Abstract
Thelebolales are globally distributed fungi with diverse ecological characteristics. The classification of Thelebolales remains controversial to date and this study introduces two new taxa, based on morphological and phylogenetic analyses. The results of phylogenetic analyses indicated that the new taxa formed distinct lineages with strong support that were separated from the other members of Thelebolales. The new taxa described herein did not form sexual structures. The phylogenetic relationships of the new taxa and the morphological differences between these taxa and the other species under Thelebolales are also discussed.
Keywords: Leotiomycetes, taxonomy, Thelebolales, two new taxa
Introduction
Eriksson and Winka (1997) established the class Leotiomycetes to accommodate the inoperculate discomycetes. Members of this class are ecologically diverse and include saprophytic fungi, endophytic fungi, plant and mammalian pathogens, aquatic and aerial filamentous fungi, mycorrhizal fungi, fungal parasites, root symbionts and wood-rotting fungi, of which the lattermost group mostly includes saprophytic fungi that grow on various substrates (Ekanayaka et al. 2019; Johnston et al. 2019). The order Thelebolales comprises important members of Leotiomycetes due to their diverse functions and potential applications (Hassan et al. 2017; Batista et al. 2020). Thelebolales was established by Haeckel in 1894; however, the classification of this order remains controversial to date (Ekanayaka et al. 2019; Johnston et al. 2019; Batista et al. 2020; Quijada et al. 2022). According to Johnston et al. (2019) and Batista et al. (2020), Thelebolales comprises Pseudeurotiaceae and Thelebolaceae. However, Ekanayaka et al. (2019) reported that Pseudeurotiaceae was nested within Thelebolaceae; thus, the former was regarded as a synonym of the latter. Recently, the work of Quijada et al. (2022) showed that Thelebolaceae is monophyletic and valid, whereas Pseudeurotiaceae is polyphyletic and includes multiple clades and established the Holwayaceae (i.e. Alatospora-Miniancora clade, Ekanayaka et al. (2019)).
The genus Pseudogymnoascus was established by Raillo in 1929; however, a type strain was formally specified during the establishment of the genus. Several years later, Samson (1972) designated Pseudogymnoascusroseus Raillo the neotype of Pseudogymnoascus, as CBS 395.65 could still be cultivated. At present, the genus Pseudogymnoascus comprises 17 valid species (Zhang et al. 2020b; Villanueva et al. 2021; Zhang et al. 2021) belonging to 13 clades (Minnis and Lindner 2013). The genus Pseudogymnoascus comprises a diverse group of fungi that are widely distributed on Earth and are highly ecologically diverse.
In this study, two new taxa belonging to the order Thelebolales were isolated in a survey on fungi from urban soil samples in China. This study provides a description, illustrations and a phylogenetic tree for the two new species isolated herein.
Materials and methods
Fungal isolation and morphology
Soil samples were collected from Cengong County (27°16'98"N, 108°81'46"E) in Kaili City, Guizhou Province, China by Zhi-Yuan Zhang in June 2020. The soil samples were collected from a depth of 3–10 cm from the soil surface. The fungi were isolated using the dilution plate method (Li et al. 2022). Briefly, 2 g of each of the collected samples was suspended in 20 ml of sterile water in a 50 ml sterile conical flask. The conical flasks were thoroughly shaken using a Vortex vibration meter. The suspension was then diluted to a concentration of 10-4. Then, 1 ml of the diluted sample was transferred to a sterile Petri dish, following which modified SDA medium (1 g dextrose, 20 g peptone, 20 g agar, and 1 litre ddH2O) containing 50 mg/l penicillin and 50 mg/l streptomycin was added and mixed. The experiment was performed in three replicates. The plates were incubated at 25 °C for 1–2 weeks and single colonies were selected from the plates and inoculated on to new potato dextrose agar (PDA) plates.
The isolates of potentially new species were transferred to a new plate containing PDA, malt extract agar (MEA), oatmeal agar (OA) and corn meal agar (CMA) and incubated in the dark at 25 °C for 14 days. Photomicrographs of the diagnostic structures were prepared using an OLYMPUS BX53 microscope, equipped with differential interference contrast (DIC) optics, an OLYMPUS DP73 high-definition colour camera and cellSens software v.1.18. The dry and living cultures were deposited at the Institute of Fungus Resources, Guizhou University, Guiyang City, Guizhou, China (GZUIFR).
DNA extraction, PCR amplification and sequencing
The total DNA was extracted using 5% chelex-100 solution. The internal transcribed spacer (ITS), nuclear large subunit (LSU) rDNA, DNA replication licensing factor (MCM7), RNA polymerase II second largest subunit (RPB2) and the translation elongation factor EF-1α (EF1A) were amplified and sequenced according to the method described by Minnis and Lindner (2013). The sequences of the primers used for amplifying these loci are listed in Suppl. material 1: table S1. The novel sequences identified in this study were deposited in GenBank (Suppl. material 1: table S2).
