8 assays. Apoptotic activity was detected using western blotting and flow cytometry. The colony formation assay showed an increased number of colonies in OV-BCKDK H1299 cells (Fig. 4G–H). The CCK-8 assay showed a higher OD 450 value for OV-BCKDK H1299 cells than for KD-BCKDK H1299 cells (Fig. 4I). As described above, BCKDK promotes the proliferation of H1299 cells. Caspase-3 expression was significantly lower in OV-BCKDK H1299 cells than in KD-BCKDK H1299 cells (Fig. 4J–K). The apoptosis rates were 2.22% and 1.60% in BCKDK-KD H1299 and BCKDK-OV H1299 cells, respectively (Fig. 4L), indicating that the apoptosis of H1299 cells was inhibited by BCKDK overexpression. Furthermore, compared to control A549 cells, BCKDK-OV A549 cells showed increased leucine and valine levels due to a reduction in BCAA catabolism. However, decreased isoleucine levels were observed in BCKDK-OV A549 cells, indicating that isoleucine may differ from leucine and valine in terms of BCAA catabolism (Fig. 4M). Thus, BCKDK promoted cell proliferation and inhibited apoptosis in NSCLC cells.
Fig. 4.
(A–B) Immunofluorescence analysis of BCKDK in A549 cells was performed by fluorescence microscope (original magnification: 200x). (C–D) The BCKDK protein level of A549 BCKDK KD and BCKDK OV stable cell lines was measured using western blotting (n = 3 for each group). (E–F) The BCKDK protein level of H1299 BCKDK KD and BCKDK OV stable cell lines was measured using western blotting (n = 3 for each group). (G–H) Colony formation assay of BCKDK OV and BCKDK KD H1299 cells. (I) CCK-8 assay of BCKDK OV and BCKDK KD H1299 cells. (J–K) The caspase-3 protein level of BCKDK OV and BCKDK KD H1299 cells was measured using western blotting (n = 3 for each group). (L) Apoptosis analysis of BCKDK OV and BCKDK KD H1299 cells. (M) BCAA assay of BCKDK OV and control A549 cells was conducted (n = 3 for each group).
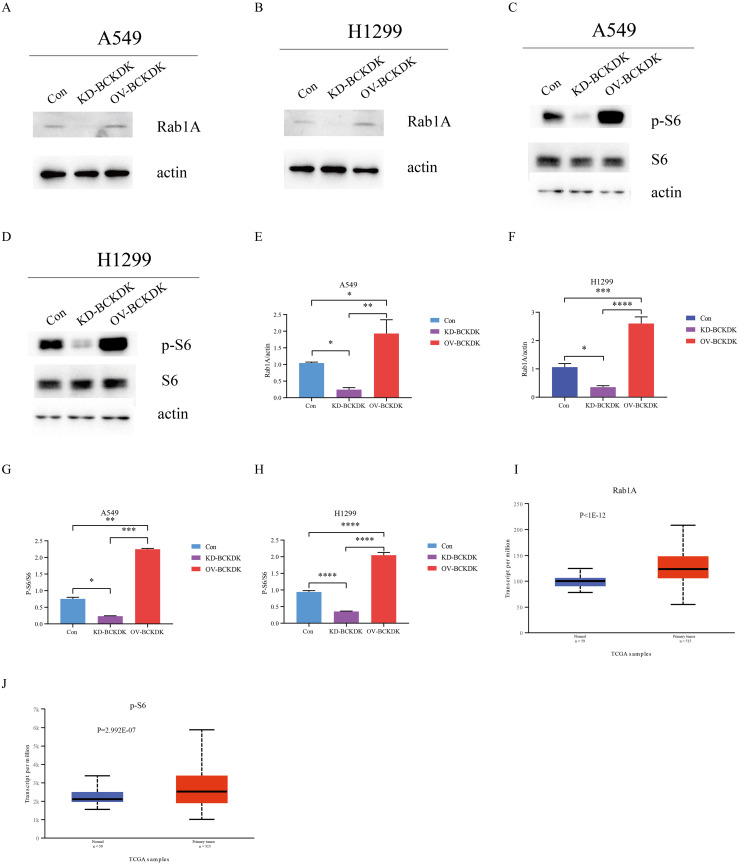
BCKDK affected Rab1A and p-S6 in NSCLC cells and tissue
To explore the detailed mechanism by which BCKDK is involved in NSCLC, S6, p-S6, and Rab1A were evaluated using western blotting in A549-KD, A549-OV, H1299-KD, and H1299-OV cell lines. Significantly increased Rab1A and p-S6 levels were observed in BCKDK-OV A549 and BCKDK-OV H1299 cells compared to those in the corresponding control cells. Reduced Rab1A and p-S6 levels were observed in BCKDK-KD A549 and BCKDK-KD H1299 cells compared to those in the corresponding control cells (Fig. 5A–H). Furthermore, the expression of Rab1A and p-S6 was higher in cancerous tissues than in adjacent tissues (http://ualcan.path.uab.edu/cgi-bin/TCGAExResultNew2.pl?genenam=RAB1A&ctype=NSCLC,http://ualcan.path.uab.edu/cgi-bin/TCGAExResultNew2.pl?genenam=RPS6 &ctype=LUAD, P <0.001, Fig. 5I–J). These results indicated that BCKDK affects Rab1A and p-S6 in NSCLC cells and tissues.
