Abstract
Vibrio parahaemolyticus is the leading cause of seafood-borne gastroenteritis worldwide. A distinctive feature of the O3:K6 pandemic clone, and its derivatives, is the presence of a second, phylogenetically distinct, type III secretion system (T3SS2) encoded within the genomic island VPaI-7. The T3SS2 allows the delivery of effector proteins directly into the cytosol of infected eukaryotic cells to subvert key host-cell processes, critical for V. parahaemolyticus to colonize and cause disease. Furthermore, the T3SS2 also increases the environmental fitness of V. parahaemolyticus in its interaction with bacterivorous protists; hence, it has been proposed that it contributed to the global oceanic spread of the pandemic clone. Several reports have identified T3SS2-related genes in Vibrio and non- Vibrio species, suggesting that the T3SS2 gene cluster is not restricted to the Vibrionaceae and can mobilize through horizontal gene transfer events. In this work, we performed a large-scale genomic analysis to determine the phylogenetic distribution of the T3SS2 gene cluster and its repertoire of effector proteins. We identified putative T3SS2 gene clusters in 1130 bacterial genomes from 8 bacterial genera, 5 bacterial families and 47 bacterial species. A hierarchical clustering analysis allowed us to define six T3SS2 subgroups (I–VI) with different repertoires of effector proteins, redefining the concepts of T3SS2 core and accessory effector proteins. Finally, we identified a subset of the T3SS2 gene clusters (subgroup VI) that lacks most T3SS2 effector proteins described to date and provided a list of 10 novel effector candidates for this subgroup through bioinformatic analysis. Collectively, our findings indicate that the T3SS2 extends beyond the family Vibrionaceae and suggest that different effector protein repertories could have a differential impact on the pathogenic potential and environmental fitness of each bacterium that has acquired the Vibrio T3SS2 gene cluster.
Keywords: effector proteins, T3SS2, type III secretion, Vibrio, Vibrio parahaemolyticus
Data Summary
All genome sequences used in this study were downloaded from the National Center for Biotechnology Information (NCBI) RefSeq or GenBank databases (see Table S1, available with the online version of this article, for accession numbers).
Files for the type III secretion system T3SS2 reconstructed phylogenetic tree (Newick tree and MSA fasta file), the hierarchical clustering data analysis file from morpheus, Table S1 with genome accession numbers and all the data of the absence/presence of T3SS2-related components, Table S2 with the prediction of novel effector proteins and Table S3 with the distribution of T3SS2s in terms of sequence types are available as part of the online supporting data at the Zenodo data repository (https://doi.org/10.5281/zenodo.7497846). The T3SS2 phylogenetic tree can be interactively visualized at: https://itol.embl.de/tree/19016190125374711626959067#.
Impact Statement.
One of the distinctive features of the O3:K6 pandemic clone of Vibrio parahaemolyticus is the presence of a phylogenetically distinct type III secretion system (T3SS) known as T3SS2. T3SSs are complex nanomachines that allow the delivery of effector proteins directly into the cytosol of eukaryotic cells to manipulate different cellular functions. The Vibrio T3SS2 is the major virulence factor of the pandemic clone and a pivotal contributor to the pathogenesis and environmental fitness of the bacterium. Several studies have identified the T3SS2 gene cluster in other bacterial species beyond the family Vibrionaceae , suggesting that this T3SS can be mobilized through horizontal gene transfer events. In this work, we performed a phylogenetic distribution analysis of the T3SS2 gene cluster in publicly available bacterial genome sequences. We identified putative T3SS2 gene clusters in 1130 bacterial genomes from 47 different bacterial species from 8 genera and 5 bacterial families. Furthermore, we classified these T3SS2s into six different subgroups (I–VI) in terms of their repertoire of effector proteins. Our findings indicate that T3SS2 is not restricted to the family Vibrionaceae and suggest that different effector protein repertories could have a differential impact on the pathogenic potential and environmental fitness of each bacterium that has acquired this T3SS2.
Introduction
Vibrio parahaemolyticus is a Gram-negative bacterium that resides in marine and estuarine environments [1, 2]. In 1996, a pandemic clone of the O3:K6 serotype emerged and spread around the globe, becoming the leading cause of seafood-borne gastroenteritis worldwide [3]. Whole-genome sequencing of the pandemic strain RIMD2210633 revealed the presence of a novel type III secretion system (T3SS2) encoded within an 80 kb genomic island on chromosome 2, known as the V. parahaemolyticus pathogenicity island 7 (VPaI-7) [4]. While all V. parahaemolyticus strains encode a T3SS on chromosome 1 (T3SS1), only V. parahaemolyticus strains derived from the pandemic clone harbour the phylogenetically distinct T3SS2 [4].
T3SSs are complex nanomachines that enable Gram-negative bacteria to deliver proteins, known as effectors, directly from the bacterial cytosol into the cytosol of eukaryotic cells [5, 6]. The translocation of effectors into target cells enables bacteria to subvert a wide variety of host cell functions [7–9]. In addition, the specific repertoire of proteins delivered to infected cells determines which cellular networks are subverted by the bacterium and ultimately influence the outcome of the infection [5, 6, 10]. In this context, even closely related T3SSs can have different repertoires of effector proteins due to horizontal gene transfer (HGT) and/or genomic rearrangement events [11, 12]. A recent global survey described that T3SSs are widely distributed among Gram-negative bacteria, identifying 174 nonredundant T3SSs from 109 distinct bacterial genera [13]. T3SSs have been classified in 13 (I–XIII) major categories based on sequence divergence of T3SS core components [13]. According to the study, the Vibrio T3SS1 is part of category Ib, and the T3SS2 is the founding member of the T3SS phylogenetic category X [13].
Currently, the T3SS2 is considered the main virulence factor of V. parahaemolyticus . Studies in animal models of infection revealed that, while the T3SS1 plays a minor role during infection, the T3SS2 is essential for the enterotoxicity and the development of the clinical signs of the disease [14–17]. Therefore, the T3SS2 is considered the major virulence factor of pandemic V. parahaemolyticus [18]. The gene expression of the T3SS2 gene cluster is induced by bile salts [19, 20] through a regulatory network involving the VtrA, VtrC and VtrB proteins [19–21], and the T3SS2 function requires host cell-surface fucosylation for efficient delivery of effector protein into infected cells [22]. Notably, the importance of the T3SS2 goes beyond increasing the pathogenic potential of pandemic strains, as the T3SS2 increases the environmental fitness of V. parahaemolyticus by promoting bacterial survival against predatory amoeba [23].
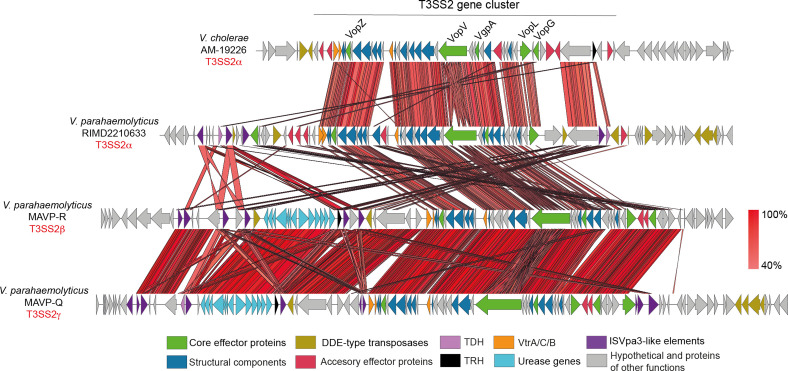
Phylogenetic analysis based on the amino acid sequence divergence of T3SS structural components has identified two distinct lineages of T3SS2, known as phylotypes T3SS2α and T3SS2β [24, 25]. T3SS2α and T3SS2β gene clusters share a similar genetic organization and a subset of effector proteins (Fig. 1) [18]. A more recent study has proposed an additional category within T3SS2β, known as T3SS2γ [26]. The T3SS2γ is very similar to the T3SS2β gene cluster but has some gene content differences, such as the presence of a tdh gene inserted within the T3SS gene cluster [26] (Fig. 1).
Fig. 1.
T3SS2 is divided into two major phylotypes T3SS2α and T3SS2β, and a subcategory within T3SS2β known as T3SS2γ. Schematic depiction of a comparison of the T3SS2 gene clusters in V. cholerae AM-19226, V. parahaemolyticus RIMD2210633, and V. parahaemolyticus MAVP-R and MAVP-Q. tblastx alignment was performed and visualized using Easyfig. Genes encoding different components are highlighted in colours according to the key.
