Abstract
Duchenne muscular dystrophy (DMD) is a debilitating genetic disorder that results in progressive muscle degeneration and premature death. DMD is caused by mutations in the gene encoding dystrophin protein, a membrane-associated protein required for maintenance of muscle structure and function. Although the genetic mutations causing the disease are well known, no curative therapies have been developed to date. The advent of genome-editing technologies provides new opportunities to correct the underlying mutations responsible for DMD. These mutations have been successfully corrected in human cells, mice, and large animal models through different strategies based on CRISPR-Cas9 gene editing. Ideally, CRISPR-editing could offer a one-time treatment for DMD by correcting the genetic mutations and enabling normal expression of the repaired gene. However, numerous challenges remain to be addressed, including optimization of gene editing, delivery of gene-editing components to all the muscles of the body, and the suppression of possible immune responses to the CRISPR-editing therapy. This review provides an overview of the recent advances toward CRISPR-editing therapy for DMD and discusses the opportunities and the remaining challenges in the path to clinical translation.
Keywords: Duchenne muscular dystrophy, CRISPR-Cas9, gene editing, AAV vectors
INTRODUCTION
Duchenne muscular dystrophy (DMD) is a X-linked recessive disease that affects 1 in 5,000 male births.1 DMD patients exhibit progressive muscle degeneration that results in muscle weakness and loss of ambulation, and despite advances in cardiac and respiratory care, almost all DMD patients succumb to premature death before the age of 30 years.2
It has been known for over 35 years that mutations in the DMD gene cause the disease.3 The DMD gene is located on the X chromosome, and it is one of the largest genes of the genome. It consists of 79 exons that encode the dystrophin protein that is located beneath the sarcolemma and connects the cytoskeleton of muscle fibers with the extracellular matrix.4 Dystrophin functions as a shock absorber, reducing the mechanical stress induced by muscle contraction, thereby maintaining sarcolemmal integrity and supporting muscle function.5
Due to the extensive length of the DMD gene, more than 7,000 mutations causing DMD have been identified.6 Approximately 70% of DMD patients harbor a large deletion ( = or >1 exon) in the DMD gene, and the other 30% of mutations include duplications, point mutations, or small insertions or deletions (INDELs). Most of the exon deletion or duplication mutations cluster into two hotspot regions spanning between exons 6 and 7, and exons 43 and 53, whereas other small mutations occur randomly through the DMD gene.6 Mutations in the DMD gene generally result in a shift in the open reading frame (ORF), rending the DMD gene out-of-frame, and ultimately introducing a premature stop codon that leads to the absence of functional dystrophin protein in skeletal muscle myofibers and in cardiomyocytes.
While the amino- (N) and carboxyl- (C) termini of the protein are essential regions, deletions in the rod-like domains of the central region that preserve the correct dystrophin ORF are tolerated and result in a Becker muscular dystrophy (BMD) phenotype that can be a relatively benign muscular dystrophy compared to DMD.7
Unfortunately, at present, there is no curative therapy for this lethal disease. Corticosteroid treatments are available for the mitigation of the secondary symptoms of DMD, such as inflammation, fibrosis, impaired angiogenesis, altered calcium homeostasis, and mitochondrial dysfunction.8,9 However, long-term use of corticosteroids causes many adverse effects, despite minimal amelioration of the DMD phenotype. An efficient strategy to treat DMD could be to express a semifunctional dystrophin to mimic the BMD-like phenotype. Truncated dystrophin lacking exons encoding the central rod domains can fit the limited packaging capacity of adeno-associated virus (AAV) for systemic delivery, and this led to the development of a wide range of mini- and microdystrophin variations for therapeutic use.10
Currently, the utilization of these truncated dystrophins is under evaluation in several clinical trials.11 Other pharmacological approaches focus on the restoration of the disrupted DMD ORF using antisense technology. Eteplirsen is the first drug specific for DMD approved by the U.S. Food and Drug Administration (FDA). It is an antisense oligonucleotide that induces the skipping of exon 51 of the DMD gene resulting in the expression of a semifunctional dystrophin and BMD-like mild symptoms in a small cohort of DMD patients.12 Two other oligonucleotides (golodirsen and viltolarsen) are under evaluation for treating DMD patients that could be therapeutically beneficial by promoting skipping of exon 53.13,14 However, all these approaches have shown only minimal clinical benefit, and none removes the underlying genetic cause of the disease to enable lifelong expression of dystrophin.
GENE-EDITING THERAPEUTIC STRATEGIES
Gene editing with CRISPR-Cas9 has the potential of permanently correcting mutations causing DMD and ameliorating the pathology of the disease. The CRISPR-Cas9 system was originally discovered in bacteria as an adaptive defense system against viruses and has been engineered for the editing of the genome of eukaryotic cells.15–17 The Cas9 endonuclease can be directed to a specific DNA sequence of the genome, using a short guide RNA (sgRNA). The target sequence is specified by the complementary of a region of 20 nucleotides of the sgRNA called the protospacer. The most studied and used Cas9 is derived from the CRISPR system of Streptococcus pyogenes (SpCas9). The SpCas9 endonuclease recognizes a protospacer adjacent motif (PAM) in DNA consisting of the sequence 5′-NGG-3′ and cuts the target DNA 3 base pairs (bp) upstream of the PAM, generating a DNA double-stranded break (DSB).
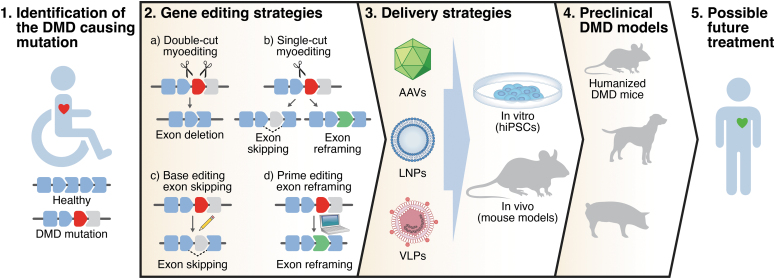
The actual gene editing can occur in two ways: (1) in the presence of a DNA donor template, homology-direct repair (HDR) can introduce the desired modification in the target locus. However, the HDR mechanism is only active in proliferative cells, with limited efficiency in postmitotic cells, such as myofibers and cardiomyocytes; (2) alternatively, nonhomologous end joining (NHEJ) works in both quiescent and proliferating cells and can imprecisely repair the DNA generating short INDELs at the site of the DNA DSB. In recent years, CRISPR-Cas9 has shown great potential for gene editing of mutations causing DMD. We use the term “myoediting” to refer to CRISPR-mediated gene editing in muscle to permanently correct genetic mutations causing DMD and restore muscle function, and here, we describe different myoediting strategies developed for treatment of DMD (Fig. 1, “Gene-editing strategies”).18–20
Figure 1.