Fig. 5.
(A, E) The Rab1A protein level of A549 BCKDK KD and BCKDK OV stable cell lines was measured by western blotting (n = 3 for each group). (B, F) The Rab1A protein level of H1299 BCKDK KD and BCKDK OV stable cell lines was measured by western blotting (n = 3 for each group). (C, G) The p-S6 and S6 protein levels of A549 BCKDK KD and BCKDK OV stable cell lines were measured using western blotting (n = 3 for each group). (D, H) The p-S6 and S6 protein levels of H1299 BCKDK KD and BCKDK OV stable cell lines were measured using western blotting (n = 3 for each group). (I–J) Boxplot showing the relative expression of Rab1A and p-S6 in normal and non-small cell lung cancer samples.
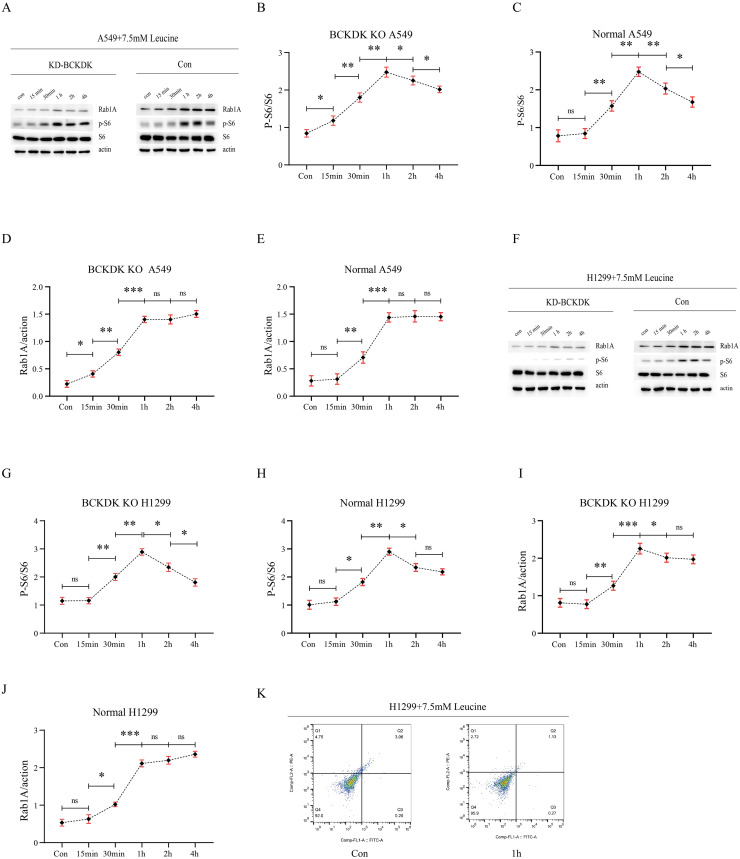
BCAA influenced Rab1A and p-S6 in A549 and H1299 cells
As previously shown, BCKDK overexpression in A549 cells increased BCAA levels. In addition, the knockdown and overexpression of BCKDK caused a decrease and increase in p-S6, and Rab1A, respectively. To determine whether BCKDK regulates Rab1A and p-S6 by modulating BCAA, we added 7.5 mM leucine to A549, BCKDK-KD A549, H1299, and BCKDK-KD H1299 cells at different time gradients, and p-S6, S6, and Rab1A were detected. Overall, there were increasing trends in p-S6 and Rab1A in A549, BCKDK-KD A549, H1299, and BCKDK-KD H1299 cells treated with 7.5 mM leucine at different time gradients. The time to start increasing and the time to reach the highest level are shown in the histogram (Fig. 6A–J). Thus, BCKDK overexpression increased BCAA levels and regulated Rab1A and p-S6.
Fig. 6.
