Abstract
The severity and progression of lung disease are highly variable across individuals with cystic fibrosis (CF) and are imperfectly predicted by mutations in the human gene CFTR, lung microbiome variation or other clinical factors. The opportunistic pathogen Pseudomonas aeruginosa (Pa) dominates airway infections in most CF adults. Here we hypothesized that within–host genetic variation of Pa populations would be associated with lung disease severity. To quantify Pa genetic variation within CF sputum samples, we used deep amplicon sequencing (AmpliSeq) of 209 Pa genes previously associated with pathogenesis or adaptation to the CF lung. We trained machine learning models using Pa single nucleotide variants (SNVs), microbiome diversity data and clinical factors to classify lung disease severity at the time of sputum sampling, and to predict lung function decline after 5 years in a cohort of 54 adult CF patients with chronic Pa infection. Models using Pa SNVs alone classified lung disease severity with good sensitivity and specificity (area under the receiver operating characteristic curve: AUROC=0.87). Models were less predictive of lung function decline after 5 years (AUROC=0.74) but still significantly better than random. The addition of clinical data, but not sputum microbiome diversity data, yielded only modest improvements in classifying baseline lung function (AUROC=0.92) and predicting lung function decline (AUROC=0.79), suggesting that Pa AmpliSeq data account for most of the predictive value. Our work provides a proof of principle that Pa genetic variation in sputum tracks lung disease severity, moderately predicts lung function decline and could serve as a disease biomarker among CF patients with chronic Pa infections.
Keywords: AmpliSeq, cystic fibrosis, genomics, machine learning, Pseudomonas aeruginosa, within–host diversity, lung function
Data Summary
All amplicon sequencing data generated in this project are deposited in NCBI GenBank under BioProject PRJNA763719: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA763719
Impact Statement.
Cystic fibrosis (CF) is among the most common, life-limiting inherited disorders, caused by mutations in the CF transmembrane conductance regulator (CFTR) gene. CFTR dysfunction causes impaired mucociliary clearance, leading to chronic airway infections, and a vicious cycle of lung inflammation and damage, resulting in progressive lung disease, the major cause of morbidity and mortality in CF patients. However, the severity of lung disease and the rate of lung function decline are highly variable across CF patients, and cannot be fully explained using existing clinical or host genetic factors. Here we employed machine learning (ML) techniques to establish a link between Pseudomonas aeruginosa (Pa) within–patient genetic diversity and lung disease severity in a cohort of CF patients with chronic Pa infections. Our study provides a proof of principle demonstrating the utility of ML tools for predictive modelling of lung function severity and decline in CF patients using Pa genetic diversity data. Our findings show the potential for ML models to identify high-risk CF patients using Pa genetic information.
Introduction
Cystic fibrosis (CF) is an autosomal recessive disorder caused by mutations in the CF transmembrane conductance regulator (CFTR) gene and is the most common lethal Mendelian disease in populations with European ancestry [1]. The resulting lung disease is the major cause of morbidity and mortality in CF patients, with lung failure the most common cause of death [2]. The severity of lung disease and the rate of lung function decline are highly variable across CF patients, and cannot be fully explained by variations in CFTR alleles or other modifier genes [3].
While CF airway infections are polymicrobial, and microbiome diversity has been associated with lung disease severity in many studies ([4–7], Pseudomonas aeruginosa (Pa) is an opportunistic pathogen found in the majority of adult CF patients and often dominates their airway microbiome [7, 8]. Infection with Pa in early life is associated with a greater decline in lung function and mortality [9–11]. Notably, Pa airway infections can persist even with highly effective CFTR-modulator therapies [12, 13].
Over the course of chronic CF lung infections, Pa undergoes genetic diversification, selection and adaptive evolution, resulting in a genetically and phenotypically diverse Pa population within each patient [14–19]. How this pathoadaptation affects the clinical course of CF lung disease remains poorly understood. We therefore focused on examining the association between Pa genetic variation and the severity and progression of lung disease in CF patients with chronic Pa infections. We hypothesized that within–host genetic variations in Pa populations during chronic CF lung infections are associated with lung disease severity (i.e baseline lung function) and subsequent progression (i.e. decline in lung function), as measured by spirometry.
While it is known that within–host mutations can significantly affect the virulence of Pa and host responses to Pa, previous studies [16, 20–23] have examined the genetic variation of Pa across cohorts of CF patients by performing whole-genome sequencing (WGS) of only one or few Pa clones isolated from CF sputum samples – an approach that fails to capture the genetic diversity of Pa within the lung and is subject to sampling bias. While shotgun metagenomic analysis of CF sputum is increasingly used for microbiome analyses [24], the overwhelming abundance of host-derived DNA in sputum samples still hampers the ability to resolve within-species genetic variation. To overcome these challenges, here we applied a novel amplicon sequencing (AmpliSeq) panel of 209 genes in the Pa genome previously known to be involved in the pathoadaptation and pathogenesis of CF infections (Dataset S1, available in the online version of this article). The AmpliSeq platform allows us to estimate single nucleotide variant (SNV) frequencies within the Pa population, directly from CF samples, without the need to culture and sequence hundreds of isolates per individual.
We then used several machine learning (ML) approaches to classify lung disease severity (at the time of sampling) and to predict disease progression (after 5 years) based on the SNV frequency data from a cohort of 54 adult CF patients with chronic Pa infection. ML has been successfully applied to predict phenotypes from genotype data in other model systems [25]. ML models can explicitly include the interactions and correlations between features (in our case, SNVs), which helps control for confounding factors such as population stratification that may exist in the dataset [26].
Our study provides a proof of principle that the population of Pa in CF sputum samples includes bacterial genetic biomarkers that are associated with disease status and could serve to identify individuals at increased risk of rapid lung function decline. Additionally, this work identified genetic variation in Pa genes that merit further investigation for their potential roles in the pathogenesis of CF lung disease.
Methods
Patient selection, sample and clinical data collection
The Calgary biobank includes frozen whole sputum samples prospectively collected from individuals with CF followed at the Calgary Adult CF clinic from 1998 to 2017, as described previously [27, 28]. A cohort of 104 individuals between the ages of 18 and 22 years with sputum available from the Calgary biobank was previously characterized [27]. For this study, we selected from this cohort all individuals with sputum cultures positive for Pa (64 out of 104 patients). Out of these 64 samples, 54 yielded AmpliSeq data of sufficient depth (>10× average depth of coverage of the targeted genes) and were retained for further analysis. Clinical data collected for each patient are outlined in Tables 1 and S1 and include age, gender, body mass index, CFTR genotype, birth cohorts and microbiology (mucoid phenotype, Pa relative abundance, microbiome diversity indices).
Table 1.
Patient clinical data
Values show the absolute count, or mean with standard deviation in parentheses where applicable. Baseline lung function is defined as severe when FEVp <60% predicted and mild otherwise. Lung function decline is defined as non-rapid when 5 year FEVp decline <5 % and rapid otherwise. The relative abundance of Pa, as well as Shannon and Simpson diversity indices, were computed based on 16S rRNA gene sequencing of the lung microbiome community. Homozygous ΔF508 indicates the counts of individuals with a ΔF508/ΔF508 genotype; others include heterozygotes or other genotypes. Test statistics are the Wilcoxon rank-sum statistic for numerical data [Pa, age, body mass index (BMI), Shannon and Simpson indices] and odds ratio for categorical data.
