ABSTRACT
The obligate intracellular human pathogen Chlamydia trachomatis (Ctr) undergoes a complex developmental cycle in which the bacterium differentiates between two functionally and morphologically distinct forms: the elementary body (EB) and the reticulate body (RB). The EB is the smaller, infectious, nondividing form which initiates infection of a susceptible host cell, whereas the RB is the larger, non-infectious form which replicates within a membrane-bound vesicle called an inclusion. The mechanism(s) which drives differentiation between these developmental forms is poorly understood. Bulk protein turnover is likely required for chlamydial differentiation given the significant differences in the protein repertoires and functions of the EB and RB. We hypothesize that periplasmic protein turnover is also critical for the reorganization of an RB into an EB, referred to as secondary differentiation. Ct441 is a periplasmic protease ortholog of tail-specific proteases (i.e., Tsp, Prc) and is expressed in Ctr during secondary differentiation. We investigated the effect of altering Tsp expression on developmental cycle progression. Through assessment of bacterial morphology and infectious progeny production, we found that both overexpression and CRISPR interference/dCas9 (CRISPRi)-mediated knockdown of Tsp negatively impacted chlamydial development through different mechanisms. We also confirmed that catalytic activity is required for the negative effect of overexpression and confirmed the effect of the mutation in in vitro assays. Electron microscopic assessments during knockdown experiments revealed a defect in EB morphology, directly linking Tsp function to secondary differentiation. These data implicate Ct441/Tsp as a critical factor in secondary differentiation.
IMPORTANCE The human pathogen Chlamydia trachomatis is the leading cause of preventable infectious blindness and bacterial sexually transmitted infections worldwide. This pathogen has a unique developmental cycle that alternates between distinct forms. However, the key processes of chlamydial development remain obscure. Uncovering the mechanisms of differentiation between its metabolically and functionally distinct developmental forms may foster the discovery of novel Chlamydia-specific therapeutics and limit development of resistant bacterial populations derived from the clinical use of broad-spectrum antibiotics. In this study, we investigate chlamydial tail-specific protease (Tsp) and its function in chlamydial growth and development. Our work implicates Tsp as essential to chlamydial developmental cycle progression and indicates that Tsp is a potential drug target for Chlamydia infections.
KEYWORDS: Chlamydia, development, differentiation, Tsp, Prc, protease, periplasm
INTRODUCTION
How a microorganism differentiates from one form to another is an intriguing question. For example, Caulobacter crescentus differentiates from swarmer to stalk cells, attack-phase Bdellovibrio cells differentiate into growth-phase cells, and Myxococcus xanthus fruiting body cells can differentiate into myxospores or peripheral rod cells (1–3). Invariably, such reorganization requires the careful control of gene expression at every level, from transcription to post-translational processes. This is also true for Chlamydia, an obligate intracellular, Gram-negative bacterium that undergoes a unique developmental cycle. During its developmental cycle, the bacterium differentiates between two functionally and morphologically distinct forms (4). The elementary body, or EB, is the smaller, infectious, non-dividing form which initiates the developmental cycle by binding to the host cell. Once bound, it induces its uptake into the host cell, where it recruits and incorporates several host cell components to form a membrane-bound vesicle called an inclusion (5–8). The EB then undergoes primary differentiation into a reticulate body, or RB, which is the larger, non-infectious, replicative form. The RB then divides via an MreB-dependent polarized budding mechanism (9–13) to increase bacterial numbers. After multiple rounds of replication, nascent RBs undergo secondary differentiation into EBs. The RBs continue to divide and differentiate asynchronously until the bacteria egress from the cell either through cell lysis or inclusion extrusion (14). Despite the known differences between EBs and RBs, the specific factors which contribute to differentiation between these forms remain unclear.
Notably, chlamydial gene expression is temporally regulated throughout development, with expression patterns falling into three broad categories: early-, mid-, and late-cycle genes (15, 16). Early-cycle genes peak shortly after infection at approximately 8 to 12 h postinfection (hpi) in Chlamydia trachomatis (Ctr) serovar L2 and are likely involved in inclusion establishment, circumvention of host innate immunity, and primary differentiation (EB to RB). Mid-cycle genes, which represent the bulk of chlamydial genes, peak in transcription around 16 hpi and likely function in “housekeeping” duties to ensure replication, division, and other growth-related tasks. Late-cycle genes peak at approximately 24 hpi and are involved in EB formation (secondary differentiation) and priming of EBs for infection and host cell exit. Coincident with these transcriptional changes is a remodeling of the bacterial proteome as developmental-specific proteins are translated. We hypothesize that protein degradation and turnover is a critical aspect of this remodeling to eliminate proteins which specify the progenitor form. For example, the EB expresses proteins that condense its nucleic acids and cross-link its outer membrane (4, 17–22). Presumably, these proteins must be eliminated or altered in some way to prevent these functions as the EB differentiates into an RB. Similarly, proteins which maintain or allow an RB state, such as cell division proteins, must be eliminated or “deactivated” to allow for EB formation.
Despite extensive morphological characterization of the chlamydial developmental cycle, the molecular mechanisms which drive differentiation in Chlamydia are poorly understood. Unlike in other bacteria, such as Caulobacter crescentus and Bacillus subtilis, which differentiate following an asymmetric division event (23, 24), there is no evidence to suggest that chlamydial differentiation is triggered by an asymmetric division event. Electron micrographic assessments from various researchers over the decades have failed to demonstrate an EB forming while attached to an RB progenitor (9, 25–28). Rather, a single RB begins a transition into an intermediate body (IB) with a condensing nucleoid, which then further condenses into a compact EB. Given the clear differences in protein repertoires between the developmental forms (29–31), we hypothesize that proteomic turnover between the developmental forms halts RB division, which is a critical first step in the reorganization of an RB into an EB during secondary differentiation. Previously, our labs established a model in which protein turnover in the Ctr cytoplasm by the ClpXP protease plays a crucial role in secondary differentiation (32). However, as a Gram-negative bacterium, Chlamydia has four compartments requiring remodeling: the cytoplasm, inner membrane, periplasm, and outer membrane. In this study, we investigated the function of a periplasmic protease in the growth and differentiation of Chlamydia.
Chlamydia encodes two putative periplasmic proteases: Ct823 (HtrA) and Ct441 (Tsp). HtrA is expressed as a mid-cycle gene in Chlamydia and is likely the major housekeeping periplasmic protease in the organism (33, 34). Ct441 is orthologous to Gram-negative tail-specific proteases (i.e., Tsp, Prc) (35–39). Typically, in Gram-negative bacteria, Tsp helps maintain peptidoglycan (PG) homeostasis in the periplasm by degrading or modifying PG-associated proteins (40–42). However, pathogenic Chlamydia does not have PG in its cell wall, and ct441/tsp has been characterized as a late-cycle gene (15, 43, 44). Therefore, we hypothesize that chlamydial Tsp has a specialized function in the transition from RB to EB. More specifically, we speculate that Tsp degrades or inactivates RB-specific periplasmic proteins during secondary differentiation, which is critical for the reorganization of an RB into an EB. To test this hypothesis, we created a series of transformant strains to evaluate the effects of overexpression or knockdown of ct441/tsp on chlamydial growth and differentiation. Overexpression of wild-type Ct441/Tsp, but not a catalytically inactive mutant of Ct441/Tsp, was highly detrimental to Ctr L2, blocking both replication and generation of infectious EBs. In contrast, knocking down Tsp expression using CRISPR interference had a limited impact on replication and inclusion size and development. However, secondary progeny were reduced even though EB-specific proteins were detected, suggesting that the EBs were non-infectious. Electron microscopy analysis indicated morphological changes in the EBs consistent with this. Overall, our data suggest that chlamydial Ct441/Tsp is a critical mediator of secondary differentiation and is essential for generation of viable EBs.
RESULTS
In vitro analysis of Ct441/Tsp proteolytic activity.
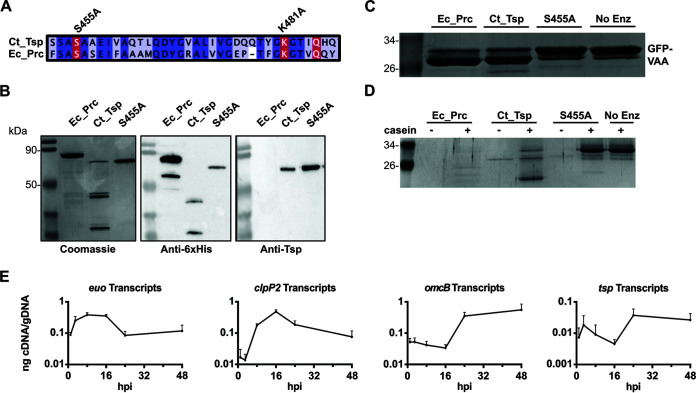
Ct441/Tsp contains a signal sequence for secretion into the periplasm, an N-terminal PDZ domain for substrate recognition (45), the conserved catalytic residues (S455 and K481) of a Tsp ortholog, and a domain of unknown function (DUF3340) at the C terminus. Ct441/Tsp is conserved across chlamydial species (Fig. S1 and 2 in the supplemental material), and key domains and active site residues are conserved with other Tsp/Prc orthologs from other Gram-negative bacteria (Fig. S3). As part of our initial studies, we wanted to validate the proteolytic activity of recombinant Ct441/Tsp (Ct_Tsp). We purified Ct_Tsp and a proteolytically inactive isoform where we mutated the essential catalytic residue, S455A (35) (Fig. 1A). Both proteins lacked the predicted secretion signal from the N terminus. In parallel, we purified a secretion-competent E. coli Prc (Ec_Prc), an ortholog of chlamydial Tsp. The 6×His affinity purification tag was cloned onto the N-terminal end for Ct_Tsp to mimic previous work characterizing Ct_Tsp function and structure (35). Ec_Prc was expressed with a C-terminal 6×His affinity tag to allow normal secretion to the periplasm prior to purification and to reflect previous Prc activity experiments (46). Recombinant proteins were purified using immobilized metal affinity chromatography. We observed that Ec_Prc, Ct_Tsp, and the S455A mutant showed banding patterns around 75 kDa, consistent with the expected molecular weight of Ec_Prc (signal peptide processed) and Ct_Tsp isoforms lacking their signal peptide (Fig. 1B). Degradation products of smaller sizes were detected below the expected bands for wild-type Ec_Prc and Ct_Tsp in both Coomassie gels and Western blots using an anti-6×His antibody (Fig. 1B), signifying potential autoproteolysis for the active proteases. Consistent with this, degradation products were not observed for the S455A mutant protein (Fig. 1B). We obtained an anti-peptide antibody directed against a region near the C terminus of Ct_Tsp (residues 611 to 626; KKTNKQDRSFGSNDLQ). Interestingly, only the full-length Ct_Tsp was detected in the blot probed with the anti-Ct_Tsp antibody (Fig. 1B). These data further suggest that wild-type Ct_Tsp undergoes C-terminal autoprocessing, at least when expressed in E. coli.
