ABSTRACT
Next to Escherichia coli, Bacillus subtilis is the most studied and best understood organism that also serves as a model for many important pathogens. Due to its ability to form heat-resistant spores that can germinate even after very long periods of time, B. subtilis has attracted much scientific interest. Another feature of B. subtilis is its genetic competence, a developmental state in which B. subtilis actively takes up exogenous DNA. This makes B. subtilis amenable to genetic manipulation and investigation. The bacterium was one of the first with a fully sequenced genome, and it has been subject to a wide variety of genome- and proteome-wide studies that give important insights into many aspects of the biology of B. subtilis. Due to its ability to secrete large amounts of proteins and to produce a wide range of commercially interesting compounds, B. subtilis has become a major workhorse in biotechnology. Here, we review the development of important aspects of the research on B. subtilis with a specific focus on its cell biology and biotechnological and practical applications from vitamin production to concrete healing. The intriguing complexity of the developmental programs of B. subtilis, paired with the availability of sophisticated tools for genetic manipulation, positions it at the leading edge for discovering new biological concepts and deepening our understanding of the organization of bacterial cells.
KEYWORDS: sporulation, cell biology, biofilm formation, probiotics, biotechnology
INTRODUCTION
In 1835, one of the founders of microbiology, Christian Gottfried Ehrenberg, described a bacterium that he named Vibrio subtilis, probably to coin the motility (“vibration”) of the thin cells (1). The microbiologist and botanist Ferdinand Julius Cohn renamed the bacterium in 1872 to Bacillus subtilis—the subtle rod—as we know it today. Cohn also discovered that B. subtilis forms heat-resistant spores as part of its life cycle (2). This finding eventually paved the way to pasteurization. Ever since, B. subtilis has attracted scientific interest and has become the best-studied and best-understood bacterium besides E. coli. This is caused by several factors. First and foremost, the sporulation cycle first documented by Cohn provides a relatively simple model for studying processes related to cell differentiation and development. Second, the nonpathogenic nature, its versatile metabolism, and ease of culturing of B. subtilis make it useful for a wide variety of applications. These include the production of traditional food in Asia by fermentation of soybeans such as Natto in Japan, or the production of vitamins, amino acids, and enzymes for washing powders. Third, B. subtilis is a relative of many important Gram-positive pathogens such as Bacillus anthracis, Staphylococcus aureus, or Listeria monocytogenes. B. subtilis serves as the model organism for these pathogens and all other Firmicutes. Finally, B. subtilis grows very fast and can easily be genetically manipulated due to the ability to take up foreign DNA and even to integrate this DNA into its own genome. Thus, B. subtilis has become extremely popular in microbiology and industry and was even named the “Microbe of the Year” by the German Association for General and Applied Microbiology in 2023, the sister organization of the ASM. Here, we will give an overview of some of the important and sometimes even spectacular discoveries that have been made with B. subtilis as the object of investigation and in the many applications of this bacterium in biotechnology, animal feeding, and concrete healing.
THE GENOME AND PROTEOME OF B. SUBTILIS
The nineties of the past century saw the beginning of the genomic revolution, and the genome sequence of B. subtilis was one of the first to be published (3). This was achieved in a huge combined European–Japanese collaboration, organized by Frank Kunst. This collaboration was the start to a long-lasting cooperation among the European B. subtilis labs, which eventually resulted in the initial identification of the set of essential genes in the determination of the expression profiles of all B. subtilis genes under 104 different conditions and in the construction of a first genome-reduced strain that lacked all prophages (4 to 6). The genome sequence was generated with individually cloned genome fragments and hand-casted sequencing gels. It is thus not surprising that the genome sequence and also the list of essential genes has undergone revisions (7 to 9).
B. subtilis is a bacterium that was subject to intensive proteome research even before the word “proteome” existed. This line of research was pioneered by Michael Hecker and was performed with the goal of getting a map of all proteins and the patterns of their abundance during different growth stages (10). In the early days, before the use of tandem mass spectrometry for protein identification, the proteins were excised from gels and identified by N-terminal sequencing. In this way, all the proteins involved in central carbon metabolism and their regulation by glucose could be studied (11), and the global regulation of protein synthesis could even by visualized in a “movie of life” (12). Today, relative quantifications are available for all proteins, and absolute numbers (i.e., copy numbers per cell) are available for many proteins (13 to 15). Recently, proteome analysis was taken even a step further, and the global protein interactome was studied by in vivo cross-linking coupled with mass spectrometry (16). This was the first time that such an approach, which is bioinformatically challenging, was applied to a complex bacterium. The study identified a large number of novel interactions, many of them involving proteins of unknown function. This is an excellent starting point for the functional identification of such proteins, and indeed, PdhI, a novel inhibitory protein of pyruvate dehydrogenase, has been identified by this approach (16).
The development of the field of synthetic biology has drawn a lot of interest in the construction of minimal cells, with the aim of understanding the roles of all remaining components of the cell. This goal can be reached by bottom-up or top-down approaches. The artificial construction of a genome and of a living cell based on this designed genome has so far only been possible for one species, Mycoplasma mycoides. The generated artificial organism M. mycoides Syn3A contains the smallest known genome that allows host-independent growth (17). B. subtilis is one of the bacteria for which genome minimization by a top-down approach was attempted. In a first step, the set of genes that are likely to be required in a minimal organism was identified. It comprises 523 and 119 genes coding for proteins and RNAs, respectively (18). Based on this blueprint, the B. subtilis genome was reduced by 40%, which is the most significant genome reduction that has so far been achieved for any complex bacterium (13, 19). Interestingly, due to the deletion of all protease-encoding genes, these strains proved to be superior for the production of otherwise difficult secreted proteins, as shown for different staphylococcal antigens (20).
The availability of a large research community and the generation of different types of data on an organism are the keys to making advances in its understanding. However, all this information is much more valuable if it is integrated in one database. The B. subtilis scientific community can make use of the database SubtiWiki, which integrates all types of information in an intuitive and interactive manner (21). This is essential for the development of novel research hypotheses and their experimental validation, which may in turn result in the identification of new functions or new regulatory or physical interactions. To the best of our knowledge, SubtiWiki is the only organism-specific database for bacteria that fully integrates all information and is completely free to use.
THE CONTROL OF SPORULATION IN B. SUBTILIS—A TALE OF THE VALUE OF SCIENTIFIC CONTROVERSY
As mentioned above, the ability of B. subtilis to form heat-resistant spores was already discovered in the 19th century by Ferdinand Cohn. The elucidation of the underlying molecular program has been a long-lasting endeavor of the B. subtilis scientific community. Jim Hoch and Richard Losick, two of the heroes of Bacillus research, discovered that sporulation is controlled by a regulated genetic program. Hoch found that a complex two-component system, later called the phosphorelay, with Spo0A as the central player was critical to the onset of sporulation (22, 23). Losick studied the RNA polymerase and discovered the involvement of several alternative sigma factors in the genetic program of sporulation (24). How was this possible? Who of them was right and who was wrong? Was the two-component system not something that is typical for E. coli, whereas alternative sigma factors are a hallmark of the B. subtilis genetic system? Later on, when the activity control of some of the alternative sigma factors was studied, Losick found that the activity of the sporulation sigma factor SigF is controlled via protein–protein interactions by an antisigma factor and an anti-antisigma factor (25). In contrast, at the same time, Michael Yudkin discovered that the antisigma factor SpoIIAB, which is encoded just upstream of SigF, is in fact a protein kinase (26). These results were the subject of a battle rather than a discussion at the International Bacillus Conference 1993 in Paris. In both controversies, it turned out that both initially contradicting findings were correct, and that they had to be assembled to get the full picture. These discussions illustrate the importance of integrating different views as well as the value of good and careful experimental work.
