Abstract
Species of Diaporthe have been reported as plant endophytes, pathogens and saprobes on a wide range of plant hosts. Strains of Diaporthe were isolated from leaf spots of Smilaxglabra and dead culms of Xanthiumstrumarium in China, and identified based on morphology and molecular phylogenetic analyses of combined internal transcribed spacer region (ITS), calmodulin (cal), histone H3 (his3), translation elongation factor 1-alpha (tef1) and β-tubulin (tub2) loci. As a result, two new species named Diaportherizhaoensis and D.smilacicola are identified, described and illustrated in the present study.
Keywords: Leaf spots, morphology, multi-gene phylogeny, taxonomy
Introduction
Diaporthe (Diaporthaceae,Diaporthales) is a species-rich genus with its asexual morph previously known as Phomopsis (Rossman et al. 2007; Udayanga et al. 2011, 2012a, 2014a, 2015; Dissanayake et al. 2017; Guarnaccia et al. 2018). The genus Diaporthe was established by Nitschke in 1870 and predates its sexual morph established in 1905, thus Diaporthe is recommended to be used for this genus following “one fungus one name” nomenclature (Nitschke 1870; Rossman et al. 2015).
The sexual morph of Diaporthe is characterized by immersed ascomata and an erumpent pseudostroma with single or multiple tapering perithecial necks. Asci are unitunicate, sessile and clavate to cylindrical. Ascospores are elliptical to fusiform, septate or aseptate, hyaline, biseriate to uniseriate in the ascus and sometimes have appendages (Udayanga et al. 2011; Senanayake et al. 2017, 2018). The asexual morph is characterized by black or dark brown conidiomata, with cylindrical phialides producing three types of aseptate and hyaline conidia (Type I: α-conidia, hyaline, fusiform, straight, guttulate or eguttulate, aseptate, smooth-walled; type II: β-conidia, hyaline, filiform, straight or hamate, aseptate, smooth-walled, eguttulate; type III: γ-conidia, rarely produced, hyaline, multiguttulate, fusiform to subcylindrical with an acute or rounded apex, while the base is sometimes truncate) (Udayanga et al. 2011; Gomes et al. 2013).
Species of Diaporthe are widely distributed, and infect a broad plant host range, e.g., agricultural crops, forest trees, vegetables, and fruits (Farr et al. 2002a, b; Crous 2005; Rossman et al. 2007; Udayanga et al. 2011, 2012a, b, 2014a, b, 2015; Gomes et al. 2013; Du et al. 2016; Dissanayake et al. 2017; Guarnaccia and Crous 2017; Fan et al. 2018). As plant pathogens, Diaporthe spp. cause severe diseases, e.g., blights, cankers, decay, dieback, leaf spots and wilt of many economically important plants in genera Castanea, Citrus, Helianthus, Macadamia, Rosa, Vaccinium and Vitis, resulting in major losses (Thompson et al. 2011; Huang et al. 2015; Guarnaccia et al. 2018, 2020; Hilário et al. 2020; Wrona et al. 2020; Caio et al. 2021; Jiang et al. 2021a).
The genus Diaporthe includes over 1000 epithets, mostly based on morphological characteristics and host associations (van der Aa et al. 1990; Santos et al. 2010; Guarnaccia et al. 2018). However, recent studies have shown that many species of Diaporthe are not host-specific, i.e., one species may infect more than one host species (Vrandecic et al. 2011; Bai et al. 2015; Zhang et al. 2018). And many Diaporthe species that are morphologically similar have proven to be genetically distinct (van Rensburg et al. 2006; Udayanga et al. 2011; Jiang et al. 2021b). Thus, polyphasic taxonomy is essential to identify and comprehensively characterize Diaporthe.
In the present study, we have analyzed five-locus dataset of combined nuclear ribosomal internal transcribed spacer (ITS), calmodulin (cal), histone (his3), translation elongation factor 1-alpha (tef1) and beta-tubulin (tub2). To aid the identification of two new species, we followed Norphanphoun et al. (2022) for the taxonomic treatments of Diaporthe. Norphanphoun et al. (2022) clustered Diaporthe into 13 workable species complexes namely D.arecae, D.biconispora, D.carpini, D.decedens, D.eres, D.oncostoma, D.pustulata, D.rudis, D.scobina, D.sojae, D.toxica, D.varians and D.vawdreyi species complexes. In addition, nine species were retained as singletons, viz., D.acerina, D.acutispora, D.crataegi, D.multiguttulata, D.ocoteae, D.perjuncta, D.pseudoalnea, D.spartinicola and D.undulata based on multilocus phylogeny.
In previous studies, Smilaxglabra and Xanthiumstrumarium have been reported as hosts of Diaporthe (Vrandecic et al. 2007, 2010; Gao et al. 2013; Thompson et al. 2018). D.eres (= D.mahothocarpi) and D.lithocarpi were identified as the cause agents of leaf spot disease based on morphology and phylogenetics on S.glabra in China (Gao et al. 2013). D.helianthi and D.longicolla, pathogens of X.strumarium, have been collected from blighted stems and branches in Croatia (Vrandecic et al. 2007, 2010). D.pseudolongicolla (= D.novem) has been reported as a branch dieback agent in X.strumarium in Australia (Thompson et al. 2018).
In this study, we introduce two new species namely Diaportherizhaoensis and D.smilacicola, collected from diseased plant tissues in China. We further provide descriptions, illustrations, and DNA sequence-based phylogeny to verify identification and placement.
Materials and methods
Isolation and morphological characterization
During 2021 and 2022, investigations were conducted to inspect for the presence of Diaporthe species associated with plant diseases in China. Leaves of Smilaxglabra and culms of Xanthiumstrumarium showing typical symptoms of Diaporthe were collected. Infected tissues were cut into 0.5 × 0.5 cm pieces using a double-edge blade, and surface sterilized as follows. These sections underwent initial immersion for 2 min in 0.5% sodium hypochlorite, followed by 1 min in sterile distilled water, 2 min in 75% ethanol, and, finally, 1 min in sterile distilled water. The disinfected fragments were then plated onto the surface of potato dextrose agar (PDA; 200 g potatoes, 20 g dextrose, 20 g agar per L) and malt extract agar (MEA; 30 g malt extract, 5 g mycological peptone, 15 g agar per L), and incubated at 25 °C to obtain the pure culture.
Species identification was based on morphological features of the new species produced on infected plant tissues and PDA plates. Conidiomata were sectioned by hand, using a double-edged blade and structures were observed under a dissecting microscope. Over 20 fruiting bodies were sectioned, and 50 conidia were selected randomly for measurement using Axio Imager 2 microscope (Zeiss, Oberkochen, Germany). Isolate characteristics incubated on PDA at 25 °C were observed and recorded at 7 days, including colony colour, texture and the arrangement of the conidiomata. The cultures were deposited in the China Forestry Culture Collection Center (CFCC; http://www.cfcc-caf.org.cn/), and the specimens in the herbarium of the Chinese Academy of Forestry (CAF; http://museum.caf.ac.cn/).
DNA extraction, amplification and sequencing
Genomic DNA was extracted from the fresh mycelium harvested from PDA plates after 7 days using a cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle 1990). For initial species confirmation, the internal transcribed spacer (ITS) region was sequenced for all isolates. The BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to compare the resulting sequences with those in GenBank. After confirmation of Diaporthe species, four additional partial loci, including calmodulin (cal), histone H3 (his3), partial translation elongation factor 1-alpha (tef1) and part of the beta-tubulin gene region (tub2) genes were amplified. The primer pairs and amplification conditions for each of the above-mentioned gene regions are provided in Table 1. A PCR reaction was conducted in a 20 µL reaction volume, and the components were as follows: 1 µL DNA template (20 ng/μl), 1 µL forward 10 µM primer, 1 µL reverse 10 µM primer, 10 µL T5 Super PCR Mix (containing Taq polymerase, dNTP and Mg2+, Beijing TisingKe Biotech Co., Ltd., Beijing, China), and 7 µL sterile water. Amplifications were performed using a T100 Thermal Cycler (Bio-Rad, Hercules, CA, USA). Strands were sequenced in both directions using PCR primers. All amplified PCR products were estimated visually 1.4% agarose gels stained with ethidium bromide and then PCR positive products were sent to Sangon Biotech (Shanghai) Co., Ltd., (Beijing, China) for sequencing.
Table 1.