FIG 1.
Purification and activity analyses of Ct_Tsp. (A) Multiple sequence alignment of the active site in Ct_Tsp and Ec_Prc denoting the conserved catalytic residues S455A (mutated in this study), K481A (mutated in this study), and Q485. (B) 6×His-tagged recombinant proteins were assessed for purity and stability on SDS-PAGE stained with Coomassie brilliant blue for total protein (top left) or probed via Western blotting for the 6×His-tag or a C-terminal region of tail-specific protease (Tsp). (C and D) To assess enzyme activity, 1 μg of enzyme and 3 μg of (C) chlamydial SsrA-tagged green fluorescent protein (GFP) (abbreviated as VAA) or (D) casein were incubated at 37°C for 3 h and the products were resolved on SDS-PAGE and Coomassie-stained. Images represent results from replicate experiments with two independently purified protein preparations. Expected sizes: Ec_Prc, 75.3 kDa after signal processing; and Ct_Tsp, 74.6 kDa. (E) Transcripts of euo, clpP2, omcB, and tsp throughout the Ctr L2 48-h developmental cycle.
Next, we tested the proteolytic activity of Ct_Tsp using either the full chlamydial SsrA tag appended to green fluorescent protein (GFP) (abbreviated as GFP_VAA) (Fig. 1C) or casein (Fig. 1D). Incubation of GFP_VAA with wild-type Ct_Tsp or Ec_Prc resulted in processing of the GFP_VAA into a smaller product (Fig. 1C), indicating their ability to recognize the SsrA tag (47). Importantly, the S455A mutant showed no processing of GFP-VAA (Fig. 1C), confirming that the S455A mutation effectively blocks Ct_Tsp activity. Using casein as a relatively unstructured substrate, we observed degradation of casein in the presence of Ec_Prc and Ct_Tsp, but not the S455A mutant (Fig. 1D), thus confirming again that the Ct_Tsp S455A mutation inhibits protease activity. Overall, these data confirm the expected in vitro protease activity of these proteins and confirm that an S455A mutation renders Ct_Tsp inactive.
ct441 is transcribed as a late-developmental cycle gene in Chlamydia.
Given that Prc orthologs are constitutively expressed in other Gram-negative bacteria, we sought to determine the transcriptional profile of ct441/tsp over the course of the chlamydial developmental cycle. Prior reports have suggested that ct441/tsp is a late-cycle gene (15). We initiated reverse transcription-quantitative PCR (RT-qPCR) studies using wild-type Ctr L2 and measured the transcript levels of known early- (e.g., euo [48]), mid- (clpP2 [49]), and late-cycle (omcB [26]) genes as well as ct441/tsp (Fig. 1E). As expected, euo transcripts peaked around 8 hpi, whereas clpP2 transcripts peaked at 16 hpi. We measured a peak in ct441/tsp transcript levels at 24 hpi. These data closely mimicked the transcriptional profile of omcB, confirming that ct441 is transcribed as a late-developmental cycle gene in Chlamydia. These data suggest that Ct_Tsp is not involved in PG homeostasis, as PG synthesis and cell division-associated genes are mid-cycle genes that function during RB growth and division (50, 51). These transcriptional data suggest that Ct_Tsp functions in the process of secondary differentiation.
Overexpression of wild-type, but not catalytically inactive, Ct_Tsp_6×His negatively impacts chlamydial growth.
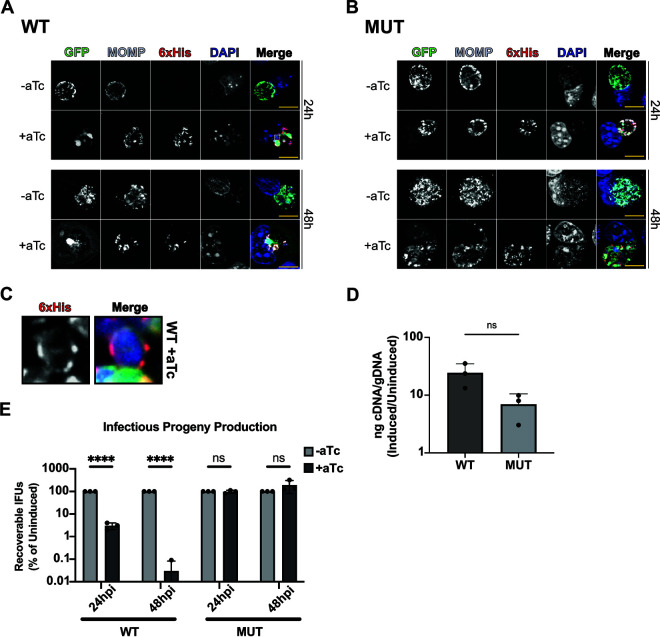
To determine whether altering Ct_Tsp levels affects developmental cycle progression, we first prepared overexpression constructs for 6×His-tagged wild-type (WT) or catalytically inactive Ct_Tsp (S455A/K481A; MUT) (Fig. 1A). Tsp was 6×His-tagged at the C terminus to allow for normal protein localization in Chlamydia. We prepared the MUT with two active site mutations to avoid the possibility that the single S455A mutant might retain some residual proteolytic activity in vivo, not detectable in vitro, which would complicate interpretation of any findings. We transformed these plasmids into Ctr L2, infected cells with the transformants, induced or did not induce overexpression at 16 hpi, and fixed samples at 24 and 48 hpi. Using indirect immunofluorescence microscopy assays (IFA), we observed a negative impact on chlamydial morphology, with a notable decrease in chlamydial numbers within the inclusion following WT, but not MUT, Ct_Tsp_6×His overexpression at both time points assessed (Fig. 2A and B). Higher magnification images revealed that WT Ct_Tsp_6×His localized to the periphery of the bacteria and overlapped the major outer membrane protein (MOMP) label (Fig. 2C). Based on our in vitro data, this localization may represent the full-length, potentially unprocessed Ct_Tsp_6×His. Nonetheless, these data are consistent with a periplasmic localization for Ct_Tsp because light microscopy cannot resolve spatial differences between a bacterial periplasm and the inner and outer membranes.
FIG 2.
Overexpression of Ct_Tsp_6×His negatively impacts Chlamydia. (A and B) Immunofluorescence assay (IFA) of wild-type (WT) Ct_Tsp_6×His overexpression at 24 and 48 hpi (A) and mutant (MUT) Ct_Tsp_6×His (S455A/K481A) overexpression at 24 and 48 hpi (B). Samples were induced at 16 hpi with 20 nM anhydrotetracycline (aTc), aldehyde-fixed at the indicated time, and stained for major outer membrane protein (MOMP; gray), 6×His (red), and DAPI (4′,6-diamidino-2-phenylindole; blue) to label DNA. GFP expressed constitutively from the overexpression plasmid is shown in green. Scale bars = 10 μm. Images captured using a Zeiss AxioImager Z.2 with Apotome2 at ×100 magnification. (C) Representative localization of induced Ct_Tsp_6×His in an individual organism. White box in panel A represents chosen bacteria for the zoomed image in panel C. (D) Transcriptional analysis of tsp using reverse transcription-quantitative PCR (RT-qPCR) following induction of WT or MUT overexpression at 16 hpi. RNA and genomic DNA (gDNA) were harvested at 24 hpi. Data are presented as a ratio of cDNA to gDNA for the induced relative to the uninduced. ns, not significant by paired Student’s t test. (E) Inclusion-forming unit (IFU) assay following overexpression of WT or MUT induced at 16 hpi and harvested at 24 or 48 hpi. IFUs were normalized to uninduced samples and plotted on a log scale. ****, P < 0.000001; ns, not significant by multiple unpaired Student’s t test. Data represent three biological replicates.
One explanation for the effects of WT Ct_Tsp_6×His overexpression may be that it is expressed at higher levels than the mutant, even though both constructs are expressed from the same plasmid backbone. We were unable to detect either 6×His-tagged or untagged Ct_Tsp by Western blotting using anti-6×His or anti-Tsp antibodies, respectively, and we speculate that either the concentration of protein within the periplasm is limiting or the C terminus is also processed in vivo in Chlamydia as observed when the protein was expressed in E. coli (Fig. 1). To address potential differences in expression of the constructs, we instead quantified ct441/tsp transcripts in the two strains. We measured no statistically significant differences in ct441/tsp transcript levels after inducing overexpression of each construct (Fig. 2D). Therefore, we conclude that differences in WT and MUT Ct_Tsp_6×His expression following induction cannot account for the observed negative phenotypes of WT overexpression.
