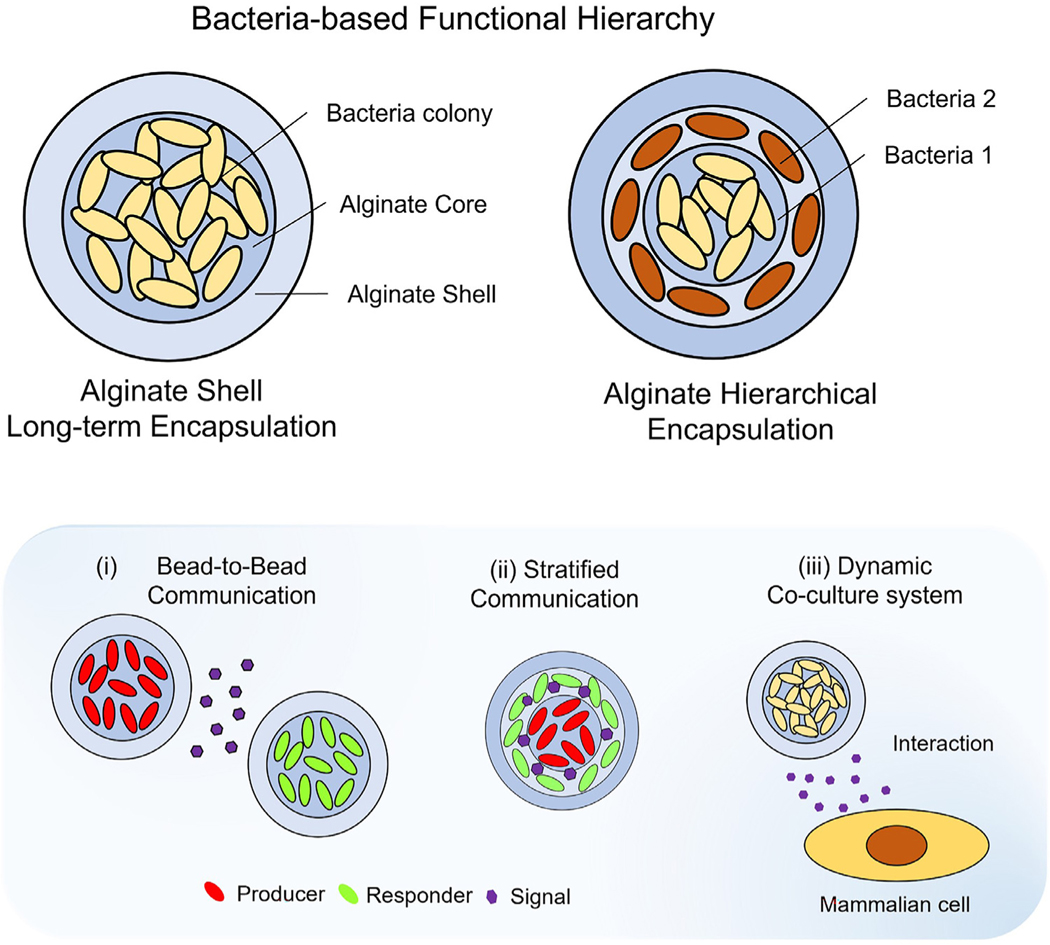
Abstract
To sequester prokaryotic cells in a biofilm-like niche, the creation of a pertinent and reliable microenvironment that reflects the heterogeneous nature of biological systems is vital for sustenance. Design of a microenvironment that is conducive for growth and survival of organisms, should account for factors such as mass transport, porosity, stability, elasticity, size, functionality, and biochemical characteristics of the organisms in the confined architecture. In this work we present an artificial long-term confinement model fabricated by natural alginate hydrogels that are structurally stable and can host organisms for over 10 days in physiologically relevant conditions. A unique feature of the confinement platform is the development of stratified habitats wherein bacterial cells can be entrapped in the core as well as in the shell layers, wherein the thickness and the number of shell layers are tunable at fabrication. We show that the hydrogel microenvironment in the beads can host complex subpopulations of organisms similar to that in a biofilm. Dynamic interaction of bacterial colonies encapsulated in different beads or within the core and stratified layers of single beads was demonstrated to show intra-species communication. Inter-species communication between probiotic bacteria and human colorectal carcinoma cells was also demonstrated to highlight a possible bidirectional communication between the organisms in the beads and the environment.
Keywords: Alginate hydrogel architectures, Bacteria encapsulation, Microbial communication, Host-microbe interaction
1. Introduction
The past two decades have seen a rapid increase in the development of three dimensional (3D) structures based on soft hydrogel materials used for the encapsulation of microorganisms given their unique properties [1–3]. Cell encapsulation based on soft hydrogel systems have been explored for over a half of century [4–7]. Recent efforts on hydrogel based modalities have been used to engineer complex hierarchical architectures [8–10]. Despite the progress, inherent complexities within the biological environment may elicit the development of relevant and robust platforms that reflects the heterogeneous nature of biological systems [11].
Bacterial cells co-exist in diverse communities in matrix/film-like habitats comprising of extracellular polymeric substances (EPS) such as polysaccharides, proteins, and DNA secreted by microorganisms [12]. To mimic the unique 3D niche in biofilms, the polysaccharide-based hydrogels originally conceptualized must meet several specifications such as elasticity of the hygroscopic milieu, biochemical properties, and biocompatibility so that the design criteria for effective encapsulation of microbial population in artificial confinements can be established [1, 13]. Nevertheless, the development of artificial encapsulation platforms based on hydrogel materials to mimic the physicochemical characteristics of the 3D biofilm environment [2] has been a challenge. A key issue is the uncontrolled leakage or rapid death of organisms in the restricted microenvironment within 1–2 days [14–19]. Microorganisms proliferate and exert substantial stress within the confined space [11] resulting in rupture of the containment system [20]. Swelling and deformation of the hydrogel inevitably occurs in a physiological environment resulting in cell leakage from these structures constructed with polymeric matrices rendering further evaluation of its function and interaction ineffective. Therefore, structurally stable and physiologically relevant 3D architectures that form biofilm-like habitats for long-term studies will be an immense asset for microbial communication studies.
Several technological solutions ( e.g., polyelectrolyte complexation [14, 21], microfluidics [22, 23], microfabricated structures [24, 25], 3D bioprinting [17, 26–28] etc.) have been suggested for the entrapment, growth, and examination of bacterial cells within confined architectures. Despite a plethora of research compounded by challenges due to proliferation, death and leakage of bacterial cells from such confined spaces, studies at the multi scale level are limited because these are laborious, time-consuming, costly, and cannot be generalized. Above all, in order to possess the ability to sequester bacterial cells in a restricted space, the system design should account for mass transport, porosity, stability, hydrodynamics, and efficient characterization ( e.g., sample collection and analysis) of the materials used in fabrication. Other design characteristics for 3D architectures are reproducibility, simplicity and cost-effective fabrication conducive to the growth of artificial biofilm-like habitats [11, 13]. None of reported work on hydrogel-based structures could fabricate a favorable 3D niche for bacterial cells in a confined volume for long-term microbial colonization and interaction studies.
Alginate hydrogels are well-known for their nanoporous structures and ionic-crosslinked networks, that permit rapid diffusion of small molecules [29]. Since the initial work on the development of alginate semipermeable barriers [4] reported nearly half a century ago, advanced core–shell architectures have emerged and concentric shell architectures were fabricated [8, 9]. Additional reports on alginate core hydrogels include the encapsulation of either eukaryotes [30, 31] or prokaryotes [16, 19, 21]. Still, the development of artificial habitats for long-term confinement in 3D biofilm-like environments have remained a challenge [11, 32]. Furthermore, its application in biological evaluation is sparse due to the lack of a relevant model for hierarchy and function assessment.
In this study, we demonstrate a rational approach for the entrapment and long-term growth of biofilm-like habitats for over 10 days and possibly longer to query microbial behavior upon confinement in alginate hydrogel-shell structures ( Fig. 1 ). A hydrated environment for hydrogel along with the buffers provides a viable microenvironment for entrapped cells against perturbations, caused by physical, chemical, or biological stimuli where cellular responses are altered dynamically, by either intrinsic or extrinsic factors [33, 34]. The rationale for this study is based on the hypothesis that biological organisms can thrive well in ideal microenvironments; therefore, our overarching goal is to construct a favorable 3D niche to provide biofilm-like microenvironments for long-term growth of microbial communities. We expect that our work based on mechanistic principles with precisely controllable features [35] has the potential to advance the development of heterogeneous 3D habitats to entrap a range of organisms for microbial communication and host-microbe interaction studies. Our work proposes a design strategy to develop layered hydrogel beads, referred to as hierarchical encapsulation wherein bacteria can be colonized in the core as well as in the stratified hydrogel-shell layers. Finally, we present three representative models to illustrate inter- and intra- species communication and function by our entrapment strategy.
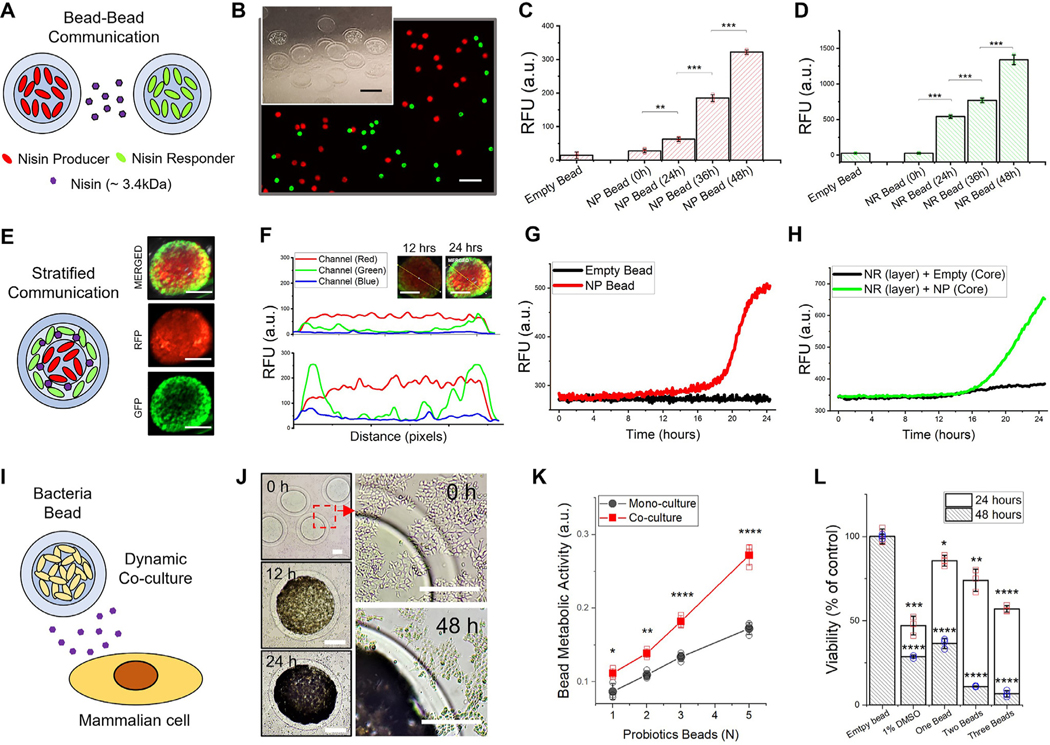
Fig. 1.
