Abstract
The hepatitis C virus (HCV) nonstructural 5A (NS5A) protein has been controversially implicated in the inherent resistance of HCV to interferon (IFN) antiviral therapy in clinical studies. In this study, the relationship between NS5A mutations and selection pressures before and during antiviral therapy and virologic response to therapy were investigated. Full-length NS5A clones were sequenced from 20 HCV genotype 1-infected patients in a prospective, randomized clinical trial of IFN induction (daily) therapy and IFN plus ribavirin combination therapy. Pretreatment NS5A nucleotide and amino acid phylogenies did not correlate with clinical IFN responses and domains involved in NS5A functions in vitro were all well conserved before and during treatment. A consensus IFN sensitivity-determining region (ISDR237–276) sequence associated with IFN resistance was not found, although the presence of Ala245 within the ISDR was associated with nonresponse to treatment in genotype 1a-infected patients (P < 0.01). There were more mutations in the 26 amino acids downstream of the ISDR required for PKR binding in pretreatment isolates from responders versus nonresponders in both HCV-1a- and HCV-1b-infected patients (P < 0.05). In HCV-1a patients, more amino acid changes were observed in isolates from IFN-sensitive patients (P < 0.001), and the mutations appeared to be concentrated in two variable regions in the C terminus of NS5A, that corresponded to the previously described V3 region and a new variable region, 310 to 330. Selection of pretreatment minor V3 quasispecies was observed within the first 2 to 6 weeks of therapy in responders but not nonresponders, whereas the ISDR and PKR binding domains did not change in either patient response group. These data suggest that host-mediated selective pressures act primarily on the C terminus of NS5A and that NS5A can perturb or evade the IFN-induced antiviral response using sequences outside of the putative ISDR. Mechanistic studies are needed to address the role of the C terminus of NS5A in HCV replication and antiviral resistance.
Hepatitis C virus (HCV) is a major cause of chronic liver disease leading to cirrhosis worldwide and is now recognized as a leading indication for orthotopic liver transplantation in the United States. The HCV genome has a high degree of genetic variability. Interindividual HCV sequence variability has led to the classification of at least six genotypes, while intraindividual variability is referred to as a quasispecies, which usually consists of a predominant viral variant and a variable mixture of highly genetically related yet distinct variants (23, 36).
Interferon (IFN) monotherapy leads to sustained virological responses in less than 20% of chronic hepatitis C cases (47). The recent introduction of IFN plus ribavirin combination therapy has significantly improved response rates (8, 38). In multivariate analyses, HCV genotype and high viral load are two important virologic factors that independently predict the likelihood of treatment failure (37, 39), stressing the importance of virologic factors in determining response to IFN therapy. Unfortunately, therapeutic intervention strategies are evolving at a faster rate than the understanding of the molecular mechanisms by which patients fail therapy. Understanding the molecular mechanisms of antiviral resistance to IFN therapy may lead to the design of better treatment strategies and/or new antiviral compounds. Furthermore, similar HCV-encoded mechanisms may be operational during manifestation of resistance to new antivirals, such as IFN plus ribavirin combination therapy and therapy with pegylated IFN.
The HCV NS5A protein is a nonstructural phosphoprotein of 56 to 58 kDa found in the cytoplasm, near either the nuclear membrane or the endoplasmic reticulum and Golgi apparatus (24, 28, 44, 50, 58). The NS5A gene product is expressed from the 3′ end of the HCV genome: the precursor NS5 protein is cleaved by the viral NS3 protein to yield NS5A and NS5B (32). Until recently, the functions of this protein were largely unknown.
NS5A has been shown to interact with a cellular kinase that is responsible for phosphorylating NS5A (28, 50, 58). A recent study identified the major phosphorylation site in the C terminus of NS5A (49). Currently, the role of NS5A phosphorylation in NS5A function is unknown, although some phosphorylation sites are well conserved (42), which suggests conservation of function. Studies in yeast indicate that the carboxy-terminal half of NS5A is capable of activating transcription when fused with the DNA-binding domain of the GAL4 protein (29, 57). The C terminus of NS5A also contains a nuclear localization signal that does not by itself direct NS5A to the nucleus, but is nonetheless functional in directing nuclear translocation when placed at the amino terminus of a reporter gene (25). The effects of antiviral therapy on mutation and selection in these functional domains of NS5A are not known.
In Japan, a consensus IFN sensitivity-determining region (ISDR) sequence in the NS5A gene was associated with lack of response to IFN in patients infected with genotype 1b, while mutations within the ISDR were associated with response to IFN therapy (2, 10, 11, 34, 52). In contrast, studies from Europe on HCV-1b-infected patients (19, 31, 42, 55, 59) and from North America on HCV-1a-infected patients (5, 22, 46) did not find such correlations. In vitro studies indicate that NS5A binds to and inhibits the IFN-induced, double-stranded RNA-activated protein kinase PKR (18). The 40-amino-acid ISDR and 26 amino acids downstream of the ISDR on NS5A constitute the PKR binding domain (17).
All of the previous clinical studies that examined NS5A mutations and response to IFN therapy have been based on retrospective analyses, and most have relied on direct sequencing of a limited region of the NS5A gene. Based on in vitro studies, it has been suggested that sequences outside the ISDR might be involved in PKR-mediated IFN resistance, particularly in the region adjacent to the C-terminal extremity of the ISDR (17). It has also been shown that NS5A lacking the ISDR can inhibit the antiviral actions of IFN in vitro (44). Furthermore, NS5A isolates from IFN-responsive patients containing multiple mutations in the ISDR, which would be predicted not to bind PKR, also inhibit the antiviral actions of IFN in vitro (41, 45). More generally, mutations in the entire carboxy-terminal region of the NS5A sequence have been associated with sensitivity to IFN (9). Because of these discrepancies, the present study examined the entire sequence of the NS5A gene in a prospective clinical trial design.
The goals of the present study were to address the following questions for a cohort of patients who received daily IFN therapy followed by IFN plus ribavirin combination therapy in a prospective randomized trial. Are mutations in the putative ISDR associated with responsiveness to IFN therapy in genotype 1a and 1b North American patients? Do pretreatment NS5A isolates from IFN-sensitive patients phylogenetically cluster separately from those from IFN-resistant patients? Do mutations in other domains of the NS5A protein correlate with IFN responsiveness? And are there selective pressures that induce changes in NS5A quasispecies during the early phase of therapy that correlate with IFN responsiveness?
MATERIALS AND METHODS
Patient population.
Serum samples were obtained from 20 treatment-naive patients infected with HCV genotype 1a or 1b. These patients were participating in a larger prospective randomized trial at the University of Washington Medical Center under written informed consent (35). Patients were stratified to three groups according to the level of HCV RNA; low (HCV RNA level below 350,000 equivalents/ml), intermediate (levels between 350,000 and 3.5 million equivalents/ml), and high (HCV RNA above 3.5 million equivalents/ml). In each group, patients were randomized to receive IFN alpha 2b in doses of 1.5, 3.0, 5.0, or 10.0 MU daily (induction) for 6 weeks. After 6 weeks of induction therapy, all patients were placed on daily doses of 3 × 106 U of IFN plus ribavirin (1 or 1.2 g/day for body weight less or more than 75 kg, respectively) for 48 weeks to complete 54 weeks of treatment. Serum was prepared from whole blood within 4 h of venipuncture and stored at −70°C.
Virological classification of response.