Most of our knowledge of T3SS2 comes from studies of the T3SS2α of V. parahaemolyticus strain RIMD2210633 and the T3SS2α of Vibrio cholerae strain AM-19226 (reviewed elsewhere [18, 27]). In this context, 15 different T3SS2 effector proteins have been identified, of which only 5 (VopZ, VopV, VopG, VopL and VgpA) are shared between V. parahaemolyticus RIMD2210633 and V. cholerae AM-19226 [28–41]. Therefore, these five effectors are called core effector proteins, while the remaining ten are known as accessory effector proteins [18, 28, 38]. While useful, this classification has the limitation of comparing the distribution of effectors of only two Vibrio strains of the same T3SS2 phylotype.
The T3SS2 gene cluster is not restricted to V. parahaemolyticus as similar T3SSs have been found in other species such as non‐O1/non‐O139 V. cholerae , Vibrio mimicus and Vibrio anguillarum [24, 25, 42–44]. Furthermore, several reports have identified the presence of one or more T3SS2-related genes in strains of bacterial genera other than Vibrio , including Aeromonas [45], Grimontia [20], Photorhabdus [46, 47], Providencia [13] and Shewanella [28, 47]. Altogether, these reports suggest that the presence of T3SS2 gene clusters extends beyond the family Vibrionaceae and could have spread through HGT events. Nevertheless, these correspond to isolated observations performed in a limited number of strains and bacterial genomes. Therefore, the overall taxonomic and phylogenetic distribution of T3SS2 and its effector proteins in bacterial genomes is currently unknown.
To address this issue, in this study, we performed a large-scale bioinformatic analysis to gain insight into the phylogenetic distribution of the T3SS2 gene clusters, including effector proteins. Our analysis identified 1130 bacterial genomes encoding putative T3SS2 gene clusters distributed over 47 different bacterial species from 8 bacterial genera. In addition, we classified these T3SS2s into six different subgroups in terms of the repertoire of effector proteins they possess. Our work provides evidence that T3SS2 extends beyond the family Vibrionaceae and that acquisition of both the T3SS2 and different repertoires of effector proteins could have an impact on the pathogenic potential and environmental fitness of these bacteria.
Methods
In silico identification and analyses of T3SS2 loci and components
To identify putative T3SS2 loci, the amino acid sequence of the VPA1338 (SctN) and VPA1339 (SctC) proteins of V. parahaemolyticus RIMD2210633 were used as queries in blastp [48], to perform analyses using publicly available bacterial genome sequences of the National Center for Biotechnology Information (NCBI) database (February 2021). A cut-off of 70 % identity and 80 % sequence coverage threshold were used to select for positive matches. All genome sequence assemblies with positive hits were downloaded from the NCBI RefSeq or GenBank databases (see Table S1 for accession numbers and metadata) and used to build local blast databases. To identify the remaining 35 T3SS2-related components in these 1130 genomes, the nucleotide and amino acid sequences of each component (Table S2) were used as queries for either tblastx or blastp analyses, respectively. A 50 % identity and 80 % sequence coverage threshold were used to select for positive matches. A threshold lower than the first screen was used to identify components that might have a higher sequence divergence. For comparative genomic analysis, nucleotide sequences were visualized and analysed by Artemis version 18.1 [49], Artemis Comparison Tool (act) release 6 [50], the multiple aligner Mauve [51] and Easyfig v2.2.2 [52]. For the hierarchical clustering analysis, a presence/absence matrix of each T3SS2 component was constructed for each bacterial genome (Table S1) and uploaded as a csv file to the online server of the versatile matrix visualization and analysis software morpheus (one minus Pearson correlation, average linkage method) (https://software.broadinstitute.org/morpheus). The hierarchical clustering data analysis file from morpheus is available as part of the online supporting data at the Zenodo data repository (https://doi.org/10.5281/zenodo.7016552). The presence and absence of each identified component was visualized as a coloured matrix generated in GraphPad Prism version 9 (GraphPad).
T3SS2 phylogenetic analyses
For T3SS phylogeny analysis, the 1130 concatenated SctN and SctC amino acid sequences were aligned with ClustalW using mega software version 7.0 [53]. The phylogenetic tree was built from the alignments obtained from mega by performing a bootstrap test of phylogeny (1000 replications) using the maximum-likelihood (ML) method with a Jones–Taylor–Thornton (JTT)+CAT correction model using FastTree2 (Galaxy version 2.1.10+galaxy1) [54] in the Galaxy platform (https://usegalaxy.org/) [55]. For the taxonomic phylogenetic analysis, a 16S RNA phylogenetic tree was reconstructed. The 16S rRNA DNA sequence of each bacterial species was obtained from the NCBI database and aligned with ClustalW using Molecular Evolutionary Genetics Analysis (mega) software. The phylogenetic tree was built from the 16S rRNA DNA alignment by performing a bootstrap test of phylogeny (ultrafast bootstrap, 1 000 replications) using the ML method using iq-tree [56] (Galaxy version 2.1.10+galaxy1). The phylogenetic trees were further visualized and edited to include important metadata with iTOL [57]. Files for the T3SS2 reconstructed phylogenetic tree (Newick file) are available as part of the online supporting data at the Zenodo data repository (https://doi.org/10.5281/zenodo.7016552). The T3SS2 phylogenetic tree can be interactively visualized at https://itol.embl.de/tree/19016190125374711626959067#.
Identification of novel T3SS2 effector candidates
Each ORF within the predicted T3SS2 gene clusters of subgroup VI (including 1 kb upstream and downstream of the first and last predicted component, respectively) was analysed by means of the EffectiveDB [58], Bastion3 [59] and bean2.0 [60] pipelines to identify potential T3SS signal sequences. ORFs with positive hits in at least one of these pipelines was further analysed to predict putative functional domains with InterProScan [61] and NCBI CDD [62]. Prediction of other secretion signals such as for the Sec or TAT systems was carried out with SignalP 6.0 [63]. Finally, each ORF was also analysed by the structure-based homology tool HHpred [64]. Multiple sequence alignments were generated by mafft [65] and T-Coffee Expresso [66], and visualized by ESPript 3.0 [67].
Results and Discussion
T3SS2 gene clusters extend beyond the family Vibrionaceae
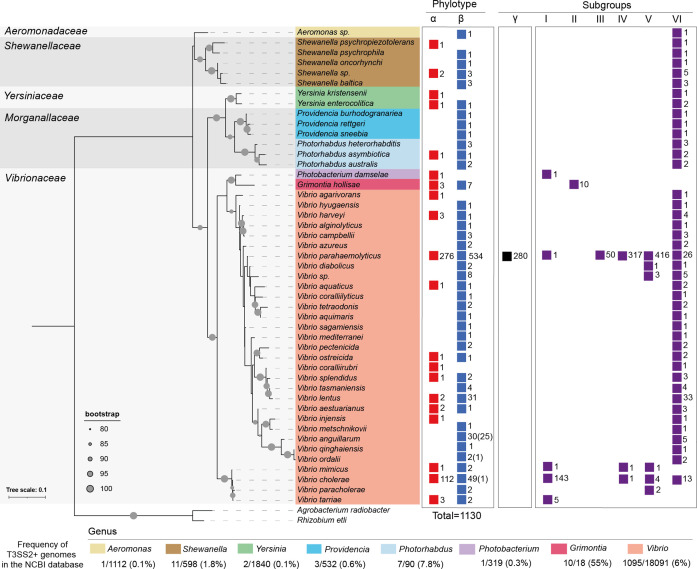
We screened all publicly available bacterial genomes in the NCBI database to identify the full breadth of bacterial species harbouring putative T3SS2 gene clusters. First, we performed blastp analysis in a search for homologues of the VPA1338 (SctN) and VPA1339 (SctC) proteins of V. parahaemolyticus RIMD2210633. SctN and SctC were chosen as they are two of the most conserved T3SS structural proteins, and are often used for phylogenetic clustering and classification of T3SSs [13, 68]. SctN is part of the ATPase complex and SctC is part of the export apparatus [18]. We used stringent cut-offs to avoid identifying homologue proteins from phylogenetically close T3SSs (≥70 % identity and ≥80 % coverage). Our analysis identified the presence of VPA1338 and VPA1339 homologues in 1130 bacterial genomes (Fig. 2, Table S1).
Fig. 2.
T3SS2 extends beyond the family Vibrionaceae . Identification and taxonomic distribution of T3SS2 gene clusters in bacterial genomes. The presence of T3SS2 gene clusters from phylotypes T3SS2α, T3SS2β and different subgroups are shown for each taxonomic category (colour coded). The numbers correspond to the total numbers of bacterial genomes within each taxonomic category that harbour a putative T3SS2 gene cluster. The number of genomes lacking most T3SS2 structural components is shown in parentheses. The ML phylogenetic tree was built from the alignments of 16S RNA DNA sequences from each representative bacterial species with a bootstrap test of phylogeny (1000 replications) with iq-tree and visualized by iTOL. For each bacterial genus, the percentage of T3SSS-positive bacterial genomes from the total NCBI database is shown. The unit of the tree scale corresponds to nucleotide substitutions per site.