Hierarchical encapsulation of microorganisms in biofilm-like heterogenous hydrogel. Schematic representation of bacteria encapsulation based on alginate core–shell structure (top left) and hierarchical encapsulation (top right) for multilayered communication. The alginate-based core–shell platform enables the development of biofilm-like stratified habitats within hierarchical multi-layered structures. The small ecological systems in a hydrogel environment act to serve as independent microbial modulators with direct or indirect activation of a response regulator by stimuli, including signaling molecules. Three representative models ( i.e., bead-to-bead interaction, stratified communication, and bacteria-mammalian cell interaction system) are presented by encapsulating specific microorganisms.
2. Materials and methods
2.1. Fabrication of alginate hydrogel-shell beads
Alginate hydrogel-shell beads were fabricated by the reported method (Fig. S1) [35]. Alginic acid polymer solutions (Sigma/71238) with concentrations ranging from 1 to 3 wt % were prepared and stored at 4 °C until further usage. Alginate core beads were fabricated using a needle-based extrusion process, in which alginate solution was dropped into a 0.1M CaCl2 solution while being gently stirred. Briefly, sterile blunt 30 gauge needles were used and connected to a syringe. A syringe-pump system (Harvard Apparatus PHD 20 0 0) was used at a constant flow infusion rate. The air outlet was positioned perpendicular to the end of the needle. A rotameter (Dwyer Instrument) was linked to a N2 gas tubing line with the desired setting at a constant air-flow rate. By adjusting the distance between the syringe needle and the air outlet, the force required to detach an alginate droplet at the end of the needle was controlled. The time taken for crosslinking of the alginate ionotropic gels was about 10 min. To remove smaller size beads less than 500 μm in diameter, the generated bead suspension was filtered using a 500-micron nylon mesh filter. The beads were placed on a nylon mesh of 200 μm size and transferred quickly to a very low concentration of alginate solution (less than 0.1 wt%). The concentration of the alginate solution increased by up to 0.5–0.6 wt % upon contact with a very low viscosity solution. The reaction container containing the core beads was then vigorously shaken to prevent aggregation of the beads. To stop interfacial gelation, the solution in the beaker was diluted more than 20 times by adding an excessive amount of deionized (DI) water. In the final step, the hydrogel-shell beads were rinsed two times with DI water and one time with 0.05M CaCl2 solution to stabilize the hydrogel core–shell structure.
2.2. Morphology observation of hydrogel beads
The morphology of the hydrogel-shell beads was observed using an inverted microscope (Leica DMI3000B) equipped with a CCD camera (Qimaging EXi Blue). The mean geometric diameter of the spherical beads was first determined using an upright microscope (Olympus BX53, 10x and 20x magnification). The size distribution of the alginate beads and the thickness of the hydrogel-shell were analyzed and measured from the optical microscopy images. ImageJ software was used to adjust the brightness of the images of the beads.
2.3. Small molecule diffusion test
Alginate is an anionic polymer. Cationic dye (Rhodamine B, MW = 479.01, Sigma) was chosen as a surrogate for cation diffusion studies. Three types of hydrogel beads (Bead 1.5, Bead 1.5/Layer and Bead 1.3/Layer) were fabricated for testing the release kinetics of hydrogel beads. The average size of beads (n = 20) was 1.51 ± 0.09 mm (Bead1.5), 1.31 ± 0.12 mm (Bead1.3). The layer thickness of the beads was tuned to be ~200 μm. The beads were immersed in 0.1M Rhodamine B solution for over 24 h at room temperature and were transferred into 5 ml DI water for evaluation. At each time interval, 100 μl volume of the solution was used for experiments. The amount of Rhodamine B released in the solution was measured by fluorescence intensity at an emission wavelength of 521 nm using a microplate reader (Synergy H1, BioTek Instruments) with a black 96 well plate. Average values of three measurements were obtained for each sample.
2.4. Bacteria encapsulation with hydrogel-shell confinement
All of the strains were used in encapsulation studies without further genetic transformation, as listed in Table S1. Wild type E. coli was grown in Luria-Bertani (LB) broth (nutritionally complete) medium or LB agar plates (BD Difco, Miller) at 37 °C unless otherwise noted for other strains. Bacterial cells were cultured overnight on LB agar plates at 37 °C. A single colony was inoculated and cultured in LB media with shaking. The bacteria concentration (OD60 0, optical density 60 0 nm) was determined using a UV/vis spectrophotometer (Eppendorf Biophotomer) and the standard agar plate count method. In the mid-exponential phase, aliquots were resuspended in fresh LB media to achieve specific bacteria densities and the required concentration before use in encapsulation studies. The alginate solution was sterilized by UV treatment (Ultraviolet lamp = 50 W, Lightexports) for 5 min in a biosafety cabinet. The alginate solution devoid of any external bacteria was pretested by incubating at 37 °C in LB media to confirm its sterility. Bacteria cultures in the mid-exponential phase were mixed with sterile alginate solution (3 wt%). A typical needle-based extrusion method (30G needles) was used to drop the cell-alginate mixture into 0.1M CaCl2 solution with mild stirring. When the size of alginate beads containing microorganisms decreased, the air extrusion method was applied without any modification to the setting configuration. All other fabrication procedures were the same to fabricate hydrogel-shell layers. All experiments commenced within 10 min after fabrication of the hydrogel-shell beads containing microorganisms.
2.5. Three-dimensional (3D) colony forming unit per bead (CFU/bead)
Turbidity was correlated with bacterial numbers determined by plate counts to measure the reproductive viability of cells (results from E. Coli are provided in Fig. S4). Bacteria aliquots for experiments at the mid-exponential phase were obtained at an optical density of 0.5 in arbitrary units (which corresponds to ~4.75 ×108 cells/ml measured by the standard agar plate count). Approximately, 106 cells were mixed with 1 ml of alginate solution in a sterilized Eppendorf tube. The number of incipient bacteria entrapped in a single core bead was calculated as a weight fraction of alginate beads (the weight of 10 droplets extruded through a 30G syringe needle (≃ 6.6 mg) divided by the total weight of the 1 ml solution containing 106 cells (≃ 1037 mg) wherein the estimated number of entrapped cells is 6 ×103 in a single bead. Bacterial aliquots were suitably diluted to control the initial number of cells to be entrapped in a bead. After the hydrogel-shell beads containing bacterial cells were cultivated in LB medium at 37 °C for 12 h, the number of colonies were counted using the ImageJ software to determine CFU count per bead (CFU/bead). To increase accuracy, customized Fiji macros by ImageJ with process parameters were used for quantifying the number of colonies from microscopy images.
2.6. Leakage test
Cell leakage tests were performed either on an agar plate or in LB media. Briefly, a conventional soft-agar overlay was employed to test cell leakage from alginate beads with/without hydrogel-shell, incorporated into an LB soft agar (0.7%) overlay, which was poured on a hard LB agar (1.5%) plate. Before the overlay completely solidified, hydrogel beads from each experimental group were mounted onto the overlay. The plates were inverted and incubated at 37 °C. The leakage was monitored and photographed under identical plate conditions. Three groups (free cell, beads with/without hydrogel-shell) were incubated in 50 ml LB media at 37 °C with mild (50 rpm) shaking. Cultured media in each Erlenmeyer beakers was collected at each time point and measured by spectrophotometry. The collected supernatant was incubated in LB media overnight and measured spectrophotometrically. Additionally, to verify cell leakage at the periphery of hydrogel, conventional hydrogel beads encapsulating GFP E. coli were monitored with confocal microscopy (Carl Zeiss LSM 780), at an excitation wavelength of 488 nm and emission at 50 0–60 0 nm.
2.7. Cell density, protein/cyclic-di-GMP quantification
All hydrogel-shell beads containing wild-type E. coli were loaded into a 96-well plate and incubated in 200 μl of nutritionally complete LB media (BD Difco, Miller) at a constant temperature of 30 °C. Before initiation of bead cultivation experiments, the CFU/bead (number of colonies per bead) was pre-determined by measuring the number of colonies formed in a single hydrogel bead. A single hydrogel bead containing bacterial colonies was collected at each time point in a 2 ml sterile Eppendorf tube. To estimate cell density in the bead, the collected samples were degraded entirely by adding 100 μl of 0.1 M sodium citrate (Sigma/S4641) in an Eppendorf tube within 90 seconds and vigorously vortexed. Then, the samples were back-diluted with 900 μl of fresh LB and measured by spectrometry. By measuring the OD600 of the samples, data set at OD600 of the bead samples was recorded. After centrifugation at 15000 rpm, for 5 min (Eppendorf 5424R), the supernatant of the sample was discarded, and the pellets used for the quantification of intracellular proteins and cyclic-di-GMP assay [36]. Degraded biomass samples were solubilized by incubation in 0.3 ml of 0.1M NaOH and 0.1% sodium dodecyl sulfate (SDS) at 95 °C for 15 min. Quantification of intracellular proteins produced by cell clusters was performed with standard bicinchoninic acid (BCA) assay (Thermo Scientific) with Bovine serum albumin as a standard. A commercial c-di-GMP Elisa assay kit (Cayman) was used for the quantification of cyclic-di-GMP (c-di-GMP). Subsequent extraction of nucleotides from the biomass pellets was performed per manufacturer’s instructions with B-Per buffer (Thermo Fisher Scientific). C-di-GMP concentrations were provided in picomoles per milligram of intracellular protein (pmol/mg of protein). To measure extracellular proteins and c-di-GMP released from bacterial bead culture, 100 μl of culture supernatant from each bead sample was separately collected and analyzed. Measurements from three biological samples were repeated in triplicate. Hydrogel bead samples containing microbial colonies were stored at −80 °C in a storage buffer (Tris-HCl 50 mM, NaCl 50 mM, EDTA 0.5 mM, pH 8.0, glycerol 5%) for long-time storage.