Response to IFN therapy was determined by quantitative analysis of HCV RNA levels by the branched DNA assay (version 2.0; Chiron Corp., Emeryville, Calif.) and by quantitative reverse transcription (RT)-PCR assay before, during, and at the end of treatment. The limit of detection of the branched DNA assay is 200,000 genome equivalents/ml. Below this limit, an endpoint dilution assay using Roche Amplicor (v.2.0 protocol) was performed to further characterize low-level viremia. The limit of detection of this assay was 100 copies/ml. Response to therapy was assessed at the end of the 6-week induction period, at the end of therapy, and 6 months after stopping therapy. End of treatment complete response was defined as the absence of HCV RNA in serum by quantitative RT-PCR at the end of treatment. Sustained response was defined as absence of HCV RNA in serum by quantitative PCR 6 months after therapy was stopped. Nonresponse was defined as continued presence of serum HCV RNA by quantitative PCR at the specified time point. Patients who were HCV RNA negative at the end of therapy but experienced a rebound in HCV viremia after stopping therapy were classified as relapsers.
Genotype analysis.
HCV genotyping was performed by restriction fragment length polymorphism analysis of the 5′ noncoding region as described by Davidson et al. (7).
RNA extraction, RT-PCR, cloning, and sequencing.
Total RNA was extracted from patient sera by the single-step guanidinium method (3). RNA was reverse transcribed in a 25-μl reaction containing 150 pmol of antisense primer 3′UTR-286 (33) (5′CAGTCATGCGGCTCACGGACCTT3′, nucleotide [nt] positions 9451 to 9481), 3 mM MgCl2, 1 mmol each of the four deoxynucleoside triphosphates, 0.6 mM dithiothreitol, 75 mM KCl, 50 mM Tris-HCl (pH 8.3), 9.4 U of RNase inhibitor (Pharmacia LKB, Piscataway, N.J.), and 130 U of Moloney murine leukemia virus reverse transcriptase (Gibco-BRL, Grand Island, N.Y.). The mixture was incubated at 37°C for 1 h and then at 95°C for 5 min. Nested PCR was then used to amplify the NS5A gene. For genotype 1a, the first-round primer set consisted of forward primer NS5A-1a-US1 (TGTTTCCCCCAC GCACTACG, nt 6125 to 6144) and antisense primer 3′UTR-286. The second-round primer set consisted of sense primer NS5A-1a 5′ NcoI (TTCCATGGGC TCCGGTTCCTGGCTAAGG, nt 6258 to 6275) and antisense primer NS5A-1a-3′ XbaI (TCTAGATTAGCAGCACACGACATCCTC, nt 7584 to 7601).
For genotype 1b, the first-round primer set included sense primer NS5A-1b-US1 (CAGCCTCACCATCACTCAGC, nt 6188 to 6210) and antisense primer NS5A-1b-DS1 (GTGGTGACGCAGCAGAGAGT, nt 7681 to 7700). The second-round primer set consisted of sense primer NS5A-1b-5′ NcoI (CCTTCCATGGGCTCCGGCTCGTGGCTAAAG, nt 6256 to 6275) and antisense primer NS5A-1b-3′ XbaI (ATCTCTACATTAGGACATTGAGCAGCAGACGA, nt 7588 to 7602). The first round of PCR was performed as follows: 10 μl of the cDNA was added to a 40-μl PCR mixture containing 50 pmol of the external, sense primer, 2.0 mmol of MgCl2, 50 mM Tris-HCl (pH 8.3), 5 mM KCl, and 8 U of Pfu polymerase (Stratagene, La Jolla, Calif.). A “hot start” nested PCR was then performed. In nested PCR, the bottom reaction mixture contained 2.0 mmol of MgCl2, 50 mM Tris-HCl (pH 8.3), 5 mM KCl, and 50 pmol of each internal primer and was separated by a wax layer from the top reaction mixture containing 50 mM Tris-HCl (pH 8.3), 5 mM KCl, 8 U of Pfu polymerase, and 2% of the first-round product. The PCRs were done in a Perkin-Elmer 9600 thermocycler, using 30 cycles with the following cycling parameters: template denaturation at 94°C for 23 s, primer annealing at 60 to 65°C for 30 s, and extension at 72°C for 3 to 6 min. A single final extension step was done at 72°C for 5 min to complete the amplification reaction. Amplified products were analyzed on 0.8% agarose gels. DNA products were purified using Qiaex (Qiagen, Valencia, Calif.). The 1.4-kb PCR products were ligated into the pCR-Blunt vector and used to transform One Shot TOP 10 competent cells (Invitrogen, San Diego, Calif.). Transformants were detected according to the manufacturer's protocol, and cloning efficiency was >80%.
For each subject, at least three clones derived from pretreatment serum were selected. Plasmid DNA was isolated from a 4.0-ml broth culture (QIAprep Miniprep; Qiagen). Recombinant pCR-Blunt-NS5A clones were sequenced by dideoxy terminator automated sequencing (ABI Prism Ready Reaction AmpliTaq Ts; Applied Biosystems, Inc., Foster City, Calif.) on an ABI Prism 377 sequencer, according to the manufacturer's instructions (Applied Biosystems Inc.). Sequences were assembled and edited with SeqEd version 1.0.3 (Applied Biosystems). The complete sequence of each NS5A clone was determined in four separate sequencing reactions per clone. Sequence analysis was performed with SeqEd and Mac Vector software version 6.5 (Oxford Molecular, Madison, Wis.). Because of higher variability in the C-terminal half of the NS5A, we also determined the amino acid sequence of this region in 10 clones for a subgroup of four patients before and following 6 weeks of therapy. Sequencing of three full-length NS5A clones was also done for nine patients (seven patients infected with genotype 1a and two infected with genotype 1b) at week 6, or earlier if the specimen titer was too low at week 6 for sequence analysis.
Phylogenetic analysis.
Prior to further analysis, primer sequences and sites containing gaps in any sequence were removed. Two cDNA clone sequences were discarded because they appeared to be artifactually defective: one had a single base insertion resulting in a shift in reading frame, and the other had a large (33 nt) deletion. Two other nonsense mutations were detected in a pretreatment sequence from subject 5, clone 1, at codon 374 of the NS5A gene and in a treatment week 6 sequence from subject 20, clone 2, at codon 157.
Sequence alignments were randomly permuted 100 times by using the SEQBOOT program from PHYLIP package version 3.572c (12, 13). DNA distance matrices were calculated by using the DNADIST program, maximum-likelihood model, with a transition-to-transversion ratio of 4.25 (54). Permuted trees were generated using the Neighbor program with random addition, and bootstrap values were obtained by using Consense. Subtype reference sequences used for phylogenetic analysis had accession numbers (and strain designations): 1a, M62321 (HCV-1) (4); 1b, D90208 (HCV-J) (30); and 1c, D14853 (HC-G9) (40). The diversity was quantified as the mean genetic distance calculated for all pairs of amino acid sequences by using the ProtDist module in the PHYLIP package, version 3.5c (14).
The VarPlot software program (version 1.2) was used to examine the ratio of nonsynonymous distance (number of nonsynonymous differences per nonsynonymous site) to synonymous distance (number of synonymous differences per synonymous site), or dN/dS, in a sliding window of nucleotide sequence (48). Briefly, a segment of defined length, in this case 150 nt (the window size), was used to determine the genetic distance, or number of substitutions per site. This process was then repeated for an overlapping segment shifted by 3 nt (the step size), with the same window size, and continued across the alignment. At each step all pairwise comparisons for a subject were performed and averaged. Windows in which dS was 0, resulting in an undefined value for dN/dS, were not included. These mean values were then averaged for all subjects in a group (nonresponse or complete response), ensuring that each subject was given equal weight. The New1 method of Ina was used to calculate the nonsynonymous and synonymous genetic distances (26).
Statistical analysis.
Clinical and biochemical characteristics of patients are expressed as mean ± standard deviation. Comparisons between the responders and nonresponders were determined by using Student's paired t test. Qualitative variables were compared using the chi-squared test. A P value of less than 0.05 was considered significant.
Nucleotide sequence accession numbers.