We next quantified the impact of overexpression on chlamydial development by measuring recoverable inclusion-forming units (IFU), a proxy for infectious EBs produced during infection of a primary cell monolayer. Cells were infected with each transformant, and expression of WT or MUT Ct_Tsp_6×His was induced or not at 16 hpi followed by collection of IFU samples at 24 and 48 hpi. IFUs were quantified after infecting fresh monolayers with serial dilutions of these samples. Here, we observed a stark decrease in infectious progeny at both 24 and 48 hpi after WT Ct_Tsp_6×His overexpression (Fig. 2E), which is consistent with the smaller inclusions and altered morphology observed (Fig. 2A). In contrast, overexpression of the MUT Ct_Tsp_6×His had no measurable effect on IFU production. We conclude from these data that overexpression of WT Ct_Tsp_6×His is detrimental to Ctr L2.
Overexpression of wild-type Ct_Tsp_6×His blocks developmental cycle progression in Chlamydia.
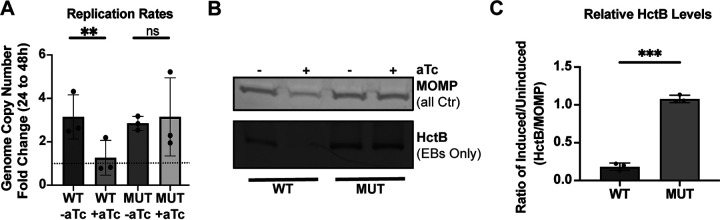
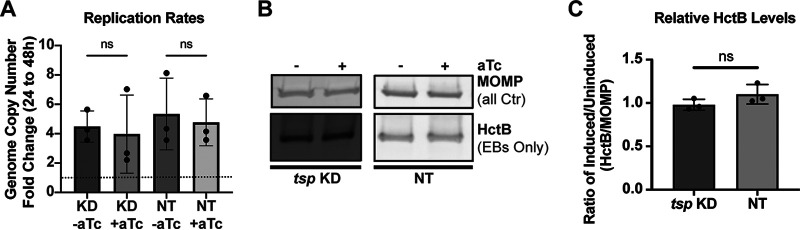
The reduction in IFUs measured after WT Ct_Tsp_6×His overexpression could be due to either a decrease in viable EBs while maintaining overall bacterial numbers (i.e., RBs and EBs) or a block in the developmental cycle (i.e., reduced RBs). To further characterize the impact on development following overexpression of Ct_Tsp_6×His, we measured genomic DNA (gDNA) levels at 24 and 48 hpi to determine effects related to genome replication as a proxy for overall bacterial numbers. We detected no increase in gDNA levels from 24 to 48 hpi after WT Ct_Tsp_6×His overexpression, indicating a block in chlamydial replication (Fig. 3A). We next probed for HctB, an EB-specific marker (22, 52, 53), by Western blotting as a proxy for progression of secondary differentiation. Here, we quantified a reduction in the HctB to MOMP ratio following induction of WT Ct_Tsp_6×His, demonstrating that secondary differentiation was blocked (Fig. 3B and C), consistent with our IFU data (Fig. 2E). In contrast, overexpression of the inactive mutant isoform had minimal effects on development with no decrease in gDNA levels and detection of HctB by Western blot. These data indicate that the increase in catalytic activity of Ct_Tsp rather than the increase in Ct_Tsp protein levels leads to the negative impact on development observed during overexpression of the WT isoform. Given the severe impact on bacterial morphology and infectious progeny production and the lack of RB replication, we conclude that overactivity of wild-type Ct_Tsp prevents developmental cycle progression by blocking RB replication.
FIG 3.
Overexpression of Ct_Tsp_6×His impairs replication of Chlamydia. (A) Quantification of genomic DNA (gDNA) determined by qPCR following overexpression of WT or MUT (S455A/K481A) Ct_Tsp. Samples were induced at 16 hpi with 20 nM aTc, and gDNA was harvested at 24 or 48 hpi. Samples were normalized to the genome copy number at 24 hpi and presented as fold change from 24 to 48 hpi. Dotted line represents the normalization of the 24-hpi sample to 1. **, P < 0.01; ns, not significant by paired Student’s t test. Individual points represent biological replicates. (B) Western blots of HctB (EBs only) and MOMP (all Ctr) levels from uninduced and induced cultures of WT or MUT overexpression strains. Samples were induced at 16 hpi with 20 nM aTc, and protein was harvested at 48 hpi. Blot is representative of three biological replicates. (C) HctB levels quantified from Western blots shown in panel B. Levels are displayed as the HctB to MOMP ratio of induced samples relative to uninduced samples. Individual points represent biological replicates. ***, P < 0.0001 by paired Student’s t test.
Knockdown of Ct_Tsp by CRISPR interference negatively impacts IFUs.
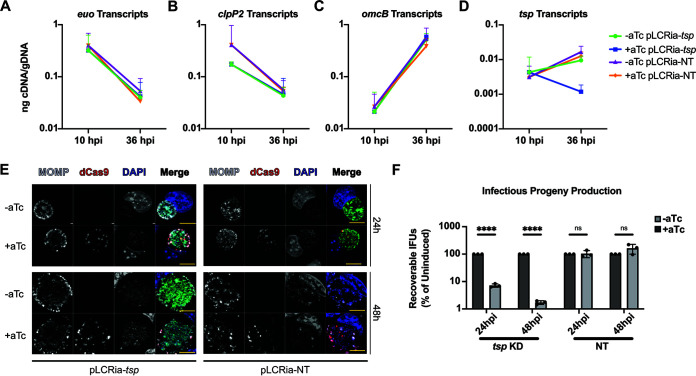
Given the effects on Ctr L2 when overexpressing Ct_Tsp, we next utilized an inducible CRISPR interference/dCas9 (CRISPRi) system to determine the effects of disrupting Ct_Tsp function in Chlamydia (54, 55). The chromosomal context of tsp indicates that it is encoded as a monocistronic unit (56, 57). We generated CRISPRi vectors that targeted either the intergenic region 5′ to ct441/tsp or no chlamydial sequence (i.e., a non-targeting [NT]) as a control. These plasmids were transformed into Ctr L2, and the transformant strains were used to infect cells. To validate the CRISPRi strains, we induced or did not induce knockdown at 4 hpi and measured transcripts at 10 and 36 hpi for both the ct441/tsp-targeting and non-targeting strains. Transcript levels for control developmental cycle genes were unaffected in either strain (Fig. 4A to C), including the late gene omcB, suggesting that overall developmental cycle progression is unaffected by ct441/tsp knockdown. Importantly, we observed a 10-fold decrease in ct441/tsp transcripts upon induction of dCas9 expression in the targeting strain, but not in the non-targeting control (Fig. 4D).
FIG 4.
Knockdown of ct441/tsp reduces infectious progeny but does not affect inclusion size. (A to D) Assessment of temporal gene regulation and confirmation of ct441/tsp knockdown by RT-qPCR. euo (A), clpP2 (B), omcB (C), and tsp (D) transcripts measured during ct441/tsp knockdown (KD) or nontargeting (NT) conditions. Ratio of cDNA normalized to gDNA is plotted on a log scale. Samples were induced at 4 hpi, and RNA or gDNA samples were taken at 10 and 36 hpi for the ct441/tsp KD and NT strains. (E) IFA of ct441/tsp KD and NT at 24 and 48 hpi. Samples were induced at 4 hpi with 20 nM aTc and stained for MOMP (gray), dCas9 (red), and DAPI (blue) to label DNA. Scale bars = 10 μm. A Zeiss AxioImager Z.2 with Apotome2 was used to take images at ×100 magnification. (F) IFU assay following ct441/tsp KD or NT induced at 4 hpi and harvested at 24 or 48 hpi. IFUs were normalized to uninduced samples and plotted on a log scale. ****, P < 0.000001; ns, not significant by multiple unpaired Student’s t test. Results are representative of three biological replicates.
After confirming knockdown, we utilized an immunofluorescence approach to visualize inclusion and bacterial morphology after ct441/tsp knockdown, which revealed no obvious impacts on these parameters (Fig. 4E). However, IFU assays revealed a substantial >1-log reduction in chlamydial growth at both 24 and 48 hpi following ct441/tsp knockdown (Fig. 4F). Importantly, the non-targeting control strain also showed no morphological abnormalities after inducing dCas9 expression and, as expected, did not affect IFU yields. These data indicate a specific negative impact of reduced ct441/tsp expression on IFU recovery without a concomitant disruption in late gene expression as represented by omcB expression (Fig. 4C).
Knockdown of Ct_Tsp by CRISPR interference negatively impacts EB viability.
Overexpression of Ct_Tsp showed congruent results between small inclusion sizes and reduced IFU yields, and this was caused by a block in developmental cycle progression. However, knocking down ct441/tsp levels resulted in the unexpected findings of normal inclusion sizes, but reduced IFU yields. To tease apart the apparent discrepancy between the IFU and inclusion size data, we again assayed replication (via gDNA levels) and EB production (via HctB production). Consistent with the normal inclusion sizes, we measured no change in gDNA levels following knockdown of ct441/tsp, which were comparable to gDNA levels in the non-targeting control (Fig. 5A). Surprisingly, HctB levels (EBs) relative to MOMP were similarly comparable between the uninduced and induced knockdown samples (Fig. 5B and C), even though ct441/tsp knockdown results in reduced IFU yields. The HctB blotting data together with the omcB transcript data suggest that secondary differentiation proceeds normally during ct441/tsp knockdown, but that the EBs produced are either non-infectious or non-viable. Taking these findings together, we conclude that Ct_Tsp likely functions in the production of viable, infectious EBs.
