Abstract
With the biological relevance of the whole cells, low cost compared with animal experiments, a wide variety of cell-based screening platforms (cell-based assay, cell-based microfluidics, cell-based biosensor, cell-based chromatography) have been developed to address the challenges of drug discovery. In this review, we conclude the current advances in cell-based screening and summary the pros and cons of the platforms for different applications. Challenges and improvement strategies associated with cell-based methods are also discussed.
Keywords: Cell-based screening, Drug candidate, Microfluidics, Biosensor, Affinity chromatography
INTRODUCTION
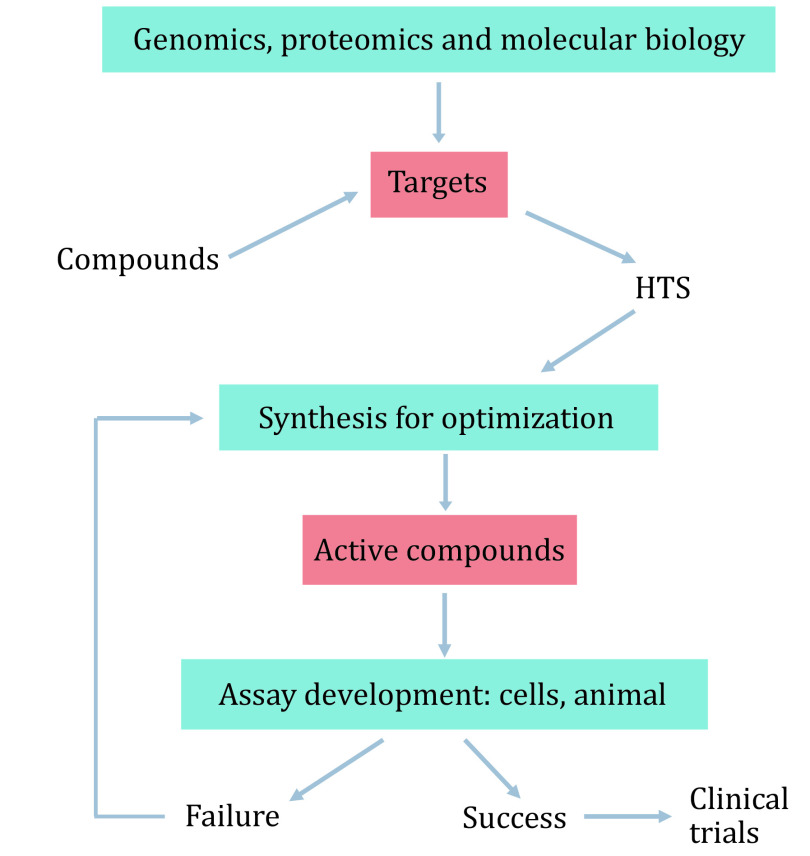
Traditional drug discovery involves a serial stage for the development of the new drug. It is expensive and can take 10–15 years. Mostly, high-throughput screening (HTS) is carried out after target confirmation, following with optimization of the compound structure, animal testing, and finally clinical trials (Fig. 1). However, it remains a high failure rate in drug discovery, which causes the tendency to discover new targets for drug repurposing for more diseases (Moridani and Harirforoosh 2014; Parvathaneni et al. 2019; Wang 2018). And the critical issue is the appropriate target (druggability of the target) that should provide an unambiguous, therapeutically significant response to improve the drug discovery (Jorgensen 2012; Roy 2019).
Figure 1.
Lead generation with HTS screening
Actually, a more important reason is the lack of biological context during the screening process. In Arduino’s study, cryopreserved mitochondria isolated from yeast strain were engineered with functional protein and then were employed as a ready-to-use screening reagent. The reduced false discovery rate was carried out by energizing mitochondria with D-lactate in a mannitol/sucrose-based medium, which indicates the significance of bionic and in vivo environment (Arduino et al. 2021). Besides, among the anti-cancer drugs, sorafenib and regorafenib show significant differences in activity, but only one difference in structure for the non-hydrogen atom (i.e., a fluorine). So there is no sufficient resolution in current methods to distinguish compounds with subtle structural changes except for animal or cell assays (Schlessinger et al. 2017). It means new tools and techniques that can better reflect the in vivo environment are required during the drug discovery process.
Currently, the demand of cell-based functional assays in HTS is increasing. One obvious advantage is that cell-based assays present more physiologically relevant systems for the screening of compounds (Fursov et al. 2005). It indicates that cell-based screening has more potential for development (Szabo et al. 2017). For example, at the beginning of the genomic era, enzyme-based biochemical screens were focused during the antibacterial drug development to replace the traditional cell-based phenotypic screens. However, after a long period of HTS practice, it was discovered that the required drugs could not be successfully provided. Thereafter, the focus in the antibiotic drug discovery field has shifted back to whole cell-based phenotypic screens directly (Yuan et al. 2021). Kumar found that the result of screening against PanC which is considered a druggable target had no significant cellular activity in a variety of biochemical screens. In contrast, traditional whole-cell screening has proven more successful. The reason may be multiple new targets can be implemented on the whole cell (Kumar et al. 2017). The discovery of antibiotics is mainly through cell-based screens, as the inhibit activity of identified novel inhibitors based on essential enzymes targets was not as expected (Datta 2021). Screening in whole cells can reveal a great deal more about the targets and action mechanism of compounds compared to in vitro screening based on enzyme or protein targets (Adamson et al. 2021).
Given the importance of biological context, preclinical models are widely used in drug discovery, including in vitro models (cell culture), ex vivo models, and in vivo models (artificial, transgenic, non-transgenic and induced) (Shi et al. 2019; Xu et al. 2021). However, more than 20,000 molecules were screened using different animal models for Alzheimer’s disease drug development during the past two decades, only Aducanumab was approved by FDA (Cacabelos et al. 2021). The challenge is that no single model faithfully reproduces all the features of human disease. So, drug discovery can integrate different important attributes in a multisystem model, which can be achieved by cell-based models (Cacabelos et al. 2021; Kumar et al. 2017; Szabo et al. 2017).
Cell-based screening in drug discovery is usually two-dimensional (2D) screening, due to that 2D cell culture models remain the accepted standard for drug screening in vitro. 2D cell culture model can provide valuable insights into biological processes and effects of new drugs with low cost and efficient workflows, which is widely used in various screening methods (Amelian et al. 2017; Thippabhotla et al. 2019). However, growing evidence indicates that 2D cell culture models often fail to represent the underlying biology of cells, such as in vivo extracellular matrix microenvironment, and therefore cannot accurately predict the in vivo drug response (Belfiore et al. 2021; Godugu and Singh 2016). This review will summarize the current state relating to different cell-based screening technologies containing 2D and 3D models. It will also provide recent perspectives about the cell-based HTS from natural herbs in drug discovery.
CELL-BASED ASSAY
The cell-based assay is usually combined with HTS, and the distinction between a cell-based assay and an in vitro screening is that the cell-based assay utilizes live cells seeded onto the floor of the well (Rajalingham 2016). Usually, cellular screening relies on different strategies ranging from reporter gene technology to protein fragment complementation assays. In order to reduce the response time, the monitoring of its first activation step can be treated as alternative approach by using fluorescence and bioluminescence resonance energy transfer (Michelini et al. 2010). Cell-based assays are used to identify the best drug candidate (Capula et al. 2019), measure proliferation (Adan et al. 2016), toxicity (Li et al. 2006), motility (Sanookpan et al. 2021), analyze cell signaling pathways (Pathe-Neuschafer-Rube et al. 2021), and changes in morphology (Rajalingham 2016). Among the cell-based assays, 2D versus 3D culture might also contribute to the results obtained.
2D screening of cell-based assay
A promising tool to bridge between species or from health to disease is in vitro cell culture. The simplest 2D models include monolayer cell culture, adding molecules or molecular libraries to the culture medium, and measuring the output with a microplate reader or microscope (Foster et al. 2021). Although they lack the sophisticated tissue structures or biophysical stimulation present in vivo, the way in which monolayer culture responds to chemical or genetic stress is largely consistent with clinical observations or primary cell data (Tu et al. 2021). In addition, a key advantage of a 2D model is the compatibility with high-throughput analysis. So, a simple 2D in vitro model may serve as a preliminary screening tool. Of course, the drawbacks of animal experiments such as extremely time-consuming and cost-intensive, a significant discrepancy between animal toxicity and human toxicity, are an aspect that promotes the development of cell-based assay (Doke and Dhawale 2015; Madden et al. 2020).
Conventionally, 2D models are performed in dishes, tubes, or well plates. The aim is to confirm the effect of the different concentrations of the candidate on cellular growth and function (Hamon et al. 2013; Hu et al. 2015). For the most widespread cell viability or cytotoxicity assays in drug discovery, 96, 384, or even 1,536 microtiter plates are most commonly used with colorimetric readouts of cell supernatants (Riss 2005; Wegener 2015). Radnai et al. presented a simple cell-based method for the discovery of novel cytokinesis inhibitors. The assay was performed in a 96-well plate format in 48 h. Then, living cells, nuclei and nuclei of dead cells are identified by a single staining step using three fluorescent dyes, followed by rapid live cell imaging (Radnai et al. 2020). Scaling up of screening systems, with the use of multiwell plates and multichannel pipettes (or even robotic liquid handling systems) is fairly commonplace. It should be noted that when using a multiwell plate, the number of cells per well and equilibration period before the assay will affect the responsiveness to compounds (Riss 2005). Heinzman et al. developed a liquid handler equipped with a 1000-μL capacity 96-tip tool for cell plating automate to minimize human error while increasing accuracy, precision, and efficiency (Heinzman et al. 2010). Soman et al. used plates that seeded with disialoganglioside (GD2) — expressing cell lines to bind and screen the anti-GD2 molecules and quantify the GD2-specific binding activities. They found that the cell-based assay showed more consistent and reproducible comparing with microtiter plate coated with purified GD2 (Soman et al. 2011). Thomas et al. developed a rotatable disc microfabricated with multichannel for performing cell growth and cell-based assays in a liquid medium. The apparatus and methods can be used to measure a variety of biochemical processes and products. Combining with non-invasive techniques does not compromise the integrity or viability of cells (Thomas 2011).