To determine whether these 1130 bacterial genomes could encode complete T3SS2 gene clusters, we further screened each genome for the presence of the 37 T3SS2-related components previously identified in V. parahaemolyticus RIMD2210633 (T3SS2α), V. parahaemolyticus MAVP-R (T3SS2β) and V. cholerae AM-19226 (T3SS2α) (Table S1). These include ORFs encoding 16 structural proteins, 15 bacterial effector proteins, 3 regulatory proteins (VtrA, VtrB and VtrC) and each of the 3 haemolysins linked to T3SS2 gene clusters (TDHVp, TRHVc and TRHVp). Most of the 1130 bacterial genomes (1106) encoded each of the T3SS structural genes, suggesting that they most likely harbour complete T3SS2 gene clusters. The exceptions were 24 V . anguillarum , 1 Vibrio ordalii and 1 V . cholerae genomes that only possess 8, 3 and 7 of the 16 T3SS2 structural components, respectively (Table S1). This was not unexpected, as it has been previously reported that some V. anguillarum strains lack most of the T3SS2 structural components due to a large internal deletion of the T3SS2 gene cluster [43], suggesting that these strains do not encode functional T3SS2s.
Taxonomic distribution analysis identified putative T3SS2 gene clusters in 47 bacterial species from 5 distinct families ( Aeromonadaceae , Shewanellaceae , Yersiniaceae , Morganellaceae and Vibrionaceae ) and 8 bacterial genera ( Aeromonas , Shewanella , Yersinia , Providencia , Photorhabdus , Photobacterium , Grimontia and Vibrio ) (Fig. 2). In some genera, we identified multiple species harbouring putative T3SS2 gene clusters, while in other genera we could identify as little as one bacterial species harbouring T3SS2 gene components (Fig. 2). In terms of bacterial species, the genus Vibrio contained the largest number of different species harbouring putative T3SS2 gene clusters (31 species), followed by Shewanella (5 species), Photorhabdus and Providencia (each with 3 species), Yersinia (2 species), and Photobacterium , Grimontia and Aeromonas (each with a single species). Finally, our analysis did not identify bacterial strains harbouring more than one T3SS2 gene cluster within their genomes. Interestingly, several of these T3SS2s were identified in recently validated or proposed Vibrio and Shewanella species, including Vibrio paracholerae [69], Vibrio tarriae [70], 'Vibrio tetraodonis' [71], Shewanella oncorhynchi [72] and 'Shewanella psychropiezotolerans' [73] (Fig. 2, Table S1).
Considering the total number of genome assemblies available in the NCBI database, putative T3SS2 gene clusters were more frequently identified in bacterial genomes from the genera Grimontia (55 %), Photorhabdus (7.8 %), Vibrio (6 %) and Shewanella (1.8 %), with a lower frequency in genomes from the genera Providencia (0.6 %), Photobacterium (0.3 %), Aeromonas (0.1 %) and Yersinia (0.1 %) (Fig. 2, Table S1). While this suggests that T3SS2 is overrepresented in some bacterial lineages, it could also reflect potential sequencing biases in terms of the diversity of bacterial genome sequences deposited in the NCBI database (e.g., clinical versus environmental and different geographical origins).
Interestingly, most of the different bacterial species identified as harbouring putative T3SS2 gene clusters can be found in aquatic habitats, suggesting that acquisition and transfer of T3SS2 actively occur in this environment. Among the exceptions was the identification of T3SS2 gene clusters in genomes of strains of the genus Photorhabdus (Fig. 2, Table S1). Bacteria from the genus Photorhabdus belonged to three species that are important insect pathogens, including some that can also cause human disease [74]. Wilkinson et al. [46] previously identified a T3SS2 gene cluster in the genome of Photorhabdus asymbiotica strain ATCC 43949. Our analysis expanded this finding by identifying complete T3SS2 gene clusters in different strains of Photorhabdus australis , Photorhabdus heterorhabditis and P. asymbiotica species (Fig. S1, Table S1). Every T3SS2 gene cluster identified is inserted in the same genome locus, which corresponded to the pauO33480 locus in the Photorhabdus laumondii strain TT01 genome, as previously reported for P. asymbiotica strain ATCC 43949 [46]. We also identified T3SS2 gene clusters in one strain of Yersinia kristensenii and two strains of Yersinia enterocolitica (Fig. 2, Table S1). Interestingly, while Y. enterocolitica can be found in aquatic environments, the two strains with putative T3SS2 gene clusters were isolated from wild ungulate carcasses and belong to the highly virulent biotype 1B [75].
Altogether, the taxonomic distribution of T3SS2 gene clusters in bacterial genomes indicates that this secretion system is not restricted to the family Vibrionaceae , suggesting that T3SS2 gene clusters can mobilize through HGT events among environmental bacteria. In support of this notion, it has been shown that under laboratory conditions naturally competent T3SS-negative V. cholerae strains can acquire and integrate the T332 gene cluster into their chromosomes, suggesting that transfer of this gene cluster might occur in natural environments [76].
T3SS2β phylotype is more abundant and divergent than the phylotype T3SS2α
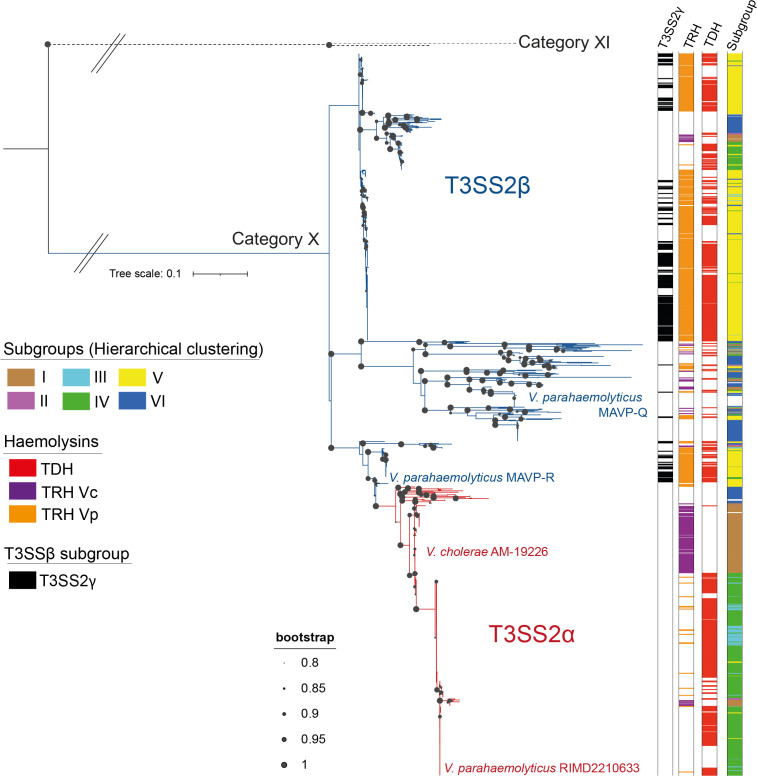
T3SS2 is divided into two major phylotypes known as T3SS2α and T3SS2β [18], and an additional subcategory within T3SS2β known as T3SS2γ [26]. To determine the distribution of T3SS2α, T3SS2β and T3SS2γ among the 1130 T3SS2s identified, we performed a phylogenetic analysis using concatenated SctN and SctC amino acid sequences. We also included the concatenated SctN and SctC sequences of the Agrobacterium radiobacter K84 and Rhizobium etli CIAT 652 T3SSs, which are representatives of T3SS category XI, the closest phylogenetic category to the Vibrio T3SS2 [13]. The phylogenetic clustering analysis showed that all the 1130 putative T3SS2s grouped closely together, while the T3SS of Agrobacterium radiobacter K84 and R. etli CIAT 652 clustered in a more distant branch (Fig. 3), supporting the notion that all the 1130 T3SSs identified in this work correspond to bona fide T3SS2s.
Fig. 3.
Phylogenetic analysis of the identified T3SS2s. ML phylogenetic tree built from the alignments of concatenated SctN and SctC amino acid sequences with a bootstrap test of phylogeny (1000 replications), with a Jones–Taylor–Thornton (JTT) correction model, and visualized by iTOL. The presence of haemolysis and subgroups are shown as coloured strips. T3SS2α and T3SS2β are shown by red and blue branches, respectively. The unit of the tree scale corresponds to nucleotide substitutions per site. The bacterial strains of representative T3SS2α and T3SS2β gene clusters are also highlighted in red and blue colours, respectively.
To classify these T3SS2s in terms of phylotypes (α or β), we used the previously classified T3SS2α of V. cholerae AM-19226 and V. parahaemolyticus RIMD2210633, and the T3SS2β of V. parahaemolyticus MAVP-Q and MAVP-R strains, to identify the major two branches that separate these two phylotypes (Fig. 3). Our analysis also revealed that 36.7 % (n=415) of the identified T3SS2s belonged to phylotype T3SS2α and 63 .3 % (n=715) to phylotype T3SS2β. The phylogenetic analysis also revealed that the greater sequence divergence was observed within the T3SS2β phylotype (Fig. 3).