FIG 5.
Knockdown of ct441/tsp does not alter replication or production of an elementary body (EB)-specific marker. (A) Quantification of gDNA determined by qPCR following ct441/tsp KD or NT control conditions. Samples were induced at 4 hpi with 20 nM aTc, and gDNA was harvested at 24 or 48 hpi. Samples were normalized to the genome copy number at 24 hpi and presented as fold change from 24 to 48 hpi. Dotted line represents the normalization of the 24-hpi sample to 1. ns, not significant by paired Student’s t test. (B) Western blot of HctB (EBs only) and MOMP (all Ctr) levels from uninduced and induced cultures of KD or NT knockdown strains. Samples were induced at 4 hpi with 20 nM aTc, and protein was harvested at 48 hpi. Blot is representative of three biological replicates. (C) HctB levels quantified from Western blots shown in panel B. Levels are displayed as the HctB to MOMP ratio of induced samples relative to uninduced samples. Individual points represent biological replicates. ns, not significant by paired Student’s t test.
Knocking down ct441/tsp levels alters EB morphology.
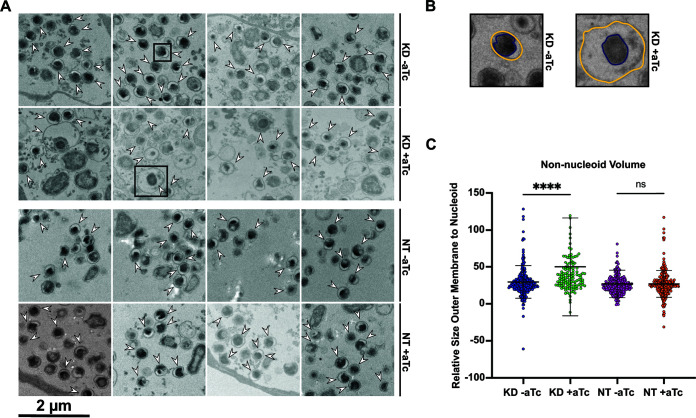
Given the apparent discrepancy between the immunofluorescence and IFU assays following knockdown of ct441/tsp, we used transmission electron microscopy (TEM) to observe EB morphology. We reasoned that the lack of infectivity may manifest in changes to EB morphology that fall below the spatial constraints of fluorescence microscopy. A defining characteristic of an RB differentiating into an EB is a condensed nucleoid due in part to the expression of the histone-like proteins HctA and HctB (17, 52, 58, 59). This condensed nucleoid is easily visualized by electron microscopy as an electron-dense black spot. We noted obvious atypical differentiating organisms in the ct441/tsp knockdown strain (Fig. 6A). During knockdown of ct441/tsp, we observed developmental forms that appeared to have a condensing or condensed nucleoid with a large, voided space between the outer membrane and nucleoid (Fig. 6A). These abnormal developmental forms were not observed in the uninduced or non-targeting controls, suggesting that these morphological impacts were not an artifact of EM processing. To quantify our observations, we measured the volume of the condensing or condensed nucleoid (purple line) and the volume of the outer membrane (yellow line) in all samples in a blinded fashion (Fig. 6B). Our analysis revealed a significant increase in the non-nucleoid volume during ct441/tsp knockdown compared to that in the uninduced and non-targeting controls (Fig. 6C). These data suggest that reducing ct441/tsp levels disrupts the morphology of differentiating developmental forms.
FIG 6.
Knockdown of ct441/tsp alters EB morphology as revealed by electron microscopy (EM) analysis. (A) Representative EM images for the ct441/tsp KD or NT control transformant strains. Scale bar = 2 μm. Samples were induced at 4 hpi with 20 nM aTc and harvested at 40 hpi. White arrows represent organisms that were identified as having condensing or condensed nucleoids (in a blinded fashion by one person) and quantified (in a blinded fashion by a different person). Black boxes show the organisms represented in panel B. (B) Representation of procedure for measuring the perimeter of the nucleoid and outer membrane for each organism. Yellow line represents the outer membrane perimeter, purple line shows the nucleoid perimeter. (C) Quantification of the non-nucleoid volume displayed as the total volume constrained by the outer membrane with the nucleoid volume subtracted. ****, P < 0.000001; ns, not significant by an ordinary one-way analysis of variance.
DISCUSSION
Understanding how pathogens like Chlamydia transition between growth states is important for many reasons, one of which is the practical application of designing strategies to interrupt or block this process. Chlamydia undergoes a unique, biphasic developmental cycle in which the organism differentiates between functional forms with distinct morphologies. The process of secondary differentiation, in which the replicating RB becomes the infectious EB, is an integral aspect of chlamydial development. Given that no studies to date have identified dividing RBs with condensing nucleoids, a key characteristic of secondary differentiating forms, we reason that secondary differentiation occurs post-division. While multiple proteomic studies have profiled the protein repertoires of EBs and RBs (29–31), the molecular mechanisms which drive the distinct proteomic profiles remain poorly understood. As such, we hypothesize that protein turnover plays a critical role during differentiation. For example, proteins that maintain an RB-like state must be eliminated or otherwise inactivated to allow an EB to form.
We have previously characterized the chlamydial Clp protease systems and proposed a model in which protein turnover in the bacterial cytosol contributes to secondary differentiation (32, 49, 60, 61). Because Chlamydia is a Gram-negative bacterium, it has four cellular compartments from which proteins may need to be eliminated in a coordinated way during differentiation events: the cytoplasm, inner membrane, periplasm, and outer membrane. Prior work from the Huston lab has evaluated the periplasmic protease HtrA suggesting its essentiality in the replicative phase of Ctr development and proposing its direct link to the production of infectious EBs (33, 34, 62). However, HtrA is expressed as a mid-cycle gene and is likely the major housekeeping protease ensuring protein homeostasis in the periplasm during RB growth and division. As such, other factors may serve a more specific function in the periplasm during secondary differentiation. Other major conserved proteases in Chlamydia include Lon, a cytoplasmic protease, FtsH, an inner membrane-anchored protease, and Tsp, another periplasmic protease. Like HtrA, Lon and FtsH are mid-cycle genes (unpublished data). However, Tsp is expressed as a late-cycle gene (15) (Fig. 1E), consistent with a proposed function in secondary differentiation. In this study, we investigated the function of the chlamydial periplasmic Tsp ortholog with the working hypothesis that it is essential for secondary differentiation.
Our findings indicate that Chlamydia is highly sensitive to disruptions in Tsp levels, with biologically significant reductions in viable EB progeny under such conditions. We demonstrated that increased levels of wild-type Tsp impaired the progression of chlamydial development such that RB replication was reduced and, accordingly, secondary differentiation was attenuated (Fig. 2 and 3). These data suggest that increased activity of wild-type Tsp causes a reduction in chlamydial viability, as overexpression of an inactive Tsp isoform exhibited little effect on Chlamydia. Interestingly, knockdown of tsp appeared to exert little effect on both replication and secondary differentiation while still reducing infectious progeny production (Fig. 4 and 5). Ultrastructural analysis suggested that the defect in infectious EB production likely arises from dysregulation of EB organization (Fig. 6). Taken together, these data indicate that endogenous levels of Tsp are important for appropriate secondary differentiation and that Chlamydia is extremely sensitive to altered Tsp levels.
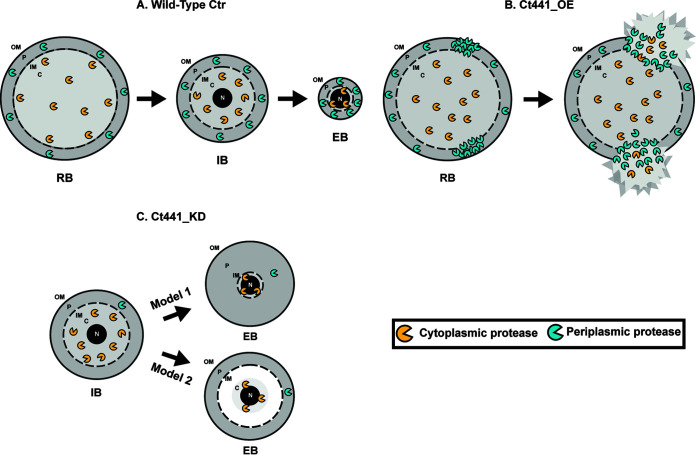
We hypothesize that chlamydial Tsp targets RB-specific periplasmic proteins for degradation or inactivation during secondary differentiation and that this is critical for the reorganization of an RB into an EB (Fig. 7A). The dysregulation of periplasmic homeostasis following the increase in Ct_Tsp_6×His levels may lead to excessive degradation of periplasmic components, resulting in membrane integrity defects and, subsequently, decreased bacterial survival (Fig. 7B). Our knockdown data suggest two possible models for why unbalanced Tsp activity disrupts secondary differentiation (Fig. 7C). The membrane separation model (Model 1) proposes that cytoplasmic proteases, such as the Clp systems, Lon, FtsH, or a combination thereof, degrade inner membrane and cytoplasmic substrates to reduce cytoplasmic volume and, concurrently, inner membrane size during secondary differentiation. In this mechanism, Tsp would reduce the outer membrane and periplasmic volumes independently of cytoplasmic volume reduction. However, the timing of these events would need to be highly coordinated to ensure the successful formation of an EB. The stationary periplasmic model (Model 2) proposes that periplasmic proteases degrade inner membrane components along with periplasmic substrates for proper secondary differentiation. Tsp may either directly degrade these substrates or cleave substrates in preparation for other periplasmic proteases, such as HtrA, to degrade them. The models differ in that during knockdown of ct441/tsp, the stationary periplasmic model indicates that turnover of periplasmic and inner membrane proteins is inhibited such that the outer membrane and inner membrane remain anchored together. The result of this is that the cytoplasm still condenses but leads to a “void” space between the condensed cytoplasm and inner membrane. The membrane separation model implies that the inner and outer membranes separate, such that the inner membrane size reduces concurrently with the cytoplasmic volume, thus resulting in separation between the inner and outer membranes. In this case, the “void” space would likely be the result of a massive increase in periplasmic volume. Both models allow for production of EB-specific proteins and virulence factors, but indicate that EBs are disorganized and therefore not infectious when Tsp expression is disrupted. Immunogold TEM to label the inner and outer membranes may help resolve these models, but it requires high-quality antibodies against relatively abundant antigens.