Loci used in this study with PCR primers and process.
| Loci | PCR primers | PCR: thermal cycles: (Annealing temp. in bold) | Reference |
|---|---|---|---|
| ITS | ITS1/ITS4 | (95 °C: 30 s, 48 °C: 30 s, 72 °C: 1 min) × 35 cycles | White et al. 1990 |
| cal | CAL228F/CAL737R | (95 °C: 15 s, 54 °C: 20 s, 72 °C: 1 min) × 35 cycles | Carbone and Kohn 1999 |
| his3 | CYLH3F/H3-1b | (95 °C: 30 s, 57 °C: 30 s, 72 °C: 1 min) × 35 cycles | Crous et al. 2004 |
| Glass and Donaldson 1995 | |||
| tef1 | EF1-728F/EF1-986R | (95 °C: 15 s, 54 °C: 20 s, 72 °C: 1 min) × 35 cycles | Carbone and Kohn 1999 |
| tub2 | T1(Bt2a)/Bt2b | (95 °C: 30 s, 55 °C: 30 s, 72 °C: 1 min) × 35 cycles | Glass and Donaldson 1995 |
| O’Donnell and Cigelnik 1997 |
Phylogenetic analyses
Sequences were edited and condensed with SeqMan v.7.1.0. The sequences generated in this study were supplemented with additional sequences obtained from GenBank (Table 2) based on blast searches and recent publications of the genus Diaporthe. The sequences were aligned with the MAFFT v.7 after which the alignments were manually corrected using MEGA v. 7.0. (Katoh and Toh 2010; Kumar et al. 2016). Phylogenetic analyses including Maximum Likelihood (ML) and Bayesian Inference (BI) methods were conducted for the single gene sequence data sets of the ITS, cal, his3, tef1 and tub2, and the combined data set of all five gene regions. ML analyses were conducted using RAxML-HPC BlackBox 8.2.10 on the CIPRES Science Gateway portal (https://www.phylo.org) (Miller et al. 2012), employing a GTRGAMMA substitution model with 1000 bootstrap replicates (Stamatakis 2014). BI analyses were conducted using a Markov Chain Monte Carlo (MCMC) algorithm in MrBayes v.3.0 (Ronquist and Huelsenbeck 2003). Two Markov chain Monte Carlo (MCMC) chains were run from a random starting tree for 1,000,000 generations, resulting in a total of 10,000 trees. The first 25% of trees sampled were discarded as burn-in and the remaining trees were used to calculate the posterior probabilities. Branches with significant Bayesian Posterior Probabilities (BPP > 0.9) were estimated in the remaining 7,500 trees. Phylogenetic trees were viewed with FigTree v. 1.4 and processed by Adobe Illustrator CS5. The nucleotide sequence data of the new taxa were deposited in GenBank, and the GenBank accession numbers of all accessions included in the phylogenetic analyses are listed in Table 2.
Table 2.
Strains and GenBank accession numbers used in this study.
| Species | Location | Host | Strain | GenBank Accession Number | ||||
|---|---|---|---|---|---|---|---|---|
| ITS | tef1 | tub2 | cal | his3 | ||||
| Diaportheabsenteum | China | Camelliasinensis | LC3429* | KP267897 | KP267971 | KP293477 | NA | KP293547 |
| D.absenteum | China | Camelliasinensis | LC3564 | KP267912 | KP267986 | KP293492 | NA | KP293559 |
| D.acaciarum | Tanzania | Acaciatortilis | CBS 138862* | KP004460 | NA | KP004509 | NA | KP004504 |
| D.acericola | Italy | Acernegundo | MFLUCC 17-0956* | KY964224 | KY964180 | KY964074 | KY964137 | NA |
| D.aceris | Japan | Acer sp. | LC8112 | KY491547 | KY491557 | KY491567 | KY491575 | NA |
| D.actinidiae | New Zealand | Actinidiadeliciosa | ICMP 13683* | KC145886 | KC145941 | NA | NA | NA |
| D.acuta | China | Pyruspyrifolia | CGMCC 3.19600* | MK626957 | MK654802 | MK691225 | MK691124 | MK726161 |
| D.alangii | China | Alangiumkurzii | CFCC 52556* | MH121491 | MH121533 | MH121573 | MH121415 | MH121451 |
| D.alangii | China | Alangiumkurzii | CFCC 52557 | MH121492 | MH121534 | MH121574 | MH121416 | MH121452 |
| D.alnea | Netherlands | Alnus sp. | CBS 146.46 | KC343008 | KC343734 | KC343976 | KC343250 | KC343492 |
| D.amaranthophila | Japan | Amaranthustricolor | MAFF 246900 | LC459575 | LC459577 | LC459579 | LC459583 | LC459581 |
| D.ambigua | South Africa | Pyruscommunis | CBS 114015* | KC343010 | KC343736 | KC343978 | KC343252 | KC343494 |
| D.angelicae | Austria | Heracleumsphondylium | CBS 111592* | KC343027 | KC343753 | KC343995 | KC343269 | KC343511 |
| D.anhuiensis | China | Cunninghamialanceolata | CNUCC 201901* | MN219718 | MN224668 | MN227008 | MN224549 | MN224556 |
| D.arctii | Austria | Arctiumlappa | CBS 139280* | KJ590736 | KJ590776 | KJ610891 | KJ612133 | KJ659218 |
| D.arecae | India | Arecacatechu | CBS 161.64* | KC343032 | KC343758 | KC344000 | KC343274 | KC343516 |
| D.arengae | Hong Kong | Arengaengleri | CBS 114979* | KC343034 | KC343760 | KC344002 | KC343276 | KC343518 |
| D.arezzoensis | Italy | Cytisus sp. | MFLUCC 15-0127 | MT185503 | NA | NA | NA | NA |
| D.aseana | Thailand | Unidentified dead leaf | MFLUCC 12-0299a* | KT459414 | KT459448 | KT459432 | KT459464 | NA |
| D.australiana | Australia | Macadamia | CBS 146457 | MN708222 | MN696522 | MN696530 | NA | NA |
| D.batatas | USA | Ipomoeabatatas | CBS 122.21* | KC343040 | KC343766 | KC344008 | KC343282 | KC343524 |
| D.beilharziae | Australia | Indigoferaaustralis | BRIP 54792* | JX862529 | JX862535 | KF170921 | NA | NA |
| D.biconispora | China | Citrusgrandis | ZJUD62 | KJ490597 | KJ490476 | KJ490418 | MT227578 | KJ490539 |
| D.biguttulata | China | Citruslimon | ZJUD47* | KJ490582 | KJ490461 | KJ490403 | NA | KJ490524 |
| D.brasiliensis | Brazil | Aspidosperma sp. | CBS 133183* | KC343042 | KC343768 | KC344010 | KC343284 | KC343526 |
| D.caatingaensis | Brazil | Tacingainamoena | CBS 141542* | KY085927 | KY115603 | KY115600 | NA | KY115605 |
| D.camelliae-oleiferae | China | Camelliaoleifera | HNZZ027* | MZ509555 | MZ504707 | MZ504718 | MZ504685 | MZ504696 |
| D.caryae | China | Caryaillinoensis | CFCC 52563* | MH121498 | MH121540 | MH121580 | MH121422 | MH121458 |
| D.caryae | China | Caryaillinoensis | CFCC 52564 | MH121499 | MH121541 | MH121581 | MH121423 | MH121459 |
| D.cercidis | China | Cercischinensis | CFCC 52565* | MH121500 | MH121542 | MH121582 | MH121424 | MH121460 |
| D.cercidis | China | Cercischinensis | CFCC 52566 | MH121501 | MH121543 | MH121583 | MH121425 | MH121461 |
| D.chiangraiensis | Thailand | Bauhinia sp. | MFLUCC 17-1669* | MF190119 | MF377598 | NA | NA | NA |
| D.chrysalidocarpi | China | Chrysalidocarpuslutescens | SAUCC194.35 | MT822563 | MT855760 | MT855876 | MT855646 | MT855532 |
| D.cichorii | Italy | Cichoriumintybus | MFLUCC 17-1023* | KY964220 | KY964176 | KY964104 | KY964133 | NA |
| D.cinmomi | China | Cinnamomum sp. | CFCC 52569* | MH121504 | MH121546 | MH121586 | NA | MH121464 |
| D.cinmomi | China | Cinnamomum sp. | CFCC 52570 | MH121505 | MH121547 | MH121587 | NA | MH121465 |
| D.citriasiana | China | Citrusunshiu | CGMCC 3.15224* | JQ954645 | JQ954663 | KC357459 | KC357491 | KJ490515 |
| D.columnaris | USA | Vacciniumvitisidaea | AR3612* | AF439625 | NA | NA | NA | NA |
| D.compacta | China | Camelliasinensis | CGMCC 3.17536* | KP267854 | KP267928 | KP293434 | NA | KP293508 |
| D.convolvuli | Turkey | Convolvulusarvensis | CBS 124654* | KC343054 | KC343780 | KC344022 | KC343296 | KC343538 |
| D.cucurbitae | Canada | Cucumis sp. | DAOM 42078* | KM453210 | KM453211 | KP118848 | NA | KM453212 |
| D.cuppatea | South Africa | Aspalathuslinearis | CBS 117499* | KC343057 | KC343783 | KC344025 | KC343299 | KC343541 |