Phylogenetic analyses
The ITS, LSU, MCM7, RPB2 and EF1A sequences were retrieved from GenBank, based on previous studies by Zhang et al. (2020b, 2021) and Villanueva et al. (2021) (Suppl. material 1: table S2). The following two datasets were used in this study: (1) the ITS + LSU dataset was used for inferring the phylogenetic placement of the two novel taxa under the order Thelebolales and (2) the ITS + LSU + MCM7 + RPB2 + EF1A dataset was used for inferring the phylogenetic placement of the new species.
The TBtools software was used for simplifying the nomenclature and renaming (Chen et al. 2020). A single-locus dataset was aligned and edited using MAFFT v.7.037b (Katoh and Standley 2013) and MEGA v.6.06 (Tamura et al. 2013). The “Concatenate Sequence” function in PhyloSuite v1.16 (Zhang et al. 2020a) was used for concatenating each locus. The best-fit substitution model was selected using the corrected Akaike Information Criterion (AICc) in ModelFinder (Kalyaanamoorthy et al. 2017). The combined loci were analysed using the Bayesian Inference (BI) and Maximum Likelihood (ML) methods. The results of ML analysis were implemented in IQ-TREE v.1.6.11 (Nguyen et al. 2015) with 104 bootstrap (BS) tests, using the ultrafast algorithm (Minh et al. 2013). BI analysis was performed with MrBayes v.3.2 (Ronquist et al. 2012) and the Markov Chain Monte Carlo (MCMC) simulations were executed for 108 generations with a sampling frequency every 103 generations and a burn-in of 25%. All the aforementioned analyses were performed in PhyloSuite v.1.16 (Zhang et al. 2020a).
Results
Phylogenetic analyses
The concatenated alignment of ITS + LSU sequences primarily from the genera under the order Thelebolales comprised 1,209 nucleotides, including inserted gaps (ITS: 433 bp, LSU: 776 bp). The concatenated ITS + LSU + MCM7 + RPB2 + EF1A dataset from Pseudogymnoascus and its related taxa comprised 2,981 nucleotides, including inserted gaps (ITS: 430 bp, LSU: 790 bp, MCM7: 475 bp, RPB2: 525 bp and EF1A: 761 bp). The best-fit evolutionary models obtained by ML and BI analyses of each locus are listed in Suppl. material 1: table S3.
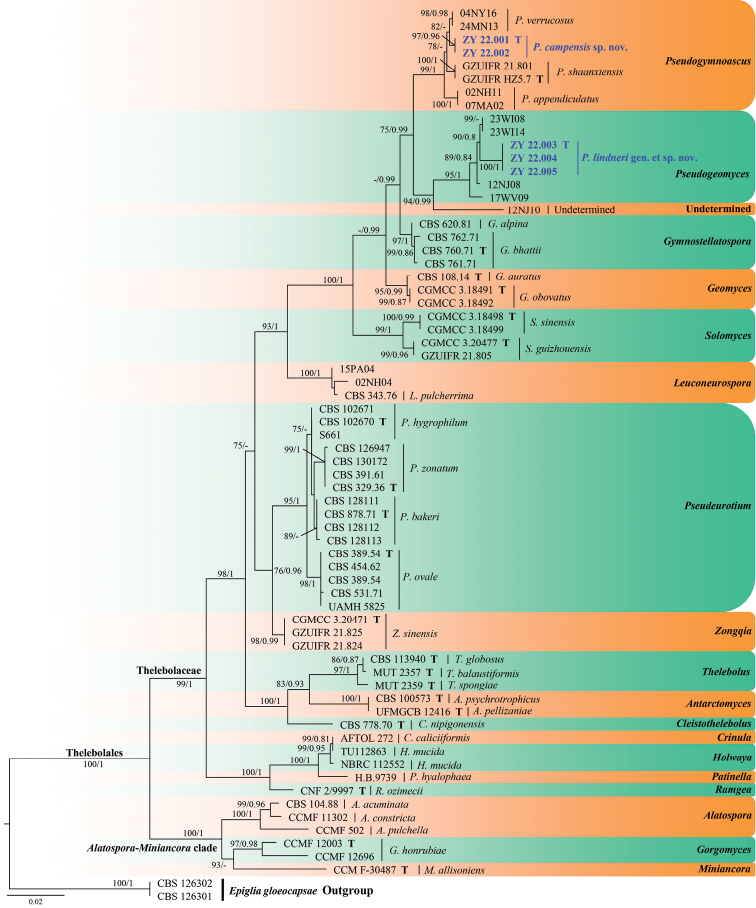
The clades formed by the genera in the first phylogenetic tree (Fig. 1) had a high support rate (Pseudogymnoascus (100% BS support [BS]/1 posterior probability [PP]), Solomyces (100% BS/1 PP), Pseudogeomyces (100% BS/1 PP), Geomyces (100% BS/1 PP), Pseudeurotium (100% BS/1 PP) and Zongqia (100% BS/1 PP)). The unidentified isolate, 12NJ10, formed a single clade (clade N; Minnis and Lindner (2013)) and was separated from the clades formed by the other genera. The new isolates identified in this study were divided into two genera, of which two isolates clustered under the genus Pseudogymnoascus and three isolates were clustered under the new genus, Pseudogeomyces.