The sequences reported herein have been assigned GenBank accession no. AF264995 to AF265165.
RESULTS
At the end of the 6-week induction period, three patients were defined as early complete responders, five were nonresponders with no significant decrease in HCV viremia, and 12 patients had a decrease in HCV RNA level of more than 10-fold; six of these patients had a decrease of more than 1,000-fold. After 12 months of IFN-ribavirin maintenance treatment, 12 of 20 (60%) patients were HCV RNA negative, while 8 of 20 patients (40%) were nonresponders. End of 12-month treatment responses occurred in 8 of 14 (57%) HCV-1a and 4 of 6 (67%) HCV-1b patients. When HCV RNA was analyzed 6 months after stopping therapy, sustained responses were observed in 5 of 13 (38%) of HCV-1a and 2 of 4 (50%) of HCV-1b patients. Due to the prospective nature of the study, long-term response data are pending for three patients. In general, patients who had end-of-treatment complete responses tended to have sustained responses, while patients who were end-of-treatment nonresponders remained nonresponders at the 6-month follow-up. The exception to this was patient 16, who experienced a relapse of viremia after therapy was discontinued. Clinical and virological data are summarized in Table 1.
TABLE 1.
Patient characteristics
| Patient no. | HCV genotype | Treatment groupa | End-of-treatment responseb | Long-term responseb | HCV RNA (log Meq)
|
No. of clones (pretherapy/on therapy) | |
|---|---|---|---|---|---|---|---|
| Pretreatment | Week 6 | ||||||
| 1 | 1b | C2 | NR | NR | 7.29 | 7.11 | 3/3 |
| 2 | 1a | B1 | NR | NR | 6.54 | 2.69 | 3 |
| 3 | 1a | C2 | CR | Pending | 7.43 | 4.69 | 11/10 |
| 4 | 1a | C1 | CR | SR | 6.98 | 4.69 | 2/3 |
| 5 | 1a | B2 | CR | Relapse | 6.31 | 4.69 | 3 |
| 6 | 1a | B4 | CR | SR | 6.2 | Negative | 3 |
| 7 | 1a | C4 | CR | SR | 7.17 | 2.69 | 3 |
| 8 | 1a | C3 | NR | NR | 7.59 | 5.53 | 3/3 |
| 9 | 1a | C2 | NR | NR | 6.12 | 6.94 | 11/10 |
| 10 | 1b | B3 | CR | Pending | 6.02 | Negative | 3 |
| 11 | 1b | C2 | CR | SR | 6.99 | 2.69 | 3/3 |
| 12 | 1b | C3 | CR | SR | 7.3 | 2.69 | 3 |
| 13 | 1b | C4 | NR | NR | 6.81 | 4.69 | 3 |
| 14 | 1a | B3 | CR | SR | 6.48 | 2.69 | 3 |
| 15 | 1a | C3 | NR | NR | 7.43 | 5.48 | 3/3 |
| 16 | 1a | C2 | CR | Relapse | 7.12 | 4.69c | 10/10 |
| 17 | 1a | A4 | CR | SR | 5.47 | Negative | 3 |
| 18 | 1a | C1 | NR | NR | 7.33 | 6.77 | 3 |
| 19 | 1b | C1 | CR | Pending | 6.78 | 4.69 | 2 |
| 20 | 1a | C2 | NR | NR | 6.55 | 6.55 | 10/10 |
Virus titers: A, <350,000; B, 350,000 to 3.5 million; C, >3.5 million. IFN dose: 1.5, 3.0, 5.0, and 10.0 MU daily for groups 1 to 4, respectively.
NR, nonresponse; CR, complete response; SR, sustained response.
Titer derived from week 2, as week 6 HCV RNA was negative.
From the 20 patients, a total of 143 full-length clones were sequenced. One hundred twenty clones represented full-length NS5A-1a, while 23 clones represented full-length NS5A-1b. Eighty-eight clones from pretreatment serum specimens (71 genotype 1a and 17 genotype 1b) and 55 clones from on-therapy serum specimens (49 genotype 1a and 6 genotype 1b) were analyzed.
We first examined the relationship between pretreatment NS5A sequences and response to therapy. Phylogenetic trees were constructed from the 143 sequences as described in Materials and Methods. The data are depicted in Fig. 1. Based on end-of-treatment responses, NS5A isolates from IFN-nonresponsive patients did not cluster separately from those from IFN-responsive patients when analyzing full-length NS5A sequences (nucleotide or protein) in HCV-1a- or 1b-infected patients. Next, we examined the sequence of regions of the NS5A protein previously described to have biological activities in vitro. Acidic regions AR1171–212 and AR2248–301, which have been shown to be critical for the transcriptional activity of the protein in yeast (16, 29, 57), were in general well conserved between isolates (Fig. 1 and data not shown). The four serine residues at positions 225, 230, 232, and 235, suggested to be important for hyperphosphorylation of NS5A (58), were also highly conserved among HCV-1a- and 1b-infected patients and within the quasispecies in each patient (Fig. 1 and data not shown). Moreover, Ser346 (position 2321 on the full-length HCV genotype 1a polyprotein), recently shown to be the major phosphorylation site on NS5A-1a (49), was absolutely conserved among all isolates, as was the putative nuclear localization signal (NLS) at positions 354 to 362 (25) (data not shown). Phylogenetic analysis limited to specific functional domains on NS5A such as AR1, AR2, ISDR, the PKR binding domain, and the NLS also did not reveal any phylogenetic clustering of responsive isolates from nonresponsive isolates (data not shown). However, in terms of the total number of mutations in pretreatment NS5A-1a isolates, we found that responsive patients had more mutations than nonresponsive patients (P < 0.001). We then focused our attention on the sequences of the putative ISDR and V3 regions that were previously associated with clinical responsiveness to IFN therapy.
FIG. 1.
Neighbor-joining phylogenetic tree constructed from 1,344-nt NS5A gene sequences (1,338 nt after stripping sites containing gaps) obtained from study subjects. The scale in genetic distance is indicated, and the number of 100 permuted trees supporting a clade is indicated when that proportion was greater than 70%. Subject identifiers next to brackets correspond to those in Table 1, and at the end of each branch tip is the cDNA clone number. All sequences were obtained from pretreatment specimens except those enclosed in boxes, which were obtained after treatment for 2 (subjects 11 and 16) or 6 (subjects 1, 3, 4, 8, 9, 15, and 20) weeks. Reference sequences for subtypes 1a, 1b, and 1c are indicated. Phylogenetic analysis was performed as described in Materials and Methods.