|
Patient data |
Baseline lung function |
Lung function decline over 5 years |
||||||
|---|---|---|---|---|---|---|---|---|
|
Severe (n=27) |
Mild (n=27) |
Test statistic |
P value |
Non-rapid (n=31) |
Rapid (n=23) |
Test statistic |
P value |
|
|
Pa relative abundance |
0.59 (0.32) |
0.35 (0.32) |
2.48 |
0.01 |
0.46 (0.33) |
0.49 (0.32) |
0.3 |
0.75 |
|
Age (years) |
19.0 (1.13) |
19.4 (1.16) |
1.40 |
0.16 |
19.2 (1.1) |
19.2 (1.25) |
0.24 |
0.80 |
|
BMI (kg m–2) |
19.0 (2.3) |
21.4 (2.3) |
3.45 |
0.0005 |
20.62 (2.9) |
19.7 (2.1) |
1.18 |
0.23 |
|
Shannon index |
1.12 (0.64) |
1.21 (0.68) |
0.42 |
0.66 |
1.31 (0.66) |
1.06 (0.65) |
1.44 |
0.24 |
|
Simpson index |
0.48 (0.26) |
0.5 (0.28) |
0.23 |
0.81 |
0.54 (0.26) |
0.45 (0.27) |
1.61 |
0.20 |
|
PES (PFGE typing) |
15 |
9 |
2.5 |
0.17 |
16 |
8 |
2.0 |
0.27 |
|
Homozygous ΔF508 |
15 |
13 |
1.34 |
0.78 |
14 |
14 |
0.52 |
0.28 |
|
Not-deceased (Death) |
22 |
25 |
0.35 |
0.42 |
27 |
20 |
1.01 |
1 |
|
Male (Gender) |
11 |
9 |
0.72 |
0.77 |
8 |
12 |
3.13 |
0.86 |
|
Mucoid |
24 |
23 |
0.71 |
1 |
27 |
20 |
0.98 |
1 |
|
Birth cohort years 1978–1984 |
11 |
8 |
0.47 |
0.49 |
12 |
7 |
1.31 |
0.25 |
|
Birth cohort years 1985–1990 |
9 |
11 |
0.2 |
0.65 |
7 |
13 |
1.8 |
0.17 |
PES, Prairie Epidemic Strain.
As a measure of lung disease severity at the time of sputum collection, we used the spirometric measure of forced expiratory volume in 1 s, percentage predicted (hereafter referred to as ‘baseline lung function’ and noted FEVp), a standard measure of lung function normalized for age, height, and self-identified sex and ethnicity. Baseline lung function was categorized as severe for FEVp <60 % predicted, and mild/moderate for FEVp ≥60 % predicted based on the European Respiratory Society/American Thoracic Society standard lung function interpretations [29]. Long-term lung function decline (hereafter noted as ‘lung function decline’) was measured using the relative rate of FEVp decline per year (determined by subject-specific constructed linear regressions over the 5 years following sputum collection as described by Acosta et al. [27]. Lung function decline was categorized as ‘rapid’ when the relative FEVp decline was >5 % per year, and ‘non-rapid’ when ≤5 % per year.
Sputum DNA extraction and microbiome analyses
Genomic DNA was extracted from a single biobanked sputum sample per patient as previously described [27], and used as template for 16S rRNA gene amplicon and Ion AmpliSeq sequencing. The Prairie Epidemic Strain (PES) genotype, a highly prevalent strain in our study population, was identified by PFGE and/or multi-locus sequence typing (MLST) . For microbiome analysis, bacterial communities in CF sputum and reagent blanks were characterized by amplification and sequencing of the V3–V4 region of the 16S rRNA gene, as previously described [27]. The sequencing reads were then processed to identify operational taxonomic units (OTUs) [28]. Relative Pa abundance was determined as the proportion of Pseudomonas reads relative to the total number of 16S rRNA gene reads.
Ion AmpliSeq panel design and sequencing
The AmpliSeq panel targeted 209 Pa genes previously implicated in pathogenicity, antimicrobial resistance and within–host pathoadaptation during chronic infection (Data S1). The AmpliSeq primer panel (generated by Life Technologies) was designed by the AmpliSeq Custom Services (White Glove, Thermo Fisher Scientific) to provide high sequencing coverage of the target genes based on the Pa PAO1 genome (NCBI accession number: GCA_000006765.1), with 100 % breadth of coverage for 205 genes and >96 % in four genes, based on the tiling of amplicons. Four additional genome assemblies of Pa clinical isolates [GCF_004375495.1, GCF_004374685.1, GCF_004374275.1 and the PES genome (NCBI BioProject: PRJNA750451)] were also evaluated along with PAO1 for the optimization of primer design, tiling and pooling to achieve maximal target coverage by the primer panel with minimal misalignments and homology with the human genome.
AmpliSeq libraries were constructed using the Ion AmpliSeq Library kit 2.0 and IonCode barcode set with the following modifications. SparQ magnetic beads (Quantabio) were used for purification, and individual libraries were quantified using the Quant-iT PicoGreen dsDNA Assay Kit (ThermoFisher). Samples were mixed in equimolar proportions and the pooled library (200 pM) was loaded on an Ion Chef for template preparation using HiQ reagents. The P1 v3 chips were sequenced using an Ion Proton sequencer (500 flows) with P1 HiQ sequencing reagents following the manufacturer’s instructions.
AmpliSeq variant calling
The quality of AmpliSeq sequencing was confirmed using TorrentSuite software (v.5.2; Thermo Fisher Scientific). Raw sequencing reads were trimmed based on a per-base phred quality score cutoff (‘q’ flag) of 18, window size of 1 bp and minimum remaining sequence length (‘l’ flag) of 19 using fastq-mcf (v.1.04.636) [30]. Reads were aligned to the PES genome (CP080405) using BWA MEM and the alignments were sorted and indexed using SAMtools (v.1.9) [31]. Samples with average sequencing depth ≤10× across the target genes were discarded, leaving 54 samples for further analysis. SNVs with minimum mapping quality of 20, minimum base quality of 18 and minimum coverage of 10× were then identified using VarScan 2 [32] and functional consequences of each SNV were inferred using snpEFF (v.2.4.2) [33]. The SNV allele frequencies (ranging from 0 to 1) at each polymorphic site covered by the AmpliSeq panel were used to generate an SNV frequency matrix, with samples as rows and nucleotide positions as columns. For baseline lung function (measured based on FEVp score) and lung function decline (disease progression) prediction analysis, all synonymous variants were filtered out and only non-synonymous variants (including nonsense and missense mutations, frameshift deletions and insertions) were used (Dataset S2). All SNVs (including synonymous sites) were included for population stratification analyses.
Heritability estimation
Here we define heritability as the variation in disease status that can be explained by genetic variation measured in the AmpliSeq SNV data. We estimated heritability as the average prediction accuracy R 2 from the elastic net model implemented in the scikit-learn Python package in held-out samples during cross-validation on continuous phenotypes (not binned into mild/severe or rapid/non-rapid). This approach is similar to that described previously by Lees et al. [26].
Bacterial population stratification
Population stratification in Pa was evaluated by calculating pairwise Pearson correlation coefficients between sputum samples based on the SNV frequency matrix followed by determination of distinct genome subgroups using hierarchical agglomerative clustering implemented in SciPy [34] and visualized using the python seaborn package [35]. This identified two major subclusters of Pa, one of which was significantly enriched in PES strains. To determine if any clinical factors were associated with these subclusters, we used t-tests for continuous variables including age, body mass index (BMI), Shannon and Simpson diversity indices and Pa abundance in the sputum sample. For binary variables including PFGE typing (PES or not), gender, host CFTR genotype, death, mucoid presence/absence status, baseline lung function and lung disease progression (lung function decline), we used a Fisher Exact test. A Chi-square test was used for the multi-categorical birth cohort factor.
Feature selection and training predictive model of lung function
In an ML context, a feature is defined as an individual measurable characteristic of an observed phenomenon. In this study, the features considered are Pa genetic variants (SNV frequencies) identified by the AmpliSeq panel and the clinical factors linked to the study patients (Table S1). To reduce the dimensionality of the dataset (i.e. to reduce the ratio of features to sample size), a feature selection approach was applied using nested cross-validation in three steps.
1. Outer loop: data were split into 80 % training (43 samples) and 20 % testing (11 samples) for 20 resamplings (folds) using a stratified shuffled split function implemented in sklearn [36]. This method ensures no overlap between train and test dataset in each fold but samples in the training dataset may overlap across folds. Stratification was performed along labels in each phenotype (i.e. baseline lung function and lung function decline) to keep the ratio of cases to controls in train and test datasets similar in each fold.