Figure 1.
Phylogram generated from a Maximum Likelihood analysis of sequences of Thelebolales, based on ITS and LSU. ML bootstrap values (≥ 75%) and Bayesian posterior probability (≥ 0.75) are indicated along branches (BP/ML). The new taxa are highlighted in bold and blue and “T” indicate ex-type cultures.
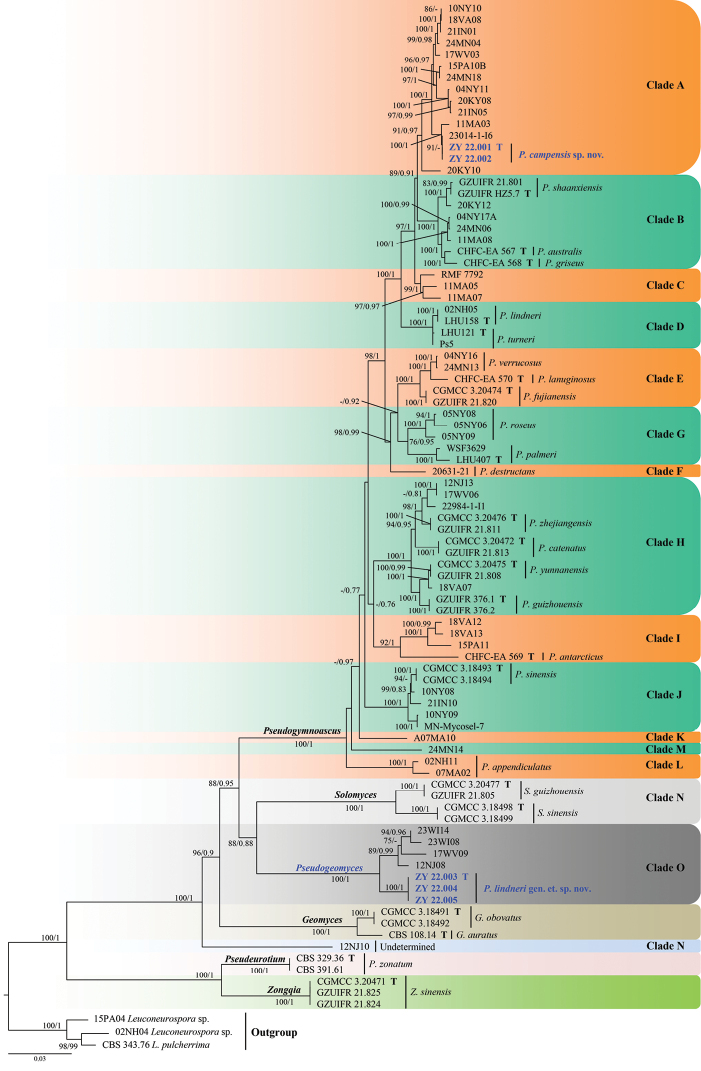
The genera in the second phylogenetic tree (Fig. 2) clustered into monophyletic clades with high support value. The new isolates (ZY 22.003, ZY 22.004 and ZY 22.005) under the new genus, Pseudogeomyces, clustered together with the other unidentified four isolates (12NJ08, 17WV09, 23WI14 and 23WI08) in a well-supported clade (100% BS /1 PP) that was separated from the other clades under Thelebolales. The new isolates, ZY 22.001 and ZY 22.002, belonging to the new species, Pseudogymnoascuscampensis, were clustered into a single clade with high support value (97% BS/0.96 PP) under the genus Pseudogymnoascus.
Figure 2.
Phylogram generated from A Maximum Likelihood analysis of sequences of Thelebolaceae, based on ITS, LSU, EF1A, RPB2 and MCM7. ML bootstrap values (≥ 75%) and Bayesian posterior probability (≥ 0.75) are indicated along branches (BP/ML). Clades are identified using clade nomenclature (A to O) formally defined by Minnis and Lindner (2013). The new taxa are highlighted in bold and blue and “T” indicate ex-type cultures.
Taxonomy
. Pseudogeomyces
Zhi.Y. Zhang & Y.F. Han gen. nov.
C875A466-442A-5911-9AB9-2804DDC232FD
846356
Etymology.
Referring to its similarity to Geomyces.
Geographical distribution.
China and the USA.
Description.
Saprobic on the soil. Sexual morph: not observed. Asexual morph: Hyphae branched, septate, smooth. Conidiophores solitary, rare branches, hyaline, smooth, arising from the erect or geniculated hyphae, usually bearing two to three branches at the tip. Conidia hyaline, rough, verrucosa, solitary, obovoid, globose to subglobose, borne on hyphae, short protrusions, side branches or in conidiophores separated by connective cells. Intercalary conidia hyaline, globose to subglobose, fusiform with both truncate. Chlamydospores not observed.