As mentioned above, recent reports have created a subcategory within T3SS2β of V. parahaemolyticus known as T3SS2γ (based mainly on the presence of both the tdh and trh genes) [26]. To determine the percentage of T3SS2γ gene clusters within the T3SS2β family, we performed a gene content analysis based on the definition of T3SS2γ gene clusters proposed by Hu et al. [13]. The analysis identified that 40 .5 % (n=280) of T3SS2β gene clusters could be classified as T3SS2γ. Notably, the presence of T3SS2γ did not cluster to any major clade of the T3SS2 phylogenetic tree (Fig. 3), supporting the notion that T3SS2γ is not a third T3SS2 phylotype but rather a subcategory within T3SS2β.
Taxonomic distribution analysis identified the presence of T3SS2α and/or T3SS2β in strains of each of the five bacterial families (Figs 2 and 3), with no clear overrepresentation of any phylotype within a particular bacterial family. Nevertheless, T3SS2β was present in a more significant number of different bacterial species (41 out of 47) in comparison to T3SS2α (20 out of 47 species) (Fig. 2). Finally, the T3SS2γ subcategory was identified solely in V. parahaemolyticus strains, which agrees with previous reports [26]. Since many V. parahaemolyticus strains that harbour T3SS2 gene clusters are related to the RIMD2210633 pandemic clone, we analysed the distribution of T3SS2 phylotypes among different V. parahaemolyticus sequence types (STs). Our analysis showed that T3SS2α was overrepresented in V. parahaemolyticus strains from ST3 (44 %), ST120 (11.5 %), ST189 (7 %) and ST332 (16.4 %), while the T3SS2β was overrepresented in strains from ST3 (11.5 %), ST36 (25.2 %) and ST631 (7.5 %) (Table S3).
T3SS2 gene clusters share a high degree of synteny
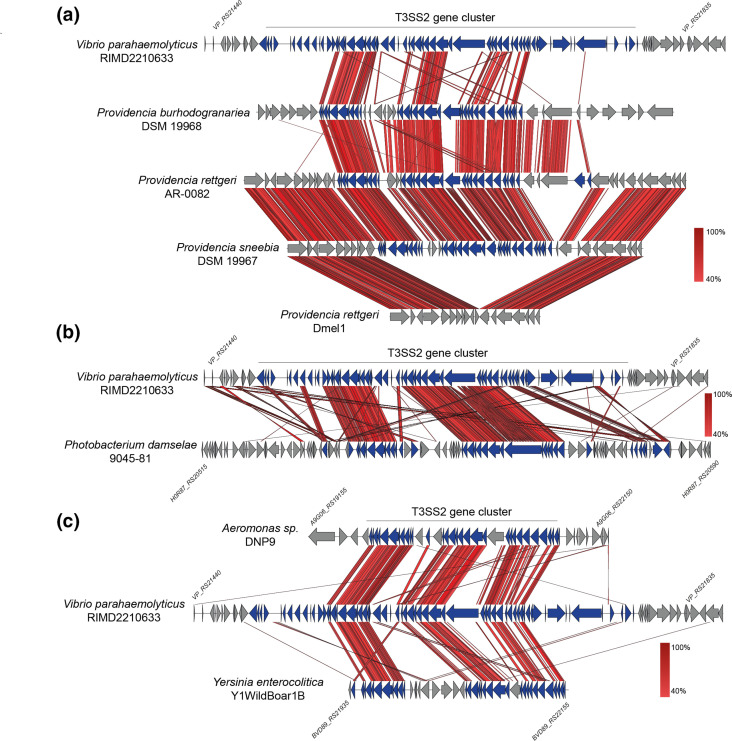
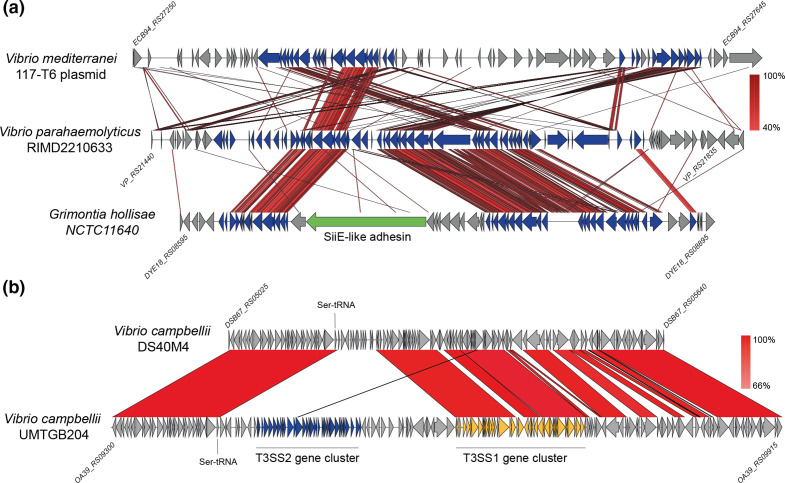
To gain insight into the genomic context of the identified T3SS2 gene clusters, we performed a comparative genomic analysis between the T3SS2 gene cluster of V. parahaemolyticus RIMD2210633 and representative strains of 74 non- Vibrio bacterial species (Table S1) with complete or almost complete bacterial genomes (large enough contigs to allow a comparative analysis). Notably, there was a high gene synteny between the T3SS2 gene cluster of V. parahaemolyticus RIMD2210633 and the gene clusters of bacterial species of different genera (e.g. Providencia , Photobacterium , Grimontia and Photorhabdus ) (Figs 4a, b, 5a and S1). In each of the T3SS2 gene clusters identified in Grimontia hollisae strains, a large gene encoding a putative adhesin (SiiE-like adhesin) is encoded in the middle of the T3SS2 gene cluster (Fig. 5a). A high gene synteny was also observed between the T3SS2 gene clusters with a lower sequence identity, such as the gene clusters of Aeromonas sp. DNP9 and Y. enterocolitica Y1Wildboar1B (Fig. 4c).
Fig. 4.
T3SS2 gene clusters share a high degree of synteny. Schematic depiction of a comparison of the T3SS2 gene clusters in V. parahaemolyticus RIMD2210633 in comparison to the gene clusters of strains of Providencia (a), Photobacterium (b), and Aeromonas and Y. enterocolitica strains (c). tblastx alignments were performed and visualized using Easyfig. Genes encoding T3SS-related genes are highlighted in blue.
Fig. 5.
Distinctive genomic locations of T3SS2 gene clusters. (a) Comparison of the T3SS2 gene cluster in V. parahaemolyticus RIMD2210633 with the T3SS2 gene cluster encoded in V. mediterranei 116-T6 plasmid and the T3SS2 gene cluster of G. hollisae NCTC11640. tblastx alignment was performed and visualized using Easyfig. (b) Schematic depiction of a comparison of the T3SS1 and T3SS2 gene clusters inserted in the vicinity of the serine tRNA gene in V. campbellii strain UMTGB204. blastn alignment was performed and visualized using Easyfig. Genes encoding T3SS-related genes are highlighted in blue.
For Vibrio species, we also analysed the genome location of these T3SS2 gene clusters. In addition to the described chromosomal location of T3SS2 within VPAI-7 of V. parahaemolyticus RIMD2210633 [4], we identified a T3SS2 gene cluster within the plasmid of a recently sequenced Vibrio mediterranei strain 117-T6 (CP033579) [77] (Fig. 5a). Another interesting observation was the presence of a T3SS2 gene cluster inserted right next to a T3SS1 gene cluster in Vibrio campbellii strain UMTGB204. In this strain, both T3SSs are inserted within the vicinity of the same serine tRNA (Fig. 5b), in which the T3SS1 of V. parahaemolyticus RIMD2210633 is located.