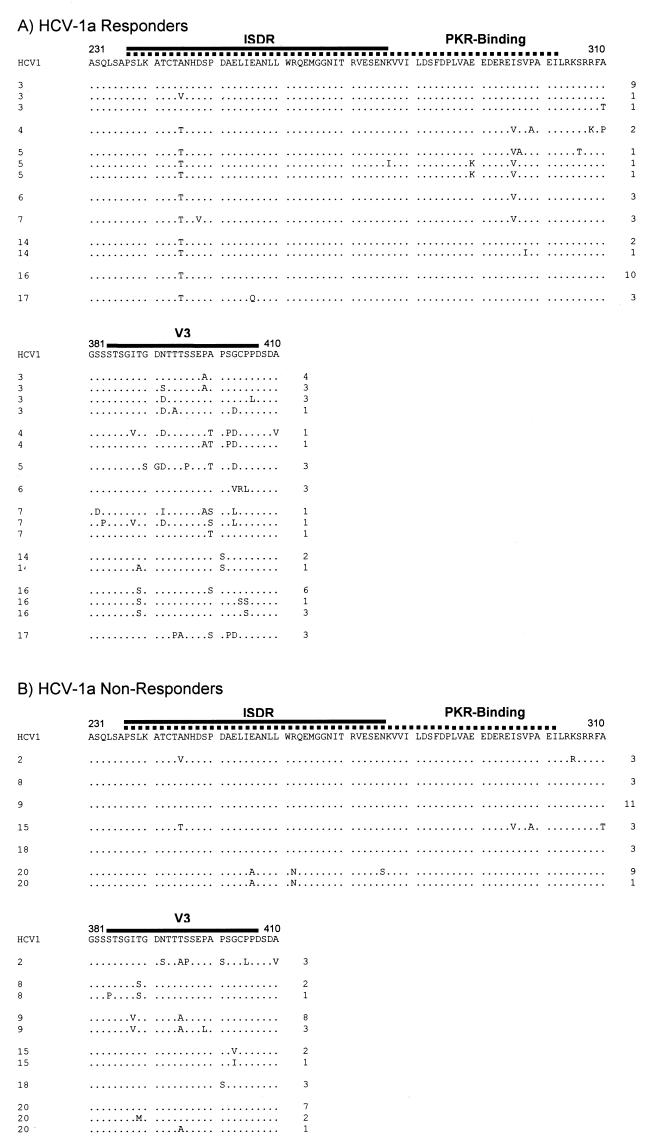
Figure 2A depicts amino acid sequence alignments of the region containing the ISDR, PKR-binding domain, and V3 region of 38 clonal isolates from the pretreatment serum of the eight patients infected with HCV genotype 1a who had end-of-treatment complete responses. Figure 2B depicts amino acid sequence alignments of the region containing the ISDR, PKR interaction domain, and V3 region of 33 clonal isolates from the pretreatment serum of the six patients infected with HCV genotype 1a who were end-of-treatment nonresponders. In all isolates, there was limited variability in the ISDR237–276 and the PKR-binding domain, which includes the ISDR plus 26 additional downstream amino acids (17). There was no significant correlation between the number of ISDR mutations and end-of-treatment responses in HCV-1a NS5A isolates. However, within the ISDR there was a preference for Thr245 in responsive patients (27 of 38 clones, 71%), while in nonresponders, Ala245 was frequently observed at this position (27 of 33 clones, 81.8%). Thus, Ala245 was more frequently observed in HCV-1a isolates from IFN-resistant patients than in HCV-1a isolates from IFN-sensitive patients (27 of 33 versus 11 of 38 clones, respectively; P < 0.001). Furthermore, responsive patients had significantly more mutations in the PKR-binding domain than nonresponders (28 total mutations relative to HCV-1 in the 38 responder clones versus 6 total mutations relative to HCV-1 in the 33 nonresponder clones; P < 0.05). There were significantly more amino acid changes in the previously designated variable 3 (V3) region (27) in HCV-1a isolates from IFN-sensitive patients than in HCV isolates from IFN-resistant patients when compared to the consensus HCV-1 sequence (P < 0.0001).
FIG. 2.
Sequence analysis of pretreatment NS5A isolates in the ISDR, PKR-binding domain, and V3 region in HCV-1a-infected patients, aligned relative to HCV-1, the prototype genotype 1a isolate. (A) Clones from patients with end-of-treatment complete response. (B) Clones from patients who were end-of-treatment nonresponders. Full-length NS5A was amplified by nested PCR using Pfu polymerase, cloned, and sequenced as described in Materials and Methods. The ISDR is highlighted by the solid line, while the PKR-binding domain is highlighted by the dotted line. The V3 region is highlighted by the solid line. The patient number is listed to the left of each sequence, while the number of clones comprising each sequence is listed to the right.
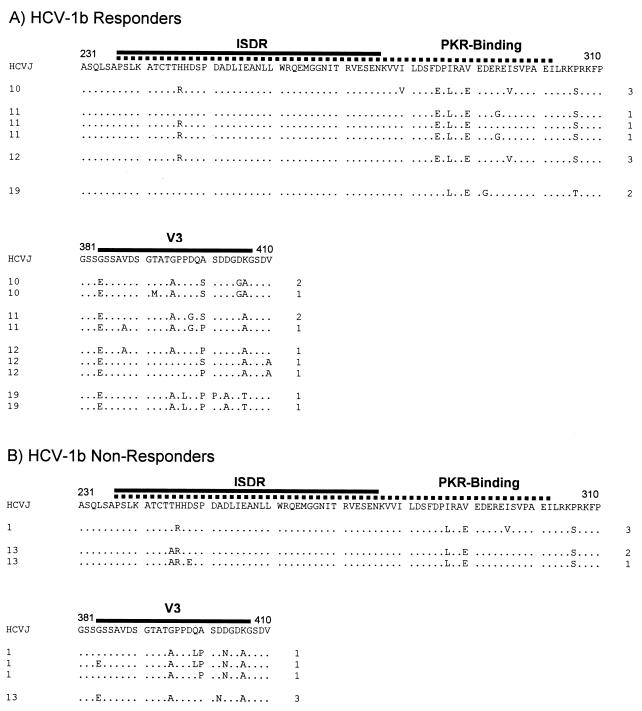
Figure 3A depicts amino acid sequence alignments of the region containing the ISDR, PKR interaction domain, and V3 region of 11 clonal isolates from the pretreatment serum of the four patients infected with HCV genotype 1b who had end-of-treatment complete responses. Figure 3B depicts amino acid sequence alignments of the region containing the ISDR, PKR interaction domain, and V3 region of six clonal isolates from the pretreatment serum of the two patients infected with HCV genotype 1a who were end-of-treatment nonresponders. In HCV-1b-infected patients, there was no correlation between the number of mutations in the ISDR and response to therapy. For the HCV-1b ISDR isolates, Arg-for-His substitutions at position 246 (2218 on the HCV polyprotein) were observed but were not different between responders and nonresponders (Fig. 3). Responsive HCV-1b patients had significantly more mutations in the PKR-binding domain compared to nonresponders (44 total mutations relative to HCV-J in the 11 responder clones versus 15 total mutations relative to HCV-J in the 6 nonresponder clones, P < 0.05). There was also a trend for more mutations in the V3 region in NS5A isolates from genotype 1b-infected responsive patients compared to nonresponsive patients (Fig. 3B).
FIG. 3.
Sequence analysis of pretreatment NS5A isolates in the ISDR, PKR-binding domain, and V3 region in HCV-1b-infected patients, aligned relative to HCV-J, the prototype genotype 1b isolate. (A) Clones from patients with end-of-treatment complete responses. (B) Clones from patients who were end-of-treatment nonresponders. Full-length NS5A was amplified by nested PCR using Pfu polymerase, cloned, and sequenced as described in Materials and Methods. The patient number is listed to the left of each sequence, while the number of clones comprising each sequence is listed to the right.
Because the previous data were derived from patients treated during the induction phase with various doses of IFN, we sequenced NS5A isolates from patients following 6 weeks of daily IFN induction therapy. We analyzed in detail 10 clones from four patients matched for pretreatment viremia (Table 1) and dose of IFN during the induction phase (3.0 × 106 U daily for 6 weeks). Two patients were end-of-treatment nonresponders (patients 9 and 20), while two patients were end-of-treatment responders (patients 3 and 16). Figure 4 depicts the changes observed in the V3 region from pretreatment to the end of the 2- to 6-week period of induction therapy. In the two nonresponder patients, the major variant at week 6 of therapy was identical to that found pretreatment. In contrast, in responder patient R1, a minor variant that accounted for 30% of the pretreatment population comprised 100% of the clones after 2 weeks of therapy. Note that for patient R1, the week 6 serum specimen was negative for HCV RNA by RT-PCR, so that we amplified the sample from the earliest positive time point (week 2). In responder patient R2, the major pretreatment variant was not detected at 6 weeks and had been replaced by a minor pretreatment variant representing 50% of the clones at 6 weeks. Furthermore, in this patient, a minor variant that represented 9.1% of the pretreatment quasispecies population increased to 30% of the quasispecies population by week 6. Thus, we observed selection of minor pretreatment V3 quasispecies variants during the early phase of IFN therapy in end-of-treatment responders compared with nonresponders. The ISDR and PKR-binding domain sequences in this same set of 10 clones from each of the four patients remained absolutely unchanged during this period (data not shown).