Type species.
Pseudogeomyceslindneri Zhi. Y. Zhang & Y. F. Han.
Notes.
Pseudogeomycesis is introduced to accommodate Pseudogeomycesislindneri obtained from urban soil in China and the four isolates (12NJ08, 17WV09, 23WI14 and 23WI08) obtained from bat hibernacular soil in New Jersey, West Virginia and Wisconsin, USA (Minnis and Lindner 2013). Unfortunately, these isolates have not been identified to species to date. Currently, the order Thelebolales consists of 24 genera (Wijayawardene et al. 2017; Ekanayaka et al. 2019; Zhang et al. 2021). The results of phylogenetic analyses (Figs 1, 2) revealed that Pseudogeomycesis formed a distinct clade with high support value. However, Ascophanus, Ascozonus, Caccobius, Coprobolus, Leptokalpion, Neelakesa and Pseudascozonus are lacking sequence data (Ekanayaka et al. 2019; https://www.ncbi.nlm.nih.gov/, retrieval in Oct 2022); thus, these genera were not included in our phylogenetic analysis. Besides, these genera were reported without asexual morphs (Wijayawardene et al. 2017). Therefore, it was not possible to compare the morphological differences of the newly-established genus, Pseudogeomycesis (sexual stage not observed), with the aforementioned genera. However, members of these genera are saprobes (involving dung and wood), terrestrial and widely distributed (Wijayawardene et al. 2017). Of the remaining genera, Pseudogeomyces were similar to Geomyces and the asexual morphs of Pseudogymnoascus. However, Pseudogeomyces differed from Geomyces and Pseudogymnoascus with the presence of two to three irregular branches at the tip of the conidiophores (Kuehn 1958; Van Oorschot 1980).
. Pseudogeomyces lindneri
Zhi. Y. Zhang & Y. F. Han sp. nov.
C915DF68-44FD-52CA-A324-3BC38B91EDB5
846365
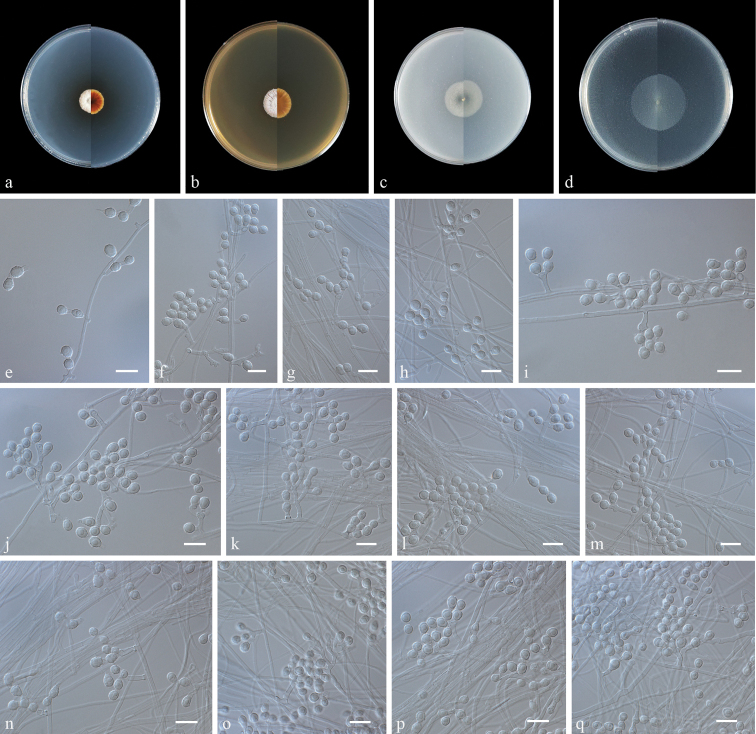
Figure 3.
Morphology of Pseudogeomyceslindneri sp. nov. a–d colony on PDA, MEA, OA and CMA after 14 d at 25 °C (upper surface and lower surface) e–q Conidiophore, Conidia and Intercalary conidia. Scale bars: 10 mm (a–d); 10 μm (e–q).
Etymology.
Named after Daniel Lindner, for acknowledging his contributions to the modern taxonomy of Pseudogymnoascus and its related taxa.
Type.
Kaili City, Guizhou Province, China 27°16'98"N, 108°81'46"E, isolated from the green belt soil, July 2022, Zhi-Yuan Zhang (holotype ZY H-22.003, ex-type ZY 22.003, ibid., ZY 22.004).
Geographical distribution.
Guizhou Province, China.
Description.