B. SUBTILIS AS A MODEL ORGANISM FOR CELL BIOLOGY
The role of B. subtilis as a major model in bacterial cell and developmental biology was greatly pushed by the discovery of its natural competence (27). The fact that B. subtilis produces endospores using a simple developmental program, further fueled the scientific interest. Here, fundamental biological principles in cell differentiation could be analyzed with the power of efficient bacterial genetics. Consequently, research on spore formation has led to a variety of tools that laid the foundations for bacterial cell biology (28). Early electron microscopy studies revealed various stages of sporulation (29). It was clear that it must be possible to dissect the molecular mechanisms behind this series of cellular events. Struck by the obvious beauty of the system, several laboratories started to engage in B. subtilis sporulation research. Sporulation of B. subtilis is initiated by nutrient limitation, and Joel Mandelstam took advantage of this in a resuspension method, by which sporulation could be timed and reliably analyzed (30). Sporulation is initiated by an asymmetric septum formation close to one cell pole. This clear subcellular differentiation was an ideal example to test protein localization in vivo. Only very shortly after the introduction of the green fluorescent protein (GFP) as a tool for cell biology (31), the labs of Richard Losick and others used the technique to generate translational fusions to sporulation genes and to localize proteins within living cells (32, 33).
B. subtilis quickly became the model organism for studying cell wall synthesis, cytokinesis, and chromosome organization. B. subtilis is, like all Firmicutes, surrounded by a thick cell wall made of peptidoglycan (PG) and teichoic acids. This cell wall acts as an exoskeleton and thus maintains the shape of the cell. It was long thought that PG synthesis relies on the transglycosylation activity of class A penicillin-binding-proteins (PBPs) that have transglycosylation activity connecting the sugar moieties of the PG scaffold and transpeptidase activity to generate the cross-links of the stem peptide (34). An unexpected finding in B. subtilis was the recent discovery of the SEDS-protein RodA being a glycosyltransferase, responsible for PG synthesis (35). PG synthesis in the rod-shaped bacilli occurs at the lateral side by a multiprotein complex termed the elongasome (36). The elongasome complex includes the bacterial actin-homologue MreB. Although the presence of actin-like proteins in bacteria was known for several years, it was only the localization of MreB-GFP fusions in B. subtilis that really pushed the idea of a bacterial actin cytoskeleton (37, 38). Initially, it was thought that MreB formed extended, helical filaments along the cell membrane. However, modern microscopy techniques rather showed that MreB forms dynamic patches that require active PG synthesis for their dynamics (39, 40; see Fig. 1A). In B. subtilis, MreB and its two paralogs Mbl and MreBH seem to modulate PG synthesis to constrain circumference, thereby helping to form the rod morphology (41).
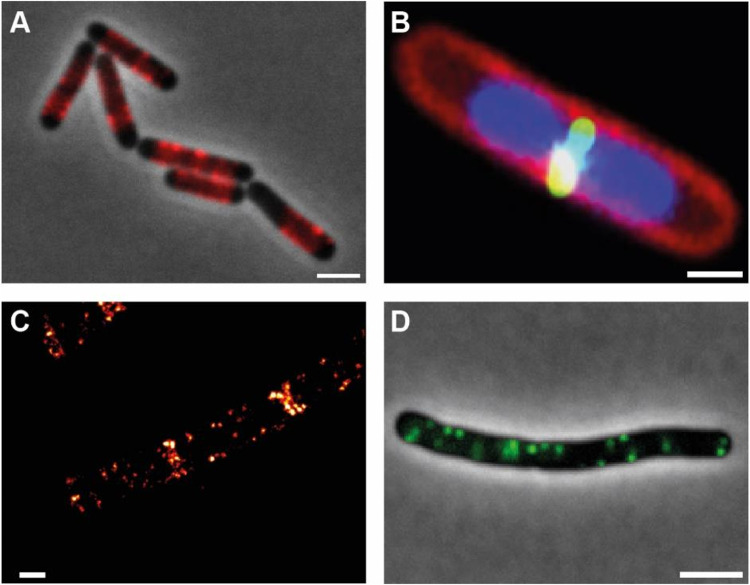
FIG 1.
Subcellular localization of proteins in B. subtilis. (A) Localization of MreB-mCherry along the lateral sides the cell. (B) FtsZ-GFP (green) localization at midcell; nucleoids (DNA) are shown in blue and the plasma membrane in red. (C) Single molecule localization microscopy of the polar scaffold protein DivIVA (DivIVA-PA-mCherry). DivIVA localizes at both sides of the division septum and in cluster along the membrane. (D) Flotillin (FloT-GFP) patches along the plasma membrane in B. subtilis. Scale bars 1 μm (A and D) and 0.5 μm (B and C).
While the elongasome is regulated by MreB, cell wall synthesis at the site of cell division is governed by the bacterial tubulin homologue FtsZ. Early immune fluorescence imaging showed that FtsZ localizes precisely at midcell (42; see Fig. 1B); however, fluorescent fusions recently revealed that dynamic treadmilling of FtsZ drives Z-ring condensation and constriction, thereby directing septal PG synthesis (43). FtsZ recruits a set of proteins collectively termed the divisome. Work on B. subtilis revealed that the divisome assembles at least in two steps, first assembling an “inner ring” on the cytoplasmic side, and in a second step recruitment of the membrane integral to PG synthesis machinery and their regulatory proteins (44). Spatial regulation of FtsZ is governed by a ParB-like nucleoid occlusion protein Noc, which binds to specific DNA sequences on the chromosome and to the plasma membrane. This membrane anchoring likely sterically hinders FtsZ ring formation across the nucleoid (45). B. subtilis uses a variation of the miniature cell (Min) system to ensure that division takes place only once per cell cycle. The polar scaffold proteins DivIVA and MinJ recruit the MinCD complex to prevent divisome reassembly at previously used division sites (46). DivIVA uses the physical cue of negative membrane curvature at constricting septa for its positioning (47; see Fig. 1C).
B. subtilis was also a prime model organism for the study of chromosome organization and segregation. Since one copy of the genome has to be transferred into the prespore during sporulation, DNA segregation mutants were identified among sporulation mutants. These included Spo0J (ParB) and Soj (ParA). Studies on Spo0J/Soj localization in the lab of Jeff Errington proposed the idea of a “mitotic like DNA segregation” system, suggesting that DNA (or at least oriC) segregation in bacteria can be an actively driven process (48). Again, work in B. subtilis revealed that Spo0J (ParB) is a CTPase and that CTP binding and hydrolysis act as a molecular switch that allows binding, spreading, and release of ParB-like proteins from DNA (49). Spo0J is required to load the structural maintenance of chromosome SMC onto the chromosome close to the origin of replication (50, 51). SMC brings the replichore arms together and is likely compacting the chromosome by a loop extrusion mechanism. Failing to load SMC leads to a block in oriC segregation (52).
In the last decade, B. subtilis has also become a model system to study plasma membrane organization and membrane fluidity (53). Several proteins that were thought to be eukaryotic inventions are actually present in B. subtilis. Among those are the flotillins FloA and FloT that are highly similar to their eukaryotic counterparts (54; Fig. 1D). Flotillins regulate membrane fluidity and membrane domain formation (55, 56). Lack of these activities leads to pleiotropic phenotypes in cell wall synthesis, biofilm formation, and signaling. Membrane surveillance and repair was shown to include the dynamin-like protein DynA (57). Again, the elaborated molecular biological toolbox allowed fast progress of membrane research in B. subtilis, likely securing B. subtilis a seat at the forefront of membrane biology research.
IMPORTANT DISCOVERIES MADE WITH B. SUBTILIS
The research with B. subtilis has resulted in important discoveries in many fields. In the early days of molecular biology, many researchers thought that what was true for E. coli was also true for all other bacteria and maybe even the elephant. The research of the past few years has shown that B. subtilis rather than E. coli can serve as a model at least for many bacteria.
The first paradigm of a regulatory mechanism was the dual control of the E. coli lac operon by lactose as the inducer and glucose that caused carbon catabolite repression. The latter regulation acts via the signal molecule cyclic AMP and a dedicated receptor protein that serves as transcription activator for the lac and many other catabolic operons (58). However, neither cAMP nor its receptor protein are present in B. subtilis, even though glucose causes catabolite repression in this bacterium as well. Pioneering work in the lab of Milton Saier identified that HPr, a small protein that is part of the PTS, a protein cascade of consecutively phosphorylated proteins that finally transport and phosphorylate a set of sugars, can be phosphorylated at a second site (Ser-46) in addition to the phosphorylation site used for sugar uptake (59). Moreover, this phosphorylation depends on the availability of glucose. Later, it could be shown in the lab of Wolfgang Hillen that this regulatory form of HPr phosphorylated on Ser-46 acts as the cofactor of the transcription repressor CcpA and that this complex in fact is responsible for catabolite repression in B. subtilis (60). Yet, the protein kinase that is responsible for the phosphorylation of HPr on Ser-46 was still unknown and was the subject of intensive research in many labs. Finally, only the availability of the B. subtilis genome sequence allowed the identification of the gene based on the N-terminal amino acid sequence of the purified protein. After a highly competitive race, two groups eventually identified the hprK gene and characterized the corresponding protein (61, 62).