| D.cyatheae | Taiwan | Cyathealepifera | YMJ 1364* | JX570889 | KC465406 | KC465403 | KC465410 | NA |
| D.discoidispora | China | Citrusunshiu | ZJUD89* | KJ490624 | KJ490503 | KJ490445 | NA | KJ490566 |
| D.drenthii | Australia | Macadamia | CBS 146453 | MN708229 | MN696526 | MN696537 | NA | NA |
| D.durionigena | Vietnam | Duriozibethinus | VTCC 930005 | MN453530 | MT276157 | MT276159 | NA | NA |
| D.endocitricola | China | Citrusmaxima | ZHKUCC 20-0012* | MT355682 | MT409336 | MT409290 | MT409312 | NA |
| D.endophytica | Brazil | Schinusterebinthifolius | CBS 133811* | KC343065 | KC343791 | KC344033 | KC343307 | KC343549 |
| D.eucalyptorum | China | Eucalyptus | CBS 132525* | MH305525 | NA | NA | NA | NA |
| D.eugeniae | Indonesia | Eugeniaaromatica | CBS 444.82* | KC343098 | KC343824 | KC344066 | KC343340 | KC343582 |
| D.fraxini-angustifoliae | Australia | Fraxinusangustifolia | BRIP 54781* | JX862528 | JX862534 | KF170920 | NA | NA |
| D.fructicola | Japan | Passifloraedulis × P.edulis f. | MAFF 246408* | LC342734 | LC342735 | LC342736 | LC342738 | LC342737 |
| D.fulvicolor | China | Pyruspyrifolia | CGMCC 3.19601* | MK626859 | MK654806 | MK691236 | MK691132 | MK726163 |
| D.ganjae | USA | Cannabissativa | CBS 180.91* | KC343112 | KC343838 | KC344080 | KC343354 | KC343596 |
| D.goulteri | Australia | Helianthusannuus | BRIP 55657a* | KJ197290 | KJ197252 | KJ197270 | NA | NA |
| D.guangdongensis | China | Citrusmaxima | ZHKUCC 20-0014* | MT355684 | MT409338 | MT409292 | MT409314 | NA |
| D.guangxiensis | China | Vitisvinifera | JZB320094* | MK335772 | MK523566 | MK500168 | MK736727 | NA |
| D.gulyae | Australia | Helianthusannuus | BRIP 54025* | JF431299 | JN645803 | KJ197271 | NA | NA |
| D.guttulata | China | Unknown | CGMCC 3.20100 | MT385950 | MT424685 | MT424705 | MW022470 | MW022491 |
| D.helianthi | Serbia | Helianthusannuus | CBS 592.81* | KC343115 | KC343841 | KC344083 | KC343357 | KC343599 |
| D.heterostemmatis | China | Heterostemmagrandiflorum | SAUCC194.85* | MT822613 | MT855925 | MT855810 | MT855692 | MT855581 |
| D.hongkongensis | China | Dichroafebrífuga | CBS 115448* | KC343119 | KC343845 | KC344087 | KC343361 | KC343603 |
| D.hordei | Norway | Hordeumvulgare | CBS 481.92* | KC343120 | KC343846 | KC344088 | KC343362 | KC343604 |
| D.huangshanensis | China | Camelliaoleifera | CNUCC 201903* | MN219729 | MN224670 | MN227010 | NA | MN224558 |
| D.hubeiensis | China | Vitisvinifera | JZB320123 | MK335809 | MK523570 | MK500148 | MK500235 | NA |
| D.hunanensis | China | Camelliaoleifera | HNZZ023* | MZ509550 | MZ504702 | MZ504713 | MZ504680 | MZ504691 |
| D.infecunda | Brazil | Schinus sp. | CBS 133812* | KC343126 | KC343852 | KC344094 | KC343368 | KC343610 |
| D.infertilis | Suriname | Camelliasinensis | CBS 230.52* | KC343052 | KC343778 | KC344020 | KC343294 | KC343536 |
| D.kochmanii | Australia | Helianthusannuus | BRIP 54033* | JF431295 | JN645809 | NA | NA | NA |
| D.kongii | Australia | Portulacagrandifla | BRIP 54031* | JF431301 | JN645797 | KJ197272 | NA | NA |
| D.krabiensis | Thailand | marine based habitats | MFLUCC 17-2481* | MN047101 | MN433215 | MN431495 | NA | NA |
| D.leucospermi | Australia | Leucospermum sp. | CBS 111980* | JN712460 | KY435632 | KY435673 | KY435663 | KY435653 |
| D.limonicola | Malta | Citruslimon | CPC 28200* | NR_154980 | MF418501 | MF418582 | MF418256 | MF418342 |
| D.litchiicola | Australia | Litchichinensis | BRIP 54900* | JX862533 | JX862539 | KF170925 | NA | NA |
| D.lithocarpi | China | Lithocarpusglabra | CGMCC 3.15175* | KC153104 | KC153095 | KF576311 | KF576235 | NA |
| D.longicolla | USA | Glycinemax | FAU599 | KJ590728 | KJ590767 | KJ610883 | KJ612124 | KJ659188 |
| D.longispora | Canada | Ribes sp. | CBS 194.36* | KC343135 | KC343861 | KC344103 | KC343377 | KC343619 |
| D.lusitanicae | Portugal | Foeniculumvulgare | CBS 123212 | KC343136 | KC343862 | KC344104 | KC343378 | KC343620 |
| D.lusitanicae | Portugal | Foeniculumvulgare | CBS 123213* | MH863280 | KC343863 | KC344105 | KC343379 | KC343621 |
| D.malorum | Portugal | Malusdomestica | CAA 734* | KY435638 | KY435627 | KY435668 | KY435658 | KY435648 |
| D.manihotia | Rwanda | Manihotutilissima | CBS 505.76 | KC343138 | KC343864 | KC344106 | KC343380 | KC343622 |
| D.masirevicii | Australia | Helianthusannuus | BRIP 57892a* | KJ197276 | KJ197239 | KJ197257 | NA | NA |
| D.mayteni | Brazil | Maytenusilicifolia | CBS 133185 | KC343139 | KC343865 | KC344107 | KC343381 | KC343623 |
| D.megalospora | Not stated | Sambucuscanadensis | CBS 143.27* | KC343140 | KC343866 | KC344108 | KC343382 | KC343624 |
| D.melitensis | Malta | Citruslimon | CPC 27873* | MF418424 | MF418503 | MF418584 | MF418258 | MF418344 |
| D.melonis | USA | Cucumismelo | CBS 507.78* | KC343142 | KC343868 | KC344110 | KC343384 | KC343626 |
| D.melonis | Indonesia | Glycinesoja | CBS 435.87 | KC343141 | KC343867 | KC344109 | KC343383 | KC343625 |
| D.middletonii | Australia | Rapistrumrugostrum | BRIP 54884e* | KJ197286 | KJ197248 | KJ197266 | NA | NA |
| D.millettiae | China | Millettiareticulata | GUCC9167* | MK398674 | MK480609 | MK502089 | MK502086 | NA |
| D.minusculata | China | saprobic on decaying wood | CGMCC 3.20098* | MT385957 | MT424692 | MT424712 | MW022475 | MW022499 |
| D.miriciae | Australia | Helianthusannuus | BRIP 54736j* | KJ197282 | KJ197244 | KJ197262 | NA | NA |
| D.musigena | Australia | Musa sp. | CBS 129519* | KC343143 | KC343869 | KC344111 | KC343385 | KC343267 |
| D.myracrodruonis | Brazil | Astroniumurundeuva | URM 7972* | MK205289 | MK213408 | MK205291 | MK205290 | 17 |
| D.nelumbonis | Taiwan | Nelumbonucifera | R. Kirschner 4114* | KT821501 | NA | LC086652 | NA | NA |
| D.neoarctii | USA | Ambrosiatrifi | CBS 109490* | KC343145 | KC343871 | KC344113 | KC343387 | KC343629 |
| D.neoraonikayaporum | Thailand | Tectonagrandis | MFLUCC 14-1136* | KU712449 | KU749369 | KU743988 | KU749356 | NA |
| D.oculi | Japan | Homosapiens | HHUF 30565* | LC373514 | LC373516 | LC373518 | NA | NA |
| D.osmanthi | China | Osmanthusfragrans | GUCC9165* | MK398675 | MK480610 | MK502091 | MK502087 | NA |
| D.ovalispora | China | Citruslimon | CGMCC 3.17256* | KJ490628 | KJ490507 | KJ490449 | NA | KJ490570 |
| D.oxe | Brazil | Maytenusilicifolia | CBS 133186* | KC343164 | KC343890 | KC344132 | KC343406 | KC343648 |
| D.pandanicola | Thailand | Pandanus sp. | MFLUCC 17-0607* | MG646974 | NA | MG646930 | NA | NA |
| D.paranensis | Brazil | Maytenusilicifolia | CBS 133184* | KC343171 | KC343897 | KC344139 | KC343413 | KC343655 |
| D.pascoei | Australia | Perseaamericana | BRIP 54847* | JX862532 | JX862538 | KF170924 | NA | NA |
| D.passiflorae | South America | Passifloraedulis | CBS 132527* | JX069860 | KY435633 | KY435674 | KY435664 | KY435654 |
| D.passifloricola | Malaysia | Passiflorafoetida | CBS 141329* | KX228292 | NA | KX228387 | NA | KX228367 |
| D.perseae | Netherlands | Perseagratissima | CBS 151.73* | KC343173 | KC343899 | KC343141 | KC343415 | KC343657 |
| D.pescicola | China | Prunuspersica | MFLUCC 16-0105* | KU557555 | KU557623 | KU557579 | KU557603 | NA |