In terms of detection on cell-based assays, improvements in various detection techniques are also promoting the development of cell-based methods. A new plate reader (Nanotaurus) was developed by Edinburgh Instruments, which has the principal features of a confocal microscope and acquires data by the technique of time correlated single photon counting. This instrument demonstrates the advantages of biochemical assays and shows strong promise for cell-based assays (Näther et al. 2006). The microscopic imaging technique is the necessary detection method for many cell-based assays, but due to the cost of equipment, it is not in general widely adopted for primary screening. So Olson et al. used enzyme complementation to provide an analytical method that uses substrates to generate luminescent signals. The principal advantage of this method is amenable to HTS using microtiter plate protocols (Olson and Eglen 2007). Mohiuddin et al. stably co-expressed target fragments tagged with luminescence probes in HEK-293FT cells and identify five compounds as lead compounds (Mohiuddin et al. 2021). Fluorescent imaging often requires the removal of background fluorescent signals to obtain robust measurements, which is challenging for high-density microplates. In view of this problem, a wash-free cell-based fluorescence assay method was proposed, which uses a laser scanning fluorescence plate cytometer. This work shows that sensitivity and efficiency are increasing while assay artifacts are reduced, and results in the development of broadly applicable cell-based fluorescence imaging assays for drug screening (Gorshkov et al. 2020).
Mainly primary animal cells, tissue specimens, and immortalized as well as tumor cell lines have been used in cell-based assays (Fritsche et al. 2021). Most cell-based screening is often engineered to overexpress targets or reporter constructs, due to that the immortalized cell lines are easy to culture and expand, which is quite suitable for HTS. For example, Spodoptera frugiperda insect cell expressed hCOX-1 and hCOX-2 proteins was used to identify the selective inhibitors of hCOX-1 and/or hCOX-2 (Zhang et al. 2004). However, the generation of cell lines involves the cell clones by proliferating ex vivo which is different from the in vivo counterparts. Its experimental condition may alter growth characteristics and signal transduction pathways. By contrast, primary cells are more closely reflect cell behaviors in human tissues and more physiologically relevant to human biology (Berg 2019; Berg et al. 2014). Tumor cell lines are another type of primary cells, and more closely reflect the genetic and clonal heterogeneity of the native tumor in vitro model system, thus providing a more accurate pre-clinical platform (Corallo et al. 2020). Wang et al. found human lactate dehydrogenase A (hLDHA) is overexpressed in osteosarcoma cells as compared to a human normal cell. So they used a cell-based phenotypic screening assay to solve the highly polar nature of hLDHA, and discovered three cellular active inhibitors (Wang et al. 2020a).
Simple 2D cell-based assays have limitations, partly due to their plate format. So a wall-less plate technology was present, which takes advantage of hydrophobic and hydrophilic surface properties of the unique liquid. This technology showed an obvious advantage when suspension cells were used in multistep experimental procedures (Quinones et al. 2013). Some groups sought to introduce an extra level of complexity to increase the physiological relevance of their 2D screening systems. Another mean was to introduce an extracellular matrix to mimic chemical and mechanical properties, which was designed for the screening models of tissue types (Foster et al. 2021). Zhang et al. first described the differentiation of hESCs into a mixed culture of neurons, astrocytes, and oligodendrocytes (Zhang et al. 2001). From 2D cell culture-based monolayers, multilayer to co-culture models, their aims were to promote physiological characters, reproducibility and mimic characteristic functionalities of disease modeling (Kutlehria and Sachdeva 2021). In order to develop in vitro models, many factors need to be considered, such as cell line type, cell culture medium, substrate roughness and stiffness. They affect the final outcome of the in vitro assay through the significantly effect of the microenvironment. Advanced technologies based on 3D models have allowed the development of more complex structures, bridging the gap between in vitro and in vivo models (Yuste et al. 2021).
Limitations of 2D format
Although simple models are easier to create and faster to reproduce, their systems present a number of limitations. Some candidate molecules often fail to perform in vivo. One reason is that the 2D models lack microenvironments, such as complex geometrical architecture, paracrine signals from neighboring cells, mechanical properties, nutrition and oxygen, to mimic the native tissue. This microenvironment will strongly influence cellular behavior and functionalities containing proliferation, differentiation and metabolism (Berg 2019; Davoudi et al. 2021; Rimann and Graf-Hausner 2012; Wollrab et al. 2016). On the other hand, enhanced drug sensitivities are proved in 2D conditions and require lower dosage ranges, resulting in ineffective in vivo (Foster et al. 2021). In cell-based assays, a main hurdle is to design a sufficiently powerful detection method with adequate signal to noise while maintaining the inherent physiology of the cells (Halim 2020).
3D screening model
Improving the success rate in the early stages of drug development requires disease models with high biological relevance for biomarker discovery and drug development. In cell-based experiments, the rapid increase in 3D cell culture technologies more closely mimics in vivo physiology, which is considered a promising step to improve the success rate of drug discovery (Langhans 2021). Especially for tumor models, 3D format is similar to in vivo tumors, which can recapitulate the complexity of the tumor microenvironment, and therefore bridge the gap between 2D monolayers and animal models (Fontana et al. 2021). The 3D cell culture models either rely on the self-organizing properties of mammalian cells or use bioengineered constructs to arrange cells like the organ. A self-assembling 3D multicellular brain model is used to mimic the complex in vivo cytoarchitecture of the brain. The data showed that the combination of 3D cell culture and bioengineering can improve reproducibility and tissue architecture (Hattori 2014; Lancaster et al. 2017). Additionally, some studies create simple 3D co-culture models by using a mixture of cell types present in the tissue microenvironment to observe the responses in vivo (Belfiore et al. 2021; Lazzari et al. 2018).
The 3D cell models include spheroids, hanging drops, scaffolds, cell sheets, hydrogels, bioreactors, and microfluidic chips (Bialkowska et al. 2020; Yuste et al. 2021). The scaffold-free 3D cell models including multicellular tumor spheroid models are better in terms of in vivo context simulation compared to 2D cell models, but they are lack of extracellular matrix recapitulation that limits their applicability in relevant drug testing (Cavo et al. 2016). Scaffolds are widely used to create 3D models, such as collagen scaffold, chitosan-alginate scaffold, nanofiber scaffold and hydrogel scaffold (Leung et al. 2010; Liu et al. 2018b; Yang and Zhao 2011). The advanced technologies such as microfluidics, biosensor and chromatography will be described later.
Successes from cell-based assay
Cell-based assays are suit to screen targets that are refractory to biochemical purification and can characterize compounds with unknown targets (Fig. 2). In physiologically relevant settings, intracellular signals can be transmitted so that agonists and antagonists can be identified. Meanwhile, different binding sites of the same receptor, especially allosteric sites, can be screened for diverse pharmacological effects of compounds (An and Tolliday 2010; Drewe and Cai 2010; Berg et al. 2014; Zaman et al. 2007).
Figure 2.