Inner loop: from the training dataset within each outer loop, samples were randomly bootstrapped (n=43, sampling with replacement) and feature importance (i.e. scores assigned to each input feature indicating the relative importance of the feature when making a prediction) were estimated using the lightGBM [37] gradient boosting ensemble method implemented in feature-selector v1.0.0 [38] for 50 folds. Feature-selector parameters were set as: n estimators=1000, learning_rate=0.05 and early_stopping=True.
Features with an importance value of zero averaged across 50 inner loops were discarded. Each of four ML models (i.e. logistic regression, SVM, random forest and XGBoost) were independently trained on a dataset of selected features and performance of the trained model was measured by estimating ROC-AUC on the test dataset ensuring no data leakage between training and testing datasets.
2. Feature importance values estimated in the 20 outer loops were averaged and the set of features required to obtain 99 % cumulative relative importance were retained to perform prediction analysis. The remaining features were filtered out.
3. ROC-AUCs of models were estimated by averaging the ROC-AUCs over the 20 outer loops.
To prevent bias or overfitting to the training data, we performed feature selection within the inner cross-validation such that no information from the training data is leaked to the held-out samples. In other words, to find the optimal coefficient values for each feature, we used inner cross-validation and set the feature importance to 0 for the features that did not contribute to the classification of the data in the test dataset, thereby preventing any leakage of information between training and testing datasets.
Four ML models were used in this study: logistic regression with l2 regularization, extreme gradient boosting implemented in XGBoost [39], ensemble decision trees implemented in random forest [40] and linear support vector machine (SVM) with linear kernel implemented in the sklearn package with default hyperparameters. Model performance was evaluated using six metrics: (1) area under the receiver operating characteristic (AUROC), accuracy (number of correct predictions/total number of predictions); (2) precision (True Positives/(True Positives+False Positives)); (3) recall (True Positives/(True Positives+False Negatives)); (4) F1 score (2*(Precision*Recall)/Precision +Recall); (5) Accuracy; and (6) balanced accuracy (bACC), the average of recall obtained on each class (i.e. severe/mild for baseline lung function and rapid/non-rapid for lung function decline).
To evaluate the statistical significance of the prediction performances (AUROC scores) obtained by ML models in comparison with random expectations, a non-parametric permutation test [36] was performed using 100 rounds of label switching followed by feature selection and model training (as done for real data) and estimating empirical P-values (i.e. the chance that the observed AUROC scores obtained using the data could be obtained by chance alone). The enrichment of predictor SNVs across functional gene categories relative to the total genes in the AmpliSeq panel was assessed using Fisher’s exact test with a family-wise error rate of 0.05 adjusted for multiple testing using the Bonferroni method.
Code availability
All computer code used to conduct the AmpliSeq data analysis, including ML methods, are available at GitHub at the following link: https://github.com/Morteza-M-Saber/Cystic_fibrosis_ML_analysis/
Results
We studied a previously described and well-characterized cohort of young adult CF patients aged 18–22 years with chronic Pa infection [27]. After filtering for AmpliSeq sequencing quality, we excluded 10 patients with low coverage of Pa, leaving 54 patients for further analysis. The clinical and demographic characteristics of all 54 patients are summarized in Table 1, and the excluded patients were not apparent outliers in their clinical profiles. From the filtered sequence data, we identified SNVs within the 209 genes represented in the AmpliSeq panel and estimated the frequency of each SNV within each patient sputum sample. In total across the 54 patient samples, we identified 7867 synonymous and 4452 non-synonymous SNVs (Dataset S2). All variants were used for population stratification analysis and only non-synonymous SNVs were used to train ML models.
We first estimated the heritability of baseline lung function (FEVp score) and lung function decline over 5 years. Baseline lung function had a heritability of 0.30 [95 % confidence interval (CI): 0.23–0.37], indicating a significant genetic component from the AmpliSeq data. In contrast, the heritability of lung function decline could not be estimated due to a poor fit of the elastic net model (mean R 2 across cross-validation folds <0). We therefore expect lung function decline to be challenging to predict from AmpliSeq data. Considering baseline and future lung disease as continuous factors, we found that AmpliSeq data explain significant variance in baseline lung function (elastic net explained variance regression score=0.41; 95 % CI: 0.37–0.46) but not in lung function decline (explained variance ~0). Due to these relatively poor model fits on continuous phenotypes, we binned each measure of disease severity into discrete, clinically relevant categories: (1) severe or mild baseline lung function, and (2) rapid or non-rapid lung function decline over 5 years (see Methods). Both measures of lung disease severity were binned based on clinically accepted threshold values [29]. As described below in detail, ML models were able to classify these discrete disease categories significantly better than random. We therefore proceeded with these discrete categories for subsequent analyses.
Stratification in the Pa population
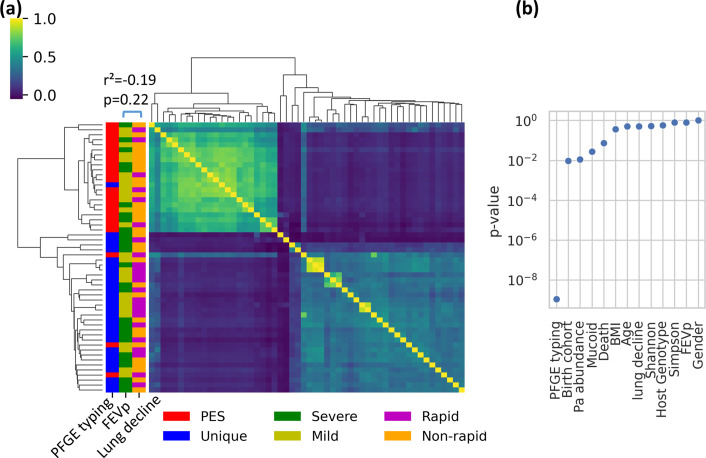
We quantified the extent of Pa population stratification, which can be problematic if there are clonally related genetic clusters that are confounded with the lung disease outcomes of interest. If a particular genetic cluster or lineage is associated with lung disease severity, it then becomes difficult to pinpoint the SNVs associated with disease because all mutations (whether related to disease or not) in a cluster are correlated with each other. We know a priori based on PFGE typing that our dataset contains a highly prevalent lineage of Pa [called Prairie Epidemic Strain or PES; sequence type (ST)-192; Table 1] suspected to be associated with severe lung disease [41]. We confirmed this by hierarchical clustering of the Pa AmpliSeq data (n=12 319 SNVs, including both synonymous and non-synonymous variants), which revealed two apparent genetic clusters (Fig. 1a), one of which was strongly associated with the PES lineage (Fisher exact test, odds ratio=168.0, P=1.1e-09; Fig. 1b). The few observed exceptions (i.e. three PES samples grouped with unique PFGE types; Fig. 1a) could be due to mixed infections or sufficient within–host diversification to obscure the genetic signal of PES ancestry. The two major Pa genetic clusters were also weakly associated with the birth cohort (Chi-square test, P=0.0095; not significant after multiple test correction; Fig. 1b) which is probably due to unequal prevalence of PES across the time periods where cohorts were recruited (Table S1). No other clinical factor was significantly associated with either genetic cluster (Fig. 1b). Importantly, neither cluster is correlated with either baseline lung function (Fisher exact test, P=0.81) or lung function decline (Fisher exact test, P= 0.51) (Fig. 1b), indicating that these disease outcomes are unlikely to be confounded by Pa population stratification, and that finer-grained predictive modelling is warranted. We also noted that lung disease progression over 5 years (lung function decline) is not significantly correlated with baseline lung function at sample collection (Fig. 1a).
Fig. 1.