Culture characteristics (14 days at 25 °C): Colonies on PDA 15–16 mm in diameter, white to pale pink, raised, fluffy, irregular, producing abundant caesious exudates; reverse: brown to cinnamon. Colonies on MEA 18–19 mm in diameter, off-white, felty, with radial grooves, nearly round, exudates and diffusible pigments absent; reverse: brown to cinnamon. Colonies on OA 25–26 mm in diameter, white, aerial mycelia sparse, flat, nearly round, exudates and diffusible pigments absent; reverse: white. Colonies on CMA 34–35 mm in diameter, white, aerial mycelia sparse, flat, nearly round, margin regular, exudates and diffusible pigments absent; reverse: white.
Hyphae hyaline, smooth, branched, septate, 1.0–2.0 μm in diameter. Conidiophores solitary, rare branches, hyaline, smooth, arising from erect or geniculated hyphae, sometimes reduced to conidiogenous cells, erect, usually bearing two to four conidiogenous cells at the tip. Conidia hyaline, rough, verrucosa, solitary, obovoid, globose to subglobose, 3.0–7.5 × 2.5–5.5 µm (av. 4.8 × 3.8, n = 50), borne on hyphae, short protrusions, side branches or in conidiophores separated by connective cells. Intercalary conidia hyaline, globose to subglobose, fusiform, with both truncate 3.5–6.5 × 3.0–4.5 µm (av. 4.9 × 4.0, n = 50). Chlamydospores not observed. Sexual morph undetermined.
Notes.
Based on multi-locus phylogenetic analyses (Figs 1, 2) and morphological characteristics, Pseudogeomyceslindneri is proposed as the type species of Pseudogeomyces. The isolates ZY 22.003, ZY 22.004 and ZY 22.005 formed a single phylogenetic clade and were separated from the other four unidentified isolates (12NJ08, 17WV09, 23WI14 and 23WI08) under Pseudogeomyces. Morphologically, Pseudoge.lindneri differed from other taxa under the family Thelebolaceae in terms of the presence of two to four irregular branches at the tip of the conidiophores and that the conidia and intercalary conidia are generally connected by connective cells in a chain (Kuehn 1958; Van Oorschot 1980).
. Pseudogymnoascus campensis
Zhi. Y. Zhang & Y. F. Han sp. nov.
EB746C2E-2B9A-5C5D-B7A5-CCD1E0A2AA33
846366
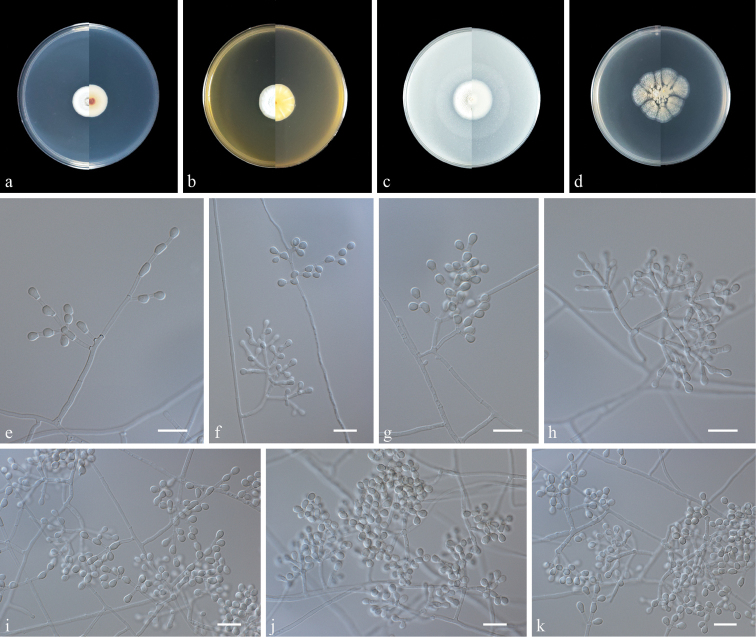
Figure 4.
Morphology of Pseudogymnoascuscampensis sp. nov. a–d colony on PDA, MEA, OA and CMA after 14 d at 25 °C (upper surface and lower surface) e, f fertile hyphae bearing arthroconidia and aleurioconidia g–k Conidiophore and Conidia. Scale bars: 10 μm (e–k).
Etymology.
Refers to Guizhou Minzu University where this fungal type was isolated.
Type.
Guizhou Minzu University, Guiyang City, Guizhou Province, China 26°37'57"N, 106°62'41"E. Colonies form on PDA as a contaminating fungus, July 2022, Zhi-Yuan Zhang (dried holotype ZY H-22.001, ex-type ZY 22.001, ibid., ZY 22.002).
Geographical distribution.
Guizhou Province, China.
Description.
Culture characteristics (14 days at 25 °C): Colonies on PDA 20–21 mm in diameter, white to light green, fluffy, nearly round, margin regular, exudates and diffusible pigments absent; reverse: claret-red to white from centre to margin. Colonies on MEA 23–24 mm in diameter, white, elevated at the centre, velvety to floccose, margin regular, exudates and diffusible pigments absent; reverse: pale yellow to white. Colonies on OA 27–28 mm in diameter, white, flat, nearly round, margin regular, exudates absent, producing a diffusible faint white pigment; reverse: white. Colonies on CMA 32–38 mm in diameter, khaki to white, radially sectored by cracks, powdery, exudates and diffusible pigments absent; reverse: khaki.
