Abstract
The new genus Pseudolepraria Kukwa, Jabłońska, Kosecka & Guzow-Krzemińska is introduced to accommodate Leprariastephaniana Elix, Flakus & Kukwa. Phylogenetic analyses of nucITS, nucLSU, mtSSU and RPB2 markers recovered the new genus in the family Ramalinaceae with strong support. The genus is characterised by its thick, unstratified thallus composed entirely of soredia-like granules, the presence of 4-O-methylleprolomin, salazinic acid, zeorin and unknown terpenoid, and its phylogenetic position. The new combination, P.stephaniana (Elix, Flakus & Kukwa) Kukwa, Jabłońska, Kosecka & Guzow-Krzemińska, is proposed.
Keywords: Lichenized fungi, morphology, Neotropics, secondary metabolites, sterile lichens, taxonomy
Introduction
During the evolution in some groups of lichenized fungi the ability to reproduce sexually has been apparently lost completely and some phylogenetic lineages are known to develop exclusively asexual lichenized propagules. This includes Lepraria Ach. (Ascomycota, Lecanoromycetes, Lecanorales, Stereocaulaceae), a well-known genus which up to quite recently comprised only crustose lichens with morphologically simple thalli consisting of soredia-like granules laying directly on substrate or on a layer of hypothalline hyphae (e.g., Ekman and Tønsberg 2002; Kukwa 2002; Sipman 2004; Flakus and Kukwa 2009; Flakus et al. 2011a; Lendemer 2011a, b, 2013a; Lendemer and Hodkinson 2013; Guzow-Krzemińska et al. 2019a). However, Lendemer and Hodkinson (2013) found, based on molecular data, that some fruticose species previously referred to Leprocaulon Nyl. also represented Lepraria s.str. and they were subsequently transferred to the latter genus. In contrast to their simplified morphology, the species produce a vast variety of secondary lichen metabolites, which are an invaluable tool, together with morphological characters that may be sparse, in the recognition of species and their identification (e.g., Laundon 1989, 1992; Tønsberg 1992; Sipman 2004; Kantvilas and Kukwa 2006; Flakus and Kukwa 2007; Saag et al. 2009; Flakus et al. 2011a; Lendemer 2011a, 2013a; Lendemer and Hodkinson 2013; Guzow-Krzemińska et al. 2019a; Kukwa 2019). It is also noteworthy that some species until recently classified as Lepraria have been shown to belong to other genera (e.g., Leprocaulon and Septotrapelia Aptroot & Chaves; Bungartz et al. 2013; Lendemer and Hodkinson 2013) or even new genera were established for some peculiar species, e.g., Andreiomyces Hodkinson & Lendemer within Arthoniomycetes (Hodkinson and Lendemer 2013), Botryolepraria Canals et al., related to Verrucariaceae in Eurotiomycetes (Kukwa and Pérez-Ortega 2010) and Lithocalla Orange in Lecanorales (probably in Ramalinaceae) in Lecanoromycetes (Orange 2020).
Lepraria includes at present c. 75 species (Wijayawardene et al. 2017; Guzow-Krzemińska et al. 2019a; Barcenas-Peña et al. 2021), most of which were described based on chemical (secondary metabolites) and morphological features and some also by molecular markers (e.g., Laundon 1989, 1992; Tønsberg 1992; Lendemer 2011a, 2012, 2013a; Lendemer and Hodkinson 2013; Guzow-Krzemińska et al. 2019a; Barcenas-Peña et al. 2021). One of the species that was placed in Lepraria based solely on morphological similarity to other members of the genus was L.stephaniana Elix, Flakus & Kukwa (Flakus et al. 2011a). This species is characterised by the thick, unstratified and non-lobed thallus composed of coarse soredia-like granules with soft appearance, and the production of 4-O-methylleprolomin, salazinic acid and terpenoids. 4-O-methylleprolomin was known only in a single Pannaria species before its discovery in L.stephaniana (Flakus et al. 2011a). Leprariastephaniana has been known until recently only from the type locality, however during field studies in 2017 in Bolivia we found two new localities of the species (one close to the type locality) (Guzow-Krzemińska et al. 2019b). Sequencing of molecular markers of those two recently collected specimens revealed that L.stephaniana is unrelated to other species of Lepraria s.str., but instead it appeared to be nested within Ramalinaceae as a previously unsequenced lineage close to Cliostomum Fr., Ramalina Ach. and allied genera. In this paper we introduce the new genus Pseudolepraria for this peculiar lineage within Ramalinaceae.
Materials and methods
Taxon sampling
The studied specimens are deposited in B, BG, KRAM, LPB, NY and UGDA herbaria. Morphology was examined by using Nikon SMZ 800N stereomicroscope. The secondary chemistry of all samples was studied by thin layer chromatography (TLC) following methods by Culberson and Kristinsson (1970) and Orange et al. (2001a).