(A–E) The p-S6, S6, and Rab1A protein levels of BCKDK KD and control A549 cells treated with 7.5 mM leucine at different time gradients were measured using western blotting (n = 3 for each group). (F–J) The p-S6, S6, and Rab1A protein levels of BCKDK KD and control H1299 cells treated with 7.5 mM leucine for different time gradients were measured using western blotting (n = 3 for each group). (K) Apoptosis assay of control H1299 cells treated with 7.5 mM leucine for 1 h.
BCAA inhibited the apoptosis of H1299 cells
Apoptotic activity of H1299 cells treated with 7.5 mM leucine was detected using flow cytometry. The apoptosis rate of H1299 cells after 7.5 mM leucine for 1 h was 1.40%, which was lower than that of H1299 cells (3.26%) that were not treated with leucine (Fig. 6K), providing evidence that BCAA inhibited the apoptosis of H1299 cells.
Discussion
Lung cancer is the second most common cancer and the leading cause of cancer-related deaths globally due to its high incidence and prevalence, imposing a heavy burden on human health worldwide. Lung cancer is divided into NSCLC and small-cell lung cancer. NSCLC accounts for approximately 85% of all lung cancers [1,2]. Compared with 20 years ago, advances in surgery, radiotherapy, chemotherapy, molecularly targeted therapy, and immunotherapy have significantly prolonged the lives of many cancer patients and greatly changed their quality of life. However, cancer is difficult to cure due to its continuous evolution, and the average 5-year survival rate for lung cancer is only 19% [20]. Thus, the discovery of new therapeutic targets through molecular biology research and development will be helpful.
Metabolic reprogramming occurs in diseases such as cancer. The metabolic characteristics of cancer cells are useful for tumor diagnosis and staging [8]. BCAA metabolism is involved in the development and progression of multiple cancers, including NSCLC, PDAC, leukemia, breast cancer, ovarian cancer, HCC, and other tumors [9], [10], [11], [12], [13], [14]. Ericksen et al. showed that suppression of BCAA degradation could promote tumor development and progression in HCC [14]. BCAA regulates multiple signaling pathways, such as protein synthesis, lipid synthesis, cell growth, and autophagy [21]. Thus, the upregulation of BCAA in patients with NSCLC could be interpreted as meeting the energetic and proliferative needs of both the host and the tumor. However, relatively few studies have investigated the role of BCAA catabolism in NSCLC. Therefore, we focused on the BCAA degradation pathway in NSCLC. In this study, we analyzed plasma metabolites in patients with NSCLC. Functional pathway analysis revealed a strong correlation between NSCLC and BCAA degradation. In addition, there is an obvious increase in BCAA levels in the plasma of patients with NSCLC. We demonstrated that NSCLC was primarily involved in the BCAA degradation. Consistently, Xiaoli Zhang et al. demonstrated that circulating BCAA levels were higher in lung cancer patients than in healthy controls [22]. BCKDH, the rate-limiting enzyme for BCAA degradation, is controlled by a pair of vital enzymes, BCKDK and PPM1K [5], [6], [7]. Thus, BCKDK is a key enzyme involved in BCAA degradation. Furthermore, studies have shown that overexpression of BCKDK promotes tumor growth and metastasis [[23], [24], [25]]. In this study, we revealed that BCKDH levels decreased and BCKDK levels increased in NSCLC through database analysis. Consistently, upregulated BCKDK expression was detected in NSCLC tissues by histochemical staining. These results correspond to an increase in the BCAA levels in NSCLC. In addition, lower BCKDH and higher BCKDK levels were correlated with shorter survival times in patients with NSCLC, showing that BCKDK is related to the prognosis of NSCLC. In this study, we discovered that BCKDK promoted cell proliferation and inhibited apoptosis in NSCLC cells. The combination of BCAA, CEA, and Cyfra21-1 is clinically useful for the differential diagnosis of NSCLC. These findings suggest that BCKDK is a potential biomarker for NSCLC diagnosis.