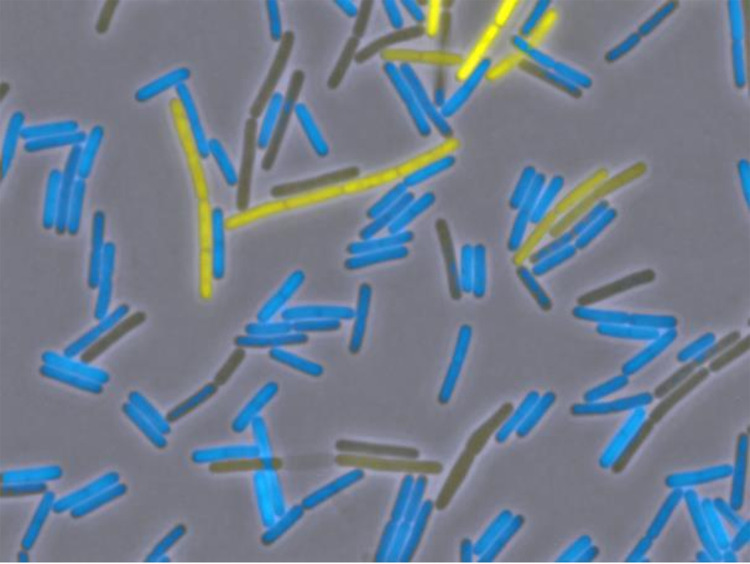
For a long time, it was generally assumed that bacterial populations grow homogeneously and that all cells in a culture have identical properties. However, we now know that differentiation of cell types, such as the development of genetic competence or of biofilms, only takes place in subpopulations within B. subtilis cultures (see Fig. 2). Indeed, each cell must decide to go its own path, and once this decision is made, the complete program is expressed until the cells are competent or form a biofilm. Pioneering work in this field was made in the labs of David Dubnau and Oscar Kuipers. Decision-making is based on so-called bistable switches that decide about the expression of key regulators, ComK and SinR in the case of genetic competence and biofilm formation (63 to 65). A B. subtilis biofilm displays features of multicellularity that we know from eukaryotes like fungi, with distinct localization of activities within the biofilm and division of labor, with some cells producing the extracellular matrix, while others sporulate (66, 67). As shown by Nicola Stanley-Wall, the B. subtilis biofilm is coated by a hydrophobic protein, BslA, which repels water more efficiently than even Teflon, providing exquisite protection from phage predation and water-soluble antimicrobials (68; see Fig. 3).
FIG 2.
Bistable expression of motility and biofilm genes. Fluorescence microscopy of cells harboring both Phag-cfp and either PtapA-yfp fusion in a wild-type strain of B. subtilis. Cells were observed using fluorescence microscopy. Phag-CFP was false, colored in blue, and PtapA-YFP in yellow. (Reproduced from reference 108).
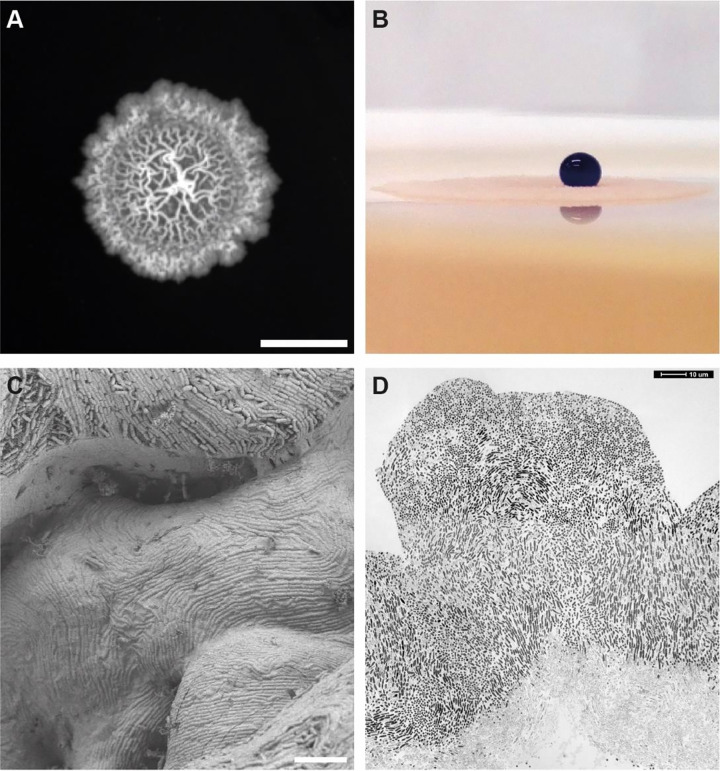
FIG 3.
Biofilm formation of B. subtilis. (A) Colony of B. subtilis on agar surface. (B) B. subtilis biofilm surfaces are highly hydrophobic. A drop of colored water (5 μL) is placed on top of the colony. (C) Scanning electron microscopy of a B. subtilis biofilm. Note the evenly covered surface of the biofilm, which occurs because of the hydrophobic protein BslA. (D) Transmission electron microscopy image of a thin section through a B. subtilis biofilm. Cells at the bottom of the biofilm tend to lyse and appear lighter. Scale bars are 0.5 cm (A) and 10 μm (C and D).
A field of research that was most strongly stimulated by discoveries with B. subtilis is the field of RNA switch-mediated regulation of gene expression. The expression of several sugar catabolic operons is controlled by mutually exclusive RNA structures that upon binding of a regulatory protein adopt a structure that allows transcription, whereas a transcription terminator is formed in the absence of the proteins (69). In the case of the tryptophan biosynthetic operon, binding of a ring-like protein in the presence of tryptophan causes transcription termination (70). While such protein-mediated regulation fits very well into the general picture of gene expression, work in the lab of Tina Henkin identified something very unexpected: there are RNA switches, the T-boxes that are controlled by interaction with tRNAs. The T-boxes are present upstream of genes involved in amino acid homeostasis, and the genes are induced by starvation for the cognate amino acid as a result of an interaction between the uncharged tRNA and the T-box RNA that leads to transcription read-through (71, 72). Even more spectacular was the discovery of RNA switches that are triggered by metabolites, the so-called riboswitches. Again, the Henkin lab pioneered in identifying a so-called S-box regulatory RNA element upstream of many genes involved in methionine biosynthesis (73). In this case, S-adenosylmethionine binds to the S-box and induces a structural change that results in transcription termination (74). In parallel, Ron Breaker identified RNA structures that bind thiamine and FMN and coined the term riboswitch for small molecule-binding RNA switches (75, 76). Bioinformatic analyses revealed that riboswitches are widespread in bacteria and that they can bind a multitude of different molecules, among them metal ions, ribonucleotide-based intermediates of metabolism, and second messengers (77). This link between riboswitches and RNA-based nucleotides can be seen as a remainder of the RNA world that preceded the current protein-based life (78).