DNA extraction, PCR amplification and DNA sequencing
DNA was extracted using a modified CTAB method (Guzow-Krzemińska and Węgrzyn 2000). We analysed four fungal markers: nucITS rDNA, mtSSU rDNA, nucLSU rDNA, and RPB2 gene. For this purpose we used the following primers: ITS1F (Gardes and Bruns 1993) and ITS4A (Kroken and Taylor 2001) for nucITS rDNA; mrSSU1 and mrSSU3R (Zoller et al. 1999) for mtSSU rDNA; ITS4A-5’ (Kroken and Taylor 2001; Nelsen et al. 2011) and LR5 (Vilgalys and Hester 1990) for nucLSU rDNA; fRPB2-5F and fRPB2-7cR (Liu et al. 1999) for RPB2 gene. Additionally, nucITS rDNA region from green algal partner was amplified using Al1500bf (Helms et al. 2001) and ITS4M primers (Guzow-Krzemińska 2006). PCR was performed in a volume of 25 µl using StartWarm HS-PCR Mix (A&A Biotechnology) following the manufacturer’s protocol. 1 µl of genomic DNA was used for amplification. The PCR cycling parameters are available in Suppl. material 1.
The efficiency of the PCR was checked by visualising the reaction products on a 1% agarose gels stained with SimplySafe (Eurx) dye in order to determine DNA fragment lengths. Afterwards, PCR products were purified using Clean-Up Concentrator (A&A Biotechnology). The sequencing was performed in Macrogen Europe (The Netherlands), using amplification primers. The newly obtained sequences were deposited in GenBank database and their accession numbers are listed in Table 1.
Table 1.
Species used in this study with their GenBank accession numbers. New sequences are marked in bold.
Sequence alignment and phylogenetic analysis
The newly generated sequences were compared to the sequences available in the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/) using BLASTn search (Altschul et al. 1990). For the phylogenetic analyses we used representatives of Ramalinaceae and Boreoplacaultrafrigida Timdal and Ropalosporalugubris (Sommerf.) Poelt were used as outgroup taxa according to previous studies (Kistenich et al. 2018; Orange 2020; van den Boom and Magain 2020). The independent alignments for each marker were generated in MAFFT using auto option and default parameters (Katoh and Standley 2013). The datasets were then subjected to Guidance2 server (Landan and Graur 2008; Penn et al. 2010; Sela et al. 2015; http://guidance.tau.ac.il/) for further analysis. The MSA algorithm was set to MAFFT and 100 bootstrap replicates were used. The Guidance confidence scores were calculated and columns with a score < 0.93 were excluded from the alignments. The terminal ends were trimmed. Single-locus matrices consisted of 61 sequences for nucITS, 62 sequences for mtSSU, 51 sequences for nucLSU, and 44 sequences for RPB2. The best ML tree was inferred for each locus using IQ-TREE with 1000 ultrafast bootstrap replicates as implemented in the IQ-TREE web server (Nguyen et al. 2015; Chernomor et al. 2016; Kalyaanamoorthy et al. 2017; Hoang et al. 2018). Congruence was examined by eye and no significant conflict between loci was observed.
For the final analysis, we concatenated four markers which resulted in a dataset of 66 terminals and 3766 positions. The concatenated dataset was subjected to IQ-TREE analysis to find best-fitting nucleotide substitution models (Nguyen et al. 2015; Chernomor et al. 2016; Kalyaanamoorthy et al. 2017; Hoang et al. 2018). The model selection was restricted to models implemented in MrBayes and the following nucleotide substitution models for the four predefined subsets were selected: GTR+F+I+G for mtSSU rDNA, K2P+I+G for nucITS, and SYM+I+G for both nucLSU rDNA and RPB2 markers. The search for maximum likelihood tree was performed in IQ-TREE and followed with 1000 standard bootstrap replicates (Nguyen et al. 2015; Chernomor et al. 2016; Kalyaanamoorthy et al. 2017; Hoang et al. 2018).
Bayesian analysis was carried out using a Markov Chain Monte Carlo (MCMC) method, in MrBayes v. 3.2.6 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) on the CIPRES Web Portal (Miller et al. 2010) using previously selected models (see above). Two parallel MCMC runs were performed, each using four independent chains and ten million generations, sampling every 1000th tree. The resulting log files were analysed using Tracer 1.7.2 (Rambaut et al. 2018). Posterior probabilities (PP) were determined by calculating a majority-rule consensus tree after discarding the initial 25% trees of each chain as the burn-in. The convergence of the chains was confirmed by the convergent diagnostic of the Potential Scale Reduction Factor (PSRF), which approached 1 and the ‘average standard deviation of split frequencies’ was < 0.01).
Phylogenetic trees were visualised using FigTree v. 1.4.3 (Rambaut 2009) and modified in Inkscape (https://inkscape.org/). Bootstrap support (BS values ≥ 70) and PP values (values ≥ 0.95) are given near the branches on the phylogenetic tree. The data were deposited at TreeBASE (Submission ID: 30149).