Furthermore, to confirm the function of BCKDK in NSCLC, stable cell lines, including BCKDK-KD A549, BCKDK-OV A549, BCKDK-KD H1299, and BCKDK-OV H1299, were used. BCKDK promotes proliferation and inhibits apoptosis in NSCLC cells. Hence, the above results support the tumor-promoting function of BCKDK and are consistent with previous studies demonstrating that BCKDK overexpression promotes cell proliferation and metastasis in colorectal cancer and HCC [[23], [24], [25]]. BCAA, particularly leucine, activates the mTORC1 pathway [15,26]. mTORC1 is a central cell growth regulator that connects cellular metabolism and growth [27] and is often activated in many types of cancer; thus, it has become an attractive cancer therapeutic target [28,29]. Rab1A, a small GTP-binding protein, regulates amino acid (AA) signaling in mTORC1 [17,18]. Rab1A overexpression promotes mTORC1 signaling and oncogenic growth in an AA- and mTORC1-dependent manner [18]. Rab1A plays a pivotal role in AA signaling and regulates mTORC1 expression in normal and cancer cells [19]. In this study, we examined whether BCKDK knockdown and overexpression modulated Rab1A and mTORC1 signaling in NSCLC. BCKDK knockdown and overexpression caused the downregulation and upregulation of p-S6, a downstream target kinase of mTORC1. The Rab1A results were consistent with those for p-S6. Additionally, Rab1A and p-S6 expression levels were higher in NSCLC tissues than in the adjacent tissues. These results suggested that BCKDK knockdown suppressed Rab1A-mTORC1 signaling in NSCLC cells. These results are consistent with the cancer-promoting functions of Rab1A and mTORC1 in other researches [19,28,29]. Xue et al. showed that BCKDK enhances the MAPK pathway through MEK rather than BCAA catabolism in colorectal cancer [23]. Therefore, whether BCKDK modulates Rab1A and mTORC1 expression through BCAA catabolism remains unclear. We added 7.5 mM leucine into A549, BCKDK-KD A549, H1299, and BCKDK-KD H1299 cells at different time gradients. As expected, Rab1A and p-S6 expressions were regulated by the addition of leucine. Thus, BCKDK affected Rab1A and p-S6 in A549 and H1299 cells through BCAA modulation. Further research revealed that Leu addition reduced the apoptosis of NSCLC cells.
Therefore, we conclude that BCKDK modulates Rab1A-mTORC1 expression by modulating BCAA degradation in NSCLC. To our knowledge, this study is the first to report that the loss of BCAA catabolism enhances Rab1A-mTORC1 signaling activity and promotes tumor proliferation in NSCLC. Additional experiments are necessary to elucidate the detailed molecular mechanisms.
Conclusions
This study showed that BCKDK enhances Rab1A-mTORC1 signaling by suppressing BCAA catabolism and promoting tumor proliferation in NSCLC. In summary, we provide new findings on the molecular mechanism of NSCLC that may suggest a new biomarker for early diagnosis or a way to identify metabolism-based targeted approaches for patients with NSCLC, resulting in more favorable outcomes.
Ethics approval and consent to participation
This study was approved by the institutional review boards of the hospitals, and written informed consent was obtained from all patients.
Ethic Committee Name: Medical Ethics Committee of Tianjin Cancer Hospital.
Approval Code: NO.bc2019114.
Approval Date: September 10, 2019.
Consent for publication
Not applicable.
Availability of data and material
This manuscript includes all data generated and analyzed in this study. More information is available from the corresponding author on request.
Funding Support
This work was funded by the National Natural Science Foundation of China (grant No. 81802432) and the Tianjin Key Medical Discipline(Specialty) Construction Project (TJYXZDXK-009A).
CRediT authorship contribution statement
Meiting Xue: Conceptualization, Methodology, Validation, Writing – original draft, Writing – review & editing. Jiawei Xiao: Conceptualization, Data curation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. Wenna Jiang: Conceptualization, Funding acquisition, Validation. Yanhui Wang: Data curation, Investigation, Resources. Duo Zuo: Resources, Supervision. Haohua An: Methodology, Visualization. Li Ren: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not applicable.