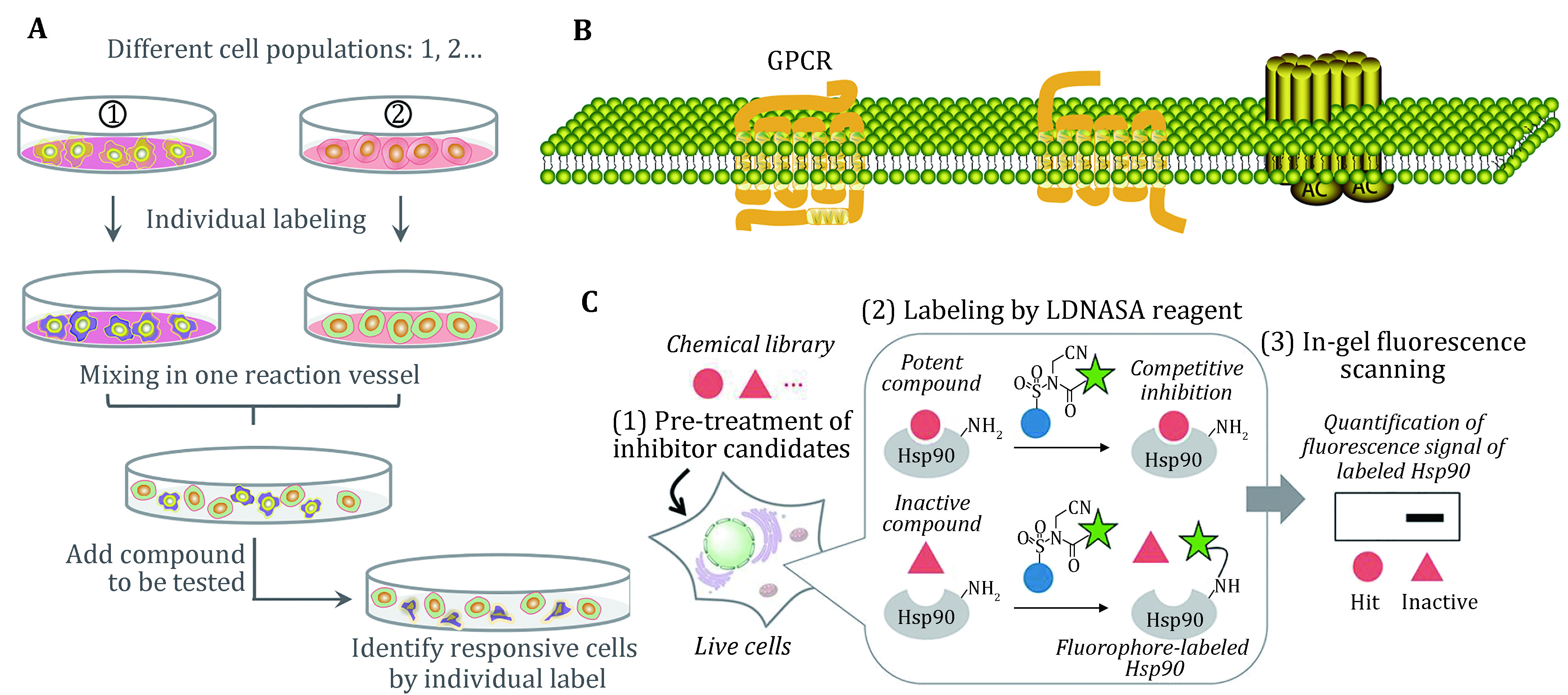
Scope of application for cell-based assay. A Unknown targets can use the whole cells for screening. B GPCRs as target that is difficult for purification. C Cell-based ligand-screening system for inhibitor or allosteric inhibitor (Ueda et al. 2020)
In the present study, some of the same compounds screened by different cell-based assays show different pharmacological activities. For example, brefeldin A can inhibit the cytotoxic effects of ricin (Wahome et al. 2010) and can also inhibit the growth of two pairs of parental and Pgp-overexpressing multidrug-resistant cell lines (Zahra et al. 2020). Apigenin stimulates hair growth through downregulation of the TGF-β1 gene (Huh et al. 2009) and is also identified as potent activators of PXR-mediated CYP3A4 promoter activation (Dong et al. 2010), activators of the JAK/STAT pathway (Tai et al. 2012). Quercetin can inhibit ABCG2 activity (Henrich et al. 2006) and prevent H. pylori adhesion and infection (Sekiguchi et al. 2008), also can be a potential IFN mimic or adjuvant in new antiviral drugs (Tai et al. 2012). Luteolin can prevent H. pylori adhesion and infection (Sekiguchi et al. 2008) and is also identified as ANO1 inhibitors as potential anticancer therapeutic agents for prostate cancer (Seo et al. 2017), besides, it is also identified as a potential IFN mimic or adjuvant in new antiviral drugs (Tai et al. 2012). In addition, Table 1 shows the active compounds screened by cell-based assay in the past five years that may be developed into promising drug candidates.
Table 1. Cell-based screening assay for candidate drugs.
|
Cell type(s) |
Model |
Active compounds |
References |
| SPR: surface plasmon resonance; MEFs: NF1-deficient mouse embryonic fibroblasts | |||
|
MCF-7 Cells/ OR6M1-expressing cell lines |
SPR chip immobilized cells. A modified carboxymethyl dextran sensor chip |
Anthraquinone, rutin |
Choi et al. 2021 |
|
DNA-PKcs and OCT4 - expressing HEK-293FT cells/ MK2 and OCT4 expressing NCI-H82 cells |
384-well plate. Two-step method. Exogenously expressing proteinsin cancer cells as first step |
A cardioglycoside and an isocarbostyril alkaloid, cholesterol-based structures (three compounds) |
Mohiuddin et al. 2021 |
|
Vero E6 cells |
96-well plates. Transfected 2-E plasmids after compounds pre-incubated |
34 hits with cell protection activity |
Wang et al. 2021 |
|
HeLa cells |
24-well plates. Treated with compounds followed by labeling of the intracellular Hsp90. Analysis with in-gel fluorescence after cell lysis |
157 compounds. Morin |
Ueda et al. 2020 |
|
COS7 cells |
Clean glass. Single-protein tracking in a living cell. Effects are evaluated by diffusion coefficient shift using fluorescence microscopy |
Hyperoside for EGFR and ErbB2. DiAB-141 and 2”-O-acetylvitexin for ErbB3 |
Kim et al. 2021 |
|
COS-7 cells |
96-well plates. Cells seeded in monolayer and molecules added to the medium |
Blebbistatin, para-aminoblebbistatin, para-nitroblebbistatin, jasplakinolide, cytochalasin D, swinholide A |
Radnai et al. 2020 |
|
HEK293:FLP-InT-REx-BiFC#20 cells |
384-well clear-bottom, black-walled microplate. Cells seeded in monolayer and molecules added to the medium |
6 compounds. Isocotoin |
Xu et al. 2020 |
|
Vero E6/A549/Huh7/ LN-18 cells |
6-well plate format and 96-well plate. Cells infected viral dilutions. Cells seeded in monolayer and antivirals in infection medium added to cells |
Sofosbuvir and ribavirin |
Vicenti et al. 2020 |
|
HEp-2/ A549/293T cells |
96-well plate. Cells seeded in monolayer and molecules and virus added to the medium |
Laby A1/A2 |
Blockus et al. 2020 |
|
Human embryonic stem cells/ Fibroblasts from healthy donors and patients |
384-well plates. Cells seeded in monolayer and molecules added to the medium |
CUDC-907 |
Kase et al. 2021 |
|
HpeG2/ HeLa cells |
Culture chamber. 3D electric cell/matrigel-substrate impedance sensing chip. Cells seeded in prechilled matrigel solution and generated 3D structure |
Taxol, cisplatin, sorafenib |
Pan et al. 2019 |
|
HeLa cells |
96-well plates and 384-well plates. Cells seeded in monolayer and molecules added to the medium followed with added viral solution |
11 compounds. Gemcitabine |
Zhang et al. 2017 |
|
MEFs |
96-well plates. Cells seeded in monolayer and molecules added to the medium |
Cantharidin, Nifedipine |
Semenova et al. 2017 |
|
HeLa cells |
384-well clear-bottomed black plates. Cells seeded in monolayer and molecules added to the medium |
18 compounds |
Hajjar et al. 2017 |
|
MDA-MB-231 breast cancer cells/ HUVECs |
96-well plates. Cells seeded in monolayer and extracts added to the medium |
Cirsimaritin, Cirsium japonicum extract, cirsimaritin |
Yeon Park et al. 2017 |
|
HepG2 ARE reporter cells |
384 well pate format. Cells seeded in monolayer and molecules added to the medium |
AZ-628, PHA-767491, SL-327, PAC-1, pifithrin-α, vitamin B12 |
Liu et al. 2018a |
|
MC3T3-E1-OSE cells |
96-well plates. Cells seeded in monolayer and molecules added to the medium |
4 compounds. T63 |
Zhao et al. 2017 |
|
ARPE-19 cells |
96-well plate. Cells seeded in monolayer and molecules added to the medium |
47 compounds |
Maruyama et al. 2018 |
|
MG-63 cells |
96-well plate. Cells seeded in monolayer and molecules added to the medium |
42 compounds |
Wang et al. 2020a |
|
A549 cells |
384-well plates. Cells seeded in monolayer and infected with DENV1-4, and treated immediately with compounds |
(S)-29 |
Kounde et al. 2017 |
ADVANCED CELL-BASED SCREENING TECHNOLOGIES
Microfluidics technologies for drug screening
Microfluidics is also known as Lab-on-a-chip, represents a technology that can precisely control and manipulate sub-millimetre scale fluids in geometry. In the last decades, microfluidic devices have gradually been used as a multi-functional tool for many types of cell-based analysis, such as in drug screening and discovery, cell culture, cell separation, intracellular signaling, toxicity and so on (Gupta et al. 2016). Microfluidic devices offer some benefits including rapid analysis, high sensitivity and reproducibility. Its key advantage is microscale dimensions that match with the cellular structures and microenvironments like the human body. Because of its nanoliter volumes samples and reagents, microfluidic technology is very cost effective. As with cell-based assay, microfluidic technology also can simulate the in vivo response. Especially, the miniaturization of microfluidics is suitable for HTS, compared with some cell-based assays (Caruso et al. 2020; Hattori et al. 2013).
In the application of high-throughput screening, three major complementary modes can be used to manipulate microfluidic. Perfusion flow mode requires a series of components to introduce reagents and samples, transferring and mixing fluids in the microchannel network. This mode manipulates the liquid flows continuously by external mechanical pumps or the capillary forces combined with electro-kinetic form (Coliaie et al. 2021; Hao et al. 2020). The liquid flows also can be driven by vacuum-driven pressure or gas-generating chemical reactions (Park et al. 2020). Gao et al. carried out one-step cell seeding and anti-cancer drug testing by a microfluidic channel combined with vacuum actuated chambers (Gao et al. 2013). Guler et al. developed a self-powered microfluidic device. The key part is a 3D-printed effervescent pump for CO2 generation from a chemical reaction. When the coagulation starts, an acid-base reaction is triggered for the gas generation that drives the fluids within the channels (Guler et al. 2018). Using gravity driven flow is another possible solution. Zhu et al. presented a gravity driven pumping system using arrays of horizontally-oriented mini-reservoirs to generate a constant flow rate across microfluidic channels (Zhu et al. 2004). The advantage of continuous-flow is easy implementation, which makes it to be the most widely accepted microfluidic platform for simple biomedical applications. However, there are some limits in the perfusion flow mode. The use of microchannels for continuous fluid delivery tends to result in higher reagent consumption. Moreover, when applied to large-scale drug screening, chip structures are often complex, involving multiple channels, liquid-controlled pump and valve designs (Liang et al. 2021).