Results and discussion
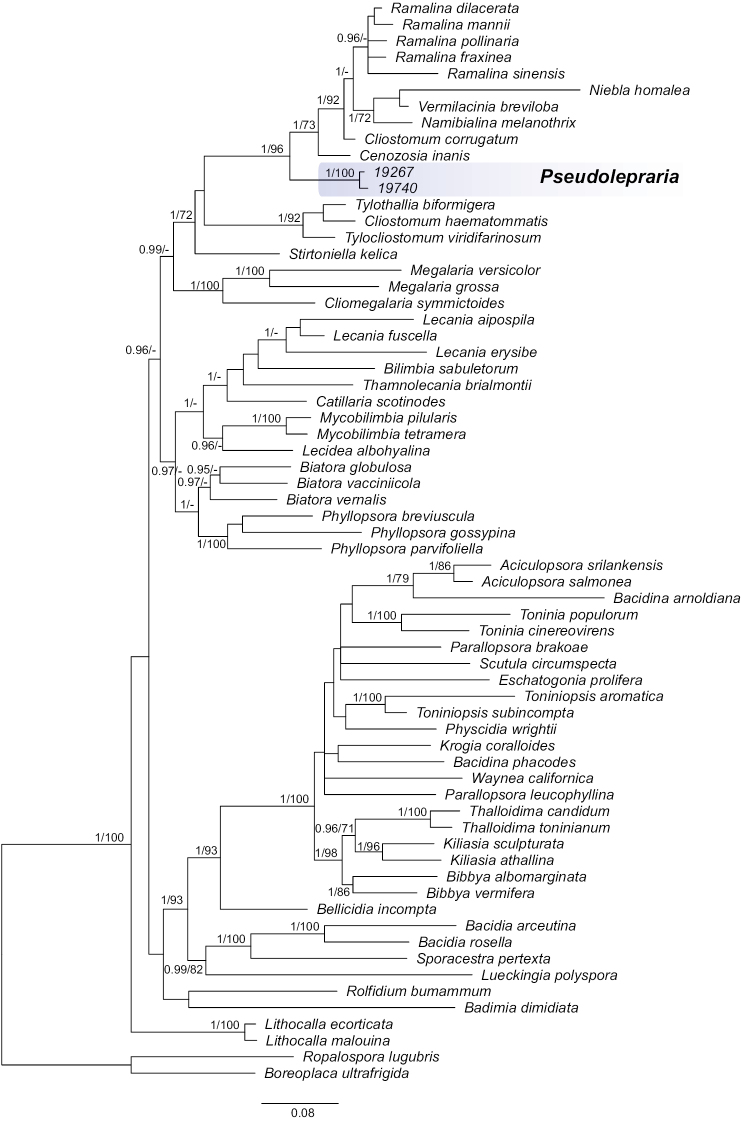
For this work we successfully sequenced nucITS, mtSSUand nucLSU from two specimens and additionally RPB2 from one specimen of Leprariastephaniana collected in Bolivia (Table 1). BLAST searches of the nucITS, nucLSU, mtSSU and RPB2 markers surprisingly showed the highest similarities to representatives of the family Ramalinaceae, i.e., the genera Cenozosia A. Massal., Cliostomum and Ramalina. Phylogenetic analysis of the concatenated dataset shows that L.stephaniana is nested inside Ramalinaceae. The newly sequenced specimens of the species were resolved in a distinct and highly supported clade sister to a clade consisting of Cliostomum s.str. represented by the type species C.corrugatum (Ach.) Fr., Cenozosiainanis (Mont.) A. Massal. and a subclade of several species of Ramalina, Namibialinamelanothrix (Laurer) Spjut & Sérus., Niebla (Ach.) Rundel & Bowler and Vermilaciniabreviloba Spjut & Sérus. (Fig. 1). A new monotypic genus, Pseudolepraria, is introduced for this lineage of Leprariastephaniana and is characterised by a thick, unstratified thallus composed of soredia-like granules, and the presence of 4-O-methylleprolomin, salazinic acid, zeorin and unknown terpenoid.
Figure 1.
Majority-rule consensus tree resulting from MrBayes analysis of the concatenated mtSSU, nucITS, nucLSU and RPB2 markers with Bayesian PP (values ≥ 0.95) and IQ-TREE bootstrap support values (BS values ≥ 70) given near the branches. Pseudolepraria is marked in blue.
Pseudolepraria is the first genus forming leprose and sterile thalli that can be placed with high support within Ramalinaceae. Orange (2020) described the genus Lithocalla and placed it with uncertainty in Ramalinaceae. In our phylogeny, Lithocalla forms the sister group to Ramalinaceae sensu Kistenich et al. (2018), but this may be an artifact of the taxon sampling. The particular placement of the genus was beyond the scope of this study. Lithocalla was introduced for two species, which were originally placed, due to morphological similarities, in Lepraria, i.e., L.ecorticata (J. R. Laundon) Kukwa and L.malouina Øvstedal (Kukwa 2006; Fryday and Øvstedal 2012). Both, Lithocallaecorticata (J. R. Laundon) Orange known from Great Britain and Norway, and L.malouina (Øvstedal) Fryday & Orange found in the Falkland Islands (Fryday and Øvstedal 2012; Orange 2020), differ from Pseudoleprariastephaniana, in their distribution in colder climates, by the production of usnic and fatty acids, the absence of zeorin and the exclusively saxicolous habitat (Orange 2020).
Species resembling Pseudolepraria in the Ramalinaceae have up until recently been included in Crocynia (Ach.) A. Massal. This genus was established for lichens with a non-corticate, byssoid, felt-like thallus and historically included several species now placed mostly in Lepraria (e.g., Laundon 1989, 1992; Kistenich et al. 2018). According to Lücking et al. (2017) Crocynia comprised three species and two of them were included in the phylogeny of Ramalinaceae by Kistenich et al. (2018), where they formed a clade nested inside Phyllopsora Müll. Arg. Consequently, Crocynia was synonymised with Phyllopsora, also because of morphological similarities (Kistenich et al. 2018). The status of the third species, C.microphyllina Aptroot (Lumbsch et al. 2011), and three species discussed by Sipman (2018) remains uncertain. The species historically placed in Crocynia differ from Pseudolepraria in the byssoid thalli not producing 4-O-methylleprolomin and in sometimes producing apothecia (Cáceres 2007; Lumbsch et al. 2011; Aptroot and Cáceres 2014; Sipman 2018).