2.8. Metabolic activity
Metabolic activity was measured based on a tetrazolium salt (MTT, Invitrogen) colorimetric assay after incubation. Single hydrogel beads containing microbial colonies were collected at each time point for evaluation. To determine the MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) reduction activity of microbial colonies, beads were transferred into U-shaped-bottom 96 well microplate (single bead per well) and 100 μl of MTT solution (25 μg/ml) mixed with fresh pre-warmed LB was dispensed in each well. The microplates, wrapped in foil, were incubated at 30 °C in a mini thermal incubator (HERATHERM, Thermo Fisher Scientific) for 2 h, and the remaining media was completely discarded. To dissolve the water-insoluble formazan crystal trapped in the bead, 200 μl of dimethylsulfoxide (DMSO, Corning) was dispensed. After 8 h, 100 μl of the formazan crystal solution released from the bead was recollected and measured. Absorbance was assessed using a Synergy H1 plate reader (BioTek) at 560 nm. A calibration process for the incubation and dissolution of the MTT-formazan assay was carried out. (Fig. S7) To calculate the activity of bacterial cells, the relative metabolic activity was evaluated, to depict the relative value of cellular activities corresponding to the cell density and the total number of cells in a single bead. The value of metabolic activities of cells in the bead obtained from the MTT-formazan assay was approximated by the value of cell density quantified in the previous step.
2.9. Pyoverdine measurement
Hydrogel beads with bacteria colonies were collected at each time point in a sterile Eppendorf tube and completely degraded by adding 100 μl of 0.1 M sodium citrate. The solution was then transferred to a 96 black well plate. Pyoverdine was quantified, according to a standard protocol [37]. Pyoverdine fluorescence (excitation 405 nm, emission 460 nm) was measured using a Synergy H1 plate reader (BioTek). Data are shown as relative fluorescence units (RFU), with arbitrary units.
2.10. Viable cell count
For viable cell counts, physical degradation of E. Coli bead samples was demonstrated using an Omni Tissue Homogenizer (Omni International). The samples were dissolved entirely by adding 100 μl of 0.1 M sodium citrate and back-diluted within 30 s in 1 ml of fresh LB media. The endpoint of a bacterial titer (10−7 to 20−10 dilutions) was then determined from the duplicate plates by standard plate count. 50 μl of the diluted samples were pipetted and dried on LB agar plates [38]. All samples on LB agar plates were incubated at 37 °C for 24 h. The average colony count between 30 and 150 colonies were considered for plate count.
2.11. Histochemical stains
Samples of the beads (100 CFU/bead, cultivated for 24 h) were prepared and fixed in 4% neutral buffered formalin solution (Sigma) for 4 h and stored at 4 °C. One bead sample was placed in a tissue cassette and the samples were immersed with 70% to 80% ethanol for 10 minutes each, 95% ethanol for 15 min, and 100% ethanol for 10 min, processed in a tissue processor (Leica Microsystems). Next, the beads were processed in xylene solution for 10 min followed by three paraffin baths; 15 min in the first bath and 25 min in the two paraffin baths, and then placed in a mold with paraffin. Bead sections cut were 5 μm thick and mounted onto hydrophilic glass slide. The hydrogel bead sections were deparaffinized in xylene and hydrated by ethanol and distilled water and stained with hematoxylin and eosin (H&E, Sigma) and Periodic acid-Schiff (PAS, Sigma).
2.12. Swollen volume ratio
Hydrogel swelling was determined at 4 °C. Hydrogel-shell beads (layer thickness; 50, 120, 200 μm, respectively) were examined by immersing the samples in 50 mL DI and LB media for 7 days. The hydrogel beads containing E. coli were prepared by the above method. The diameter of the swollen hydrogel beads was observed and recorded with an inverted microscope (Zeiss Primovert) installed with a charged-coupled device (CCD) video camera (5-megapixel Swiftcam, Swift Optical Instruments). The spherical volume (Vt) was calculated from the diameter of the swollen beads measured at each time point and compared with the incipient volume (V0) before swelling. The volume ratio (Vt/V0) of the swollen samples was presented.
2.13. Compressive rheological test
Rheological characterization of hydrogel-shell beads before/after bacterial colonization was performed using a TA-XT2 texture analyzer (Texture Technologies Corp) equipped with a 35-mm diameter probe in an acrylic cylinder (TA-11). The hydrogel beads were mounted on the plate-plate cylinder and compressed at a rate of 0.5 mm/s, force-strain curves were obtained with the following test settings: 70–90% strain deformation, relaxation time of 5 s, and force of 20 g. In the pre-characterization step, alginate hydrogel samples (2 mm of core diameter, 300 μm of hydrogel-shell thickness) stored in DI water were measured as control for calibration. For comparison, the hydrogel beads containing E. coli (600 CFU/bead, 300 μm of hydrogel-shell thickness) incubated in LB media (for 0, 24, 72, and 120 h, respectively) were measured at 70% strain deformation calibrated in the premeasurement step.
2.14. GFP and RFP E. coli encapsulation
GFP E. coli Dh5 α was grown in LB medium containing kanamycin at 25 μg/mL, and RFP E. coli EPI 300 was grown at 37 °C in LB medium without antibiotics. Alginate core matrices (0.5 and 2 mm in diameter) entrapping either GFP or RFP expressing E. coli in the mid-exponential phase were produced by the same extrusion/gelation and layer fabrication method. Hydrogel-shell beads containing GFP and RFP E. coli cells were cultivated in 35 mm Petri dishes at 37 °C for seven days in 5 ml LB medium. 50 μl of growth media was aliquoted into 10 ml of fresh media every day and incubated overnight to evaluate cell leakage. Digital images were acquired by a smartphone camera (Samsung Galaxy Note 20). A hand-held UV-lamp (Schleicher & Schuell) at 365 nm was used to illuminate GFP and RFP expression by E. coli in a dark room. Hydrogel beads encapsulating GFP and RFP E. coli were monitored with fluorescence microscopy (Carl Zeiss Observer Z1). Fluorescence signals were measured using a Synergy H1 microplate reader (BioTek Instruments).
2.15. Hierarchical architectures and immobilization in a layered geometry
Multi-layered hydrogel beads were fabricated by alternating the procedures of hydrogel bead fabrication utilizing similar principles [35]. The beads with ‘n’ layers were mounted on a nylon mesh, and the excessive CaCl2 solution was carefully removed at the bottom of the nylon mesh. Prior to drying, the beads were transferred into a reaction vessel containing a low viscous alginate solution (less 0.1%wt). The subsequent procedures were the same as described above, but minor modifications were required to entrap and colonize cells in a layered geometry. Before transferring the core beads into a reaction vessel, alginate solution (less 0.1 %wt) and a bacterial aliquot of known concentration in mid-exponential phase were mixed to attain the desired density and uniform distribution of the cells. Vigorous agitation was initiated after dropping the beads into the low viscous alginate solution mixed with the bacterial aliquots of known concentration. The final concentration of alginate solution rapidly increased up to 0.5 %wt. By varying bacterial aliquots (E. coli, B subtilis, GFP E. Coli. and RFP E. coli), different types of stratified strains could be immobilized in each of the layered geometries in a multi-layered hieratical system and imaged under UV illumination by a hand-held UV lamp (365 nm).
2.16. Bead–Bead and stratified communication
For nisin-controlled gene expression (NICE) system [39], L. lactis strains, as listed in Table S1, were grown in M17 medium (HiMedia) containing chloramphenicol (Sigma/C0378) at 25 μg/mL at 30 °C. Alginate matrices entrapping the strains in the mid-exponential phase were produced by the same extrusion/gelation and layer fabrication method. Hydrogel-shell beads containing nisin producer (NP) and nisin responder (NR) cells were cultivated in either 150 mm Petri dishes or 24 well plate at 30 °C for 48 h in M17 media. The colonized beads were collected every 12 h and transferred into a black 96 well plate. Fluorescence signals induced by nisin molecules secreted from NP bead was measured using a Synergy H1 microplate reader (BioTek Instruments). To verify secretion of nisin molecules produced from NP beads, supernatants cultured with NP beads were collected at 12 and 24 h. NR bead was incubated in 200 μl of the collected supernatant at a constant temperature of 30 °C for 12 h using a Synergy H1 microplate reader (Biotek). Fluorescent signal emitted from the NR beads was measured at an emission wavelength of 521 nm every 5 min.
NP and NR cells (inner core: Nisin Producer, outer layer: Nisin Responder) were immobilized in the core and layered geometries in a multi-layered system. To entrap and colonize cells in a layered geometry, subsequent procedures similar to the one described above was used. The core has an average diameter of 1 mm with 150 μm of the 1st layer thickness and 100 μm of the 2nd layer thickness. After incubation for 12 and 24 h, fluorescence images and multi-channel intensity profiling of hydrogel samples (RFP and GFP expressing L. Lactis ) were characterized using a Chemidoc XRS system (Bio-rad). Fluorescence emission of the beads containing GFP and RFP expressing L. Lactis cells was detected using the 530/28 and 605/50 standard filter with excitation in the range of blue/green epi-illumination. Images acquired from Image Lab software (Bio-rad) were used for fluorescence intensity profiling by ImageJ software.
2.17. Dynamic Co-culture with mammalian cells
Human colorectal carcinoma (HCT116, ATCC® CCL-247 ™) was maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Corning) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% antibiotics (100 IU/ml penicillin and 100 μg/ml streptomycin, Lonza). Wild-type Lactococcus lactis subsp. lactis (ATCC 11454) were inoculated in brain–heart infusion (BHI) broth (BD Difco) at 37 °C. Growth of L. lactis ATCC 11454 was pre-examined in DMEM media containing 10% FBS with/without antibiotics. Co-culture experiments with bacteria bead and mammalian cells were conducted without antibiotics, which inhibit growth of microorganisms in DMEM media.
Alginate beads entrapping L. lactis ATCC 11454 strain in the mid-exponential phase were produced by the same extrusion/gelation and layer fabrication method. Before initiation of co-cultivation experiments utilizing the beads, the CFU/bead of L. lactis ATCC 11454 was pre-determined for an average bead diameter of 500 μm and 100 μm of the layer thickness. Probiotic L. lactis beads were loaded into a 48 or 96 well plate where cancer cells were pre-incubated for 12 h at 37 °C in a humidified incubator. The bead samples were harvested at each time point after 12 h of co-culturing with mammalian cells. Beads were then transferred into U-shaped-bottom 96 well microplate (at three beads per well) and a tetrazolium salt (MTT) colorimetric assay was performed to measure the metabolic status. To estimate the density of L. lactis in the bead, the harvested bead was degraded entirely by the addition of 100 μl of 0.1 M sodium citrate in an Eppendorf tube and the samples were back diluted with 900 μl of DI water and the OD600 of the samples was measured by spectrophotometry. After harvesting the hydrogel beads, WST-1 assay (Roche Applied Science) was used to assess in vitro proliferation of HTC116 cells. Additionally, flow cytometric analyses were conducted to assess Live/Dead and apoptosis assay for HTC 116 cells co-cultured with probiotics beads was performed. Calcein-AM (Invitrogen) and Propidium Iodine (Invitrogen) were used for Live/Dead assay. Annexin V-FITC (eBiosciences) and Propidium Iodine were used for cell apoptosis studies. Control and experimental groups of HTC116 cells were doubled-stained for flow cytometry analysis and subsequently analyzed with a BD LSR II flow cytometer (BD Biosciences). Data was collected with the FACSDiva 6.1.1. software (BD Biosciences) and analyzed with the FCS Express 6 software (De Novo).