Droplet mode always uses water-in-oil emulsion droplets to compartmentalize reagents into nanoliter to picoliter volumes. It will create unavoidable interface fluctuation during emulsification. It can encapsulate biomolecules into discrete droplets and uses the generated units for analysis. The droplets are usually generated by pressure-driven flow (Shembekar et al. 2016), containing hydrodynamics and pneumatic pressure. Electrowetting can generate droplets by surface tension drive (Lian 2019; Liu et al. 2021). Gravity-driven overflow microfluidic system can infuse fluids steadily and continuously, which requires less manual power (Gao et al. 2019). The hanging-drop platform used in the tissue model enables continuous inter tissue communication, constant medium turnover, and immediate exchange of metabolites by gravity-driven flow through the network (Boos et al. 2019). Droplets encapsulation can exclude sample loss on the surface wall by preventing the contact between the sample and the droplet wall. Comparing with continuous microfluidics, droplet-based microfluidic overcomes complex fluidic control, does not require separated channels for each sample, and minimizes dilution and contamination issues (Damiati et al. 2018). Its key characteristics are using a few microliters of samples and requiring few cells. Furthermore, a high degree of automation and ease of integration with HTS makes it very promising in drug discovery (Shembekar et al. 2016; Wang et al. 2020b). When droplet-based microfluidics is used to generate microcarriers, they exhibit the advantages of high drug loading and relatively long drug release. However, the formation of monodispersed carriers is not constant or repeatable due to the solvent evaporation and droplet solidification step. In particular, the formation of nano-sized carriers is limited by droplet-based microfluidic systems. Moreover, mechanical stirring will destroy the shape, morphology, size uniformity and loading efficiency of the droplets (Damiati et al. 2018).
Cell microarrays mode has been well established for cellular phenotypes investigation and offers invaluable advantages of HTS. This screening mode needs the generation of cell microarrays on a 2D solid substrate, and then applies drug combinations or drug libraries to those arrays (Li et al. 2018). Arrays can be composed of single cells, cell monolayers, aggregates or spheroids. Microarrays can screen for thousands of different samples simultaneously in one single experiment with low reagent consumption and high-content readouts. Although effective, their high cost and the requirement of specialized equipment for their manufacture limit their scope of application. Besides, cells cultured on the microarray can cause neighboring effects and cross-contamination (Du et al. 2016; Jonczyk et al. 2016; Zhang et al. 2016).
Microfluidic technology is an effective tool for the enhancement of drug discovery. But single cell analysis is mostly used for cell function research. The heterogeneous responses from individual cells can provide information at both the individual and population levels (Seah et al. 2018; Yin and Marshall 2012). As mentioned before, the 2D monolayer cell lacks the microenvironment, leading to the ineffective for disease. So, the combination of microfluidic technology with the 3D cell culture offers great potential for drug discovery (Liu et al. 2019). A microfluidic platform was developed for anticancer compound screening by using multicellular spheroids as a 3D model derived from tumor biopsies. The characters of this lab-on-a-chip platform are self-generating nutrients, drug concentration gradients perfusion and equipment-free (Mulholland et al. 2018). The supporting matrix or carrier for the 3D cell culture is an important factor in microdevices. It can be summed up as gel-supported 3D cell culture, gel-free 3D cell culture and gel-coated 3D cell culture. Gel-supported 3D cell culture allows the encapsulation of cells into the hydrogel, and permits oxygen permeability and nutrient transport. In order to mimic in vivo microenvironment, native extracellular matrix proteins are always used as the basis of hydrogel scaffolding, such as collagen, fibrin, fibronectin, hyaluronic acid, matrigel, agarose, poly(ethylene glycol) diacrylate, or a mixture of both. While for gel-free 3D cell culture, intercellular polymeric linker polyethylenimine-hydrazide, microwells, suspension or spheroids model can be selected to supplement the gel-supported 3D cell culture (Li et al. 2012).
Cell-based sensor for drug screening
Cell-based biosensor systems consist of three components. The sensing unit contains cells for target identification. A transducer is used for converting biological reactions to chemical/electrical/optical signals, and the output system can amplify and readout signals (Zhou et al. 2011). It plays an outstanding role in drug discovery, cancer research and immunology. Cell-based biosensor systems that use whole cells as a living model have an obvious advantage, which is responding in a manner that can offer insight into the physiological effect of an analyte. The advantages include the detection of unknown compounds and toxins, readily coupling with HTS for drug candidates screening, and reducing the need for animal testing (Ozsoylu et al. 2021). In cell-based sensor detection, the key factors of cell function affected by the analytes can be singled-out without being disturbed by more complex, whole organism or whole organ responses. Cells grown in a thin layer have advantages in cell-based sensors, that is, they can be observed under a microscope or other optical equipment. Different cell types of cell-based sensors also show different advantages. For example, microorganism cells can be cultured easily and grow rapidly. It is less expensive to culture compared with mammalian cells. However, the mammalian cells can provide bioavailability and physiologic responses relevant to humans (Banerjee et al. 2010).
Since the cell-based biosensor uses living cells, its limitations are stability and robustness. On the one hand, researchers are trying to develop label-free biosensor technologies, which monitor the behavior of cells without stains damage or photobleaching effects (Shamah and Cunningham 2011). Due to the non-invasive nature of this technology, living cells can be continuously investigated, so real-time kinetic measurement can be achieved (Ona and Shibata 2010; Xi et al. 2008). Cryopreservation is another solution to maintain certain vital parameters of cells inside the sensor system. Özsoylu et al. proposed an on-sensor cryopreservation strategy with the modified chip surface. It can be effective for keeping cells viable on a biosensor chip (Ozsoylu et al. 2021). Due to the demand for high-throughput cellular assays, miniaturization of cell-based biosensors needs to be achieved by preparing cell microarrays. Flat substrates (positioning arrays) or particles (solution or suspension arrays) are used to immobilize different cells using various microfabrication technologies to achieve multiplexing and high-throughput cell-based sensing (Hong et al. 2017).
Despite the advantages of cell-based biosensors, some limitations are associated with the existing systems. Most cells used in the sensor are cultured on hard 2D glass or plastic matrix, which cannot mimic in vivo counterparts. Its weak cell-substrate attachment greatly shortens the effectiveness and life of cell-based biosensors (Mao and Kisaalita 2004). Advances in novel biomaterials and nano/micro engineering technologies have enabled to immobilize cells using scaffold-free 3D methods. So it is promising to address the limitation of 2D cell-based biosensors (Zhou et al. 2011). Dipeptide-derived hydrogel matrix was employed to encapsulate cells and enzymes that are used as sensing elements. This method is based on the self-assembly function of a small molecular hydrogel. An established 3D culture model based cellular biosensing system is useful for cellular function and drug discovery (Lian et al. 2017).
Cell-based chromatography for drug screening
The technologies mentioned are not suitable for the HTS of complex systems like natural herbs. Natural products can be used to treat various diseases. For many years, plant-derived products have been recognized as sources of therapeutic agents and structural diversity (Chopra and Dhingra 2021). Nevertheless, natural products also present challenges for drug discovery, now we will introduce several improved analytical tools to open up the new opportunity (Atanasov et al. 2021). The chromatographic methods established by adsorbing cell membrane on the surface of silica gel to screen bioactive compounds from traditional medicines are lack of stability. So a new strategy was designed for attaching cells onto amino microspheres. The microspheres were prepared by coating poly (oligo (ethylene glycol) methacrylate) with RGD peptide using atom transfer radical polymerization. Then the cells were immobilized to the microspheres based on the specific affinity between integrin on the cells and the RGD peptide. This method can increase the density of cells in the stationary phase at the same time. As a result, three bioactive compounds were screened from Ligusticum chuanxiong using the established cell column (Li et al. 2015). Liu et al. developed a novel hollow fiber cell fishing procedure with high-performance liquid chromatography. These methods were used for rapid screening, fishing, and analysis of bioactive compounds from traditional Chinese medicines. Firstly, the cells were seeded on the internal surface of the fibers, followed by inserting into the extracts of herbs. The active compounds can be screened by cells inside the fibers. Finally, the active compounds were dissociated and analyzed using HPLC/MS (Liu et al. 2014). Although the screening process approximates the interaction between the bioactive component and the cells in vivo, the stationary phase cannot be reused due to the sensitivity of live cells inoculated on the fiber. Recently, we reported an innovative cell-based microcarrier chromatography to simulate in vivo drug-receptor interaction. Cells firstly grow on the microcarriers, then the attachment can be improved using paraformaldehyde. The success of paraformaldehyde fixation is based on a layer of denatured collagen on the surface of the microcarrier. Due to the use of microcarriers for 3D cell culture, the stationary phase loaded into the column also presents 3D characteristics. Combing with HPLC/MS, active compounds can be bionically screened and identified successfully (Wei et al. 2021). Although cell-based chromatography can more likely screen active lead drugs, it lacks the function of predicting cellular effects after screening and identification, and needs to combine with the cell-based assay for further activity verification.
CONCLUSIONS
The need to increase clinically available drugs while reducing development costs is continuing to drive the development of cell-based screening methods. Each platform described in this review for drug discovery has associated strengths and limitations. In general, cell-based screening methods can build a bridge between animal experiments and human diseases. They are suitable to screen targets that are refractory to biochemical purification and characterize compounds with unknown targets. The screening results can be more physiologically relevant. Compared with animal experiments, cell-based screening methods are more efficient and less expensive. In addition, among these screening platforms, 3D models have more potential for drug development compared to 2D cell-based screening methods. Although numerous approaches exist today, it is very likely that a new strategy can combine several advantages of each approach in the future.