FIG. 4.
Selection of minor pretreatment V3 quasispecies variants during the early phase of IFN therapy in responsive patients. The sequences of 10 clones from each of four patients infected with HCV-1a were generated before and following 6 weeks of IFN induction therapy (onRx). For patient 16, HCV RNA was RT-PCR negative at 6 weeks, so on-therapy sequences were obtained following 2 weeks of therapy. Based on end-of-treatment responses, patients 9 and 20 were nonresponders, while patients 16 and 3 responded to treatment. All four patients were matched for pretreatment viremia levels and IFN dose. The major V3 quasispecies variant after 6 weeks of therapy in the two nonresponsive patients was identical to the pretreatment major V3 quasispecies variant and is highlighted with an asterisk (∗). In contrast, the major V3 quasispecies variant after 6 weeks of therapy in the two responsive patients was derived from a minor pretreatment V3 quasispecies variant and is highlighted with a pound sign (#). Bracketed numbers represent the relative proportion of each variant in the population.
One approach to quantitating genetic variability between clinical isolates is to determine the mean genetic distance, which provides an estimate of the genetic diversity of the population. In Figure 5, mean genetic distances from HCV-1a (panel A) and HCV-1b (panel B) pretreatment isolates for full-length NS5A and the V3 region are plotted. In genotype 1a-infected patients, the mean distance between full-length NS5A clones obtained from responders was higher than in nonresponders, although the difference was not statistically significant (0.0087 ± 0.0073 versus 0.0083 ± 0.0029, P = 0.07). In the V3 region, the mean genetic distance for pretreatment isolates was significantly higher for responders than for nonresponders (0.0457 ± 0.0506 versus 0.0185 ± 0.0216, P < 0.001). For genotype 1b-infected patients (Fig. 5B), there was a trend for increased genetic distance in the V3 region from responders compared to nonresponders (0.0521 ± 0.0274 versus 0.0291 ± 0.0290, P = 0.24). The mean genetic distances for sequences obtained following 6 weeks of IFN therapy for full-length NS5A and the V3 region were also higher in responders than nonresponders (data not shown). Collectively, the data in Fig. 2 to 5 indicate that there were significantly more mutations in the NS5A gene from patients who responded to IFN therapy than in patients who failed to respond. Most of the mutations were concentrated downstream of the ISDR, primarily in the V3 region.
FIG. 5.
Comparison of pretreatment genetic diversity in various regions of the NS5A protein. Genetic diversity was estimated by calculating the mean genetic distance as described in Materials and Methods. (A) Mean genetic diversity in responders (open bars) versus nonresponders (solid bars) for full-length NS5A and the V3 region for genotype 1a-infected patients. (B) Mean genetic diversity in responders (open bars) versus nonresponders (solid bars) for full-length NS5A and the V3 region for genotype 1b-infected patients.
We then calculated the nonsynonymous-to-synonymous mutation ratio (dN/dS) along the entire NS5A gene to determine if there was any evidence of selective forces acting on NS5A. For this, we employed the software VarPlot (48), which scans nucleotide sequences with a sliding window to provide a plot of dN/dS for the entire protein. Figure 6 depicts VarPlot analyses for all patients, grouped according to end-of-treatment responses. Figure 6A depicts VarPlot analysis on pretreatment NS5A isolates, while Fig. 6B depicts the results after 2 or 6 weeks of induction therapy. Both figures indicate that there is greater variability of the C terminus of NS5A in responders before and during antiviral therapy. The amino terminus of NS5A also appears to have elevated dN/dS ratios in both responders and nonresponders before and during antiviral therapy, centered on amino acid position 60 (Fig. 6A and B). The ISDR had virtually zero nonsynonymous changes both before and during induction therapy. The dN/dS ratio in the V3 region in end-of-treatment responders was significantly elevated both before and during therapy compared to end-of-treatment nonresponders. During IFN therapy, the dN/dS ratio in nonresponders appeared to decrease. Elevated dN/dS ratios centered on position 320 were also noted during induction therapy (Fig. 5B). The data indicate the presence of two variable regions in the C terminus of NS5A, the previously described V3 region centered around amino acid 400, and a new variable region centered around amino acid 320. In both variable regions, the dN/dS ratio is higher in end-of-treatment responders than nonresponders both before and during antiviral therapy, suggesting that the two variable regions may be under positive selection pressure.
FIG. 6.
VarPlot analysis of the entire NS5A open reading frame. VarPlot analysis was performed as described in Materials and Methods. The x axis represents the amino acid positions on the NS5A protein, while the y axis represents the ratio of nonsynonymous to synonymous nucleotide changes (dN/dS). The relative positions of the ISDR, PKR-binding domain, and the V3 region are indicated above the plots. (A) Results based on analysis of NS5A clones before therapy. (B) Results based on analysis of NS5A clones following 6 weeks of daily induction therapy. Dotted lines represent VarPlot results for complete responders (CR), while solid lines represent VarPlot results for nonresponders (NR).
DISCUSSION
Whether or not antiviral resistance influences therapeutic outcomes in chronic hepatitis C remains an important and controversial issue. The present study addressed HCV genetic sequences over the entire NS5A gene in a prospective longitudinal treatment trial involving both induction and combination therapy.
This is the first study to examine the entire NS5A coding region both before and during treatment in patients in a prospective trial of induction and combination therapy. We found that pretreatment NS5A nucleotide or amino acid phylogenies did not correlate with clinical IFN responses, and domains on NS5A involved in NS5A functions in vitro were all well conserved during treatment. This study also found no evidence of a consensus ISDR sequence associated with IFN resistance, although the presence of Ala245 within the ISDR was associated with nonresponse to treatment. There were more mutations in the PKR-binding domain in pretreatment isolates from responders versus nonresponders in both HCV-1a- and HCV-1b-infected patients. In HCV-1a patients, more amino acid changes were observed in isolates from IFN-sensitive patients, and these mutations appeared to be concentrated in two variable regions in the C terminus of NS5A. The two variable regions corresponded to the previously described V3 region and a new variable region from 310 to 330. These variable regions appeared to be under positive selective pressure. Selection of minor pretreatment V3 quasispecies was observed during the first 2 to 6 weeks of IFN therapy in responsive but not nonresponsive patients. During this time, the ISDR and PKR-binding domains on NS5A remained unchanged in both patient groups.
There are potential limitations in the current study. Results might be complicated by the fact that patients were initially treated with various doses of IFN during the induction phase. However, we performed extensive analysis of 10 clones from each of four patients matched for pretreatment viremia, genotype, and IFN dose during the induction period. Furthermore, as this is a prospective study, sustained response data are still pending for some patients. As a result, all comparisons were based on end-of-treatment responses. For patients for whom sustained response data were available, however, we found that in general, end-of-treatment responses reflected sustained responses. Moreover, relatively small numbers of patients were studied. Although we did not examine the changes in the NS5A gene over time in patients not treated with IFN, several studies have previously demonstrated that IFN places pressure on the HCV genome, resulting in an increase in the rate of fixation of mutations (21, 43, 46). The data therefore suggest that the mutational patterns that we observed in the NS5A gene were a direct consequence of the effect of IFN on HCV replication and quasispecies dynamics. Thus, additional studies on larger patient cohorts are warranted.