Pseudolepraria is very similar to Lepraria s.str. in sharing the same thallus morphology and, to a certain extent, secondary chemistry (presence of salazinic acid and terpenoids) (e.g., Aptroot 2002; Sipman 2004; Saag et al. 2009; Flakus et al. 2011a; Lendemer 2011a, b). They differ, apart from the phylogenetic position, only in the presence of the very rarely reported 4-O-methylleprolomin, a diphenyl ether previously found only in one Pannaria species (Flakus et al. 2011a). Pseudolepraria differs also in the habitat preferences. It was found only in tropical forests at low elevations (300–470 m a.s.l.), whereas Lepraria in tropical South America, including Bolivian ecosystems, are found mostly above 1000 m above sea level (only one locality of L.finkii (B. de Lesd.) R.C. Harris found at the elevation of 890 m), in montane forests and open high Andean vegetation (Flakus and Kukwa 2007; Flakus et al. 2011a, b, 2012, 2015; Guzow-Krzemińska et al. 2019a). This is in agreement with the statement presented by Orange et al (2001b), who considered Lepraria to be restricted to montane habitats in the tropics.
Poelt (1987) considered the genus Lepraria as a ‘box of analogous groups of lichens of completely different origin, held together by the same highly specialized thallus type’. Poelt (1987) also stated that the leprarioid thallus type and the loss of generative reproduction developed in evolution through the reduction of the thallus structures as an adaptation for growing in bark crevices and on over-hanging rocks in ecologically specialised group of lichenized fungi, which includes Lepraria, but also, as Poelt (1987) mentioned, Leproplaca (Nyl.) Nyl. and some species of the genus Chrysothrix Mont. (Poelt 1987). However, this is only partly true, as some lichen groups with this type of thallus (e.g., species of Leprarianeglecta group) can grow also in other habitats (e.g., Laundon 1992; Lendemer 2013b). Nevertheless, the statement of Poelt (1987) was true and innovative at this time and it was later shown that the leprarioid thallus indeed originated in several unrelated lichen lineages (e.g., Ekman and Tønsberg 2002; Kukwa and Pérez-Ortega 2010; Hodkinson and Lendemer 2013; Malíček et al. 2018; Guzow-Krzemińska et al. 2017; Orange 2020). Furthermore, some leprarioid genera are known exclusively in sterile state, like Andreiomyces (Arthoniales, Arthoniomycetes), Botryolepraria (Verrucariales, Eurotiomycetes), Lepraria and Lithocalla (both in Lecanorales, Lecanoromycetes) (Ekman and Tønsberg 2002; Kukwa and Pérez-Ortega 2010; Hodkinson and Lendemer 2013; Orange 2020). Pseudolepraria is another addition to this group, however, as only a few collections are available, it may eventually be found with ascomata.
Buschbom and Mueller (2006) suggested that the asexual way of reproduction is advantageous because the symbiosis with the optimal photobiont for a given environment allows the rapid dissemination of both partners. Therefore, it is more important for the mycobiont to keep the relationship with suitable algal species; however this does not mean that the symbiosis in asexually reproducing species cannot be broken. Kosecka et al. (2021) showed for some Lepraria species that the mycobiont can form thalli with different, locally adapted algal strains. We partially sequenced the nucITS region of the photobiont of Pseudoleprariastephaniana (Table 1) and found that both thalli associate with the same green algal partner (100% of identity). BLAST hits were closest to Symbiochloris, Dictyochloropsis, Massjukichlorella and Watanabea spp., all of which were quite dissimilar to the photobiont sequences of Pseudoleprariastephaniana.
Taxonomy
. Pseudolepraria
Kukwa, Jabłońska, Kosecka & Guzow-Krzemińska gen. nov.
36CF85FA-ED27-57CF-A739-A95AF89D9AF5
847408
Diagnosis.
Characterised by thick, unstratified thallus composed of soredia-like granules, the presence of 4-O-methylleprolomin, salazinic acid, zeorin, and unknown terpenoid, and the phylogenetic position within Ramalinaceae.
Generic type.
Pseudoleprariastephaniana (Elix, Flakus & Kukwa) Kukwa, Jabłońska, Kosecka & Guzow-Krzemińska
Description.
As this is a monotypic genus the description below constitutes the generic description.
Etymology.
The new name refers to the similarity to the genus Lepraria, in which this particular species was originally placed.
. Pseudolepraria stephaniana
(Elix, Flakus & Kukwa) Kukwa, Jabłońska, Kosecka & Guzow-Krzemińska comb. nov.
E28E76E4-5CFC-51ED-95F9-BFB3CEC406BB
847409
Figure 2.
Morphology of Pseudoleprariastephaniana (type). Scale bar: 0.5 mm.
Lepraria stephaniana Elix, Flakus & Kukwa, in Flakus et al., Lichenologist 43: 64, 2011 (2010). Basionym.
Type.
Bolivia. Dept. La Paz: Prov. Iturralde, between Ixiamas and Santa Rosa de Maravillas villages, elev. 305 m, 13°49'16"S, 68°07'18"W, preandean Amazon forest, on bark of tree, 28 July 2008, M. Kukwa 6828 (holotype: UGDA L!; isotypes: B!, BG!, KRAM!, LPB!, NY!).