2.18. Statistical analysis
Experiments were performed in triplicate. The data for the experiments was expressed as mean ± standard deviation (s.d.) and analyzed using unpaired Student’s t-test and one-way ANOVA with a post hoc test (Graphpad prism 9.0.), where post-hoc comparisons were conducted using the Tukey’s method. P values < 0.05 were considered statistically significant, and the details of the statistical tests are presented in each figure legends. No statistical analysis was performed to pre-determine a required effect size.
3. Results
3.1. Bioconfinement in hydrogel-shell beads
Our initial goal was to confirm whether the microenvironment in the alginate hydrogel layers allow effective cell growth to prevent cell leakage due to core–shell disintegration (Fig. 2A, Fig. S1). A representative strain, wild-type E. coli K-12 strain MG1655 (denoted below by E. coli), was chosen as a surrogate model to demonstrate these concepts. Besides, bacterial cells typically develop communities, forming biofilms. A biofilm can range in thickness from a few micrometers to several hundred micrometers, depending on environmental factors including nutrient supply [12]. Our experiments will utilize hydrogel-shell beads with an average diameter of 2 mm instead of a scaled-down size to simulate a diffusion gradient in a biofilm-like environment.
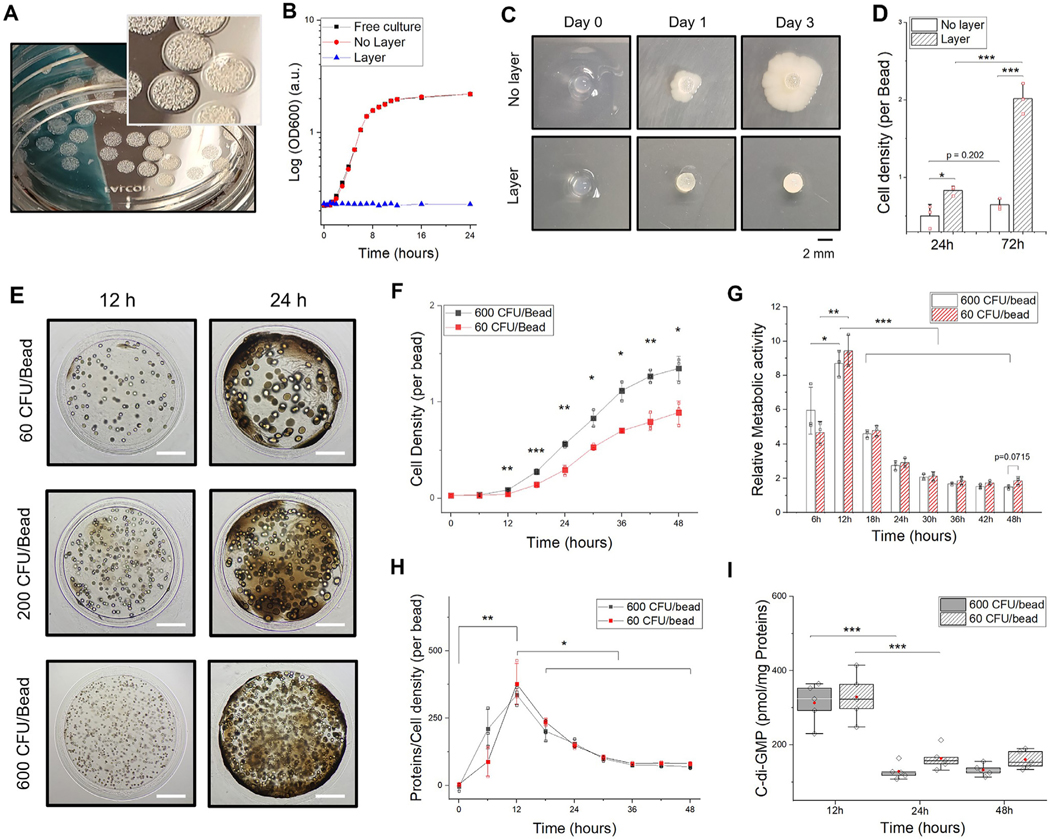
Fig. 2.
Encapsulation of microorganisms in biofilm-like heterogenous hydrogel. A. Representative image of hydrogel-shell beads with E. coli colonization after 24 h. The core has a diameter of 2 mm with 150 μm of outer layer thickness. B. Optical density (OD600) measurements of culture supernatant for leakage test. More than ten hydrogel beads and planktonic cells were cultured in nutritionally complete LB medium at 37 °C. C. Comparison of results from bacteria growth inside alginate hydrogel with/without the shell layer on 2D agar plates. Soft agar overlay technique was used to test cell leakage. D. E. coli densities (per bead) measured by spectrophotometry in 1 ml of LB medium after complete degradation of a single alginate bead. E. Microscopic images of hydrogel-shell beads at different time points. Colony forming units per bead (CFU/bead) of E. coli colonies after 12 h (upper: 60 CFU/bead, middle: 200 CFU/bead, lower: 600 CFU/bead). Scale bar is 500 μm. F. Measurements of E. coli density per bead (60 and 600 CFU/bead, respectively) every 6 h for 2 days. G. Relative metabolic activity of whole cell population in a bead; Calibration of the assay was shown in Fig S7. H. Relative quantification of proteins estimated by cell density per bead at each time point. I. Intracellular c-di-GMP components of E. Coli (600 and 60 CFU/Bead) (n = 5, independent experiments; biological replicates per condition). All data represent mean ± standard deviation (n = 3, independent experiments; biological replicates per condition); *p < 0.05; **p < 0.01; and ***p < 0.001 Significance by student’s t-test and one-way ANOVA, unless otherwise noted.
Typically, planktonic (free-swimming) cells in a biocompatible medium reach stationary phase within a few hours ( Fig. 2B). No substantial difference between the cells in free liquid culture and those grown in conventional alginate beads was noted because the leakage of E. coli from the conventional beads into the medium occurred almost instantly at the peripheral region of the beads (Fig. S2). When a small number of bacterial cells leak out of the polymeric encapsulants, cells outside the encapsulating matrix proliferate rapidly to distort the entire system. For example, stochastic entrapment techniques based on microfluidics [40] or coaxial encapsulation [41] strategies are known to generate numerous hydrogel microbial beads in a small volume, but these methods are not efficient and often result in the leakage of a significant number of unentrapped cells by the beads leading to contamination of the surrounding environment [22]. In our design, upon hydrogel confinement, bacteria replicate and initiate the production of various cellular substances within the bead. (Video S1, 2) Further, hydrogel beads with E. coli were examined on 2D agar plates, where a modified agar overlay technique was utilized to show that there is no cell leakage ( Fig. 2C). We show that E. coli could be stably entrapped inside the hydrogel-shell beads for several days and the organisms continued to grow in the bead until saturation (Fig. 2D).
Not surprisingly, a single E. coli cell in the bead can replicate and grow into a colony of cells when placed in a desirable medium. (Fig. S3) The hydrogel-shell beads function as a structural scaffold and form colonies within 12 h of its residency in the core. (Video S3) To estimate the behavior of cellular aggregates in a 3D hydrogel environment, bacteria counts were provided in colony forming units per bead (CFU/bead), by precise tuning of the initial number of entrapped bacterial cells ( Fig. 2E, Fig. S4). The colonies were tens to hundreds of micrometers in size and can be monitored throughout the transparent hydrogel layers. Eventually, the colonies develop into large cellular aggregates and occupy the available inner space. We also show that other wild-type strains ( Bacillus subtilis 168, Lactococcus lactis ATCC 11454, Pseudomonas aeruginosa PAO-1) could also be cultivated into distinguishable microcolonies within the beads (Fig. S5). The outer shell formation with entrapped cells in the core can create application-tailored artificial microenvironments to query bacteria behavior.
3.2. Heterogeneous micro-niches
Bacteria colonization in the hydrogel environment could result in dynamic alteration of the intricate physiological processes resulting in the production of secondary metabolites by either intrinsic or extrinsic factors or both. The concomitant production of the components of EPS and metabolic activity were further evaluated and compared between a group containing 60 and 600 CFU/bead. While the total number of bacterial cells increased in the core as expected (Fig. 2F), the amount of total protein produced by cell aggregates increased proportionally (Fig. S6). The initial number of cells ( i.e., CFU/bead) entrapped could influence the production of cellular components by reaching a saturation point given the space constraint and nutrient availability in the bead environment. Evaluation of the relative metabolic activity of the cells entrapped in a single bead revealed a rapid increase in cellular activity (Fig. 2G, Fig. S7). A higher level of metabolic activity was noted till the density of cells increased beyond the threshold, reflected by the production of proteins in the initial stages of development (Fig. 2H). Since the availability of nutrients influence cell proliferation, we noted that the cells at the periphery of the hydrogel beads consume most of the nutritional resources. Our results indicate varying levels of interaction among cells that are in different metabolic states [12, 42] even though the total viable cell count increased (Fig. S8).
There has been a growing consensus on the role of high population density and collective behavior of microorganisms, associated with the production, release, exchange, and detection of small molecules [43]. Cyclic-di-GMP (c-di-GMP), is one of the most common second messenger molecule known to regulate biofilm formation, motility, virulence, differentiation, and other processes. Higher intracellular levels of c-di-GMP was observed during the early growth phase ( Fig. 2I, Fig. S9) [36]. High intracellular c-di-GMP levels have been shown to be associated with biofilm formation in the early stages of development, whereas a low intracellular c-di-GMP level could promote a planktonic lifestyle [44]. In addition, similar to subpopulations in stratified biofilm structures [20], preferential growth behavior of cells in favorable environments induce heterogeneous communities ( i.e., high/low metabolically active cells) with different growth rates (Fig. S10). Metabolically active cells were positioned mostly around the periphery of the hy drogel bead due to the diffusion gradient of nutrients in the environment. Histological evaluation of the core-shell beads with microorganisms exhibited heterogeneous distributions of bacteria aggregates at the center and periphery (Fig. S11). These distributions highlight a self-generated matrix within the hydrogel which renders heterogeneous subpopulations as in a biofilm.
3.3. Hydrogel long-term entrapment
To query bacteria behavior during long-term confinement, key factors considered were disruption of the bead structures and leakage of the cells due to the growth of microorganisms. We assessed whether constructing thicker shell layers can sustain bacteria proliferation and colonization, while maintaining the integrity of the core–shell structures. Our initial studies show that thinner outer layers (< 100 μm shell thickness) can easily rupture within 48 h in a nutrition rich media (Fig. 3A ). By contrast, thicker layered beads (~300 μm shell thickness) can provide adequate stiffness without rupturing (Fig. 3B), unless denatured by external factors. Enzymatic degradation studies of hydrogel layers by alginate lyase (Video S4, 5) shows that the structures are stable without undergoing any enzymatic hydrolysis.