References
- 1.Lindsey A., Torre MSPH, et al. Global cancer statistics, 2012. CA-Cancer J. Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Li C, Wang H, Jiang Y, Fu W, Liu X, Zhong R, Cheng B, Zhu F, Xiang Y, He J, Liang W. Advances in lung cancer screening and early detection. Cancer Biol. Med. 2022;19(5):591–608. doi: 10.20892/j.issn.2095-3941.2021.0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel Rebecca L, et al. Cancer statistics, 2021. CA-Cancer J. Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 4.Harper A, Miller R, Block K. Branched-chain amino acid metabolism. Annu. Rev. Nutr. 1984;4:409–454. doi: 10.1146/annurev.nu.04.070184.002205. [DOI] [PubMed] [Google Scholar]
- 5.Brosnan JT, Brosnan ME. Branched-chain amino acids: enzyme and substrate regulation. J. Nutr. 2006;136(1 Suppl):207S–211S. doi: 10.1093/jn/136.1.207S. [DOI] [PubMed] [Google Scholar]
- 6.Adeva María M., et al. Insulin resistance and the metabolism of branched-chain amino acids in humans. Amino Acids. 2012;43(1):171–181. doi: 10.1007/s00726-011-1088-7. [DOI] [PubMed] [Google Scholar]
- 7.Lu G, Sun H, She P, et al. Protein phosphatase 2Cm is a critical regulator of branched-chain amino acid catabolism in mice and cultured cells. J. Clin. Invest. 2009;119(6):1678–1687. doi: 10.1172/JCI38151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu J, Thompson CB. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 2019;20(7):436–450. doi: 10.1038/s41580-019-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayers JR, Torrence ME, Danai LV, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. 2016;353:1161–1165. doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Z, Achreja A, Meurs N, et al. Tumour-reprogrammed stromal BCAT1 fuels branchedchain ketoacid dependency in stromal-rich PDAC tumours. Nat. Metab. 2020;2:775–792. doi: 10.1038/s42255-020-0226-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu Z, Liu Y, Cai F, et al. Loss of EZH2 reprograms BCAA metabolism to drive leukemic transformation. Cancer Discov. 2019;9:1228–1247. doi: 10.1158/2159-8290.CD-19-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, Han J. Branched-chain amino acid transaminase 1 (BCAT1) promotes the growth of breast cancer cells through improving mTOR-mediated mitochondrial biogenesis and function. Biochem. Biophys. Res. Commun. 2017;486:224–231. doi: 10.1016/j.bbrc.2017.02.101. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Faddaoui A, Bachvarova M, et al. BCAT1 expression associates with ovarian cancer progression: possible implications in altered disease metabolism. Oncotarget. 2015;6:31522–31543. doi: 10.18632/oncotarget.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ericksen RE, Lim SL, McDonnell E, et al. Loss of BCAA catabolism during carcinogenesis enhances mTORC1 activity and promotes Tumor development and progression. Cell Metab. 2019;29(5):1151–1165. doi: 10.1016/j.cmet.2018.12.020. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfson RL, Chantranupong L, Saxton RA, et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351:43–48. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mossmann D, Park S, Hall MN. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer. 2018;18:744–757. doi: 10.1038/s41568-018-0074-8. [DOI] [PubMed] [Google Scholar]
- 17.Saraste J, Lahtinen U, Goud B. Localization of the small GTP-binding protein rab1p to early compartments of the secretory pathway. J. Cell Sci. 1995;108:1541–1552. doi: 10.1242/jcs.108.4.1541. [DOI] [PubMed] [Google Scholar]
- 18.Thomas JD, Zhang YJ, Wei YH, et al. Rab1A is an mTORC1 activator and a colorectal oncogene. Cancer Cell. 2014;26:754–769. doi: 10.1016/j.ccell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Wang X, Yuan Z, et al. Amino acids-Rab1A-mTORC1 signaling controls whole-body glucose homeostasis. Cell Rep. 2021;34(11) doi: 10.1016/j.celrep.2021.108830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 21.O'Connell TM. The complex role of branched chain amino acids in diabetes and cancer. Metabolites. 2013;3:931–945. doi: 10.3390/metabo3040931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Zhu X, Wang C, et al. Non-targeted and targeted metabolomics approaches to diagnosing lung cancer and predicting patient prognosis. Oncotarget. 2016;7(39):63437–63448. doi: 10.18632/oncotarget.11521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue P, Zeng F, Duan Q, et al. BCKDK of BCAA catabolism cross-talking with the MAPK pathway promotes tumorigenesis of colorectal cancer. EBioMedicine. 2017;20:50–60. doi: 10.1016/j.ebiom.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian Q, Yuan P, Quan C, et al. Phosphorylation of BCKDK of BCAA catabolism at Y246 by Src promotes metastasis of colorectal cancer. Oncogene. 2020;39(20):3980–3996. doi: 10.1038/s41388-020-1262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhai M, Yang Z, Zhang C, et al. APN-mediated phosphorylation of BCKDK promotes hepatocellular carcinoma metastasis and proliferation via the ERK signaling pathway. Cell Death. Dis. 2020;11(5):396. doi: 10.1038/s41419-020-2610-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 2013;14(3):133–139. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Guan K. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 2019;21(1):63–71. doi: 10.1038/s41556-018-0205-1. [DOI] [PubMed] [Google Scholar]
- 28.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 29.Sun S. mTOR kinase inhibitors as potential cancer therapeutic drugs. Cancer Lett. 2013;340(1):1–8. doi: 10.1016/j.canlet.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This manuscript includes all data generated and analyzed in this study. More information is available from the corresponding author on request.