Pa populations are stratified into two genetic clusters, neither of which is associated with baseline lung function (FEVp) or lung function decline. (a) Heatmap showing correlations in within–host Pa SNV frequencies between pairs of sputum samples. Strong correlations are in yellow; weak correlations in blue. Rows and columns (samples) are ordered by hierarchical clustering. Distribution of baseline lung function measured by FEVp score (27 Severe and 27 Mild individuals), lung function decline (23 Rapid and 31 Non-rapid individuals) and PFGE typing (25 PES and 29 Unique) are presented on the y-axis. Baseline lung function and lung function decline over 5 years are not significantly correlated (Pearson R 2 score=−0.19, P=0.22). (b) P-values for the association between clinical data and genetic clusters are determined by t-tests for numerical data and Chi-square tests for categorical data (Methods). Only the association between PFGE type (PES or non-PES) is significantly associated with the genetic clusters in (a) (P<0.0045 after Bonferroni correction for multiple tests).
Genetic and clinical features associated with baseline lung function and lung function decline in CF patients
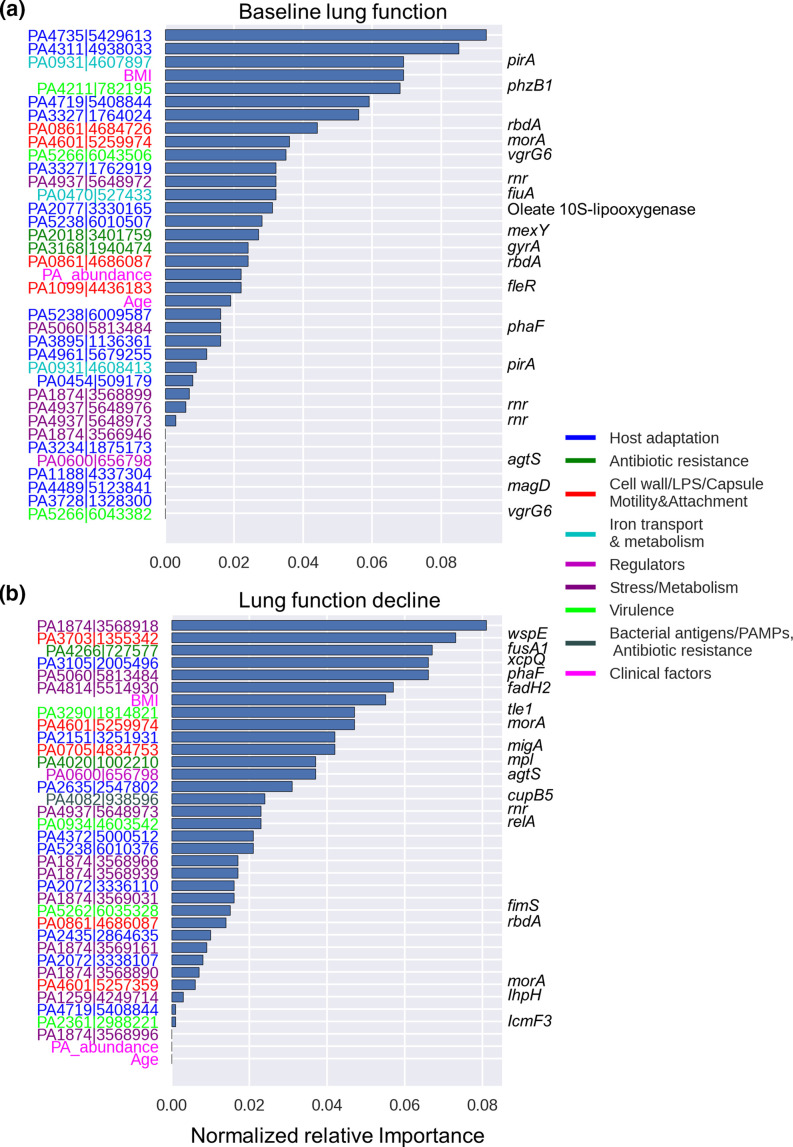
A common challenge in predicting outcomes from sequence data is the sparsity of the data, that is the relatively few available samples compared to the large number of genetic markers (called ‘features’ in ML context). To resolve this problem, feature selection can be used to remove non-informative features (i.e. SNVs and clinical factors) and focus only on the most predictive ones [42–45]. We used a nested cross-validation approach for feature selection based on ensemble gradient boosting (see Methods). Out of 4452 non-synonymous SNVs and 11 clinical factors considered, our model selected only 34 SNVs (hereafter called predictor SNVs) and three clinical factors (age, BMI and Pa relative abundance in the sputum microbiome) that account for 99 % of the cumulative feature importance (Fig. 2). This means that a minimal set of SNVs and clinical factors provides 99 % of the information used in predicting baseline lung function at the time of sampling (Fig. 2a). An equivalent analysis for lung function decline after 5 years identified 33 predictor SNVs and the same three clinical factors that contributed to 99 % of the cumulative feature importance (Fig. 2b). Including the number of polymorphic SNVs (within–sample frequency >0 and <1) to the models did not improve predictions nor was it selected as an important feature, suggesting that simple measures of within–patient diversity have limited predicted value. For both baseline lung function and future lung function decline, the phenotype is not simply predicted based on the presence/absence of each SNV, but rather on more subtle information about SNV allele frequencies within patients. In other words, predictive SNVs occur at a range of frequencies, with only 223 being clustered around 0 or 1 (Fig. S1).
Fig. 2.
Pa genes and clinical factors selected as predictive features of baseline lung function and lung function decline. Normalized importance of genomic and clinical data that contribute to 99 % cumulative relative importance for the prediction of (a) baseline lung function at time of sample collection and (b) risk of 5 year progression (lung function decline). On the y-axis, gene identifiers (locus tag|chromosome location based on PES genome) are colour-coded based on their functional classification. Named genes are shown on the right, when available.
The three selected clinical factors associated with both baseline lung function and lung function decline are BMI, Pa relative abundance from 16S rRNA gene amplicon sequence data from a previous study of the same cohort [27] and age (Fig. 2). Multiple studies have shown an association between poor lung function and low BMI [13, 46, 47], high abundance of Pa [5] and age [5, 48]. As expected, Pa relative abundance also showed a strong negative correlation with Shannon and Simpson microbiome diversity indices (Fig. S2), indicating that Pa abundance can be considered as a proxy for lung microbiome diversity in our dataset. However, Shannon and Simpson diversity indices were not selected as predictive features in our model, consistent with a previous study [48]. This suggests that, even if low microbiome diversity indices are associated with CF disease progression, the low diversity is probably driven by the dominance of key pathogens such as Pa. The recovery of previously known clinical determinants of lung function in CF patients supports the reliability of our feature selection approach.