| D.phaseolorum | USA | Phaseolusvulgaris | AR4203* | KJ590738 | KJ590739 | KJ610893 | KJ612135 | KJ659220 |
| D.phoenicicola | India | Arecacatechu | CBS 161.64* | MH858400 | GQ250349 | JX275440 | JX197432 | NA |
| D.podocarpi-macrophylli | China | Podocarpusmacrophyllus | CGMCC 3.18281* | KX986774 | KX999167 | KX999207 | KX999278 | KX999246 |
| D.pseudolongicolla | Serbia | Glycinemax | PL42* | JQ697843 | JQ697856 | NA | NA | NA |
| D.pseudolongicolla | Croatia | Glycinemax | CBS 127269 | KC343155 | KC343881 | KC344123 | KC343397 | KC343639 |
| D.pseudomangiferae | Dominican Republic | Mangiferaindica | CBS 101339* | KC343181 | KC343907 | KC344149 | KC343423 | KC343665 |
| D.pseudooculi | Japan | Homosapiens | HHUF 30617* | NR_161019 | LC373517 | LC373519 | NA | NA |
| D.pseudophoenicicola | Spain | Phoenixdactylifera | CBS 462.69* | KC343184 | KC343910 | KC344152 | KC343426 | KC343668 |
| D.pseudophoenicicola | Iraq | Mangiferaindica | CBS 176.77 | KC343183 | KC343909 | KC344151 | KC343425 | KC343667 |
| D.pterocarpicola | Thailand | Pterocarpusindicus | MFLUCC 10-0580a* | JQ619887 | JX275403 | JX275441 | JX197433 | NA |
| D.pyracanthae | Portugal | Pyracanthacoccinea | CBS 142384* | KY435635 | KY435625 | KY435666 | KY435656 | KY435646 |
| D.racemosae | South Africa | Euclearacemosa | CPC 26646* | MG600223 | MG600225 | MG600227 | MG600219 | MG600221 |
| D.raonikayaporum | Brazil | Spondiasmombin | CBS 133182* | KC343188 | KC343914 | KC344156 | KC343430 | KC343672 |
| D.rhodomyrti | China | Rhodomyrtustomentosa | CFCC 53101 | MK432643 | MK578119 | MK578046 | MK442965 | MK442990 |
| D.rhodomyrti | China | Rhodomyrtustomentosa | CFCC 53102 | MK432644 | MK578120 | MK578047 | MK442966 | MK442991 |
| D.rizhaoensis | China | Xanthiumstrumarium | CFCC 57562* | OP955930 | OP959767 | OP959773 | OP959782 | OP959785 |
| D.rizhaoensis | China | Xanthiumstrumarium | CFCC 57563 | OP955931 | OP959766 | OP959772 | OP959781 | OP959784 |
| D.rizhaoensis | China | Xanthiumstrumarium | CFCC 57564 | OP955932 | OP959765 | OP959771 | OP959780 | OP959783 |
| D.rosae | Thailand | Rosa sp. | MFLUCC 17-2658* | MG828894 | NA | MG843878 | MG829273 | NA |
| D.rosiphthora | Brazil | Rosa sp. | COAD 2914* | MT311197 | MT313693 | NA | MT313691 | NA |
| D.rossmaniae | Portugal | Vacciniumcorymbosum | CAA762* | MK792290 | MK828063 | MK837914 | MK883822 | MK871432 |
| D.sackstonii | Australia | Helianthusannuus | BRIP 54669b* | KJ197287 | KJ197249 | KJ197267 | NA | NA |
| D.salinicola | Thailand | Xylocarpus sp. | MFLU 18-0553* | MN047098 | MN077073 | NA | NA | NA |
| D.sambucusii | China | Sambucuswilliamsii | CFCC 51986* | KY852495 | KY852507 | KY852511 | KY852499 | KY852503 |
| D.sambucusii | China | Sambucuswilliamsii | CFCC 51987 | KY852496 | KY852508 | KY852512 | KY852500 | KY852504 |
| D.schimae | China | Schimasuperba | CFCC 53103* | MK432640 | MK578116 | MK578043 | MK442962 | MK442987 |
| D.schimae | China | Schimasuperba | CFCC 53104 | MK432641 | MK578117 | MK578044 | MK442963 | MK442988 |
| D.schini | Brazil | Schinusterebinthifolius | CBS 133181* | KC343191 | KC343917 | KC344159 | KC343433 | KC343675 |
| D.schoeni | Italy | Schoenusnigricans | MFLU 15-1279* | KY964226 | KY964182 | KY964109 | KY964139 | |
| D.sclerotioides | Netherlands | Cucumissativus | CBS 296.67* | KC343193 | KC343919 | KC344161 | KC343435 | KC343677 |
| D.searlei | Australia | Macadamia | CBS 146456* | MN708231 | NA | MN696540 | NA | NA |
| D.sennae | China | Sennabicapsularis | CFCC 51636* | KY203724 | KY228885 | KY228891 | KY228875 | NA |
| D.sennae | China | Sennabicapsularis | CFCC 51637 | KY203725 | KY228886 | KY228892 | KY228876 | NA |
| D.serafiniae | Australia | Helianthusannuus | BRIP 55665a* | KJ197274 | KJ197236 | KJ197254 | NA | NA |
| D.siamensis | Thailand | Dasymaschalon sp. | MFLUCC 10-0573a* | JQ619879 | JX275393 | JX275429 | JX197423 | NA |
| D.sinensis | China | Amaranthus sp. | ZJUP0033-4* | MK637451 | MK660449 | MK660447 | NA | MK660451 |
| D.smilacicola | China | Smilaxglabra | CFCC 54582* | OP955933 | OP959770 | OP959776 | OP959779 | OP959788 |
| D.smilacicola | China | Smilaxglabra | CFCC 58764 | OP955934 | OP959769 | OP959775 | OP959778 | OP959787 |
| D.smilacicola | China | Smilaxglabra | CFCC 58765 | OP955935 | OP959768 | OP959774 | OP959776 | OP959786 |
| D.sojae | USA | Glycinemax | FAU635* | KJ590719 | KJ590762 | KJ610875 | KJ612116 | KJ659208 |
| D.spinosa | China | Pyruspyrifolia | CGMCC 3.19602* | MK626849 | MK654811 | MK691234 | MK691129 | MK726156 |
| D.stewartii | Not stated | Cosmosbipinnatus | CBS 193.36* | MH867279 | GQ250324 | JX275421 | JX197415 | NA |
| D.subellipicola | China | on dead wood | KUMCC 17-0153* | MG746632 | MG746633 | MG746634 | NA | NA |
| D.subordinaria | New Zealand | Plantagolanceolata | CBS 464.90* | KC343214 | KC343940 | KC344182 | KC343456 | KC343698 |
| D.taiwanensis | Taiwan | Ixorachinensis | NTUCC 18-105-1* | MT241257 | MT251199 | MT251202 | MT251196 | NA |
| D.taoicola | China | Prunuspersica | MFLUCC 16-0117* | KU557567 | KU557635 | KU557591 | NA | NA |
| D.tarchonanthi | South Africa | Tarchonanthuslittoralis | CBS 146073* | MT223794 | NA | MT223733 | NA | MT223759 |
| D.tecomae | Brazil | Tabebuia sp. | CBS 100547* | KC343215 | KC343941 | KC344183 | KC343457 | KC343699 |
| D.tectonae | Thailand | Tectonagrandis | MFLUCC 12-0777* | KU712430 | KU749359 | KU743977 | KU749345 | NA |
| D.tectonendophytica | Thailand | Tectonagrandis | MFLUCC 13-0471* | KU712439 | KU749367 | KU743986 | KU749354 | NA |
| D.tectonigena | China | Tectonagrandis | MFLUCC 12-0767* | KU712429 | KU749371 | KU743976 | KU749358 | NA |
| D.tectonigena | China | Camelliasinensis | LC6512 | KX986782 | KX999174 | KX999214 | KX999284 | KX999254 |
| D.terebinthifolii | Brazil | Schinusterebinthifolius | CBS 133180* | KC343216 | KC343942 | KC344184 | KC343458 | KC343700 |
| D.thunbergiicola | Thailand | Thunbergialaurifolia | MFLUCC 12-0033* | KP715097 | KP715098 | NA | NA | NA |
| D.tulliensis | Australia | Theobromacacao | BRIP 62248a* | KR936130 | KR936133 | KR936132 | NA | NA |
| D.ueckeri | USA | Cucumismelo | FAU656* | KJ590726 | KJ590747 | KJ610881 | KJ612122 | KJ659215 |
| D.unshiuensis | China | Fortunellamargarita | CGMCC 3.17566* | KJ490584 | KJ490463 | KJ490405 | NA | KJ490526 |
| D.unshiuensis | China | Caryaillinoensis | CFCC 52594 | MH121529 | MH121571 | MH121606 | MH121447 | MH121487 |
| D.unshiuensis | China | Caryaillinoensis | CFCC 52595 | MH121530 | MH121572 | MH121607 | MH121448 | MH121488 |
| D.vawdreyi | Australia | Psidiumguajava | BRIP 57887a | KR936126 | KR936129 | KR936128 | NA | NA |
| D.vexans | USA | Solanummelongena | CBS 127.14 | KC343229 | KC343955 | KC344197 | KC343471 | KC343713 |
| D.viniferae | China | Vitisvinifera | JZB320071* | MK341550 | MK500107 | MK500112 | MK500119 | NA |
| D.vochysiae | Brazil | Vochysiadivergens | LGMF1583* | MG976391 | MK007526 | MK007527 | MK007528 | MK033323 |
| D.xishuangbanica | China | Camelliasinensis | CGMCC 3.18283* | KX986784 | KX999176 | KX999217 | NA | NA |
| D.xishuangbanica | China | Camelliasinensis | LC6707 | KX986783 | KX999175 | KX999216 | NA | KX999255 |
Notes: NA, not applicable. * ex-type strains.