Hierarchical clustering analysis identifies six subgroups with different repertoires of effector proteins
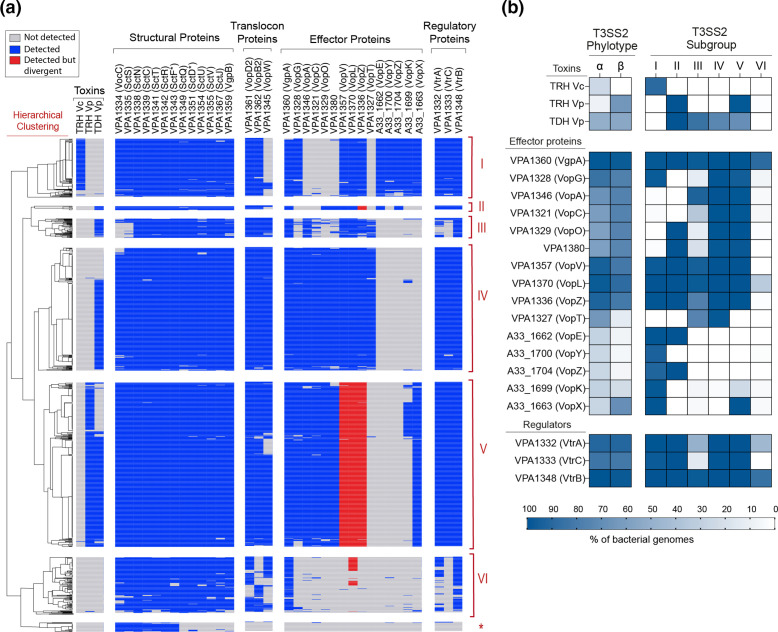
While phylogenetic analysis of T3SSs using core structural components provides insight into the evolution of T3SSs, the acquisition, transfer or loss of accessory and effector encoding genes provides an additional layer of complexity to the evolution of these systems [11, 13, 68]. To gain further insight into this diversity, we performed a hierarchical clustering analysis of 37 T3SS2-related genes in the 1130 bacterial genomes (Fig. 6). For the analysis of the effector proteins, we included each T3SS2 effector described for either V. parahaemolyticus RIMD2210633 or V. cholerae AM-19226 [18]. The hierarchical clustering analysis allowed us to identify six major subgroups (I to VI) with similar gene content profiles (Fig. 6a, Table S1).
Fig. 6.
Hierarchical clustering analysis of T3SS2-related genes identifies six distinct subgroups. (a) Clustering analysis was performed using versatile matrix visualization and analysis software morpheus (one minus Pearson correlation, average linkage method). Boxes representing genes not detected, detected or detected but divergent (50–70 % amino acid sequence identity with at least 80 % sequence coverage) are coloured in grey, blue or red, respectively. An asterisk highlights the group of V. anguillarum genomes that only harboured a limited number of T3SS structural components. (b) Comparison of the frequency of detection of T3SS2 effector proteins, toxins and regulators between each T3SS2 phylotype and subgroup. Frequencies are colour coded and expressed as percentages.
Most structural components were highly conserved in all subgroups, except for the hydrophilic translocator VopW of subgroups I and VI, the structural component SctS and the VocC chaperone of subgroup III, and the VopB2 and VopD2 translocon proteins of subgroup VI (Fig. 6a). While it is possible that these gene clusters do not encode such components, the inability to identify these components could be due to the lower sequencing coverage (<68×) of most of these bacterial genomes. The vtrA, vtrB and vtrC genes, which encode the regulatory complex that induces the expression of T3SS2 gene clusters in response to bile [19, 20], were also highly conserved, except for subgroups III and VI, where many bacterial strains harboured divergent homologues of VtrA and VtrC (Fig. 6a).
Since most structural genes were highly conserved across all genomes, the hierarchical clustering analysis mainly classified each T3SS2 based on the repertoire of effector proteins identified. As shown in Fig. 6(b), each T3SS2 subgroup was enriched in a distinct set of effector proteins and associated haemolysins. While most subgroups were enriched for a combination of 10–12 different T3SS2 effectors (subgroups I to V), subgroup VI was vastly underrepresented in terms of the presence of known effector proteins. This subgroup was primarily enriched in the VgpA effector protein, and lacking most other known T3SS2 effector proteins. The presence of VgpA homologues in each subgroup supports this effector's critical role in T3SS2 function [37, 38]. As expected, the current phylotype classification (T3SS2α versus T3SS2β) was a poor predictor of the presence/absence of individual effector proteins (Fig. 6a, b), highlighting the utility of a gene-content classification scheme.
Interestingly, most T3SS2 subgroups were enriched for genomes of particular bacterial species (Fig. 2, Table S1): (i) subgroup I included strains from five different bacterial species (Photorhabdus damselae, V. cholerae , V. mimicus , V. parahaemolyticus and V. tarriae); (ii) subgroup II included strains only from G. hollisae species; (iii) subgroup III included V. parahaemolyticus strains exclusively; (iv) subgroup IV included strains from three species ( V. cholerae , V. mimicus and V. parahaemolyticus ); (v) subgroup V included strains from six species ( V. parahaemolyticus , V. cholerae , Vibrio diabolicus , V. mimicus, V. paracholerae and Vibrio sp.); (vi) subgroup VI harboured the most diverse set of bacterial species with a total of 42 bacterial species (Fig. 2). Subgroup VI was also the only subgroup with bacterial strains beyond the family Vibrionaceae . Finally, while no T3SS2 subgroup was exclusive to a particular T3SS2 phylotype, some subgroups were indeed overrepresented by different phylotypes. Subgroups I, III and IV were overrepresented in strains from the T3SS2α phylotype, while subgroups II, V and VI were overrepresented in strains from the T3SS2β phylotype (Fig. 3, Table S1). Finally, every V. parahaemolyticus strain of the T3SS2γ subcategory clustered within subgroup V (Table S1).
Altogether, the hierarchical clustering analysis suggests that T3SS2s have acquired a diverse set of effector proteins that could ultimately impact on the contribution of each T3SS2 to the pathogenic potential and environmental fitness of different bacterial strains.
T3SS2 subgroup VI encodes 10 novel effector candidates
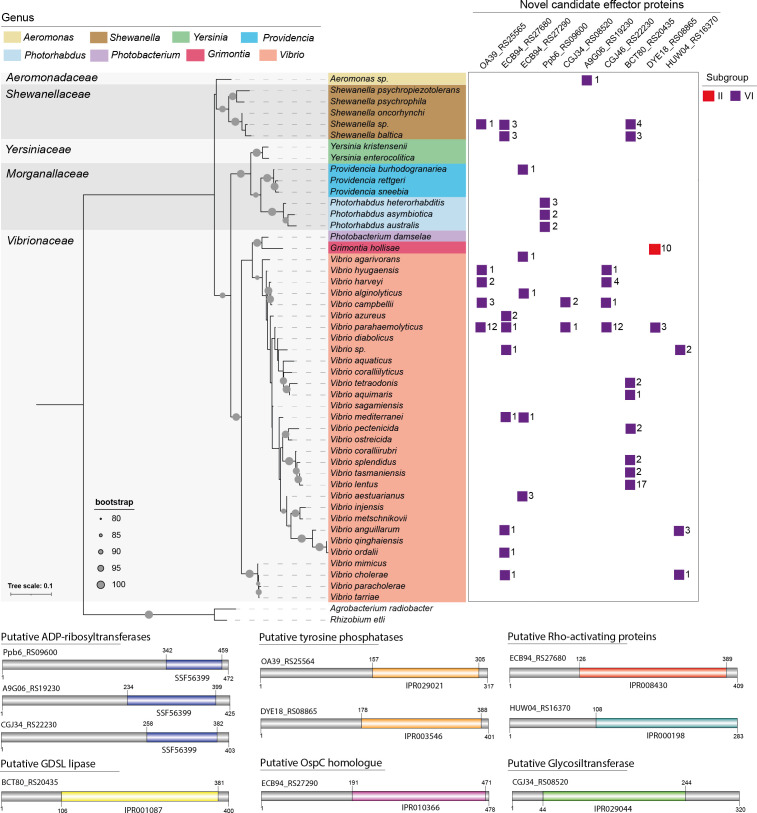
As mentioned above, the T3SS2 gene clusters of subgroup VI stand out as they are the most taxonomically diverse (Fig. 2) and lack most of the known T3SS2 effector proteins (Fig. 6). The high degree of synteny of these gene clusters to the T3SS2 gene cluster of V. parahaemolyticus RIMD2210633 (Figs 4 and S1) and their distribution in the T3SS2 phylogenetic tree (Fig. 3) suggests that these are not part of a phylogenetically distinct T3SS2, but rather have a different set of effector proteins. To test this hypothesis, we analysed the vicinity of the T3SS2 gene cluster from one strain of each species of subgroup VI (Table S1) and performed bioinformatic analysis to identify putative T3SS2 effector candidates. We analysed each ORF of these T3SS2 clusters based on four approaches, including: (i) identification of putative T3SS secretion signals by T3SS effector prediction pipelines (bean2.0, Bastion3 and EffectiveDB); (ii) bioinformatic analysis to identify conserved functional domains (InterProScan, NCBI CDD); (iii) sequence identity to known T3SS effector proteins of other bacterial species (blastp, amino acid sequence identity >40 %); and (iv) functional predictions via remote homology searches using the HHpred pipeline.