The putative ISDR was highly conserved in both genotype 1a and genotype 1b sequences. Most of the previous studies have compared HCV-1b sequences. The current study showed that in all genotype 1b isolates, substitutions at position His245 were not different between responders and nonresponders, while Ala245 was associated with nonresponse to treatment. Thus, before treatment, no specific sequence was characteristic for either responsive or nonresponsive clones. Gale and colleagues have proposed redefining the ISDR as a larger sequence element comprising amino acids 237 to 302 (2209 to 2274 of the HCV open reading frame) (17). When considering the additional 26 amino acids, we found that there were more mutations in this region in responders than in nonresponders, in agreement with a recent study (53). The relevance of this finding to NS5A-mediated inhibition of PKR in vitro is not clear, but one prediction is that NS5A isolates containing mutations in the 26-amino-acid region should bind less efficiently or not at all to PKR.
It has been demonstrated that NS5A isolates from patients who were complete responders to IFN therapy antagonize the antiviral actions of IFN against encephalomyocarditis virus in vitro (41, 45). The NS5A proteins had ISDR sequences with multiple mutations relative to the consensus IFN-“resistant” ISDR sequence. Based on studies in the Katze lab, these NS5A proteins should not be able to bind and inhibit PKR, yet they still partially inhibited IFN action. These data suggest that although the NS5A protein clearly inhibits the IFN system, sequences outside the ISDR may be involved. Furthermore, although NS5A can partially inhibit IFN activity, the inhibitory effects in vitro do not correlate with clinical response to therapy (41). That is, NS5A isolates from nonresponder patients do not inhibit the IFN system in vitro more than NS5A isolates from patients who respond to therapy. These data suggest that other mechanisms of NS5A-mediated IFN resistance may be at work. However, there is one caveat with this argument, based on data presented in this study. Despite the fact that we found no correlation between ISDR sequences and clinical response to therapy, each NS5A had a different profile of mutations in the rest of the NS5A protein. Thus, lack of correlation between the ISDR and IFN response does not necessarily negate the PKR-ISDR hypothesis, since mutations in other regions of the NS5A protein may affect protein conformation and biological activity.
HCV-1a is the most common cause of chronic hepatitis C in the United States. Little is known about mutations in the NS5A gene in genotype 1a-infected patients. Previous analysis of HCV genotype 1a clones did not reveal any specific amino acid substitution pattern related to the response to IFN therapy (5, 15, 22, 46, 59). In the present study, Ala245 was more frequently observed in nonresponders than in responders. Pawlotsky and colleagues also showed a hot spot for amino acid changes at this position when analyzing HCV-1b quasispecies (42) and suggested that this region might interact with host proteins or could be involved in the phosphorylation of NS5A. These hypotheses require further investigation.
The evolution of NS5A during therapy is controversial. A relative stability over time of NS5A-ISDR has been described by some groups (2, 59), suggesting that selection of the resistant ISDR phenotype does not usually occur during IFN therapy. However, in a subset of genotype 1b-infected patients, evolution or selection of the ISDR to the prototype “resistant” ISDR sequence during IFN therapy is associated with nonresponse (20, 46), but it is not clear when selection of NS5A quasispecies occurs. In the present study, the ISDR and PKR-binding domains were relatively conserved before and during therapy. By contrast, genetic variability was detected in two variable regions in the C terminus of NS5A. Interestingly, in the group of HCV-1a-infected patients, no significant changes were observed in the variable regions in nonresponders, whereas more mutations were observed in responders. The average within-sample genetic distance within the quasispecies was significantly higher and increased significantly during therapy in responders compared to nonresponders. Detailed analysis of a subset of four patients matched for viremia and dose of IFN showed no amino acid change in the V3 region in two nonresponders, whereas an enrichment of minor pretreatment quasispecies was observed after 2 to 6 weeks of therapy in responders. VarPlot analysis suggested that positive selective forces act on the V3 variable region and also on a second variable region centered on amino acid position 320. These data suggest that in HCV-1a-infected patients, selection of NS5A quasispecies may occur during the first few weeks of IFN therapy, and the selective forces appear to target the C terminus of the protein. We also found evidence for elevated dN/dS ratios in all patients in the amino terminus of NS5A, centered on amino acid 70. Additional studies are required before the potential effect of selection at this site on NS5A function and the selective forces driving the changes can be characterized.
Currently, the selective forces that drive changes in the NS5A protein are not known. It is possible that the immunological pressure drives the fixation of mutations in the C-terminal variable regions. In this respect, secondary-structure predictions indicate that the V3 region has a hydrophilic character, predicted to be accessible to at least humoral responses (data not shown). It is possible that humoral and cytotoxic responses against these regions drive the changes observed. Indeed, it has been shown that immune responses directed against NS5A correlate with response to IFN therapy (15). Since the V3 region was found to change in responders to a greater extent than in nonresponders, this hypothesis fits with recent data which suggest that strong multispecific immune responses to HCV result in clearance of virus (6). However, as of the writing of the manuscript, no cytotoxic T-lymphocyte responses against this region of NS5A have been documented (51; M. Koziel, personal communication). The V3 region is situated very close to the NLS (354 to 362) and the proline-rich region (PRR310–354) of NS5A. In fact, the new variable region that we found in this study centered at position 320 lies directly within the PRR310–354. Interestingly, amino acid positions 350 to 356, which include a portion of the PRR, have recently been shown to mediate binding of NS5A to growth factor receptor-bound protein 2 (Grb-2), an interaction that subsequently inhibits mitogenic signaling (56). Thus, the variable regions in the C terminus of NS5A may affect secondary and tertiary folding of the NS5A protein, thereby affecting various biologic functions, such as inhibition of signal transduction cascades. These, in turn, might affect the IFN-induced antiviral response. These possibilities will remain speculative until they can be addressed in model in vitro systems.
In our study, the rate of fixation of mutations in the ISDR region and also in the entire NS5A gene was lower than found in previous studies. A possible explanation might be the use of Pfu polymerase instead of Taq. To limit the number of artifacts during the amplification of virus genomes, the use of thermostable polymerases that have lower error rates than Taq has been suggested. Pfu polymerase contains a 3′-5′ exonuclease activity and has been shown to have a fidelity nine times higher than that for Taq polymerase (1). This suggests that proofreading polymerases should be used to investigate the quasispecies and their potential influence on the outcome of antiviral therapy.
In summary, this genetic study provided evidence of positive selective forces acting on the NS5A gene during IFN therapy, especially in the C terminus. The data imply that NS5A-mediated antiviral resistance may involve sequences outside of the putative ISDR. Mechanistic studies are needed to address the role of NS5A C-terminal variable regions in HCV replication and antiviral resistance.
ACKNOWLEDGMENTS
We thank Paula Cox and Nathan Comsia for patient and sample management and Minjun Chung, Anthony Marquardt, Maureen Guajardo, Ka Wing Ng, Sharon Wendt, and Cassidy Patriarca for HCV RNA determinations.
This research was partially supported by educational grants from Schering-Plough to S.J.P. and R.L.C. and by NIH grant DK-057998 to S.C.R. S.J.P. is a Liver Scholar of the American Liver Foundation.