Results
Phylogeny
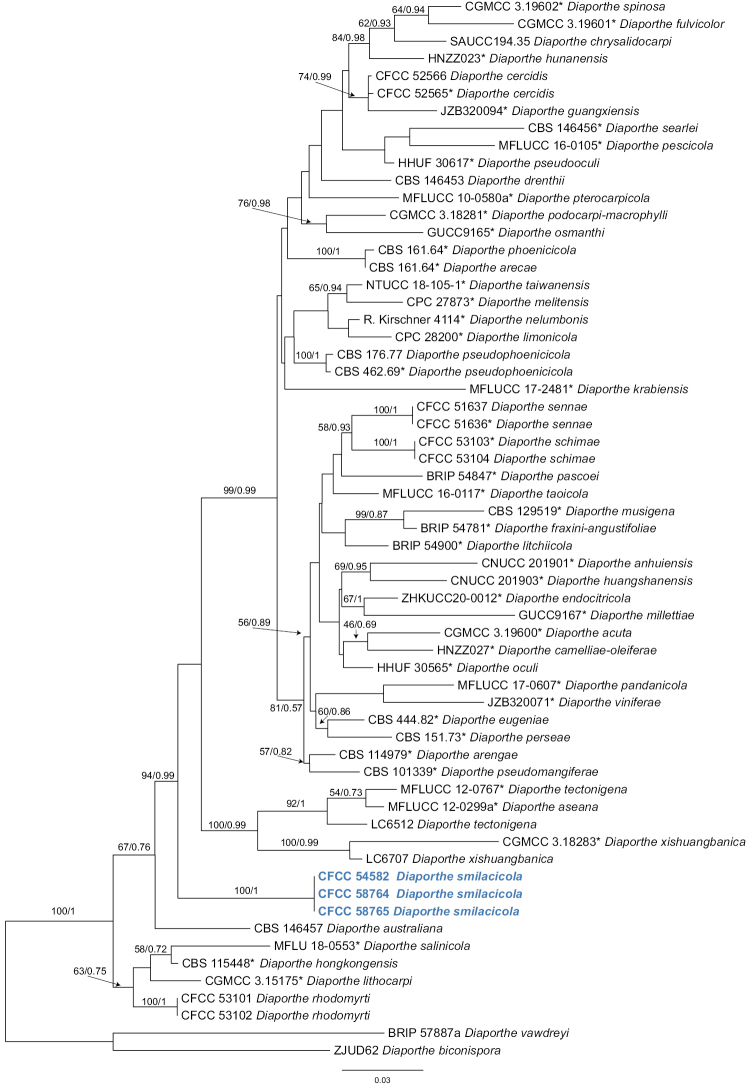
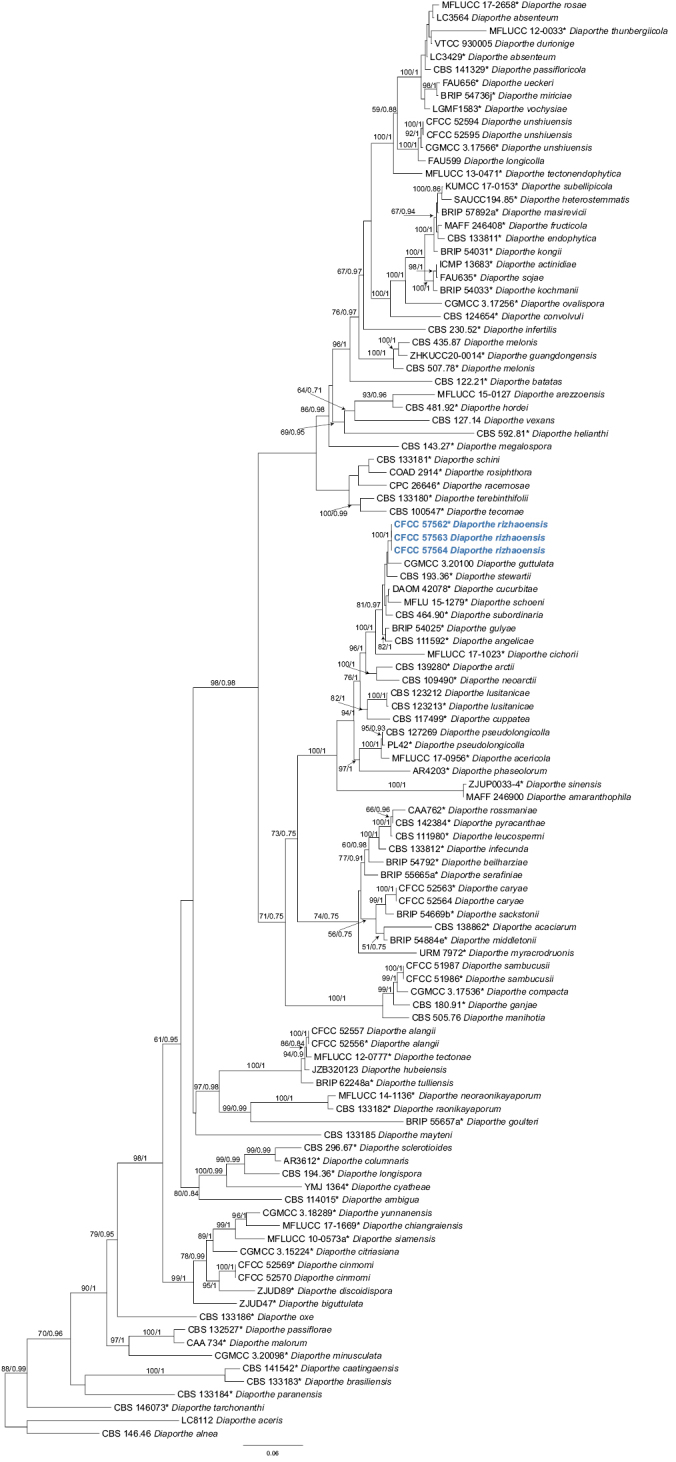
In the present study, we followed Norphanphoun et al. (2022) for the species complexes treatments of Diaporthe. Firstly, we conducted a genus tree including all species belonging to this genus according to Norphanphoun et al. (2022). After that, the phylogenetic analysis revealed that three isolates (CFCC 57562, CFCC 57563 and CFCC 57564) clustered in a distinct clade in the D.sojae species complex, and three isolates (CFCC 54582, CFCC 58764 and CFCC 58765) clustered in a distinct clade in the D.arecae species complex (Figs 1, 2). The combined sequence alignments of D.arecae species complex comprised 62 strains, with D.vawdreyi (BRIP 57887a) and D.biconispora (ZJUD62) as the outgroup taxa. The dataset comprised 2791 characters including alignment gaps (634 for ITS, 381 for tef1, 791 for tub2, 499 for cal and 486 for his3). The combined sequence alignments of D.sojae species complex comprised 111 strains, with D.aceris (LC8112) and D.alnea (CBS 146.46) as the outgroup taxa. The dataset comprised 2799 characters including alignment gaps (671 for ITS, 483 for tef1, 483 for tub2, 593 for cal and 569 for his3). The final maximum likelihood tree topology was similar to Bayesian analysis.
Figure 1.
Phylogram of Diaporthesojae species complex resulting from a maximum likelihood analysis based on a combined matrix of ITS, cal, his3, tef1 and tub2 loci. Numbers above the branches indicate ML bootstrap values (left, ML BS ≥ 50%) and Bayesian posterior probabilities (right, BPP ≥ 0.9). Isolates from the present study are marked in bold blue.
Figure 2.
Phylogram of Diaporthearecae species complex resulting from a maximum likelihood analysis based on a combined matrix of ITS, cal, his3, tef1 and tub2 loci. Numbers above the branches indicate ML bootstrap values (left, ML BS ≥ 50%) and Bayesian posterior probabilities (right, BPP ≥ 0.9). Isolates from the present study are marked in bold blue.
Taxonomy
. Diaporthe rizhaoensis
Y.Q. Zhu & Ning Jiang sp. nov.
B0CFC039-E2A1-5035-8608-3273A2023E46
846816
Figure 3.
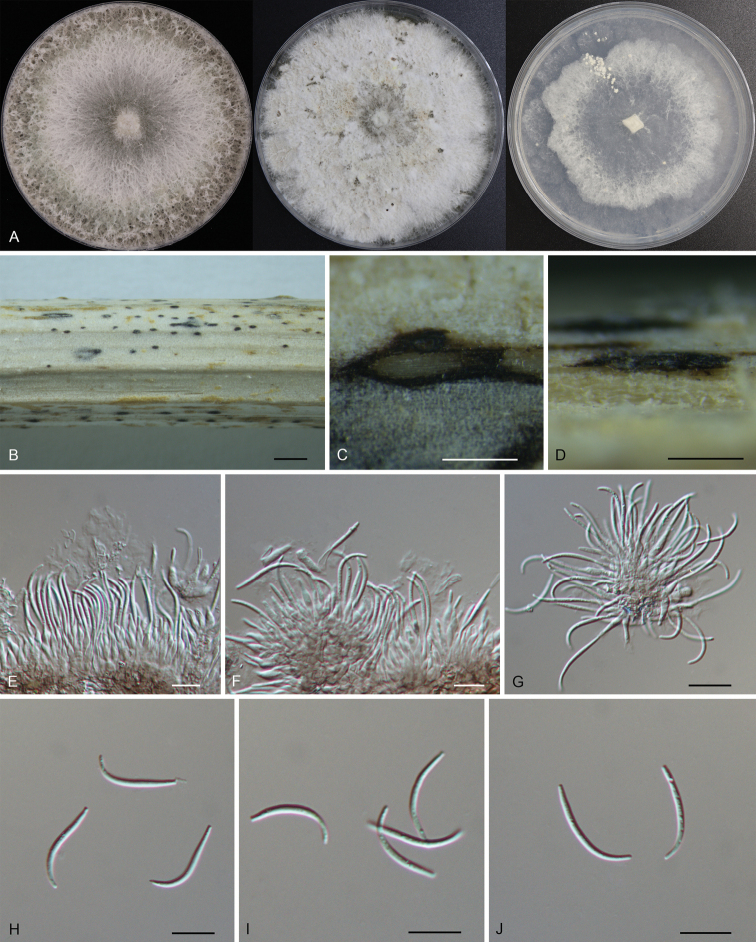
Morphology of DiaportherizhaoensisA colonies on PDA, MEA and SNA at 25 °C after 2 weeks B habit of conidiomata on the host C transverse section of the conidioma D longitudinal section through the conidioma E–G conidiogenous cells with attached beta conidia H–J beta conidia. Scale bars: 500 µm (B); 100 µm (C, D); 10 µm (E–J).