Description
(adopted from Flakus et al. 2011a). Thallus crustose, thick, usually not delimited nor lobed, green-grey to creamy-white, not stratified, but sometimes with a poorly differentiated, pseudo-medullary layer of decaying granules. Hypothallus indistinct. Granules coarse with soft appearance, irregularly rounded, up to 100(–200) μm in diam., composed of very lax hyphae mixed with algal cells, usually with projecting hyphae up to c. 30(–50) μm long. Photobiont green, cells globose, up to 12 μm in diam.
Chemistry.
Substances detected: 4-O-methylleprolomin (major), salazinic acid (minor), zeorin (minor) and an unknown terpenoid (minor) with Rf class values A6, B6, C6. Thallus reactions: K+ yellow turning brownish to red, P+ yellow, C–, KC– (see also Flakus et al. 2011a).
Distribution and habitat.
The species is known only from three localities in Bolivia. It was found on bark of trees in transition Chaqueño-Amazon or preandean Amazon forests at elevation between c. 300 to 470 m a.s.l. (Flakus et al. 2011a; Guzow-Krzemińska et al. 2019b).
Specimens used for DNA extraction
(Table 1). Bolivia. Dept. La Paz: Prov. Abel Iturralde, between Santa Rosa de Maravillas and Tumupasa, 13°58'43"S, 67°58'14"W, elev. 300 m, natural Preandean Amazon forest, corticolous, 25 May 2017, M. Kukwa 19740 (LPB, UGDA). Dept. Santa Cruz: Prov. Ichilo, Parque Nacional y Área Natural de Manejo Integrado Amboró, Sendero a la Cascada, near Villa Amboró, 17°44'02"S, 63°35'05"W, elev. 470 m, transition Chaqueño-Amazon forest, in the valley, corticolous, 11 May 2017, M. Kukwa 19267 (LPB, UGDA).
Supplementary Material
Acknowledgements
We are very grateful to James C. Lendemer (New York Botanical Garden), Tor Tønsberg (University of Bergen) and an anonymous reviewer for invaluable comments, and the members of Herbario Nacional de Bolivia, Instituto de Ecología, Universidad Mayor de San Andrés, La Paz, for their generous cooperation. This research received funding from the National Science Centre (project no 2015/17/B/NZ8/02441).
Citation
Kukwa M, Kosecka M, Jabłońska A, Flakus A, Rodriguez-Flakus P, Guzow-Krzemińska B (2023) Pseudolepraria, a new leprose genus revealed in Ramalinaceae (Ascomycota, Lecanoromycetes, Lecanorales) to accommodate Lepraria stephaniana. MycoKeys 96: 97–112. https://doi.org/10.3897/mycokeys.96.98029
Contributor Information
Martin Kukwa, Email: martin.kukwa@ug.edu.pl.
Beata Guzow-Krzemińska, Email: beata.guzow@biol.ug.edu.pl.
Supplementary materials
The PCR parameters
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Beata Guzow-Krzemińska, Magdalena Kosecka
Data type
PCR parameters (word document)
Explanation note
The PCR parameters are presented for each marker.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. Journal of Molecular Biology 215(3): 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Aptroot A. (2002) New and interesting lichens and lichenicolous fungi in Brazil. Fungal Diversity 9: 15–45. [Google Scholar]
- Aptroot A, Cáceres MES. (2014) A key to the corticolous microfoliose, foliose and related crustose lichens from Rondônia, Brazil, with the description of four new species. Lichenologist 46(6): 783–799. 10.1017/S0024282914000358 [DOI] [Google Scholar]
- Barcenas-Peña A, Diaz R, Grewe F, Widhelm T, Lumbsch HT. (2021) Contributions to the phylogeny of Lepraria (Stereocaulaceae) species from the Southern Hemisphere, including three new species. The Bryologist 124(4): 494–505. 10.1639/0007-2745-124.4.494 [DOI] [Google Scholar]
- Bungartz F, Hillmann G, Kalb K, Elix JA. (2013) Leprose and leproid lichens of the Galapagos, with a particular focus on Lepraria (Stereocaulaceae) and Septotrapelia (Pilocarpaceae). Phytotaxa 150(1): 1–28. 10.11646/phytotaxa.150.1.1 [DOI] [Google Scholar]
- Buschbom J, Mueller GM. (2006) Testing “species pair” hypotheses: Evolutionary processes in the lichen-forming species complex Porpidiaflavocoerulescens and Porpidiamelinodes. Molecular Biology and Evolution 23(3): 574–586. 10.1093/molbev/msj063 [DOI] [PubMed] [Google Scholar]
- Cáceres MES. (2007) Corticolous crustose and microfoliose lichens of northeastern Brazil. Libri Botanici 22: 1–168. [Google Scholar]
- Chernomor O, von Haeseler A, Minh BQ. (2016) Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology 65(6): 997–1008. 10.1093/sysbio/syw037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culberson CF, Kristinsson H. (1970) A standardized method for the identification of lichen products. Journal of Chromatography A 46: 85–93. 10.1016/S0021-9673(00)83967-9 [DOI] [PubMed] [Google Scholar]