REFERENCES
- 1.Bracho M A, Moya A, Barrio E. Contribution of Taq polymerase-induced errors to the estimation of RNA virus diversity. J Gen Virol. 1998;79:2921–2928. doi: 10.1099/0022-1317-79-12-2921. [DOI] [PubMed] [Google Scholar]
- 2.Chayama K, Tsubota A, Kobayashi M, Okamoto K, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase H, Saitoh S, Suzuki Y, Murashima N, Ikeda K, Kumada H. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology. 1997;25:745–749. doi: 10.1002/hep.510250342. [DOI] [PubMed] [Google Scholar]
- 3.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 4.Choo Q L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina S R, Barr P J. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung R T, Monto A, Dienstag J L, Kaplan L M. Mutations in the NS5A region do not predict interferon-responsiveness in American patients infected with genotype 1b hepatitis C virus. J Med Virol. 1999;58:353–358. [PubMed] [Google Scholar]
- 6.Cooper S, Erickson A L, Adams E J, Kansopon J, Weiner A J, Chien D Y, Houghton M, Parham P, Walker C M. Analysis of a successful immune response against hepatitis C virus. Immunity. 1999;10:439–449. doi: 10.1016/s1074-7613(00)80044-8. [DOI] [PubMed] [Google Scholar]
- 7.Davidson F, Simmonds P, Ferguson J C, Jarvis L M, Dow B C, Follett E A, Seed C R, Krusius T, Lin C, Medgyesi G A, et al. Survey of major genotypes and subtypes of hepatitis C virus using RFLP of sequences amplified from the 5′ non-coding region. J Gen Virol. 1995;76:1197–1204. doi: 10.1099/0022-1317-76-5-1197. [DOI] [PubMed] [Google Scholar]
- 8.Davis G L, Esteban-Mur R, Rustgi V, Hoefs J, Gordon S C, Trepo C, Shiffman M L, Zeuzem S, Craxi A, Ling M H, Albrecht J. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. N Engl J Med. 1998;339:1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 9.Duverlie G, Khorsi H, Castelain S, Jaillon O, Izopet J, Lunel F, Eb F, Penin F, Wychowski C. Sequence analysis of the NS5A protein of European hepatitis C virus 1b isolates and relation to interferon sensitivity. J Gen Virol. 1998;79:1373–1381. doi: 10.1099/0022-1317-79-6-1373. [DOI] [PubMed] [Google Scholar]
- 10.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A gene. J Clin Investig. 1995;96:224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein J. PHYLIP—phylogeny inference package. Cladistics. 1989;5:164–166. [Google Scholar]
- 13.Felsenstein J. Confidence limits on phylogenesis: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein J. PHYLIP (phylogeny inference package), version 3.5c. Seattle, Wash: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 15.Frangeul L, Cresta P, Perrin M, Duverlie G, Khorsi H, Musset L, Opolon P, Huraux J M, Lunel F. Pattern of HCV antibodies with special reference to NS5A reactivity in HCV-infected patients: relation to viral genotype, cryoglobulinemia and response to interferon. J Hepatol. 1998;28:538–543. doi: 10.1016/s0168-8278(98)80275-4. [DOI] [PubMed] [Google Scholar]
- 16.Fukuma T, Enomoto N, Marumo F, Sato C. Mutations in the interferon-sensitivity determining region of hepatitis C virus and transcriptional activity of the nonstructural region 5A protein. Hepatology. 1998;28:1147–1153. doi: 10.1002/hep.510280433. [DOI] [PubMed] [Google Scholar]
- 17.Gale M, Blakely C M, Kwieciszewski B, Tan S L, Dossett M, Tang N M, Korth M J, Polyak S J, Gretch D R, Katze M G. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gale M J, Korth M J, Tang N M, Tan S L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 19.Germanidis G, Pellerin M, Bastie A, Stuyver L, Duverlie G, Darthy F, Remire J, Duval J, Dhumeaux D, Pawlotsky J M. Study of the genetic heterogeneity of the NS5A region of the HCV-1b genome and evolution under interferon alfa therapy. Hepatology. 1996;24:264A. [Google Scholar]
- 20.Gerotto M, Dal P-F, Sullivan D G, Chemello L, Cavalletto L, Polyak S J, Pontisso P, Gretch D R, Alberti A. Evidence for sequence selection within the non-structural 5A gene of hepatitis C virus type 1b during unsuccessful treatment with interferon-alpha. J Viral Hepatitis. 1999;6:367–372. doi: 10.1046/j.1365-2893.1999.00166.x. [DOI] [PubMed] [Google Scholar]
- 21.Gerotto M, Sullivan D C, Polyak S J, Chemello L, Cavalletto L, Pontisso P, Alberti A, Gretch D R. Effect of retreatment with interferon alone or interferon plus ribavirin on hepatitis C virus quasispecies diversification in nonresponder patients with chronic hepatitis C. J Virol. 1999;73:7241–7247. doi: 10.1128/jvi.73.9.7241-7247.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofgärtner W T, Polyak S J, Sullivan D G, Carithers R L, Gretch D R. Mutations in the NS5A gene of hepatitis C virus in North American patients infected with HCV genotype 1a or 1b. J Med Virol. 1997;53:118–126. doi: 10.1002/(sici)1096-9071(199710)53:2<118::aid-jmv3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 23.Holland J J, De La Torre J C, Steinhauer D A. RNA virus populations as quasispecies. Curr Top Microbiol Immunol. 1992;176:1–20. doi: 10.1007/978-3-642-77011-1_1. [DOI] [PubMed] [Google Scholar]
- 24.Ide Y, Tanimoto A, Sasaguri Y, Padmanabhan R. Hepatitis C virus NS5A protein is phosphorylated in vitro by a stably bound protein kinase from HeLa cells and by cAMP-dependent protein kinase A-alpha catalytic subunit. Gene. 1997;201:151–158. doi: 10.1016/s0378-1119(97)00440-x. [DOI] [PubMed] [Google Scholar]
- 25.Ide Y, Zhang L W, Chen M, Inchauspe G, Bahl C, Sasaguri Y, Padmanabhan R. Characterization of the nuclear localization signal and subcellular distribution of hepatitis C virus nonstructural protein NS5A. Gene. 1996;182:203–211. doi: 10.1016/s0378-1119(96)00555-0. [DOI] [PubMed] [Google Scholar]
- 26.Ina Y. New methods for estimating the numbers of synonymous and nonsynonymous substitutions. J Mol Evol. 1995;40:190–226. doi: 10.1007/BF00167113. [DOI] [PubMed] [Google Scholar]
- 27.Inchauspe G, Zebedee S, Lee D H, Sugitani M, Nasoff M, Prince A M. Genomic structure of the human prototype strain H of hepatitis C virus: comparison with American and Japanese isolates. Proc Natl Acad Sci USA. 1991;88:10292–10296. doi: 10.1073/pnas.88.22.10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaneko T, Tanji Y, Satoh S, Hijikata M, Asabe S, Kimura K, Shimotohno K. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem Biophys Res Commun. 1994;205:320–326. doi: 10.1006/bbrc.1994.2667. [DOI] [PubMed] [Google Scholar]
- 29.Kato N, Lan K H, OnoNita S K, Shiratori Y, Omata M. Hepatitis C virus nonstructural region 5A protein is a potent transcriptional activator. J Virol. 1997;71:8856–8859. doi: 10.1128/jvi.71.11.8856-8859.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khorsi H, Castelain S, Wyseur A, Izopet J, Canva V, Rombout A, Capron D, Capron J P, Lunel F, Stuyver L, Duverlie G. Mutations of hepatitis C virus 1b NS5A 2209-2248 amino acid sequence do not predict the response to recombinant interferon-alfa therapy in French patients. J Hepatol. 1997;27:72–77. doi: 10.1016/s0168-8278(97)80282-6. [DOI] [PubMed] [Google Scholar]