Conflict of interest
Fen Wei, Sicen Wang and Xilan Gou declare that they have no conflict of interest.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82003709, 81973277).
Compliance with Ethical Standards
Human and animal rights and informed consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Adamson CS, Chibale K, Goss RJM, Jaspars M, Newman DJ, Dorrington RA Antiviral drug discovery: preparing for the next pandemic. Chem Soc Rev. 2021;50(6):3647–3655. doi: 10.1039/D0CS01118E. [DOI] [PubMed] [Google Scholar]
- Adan A, Kiraz Y, Baran Y Cell proliferation and cytotoxicity assays. Curr Pharm Biotechnol. 2016;17(14):1213–1221. doi: 10.2174/1389201017666160808160513. [DOI] [PubMed] [Google Scholar]
- Amelian A, Wasilewska K, Megias D, Winnicka K Application of standard cell cultures and 3D in vitro tissue models as an effective tool in drug design and development. Pharmacol Rep. 2017;69(5):861–870. doi: 10.1016/j.pharep.2017.03.014. [DOI] [PubMed] [Google Scholar]
- An WF, Tolliday N Cell-based assays for high-throughput screening. Mol Biotechnol. 2010;45(2):180–186. doi: 10.1007/s12033-010-9251-z. [DOI] [PubMed] [Google Scholar]
- Arduino DM, Goh V, Mokranjac D, Perocchi F Drug discovery assay to identify modulators of the mitochondrial Ca(2+) uniporter. Methods Mol Biol. 2021;2277:69–89. doi: 10.1007/978-1-0716-1270-5_5. [DOI] [PubMed] [Google Scholar]
- Atanasov AG, Zotchev SB, Dirsch VM, International Natural Product Sciences T, Supuran CT Natural products in drug discovery: advances and opportunities. Nat Rev Drug Discov. 2021;20(3):200–216. doi: 10.1038/s41573-020-00114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee P, Franz B, Bhunia AK Mammalian cell-based sensor system. Adv Biochem Eng Biotechnol. 2010;117:21–55. doi: 10.1007/10_2009_21. [DOI] [PubMed] [Google Scholar]
- Belfiore L, Aghaei B, Law AMK, Dobrowolski JC, Raftery LJ, Tjandra AD, Yee C, Piloni A, Volkerling A, Ferris CJ, Engel M Generation and analysis of 3D cell culture models for drug discovery. Eur J Pharm Sci. 2021;163:105876. doi: 10.1016/j.ejps.2021.105876. [DOI] [PubMed] [Google Scholar]
- Berg EL Human cell-based in vitro phenotypic profiling for drug safety-related attrition. Front Big Data. 2019;2:47. doi: 10.3389/fdata.2019.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg EL, Hsu YC, Lee JA Consideration of the cellular microenvironment: physiologically relevant co-culture systems in drug discovery. Adv Drug Deliv Rev. 2014;69−70:190–204. doi: 10.1016/j.addr.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Bialkowska K, Komorowski P, Bryszewska M, Milowska K Spheroids as a type of three-dimensional cell cultures-examples of methods of preparation and the most important application. Int J Mol Sci. 2020;21(17):6225. doi: 10.3390/ijms21176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blockus S, Sake SM, Wetzke M, Grethe C, Graalmann T, Pils M, Le Goffic R, Galloux M, Prochnow H, Rox K, Huttel S, Rupcic Z, Wiegmann B, Dijkman R, Rameix-Welti MA, Eleouet JF, Duprex WP, Thiel V, Hansen G, Bronstrup M, Haid S, Pietschmann T Labyrinthopeptins as virolytic inhibitors of respiratory syncytial virus cell entry. Antiviral Res. 2020;177:104774. doi: 10.1016/j.antiviral.2020.104774. [DOI] [PubMed] [Google Scholar]
- Boos JA, Misun PM, Michlmayr A, Hierlemann A, Frey O Microfluidic multitissue platform for advanced embryotoxicity testing in vitro. Adv Sci (Weinh) 2019;6(13):1900294. doi: 10.1002/advs.201900294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacabelos R, Carrera I, Martinez-Iglesias O, Cacabelos N, Naidoo V (2021) What is the gold standard model for Alzheimer's disease drug discovery and development? Expert Opin Drug Discov 16(12): 1415−1440
- Capula M, Corno C, El Hassouni B, Li Petri G, Arandelovic S, Group EP A brief guide to performing pharmacological studies in vitro: reflections from the EORTC-PAMM course "preclinical and early-phase clinical pharmacology". Anticancer Res. 2019;39(7):3413–3418. doi: 10.21873/anticanres.13485. [DOI] [PubMed] [Google Scholar]
- Caruso G, Musso N, Grasso M, Costantino A, Lazzarino G, Tascedda F, Gulisano M, Lunte SM, Caraci F Microfluidics as a novel tool for biological and toxicological assays in drug discovery processes: focus on microchip electrophoresis. Micromachines (Basel) 2020;11(6):593. doi: 10.3390/mi11060593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavo M, Fato M, Penuela L, Beltrame F, Raiteri R, Scaglione S Microenvironment complexity and matrix stiffness regulate breast cancer cell activity in a 3D in vitro model. Sci Rep. 2016;6:35367. doi: 10.1038/srep35367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YR, Shim J, Park JH, Kim YS, Kim MJ Discovery of orphan olfactory receptor 6M1 as a new anticancer target in MCF-7 cells by a combination of surface plasmon resonance-based and cell-based systems. Sensors (Basel) 2021;21(10):3468. doi: 10.3390/s21103468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra B, Dhingra AK Natural products: a lead for drug discovery and development. Phytother Res. 2021;35(9):4660–4702. doi: 10.1002/ptr.7099. [DOI] [PubMed] [Google Scholar]
- Coliaie P, Kelkar MS, Langston M, Liu C, Nazemifard N, Patience D, Skliar D, Nere NK, Singh MR Advanced continuous-flow microfluidic device for parallel screening of crystal polymorphs, morphology, and kinetics at controlled supersaturation. Lab Chip. 2021;21(12):2333–2342. doi: 10.1039/D1LC00218J. [DOI] [PubMed] [Google Scholar]
- Corallo D, Frabetti S, Candini O, Gregianin E, Dominici M, Fischer H, Aveic S Emerging neuroblastoma 3D in vitro models for pre-clinical assessments. Front Immunol. 2020;11:584214. doi: 10.3389/fimmu.2020.584214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiati S, Kompella UB, Damiati SA, Kodzius R Microfluidic devices for drug delivery systems and drug screening. Genes (Basel) 2018;9(2):103. doi: 10.3390/genes9020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S Learnings from past failures: future routes of antimicrobial drug discovery. Drug Discov Today. 2021;26(9):2105–2107. doi: 10.1016/j.drudis.2021.07.017. [DOI] [PubMed] [Google Scholar]
- Davoudi F, Ghorbanpoor S, Yoda S, Pan X, Crowther GS, Yin X, Murchie E, Hata AN, Willers H, Benes CH Alginate-based 3D cancer cell culture for therapeutic response modeling. STAR Protoc. 2021;2(2):100391. doi: 10.1016/j.xpro.2021.100391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doke SK, Dhawale SC Alternatives to animal testing: a review. Saudi Pharm J. 2015;23(3):223–229. doi: 10.1016/j.jsps.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Lin W, Wu J, Chen T Flavonoids activate pregnane x receptor-mediated CYP3A4 gene expression by inhibiting cyclin-dependent kinases in HepG2 liver carcinoma cells. BMC Biochem. 2010;11:23. doi: 10.1186/1471-2091-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewe J, Cai SX Cell-based apoptosis assays in oncology drug discovery. Expert Opin Drug Discov. 2010;5(6):583–596. doi: 10.1517/17460441.2010.486829. [DOI] [PubMed] [Google Scholar]
- Du G, Fang Q, den Toonder JM Microfluidics for cell-based high throughput screening platforms — a review. Anal Chim Acta. 2016;903:36–50. doi: 10.1016/j.aca.2015.11.023. [DOI] [PubMed] [Google Scholar]
- Fontana F, Marzagalli M, Sommariva M, Gagliano N, Limonta P In vitro 3D cultures to model the tumor microenvironment. Cancers (Basel) 2021;13(12):2970. doi: 10.3390/cancers13122970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster NC, Hall NM, El Haj AJ Two-dimensional and three-dimensional cartilage model platforms for drug evaluation and high-throughput screening assays. Tissue Eng Part B Rev. 2021 doi: 10.1089/ten.teb.2020.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche E, Haarmann-Stemmann T, Kapr J, Galanjuk S, Hartmann J, Mertens PR, Kampfer AAM, Schins RPF, Tigges J, Koch K Stem cells for next level toxicity testing in the 21st century. Small. 2021;17(15):e2006252. doi: 10.1002/smll.202006252. [DOI] [PubMed] [Google Scholar]
- Fursov N, Cong M, Federici M, Platchek M, Haytko P, Tacke R, Livelli T, Zhong Z Improving consistency of cell-based assays by using division-arrested cells. Assay Drug Dev Technol. 2005;3(1):7–15. doi: 10.1089/adt.2005.3.7. [DOI] [PubMed] [Google Scholar]