- Ekman S, Tønsberg T. (2002) Most species of Lepraria and Leproloma form a monophyletic group closely related to Stereocaulon. Mycological Research 106(11): 1262–1276. 10.1017/S0953756202006718 [DOI] [Google Scholar]
- Flakus A, Kukwa M. (2007) New species and records of Lepraria (Stereocaulaceae, lichenized Ascomycota) from South America. Lichenologist 39(5): 463–474. 10.1017/S0024282907007116 [DOI] [Google Scholar]
- Flakus A, Kukwa M. (2009) Leprariaglaucosorediata sp. nov. (Stereocaulaceae, lichenized Ascomycota) and other interesting records of Lepraria. Mycotaxon 108(1): 353–364. 10.5248/108.353 [DOI] [Google Scholar]
- Flakus A, Elix JA, Rodriguez P, Kukwa M. (2011a) New species and records of Lepraria (Stereocaulaceae, lichenized Ascomycota) from South America. Lichenologist 43(1): 57–66. 10.1017/S0024282910000502 [DOI] [Google Scholar]
- Flakus A, Oset M, Jabłońska A, Rodriguez Saavedra P, Kukwa M. (2011b) Contribution to the knowledge of the lichen biota of Bolivia. 3. Polish Botanical Journal 56(2): 159–183. [Google Scholar]
- Flakus A, Etayo J, Schiefelbein U, Ahti T, Jabłońska A, Oset M, Bach K, Rodriguez Saavedra P, Kukwa M. (2012) Contribution to the knowledge of the lichen biota of Bolivia. 4. Polish Botanical Journal 7(2): 427–461. [Google Scholar]
- Flakus A, Sipman HJM, Rodriguez Flakus P, Jabłońska A, Oset M, Meneses QRI, Kukwa M. (2015) Contribution to the knowledge of the lichen biota of Bolivia. 7. Polish Botanical Journal 60(1): 81–98. 10.1515/pbj-2015-0001 [DOI] [Google Scholar]
- Fryday AM, Øvstedal DO. (2012) New species, combinations and records of lichenized fungi from the Falkland Islands (Islas Malvinas). Lichenologist 44(4): 483–500. 10.1017/S0024282912000163 [DOI] [Google Scholar]
- Gardes M, Bruns TD. (1993) ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Molecular Ecology 2(2): 113–118. 10.1111/j.1365-294X.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- Guzow-Krzemińska B. (2006) Photobiont flexibility in the lichen Protoparmeliopsismuralis as revealed by ITS rDNA analyses. Lichenologist 38(5): 469–476. 10.1017/S0024282906005068 [DOI] [Google Scholar]
- Guzow-Krzemińska B, Węgrzyn G. (2000) Potential use of restriction analysis of PCR-amplified DNA fragments in taxonomy of lichens. Mycotaxon 76: 305–313. [Google Scholar]
- Guzow-Krzemińska B, Malíček J, Tønsberg T, Oset M, Łubek A, Kukwa M. (2017) Lecanorastanislai, a new, sterile, usnic acid containing lichen species from Eurasia and North America. Phytotaxa 329(3): 201–211. 10.11646/phytotaxa.329.3.1 [DOI] [Google Scholar]
- Guzow-Krzemińska B, Jabłońska A, Flakus A, Rodriguez-Flakus P, Kosecka M, Kukwa M. (2019a) Phylogenetic placement of Leprariacryptovouauxii sp. nov. (Lecanorales, Lecanoromycetes, Ascomycota) with notes on other Lepraria species from South America. MycoKeys 53: 1–22. 10.3897/mycokeys.53.33508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzow-Krzemińska B, Flakus A, Kosecka M, Jabłońska A, Rodriguez-Flakus P, Kukwa M. (2019b) New species and records of lichens from Bolivia. Phytotaxa 397(4): 257–279. 10.11646/phytotaxa.397.4.1 [DOI] [Google Scholar]
- Helms G, Friedl T, Rambold G, Mayrhofer H. (2001) Identification of photobionts from the lichen family Physciaceae using algal-specific ITS rDNA sequencing. Lichenologist 33(1): 73–86. 10.1006/lich.2000.0298 [DOI] [Google Scholar]
- Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. (2018) UFBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35(2): 518–522. 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodkinson BP, Lendemer JC. (2013) Next-generation sequencing reveals sterile crustose lichen phylogeny. Mycosphere 4(6): 1028–1039. 10.5943/mycosphere/4/6/1 [DOI] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17(8): 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. (2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14(6): 587–589. 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantvilas G, Kukwa M. (2006) A new species of Lepraria (lichenized Ascomycetes) from Tasmania’s wet forests. Muelleria 23: 3–6. 10.5962/p.291578 [DOI] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT: multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistenich S, Timdal E, Bendiksby M, Ekman S. (2018) Molecular systematics and character evolution in the lichen family Ramalinaceae (Ascomycota: Lecanorales). Taxon 67(5): 871–904. 10.12705/675.1 [DOI] [Google Scholar]
- Kosecka M, Guzow-Krzemińska B, Černajová I, Škaloud P, Jabłońska A, Kukwa M. (2021) New lineages of photobionts in Bolivian lichens expand our knowledge on habitat preferences and distribution of Asterochloris algae. Scientific Reports 11(1): e8701. 10.1038/s41598-021-88110-0 [DOI] [PMC free article] [PubMed]