Our analysis identified a set of 10 novel effector candidates that harboured putative T3SS secretion signal sequences and putative functional domains present in known T3SS effector proteins. These included putative ADP-ribosyltransferases, tyrosine phosphatases, glycosyltransferases and Rho-activating proteins (Table 1, Figs 7 and S2). In addition, five of these ORFs encode homologues of known T3SS effector proteins or toxins such as the effector proteins OspC1, AoPH and VopC of Shigella flexneri , Aeromonas hydrophila and V. parahaemolyticus , respectively (Tables 1 and S2, Fig. 7). The 10 effector candidates identified were present in diverse bacterial species of the T3SS2 subgroup VI, except for the putative tyrosine phosphatases (DYE18_RS08865) that also was identified in G. hollisae strains of T3SS2 subgroup II (Fig. 7, Table S2).
Table 1.
Prediction of T3SS effector candidates within the T3SS2 subgroup VI
Representative ORFs for each prediction are shown with the results of different prediction software (1, bean2.0; 2, BastionX; 3, EffectiveT3; 4, EffectiveCCBD; 5, EffectiveELD), the identification of putative functional domains (InterproScan), and the amino acid sequence identity to known T3SS effector proteins from other organisms. ND, Not determined.
|
Effector candidate |
Length (aa) |
T3SS effector prediction |
Functional domain prediction |
Similarity to known T3SS effectors or toxins |
|---|---|---|---|---|
|
OA39_RS25565 |
317 |
1, 2, 3, 4 |
Tyrosine phosphatase (IPR029021) |
nd |
|
ECB94_RS27680 |
409 |
1, 2, 3, 4 |
CNF1 Rho-activating domain (IPR008430) |
27.7 % identity to VopC (blastp, 3e-48 E value) |
|
ECB94_RS27290 |
478 |
1, 2, 3, 4 |
Shigella flexneri OspC (IPR010366) |
49.9 % identity to OspC1 (blastp, 5e-153 E value) |
|
Ppb6_RS09600 |
472 |
1, 3, 4 |
ADP-ribosylation (SSF56399) |
nd |
|
CGJ34_RS08520 |
320 |
1, 3, 4 |
Glycosyltransferase (IPR029044) |
Similarity to Lgt1 toxin (HHpred, 2.4e-16 E value) |
|
A9G06_RS19230 |
425 |
1, 2 |
ADP-ribosylation (SSF56399) |
nd |
|
CGJ46_RS22230 |
403 |
1, 4 |
ADP-ribosylation (SSF56399) |
nd |
|
BCT80_RS20435 |
400 |
1 |
GDSL lipase (IPR001087) |
22.4 % identity to VPA0226 (blastp, 3e-18 E value) |
|
DYE18_RS08865 |
401 |
1, 5 |
Tyrosine phosphatase SptP/YopH (IPR003546) |
61.2 % identity to AopH (blastp, 1e-119 E value) |
|
HUW04_RS16370 |
283 |
1, 5 |
Rho GTPase-activating domain (IPR000198) |
nd |
Fig. 7.
T3SS2 subgroup VI encodes 10 novel effector candidates. The figure shows the taxonomic distribution of each predicted effector protein identified in bacterial genomes. The numbers correspond to the total numbers of bacterial genomes within each taxonomic category that harbour novel effector candidates. A schematic representation of novel candidates highlighting the presence of predicted functional domains in different colours is shown at the bottom. The unit of the tree scale corresponds to nucleotide substitutions per site.
Interestingly, one candidate (BCT80_RS20435) harboured a predicted GDSL lipase domain (IPR001087) and a 22.5 % amino acid sequence identity to the VPA0226 protein of V. parahaemolyticus (Fig. S3). VPA0226 is a secreted lipase required for V. parahaemolyticus RIMD2210633 to egress from the infected cell. Notably, in V. parahaemolyticus , VPA0226 is not secreted by the T3SS2 but rather by the T2SS (type II secretion system) [78]. Unlike VPA0226, analysis of BCT80_RS20435 showed no detectable T2SS or TAT secretion signal (Fig. S3).
Overall, our analysis suggests that subgroup VI encodes a subset of novel T3SS2 effector proteins. Further work is needed to confirm that each candidate corresponds to T3SS2 effector proteins.
Revisiting the current classification of core and accessory T3SS2 effector proteins
Recently, Matsuda et al. [18] proposed a classification of T3SS2 effector proteins based on the presence of these effectors in V. parahaemolyticus and V. cholerae . Effectors present in both V. cholerae and V. parahaemolyticus are known as core effectors, and those present in either V. cholerae or V. parahaemolyticus have been called accessory effector proteins. While useful, this classification relies on analysing only two strains of two bacterial species.
To revisit this classification, we analysed the presence/absence of each T3SS2 effector protein in the 47 bacterial species in which we have identified putative T3SS2 gene clusters. As shown in Fig. S4, only VgpA was present in at least one strain of most bacterial species (except for Y. enterocolitica and Y. kristensenii strains). The high prevalence of this effector protein could be explained by its additional structural role [38]. Another effector, VopT, was identified exclusively in V. parahaemolyticus strains. The remaining 13 T3SS2 effector proteins were present at various degrees with no overrepresentation in any bacterial species. Altogether, our data show a wide distribution of T3SS2 effector proteins in different bacterial species and that there is no clear correlation between the number of bacterial species that harbour a particular effector protein, which is the basis of the current classification scheme. We argue that a better alternative is to classify T3SS2s based on the repertoire of their effector proteins in the context of all identified T3SS2 loci in bacterial genomes (subgroups I–VI).
Nevertheless, such a classification scheme is not without limitations. First, we believe that for such a classification scheme to be useful a larger dataset of T3SS2-positive bacterial genomes beyond the family Vibrionaceae are needed, to fully grasp the diversity of this system. Second, a large fraction of these genomes would need to be of high quality to avoid sequence coverage bias. And finally, an inherent limitation is that this classification scheme could turn out to be dynamic, as it may change over time if novel effector encoding genes are acquired through HGT or if some are lost due to genomic rearrangements.
It has been shown that genes encoding bacterial effector proteins can be acquired through HGT, and can also further evolve through terminal reassortment and pseudogenization events [11, 12, 79–81]. This supports the notion that different repertoires of effector proteins will be selected by different bacterial species in terms of their contribution to their adaptation to different ecological niches. In this context, it could be possible that different T3SS2-positive bacterial strains use different effector protein repertoires (subgroups I–VI) to differentially interact with environmental eukaryotic hosts which ultimately would impact on their pathogenic potential.
Conclusion
The acquisition and transfer of T3SS gene clusters can potentially increase the virulence and the environmental fitness of the recipient bacteria. In this context, it has been proposed that the acquisition of the T3SS2 gene cluster by V. parahaemolyticus was a crucial factor in the global spread, and increased pathogenic potential of the O3:K6 pandemic clone and its derivatives. In this study, we performed a genome-wide analysis to determine the phylogenetic distribution of the Vibrio T3SS2 gene cluster in public genome databases. Our analysis revealed that T3SS2 gene clusters extend beyond the family Vibrionaceae , share a high degree of synteny and that it is possible to categorize T3SS2 in six different subgroups (I–VI) in terms of their repertoire of effector proteins. Notably, it is plausible to think that acquisition of the T3SS2 gene cluster by HGT events in each of the bacteria identified in this study could increase their human pathogenic potential and environmental fitness, just as it has been proposed for the V. parahaemolyticus pandemic clone. Finally, since the repertoire of T3SS effector proteins delivered to the target cells ultimately defines the cellular outcome of an infection, we believe that future studies should focus on testing the contribution of each repertoire of T3SS2 effector proteins to the pathogenic potential and environmental fitness of these bacterial species.
Supplementary Data
Funding information
This work was funded by the Howard Hughes Medical Institute (HHMI)–Gulbenkian International Research Scholar grant number 55008749 and FONDECYT grant 1201805 (Agencia Nacional de Investigación y Desarrollo; ANID) (to C.J.B.). S.A.J. was supported by national PhD grant 21210879 (ANID). I.M.U. was supported by FONDECYT grant 3200874 (ANID). V.B. was supported by DICYT (Dirección de Investigación Científica y Tecnológica) grant 022001BZ Universidad de Santiago de Chile (USACH).
Acknowledgements
We thank members of the Blondel Laboratory for helpful discussions on all aspects of this project and for their comments on the manuscript.
Author contributions
S.A.J.: investigation, visualization, review and editing final manuscript. N.P.: investigation, visualization, review and editing final manuscript. I.M.U.: investigation, visualization, review and editing final manuscript. V. B.: investigation, visualization, review and editing final manuscript. C.J.B.: conceptualization, methodology, formal analysis, investigation, visualization, resources, data curation, writing – original draft and review and editing, funding acquisition, supervision and project administration.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: HGT, horizontal gene transfer; ML, maximum likelihood; NCBI, National Center for Biotechnology Information; ST, sequence type; T3SS, type III secretion system.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Supporting data are available via Zenodo (https://doi.org/10.5281/zenodo.7016552). Four supplementary figures and three supplementary tables are available with the online version of this article.