Ground-breaking discoveries have been made with B. subtilis in many fields of research. Several metal ions are essential for life, but they may become toxic at high intracellular concentrations. The lab of John Helmann has made significant contributions to this research area (79). Regulation of amino acid homeostasis has for a long time been studied by Boris Belitsky and Linc Sonenshein. They have discovered that B. subtilis contains an active glutamate dehydrogenase, RocG, that converts glutamate to 2-oxoglutarate. This enzyme is part of the arginine degradative pathway. In contrast, a second, constitutively expressed glutamate dehydrogenase, GudB, is cryptic in laboratory strains in B. subtilis (80, 81). However, in undomesticated strains, GudB is active and is the major player in glutamate utilization. Recently it was discovered that the activity of GudB is inhibited in wild-type strains by direct interaction of the enzyme with the biosynthetic enzyme glutamate dehydrogenase GltAB. In this way, the formation of a futile cycle that would result in a waste of glutamate can be prevented. This kind of interaction of opposing enzymes has been coined counterenzyme complex (82). A huge variety of regulatory systems that respond to distinct external stimuli have been discovered in B. subtilis and other bacteria. Recently, it has been shown that electrochemical signaling using potassium ions plays an important role in spore germination and biofilm formation in B. subtilis (83, 84). Glyphosate is an herbicide that is used worldwide. The weed killer inhibits the 5-enolpyruvyl-shikimate-3-phosphate synthase, which is essential for the synthesis of aromatic amino acids. However, until recently, no transporter for this important molecule has been identified. Using B. subtilis as the model organism, it could be demonstrated that glyphosate enters the cell via a glutamate transporter, GltT (85). Most phototrophic organisms as well as animals possess an internal circadian clock. In humans, this clock is responsible for the control of the sleep–wake cycle. The recent discovery of a circadian clock in the nonphotosynthetic bacterium B. subtilis (86) came as quite a surprise and indicates that there is likely much more hidden in the biology of this bacterium that deserves further research. The large body of knowledge that we have about this bacterium will now also allow us to study proteins and functions for which our knowledge is still limited (87).
B. SUBTILIS: AN ESSENTIAL WORKHORSE FOR INDUSTRIAL BIOTECHNOLOGY
There are multiple reasons why B. subtilis has become established as an important expression host in the biotech industry. It is a robust, rapidly growing microorganism that efficiently and rapidly converts organic substrates into biotechnological products in short fermentation cycles. In addition, it also possesses the extraordinary ability to secrete large quantities of protein (20 to 25 g/L) into the culture medium, making it a frequent choice as an industrial platform organism for large-scale production of degradative enzymes and proteins (88). Numerous molecular biological tools for selective metabolic engineering have been developed in recent decades, including efficient, simple CRISPR Cas9 methods that allow researchers to edit the genome with base-by-base precision (89, 90).
Of all industrial processes utilizing B. subtilis to generate an organic molecule via fermentation, vitamin B2 (riboflavin) production is likely the most significant. As a precursor of flavin coenzymes (FAD, FMN), vitamin B2 is essential for metabolism in all living cells. For their growth and reproduction, animals and humans in particular depend on riboflavin intake in the form of nutritional supplements and feed additives. As such, research on microbial production methods began as early as the 1940s (91). According to more recent market estimates, approximately 12,700 metric tons of riboflavin were produced in 2021 at a value of nearly $400 million (USD) (https://www.mordorintelligence.com/industry-reports/riboflavin-market). Roughly 70% of this volume has traditionally been used for animal feeds, with the other 30% going to human nutritional supplements and/or pharmaceuticals.
From a biotechnology perspective, fermentative vitamin B2 production is the classic example of a biotech process replacing a chemical production process, having done so within just 15 years. The microbial production process delivers more than just economic benefits, however—it is especially valuable in many aspects of sustainability as well. Establishing today’s highly efficient processes required the use of all available methods of efficient strain and process development (see reference 92 for review). It is worth noting that the Russian Institute for Genetics and Selection of Industrial Microorganisms, Moscow, used riboflavin in 1983 in an example of what was presumably the first genetically modified production strain created for a small organic molecule (88).
Riboflavin is not the only organic molecule, however, that can be produced using B. subtilis. The patent literature describes processes for producing pantothenic acid (vitamin B5) (88). In addition, B. subtilis is also known for producing a huge diversity of secondary metabolites such as surfactin and other lipopeptides such as fengycin (93). Another interesting, industrially significant substance produced by B. subtilis is γ-polyglutamic acid, an anionic homopolyamide consisting of D- and L-glutamic acid units that is used as a thickener, moisturizer, or antifreeze in the food and cosmetics industries (94).
In addition to their many uses in the food, beverage, textile, leather, detergent, and cleaning industries, large quantities of industrial enzymes are also needed in animal nutrition and various medical applications. The market for industrial enzymes was valued at approximately $6 billion in 2021 (https://www.mordorintelligence.com/industry-reports/industrial-enzymes-market). Here again, B. subtilis plays a critical role as an efficient, heterologous expression host for hydrolytic enzymes such as proteases, amylases, and lipases (95). Protein secretion in B. subtilis and possibilities for its improvement have been extensively studied in the group of Jan Maarten van Dijl (20, 96, 97). In fact, enzymes produced with B. subtilis or close relatives such as Bacillus licheniformis are the major active compounds in all commercially available washing powders. In addition, B. subtilis is also the natural source of neutral and alkaline proteases, whose biological function guarantees access to organic materials in the soil. Because these intrinsic proteases negatively affect heterologous expression of hydrolytic enzymes, however, the WB800N strain was produced, in which all 8 of the known B. subtilis proteases were inactivated (98). Recently, genome-reduced strains of B. subtilis were suggested to be superior hosts for the secretion of heterologous proteins (20).
Despite its wide variety of potential applications in industrial biotechnology, B. subtilis is often a source of unwelcome contamination problems in production facilities, as its biofilms can adhere to tubing and conduits, and its resilient spores are very difficult to remove.
APPLICATION OF B. SUBTILIS AS A PROBIOTIC
The term “probiotic” was first coined in 1954 to describe substances crucial to a healthy life. In 2001, a WHO expert committee proposed the following definition of probiotics, which remains valid today: “live microorganisms which, when consumed in appropriate amounts in food, confer a health benefit on the host” (99). In addition to representatives of the genus Lactobacillus, many Bacillus species, and especially strains of B. subtilis, have also been used in a variety of ways as probiotics for animals and humans.
Among its various metabolic properties, its ability to form spores in particular makes B. subtilis attractive as a probiotic. The heat resistance of Bacillus spores does more than ensure shelf-life stability of corresponding products at temperatures exceeding room temperature—it also means that manufacturers can mix the spores directly into animal feeds, which are pelletized at 80 to 85°C. In addition, many spores also survive the low-pH environment of the gastric passage and the bile acids of the small intestine. Once the spores germinate, the effects of the probiotics can be shown in the upper and, most especially, in the lower intestinal tract. Depending on the strain, Bacillus-based probiotics in particular exhibit a variety of different modes of action such as direct or indirect inhibition of pathogens. Certain probiotics also strengthen the intestinal barrier and the immune system, produce metabolites that other microbiota selectively metabolize (“cross-feeding”), or secrete enzymes that make indigestible food available to the host and to microbiota.
Based on the collected genetic information and the physiological properties of B. subtilis strains, the European Food Safety Authority (EFSA) has included the microbe on its Qualified Presumption of Safety (QPS) list, provided the strains used are not resistant to antibiotics and do not produce other toxic substances. Some of the enzymes produced by B. subtilis and used in industry, such as nattokinase, also have the “Generally Regarded as Safe” (GRAS) status with the U.S. Food and Drug Administration (FDA).
B. SUBTILIS IN LIVESTOCK FARMING AND HUMAN USE
Bacillus-based probiotics for poultry and pig farming have been developed and launched as spore products as early as the late 1980s. Even at that time, the primary motivation was preventive control of pathogens and support for intestinal health as an alternative to the use of antibiotics. It should be emphasized that, at the time, the use of antibiotics went beyond medical justification—the lion’s share of these were what are known as antibiotic growth promoters (AGPs), which were used in subtherapeutic doses because they improved animals’ performance. The WHO sees a clear correlation between the growing use of antibiotics in livestock farming and in human and veterinary medicine, on the one hand, and the ever-growing spread of antibiotic-resistant bacteria limiting treatment options in hospitals, on the other. AGPs were therefore banned entirely in the EU starting in 2006. Other countries followed suit; nevertheless, quantities have gone up worldwide, with 73% of all antibiotic use in 2021 still attributable to meat production (https://www.fairr.org/index/key-findings/risk-opportunity-factors/antibiotics/).
The EU’s ban on AGPs, along with more restrictive prescription practices, have led to increased interest in alternative feed additives, whereby probiotics are considered to have the greatest potential. According to various market studies, the current size of the market for animal feed probiotics was approximately $2.7 billion in 2021 (https://www.feedandadditive.com/global-feed-probiotics-market/). Fragmented among hundreds of manufacturers, this market encompasses Bacillus as well as lactic acid bacteria and certain yeast products.