- Gao Y, Li P, Pappas D A microfluidic localized, multiple cell culture array using vacuum actuated cell seeding: integrated anticancer drug testing. Biomed Microdevices. 2013;15(6):907–915. doi: 10.1007/s10544-013-9779-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GaoW, Liu M, Chen S, Zhang C, Zhao Y Droplet microfluidics with gravity-driven overflow system. Chem Eng J. 2019;362:169–175. doi: 10.1016/j.cej.2019.01.026. [DOI] [Google Scholar]
- Godugu C, Singh M AlgiMatrix-based 3D cell culture system as an in vitro tumor model: an important tool in cancer research. Methods Mol Biol. 2016;1379:117–128. doi: 10.1007/978-1-4939-3191-0_11. [DOI] [PubMed] [Google Scholar]
- Gorshkov K, Pradhan M, Xu M, Yang S, Lee EM, Chen CZ, Shen M, Zheng W Cell-based no-wash fluorescence assays for compound screens using a fluorescence cytometry plate reader. J Pharmacol Exp Ther. 2020;374(3):500–511. doi: 10.1124/jpet.120.265207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N, Liu JR, Patel B, Solomon DE, Vaidya B, Gupta V Microfluidics-based 3D cell culture models: utility in novel drug discovery and delivery research. Bioeng Transl Med. 2016;1(1):63–81. doi: 10.1002/btm2.10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guler MT, Isiksacan Z, Serhatlioglu M, Elbuken C Self-powered disposable prothrombin time measurement device with an integrated effervescent pump. Sensor Actuat B-Chem. 2018;273:350–357. doi: 10.1016/j.snb.2018.06.042. [DOI] [Google Scholar]
- Hajjar D, Kremb S, Sioud S, Emwas AH, Voolstra CR, Ravasi T Anti-cancer agents in Saudi Arabian herbals revealed by automated high-content imaging. PLoS One. 2017;12(6):e0177316. doi: 10.1371/journal.pone.0177316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim AB Do we have a satisfactory cell viability assay? Review of the currently commercially-available assays. Curr Drug Discov Technol. 2020;17(1):2–22. doi: 10.2174/1570163815666180925095433. [DOI] [PubMed] [Google Scholar]
- Hamon M, Jambovane S, Bradley L, Khademhosseini A, Hong JW Cell-based dose responses from open-well microchambers. Anal Chem. 2013;85(10):5249–5254. doi: 10.1021/ac400743w. [DOI] [PubMed] [Google Scholar]
- Hao Z, Huang W, Li Y, Tong H, Liu G, Wang Z Flow modeling and experimental verification of flow resistors used in microfluidic chips driven by capillary force. J Micromech Microeng. 2020;30(11):115015. doi: 10.1088/1361-6439/abb52c. [DOI] [Google Scholar]
- Hattori K, Sugiura S, Kanamori T Pressure-driven microfluidic perfusion culture device for integrated dose-response assays. J Lab Autom. 2013;18(6):437–445. doi: 10.1177/2211068213503155. [DOI] [PubMed] [Google Scholar]
- Hattori N Cerebral organoids model human brain development and microcephaly. Mov Disord. 2014;29(2):185. doi: 10.1002/mds.25740. [DOI] [PubMed] [Google Scholar]
- Henrich CJ, Bokesch HR, Dean M, Bates SE, Robey RW, Goncharova EI, Wilson JA, McMahon JB A high-throughput cell-based assay for inhibitors of ABCG2 activity. J Biomol Screen. 2006;11(2):176–183. doi: 10.1177/1087057105284576. [DOI] [PubMed] [Google Scholar]
- Hong HJ, Koom WS, Koh WG Cell microarray technologies for high-throughput cell-based biosensors. Sensors (Basel) 2017;17(6):1293. doi: 10.3390/s17061293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Wala I, Han H, Nagatani J, Barger T, Civoli F, Kaliyaperumal A, Zhuang Y, Gupta S Comparison of cell-based and non-cell-based assay platforms for the detection of clinically relevant anti-drug neutralizing antibodies for immunogenicity assessment of therapeutic proteins. J Immunol Methods. 2015;419:1–8. doi: 10.1016/j.jim.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Huh S, Lee J, Jung E, Kim SC, Kang JI, Lee J, Kim YW, Sung YK, Kang HK, Park D A cell-based system for screening hair growth-promoting agents. Arch Dermatol Res. 2009;301(5):381–385. doi: 10.1007/s00403-009-0931-0. [DOI] [PubMed] [Google Scholar]
- Heinzman JM, Rice SD, Corkan LA Robotic liquid handlers and semiautomated cell quantification systems increase consistency and reproducibility in high-throughput, cell-based assay. J AssocLab Autom. 2010;15(1):7–14. doi: 10.1016/j.jala.2009.08.010. [DOI] [Google Scholar]
- Jonczyk R, Kurth T, Lavrentieva A, Walter JG, Scheper T, Stahl F Living cell microarrays: an overview of concepts. Microarrays (Basel) 2016;5(2):11. doi: 10.3390/microarrays5020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL Challenges for academic drug discovery. Angew Chem Int Ed Engl. 2012;51(47):11680–11684. doi: 10.1002/anie.201204625. [DOI] [PubMed] [Google Scholar]
- Kase N, Terashima M, Ohta A, Niwa A, Honda-Ozaki F, Kawasaki Y, Nakahata T, Kanazawa N, Saito MK Pluripotent stem cell-based screening identifies CUDC-907 as an effective compound for restoring the in vitro phenotype of Nakajo-Nishimura syndrome. Stem Cells Transl Med. 2021;10(3):455–464. doi: 10.1002/sctm.20-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-K, Kim YS, Kim CS, Lee NK Method for the rapid screening of drug candidates using single-protein tracking in a living cell. Bull Korean Chem Soc. 2021;42(3):393–397. doi: 10.1002/bkcs.12198. [DOI] [Google Scholar]
- Kounde CS, Yeo HQ, Wang QY, Wan KF, Dong H, Karuna R, Dix I, Wagner T, Zou B, Simon O, Bonamy GMC, Yeung BKS, Yokokawa F Discovery of 2-oxopiperazine dengue inhibitors by scaffold morphing of a phenotypic high-throughput screening hit. Bioorg Med Chem Lett. 2017;27(6):1385–1389. doi: 10.1016/j.bmcl.2017.02.005. [DOI] [PubMed] [Google Scholar]
- Kumar A, Chettiar S, Parish T Current challenges in drug discovery for tuberculosis. Expert Opin Drug Discov. 2017;12(1):1–4. doi: 10.1080/17460441.2017.1255604. [DOI] [PubMed] [Google Scholar]
- Kutlehria S, Sachdeva MS Role of in vitro models for development of ophthalmic delivery systems. Crit Rev Ther Drug Carrier Syst. 2021;38(3):1–31. doi: 10.1615/CritRevTherDrugCarrierSyst.2021035222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Corsini NS, Wolfinger S, Gustafson EH, Phillips AW, Burkard TR, Otani T, Livesey FJ, Knoblich JA Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol. 2017;35(7):659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhans SA Using 3D in vitro cell culture models in anti-cancer drug discovery. Expert Opin Drug Discov. 2021;16(8):841–850. doi: 10.1080/17460441.2021.1912731. [DOI] [PubMed] [Google Scholar]
- Lazzari G, Nicolas V, Matsusaki M, Akashi M, Couvreur P, Mura S Multicellular spheroid based on a triple co-culture: a novel 3D model to mimic pancreatic tumor complexity. Acta Biomater. 2018;78:296–307. doi: 10.1016/j.actbio.2018.08.008. [DOI] [PubMed] [Google Scholar]
- Leung M, Kievit FM, Florczyk SJ, Veiseh O, Wu J, Park JO, Zhang M Chitosan-alginate scaffold culture system for hepatocellular carcinoma increases malignancy and drug resistance. Pharm Res. 2010;27(9):1939–1948. doi: 10.1007/s11095-010-0198-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tan W, Xiao W, Carney RP, Men Y, Li Y, Quon G, Ajena Y, Lam KS, Pan T A plug-and-play, drug-on-pillar platform for combination drug screening implemented by microfluidic adaptive printing. Anal Chem. 2018;90(23):13969–13977. doi: 10.1021/acs.analchem.8b03456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wang J, Liu G, Sun H, Bian L, Zhao X, Zheng X Screening bioactive compounds from Ligusticum chuanxiong by high density immobilized human umbilical vein endothelial cells. Anal Bioanal Chem. 2015;407(19):5783–5792. doi: 10.1007/s00216-015-8764-5. [DOI] [PubMed] [Google Scholar]
- Li W, Lam MS, Birkeland A, Riffel A, Montana L, Sullivan ME, Post JM Cell-based assays for profiling activity and safety properties of cancer drugs. J Pharmacol Toxicol Methods. 2006;54(3):313–319. doi: 10.1016/j.vascn.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Li XJ, Valadez AV, Zuo P, Nie Z Microfluidic 3D cell culture: potential application for tissue-based bioassays. Bioanalysis. 2012;4(12):1509–1525. doi: 10.4155/bio.12.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian J, Luo X, Huang X, Wang Y, Xu Z, Ruan X Investigation of microfluidic co-flow effects on step emulsification: interfacial tension and flow velocities. Colloid Surface A. 2019;568:381–390. doi: 10.1016/j.colsurfa.2019.02.040. [DOI] [Google Scholar]
- Lian M, Xu L, Zhu X, Chen X, Yang W, Wang T Seamless signal transduction from three-dimensional cultured cells to a superoxide anions biosensor via in situ self-assembly of dipeptide hydrogel. Anal Chem. 2017;89(23):12843–12849. doi: 10.1021/acs.analchem.7b03371. [DOI] [PubMed] [Google Scholar]