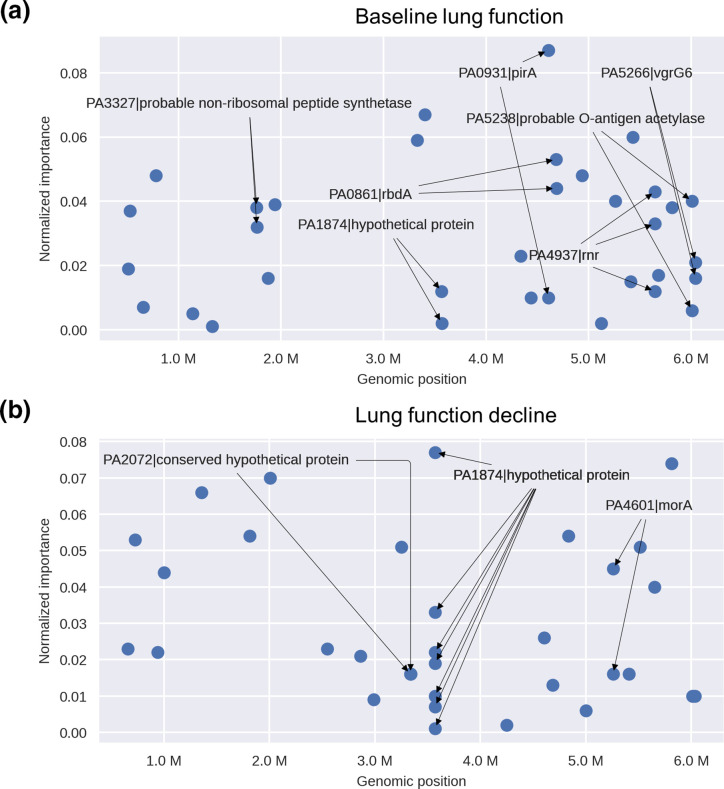
To interpret the possible roles of Pa SNVs in CF lung disease, we classified the known or predicted function of genes containing predictor SNVs (hereafter called predictor genes) into functional categories manually curated based on existing literature. The predictor SNVs with the highest weighted importance for both baseline lung function and lung function decline outcomes are located within genes that play a role in seven functional categories (Table 2). The distribution of predictor genes is generally similar to the distribution of gene functions included in the AmpliSeq panel (Fig. S3). However, the predictor genes for baseline lung function are enriched in iron transport and metabolism (13.4 % in baseline lung function predictor genes vs. 1.4 % in the AmpliSeq panel, P=0.00018; Fig. S3). The genes encoding the ferric enterobactin receptor (PirA) and the ferrichrome receptor (FiuA) respectively account for 8.9 and 3.7 % of the total normalized importance for baseline lung function (Fig. 2a), and pirA contains multiple predictor SNVs (Fig. 3a). In contrast, the predictor genes for lung function decline are enriched in stress/metabolism functions (33.6 % in lung function decline predictor genes vs. 13.4 % in the AmpliSeq panel, P=0.002; Fig. S3). Notably, the hypothetical protein PA1874 accounts for 16.9 % of the total normalized importance for prediction of lung function decline and includes seven out of 33 predictor SNVs (Figs 2b and 3b), as well as two predictor SNVs for baseline lung function (Fig. 3). This hypothetical protein has also been shown to play a role in resistance of Pa to multiple antibiotics [49]. The PA4937 gene, which encodes an RNase R exoribonuclease, also contains multiple SNVs predictor genes of both baseline lung function and lung function decline (Figs 2a and 3a). The two genes PA0861 (rbdA) and PA4601 (morA), which encode regulators involved in bacterial motility and biofilm formation, are also predictor genes for both baseline lung function and lung function decline (Fig. 2).
Table 2.
Functional classification of predictor genes used for prediction of baseline lung function and lung function decline
|
Baseline lung function predictor genes |
Lung function decline predictor genes |
Shared genes |
|
|---|---|---|---|
|
Host adaptation |
PA0454|conserved hypothetical protein PA1188|hypothetical protein PA2077|oleate 10S-lipoxygenase PA3234|probable sodium:solute symporter PA3327|probable non-ribosomal peptide synthetase PA3728|hypothetical protein PA3895|probable transcriptional regulator PA4311|conserved hypothetical protein PA4489|magD PA4735|hypothetical protein PA4961|hypothetical protein |
PA2072|conserved hypothetical protein PA2151|conserved hypothetical protein PA2435|probable cation-transporting P-type ATPase PA2635|hypothetical protein PA3105|xcpQ PA4372|hypothetical protein |
PA4719|probable transporter PA5238|probable O-antigen acetylase |
|
Antibiotic resistance |
PA2018|mexY PA3168|gyrA |
PA4020|mpl PA4082|cupB5 PA4266|fusA1 |
|
|
Cell wall, LPS, capsule, motility and attachment |
PA1099|fleR |
PA0705|migA PA3704|wspE PA4082|cupB5 |
PA0861|rbdA PA4601|morA |
|
Iron transport and metabolism |
PA0470|fiuA PA0931|pirA |
||
|
Regulators |
PA0600|agtS |
||
|
Stress/metabolism |
PA1259|lhpH PA4814|fadH2 |
PA1874|hypothetical protein PA4937|rnr PA5060|phaF |
|
|
Virulence |
PA4211|phzB1 PA5266|vgrG6 |
PA0934|relA PA2361|icmF3 PA3290|tle1 PA5262|fimS |
LPS, lipopolysaccharide.
Fig. 3.
Genomic locations and importance of genes containing predictor SNVs. (a) Genes containing SNVs predictive of baseline lung function. The x-axis shows nucleotide positions across the Pa reference genome. (b) Genes containing SNVs predictive of lung function decline. Genes including multiple SNVs are shown with arrows. Note that the two arrows from PA2072 point to two different nearby SNVs in the same gene.
Predicting lung disease severity and progression in individuals with CF using genetic and clinical factors
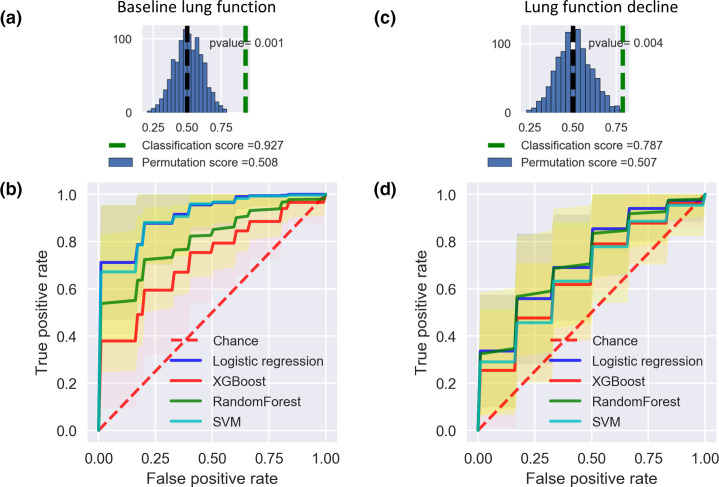
We used a nested cross-validation approach to compare the performance of four L models including l2-regularized logistic regression, support vector machines (SVM), random forests, and extreme gradient boosting (XGBoost) using the AUROC curve and other standard metrics (see Methods). To prevent data leakage (i.e. information from outside the training dataset being used to create the prediction model), feature selection was performed on a training dataset and the performance of the trained model was estimated on a completely independent test dataset. It should be noted that while an ensemble light gradient boosting model was used for feature selection (see Methods), we did not use it for predictive modelling to avoid overfitting. Of the ML methods tested, logistic regression had the best predictive performance for both phenotypes (Table 3, Fig. 4). Logistic regression is a simple classification model, which makes it reasonably robust against overfitting [50]. Cross-validation showed that logistic regression was the most accurate and precise model for both baseline lung function, with an average AUROC score of 0.87 (95 % CI, 0.84–0.90), and for lung function decline, with a score of 0.74 (95 % CI: 0.71–0.78; Tables 3 and S2). The second best method was SVM, another type of linear model (Table S2). Across all models, the baseline lung function phenotype was more accurately predicted than lung function decline, consistent with predictions becoming more uncertain further into the future (Tables 3 and S2). Importantly, all models could predict both phenotypes significantly better than expected by chance (compared to a permutation test using data with shuffled outcome labels; Figs 4a, c and S4).
Table 3.
Performance of logistic regression in predicting baseline lung function and lung function decline using genomic data only, or a combination of genomic and clinical data
See Methods for descriptions of the performance metrics.
|
Genomic data (95 % CI) |
Genomic and clinical data (95 % CI) |
||
|---|---|---|---|
|
Baseline lung function |
AUROC |
0.87 (0.84,0.9) |
0.92 (0.84,1) |
|
bACC |
0.81 (0.78,0.84) |
0.83 (0.72,0.94) |
|
|
Accuracy |
0.81 (0.78,0.84) |
0.83 (0.72,0.94) |
|
|
F1 |
0.81 (0.78,0.83) |
0.83 (0.72,0.94) |
|
|
Precision |
0.83 (0.81,0.86) |
0.84 (0.73,0.94) |
|
|
Recall |
0.81 (0.78,0.84) |
0.83 (0.72,0.94) |
|
|
Lung function decline |
AUROC |
0.74 (0.71,0.78) |
0.79 (0.7,0.88) |
|
bACC |
0.63 (0.59,0.66) |
0.66 (0.59,0.74) |
|
|
Accuracy |
0.64 (0.6,0.67) |
0.67 (0.6,0.75) |
|
|
F1 |
0.62 (0.58,0.65) |
0.66 (0.58,0.74) |
|
|
Precision |
0.65 (0.61,0.69) |
0.69 (0.6,0.78) |
|
|
Recall |
0.64 (0.6,0.67) |
0.67 (0.6,0.75) |
Fig. 4.