Overview of strategy to correct DMD by myoediting. DMD mutations are identified in DMD patients. Different gene-editing strategies and delivery systems are tested in vitro (in human hiPSCs from patients) and in vivo (in DMD mouse models) to assess myoediting by restoration of dystrophin. Efficacy and safety of myoediting is tested in the appropriate preclinical models with the goal of proposing a therapeutic approach for DMD patients. DMD, Duchenne muscular dystrophy; hiPSCs, human induced pluripotent stem cells.
Double-cut myoediting
Deletion of one or more exons can be used to restore the ORF of DMD. In the double-cut strategy, removal of a target exon(s) is accomplished using two sgRNAs to simultaneously target the introns flanking the exon(s) to be deleted. This approach was extensively applied to restore the DMD ORF in human induced pluripotent stem cells (hiPSCs) from patients and human myoblasts harboring different mutations.21–28 Exon deletion is especially suitable to correct exon duplication mutations, as it can restore full-length dystrophin.25,29–31 Moreover, as a general approach, double-cut myoediting can excise a multiexonic genomic region, deleting the mutational hotspot of the central rod domain (exons 45–55) and producing a truncated but functional form of dystrophin.21,32
However, the deletion of numerous exons can impact dystrophin function, and when two sgRNAs are used, exon deletions can generate diverse and unpredictable genome modifications, including exogenous DNA integration or aberrant splicing at the DSB cut sites.33 Another weakness of the double-cut strategy is the low editing efficiency, due to the necessity of simultaneous cutting using two sgRNAs and the subsequent joining of distant free DNA ends. Two sgRNAs also increases the probability of additional off-target effects.33
Single-cut myoediting
Limitations of the double-cut myoediting approach can be partially overcome by single-cut gene editing. In this strategy, only one sgRNA generates a single DSB in the proximity of the splice site of the target exon. Repair of the DSB through the NHEJ pathway generates INDELs in the target locus and reconstitution of the correct ORF can occur: (1) by exon skipping if the INDELs destroy the splice consensus site of the exon to be skipped, or (2) by exon reframing if the appropriate number of INDELs occurs in the exonic region. Based on the Leiden DMD mutation database, almost 80% of the DMD patients could benefit from exon skipping, so designing a relatively small number of optimized sgRNA allows efficient correction of most DMD mutations.34
Indeed, several studies demonstrated high editing efficiency using the single-cut myoediting strategy to restore dystrophin expression in muscle cells derived from hiPSC of DMD patients.25,35–39 Another advantage of the single-cut approach is that it decreases the likelihood of off-target mutations since only one sgRNA is used. However, this strategy relies on the endonuclease activity of the Cas9, and generation of DNA DSBs has been shown to be deleterious to cells and can lead to integration events in the genome, if AAV is used as a delivery vector.33,40–43
Base editing myoediting
Base editing allows the permanent and precise modification of the genome, without generating a DNA DSB.44 At the moment, two major classes of base editors are available: cytosine base editors that convert DNA C•G base pairs to T•A base pairs, and adenine base editors (ABEs) that convert DNA A•T base pairs to G•C base pairs.45,46 Base editors can be used to correct not only point mutations in the DMD gene, but also to induce beneficial exon skipping via an approach termed “single-swap” editing of splice sites.47–49 Importantly, while moderate bystander editing is an inherent downside of base editing, this is negated in the single-swap approach as the bystander edits occur in the intron or to-be-skipped exon and not in the mature transcript.
However, as discussed further below, a current major weakness of base editors is their large size that impinges on the limited cargo capacity of the AAV vectors commonly used as delivery systems for myoediting strategies.
Prime editing reframing myoediting
Another nucleotide editing technology, prime editing, can permanently modify the genome without cutting DNA.50 It consists of a Cas9 nickase fused with an engineered reverse transcriptase. This, in combination with a prime editing guide RNA, can perform targeted and precise small insertions, deletions, or base changing. This technology has been used to correct point mutations in myoblasts of DMD patients.51,52 In a more general approach, prime editing was recently used to precisely insert the correct number of nucleotides to reframe the DMD ORF in human cardiomyocytes harboring an exon deletion mutation in the DMD gene and restoring dystrophin expression.48 However, despite the great potential of prime editing to correct DMD causing mutations, up to now, there is no report of its applicability in vivo.
IN VIVO DYSTROPHIN RESTORATION BY GENE EDITING
To test the therapeutic potential of DMD gene editing in vivo, several DMD mouse models representing the most commonly deleted exons in DMD patients (including deletion of exon 43, 44, 45, 50, 51, or 52) were generated by CRISPR technology.36,37,48,53 These animal models permitted validation of the myoediting strategies initially described in quiescent cells. Plus, they provided an estimate of the in vivo recovery of dystrophin, as well as, evaluation of the safety of gene editing, and optimization of delivery of the gene-editing components.