- Liang Y, Pan J, Fang Q Research advances of high-throughput cell-based drug screening systems based on microfluidic technique. Se Pu. 2021;39(6):567–577. doi: 10.3724/SP.J.1123.2020.07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HY, Tuckett AZ, Fennell M, Garippa R, Zakrzewski JL Repurposing of the CDK inhibitor PHA-767491 as a NRF2 inhibitor drug candidate for cancer therapy via redox modulation. Invest New Drugs. 2018a;36(4):590–600. doi: 10.1007/s10637-017-0557-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhang Z, Liu Y, Cui Z, Zhang T, Li Z, Ma W Cancer cells growing on perfused 3D collagen model produced higher reactive oxygen species level and were more resistant to cisplatin compared to the 2D model. J Appl Biomater Funct Mater. 2018b;16(3):144–150. doi: 10.1177/2280800018764763. [DOI] [PubMed] [Google Scholar]
- Liu W, Tao Y, Ge Z, Zhou J, Xu R, Ren Y Pumping of electrolyte with mobile liquid metal droplets driven by continuous electrowetting: a full-scaled simulation study considering surface-coupled electrocapillary two-phase flow. Electrophoresis. 2021;42(7-8):950–966. doi: 10.1002/elps.202000237. [DOI] [PubMed] [Google Scholar]
- Liu X, Hu S, Chen X, Bai X Hollow fiber cell fishing with high-performance liquid chromatography for rapid screening and analysis of an antitumor-active protoberberine alkaloid group from Coptis chinensis. J Pharm Biomed Anal. 2014;98:463–475. doi: 10.1016/j.jpba.2014.06.030. [DOI] [PubMed] [Google Scholar]
- Liu X, Zheng W, Jiang X Cell-based assays on microfluidics for drug screening. ACS Sens. 2019;4(6):1465–1475. doi: 10.1021/acssensors.9b00479. [DOI] [PubMed] [Google Scholar]
- Madden JC, Enoch SJ, Paini A, Cronin MTD A review of in silico tools as alternatives to animal testing: principles, resources and applications. Altern Lab Anim. 2020;48(4):146–172. doi: 10.1177/0261192920965977. [DOI] [PubMed] [Google Scholar]
- Mao C, Kisaalita WS Characterization of 3-D collagen hydrogels for functional cell-based biosensing. Biosens Bioelectron. 2004;19(9):1075–1088. doi: 10.1016/j.bios.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Maruyama J, Inami K, Michishita F, Jiang X, Iwasa H, Nakagawa K, Ishigami-Yuasa M, Kagechika H, Miyamura N, Hirayama J, Nishina H, Nogawa D, Yamamoto K, Hata Y Novel YAP1 activator, identified by transcription-based functional screen, limits multiple myeloma growth. Mol Cancer Res. 2018;16(2):197–211. doi: 10.1158/1541-7786.MCR-17-0382. [DOI] [PubMed] [Google Scholar]
- Michelini E, Cevenini L, Mezzanotte L, Coppa A, Roda A Cell-based assays: fuelling drug discovery. Anal Bioanal Chem. 2010;398(1):227–238. doi: 10.1007/s00216-010-3933-z. [DOI] [PubMed] [Google Scholar]
- Mohiuddin IS, Wei SJ, Yang IH, Martinez GM, Yang S, Cho EJ, Dalby KN, Kang MH Development of cell-based high throughput luminescence assay for drug discovery in inhibiting OCT4/DNA-PKcs and OCT4-MK2 interactions. Biotechnol Bioeng. 2021;118(5):1987–2000. doi: 10.1002/bit.27712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moridani M, Harirforoosh S Drug development and discovery: challenges and opportunities. Drug Discov Today. 2014;19(11):1679–1681. doi: 10.1016/j.drudis.2014.06.003. [DOI] [PubMed] [Google Scholar]
- Mulholland T, McAllister M, Patek S, Flint D, Underwood M, Sim A, Edwards J, Zagnoni M Drug screening of biopsy-derived spheroids using a self-generated microfluidic concentration gradient. Sci Rep. 2018;8(1):14672. doi: 10.1038/s41598-018-33055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näther DU, Fenske R, Hurteaux R, Majno S, Smith SD Time correlated single photon counting — an advancing technique in a plate reader for assay development and high throughput screening. Proceedings of SPIE. 2006;6372(1):637208. doi: 10.1117/12.688419. [DOI] [Google Scholar]
- Olson KR, Eglen RM Beta galactosidase complementation: a cell-based luminescent assay platform for drug discovery. Assay Drug Dev Technol. 2007;5(1):137–144. doi: 10.1089/adt.2006.052. [DOI] [PubMed] [Google Scholar]
- Ona T, Shibata J Advanced dynamic monitoring of cellular status using label-free and non-invasive cell-based sensing technology for the prediction of anticancer drug efficacy. Anal Bioanal Chem. 2010;398(6):2505–2533. doi: 10.1007/s00216-010-4223-5. [DOI] [PubMed] [Google Scholar]
- Ozsoylu D, Isik T, Demir MM, Schoning MJ, Wagner T Cryopreservation of a cell-based biosensor chip modified with elastic polymer fibers enabling ready-to-use on-site applications. Biosens Bioelectron. 2021;177:112983. doi: 10.1016/j.bios.2021.112983. [DOI] [PubMed] [Google Scholar]
- Pan Y, Hu N, Wei X, Gong L, Zhang B, Wan H, Wang P 3D cell-based biosensor for cell viability and drug assessment by 3D electric cell/matrigel-substrate impedance sensing. Biosens Bioelectron. 2019;130:344–351. doi: 10.1016/j.bios.2018.09.046. [DOI] [PubMed] [Google Scholar]
- Park J, Han DH, Park JK Towards practical sample preparation in point-of-care testing: user-friendly microfluidic devices. Lab Chip. 2020;20(7):1191–1203. doi: 10.1039/D0LC00047G. [DOI] [PubMed] [Google Scholar]
- Parvathaneni V, Kulkarni NS, Muth A, Gupta V Drug repurposing: a promising tool to accelerate the drug discovery process. Drug Discov Today. 2019;24(10):2076–2085. doi: 10.1016/j.drudis.2019.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathe-Neuschafer-Rube A, Neuschafer-Rube F, Puschel GP Cell-based reporter release assay to determine the activity of calcium-dependent neurotoxins and neuroactive pharmaceuticals. Toxins (Basel) 2021;13(4):247. doi: 10.3390/toxins13040247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones GA, Moore TI, Nicholes K, Lee H, Kim S, Sun L, Jeon NL, Stephan JP Application of a new wall-less plate technology to complex multistep cell-based investigations using suspension cells. Blood. 2013;121(7):e25–e33. doi: 10.1182/blood-2012-07-446294. [DOI] [PubMed] [Google Scholar]
- Radnai L, Stremel RF, Vaissiere T, Lin L, Cameron M, Martin WH, Rumbaugh G, Kamenecka TM, Griffin PR, Miller CA A simple and robust cell-based assay for the discovery of novel cytokinesis inhibitors. J Biol Methods. 2020;7(3):e136. doi: 10.14440/jbm.2020.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajalingham K Cell-based assays in high-throughput mode (HTS) BioTechnologia. 2016;97(3):227–234. [Google Scholar]
- Rimann M, Graf-Hausner U Synthetic 3D multicellular systems for drug development. Curr Opin Biotechnol. 2012;23(5):803–809. doi: 10.1016/j.copbio.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Riss T Selecting cell-based assays for drug discovery screening. Cell Notes. 2005;13:16–21. [Google Scholar]
- Roy A Challenges with risk mitigation in academic drug discovery: finding the best solution. Expert Opin Drug Discov. 2019;14(2):95–100. doi: 10.1080/17460441.2019.1553952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanookpan K, Nonpanya N, Sritularak B, Chanvorachote P Ovalitenone inhibits the migration of lung cancer cells via the suppression of AKT/mTOR and epithelial-to-mesenchymal transition. Molecules. 2021;26(3):638. doi: 10.3390/molecules26030638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger A, Abagyan R, Carlson HA, Dang KK, Guinney J, Cagan RL Multi-targeting drug community challenge. Cell Chem Biol. 2017;24(12):1434–1435. doi: 10.1016/j.chembiol.2017.12.006. [DOI] [PubMed] [Google Scholar]
- Seah YFS, Hu H, Merten CA Microfluidic single-cell technology in immunology and antibody screening. Mol Aspects Med. 2018;59:47–61. doi: 10.1016/j.mam.2017.09.004. [DOI] [PubMed] [Google Scholar]
- Sekiguchi H, Washida K, Murakami A Suppressive effects of selected food phytochemicals on CD74 expression in NCI-N87 gastric carcinoma cells. J Clin Biochem Nutr. 2008;43(2):109–117. doi: 10.3164/jcbn.2008054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova G, Stepanova DS, Deyev SM, Chernoff J Medium throughput biochemical compound screening identifies novel agents for pharmacotherapy of neurofibromatosis type 1. Biochimie. 2017;135:1–5. doi: 10.1016/j.biochi.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y, Ryu K, Park J, Jeon DK, Jo S, Lee HK, Namkung W Inhibition of ANO1 by luteolin and its cytotoxicity in human prostate cancer PC-3 cells. PLoS One. 2017;12(3):e0174935. doi: 10.1371/journal.pone.0174935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamah SM, Cunningham BT Label-free cell-based assays using photonic crystal optical biosensors. Analyst. 2011;136(6):1090–1102. doi: 10.1039/c0an00899k. [DOI] [PubMed] [Google Scholar]