FIG 7.
Proposed model for Ct441/Tsp function in secondary differentiation. (A) Hypothesized model of Ct441/Tsp targeting reticulate body (RB)-specific proteins during secondary differentiation. During differentiation, Tsp degrades periplasmic proteins to facilitate reduction of periplasmic volume concurrent with reduction of cytoplasmic volume. (B) Depiction of effects of increased Ct441/Tsp activity on RBs. Excess active Tsp degrades periplasmically exposed proteins (either specifically or nonspecifically), leading to compromised membrane integrity. (C) Two potential models in which nonfunctional EBs are produced during ct441/tsp knockdown: the membrane separation model (Model 1) and the stationary periplasmic model (Model 2). In Model 1, the inner membrane separates from the outer membrane as the cytoplasmic volume decreases, resulting in a large increase in periplasmic volume. The separation of the membranes results in a decrease in protein density, leaving only diffusible molecules to fill the space. In Model 2, the inner and outer membranes remain anchored and do not reduce in size with the cytoplasm, resulting in a “void” space between the condensing nucleoid and the inner membrane. This space likely contains readily diffusible metabolites and ions with few (if any) proteins or nucleic acids. OM, outer membrane; P, periplasm; IM, inner membrane.
In model organisms such as E. coli and Pseudomonas, Tsp orthologs have been characterized as C-terminal processing proteases (39, 63). Structural homology of chlamydial Tsp to orthologous proteases of other Gram-negative bacterial species shows conservation of key motifs, including the signal sequence, PDZ domain, and catalytic residues (Fig. S3). However, several regions do not align well with Tsp homologs, including a domain of unknown function, annotated as DUF3340. DUF3340 has been shown to be critical to the catalytic activity of Ct_Tsp (35) and may contribute to a novel function of chlamydial Tsp. Most studies indicate these C-terminal processing proteases function in peptidoglycan homeostasis and regulate cell wall synthesis by targeting PG synthases or hydrolases (40–42). Such proteins are typically secreted into the periplasm or are membrane-anchored with domains exposed to the periplasm. Interestingly, pathogenic Chlamydia species lack PG in their cell walls and only synthesize a transient PG ring during their MreB-dependent polarized division process (9–13, 44). Because only the RB is capable of dividing, and division and PG-related genes are expressed during the RB phase of the development (i.e., mid-cycle [50, 51]), the late developmental cycle expression of the chlamydial Tsp ortholog is incongruent with a homeostatic function in cell wall synthesis. Nonetheless, it is possible that Tsp degrades, or facilitates degradation of, chlamydial PG-associated proteins to ensure that cell division ceases so that the EB can begin to form. Interestingly, non-pathogenic Chlamydia species such as Protochlamydia amoebophila do contain a PG sacculus (64). We speculate that in these organisms, Tsp activity may be required constitutively throughout the developmental cycle. Indeed, these organisms also lack the alternative sigma factor σ28, which regulates late-cycle tsp expression in C. trachomatis (Hatch and Ouellette in prep, Domman and Horn [65]). Therefore, it is expected that P. amoebophila tsp transcription peaks mid-cycle and that it is not a critical mediator of secondary differentiation. Further work is required to test this prediction.
In E. coli, Prc can cleave substrates in the periplasm that are labeled with an SsrA tag for degradation (47). Furthermore, the PDZ domain has been characterized as facilitating the interaction of E. coli Prc with the targeted SsrA-tagged substrate (66). Our in vitro studies confirmed that recombinant Ct_Tsp can recognize the SsrA tag (Fig. 1C). Previously, our lab described a role of trans-translation during secondary differentiation and proposed that the accumulation of SsrA-tagged proteins may serve as an initiating mechanism for secondary differentiation in the cytoplasm of Chlamydia by switching the substrate preference of ClpX (32). Intriguingly, we observed SsrA-tagged proteins throughout the bacterium and on the inclusion membrane, suggesting that SsrA-tagged proteins are also present in the periplasm and accumulate over the course of the developmental cycle. We hypothesize that Tsp, in addition to potential functions in degrading PG/division proteins, may be needed to specifically eliminate SsrA-tagged proteins in the periplasm during secondary differentiation to prevent the accumulation of potentially toxic partial peptides in the EB. This is under investigation.
In conclusion, we have shown that Tsp is essential for chlamydial development. Given that ct441/tsp is a late gene and that knocking down its expression reduces EB viability through dysregulation of size and membrane organization, we conclude that Tsp is a critical mediator of secondary differentiation. While the molecular mechanism of proteolysis of chlamydial Tsp is likely similar to that of other Prc homologs, we speculate that the unique gene repertoire of Chlamydia will reveal novel substrates for this protease. We are currently working to develop the strategies necessary to identify substrates, but these studies are complicated by the obligate intracellular growth of Chlamydia, which precludes large-scale bacterial cultures. Overall, this work contributes to the understanding of periplasmic condensation during chlamydial differentiation and further defines another key mediator of this process.
MATERIALS AND METHODS
Primers, plasmids, and strains.
A list of all primers, plasmids, and strains used in these studies can be found in Table S1 in the supplemental material.
Strains and cell culture.
McCoy mouse fibroblast cells were used for transformation, overexpression, knockdown studies, and protein extractions. McCoy cells were also used to harvest DNA and RNA samples from knockdown or overexpression strains. During our transcriptional profiling experimentation, human epithelial HEp-2 cells were used to harvest DNA and RNA samples from wild-type CtrL2. HeLa cells were utilized for plaque purification. These lines were expanded regularly in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO). Cells were regularly tested for Mycoplasma using the LookOut Mycoplasma PCR detection kit from Sigma-Aldrich. For plasmid transformation into Chlamydia, we used C. trachomatis serovar L2 EBs which lacked the endogenous plasmid (-pL2). Wild-type, density gradient-purified C. trachomatis L2/434/Bu EBs were used for transcriptional profiling studies.
Bioinformatics.
Gene sequences from all organisms were obtained from the KEGG Genome Browser (67–69). Multiple sequence alignments were generated using Clustal Omega (70) with default settings and were shown using Jalview version 2 (71). Pairwise alignments to find percent identity for Ct441 and other Gram-negative Tsp orthologs were performed using the NCBI Protein BLAST Function (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (72).
Purification and activity analysis of recombinant Tsp.
The ct_tsp and ec_prc genes were PCR-amplified from C. trachomatis L2/434/Bu and E. coli K-12, respectively. PCR products were then cloned into pLATE52 (N-ter 6×His tag, tsp) or pLATE31 (C-ter 6×His tag, prc) (Thermo Fisher Scientific) and vectors were transformed into E. coli DH5α. Transformants were selected on LB agar with ampicillin (100 μg/mL) at 37°C. Plasmids were purified and then sequence-verified by PSOMAGEN. Site-directed mutagenesis was performed on pLATE52-tsp to construct the S455A catalytic mutant.
For protein purification, plasmids were transformed into E. coli BL21(DE3) C43 (35). The recombinant proteins were purified as described by Kohlmann et al. (73). Briefly, bacteria were grown at 37°C in 2×YT medium with ampicillin until reaching an optical density at 600 nm (OD600) of 0.8 to 1. Cultures were then shifted to 20°C and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 16 h. Bacteria were harvested and then lysed via sonication in 50 mM Na2HPO4, 500 mM NaCl, 20 mM imidazole, 5% vol/vol glycerol (pH 8.0). Clarified supernatants were then bound to HisPur cobalt resin (Thermo Fisher Scientific) for immobilized metal affinity chromatography. Proteins were eluted in modified lysis buffer (500 mM imidazole) and then exchanged into storage buffer (20 mM Tris, 150 mM NaCl [pH 7.4]) using Amicon centrifugal concentration devices (Millipore).
Laemmli-treated purified recombinant proteins (1 μg) were run on 10% SDS-PAGE gels followed by staining with Coomassie brilliant blue to detect protein. For Western blotting, 500 ng of recombinant protein was used. Proteins were transferred from SDS-PAGE to nitrocellulose and blotted overnight at 4°C with a mouse anti-6×His-antibody (1:1,000, HIS.H8, Invitrogen, Waltham, MA) or rabbit anti-Tsp-antibody (1:200, produced by Pacific Immunology [St. Ramona, CA] against KKTNKQDRSFGSNDLQ) diluted in 5% milk Tris-buffered saline (TBS). Blots were then washed with 0.05% Tween-TBS (TTBS) and probed with either a goat anti-mouse IgG HRP (horseradish peroxidase)-conjugated antibody (1:2,000, AP124P, Millipore) or a goat anti-rabbit IgG HRP conjugated antibody (1:2,000, 32260, Invitrogen) for 1 h at room temperature. Blots were subsequently washed in TTBS followed by TBS and then incubated with Immobulin chemiluminescent substrate (Millipore). Blot and gel images were acquired using a Bio-Rad Chemidoc MP.