Inarguably, one of the major challenges of postnatal myoediting is the efficient delivery of the CRISPR editor and the sgRNA to the target tissues: the skeletal and cardiac muscles. Both viral and nonviral delivery systems have been developed for this aim (Fig. 1, “Delivery strategies”). Among the viral systems, AAV is the most commonly used vector for myoediting as it has the following features: (1) low pathogenicity, (2) low immunogenicity, and (3) several serotypes that show tropism for skeletal and cardiac muscles (serotypes1, 6, 8, 9, rh10, rh74).54 However, AAVs have a packaging cargo size limitation (<4.7 kb) close to the SpCas9 cDNA size (∼4.2 kb). Several groups used a dual-AAV system (one encoding the Cas, the other the sgRNAs), or a single-AAV system with a smaller Cas9 (i.e., Staphylococcus aureus Cas9, SaCas9, or Campylobacter jejuni Cas9, CjCas9) to restore dystrophin expression in different DMD mouse models by double- or single-cut myoediting strategies.36–39,53,55–62
Importantly, AAV delivery permits the utilization of specific promoters (e.g., muscle creatine kinase promoter) to direct the expression of Cas9 only in muscle cells, adding an additional level of cell-specificity editing in addition to viral tropism.53 One major disadvantage of AAV delivery is the high dosage of the AAV vector(s) necessary for efficient gene myoediting. As an optimization strategy to reduce viral dose, utilization of a self-complementary AAV has been used for expression of the sgRNAs, providing a 20-fold reduction of dose, compared to single-stranded AAV, to achieve efficient gene editing.63,64
Despite their large size, base editors have been used effectively to restore dystrophin expression in different DMD mouse models.48,65–67 To correct nonsense point mutations, ABEs were split into two trans-splicing AAV vectors to overcome the packaging limitation of AAV and then delivered into muscle, or systemically, demonstrating the therapeutic potential of base editing in adult DMD animal models.65,67 The same packaging strategy was used to demonstrate the efficient recovery of dystrophin in vivo by single-swap base editing of splice sites.48 In another study, the utilization of the small SaCas9 permitted packaging of a functional base editor in a single AAV and sgRNAs in another AAV (similarly described for the dual-AAV system for single- and double-cut myoediting strategies).66 However, the development and utilization of an efficient single AAV delivery solution for expression of both base editor and sgRNA (“all-in-one”) is desirable for meaningful and consequential preclinical studies.
As a nonviral delivery system, lipid nanoparticles (LNPs) were used to deliver in vivo gene-editing components to muscle cells to correct DMD causing mutations. Ribonucleoprotein complexes of CRISPR editing components for single-cut editing were encapsulated into LNPs and delivered to skeletal muscle, showing effective, although less efficient, gene editing to restore dystrophin expression.68 Importantly, it was demonstrated that LNP delivery shows low immunogenicity, allowing repeated local administration into skeletal muscles to induce stable genomic exon skipping and restore dystrophin protein in DMD mice.69 However, systemic gene-editing correction in skeletal muscles and heart of DMD mice using LNPs has not yet been demonstrated.
FUTURE CHALLENGES
CRISPR gene-editing technologies provide powerful tools for treating genomic mutations of DMD patients that cause the disease, theoretically enabling permanent restoration of dystrophin expression and function of muscle cells. Unlike replacement gene therapy approaches, the gene myoediting strategies enable endogenous regulation of dystrophin, thereby expressing appropriate amounts of protein in the correct tissues. Although myoediting strategies have been successfully applied in human iPSCs and animal models, there are several key obstacles that need to be overcome before considering clinical application. These issues include the following: (1) developing new editing strategies to improve efficiency and reduce dose; (2) affirming safety features of the gene-editing components; and (3) providing testing of myoediting in meaningful preclinical models.
New gene-editing strategies
With the exception of correcting point or duplication DMD mutations, all gene-editing strategies attempt to restore a functional, although truncated, form of dystrophin. These myoediting approaches result in converting a DMD lethal phenotype to a milder Becker phenotype of muscle cells. Fortuitously, in vitro studies report similar protein stability between full-length dystrophin and all in-frame exon-skipped isoforms in the mutational hotspot regions encoding the central rod domain.70 However, when designing new exon skipping strategies for DMD, the functionality of the resulting Becker-type isoform must be taken into consideration.
Other gene-editing strategies for DMD target overexpression of utrophin, a paralog of dystrophin that can compensate for its functional deficiency.71,72 In a recent study, a double-cut gene-editing strategy was applied to delete several microRNA binding sites of utrophin in DMD stem cells, thereby inducing overexpression of utrophin to compensate dystrophin deficiency.73 The main advantage of this approach is that it is applicable to all DMD mutations. However, the level of utrophin overexpression has not been measured in vivo, and due to sequence differences between dystrophin and utrophin, this strategy may lead to a mild Becker phenotype.74
In the future, gene-editing strategies that can efficiently and effectively restore full-length dystrophin are desirable. Homology-independent targeted integration (HITI) and single homology arm donor mediated intron-targeting integration (SATI) strategies were developed to perform robust DNA knock-in, in proliferating and quiescent cells both for in vitro and in vivo applications.75,76 HITI technology was shown to edit the genome and restore full-length dystrophin protein in a DMD mouse model with an exon deletion mutation.77 However, the efficiency was low, and minimal total dystrophin recovery was reported.77
In addition, several groups have developed new tools and approaches for integration of large DNA fragments into the genome without introducing DNA DSBs, such as CRISPR-associated transposase, insert transposable elements by guide RNA-assisted targeting, twin prime editor, and programmable addition via site-specific targeting elements.78–81 Future developments of these technologies to improve their gene-editing efficiency and efficacy in vivo could lead to new potential therapies that restore the full-length dystrophin protein.
Safety
Up to now, the biggest obstacle of translating gene-editing therapies for DMD to the clinic is efficient and safe delivery of the gene-editing components to muscles. AAV has provided the most effective means of systemic delivery of gene-editing components; however, it is crucial to further examine the safety features of this viral vector for application of CRISPR for clinical application for DMD patients. In fact, a high dose of AAV is required for editing all the muscles of the human body. However, high dose of AAV can trigger adverse events in multiple organs, such as liver toxicity.82 To lower viral dose, new AAV capsid variants with enhanced muscle tropism, such as AAVMYO and MyoAAV, are being developed. These AAV variants have the potential to lower the viral dose by at least 10-fold, offering a safer therapeutic dose.83–85
Moreover, as previously described, the dual-AAV system, which is the most popular method for in vivo delivery of the gene-editing components, necessitates using a high dose of AAV for efficient gene editing.36,37,39,53,55–60,62 The development of new “all-in-one” AAV vectors that combine small Cas9 variants and sgRNAs in the same vector would not only reduce the amount of viral dose but would also address the problem related to manufacturing different clinical-grade AAV vectors in high quantity.38,61 However, several studies with the dual-AAV system have demonstrated that efficient systemic editing requires higher amounts of the AAV encoding the sgRNAs, and this ratio difference can only be achieved with the dual-AAV approach.36,86,87
Besides AAV vectors, virus-like particles (VLPs) have recently been created to deliver mRNA of gene-editing components to correct mutations causing DMD in vivo.88 The possibility of modifying the viral envelope to target specific cell types will allow further development of new delivery strategies based on VLPs.89,90 However, due to the limited publications using this new delivery strategy, more studies are necessary to assess its safety.