Hyphae hyaline, smooth, branched, septate, 1.0–2.5 μm in diameter. Sometimes lateral hyphae end in barrel-, reniform- or pyriform-shaped chains with blunt-ended arthroconidia, sometimes bearing aleurioconidia, sessile or stalked. Conidiophores abundant, solitary, erect, arising in acute angles with the main axis, hyaline, smooth, usually bearing verticils of two to three branches arising from the stipe at an acute angle. Aleurioconidia pyriform or obovoid, with a broad truncated basal scar, 3.0–5.0 × 2.0–2.5 µm (av. 3.6 × 2.7, n = 50), in conidiophores separated by connective cells, smooth or rough. Intercalary conidia barrel, reniform, pyriform to elongated or irregular, with a broad truncated scar at the base or both ends, 3.5–5.5 × 2.0–3.0 µm (av. 4.0 × 2.6, n = 50), smooth or rough. Arthroconidia not observed. Sexual morph unknown.
Notes.
Minnis and Lindner (2013) proposed multiple clades of Pseudogymnoascus and allies (clades A to O), based on phylogenetic analyses using North American isolates. In this study, Pseudogymnoascuscampensis was placed in clade A (Fig. 1). Clade A harbours 13 isolates for which no morphological data are yet available and remain as unidentified species to date (Minnis and Lindner 2013; Leushkin et al. 2015). These isolates were obtained from bat hibernacular soil in the USA (Minnis and Lindner 2013). Pseudogymnoascuscampensis (ZY 22.001 and ZY 22.002), 23014-1-I6 and 11MA03 formed an independent lineage with strong support (MLBS 100/PP 1, Fig. 1). The closest known species to Pseudogy.campensis are Pseudogy.shaanxiensis, Pseudogy.australis and Pseudogy.griseus, which are members of the neighbouring clade B (Zhang et al. 2020b, Villanueva et al. 2021). However, Pseudogy.campensis can be distinguished from Pseudogy.shaanxiensis, Pseudogy.australis and Pseudogy.griseus by the absence of exudates on PDA, MEA and CMA media and lack of arthroconidia (Zhang et al. 2020b; Villanueva et al. 2021).
Discussion
Previously, Minnis and Lindner (2013) performed a phylogenetic analysis, based on numerous multi-loci sequences of Pseudogymnoascus and its allies isolated from North American cave soils and obtained very robust results. However, many of the isolates obtained in the study were not identified as species. Based on their work, we subsequently defined Pseudogymnoascus and its allies isolated from China and reported two new genera and several new species (Zhang et al. 2020b, 2021). Similarly, Villanueva et al. (2021) identified four strains isolated from Antarctica, based on the above study and found that they were all previously undescribed species. In this study, one new genus and one new species are being proposed, based on the aforementioned study.
The classification of Thelebolales remains controversial to date (Ekanayaka et al. 2019; Johnston et al. 2019; Batista et al. 2020; Quijada et al. 2022). In contrast, however, the work of Ekanayaka et al. (2019) contained more genera in Thelebolales; therefore, we continued the phylogenetic analysis in Thelebolales, based on this study. This study, based on ITS+LSU phylogenetic analyses, showed that Thelebolales consisted of Thelebolaceae and Alatospora-Miniancora clade (Fig. 1), which is consistent with Ekanayaka et al. (2019). Our proposed new genus Pseudogeomyces was nested in Thelebolaceae and is well supported (Fig. 1).
The ITS region is the most frequently used molecular marker in fungal classification studies, primarily due to its suitable variability. Additionally, Vu et al. (2019) demonstrated the high efficacy of ITS and LSU concatenation in discriminating filamentous fungal species. Numerous fungal ITS and LSU sequences are presently available in public databases (Zhang et al. 2022). Additionally, some fungal taxa, including the majority of genera under Thelebolales, have only ITS and/or LSU regions. Therefore, we only explored the position of the new genus, Pseudogeomyces, in Thelebolales, based on the phylogenetic analysis of ITS + LSU sequences.
In accordance with the most recent revision to the rules governing fungal nomenclature, presently referred to as the “International Code of Nomenclature for algae, fungi and plants”, the system of dual nomenclature sanctioned by Article 59 has been modified to ‘‘One Fungus, One Name’’ (McNeill et al. 2012), where a single name is applied, regardless of the life stage considered. Most of the new taxa erected in recent years under Pseudogymnoascus and allies are based on asexual structures rather than sexual structures (Zhang et al. 2020b; Villanueva et al. 2021; Zhang et al. 2021). In this study, the new isolates were separately cultured in four media for observing the sexual structures, but the approach proved unsuccessful. The sexual structures of fungi appear when grown in nature rather than under laboratory conditions. Therefore, studying the production of sexual structures by these fungi under laboratory conditions is highly necessary.