References
- 1.Brumfield KD, Usmani M, Chen KM, Gangwar M, Jutla AS, et al. Environmental parameters associated with incidence and transmission of pathogenic Vibrio spp. Environ Microbiol. 2021;23:7314–7340. doi: 10.1111/1462-2920.15716. [DOI] [PubMed] [Google Scholar]
- 2.Letchumanan V, Chan K-G, Lee L-H. Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front Microbiol. 2014;5:705. doi: 10.3389/fmicb.2014.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Velazquez-Roman J, León-Sicairos N, de Jesus Hernández-Díaz L, Canizalez-Roman A. Pandemic Vibrio parahaemolyticus O3:K6 on the American continent. Front Cell Infect Microbiol. 2014;3:110. doi: 10.3389/fcimb.2013.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae . Lancet. 2003;361:743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 5.Galán JE, Lara-Tejero M, Marlovits TC, Wagner S. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol. 2014;68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lara-Tejero M, Galán JE. Protein secretion in bacteria. EcoSal Plus. 2019;8:245–259. doi: 10.1128/9781683670285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Portaliou AG, Tsolis KC, Loos MS, Zorzini V, Economou A. Type III secretion: building and operating a remarkable nanomachine. Trends Biochem Sci. 2016;41:175–189. doi: 10.1016/j.tibs.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Dean P. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol Rev. 2011;35:1100–1125. doi: 10.1111/j.1574-6976.2011.00271.x. [DOI] [PubMed] [Google Scholar]
- 9.Scott NE, Hartland EL. Post-translational mechanisms of host subversion by bacterial effectors. Trends Mol Med. 2017;23:1088–1102. doi: 10.1016/j.molmed.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Ruano-Gallego D, Sanchez-Garrido J, Kozik Z, Núñez-Berrueco E, Cepeda-Molero M, et al. Type III secretion system effectors form robust and flexible intracellular virulence networks. Science. 2021;371:eabc9531. doi: 10.1126/science.abc9531. [DOI] [PubMed] [Google Scholar]
- 11.Brown NF, Finlay BB. Potential origins and horizontal transfer of type III secretion systems and effectors. Mob Genet Elements. 2011;1:118–121. doi: 10.4161/mge.1.2.16733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stavrinides J, Ma W, Guttman DS. Terminal reassortment drives the quantum evolution of type III effectors in bacterial pathogens. PLoS Pathog. 2006;2:e104. doi: 10.1371/journal.ppat.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Huang H, Cheng X, Shu X, White AP, et al. A global survey of bacterial type III secretion systems and their effectors. Environ Microbiol. 2017;19:3879–3895. doi: 10.1111/1462-2920.13755. [DOI] [PubMed] [Google Scholar]
- 14.Yang H, de Souza Santos M, Lee J, Law HT, Chimalapati S, et al. A novel mouse model of enteric Vibrio parahaemolyticus infection reveals that the type III secretion system 2 effector VopC plays a key role in tissue invasion and gastroenteritis. mBio. 2019;10:e02608-19. doi: 10.1128/mBio.02608-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritchie JM, Rui H, Zhou X, Iida T, Kodoma T, et al. Inflammation and disintegration of intestinal villi in an experimental model for Vibrio parahaemolyticus-induced diarrhea. PLoS Pathog. 2012;8:e1002593. doi: 10.1371/journal.ppat.1002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piñeyro P, Zhou X, Orfe LH, Friel PJ, Lahmers K, et al. Development of two animal models to study the function of Vibrio parahaemolyticus type III secretion systems. Infect Immun. 2010;78:4551–4559. doi: 10.1128/IAI.00461-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubbard TP, Chao MC, Abel S, Blondel CJ, Abel Zur Wiesch P, et al. Genetic analysis of Vibrio parahaemolyticus intestinal colonization. Proc Natl Acad Sci USA. 2016;113:6283–6288. doi: 10.1073/pnas.1601718113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuda S, Hiyoshi H, Tandhavanant S, Kodama T. Advances on Vibrio parahaemolyticus research in the postgenomic era. Microbiol Immunol. 2020;64:167–181. doi: 10.1111/1348-0421.12767. [DOI] [PubMed] [Google Scholar]
- 19.Gotoh K, Kodama T, Hiyoshi H, Izutsu K, Park K-S, et al. Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants. PLoS One. 2010;5:e13365. doi: 10.1371/journal.pone.0013365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P, Rivera-Cancel G, Kinch LN, Salomon D, Tomchick DR, et al. Bile salt receptor complex activates a pathogenic type III secretion system. Elife. 2016;5:e15718. doi: 10.7554/eLife.15718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kodama T, Gotoh K, Hiyoshi H, Morita M, Izutsu K, et al. Two regulators of Vibrio parahaemolyticus play important roles in enterotoxicity by controlling the expression of genes in the Vp-PAI region. PLoS One. 2010;5:e8678. doi: 10.1371/journal.pone.0008678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blondel CJ, Park JS, Hubbard TP, Pacheco AR, Kuehl CJ, et al. CRISPR/Cas9 screens reveal requirements for host cell sulfation and fucosylation in bacterial type III secretion system-mediated cytotoxicity. Cell Host Microbe. 2016;20:226–237. doi: 10.1016/j.chom.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matz C, Nouri B, McCarter L, Martinez-Urtaza J. Acquired type III secretion system determines environmental fitness of epidemic Vibrio parahaemolyticus in the interaction with bacterivorous protists. PLoS One. 2011;6:e20275. doi: 10.1371/journal.pone.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okada N, Iida T, Park K-S, Goto N, Yasunaga T, et al. Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect Immun. 2009;77:904–913. doi: 10.1128/IAI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okada N, Matsuda S, Matsuyama J, Park K-S, de los Reyes C, et al. Presence of genes for type III secretion system 2 in Vibrio mimicus strains. BMC Microbiol. 2010;10:302. doi: 10.1186/1471-2180-10-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu F, Gonzalez-Escalona N, Drees KP, Sebra RP, Cooper VS, et al. Parallel evolution of two clades of an Atlantic-endemic pathogenic lineage of Vibrio parahaemolyticus by independent acquisition of related pathogenicity islands. Appl Environ Microbiol. 2017;83:e01168-17. doi: 10.1128/AEM.01168-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller KA, Tomberlin KF, Dziejman M. Vibrio variations on a type three theme. Curr Opin Microbiol. 2019;47:66–73. doi: 10.1016/j.mib.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plaza N, Urrutia IM, Garcia K, Waldor MK, Blondel CJ. Identification of a family of Vibrio type III secretion system effectors that contain a conserved serine/threonine kinase domain. mSphere. 2021;6:e00599-21. doi: 10.1128/mSphere.00599-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Krachler AM, Broberg CA, Li Y, Mirzaei H, et al. Type III effector VopC mediates invasion for Vibrio species. Cell Rep. 2012;1:453–460. doi: 10.1016/j.celrep.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kodama T, Rokuda M, Park K-S, Cantarelli VV, Matsuda S, et al. Identification and characterization of VopT, a novel ADP-ribosyltransferase effector protein secreted via the Vibrio parahaemolyticus type III secretion system 2. Cell Microbiol. 2007;9:2598–2609. doi: 10.1111/j.1462-5822.2007.00980.x. [DOI] [PubMed] [Google Scholar]
- 31.Hiyoshi H, Okada R, Matsuda S, Gotoh K, Akeda Y, et al. Interaction between the type III effector VopO and GEF-H1 activates the RhoA-ROCK pathway. PLoS Pathog. 2015;11:e1004694. doi: 10.1371/journal.ppat.1004694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X, Gewurz BE, Ritchie JM, Takasaki K, Greenfeld H, et al. A Vibrio parahaemolyticus T3SS effector mediates pathogenesis by independently enabling intestinal colonization and inhibiting TAK1 activation. Cell Rep. 2013;3:1690–1702. doi: 10.1016/j.celrep.2013.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trosky JE, Mukherjee S, Burdette DL, Roberts M, McCarter L, et al. Inhibition of MAPK signaling pathways by VopA from Vibrio parahaemolyticus . J Biol Chem. 2004;279:51953–51957. doi: 10.1074/jbc.M407001200. [DOI] [PubMed] [Google Scholar]