Subclinical necrotic enteritis, an illness caused by toxin-producing Clostridium perfringens strains, represents a huge challenge in the poultry industry, especially when producers need to do away with antibiotics. The disease produces lesions in the intestinal wall, resulting in losses on the order of $6 billion annually (100). Identifying and developing a Bacillus-based probiotic involved a complex screening process carried out on an extensive collection of 500 Bacillus strains. Covering over 20 parameters—including sporulation efficiency, heat resistance, survival under intestinal tract conditions, inhibition of pathogens such as C. perfringens (Fig. 4), and safety evaluation and production efficiency—the analysis led to, among other results, the identification of the strain B. subtilis DSM 32315 (101). The poultry product based on this strain was subjected to numerous animal studies in 2017 that successfully demonstrated its efficiency, particularly in C. perfringens models (102).
FIG 4.
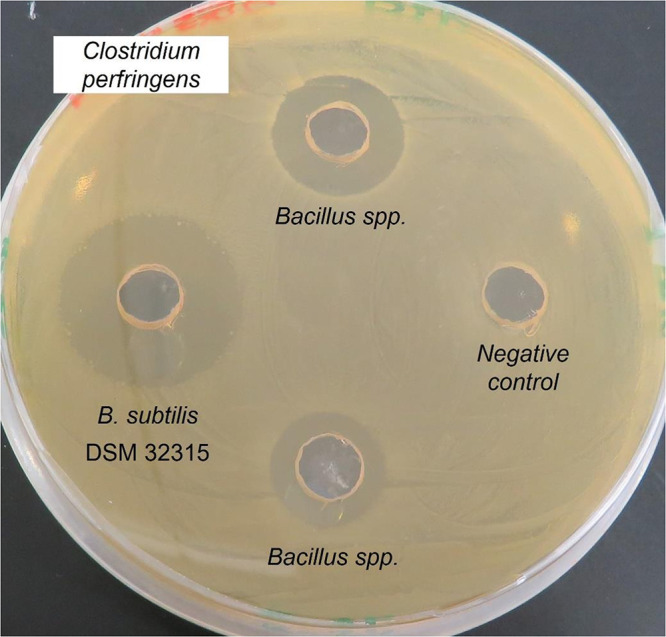
Pathogen Inhibition assay. To analyze the effect of B. subtilis on the pathogen Clostridium perfringens, the pathogenic strain was plated onto a Caso-Yeast agar plate. Small holes were punched into the agar and similar amounts of liquid Bacillus cultures or only medium as negative control were filled into them. The plates were incubated for 24 h under anaerobic conditions. The culture diffuses into the agar and inhibits the growth of C. perfringens around the holes. In comparison to the other two tested Bacillus strains, the culture of B. subtilis DSM 32315 shows the biggest inhibition halo and therefore the highest inhibition of C. perfringens.
The market for probiotic food supplements has traditionally been dominated by Lactobacillus strains and primarily addresses intestinal health. Market studies have shown that end-consumer sales of food supplements came to roughly $8.9 billion in 2019 (103). Enterogermina, registered in Italy in 1958, was one of the first over-the-counter medicinal supplements containing Bacillus-based probiotics. The product, claiming to strengthen the immune system, was marketed as containing four B. subtilis strains that were later reclassified as B. clausii. Recent years have also seen an increased focus on developing Bacillus-containing products to improve intestinal health.
Synbiotic concepts for improving intestinal health have shown particular innovation potential. A synbiotic is a combination of a living microorganism and a substrate selectively metabolized by host microorganisms that confers a health benefit on the host. When combined here with the L-alanyl-L-glutamine dipeptide, the B. subtilis DSM 32315 strain indicated above again produces surprising effects. Extensive studies have demonstrated that the presence of this synbiotic not only prompts microbiota to increase butyrate production in vitro—the same observation could even be confirmed via feces analyses in a human pilot study. In addition to serving as an important source of energy for enterocytes, butyrate has also been shown in the literature to have many benefits, which include strengthening intestinal integrity, improving the immune system and metabolic health, and producing anti-inflammatory effects. Surprisingly, blood analyses also revealed a positive impact on lipid and glucose metabolism in test subjects who had taken the synbiotic (104).
B. SUBTILIS-BASED SELF-HEALING CONCRETE
With an annual global demand of around 10 billion metric tons, concrete is the world’s most important construction material. Concrete is primarily a blend of cement, an inorganic binder, water, and aggregates such as sand, gravel, or crushed limestone. Producing some four billion metric tons of cement worldwide requires burning limestone, a process that contributes greatly to the global CO2 emissions. Being the source of 8 to 10% of the global anthropogenic emissions of CO2 makes cement production one of the world’s most emissions-intense industrial processes (105). In addition to establishing methods that produce less CO2, one particularly effective way of reducing emissions is to make concrete that lasts longer.
As concrete ages, small cracks arise that allow water and salt ions to penetrate; this process can cause steel reinforcements to rust and to corrode. A microbiological approach to close these cracks is biomineralization, specifically microbiologically induced calcium carbonate precipitation (MICP) (105). This process was described in the late 20th century and can be achieved in autotrophic and, especially, heterotrophic bacteria via various metabolic pathways. To put it simply, MICP is a process by which the metabolic activity of microorganisms results in the production of CO32– ions in an alkaline environment. The Ca2+ ions present in solution during cement hydration bind to negatively charged groups on the microbial cell wall and then react with the carbonate ions, resulting in extracellular formation of insoluble CaCO3. The conditions under which these microorganisms have to produce these effects are very challenging: the environment inside concrete is highly alkaline (pH 13), there is little oxygen available, and the curing process produces temperatures of 60°C.
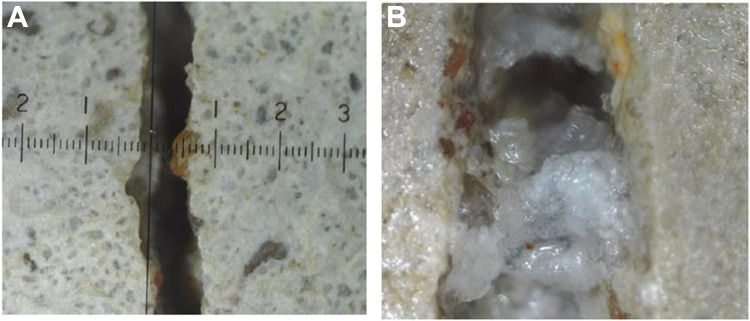
These conditions once again greatly favor Bacillus spores, which are used in MICP. Here, spores of B. subtilis DSM 32315 achieved yet another success, in this case as an additive for self-healing concrete, whereby the water that penetrates cracks causes the spores to germinate and then close those cracks via CaCO3 precipitation (106; Fig. 5).
FIG 5.
Use of B. subtilis DSM 32315 for concrete healing. (A) Picture of a concrete test object after mechanical introduction of a 0.5- to 1-mm crack. (B) Picture of the test object after 28 days of incubation in a cloud chamber at 20°C/65% relative humidity, followed by 14 days of incubation in a cloud chamber (20°C/100% relative humidity). The presence of B. subtilis spores induced biomineralization and filling of the crack via CaCO3 formation. (Photos courtesy of Anke Reinschmidt, used with permission).
FUTURE APPLICATIONS
The examples shown here reinforce the significance of B. subtilis for industrial biotechnology. In addition, numerous B. subtilis products have already been introduced for biomining, as additives in household cleansers and as microbial biostimulants, a class of bacteria that support plant growth by protecting plants from biotic and abiotic stress. Because its surface structure interacts with metals and rare earth elements, B. subtilis is also employed in biomining, the use of microorganisms for binding and extracting metals. Another interesting potential application of B. subtilis for human health could be Alzheimer’s disease. In the corresponding research, it was observed that the gut-associated biofilm in the Alzheimer’s model Caenorhabditis elegans had a protective effect on nerve cells (107).
Various properties, such as microbiome modulation and enzyme production, make B. subtilis ideal for use in innovative cleansers for hard surfaces, especially in hospitals, where it inhibits colonization by pathogenic organisms. In terms of industrial applications, B. subtilis is already indispensable, and many more innovations are anticipated.
ACKNOWLEDGMENTS
We are grateful to Urśka Repnik (central microscopy facility Kiel), Sarah Baur, and Helge Feddersen (Kiel) for help with the images. We thank all the employees of the Evonik Biotech Hub who contributed to these results, as well as all group members of the Bramkamp and Stülke labs for their contributions to Bacillus research. Anke Reinschmidt is acknowledged for providing photos in Fig. 5.