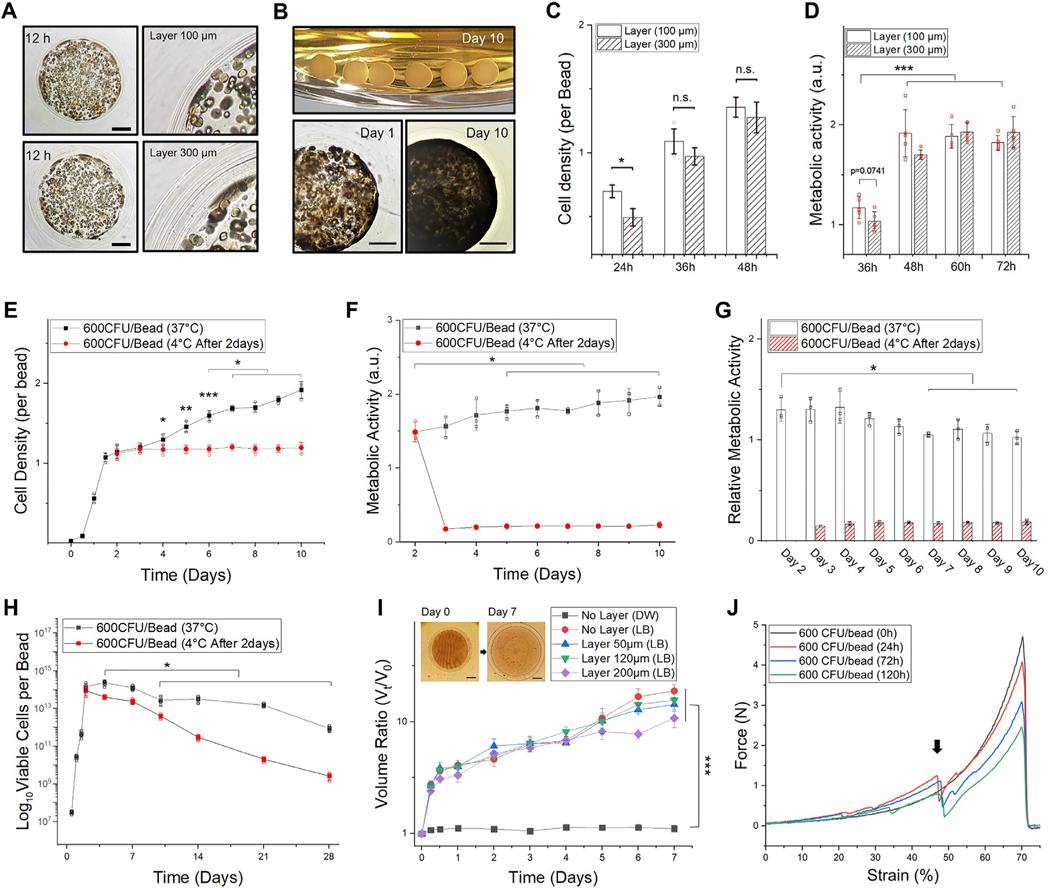
Fig. 3.
Thick hydrogel layer for long-term confinement. The core has an average diameter of 2 mm (WT E. coli 600 CFU/bead). A. A representative microscopic image (left) and magnified view (right) of hydrogel-shell beads with 100 and 300 μm of layer thickness. Scale bar is 500 μm. B. Image of hydrogel-shell beads cultured for 10 days (upper), microscopic images of hydrogel-shell beads with 300 μm of layer thickness in day 1 (lower-left) and day 10 (lower-right). Scale bar is 500 μm. C. Comparison of E. coli cell densities per bead. D. Comparison of metabolic activity with different layer thickness (n = 5, independent experiments per condition). E. Measurements of E. coli densities per bead for 10 days. F. Measurement of metabolic activities. G. Relative metabolic activity in cells. H. Viable cell counts of hydrogel beads cultivated for 4 weeks, determined by the standard plate method. The total viable cells per bead (y-axis) was plotted in the range of 107 to 1017 at the log scale. (E-H) Hydrogel-shell beads with 300 μm of layer thickness; after culturing the beads for 2 days the same experimental groups were placed in a cold room (4 °C) as a comparison group. I. Hydrogel bead swollen volume ratio ( V t / V 0 ). Volume of the beads at each time point (Vt) relative to initial volume ( V 0 ). J. Plot of force–deformation (as % strain) curves show the mechanical behavior of layered hydrogel beads after microbial colonization (0, 24, 72, and 120 h, respectively). The black arrow indicates layer breaking point. Data represent mean ± standard deviation ( n = 3, biological replicates per condition); *p < 0.05; **p < 0.01; and ***p < 0.001 Significance by student’s t-test and one-way ANOVA, unless otherwise noted.
Continued proliferation of cell clusters into colonies containing millions of tightly packed cells can exert substantial stress at the outer layers. When the hydrogel layer is thicker (> 300 μm of shell thickness), the growth of bacteria is slightly diminished due to decreased diffusion of nutrient components (Fig. 3C). After reaching a saturation point, the metabolic activity of cell aggregates was found to be similar to that of beads with relatively thinner shell layers (Fig. 3D). Since the shell of the hydrogel beads comprise of natural alginate polymers crosslinked with Ca 2+, the permeability of alginate polymer networks is mostly affected by gelation (Fig. S12). Consequently, the thicker layers of the outer shell does not significantly alter the growth of E. coli, demonstrating that our design could sustain long-term (10 days or more) entrapment.
E. coli inside the beads proliferated even after reaching a near saturation point within the bead microenvironment ( Fig. 3E ). Although cells aggregated considerably and occupied the inner regions, the metabolic activities of the microbial population in a single bead increased ( Fig. 3F ) with time for up to 10 days as the volume of the bead increased by ~400%. However, relative metabolic activities of the entrapped cells decreased gradually (~75%) compared to day 2 (Fig. 3G). Bacteria components produced by the entrapped cells could create a dense environment to inhibit effective diffusion of substrates from/into the core region. There is a possibility of accumulation of byproducts, which should be taken into account due to the increased diffusion barrier. As a result, a large portion of the cells began to lose their viability under unfavorable circumstances for survival (Fig. 3H). However, it is known that the cells enter a differentiated state in the beads to tolerate exogenous stress in adverse environments [45]. These stressed cells are stochastically formed in a subpopulation of less active cells within confined structures and enter a dormant, nondividing state for long-term survival [42].
3.4. Expansion of hydrogel environment
Alginic acid is highly hygroscopic and alginate hydrogels tend to swell without being dissolved within a short period of time while retaining the moisture content. The swollen bead in the biological medium facilitates the formation of a more porous and less dense gel compared to the non-swollen gel matrix [29,46]. To support this behavior, swelling properties of alginate beads either with a shell layer of defined thickness or without the layer with E. Coli in the core were examined in a cold room by minimizing the influence of other factors such as thermal expansion, cell growth, metabolic activities, and EPS generation. In our system, the volume of beads increased ~5X in the growth medium in 4 h reaching a maximum of 10X the initial volume in 7 days (Fig. 3I, Fig. S13). The initial increase in volume associated with the total number of microbes entrapped in the core of the hydrogel was due to the tonicity of the solution and osmotic imbalance (Fig. S14). Furthermore, after bacterial cells were colonized, the mechanical properties of the hydrogel core was gradually altered due to the production of various byproducts of bacteria (Fig. 3J, Fig. S15).
This unique environment, as depicted in Fig. 4, along with the swelling of the core and shell favors hydration, proliferation, and colonization of organisms affirming a biofilm-like microenvironment in the swollen core. Transport of essential substrates such as nutrients would not be a limiting factor in cell-free alginate hydrogels [16]. Spatial heterogeneity (i.e., distribution of cell populations) is an important feature of biofilm environments given the presence of concentration gradient of nutrients, bacterial waste, and signaling molecules [45,47]. For spatial organization, physiological heterogeneity will be accompanied by spatial heterogeneity. The chemical/biological gradient within the hydrogel environment was gradually developed because of the diffusion properties of alginate polymer matrixes and substrates generated by cells. Consequently, the heterogeneous micro-niche were molded throughout the cross-section of hydrogel in contrast to homogenous growth environments of planktonic cells in growth-free condition.
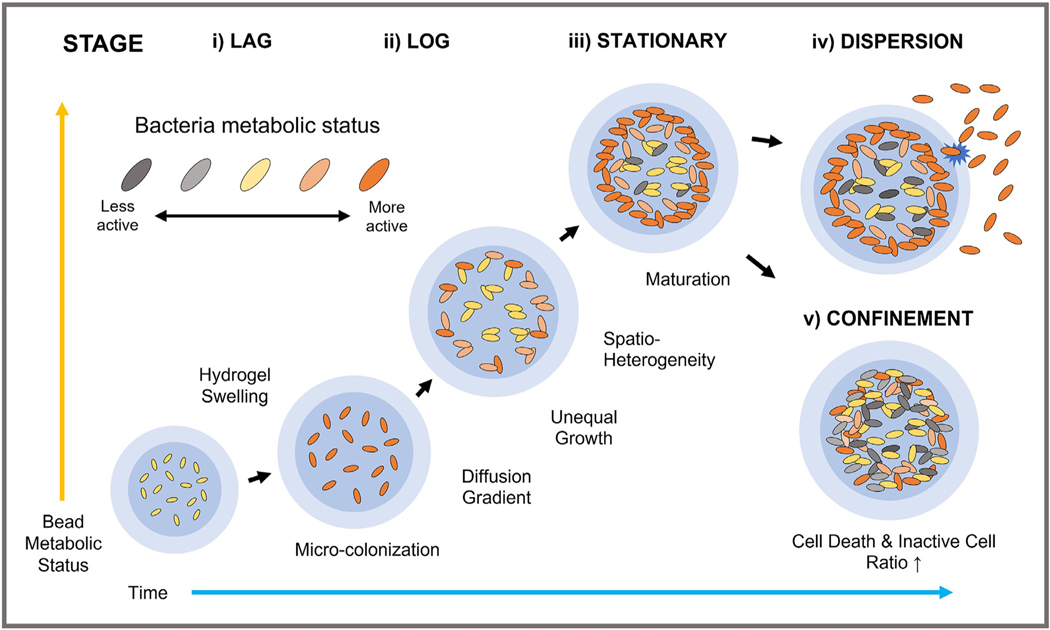
Fig. 4.