Predictive models of baseline lung function and lung function decline perform significantly better than expected by chance. (a) Classification score of baseline lung function using logistic regression (green dashed line) is significantly higher than expected based on permuted data (mean shown in black dashed line). (b) Average AUROC scores of different ML models to predict baseline lung function, compared to the random expectation (permuted sample labels). Shading indicates the 95 % confidence interval. (c) Classification score of lung function decline using logistic regression compared to permuted data. (d) Average AUROC scores for lung function decline prediction.
Clinical factors have been previously used to predict lung disease progression in CF patients [51]. We therefore assessed if integrating clinical factors could improve upon the predictions based on Pa AmpliSeq data alone. Including the three clinical factors identified by feature selection (BMI, age and Pa relative abundance) in our predictive models led to modest performance increases (~5 % increase in AUROC) for both baseline lung function and lung function decline outcomes across the four ML models considered (Tables 3 and S2). Using clinical factors alone was always inferior to AmpliSeq data to classify baseline lung function, and was unable to predict lung function decline better than a random expectation (Fig. S5). We conclude that, while these clinical factors are useful, most of the predictive power comes from the Pa genetic data.
Lack of generalizability is one of the main limiting factors for the translation of prediction models into clinically useful diagnostics. ML models often have low generalizability (i.e. ‘overfit’) in scenarios where the model performs well on the dataset used to train the model but fails to achieve similar prediction accuracy on new data. We plotted learning curves to assess how ML (using logistic regression) predictions improved by training on more data [52]. We found that the performance difference between training and testing data decreases as sample size increases (Fig. S6). There were no major differences in prediction accuracy of training and testing datasets (Fig. S6), which suggests the model does not suffer from significant overfitting. We also noted that cross-validation scores for both baseline lung function and lung function decline models continued to increase for the testing dataset as more data were used for model training (Fig. S6), which suggests the model could be further improved with more data.
Discussion
Considering the critical role of Pa in CF-related morbidity and mortality, here we established a link between Pa within–host genetic diversity and CF lung disease severity in a cohort of young adults with chronic Pa infections. Despite a modest sample size, our study provides a proof of principle demonstrating the utility of ML models for predictive modelling of lung function severity and decline in CF patients using bacterial genetic and clinical data. Although our models do not appear to be significantly overfitted, fully validating their predictive performance will require independent cohorts. We also identified potential genetic biomarkers associated with lung disease severity. Overall, our findings provide evidence that ML models can identify CF individuals at high risk for poor Pa infection outcomes using Pa genetic data.
Our work is based on a subset of samples from a previously described cohort study that identified dominance of Pa in the sputum microbiome (and the resulting reduction of community diversity) as a predictor of lung function decline in a cohort of young CF adults [27]. Here we focused on a subset of patients with a lung microbiome dominated by Pa. While these patients are already at increased risk of lung disease, we found that the severity of disease at the time of sampling and 5 years into the future could be predicted based on genetic variation within the infecting Pa population. Even in this patient cohort in which Pa was always present, we confirmed that Pa relative abundance is associated with disease severity and progression – although it is a less important predictor than many SNVs within the Pa genome. This suggests that genetic variation in dominant pathogens can significantly complement and improve upon predictions of disease status based on the microbiome. Along these lines, another recent study showed that the Pa genomic data could predict pathogenicity in mouse models [53].
In addition to variation in the host genome, the polymicrobial community inhabiting the CF lung has been identified as an important modifier of disease progression. Numerous studies of the lung microbiome have shown an association between decreasing microbial community diversity and worsening lung function [4–7, 54], as well as progression to end-stage lung disease [27]. However, microbiome diversity may have limited predictive value as there is high interpersonal variability in lung microbiomes [6], and a large number of adult CF individuals have microbiomes dominated by pathogens such as PA. Zhao et al. [55] recently showed that a combination of microbiome data and clinical metadata improved predictive performance compared to either data type alone. Our results suggest that genetic diversity within key pathogens such as Pa could complement or even supersede microbiome community diversity for predicting clinical outcomes in specific patient subsets.
Limitations of our study include a relatively small sample size of patients (N=54) from a single cohort. As such, we consider our work a proof of concept that could be improved upon in larger cohorts. Indeed, learning curves showed that predictive accuracy is likely to improve with more samples. Although we performed nested cross-validation by subsampling our 54 patients for model training and testing, the model should ideally be tested on a completely independent cohort to assess its real-world predictive value. Other recent studies have suggested that standard cross-validation techniques can overestimate predictive accuracy due to strong genetic linkage across bacterial genomes [26, 56, 57]. Pa has a relatively high recombination rate [58] which should reduce the confounding effects of linkage. Importantly, the AmpliSeq data quantify within–patient genetic diversity which does contain traces of ancestry (e.g. PES vs. non-PES lineages) but should also be enriched in de novo mutations which are unlinked to deep-branching genomic backgrounds. Future work could attempt to disentangle de novo mutations from co-infection with different lineages, thereby determining which predictive features are lineage-associated. Nevertheless, we used a high number of cross-validation folds (k=20 for the outer loop and k=50 for the inner loop) relative to the sample size of 54 to help reduce the overestimation of accuracy. Using high values of k is similar to ‘leave-one-strain-out’ validation, which can be less prone to accuracy inflation [56]. Regardless, the accuracy and generality of our results will require replication in independent cohorts. Despite these limitations, our models made significantly better predictions than expected by chance. As expected, predicting lung function decline 5 years into the future proved more challenging than doing so at the time of sampling. These results provide a key first step toward clinical diagnostics of patients most at risk of lung function decline.
As with any genotype–phenotype association method, our approach does not fully guarantee causal relationships, and rather points to candidate genes. Further experimental testing is therefore required to determine whether Pa SNVs play a causal role in lung function decline, or simply serve as useful biomarkers. Regardless, we were able to pinpoint SNVs in several genes of interest. This was feasible because the strong population stratification of Pa into PES and non-PES lineages was fortunately not associated with the disease outcomes of interest. This allowed us to identify SNVs in several genes that provided independent biomarkers of disease.
Several genes containing SNVs predictive of disease status and progression were identified as candidates for further investigation. For example, baseline lung function predictor SNVs are enriched in genes involved in iron transport and metabolism. The AmpliSeq panel only included three iron-related genes, of which two (pirA and fiuA) contained SNVs associated with baseline lung function. Updated AmpliSeq panels or whole-genome sequencing, along with targeted experimental studies, could be used to test the hypothesis that variation in these genes plays a role in disease progression. Multiple studies have shown competition for iron to be key for the survival and virulence of many of the pathogens that reside in the CF lung, including Pa [59, 60]. We also found that SNVs predictive of lung function decline are enriched in genes involved in stress/metabolism. Notably, the gene PA1874 includes seven predictor SNVs comprising 16.9 % of the total feature importance for lung function decline, and two predictor SNVs for baseline lung function prediction, suggesting its general importance in disease severity and progression in CF patients. PA1874 encodes a multidrug efflux pump involved in biofilm-dependent resistance to antibiotics including tobramycin, gentamicin and ciprofloxacin [49, 61] and could be a potentially promising biomarker of CF disease severity, which merits further investigation.