Etymology.
Named after the collection site of the type specimen, Rizhao City.
Description.
Conidiomata pycnidial, small, scattered, slightly erumpent through bark surface, nearly flat, discoid, with a solitary undivided locule, 150–400 μm diam. Conidiogenous cells 6.7–11.4 × 1.6–3.0 μm, hyaline, unbranched, densely aggregated, mostly ampulliform, guttulate, aseptate, straight or slightly curved, swelling at base, tapering towards apex. Beta conidia 12.9–23.4 × 1.1–2.1 μm (mean = 18.7 × 1.4 μm, n = 50), hyaline, filiform, straight or slightly curved, aseptate, base subtruncate, tapering towards the base. Alpha conidia and gamma conidia not observed. Sexual morph not observed.
Culture characters.
Colonies on potato dextrose agar (PDA) flat, spreading, with flocculent aerial mycelium and entire edge, white, reaching a 90 mm diameter after 14 days at 25 °C; on malt extract agar (MEA) flat, spreading, with flocculent aerial mycelium and crenate edge, white, reaching a 90 mm diameter after 14 days at 25 °C, forming black conidiomata with black conidial masses; on synthetic low nutrient agar (SNA) flat, spreading, with flocculent aerial mycelium forming concentric rings and entire edge, white, reaching a 90 mm diameter after 14 days at 25 °C.
Materials examined.
China, Shandong Province, Rizhao City, Wulian County, Zhongzhi Town, on dead culms of Xanthiumstrumarium, 5 May 2022, Ning Jiang & Chengbin Wang (holotype CAF 800069; ex-holotype culture CFCC 57562). Shandong Province, Rizhao City, Wulian County, Xumeng Town, on dead culms of Xanthiumstrumarium, 5 May 2022, Ning Jiang & Chengbin Wang (cultures CFCC 57563 and CFCC 57564).
Notes.
Diaportherizhaoensis formed a distinct clade with high support (ML/BI = 100/1), and was close to D.guttulata and D.stewartia (Fig. 1). Diaportherizhaoensis is different from D.stewartia by host association (D.rizhaoensis on Xanthiumstrumarium vs. D.stewartia on Cosmosbipinnatus) (Harrison 1935; Dissanayake et al. 2020). In addition, D.guttulata and D.stewartia are only known in sexual morph. Moreover, Diaportherizhaoensis can be distinguished from D.guttulata (15/364 in cal, 5/428 in his3, 5/313 in tef1, and 1/408 in tub2) and D.stewartii (3/532 in ITS, 7/451 in cal, and 7/369 in tub2) by sequence data. Diaporthehelianthi, D.longicolla, D.pseudolongicolla (= D.novem) and D.rizhaoensis have been reported form the host Xanthiumstrumarium (Vrandecic et al. 2007, 2010; Petrović et al. 2018; Thompson et al. 2018). Morphologically, Diaporthehelianthi is a bit longer than D.rizhaoensis in the beta conidia, but not fully distinguished (Vrandecic et al. 2007, 2010). Morphology of D.longicolla and D.pseudolongicolla on Xanthiumstrumarium were not available. However, these four species are phylogenetically distinguished in the phylogram of D.sojae species complex (Fig. 1).
. Diaporthe smilacicola
Y.Q. Zhu & Ning Jiang sp. nov.
1BC54B14-2E2C-5952-8A5A-3DC760DCB39F
846818
Figure 4.
Morphology of DiaporthesmilacicolaA colonies on PDA, MEA and SNA at 25 °C after 2 weeks B leaf spots on the host surface C conidiomata formed on the PDAD, E conidiogenous cells with attached alpha conidia F–G alpha conidia. Scale bars: 200 µm (C); 10 µm (D–G).
Etymology.
Named after the host genus, Smilax.
Description.
Leaf spots subcircular to irregular, pale brown to brown, with dark brown margin. Conidiomata pycnidial, scattered, subglobose to globose, black, erumpent, exuding faint yellow translucent conidial droplets from central ostioles, 150–350 μm diam. Conidiogenous cells 11–16.2 × 1.8–2.4 μm, hyaline, phialidic, cylindrical, terminal, slightly tapering towards the apex. Alpha conidia 5.7–9.7 × 2.0–3.5 μm (mean = 7.8 × 2.6 μm, n = 50), hyaline, aseptate, smooth, guttulate, ellipsoidal to oblong ellipsoidal, with both ends obtuse. Beta conidia and gamma conidia not observed. Sexual morph not observed.
Culture characters.
Colonies on PDA flat, with flocculent aerial mycelium and crenate edge, white to gray, reaching a 90 mm diameter after 14 days at 25 °C, forming black conidiomata with black conidial masses; on MEA flat, spreading, with flocculent aerial mycelium forming concentric rings, off-white to luteous, reaching a 90 mm diameter after 14 days at 25 °C; on SNA flat, spreading, with flocculent aerial mycelium forming concentric rings and entire edge, white, reaching a 90 mm diameter after 14 days at 25 °C.
Materials examined.
China, Hunan Province, Changsha City, Changsha County, Kaihui Town, on leaf spots of Smilaxglabra, 2 November 2020, Ning Jiang (holotype CAF 800070; ex-holotype culture CFCC 54582). Hunan Province, Shaoshan City, on leaf spots of Smilaxglabra, 2 November 2020, Ning Jiang (cultures CFCC 58764 and CFCC 58765).
Notes.
Three Diaporthe isolates representing D.smilacicola formed a well-supported clade (ML/BI = 100/1), and appear to be distinct from the other Diaporthe species phylogenetically (Fig. 2). Diaportheeres (= D.mahothocarpi), D.lithocarpi and D.smilacicola have been reported from the host S.glabra (Gao et al. 2013; Chaisiri et al. 2021). Morphologically, these three species are similar in conidial shape and size. However, Diaportheeres belongs to D.eres species complex, which is different from D.lithocarpi and D.smilacicola in D.arecae species complex. D.smilacicola is obviously different from D.lithocarpi based on sequence data (22/467 in ITS, 31/393 in cal, 52/317 in tef1, 19/420 in tub2) (Fig. 2).
Discussion
Based on the morphology and the multi-locus phylogeny, six isolates from the present study can be recognized as two new species of Diaporthe, viz. D.rizhaoensis from dead culms of Xanthiumstrumarium and D.smilacicola from leaf spots of Smilaxglabra.
Species identification in Diaporthe was primarily based on the assumption of host-specificity, which has largely impeded the progress of establishing a proper taxonomy of Diaporthe (Gomes et al. 2013). More than one species of Diaporthe can often be recovered from a single host and one species was found to be associated with different host plants (Gomes et al. 2013; Gao et al. 2017; Guarnaccia and Crous 2017; Guarnaccia et al. 2018; Guo et al. 2020). For example, D.eres can infect blackberry (Vrandecic et al. 2011), pear (Bai et al. 2015), and jujube (Zhang et al. 2018); D.pometiae was isolated from Heliconiametallica and Perseaamericana (Huang et al 2021); D.melastomatis was collected from three hosts namely Camelliasinensis, Melastomamalabathricum and Millettiareticulata (Sun et al. 2021); D.australiana, D.drenthii, D.macadamiae and D.searlei can cause diseases on macadamia in Australia and South Africa (Wrona et al. 2020) and seven endophytic Diaporthe species were discovered on Citrus trees (Huang et al. 2015). As was revealed in the present study, two additional species of Diaporthe were proposed from the host Smilaxglabra and Xanthiumstrumarium. This study further demonstrates that host association is not a robust character to distinguish members of Diaporthe.
Recently, the species classification of Diaporthe has become more dependent on DNA sequence-based methods rather than traditional morphological characterization. (Udayanga et al. 2014a, b, 2015; Fan et al. 2015; Gao et al. 2017; Guarnaccia and Crous 2017; Guarnaccia et al. 2018; Hyde et al. 2018, 2020; Yang et al. 2018, 2020, 2021; Long et al. 2019; Cao et al. 2022). The ITS sequence offers convincing proof for species demarcation and is recommended for identifying species boundaries in the genus Diaporthe (Santos and Phillips 2009, 2011; Thompson et al. 2011). However, the intraspecific variation is even greater than the interspecific variation, which makes it difficult to identify Diaporthe species using the ITS sequence alone (Crouch et al. 2009). Considering this, concatenation of a five-loci dataset (ITS-tef1-tub2-cal-his3) was recommended as the best combination for species identification within the genus (Udayanga et al. 2014; Fan et al. 2018; Yang et al. 2018; Guo et al. 2020). Two phylograms resulted from the present study also support the feasibility of the five loci data to separate species of Diaporthe.