Concerns over the immune response of humans to CRISPR gene-editing therapy components remain a safety issue. An immune response can be evoked not only by the AAV vectors but also by the Cas proteins. Cas9 is a bacterial protein and the presence of Cas9-specific antibodies has been detected in human plasma.91 It was also reported that sgRNA may trigger an innate immune response within human cells in vitro, but it has not been demonstrated whether sgRNAs induce an immune response in vivo.92,93 Furthermore, more studies are needed to assess specific post-transcriptional modifications of sgRNAs that were administered systemically to determine if these modifications could induce an immune response in vivo.93 Importantly, an immune response against AAV and Cas9 has not been detected in neonatal mice, suggesting that an immune response may be avoided by treating humans at early ages.33
Another safety concern of gene-editing therapy is the potential of off-target activity. Different bioinformatic investigative methods have been developed to detect off-target effects, such as Digenome-seq, GUIDE-seq, and CIRCLE-seq.94–96 Although off-target activity has been observed with gene editing in proliferating cells in tissue culture, it is documented to be minimal in animal models by in vivo studies, especially in postmitotic muscle cells.97 Nonetheless, off-target activity by CRISPR gene editing can be further reduced using high fidelity Cas9 or by optimization of the sgRNA.98–105
Preclinical animal models
Generating and using appropriate preclinical DMD animal models will allow optimization of gene-editing strategies by assessing delivery of the gene-editing components, and evaluating the outcome of DMD myoediting in vivo (Fig. 1, “Preclinical DMD models”).
Mouse and human dystrophin proteins are highly conserved in exon composition and amino acid sequence. However, the mouse and human dystrophin genes vary at the genomic level and the differences in nucleotide sequence impede testing of human-specific sgRNAs in DMD mouse models. Creation of humanized DMD mouse models allows for preclinical trials to evaluate the efficiency of genome editing of human-specific sgRNAs in an animal model for eventual use of the sgRNA in clinical trials. A humanized mouse was generated, in which the entire human DMD sequence was integrated into mouse chromosome 5 and subsequently mutations were introduced to recapitulate the dystrophic phenotype.60,106,107 However, a recent study revealed that this humanized DMD mouse model carries two copies of the human DMD transgene at the integration locus.108
A new generation of humanized DMD mouse models were generated in which mouse exon(s) were replaced by human ortholog(s) within the endogenous genomic location on the X chromosome.39,69,109 These chimeric mouse models containing human DMD genome sequence allow testing of gene-editing efficiency of human-specific sgRNAs for single-cut or base editing correction strategies.39,69,109 In the future, it would be useful for preclinical studies to establish additional humanized DMD mouse models by substituting all the therapeutically relevant exons with human sequence.
DMD mouse models are helpful tools to test gene-editing strategies as they mimic the pathological features of the diseases. However, due to the compensatory effects of utrophin and muscle regeneration, they do not fully recapitulate the phenotype of DMD patients.110 For this reason, therapeutic (and also delivery) strategies need to be optimized in larger animals and more pathologically severe DMD mammalian models, before clinical application.111 Although one study suggested that Cas9 immunity was induced by AAV delivery vectors in large mammals, encouragingly two other large animal applications of gene editing, one in a DMD dog model and another in an induced DMD pig model, efficiently restored dystrophin protein after systemic administration of AAV9, validating therapeutic gene editing in large animals.27,112,113
Another important variable to consider before moving DMD gene myoediting into the clinic is the timing of gene-editing correction. All in vivo DMD editing studies that show efficient dystrophin restoration have been performed by administering the editing components to young animals, suggesting that early intervention is effective and most likely better than delivery of AAV CRISPR to older animals. In fact, administration of AAV CRISPR to younger patients is preferable because (1) a lower amount of AAV CRISPR is needed for younger animals due to the smaller size; (2) young patients are less likely to have pre-existing immunity against AAV9; and (3) younger muscles are better preserved, showing minimal pathological features of dystrophic muscles. Following administration, the durability and longevity of the gene-editing correction has to be monitored.
Long-term studies after double-cut gene editing showed skeletal muscle editing at 12 or 18 months after editing.33,86 Recently, a study using single-cut myoediting reported lifelong expression of dystrophin in myoedited mice and corrected skeletal muscle was highly resistant to necrosis and fibrosis, plus showed sustained dystrophin expression in response to chronic injury.114
Due to the high turnover rate of DMD muscles, the ability to deliver the gene-editing components to satellite cells, the muscle stem cells, would likely improve and sustain dystrophin expression in myoedited muscle. In this regard, AAV infectivity of satellite cells has been controversial and needs further investigation and development.56,115–118 Encouragingly, MyoAAV transduces satellite cells with about three times higher efficiency compared to AAV9, and in the future, the utilization of a satellite cell-specific promoter to drive the gene-editing components could improve and advance gene editing of DMD.84
CONCLUDING REMARKS
Different CRISPR myoediting strategies have been successfully applied in DMD human iPSCs and DMD animal models, providing new, promising, effective treatments for DMD. The efficacy of gene-editing technologies is no longer an obstacle for the treatment of DMD in clinical applications. Considering all the recent preclinical successes obtained with CRISPR systems, it seems reasonable that the remaining safety and delivery challenges will be overcome in the next few years.
ACKNOWLEDGMENTS
We thank J. Cabrera for his creative assistance with the graphics.
AUTHORs' CONTRIBUTIONS
F.C. wrote the initial draft, and E.N.O. and R.B.D. edited the article. All authors revised the article and approved it for publication.