Among the set of clinical factors studied, BMI, Pa abundance and age were identified as important predictors of both baseline lung function and lung function decline. These are all known risk factors for CF disease severity and progression [5, 46–48]. By including these features in our prediction models, we noted a moderate increase across all the measured metrics relative to using only AmpliSeq data. These results are in line with previous studies showing the improvement of ML-based phenotype prediction by adding relevant clinical data [53, 62]. We note that clinical factors only modestly improved the performance of the models (~5 %), highlighting the rich information and predictive value of the Pa AmpliSeq data alone.
In summary, our study demonstrates that SNVs in the Pa genome, identified by ML models, can be powerful predictors of lung disease severity and progression in CF patients with chronic Pa infections. Even though this disease outcome is affected by multiple microbial, host genetic and environmental factors, Pa SNVs add complementary predictive value. With additional genetic and clinical data, our ML model could be further fine-tuned and eventually used as a biomarker to pre-emptively identify individuals with CF at high risk for more aggressive observation and treatment.
Supplementary Data
Funding information
The project was supported by funding from CIHR (PJT-148827 to D.N.) and a Vertex Research Innovation Award (D.N.), and salary support from the Cystic Fibrosis Canada Research Fellowship (Award ID 558850 to J.D.), the Leopoldina Foundation (German National Academy of Sciences Leopoldina, Award ID LPDS 2017–17), the Reseau en Santé respiratoire (I.L.), and the Fonds de Recherche en Santé Quebec (I.L., D.N.). M.M.S. and B.J.S. were supported by a Genome Canada and Genome Quebec Bioinformatics and Computational Biology grant.
Acknowledgements
We would like to acknowledge Michael Surette for providing the PES genome sequence and Pradeep K. Singh for input in the AmpliSeq design.
Conflicts of interest
No reported conflicts.
Ethical statement
The study was carried out with the approval from the Research Ethics Boards from the University of Calgary (15-0854) and McGill University Health Centre (15-623).
Footnotes
Abbreviations: AUROC, area under the receiver operating characteristic; AUROC, area under the receiver operating characteristic; BMI, body mass index; CF, cystic fibrosis; CFTR, CF transmembrane conductance regulator; LightGBM, Light Gradient Boosting Method; ML, maching learning; Pa, Pseudomonas aeruginosa; PES, Prairie Epidemic Strain; SNV, single nucleotide variant.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Six supplementary figures, two supplementary tables and two datasets are available with the online version of this article.
References
- 1.Welsh MJ, Ramsey BW, Accurso F, Cutting GR. In: Metabolic & Molecular Bases of Inherited Disease. 8th. Scriver CR, Beaudet AL, Sly WS, Valle D, editors. Vol. 3. New York: McGraw-Hill; 2001. Cystic fibrosis; pp. 5121–5188. edn. vol. [Google Scholar]
- 2.Turcios NL. Cystic fibrosis lung disease: an overview. Respir Care. 2020;65:233–251. doi: 10.4187/respcare.06697. [DOI] [PubMed] [Google Scholar]
- 3.Shanthikumar S, Neeland MN, Saffery R, Ranganathan S. Gene modifiers of cystic fibrosis lung disease: a systematic review. Pediatr Pulmonol. 2019;54:1356–1366. doi: 10.1002/ppul.24366. [DOI] [PubMed] [Google Scholar]
- 4.Coburn B, Wang PW, Diaz Caballero J, Clark ST, Brahma V, et al. Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep. 2015;5:10241. doi: 10.1038/srep10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One. 2010;5:e11044. doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuthbertson L, Walker AW, Oliver AE, Rogers GB, Rivett DW, et al. Lung function and microbiota diversity in cystic fibrosis. Microbiome. 2020;8:45. doi: 10.1186/s40168-020-00810-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci. 2012;109:5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goddard AF, Staudinger BJ, Dowd SE, Joshi-Datar A, Wolcott RD, et al. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc Natl Acad Sci. 2012;109:13769–13774. doi: 10.1073/pnas.1107435109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emerson J, Rosenfeld M, McNamara S, Ramsey B, Gibson RL. Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr Pulmonol. 2002;34:91–100. doi: 10.1002/ppul.10127. [DOI] [PubMed] [Google Scholar]
- 10.Fothergill JL, Walshaw MJ, Winstanley C. Transmissible strains of Pseudomonas aeruginosa in cystic fibrosis lung infections. Eur Respir J. 2012;40:227–238. doi: 10.1183/09031936.00204411. [DOI] [PubMed] [Google Scholar]
- 11.Kosorok MR, Zeng L, West SE, Rock MJ, Splaingard ML, et al. Acceleration of lung disease in children with cystic fibrosis after Pseudomonas aeruginosa acquisition. Pediatr Pulmonol. 2001;32:277–287. doi: 10.1002/ppul.2009.abs. [DOI] [PubMed] [Google Scholar]
- 12.Harris JK, Wagner BD, Zemanick ET, Robertson CE, Stevens MJ, et al. Changes in airway microbiome and inflammation with ivacaftor treatment in patients with cystic fibrosis and the G551D mutation. Ann Am Thorac Soc. 2020;17:212–220. doi: 10.1513/AnnalsATS.201907-493OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hisert KB, Heltshe SL, Pope C, Jorth P, Wu X, et al. Restoring cystic fibrosis transmembrane conductance regulator function reduces airway bacteria and inflammation in people with cystic fibrosis and chronic lung infections. Am J Respir Crit Care Med. 2017;195:1617–1628. doi: 10.1164/rccm.201609-1954OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bragonzi A, Paroni M, Nonis A, Cramer N, Montanari S, et al. Pseudomonas aeruginosa microevolution during cystic fibrosis lung infection establishes clones with adapted virulence. Am J Respir Crit Care Med. 2009;180:138–145. doi: 10.1164/rccm.200812-1943OC. [DOI] [PubMed] [Google Scholar]
- 15.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol. 2012;10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 16.Marvig RL, Sommer LM, Molin S, Johansen HK. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet. 2015;47:57–64. doi: 10.1038/ng.3148. [DOI] [PubMed] [Google Scholar]
- 17.Mowat E, Paterson S, Fothergill JL, Wright EA, Ledson MJ, et al. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am J Respir Crit Care Med. 2011;183:1674–1679. doi: 10.1164/rccm.201009-1430OC. [DOI] [PubMed] [Google Scholar]
- 18.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tümmler B. Pseudomonas. Boston, MA: Springer; 2006. Clonal variations in Pseudomonas aeruginosa ; pp. 35–68. [Google Scholar]
- 20.Dettman JR, Kassen R. Evolutionary genomics of niche-specific adaptation to the cystic fibrosis lung in Pseudomonas aeruginosa . Mol Biol Evol. 2021;38:663–675. doi: 10.1093/molbev/msaa226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klockgether J, Cramer N, Fischer S, Wiehlmann L, Tümmler B. Long-term microevolution of Pseudomonas aeruginosa differs between mildly and severely affected cystic fibrosis lungs. Am J Respir Cell Mol Biol. 2018;59:246–256. doi: 10.1165/rcmb.2017-0356OC. [DOI] [PubMed] [Google Scholar]
- 22.Marvig RL, Johansen HK, Molin S, Jelsbak L. Genome analysis of a transmissible lineage of Pseudomonas aeruginosa reveals pathoadaptive mutations and distinct evolutionary paths of hypermutators. PLoS Genet. 2013;9:e1003741. doi: 10.1371/journal.pgen.1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams D, Evans B, Haldenby S, Walshaw MJ, Brockhurst MA, et al. Divergent, coexisting Pseudomonas aeruginosa lineages in chronic cystic fibrosis lung infections. Am J Respir Crit Care Med. 2015;191:775–785. doi: 10.1164/rccm.201409-1646OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim YW, Evangelista JS, Schmieder R, Bailey B, Haynes M, et al. Clinical insights from metagenomic analysis of sputum samples from patients with cystic fibrosis. J Clin Microbiol. 2014;52:425–437. doi: 10.1128/JCM.02204-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dias R, Torkamani A. Artificial intelligence in clinical and genomic diagnostics. Genome Med. 2019;11:1–12. doi: 10.1186/s13073-019-0689-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lees JA, Mai TT, Galardini M, Wheeler NE, Horsfield ST, et al. Improved prediction of bacterial genotype-phenotype associations using interpretable pangenome-spanning regressions. mBio. 2020;11:e01344-20. doi: 10.1128/mBio.01344-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Acosta N, Heirali A, Somayaji R, Surette MG, Workentine ML, et al. Sputum microbiota is predictive of long-term clinical outcomes in young adults with cystic fibrosis. Thorax. 2018;73:1016–1025. doi: 10.1136/thoraxjnl-2018-211510. [DOI] [PubMed] [Google Scholar]
- 28.Acosta N, Whelan FJ, Somayaji R, Poonja A, Surette MG, et al. The evolving cystic fibrosis microbiome: a comparative cohort study spanning 16 years. Ann Am Thorac Soc. 2017;14:1288–1297. doi: 10.1513/AnnalsATS.201609-668OC. [DOI] [PubMed] [Google Scholar]
- 29.Pellegrino R, Viegi G, Brusasco V, Crapo RO, Burgos F, et al. Interpretative strategies for lung function tests. Eur Respir J. 2005;26:948–968. doi: 10.1183/09031936.05.00035205. [DOI] [PubMed] [Google Scholar]
- 30.Aronesty E. Fastq-mcf: sequence quality filter, clipping and processor. 2013.