- Kroken S, Taylor JW. (2001) A gene genealogical approach to recognize phylogenetic species boundaries in the lichenized fungus Letharia. Mycologia 93(1): 38–53. 10.1080/00275514.2001.12061278 [DOI] [Google Scholar]
- Kukwa M. (2002) Taxonomic notes on the lichen genera Lepraria and Leproloma. Annales Botanici Fennici 39: 225–226. [Google Scholar]
- Kukwa M. (2006) Notes on taxonomy and distribution of the lichen species Leprariaecorticata comb. nov. Mycotaxon 97: 63–66. [Google Scholar]
- Kukwa M. (2019) Leprariajuanfernandezii, a new lichen species from the Southern Hemisphere. Plant and Fungal Systematics 64(2): 233–235. 10.2478/pfs-2019-0019 [DOI] [Google Scholar]
- Kukwa M, Pérez-Ortega S. (2010) A second species of Botryolepraria from the Neotropics and the phylogenetic placement of the genus within Ascomycota. Mycological Progress 9(3): 345–351. 10.1007/s11557-009-0642-0 [DOI] [Google Scholar]
- Landan G, Graur D. (2008) Local reliability measures from sets of co-optimal multiple sequence alignments. Pacific Symposium on Biocomputing 13: 15–24. [PubMed] [Google Scholar]
- Laundon JR. (1989) The species of Leproloma-the name for the Leprariamembranacea group. Lichenologist 21(1): 1–22. 10.1017/S0024282989000034 [DOI] [Google Scholar]
- Laundon JR. (1992) Lepraria in the British Isles. Lichenologist 24(4): 315–350. 10.1017/S002428299200046X [DOI] [Google Scholar]
- Lendemer JC. (2011a) A taxonomic revision of the North American species of Lepraria s.l. that produce divaricatic acid, with notes on the type species of the genus L.incana. Mycologia 103(6): 1216–1229. 10.3852/11-032 [DOI] [PubMed] [Google Scholar]
- Lendemer JC. (2011b) A standardized morphological terminology and descriptive scheme for Lepraria (Stereocaulaceae). Lichenologist 43(5): 379–399. 10.1017/S0024282911000326 [DOI] [Google Scholar]
- Lendemer JC. (2012) Perspectives on chemotaxonomy: Molecular data confirm the existence of two morphologically distinct species within a chemically defined Leprariacaesiella (Stereocaulaceae). Castanea 77(1): 89–105. 10.2179/11-042 [DOI] [Google Scholar]
- Lendemer JC. (2013a) A monograph of the crustose members of the genus Lepraria Ach. s. str. (Stereocaulaceae, Lichenized Ascomycetes) in North America north of Mexico. Opuscula Philolichenum 12(1): 27–141. [Google Scholar]
- Lendemer JC. (2013b) Shifting paradigms in the taxonomy of lichenized fungi: Molecular phylogenetic evidence corroborates morphology but not chemistry in the Leprarianeglecta group. Memoirs of the New York Botanical Garden 108: 127–153. [Google Scholar]
- Lendemer JC, Hodkinson BP. (2013) A radical shift in the taxonomy of Lepraria s.l.: Molecular and morphological studies shed new light on the evolution of asexuality and lichen growth form diversification. Mycologia 105(4): 994–1018. 10.3852/12-338 [DOI] [PubMed] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: Evidence from an RNA Polymerase II Subunit. Molecular Biology and Evolution 16(12): 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Lücking R, Hodkinson BP, Leavitt SD. (2017 [2016]) The 2016 classification of lichenized fungi in the Ascomycota and Basidiomycota – Approaching one thousand genera. The Bryologist 119(4): 361–416. 10.1639/0007-2745-119.4.361 [DOI] [Google Scholar]
- Lumbsch HT, Ahti T, Altermann S, Amo De Paz G, Aptroot A, Arup U, Bárcenas Peña A, Bawingan PA, Benatti MN, Betancourt L, Björk CR, Boonpragob K, Brand M, Bungartz F, Cáceres MES, Candan M, Chaves JL, Clerc P, Common R, Coppins BJ, Crespo A, Dal-Forno M, Divakar PK, Duya MV, Elix JA, Elvebakk A, Fankhauser JD, Farkas E, Itatí Ferraro L, Fischer E, Galloway DJ, Gaya E, Giralt M, Goward T, Grube M, Hafellner J, Hernández M JE, Herrera Campos MA, Kalb K, Kärnefelt I, Kantvilas G, Killmann D, Kirika P, Knudsen K, Komposch H, Kondratyuk S, Lawrey JD, Mangold A, Marcelli MP, McCune B, Messuti MI, Michlig A, Miranda González R, Moncada B, Naikatini A, Nelsen MP, Øvstedal DO, Palice Z, Papong K, Parnmen S, Pérez-Ortega S, Printzen C, Rico VJ, Rivas Plata E, Robayo J, Rosabal D, Ruprecht U, Salazar Allen N, Sancho L, Santos De Jesus L, Santos Vieira T, Schultz M, Seaward MRD, Sérusiaux E, Schmitt I, Sipman HJM, Sohrabi M, Søchting U, Søgaard MZ, Sparrius LB, Spielmann A, Spribille T, Sutjaritturakan J, Thammathaworn A, Thell A, Thor G, Thüs H, Timdal E, Truong C, Türk R, Umaña Tenorio L, Upreti DK, van den Boom P, Vivas Rebuelta M, Wedin M, Will-Wolf S, Wirth V, Wirtz N, Yahr R, Yeshitela K, Ziemmeck F, Wheeler T, Lücking R. (2011) One hundred new species of lichenized fungi: a signature of undiscovered global diversity. Phytotaxa 18: 1–127. 10.11646/phytotaxa.18.1.1 [DOI] [Google Scholar]