Supplementary Material
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (no. 32060011, 31860520) and the National Key R&D Program of China (Grant 2019YFD1002003). We appreciate Charlesworth for English language editing to the whole manuscript.
Citation
Zhang Z-Y, Han Y-F, Chen W-H, Tao G (2023) Additions to Thelebolales (Leotiomycetes, Ascomycota): Pseudogeomyces lindneri gen. et sp. nov. and Pseudogymnoascus campensis sp. nov. MycoKeys 95: 47–60. https://doi.org/10.3897/mycokeys.95.97474
Contributor Information
Zhi-Yuan Zhang, Email: zhangzhiyuan_16@163.com.
Gang Tao, Email: ttg729@sina.com.
Supplementary materials
Sequences of primers used for the amplification of molecular markers in this study. GenBank accession numbers of the sequences used in this study. The best-fit evolutionary model in the phylogenetic analyses
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Zhi-Yuan Zhang, Yan-Feng Han, Wan-Hao Chen, Gang Tao
Data type
table (word file)
References
- Batista TM, Hilário HO, de Brito GAM, Moreira RG, Furtado C, De Menezes GCA, Rosa CA, Rosa LH, Franco GR. (2020) Whole-genome sequencing of the endemic Antarctic fungus Antarctomycespellizariae reveals an ice-binding protein, a scarce set of secondary metabolites gene clusters and provides insights on Thelebolales phylogeny. Genomics 112(5): 2915–2921. 10.1016/j.ygeno.2020.05.004 [DOI] [PubMed] [Google Scholar]
- Chen CJ, Chen H, Zhang Y, Thomas HR, Frank MH, He YH, Xia R. (2020) TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant 13(8): 1194–1202. 10.1016/j.molp.2020.06.009 [DOI] [PubMed] [Google Scholar]
- Ekanayaka AH, Hyde KD, Gentekaki E, McKenzie EHC, Zhao Q, Bulgakov TS, Camporesi E. (2019) Preliminary classification of Leotiomycetes. Mycosphere 10(1): 310–489. 10.5943/mycosphere/10/1/7 [DOI] [Google Scholar]
- Eriksson OE, Winka K. (1997) Supraordinal taxa of Ascomycota. Myconet 1: 1–16. [Google Scholar]
- Hassan N, Rafiq M, Hayat M, Nadeem S, Shah AA, Hasan F. (2017) Potential of psychrotrophic fungi isolated from Siachen glacier, Pakistan, to produce antimicrobial metabolites. Applied Ecology and Environmental Research 15(3): 1157–1171. 10.15666/aeer/1503_11571171 [DOI] [Google Scholar]
- Johnston PR, Quijada L, Smith CA, Baral HO, Hosoya T, Baschien C, Pärtel K, Zhuang WY, Haelewaters D, Park D, Carl S, López-Giráldez F, Wang Z, Townsend JP. (2019) A multigene phylogeny toward a new phylogenetic classification of Leotiomycetes. IMA Fungus 10(1): 1. 10.1186/s43008-019-0002-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. (2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14(6): 587–589. 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn HH. (1958) A preliminary survey of the Gymnoascaceae. I. Mycologia 50(3): 417–439. 10.1080/00275514.1958.12024739 [DOI] [Google Scholar]
- Leushkin EV, Logacheva MD, Penin AA, Sutormin RA, Gerasimov ES, Kochkina GA, Ivanushkina NE, Vasilenko OV, Kondrashov AS, Ozerskaya SM. (2015) Comparative genome analysis of Pseudogymnoascus spp. reveals primarily clonal evolution with small genome fragments exchanged between lineages. BMC Genomics 16(1): 1. 10.1186/s12864-015-1570-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang ZY, Ren YL, Chen WH, Liang JD, Pan JM, Huang JZ, Liang ZQ, Han YF. (2022) Morphological characteristics and phylogenetic evidence reveal two new species of Acremonium (Hypocreales, Sordariomycetes). MycoKeys 91: 85–96. 10.3897/mycokeys.91.86257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill J, Buck WR, Demoulin V, Greuter W, Hawkworth DL, Herendeen PS, Knapp S, Marhold K, Prado J, Prud’homme van Reine WF, Smith GF, Wiersema JH, Turland NJ. (2012) International code of nomenclature for algae, fungi and plants (Melbourne code); adopted by the 18th International Botanical Congress, Melbourne, Australia, Jul 2011. Koenigstein, Germany: Koeltz Scientific Books.