AUTHOR DISCLOSURE
E.N.O. is a consultant for Vertex Pharmaceuticals and Tenaya Therapeutics. The other authors declare no competing interests.
FUNDING INFORMATION
This work was supported by grants from the National Institutes of Health (HL-130253, HD-087351, HL-157281) and the Robert A. Welch Foundation (grant 1-0025 to E.N.O.).
REFERENCES
- 1. O'Brien KF, Kunkel LM. Dystrophin and muscular dystrophy: Past, present, and future. Mol Genet Metab 2001;74(1–2):75–88; doi: 10.1006/mgme.2001.3220 [DOI] [PubMed] [Google Scholar]
- 2. Passamano L, Taglia A, Palladino A, et al. Improvement of survival in Duchenne Muscular Dystrophy: Retrospective analysis of 835 patients. Acta Myol 2012;31(2):121–125. [PMC free article] [PubMed] [Google Scholar]
- 3. Aartsma-Rus A, Van Deutekom JC, Fokkema IF, et al. Entries in the Leiden Duchenne muscular dystrophy mutation database: An overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve 2006;34(2):135–144; doi: 10.1002/mus.20586 [DOI] [PubMed] [Google Scholar]
- 4. Hoffman EP, Brown RH Jr., Kunkel LM. Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell 1987;51(6):919–928; doi: 10.1016/0092-8674(87)90579-4 [DOI] [PubMed] [Google Scholar]
- 5. Mercuri E, Muntoni F. Muscular dystrophies. Lancet 2013;381(9869):845–860; doi: 10.1016/S0140-6736(12)61897-2 [DOI] [PubMed] [Google Scholar]
- 6. Bladen CL, Salgado D, Monges S, et al. The TREAT-NMD DMD Global Database: Analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum Mutat 2015;36(4):395–402; doi: 10.1002/humu.22758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Monaco AP, Bertelson CJ, Liechti-Gallati S, et al. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 1988;2(1):90–95; doi: 10.1016/0888-7543(88)90113-9 [DOI] [PubMed] [Google Scholar]
- 8. Goemans N, Buyse G. Current treatment and management of dystrophinopathies. Curr Treat Options Neurol 2014;16(5):287; doi: 10.1007/s11940-014-0287-4 [DOI] [PubMed] [Google Scholar]
- 9. Zhang T, Kong X. Recent advances of glucocorticoids in the treatment of Duchenne muscular dystrophy (Review). Exp Ther Med 2021;21(5):447; doi: 10.3892/etm.2021.9875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duan D. Systemic AAV micro-dystrophin gene therapy for duchenne muscular dystrophy. Mol Ther 2018;26(10):2337–2356; doi: 10.1016/j.ymthe.2018.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Asher DR, Thapa K, Dharia SD, et al. Clinical development on the frontier: Gene therapy for duchenne muscular dystrophy. Expert Opin Biol Ther 2020;20(3):263–274; doi: 10.1080/14712598.2020.1725469 [DOI] [PubMed] [Google Scholar]
- 12. Lim KR, Maruyama R, Yokota T. Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des Devel Ther 2017;11:533–545; doi: 10.2147/DDDT.S97635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Komaki H, Nagata T, Saito T, et al. Systemic administration of the antisense oligonucleotide NS-065/NCNP-01 for skipping of exon 53 in patients with Duchenne muscular dystrophy. Sci Transl Med 2018;10(437):eaan0713; doi: 10.1126/scitranslmed.aan0713 [DOI] [PubMed] [Google Scholar]
- 14. Clemens PR, Rao VK, Connolly AM, et al. Safety, tolerability, and efficacy of viltolarsen in boys with duchenne muscular dystrophy amenable to exon 53 skipping: A phase 2 randomized clinical trial. JAMA Neurol 2020;77(8):982–991; doi: 10.1001/jamaneurol.2020.1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012;337(6096):816–821; doi: 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339(6121):819–823; doi: 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science 2013;339(6121):823–826; doi: 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y, Long C, Bassel-Duby R, et al. Myoediting: Toward prevention of muscular dystrophy by therapeutic genome editing. Physiol Rev 2018;98(3):1205–1240; doi: 10.1152/physrev.00046.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Min YL, Bassel-Duby R, Olson EN. CRISPR correction of duchenne muscular dystrophy. Annu Rev Med 2019;70:239–255; doi: 10.1146/annurev-med-081117-010451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chemello F, Bassel-Duby R, Olson EN. Correction of muscular dystrophies by CRISPR gene editing. J Clin Invest 2020;130(6):2766–2776; doi: 10.1172/JCI136873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ousterout DG, Kabadi AM, Thakore PI, et al. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun 2015;6:6244; doi: 10.1038/ncomms7244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maggio I, Liu J, Janssen JM, et al. Adenoviral vectors encoding CRISPR/Cas9 multiplexes rescue dystrophin synthesis in unselected populations of DMD muscle cells. Sci Rep 2016;6:37051; doi: 10.1038/srep37051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kyrychenko V, Kyrychenko S, Tiburcy M, et al. Functional correction of dystrophin actin binding domain mutations by genome editing. JCI Insight 2017;2(18):e95918; doi: 10.1172/jci.insight.95918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Duchene BL, Cherif K, Iyombe-Engembe JP, et al. CRISPR-induced deletion with SaCas9 restores dystrophin expression in dystrophic models in vitro and in vivo. Mol Ther 2018;26(11):2604–2616; doi: 10.1016/j.ymthe.2018.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Long C, Li H, Tiburcy M, et al. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci Adv 2018;4(1):eaap9004; doi: 10.1126/sciadv.aap9004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brescia M, Janssen JM, Liu J, et al. High-capacity adenoviral vectors permit robust and versatile testing of DMD gene repair tools and strategies in human cells. Cells 2020;9(4):869; doi: 10.3390/cells9040869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moretti A, Fonteyne L, Giesert F, et al. Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat Med 2020;26(2):207–214; doi: 10.1038/s41591-019-0738-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen M, Shi H, Gou S, et al. In vivo genome editing in mouse restores dystrophin expression in Duchenne muscular dystrophy patient muscle fibers. Genome Med 2021;13(1):57; doi: 10.1186/s13073-021-00876-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lattanzi A, Duguez S, Moiani A, et al. Correction of the exon 2 duplication in DMD myoblasts by a single CRISPR/Cas9 system. Mol Ther Nucleic Acids 2017;7:11–19; doi: 10.1016/j.omtn.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pini V, Mariot V, Dumonceaux J, et al. Transiently expressed CRISPR/Cas9 induces wild-type dystrophin in vitro in DMD patient myoblasts carrying duplications. Sci Rep 2022;12(1):3756; doi: 10.1038/s41598-022-07671-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang DN, Wang ZQ, Jin M, et al. CRISPR/Cas9-based genome editing for the modification of multiple duplications that cause Duchenne muscular dystrophy. Gene Ther 2022;29(12):730–737; doi: 10.1038/s41434-022-00336-3 [DOI] [PubMed] [Google Scholar]