Process of bacteria confinement in 3D hydrogel beads with desired shell thickness. Alginate polymer networks with hydrogel swelling enhance diffusion of essential substrates. Microcolony formation is noted at all depths of the core, and chemical gradients lead to unequal growth from the center to the periphery of hydrogel beads. Small substrates to support growth and viability would be preferentially consumed by the cells located at the periphery regions. The EPS components including biomass and proteins accumulate to inhibit oxygen and nutrient diffusion towards the center. The heterogenous growth leads to high local cell density contributing to considerable compression, or rupture of the hydrogel matrix. Bacteria dispersion from the beads could occur due to complex interactions with the surroundings and degradation of hydrogel-shell layer. Pattern of bacteria dispersion varies, depending on both species and environmental factors. Such lifestyle of the formed structures could be altered to tolerate exogenous stresses and adverse environments for long-term survival.
3.5. Bacteria-derived functional bead
Popular strategies to create bacteria-derived functional biomaterials are developed by entrapment of genetically modified microorganisms expressing recombinant proteins (Fig. 5A) [19, 27, 28, 48] but unfavorable conditions or procedures might lead to rapid cell death or leakage. When a hydrogel barrier was constructed to confine GFP and RFP expressing E. coli within a bead environment, substantial differences were noted compared to conventional encapsulation strategies (Fig. 5B). Bacterial cells tend to grow at the periphery of the encapsulant. Given the transparent optical properties of alginate hydrogels, monitoring bacterial cells was possible not only with naked eyes but also by fluorescence (photoluminescence) spectroscopy (Fig. 5C,D, Fig. S16). Under confined conditions, accumulation of recombinant proteins produced by E. coli would be proportional to the cell density. It is worth noting that a complete biofilm-like confinement is highly desirable to design effective bacteria-derived functional hydrogels.
Fig. 5.
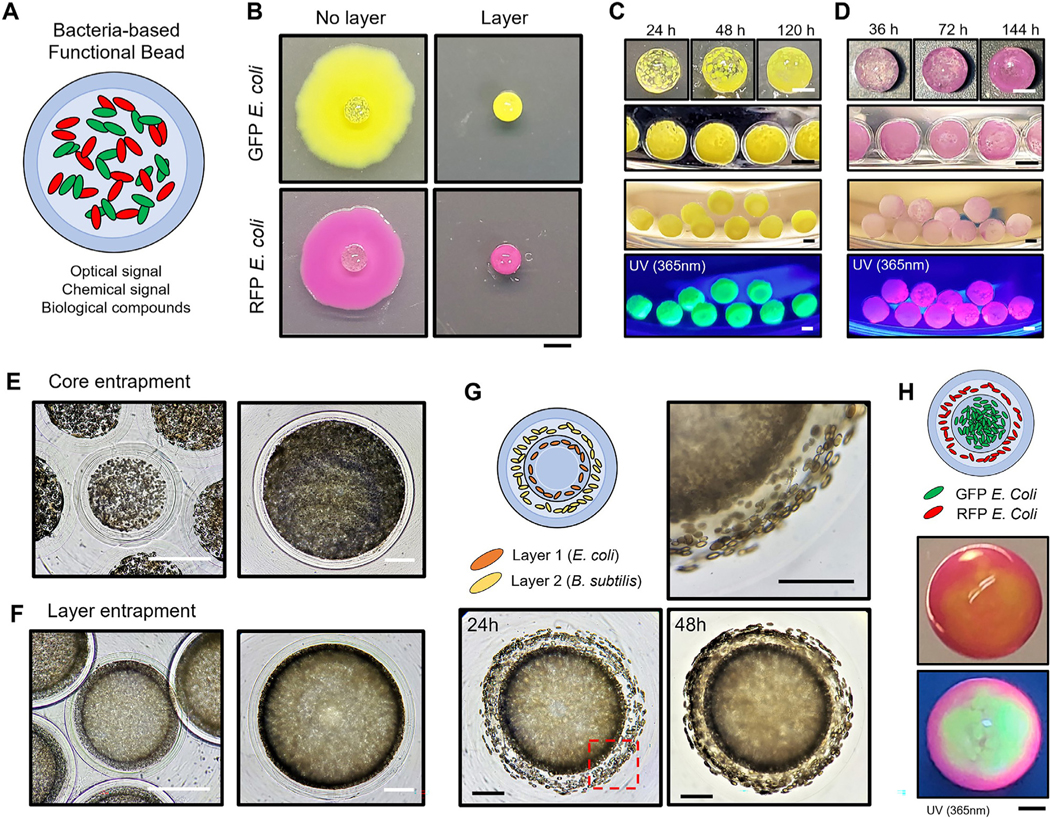
Bacteria-derived functional beads and hierarchical immobilization A. An illustration of bacteria-derived functional beads for multi-purpose. B. GFP and RFP E. coli encapsulation with/without hydrogel-shell. Soft agar overlay onto 2D agar plates was used to observe cell leakage at day 7. Scale bar is 2 mm. C-D. GFP and RFP E. coli encapsulation at different time points. Images of GFP and RFP E. coli hydrogel-shell beads illuminated by a hand-held 365 nm UV light. The core has an average diameter of 1.5 mm with 200 μm of layer thickness. Scale bar is 1 mm. E-F. Comparison of microscopic images of E. coli core and layer entrapment. Scale bar is 500 μm. G. Hierarchical encapsulation of two different type of microorganisms (E. coli and B. subtilis). The empty core has an average diameter of 1.5 mm. (1st layer, E. coli: 200 μm; 2nd layer, B. subtilis: 300 μm; and 3rd layer: 200 μm) The red dash box (lower-left) indicates the magnified image (upper-right) of the layered regions. Scale bar is 500 μm. H. Multilayered hydrogel beads for functional hierarchy. A representative image of stratified GFP and RFP E. coli immobilization, under room light (middle) and 365 nm UV light (lower). Images were taken under room light and 365 nm UV light. Scale bar is 500 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
One of the unique characteristics of the fabricated multi-layered architectures is the precise tuning of dimensions to entrap a specified number of microorganisms in the core or shell layers ( Fig. 5E,F, Video S6, 7). The outer-most hydrogel layer offers a degree of stability to sustain growth within each layer without leakage. By expanding the multi-layered fabrication concept, we show that various microorganisms could be sequentially encapsulated in each layer ( Fig. 5G,H, Video S8). The stratified immobilization reflects the hierarchical and heterogeneous nature of biological structures enabling the development of complex multi-functional hydrogel architectures derived from bacteria.
3.6. Hydrogel modulator and communication systems
Cell communication typically uses chemical signals mediated by intra/extracellular messenger molecules that not only governs the basic activities of cells but also regulates the physiological functions in a milieu. In Fig. 6, we demonstrate three representative cell communication modules enabled by the hydrogel core–shell beads. Hydrogel beads encapsulating specific bacteria could be co-cultured with other hydrogel modulators or mammalian cells.
Fig. 6.
Hydrogel bead modulators for inter- and intra- species communication. A. Bead-to-Bead communication model mediated by nisin (MW = 3,354.07 g/mol) to modulate the expression system. RFP expressing L. Lactis (NP, Nisin Producer) and GFP expressing L. Lactis (NR, Nisin Responder) encapsulated in hydrogel-shell beads. B. A representative fluorescence image and magnified view of nisin-producing and responding L. lactis beads for communication. The core has an average diameter of 500 μm with 100 μm of layer thickness. Scale bar is 2 mm. (inset image, Scale bar is 500 μm) C. Fluorescence signal detection of NP bead D. Fluorescence signal detection of NR beads at each time point (0, 24, 36 and 48 h). NP and NR beads were incubated together for 48 h. E. Stratified communication model in a single hierarchical bead. Fluorescence images of stratified beads (inner core: NP, outer layer: NR) incubated for 24 h. Scale bar is 500 μm F. Multi-channel fluorescent intensity profiling of stratified hydrogel beads containing NP and NR cells at different time points (12 and 24 h, respectively). Fluorescence intensity profile across the bead diameter (denoted by the yellow line) is provided in the inset images (Scale bar is 500 μm). G. Real-time RFP signal (at 5 min interval) from empty bead and NP bead for 24 h. H. Real-time GFP signal from hydrogel bead encapsulating NP (core) and NR (layer) strains in hierarchical hydrogel structures. I. in vitro co-culture model of bacteria bead and cancer cells to elucidate dynamic cellular response in relation to metabolites secreted from live bacteria. J. Microscopic images (left) and magnified view (right) at each time point of HCT-116 cells co-cultured with micro-colonized beads. The red dash box indicates the magnified image of the layered region of probiotic L. Lactis bead and HCT-116 cells. Scale bar is 250 μm. K. Comparative metabolic activities of L. Lactis colonized in a hydrogel environment after co-cultivation with HCT-116 cells for 12 h. L. Cell viability of HCT-116 cells co-cultured with probiotic L. Lactis beads for 24 and 48 h. An aqueous solution of DMSO (1% v/v) as a positive control and an empty bead as a negative control were used for the assay. The results were statistically compared with the negative control group. All data represent mean ± standard deviation ( n = 3, biological replicates per condition); *p < 0.05; **p < 0.01; ***p < 0.001 and ****p < 0.0001 Significance by student’s t-test and one-way ANOVA, unless otherwise noted. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
First, a density-dependent microenvironment for bacteria communication such as quorum sensing (QS) is triggered when the cell density increased above a threshold [12, 49]. For example, a greater proportion of extracellular factors such as acyl homoserine lactones (AHLs) and autoinducer peptides (AIPs) would regulate numerous processes [45]. To substantiate QS of bacteria, we examined nisin-controlled hydrogel beads encapsulating a nisin producer (NP, RFP expressing) and a nisin responder (NR, GFP expressing) L. lactis strain (Fig. 6A, Fig. S17). Both RFP and GFP signals could be detected from each beads when co-cultured, after 24 h (Fig. 6B). The NP beads showed RFP signal due to the autoregulation mechanism of nisin biosynthesis (Fig. 6C). The NR beads expressed GFP signal upon receiving the signaling molecules produced from the NP bead (Fig. 6D). This communicative mode could be in accordance with the population density through the use of signaling molecules [33].
Next, a stratified communication model for developing hierarchical hydrogel structures was shown to exemplify bacteria interactions in a heterogeneous environment [50] ( Fig. 6E, Fig. S18). In this system, RFP signals at the core and GFP signal at the periphery of the hydrogel core-shell bead were detected after 24 h of incubation, respectively ( Fig. 6F). Fluorescent signal enhancement was monitored in real-time from the hierarchical hydrogel beads encapsulated with NP L. Lactis in the core and NR L. Lactis in the shell layer of the bead (Fig. 6G,H). Our results support the fact that hydrogel beads serve as an excellent platform to mediate transport of signaling molecules, depicting a dynamic pairwise interaction.