Ct_Tsp and Ec_Prc activity assays.
β-Casein (Sigma-Aldrich) and SsrA-tagged GFP (VAA C terminus), purified as described by Wood et al. (32). were used to assess the protease activity of Ec_Prc and Ct_Tsp/Ct_Tsp S455A. Here, 1 μg of enzyme was incubated with 3 μg of substrate in 50 mM Tris, 150 mM NaCl (pH 8) for 3 h at 37°C. Reactions were terminated through the addition of Laemmli buffer and heating at 95°C for 5 min. Samples were then separated on 12% SDS-PAGE gels and stained with Coomassie brilliant blue to detect protein.
Chlamydial plasmid construction.
The Hifi Cloning System (New England Biolabs [NEB], Ipswich, MA) was utilized to create constructs for chlamydial transformation. The NEBuilder assembly tool from NEB was used to design primers. Primers were created with overlap to the desired vector. We added a 6×His tag at the C-terminal end of our gene of interest to detect our overexpressed proteins. Purified chlamydial genomic DNA was used as a template to amplify our gene of interest using Phusion DNA polymerase (Thermo Fisher Scientific). Once assembled, the product was ligated into the respective vector and transformed into DH10β E. coli (NEB) for expansion. The isolated plasmid was verified by restriction enzyme digest and sequencing before transformation into C. trachomatis serovar L2 (-pL2). The Ct_Tsp_6×His overexpression strains utilized the pBOMBL expression backbone (55). A CRISPR interference approach using the pBOMBLCRia vector encoding an inducible catalytically dead Cas9 protein and guide RNA was engineered by our lab and used to obtain the ct441/tsp knockdown (54, 55).
Chlamydial transformation.
Prior to the transformation, a 6-well plate was seeded with McCoy cells. Two wells were utilized per plasmid transformation. For transformation into C. trachomatis, ~2 μg of sequence-verified plasmid was incubated at room temperature with 106 CtrL2 EBs that lacked the endogenous plasmid (-pL2; ATCC 25667R) suspended in 50 μL CaCl2 for 30 min. The transformation mixture was diluted with 1 mL of Hanks’ balanced salt solution (HBSS) and added dropwise to one well of the 6-well plate containing 1 mL of HBSS. The cells were centrifuged at 400 × g for 15 min at room temperature and incubated at 37°C for 15 min. After incubation, the inoculum was replaced with DMEM/10% FBS. At 8 hpi, the medium was removed and replaced with DMEM/10% FBS containing 1 μg/mL of cycloheximide and either 1 or 2 U/mL of penicillin. Cells were monitored and passaged every 48 h until a population of GFP-positive, penicillin-resistant inclusions was observed. The transformants were then harvested and frozen at −80°C in a solution of sucrose-phosphate (2SP). Samples were expanded to make stocks and titrated to calculate the multiplicity of infection (MOI).
IFA imaging and IFU assays to measure the effects of overexpression of Ct_Tsp isoforms or knockdown of tsp transcripts on Chlamydia.
A confluent monolayer of McCoy cells was infected with Ctr transformed with plasmids encoding the various Tsp isoforms fused to a 6×His affinity tag or CRISPRi vectors targeting tsp or a non-targeting sequence control. Penicillin and cycloheximide treatment were maintained in the medium for the duration of the experiment. For overexpression experiments, at 16 hpi, 20 nM anhydrotetracycline (aTc) was used to induce samples. At 24 or 48 hpi, three wells of a 24-well plate were scraped in 250 μL 2SP, pooled, vortexed with three 1-mm glass beads, and frozen at −80°C for IFU determination. For knockdown experiments, at 4 hpi, samples were either induced or not induced with 20 nM aTc. RNA and DNA samples were collected at 10 and 36 hpi as described below. Transcript data were analyzed as a ratio of cDNA to gDNA. Alongside the experimental time points for both studies, coverslips were fixed with a formaldehyde-glutaraldehyde solution followed by methanol to assess expression of the constructs in the primary infection. Coverslips were stained with primary antibodies, goat anti-Ctr MOMP (Meridian Biosciences, Memphis, TN) and rabbit anti-6×His (Abcam; Cambridge, MA) for overexpression studies or rabbit anti-dCas9 (Abcam) for knockdown studies, and DNA was labeled with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen). Donkey secondary antibodies were used against the goat and rabbit primary antibodies. Coverslips were then mounted on glass slides using ProLong Glass mounting medium (Invitrogen). Images were acquired from a ×100 objective on a Zeiss AxioImager Z.2 equipped with Apotome.2 optical sectioning hardware and X-Cite Series 120PC illumination lamp using a 2MP Axiocam 506 monochrome camera. To quantify IFUs, the thawed IFU samples were serially diluted at 1:10 in 2SP buffer and used to infect a fresh monolayer of McCoy cells. At 24 hpi, these titrated samples were fixed with a formaldehyde-glutaraldehyde mix to preserve the GFP signal expressed from the plasmid and then permeabilized using methanol. Primary goat anti-Ctr MOMP followed by donkey anti-goat 594 (Invitrogen) was used to label inclusions. Inclusions per field of view were systematically counted. Data were plotted on a log scale and are presented as a percentage of the uninduced sample.
Isolation of genomic DNA and qPCR readouts.
At the indicated time points, cells were trypsinized and spun at 400 × g for 5 min at 4°C. The pellet was resuspended in 500 μL phosphate-buffered saline (PBS) and frozen at −80°C. Samples were freeze-thawed three times, and DNA was extracted using the Qiagen DNeasy kit for processing. DNA concentrations were measured on a Nanodrop spectrophotometer, and samples were diluted to 5 ng/μL. A master mix was created using gene-specific primers and SYBR Green, which was added into wells of a 96-well optical plate containing 5 μL of the diluted gDNA. Blank wells of the master mix alone were used to assess contamination. The plate was analyzed on a QuantStudio 3 thermal cycler with standard amplification conditions followed by a melting curve analysis. Each sample was analyzed in triplicate. A standard curve was created using Ctr L2 genomic DNA to quantify DNA levels. These data were normalized as genome copy numbers, where all data points were multiplied by 888,676 (number of chromosomal copies per 1 ng DNA) prior to plotting on a log scale.
Isolation of RNA and generation of cDNA.
To collect RNA, 1 mL of TRIzol (Invitrogen) was used to harvest infected cells from one well of a 6-well plate at the indicated time points, and 200 μL of chloroform was added to extract the RNA following the manufacturer’s guidelines. Once spun at 3,000 × g for 5 min at 4°C, 500 μL of the aqueous layer was extracted without disturbance and precipitated with an equal volume of isopropanol. DNase was used to treat 10 μg of total RNA and eliminate contaminating DNA. cDNA was then generated from 1 μg of the DNase-treated RNA using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions in a total volume of 20 μL. For the control conditions, no reverse transcriptase was added (i.e., no RT). After the reaction was complete, samples were diluted with 200 μL nuclease-free H2O, and cDNA was aliquoted to avoid freeze-thawing. 5 μL of cDNA per well was loaded in a qPCR plate with gene-specific primers and SYBR green for detection. Three wells were loaded for each sample as internal replicates. Blank wells and no-RT samples were controlled for any contamination. A standard curve was generated using Ctr L2 isolated genomic DNA. The qPCR plate was analyzed on a QuantStudio 3 thermal cycler with standard amplification conditions followed by a melting curve analysis as described above for DNA quantification.
Determining the transcriptional profile of ct441/tsp during chlamydial development.
HEp2 cells were seeded in 6-well plates, and two wells were used per condition. No antibiotics were added during this experiment. To collect RNA at the time of infection, the protocol described above was followed with wild-type Ctr L2 being used to infect the cells. DNA was harvested as described above. Samples of RNA and DNA were harvested, as described above, throughout the developmental cycle, specifically at 1, 3, 8, 16, 24, and 48 hpi. Primers against tsp and representative early- (euo), mid- (clpP2), and late-cycle (omcB) genes were used to quantify their transcript levels using qPCR. The ratio of cDNA to gDNA was calculated and plotted on a log scale.
Observation of HctB during ct441/tsp knockdown or overexpression of Ct_Tsp_6×His.
McCoys were seeded in a 6-well plate before infection. The respective wells were infected and then either induced or not induced with 20 nM aTc. Throughout the duration of the experiment, penicillin and cycloheximide were maintained in the medium. The Ct_Tsp_6×His overexpression strains were induced at 16 hpi. The knockdown and non-targeting strains were induced at 4 hpi. At 48 hpi, protein lysate was harvested using 8 M urea-containing buffer which had 0.1% SDS, 2.5% beta-mercaptoethanol, 10 mM Tris, and nuclease added immediately before use. The protein samples were quantified using the EZQ protein assay kit (Thermo Fisher Scientific). Next, 30 μg of protein was loaded per well for SDS-PAGE. Protein was transferred to a polyvinylidene difluoride membrane which was then stained with primary goat anti-MOMP and rabbit anti-HctB (kind gift from T. Hackstadt; Rocky Mountain Labs/NIH). Donkey anti-goat 680 and donkey anti-rabbit 800 secondary antibodies (LI-COR Biosciences; Lincoln, NE) were utilized. The FIJI is Just ImageJ (FIJI) program was used for densitometric quantification. Ratios were displayed as uninduced divided by induced and HctB to MOMP.
Morphology analysis using transmission electron microscopy to observe ct441/tsp knockdown samples.