The two newly introducing species could potentially be pathogens, because they were isolated from diseased plant tissues, and their pathogenicity should be evaluated in further studies. And, it is necessary to evaluate the effects of environmental conditions, such as temperature, pH, and carbon sources, on mycelium growth and pathogenicity.
Supplementary Material
Acknowledgements
This research was funded by Fundamental Research Funds for the Central Non-profit Research Institution of Chinese Academy of Forestry (grant CAFYBB2018ZB001), and National Microbial Resource Center of the Ministry of Science and Technology of the People’s Republic of China (grant NMRC-2021-7).
Citation
Zhu Y-Q, Ma C-Y, Xue H, Piao C-G, Li Y, Jiang N (2023) Two new species of Diaporthe (Diaporthaceae, Diaporthales) in China. MycoKeys 95: 209–228. https://doi.org/10.3897/mycokeys.95.98969
Funding Statement
This research was funded by Fundamental Research Funds for the Central Non-profit Research Institution of Chinese Academy of Forestry (grant CAFYBB2018ZB001), and National Microbial Resource Center of the Ministry of Science and Technology of the People’s Republic of China (grant NMRC-2021-7).
Reference
- Bai Q, Zhai LF, Chen XR, Hong N, Xu WX, Wang GP. (2015) Biological and molecular characterization of five Phomopsis species associated with pear shoot canker in China. Plant Disease 99(12): 1704–1712. 10.1094/PDIS-03-15-0259-RE [DOI] [PubMed] [Google Scholar]
- Caio P, Bruno F, Carlos AP, Robert B. (2021) Diaportherosiphthora sp. nov.: Yet another rose dieback fungus. Crop Protection (Guildford, Surrey) 139: 105365. 10.1016/j.cropro.2020.105365 [DOI]
- Cao L, Luo D, Lin W, Yang Q, Deng X. (2022) Four new species of Diaporthe (Diaporthaceae,Diaporthales) from forest plants in China. MycoKeys 91: 25–47. 10.3897/mycokeys.91.84970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. (1999) A Method for designing primer sets for speciation studies in filamentous Ascomycetes. Mycologia 91(3): 553–556. 10.1080/00275514.1999.12061051 [DOI] [Google Scholar]
- Chaisiri C, Liu X, Lin Y, Fu Y, Zhu F, Luo C. (2021) Phylogenetic and haplotype network analyses of Diaportheeres species in China based on sequences of multiple loci. Biology (Basel) 10(3): 179. 10.3390/biology10030179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch JA, Clarke BB, Hillman BI. (2009) What is the value of ITS sequence data in Colletotrichum systematics and species diagnosis? A case study using the falcate-spored graminicolous Colletotrichum group. Mycologia 101(5): 648–656. 10.3852/08-231 [DOI] [PubMed] [Google Scholar]
- Crous PW. (2005) Impact of molecular phylogenetics on the taxonomy and diagnostics of fungi. Bulletin OEPP. EPPO Bulletin. European and Mediterranean Plant Protection Organisation 35(1): 47–51. 10.1111/j.1365-2338.2005.00811.x [DOI] [Google Scholar]
- Crous PW, Groenewald JZ, Risède JM, Simoneau P, Hywel-Jones NL. (2004) Calonectria species and their Cylindrocladium anamorphs: Species with sphaeropedunculate vesicles. Studies in Mycology 50: 415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissanayake AJ, Phillips AJL, Hyde KD, Yan JY, Li XH. (2017) The current status of species in Diaporthe. Mycosphere 8(5): 1106–1156. 10.5943/mycosphere/8/5/5 [DOI] [Google Scholar]
- Dissanayake AJ, Chen YY, Liu JK. (2020) Unravelling Diaporthe species associated with woody hosts from karst formations (Guizhou) in China. Journal of Fungi (Basel, Switzerland) 6(4): 251. 10.3390/jof6040251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. (1990) Isolation of plant DNA from fresh tissue. Focus (San Francisco, Calif. ) 12: 13–15. [Google Scholar]
- Du Z, Fan XL, Hyde KD, Yang Q, Liang YM, Tian CM. (2016) Phylogeny and morphology reveal two new species of Diaporthe from Betula spp. in China. Phytotaxa 269(2): 90–102. 10.11646/phytotaxa.269.2.2 [DOI] [Google Scholar]
- Fan XL, Hyde KD, Udayanga D, Wu XY, Tian CM. (2015) Diaportherostrata, a novel ascomycete from Juglansmandshurica associated with walnut dieback. Mycological Progress 14(10): 1–8. 10.1007/s11557-015-1104-5 [DOI] [Google Scholar]
- Fan XL, Yang Q, Bezerra JDP, Alvarez LV, Tian CM. (2018) Diaporthe from walnut tree (Juglansregia) in China, with insight of Diaportheeres complex. Mycological Progress 17(7): 841–853. 10.1007/s11557-018-1395-4 [DOI] [Google Scholar]
- Farr DF, Castlebury LA, Rossman AY. (2002a) Morphological and molecular characterization of Phomopsisvaccinii and additional isolates of Phomopsis from blueberry and cranberry in the eastern United States. Mycologia 94(3): 494–504. 10.1080/15572536.2003.11833214 [DOI] [PubMed] [Google Scholar]
- Farr DF, Castlebury LA, Rossman AY, Putnam ML. (2002b) A new species of Phomopsis causing twig dieback of Vacciniumvitisidaea (lingonberry). Mycological Research 106(6): 745–752. 10.1017/S095375620200583X [DOI] [Google Scholar]
- Gao YH, Sun W, Su YY, Cai L. (2013) Three new species of Phomopsis in Gutianshan nature reserve in China. Mycological Progress 13(1): 111–121. 10.1007/s11557-013-0898-2 [DOI] [Google Scholar]
- Gao YH, Liu F, Duan W, Crous PW, Cai L. (2017) Diaporthe is paraphyletic. IMA Fungus 8(1): 153–187. 10.5598/imafungus.2017.08.01.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61(4): 1323–1330. 10.1128/aem.61.4.1323-1330.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes RR, Glienke C, Videira SIR, Lombard L, Groenewald JZ, Crous PW. (2013) Diaporthe: A genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 31(1): 1–41. 10.3767/003158513X666844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Crous PW. (2017) Emerging citrus diseases in Europe caused by species of Diaporthe. IMA Fungus 8(2): 317–334. 10.5598/imafungus.2017.08.02.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Groenewald JZ, Woodhall J, Armengol J, Cinelli T, Eichmeier A, Ezra D, Fontaine F, Gramaje D, Gutierrez-Aguirregabiria A, Kaliterna J, Kiss L, Larignon P, Luque J, Mugnai L, Naor V, Raposo R, Sándor E, Váczy KZ, Crous PW. (2018) Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in Europe. Persoonia 40(1): 135–153. 10.3767/persoonia.2018.40.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Martino I, Tabone G, Brondino L, Gullino ML. (2020) Fungal pathogens associated with stem blight and dieback of blueberry in northern Italy. Phytopathologia Mediterranea 59(2): 229–245. 10.14601/Phyto-11278 [DOI] [Google Scholar]
- Guo YS, Crous PW, Bai Q, Fu M, Yang MM, Wang XH, Du YM, Hong N, Xu WX, Wang GP. (2020) High diversity of Diaporthe species associated with pear shoot canker in China. Persoonia 45(1): 132–162. 10.3767/persoonia.2020.45.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison AL. (1935) The perfect stage of Phomopsisstewartii on Cosmos. Mycologia 27(5): 521–526. 10.1080/00275514.1935.12017096 [DOI] [Google Scholar]
- Hilário S, Amaral IA, Gonçalves MF, Lopes A, Santos L, Alves A. (2020) Diaporthe species associated with twig blight and dieback of Vacciniumcorymbosum in Portugal, with description of four new species. Mycologia 112(2): 293–308. 10.1080/00275514.2019.1698926 [DOI] [PubMed] [Google Scholar]
- Huang F, Udayanga D, Wang X, Hou X, Mei X, Fu Y, Li H. (2015) Endophytic Diaporthe associated with Citrus: A phylogenetic reassessment with seven new species from China. Fungal Biology 119(5): 331–347. 10.1016/j.funbio.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Huang ST, Xia JW, Zhang XG, Sun WX. (2021) Morphological and phylogenetic analyses reveal three new species of Diaporthe from Yunnan, China. MycoKeys 78: 49–77. 10.3897/mycokeys.78.60878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde KD, Chaiwan N, Norphanphoun C, Boonmee S, Camporesi E, Chethana KWT, Dayarathne MC, de Silva IN, Dissanayake AJ, Ekanayaka AH. (2018) Mycosphere notes 169–224. Mycosphere 9(2): 271–430. 10.5943/mycosphere/9/2/8 [DOI] [Google Scholar]
- Hyde KD, Dong Y, Phookamsak R, Jeewon R, Bhat DJ, Gareth Jones EB, Liu NG, Abeywickrama PD, Mapook A, Wei D. (2020) Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100(1): 1–273. 10.1007/s13225-020-00439-5 [DOI] [Google Scholar]
- Jiang N, Fan XL, Tian CM. (2021a) Identification and characterization of leaf-inhabiting fungi from Castanea plantations in China. Journal of Fungi (Basel, Switzerland) 7(1): 64. 10.3390/jof7010064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Voglmayr H, Piao CG, Li Y. (2021b) Two new species of Diaporthe (Diaporthaceae,Diaporthales) associated with tree cankers in the Netherlands. MycoKeys 85: 31–56. 10.3897/mycokeys.85.73107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. (2010) Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26(15): 1899–1900. 10.1093/bioinformatics/btq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33(7): 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H, Zhang Q, Hao YY, Shao XQ, Wei XX, Hyde KD, Wang Y, Zhao DG. (2019) Diaporthe species in south-western China. MycoKeys 57: 113–127. 10.3897/mycokeys.57.35448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment. Bridging from the extreme to the campus and beyond. Association for Computing Machinery, USA, 8 pp. 10.1145/2335755.2335836 [DOI] [Google Scholar]
- Nitschke T. (1870) Pyrenomycetes Germanici (2nd edn.). Eduard T rewendt, Breslau, 161–320.