- Malíček J, Palice Z, Vondrák J, Łubek A, Kukwa M. (2018) Bacidiaalbogranulosa (Ramalinaceae, lichenized Ascomycota), a new sorediate lichen from European old-growth forests. MycoKeys 44: 51–62. 10.3897/mycokeys.44.30199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environments Workshop (GCE). 14 Nov. 2010. New Orleans Convention Center, New Orleans, 8 pp. 10.1109/GCE.2010.5676129 [DOI] [Google Scholar]
- Nelsen MP, Lücking R, Mbatchou JS, Andrew CJ, Spielmann AA, Lumbsch TH. (2011) New insights into relationships of lichen-forming Dothideomycetes. Fungal Diversity 51(1): 155–162. 10.1007/s13225-011-0144-7 [DOI] [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32(1): 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orange A. (2020) Lithocalla (Ascomycota, Lecanorales), a new genus of leprose lichens containing usnic acid. Lichenologist 52(6): 425–435. 10.1017/S0024282920000419 [DOI] [Google Scholar]
- Orange A, James PW, White FJ. (2001a) Microchemical Methods for the Identification of Lichens. British Lichen Society, London, 101 pp. [Google Scholar]
- Orange A, Wolseley P, Karunaratne V, Bombuwala K. (2001b) Two leprarioid lichens new to Sri Lanka. Bibliotheca Lichenologica 78: 327–333. [Google Scholar]
- Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T. (2010) GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Research 38: W23–W28. 10.1093/nar/gkq443 [DOI] [PMC free article] [PubMed]
- Poelt J. (1987) On the reduction of morphological structures in lichens. Bibliotheca Lichenologica 25: 35–45. [Google Scholar]
- Rambaut A. (2009) FigTree ver. 1.4.3. http://tree.bio.ed.ac.uk/software/figtree [Accessed 4 Oct. 2016]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. (2018) Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Systematic Biology 67(5): 901–904. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12): 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Saag L, Saag A, Randlane T. (2009) World survey of the genus Lepraria (Stereocaulaceae, lichenized Ascomycota). Lichenologist 41(1): 25–60. 10.1017/S0024282909007993 [DOI] [Google Scholar]
- Sela I, Ashkenazy H, Katoh K, Pupko T. (2015) GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Research 43: W7–W14. 10.1093/nar/gkv318 [DOI] [PMC free article] [PubMed]
- Sipman HJM. (2004) Survey of Lepraria species with lobed thallus margins in the tropics. Herzogia 17: 23–35. [Google Scholar]
- Sipman HJM. (2018) Three new lichen species and 48 new records from Vanuatu. Australasian Lichenology 82: 106–129. [Google Scholar]
- Tønsberg T. (1992) The sorediate and isidiate, corticolous, crustose lichens in Norway. Sommerfeltia 14(1): 1–331. 10.2478/som-1992-0002 [DOI] [Google Scholar]
- van den Boom P, Magain N. (2020) Three new lichen species from Macaronesia belonging in Ramalinaceae, with the description of a new genus. Plant and Fungal Systematics 65(1): 167–175. 10.35535/pfsyst-2020-0011 [DOI] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayawardene NN, Hyde KD, Rajeshkumar KC, Hawksworth DL, Madrid H, Kirk PM, Braun U, Singh RV, Crous PW, Kukwa M, Lücking R, Kurtzman CP, Yurkov A, Haelewaters D, Aptroot A, Lumbsch HT, Timdal E, Ertz D, Etayo J, Phillips AJL, Groenewald JZ, Papizadeh M, Selbmann L, Dayarathne MC, Weerakoon G, Jones EBG, Suetrong S, Tian Q, Castañeda-Ruiz RF, Bahkali AH, Pang K-L, Tanaka K, Qin DD, Sakayaroj J, Hujslová M, Lombard L, Shenoy BD, Suija A, Maharachchikumbura SSN, Thambugala KM, Wanasinghe DN, Sharma BO, Gaikwad S, Pandit G, Zucconi L, Onofri S, Egidi E, Raja HA, Kodsueb R, Cáceres MES, Pérez-Ortega S, Fiuza PO, Monteiro SJ, Vasilyeva LN, Shivas RG, Prieto M, Wedin M, Olariaga I, Lateef AA, Agrawal Y, Fazeli SAS, Amoozegar MA, Zhao GZ, Pfliegler WP, Sharma G, Oset M, Abdel-Wahab MA, Takamatsu S, Bensch K, de Silva NI, De Kesel A, Karunarathna A, Boonmee S, Pfister DH, Lu Y-Z, Luo Z-L, Boonyuen N, Daranagama DA, Senanayake IC, Jayasiri SC, Samarakoon MC, Zeng X-Y, Doilom M, Quijada L, Rampadarath S, Heredia G, Dissanayake AJ, Jayawardana RS, Perera RH, Tang LZ, Phukhamsakda C, Hernández-Restrepo M, Ma X, Tibpromma S, Gusmao LFP, Weerahewa D, Karunarathna SC. (2017) Notes for genera: Ascomycota. Fungal Diversity 86(1): 1–594. 10.1007/s13225-017-0386-0 [DOI] [Google Scholar]
- Zoller S, Scheidegger C, Sperisen C. (1999) PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. Lichenologist 31(5): 511–516. 10.1006/lich.1999.0220 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The PCR parameters
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Beata Guzow-Krzemińska, Magdalena Kosecka
Data type
PCR parameters (word document)
Explanation note
The PCR parameters are presented for each marker.