- 32. Young CS, Hicks MR, Ermolova NV, et al. A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell Stem Cell 2016;18(4):533–540; doi: 10.1016/j.stem.2016.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nelson CE, Wu Y, Gemberling MP, et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat Med 2019;25(3):427–432; doi: 10.1038/s41591-019-0344-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fletcher S, Adams AM, Johnsen RD, et al. Dystrophin isoform induction in vivo by antisense-mediated alternative splicing. Mol Ther 2010;18(6):1218–1223; doi: 10.1038/mt.2010.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Y, Long C, Li H, et al. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci Adv 2017;3(4):e1602814; doi: 10.1126/sciadv.1602814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Min YL, Li H, Rodriguez-Caycedo C, et al. CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci Adv 2019;5(3):eaav4324; doi: 10.1126/sciadv.aav4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Min YL, Chemello F, Li H, et al. Correction of three prominent mutations in mouse and human models of duchenne muscular dystrophy by single-cut genome editing. Mol Ther 2020;28(9):2044–2055; doi: 10.1016/j.ymthe.2020.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Y, Nishiyama T, Li H, et al. A consolidated AAV system for single-cut CRISPR correction of a common Duchenne muscular dystrophy mutation. Mol Ther Methods Clin Dev 2021;22:122–132; doi: 10.1016/j.omtm.2021.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y, Li H, Nishiyama T, et al. A humanized knockin mouse model of Duchenne muscular dystrophy and its correction by CRISPR-Cas9 therapeutic gene editing. Mol Ther Nucleic Acids 2022;29:525–537; doi: 10.1016/j.omtn.2022.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haapaniemi E, Botla S, Persson J, et al. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med 2018;24(7):927–930; doi: 10.1038/s41591-018-0049-z [DOI] [PubMed] [Google Scholar]
- 41. Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 2018;36(8):765–771; doi: 10.1038/nbt.4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hanlon KS, Kleinstiver BP, Garcia SP, et al. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat Commun 2019;10(1):4439; doi: 10.1038/s41467-019-12449-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hanson B, Stenler S, Ahlskog N, et al. Non-uniform dystrophin re-expression after CRISPR-mediated exon excision in the dystrophin/utrophin double-knockout mouse model of DMD. Mol Ther Nucleic Acids 2022;30:379–397; doi: 10.1016/j.omtn.2022.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 2020;38(7):824–844; doi: 10.1038/s41587-020-0561-9 [DOI] [PubMed] [Google Scholar]
- 45. Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016;533(7603):420–424; doi: 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017;551(7681):464–471; doi: 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yuan J, Ma Y, Huang T, et al. Genetic modulation of RNA splicing with a CRISPR-guided cytidine deaminase. Mol Cell 2018;72(2):380.e7–394.e7; doi: 10.1016/j.molcel.2018.09.002 [DOI] [PubMed] [Google Scholar]
- 48. Chemello F, Chai AC, Li H, et al. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Sci Adv 2021;7(18):eabg4910; doi: 10.1126/sciadv.abg4910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang P, Li H, Zhu M, et al. Correction of DMD in human iPSC-derived cardiomyocytes by base-editing-induced exon skipping. Mol Ther Methods Clin Dev 2023;28:40–50; doi: 10.1016/j.omtm.2022.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019;576(7785):149–157; doi: 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Happi Mbakam C, Rousseau J, Tremblay G, et al. Prime editing permits the introduction of specific mutations in the gene responsible for duchenne muscular dystrophy. Int J Mol Sci 2022;23(11):6160; doi: 10.3390/ijms23116160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Happi Mbakam C, Rousseau J, Lu Y, et al. Prime editing optimized RTT permits the correction of the c.8713C>T mutation in DMD gene. Mol Ther Nucleic Acids 2022;30:272–285; doi: 10.1016/j.omtn.2022.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Amoasii L, Long C, Li H, et al. Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci Transl Med 2017;9(418):eaan8081; doi: 10.1126/scitranslmed.aan8081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zincarelli C, Soltys S, Rengo G, et al. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther 2008;16(6):1073–1080; doi: 10.1038/mt.2008.76 [DOI] [PubMed] [Google Scholar]
- 55. Long C, Amoasii L, Mireault AA, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 2016;351(6271):400–403; doi: 10.1126/science.aad5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tabebordbar M, Zhu K, Cheng JKW, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016;351(6271):407–411; doi: 10.1126/science.aad5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nelson CE, Hakim CH, Ousterout DG, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 2016;351(6271):403–407; doi: 10.1126/science.aad5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bengtsson NE, Hall JK, Odom GL, et al. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun 2017;8:14454; doi: 10.1038/ncomms14454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. El Refaey M, Xu L, Gao Y, et al. In vivo genome editing restores dystrophin expression and cardiac function in dystrophic mice. Circ Res 2017;121(8):923–929; doi: 10.1161/CIRCRESAHA.117.310996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Young CS, Mokhonova E, Quinonez M, et al. Creation of a novel humanized dystrophic mouse model of duchenne muscular dystrophy and application of a CRISPR/Cas9 gene editing therapy. J Neuromuscul Dis 2017;4(2):139–145; doi: 10.3233/JND-170218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Koo T, Lu-Nguyen NB, Malerba A, et al. Functional rescue of dystrophin deficiency in mice caused by frameshift mutations using campylobacter jejuni Cas9. Mol Ther 2018;26(6):1529–1538; doi: 10.1016/j.ymthe.2018.