- Minh Q, Nguyen M, von Haeseler AA. (2013) Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution 30(5): 1188–1195. 10.1093/molbev/mst024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnis AM, Lindner DL. (2013) Phylogenetic evaluation of Geomyces and allies reveals no close relatives of Pseudogymnoascusdestructans, comb. nov., in bat hibernacula of eastern North America. Fungal Biology 117(9): 638–649. 10.1016/j.funbio.2013.07.001 [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32(1): 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijada L, Matočec N, Kušan I, Tanney JB, Johnston PR, Mešić A, Pfister DH. (2022) Apothecial ancestry, evolution, and re-evolution in Thelebolales (Leotiomycetes, Fungi). Biology 11(4): 583. 10.3390/biology11040583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA. (1972) Notes on Pseudogymnoascus, Gymnoascus and related genera. Acta Botanica Neerlandica 21(5): 517–527. 10.1111/j.1438-8677.1972.tb00804.x [DOI] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30(12): 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oorschot CA. (1980) A revision of Chrysosporium and allied genera. Studies in Mycology 20: 1–89. [Google Scholar]
- Villanueva P, Vásquez G, Gil-Durán C, Oliva V, Díaz A, Henríquez M, Álvarez E, Laich F, Chávez R, Vaca I. (2021) Description of the first four species of the genus Pseudogymnoascus from Antarctica. Frontiers in Microbiology 12: 713189. 10.3389/fmicb.2021.713189 [DOI] [PMC free article] [PubMed]
- Vu D, Groenewald M, de Vries M, Gehrmann T, Stielow B, Eberhardt U, Al-Hatmi A, Groenewald JZ, Cardinali G, Houbraken J, Boekhout T, Crous PW, Robert V, Verkley GJM. (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology 92(1): 135–154. 10.1016/j.simyco.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayawardene NN, Hyde KD, Rajeshkumar KC, Hawksworth DL, Madrid H, Kirk PM, Braun U, Singh RV, Crous PW, Kukwa M, Lücking R, Kurtzman CP, Yurkov A, Haelewaters D, Aptroot A, Lumbsch HT, Timdal E, Ertz D, Etayo J, Phillips AJL, Groenewald JZ, Papizadeh M, Selbmann L, Dayarathne MC, Weerakoon G, Jones EBG, Suetrong S, Tian Q, Castañeda-Ruiz RF, Bahkali AH, Pang KL, Tanaka K, Dai DQ, Sakayaroj J, Hujslová M, Lombard L, Shenoy BD, Suija A, Maharachchikumbura SSN, Thambugala KM, Wanasinghe DN, Sharma BO, Gaikwad S, Pandit G, Zucconi L, Onofri S, Egidi E, Raja HA, Kodsueb R, Cáceres MES, Pérez-Ortega S, Fiuza PO, Monteiro JS, Vasilyeva LN, Shivas RG, Prieto M, Wedin M, Olariaga I, Lateef AA, Agrawal Y, Fazeli SAS, Amoozegar MA, Zhao GZ, Pfliegler WP, Sharma G, Oset M, Abdel-Wahab MA, Takamatsu S, Bensch K, de Silva NI, de Kesel A, Karunarathna A, Boonmee S, Pfister DH, Lu YZ, Luo ZL, Boonyuen N, Daranagama DA, Senanayake IC, Jayasiri SC, Samarakoon MC, Zeng XY, Doilom M, Quijada L, Rampadarath S, Heredia G, Dissanayake AJ, Jayawardana RS, Perera RH, Tang LZ, Phukhamsakda C, Hernández-Restrepo M, Ma X, Tibpromma S, Gusmao LFP, Weerahewa D, Karunarathna SC. (2017) Notes for genera: Ascomycota. Fungal Diversity 86(1): 1–594. 10.1007/s13225-017-0386-0 [DOI] [Google Scholar]
- Zhang D, Gao FL, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. (2020a) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources 20(1): 348–355. 10.1111/1755-0998.13096 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Dong C, Chen W, Mou Q, Lu X, Han Y, Huang J, Liang Z. (2020b) The enigmatic Thelebolaceae (Thelebolales, Leotiomycetes): One new genus Solomyces and five new species. Frontiers in Microbiology 11: 572596. 10.3389/fmicb.2020.572596 [DOI] [PMC free article] [PubMed]
- Zhang ZY, Shao QY, Li X, Chen WH, Liang JD, Han YF, Huang JZ, Liang ZQ. (2021) Culturable fungi from urban soils in China I: Description of 10 new taxa. Microbiology Spectrum 9(2): e00867–e21. 10.1128/Spectrum.00867-21 [DOI] [PMC free article] [PubMed]
- Zhang Z, Chen W, Liang Z, Zhang L, Han Y, Huang J, Liang Z. (2022) Revealing the non-overlapping characteristics between original centers and genetic diversity of Purpureocilliumlilacinum. Fungal Ecology 60: 101179. 10.1016/j.funeco.2022.101179 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of primers used for the amplification of molecular markers in this study. GenBank accession numbers of the sequences used in this study. The best-fit evolutionary model in the phylogenetic analyses
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Zhi-Yuan Zhang, Yan-Feng Han, Wan-Hao Chen, Gang Tao
Data type
table (word file)