McCoy cells were infected with the ct441/tsp knockdown or nontargeting strains. Induction with 20 nM aTc occurred or did not occur at 4 hpi for the knockdown or non-targeting strains. Samples for TEM imaging were fixed by immersion in a solution of 2% glutaraldehyde, 2% paraformaldehyde in 0.1 M Sorenson’s phosphate buffer (pH 7.2) for a minimum of 24 h at 4°C. Samples were then washed three times with PBS to clear excess fixative. During processing, samples were post-fixed in a 1% aqueous solution of osmium tetroxide for 30 min. Subsequently, samples were dehydrated in a graded ethanol series (50, 70, 90, 95, 100%), and propylene oxide was used as a transition solvent between the ethanol and Embed 812 resin. Samples were allowed to sit overnight in a 50:50 propylene oxide:resin solution until all the propylene oxide had evaporated. Samples were then incubated in fresh resin for 2 h at room temperature before final embedding. Polymerization took place at 65°C for 24 h. Thin sections (90 nm) made with a Leica UC7 Ultracut ultramicrotome were placed on 200-mesh copper grids, post-stained with 2% uranyl acetate followed by Reynolds lead citrate, and examined on a Tecnai G2 Spirit TWIN (FEI) operating at an accelerating voltage of 80 kV. Images were acquired digitally with an AMT digital imaging system.
Organisms which had a condensing or condensed nucleoid were identified and labeled by one individual in a blind fashion. Using FIJI, the perimeter of the nucleoid and the outer membrane were measured for each organism, also in a blinded fashion, by a different individual. Using the perimeter values, radius was calculated using the formula . To account for the electron microscopy images being two-dimensional, we adjusted for three-dimensional space by calculating volume from the calculated radii using the equation . We used these volumes of the outer membrane and nucleoid to infer the difference between these boundaries.
ACKNOWLEDGMENTS
This work was supported in part by a National Institutes of Health (NIAID) award to S.P.O. and D.J.F. (R21AI171228) and by university start-up funds to both from their respective institutions. Additional support for A.R.S. was provided by an INBRE award from the National Institute of General Medical Sciences of the National Institutes of Health (5P20GM103427).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank Nathan Hatch for helping with the blinded analysis of the EM data. We thank Tom Bargar and Nicholas Conoan of the Electron Microscopy Core Facility (EMCF) at the University of Nebraska Medical Center for technical assistance. The EMCF is supported by state funds from the Nebraska Research Initiative (NRI) and the University of Nebraska Foundation, and institutionally by the Office of the Vice Chancellor for Research.
Footnotes
Supplemental material is available online only.
Contributor Information
Scot P. Ouellette, Email: scot.ouellette@unmc.edu.
Laurie E. Comstock, University of Chicago
REFERENCES
- 1.Gober JW, Marques MV. 1995. Regulation of cellular differentiation in Caulobacter crescentus. Microbiol Rev 59:31–47. doi: 10.1128/mr.59.1.31-47.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomashow MF, Cotter TW. 1992. Bdellovibrio host dependence: the search for signal molecules and genes that regulate the intraperiplasmic growth cycle. J Bacteriol 174:5767–5771. doi: 10.1128/jb.174.18.5767-5771.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Julien B, Kaiser AD, Garza A. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc Natl Acad Sci USA 97:9098–9103. doi: 10.1073/pnas.97.16.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdelrahman YM, Belland RJ. 2005. The chlamydial developmental cycle. FEMS Microbiol Rev 29:949–959. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Clifton DR, Dooley CA, Grieshaber SS, Carabeo RA, Fields KA, Hackstadt T. 2005. Tyrosine phosphorylation of the chlamydial effector protein Tarp is species specific and not required for recruitment of actin. Infect Immun 73:3860–3868. doi: 10.1128/IAI.73.7.3860-3868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci USA 101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lane BJ, Mutchler C, Al Khodor S, Grieshaber SS, Carabeo RA. 2008. Chlamydial entry involves TARP binding of guanine nucleotide exchange factors. PLoS Pathog 4:e1000014. doi: 10.1371/journal.ppat.1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keb G, Ferrell J, Scanlon KR, Jewett TJ, Fields KA. 2021. Chlamydia trachomatis TmeA directly activates N-WASP to promote actin polymerization and functions synergistically with TarP during invasion. mBio 12:e02861-20. doi: 10.1128/mBio.02861-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdelrahman Y, Ouellette SP, Belland RJ, Cox JV. 2016. Polarized cell division of Chlamydia trachomatis. PLoS Pathog 12:e1005822. doi: 10.1371/journal.ppat.1005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ouellette SP, Karimova G, Subtil A, Ladant D. 2012. Chlamydia co-opts the rod shape-determining proteins MreB and Pbp2 for cell division. Mol Microbiol 85:164–178. doi: 10.1111/j.1365-2958.2012.08100.x. [DOI] [PubMed] [Google Scholar]
- 11.Ouellette SP, Lee J, Cox JV. 2020. Division without binary fission: cell division in the FtsZ-less Chlamydia. J Bacteriol 202:e00252-20. doi: 10.1128/JB.00252-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouellette SP, Fisher-Marvin LA, Harpring M, Lee J, Rucks EA, Cox JV. 2022. Localized cardiolipin synthesis is required for the assembly of MreB during the polarized cell division of Chlamydia trachomatis. PLoS Pathog 18:e1010836. doi: 10.1371/journal.ppat.1010836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, Cox JV, Ouellette SP. 2020. Critical role for the extended N terminus of Chlamydial MreB in directing its membrane association and potential interaction with divisome proteins. J Bacteriol 202:e00034-20. doi: 10.1128/JB.00034-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hybiske K, Stephens RS. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc Natl Acad Sci USA 104:11430–11435. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, Beatty WL, Caldwell HD. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci USA 100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw EI, Dooley CA, Fischer ER, Scidmore MA, Fields KA, Hackstadt T. 2000. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol 37:913–925. doi: 10.1046/j.1365-2958.2000.02057.x. [DOI] [PubMed] [Google Scholar]
- 17.Barry CE 3rd, Hayes SF, Hackstadt T. 1992. Nucleoid condensation in Escherichia coli that express a chlamydial histone homolog. Science 256:377–379. doi: 10.1126/science.256.5055.377. [DOI] [PubMed] [Google Scholar]
- 18.Everett KD, Hatch TP. 1995. Architecture of the cell envelope of Chlamydia psittaci 6BC. J Bacteriol 177:877–882. doi: 10.1128/jb.177.4.877-882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatch TP, Allan I, Pearce JH. 1984. Structural and polypeptide differences between envelopes of infective and reproductive life cycle forms of Chlamydia spp. J Bacteriol 157:13–20. doi: 10.1128/jb.157.1.13-20.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatch TP, Miceli M, Sublett JE. 1986. Synthesis of disulfide-bonded outer membrane proteins during the developmental cycle of Chlamydia psittaci and Chlamydia trachomatis. J Bacteriol 165:379–385. doi: 10.1128/jb.165.2.379-385.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackstadt T, Todd WJ, Caldwell HD. 1985. Disulfide-mediated interactions of the chlamydial major outer membrane protein: role in the differentiation of chlamydiae? J Bacteriol 161:25–31. doi: 10.1128/jb.161.1.25-31.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brickman TJ, Barry CE, Hackstadt T. 1993. Molecular cloning and expression of hctB encoding a strain-variant chlamydial histone-like protein with DNA-binding activity. J Bacteriol 175:4274–4281. doi: 10.1128/jb.175.14.4274-4281.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsokos CG, Laub MT. 2012. Polarity and cell fate asymmetry in Caulobacter crescentus. Curr Opin Microbiol 15:744–750. doi: 10.1016/j.mib.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Errington J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol 1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- 25.Matsumoto A, Manire GP. 1970. Electron microscopic observations on the effects of penicillin on the morphology of Chlamydia psittaci. J Bacteriol 101:278–285. doi: 10.1128/jb.101.1.278-285.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouellette SP, Hatch TP, AbdelRahman YM, Rose LA, Belland RJ, Byrne GI. 2006. Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNγ-mediated host cell tryptophan starvation. Mol Microbiol 62:1387–1401. doi: 10.1111/j.1365-2958.2006.05465.x. [DOI] [PubMed] [Google Scholar]