Work in the Bramkamp and Stülke laboratories is funded by the Deutsche Forschungsgemeinschaft (DFG) (BR2915/7 and BR2915/10 to M.B., Priority Program SPP1879 [STU 214/16] and SFB 1565 [Projektnummer 469281184 (P11)] to J.S.).
Contributor Information
Jörg Stülke, Email: jstuelk@gwdg.de.
Stefan Pelzer, Email: stefan.pelzer@evonik.com.
George O'Toole, Geisel School of Medicine at Dartmouth.
REFERENCES
- 1.Ehrenberg CG. 1835. Dritter Beitrag zur Erkenntnis grosser Organisation in der Richtung des kleinsten Raumes. Physikal Abh Königl Akad Wiss Berlin 1833:143–336. [Google Scholar]
- 2.Cohn F. 1877. Untersuchungen über Bakterien. IV. Beiträge zur Biologie der Bacillen. Beitr Biol Pflanz 2:249–276. [Google Scholar]
- 3.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessières P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Cordani JJ, Connerton IF, Cummings NJ, Daniel RA, Denziot F, Devine KM, Düsterhöft A, Ehrlich SD, Emmerson PT, Entian KD, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim SY, Glaser P, Goffeau A, Golightly EJ, Grandi G, Guiseppi G, Guy BJ, Haga K, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Débarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauël C, et al. 2003. Essential Bacillus subtilis genes. Proc Natl Acad Sci USA 100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Härtig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Chat LL, Lecointe F, Lewis P, Liebermeister W, March A, Mars RAT, Nannapaneni P, Noone D, Pohl S, Rinn B, Rügheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stülke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Völker U, Bessières P, et al. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 6.Westers H, Dorenbos R, van Dijl JM, Kabel J, Flanagan T, Devine KM, Jude F, Seror SJ, Beekman AC, Darmon E, Eschevins C, de Jong A, Bron S, Kuipers OP, Albertini AM, Antelmann H, Hecker M, Zamboni N, Sauer U, Bruand C, Ehrlich DS, Alonso JC, Salas M, Quax WJ. 2003. Genome engineering reveals large dispensable regions in Bacillus subtilis. Mol Biol Evol 20:2076–2090. doi: 10.1093/molbev/msg219. [DOI] [PubMed] [Google Scholar]
- 7.Barbe V, Cruveiller S, Kunst F, Lenoble P, Meurice G, Sekowska A, Vallenet D, Wang T, Moszer I, Médigue C, Danchin A. 2009. From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology (Reading) 155:1758–1775. doi: 10.1099/mic.0.027839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Commichau FM, Pietack N, Stülke J. 2013. Essential genes in Bacillus subtilis: a re-evaluation after ten years. Mol Biosyst 9:1068–1075. doi: 10.1039/c3mb25595f. [DOI] [PubMed] [Google Scholar]
- 9.Koo BM, Kritikos G, Farelli JD, Todor H, Tong K, Kimsey H, Wapinski I, Galardini M, Cabal A, Peters JM, Hachmann AB, Rudner DZ, Allen KN, Typas A, Gross CA. 2017. Construction and analysis of two-genome-scale deletion libraries for Bacillus subtilis. Cell Syst 4:291–305.e7. doi: 10.1016/j.cels.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antelmann H, Bernhardt J, Schmid R, Mach H, Völker U, Hecker M. 1997. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis 18:1451–1463. doi: 10.1002/elps.1150180820. [DOI] [PubMed] [Google Scholar]
- 11.Tobisch S, Zühlke D, Bernhardt J, Stülke J, Hecker M. 1999. Role of CcpA in regulation of the central pathways of carbon catabolism in Bacillus subtilis. J Bacteriol 181:6996–7004. doi: 10.1128/JB.181.22.6996-7004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernhardt J, Weibezahn J, Scharf C, Hecker M. 2003. Bacillus subtilis during feast and famine: visualization of the overall regulation of protein synthesis during glucose starvation by proteome analysis. Genome Res 13:224–237. doi: 10.1101/gr.905003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reuß DR, Altenbuchner J, Mäder U, Rath H, Ischebeck T, Sappa PK, Thürmer A, Guérin C, Nicolas P, Steil L, Zhu B, Feussner I, Klumpp S, Daniel R, Commichau FM, Völker U, Stülke J. 2017. Large-scale reduction of the Bacillus subtilis genome: consequences for the transcriptional network, resource allocation, and metabolism. Genome Res 27:289–299. doi: 10.1101/gr.215293.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maass S, Sievers S, Zühlke D, Kuzinski J, Sappa PK, Muntel J, Hessling B, Bernhardt J, Sietmann R, Völker U, Hecker M, Becher D. 2011. Efficient, global-scale quantification of absolute protein amounts by integration of targeted mass spectrometry and two-dimensional gel-based proteomics. Anal Chem 83:2677–2684. doi: 10.1021/ac1031836. [DOI] [PubMed] [Google Scholar]
- 15.Muntel J, Fromion V, Goelzer A, Maaβ S, Mäder U, Büttner K, Hecker M, Becher D. 2014. Comprehensive absolute quantification of the cytosolic proteome of Bacillus subtilis by data independent, parallel fragmentation in liquid chromatography/mass spectrometry (LC/MSE). Mol Cell Proteomics 13:1008–1019. doi: 10.1074/mcp.M113.032631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Reilly FJ, Graziadei A, Forbrig C, Bremenkamp R, Charles K, Lenz S, Elfmann C, Fischer L, Stülke J, Rappsilber J. 2023. Protein complexes in cells by AI-assisted structural proteomics. Mol Syst Biol 19:e11544. doi: 10.15252/msb.202311544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchison CA, III, Chuang RY, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, Pelletier JF, Qi ZQ, Richter RA, Strychalski EA, Sun L, Suzuki Y, Tsvetanova B, Wise KS, Smith HO, Glass JI, Merryman C, Gibson DG, Venter JC. 2016. Design and synthesis of a minimal bacterial genome. Science 351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 18.Reuß DR, Commichau FM, Gundlach J, Zhu B, Stülke J. 2016. The blueprint of a minimal cell: MiniBacillus. Microbiol Mol Biol Rev 80:955–987. doi: 10.1128/MMBR.00029-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michalik S, Reder A, Richts B, Faßhauer P, Mäder U, Pedreira T, Poehlein A, van Heel AJ, van Tilburg AY, Altenbuchner J, Klewing A, Reuß DR, Daniel R, Commichau FM, Kuipers OP, Hamoen LW, Völker U, Stülke J. 2021. The Bacillus subtilis minimal genome compendium. ACS Synth Biol 10:2767–2771. doi: 10.1021/acssynbio.1c00339. [DOI] [PubMed] [Google Scholar]
- 20.Aguilar Suárez R, Stülke J, van Dijl JM. 2019. Less is more: toward a genome-reduced Bacillus cell factory for “difficult proteins.” ACS Synth Biol 8:99–108. doi: 10.1021/acssynbio.8b00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedreira T, Elfmann C, Stülke J. 2022. The current state of SubtiWiki, the database for the model organism Bacillus subtilis. Nucleic Acids Res 50:D875–D882. doi: 10.1093/nar/gkab943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrari FA, Trach KA, LeCoq D, Spence J, Ferrari E, Hoch JA. 1985. Characterization of the spo0A locus and its deduced product. Proc Natl Acad Sci USA 82:2647–2651. doi: 10.1073/pnas.82.9.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burbulys D, Trach KA, Hoch JA. 1991. The initiation of sporulation in Bacillus subtilis is controlled by a multicomponent phosphorelay. Cell 64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 24.Haldenwang WG, Lang N, Losick R. 1981. A sporulation-induced sigma-like regulatory protein from Bacillus subtilis. Cell 23:615–624. doi: 10.1016/0092-8674(81)90157-4. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt R, Margolis P, Duncan L, Coppolecchia R, Moran CP, Jr, Losick R. 1990. Control of developmental transcription factor sigma F by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis. Proc Natl Acad Sci USA 87:9221–9225. doi: 10.1073/pnas.87.23.9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Min KT, Hilditch CM, Diederich B, Errington J, Yudkin MD. 1993. σF, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-σ factor that is also a protein kinase. Cell 74:735–742. doi: 10.1016/0092-8674(93)90520-z. [DOI] [PubMed] [Google Scholar]
- 27.Anagnostopoulos C, Spizizen J. 1961. Requirements for transformation in Bacillus subtilis. J Bacteriol 81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Errington J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol 1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- 29.Ryter A. 1965. Morphologic study of the sporulation of Bacillus subtilis. Ann Inst Pasteur (Paris) 108:40–60. [PubMed] [Google Scholar]