- 34.Hiyoshi H, Kodama T, Saito K, Gotoh K, Matsuda S, et al. VopV, an F-actin-binding type III secretion effector, is required for Vibrio parahaemolyticus-induced enterotoxicity. Cell Host Microbe. 2011;10:401–409. doi: 10.1016/j.chom.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 35.Liverman ADB, Cheng H-C, Trosky JE, Leung DW, Yarbrough ML, et al. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc Natl Acad Sci USA. 2007;104:17117–17122. doi: 10.1073/pnas.0703196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calder T, Kinch LN, Fernandez J, Salomon D, Grishin NV, et al. Vibrio type III effector VPA1380 is related to the cysteine protease domain of large bacterial toxins. PLoS One. 2014;9:e104387. doi: 10.1371/journal.pone.0104387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu M, Zhang Y, Gu D, Chen X, Waldor MK, et al. Nucleolar c-Myc recruitment by a Vibrio T3SS effector promotes host cell proliferation and bacterial virulence. EMBO J. 2021;40:e105699. doi: 10.15252/embj.2020105699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tandhavanant S, Matsuda S, Hiyoshi H, Iida T, Kodama T. Vibrio parahaemolyticus senses intracellular K+ to translocate type III secretion system 2 effectors effectively. mBio. 2018;9:e01366-18. doi: 10.1128/mBio.01366-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki M, Danilchanka O, Mekalanos JJ. Vibrio cholerae T3SS effector VopE modulates mitochondrial dynamics and innate immune signaling by targeting Miro GTPases. Cell Host Microbe. 2014;16:581–591. doi: 10.1016/j.chom.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaand M, Miller KA, Sofia MK, Schlesener C, Weaver JWA, et al. Type three secretion system island encoded proteins required for colonization by non-O1/non-O139 serogroup Vibrio cholerae . Infect Immun. 2015;83:2862–2869. doi: 10.1128/IAI.03020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alam A, Miller KA, Chaand M, Butler JS, Dziejman M. Identification of Vibrio cholerae type III secretion system effector proteins. Infect Immun. 2011;79:1728–1740. doi: 10.1128/IAI.01194-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dziejman M, Serruto D, Tam VC, Sturtevant D, Diraphat P, et al. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc Natl Acad Sci USA. 2005;102:3465–3470. doi: 10.1073/pnas.0409918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naka H, Dias GM, Thompson CC, Dubay C, Thompson FL, et al. Complete genome sequence of the marine fish pathogen Vibrio anguillarum harboring the pJM1 virulence plasmid and genomic comparison with other virulent strains of V. anguillarum and V. ordalii . Infect Immun. 2011;79:2889–2900. doi: 10.1128/IAI.05138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arteaga M, Velasco J, Rodriguez S, Vidal M, Arellano C, et al. Genomic characterization of the non-O1/non-O139 Vibrio cholerae strain that caused a gastroenteritis outbreak in Santiago, Chile, 2018. Microb Genom. 2020;6:e000340. doi: 10.1099/mgen.0.000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vázquez-Rosas-Landa M, Ponce-Soto GY, Eguiarte LE, Souza V. Comparative genomics of free-living Gammaproteobacteria: pathogenesis-related genes or interaction-related genes? Pathog Dis. 2017;75:ftx059. doi: 10.1093/femspd/ftx059. [DOI] [PubMed] [Google Scholar]
- 46.Wilkinson P, Waterfield NR, Crossman L, Corton C, Sanchez-Contreras M, et al. Comparative genomics of the emerging human pathogen Photorhabdus asymbiotica with the insect pathogen Photorhabdus luminescens . BMC Genomics. 2009;10:302. doi: 10.1186/1471-2164-10-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akeda Y, Kodama T, Saito K, Iida T, Oishi K, et al. Identification of the Vibrio parahaemolyticus type III secretion system 2-associated chaperone VocC for the T3SS2-specific effector VopC. FEMS Microbiol Lett. 2011;324:156–164. doi: 10.1111/j.1574-6968.2011.02399.x. [DOI] [PubMed] [Google Scholar]
- 48.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 49.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, et al. ACT: the artemis comparison tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 51.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eichinger V, Nussbaumer T, Platzer A, Jehl M-A, Arnold R, et al. EffectiveDB – updates and novel features for a better annotation of bacterial secreted proteins and type III, IV, VI secretion systems. Nucleic Acids Res. 2016;44:D669–D674. doi: 10.1093/nar/gkv1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J, Li J, Yang B, Xie R, Marquez-Lago TT, et al. Bastion3: a two-layer ensemble predictor of type III secreted effectors. Bioinformatics. 2019;35:2017–2028. doi: 10.1093/bioinformatics/bty914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong X, Lu X, Zhang Z. BEAN 2.0: an integrated web resource for the identification and functional analysis of type III secreted effectors. Database. 2015;2015:bav064. doi: 10.1093/database/bav064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blum M, Chang H-Y, Chuguransky S, Grego T, Kandasaamy S, et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021;49:D344–D354. doi: 10.1093/nar/gkaa977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48:D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teufel F, Almagro Armenteros JJ, Johansen AR, Gíslason MH, Pihl SI, et al. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol. 2022;40:1023–1025. doi: 10.1038/s41587-021-01156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, et al. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol. 2018;430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 65.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 67.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abby SS, Rocha EPC. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 2012;8:e1002983. doi: 10.1371/journal.pgen.1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Islam MT, Nasreen T, Kirchberger PC, Liang KYH, Orata FD, et al. Population analysis of Vibrio cholerae in aquatic reservoirs reveals a novel sister species (Vibrio paracholerae sp. nov.) with a history of association with humans. Appl Environ Microbiol. 2021;87:e0042221. doi: 10.1128/AEM.00422-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Islam MT, Liang K, Orata FD, Im MS, Alam M, et al. Vibrio tarriae sp. nov., a novel member of the Cholerae clade. Int J Syst Evol Microbiol. 2022;72:005571. doi: 10.1099/ijsem.0.005571. [DOI] [PubMed] [Google Scholar]
- 71.Loughran RM, Emsley SA, Jefferson T, Wasson BJ, Deadmond MC, et al. Vibrio tetraodonis subsp. pristinus subsp. nov., isolated from the coral Acropora cytherea at Palmyra Atoll, and creation and emended description of Vibrio tetraodonis subsp. tetraodonis subsp. nov. Antonie van Leeuwenhoek. 2022;115:1215–1228. doi: 10.1007/s10482-022-01766-0. [DOI] [PubMed] [Google Scholar]
- 72.Altun S, Duman M, Ay H, Saticioglu IB. Shewanella oncorhynchi sp. nov., a novel member of the genus Shewanella, isolated from rainbow trout (Oncorhynchus mykiss) Int J Syst Evol Microbiol. 2022;72:005460. doi: 10.1099/ijsem.0.005460. [DOI] [PubMed] [Google Scholar]
- 73.Yu L, Jian H, Gai Y, Yi Z, Feng Y, et al. Characterization of two novel psychrophilic and piezotolerant strains, Shewanella psychropiezotolerans sp. nov. and Shewanella eurypsychrophilus sp. nov, adapted to an extreme deep-sea environment. Syst Appl Microbiol. 2021;44:126266. doi: 10.1016/j.syapm.2021.126266. [DOI] [PubMed] [Google Scholar]
- 74.Gerrard JG, McNevin S, Alfredson D, Forgan-Smith R, Fraser N. Photorhabdus species: bioluminescent bacteria as emerging human pathogens? Emerg Infect Dis. 2003;9:251–254. doi: 10.3201/eid0902.020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Macori G, Romano A, Adriano D, Razzuoli E, Bianchi DM, et al. Draft genome sequences of four Yersinia enterocolitica strains, isolated from wild ungulate carcasses. Genome Announc. 2017;5:e00192-17. doi: 10.1128/genomeA.00192-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morita M, Yamamoto S, Hiyoshi H, Kodama T, Okura M, et al. Horizontal gene transfer of a genetic island encoding a type III secretion system distributed in Vibrio cholerae . Microbiol Immunol. 2013;57:334–339. doi: 10.1111/1348-0421.12039. [DOI] [PubMed] [Google Scholar]
- 77.Liu Q, Xu M, He Y, Tao Z, Yang R, et al. Complete genome sequence of Vibrio mediterranei 117-T6, a potentially pathogenic bacterium isolated from the conchocelis of Pyropia spp. Microbiol Resour Announc. 2019;8:e01569-18. doi: 10.1128/MRA.01569-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chimalapati S, de Souza Santos M, Lafrance AE, Ray A, Lee W-R, et al. Vibrio deploys type 2 secreted lipase to esterify cholesterol with host fatty acids and mediate cell egress. Elife. 2020;9:e58057. doi: 10.7554/eLife.58057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma W, Dong FFT, Stavrinides J, Guttman DS. Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS Genet. 2006;2:e209. doi: 10.1371/journal.pgen.0020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ortega AP, Villagra NA, Urrutia IM, Valenzuela LM, Talamilla-Espinoza A, et al. Lose to win: marT pseudogenization in Salmonella enterica serovar Typhi contributed to the surV-dependent survival to H2O2, and inside human macrophage-like cells. Infect Genet Evol. 2016;45:111–121. doi: 10.1016/j.meegid.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 81.Rohmer L, Guttman DS, Dangl JL. Diverse evolutionary mechanisms shape the type III effector virulence factor repertoire in the plant pathogen Pseudomonas syringae . Genetics. 2004;167:1341–1360. doi: 10.1534/genetics.103.019638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.