- Shembekar N, Chaipan C, Utharala R, Merten CA Droplet-based microfluidics in drug discovery, transcriptomics and high-throughput molecular genetics. Lab Chip. 2016;16(8):1314–1331. doi: 10.1039/C6LC00249H. [DOI] [PubMed] [Google Scholar]
- Shi D, Mi G, Wang M, Webster TJ In vitro and ex vivo systems at the forefront of infection modeling and drug discovery. Biomaterials. 2019;198:228–249. doi: 10.1016/j.biomaterials.2018.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soman G, Yang X, Jiang H, Giardina S, Mitra G Comparison of GD2 binding capture ELISA assays for anti-GD2-antibodies using GD2-coated plates and a GD2-expressing cell-based ELISA. J Immunol Methods. 2011;373(1-2):181–191. doi: 10.1016/j.jim.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo M, Svensson Akusjarvi S, Saxena A, Liu J, Chandrasekar G, Kitambi SS Cell and small animal models for phenotypic drug discovery. Drug Des Devel Ther. 2017;11:1957–1967. doi: 10.2147/DDDT.S129447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai ZF, Zhang GL, Wang F Identification of small molecule activators of the janus kinase/signal transducer and activator of transcription pathway using a cell-based screen. Biol Pharm Bull. 2012;35(1):65–71. doi: 10.1248/bpb.35.65. [DOI] [PubMed] [Google Scholar]
- Thippabhotla S, Zhong C, He M 3D cell culture stimulates the secretion of in vivo like extracellular vesicles. Sci Rep. 2019;9(1):13012. doi: 10.1038/s41598-019-49671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas N (2011) Microfabricated apparatus for cell based assays. United States, US7935522B2
- Tu C, Cunningham NJ, Zhang M, Wu JC Human induced pluripotent stem cells as a screening platform for drug-induced vascular toxicity. Front Pharmacol. 2021;12:613837. doi: 10.3389/fphar.2021.613837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Tamura T, Hamachi I Development of a cell-based ligand-screening system for identifying Hsp90 inhibitors. Biochemistry. 2020;59(2):179–182. doi: 10.1021/acs.biochem.9b00781. [DOI] [PubMed] [Google Scholar]
- Vicenti I, Dragoni F, Giannini A, Giammarino F, Spinicci M, Saladini F, Boccuto A, Zazzi M Development of a cell-based immunodetection assay for simultaneous screening of antiviral compounds inhibiting Zika and Dengue virus replication. SLAS Discov. 2020;25(5):506–514. doi: 10.1177/2472555220911456. [DOI] [PubMed] [Google Scholar]
- Wahome PG, Bai Y, Neal LM, Robertus JD, Mantis NJ Identification of small-molecule inhibitors of ricin and shiga toxin using a cell-based high-throughput screen. Toxicon. 2010;56(3):313–323. doi: 10.1016/j.toxicon.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhao Q, Liu J, Wang Z, Kong D Identification of human lactate dehydrogenase A inhibitors with anti-osteosarcoma activity through cell-based phenotypic screening. Bioorg Med Chem Lett. 2020a;30(4):126909. doi: 10.1016/j.bmcl.2019.126909. [DOI] [PubMed] [Google Scholar]
- Wang L Drug discovery in China: challenges and opportunities. Natl Sci Rev. 2018;5(5):768–773. doi: 10.1093/nsr/nwy085. [DOI] [Google Scholar]
- Wang Y, Chen Z, Bian F, Shang L, Zhu K, Zhao Y Advances of droplet-based microfluidics in drug discovery. Expert Opin Drug Discov. 2020b;15(8):969–979. doi: 10.1080/17460441.2020.1758663. [DOI] [PubMed] [Google Scholar]
- Wang Y, Fang S, Wu Y, Cheng X, Zhang LK, Shen XR, Li SQ, Xu JR, Shang WJ, Gao ZB, Xia BQ (2021) Discovery of SARS-CoV-2-E channel inhibitors as antiviral candidates. Acta Pharmacol Sin. https://doi.org/10.1038/s41401-021-00732-2
- Wegener J Cell-based microarrays for in vitro toxicology. Annu Rev Anal Chem (Palo Alto Calif) 2015;8:335–358. doi: 10.1146/annurev-anchem-071213-020051. [DOI] [PubMed] [Google Scholar]
- Wei F, Zhang X, Cui P, Gou X, Wang S Cell-based 3D bionic screening by mimicking the drug-receptor interaction environment in vivo. J Mater Chem B. 2021;9(3):683–693. doi: 10.1039/D0TB02661A. [DOI] [PubMed] [Google Scholar]
- Wollrab V, Caballero D, Thiagarajan R, Riveline D (2016) Ordering single cells and single embryos in 3D confinement: a new device for high content screening. J Vis Exp (115): 51880. https://doi.org/10.3791/51880
- Xi B, Yu N, Wang X, Xu X, Abassi YA The application of cell-based label-free technology in drug discovery. Biotechnol J. 2008;3(4):484–495. doi: 10.1002/biot.200800020. [DOI] [PubMed] [Google Scholar]
- Xu Y, Liu R, Leu NA, Zhang L, Ibragmova I, Schultz DC, Wang PJ A cell-based high-content screen identifies isocotoin as a small molecule inhibitor of the meiosis-specific MEIOB-SPATA22 complexdagger. Biol Reprod. 2020;103(2):333–342. doi: 10.1093/biolre/ioaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Shrestha N, Preat V, Beloqui A An overview of in vitro, ex vivo and in vivo models for studying the transport of drugs across intestinal barriers. Adv Drug Deliv Rev. 2021;175:113795. doi: 10.1016/j.addr.2021.05.005. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhao X A 3D model of ovarian cancer cell lines on peptide nanofiber scaffold to explore the cell-scaffold interaction and chemotherapeutic resistance of anticancer drugs. Int J Nanomedicine. 2011;6:303–310. doi: 10.2217/nnm.10.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeon Park J, Young Kim H, Shibamoto T, Su Jang T, Cheon Lee S, Suk Shim J, Hahm DH, Lee HJ, Lee S, Sung Kang K Beneficial effects of a medicinal herb, Cirsium japonicum var. maackii, extract and its major component, cirsimaritin on breast cancer metastasis in MDA-MB-231 breast cancer cells. Bioorg Med Chem Lett. 2017;27(17):3968–3973. doi: 10.1016/j.bmcl.2017.07.070. [DOI] [PubMed] [Google Scholar]
- Yin H, Marshall D Microfluidics for single cell analysis. Curr Opin Biotechnol. 2012;23(1):110–119. doi: 10.1016/j.copbio.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Yuan T, Werman JM, Sampson NS The pursuit of mechanism of action: uncovering drug complexity in TB drug discovery. RSC Chem Biol. 2021;2(2):423–440. doi: 10.1039/D0CB00226G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste I, Luciano FC, Gonzalez-Burgos E, Lalatsa A, Serrano DR Mimicking bone microenvironment: 2D and 3D in vitro models of human osteoblasts. Pharmacol Res. 2021;169:105626. doi: 10.1016/j.phrs.2021.105626. [DOI] [PubMed] [Google Scholar]
- Zahra R, Furqan M, Ullah R, Mithani A, Saleem RSZ, Faisal A A cell-based high-throughput screen identifies inhibitors that overcome P-glycoprotein (Pgp)-mediated multidrug resistance. PLoS One. 2020;15(6):e0233993. doi: 10.1371/journal.pone.0233993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman GJ, de Roos JA, Blomenrohr M, van Koppen CJ, Oosterom J Cryopreserved cells facilitate cell-based drug discovery. Drug Discov Today. 2007;12(13-14):521–526. doi: 10.1016/j.drudis.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Zhang P, Zhang J, Bian S, Chen Z, Hu Y, Hu R, Li J, Cheng Y, Zhang X, Zhou Y, Chen X, Liu P High-throughput superhydrophobic microwell arrays for investigating multifactorial stem cell niches. Lab Chip. 2016;16(16):2996–3006. doi: 10.1039/C6LC00331A. [DOI] [PubMed] [Google Scholar]
- Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19(12):1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- Zhang WY, Yang XN, Jin DZ, Zhu XZ Expression and enzyme activity determination of human cyclooxygenase-1 and -2 in a baculovirus-insect cell system. Acta Pharmacol Sin. 2004;25(8):1000–1006. [PubMed] [Google Scholar]
- Zhang Z, Yang E, Hu C, Cheng H, Chen CY, Huang D, Wang R, Zhao Y, Rong L, Vignuzzi M, Shen H, Shen L, Chen ZW Cell-based high-throughput screening assay identifies 2',2'-Difluoro-2'-deoxycytidine gemcitabine as a potential antipoliovirus agent. ACS Infect Dis. 2017;3(1):45–53. doi: 10.1021/acsinfecdis.6b00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XL, Chen JJ, Zhang GN, Wang YC, Si SY, Chen LF, Wang Z Small molecule T63 suppresses osteoporosis by modulating osteoblast differentiation via BMP and WNT signaling pathways. Sci Rep. 2017;7(1):10397. doi: 10.1038/s41598-017-10929-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Huang G, Wang S, Wu J, Lee WG, Chen Y, Xu F, Lu T Advances in cell-based biosensors using three-dimensional cell-encapsulating hydrogels. Biotechnol J. 2011;6(12):1466–1476. doi: 10.1002/biot.201100098. [DOI] [PubMed] [Google Scholar]
- Zhu X, Yi Chu L, Chueh BH, Shen M, Hazarika B, Phadke N, Takayama S Arrays of horizontally-oriented mini-reservoirs generate steady microfluidic flows for continuous perfusion cell culture and gradient generation. Analyst. 2004;129(11):1026–1031. doi: 10.1039/b407623k. [DOI] [PubMed] [Google Scholar]