- 27.Gaylord WH, Jr. 1954. Intracellular forms of meningopneumonitis virus. J Exp Med 100:575–580. doi: 10.1084/jem.100.6.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JK, Enciso GA, Boassa D, Chander CN, Lou TH, Pairawan SS, Guo MC, Wan FYM, Ellisman MH, Sütterlin C, Tan M. 2018. Replication-dependent size reduction precedes differentiation in Chlamydia trachomatis. Nat Commun 9:45. doi: 10.1038/s41467-017-02432-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skipp PJ, Hughes C, McKenna T, Edwards R, Langridge J, Thomson NR, Clarke IN. 2016. Quantitative proteomics of the infectious and replicative forms of Chlamydia trachomatis. PLoS One 11:e0149011. doi: 10.1371/journal.pone.0149011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Østergaard O, Follmann F, Olsen AW, Heegaard NH, Andersen P, Rosenkrands I. 2016. Quantitative protein profiling of Chlamydia trachomatis growth forms reveals defense strategies against tryptophan starvation. Mol Cell Proteomics 15:3540–3550. doi: 10.1074/mcp.M116.061986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saka HA, Thompson JW, Chen Y-S, Kumar Y, Dubois LG, Moseley MA, Valdivia RH. 2011. Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms. Mol Microbiol 82:1185–1203. doi: 10.1111/j.1365-2958.2011.07877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood NA, Swoboda AR, Blocker AM, Fisher DJ, Ouellette SP. 2022. Tag-dependent substrate selection of ClpX underlies secondary differentiation of Chlamydia trachomatis. mBio 13:e01858-22. doi: 10.1128/mbio.01858-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huston WM, Swedberg JE, Harris JM, Walsh TP, Mathews SA, Timms P. 2007. The temperature activated HtrA protease from pathogen Chlamydia trachomatis acts as both a chaperone and protease at 37 degrees C. FEBS Lett 581:3382–3386. doi: 10.1016/j.febslet.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 34.Huston WM, Theodoropoulos C, Mathews SA, Timms P. 2008. Chlamydia trachomatis responds to heat shock, penicillin induced persistence, and IFN-gamma persistence by altering levels of the extracytoplasmic stress response protease HtrA. BMC Microbiol 8:190. doi: 10.1186/1471-2180-8-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohlmann F, Shima K, Hilgenfeld R, Solbach W, Rupp J, Hansen G. 2015. Structural basis of the proteolytic and chaperone activity of Chlamydia trachomatis CT441. J Bacteriol 197:211–218. doi: 10.1128/JB.02140-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silber KR, Keiler KC, Sauer RT. 1992. Tsp: a tail-specific protease that selectively degrades proteins with nonpolar C termini. Proc Natl Acad Sci USA 89:295–299. doi: 10.1073/pnas.89.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keiler KC, Sauer RT. 1995. Identification of active site residues of the Tsp protease. J Biol Chem 270:28864–28868. doi: 10.1074/jbc.270.48.28864. [DOI] [PubMed] [Google Scholar]
- 38.Keiler KC, Sauer RT. 1996. Sequence determinants of C-terminal substrate recognition by the Tsp protease. J Biol Chem 271:2589–2593. doi: 10.1074/jbc.271.5.2589. [DOI] [PubMed] [Google Scholar]
- 39.Keiler KC, Silber KR, Sauer RT, Downard KM, Papayannopoulos IA, Biemann K. 1995. C-terminal specific protein degradation: activity and substrate specificity of the Tsp protease. Protein Sci 4:1507–1515. doi: 10.1002/pro.5560040808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh SK, Parveen S, SaiSree L, Reddy M. 2015. Regulated proteolysis of a cross-link–specific peptidoglycan hydrolase contributes to bacterial morphogenesis. Proc Natl Acad Sci USA 112:10956–10961. doi: 10.1073/pnas.1507760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sommerfield AG, Darwin AJ. 2022. Bacterial carboxyl-terminal processing proteases play critical roles in the cell envelope and beyond. J Bacteriol 204:e0062821. doi: 10.1128/jb.00628-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeon WJ, Cho H. 2022. A cell wall hydrolase MepH is negatively regulated by proteolysis involving Prc and NlpI in Escherichia coli. Front Microbiol 13:878049. doi: 10.3389/fmicb.2022.878049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liechti G, Kuru E, Packiam M, Hsu YP, Tekkam S, Hall E, Rittichier JT, VanNieuwenhze M, Brun YV, Maurelli AT. 2016. Pathogenic Chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. PLoS Pathog 12:e1005590. doi: 10.1371/journal.ppat.1005590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, VanNieuwenhze M, Maurelli AT. 2014. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506:507–510. doi: 10.1038/nature12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beebe KD, Shin J, Peng J, Chaudhury C, Khera J, Pei D. 2000. Substrate recognition through a PDZ domain in tail-specific protease. Biochemistry 39:3149–3155. doi: 10.1021/bi992709s. [DOI] [PubMed] [Google Scholar]
- 46.Su M-Y, Som N, Wu C-Y, Su S-C, Kuo Y-T, Ke L-C, Ho M-R, Tzeng S-R, Teng C-H, Mengin-Lecreulx D, Reddy M, Chang C-I. 2017. Structural basis of adaptor-mediated protein degradation by the tail-specific PDZ-protease Prc. Nat Commun 8:1516. doi: 10.1038/s41467-017-01697-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keiler KC, Waller PRH, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 48.Wichlan DG, Hatch TP. 1993. Identification of an early-stage gene of Chlamydia psittaci 6BC. J Bacteriol 175:2936–2942. doi: 10.1128/jb.175.10.2936-2942.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood NA, Chung KY, Blocker AM, Rodrigues de Almeida N, Conda-Sheridan M, Fisher DJ, Ouellette SP. 2019. Initial characterization of the two ClpP Paralogs of Chlamydia trachomatis suggests unique functionality for each. J Bacteriol 201:e00613-19. doi: 10.1128/JB.00613-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ouellette SP, Rueden KJ, AbdelRahman YM, Cox JV, Belland RJ. 2015. Identification and partial characterization of potential FtsL and FtsQ homologs of Chlamydia. Front Microbiol 6:1264. doi: 10.3389/fmicb.2015.01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ouellette SP, Rueden KJ, Gauliard E, Persons L, de Boer PA, Ladant D. 2014. Analysis of MreB interactors in Chlamydia reveals a RodZ homolog but fails to detect an interaction with MraY. Front Microbiol 5:279. doi: 10.3389/fmicb.2014.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barry CE 3rd, Brickman TJ, Hackstadt T. 1993. Hc1-mediated effects on DNA structure: a potential regulator of chlamydial development. Mol Microbiol 9:273–283. doi: 10.1111/j.1365-2958.1993.tb01689.x. [DOI] [PubMed] [Google Scholar]
- 53.Fahr MJ, Douglas AL, Xia W, Hatch TP. 1995. Characterization of late gene promoters of Chlamydia trachomatis. J Bacteriol 177:4252–4260. doi: 10.1128/jb.177.15.4252-4260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ouellette SP. 2018. Feasibility of a conditional knockout system for Chlamydia Based on CRISPR interference. Front Cell Infect Microbiol 8:59. doi: 10.3389/fcimb.2018.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ouellette SP, Blay EA, Hatch ND, Fisher-Marvin LA. 2021. CRISPR interference to inducibly repress gene expression in Chlamydia trachomatis. Infect Immun 89:e0010821. doi: 10.1128/IAI.00108-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 57.Price MN, Alm EJ, Arkin AP. 2005. Interruptions in gene expression drive highly expressed operons to the leading strand of DNA replication. Nucleic Acids Res 33:3224–3234. doi: 10.1093/nar/gki638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hackstadt T, Baehr W, Ying Y. 1991. Chlamydia trachomatis developmentally regulated protein is homologous to eukaryotic histone H1. Proc Natl Acad Sci USA 88:3937–3941. doi: 10.1073/pnas.88.9.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hackstadt T, Brickman TJ, Barry CE 3rd, Sager J. 1993. Diversity in the Chlamydia trachomatis histone homologue Hc2. Gene 132:137–141. doi: 10.1016/0378-1119(93)90526-9. [DOI] [PubMed] [Google Scholar]
- 60.Wood NA, Blocker AM, Seleem MA, Conda-Sheridan M, Fisher DJ, Ouellette SP. 2020. The ClpX and ClpP2 orthologs of Chlamydia trachomatis perform discrete and essential functions in organism growth and development. mBio 11:e02016-20. doi: 10.1128/mBio.02016-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pan S, Jensen AA, Wood NA, Henrichfreise B, Brötz-Oesterhelt H, Fisher DJ, Sass P, Ouellette SP. 2022. In vivo and in vitro characterization of the ClpC AAA+ ATPase of Chlamydia trachomatis. mBio e0007523. doi: 10.1128/mbio.00075-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marsh JW, Ong VA, Lott WB, Timms P, Tyndall JD, Huston WM. 2017. CtHtrA: the lynchpin of the chlamydial surface and a promising therapeutic target. Future Microbiol 12:817–829. doi: 10.2217/fmb-2017-0017. [DOI] [PubMed] [Google Scholar]
- 63.Seo J, Darwin AJ. 2013. The Pseudomonas aeruginosa periplasmic protease CtpA can affect systems that impact its ability to mount both acute and chronic infections. Infect Immun 81:4561–4570. doi: 10.1128/IAI.01035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pilhofer M, Aistleitner K, Biboy J, Gray J, Kuru E, Hall E, Brun YV, VanNieuwenhze MS, Vollmer W, Horn M, Jensen GJ. 2013. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat Commun 4:2856. doi: 10.1038/ncomms3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Domman D, Horn M. 2015. Following the footsteps of chlamydial gene regulation. Mol Biol Evol 32:3035–3046. doi: 10.1093/molbev/msv193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spiers A, Lamb HK, Cocklin S, Wheeler KA, Budworth J, Dodds AL, Pallen MJ, Maskell DJ, Charles IG, Hawkins AR. 2002. PDZ domains facilitate binding of high temperature requirement protease A (HtrA) and tail-specific protease (Tsp) to heterologous substrates through recognition of the small stable RNA A (ssrA)-encoded peptide. J Biol Chem 277:39443–39449. doi: 10.1074/jbc.M202790200. [DOI] [PubMed] [Google Scholar]
- 67.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Troshin PV, Procter JB, Barton GJ. 2011. Java bioinformatics analysis web services for multiple sequence alignment: JABAWS:MSA. Bioinformatics 27:2001–2002. doi: 10.1093/bioinformatics/btr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schäffer AA, Yu Y-K. 2005. Protein database searches using compositionally adjusted substitution matrices. FEBS J 272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kohlmann F, Shima K, Rupp J, Solbach W, Hilgenfeld R, Hansen G. 2015. Production, crystallization and X-ray diffraction analysis of the protease CT441 from Chlamydia trachomatis. Acta Crystallogr F Struct Biol Commun 71:1454–1458. doi: 10.1107/S2053230X15020518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S3 and Table S1. Download jb.00099-23-s0001.pdf, PDF file, 11.1 MB (11.1MB, pdf)