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Maino E, Wojtal D, Evagelou SL, et al. Targeted genome editing in vivo corrects a Dmd duplication restoring wild-type dystrophin expression. EMBO Mol Med 2021;13(5):e13228; doi: 10.15252/emmm.202013228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang Y, Li H, Min YL, et al. Enhanced CRISPR-Cas9 correction of Duchenne muscular dystrophy in mice by a self-complementary AAV delivery system. Sci Adv 2020;6(8):eaay6812; doi: 10.1126/sciadv.aay6812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang Y, Bassel-Duby R, Olson EN. CRISPR-Cas9 correction of duchenne muscular dystrophy in mice by a self-complementary AAV delivery system. Methods Mol Biol 2023;2587:411–425; doi: 10.1007/978-1-0716-2772-3_21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ryu SM, Koo T, Kim K, et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol 2018;36(6):536–539; doi: 10.1038/nbt.4148 [DOI] [PubMed] [Google Scholar]
- 66. Li J, Wang K, Zhang Y, et al. Therapeutic exon skipping through a CRISPR-guided cytidine deaminase rescues dystrophic cardiomyopathy in vivo. Circulation 2021;144(22):1760–1776; doi: 10.1161/CIRCULATIONAHA.121.054628 [DOI] [PubMed] [Google Scholar]
- 67. Xu L, Zhang C, Li H, et al. Efficient precise in vivo base editing in adult dystrophic mice. Nat Commun 2021;12(1):3719; doi: 10.1038/s41467-021-23996-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wei T, Cheng Q, Min YL, et al. Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat Commun 2020;11(1):3232; doi: 10.1038/s41467-020-17029-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kenjo E, Hozumi H, Makita Y, et al. Low immunogenicity of LNP allows repeated administrations of CRISPR-Cas9 mRNA into skeletal muscle in mice. Nat Commun 2021;12(1):7101; doi: 10.1038/s41467-021-26714-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McCourt JL, Rhett KK, Jaeger MA, et al. In vitro stability of therapeutically relevant, internally truncated dystrophins. Skelet Muscle 2015;5:13; doi: 10.1186/s13395-015-0040-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Love DR, Hill DF, Dickson G, et al. An autosomal transcript in skeletal muscle with homology to dystrophin. Nature 1989;339(6219):55–58; doi: 10.1038/339055a0 [DOI] [PubMed] [Google Scholar]
- 72. Khurana TS, Hoffman EP, Kunkel LM. Identification of a chromosome 6-encoded dystrophin-related protein. J Biol Chem 1990;265(28):16717–16720. [PubMed] [Google Scholar]
- 73. Sengupta K, Mishra MK, Loro E, et al. Genome editing-mediated utrophin upregulation in duchenne muscular dystrophy stem cells. Mol Ther Nucleic Acids 2020;22:500–509; doi: 10.1016/j.omtn.2020.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Guiraud S, Edwards B, Babbs A, et al. The potential of utrophin and dystrophin combination therapies for Duchenne muscular dystrophy. Hum Mol Genet 2019;28(13):2189–2200; doi: 10.1093/hmg/ddz049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Suzuki K, Tsunekawa Y, Hernandez-Benitez R, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 2016;540(7631):144–149; doi: 10.1038/nature20565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Suzuki K, Yamamoto M, Hernandez-Benitez R, et al. Precise in vivo genome editing via single homology arm donor mediated intron-targeting gene integration for genetic disease correction. Cell Res 2019;29(10):804–819; doi: 10.1038/s41422-019-0213-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pickar-Oliver A, Gough V, Bohning JD, et al. Full-length dystrophin restoration via targeted exon integration by AAV-CRISPR in a humanized mouse model of Duchenne muscular dystrophy. Mol Ther 2021;29(11):3243–3257; doi: 10.1016/j.ymthe.2021.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Klompe SE, Vo PLH, Halpin-Healy TS, et al. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature 2019;571(7764):219–225; doi: 10.1038/s41586-019-1323-z [DOI] [PubMed] [Google Scholar]
- 79. Strecker J, Ladha A, Gardner Z, et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science 2019;365(6448):48–53; doi: 10.1126/science.aax9181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Anzalone AV, Gao XD, Podracky CJ, et al. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat Biotechnol 2022;40(5):731–740; doi: 10.1038/s41587-021-01133-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yarnall MTN, Ioannidi EI, Schmitt-Ulms C, et al. Drag-and-drop genome insertion of large sequences without double-strand DNA cleavage using CRISPR-directed integrases. Nat Biotechnol 2023;41:500–512; doi: 10.1038/s41587-022-01527-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hinderer C, Katz N, Buza EL, et al. Severe toxicity in nonhuman primates and piglets following high-dose intravenous administration of an adeno-associated virus vector expressing human SMN. Hum Gene Ther 2018;29(3):285–298; doi: 10.1089/hum.2018.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Weinmann J, Weis S, Sippel J, et al. Identification of a myotropic AAV by massively parallel in vivo evaluation of barcoded capsid variants. Nat Commun 2020;11(1):5432; doi: 10.1038/s41467-020-19230-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tabebordbar M, Lagerborg KA, Stanton A, et al. Directed evolution of a family of AAV capsid variants enabling potent muscle-directed gene delivery across species. Cell 2021;184(19):4919.e22–4938.e22; doi: 10.1016/j.cell.2021.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. El Andari J, Renaud-Gabardos E, Tulalamba W, et al. Semirational bioengineering of AAV vectors with increased potency and specificity for systemic gene therapy of muscle disorders. Sci Adv 2022;8(38):eabn4704; doi: 10.1126/sciadv.abn4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hakim CH, Wasala NB, Nelson CE, et al. AAV CRISPR editing rescues cardiac and muscle function for 18months in dystrophic mice. JCI Insight 2018;3(23); doi: 10.1172/jci.insight.124297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wasala NB, Million ED, Watkins TB, et al. The gRNA vector level determines the outcome of systemic AAV CRISPR therapy for duchenne muscular dystrophy. Hum Gene Ther 2022;33(9–10):518–528; doi: 10.1089/hum.2021.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gee P, Lung MSY, Okuzaki Y, et al. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat Commun 2020;11(1):1334; doi: 10.1038/s41467-020-14957-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Segel M, Lash B, Song J, et al. Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery. Science 2021;373(6557):882–889; doi: 10.1126/science.abg6155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Banskota S, Raguram A, Suh S, et al. Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell 2022;185(2):250.e16–265.e16; doi: 10.1016/j.cell.2021.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Charlesworth CT, Deshpande PS, Dever DP, et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med 2019;25(2):249–254; doi: 10.1038/s41591-018-0326-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wienert B, Shin J, Zelin E, et al. In vitro-transcribed guide RNAs trigger an innate immune response via the RIG-I pathway. PLoS Biol 2018;16(7):e2005840; doi: 10.1371/journal.pbio.2005840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kim S, Koo T, Jee HG, et al. CRISPR RNAs trigger innate immune responses in human cells. Genome Res 2018;28(3):367–373; doi: 10.1101/gr.231936.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kim D, Bae S, Park J, et al. Digenome-seq: Genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods 2015;12(3):237–243, 1 p following 243; doi: 10.1038/nmeth.3284 [DOI] [PubMed] [Google Scholar]
- 95. Tsai SQ, Zheng Z, Nguyen NT, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 2015;33(2):187–197; doi: 10.1038/nbt.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Tsai SQ, Nguyen NT, Malagon-Lopez J, et al. CIRCLE-seq: A highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods 2017;14(6):607–614; doi: 10.1038/nmeth.4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Olson EN. Toward the correction of muscular dystrophy by gene editing. Proc Natl Acad Sci U S A 2021;118(22):e2004840117; doi: 10.1073/pnas.2004840117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Fu Y, Sander JD, Reyon D, et al. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 2014;32(3):279–284; doi: 10.1038/nbt.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Doench JG, Fusi N, Sullender M, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016;34(2):184–191; doi: 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016;529(7587):490–495; doi: 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Slaymaker IM, Gao L, Zetsche B, et al. Rationally engineered Cas9 nucleases with improved specificity. Science 2016;351(6268):84–88; doi: 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Chen JS, Dagdas YS, Kleinstiver BP, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 2017;550(7676):407–410; doi: 10.1038/nature24268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kulcsar PI, Talas A, Huszar K, et al. Crossing enhanced and high fidelity SpCas9 nucleases to optimize specificity and cleavage. Genome Biol 2017;18(1):190; doi: 10.1186/s13059-017-1318-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Casini A, Olivieri M, Petris G, et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol 2018;36(3):265–271; doi: 10.1038/nbt.4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kocak DD, Josephs EA, Bhandarkar V, et al. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat Biotechnol 2019;37(6):657–666; doi: 10.1038/s41587-019-0095-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. t Hoen PA, de Meijer EJ, Boer JM, et al. Generation and characterization of transgenic mice with the full-length human DMD gene. J Biol Chem 2008;283(9):5899–5907; doi: 10.1074/jbc.M709410200 [DOI] [PubMed] [Google Scholar]
- 107. Veltrop M, van Vliet L, Hulsker M, et al. A dystrophic Duchenne mouse model for testing human antisense oligonucleotides. PLoS One 2018;13(2):e0193289; doi: 10.1371/journal.pone.0193289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Yavas A, Weij R, van Putten M, et al. Detailed genetic and functional analysis of the hDMDdel52/mdx mouse model. PLoS One 2020;15(12):e0244215; doi: 10.1371/journal.pone.0244215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Li G, Jin M, Li Z, et al. Mini-dCas13X-mediated RNA editing restores dystrophin expression in a humanized mouse model of Duchenne muscular dystrophy. J Clin Invest 2023;133(3):e162809; doi: 10.1172/JCI162809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Deconinck N, Tinsley J, De Backer F, et al. Expression of truncated utrophin leads to major functional improvements in dystrophin-deficient muscles of mice. Nat Med 1997;3(11):1216–1221; doi: 10.1038/nm1197-1216 [DOI] [PubMed] [Google Scholar]
- 111. Wasala NB, Chen SJ, Duan D. Duchenne muscular dystrophy animal models for high-throughput drug discovery and precision medicine. Expert Opin Drug Discov 2020;15(4):443–456; doi: 10.1080/17460441.2020.1718100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hakim CH, Kumar SRP, Perez-Lopez DO, et al. Cas9-specific immune responses compromise local and systemic AAV CRISPR therapy in multiple dystrophic canine models. Nat Commun 2021;12(1):6769; doi: 10.1038/s41467-021-26830-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Amoasii L, Hildyard JCW, Li H, et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 2018;362(6410):86–91; doi: 10.1126/science.aau1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Karri DR, Zhang Y, Chemello F, et al. Long-term maintenance of dystrophin expression and resistance to injury of skeletal muscle in gene edited DMD mice. Mol Ther Nucleic Acids 2022;28:154–167; doi: 10.1016/j.omtn.2022.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Arnett AL, Konieczny P, Ramos JN, et al. Adeno-associated viral (AAV) vectors do not efficiently target muscle satellite cells. Mol Ther Methods Clin Dev 2014;1:14038; doi: 10.1038/mtm.2014.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Goldstein JM, Tabebordbar M, Zhu K, et al. In situ modification of tissue stem and progenitor cell genomes. Cell Rep 2019;27(4):1254.e7–1264.e7; doi: 10.1016/j.celrep.2019.03.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Nance ME, Shi R, Hakim CH, et al. AAV9 edits muscle stem cells in normal and dystrophic adult mice. Mol Ther 2019;27(9):1568–1585; doi: 10.1016/j.ymthe.2019.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Kwon JB, Ettyreddy AR, Vankara A, et al. In vivo gene editing of muscle stem cells with adeno-associated viral vectors in a mouse model of duchenne muscular dystrophy. Mol Ther Methods Clin Dev 2020;19:320–329; doi: 10.1016/j.omtm.2020.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]