- 31.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waskom ML. seaborn: statistical data visualization. J Open Source Software. 2021;6:3021. doi: 10.21105/joss.03021. [DOI] [Google Scholar]
- 36.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 37.Ke G, Meng Q, Finley T, Wang T, Chen W, et al. Lightgbm: a highly efficient gradient boosting decision tree. Adv Neural Inf Process Syst. 2017;30:3146–3154. [Google Scholar]
- 38.Koehrsen W. Feature Selector: Feature Selection in Python. 2019. https://github.com/WillKoehrsen/feature-selector
- 39.Chen T, Guestrin C. Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining. 2016. Xgboost: A scalable tree boosting system; pp. 785–794. [Google Scholar]
- 40.Svetnik V, Liaw A, Tong C, Culberson JC, Sheridan RP, et al. Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci. 2003;43:1947–1958. doi: 10.1021/ci034160g. [DOI] [PubMed] [Google Scholar]
- 41.Somayaji R, Lam JC, Surette MG, Waddell B, Rabin HR, et al. Long-term clinical outcomes of “Prairie Epidemic Strain” Pseudomonas aeruginosa infection in adults with cystic fibrosis. Thorax. 2017;72:333–339. doi: 10.1136/thoraxjnl-2015-208083. [DOI] [PubMed] [Google Scholar]
- 42.Macesic N, Bear Don’t Walk OJ, Pe’er I, Tatonetti NP, Peleg AY, et al. Predicting phenotypic polymyxin resistance in Klebsiella pneumoniae through machine learning analysis of genomic data. mSystems. 2020;5 doi: 10.1128/mSystems.00656-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Méric G, Mageiros L, Pensar J, Laabei M, Yahara K, et al. Disease-associated genotypes of the commensal skin bacterium Staphylococcus epidermidis . Nat Commun. 2018;9:5034. doi: 10.1038/s41467-018-07368-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mobegi FM, Cremers AJH, de Jonge MI, Bentley SD, van Hijum SAFT, et al. Deciphering the distance to antibiotic resistance for the pneumococcus using genome sequencing data. Sci Rep. 2017;7:1–13. doi: 10.1038/srep42808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Recker M, Laabei M, Toleman MS, Reuter S, Saunderson RB, et al. Clonal differences in Staphylococcus aureus bacteraemia-associated mortality. Nat Microbiol. 2017;2:1381–1388. doi: 10.1038/s41564-017-0001-x. [DOI] [PubMed] [Google Scholar]
- 46.Kumru B, Emiralioğlu N, Gökmen Ozel H. Does body mass index affect lung function in patients with cystic fibrosis? Clinical Nutrition. 2018;37:S91. doi: 10.1016/j.clnu.2018.06.1355. [DOI] [Google Scholar]
- 47.Snell GI, Bennetts K, Bartolo J, Levvey B, Griffiths A, et al. Body mass index as a predictor of survival in adults with cystic fibrosis referred for lung transplantation. J Heart Lung Transplant. 1998;17:1097–1103. [PubMed] [Google Scholar]
- 48.Zhao CY, Hao Y, Wang Y, Varga JJ, Stecenko AA, et al. Microbiome data enhances predictive models of lung function in people with cystic fibrosis. J Infect Dis. 2020:jiaa655. doi: 10.1093/infdis/jiaa655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L, Mah TF. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J Bacteriol. 2008;190:4447–4452. doi: 10.1128/JB.01655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuhn M, Johnson K. Applied Predictive Modeling. New York, NY: Springer New York; 2013. Over-fitting and model tuning; pp. 61–92. [Google Scholar]
- 51.Alaa AM, van der Schaar M. Prognostication and risk factors for cystic fibrosis via automated machine learning. Sci Rep. 2018;8:11242. doi: 10.1038/s41598-018-29523-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raschka S. MLxtend: Providing machine learning and data science utilities and extensions to Python’s scientific computing stack. J Open Source Software. 2018;3:638. doi: 10.21105/joss.00638. [DOI] [Google Scholar]
- 53.Pincus NB, Ozer EA, Allen JP, Nguyen M, Davis JJ, et al. A genome-based model to predict the virulence of Pseudomonas aeruginosa isolates. mBio. 2020;11:e01527-20. doi: 10.1128/mBio.01527-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Gast CJ, Walker AW, Stressmann FA, Rogers GB, Scott P, et al. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J. 2011;5:780–791. doi: 10.1038/ismej.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao CY, Hao Y, Wang Y, Varga JJ, Stecenko AA, et al. Microbiome data enhances predictive models of lung function in people with cystic fibrosis. J Infect Dis. 2021;223:S246–S256. doi: 10.1093/infdis/jiaa655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mallawaarachchi S, Tonkin-Hill G, Croucher NJ, Turner P, Speed D, et al. Genome-wide association, prediction and heritability in bacteria with application to Streptococcus pneumoniae . NAR Genom Bioinforma. 2022;4:lqac011. doi: 10.1093/nargab/lqac011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saber MM, Shapiro BJ. Benchmarking bacterial genome-wide association study methods using simulated genomes and phenotypes. Microb Genom. 2020;6 doi: 10.1099/mgen.0.000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maatallah M, Cheriaa J, Backhrouf A, Iversen A, Grundmann H, et al. Population structure of Pseudomonas aeruginosa from five Mediterranean countries: evidence for frequent recombination and epidemic occurrence of CC235. PLoS One. 2011;6:e25617. doi: 10.1371/journal.pone.0025617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bouvier NM. Cystic fibrosis and the war for iron at the host-pathogen battlefront. Proc Natl Acad Sci. 2016;113:1480–1482. doi: 10.1073/pnas.1525101113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Firoz A, Haris M, Hussain K, Raza M, Verma D, et al. Can targeting iron help in combating chronic Pseudomonas infection? A systematic review. Cureus. 2021;13 doi: 10.7759/cureus.13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poudyal B, Sauer K. The ABC of biofilm drug tolerance: the MerR-like regulator BrlR is an activator of ABC transport systems, with PA1874-77 contributing to the tolerance of Pseudomonas aeruginosa biofilms to tobramycin. Antimicrob Agents Chemother. 2018;62 doi: 10.1128/AAC.01981-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MacFadden DR, Melano RG, Coburn B, Tijet N, Hanage WP, et al. Comparing patient risk factor-, sequence type-, and resistance locus identification-based approaches for predicting antibiotic resistance in Escherichia coli bloodstream infections. J Clin Microbiol. 2019;57 doi: 10.1128/JCM.01780-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.