Co-culture studies could provide intuitive options to reveal the relationship between different organisms to assess host-microbe communication. Very few systems [51–53] to modify expression at the genetic level to control excessive growth of microorgan isms (Fig. S19) exist in response to external cues. A robust in vitro co-culture system was developed to demonstrate dynamic alteration and reciprocal interaction between bacteria and mammalian cells. To conceptualize dynamic host-microbe interactions in vitro, bacteria colonies in hydrogel-shell beads was co-cultured with mammalian cells (Fig. 6I, Fig. S20). A natural probiotic strain, L. Lactis ATCC11454, was recapitulated to demonstrate the production and response of bacteriocin nisin against colorectal carcinoma cells (Fig. 6J). Interestingly, the metabolic status of the probiotic bacteria in forming colonies was pronounced when co-cultured with carcinoma cells (Fig. 6K, Fig. S21). The bacteriocin secreted through the hydrogel barriers, as hypothesized, inhibited the proliferation of HTC116 cells (Fig. 6L) and induced cell apoptosis (Fig. S22) [54]. Our results imply a possible bidirectional alteration of the configuration as mediated by soluble factors (e.g., metabolite, chemokines or cytokines etc.). The co-culture model system provides unprecedented tools for in vitro tuning to not only assess host-microbe interactions but a myriad of other specific applications.
4. Discussion
We show biofilm-like entrapment of microorganisms for long-term colonization in heterogeneous environment which comprise of alginate hydrogel core with concentric shell layers. A comprehensive understanding of the diffusion characteristics of the core-shell layers is critical for cell encapsulation to maintain the rate of influx and efflux of small molecules and metabolites that facilitate interaction, communication, and viability of organisms. The fabricated hydrogel-shell beads of different scales comprise of natural alginate polymers crosslinked with Ca2+ ions without any auxiliary crosslinking agents. In general, diffusion rate of substances into/from alginate gel networks would be varied depending on factors such as molecular weight, polarity, and structure [55]. The release kinetics of a cationic dye is shown in Fig. S12. Our results indicate that the diffusion of small molecules was unaffected either with or without the hydrogel layer during the time course of this study. The parameters governing solute diffusion in alginate hydrogels were investigated wherein, molecules (Molecular Weight less than 105 g/mol, 100 kDa) such as glucose, L-tryptophan, vitamin b12, α-lactalbumin, and transferrin, could freely diffuse into and out of the designed alginate hydrogel structures [30,55,56]. Ideally, the polysaccharide-based core-shell beads facilitate diffusion and transport of nutrients and metabolites within the hydrogel polymer networks.
Characteristics of alginate hydrogel-shell and core primarily depend on the properties of alginic acid gelling and non-gelling ions. The concentration of gelling and non-gelling ions affect the rearrangement of alginate polymers within the hydrogel structures. During gelation, excessive Ca2+ ions are involved in more than two alginate polymer chains in the crosslinking zone, called “egg-box” junctions [57]. According to a modified egg-box model, this may have a significant impact on the porosity of alginate gel. Besides, the swelling properties of the alginate shell and core is influenced by the gelation kinetics under different conditions. Monovalent ions (e.g., K+, Na+) in the medium diffuse into the hydrogel matrix and then Ca2+ exchange occurs, which are attached to carboxyl groups of β-D-mannuronic acid chains (M blocks) [56]. This leads to more hydrated structures with the relaxation of M block chains, while maintaining the structural integrity due to the interaction between α-L-guluronic acid chains (G blocks) and Ca2+ ions. The swollen gel networks can alter the diffusion properties of both the core and shell. Gradually, the EPS components, including biomass accumulate to inhibit oxygen and nutrient diffusion towards the center. As a result, the high porosity and large pore size of alginate networks could significantly contribute to the development of unique microenvironments for microbial colonization that promote the formation of an EPS matrix-like biofilm.
In soft hydrogel research, one of the extensively attempted methods to date is thin layer deposition with polycationic polymers such as poly-L-lycine, polyornithine or a covalent crosslinking of deposited layers by cytotoxic processes and/or chemicals [14,19,58]. However, such methods could considerably inhibit free diffusion of small substrates not only by blocking the nanoporous structures of alginate polymer networks, but also by increasing the electrostatic interactions at the layered regions. Even if bacterial cells are initially able to survive the environmental adversities, many inconsistent results on bacteria encapsulation [7] are reported. Unfavorable methodologies can only temporarily confine bacterial cells to ultimately result in either cell leakage or rapid cell death from such confined environments at later stages. To cultivate bacterial cells with spatial confinement, it is essential to evaluate whether the restricted space would be favorable in simulating an appropriate milieu for growth. The conceived beads render a biofilm-like microenvironment in the swollen hydrogel networks with enhanced diffusion, to facilitate hydration, proliferation, colonization and interaction of organisms compared to the existing entrapment technologies [16, 17,19,59].
Confined cells physiologically would be distinct from planktonic cells in free cultures, and the activities of aggregate cells in biofilms are resolved spatially and temporally. Further, a generalized perspective of hydrogel design parameters for encapsulating prokaryotic cells is necessary because of the diverse growth stages involved in the cell cycle of prokaryotes. For example, the presence of hypoxic conditions [60, 61] in the inner regions of hydrogel beads would be indispensable and beneficial for obligate anaerobes. In the absence of oxygen, nutritional interactions such as cross-feeding may orchestrate biochemical cycles among bacterial communities. The 3D hydrogel environments with complex heterogeneities could serve as a structured biofilm platform wherein metabolic activities could be promoted and regulated by nutrient gradients and spatial-temporal constraints. Yet, considerably less is known on the physiological characteristics of bacteria in biofilms. Quantative evaluation of confinement, physiological regulation, and heterogeneity of microorganisms with varied population density (e.g., allee effect) [62] is beyond the scope of our study. However, this study provides basic insights on the characteristics of various microorganisms in restricted and stratified environments and how confinement alters the physiological characteristics of organisms in biofilms.
5. Conclusion
In summary, we present a strategy to develop tunable alginate hydrogel core-shell beads for bacteria encapsulation. The core-shell beads with hierarchical architecture can host bacterial cells for more than ten days in physiologically relevant conditions. Further, the hydrogel microenvironment can support stratified subpopulations of bacteria as in a biofilm. Bacterial cells can be entrapped in the core as well as in outer shell layers which can be tuned for thickness and number of layers as well as cell density. Further, we have demonstrated intra- and inter- species communication to show dynamic interaction of bacterial colonies encapsulated in independent microenvironments upon complete confinement. We believe that a myriad of biological scenarios can be examined in the future to reveal various microbial functions at the relevant spatial scales in situ. Along with the biofilm-like confinement, our findings provide new insights on the rational design of soft hydrogel architectures with functional architectures. Future experiments could constitute engineering hydrogel materials to promote specific microenvironments to understand multi-organism (host-microbe) interactions to trigger or evaluate specific response as end points.
Supplementary Material
Statement of significance.
Bacteria confinement in a natural soft hydrogel structure has always been a challenge due to the collapse of hydrogel architectures. Alternative methods have been attempted to encapsulate microorganisms by employing various processes to avoid/minimize rupturing of hydrogel structures. However, most of the past approaches have been unfavorable in balancing cell proliferation and functionality upon confinement. Our study addresses the fundamental gap in knowledge necessary to create favorable and complex 3D biofilm mimics utilizing natural hydrogel for microbial colonization for long-term studies. Our approach represents a cornerstone in the development of 3D functional architectures not only to advance studies in microbial communication, host-microbe interaction but also to address basic and fundamental questions in biology.
Acknowledgments
We thank Dr. James Imaly, Dr. Rachel Whitaker, Dr. Cari Vanderpool and Dr. Ting Lu (University of Illinois at Urbana-Champaign, Urbana, IL, USA) for providing microbial strains used in this study. This work was supported by the UIUC startup funds to J. Irudayaraj. Partial fellowship support to Yoon Jeong was provided by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number T32EB019944. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.actbio.2023.01.003.
References
- [1].Green JJ, Elisseeff JH, Mimicking biological functionality with polymers for biomedical applications, Nature 540 (7633) (2016) 386–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Koo H, Yamada KM, Dynamic cell–matrix interactions modulate microbial biofilm and tissue 3D microenvironments, Curr. Opin. Cell Biol. 42 (2016) 102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tibbitt MW, Anseth KS, Hydrogels as extracellular matrix mimics for 3D cell culture, Biotechnol. Bioeng. 103 (4) (2009) 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chang TM, Semipermeable microcapsules, Science 146 (3643) (1964) 524–525. [DOI] [PubMed] [Google Scholar]
- [5].Lim F, Sun AM, Microencapsulated islets as bioartificial endocrine pancreas, Science 210 (4472) (1980) 908–910. [DOI] [PubMed] [Google Scholar]
- [6].Hackel U, Klein J, Megnet R, Wagner F, Immobilisation of microbial cells in polymeric matrices, Eur. J. Appl. Microbiol. Biotechnol. 1 (4) (1975) 291–293. [Google Scholar]
- [7].Cassidy M, Lee H, Trevors J, Environmental applications of immobilized microbial cells: a review, J. Ind. Microbiol. Biotechnol. 16 (2) (1996) 79–101. [Google Scholar]
- [8].Ladet S, David L, Domard A, Multi-membrane hydrogels, Nature 452 (7183) (2008) 76–79. [DOI] [PubMed] [Google Scholar]