- 30.Sterlini JM, Mandelstam J. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J 113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. 1994. Green fluorescent protein as a marker for gene expression. Science 263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 32.Webb CD, Decatur A, Teleman A, Losick R. 1995. Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. J Bacteriol 177:5906–5911. doi: 10.1128/jb.177.20.5906-5911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shapiro L, Losick R. 2000. Dynamic spatial regulation in the bacterial cell. Cell 100:89–98. doi: 10.1016/s0092-8674(00)81686-4. [DOI] [PubMed] [Google Scholar]
- 34.Angeles DM, Scheffers DJ. 2021. The cell wall of Bacillus subtilis. Curr Issues Mol Biol 41:539–596. doi: 10.21775/cimb.041.539. [DOI] [PubMed] [Google Scholar]
- 35.Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, Kahne D, Walker S, Kruse AC, Bernhardt TG, Rudner DZ. 2016. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537:634–638. doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daniel RA, Errington J. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 37.Jones LJ, Carballido-Lopez R, Errington J. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 38.Graumann PL. 2007. Cytoskeletal elements in bacteria. Annu Rev Microbiol 61:589–618. doi: 10.1146/annurev.micro.61.080706.093236. [DOI] [PubMed] [Google Scholar]
- 39.Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, Carballido-Lopez R. 2011. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 40.Defeu Soufo HJ, Graumann PL. 2004. Dynamic movement of actin-like proteins within bacterial cells. EMBO Rep 5:789–794. doi: 10.1038/sj.embor.7400209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hussain S, Wivagg CN, Szwedziak P, Wong F, Schaefer K, Izore T, Renner LD, Holmes MJ, Sun Y, Bisson-Filho AW, Walker S, Amir A, Lowe J, Garner EC. 2018. MreB filaments align along greatest principal membrane curvature to orient cell wall synthesis. Elife 7:e32471. doi: 10.7554/eLife.32471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Lutkenhaus J. 1993. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol Microbiol 9:435–442. doi: 10.1111/j.1365-2958.1993.tb01705.x. [DOI] [PubMed] [Google Scholar]
- 43.Whitley KD, Jukes C, Tregidgo N, Karinou E, Almada P, Cesbron Y, Henriques R, Dekker C, Holden S. 2021. FtsZ treadmilling is essential for Z-ring condensation and septal constriction initiation in Bacillus subtilis cell division. Nat Commun 12:2448. doi: 10.1038/s41467-021-22526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. 2009. Two-step assembly dynamics of the Bacillus subtilis divisome. J Bacteriol 191:4186–4194. doi: 10.1128/JB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams DW, Wu LJ, Errington J. 2015. Nucleoid occlusion protein Noc recruits DNA to the bacterial cell membrane. EMBO J 34:491–501. doi: 10.15252/embj.201490177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bramkamp M, van Baarle S. 2009. Division site selection in rod-shaped bacteria. Curr Opin Microbiol 12:683–688. doi: 10.1016/j.mib.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 47.Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, Errington J, Marenduzzo D, Hamoen LW. 2009. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J 28:2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glaser P, Sharpe ME, Raether B, Perego M, Ohlsen K, Errington J. 1997. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev 11:1160–1168. doi: 10.1101/gad.11.9.1160. [DOI] [PubMed] [Google Scholar]
- 49.Soh YM, Davidson IF, Zamuner S, Basquin J, Bock FP, Taschner M, Veening JW, De Los Rios P, Peters JM, Gruber S. 2019. Self-organization of parS centromeres by the ParB CTP hydrolase. Science 366:1129–1133. doi: 10.1126/science.aay3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan NL, Marquis KA, Rudner DZ. 2009. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 137:697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gruber S, Errington J. 2009. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 52.Gruber S, Veening JW, Bach J, Blettinger M, Bramkamp M, Errington J. 2014. Interlinked sister chromosomes arise in the absence of condensin during fast replication in B. subtilis. Curr Biol 24:293–298. doi: 10.1016/j.cub.2013.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gohrbandt M, Lipski A, Grimshaw JW, Buttress JA, Baig Z, Herkenhoff B, Walter S, Kurre R, Deckers-Hebestreit G, Strahl H. 2022. Low membrane fluidity triggers lipid phase separation and protein segregation in living bacteria. EMBO J 41:e109800. doi: 10.15252/embj.2021109800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bramkamp M, Lopez D. 2015. Exploring the existence of lipid rafts in bacteria. Microbiol Mol Biol Rev 79:81–100. doi: 10.1128/MMBR.00036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopez D, Kolter R. 2010. Functional microdomains in bacterial membranes. Genes Dev 24:1893–1902. doi: 10.1101/gad.1945010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zielińska A, Savietto A, de Sousa Borges A, Martinez D, Berbon M, Roelofsen JR, Hartman AM, de Boer R, Van der Klei IJ, Hirsch AK, Habenstein B, Bramkamp M, Scheffers D-J. 2020. Flotillin-mediated membrane fluidity controls peptidoglycan synthesis and MreB movement. Elife 9:e57179. doi: 10.7554/eLife.57179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sawant P, Eissenberger K, Karier L, Mascher T, Bramkamp M. 2016. A dynamin-like protein involved in bacterial cell membrane surveillance under environmental stress. Environ Microbiol 18:2705–2720. doi: 10.1111/1462-2920.13110. [DOI] [PubMed] [Google Scholar]
- 58.Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 59.Deutscher J, Saier MH. Jr, 1983. ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc Natl Acad Sci USA 80:6790–6794. doi: 10.1073/pnas.80.22.6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. 1995. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol Microbiol 15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 61.Galinier A, Kravanja M, Engelmann R, Hengstenberg W, Kilhoffer MC, Deutscher J, Haiech J. 1998. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc Natl Acad Sci USA 95:1823–1828. doi: 10.1073/pnas.95.4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reizer J, Hoischen C, Titgemeyer F, Rivolta C, Rabus R, Stülke J, Karamata D, Saier MH, Jr, Hillen W. 1998. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol Microbiol 27:1157–1169. doi: 10.1046/j.1365-2958.1998.00747.x. [DOI] [PubMed] [Google Scholar]
- 63.Maamar H, Dubnau D. 2005. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol 56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chai Y, Chu F, Kolter R, Losick R. 2008. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol 67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Veening JW, Smits WK, Kuipers P. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- 66.Aguilar C, Vlamakis H, Losick R, Kolter R. 2007. Thinking about Bacillus subtilis as a multicellular organism. Curr Opin Microbiol 10:638–643. doi: 10.1016/j.mib.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hobley L, Ostrowski A, Rao FV, Bromley KM, Porter M, Prescott AR, MacPhee CE, van Aalten DM, Stanley-Wall NR. 2013. BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc Natl Acad Sci USA 110:13600–13605. doi: 10.1073/pnas.1306390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stülke J. 2002. Control of transcription termination in bacteria by RNA-binding proteins that modulate RNA structures. Arch Microbiol 177:433–440. doi: 10.1007/s00203-002-0407-5. [DOI] [PubMed] [Google Scholar]
- 70.Babitzke P. 2004. Regulation of transcription attenuation and translation initiation by allosteric control of an RNA-binding protein: the Bacillus subtilis TRAP protein. Curr Opin Microbiol 7:132–139. doi: 10.1016/j.mib.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 71.Henkin TM. 1994. tRNA-directed transcription antitermination. Mol Microbiol 13:381–387. doi: 10.1111/j.1365-2958.1994.tb00432.x. [DOI] [PubMed] [Google Scholar]