- 32.Kolykhalov A A, Agapov E V, Rice C M. Specificity of the hepatitis C virus NS3 serine protease: effects of substitutions at the 3/4A, 4A/4B, 4B/5A, and 5A/5B cleavage sites on polyprotein processing. J Virol. 1994;68:7525–7533. doi: 10.1128/jvi.68.11.7525-7533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolykhalov A A, Feinstone S M, Rice C M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurosaki M, Enomoto N, Murakami T, Sakuma I, Asahina Y, Yamamoto C, Ikeda T, Tozuka S, Izumi N, Marumo F, Sato C. Analysis of genotypes and amino acid residues 2209–2248 of the NS5A region of hepatitis C virus in relation to the response to interferon-beta therapy. Hepatology. 1997;25:750–753. doi: 10.1002/hep.510250343. [DOI] [PubMed] [Google Scholar]
- 35.Larson A M, Schwartz J M, Cox P P, Polyak S J, Nousbaum J B, Gretch D R, Carithers R L. High dose induction interferon therapy improves early response rate in naive genotype 1 patients. Hepatology. 1999;30:A1897. [Google Scholar]
- 36.Martell M, Esteban J I, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinot Peignoux M, Marcellin P, Pouteau M, Castelnau C, Boyer N, Poliquin M, Degott C, Descombes I, Le Breton V, Milotova V, et al. Pretreatment serum hepatitis C virus RNA levels and hepatitis C virus genotype are the main and independent prognostic factors of sustained response to interferon alfa therapy in chronic hepatitis C. Hepatology. 1995;22:1050–1056. [PubMed] [Google Scholar]
- 38.McHutchison J G, Gordon S C, Schiff E R, Shiffman M L, Lee W M, Rustgi V K, Goodman Z D, Ling M H, Cort S, Albrecht J K. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 39.Nousbaum J B, Pol S, Nalpas B, Landais P, Berthelot P, Brechot C. Hepatitis C virus type 1b (II) infection in France and Italy. Collaborative Study Group. Ann Intern Med. 1995;122:161–168. doi: 10.7326/0003-4819-122-3-199502010-00001. [DOI] [PubMed] [Google Scholar]
- 40.Okamoto H, Kojima M, Sakamoto M, Iizuka H, Hadiwandowo S, Suwignyo S, Miyakawa Y, Mayumi M. The entire nucleotide sequence and classification of a hepatitis C virus isolate of a novel genotype from an Indonesian patient with chronic liver disease. J Gen Virol. 1994;75:629–635. doi: 10.1099/0022-1317-75-3-629. [DOI] [PubMed] [Google Scholar]
- 41.Paterson M, Laxton C D, Thomas H C, Ackrill A M, Foster G R. Hepatitis C virus NS5A protein inhibits interferon antiviral activity, but the effects do not correlate with clinical response. Gastroenterology. 1999;117:1187–1197. doi: 10.1016/s0016-5085(99)70405-1. [DOI] [PubMed] [Google Scholar]
- 42.Pawlotsky J M, Germanidis G, Neumann A U, Pellerin M, Frainais P O, Dhumeaux D. Interferon resistance of hepatitis C virus genotype 1b: Relationship to nonstructural 5A gene quasispecies mutations. J Virol. 1998;72:2795–2805. doi: 10.1128/jvi.72.4.2795-2805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pawlotsky J M, Pellerin M, Bouvier M, RoudotThoraval F, Germanidis G, Bastie A, Darthuy F, Remire J, Soussy C J, Dhumeaux D. Genetic complexity of the hypervariable region 1 (HVR1) of hepatitis C virus (HCV): influence on the characteristics of the infection and responses to interferon alfa therapy in patients with chronic hepatitis C. J Med Virol. 1998;54:256–264. [PubMed] [Google Scholar]
- 44.Polyak S J, Paschal D, McArdle S, Gale M, Moradpour D, Gretch D R. Characterization of the effects of hepatitis C virus non-structural 5A protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology. 1999;29:1262–1271. doi: 10.1002/hep.510290438. [DOI] [PubMed] [Google Scholar]
- 45.Polyak S J, Paschal D M, McArdle S, Duverlie G, Gretch D R. Antiviral resistance in hepatitis C: further evidence for ISDR-independent NS5A-mediated interferon resistance in vitro. Hepatology. 1999;30:A591. [Google Scholar]
- 46.Polyak S J, McArdle S, Liu S L, Sullivan D G, Chung M J, Hofgartner W T, Carithers R L, McMahon B J, Mullins J I, Corey L, Gretch D R. Evolution of hepatitis C virus quasispecies in hypervariable region 1 and the putative interferon sensitivity-determining region during interferon therapy and natural infection. J Virol. 1998;72:4288–4296. doi: 10.1128/jvi.72.5.4288-4296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poynard T, Leroy V, Cohard M, Thevenot T, Mathurin P, Opolon P, Zarski J P. Meta-analysis of interferon randomized trials in the treatment of viral hepatitis C: effects of dose and duration. Hepatology. 1996;24:778–789. doi: 10.1002/hep.510240405. [DOI] [PubMed] [Google Scholar]
- 48.Ray S C, Wang Y M, Laeyendecker O, Ticehurst J R, Villano S A, Thomas D L. Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: hypervariable region 1 as a decoy. J Virol. 1999;73:2938–2946. doi: 10.1128/jvi.73.4.2938-2946.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reed K E, Rice C M. Identification of the major phosphorylation site of the hepatitis C virus H strain NS5A protein as serine 2321. J Biol Chem. 1999;274:28011–28018. doi: 10.1074/jbc.274.39.28011. [DOI] [PubMed] [Google Scholar]
- 50.Reed K E, Xu J, Rice C M. Phosphorylation of the Hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J Virol. 1997;71:7187–7197. doi: 10.1128/jvi.71.10.7187-7197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rehermann B, Chisari F V. Cell mediated immune response to the hepatitis C virus. Curr Top Microbiol Immunol. 2000;242:299–325. doi: 10.1007/978-3-642-59605-6_14. [DOI] [PubMed] [Google Scholar]
- 52.Saiz J C, Lopez Labrador F X, Ampurdanes S, Dopazo J, Forns X, SanchezTapias J M, Rodes J. The prognostic relevance of the nonstructural 5A gene interferon sensitivity determining region is different in infections with genotype 1b and 3a isolates of hepatitis C virus. J Infect Dis. 1998;177:839–847. doi: 10.1086/515243. [DOI] [PubMed] [Google Scholar]
- 53.Sarrazin C, Berg T, Lee J H, Ruster B, Kronenberger B, Roth W K, Zeuzem S. Mutations in the protein kinase-binding domain of the NS5A protein in patients infected with hepatitis C virus type 1a are associated with treatment response. J Infect Dis. 2000;181:432–441. doi: 10.1086/315263. [DOI] [PubMed] [Google Scholar]
- 54.Smith D B, Simmonds P. Characteristics of nucleotide substitution in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J Mol Evol. 1997;45:238–246. doi: 10.1007/pl00006226. [DOI] [PubMed] [Google Scholar]
- 55.Squadrito G, Leone F, Sartori M, Nalpas B, Berthelot P, Raimondo G, Pol S, Brechot C. Mutations in the nonstructural 5A region of hepatitis C virus and response of chronic hepatitis C to interferon alfa. Gastroenterology. 1997;113:567–572. doi: 10.1053/gast.1997.v113.pm9247477. [DOI] [PubMed] [Google Scholar]
- 56.Tan S L, Nakao H, He Y P, Vijaysri S, Neddermann P, Jacobs B L, Mayer B J, Katze M G. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc Natl Acad Sci USA. 1999;96:5533–5538. doi: 10.1073/pnas.96.10.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanimoto A, Ide Y, Arima N, Sasaguri Y, Padmanabhan R. The amino terminal deletion mutants of hepatitis C virus nonstructural protein NS5A function as transcriptional activators in yeast. Biochem Biophys Res Commun. 1997;236:360–364. doi: 10.1006/bbrc.1997.6967. [DOI] [PubMed] [Google Scholar]
- 58.Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995;69:3980–3986. doi: 10.1128/jvi.69.7.3980-3986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeuzem S, Lee J H, Roth W K. Mutations in the nonstructural 5A gene of European hepatitis C virus isolates and response to interferon alfa. Hepatology. 1997;25:740–744. doi: 10.1002/hep.510250341. [DOI] [PubMed] [Google Scholar]