- [9].Zarket BC, Raghavan SR, Onion-like multilayered polymer capsules synthesized by a bioinspired inside-out technique, Nat. Commun. 8 (2017) 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Liu G, Ding Z, Yuan Q, Xie H, Gu Z, Multi-layered hydrogels for biomedical applications, Front. Chem. 6 (2018) 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wondraczek L, Pohnert G, Schacher FH, Köhler A, Gottschaldt M, Schubert US, Küsel K, A.A. Brakhage, Artificial microbial arenas: materials for observing and manipulating microbial consortia, Adv. Mater. 31 (24) (2019) 1900284. [DOI] [PubMed] [Google Scholar]
- [12].Flemming H-C, Wingender J, The biofilm matrix, Nat. Rev. Microbiol. 8 (9) (2010) 623–633. [DOI] [PubMed] [Google Scholar]
- [13].Strathmann M, Griebe T, Flemming H-C, Artificial biofilm model–a useful tool for biofilm research, Appl. Microbiol. Biotechnol. 54 (2) (2000) 231–237. [DOI] [PubMed] [Google Scholar]
- [14].Kim BJ, Park T, Moon HC, Park SY, Hong D, Ko EH, Kim JY, Hong JW, Han SW, Kim YG, Cytoprotective alginate/polydopamine core/shell microcapsules in microbial encapsulation, Angew. Chem. Int. Ed. 53 (52) (2014) 14 4 43–14 4 46. [DOI] [PubMed] [Google Scholar]
- [15].Li P, Müller M, Chang MW, Frettlöh M, Schönherr H, Encapsulation of autoinducer sensing reporter bacteria in reinforced alginate-based microbeads, ACS Appl. Mater. Interfaces 9 (27) (2017) 22321–22331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li Z, Behrens AM, Ginat N, Tzeng SY, Lu X, Sivan S, Langer R, Jaklenec A, Biofilm-inspired encapsulation of probiotics for the treatment of complex infections, Adv. Mater. 30 (51) (2018) 1803925. [DOI] [PubMed] [Google Scholar]
- [17].Connell JL, Ritschdorff ET, Whiteley M, Shear JB, 3D printing of microscopic bacterial communities, Proc. Natl. Acad. Sci. 110 (46) (2013) 18380–18385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chu EK, Kilic O, Cho H, Groisman A, Levchenko A, Self-induced mechanical stress can trigger biofilm formation in uropathogenic Escherichia coli, Nat. Commun. 9 (2018) 4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tang T-C, Tham E, Liu X, Yehl K, Rovner AJ, Yuk H, de la Fuente-Nunez C, Isaacs FJ, Zhao X, Lu TK, Hydrogel-based biocontainment of bacteria for continuous sensing and computation, Nat. Chem. Biol. 17 (6) (2021) 724–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S, Biofilms: an emergent form of bacterial life, Nat. Rev. Microbiol. 14 (9) (2016) 563. [DOI] [PubMed] [Google Scholar]
- [21].Zheng D-W, Pan P, Chen K-W, Fan J-X, Li C-X, Cheng H, Zhang X-Z, An orally delivered microbial cocktail for the removal of nitrogenous metabolic waste in animal models of kidney failure, Nat. Biomed. Eng. 4 (9) (2020) 853–862. [DOI] [PubMed] [Google Scholar]
- [22].Hol FJ, Dekker C, Zooming in to see the bigger picture: Microfluidic and nanofabrication tools to study bacteria, Science 346 (6208) (2014) 1251821. [DOI] [PubMed] [Google Scholar]
- [23].Vincent ME, Liu W, Haney EB, Ismagilov RF, Microfluidic stochastic confinement enhances analysis of rare cells by isolating cells and creating high density environments for control of diffusible signals, Chem. Soc. Rev. 39 (3) (2010) 974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Weibel DB, DiLuzio WR, Whitesides GM, Microfabrication meets microbiology, Nat. Rev. Microbiol. 5 (3) (2007) 209–218. [DOI] [PubMed] [Google Scholar]
- [25].You Z, Pearce DJ, Giomi L, Confinement-induced self-organization in growing bacterial colonies, Sci. Adv. 7 (4) (2021) eabc8685. [DOI] [PubMed] [Google Scholar]
- [26].Schaffner M, Rühs PA, Coulter F, Kilcher S, Studart AR, 3D printing of bacteria into functional complex materials, Sci. Adv. 3 (12) (2017) eaao6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Huang J, Liu S, Zhang C, Wang X, Pu J, Ba F, Xue S, Ye H, Zhao T, Li K, Programmable and printable Bacillus subtilis biofilms as engineered living materials, Nat. Chem. Biol. 15 (1) (2019) 34–41. [DOI] [PubMed] [Google Scholar]
- [28].Johnston TG, Yuan S-F, Wagner JM, Yi X, Saha A, Smith P, Nelson A, Alper HS, Compartmentalized microbes and co-cultures in hydrogels for on-demand bioproduction and preservation, Nat. Commun. 11 (2020) 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lee KY, Mooney DJ, Hydrogels for tissue engineering, Chem. Rev. 101 (7) (2001) 1869–1880. [DOI] [PubMed] [Google Scholar]
- [30].Bochenek MA, Veiseh O, Vegas AJ, McGarrigle JJ, Qi M, Marchese E, Omami M, Doloff JC, Mendoza-Elias J, Nourmohammadzadeh M, Alginate encapsulation as long-term immune protection of allogeneic pancreatic islet cells transplanted into the omental bursa of macaques, Nat. Biomed. Eng. 2 (11) (2018) 810–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kang S-M, Lee J-H, Huh YS, Takayama S, Alginate microencapsulation for three-dimensional in vitro cell culture, ACS Biomater. Sci. Eng. 7 (7) (2020) 2864–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang L, Zhang X, Tang C, Li P, Zhu R, Sun J, Zhang Y, Cui H, Ma J, Song X, Engineering consortia by polymeric microbial swarmbots, Nat. Commun. 13 (2022) 3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jeong Y, Kong W, Lu T, Irudayaraj J, Soft hydrogel-shell confinement systems as bacteria-based bioactuators and biosensors, Biosens. Bioelectron. 219 (2023) 114809. [DOI] [PubMed] [Google Scholar]
- [34].Yuk H, Wu J, Zhao X, Hydrogel interfaces for merging humans and machines, Nat. Rev. Mater. 7 (2022) 935–952. [Google Scholar]
- [35].Jeong Y, Irudayaraj J, Multi-layered alginate hydrogel structures and bacteria encapsulation, Chem. Commun. 58 (61) (2022) 8584–8587. [DOI] [PubMed] [Google Scholar]
- [36].Chua SL, Hultqvist LD, Yuan M, Rybtke M, Nielsen TE, Givskov M, Tolker-Nielsen T, Yang L, In vitro and in vivo generation and characterization of Pseudomonas aeruginosa biofilm–dispersed cells via c-di-GMP manipulation, Nat. Prot. 10 (8) (2015) 1165. [DOI] [PubMed] [Google Scholar]
- [37].Das MC, Sandhu P, Gupta P, Rudrapaul P, De UC, Tribedi P, Akhter Y, Bhattacharjee S, Attenuation of Pseudomonas aeruginosa biofilm formation by Vitexin: a combinatorial study with azithromycin and gentamicin, Sci. Rep. 6 (1) (2016) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chen C-Y, Nace GW, Irwin PL, A 6 × 6 drop plate method for simultaneous colony counting and MPN enumeration of Campylobacter jejuni, Listeria monocytogenes, and Escherichia coli, J. Microbiol. Methods 55 (2) (2003) 475–479. [DOI] [PubMed] [Google Scholar]
- [39].Mierau I, Kleerebezem M, 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis, Appl. Microbiol. Biotechnol. 68 (6) (2005) 705–717. [DOI] [PubMed] [Google Scholar]
- [40].Kaminski TS, Scheler O, Garstecki P, Droplet microfluidics for microbiology: techniques, applications and challenges, Lab Chip 16 (12) (2016) 2168–2187. [DOI] [PubMed] [Google Scholar]
- [41].Loscertales IG, Barrero A, Guerrero I, Cortijo R, Marquez M, Ganan-Calvo A, Micro/nano encapsulation via electrified coaxial liquid jets, Science 295 (5560) (2002) 1695–1698. [DOI] [PubMed] [Google Scholar]
- [42].Lewis K, Persister cells, Annu. Rev. Microbiol. 64 (2010) 357–372. [DOI] [PubMed] [Google Scholar]
- [43].McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S, Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal, Nat. Rev. Microbiol. 10 (1) (2012) 39–50. [DOI] [PubMed] [Google Scholar]
- [44].Blanka A, Düvel J, Dötsch A, Klinkert B, Abraham W-R, Kaever V, Ritter C, Narberhaus F, Häussler S, Constitutive production of c-di-GMP is associated with mutations in a variant of Pseudomonas aeruginosa with altered membrane composition, Sci. Signal. 8 (372) (2015) ra36-ra36. [DOI] [PubMed] [Google Scholar]
- [45].Stewart PS, Franklin MJ, Physiological heterogeneity in biofilms, Nat. Rev. Microbiol. 6 (3) (2008) 199–210. [DOI] [PubMed] [Google Scholar]
- [46].Li J, Mooney DJ, Designing hydrogels for controlled drug delivery, Nat. Rev. Mater. 1 (12) (2016) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wimpenny J, Manz W, Szewzyk U, Heterogeneity in biofilms, FEMS Microbiol. Rev. 24 (5) (2000) 661–671. [DOI] [PubMed] [Google Scholar]
- [48].Liu X, Inda ME, Lai Y, Lu TK, Zhao X, Engineered living hydrogels, Adv. Mater. 34 (2022) 2201326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Flemming H-C, Neu TR, Wozniak DJ, The EPS matrix: the “house of biofilm cells, J. Bacteriol. 189 (22) (2007) 7945–7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Seth EC, Taga ME, Nutrient cross-feeding in the microbial world, Front. Microbiol. 5 (2014) 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kim J, Hegde M, Jayaraman A, Co-culture of epithelial cells and bacteria for investigating host–pathogen interactions, Lab Chip 10 (1) (2010) 43–50. [DOI] [PubMed] [Google Scholar]
- [52].Din MO, Danino T, Prindle A, Skalak M, Selimkhanov J, Allen K, Julio E, Atolia E, Tsimring LS, Bhatia SN, Synchronized cycles of bacterial lysis for in vivo delivery, Nature 536 (7614) (2016) 81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, Camacho DM, Fadel CW, Bein A, Swenor B, Nestor B, Cronce MJ, Tovaglieri A, A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip, Nat. Biomed. Eng. 3 (7) (2019) 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ahmadi S, Ghollasi M, Hosseini HM, The apoptotic impact of nisin as a potent bacteriocin on the colon cancer cells, Microbial Pathogen. 111 (2017) 193–197. [DOI] [PubMed] [Google Scholar]
- [55].Lee BB, Ravindra P, Chan ES, Size and shape of calcium alginate beads produced by extrusion dripping, Chem. Eng. Technol. 36 (10) (2013) 1627–1642. [Google Scholar]
- [56].Bajpai S, Sharma S, Investigation of swelling/degradation behaviour of alginate beads crosslinked with Ca2 + and Ba2 + ions, Reactive Funct. Polym. 59 (2) (2004) 129–140. [Google Scholar]
- [57].Braccini I, Pérez S, Molecular basis of Ca2 + -induced gelation in alginates and pectins: the egg-box model revisited, Biomacromolecules 2 (4) (2001) 1089–1096. [DOI] [PubMed] [Google Scholar]
- [58].Larrañaga A, Lomora M, Sarasua J-R, Palivan CG, Pandit A, Polymer capsules as micro-/nanoreactors for therapeutic applications: Current strategies to control membrane permeability, Progr. Mater. Sci. 90 (2017) 325–357. [Google Scholar]
- [59].Berdy B, Spoering AL, Ling LL, Epstein SS, In situ cultivation of previously uncultivable microorganisms using the ichip, Nat. Protocols 12 (10) (2017) 2232–2242. [DOI] [PubMed] [Google Scholar]
- [60].Omar SH, Oxygen diffusion through gels employed for immobilization, Appl. Microbiol. Biotechnol. 40 (2) (1993) 173–181. [Google Scholar]
- [61].Shishido M, Toda M, Simulation of oxygen concentration profile in calcium alginate gel beads entrapping microbes during biological phenol degradation, Chem. Eng. Sci. 51 (6) (1996) 859–872. [Google Scholar]
- [62].Stephens PA, Sutherland WJ, Freckleton RP, What is the Allee effect? Oikos 87 (1) (1999) 185–190. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.