- 72.Gutiérrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. 2009. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol Mol Biol Rev 73:36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grundy FJ, Henkin TM. 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in Gram-positive bacteria. Mol Microbiol 30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 74.McDaniel BAM, Grundy FJ, Artsimovitch I, Henkin TM. 2003. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci USA 100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Winkler W, Nahvi A, Breaker RR. 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 76.Winkler WC, Cohen-Chalamish S, Breaker RR. 2002. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA 99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roth A, Breaker RR. 2009. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem 78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nelson JW, Breaker RR. 2017. The lost language of the RNA world. Sci Signal 10:eaam8812. doi: 10.1126/scisignal.aam8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chandrangsu P, Rensing C, Helmann JD. 2017. Metal homeostasis and resistance in bacteria. Nat Rev Microbiol 15:338–350. doi: 10.1038/nrmicro.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Belitsky BR, Sonenshein AL. 1998. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J Bacteriol 180:6298–6305. doi: 10.1128/JB.180.23.6298-6305.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sonenshein AL. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol 5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- 82.Jayaraman V, Lee JD, Elad N, Vimer S, Sharon M, Fraser JS, Tawfik DS. 2022. A counter-enzyme complex regulates glutamate metabolism in Bacillus subtilis. Nat Chem Biol 18:161–170. doi: 10.1038/s41589-021-00919-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Prindle A, Liu J, Asally M, Ly S, Garcia-Ojalvo J, Süel GM. 2015. Ion channels enable electrical communication in bacterial communities. Nature 527:59–63. doi: 10.1038/nature15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kikuchi K, Galera-Laporta L, Weatherwax C, Lam JY, Moon EC, Theodorakis EA, Garcia-Ojalvo J, Süel GM. 2022. Electrochemical potential enables dormant spores to integrate environmental signals. Science 378:43–49. doi: 10.1126/science.abl7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wicke D, Schulz LM, Lentes S, Scholz P, Poehlein A, Gibhardt J, Daniel R, Ischebeck T, Commichau FM. 2019. Identification of the first glyphosate transporter by genomic adaptation. Environ Microbiol 21:1287–1305. doi: 10.1111/1462-2920.14534. [DOI] [PubMed] [Google Scholar]
- 86.Eelderink-Chen Z, Bosman J, Sartor F, Dodd AN, Kovács AT, Merrow M. 2021. A circadian clock in a nonphotosynthetic prokaryote. Sci Adv 7:eabe2086. doi: 10.1126/sciadv.abe2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wicke D, Meißner J, Warneke R, Elfmann C, Stülke K. 2023. Understudied proteins and understudied functions in the model bacterium Bacillus subtilis—a major challenge in current research. Mol Microbiol doi: 10.1111/mmi.15053. [DOI] [PubMed] [Google Scholar]
- 88.Hohmann HP, van Dijl JM, Krishnappa L, Prágai Z. 2017. Host organisms: Bacillus subtilis, p. 221–297. In Wittmann C, Liao JC (ed), Industrial biotechnology. Wiley, New York, NY. [Google Scholar]
- 89.Altenbuchner J. 2016. Editing of the Bacillus subtilis genome by the CRISPR-Cas9 system. Appl Environ Microbiol 82:5421–5427. doi: 10.1128/AEM.01453-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sachla AJ, Alfonso AJ, Helmann JD. 2021. A simplified method for CRISPR-Cas9 engineering of Bacillus subtilis. Microbiol Spectr 9:e00754-21. doi: 10.1128/Spectrum.00754-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wickerham L, Flickinger M, Johnston RM. 1946. The production of riboflavin by Ashbya gossypii. Arch Biochem 9:95–98. [PubMed] [Google Scholar]
- 92.Schwechheimer SK, Park EY, Revuelta JL, Becker J, Wittmann C. 2016. Biotechnology of riboflavin. Appl Microbiol Biotechnol 100:2107–2119. doi: 10.1007/s00253-015-7256-z. [DOI] [PubMed] [Google Scholar]
- 93.Kaspar F, Neubauer P, Gimpel M. 2019. Bioactive secondary metabolites from Bacillus subtilis: A comprehensive review. J Nat Prod 82:2038–2053. doi: 10.1021/acs.jnatprod.9b00110. [DOI] [PubMed] [Google Scholar]
- 94.Li D, Hou L, Gao Y, Tian Z, Fan B, Wang LF, Li S. 2022. Recent advances in microbial synthesis of poly-γ-glutamic acid: A review. Foods 11:739. doi: 10.3390/foods11050739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Danilova I, Sharipova M. 2020. The practical potential of Bacilli and their enzymes for industrial production. Front Microbiol 11:1782. doi: 10.3389/fmicb.2020.01782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tjalsma H, Antelmann H, Jongbloed JD, Braun PG, Darmon E, Dorenbos R, Dubois JY, Westers H, Zanen G, Quax WJ, Kuipers OP, Bron S, Hecker M, van Dijl JM. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol Mol Biol Rev 68:207–233. doi: 10.1128/MMBR.68.2.207-233.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.van Dijl JM, Hecker M. 2013. Bacillus subtilis: from soil bacterium to super-secreting cell factory. Microb Cell Fact 12:3. doi: 10.1186/1475-2859-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jeong H, Jeong DE, Park SH, Kim SJ, Choi SK. 2018. Complete genome sequence of Bacillus subtilis strain WB800N, an extracellular protease-deficient derivative of strain 168. Microbiol Resour Announc 7:e01380-18. doi: 10.1128/MRA.01380-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen O, Heyndrickx M, Meynier A, Ouwehand A, Pot B, Stahl B, Theis S, Vaughan E, Miani M. 2023. Dietary probiotics, prebiotics and the gut microbiota in human health. Zenodo doi: 10.5281/zenodo.7590106. [DOI] [Google Scholar]
- 100.Fathima S, Hakeem WGA, Shanmugasundaram R, Selvaraj RK. 2022. Necrotic enteritis in broiler chickens: a review on the pathogen, pathogenesis, and prevention. Microorganisms 10:1958. doi: 10.3390/microorganisms10101958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pelzer S, Petri D, Giatsis C, Molck S, Kipker M, Kleinboelting J, Stannek-Göbel L, Doranalli K, Htoo JKK, Borgmeier C, Herbold S, Meurer G. 2017. Bacillus subtilis strain with probiotic activity. PCT Patent WO2017207372A1.
- 102.Whelan RA, Doranalli K, Rinttilä T, Vienola K, Jurgens G, Apajalahti J. 2019. The impact of Bacillus subtilis DSM 32315 on the pathology, performance, and intestinal microbiome of broiler chickens in a necrotic enteritis challenge. Poult Sci 98:3450–3463. doi: 10.3382/ps/pey500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.BCC Research LLC. 2022. Probiotics in food, beverages, dietary supplements and animal feed. Report Code: FOD035H.
- 104.Tom Dieck H, Schön C, Wagner T, Pankoke HC, Fluegel M, Speckmann B. 2021. A synbiotic formulation comprising Bacillus subtilis DSM 32315 and L-alanyl-L-glutamine improves intestinal butyrate levels and lipid metabolism in healthy humans. Nutrients 14:143. doi: 10.3390/nu14010143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Castro-Alonso MJ, Montañez-Hernandez LE, Sanchez-Muñoz MA, Macias Franco MR, Narayanasamy R, Balagurusamy N. 2019. Microbially induced calcium carbonate precipitation (MICP) and its potential in bioconcrete: microbiological and molecular concepts. Front Mater 6:126. doi: 10.3389/fmats.2019.00126. [DOI] [Google Scholar]
- 106.Müller T, Hintermayer S, Hellriegel J, Martens C, Haas I. 2021. Composition comprising at least one microorganism and use thereof. US Patent Application Publication US 2021/0163357 A1. [Google Scholar]
- 107.Cogliati S, Clementi V, Francisco M, Crespo C, Arganaraz F, Grau R. 2020. Bacillus subtilis delays neurodegeneration and behavioral impairment in the Alzheimer's disease model Caenorhabditis elegans. J Alzheimers Dis 73:1035–1052. doi: 10.3233/JAD-190837. [DOI] [PubMed] [Google Scholar]
- 108.Diethmaier C, Pietack N, Gunka K, Wrede C, Lehnik-Habrink M, Herzberg C, Hübner S, Stülke J. 2011. A novel factor controlling bistability in Bacillus subtilis: the YmdB protein affects flagellin expression and biofilm formation. J Bacteriol 193:5997–6007. doi: 10.1128/JB.05360-11. [DOI] [PMC free article] [PubMed] [Google Scholar]