- Norphanphoun C, Gentekaki E, Hongsanan S, Jayawardena R, Senanayake C, Manawasinghe I, Abeywickrama P, Bhunjun CS, Hyde KD. (2022) Diaporthe: Formalizing species-group concepts. Mycosphere 13(1): 752–819. 10.5943/mycosphere/13/1/9 [DOI] [Google Scholar]
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7(1): 103–116. 10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- Petrović K, Riccioni L, Đorđević V, Balešević-Tubić S, Miladinović J, Ćeran M, Rajković D. (2018) Diaporthepseudolongicolla: The new pathogen on soybean seed in Serbia. Ratarstvo i Povrtarstvo 55(2): 103–109. 10.5937/ratpov55-18582 [DOI] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12): 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Rossman AY, Farr DF, Castlebury LA. (2007) A review of the phylogeny and biology of the Diaporthales. Mycoscience 48(3): 135–144. 10.1007/S10267-007-0347-7 [DOI] [Google Scholar]
- Rossman AY, Adams GC, Cannon PF, Castlebury LA, Crous PW, Gryzenhout M, Jaklitsch WM, Mejia LC, Stoykov D, Udayanga D, Voglmayr H, Walker DM. (2015) Recommendations of generic names in Diaporthales competing for protection or use. IMA Fungus 6(1): 145–154. 10.5598/imafungus.2015.06.01.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Phillips AJL. (2009) Resolving the complex of Diaporthe (Phomopsis) species occurring on Foeniculumvulgare in Portugal. Fungal Diversity 34(11): 111–125. [Google Scholar]
- Santos JM, Correia VG, Phillips AJL. (2010) Primers for mating-type diagnosis in Diaporthe and Phomopsis, their use in teleomorph induction in vitro and biological species definition. Fungal Biology 114(2–3): 255–270. 10.1016/j.funbio.2010.01.007 [DOI] [PubMed] [Google Scholar]
- Santos JM, Vrandečić K, Ćosić J, Duvnjak T, Phillips AJL. (2011) Resolving the Diaporthe species occurring on soybean in Croatia. Persoonia 27(1): 9–19. 10.3767/003158511X603719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake IC, Crous PW, Groenewald JZ, Maharachchikumbura SSN, Jeewon R, Phillips AJL, Bhat DJ, Perera RH, Li QR, Li WJ, Tangthirasunun N, Norphanphoun C, Karunarathna SC, Camporesi E, Manawasighe IS, Al-Sadi AM, Hyde KD. (2017) Families of Diaporthales based on morphological and phylogenetic evidence. Studies in Mycology 86(1): 217–296. 10.1016/j.simyco.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake IC, Jeewon R, Chomnunti P, Wanasinghe DN, Norphanphoun C, Karunarathna A, Pem D, Perera RH, Camporesi E, McKenzie EHC, Hyde KD, Karunarathna SC. (2018) Taxonomic circumscription of Diaporthales based on multigene phylogeny and morphology. Fungal Diversity 93(1): 241–443. 10.1007/s13225-018-0410-z [DOI] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9): 312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Huang S, Xia J, Zhang X, Li Z. (2021) Morphological and molecular identification of Diaporthe species in south-western China, with description of eight new species. MycoKeys 77: 65–95. 10.3897/mycokeys.77.59852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SM, Tan YP, Young AJ, Neate SM, Aitken EAB, Shivas RG. (2011) Stem cankers on sunflower (Helianthusannuus) in Australia reveal a complex of pathogenic Diaporthe (Phomopsis) species. Persoonia 27(1): 80–89. 10.3767/003158511X617110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SM, Tan YP, Neate SM, Grams RM, Shivas RG, Lindbeck K, Aitken EAB. (2018) Diaporthenovem isolated from sunflower (Helianthusannuus) and other crop and weed hosts in Australia. European Journal of Plant Pathology 152(3): 823–831. 10.1007/s10658-018-1515-7 [DOI] [Google Scholar]
- Udayanga D, Liu X, McKenzie EH, Chukeatirote E, Bahkali AH, Hyde KD. (2011) The genus Phomopsis: Biology, applications, species concepts and names of common phytopathogens. Fungal Diversity 50(1): 189–225. 10.1007/s13225-011-0126-9 [DOI] [Google Scholar]
- Udayanga D, Liu X, Crous PW, McKenzie EH, Chukeatirote E, Chukeatirote E, Hyde KD. (2012a) A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal Diversity 56(1): 157–171. 10.1007/s13225-012-0190-9 [DOI] [Google Scholar]
- Udayanga D, Liu X, McKenzie EH, Chukeatirote E, Hyde KD. (2012b) Multi-locus phylogeny reveals three new species of Diaporthe from Thailand. Cryptogamie. Mycologie 33(3): 295–309. 10.7872/crym.v33.iss3.2012.295 [DOI] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2014) Insights into the genus Diaporthe: Phylogenetic species delimitation in the D.eres species complex. Fungal Diversity 67(1): 203–229. 10.1007/s13225-014-0297-2 [DOI] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Hyde KD. (2014a) Species limits in Diaporthe: Molecular re-assessment of D.citri, D.cytosporella, D.foeniculina and D.rudis. Persoonia 32(1): 83–101. 10.3767/003158514X679984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2014b) Insights into the genus Diaporthe: Phylogenetic species delimitation in the D.eres species complex. Fungal Diversity 67(1): 203–229. 10.1007/s13225-014-0297-2 [DOI] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2015) The Diaporthesojae species complex: Phylogenetic re-assessment of pathogens associated with soybean, cucurbits and other field crops. Fungal Biology 119(5): 383–407. 10.1016/j.funbio.2014.10.009 [DOI] [PubMed] [Google Scholar]
- van der Aa HA, Noordeloos ME, de Gruyter J. (1990) Species concepts in some larger genera of the Coelomycetes. Studies in Mycology 32: 3–19. [Google Scholar]
- van Rensburg JCJ, Lamprecht SC, Groenewald JZ, Castlebury LA, Crous PW. (2006) Characterization of Phomopsis spp. associated with dieback of rooibos (Aspalathuslinearis) in South Africa. Studies in Mycology 55: 65–74. 10.3114/sim.55.1.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrandecic K, Cosic J, Jurkovic D, Riccioni L, Duvnjak T. (2007) First report of Phomopsislongicolla on cocklebur (Xanthiumstrumarium) in Croatia. Plant Disease 91(12): 1687–1687. 10.1094/PDIS-91-12-1687B [DOI] [PubMed] [Google Scholar]
- Vrandecic K, Jurkovic D, Riccioni L, Cosic J, Duvnjak T. (2010) Xanthiumitalicum, Xanthiumstrumarium and Arctiumlappa as new hosts for Diaporthehelianthi. Mycopathologia 170(1): 51–60. 10.1007/s11046-010-9289-2 [DOI] [PubMed] [Google Scholar]
- Vrandecic K, Jurkovic D, Cosic J, Postic J, Riccioni L. (2011) First report of cane blight on blackberry caused by Diaportheeres in Croatia. Plant Disease 95(5): 612–612. 10.1094/PDIS-11-10-0860 [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 18: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Wrona CJ, Mohankumar V, Schoeman MH, Tan YP, Shivas RG, Jeff‐Ego OS, Akinsanmi OA. (2020) Phomopsis husk rot of macadamia in Australia and South Africa caused by novel Diaporthe species. Plant Pathology 69(5): 911–921. 10.1111/ppa.13170 [DOI] [Google Scholar]
- Yang Q, Fan XL, Guarnaccia V, Tian CM. (2018) High diversity of Diaporthe species associated with dieback diseases in China, with twelve new species described. MycoKeys 39: 97–149. 10.3897/mycokeys.39.26914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Jiang N, Tian CM. (2020) Three new Diaporthe species from Shaanxi Province, China. MycoKeys 67: 1–18. 10.3897/mycokeys.67.49483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Jiang N, Tian CM. (2021) New species and records of Diaporthe from Jiangxi Province, China. MycoKeys 77: 41–64. 10.3897/mycokeys.77.59999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QM, Yu CL, Li GF, Wang CX. (2018) First report of Diaportheeres causing twig canker on Zizyphusjujuba (Jujube) in China. Plant Disease 102(7): 1458. 10.1094/PDIS-12-17-1910-PDN [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.